Abstract
PDZ (PSD95/Discs large/ZO-1) domains are ubiquitous protein interaction motifs found in scaffolding proteins involved in signal transduction. Despite the fact that many PDZs show a limited tendency to undergo structural change, the PDZ family has been associated with long-range communication and allostery. One of the PDZ domains studied most in terms of structure and biophysical properties is the second PDZ (“PDZ2”) domain from protein tyrosine phophatase 1E (PTP1E, also known as PTPL1). Previously we showed through NMR relaxation studies that binding of the RA-GEF2 C-terminal peptide substrate results in long-range propagation of side-chain dynamic changes in human PDZ2 [Fuentes, et al., J. Mol. Biol. (2004), 335, 1105-1115]. Here, we present the first X-ray crystal structures of PDZ2 in the absence and presence of RA-GEF2 ligand, solved to resolutions of 1.65 and 1.3 Å, respectively. These structures deviate somewhat from previously determined NMR structures, and indicate that very minor structural changes in PDZ2 accompany peptide binding. NMR residual dipolar couplings confirm the crystal structures to be accurate models of the time-averaged atomic coordinates of PDZ2. The impact on side-chain dynamics was further tested with a C-terminal peptide from APC, which showed near-identical results to that of RA-GEF2. Thus, allosteric transmission in PDZ2 induced by peptide binding is conveyed purely and robustly by dynamics. 15N relaxation dispersion measurements did not detect appreciable populations of a kinetic structural intermediate. Collectively, for ligand binding to PDZ2, these data support a lock-and-key binding model from a structural perspective and an allosteric model from a dynamical perspective, which together suggest a complex energy landscape for functional transitions within the ensemble.
Keywords: PTP-1E, PDZ2, NMR, crystallography, dynamics, relaxation dispersion, residual dipolar couplings, 2H-methyl relaxation
The PDZ (PSD95/Discs large/ZO-1) domain family is one of the most abundant protein interacting modules found from bacteria to humans, with over 200 PDZ domains encoded in the human genome (1-3). While they influence diverse functions in the cell, they are typically involved in targeting and assembly of multiprotein signaling complexes at synapses or other membrane proximal loci. PDZ domains fulfill this function through their facility in binding C-termini sequences (4-7 amino acids) of target proteins. They are often found in tandem arrays within a PDZ-containing protein, consistant with their role as scaffolds for association with membrane receptors, enzymes and ion channels (1). They share a common fold, consisting of 2 α-helices and 6 β-strands, with the second α-helix (α2) and second β-strand (β2) forming the canonical peptide binding groove (4).
In addition to scaffolding, numerous studies indicate that PDZ domains can have more direct regulatory functions. In particular, a subset of PDZs has now been characterized as displaying allostery (5-10). This is exemplified by the PDZ domain from Par6, which, upon binding CDC42 to the adjacent semi-CRIB motif contacting the PDZ at an interface away from the peptide binding groove, undergoes a conformational change at the binding groove (5). There is also recent evidence for interdomain allostery with PDZs (8, 11-12). Thus, while all PDZs have the capacity to serve as “passive” scaffolds, at least a subset appear to possess higher-order functional roles (13). A central question in the PDZ field is, what distinguishes allosteric PDZs from simple scaffold PDZs, and to what degree are allosteric properties conserved? Further, although only some PDZs have “active” functions, are some properties related to these functions found in all PDZs because they either derive from a common descendent or those properties are intrinsic to the PDZ fold? Interestingly, although many PDZ structures have been determined in the absence and presence of ligands, observations of large conformational changes in PDZ domains have been rare (14). Thus, much of the exploration of potential allosteric effects in PDZs have focused on more subtle origins than gross conformational change (see below).
As a result of such questions, during the last decade PDZ domains have been selected for biophysical study of their internal signaling properties. In 1999, Ranganathan and coworkers used sequence covariation analysis to reveal an evolutionarily conserved energy transmission pathway that connected to a key residue in the peptide binding site (15). Specific PDZ domains were subsequently tested for intramolecular energy propagation using perturbation-response approaches (16-18), and analogous computational methods were developed that revealed PDZ-specific communication pathways (19-23). These studies demonstrated that perturbation at localized positions in PDZ domains cause changes in dynamic fluctuations that propagate to more distal regions of the domain. They also have typically focused on two specific PDZ domains: PDZ3 from postsynaptic density-95 (PSD-95), and PDZ2 from the protein tyrosine phosphatase PTP1E/PTPL1. Hence, “PDZ3” and “PDZ2” have emerged as the preferred PDZ domains for biophysical studies. Because of their representative status, gaining complete structural, dynamic, and biochemical information on these systems is highly desirable for fundamental understanding of PDZ domain function.
Historically, long-range effects (e.g. allostery) have been associated with conformational change. Thus, to understand how certain PDZ domains carry out their active functions, it is necessary to evaluate both structural and dynamic features of these systems. The archetypal PDZ domain is the third PDZ domain (“PDZ3”) from PSD-95. Early structural studies demonstrated a lack of significant structural change upon binding C-terminal peptide ligand (24). Recently, Petit et al. showed that PDZ3 is indeed allosteric and that the mechanism of allostery is not structural, but resides in the conformational entropy of side-chain dynamics (9, 25). In the case of PDZ2 (second PDZ from PTP1E/PTPL1, human form), the issue of structural change upon ligand binding is less clear. Several NMR structures have been reported for PDZ2. Human PDZ2 was reported for the apo state (26) and bound to RA-GEF2 peptide (27). Although the backbone RMSD (using mean structures) between these two structures is 1.3 Å, with some subtle shifting of α2 upon peptide binding, clear conformational changes were not mentioned (27). Mouse PDZ2, which differs by 6 amino acid substitutions (mostly in loops), was reported for the apo state (28) and bound to the APC peptide (18). Subtle but significant structural changes were found upon APC binding, with a change in the tilt of α2 of 10° (18). One complication in interpreting these NMR structures is that the free mouse and human do not agree very well and there appear to be some statistical problems with human PDZ2, as pointed out (28). In addition, none of these structures agree well with residual dipolar coupling (RDC) measurements reported here. Thus, at least for human PDZ2 binding the RA-GEF2 peptide, the question of conformational change has remained unresolved. As a result, in our previous study of side-chain dynamics in PDZ2 we concluded that a substantial role of structural changes in dynamic propagation could not be excluded (16).
In addition to the role of dynamics in intramolecular signaling in PDZ domains, dynamics has also been proposed to be important for PDZ domains' binding promiscuity and specificity (29-32). Specific PDZ domains can bind to different classes of peptide ligands, and conversely, different PDZs are known in some cases to bind the same ligand (33). Still unknown is how specific PDZ domains achieve the optimal balance between promiscuity and specificity, an issue also important for PDZ targeted drug design (34-35). The origin of PDZ binding promiscuity is an active area of research.
Because of the popularity of PDZ2 for structure-based biophysical studies of folding (36-40), binding (8, 18, 31, 39), and energy transmission (16-17, 21-22, 41-42), the lack of reliable structural models for free and peptide-bound PDZ2 has compromised the interpretations of these studies and threatens to discourage future work on this model system. Without good structural information, it is impossible to weigh the balance of structure and dynamics in PDZ2, and, by extension, in PDZ domains. Here, we have determined the structural coordinates of apo and RA-GEF2 bound human PDZ2 using X-ray crystallography to resolutions of 1.65 and 1.3 Å, respectively. The coordinates were found to be consistent with solution NMR RDC measurements, thus indicating that the structures also represent (time-averaged) PDZ2 faithfully in solution. Overall, changes in PDZ2 structure upon binding RA-GEF2 peptide are very small with RMSD of 0.3 Å. In addition, to test the robustness of our previous finding of propagation of dynamic changes in PDZ2 and to gain insight into binding specificity, we also characterized dynamic propagation upon binding a C-terminal peptide from APC, using 2H methyl relaxation. These results show that both RA-GEF2 and APC peptide binding induce highly similar long-range perturbative effects to ps-ns side-chain dynamics, and this propagation is not driven by structural changes. Finally, to gain insight into the mechanism of peptide binding, both RA-GEF2 and APC peptides were investigated for their binding kinetics at the site-specific level using NMR relaxation dispersion methods.
Experimental Procedures
Protein expression and purification
The second PDZ domain (1361-1456) from human PTP1E/PTPL1 was sub-cloned into pET21 vector as described (16). Protein was overexpressed in the BL21(DE3) cell line in LB or M9 minimal media. Cells transformed with PDZ2 vector were induced with 1 mM IPTG and grown at either 22 or 37 °C overnight for protein expression. PDZ2 was purified using the same procedure as reported (16) and verified by mass spectroscopy. For crystallization, protein was exchanged into buffer containing 50 mM NaCl and 20 mM Tris-HCl pH 6.8. For NMR study, protein was dissolved in 150 mM NaCl and 50 mM sodium phosphate pH 6.8, and 10% D2O. To prepare isotope-labeled samples for NMR, isotopically enriched chemicals (15NH4Cl, U-13C6 (99%) D-glucose, and D2O) were used in the minimal media.
Peptide preparation
RA-GEF2 peptide (Ac-ENEQVSAV) was a product of GenScript (Piscataway, NJ). The peptide concentrations were determined by PULCON (43-44). APC peptide (GSYLVTSV) was chemically synthesized with F-MOC modified amino acids using solid phase methods (45). The peptide product was purified by HPLC using a reversed-phase C18 column and acetonitrile gradient. The identity and purity of the resultant peptide was checked by mass spectrometry. The APC peptide stock concentration was determined by UV absorbance with an extinction coefficient of 1490 cm−1M−1 at 280 nm.
Crystallization
The apo and RA-GEF2 bound PDZ2 crystals were obtained using the hanging drop diffusion method. Apo-PDZ2 was crystallized via mixing 1.5 μl of 60 mg/ml protein and 1.5 μl of well buffer containing 28% PEG 3350, 0.2 M KI, 0.2 M NaSCN, 0.1 M sodium acetic acid pH 4.5, and 5% 2-propanol at room temperature. RA-GEF2 bound PDZ2 co-crystals were obtained in 20% PEG 3350, 0.2 M NaSCN, 0.8 M (NH4)2SO4 and 0.1 M sodium citrate pH 5.5 in the presence of 10 mM RA-GEF2 peptide at 4 °C. It should be noted that these crystallization solutions served as effective cryoprotectants. In the case of the complex, incomplete mixing of PEG and (NH4)2SO4 likely led to high local concentrations of PEG. Therefore, the crystals of free and peptide-bound PDZ2 were directly flash-frozen using liquid nitrogen for storage without additional cryo protection step.
Structure determination and refinement
The apo and peptide bound PDZ2 domain crystal diffraction data were collected in beamline X29A of the National Synchrotron Light Source at Brookhaven National Laboratory. Both data sets were collected with X-ray wavelength 1.0809 Å at 100 K (Table 1). Space groups were determined using xtriage (46). The integrated and scaled data by HKL2000 (47) were applied to AMoRe integrated in the CCP4 package for molecular replacement (48). To build the initial apo structural model, PDZ2 from SAP97 (49) (PDB ID: 2AWX) was used as a search model. The apo structure was processed further with alternating rounds of refinement by REFMAC (50) and phenix.refine (51) and manual model building by Coot (52). TLS refinement with phenix was applied with TLS parameters from the TLSMD server (53). Densities for the iodine ions, which were added during the crystallization process, were characterized utilizing Bijvoet difference maps. For the peptide-bound structure model, the apo PDZ structure was utilized as a search model for molecular replacement, but without peptide coordinates to rule out phase bias. Peptide electron density was clearly visible after the first cycle of refinement and then filled with peptide model. The peptide-bound structure was also processed with alternating rounds of refinement by REFMAC (50) and phenix.refine (51) and manual model building by Coot (52). During refinement with phenix.refine, the individual anisotropic ADP refinement option was utilized. Both structures have weak additional electron densities occupying the non-protein space that are modeled with water molecules.
Table 1.
Data Collection Statistics
| Apo PDZ2a | RA-GEF2 PDZ2a | |
|---|---|---|
| Wavelength (Å) | 1.0809 | 1.0809 |
| Resolution Range (Å) | 50-1.65 | 50-1.3 |
| no. of reflections | 1216653 | 460997 |
| no. of unique reflections | 74902 | 34689 |
| Completeness (%) | 98.6 (73.6)a | 99.1 (97.7) |
| Space group | P212121 | H32 |
| Cell parameters (Å) | 63.023 95.148 101.989 | 73.965 73.965 134.056 |
| Cell angles (deg) | 90 90 90 | 90 90 120 |
| Average redundancy | 16.2 (10.8) | 13.3 (10.7) |
| Rmerge (%) | 11.4 (85.6)b | 5.6 (18.0) |
| <I/σ> | 19.232 | 47.456 |
The values in parentheses are for the highest resolution shell.
Rmerge = Σh Σi | Ii (h) - <I(h)>|/ΣhΣiIi(h), where Ii(h) is the intensity of an individual measurement of the reflection and <I(h)> is the mean intensity of the reflection.
NMR spectroscopy
All NMR experiments were carried out at 25 °C (calibrated using methanol) on 500 and/or 600 MHz Varian Inova spectrometers. The protein concentration used was 1 mM. To prepare peptide saturated protein samples, RA-GEF2 or APC peptide was added to a peptide:protein ratio of 1.8:1. Protein concentrations were determined by UV absorbance with ε280=1490 cm−1M−1. All NMR spectra were initially processed by NMRPipe (54) and subsequently applied to NMRView (55) or in-house programs lab for further analysis.
RDC data collection and analysis
Using the IPAP-HSQC experiment (56), 15N-1H RDC data were collected for isotropic and anisotropic samples on a 500-MHz magnet. Proteins were aligned by axial stretching of a 6-mm polyacrylamide gel (6%) into a 5-mm NMR tube (New Era Enterprises, Inc., Vineland, NJ) (57). The residual dipolar couplings were extracted using the RDC module of NMRPipe. Q-factors of RDC data were calculated by REDCAT (58). The residues in flexible loops, termini together with overlapping resonances were excluded in RDC data analysis.
Binding affinities and populations
The binding affinity between the RA-GEF2 peptide and PDZ2 was determined by fluorescence and further confirmed by NMR titration. The two methods produced the same Kd of 10 μM. The APC-PDZ2 binding affinity was also measured by NMR titration, yielding a Kd of 10 μM (Fig. S3). With Kd, [peptide], and [PDZ2] known, the populations of peptide-bound and unbound PDZ2 can be calculated as:
| (eq. 1) |
, and
| (eq. 2) |
where [PB] is the peptide-bound PDZ2 population and [PA] is the unbound PDZ2 populations.
15N Relaxation dispersion
15N Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion experiments were carried out using compensated CPMG pulse sequence (59). For all PDZ-peptide complexes, time delays between consecutive 180° CPMG pulses were set as 0.556, 0.652, 0.75, 0.936, 1.25, 1.5, 1.875, 2.5, 3, 3.75, 5, 7.5, and 15 ms. The total relaxation time in CPMG train was 60 ms. The RA-GEF2 bound PDZ2 relaxation dispersion data were acquired at two sub-saturated states with peptide:protein molar ratios of 1:19.6 and 1:1.97, respectively. APC bound relaxation dispersion data were collected at a single peptide:protein ratio of 1:19.6. The relaxation dispersion data were collected on 500 and 600 MHz spectrometers in an interleaved manner.
Relaxation dispersion curves were fitted both locally (residue-specific fits) and globally using the in-house program exrate (60). For global fitting, a single exchange rate kex, a single [PA], and residue-dependent Δω and R20 values were fitted using the general Carver-Richards expression (61). For the sample with ∼5% saturation, [PA] obtained from global fitting was 94.7%, in excellent agreement with 94.8% based on known Kd and concentrations. We found that fitted Δωs agreed very well with the directly observed chemical shift differences between free and fully saturated PDZ2 (Δωtitration). For local fitting of individual residues, the [PA] [PB] product was set as a known constant (based on the global fit). In the local fits, the better of the two fits between use of the general or “fast” models was determined based on agreement of ΔωCPMG with Δωtitration.
ps-ns dynamics
Backbone and side-chain dynamics of APC-bound PDZ2 was studied in the same manner as for the RA-GEF2 complex reported previously (16). Briefly, 15N backbone relaxation experiments were used to collect 15N T1, T2 and {1H}-15N nuclear Overhauser enhancement (NOE) (62) at 500 MHz and 600 MHz. Methyl bearing side-chain dynamics was extracted from 2H relaxation within CH2D isotopomers. IzCz, IzCzDz and IzCzDy relaxation experiments were collected at 500 and 600 MHz and analyzed as described previously (16).
Results and Discussion
Crystal structures of apo and peptide-bound PDZ2
In order to detect conformational changes resulting from peptide binding, crystallography was employed to determine structures of PDZ2 in the absence and presence of an 8-mer C-terminal peptide ligand from RA-GEF2 (27, 63). Crystals in both forms diffracted X-rays to reasonably high resolution, 1.65 Å for apo PDZ2 and 1.3 Å for RA-GEF2 bound PDZ2, respectively. The final R-factors for apo and peptide-bound PDZ2 are 19.7% and 16.4% respectively (Table 2).
Table 2.
Structure Refinement Statistics
| Apo PDZ2 | RA-GEF2 PDZ2 | |
|---|---|---|
| Resolution Range (Å) | 39.6-1.6 | 29.7-1.3 |
| no. of reflections | 133400 | 34191 |
| R-factor (%)a | 19.7 | 16.4 |
| R-free (%)b | 23.7 | 18.9 |
| no. of non-H atoms | 4167 | 1040 |
| no. of water molecules | 433 | 238 |
| Ramachandranc | ||
| In most favoured regions | 98.0 | 98.3 |
| In additional allowed regions | 2.0 | 1.7 |
| In disallowed regions | 0.0 | 0.0 |
Rcryst = Σh ‖Fobs| - |Fcalc‖/Σh |Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively.
Rfree was calculated as Rcryst using approximately 5% of randomly selected unique reflections that were omitted from the structure refinement. Values in parentheses are for the highest resolution shell.
The Ramachandran analysis is performed using Molprobity.
Apo PDZ2 crystals belong to the P212121 space group. In the asymmetric unit, six monomers are packed to form two layers of three-blade propeller like structures (supporting materials, Fig. S1A and S1B). The average pair-wise Cα RMSD of the monomers is 0.18 Å, indicating all monomers are essentially identical. As expected, the crystal structure solved here conforms to the canonical PDZ domain fold, comprising 6 β-strands and 2 α-helices (Fig. 1A). The second β-strand (β2) and the second α-helix (α2) constitute the peptide binding groove. The RA-GEF2:PDZ2 complex crystals belong to space group R32 (H32). One molecule appears in each asymmetric unit. A hexamer conformation (32 symmetry), generated by crystallographic symmetry, is identical to the hexamer structure in the apo form. Based on calculation of the buried surface area in the hexamer interface by PISA (64), this PDZ domain molecule is expected to exist as a hexamer in solution; however, there is no evidence of this from NMR relaxation (16), which is sensitive to the rate of molecular tumbling, nor are higher-order oligomeric species evident from size exclusion chromatography. In the peptide-bound PDZ2 structure (Fig. 1B), hydrogen atoms were also modeled. In the RA-GEF2 peptide, the five C-terminal residues (QVSAV) show electron density. Using PDZ ligand numbering, counting backwards from the C-terminus, these are residues (0) to (−4). The RA-GEF2 peptide fitted into the binding groove forms an anti-parallel β-strand with protein strand β2. The interaction is further strengthened by packing of the most C-terminal valine side chain with the surrounding hydrophobic patch. The interaction is also stabilized by hydrogen bonding between Ser(−2) and the conserved H71 sidechain, as well as between the backbone of Ala(−1) and R79. In the apo state, the side chain of R79 adopts different conformations in the six different monomers. Upon binding peptide, this apparent flexibility is lost by hydrogen bonding to the carbonyl of Ala(−1). Consistent with previous studies, RA-GEF2 residues back to (−4) are hydrogen bonded with the protein (Fig. 1C) (26). In establishing this intricate hydrogen bonding network, several bound water molecules are also involved (Fig. 1C).
Fig. 1.
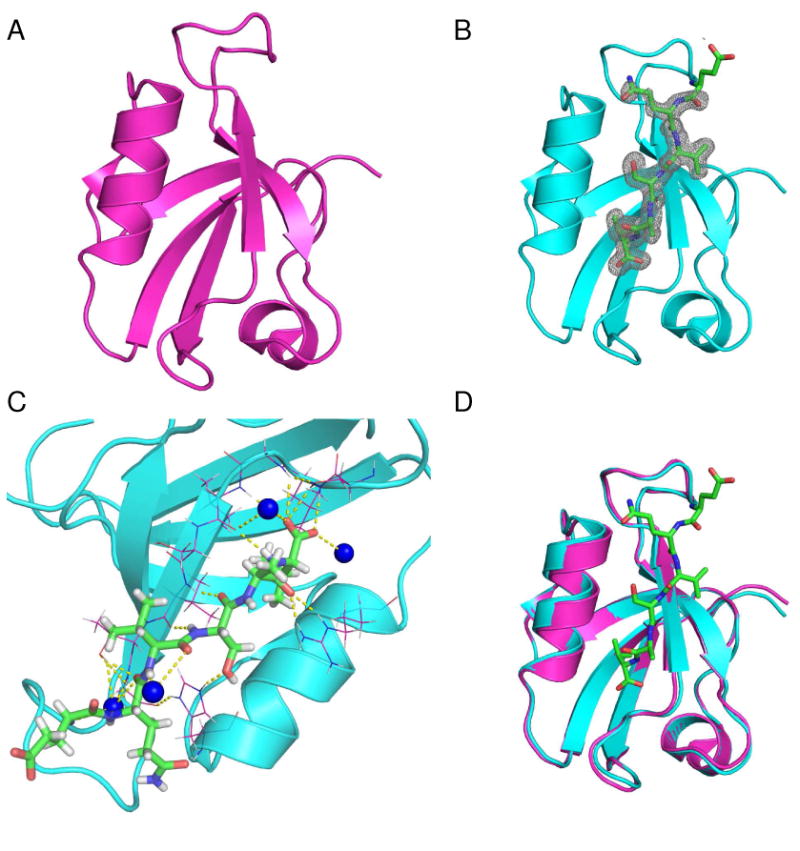
Cartoon representation of PDZ2 crystal structures. Apo (A) and RA-GEF2 bound (B) PDZ2 structures. Peptide is shown as stick model and electron density is shown as gray mesh. The density contour level is 1.5 σ. Peptide residues QVSAV have visible electron density. (C) RA-GEF2 and PDZ2 interaction network. RA-GEF2 peptide is shown by stick model and surrounding PDZ2 residues involved in peptide interaction are shown as lines. Bound water molecules involved in PDZ2 peptide interaction are shown as blue balls. The hydrogen bonds relevant to peptide binding are shown by yellow dotted lines. (D) Structural Superposition of apo (magenta) and RA-GEF2 bound PDZ2 (cyan). The RAGEF peptide is shown by green stick model. All structural graphics were prepared using PyMOL.
All atoms of apo and bound structures have very distinct electron densities, except side-chain atoms of loop residues S29-G33 and terminal residues Q93 and S94. Intriguingly, an irregular 310-helix is identified for residue fragment 30-33 (VRHGG), which is usually characterized as a partially structured loop in NMR structures or other PDZ2 homologues. Compared to the average temperature factor of the free protein (38 Å2), high temperature factors (68 Å2 on average) are observed for this fragment, suggesting high flexibility. Even though B-factors are high, the backbone traces are very similar for all PDZ molecules. This fragment is also involved in crystal packing for both apo and peptide-bound PDZ2, as revealed by crystal lattice packing (Fig. S2). It is thus possible that the 310-helix of residues 30-34 is stabilized in part by the crystal lattice. Nevertheless, in PDZ domains from HtrA proteases, non-canonical helices have been observed in the intervening residues between β2 and β3 (65). Furthermore, 13Cα chemical shifts are consistent with some degree of helicity in solution for residues 31-33 (in both free and RA-GEF2 bound states), with an average (positive) deviation from random coil values of 1.6 ± 0.4 ppm.
These high-resolution structures enable a new assessment of ligand induced conformational changes in PDZ2. As shown in Fig. 1D, no substantial conformational changes are observed: the Cα RMSD of apo and peptide-bound structures is 0.29 Å (0.21 Å if loop residues 26-32 are excluded). This is reminiscent of peptide binding to PDZ3 of PSD95, for which no structural change was found (24), but distinct from the previously published NMR model of APC-bound mouse PDZ2, for which a 10° rotation of α2 was reported (18). The differences between the crystal structures and previously published PTP1E PDZ2 NMR structures are compared quantitatively in Table 3. The RMSDs between crystal and NMR structures range from 0.9 – 2.0 Å. Upon superposition of the apo and bound crystal structures here (excluding α2), RA-GEF2 binding induces a reorientation of α2 of only 2.8°. Thus, based on RMSDs, our crystal structures appear very similar to each other, yet show significant differences from the other PDZ structures. Significant discrepancies are also found among the NMR structures (Table 3), which are either human or mouse forms, even though human PDZ2 (3PDZ and 1D5G) (26, 28) differs from mouse homologue (1GM1 and 1VJ6) (18, 27) by only 6 residues (including 2 conservative mutations). One possible source for these discrepancies is the different methodologies in structure determination. Despite the apparent high resolution, the crystal structures may be influenced by crystal packing effects that introduce structural artifacts and conformational trapping (66-67). Similarly, the NMR structures may suffer from inadequate NOE's to fully define the structure in all regions. Thus a question arises: Are the crystal structures solved here good models for PDZ2 in solution? This prompted us to employ a solution-based approach, residual dipolar couplings (RDC), to further assess the crystal structures.
Table 3.
RMSDs of published PTP1E PDZ2 structures and crystal structures
| RMSD (Å) | b3LNX | 3PDZ | a1GM1 | b3LNYRA-GEF2 | 1D5GRA-GEF2 | a1VJ6APC |
|---|---|---|---|---|---|---|
| b3LNX | - | 1.78c | 0.85 | 0.29 | ||
| 3PDZ | 1.99 | - | 1.90 | 2.09 | ||
| a1GM1 | 0.98 | 2.28 | - | 1.39 | ||
| b3LNYRA-GEF2 | 0.30 | - | 1.32 | 1.21 | ||
| 1D5GRA-GEF2 | 2.28 | 1.83 | - | 1.60 | ||
| a1VJ6APC | 1.69 | 1.54 | 2.05 | - |
Mouse PTP PDZ2, which has 92% sequence identity to human PDZ2.
Crystal structures solved in this research.
The RMSD values above diagonal were calculated based on Cα structure alignments; values below the diagonal were calculated based on all heavy atoms.
Structure validation through solution RDCs
Residual dipolar couplings (RDCs) provide orientation information on internuclear vectors in biomolecules and are widely used in NMR structure calculations and domain-domain docking (68). Alternatively, solution RDCs can be used as a powerful tool to assess the quality of structural models generated without RDC information, which includes, for example, crystal structures. Similar to the R-factor in crystallography, a quality metric called the Q-factor is calculated by fitting experimental RDC data to a structural model (69). The Q-factor varies between 0 and 1, with low Q values indicating high consistency between RDCs and the model, and high values indicating low consistency. Thus, high Q values (> 0.3-0.4) are generally suggestive of low structural quality, assuming that there are no problems/artifacts in the RDCs. Due to intrinsic errors in RDC data collection, the lower limit for Q-factors in practice is around 0.1 (70).
To evaluate all deposited PDZ2 structures (none of which used RDCs in refinement), amide 1H-15N RDC data were collected for PDZ2 in apo, RA-GEF2 bound, and APC bound states. The crystal structures of apo and RA-GEF2 bound forms fitted to their respective RDCs yield low Q values of 0.22 and 0.21 respectively (Table 4). This good agreement suggests that the crystalline PDZ2 structures are not significantly affected by crystal packing and conformational trapping. By contrast, the NMR structures generate significantly higher Q values (from 0.39 to 0.82, Table 4). We note that many of the RDCs were also collected using lipid bicelles, and the Q-factors were very similar (data not shown). Overall, based on the computed Q-factors, the crystal structures reported here represent the average solution features of PDZ2 (apo or bound) significantly better than the existing NMR structures. We therefore expect that these crystal structures will provide more accurate coordinates for molecular dynamics simulation starting structures or structure-based studies of PDZ2.
Table 4.
Q-factors calculated by fitting RDC data to structural models
| RDC datab | Structurea | |||||
|---|---|---|---|---|---|---|
| 3LNX | 3PDZ | 1GM1 | 3LNYRA-GEF2 | 1D5GRA-GEF2 | 1VJ6APC | |
| Apo PDZ | 0.22 | 0.82(0.82) | 0.39(0.56) | 0.29 | - | - |
| PDZ2RA-GEF2 | 0.26 | - | - | 0.21 | 0.67(0.71) | 0.56(0.58) |
| PDZ2APC | 0.20 | - | - | 0.23 | 0.81(0.77) | 0.69(0.75) |
The apo and RA-GEF2 bound PDZ2 structures solved here are 3LNX and 3LNY. For NMR structures, Q-factors were obtained by fitting against the best representative structure of the ensemble. Alternatively, RDCs were fit to bond vector orientations that represent averaging over the NMR ensemble (values in parentheses). All Q-factors were calculated using REDCAT.
In addition, the RDC analysis suggests that an overall lack of change in the time-averaged conformations of PDZ2 in response to peptide binding also holds true in solution. This is evident from the low Q-factors of 0.22 and 0.21 for apo and bound PDZ2 (Table 4). It is also evident upon considering that the apo PDZ2 RDCs are nearly as consistent with PDZ2RA-GEF2 structure as with apo PDZ2 (Q-factors of 0.29 versus 0.22). Conversely, the PDZ2RA-GEF2 RDCs are nearly as consistent with the apo structure as with the PDZ2RA-GEF2 structure (Q-factors of 0.26 versus 0.21). These relatively small differences in Q-factors (0.05, 0.07) are suggestive of subtle structural and/or dynamic differences that exist between free and RA-GEF2 bound forms, although a significant portion of the differences may be due to experimental uncertainty in the RDCs. It is interesting to note that RDCs from the APC-PDZ2 complex fit slightly better to apo-PDZ2 than the RA-GEF2 bound structure (Table 4). We note that the Q-factor fitting included RDCs from α2 and β2 (Fig. S4), which form critical hydrogen bonds with peptide and should report on any structural change. The RDC data here appear to contradict a previous report of a 10° change in α2 orientation, in solution, upon binding the APC peptide (18). However, that was carried out on mouse PDZ2, and it remains possible that mouse and human PDZ2s differ in this respect. We also note that there may be dynamic aspects to α2 in human PDZ2, as suggested from a slightly increased 15N R2 at R79 relative to the other structured regions (in the apo form, data not shown). We speculate that α2 may undergo segmental motion on the ns-μs timescale. In summary, the RDC data are highly consistent with the crystal structures and show that neither RA-GEF2 nor APC peptides induce significant conformational changes to human PDZ2 in solution.
Long-range “pure” dynamic propagation in PDZ2 also results from APC peptide binding
In our previous study of the RA-GEF2 peptide binding to PDZ2 (16), binding was observed to perturb ps-ns dynamics of methyl-bearing side chains not only at the binding site, but also at two surfaces of PDZ2 distal to the peptide binding pocket. At that time, it was unclear to what extent the dynamic propagation was due to changes in peptide-induced structural changes in PDZ2. The combined crystallographic and NMR results here strongly suggest that conformational change does not drive the dynamic changes and that PDZ2 channels the impact of peptide binding as a relatively “pure” dynamic response to distal surfaces 1 and 2 (16). The emerging picture appears to be that a network of residues extends through much of PDZ2. Atom fluctuations around mean positions of the network confer variable force patterns that can transmit perturbations over distances. We note that such behavior has recently been used as a perturbation-response tool in the context of molecular dynamics simulations (20-21, 71-72). Thus, a major event such as peptide binding in the PDZ active site can alter fluctuation patterns well beyond the binding site without significant changes in mean structural positions. The patterns may in some cases manifest as correlated motions, as demonstrated recently for PDZ2 (42). This qualitative model is consistent with the ease of dynamic perturbation by both mutation and ligand binding (73).
To further test this model and potentially increase confidence in the long-range dynamic propagation observed for RA-GEF2 binding, we characterized the methyl side-chain dynamics of PDZ2 bound to a C-terminal peptide derived from the APC protein (74) using 2H relaxation. This peptide (GSYLVTSV) binds with Kd ∼10 μM, similar to RA-GEF2 (Fig. S3). The changes in S2axis and τe upon APC peptide binding are very similar to those in RA-GEF2 (Fig. 2 for S2axis and Fig. S5 for τe). The patterns of changes in S2axis in PDZ2 upon binding either peptide are shown in Fig. 3. In the case of RA-GEF2 binding, propagation was previously observed out to “distal surfaces 1 and 2”, although distal surface 1 is less apparent in Fig. 3A because changes in τe are not shown. In the APC complex, only propagation to distal surface 1 is observed, but the pattern is near identical to that from RA-GEF2, both in terms of residues in the dynamic network and the magnitude of the dynamic response. One residue that shows a different response from the RA-GEF2 complex is at I6 at the N-terminal region of beta strand 1. Construction of a 2-way contingency table based on the presence or absence of significant ΔS2axis values in specific methyl groups in both complexes resulted in a high level of pattern matching, with the Fisher's exact test p-value of 7.4 × 10-4 (Table 5). This high degree of similarity in dynamic responses to RA-GEF2 and APC peptides demonstrates that the propagated dynamic responses are indeed real, reproducible, and more indicative of PDZ2 than ligand sequence (at least in these two cases). In addition, because the 1H-15N RDCs measured for APC-bound PDZ2 agree equally well with the crystal structure of RA-GEF2/PDZ2 (Q = 0.23, Table 4), these data also support pure dynamic propagation. We suggest that these data represent one of the best examples of dynamic propagation – or dynamic signal transduction (75) – detected experimentally and site-specifically, in the absence of conformational changes (25, 76-77).
Fig. 2.
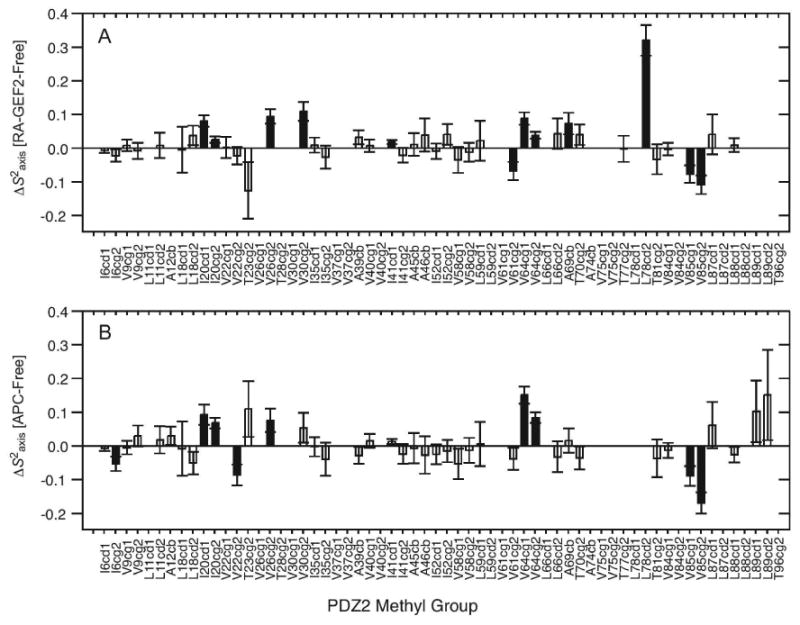
Methyl-bearing side-chain dynamics changes (ΔS2axis) induced by RA-GEF2 (A) and APC (B) binding, with respect to free PDZ2. The methyl groups with significant changes in S2axis (ΔS2axis>2σ) are shown in filled bars. Fig 2A was adapted from Fuentes et al. (16)
Fig. 3.
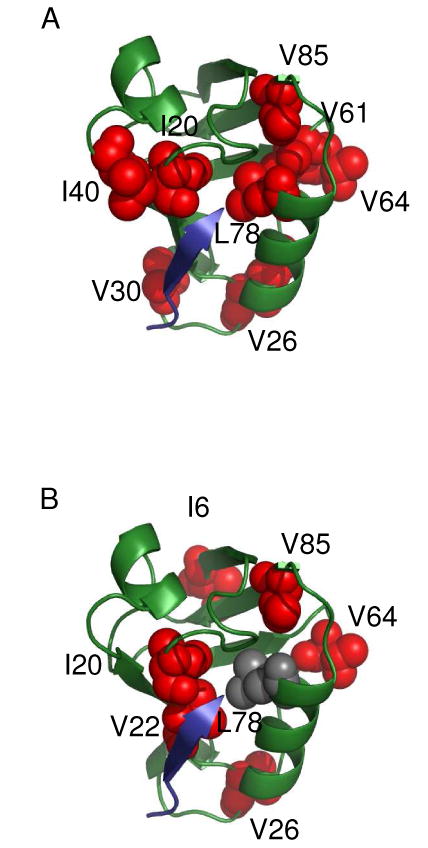
Graphical comparison of side-chain dynamic changes induced by RA-GEF2 (A) and APC binding (B). Red spheres represent residues experiencing significant (ΔS2axis>2σ) side-chain dynamic changes and peptide is shown as blue cartoon. The figures were prepared by PyMOL.
Table 5.
Contingency table showing a correlation between APC and RA-GEF2 induced ΔS2axis
| APC | RA-GEF2 | ||
|---|---|---|---|
| Significant | Insignificant | Total | |
| Significant | 7 | 2 | 9 |
| Insignificant | 4 | 25 | 29 |
| Total | 11 | 27 | 38 |
The p value based on Fisher's exact test is 0.00074.
μs-ms timescale peptide binding dynamics
A previous pre-steady state kinetic study of mouse PDZ2 binding to RA-GEF2 peptide showed that peptide association proceeds through an induced-fit mechanism (18). These kinetic data suggested that PDZ2 undergoes a ligand induced conformational change with kobs of ∼7000 s−1. While the X-ray and RDC data presented above (on human PDZ2) do not support the existence of overall conformational change, it remains possible that conformational changes take place at low populations. To probe this possibility, we investigated μs-ms motions in PDZ2 using Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion methods (61). In principle, this strategy allows monitoring of the kinetics (kex) and structural effects (as interpreted through the difference in chemical shift between states, Δω) of conformational events at the residue level and can detect minor populations as low as 0.5-1% (78).
Microsecond-millisecond timescale dynamics are frequently associated with conformational change, enzyme catalysis, and protein folding (79). 15N CPMG relaxation dispersion experiments revealed that neither apo PDZ2 nor RA-GEF2 saturated PDZ2 exhibit significant μs-ms motion (data not shown). However, for binding interactions of moderate strength (∼micromolar), ligand binding and dissociation can occur on this timescale and are amenable to characterization by relaxation dispersion using sub-saturated complexes (80-83). More specifically, there is the potential for identification of dynamic events that occur during binding. Of interest here, non-two-state behavior was reported recently for peptide binding to the PDZ domain of AF-6, based on relaxation dispersion data (84). To gain insight into the kinetics of binding and ligand specificity with site-specific resolution for PDZ2, 15N CPMG relaxation dispersion experiments were carried out on both RA-GEF2 and APC peptide complexes with 5% or 50% molar amounts of peptide. The lack of μs-ms exchange in the end states of the binding reaction is ideal for interpretation of line-broadening (i.e. relaxation dispersion) due to dynamic cycling of ligand binding and release.
To bring the peptide binding kinetics into an exchange window suitable for characterization by CPMG relaxation dispersion, PDZ2 protein was mixed with substoichiometric amounts of peptide. For dispersion curve analysis, we employed two-site exchange since the above structural studies indicated no evidence for conformational change. A two-site exchange binding process can be described by the following:
| (eq. 3) |
where kon and k off are the on-rate and off-rate of peptide binding, respectively. The exchange rate (kex) is modulated by the free peptide concentration based on the following expression:
| (eq. 4) |
Upon addition of 5% RA-GEF2 or APC peptide, relaxation dispersion was observed for residues along the binding groove and some distal regions. The high quality of the fits in Figure 4 is typical of the entire data sets for both peptide complexes. Local kex and Δω were fit assuming a fixed pA value of 0.95. Fits were carried out using the full Carver-Richards equation, as well as the simplified form for fast exchange (61), and we report the parameters which yielded better agreement with Δω determined from titration. Individually fitted exchange rates (kex), chemical shift changes (Δω) and intrinsic spin-spin relaxation rates (R20) for PDZ2 residues in complex with RA-GEF2 and APC are provided in Tables 6 and 7, respectively. The distribution of kex values were quite uniform, with average kex values of 408 ±127 s and 663±158 s for PDZ2 bound to 5% RAGEF or 5% APC peptides, respectively. Given the similarity of the locally fitted exchange rates, the data for each complex were globally fit to a model in which all residues share the same exchange rate and population, but Δω and R20 are allowed to vary for each residue. The global fitting results are very similar to the local results (Table S1, S2). Importantly, globally fitted PA values were determined to be 94.7% and 94.5% for RA-GEF2 and APC, respectively, in excellent agreement with the predicted fraction of free protein (95%) based on measured Kd values and reactant concentrations. In addition, fitted Δω values (ΔωCPMG) for both peptides are remarkably consistent with the Δω values based on peptide titrations (Δωtitration) (Fig. 4C and 4D). This strongly suggests PDZ2 samples two states (apo and fully bound) in the presence of peptide and these alone are responsible for dispersion. We note however, that there are a few resonances in each system for which we observe divergence between ΔωCPMG and Δωtitration. In the case of the APC complex, all of these outliers have very small Δω values. These discrepancies do not warrant further consideration since it is known that fitting relaxation dispersion with small chemical shift changes is error prone (85). In the case of the RA-GEF2 complex, we find Δω divergence for three residues with significant titration Δω values: G19, S21, and G34. Interestingly, G19 and S21 exhibit the smallest values of kex (255 and 204 s−1) in PDZ2. G19 and S21 are located at the binding pocket (in or near β2), and G34 lies at the end of the β2-β3 loop. Thus, although the majority of resonances in PDZ2 indicate simple two-state binding in the sensitivity regime for 15N CPMG relaxation dispersion, these few residues appear to hint at the existence of a RA-GEF2 binding intermediate localized to the vicinity of the peptide site. The behavior of G19, S21, and G34 is reminiscent of previously observed non-two-state behavior in ligand binding as observed from NMR relaxation dispersion (80, 82, 86). The divergence from two-state behavior here appears to be smaller than in those studies, yet larger than in the case of an SH3-ligand interaction (83). Fits of the dispersion data to a 3-site exchange model was not carried out since this is advised only for when an abundance of dispersion curves is available (87).
Fig. 4.
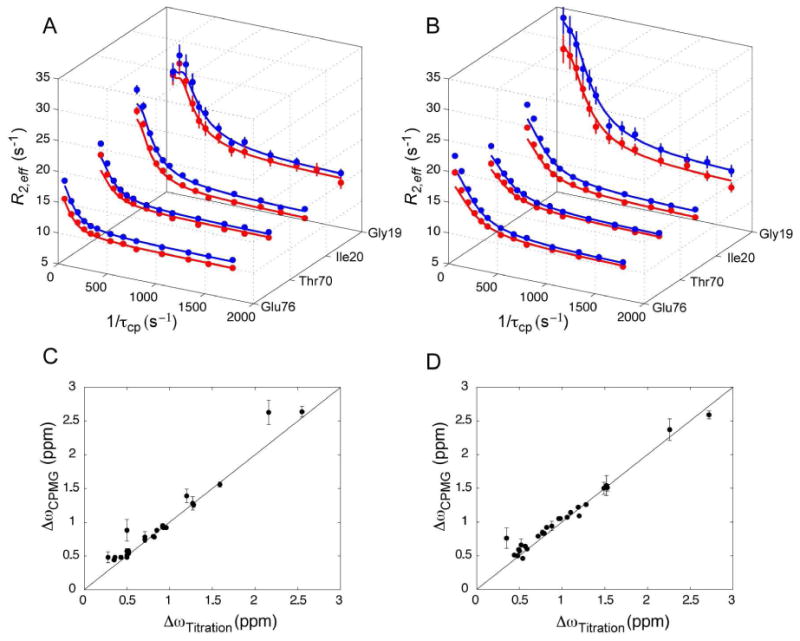
Two-state binding of RA-GEF2 and APC peptides based on 15N relaxation dispersion. Relaxation dispersion curves for select resonances in PDZ2 5% saturated with RA-GEF2 (A) and APC (B) peptides. Data acquired at 500 and 600 MHz (1H Larmor frequency) are shown in red and blue respectively. Data quality for these residues is typical of the entire dataset. In (C) and (D), correlation plots of fitted Δω values from relaxation dispersion and 15N Δω values from peptide titration. ΔωCPMG values are from global fits, as described in the main text. Data for RA-GEF2 and APC are in (C) and (D) respectively. The line is y = x.
Table 6.
Local fitting results of 5% RA-GEF2 bound PDZ2 relaxation data
| Residue | kex (s-1) | ΔωCPMG (ppm) | aR20 (s-1) | bR20 (s-1) | cΔωtitration (ppm) |
|---|---|---|---|---|---|
| 17 | 385±40 | 0.93±0.02 | 9.96 ±0.09 | 10.46 ±0.10 | 0.92 |
| 19 | 255±22 | 2.63±0.18 | 12.80 ±0.38 | 13.44 ±0.31 | 2.16 |
| 20 | 357±21 | 1.56±0.04 | 10.39 ±0.14 | 11.37 ±0.13 | 1.59 |
| 21 | 204±28 | 1.39±0.10 | 10.59 ±0.14 | 11.36 ±0.12 | 1.20 |
| 22 | 478±369 | 0.48±0.08 | 14.69 ±0.26 | 14.41 ±0.22 | 0.28 |
| 23 | 438±87 | 0.48±0.02 | 10.76 ±0.09 | 11.70 ±0.09 | 0.50 |
| 24 | 336±98 | 0.57±0.04 | 12.49 ±0.19 | 12.67 ±0.18 | 0.50 |
| 27 | 446±168 | 1.28±0.10 | 17.09 ±0.52 | 19.95 ±0.57 | 1.27 |
| 31 | 432±154 | 0.78±0.08 | 19.42 ±0.45 | 20.70 ±0.38 | 0.71 |
| 34 | 926±465 | 0.88±0.16 | 14.64 ±0.42 | 17.68 ±0.58 | 0.50 |
| 35 | 480±48 | 0.95±0.03 | 12.72 ±0.14 | 13.88 ±0.16 | 0.92 |
| 40 | 358±43 | 0.51±0.02 | 9.59 ±0.07 | 10.04 ±0.07 | 0.50 |
| 45 | 440±39 | 0.74±0.02 | 10.43 ±0.10 | 11.11 ±0.10 | 0.71 |
| 66 | 482±82 | 0.48±0.03 | 10.85 ±0.08 | 11.49 ±0.09 | 0.43 |
| 67 | 348±29 | 0.54±0.01 | 9.83 ±0.05 | 10.53 ±0.05 | 0.52 |
| 70 | 434±29 | 0.92±0.02 | 11.17 ±0.10 | 11.67 ±0.10 | 0.93 |
| 71 | 386±67 | 0.48±0.02 | 12.94 ±0.10 | 13.38 ±0.09 | 0.36 |
| 72 | 319±33 | 1.26±0.04 | 10.90 ±0.13 | 11.19 ±0.12 | 1.28 |
| 74 | 448±20 | 0.92±0.01 | 10.07 ±0.07 | 10.65 ±0.07 | 0.96 |
| 75 | 412±19 | 0.79±0.01 | 10.53 ±0.05 | 10.98 ±0.06 | 0.81 |
| 76 | 326±32 | 0.88±0.02 | 10.06 ±0.08 | 11.10 ±0.08 | 0.85 |
| 79 | 285±10 | 2.64±0.08 | 11.19 ±0.16 | 11.84 ±0.15 | 2.55 |
| 80 | 407±27 | 0.78±0.02 | 9.43 ±0.08 | 9.85 ±0.09 | 0.82 |
| 81 | 445±44 | 0.48±0.02 | 7.79 ±0.05 | 7.83 ±0.05 | 0.36 |
| 82 | 394±109 | 0.57±0.04 | 14.01 ±0.17 | 13.67 ±0.16 | 0.52 |
| 86 | 384±73 | 0.44±0.02 | 10.73 ±0.07 | 11.30 ±0.08 | 0.35 |
Values at 500 MHz.
Values at 600 MHz.
15N Δωtitration values were calculated as the difference between apo and RA-GEF2 saturated PDZ2.
Table 7.
Local fitting results of 5% APC bound PDZ2 relaxation data
| Residue | kex (s-1) | ΔωCPMG (ppm) | aR20 (s-1) | bR20 (s-1) | cΔωtitration (ppm) |
|---|---|---|---|---|---|
| 11 | 480 ±54 | 0.46 ±0.02 | 10.07 ±0.04 | 10.52 ±0.05 | 0.54 |
| 17 | 690 ±25 | 1.14 ±0.02 | 9.84 ±0.08 | 10.28 ±0.09 | 1.10 |
| 19 | 550 ±110 | 2.37 ±0.16 | 12.16 ±0.41 | 13.4 ±0.5 | 2.26 |
| 20 | 640 ±30 | 1.22 ±0.02 | 10.48 ±0.1 | 11.4 ±0.12 | 1.19 |
| 21 | 620 ±36 | 1.09 ±0.02 | 10.71 ±0.1 | 11.17 ±0.15 | 1.20 |
| 22 | 610 ±92 | 0.84 ±0.04 | 14.02 ±0.16 | 14.24 ±0.21 | 0.77 |
| 23 | 570 ±50 | 0.64 ±0.02 | 11.39 ±0.06 | 11.79 ±0.07 | 0.57 |
| 24 | 765 ±65 | 1.51 ±0.05 | 12.47 ±0.23 | 11.86 ±0.3 | 1.53 |
| 25 | 960 ±260 | 1.54 ±0.15 | 13.32 ±0.61 | 13.01 ±0.92 | 1.52 |
| 27 | 720 ±130 | 1.50 ±0.09 | 16.89 ±0.46 | 18.78 ±0.66 | 1.49 |
| 28 | 710 ± 270 | 0.66 ±0.09 | 16.61 ±0.25 | 18.23 ±0.31 | 0.52 |
| 31 | 620 ±140 | 0.94 ±0.07 | 19.2 ±0.28 | 21.09 ±0.41 | 0.88 |
| 34 | 1280 ±580 | 0.76 ±0.15 | 15.49 ±0.26 | 18.32 ±0.4 | 0.35 |
| 35 | 610 ±47 | 0.92 ±0.02 | 13.16 ±0.11 | 14.26 ±0.14 | 0.82 |
| 45 | 610 ±46 | 0.79 ±0.02 | 10.67 ±0.07 | 11.24 ±0.09 | 0.72 |
| 46 | 510 ±57 | 0.60 ±0.02 | 10.36 ±0.07 | 10.87 ±0.09 | 0.59 |
| 54 | 660 ±56 | 0.51 ±0.02 | 10.94 ±0.05 | 11.19 ±0.06 | 0.44 |
| 66 | 490 ±70 | 0.50 ±0.03 | 11.34 ±0.07 | 11.6 ±0.08 | 0.48 |
| 70 | 690 ±33 | 1.05 ±0.02 | 11.2 ±0.08 | 11.56 ±0.11 | 0.96 |
| 72 | 620 ±29 | 1.07 ±0.02 | 10.75 ±0.08 | 11.03 ±0.1 | 1.06 |
| 74 | 570 ±20 | 0.83 ±0.01 | 10.38 ±0.04 | 10.8 ±0.05 | 0.79 |
| 76 | 680 ±26 | 1.26 ±0.02 | 10.33 ±0.09 | 10.73 ±0.1 | 1.28 |
| 78 | 650 ±69 | 0.59 ±0.02 | 10.54 ±0.06 | 11.06 ±0.07 | 0.49 |
| 79 | 690 ±59 | 2.59 ±0.06 | 10.6 ±0.19 | 10.86 ±0.24 | 2.72 |
| 80 | 619 ±23 | 1.05 ±0.01 | 9.71 ±0.06 | 9.86 ±0.08 | 0.98 |
| 81 | 616 ±46 | 0.57 ±0.02 | 7.52 ±0.04 | 8.05 ±0.06 | 0.50 |
Values at 500 MHz.
Values at 600 MHz.
15N Δωtitration values were calculated as the difference between apo and RA-GEF2 saturated PDZ2.
The primarily two-state relaxation dispersion behavior reported here contrasts with the CPMG-derived ligand binding dynamics in the AF-6 PDZ domain, which showed extensive discrepancies between ΔωCPMG and Δωtitration and hence is suggestive of an intermediate state during the binding process (84). However, in the AF-6 PDZ, the apo protein samples different conformations on the millisecond timescale, which complicates the interpretation of peptide binding dynamics. In the sub-saturated complexes of PDZ2, chemical exchange only arises from peptide binding dynamics, leading to tight correlations between ΔωCPMG and Δωtitration (Figure 4C,D). As some PDZ domains are known to change their shape (5, 14), future studies of apo dynamics and ligand binding dynamics on the μs-ms timescale should help to determine how common alternative conformational states in PDZ domains are.
For clean two-site exchange, it is reasonable to expect the on-rate for peptide binding (kon) to approach the diffusion limit. To test this, we calculated kon and k off from the dependence of kex on peptide concentration. To this end, an additional set of relaxation dispersion data were collected with 50% RA-GEF2. The higher ligand concentration pushed exchange rates into the intermediate regime and hence many resonances disappeared. Nevertheless, enough relaxation dispersion curves were obtained to perform global fitting (Table 8). Solving the two linear equations (eq. 4) at the two peptide concentrations (using globally determined kex), the on-rate was determined to be 3.6×107 s−1 M−1, which is approaching the diffusion limit, and the off-rate is 307 s−1, which is very similar to the previously reported value, 270±20 s-1.(18).
Table 8.
Global fitting results of 50% RA-GEF2 bound PDZ2 relaxation data
| Residue | kex (s-1) | ΔωCPMG (ppm) | [PA] (%) | aR20 (s-1) | cΔωtitration (ppm) |
|---|---|---|---|---|---|
| 11 | 554±42 | 0.40±0.03 | 56.5±0.13 | 10±0.13 | 0.38 |
| 17 | 554±42 | 0.89±0.03 | 56.5±0.13 | 0.48±0.04 | 0.92 |
| 23 | 554±42 | 0.92±0.07 | 56.5±0.13 | 12.38±0.56 | 0.50 |
| 35 | 554±42 | 0.98±0.02 | 56.5±0.13 | 0.46±0.03 | 0.92 |
| 39 | 554±42 | 0.30±0.03 | 56.5±0.13 | 11.19±0.12 | 0.27 |
| 66 | 554±42 | 0.99±0.05 | 56.5±0.13 | nc | 0.43 |
| 74 | 554±42 | 0.25±n | 56.5±0.13 | nc | 0.96 |
Values for 500 MHz field.
The experimental δωs were calculated from apo and RA-GEF2 saturated PDZ2.
No reasonable fitting values can be obtained.
Summary
Taken together, the X-ray and NMR results reported here on RA-GEF2 and APC peptides are inconsistent with an induced-fit or conformational selection mechanism of binding to PDZ2, and highly consistent with binding via “lock-and-key”. No significant changes in PDZ2 coordinates are observed between the apo and RA-GEF2 peptide bound crystal structures, which is supported further by 1H-15N RDCs. The absence of significant CPMG relaxation dispersion for apo (or peptide bound) PDZ2 is consistent with lack of conformational change in the crystal structures. We note, however, that a caveat of the relaxation dispersion experiments is that processes faster than ∼100 μs are not detected and hence sampling of intermediate binding states on a timescale faster than this cannot be excluded. In the context of this “rigid” PDZ2 domain, binding of both RA-GEF2 and APC peptides induce very similar patterns of changes in ps-ns side-chain dynamic fluctuations that propagate away from the binding site, forming apparent allosteric pathways. Thus, the primary physical impact of peptide binding to PDZ2 is dynamic and not structural in nature. This has implications for understanding the physical basis for long-range communication and allostery in proteins.
Supplementary Material
Acknowledgments
The authors of the paper thank Karl Koshlap of the UNC School of Pharmacy NMR facility and Greg Young of the UNC Biomolecular NMR Laboratory for technical assistance. We also thank Dan Cline for assistance with peptide synthesis and purification, and Howard Robinson (at BNL) for X-ray diffraction data collection. Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886.
Abbreviations
- NMR
nuclear magnetic resonance
- RDCs
residual dipolar couplings
- S2axis
order parameter characterizing amplitude of methyl symmetry axis motions
- PDZ
PSD95/Discs large/ZO-1
- APC
Adenomatous Polyposis Coli-protein
- RA-GEF2
Ras guanine nucleotide exchange factor 2
- PTP-1E
protein tyrosine phosphatase 1E
Footnotes
This work was funded by NSF grant 0344354 (A.L.L.), NIH Protein Structure initiative grant GM 07491, NIH grant GM059791 (to HK) and internal bridge funding from UNC-Chapel Hill (A.L.L.)
SUPPORTING INFORMATION AVAILABLE
Figures showing asymmetric unit of apo PDZ2, crystal packing, effective correlation time (τe), residues used in RDC fitting. Tables showing global fitting results of RA-GEF2 and APC induced relaxation dispersion. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- 2.Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Ponting CP. Evidence for PDZ domains in bacteria, yeast, and plants. Protein Sci. 1997;6:464–468. doi: 10.1002/pro.5560060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Ham M, Hendriks W. PDZ domains-glue and guide. Mol Biol Rep. 2003;30:69–82. doi: 10.1023/a:1023941703493. [DOI] [PubMed] [Google Scholar]
- 5.Peterson FC, Penkert RR, Volkman BF, Prehoda KE. Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition. Mol Cell. 2004;13:665–676. doi: 10.1016/s1097-2765(04)00086-3. [DOI] [PubMed] [Google Scholar]
- 6.Sohn J, Grant RA, Sauer RT. Allosteric activation of DegS, a stress sensor PDZ protease. Cell. 2007;131:572–583. doi: 10.1016/j.cell.2007.08.044. [DOI] [PubMed] [Google Scholar]
- 7.Zhang M. Scaffold proteins as dynamic switches. Nat Chem Biol. 2007;3:756–757. doi: 10.1038/nchembio1207-756. [DOI] [PubMed] [Google Scholar]
- 8.van den Berk LC, Landi E, Walma T, Vuister GW, Dente L, Hendriks WJ. An allosteric intramolecular PDZ-PDZ interaction modulates PTP-BL PDZ2 binding specificity. Biochemistry. 2007;46:13629–13637. doi: 10.1021/bi700954e. [DOI] [PubMed] [Google Scholar]
- 9.Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Hidden dynamic allostery in a PDZ domain. Proc Natl Acad Sci U S A. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan J, Pan L, Chen X, Wu L, Zhang M. The structure of the harmonin/sans complex reveals an unexpected interaction mode of the two Usher syndrome proteins. Proc Natl Acad Sci U S A. 2010;107:4040–4045. doi: 10.1073/pnas.0911385107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian Y, Prehoda KE. Interdomain interactions in the tumor suppressor discs large regulate binding to the synaptic protein GukHolder. J Biol Chem. 2006;281:35757–35763. doi: 10.1074/jbc.M607057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Callaway DJ, Bu Z. Ezrin induces long-range interdomain allostery in the scaffolding protein NHERF1. J Mol Biol. 2009;392:166–180. doi: 10.1016/j.jmb.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bezprozvanny I, Maximov A. PDZ domains: More than just a glue. Proc Natl Acad Sci U S A. 2001;98:787–789. doi: 10.1073/pnas.98.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mishra P, Socolich M, Wall MA, Graves J, Wang Z, Ranganathan R. Dynamic scaffolding in a G protein-coupled signaling system. Cell. 2007;131:80–92. doi: 10.1016/j.cell.2007.07.037. [DOI] [PubMed] [Google Scholar]
- 15.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 16.Fuentes EJ, Der CJ, Lee AL. Ligand-dependent dynamics and intramolecular signaling in a PDZ domain. J Mol Biol. 2004;335:1105–1115. doi: 10.1016/j.jmb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Fuentes EJ, Gilmore SA, Mauldin RV, Lee AL. Evaluation of energetic and dynamic coupling networks in a PDZ domain protein. J Mol Biol. 2006;364:337–351. doi: 10.1016/j.jmb.2006.08.076. [DOI] [PubMed] [Google Scholar]
- 18.Gianni S, Walma T, Arcovito A, Calosci N, Bellelli A, Engstrom A, Travaglini-Allocatelli C, Brunori M, Jemth P, Vuister GW. Demonstration of long-range interactions in a PDZ domain by NMR, kinetics, and protein engineering. Structure. 2006;14:1801–1809. doi: 10.1016/j.str.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Ota N, Agard DA. Intramolecular signaling pathways revealed by modeling anisotropic thermal diffusion. J Mol Biol. 2005;351:345–354. doi: 10.1016/j.jmb.2005.05.043. [DOI] [PubMed] [Google Scholar]
- 20.Sharp K, Skinner JJ. Pump-probe molecular dynamics as a tool for studying protein motion and long range coupling. Proteins. 2006;65:347–361. doi: 10.1002/prot.21146. [DOI] [PubMed] [Google Scholar]
- 21.Ho BK, Agard DA. Probing the flexibility of large conformational changes in protein structures through local perturbations. PLoS Comput Biol. 2009;5:e1000343. doi: 10.1371/journal.pcbi.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong Y, Karplus M. Signaling pathways of PDZ2 domain: a molecular dynamics interaction correlation analysis. Proteins. 2009;74:145–154. doi: 10.1002/prot.22139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho BK, Agard DA. Conserved tertiary couplings stabilize elements in the PDZ fold, leading to characteristic patterns of domain conformational flexibility. Protein Sci. 2010;19:398–411. doi: 10.1002/pro.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 25.Cooper A, Dryden DT. Allostery without conformational change. A plausible model. Eur Biophys J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- 26.Kozlov G, Gehring K, Ekiel I. Solution structure of the PDZ2 domain from human phosphatase hPTP1E and its interactions with C-terminal peptides from the Fas receptor. Biochemistry. 2000;39:2572–2580. doi: 10.1021/bi991913c. [DOI] [PubMed] [Google Scholar]
- 27.Kozlov G, Banville D, Gehring K, Ekiel I. Solution structure of the PDZ2 domain from cytosolic human phosphatase hPTP1E complexed with a peptide reveals contribution of the beta2-beta3 loop to PDZ domain-ligand interactions. J Mol Biol. 2002;320:813–820. doi: 10.1016/s0022-2836(02)00544-2. [DOI] [PubMed] [Google Scholar]
- 28.Walma T, Spronk CA, Tessari M, Aelen J, Schepens J, Hendriks W, Vuister GW. Structure, dynamics and binding characteristics of the second PDZ domain of PTP-BL. J Mol Biol. 2002;316:1101–1110. doi: 10.1006/jmbi.2002.5402. [DOI] [PubMed] [Google Scholar]
- 29.Niv MY, Weinstein H. A flexible docking procedure for the exploration of peptide binding selectivity to known structures and homology models of PDZ domains. J Am Chem Soc. 2005;127:14072–14079. doi: 10.1021/ja054195s. [DOI] [PubMed] [Google Scholar]
- 30.Basdevant N, Weinstein H, Ceruso M. Thermodynamic basis for promiscuity and selectivity in protein-protein interactions: PDZ domains, a case study. J Am Chem Soc. 2006;128:12766–12777. doi: 10.1021/ja060830y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerek ZN, Keskin O, Ozkan SB. Identification of specificity and promiscuity of PDZ domain interactions through their dynamic behavior. Proteins. 2009;77:796–811. doi: 10.1002/prot.22492. [DOI] [PubMed] [Google Scholar]
- 32.Gerek ZN, Ozkan SB. A flexible docking scheme to explore the binding selectivity of PDZ domains. Protein Sci. 2010;19:914–928. doi: 10.1002/pro.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stiffler MA, Chen JR, Grantcharova VP, Lei Y, Fuchs D, Allen JE, Zaslavskaia LA, MacBeath G. PDZ domain binding selectivity is optimized across the mouse proteome. Science. 2007;317:364–369. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dev KK. Making protein interactions druggable: targeting PDZ domains. Nat Rev Drug Discov. 2004;3:1047–1056. doi: 10.1038/nrd1578. [DOI] [PubMed] [Google Scholar]
- 35.Wang NX, Lee HJ, Zheng JJ. Therapeutic use of PDZ protein-protein interaction antagonism. Drug News Perspect. 2008;21:137–141. [PMC free article] [PubMed] [Google Scholar]
- 36.Jemth P, Gianni S. PDZ domains: folding and binding. Biochemistry. 2007;46:8701–8708. doi: 10.1021/bi7008618. [DOI] [PubMed] [Google Scholar]
- 37.Gianni S, Geierhaas CD, Calosci N, Jemth P, Vuister GW, Travaglini-Allocatelli C, Vendruscolo M, Brunori M. A PDZ domain recapitulates a unifying mechanism for protein folding. Proc Natl Acad Sci U S A. 2007;104:128–133. doi: 10.1073/pnas.0602770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calosci N, Chi CN, Richter B, Camilloni C, Engstrom A, Eklund L, Travaglini-Allocatelli C, Gianni S, Vendruscolo M, Jemth P. Comparison of successive transition states for folding reveals alternative early folding pathways of two homologous proteins. Proc Natl Acad Sci U S A. 2008;105:19241–19246. doi: 10.1073/pnas.0804774105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milev S, Bjelic S, Georgiev O, Jelesarov I. Energetics of peptide recognition by the second PDZ domain of human protein tyrosine phosphatase 1E. Biochemistry. 2007;46:1064–1078. doi: 10.1021/bi061869i. [DOI] [PubMed] [Google Scholar]
- 40.Rao F, Karplus M. Protein dynamics investigated by inherent structure analysis. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0915087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Los Rios P, Cecconi F, Pretre A, Dietler G, Michielin O, Piazza F, Juanico B. Functional dynamics of PDZ binding domains: a normal-mode analysis. Biophys J. 2005;89:14–21. doi: 10.1529/biophysj.104.055004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhulesia A, Gsponer J, Vendruscolo M. Mapping of two networks of residues that exhibit structural and dynamical changes upon binding in a PDZ domain protein. J Am Chem Soc. 2008;130:8931–8939. doi: 10.1021/ja0752080. [DOI] [PubMed] [Google Scholar]
- 43.Dreier L, Wider G. Concentration measurements by PULCON using X-filtered or 2D NMR spectra. Magn Reson Chem. 2006;44:S206–212. doi: 10.1002/mrc.1838. Spec No. [DOI] [PubMed] [Google Scholar]
- 44.Wider G, Dreier L. Measuring protein concentrations by NMR spectroscopy. J Am Chem Soc. 2006;128:2571–2576. doi: 10.1021/ja055336t. [DOI] [PubMed] [Google Scholar]
- 45.Dick F. Acid cleavage/deprotection in Fmoc/tBu solid-phase peptide synthesis. Methods Mol Biol. 1994;35:63–72. doi: 10.1385/0-89603-273-6:63. [DOI] [PubMed] [Google Scholar]
- 46.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 48.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 49.von Ossowski I, Oksanen E, von Ossowski L, Cai C, Sundberg M, Goldman A, Keinanen K. Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide. Febs J. 2006;273:5219–5229. doi: 10.1111/j.1742-4658.2006.05521.x. [DOI] [PubMed] [Google Scholar]
- 50.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 51.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. Journal of Applied Crystallography. 2006;39:109–111. [Google Scholar]
- 54.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of Biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 55.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. Journal of Biomolecular NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 56.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 57.Sass HJ, Musco G, Stahl SJ, Wingfield PT, Grzesiek S. Solution NMR of proteins within polyacrylamide gels: diffusional properties and residual alignment by mechanical stress or embedding of oriented purple membranes. Journal of Biomolecular NMR. 2000;18:303–309. doi: 10.1023/a:1026703605147. [DOI] [PubMed] [Google Scholar]
- 58.Valafar H, Prestegard JH. REDCAT: a residual dipolar coupling analysis tool. J Magn Reson. 2004;167:228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 59.Loria JP, Rance M, Palmer AG., 3rd A TROSY CPMG sequence for characterizing chemical exchange in large proteins. Journal of Biomolecular NMR. 1999;15:151–155. doi: 10.1023/a:1008355631073. [DOI] [PubMed] [Google Scholar]
- 60.Mauldin RV, Carroll MJ, Lee AL. Dynamic dysfunction in dihydrofolate reductase results from antifolate drug binding: modulation of dynamics within a structural state. Structure. 2009;17:386–394. doi: 10.1016/j.str.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palmer AG, 3rd, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 62.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 63.Gao X, Satoh T, Liao Y, Song C, Hu CD, Kariya Ki K, Kataoka T. Identification and characterization of RA-GEF-2, a Rap guanine nucleotide exchange factor that serves as a downstream target of M-Ras. J Biol Chem. 2001;276:42219–42225. doi: 10.1074/jbc.M105760200. [DOI] [PubMed] [Google Scholar]
- 64.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 65.Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature. 2002;416:455–459. doi: 10.1038/416455a. [DOI] [PubMed] [Google Scholar]
- 66.Andrec M, Snyder DA, Zhou Z, Young J, Montelione GT, Levy RM. A large data set comparison of protein structures determined by crystallography and NMR: statistical test for structural differences and the effect of crystal packing. Proteins. 2007;69:449–465. doi: 10.1002/prot.21507. [DOI] [PubMed] [Google Scholar]
- 67.Wang J, Zuo X, Yu P, Byeon IJ, Jung J, Wang X, Dyba M, Seifert S, Schwieters CD, Qin J, Gronenborn AM, Wang YX. Determination of multicomponent protein structures in solution using global orientation and shape restraints. J Am Chem Soc. 2009;131:10507–10515. doi: 10.1021/ja902528f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bax A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci. 2003;12:1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cornilescu G, Bax A. Validation of Protein Structure from Anisotropic Carbonyl Chemical Shifts in a Dilute Liquid Crystalline Phase. J Am Chem Soc. 1998;120:6836–6837. [Google Scholar]
- 70.Cornilescu G, Bax A. Measurement of Proton, Nitrogen, and Carbonyl Chemical Shielding Anisotropies in a Protein Dissolved in a Dilute Liquid Crystalline Phase. J Am Chem Soc. 2000;122:10143–10154. [Google Scholar]
- 71.Stacklies W, Vega MC, Wilmanns M, Grater F. Mechanical network in titin immunoglobulin from force distribution analysis. PLoS Comput Biol. 2009;5:e1000306. doi: 10.1371/journal.pcbi.1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stacklies W, Xia F, Grater F. Dynamic allostery in the methionine repressor revealed by force distribution analysis. PLoS Comput Biol. 2009;5:e1000574. doi: 10.1371/journal.pcbi.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whitley MJ, Lee AL. Frameworks for understanding long-range intra-protein communication. Curr Protein Pept Sci. 2009;10:116–127. doi: 10.2174/138920309787847563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Erdmann KS, Kuhlmann J, Lessmann V, Herrmann L, Eulenburg V, Muller O, Heumann R. The Adenomatous Polyposis Coli-protein (APC) interacts with the protein tyrosine phosphatase PTP-BL via an alternatively spliced PDZ domain. Oncogene. 2000;19:3894–3901. doi: 10.1038/sj.onc.1203725. [DOI] [PubMed] [Google Scholar]
- 75.Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wand AJ. Dynamic activation of protein function: a view emerging from NMR spectroscopy. Nat Struct Biol. 2001;8:926–931. doi: 10.1038/nsb1101-926. [DOI] [PubMed] [Google Scholar]
- 77.Tsai CJ, del Sol A, Nussinov R. Allostery: absence of a change in shape does not imply that allostery is not at play. J Mol Biol. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mittermaier A, Kay LE. New tools provide new insights in NMR studies of protein dynamics. Science. 2006;312:224–228. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 79.Loria JP, Berlow RB, Watt ED. Characterization of enzyme motions by solution NMR relaxation dispersion. Acc Chem Res. 2008;41:214–221. doi: 10.1021/ar700132n. [DOI] [PubMed] [Google Scholar]
- 80.Mittag T, Schaffhausen B, Gunther UL. Direct observation of protein-ligand interaction kinetics. Biochemistry. 2003;42:11128–11136. doi: 10.1021/bi0347499. [DOI] [PubMed] [Google Scholar]
- 81.Mittag T, Schaffhausen B, Gunther UL. Tracing kinetic intermediates during ligand binding. J Am Chem Soc. 2004;126:9017–9023. doi: 10.1021/ja0392519. [DOI] [PubMed] [Google Scholar]
- 82.Tolkatchev D, Xu P, Ni F. Probing the kinetic landscape of transient peptide-protein interactions by use of peptide (15)n NMR relaxation dispersion spectroscopy: binding of an antithrombin peptide to human prothrombin. J Am Chem Soc. 2003;125:12432–12442. doi: 10.1021/ja021238l. [DOI] [PubMed] [Google Scholar]
- 83.Hansen DF, Vallurupalli P, Lundstrom P, Neudecker P, Kay LE. Probing chemical shifts of invisible states of proteins with relaxation dispersion NMR spectroscopy: how well can we do? J Am Chem Soc. 2008;130:2667–2675. doi: 10.1021/ja078337p. [DOI] [PubMed] [Google Scholar]
- 84.Niu X, Chen Q, Zhang J, Shen W, Shi Y, Wu J. Interesting structural and dynamical behaviors exhibited by the AF-6 PDZ domain upon Bcr peptide binding. Biochemistry. 2007;46:15042–15053. doi: 10.1021/bi701303p. [DOI] [PubMed] [Google Scholar]
- 85.McElheny D, Schnell JR, Lansing JC, Dyson HJ, Wright PE. Defining the role of active-site loop fluctuations in dihydrofolate reductase catalysis. Proc Natl Acad Sci U S A. 2005;102:5032–5037. doi: 10.1073/pnas.0500699102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Namanja AT, Wang XJ, Xu B, Mercedes-Camacho AY, Wilson BD, Wilson KA, Etzkorn FA, Peng JW. Toward flexibility-activity relationships by NMR spectroscopy: dynamics of Pin1 ligands. J Am Chem Soc. 2010;132:5607–5609. doi: 10.1021/ja9096779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Korzhnev DM, Neudecker P, Mittermaier A, Orekhov VY, Kay LE. Multiple-site exchange in proteins studied with a suite of six NMR relaxation dispersion experiments: an application to the folding of a Fyn SH3 domain mutant. J Am Chem Soc. 2005;127:15602–15611. doi: 10.1021/ja054550e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


