Abstract
High-resolution structures reveal that yeast ribosomal protein L11 and its bacterial/archael homologs called L5 contain a highly conserved, basically charged internal loop that interacts with the peptidyl-transfer RNA (tRNA) T-loop. We call this the L11 ‘P-site loop’. Chemical protection of wild-type ribosome shows that that the P-site loop is inherently flexible, i.e. it is extended into the ribosomal P-site when this is unoccupied by tRNA, while it is retracted into the terminal loop of 25S rRNA Helix 84 when the P-site is occupied. To further analyze the function of this structure, a series of mutants within the P-site loop were created and analyzed. A mutant that favors interaction of the P-site loop with the terminal loop of Helix 84 promoted increased affinity for peptidyl-tRNA, while another that favors its extension into the ribosomal P-site had the opposite effect. The two mutants also had opposing effects on binding of aa-tRNA to the ribosomal A-site, and downstream functional effects were observed on translational fidelity, drug resistance/hypersensitivity, virus maintenance and overall cell growth. These analyses suggest that the L11 P-site loop normally helps to optimize ribosome function by monitoring the occupancy status of the ribosomal P-site.
INTRODUCTION
Over the past decade, atomic resolution ribosome structures have revealed the locations of critical elements. However, these static images do not reveal the dynamic movements within this complex macromolecule. The ribosome must coordinate multiple activities between spatially and functionally different sites in two subunits. These include three transfer RNA (tRNA)-binding sites, the peptidyltransferase and decoding centers and the elongation factor interacting regions. Events occurring in these regions must be carefully coordinated to assure rapid and accurate decoding of messenger RNAs (mRNAs). Current efforts in the field are focusing on determining the mechanisms by which these functional centers synchronize their actions and communicate with each other.
The eukaryotic ribosome contains nearly 80 intrinsic proteins. The high degree of similarity across species, from the primary amino acid sequences to their tertiary structures, suggests conserved functional roles beyond serving as mere scaffolding for the rRNAs. Ribosomal protein L11 of Saccharomyces cerevisiae is an essential, highly conserved component of the 60S subunit (in bacteria and archaea, the homologous protein is named L5; the yeast nomenclature is used throughout this text to minimize confusion). At the primary amino acid sequence level, L11 is well conserved among eukaryotes (∼67–97% identity), while bacterial and archaeal L5 proteins are less well conserved (27–55% identical) (Supplementary Figure S1A). L11 is uniquely positioned at the interface between the large subunit central protuberance (Figure 1A and 1B) and the head of the small subunit (Figure 1A) (1–5). In the small subunit, the head region undergoes significant rotational movement relative to the central protuberance between the pre- and post-translocational states (6), and the protein–protein interactions between L11 and S18 (S13 in bacteria and archaea) on the small subunit (the B1b and B1c intersubunit bridges) undergo the largest intersubunit structural rearrangements between these two states (7–9). These observations suggest that L11 may play a central role as an informational conduit between the two subunits. Detailed analysis of X-ray crystallographic and cryo-EM structures (Figure 1B) reveals that the concave surface of the β-sheet portion of L11 interacts with specific nucleotides in the minor groove of 23S rRNA helix 84 (10). L11 also makes contacts with the helix III and loop C regions of 5S rRNA; these connections have been hypothesized to help stabilize 5S rRNA interactions and may participate in an information signal transmission network linking functional centers within the ribosome (1,11). Importantly, the B1b and B1c intersubunit bridges with S18 are the only protein–protein interactions between the two subunits (2,3,5,8). Analyses of these structures indicate that contacts involving L11 and S18 through the B1b and B1c bridges break and rearrange after eEF-2 binding and ribosome ratcheting, controlled in part by differentially charged amino acid side chains between the two proteins (2,4,5,7,9,12). An internal loop of L11 that we denote the ‘L11 P-site loop’, which is roughly formed by amino acid residues 48–68, also directly contacts the T-loop of the peptidyl-tRNA in the P-site through tRNA nucleotide 56 (3–5). At the level of primary amino acid sequence, the P-site loop is highly conserved among eukaryotes (85–100% identity), while it is less well conserved among bacteria and archaea (42–57% identity) (Supplementary Figure S1B). At the biochemical level, however, the P-site loop is significantly more homogeneous, containing a large number of well-aligned charged and aromatic amino acids. In particular, A50, F57, R60 and I65 (yeast numbering) are universally conserved. An alignment of the P-site loop structures from yeast, Haloarcula marismortui, Thermus thermophilus and Escherichia coli reveals that the P-site loop is extremely well conserved at the structural level (Supplementary Figure S1C).
Figure 1.
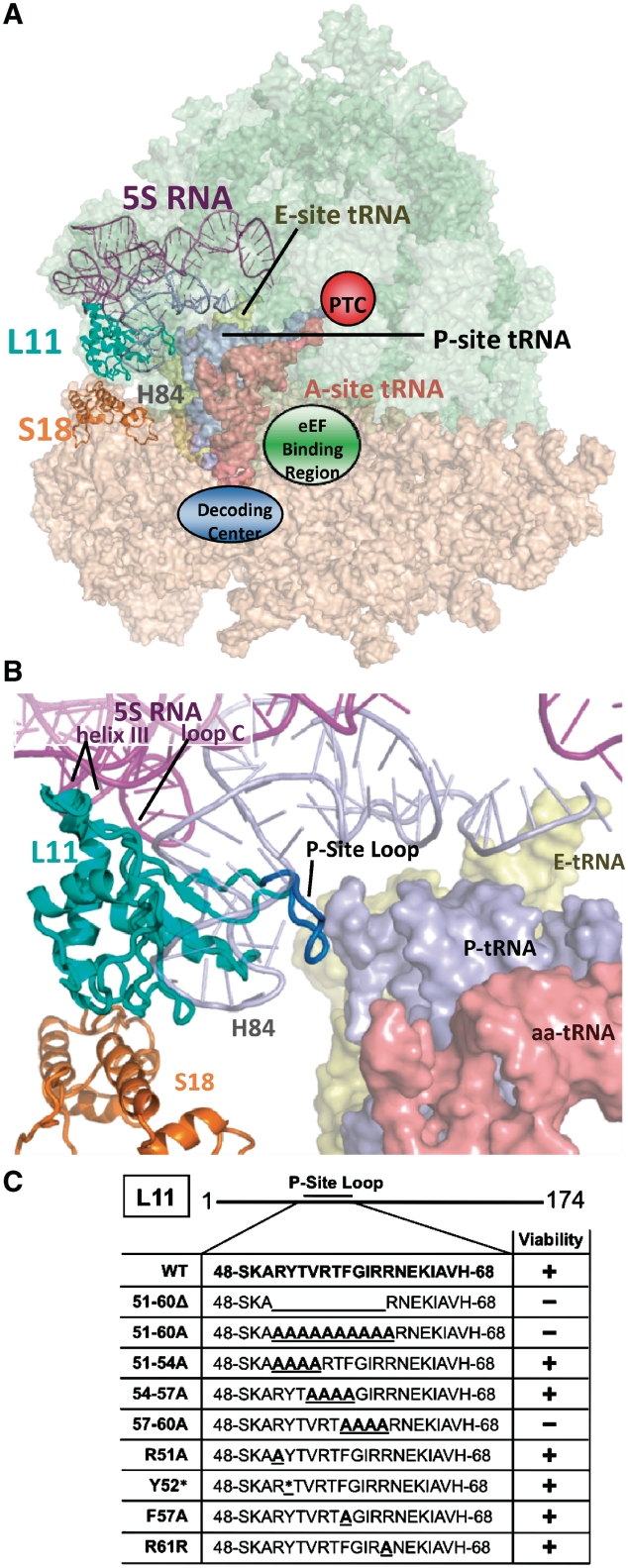
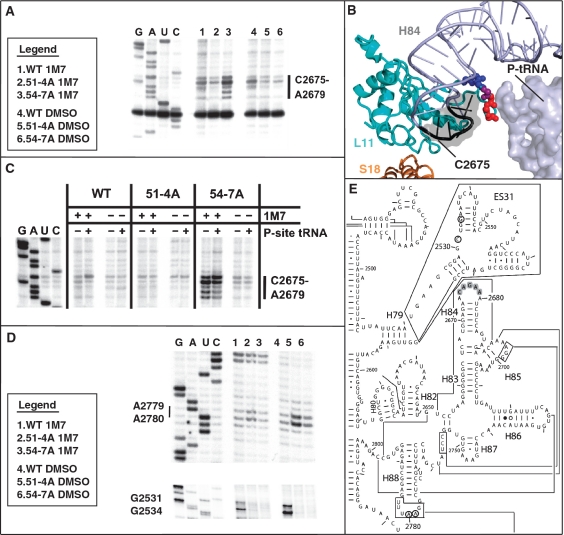
Localization of L11 within the ribosome. (A) Image of the yeast ribosome. The large subunit is colored green, and the small subunit is pink. L11 (cyan) is located in the central protuberance of the large subunit where it interacts with 5S rRNA, Helix 84 of 25S rRNA, the T-loop of the peptidyl-tRNA, and the small subunit protein S18 via the B1b and B1c intersubunit bridges. (B) Close-up view of L11 and neighboring structures. Amino acids of L11′s P-site loop targeted for mutation (R51-R61) are colored deep blue. (C) L11 P-site loop amino acids, mutations analyzed in the current study, and their viabilities as the sole form of L11. Y52* represents multiple mutations: Δ, A, R, E, S, I, Q, N, H and F. Ribosomal structures generated in PyMol using yeast cryo-EM (4) with tRNAs from T. thermophilus (5).
In yeast, L11 is encoded by the paralogous genes RPL11A and RPL11B located on chromosomes 16 and 7, respectively (13). The 19-kDa proteins are 174 amino acids long and are identical except for an alanine (L11A) to threonine (L11B) difference at the third amino acid position. Analysis of L11 in the late 1980s (a.k.a. L16) showed that expression of either isoform was sufficient for cell viability (14). However, when expressed as the sole form of L11, RPL11A mRNA transcripts accumulated to only 33–40% of wild-type levels as compared to cells expressing both isogenes, while RPL11B mRNAs accumulated to 60–66%. Expression of either isogene alone also affected 60S subunit assembly: a strain expressing only L11B grew at wild-type rates but synthesized fewer 60S subunits than wild-type cells (although apparently not below a threshold necessary for wild-type growth rates), while strains expressing only L11A grew more slowly than wild-type, and synthesized only 33–40% of wild-type levels of total L11 and 60S subunits (14). A random mutagenesis screen of RPL11B for cold-sensitive mutants identified alleles that promoted 25S pre-rRNA processing and initiation defects (15). The specific mutants identified in that study were S34P, S41P, S97F, A98V, S119C and G135D. In Arabidopsis, divergent 5′ untranslated regions (UTRs) between the two isogenes were found to result in differential expression among plant tissues (16). In addition to its function as a ribosomal protein, L11 has been implicated in p53 activation through its interactions with HDM2 in the nucleus of human fibroblast cells (17), and mutant forms of L11 have been linked to Daimond–Blackfan anemia in humans (18).
Although the structural information suggests that L11 should play a significant role in translation, functional analyses of the protein in this role have not been performed. In this report, a series of mutants were generated using a reverse genetics approach to parse the role of the L11 P-site loop. Detailed biochemical and structural analyses focused on two multi-amino acid mutants with opposing effects on rRNA structure and tRNA binding. We propose that prior to peptidyltransfer, the presence of peptidyl-tRNA in the large subunit P-site positions the L11 P-site loop to interact with the Helix 84 of the large subunit rRNA. After peptidyltransfer, spontaneous translocation of the deacylated tRNA to the large subunit E-site allows the L11 P-site loop to extend into the P-site, breaking contact with Helix 84. By this model, we hypothesize that the L11 P-site loop functions locally as a sensor of the occupancy status of the ribosomal P-site.
MATERIALS AND METHODS
Strains, plasmids and media
Restriction enzymes were obtained from Promega (Madison, WI, USA), MBI Fermentas (Vilnius, Lithuania) and Roche Applied Science (Indianapolis, IN, USA). The QuikChange XL II site-directed specific mutagenesis kit was purchased from Stratagene (La Jolla, CA, USA). DNA sequencing was performed by Genewiz (Germantown, MD, USA). Escherichia coli DH5α was used to amplify plasmid DNA. Transformation of yeast and E. coli and were performed as previously described (19). YPAD, SD and 4.7 MB plates for testing the killer phenotype were as previously reported (19). Plasmids for expression of dual luciferase reporters were described previously (20).
Saccharomyces cerevisiae strain PSY2088 (MATα rpl11a::HIS3 rpl11b::HIS3 ura3-52 leu2Δ1 trp1Δ63 his3Δ200 + YCpL11B URA3), an rpl11a/rpl11b gene deletion strain in which L11 is supplied by a URA3-CEN6 based RPL11B clone, was a generous gift from Dr. Pamela Silver (21). The L-A and M1 viruses were introduced into PSY2088 by cytoplasmic mixing (cytoduction) through nonproductive mating with JD758 [MATa kar1-1 arg1 (L-AHN M1)] to produce the Killer+ strain JD1313 as previously described (19). Wild-type RPL11B was isolated from yeast strain PSY2088 plasmid (pYCP50L11B URA3). Using flanking BamHI restriction sites, a 2.2-kb fragment of DNA containing both the 525-bp wild-type RPLL11B ORF plus the native 5′ and 3′ UTR regions (1228 bp and 485 bp, respectively) was purified by agarose gel electrophoresis. This 2238-bp fragment was ligated into BamHI digested pRS314, a low copy TRP1-selectable plasmid (purchased from ATCC, Manassas, VA, USA) (22) to create pRS314L11B-TRP1. This plasmid served as the template for generation of rpl11B mutants by site directed mutagenesis using the primers listed in Supplementary Table S1. Wild-type and mutant pRS314L11B-TRP1 clones were transformed into JD1313, selected for growth on –trp medium, and cells having lost the URA3-based plasmid were identified by their ability to grow in the presence of 5-fluoroorotic acid (5-FOA) (23).
Cell growth and drug resistance/sensitivity phenotypes
The effects of temperature and translational inhibitors were assessed by standard 10-fold dilution spot assays. Yeast were grown in H-tryptophan synthetic deletion (SD) media (–Trp) to mid log phase. OD595 values were obtained, and cells were serially diluted 10-fold from 105 to 1 CFU per 2.2 µl and spotted on –Trp plates. Growth was monitored at 20°C, 30°C and 37°C, and pharmacogenetic assays utilized 2 mg/ml paromomycin, 40 µg/ml anisomycin or 30 µg/ml sparsomycin incubated at 30°C for 3–5 days. Killer virus assays were performed as previously described (19).
Translational fidelity assays
The dual luciferase reporter plasmids pYDL-control, pYDL-LA, pYDL-Ty1, pYDL-UAA (20) and pYDL-AGC218 (24) were employed to quantitatively monitor programmed −1 ribosomal frameshifting, programmed +1 ribosomal frameshifting, suppression of a UAA codon and suppression of an AGC serine codon in place of an AGA argine codon in the firefly luciferase catalytic site respectively. In this study, the reporters were housed in LEU2-based reporters: the 0 frame dual luciferase reporter was pJD419, the L-A dsRNA virus −1 PRF containing reporter was pJD420 and the Ty1 containing +1 PRF reporter was pJD421. Cells were grown overnight in 5-ml volumes of –leu synthetic depletion media to mid log phase (A595 = 0.8–1.5). Cells were washed, resuspended in lysis buffer (1X PBS pH 7.4, 1 mM PMSF) and lysed using 0.5-mm glass beads with a vortex mixer for 3–5 min at 4°C. Lysates were clarified by centrifugation for 5 min. at 8000 r.p.m. at 4°C. Samples were maintained on ice, and 5 μl of clarified lysate was added to 50 μl of pre-aliquoted Promega LARII reagent, mixed by pipetting, and read in a TD20/20 luminometer. Immediately upon completion of this read, 50 µl of Promega Stop and Glo buffer was added to the tube, pipetted to mix and read again. This was repeated 6–12 times per strain per reporter depending on the consistency of the data. Frameshifting rates were determined by taking the ratio of firefly to Renilla luciferases for each sample, and then taking the ratio of the average ratios of the 0 frame samples to that of test reporter ratios to obtain the rates for both −1 and +1 PRF. These results were then analyzed by t-test to determine statistical significance compared to wild-type levels as previously described (25). Prior to determining rates of UAA readthrough (nonsense suppression), strains were cured of the endogenous yeast prion [PSI+] by daily serial passage of cells in -trp liquid media containing 5 mM guanidine hydrochloride for 10 days. Rates of nonsense suppression were determined as previously described (20) using the LEU2-selectable 0-frame control pJD419 and in-frame UAA containing reporter pJD702. Missense reporters were based on URA3 plasmids previously described for the sense reporter (20) and for the firefly luciferase 218 arginine codon (AGA) to serine (AGC) missense reporter plasmid pYDL-AGC (24). Methodologies were the same as those for other dual luciferase assays described above.
Ribosome preparation
Cells were grown overnight in a 30°C shaker in 500 ml of YPAD media to mid-log phase (OD595 0.8–1.5), cooled to 4°C for 1 h to allow ribosomes to run off of transcripts while remaining tightly coupled. Cells were harvested by centrifugation and washed three times with 40 ml 0.9% KCl solution. Cell pellets were stored at −80°C until needed, at which time they were thawed and resuspended in 1 ml binding buffer (10 mM Tris–HCl pH 7.5, 10 mM MgCl2, 60 mM NH4Cl, 2 mM DTT, 1 mM PMSF) per gram of cells. Cells were lysed with a 1:1 vol of Zirconian beads (BioSpec, Bartlesville, OK, USA) and disrupted using two 2-min pulses of a minibead beater. Lysates were clarified by centrifugation at 20 000 r.p.m. (50 000 g) using an MSL-50 rotor at 4°C for 25 min. Ribosomes were chromatographically purified using Sulfolink beads (Pierce, Rockford, IL, USA) as previously described (26), and eluted from the resin in 8 ml of elution buffer (10 mM Tris–HCl pH 7.5, 10 mM MgCl2, 500 mM KCl, 2 mM DTT, 0.5 mg/ml heparin). Eluted ribosomes were treated with 2 mM puromycin and 1 mM GTP for 30 min at 30°C and were layered on top of a 22-ml glycerol cushion [50 mM HEPES-KOH pH 7.6, 10 mM Mg(CH3COO)2, 50 mM NH4Cl, 1 mM DTT, 25% glycerol] and pelleted by centrifugation at 30 000 r.p.m. at 4°C for 18–20 h. Pellets were washed with 1 ml of storage buffer [50 mM HEPES-KOH pH 7.6, 10 mM Mg(CH3COO)2, 50 mM NH4Cl, 1 mM DTT, 25% glycerol], and resuspended in 200–400 μl of storage buffer. Concentrations were determined spectrophotometrically (1 OD260 = 20 pmol ribosomes). The salt-washed ribosomes were aliquoted and stored at −80°C for up to 3 months. Ribosomal rRNA quality was checked on 1.3% agarose gels and rRNA to protein ratios were monitored by determining OD260 to OD280 ratios. Polysome profiles were obtained by sucrose density gradient centrifugation as previously described (27).
Assays of ribosome/tRNA interactions
[14C]Phe-tRNAPhe and Ac-[14C]Phe-tRNAPhe species were generated and purified by high-performance liquid chromatography (HPLC) as previously described (28). Salt-washed ribosomes were thawed and activated at 30°C for 5 min. To monitor binding of [14C]Phe-tRNAPhe to the A-site, 125 pmol of ribosomes were pre-incubated at 30°C for 30 min in 150 µl of A-Site Binding Buffer [80 mM Tris pH 7.4 160 mM NH4Cl, 15 mM Mg(CH3COO)2, 2 mM spermidine, 6 mM β-mercaptoethanol] containing 1.33 mM GTP, 0.25 mg polyU, 51 µg soluble binding factors generated as described (29) and 0.1 mg uncharged tRNAPhe. Subsequently, 12.5 pmol of ribosomes were added to 2-fold serial dilutions of [14C]tRNAPhe to a total volume of 30 μl and incubated at 30°C for 35 min. Samples were passed through Millipore (Billerica, MA, USA) nitrocellulose filters using a multi-sample vacuum manifold, filters were washed three times with 3 ml of A-site binding buffer and amounts of bound [14C]Phe-tRNAPhe were determined by scintillation counting. Background values (no ribosome controls) were subtracted from all samples. All strains were tested at least twice in triplicate. Kd values were calculated using GraphPad Prism Software fitted to 1-site binding with ligand depletion formula. To examine binding of Ac-[14C]Phe-tRNAPhe to the P-site, 150 pmol of ribosomes were resuspended in 150 µl of P-Site Binding Buffer [80 mM Tris pH 7.4 160 mM NH4Cl, 11 mM Mg(CH3COO)2, 2 mM spermidine, 6 mM β-me] plus 0.25 mg polyU. Ribosomes (17 pmol) were added to 2-fold serial dilutions of Ac-[14C]Phe-tRNAPhe to a total volume of 30 μl and incubated at 30°C for 40 min. Filter binding assays and determination of steady-state Kd values were performed as described above. Multiple turnover peptidyltransfer reactions utilized 250 pmol of ribosomes pre-loaded with Ac-Phe-tRNAPhe by incubating for 40 min at 30°C with 256 pmol of Ac-[14C]Phe-tRNAPhe in 300 μl of binding buffer (BB) [80 mM Tris pH 7.4 160 mM NH4Cl, 11 mM Mg(CH3COO)2, 2 mM spermidine, 6 mM β-mercaptoethanol] including 0.17 mg of polyuridine. Puromycin was added to each 100-ml reaction to a final concentration of 25 mM, samples were incubated at 30°C and 10 μl samples were taken at 2-, 5-, 10-, 20-, 40- and 60-min time points, added to 90 μl binding buffer plus 100 μl of 1 M NaOH to stop the puromycin reactions. Two 5-μl aliquots were taken from each sample and added directly to 5 ml of scintillation fluid to determine total possible counts. Peptidylpuromycin products were extracted into 400 μl of ethyl acetate, half of which were added to 5 ml of scintillation fluid for counting. Data were plotted by counts per minute versus the percentage of N-Ac-[14C]Phe-tRNAphe reacted. Kobs was determined by plotting first-order time plots using the formula: ln[100/(100 – x)] where ‘x’ is the percentage of tRNA reacted. The first three time points yielded straight lines, the slopes of which are equal to the Kobs values.
rRNA structural analysis using selective 2′-hydroxyl acylation analyzed by primer extension
Empty, polyU loaded, or polyU and deacelated P-site tRNA loaded salt-washed ribosomes (50 pmol) were resuspended in 200 µl of selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) buffer [80 mM Tris–HCl pH 7.4, 100 mM NaCl 15 mM Mg(CHCOOH)2] and incubated for 10 min at 30°C. Samples were split, and 10 µl of dimethyl sulfoxide (DMSO) was added to half of the samples, while 10 µl of 60 mM 1M7 was added to the other half. Samples were incubated at 30°C for 20 min. Ribosomes were precipitated by the addition of 275 µl of ice-cold 100% ethanol and stored at −20°C for 1–2 h. Ribosomes were pelleted by centrifugation at 10 000 r.p.m. for 2 min and resuspended in lysis buffer and rRNAs were isolated using an Ambion (Austin, TX, USA) RNAqueous®-Micro RNA isolation kit. Optical densities were taken at 260 nm and 280 nm to monitor the quantity and quality of RNA, and samples were resuspended at a concentration of 1 µg rRNA/7 µl in pure water. HPLC purified oligonucleotide primers purchased from IDT (Coralville, IA, USA) are listed in Supplementary Table 2. Oligonucleotides were resuspended to 25 pmol/µl, 3′ end labeled with γ[32P]ATP with T4 polynucleotide kinase (Roche, Indianapolis, IN, USA), and purified from free radiolabeled nucleotide by passage through a MicroSpin G-25 column (GE Healthcare, Piscataway, NJ, USA). Annealing reactions utilized 1 µg of modified rRNAs and 3 µl of labeled oligonucleotide heated at 80°C for 1 min, followed by a 4–7-min incubation at 5–10°C below the Tm of each oligonucleotide. Annealed rRNA/primers (2 µl each) were added to 3 µl of cold enzyme mix [0.25 µl 10 mM dNTP, 0.25 µl 100 mM DTT, 1 µl 5X Superscript III Buffer, 0.25 µl Superscript III (Invitrogen Life Technologies, Carlsbad, CA, USA), 1.25 µl H2O]. For sequencing samples, an additional 1 µl of each ddNTP was added to each C, T, A, G, sample, respectively. Primer extension reactions were performed at 52°C for 25 min, with potential 2-min-long extensions preceding the 52°C at lower temperatures depending on the individual Tm values of the primers. Denaturing RNA loading dye (2 µl) was added to each sample, heated to 94°C for 2.5 min, and samples were resolved through 6% urea-acrylamide denaturing gels. Gels were dried and radiolabeled samples were visualized by phosphorimagery.
Computational analyses and ribosome structure visualization
The published structures for the 70S ribosome from E. coli [PDB accession numbers: 2AVY, 2AW4; (2)], as well as yeast 80S structures from yeast (1S1I, 1S1H, 3JYV, 3JYW, 3JYX; (4,12)] were used in the analysis of this work and the generation of figures. Published T. thermophilus 70S subunits containing A-site, P-site and E-site Phe-tRNA were also employed (1G1X, (5). All structures were visualized and manipulated using MacPyMol software (30).
RESULTS
Generation and initial characterization of rpl11b alleles
The visualization of a single salient loop of L11 interacting with peptidyl-tRNA indicated that it might play a vital role in sensing peptidyl-tRNA occupancy status and transmitting this information to other functional centers of the ribosome. As cells expressing RPL11B alone were healthier than those solely expressing RPL11A, genetic manipulations began with the yeast rpl11AΔ rpl11BΔ double knockout strain JD1313 expressing wild-type (WT) RPL11B from a low-copy, URA3-selectable episomal plasmid (pRPL11B-URA3). Oligonucleotide site-directed mutagenesis was used to construct a series of mutants, each containing changes of 1, 4 or 10 sequential amino acids (Figure 1C). Stretches of amino acids from arginine 51 to arginine 61 in the L11 P-site loop were targeted for site-directed mutagenesis expressed from a low-copy, TRP1-selectable episomal plasmid under control of the endogenous RPL11B promoter (pRPL11B-TRP1). After transformation and selection on SD medium lacking tryptophan (–trp), cells expressing only mutant rpl11B alleles were identified by their ability to grow on SD–trp medium containing 5-flouroorotic acid (5-FOA). Three of the multiple substitution mutants were inviable as the sole forms of L11B. These were R51YTVRTFGIR60→alanine (i.e. 51-60A); deletion of residues 51-60 (51-60Δ); and F57GIR60→alanine (57-60A). Viable mutants, R51YTV54→alanine (51-4A), V54RTF57→alanine (54-7A), R51A, Y52* (mutations including Δ, A, R, E, S, I, Q, N, H and F), F57A and R61A were rescued from yeast into E. coli, and the mutations were confirmed by DNA sequencing.
The L11 P-site loop mutants confer temperature- and drug-specific growth phenotypes
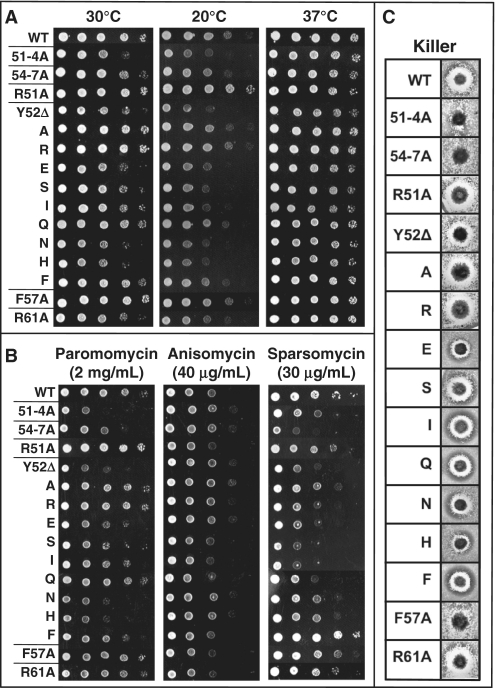
Standard 10-fold dilution spot assays were performed multiple times to measure the effects of the L11 P-site loop mutants on cell growth across a range of temperatures (30°C, 20°C and 37°C) (Figure 2A). Growth at 30°C represents the ideal temperature, and thus all other conditions were compared to this baseline. At 30°C, the 51-4A mutation conferred depressed growth while 54-7A displayed roughly wild-type growth. Cold sensitivity was assessed at 20°C and both mutants grew at wild-type rates. 51-4A showed enhanced growth at 37°C relative to itself at 30°C, while mutant 54-7A was similar to wild type. R51A grew at wild-type rates at 30°C and 37°C but showed enhanced growth at 20°C. The Y52* mutants displayed mutant-specific effects on growth rates at 30°C, but did not confer significant phenotypes at either 20°C or 37°C. F57A had wild-type growth rates at all temperatures, while R61A showed depressed growth at 30°C, which was rescued at 37°C.
Figure 2.
Phenotypic analyses of the viable L11 mutants. (A) 10-fold dilutions of indicated yeast strains were spotted onto SD–Trp media and incubated at temperatures indicated, or (B) on SD–Trp media containing paromomycin, anisomycin or sparsomycin at the indicated concentration and grown at 30°C. (C) Killer virus assays. Wild-type (WT) Killer+ cells are identified by a zone of growth inhibition. 51-4A, 54-7A and F57A mutants lack this halo, indicating the Killer− phenotype.
Small molecule inhibitors of protein translation are useful probes for identifying changes in ribosome function. This study utilized three such molecules: paromomycin, anisomycin and sparsomycin. The effects of all three drugs were monitored using dilution spot assays at 30°C on SD–trp media containing various drug concentrations. Paromomycin is an aminoglycoside antibiotic that increases translational error rates by artificially stabilizing codon:anticodon interactions at the decoding center in the small ribosomal subunit (31). As compared to their intrinsic growth in the absence of drug, both the 51-4A and 54-7A mutants were slightly hypersensitive to 2 mg/ml paromomycin, as were Y52Δ, Y52N and Y52H. In contrast, R51A, F57A and R61A were all paromomycin resistant (Figure 2B). Anisomycin competes with the 3′ end of the aa-tRNA for binding to the A-site pocket of the ribosome (32,33). Both 51-4A and 54-7A showed anisomycin resistance at 40 µg/ml, as did several Y52* mutants, and R61A (Figure 2B). Sparsomycin binds to the P-site and interferes with peptidyl-tRNA binding and peptidyl transfer (33,34). 51-4A and 54-7A mutants were hypersensitive to 30 µg/ml sparsomycin, as were most of the Y52* mutants, with the exception of Y52F, which conferred slight resistance to this drug (Figure 2B).
The yeast ‘killer’ system is composed of the L-A helper and M1 satellite dsRNA viruses (35). The L-A dsRNA viral genome encodes a capsid protein (Gag), and an RNA-dependent RNA polymerase (Pol) that is synthesized as a Gag-pol fusion protein consequent to a −1 Programmed Ribosomal Frameshifting (PRF) event (36). The M1 satellite dsRNA is encapsidated and replicated in L-A encoded viral particles, and the M1 (+) strand encodes a secreted toxin that kills uninfected yeast through its interactions with the GPI-anchored Kre1p cell wall assembly protein (37). Changes in −1 PRF efficiency alter the ratio of Gag to Gag-pol, and inhibit the ability of cells to maintain M1 (19). To monitor the effects of the mutants on Killer virus maintenance, colonies of JD1313 cells expressing either wild-type or mutant rpl11B alleles were spotted onto a lawn of diploid, Killer− indicator cells. Cells expressing wild-type RPL11B were Killer+ as demonstrated by their ability to inhibit growth of the indicator cells (Figure 2C). In contrast, isogenic cells expressing the 51-4A, 54-7A and F57A mutants were Killer−. A weak killer phenotype, defined by decreased zones of growth inhibition, was observed in mutants Y52E, Y52N, Y52H and F57A.
The rpL11B mutants affect translational fidelity
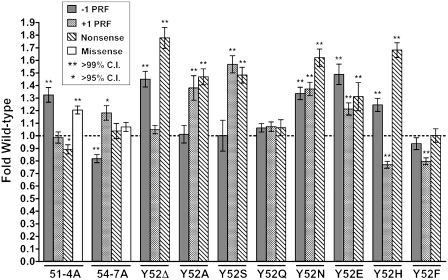
‘Translational fidelity’ is generically used to describe the accuracy of protein synthesis. A series of bicistronic reporter plasmids were used to quantitatively monitor the effects of the L11B mutants on four aspects of translational fidelity: −1 PRF, +1 PRF, suppression of a UAA nonsense codon and incorporation of a missense near-cognate amino acid. In JD1313 cells expressing wild-type RPL11B, −1 PRF directed by the L-A dsRNA viral signal was 6.07% ± 0.16%. This compares favorably with other ‘wild-type’ strains in our laboratory (normal range from 4% to 8% (20,25). The 51-4A mutant promoted increased −1 PRF (1.33 ± 0.06-fold relative to wild type), while 54-7A trended in the opposite direction (0.84 ± 0.03-fold relative to wild type) (Figure 3, and Table 1). Both these values were statistically significant and correlate well with the Killer− phenotypes. Y52Δ, Y52N, Y52E and Y52H mutants also showed increased rates of -1 PRF, with statistically significant rates ranging from Y52H at 1.20-fold wild type to Y52E at 1.49-fold wild-type. Y52A, Y52S, Y52Q and Y52F all had wild-type rates of −1 PRF.
Figure 3.
The L11B mutants promote defects in translational fidelity. Isogenic yeast cells expressing either wild-type or mutant forms of L11B were transformed with dual luciferase reporters and control plasmids and rates of translational recoding were determined. All results are graphed as fold wild type. −1 PRF was measured using the yeast L-A virus frameshift signal. +1 PRF was directed by the frameshift signal derived from the Ty1 retrotransposable element. Nonsense suppression denotes the percentage of ribosomes able to suppress an in-frame UAA termination codon positioned between the Renilla and firefly luciferase reporter genes. Missense suppression rates were evaluated by incorporation of an arginine (AGA) near-cognate amino acid instead of a cognate serine (AGC) at the catalytic codon 218 within the firefly luciferase gene. Error bars denote standard error. P-values are indicated above samples showing statistically significant changes.
Table 1.
Summary of experimental findings for the 51-4A and 54-7A mutants of yeast L11
| Mutant | 51-4A | 54-7A |
| H84 SHAPE | Protected | Deprotected |
| P-tRNA binding | ↓ Kd | ↑ Kd |
| Sparsomycin (P-site) | Hypersensitive | Hypersensitive |
| aa-tRNA binding | ↑ Kd | ↓ Kd |
| Anisomycin (A-site) | Resistant | Resistant |
| Missense | Increased | No effect |
| Paromomycin (decoding center) | Hypersensitive | Slightly Hypersensitive |
| –1 PRF | Increased | Decreased |
| Killer | Killer− | Killer− |
| +1 PRF | No effect | Increased |
While both −1 and +1 PRF are kinetically driven events, the substrates for the slippage are distinct: −1 PRF requires that both the ribosomal A- and P-sites are occupied by tRNAs, while +1 PRF occurs while the A-site is empty (38). Rates of +1 PRF were monitored using a cis-acting signal derived from the Ty1 retrotransposable element using pYDL-Ty1. Baseline +1 PRF efficiencies in cells expressing wild-type RPL11B were 10.98% ± 0.30%. 51-4A had no effects on +1 PRF, while 54-7A promoted a small but statistically significant increase (1.18 ± 0.06-fold of wild type; Figure 3). Significant changes in +1 PRF were also observed in the Y52A, Y52S, Y52N, Y52E, Y52H and Y52F mutants. mRNA decoding occurs in the small subunit decoding center, and changes in termination codon recognition (nonsense suppression) is another indicator of altered translational fidelity. pYDL-UAA (39), which contains an in-frame termination codon immediately 5′ of the firefly luciferase gene, was used to monitor this parameter. The baseline rate of nonsense suppression in cells expressing RPL11B was 0.137% ± 0.003%. The 51-4A mutant slightly improved this aspect of translational fidelity, with nonsense suppression levels decreasing to 0.88 ± 0.04-fold of wild-type levels. 54-7A did not affect UAA recognition (Figure 3). Y52Δ, Y52A, Y52S, Y52N, Y52E and Y52H all promoted increased rates of nonsense suppression ranging from 1.31- to 1.78-fold wild type. pYDL-AGC218 tests missense suppression levels by monitoring rates of incorporation of an arginine (AGA) near-cognate amino acid instead of a cognate serine (AGC) at the catalytic codon 218 within the firefly luciferase gene as previously described (24). Thus, in this assay, mis-utilization of near-cognate tRNAArg at the Ser AGC codon restores firefly luciferase activity. Wild-type missense levels were measured at 0.074% ± 0.002, comparable to previous studies (24). Mutant 51-4A had significantly higher levels of missense suppression (measured at 1.21 ± 0.03-fold wild-type), while 54-7A did not significantly affect this phenomenon (1.07 ± 0.04 fold wild type) (Figure 3). Missense suppression was not assayed for the single amino acid mutants.
The mutant rpl11b alleles promote opposing effects on tRNA binding to the ribosomal A- and P-sites
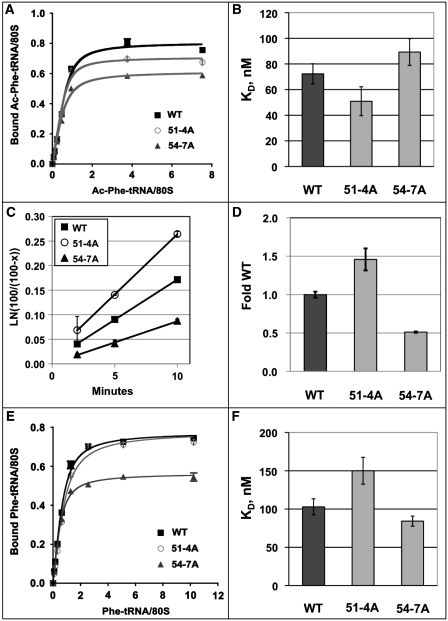
Sucrose gradient analyses were employed to fractionate cycloheximide arrested elongating ribosomes on mRNAs in lysates generated from JD1313 cells expressing wild-type L11B, 51-4A, and 54-7A. In all strains the 60S peak was smaller than that of the 40S fraction which can be attributed to the presence of only a single copy of RPL11B, which has previously been shown to effectively reduce the number of 60S subunits produced by the cell to 60–66% of true wild-type levels while having no visible phenotypic effect on growth (14). No significant differences were observed among the samples (data not shown). Phenotypic variation in PRF and in the presence of anisomycin and sparsomycin are indicative of altered interactions between the ribosome and tRNAs. P-site tRNA Kd values were determined in vitro by binding 2-fold serial dilutions of N-acetylated-[14C]Phe-tRNA to ribosomes until saturation was achieved (Figure 4A), and the resulting data were used to determine steady-state single site binding Kd values (Figure 4B). Wild-type ribosomes bound this P-site substrate with a Kd of 72.3 ± 7.9 nM. The 51-4A mutants promoted a slight increase in affinity for P-site substrate (Kd = 50.9 ± 11.2 nM), while 54-7A had the opposite effect (Kd = 89.3 ± 10.4 nM). Given the physical interaction between the L11 P-site loop and peptidyl-tRNA, it was imperative to determine whether the observed small changes in P-site affinities promoted by the mutants were biochemically significant. To this end, multiple turnover puromycin reactions were performed. In these experiments, puromycin was added to ribosomes pre-incubated with excess P-site substrate, i.e. Ac-[14C]Phe-tRNAPhe, and accumulation of the peptidylpuromycin product was monitored over time. In these reactions, the first round of peptidylpuromycin synthesis is very rapid. Next, in a slow step, the ribosome intrinsically translocates the deacylated tRNAPhe into the E-site (40), followed by the slow diffusion of Ac-[14C]Phe-tRNAPhe into the P-site where it can react with puromycin. Repetition of this cycle results in slow multiple rounds of product synthesis (Figure 4C). Assuming that the L11 mutants do not affect either rates of intrinsic translocation or of Ac-[14C]Phe-tRNAPhe diffusion into the P-site, changes in product accumulation, i.e. Kobs, should be due to differences in binding affinities for the P-site substrate. Consistent with this model 51-4A promoted 1.46 ± 0.14-fold increased Kobs relative to wild-type ribosomes, while 54-7A decreased Kobs to 0.51 ± 0.01-fold that of wild type (Figure 4D). To assay for changes in tRNA binding to the A-site, steady-state Kd values were determined in vitro by binding 2-fold serial dilutions of [14C]Phe-tRNAPhe to ribosomes pre-loaded with uncharged tRNAPhe in the P-site. Wild-type Kd values were 103 ± 11 nM. 51-4A promoted decreased affinity for the substrate with a value of 150 ± 19 nM, while 54-7A promoted a small increase in affinity (84 ± 7 nM) (Figure 4E and F).
Figure 4.
The L11B mutants promote opposing affinities for tRNAs. (A) Binding of tRNA to the P-site. Seventeen picomoles of salt-washed ribosomes were incubated for 40 min at 30°C with 2-fold dilutions of N-acetylated-[14C]Phe-tRNA and poly(U). 80S-tRNA-poly(U) complexes were bound to nitrocellulose filters and washed with binding buffer. Samples were read by radioactive scintillation counting. Curves were generated using GraphPad Prism 4. (B) P-site tRNA binding Kd values were determined using one site binding with ligand depletions equation. Error bars show standard errors. (C) First-order time plots of multiple turnover peptidylpuromycin reaction. Ribosomes pre-bound with N-acetylated-[14C]Phe-tRNA and poly(U) were incubated with puromycin. ‘x’ equals percentage of tRNA reacted. (D) Fold wild-type Kobs rates of peptidylpuromycin product formation. Error bars show standard deviations. (E) Binding of tRNA to the A-site. Salt-washed ribosomes were pre-incubated at 30°C with tRNAPhe to block the P-site, then incubated for 35 min with [14C]Phe-tRNA plus binding factors and poly(U) as described for P-site binding. (F) Kd values for A-site tRNA binding. Error bars show standard errors.
The P-site loop is flexible depending on the occupancy status of the P-site
The highly basic nature of the P-site loop, its interaction with peptidyl-tRNA, and its proximity to 25S rRNA Helix 84 (H84) suggested that it might interact with either of these two RNA components depending on the occupancy status of the P-site. Changes in interactions between the P-site loop and local rRNA structures may in turn propagate outward to more distant regions of the ribosome. To test this, SHAPE (41–43) was employed to probe for structural alterations in selected regions of the 25S, 18S and 5S rRNAs due to either the L11B mutants or in wild-type ribosomes with occupied or unoccupied P-sites. Due to the large size and complex three-dimensional structure of the ribosome, the entire rRNA content was not examined. Rather, approximately one-third of the rRNA bases were interrogated, focusing on those bases closest to L11, the A- and P-sites, and the decoding center.
In the first series of experiments, salt-washed wild-type and 51-4A, 54-7A, Y52Q and Y52F mutant ribosomes (chosen for structural analyses because they had the most pronounced genetic phenotypes) were treated with 1M7, an electrophile that adds an adduct onto the 2′OH groups of solvent exposed base sugars. Modifications were performed on salt-washed ribosomes because they represent the thermodynamic ‘ground state’ of the ribosome. Thus, the structural changes observed are indicative of changes in the full ‘dynamic potential’ of the ribosome as opposed to conformations locked in by e.g. occupation of binding sites by tRNAs or ribosome-associated factors. rRNAs were extracted, hybridized with 5′ [32P]-labeled oligonucleotide primers and reverse transcriptase primer extension reactions were performed. The products were separated through urea-acylamide denaturing gels, and visualized using a phosphorimager. 2′-OH ribose modification results in a strong stop 1-nt 3′ of modified bases, and the intensity of the stops are proportional to the solvent accessibility and flexibility of riboses. Comparison of the protection patterns between wild-type and mutant ribosomes enables identification of specific bases which became protected or deprotected relative to WT.
In all areas examined, rpl11b ribosomes Y52Q and Y52F matched the wild-type rRNA base modification profile (data not shown), while 51-4A and 54-7A ribosomes revealed consistently reproducible differences. The most significant changes in rRNA structure were observed in bases C2675-A2679 (E. coli numbering: C2306-2310) located in the terminal loop of 25S rRNA H84 (Figure 5A and E). The two mutants promoted opposing patterns of base protection/deprotection in this structure. Specifically, as compared to wild-type ribosomes, 51-4A promoted enhanced protection of this loop, while the loop was deprotected in the 54-7A mutants. Analysis of the recent cryo-EM yeast ribosome structure (4) revealed that these H84 loop bases are located within 3 Å of the stretches of amino acids changed to alanines in both the 51-4A and 54-7A mutants (Figure 5B). These findings suggested that the two mutants had the effects of displacing the P-site loop into two opposing conformational states: extended toward the P-site (54-57A), or retracted into H84 (51-54A). To test whether these two states are naturally dependent on P-site occupancy, the experiments were repeated with wild-type and mutant ribosomes with or without tRNAPhe in their P-sites. Consistent with this model, addition of tRNA to the P-site of wild-type ribosomes resulted in slightly enhanced protection of the H84 terminal loop bases closest to the P-site loop (A2676-A2679). Interestingly, C2675 showed significant deprotection when the P-site was occupied by tRNA. This base is on the far side of the terminal end of H84 from the P-site loop, suggesting that H84 itself alters its conformation upon tRNA occupancy of the P-site (Figure 5C). 51-4A’s H84 bases were unchanged between P-site bound and unbound ribosomes, consistent with the P-site loop positioned in the ‘retracted’ state in this mutant, although small differences in the protection patterns suggest that the P-site loop is in a slightly different orientation in this mutant. In contrast, while 54-7A ribosomes, i.e. the P-site loop ‘extended’ state, showed deprotection at all bases (C2675-A2679) for both P-site bound and salt washed ribosomes, bases A2676-A2679 were less deprotected when tRNA was in the P-site and C2675 was even more reactive, consistent with the notion that the P-site loop interacts with H84 when peptidyl-tRNA is in the P-site.
Figure 5.
L11 mutants promote local and distant changes in rRNA structure. (A) 1M7 SHAPE modification of wild-type and mutant salt washed ribosomes show opposite effects of solvent accessibility on H84 bases C2675-A2679 in mutants 51-4A and 54-7A relative to wild type. DMSO lanes are unmodified controls. Sequencing ladder is shown to the left. (B) PyMol generated image of H84′s protected/deprotected bases (black with gray surface) in mutants 51-4A and 54-7A respectively, for salt-washed empty ribosomes. P-site tRNA is added for reference. Blue spheres mark amino acids changed to alanines in 51-4A, red for 54-7A, purple is mutated in both. (C) Changes in H84 accessibility upon binding of P-site tRNA. Ac-Phe-tRNAPhe was pre-bound to salt washed ribosomes along with poly(U) and complexes were probed with 1M7 or DMSO controls. In both wild type and 54-7A base C2675 became more deprotected with tRNA bound to the P-site while bases A2676-A2679 show increased protection. 51-4A’s level of protection is unchanged. (D) Differences observed, in both treated and untreated lanes, of natural stops between wild-type and mutant strains in the terminal loop of 25S rRNA Helix 88 (A2779, A2780), and in Expansion Segment 31 (G2531, G2534). (E) Two-dimensional structure of yeast 25S rRNA showing locations of bases showing changes in reactivity. H84 gray highlighted bases protected/deprotected relative to wild-type in empty salt-washed ribosomes in the 51-4A and 54-7A mutants, respectively. Open circled bases indicate sites of decreased innate lability (C2531, G2534) in ES31 for both 51-4A and 54-7A, or increased natural lability (A2779-A2780) in H88 for the 51-4A mutant.
Although no other SHAPE-specific changes were observed, several other phosphodiester bonds 3′ of specific 25S rRNA bases were reproducibly more, or less, intrinsically labile as compared to wild type (Figure 5D). In both mutants, G2531 and G2534 located in expansion segment 31 (ES31) were more stable than in wild-type ribosomes as evidenced by reduced intensity of strong reverse transcriptase stops 1-nt 5′ of these bases. Additionally, bases A2779-A2780 (E. coli A2407-U2408) located in the terminal loop of Helix 88 were hyper-labile in 51-4A mutant ribosomes as compared to WT, as shown by the presence of strong stops with increased intensity 1-nt 5′ of these bases. These are mapped onto the two-dimensional structure of yeast 25S rRNA (Figure 5E).
DISCUSSION
The L11 P-site loop is largely comprised of polar amino acids and carries a net positive charge, making it ideal for interactions with the phosphate backbones of nucleic acids, e.g. rRNA and tRNA. Positioned between H84 and the peptidyl-tRNA T-loop, several of its amino acids are within H-bonding distance of H84 (∼3.3 Å), while C56 of the peptidyl tRNA T-loop comes within 2.1 Å of G58 in the L11 P-site loop (4,5), suggesting that the L11 P-site loop can directly interact with both of the RNA-based structures. While currently available X-ray crystal structures are unavailable for ratchet-state ribosomes, a recently published examination of tRNA movement through the E. coli ribosome using large-scale analysis of cryo-EM images implicates the P-site loop as a dynamic arm interacting with and moving in relation to tRNAs passing across the P-site (44). Although these studies were performed at resolutions of 9–20 Å, leaving considerable ambiguity regarding the precise residues involved, they clearly reveal highly dynamic interactions between the P-site loop and both P-site, and E-site tRNAs.
Although death is not a phenotype per se, the inviable mutants are informative nonetheless in so far as they demonstrate that the amino acids F57GIR60 are absolutely required for viability. While F57 is universally conserved, it does not appear to be essential on its own for viability, as witnessed in the mild phenotypes of the F57A mutant. Similarly, all single amino acid changes explored here resulted in viable cells, suggesting a certain degree of biochemical/biophysical redundancy within this essential loop. In support of this notion, the strongest growth phenotypes observed across a range of temperatures and small molecule translational inhibitors were concentrated in the multiple alanine substitutions, i.e. 51-4A and 54-7A, thus directing the bulk of the biochemical and structural analyses to these two mutants.
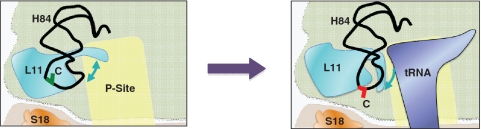
Analysis of the results of the assays performed on the viable multiple alanine substitution mutants (summarized in Table 1) provoke the hypothesis that the L11 P-site loop may dynamically function to help the ribosome sense the occupancy status of the large ribosomal subunit P-site. This is modeled in Figure 6. When the P-site is unoccupied, the P-site loop can extend into this space, moving away from the terminal loop of H84. Upon occupation of the P-site, the peptidyl-tRNA T-loop displaces the L11 P-site loop, causing its retraction into H84. By this model, the rRNA SHAPE analyses depicting increased protection of Helix 84 by the 51-4A mutant show that this mutant drives the L11 P-site loop equilibrium toward the ‘retracted’ state. Conversely, increased deprotection of Helix 84 in the 54-7A mutant suggests that this more mimics the P-site unoccupied state, i.e. the ‘extended’ P-site loop state. This analysis directly explains the P-site binding data. Retraction of the P-site loop from the P-site results in 51-4A ribosomes having higher intrinsic affinity for this substrate while extension of this structure into the P-site creates a steric clash with the peptidyl-tRNA T-loop, resulting in decreased affinity for this substrate. That neither mutant conferred optimal peptidyl-tRNA P-site occupancy may account for their hypersensitivity to sparsomycin, especially for 54-7A in which the P-site loop is already competing with the tRNA for the P-site. Mutants 57-60A, 51-60A and 51-60Δ appear to disrupt the normal function of the P-site loop to a lethal level. In addition, the observation that tRNA binding to the P-site results in deprotection of C2675 implicates H84 itself as a structurally dynamic unit. The functional consequences of this are not clear, although it is tempting to speculate that this conformational change may play a role in the structural rearrangements of the B1b and B1c bridges between the pre- and post-translocational states.
Figure 6.
Model: the P-site loop acts as a sensor of the occupancy status of the P-site. (Left) When the large subunit P-site is unoccupied by tRNA, the L11 P-site loop is able to extend into this space leaving the distal loop of H84 partially deprotected from chemical attack. This conformation is favored by the 54-7A mutant of L11B. (Right panel) Occupation of the P-site by peptidyl-tRNA displaces the L11 P-site loop, causing it to tightly retract from the P-site and interact with H84, resulting in increased protection of the H84 terminal loop from chemical attack. H84 likely moves toward the P-site loop slightly, increasing the exposure of C2675 to the surrounding solvent. This conformation is favored by the L11B 51-4A mutant.
The lack of rRNA structural changes in the A-site or in the decoding center suggest that the biochemical and phenotypic effects observed are indirectly due to the changes described above. The reciprocal effects between Ac-aa–tRNA binding with the P-site and aa–tRNA interactions with the A-site are intriguing. In the aa–tRNA binding reactions, the ribosomal P-sites were occupied with daeacylated tRNA. We suggest that in the 51-4A mutant, the P-site ligand is more ‘locked’ into a suboptimal conformation, which in turn feeds back to the A-site, resulting in decreased affinity for its ligand. Conversely, the lessened ability of 54-7A mutant ribosomes to lock P-site ligand in a suboptimal conformation may account for the increased affinity of these ribosomes for A-site ligand. Anisomycin resistance by both mutants also followed the reciprocal P-site/A-site pattern, i.e. both mutants were sparsomycin hypersensitive. Paromomycin interacts with the decoding center in the small subunit, where it promotes misreading of near-cognate codons in the A-site by stabilizing codon–anticodon interactions (45). This sensitivity may be attributable to an observed increase in missense incorporation of a near cognate arginine (AGA) over that of the sense serine codon (AGC) in mutant 51-4A. Intriguingly, 54-7A had wild-type levels of missense incorporation suggesting that its sensitivity to paromomycin was indirect. The reciprocal anisomycin/paromomycin phenotypes of the L11 mutants demonstrate the effects of this protein on A-site ligand based ribosomal functions over very long distances. Similar phenotypic patterns were previously observed with mutants of other large subunit components (46,47).
The observed effects on -1 PRF are consistent with a recent kinetic analysis demonstrating that aa–tRNA slippage is the most highly weighted parameter in determining the rate at which this process occurs (Liao,P.Y. et al., submitted for publication). Here, increased affinity for aa–tRNA by the large subunit suggests that the 51-4A ribosomes stabilize the frameshifted (i.e. near-cognate) tRNAs, reducing their ability to be proofread, thus promoting increased rates of −1 PRF. This is consistent with the observed increased rates of missense decoding in this mutant. Conversely, post-slippage A-site tRNAs are even less stable in the 54-7A mutants, leading these to be more efficiently proofread, and thus promoting decreased −1 PRF efficiency. In both cases, altering −1 PRF from the optimum ‘golden mean’ precludes these cells from maintaining the yeast killer virus (19,48). Programmed +1 frameshifting is completely dependent on peptidyl-tRNA slippage. Increased +1 PRF in the 54-57A mutant is consistent with decreased affinity for this substrate. The failure to observe decreased +1 PRF in the 51-54A mutant, despite its increased affinity for peptidyl-tRNA, is not entirely clear, although this may be due to the inability of these ribosomes to achieve a threshold beyond which +1 PRF effects can be observed.
The changes in rRNA stability observed in the terminal loop of Helix 88 and in ES31 are intriguing. Chemical protection experiments revealed the terminal loop of Helix 88 is involved in a kissing loop interaction with the terminal loop of Helix 22, and this interaction is apparent in the X-ray crystal and cryo-EM structures (4,49). Increased lability at A2779 and A2780 was previously observed in the Y11C mutant of ribosomal protein L10 (homolog of E. coli L16) located at the base of the aa–tRNA accommodation corridor, and in the Ψ2922C (E. coli U2554) 25S rRNA mutant located in the peptidyltransferase center (50,51). The observation that mutations located in three very different and topologically distinct regions of the large subunit conferred similar structural effects suggest that this kissing loop interaction plays an important role in ribosome function. Its location on the cytoplasmic face of the ribosome where deacylated tRNA leaves the molecule implies that the interaction between the terminal loops of Helices 88 and 22 may be involved in gating this deacylated tRNA exit corridor open and closed. This is consistent with the model of allosteric coordination between the A- and E-sites (52,53), which would indicate that the defects conferred by all of these mutants on aa–tRNA binding might impair this E-site gating function. The decreased lability of C2531 and G2534 in ES31 is similarly intriguing, raising more questions than answers. No function is currently associated with this expansion segment, but recent cryo-EM analysis shows it to be located on a solvent accessible surface of the large subunit (4). Perhaps this site is also involved in A-site/E-site coordination. Alternatively, it may be a site for recognition of defective ribosomes by the nonfunctional ribosome decay apparatus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (R01 GM058859 to J.D.D.). Funding for open access charge: The National Institutes of Health (R01 GM058859).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr. Rasa Rakauskait ė for assistance training, offering technical support and advice above and beyond the call of duty. Further thanks as well to Dr. Arturas Meskauskas, Dr. Karen Jack, Hamid-Reza Shahshahan, Ashton Trey Belew, Dr. Jonathan Leshin and the rest of our laboratory for help and support. We thank Dr. Pamela Silver for providing us with strain PSY2088.
REFERENCES
- 1.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 2.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 3.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae–tRNA- ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 4.Taylor DJ, Devkota B, Huang AD, Topf M, Narayanan E, Sali A, Harvey SC, Frank J. Comprehensive molecular structure of the eukaryotic ribosome. Structure. 2009;17:1591–1604. doi: 10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc. Natl Acad. Sci. USA. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sergiev PV, Kiparisov SV, Burakovsky DE, Lesnyak DV, Leonov AA, Bogdanov AA, Dontsova OA. The conserved A-site finger of the 23S rRNA: just one of the intersubunit bridges or a part of the allosteric communication pathway? J. Mol. Biol. 2005;353:116–123. doi: 10.1016/j.jmb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 10.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 11.Dontsova OA, Dinman JD. 5S rRNA: structure and function from head to toe. Int. J. Biomed. Sci. 2005;1:2–7. [PMC free article] [PubMed] [Google Scholar]
- 12.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leer RJ, Raamsdonk-Duin MM, Mager WH, Planta RJ. The primary structure of the gene encoding yeast ribosomal protein L16. FEBS Lett. 1984;175:371–376. doi: 10.1016/0014-5793(84)80771-1. [DOI] [PubMed] [Google Scholar]
- 14.Rotenberg MO, Moritz M, Woolford JL., Jr Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 1988;2:160–172. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- 15.Moritz M, Pulaski BA, Woolford JL., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol. Cell Biol. 1991;11:5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams ME, Sussex IM. Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. Plant J. 1995;8:65–76. doi: 10.1046/j.1365-313x.1995.08010065.x. [DOI] [PubMed] [Google Scholar]
- 17.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gazda HT, Sheen MR, Vlachos A, Choesmel V, O'Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am. J. Hum. Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinman JD, Wickner RB. Ribosomal frameshifting efficiency and Gag/Gag-pol ratio are critical for yeast M 1 double-stranded RNA virus propagation. J. Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stage-Zimmermann T, Schmidt U, Silver PA. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell. 2000;11:3777–3789. doi: 10.1091/mbc.11.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1990. [Google Scholar]
- 24.Plant EP, Nguyen P, Russ JR, Pittman YR, Nguyen T, Quesinberry JT, Kinzy TG, Dinman JD. Differentiating between near- and non-cognate codons in Saccharomyces cerevisiae. PLoS ONE. 2007;2:e517. doi: 10.1371/journal.pone.0000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs JL, Dinman JD. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 2004;32:e160–e170. doi: 10.1093/nar/gnh157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leshin JA, Rakauskaite R, Dinman JD, Meskauskas A. Enhanced purity, activity and structural integrity of yeast ribosomes purified using a general chromatographic method. RNA Biol. 2010;7:1–7. doi: 10.4161/rna.7.3.11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foiani M, Cigan AM, Paddon CJ, Harashima S, Hinnebusch AG. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell Biol. 1991;11:3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Triana-Alonso FJ, Spahn CM, Burkhardt N, Rohrdanz B, Nierhaus KH. Experimental prerequisites for determination of tRNA binding to ribosomes from Escherichia coli. Methods Enzymol. 2000;317:261–276. doi: 10.1016/s0076-6879(00)17019-3. [DOI] [PubMed] [Google Scholar]
- 29.Dresios J, Derkatch IL, Liebman SW, Synetos D. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry. 2000;39:7236–7244. doi: 10.1021/bi9925266. [DOI] [PubMed] [Google Scholar]
- 30.DeLano WL. The PyMOL molecular graphics system. 2006. http://www.pymol.org (2 August 2010, date last accessed) [Google Scholar]
- 31.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 32.Grollman AP. Inhibitors of protein biosynthesis. II. Mode of action of anisomycin. J. Biol. Chem. 1967;242:3226–3233. [PubMed] [Google Scholar]
- 33.Hansen JL, Moore PB, Steitz TA. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- 34.Schlunzen F, Zarivach R, Harms R, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 35.Wickner RB. Double-stranded RNA viruses of Saccharomyces cerevisiae Microbiol. Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dinman JD, Icho T, Wickner RB. A -1 ribosomal frameshift in a double-stranded RNA virus forms a Gag-pol fusion protein. Proc. Natl Acad. Sci. USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breinig F, Tipper DJ, Schmitt MJ. Kre1 p, the plasma membrane receptor for the yeast K1 viral toxin. Cell. 2002;108:395–405. doi: 10.1016/s0092-8674(02)00634-7. [DOI] [PubMed] [Google Scholar]
- 38.Harger JW, Meskauskas A, Dinman JD. An ‘integrated model' of programmed ribosomal frameshifting and post-transcriptional surveillance. TIBS. 2002;27:448–454. doi: 10.1016/s0968-0004(02)02149-7. [DOI] [PubMed] [Google Scholar]
- 39.Harger JW, Dinman JD. Evidence against a direct role for the Upf proteins in frameshfiting or nonsense codon readthrough. RNA. 2004;10:1721–1729. doi: 10.1261/rna.7120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spirin AS. Translocation mechanism of ribosomes. Mol. Biol. 1977;11:1335–1343. [PubMed] [Google Scholar]
- 41.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 42.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 43.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 44.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 45.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 46.Meskauskas A, Russ JR, Dinman JD. Structure/function analysis of yeast ribosomal protein L2. Nucleic Acids Res. 2008;36:1826–1835. doi: 10.1093/nar/gkn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rakauskaite R, Dinman JD. An arc of unpaired “hinge bases” facilitates information exchange among functional centers of the ribosome. Mol. Cell Biol. 2006;26:8992–9002. doi: 10.1128/MCB.01311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plant EP, Rakauskaite R, Taylor DR, Dinman JD. Achieving a golden mean: mechanisms by which coronaviruses ensure synthesis of the correct stoichiometric ratios of viral proteins. J. Virol. 2010;84:4330–4440. doi: 10.1128/JVI.02480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leffers H, Kjems J, Ostergaard L, Larsen N, Garrett RA. Evolutionary relationships amongst archaebacteria. A comparative study of 23 S ribosomal RNAs of a sulphur-dependent extreme thermophile, an extreme halophile and a thermophilic methanogen. J. Mol. Biol. 1987;195:43–61. doi: 10.1016/0022-2836(87)90326-3. [DOI] [PubMed] [Google Scholar]
- 50.Petrov AN, Meskauskas A, Roshwalb SC, Dinman JD. Yeast ribosomal protein L10 helps coordinate tRNA movement through the large subunit. Nucleic Acids Res. 2008;36:6187–6198. doi: 10.1093/nar/gkn643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rakauskaite R, Dinman JD. rRNA mutants in the yeast peptidyltransferase center reveal allosteric information networks and mechanisms of drug resistance. Nucleic Acids Res. 2008;36:1497–1507. doi: 10.1093/nar/gkm1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Budkevich TV, El'skaya AV, Nierhaus KH. Features of 80S mammalian ribosome and its subunits. Nucleic Acids Res. 2008;36:4736–4744. doi: 10.1093/nar/gkn424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dinos G, Kalpaxis DL, Wilson DN, Nierhaus KH. Deacylated tRNA is released from the E site upon A site occupation but before GTP is hydrolyzed by EF-Tu. Nucleic Acids Res. 2005;33:5291–5296. doi: 10.1093/nar/gki833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.