Abstract
Shigella dysenteriae serotype 1 (SD1) causes the most severe form of epidemic bacillary dysentery. We present the first comprehensive proteome analysis of this pathogen, profiling proteins from bacteria cultured in vitro and bacterial isolates from the large bowel of infected gnotobiotic piglets (in vivo). Overall, 1061 distinct gene products were identified. Differential display analysis revealed that SD1 cells switched to an anaerobic energy metabolism in vivo. High in vivo abundances of amino acid decarboxylases (GadB and AdiA) which enhance pH homeostasis in the cytoplasm and protein disaggregation chaperones (HdeA, HdeB and ClpB) were indicative of a coordinated bacterial survival response to acid stress. Several type III secretion system (T3SS) effectors were increased in abundance in vivo, including OspF, IpaC and IpaD. These proteins are implicated in invasion of colonocytes and subversion of the host immune response in S. flexneri. These observations likely reflect an adaptive response of SD1 to the hostile host environment. Seven proteins, among them the T3SS effectors OspC2 and IpaB, were detected as antigens in western blots using piglet antisera. The outer membrane protein OmpA, the heat shock protein HtpG and OspC2 represent novel SD1 subunit vaccine candidates and drug targets.
Keywords: acid stress, bacillary dysentery, proteome analysis, Shigella dysenteriae
1. Introduction
Shigella dysenteriae serotype 1 (SD1), a serious cause of bacillary dysentery or shigellosis, is a disease essentially limited to the human host and primates and transmitted via the oral-fecal route [1, 2]. SD1 survives in contaminated food products or water and has an extremely low infectious dose (10–100 organisms). Shigellosis starts with an acute infection of the cecum and is followed by bacterial invasion of the colonic mucosa causing symptoms such as cramps, diarrhea and fever. Destruction of the epithelial cell barrier and infiltration of inflammatory cells into the gut mucosa lead to colonic bleeding. When untreated, particularly in young children and immune-deficient patients, 10–15% of the cases result in death. In relation to the magnitude of the efforts aimed at prevention and effective drug treatment of shigellosis, the estimated 1.1 million deaths per annum are staggering [2]. The rapid emergence of drug resistance of Shigella species towards β-lactams, tetracyclines and aminoglycosides raises additional concerns [2, 3]. A more comprehensive understanding of the pathogenesis of shigellosis and the development of an effective vaccine are therefore of utmost importance. All four Shigella species are Gram-negative, non-spore forming, non-motile, facultative anaerobes. Unlike most S. dysenteriae serotypes and the other Shigella species (S. bodeii, S. flexneri and S. sonnei), SD1 and some Shiga toxin-producing Escherichia (E.) coli express a potent toxin called Shiga toxin 1 (Stx1) [4]. This toxin causes cell death, primarily in the microvascular endothelium, which contribute to GI bleeding.
The involvement of additional virulence factors in the pathogenesis of shigellosis has been investigated. It is clear that the type III secretion system (T3SS), also called Mix-Spa in Shigella, and the effector proteins secreted through it have important functions in invasion and cell-to-cell spreading of bacterial cells in the intestinal epithelium [5, 6]. The proteins forming the S. dysenteriae T3SS and all of its effectors are encoded on a large virulence plasmid [7]. Upon host cell contact, the invasion plasmid antigens IpaB and IpaC integrate into host cell membranes, thus forming a translocator pore that induces the injection of effectors into host cells [8, 9]. Numerous effector proteins have been characterized, particularly in studies of S. flexneri strains 5 and 2a. They induce changes in the cytoskeleton of phagocytic and epithelial cells, activate or deactivate signaling pathways that affect the secretion of inflammatory chemokines and eventually cause massive infiltration of the mucosa by polymorphonuclear leukocytes [10, 11]. Also important for virulence are the heterogeneous Shigella O-antigens, outer components of the cell surface lipopolysaccharide (LPS) [12]. The SD1 O-antigen consists of →3-α-L-rhamnosyl-1→3-α-L-rhamnosyl-1→2-α-D-galactosyl-1→3-α-D-N-acetylglucosaminyl-1→ repeat units [13]. While the small SD1 plasmid encodes the galactosyl transferase Rfp, other enzymes essential for O-antigen biosynthesis are encoded by chromosomal genes shared among Shigella spp., such as galETKM and rfbBDACX [14]. The diversity of O-antigen structures may have had an impact on the adaptation and survival of Shigella clones to and the survival chances in specific host environmental niches. O-antigens are strongly immunogenic in humans. Therefore, attenuated Shigella strains expressing genetically engineered LPS O-antigens have been tested as vaccine candidates. However, an effective vaccine against S. dysenteriae remains elusive [2].
The first completed S. dysenteriae genome was reported for the strain Sd197 in 2007 and included sequences of the chromosome, a large virulence-associated plasmid (pSD1_197) and a small plasmid (pSD197_Spa) [7]. The focus of Shigella proteomic surveys has been S. flexneri 2a, which causes endemic shigellosis. Nearly 170 S. flexneri gene products including several extracellular and OM proteins were identified in 2D gels [15]. Extensive 2D-LC-MS/MS-based profiling of membrane fractions yielded 666 S. flexneri proteins of which at least 200 proteins were predicted to be integrated in the inner membrane (IM) or outer membrane (OM). Examining exponential vs. stationary phase cultures, strong abundance changes of S. flexneri proteins involved in energy metabolism and the oxidative stress response were observed [16]. Additional reports described the immunogenicity of S. flexneri proteins using sera from either infected animals or patients. Strong antigens appeared to localize to the OM (OmpA, IpaD, TolC, YaeT) and the periplasm (MglB, TolB and AnsB) [17–19]. To our knowledge, neither a comprehensive survey of the S. dysenteriae proteome nor a proteome analysis of any Shigella spp. pertaining to the mammalian host environment has been published to date. Here, we present a proteomic survey of SD1 cells (strain Sd1617), which were collected from infected gnotobiotic piglets, and subsequently analyzed the data in the context of bacterial invasion of colonocytes and survival in the host.
2. Materials and methods
2.1. Materials
Reagents for protein extraction from cell lysates, protein analysis in 2D gels and by MS, were used as previously described [20]. RNase and DNase I (bovine pancreas) were from Sigma-Aldrich (St. Louis, MO). Sequencing grade porcine trypsin was obtained from Promega (Madison, WI). Triton X-100, tetradecanoylamidopropyl-dimethylammonio-butanesulfonate (ASB-14) and CHAPS were purchased from Calbiochem (LaJolla, CA). IPG strips were from GE Healthcare (Piscataway, NJ). The five serum samples isolated from gnotobiotic piglets included a control serum, serum P1 and P2 (21 and 9 days post-infection with SD1 strain Sd1617) and serum P3 and P4 (21 and 8 days post-infection with strain Sd1617 ΔstxAB).
2.2. Bacterial strains and culture conditions
Shigella dysenteriae serotype 1 Sd1617 [21], kindly provided by Dr. Malabi M. Venkatesan of Walter Reed Army Institute of Research, was routinely grown on Tryptic Soy Agar plates (TSA) containing 0.05% Congo Red (w/v). The inoculum for animal experiments was prepared by selecting a typical colony from TSA plates and growth of the bacteria in Luria-Bertani (LB) media overnight. A single colony was inoculated into LB media and grown in shaker flasks to stationary phase for 18 h to generate the in vitro bacterial sample. Cell pellets were harvested by centrifugation at 5,000 × g, washed once with ice-cold PBS, suspended in hypotonic lysis buffer (buffer HL) composed of 25 mM Tris-OAc (pH 7.8), 5 mM EDTA, 150 μg/ml lysozyme, 1 mM AEBSF, 1 mM BAM and 0.05% Triton X-100 and frozen at −80°C.
2.3. Animal experiments and isolation of Sd1617 bacterial cells from the intestine of gnotobiotic piglets
Piglets were delivered by cesarian section and housed at the Division of Infectious Disease of Tufts University Cummings School of Veterinary Medicine in accordance with approved procedures of the Institutional Animal Care and Use Committee at Tufts University. Of several animals assigned to this project two fulfilled the selection criteria. One piglet was inoculated with 1×108 Sd1617 cells, in which diarrhea started 24 h later and lasted until it was euthanized four days later. The second piglet was inoculated with 5×108 Sd1617 cells, developed diarrhea within 18 h and was euthanized three days after inoculation. Piglets ate well but appeared chilled when euthanized. To isolate and purify bacteria from piglets’ guts, contents of the cecum and colon were collected into sterile histological cups on ice immediately after euthanasia, suspended in PBS (4°C) and pelleted at 5,000 × g. The pellet was re-suspended in 65% isotonic Percoll (1 g pellet to 30 ml 65% Percoll solution) and centrifuged for 30 min at 14,500 × g at 4°C. The near-bottom bacterial gradient layer was collected using a 3–5 ml syringe with needle (0.9 × 25 mm), suspended in PBS and pelleted at 16,100 × g. Bacteria were washed once more with PBS, suspended in ice-cold buffer HL and frozen at −80°C.
2.4. Preparation of Sd1617 cell lysates and subcellular fractionation
SD1 bacteria were thawed and agitated for 30 minutes at 20°C to complete the enzymatic cell lysis in the presence of Triton X-100 and EDTA. The lysate was adjusted to concentrations of 1 mM AEBSF, 1 mM benzamidine, 10 μg/ml leupeptin and 20 mM MgCl2. DNAse I and RNAse (10 μg/mL each) were added to digest nucleic acids while gently agitating for 1 h at 20°C. The lysate was centrifuged at 20,000 × g for 30 min at 4°C. Supernatant and pellet fractions were recovered. Total protein in the supernatant was measured using the bicinchoninic acid assay. Using membrane filtration units (10 kDa NMWL), the supernatant was desalted into 25 mM NH4HCO3 (pH 7.8), 1 mM EDTA and 1 mM benzamidine and concentrated to 1.5–5 mg/mL protein. The pellet fraction, enriched in mixed membranes, was homogenized in a ca. 7.5-fold volume of 10 mM Tris-HCl (pH 7.8), 5 mM EDTA, 0.2 mM DTT, 10 μg/ml Leupeptin, 5 μg/ml Pepstatin, 10 μg/ml Nα-p-Tosyl-L-arginine methyl ester and 2 mM PMSF. Sodium bromide was added at a 2.5 M concentration, and the suspension was stirred for 1 h at 20°C followed by centrifugation at 20,000 × g for 30 min. The high salt-extracted supernatant (hs-MBR) was desalted, lyophilized and solubilized in gel rehydration buffer (buffer GR) composed of 8 M urea, 2 M thiourea, 4% (w/v) CHAPS, 18 mM DTT and 0.5% (v/v) Bio-Lyte pH 3–10 carrier ampholytes at 1–2 mg/mL protein. The pellet was re-homogenized in ice-cold 0.18 M Na2CO3 (pH 11.3), 50 mM DTT, 1 mM CaCl2, 1 mM MgCl2 and 1 mM MnCl, stirred for 1 h at 20°C and centrifuged at 20,000 × g for 30 min. The supernatant was discarded, while the insoluble pellet was recovered and frozen at −80°C, or directly re-suspended in 8 M urea, 2 M thiourea, 1% (w/v) ASB-14, 2 mM tributylphosphine and 0.5% (v/v) Bio-Lyte pH 3–10 carrier ampholytes. After protein solubilization for 30 min at 20°C and centrifugation at 16,100 × g for 15 min, the supernatant (usb-MBR) fraction was applied to SDS-PAGE to estimate the protein concentration from CBB-stained gel bands.
2.5. Fractionation of Sd1617 cell lysates via microscale in-solution isoelectric focusing
Proteins present in the cell lysate supernatant were fractionated on the basis of their isoelectric points using the ZOOM IEF fractionator (Invitrogen). IEF fractionator assembly, sample loading and electrophoretic separation parameters were selected as suggested in the instrument’s user manual. Briefly, 0.75–1 mg protein was lyophilized and subsequently re-suspended in ca. 3 ml of the buffer GR. The suspension was incubated for 30 min at 20°C and then spun at 16,100 × g for 15 min to remove any insoluble material. The supernatant was subjected to IEF for a total of ca. 1,100 V-h, yielding five separate IEF fractions with the pI boundaries 3.0–4.6, 4.6–5.4, 5.4–6.2, 6.2–7.0 and 7.0–10.0 in volumes of 0.2–0.7 ml. Protein concentrations of the IEF fractions were estimated from the intensity of CBB-stained bands in SDS-PAGE gels.
2.6. Protein separation and analysis in 2D gels
Solubilized hs-MBR and usb-MBR fractions were loaded onto 24 cm IPG gel strips (pH range 4–7) in a 100–150 μg protein quantity using the anode cup-loading method. The cell lysate ZOOM IEF fractions were applied to IPG gels in volumes of ca. 150 μl using the anode cup-loading method, with the exception of the 4.6–5.4 IEF fraction, which was applied in a volume of ca. 450 μL using in-gel rehydration due to the low protein concentration of this fraction. Separation of narrow pH range IEF fractions in the 1st 2D gel dimension was performed as follows: (1) pI 4.6–5.4 fraction in IPG strips in the pH range 4.5–5.5, (2) pI 5.4–6.2 fraction in IPG strips in the pH range 5–6, (3) pI 6.2–7.0 fraction in IPG strips in the pH range 5–8, and (4) pI 7.0–10.0 fraction in IPG strips in the pH range 6–9. Proteins in acidic IPG strips (pH 4.5–5.5 and 5–6), medium pH IPG strips (pH 4–7 and 5–8), and basic IPG strips (pH 6–9) were focused for a total of ca. 95,000, 60,000 and 75,000 V-h, respectively. The 2nd dimension separation via SDS-PAGE (25 × 19.5 × 0.15 cm slab gels), gel staining and conversion of scanned gels into 16-bit TIFF images were performed as described previously [22]. Protein spot matching and the determination of normalized volumetric spot intensities in 2D gel images were facilitated by use of the software tool Proteomweaver v.4.0. To assess the reproducibility of spot quantitation, two to four gel images per group from one in vitro and two in vivo samples were analyzed. If feasible, coefficients of correlation were determined. Matching of spots and spot trains in gels was confirmed by extensive MS analysis efforts. The positions of 25 cytoplasmic protein spots were used as landmarks for Mr and pI calibrations in 2D gels.
2.7. Mass spectrometry analysis
Methods for spot picking, peptide digestion and MS techniques were previously described in detail [20, 22]. Briefly, peptide digests were analyzed using a MALDI-TOFTOF mass spectrometer (4700 Proteomics Analyzer, Applied Biosystems) and a nano-electrospray LC-MS/MS system (LTQ-IT mass spectrometer, Thermo-Finnigan, San Jose, CA) equipped with an Agilent 1100 series solvent delivery system (Agilent, Palo Alto, CA). Peptide separation for LC-MS/MS analysis was performed using a PicoTip microcapillary reversed-phase column (BioBasic C18, 75 μm X 10 cm, New Objective, Woburn, MA) at a flow rate of 350 nL/min. MS and MS/MS data were searched against the latest release of the S. dysenteriae strain Sd197 genome database in NCBInr using the Mascot searching engine v.2.2 (Matrix Science, London, UK). Carbamidomethyl was invariably selected as a fixed modification. One missed tryptic cleavage was allowed. MALDI search parameters (+1 ions) included mass error tolerances of ±100 ppm for peptide ions and ±0.2 Da for fragment ions. LTQ-IT search parameters (+1, +2 and +3 ions) included mass error tolerances of ±1.4 Da for peptide ions and ±0.5 Da for fragment ions. MALDI peak lists were created using the Peaks-to-Mascot function in the 4000 series Explorer (Applied Biosystems). The .txt files were searched with Mascot. LTQ peak lists were created with Mascot Daemon using the data import filter lcq_dta.exe from XCalibur v2.2 (Thermo Electron), which converts binary .raw files into peak list .dta files. Protein identifications were considered as significant based on a previously reported decision tree [20, 22], with the additional requirement of at least two peptides (e-value <0.1) from a single protein spot. Applying these criteria to a Mascot decoy database search option, searches with a randomized S. dysenteriae Sd197 protein decoy database did not yield a single identification on the protein level. This was indicative of an estimated protein false discovery rate of less than 0.1%.
2.8. 2D western blotting experiments using antisera from infected piglets
Mini-2D gels were run in the pH range 4–7 and the Mr range of ca. 200-10 kDa loading ca. 50 μg protein per sample, including cell lysate supernatants and usb-MBR fractions each derived from in vitro and in vivo Sd1617 cells. IEF analysis was performed in the ZOOM IPG-Runner system (Invitrogen Inc.) for 2,000 V-h in 7 cm IPG gel strips and SDS-PAGE analysis in 4–12% Bis-Tris gels (8 × 6.5 × 0.1 cm). SD1 lysate supernatants were also fractionated by anion exchange LC on a 0.8 mL POROS HQ column using a 0.02–1 M NaCl gradient in Tris-HCl at pH 8.1. All fourteen eluted fractions were separated via SDS-PAGE. Replicates of SDS-PAGE and mini-2D gels were stained in CBB and or subjected to electroblotting onto PVDF membrane sheets at a constant voltage of 22 V for 2 h at 4°C using Nu-PAGE transfer buffer (Invitrogen Inc.). Blotted proteins were stained with Ponceau S in 5% AcOH followed by membrane blocking with 5% dry milk powder in TBS/0.05% Tween-20 overnight at 4°C. The PVDF membrane was incubated with a 1:1,000 piglet antiserum dilution in milk blocking buffer for 1 h at 20°C. Incubation with a conjugate of alkaline phosphatase to goat anti-porcine IgG (Southern Biotech) diluted to 1:5,000 in milk blocking buffer for 1 h at 20°C followed. TBS/0.05% Tween-20 solutions were used to wash the membrane three times for 5 min following each of the antibody incubation steps. Immune-reactive spots (mini-2D gels) and bands (SDS-PAGE) were visualized with BCIP/NBT phosphatase substrates, and matched to the equivalent CBB-stained gel spots/bands. Following comprehensive MS analysis, we cross-validated the antigen identities based on the protein identification data, spot shapes in 2D gels vs. blots and overlaps of protein identifications derived from both separation approaches.
2.9. Bioinformatic analysis tools
To predict motifs for lipoproteins, TMD proteins and export signal sequences in SD1 proteins, queries of the entire S. dysenteriae Sd197 genome were performed using the CMR database at http://cmr.jcvi.org/. The algorithms LipoP, SignalP, TatP and TMHMM were accessed at www.cbs.dtu.dk. In silico predictions of subcellular protein localizations were obtained using PSORTb v.2.0 searches (www.psort.org/psortb/).
3. Results and Discussion
3.1. S. dysenteriae Sd1617 proteome derived from bacteria isolated from piglets’ guts
Proteomic analysis of S. dysenteriae is urgently needed to identify novel targets for disease prevention and therapeutic intervention. Sequencing and annotation of the first S. dysenteriae serotype 1 (SD1) genome, from strain Sd197, suggested 4274 chromosomal ORFs, 224 plasmid pSD1_197-encoded ORFs and eight plasmid pSD197_spA-encoded ORFs [7]. Comparative analysis of Shigella and E. coli genomes resulted in the discovery of 955 gene deletions in the Sd197 strain compared to E. coli strain MG1655. This is a remarkable case of reductive evolution which apparently resulted from the adaptation to an intracellular milieu, in a highly host-specific environmental niche in the large bowel of humans and primates [23]. Recently, a gnotobiotic piglet model was developed (K. Jeong, Q. Zhang, J. Nunnari and S. Tzipori, unpublished) to serve as an alternative to a primate model, which apart from serious ethical considerations is inconsistent, expensive and difficult to work with. While a normal piglet develops no symptoms, a gnotobiotic piglet delivered via caesarian section and devoid of a natural gut microbial flora develops severe diarrhea after oral challenge. The gut becomes heavily colonized by S. dysenteriae with evidence of cellular invasion and mucosal ulceration as observed in humans. The ability to recover more than 109 purified SD1 cells from the gut allowed a unique study of the pathogen’s adaptation to the host environment. We compared the in vivo proteome state with that derived from stationary phase bacteria cultured in LB media (in vitro). We acknowledge that some of the detected protein changes may pertain only to the selected in vitro growth condition and not to other growth stages, such as the exponential phase. Furthermore, the human and primate gut where SD1 infection normally occurs have a complex microbial flora unlike the gnotobiotic piglets. A complex microbial environment in the gut likely results in competitive and adaptive processes affecting the SD1 proteome. Nonetheless, this study promised to yield proteomic data indicative of how this pathogen adapted to the gastro-intestinal tract. We benefitted from the similarity of E. coli and SD1 genomes, which allowed us to examine the data in the context of characterized E. coli metabolism, stress response and communication systems.
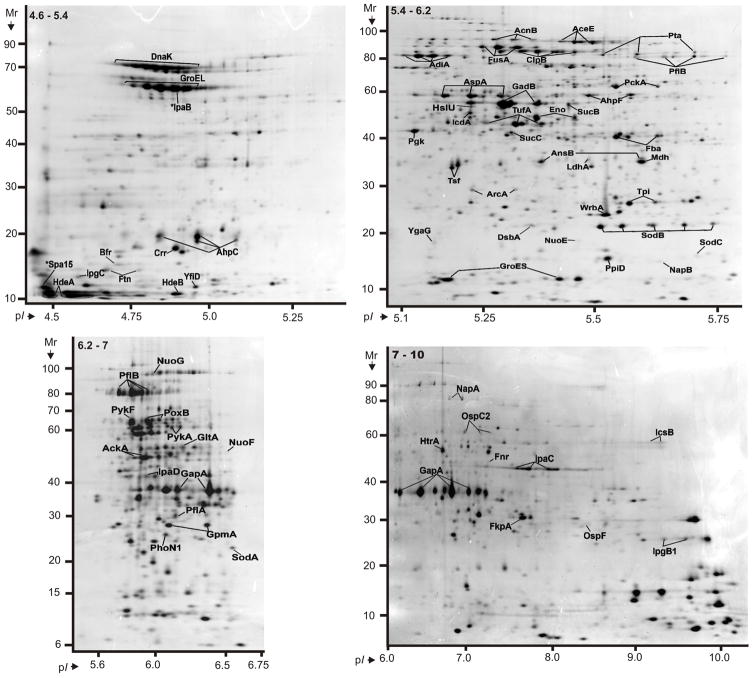
Soluble proteins were recovered from SD1 cell lysates, pre-fractionated by in-solution IEF and resolved in four narrow-to-medium pH range IPG gels (pH 4.5–5.5, 5–6, 5–8 and 6–9). Insoluble pellets were also recovered and first extracted with a 2.5 M NaBr solution, yielding the hs-MBR fraction, followed by protein solubilization with denaturants (8 M urea, 2 M thiourea and 1% detergent ASB-14). The latter step yielded the usb-MBR fraction. Both fractions were subjected to analysis in 2D gels using the IPG range 4–7. Including protein display and MS analysis data from in vitro and in vivo growth conditions, 3000 2D gel spots were identified. Due to protein appearance in the form of spot trains and spot overlaps derived from different subcellular fractions, pH boundaries and replicates of 2D gels, the 3000 spot identifications collapsed into 1061 unique gene products, corresponding to ca. 24% of the theoretical SD1 proteome. Genome annotation predicts 4505 ORFs. Spot trains in 2D gels often represent chemical protein artifacts derived from sample preparation, in particular deamidation of N or Q amino acid side chains that results in acidic shifts of spot pI values. Therefore, the differential display analysis was not performed for individual protein spots part of spot trains. Circa 66% of all identified gene products were assigned to distinct 2D gel spots, as shown in Figures S1-S6 (Suppl. Information). The denoted spots are listed in Table S1 with a description for each protein pertaining to MS data, the function and localization in the cell.
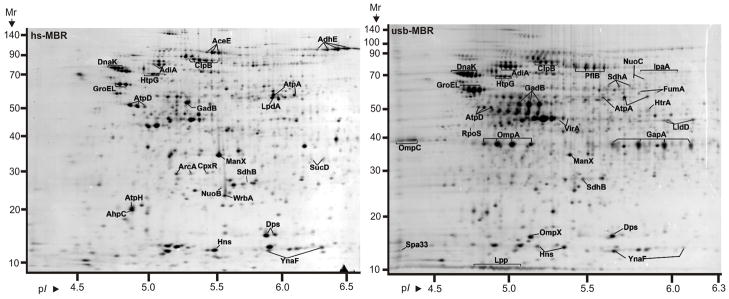
PSORTb search results predicted 599 cytoplasmic proteins, 38 integral IM proteins, 23 OM proteins and 65 periplasmic proteins. Subcellular localizations of 348 proteins were not definitively assigned. The chart in Figure 1 displays protein functional role categories according to the CMR database (http://cmr.jcvi.org). Proteins involved in energy metabolism, protein synthesis and transport and substrate-binding functions were numerically the largest categories (15.4%, 8.7% and 8.4%, respectively). A recent membrane proteome survey of S. flexneri 2a [24] reported identifications of more than 600 proteins, including 159 proteins assigned to the IM and 35 proteins assigned to the OM. For technical reasons, 2D gels are not suitable for in depth profiling of integral IM proteins. The overlap of OM-localized protein orthologs comparing our and their dataset was considerable with ca. 65%. We identified 31 plasmid pSD1-197-encoded proteins (Figure 2). Wei et al. [24] identified 80% of the respective S. flexneri proteins expressed from the plasmid pCP301, including eight Mxi-Spa proteins (structural components of the T3SS) and eleven effectors and chaperones clustered in the ipa gene locus of each virulence plasmid. Many highly abundant SD1 proteins had functions in energy and carbon metabolism, protein synthesis and the response to oxidative and heat stress. This included glycolytic enzymes (e.g. PckA, GapA, Tpi, Fba, Pgk, GpmA and Eno), elongation factors (e.g. FusA, TufA and Tsf), chaperones (e.g. GroEL, GroES, DnaK, PpiD and DsbA) and various stress response proteins (e.g. WrbA, AhpC, AhpF, SodB and HtrA). The proteins are displayed in the gel images of Figure 3. Gel images representing membrane fractions (Figure 4) featured other stress response proteins (Dps and YnaF), subunits of protein complexes required for energy metabolism (AtpA, AtpD, AtpH, AceE, LpdA and ManX), β-barrel OM proteins (OmpA, OmpC and OmpX) and Braun’s lipoprotein (Lpp), which is responsible for covalent linkage of the peptidoglycan to the OM. Proteins with regulatory functions were usually less abundant (Hns, RpoS, CpxR, ArcA, Fnr; Figures 3 and 4).
Fig. 1.
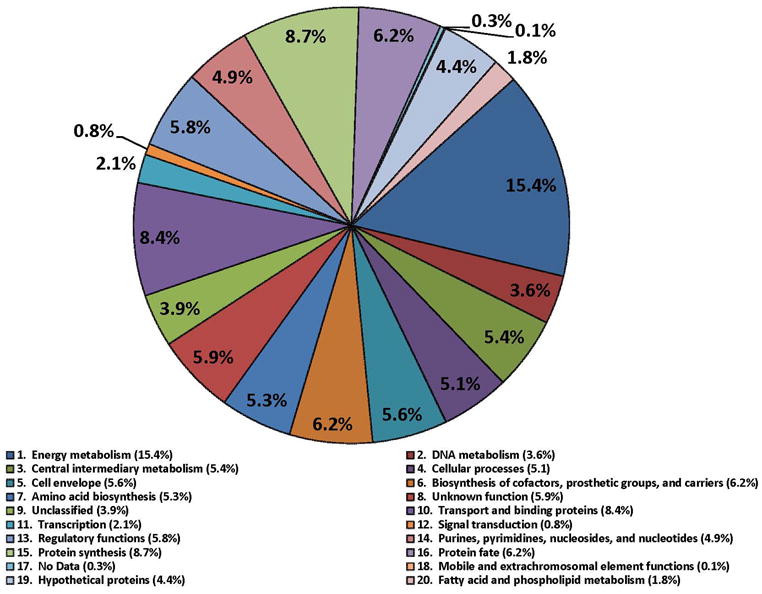
Representation of functional role categories of proteins identified in a 2D gel-based proteome survey from cell lysates of S. dysenteriae serotype 1. The pie chart is based on functional assignments derived from strain SD1 strain 197 genome in the CMR database. All of the 1061 proteins were identified in 2D gel/MS experiments.
Fig. 2.
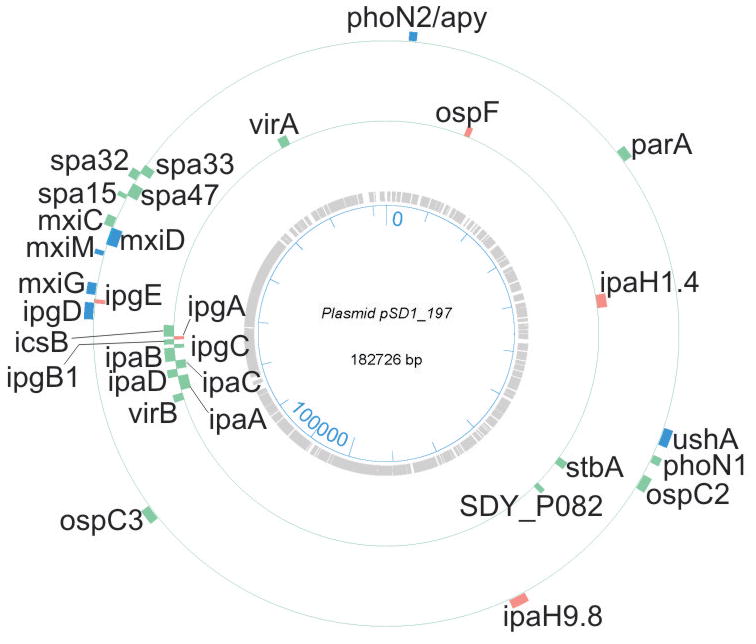
A map of the plasmid pSD1_197 and the gene locations corresponding to proteins identified in the S. dysenteriae serotype 1 (strain Sd1617) proteome analysis. Genes are listed with short names. The bar length reflects the size of the gene. Colorations: green, proteins identified in vivo and in vitro; blue, proteins identified only in vitro; orange, proteins identified only in vivo.
Fig. 3.
Proteins derived from a strain Sd1617 cell lysate and separated in 2D gels representing four pH ranges. Cells were isolated from the large bowel of the infected piglet #1. The IPG strips for the separation according to the proteins’ pI values were 4.5–5.5 (top left), 5–6.2 (top right), 5–8 (bottom left) and 6–10 (bottom right). The denoted pH ranges (top left insert in each gel image) pertain to protein pre-focusing via in-solution IEF. Experimental conditions for 2D gel electrophoresis and staining are described in the Methods. Many displayed proteins are listed in Table 1 with further descriptions.
Fig. 4.
Proteins derived from the insoluble fraction of a strain Sd1617 cell lysate separated in 2D gels in the pH range 4–7. Cells were isolated from the large bowel of infected piglet #1. The insoluble fraction was extracted with 2.5 M NaBr (hs-MBR fraction, left), followed by solubilization with 8 M urea, 2 M thiourea and 1% amidosulfobetaine-14 (usb-MBR fraction, right). Experimental conditions for 2D gel electrophoresis and staining are described in the Methods. Several proteins are listed in Table 1 with further descriptions. Integral OM proteins (e.g. OmpA and OmpX) were enriched in the usb-MBR fraction. Proteins responding to various stresses (e.g. AdiA, GadB, GroEL, DnaK, HtpG and ClpB) were relatively abundant in both fractions.
3.2. Transition of SD1 from aerobic to anaerobic respiration in vivo
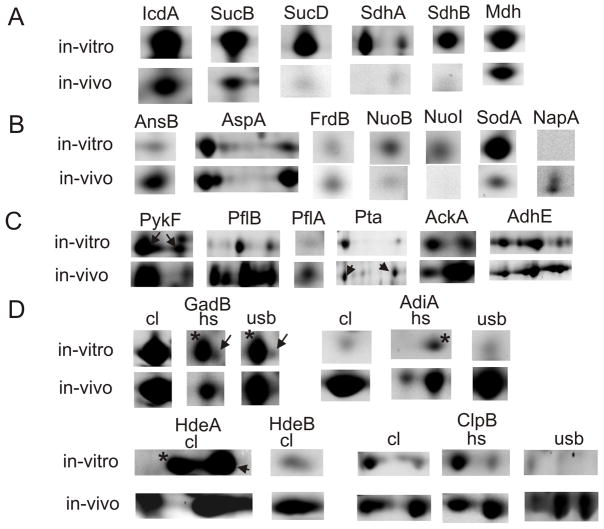
Enterobacteria are facultative anaerobes and, when starved of oxygen, switch to anaerobic/ microaerobic energy metabolisms in the host’s intestine [25]. We examined quantitative changes of SD1 proteins (in vitro vs. in vivo) as it pertained to the pathways of respiration and mixed acid fermentation. Data were available for bacterial isolates from two piglets and one cell culture experiment (Table 1). Differential display data for some proteins are illustrated in 2D gel montage views in Figure 5. The bacterial tricarboxylic acid (TCA) cycle is most active under fully aerobic conditions. We observed that many TCA cycle enzymes (e.g., IcdA, SucB, SucD, SdhA, SdhB and Mdh; Figure 5A) were strongly decreased in SD1 cells in vivo. NADH:ubiquinone oxidoreductase (Nuo) is a large IM-associated complex transferring electrons from NADH to the quinone pool. Six peripheral membrane subunits of Nuo were profiled, two of which were markedly decreased in abundance in vivo (e.g., NuoB and NuoI; Figure 5B). Nuo may play a less important role in electron transport in vivo due to the apparently decreased TCA cycle activity. However, Nuo is also active under anaerobic conditions. A few subunits of protein complexes utilized for electron transport chains under anaerobic conditions and localized in the IM were reliably quantitated (e.g., FrdB of fumarate reductase and NapA of the periplasmic nitrate reductase; Figure 5B). The NapB and FrdA subunits were also identified from 2D gels. The data were not sufficient to suggest increased functional roles of such anaerobic respiration enzymes in vivo. The E. coli periplasmic nitrate reductase (Nap) has been described as an electron acceptor induced under low nitrate concentrations in a process mediated by Fnr, one of the main global regulators at aerobic/anaerobic interface [26]. Interestingly, enzymes involved in fumarate mobilization, periplasmic asparaginase II (AnsB) and cytoplasmic aspartate ammonia lyase (AspA), were markedly increased in abundance in vivo (Figure 5B). Higher fumarate concentrations may enhance substrate turnover by the Frd complex and lead to increased menaquinol production under anaerobic conditions. In a recent proteomic study on growth phase transition in S. flexneri 2a, abundance increases of AnsB, AspA, FrdA and FrdB were observed upon entry into the stationary phase [16].
Table 1.
Differential 2D gel display data for S. dysenteriae serotype 1 proteins comparing in vitro and in vivo spot profiles
| Fractiona) | Protein name | Protein description | Mr (kDa)b) | pIb) | Vs cell cult.c) | No. gelsd) | CVe) | Vs pig #1c) | No. gelsd) | CVe) | Vs pig #2c) | No. gelsd) | Ratio pig #1/ccf) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Energy metabolism | |||||||||||||
| CL 4.5 – 5.4 | YfiD | putative formate acetyltransferase | 14374 | 4.96 | NA | 1.9 | 2 | 1.399 | 1 | ||||
| CL 5.4 – 6.2 | AcnB | aconitate hydrase B | 94038 | 5.27 | 2.891 | 3 | 27.53% | 0.72 | 4 | 16.98% | 1.393 | 2 | 0.249 |
| CL 5.4 – 6.2 | Pta | phosphotransacetylase | 77480 | 5.52 | 1.61 | 3 | 23.34% | 0.959 | 4 | 33.35% | 0.762 | 2 | 0.596 |
| CL 5.4 – 6.2 | AspA | aspartate ammonia-lyase | 54714 | 5.54 | 3.279 | 3 | 35.45% | 6.127 | 2 | 5.039 | 2 | 1.869 | |
| CL 5.4 – 6.2 | IcdA | isocitrate dehydrogenase | 46069 | 5.28 | 4.306 | 3 | 21.87% | 0.607 | 4 | 43.48% | 0.619 | 2 | 0.141 |
| CL 5.4 – 6.2 | SucC | succinyl-CoA synthetase, beta subunit | 41610 | 5.37 | 1.999 | 3 | 36.57% | 0.519 | 4 | 37.10% | 0.369 | 2 | 0.260 |
| CL 5.4 – 6.2 | SucB | dihydrolipoamide acetyltransferanse | 43990 | 5.58 | 4.125 | 3 | 31.42% | 2.31 | 4 | 78.41% | 1.018 | 2 | 0.560 |
| CL 5.4 – 6.2 | AnsB | periplasmic L-asparaginase II | 36942 | 5.68 | 0.581 | 3 | 6.70% | 1.41 | 4 | 9.90% | 0.862 | 2 | 2.427 |
| CL 5.4 – 6.2 | LdhA | fermentative D-lactate dehydrogenase, NAD-dependent | 36869 | 5.37 | 0.554 | 3 | 33.63% | 0.13 | 4 | 48.35% | NA | 0.235 | |
| CL 5.4 – 6.2 | Mdh | malate dehydrogenase | 32477 | 5.61 | 3.907 | 3 | 28.69% | 2.174 | 4 | 34.52% | 1.473 | 2 | 0.556 |
| CL 5.4 – 6.2 | NuoE | NADH dehydrogenase I chain E | 18879 | 5.4 | 0.24 | 3 | 38.36% | 0.078 | 4 | 30.36% | 0.019 | 2 | 0.325 |
| CL 6.2 – 7 | NuoG | NADH dehydrogenase I chain G | 101514 | 5.89 | 0.757 | 4 | 8.11% | 0.927 | 2 | NA | 1.225 | ||
| CL 6.2 – 7 | PflB | formate acetyltransferase 1, pyruvate formate lyase | 85581 | 5.69 | 1.551 | 4 | 17.48% | 8.5 | 2 | NA | 5.480 | ||
| CL 6.2 – 7 | PykA | pyruvate kinase II | 51583 | 6.24 | 0.679 | 4 | 19.62% | 0.998 | 2 | NA | 1.470 | ||
| CL 6.2 – 7 | GltA | citrate synthase | 48384 | 6.06 | 5.938 | 4 | 16.23% | 1.328 | 2 | NA | 0.224 | ||
| CL 6.2 – 7 | AckA | acetate kinase | 43593 | 5.85 | 0.745 | 4 | 33.93% | 2.394 | 2 | NA | 3.213 | ||
| CL 6.2 – 7 | PflA | pyruvate formate lyase activating enzyme 1 | 28528 | 6 | 0.18 | 4 | 31.76% | 0.689 | 2 | NA | 3.828 | ||
| CL 6.2 – 7 | PykF | pyruvate kinase I, fructose stimulated | 51027 | 5.77 | 2.966 | 4 | 6.19% | 3.89 | 2 | NA | 1.312 | ||
| CL 6.2 – 7 | PoxB | pyruvate oxidase | 62541 | 5.89 | 3.722 | 4 | 12.92% | 2.451 | 2 | NA | 0.659 | ||
| CL 7 – 10 | NapA | probable nitrate reductase 3, periplasmic | 93682 | 7.85 | NA | 2.551 | 2 | 1.634 | 1 | ||||
| CL 7 – 10 | NuoF | NADH dehydrogenase I chain F | 49774 | 6.44 | 1.404 | 3 | 9.86% | 0.16 | 2 | NA | 0.114 | ||
| hs-MBR | AdhE | bifunctional acetaldehyde-CoA/alcohol dehydrogenase | 96580 | 6.32 | 7.387 | 2 | 10.56 | 2 | 12.82 | 1 | 1.429 | ||
| hs-MBR | SucD | succinyl-CoA synthetase, alpha subunit | 30128 | 6.18 | 12.164 | 2 | 0.45 | 2 | 0.615 | 1 | 0.037 | ||
| hs-MBR | NuoB | NADH dehydrogenase I chain B | 25339 | 5.58 | 1.338 | 2 | 0.398 | 2 | 0.562 | 1 | 0.297 | ||
| hs-MBR | FrdB | fumarate reductase, iron-sulfur protein subunit | 27735 | 6.07 | 1.177 | 2 | 0.958 | 2 | 0.913 | 1 | 0.814 | ||
| hs-MBR | SdhB | succinate dehydrogenase, iron sulfur protein | 27452 | 5.97 | 7.432 | 2 | 0.198 | 2 | 0.248 | 1 | 0.027 | ||
| hs-MBR | NuoI | NADH dehydrogenase I chain I | 21067 | 5.4 | 1.264 | 2 | NA | NA | |||||
| usb-MBR | SdhA | succinate dehydrogenase, flavoprotein subunit | 65024 | 5.85 | 8.067 | 2 | 0.389 | 2 | 0.743 | 1 | 0.048 | ||
| usb-MBR | NuoC | NADH dehydrogenase I chain C, D | 68909 | 5.98 | 0.739 | 4 | 6.54% | 0.293 | 2 | NA | 0.396 | ||
| usb-MBR | FumA | fumarase A | 60744 | 6.11 | 3.497 | 4 | 3.41% | 0.581 | 2 | NA | 0.166 | ||
| usb-MBR | LldD | L-lactate dehydrogenase | 42899 | 6.33 | 4.661 | 4 | 4.19% | 2.402 | 2 | NA | 0.515 | ||
| Acid resistance | |||||||||||||
| CL 4.5 – 5.4 | HdeA | Hypoth. protein SDY_3543, acid resistance chaperone | 11964 | 5.06 | 23.858 | 2 | 48.96 | 2 | 65.16 | 1 | 2.052 | ||
| CL 4.5 – 5.4 | HdeB | Hypoth. protein SDY_3544, acid resistance chaperone | 12610 | 6.55 | 2.206 | 2 | 5.929 | 2 | 2.773 | 1 | 2.688 | ||
| CL 5.4 – 6.2 | AdiA | biodegradative arginine decarboxylase | 85071 | 5.09 | 0.128 | 3 | 50.89% | 9.111 | 2 | NA | 71.180 | ||
| CL 5.4 – 6.2 | ClpB | protein disaggregation chaperone | 95725 | 5.37 | 1.686 | 3 | 25.34% | 7.259 | 4 | 25.31% | 5.716 | 2 | 4.305 |
| CL 5.4 – 6.2 | GadB | glutamate decarboxylase isozyme | 53220 | 5.28 | 9.42 | 3 | 28.92% | 14.74 | 4 | 5.14% | 17.01 | 2 | 1.564 |
| hs-MBR | AdiA | biodegradative arginine decarboxylase | 85071 | 5.09 | NA | 2 | 3.618 | 2 | 0.575 | 1 | |||
| hs-MBR | ClpB | protein disaggregation chaperone | 95725 | 5.37 | 0.992 | 2 | 6.245 | 2 | 4.791 | 1 | 6.295 | ||
| hs-MBR | GadB | glutamate decarboxylase isozyme | 53220 | 5.28 | 0.254 | 2 | 5.082 | 2 | 7.561 | 1 | 20.008 | ||
| usb-MBR | AdiA | biodegradative arginine decarboxylase | 85071 | 5.09 | NA | 4 | 11.85% | 11.48 | 2 | NA | |||
| usb-MBR | ClpB | protein disaggregation chaperone | 95725 | 5.37 | 0.165 | 4 | 29.63% | 8.74 | 2 | NA | 52.970 | ||
| usb-MBR | GadB | glutamate decarboxylase isozyme | 53220 | 5.28 | 0.641 | 4 | 21.81% | 10.29 | 2 | NA | 16.055 | ||
| Host cell invasion and subversion of host immune response | |||||||||||||
| CL 4.5 – 5.4 | IpgC | cytoplasmic chaperone for IpaB and IpaC | 17916 | 4.58 | NA | 1.324 | 2 | 4.156 | 1 | ||||
| CL 5.4 – 6.2 | IpaD | invasion plasmid antigen IpaD | 36738 | 5.69 | 0.08 | 2 | 0.199 | 4 | 19.49% | 0.012 | 1 | 2.488 | |
| CL 5.4 – 6.2 | PhoN2 | periplasmic ATP diphosphohydrolase PhoN2/Apy | 27728 | 5.46 | 0.377 | 3 | 36.02% | NA | NA | ||||
| CL 5.4 – 6.2 | StxB | Shiga toxin subunit B precursor | 9851 | 7.77 | 0.267 | 3 | 6.74% | NA | NA | ||||
| CL 6.2 – 7 | IpaD | invasion plasmid antigen IpaD | 36738 | 5.69 | NA | 0.634 | 2 | NA | |||||
| CL 6.2 – 7 | PhoN1 | periplasmic non-specific acid phosphatase PhoN1 | 27266 | 6.43 | 2.28 | 4 | 20.56% | 0.948 | 2 | NA | 0.416 | ||
| CL 7 – 10 | OspC2 | outer Shigella protein family protein OspC2 | 55775 | 6.58 | 0.467 | 4 | 28.58% | 2.262 | 2 | NA | 4.844 | ||
| CL 7 – 10 | IpaC | invasion plasmid antigen IpaC | 38731 | 7.85 | 1.252 | 3 | 39.25% | 18.38 | 2 | 3.453 | 2 | 14.681 | |
| CL 7 – 10 | IcsB | invasive protein IcsB | 56540 | 9.34 | NA | 2.951 | 2 | NA | |||||
| CL 7 – 10 | OspF | mouse killing factor, phosphothreonine lyase | 28167 | 8.2 | NA | 1.493 | 2 | NA | |||||
| CL 7 – 10 | IpgB1 | membrane ruffles-inducing protein IpgB1 | 23731 | 9.45 | 3.906 | 2 | 5.383 | 2 | 3.143 | 2 | 1.378 | ||
| CL 7 – 10 | StxA | Shiga toxin subunit A precursor | 34906 | 9.63 | 1.454 | 2 | NA | NA | |||||
| usb-MBR | IpaA | invasion plasmid antigen IpaA | 70223 | 6.27 | NA | 0.932 | 2 | NA | |||||
| Oxidative stress response, iron storage, quorum sensing | |||||||||||||
| CL 4.5 – 5.4 | Ftn | cytoplasmic ferritin | 19468 | 4.77 | 9.182 | 2 | 1.733 | 2 | 2.389 | 1 | 0.189 | ||
| CL 4.5 – 5.4 | Bfr | iron storage and detoxification protein, bacterioferrin | 18326 | 4.64 | 12.636 | 2 | 1.713 | 2 | 1.155 | 1 | 0.136 | ||
| CL 5.4 – 6.2 | YgaG | S-ribosylhomocysteinase, LuxS ortholog | 19575 | 5.18 | 1.491 | 3 | 16.03% | 0.209 | 4 | 77.25% | 0.117 | 2 | 0.140 |
| CL 5.4 – 6.2 | SodB | superoxide dismutase, iron | 21310 | 5.58 | 3.337 | 3 | 32.21% | 6.554 | 4 | 13.95% | 5.134 | 2 | 1.964 |
| CL 5.4 – 6.2 | WrbA | putative NAD(P)H:quinone oxidoreductase | 20862 | 5.59 | 1.682 | 3 | 14.05% | 3.675 | 4 | 4.96% | 3.394 | 2 | 2.185 |
| CL 5.4 – 6.2 | SodC | superoxide dismutase (Cu-Zn) | 17764 | 6.04 | 1.697 | 3 | 1.11% | 0.181 | 4 | 36.38% | 0.125 | 2 | 0.107 |
| CL 6.2 – 7 | SodA | superoxide dismutase, manganese | 23079 | 6.45 | 3.279 | 4 | 9.47% | 0.401 | 2 | NA | 0.122 | ||
| CL 7 – 10 | SitA | iron transport protein, periplasmic-binding protein | 33565 | 8.34 | 2.108 | 3 | 10.84% | NA | NA | ||||
| hs-MBR | Dps | stationary phase protection protein Dps | 18684 | 5.72 | 26.394 | 2 | 6.843 | 2 | 12.9 | 1 | 0.259 | ||
subcellular fraction in which the protein spot was identified; CL, cell lysate listed with the pI range for protein detection; hs-MBR, high salt-extracted membrane fraction; usb-MBR, urea/thiourea/amidosulfobetaine-14-extracted membrane fraction;
predicted molecular weight (Mr) and pI values;
normalized relative spot intensity volume (or sum of spot volumes) for a S. dysenteriae protein measured using the software Proteomweaver v.4.0; cell cult. (in vitro cell culture, stationary phase); pig #1, cells isolated from the gut of piglet #1; pig# 2, cells isolated from the gut of piglet #2;
number of gels in which the protein was quantitated;
CV, if at least 3 spot intensity measurements per group were available;
ratio of protein spot quantities: in vivo piglet #1 vs. in vitro cell culture; NA: protein was not detected in 2D gels.
Fig. 5.
Proteins altered in abundance comparing in vitro and in vivo 2D spot profiles. A: tricarboxylic acid cycle enzymes; B: enzymes or enzyme subunits with known or putative functions in anaerobic/ microaerobic respiration: fumarate as electron acceptor (AnsB, AspA and FrdB), NADH as electron donor (NuoB and NuoI), nitrate as electron acceptor (NapA); C: mixed acid fermentation enzymes; D: acid stress response proteins: amino acid decarboxylases (GadB and AdiA), protein disaggregation chaperones (HdeA, HdeB and ClpB); in row D, the comparison of spot quantities is shown for two or three fractions (cl, cell lysate; hs, hs-MBR fraction; usb, usb-MBR fraction). ▶ indicates a correctly matched spot, if a neighboring spot (*) represented a different protein. N-fold abundance differences are reported in Table 1.
Enteric bacteria starved of oxygen also generate energy via substrate level phosphorylation such as mixed acid fermentation. Several fermentative enzymes were markedly increased in vivo (Figure 5C). This included a pyruvate-formate lyase activating enzyme (PflA), two pyruvate-formate lyases (PflB and YfiD), and acetate kinase (AckA). Other enzymes were moderately or not increased in vivo, such as phosphate acetyltransferase (Pta), acetaldehyde dehydrogenase (AdhE) and pyruvate kinase (PykF), all of which are shown in Figure 5C, pyruvate oxidase (PoxB) and fermentative D-lactate dehydrogenase (LdhA). Fermentation pathways and associated stress responses have been characterized extensively in E. coli [27, 28]. The pyruvate-formate lyase branch is responsible for the catabolism of pyruvate to acetate and produces ATP in two of the four enzymatic steps. This pathway relies on the activities of PykF, PflA, PflB, Pta and AckA and seemed to be utilized more in SD1 cells in vivo. YfiD is a stress-induced pyruvate-formate lyase and replaces PflB upon inactivation of the latter enzyme during oxidative stress in E. coli [29]. This was indicative of high oxidative stress in SD1 cells in vivo and a critical metabolic function of YfiD. Other mixed acid fermentation branches appeared to be less influenced by the in vivo vs. in vitro environments, such as the reductive pathways for lactate (LdhA) and ethanol (AdhE). Both enzymes generate NAD+ from NADH. In summary, significant abundance changes were observed for enzymes that are part of the TCA cycle (down-regulation in vivo) and mixed acid fermentation via PflA/PflB/YfiD (up-regulation in vivo), suggesting a shift from anaerobic energy generation to fermentation in SD1 cells in the host environment.
Fnr and the two-component regulator ArcA/ArcB have been characterized as the main global regulators responsible for the switch from aerobic to microaerobic/ anaerobic respiration in Gram-negative bacteria. While Fnr was less abundant in 2D gels than the transcription factor ArcA (Figure 3), the integral IM protein ArcB was not detected. The pI values of ArcA (5.2 and 5.3) matched the predicted pI values of singly (Asp54-) phosphorylated and unphosphorylated ArcA, respectively. ArcA activation involves its phosporylation by the histidine sensor kinase ArcB. E. coli ArcA regulates the expression of genes encoding TCA cycle and fermentation enzymes, as summarized in the EcoCyc database [27]. The ArcA/ArcB system also influences the expression of proteins implicated in oxidative stress such as SodA [30]. SodA was strongly decreased in abundance in SD1 cells in vivo (Figure 5B). Another cytoplasmic superoxide dismutase, SodB (Figure 2), was very abundant in vivo and in vitro. In contrast to SodA, E. coli SodB has been described as a superoxide detoxifying enzyme under aerobic and anaerobic conditions [27]. In conclusion, this data supports regulatory roles of ArcA and ArcB in SD1 cells encountering an anaerobic environment in the piglet’s intestine.
3.3. S. dysenteriae acid resistance systems
Dedicated acid resistance systems contribute to the survival of commensal and pathogenic E. coli strains while passing through the acidic stomach (pH 2–3) and moderately acidic parts of the intestine (pH 4.5–6.5). They also play a role in the neutralization of intracellular acidic fermentation products [31, 32]. Among the three E. coli acid resistance systems (ARs), are AR-1, whose components are not entirely known but include the F0F1 proton-translocating ATPase, AR-2 and AR-3, each of which is an amino acid decarboxylase/ antiporter-dependent system. AR-2 results in glutamate decarboxylation and the export of the product γ–aminobutyrate. AR-3 results in arginine decarboxylation and the export of the product agmatine. Via consumption of a proton and release of CO2, AR-2 and AR-3 directly influence pH homeostasis in the cytoplasm. There are two E. coli glutamate decarboxylases (GadA and GadB), which are most active at a pH of ca. 4, and one arginine decarboxylase (AdiA), which is most active at a pH of ca. 5. The antiporters (GadC and AdiC, respectively) expel the decarboxylation products and simultaneously import amino acid substrates from the periplasm. In order to explain the absence of strong changes in the membrane potential when the AR-2 and AR-3 systems are active, a more complex model has coupled chloride export via the ion channel ClC to amino acid antiport. Chloride export would counteract membrane hyper-polarization, arising as a result of the AR-2/AR-3 activities [33].
SD1 acid resistance systems have not been studied to date. The E. coli GadB ortholog was one of the most abundant cytoplasmic proteins (Figure 3), not only in bacteria isolated from piglets’ guts but also from stationary phase cells (in vitro). The enzyme’s high expression level in SD1 cells seems not to be limited to acidic pH environments. Indeed, induction of gadA/BC was linked to fermentative and stationary phase growth conditions in S. flexneri and E. coli [32, 34]. Acetate, D-lactate and formate are fermentation products of enzymes referred to in section 3.2 and likely contribute to decreased intracellular pH values in SD1 cells under anaerobic conditions. There was no evidence for expression of GadA in SD1 cells, although an ORF annotated as gadA is present in the Sd197 genome. We conclude that GadB is the functional glutamate decarboxylase of the S. dysenteriae AR-2 system. The enzyme was surprisingly abundant in the usb-MBR fraction in vivo (Figure 4). Recently, E. coli GadB was shown to be recruited to the IM when the intracellular pH fell [35]. A triple helix bundle near the protein’s N-terminus was determined to be important for this process. The authors argued that local pH changes first occur in proximity to the IM, where a proton enters the cytoplasm, and result in signaling events that move GadB towards the IM to neutralize the pH. This motive process could moderate detrimental effects of widespread cytoplasmic acidification [35]. Our data suggest that GadB is also recruited to the SD1 membrane, apparently to a greater extent in vivo than in vitro (Figure 5D and Table 1). Neither the antiporter GadC nor the ion channel ClC were profiled in SD1 membrane fractions. These proteins, annotated as XasS and SDY_1564 in the Sd197 genome, have 12 and 10 transmembrane domains (TMDs), respectively. Hydrophobic TMDs explain why GadC and CIC were not detected in 2D gels. AR-2 regulation in E. coli is a complex process. Studies have implicated the alternative sigma factor σs and CRP [31], but also a network of more specific transcription factors (GadX, EvgA and YdeO). These three regulatory proteins influence the expression of many acid resistance genes [32, 36] and were not detected in the survey of the SD1 proteome.
Arginine decarboxylase (AdiA) was not identified in 2D gel profiles of SD1 fractions derived from the in vitro growth state. Its abundance varied markedly in the in vivo proteome profiles. Bacteria isolated from piglet #1, fed directly before euthanasia, expressed AdiA in higher concentrations than bacteria isolated from piglet #2, fed 3 h before euthanasia (Table 1). Food intake increases the piglets’ gastric acid production, possibly resulting in a falling pH in the intestine and increased expression of AdiA. In a study on E. coli O157:H7, cells cultured in the presence of volatile fatty acids showed enhanced expression of both AdiA and GadB [37]. We hypothesize that volatile fatty acid uptake by SD1 cells results in cytoplasmic acidification, requiring the activity of GadB and AdiA to counteract the intracellular pH decrease. AdiA was also quite abundant in usb-MBR fractions in vivo (Figure 4), suggesting recruitment to the IM as previously described for GadB. Neither the antiporter AdiC nor the F0F1 proton-translocating ATPase of a putative AR-1 system were profiled in this survey.
We also examined SD1 protein profiles with respect to acid stress responses. Two periplasmic proteins, HdeA and HdeB, have been characterized as acid stress chaperones in E. coli [38, 39]. While HdeA protects periplasmic proteins from aggregation and denaturation at a pH optimum of 2, HdeB is more protective at pH 3 [38]. The ability of HdeA and HdeB to solubilize mixed protein aggregates was also demonstrated [39]. Both protein orthologs of SD1 were highly expressed in vivo, comparable to the abundant periplasmic isomerase FkpA (Figure 3), and featured low Mr and acidic pI values. HdeA was also highly abundant in vitro. HdeB was markedly decreased in vitro, suggesting that the latter chaperone was specifically induced by acid stress in the piglet’s intestine (Figure 5D). High expression levels of the chaperones supported the notion of their association with periplasmic protein aggregates in stoichiometric proportions. The expression of adiA, hdeA and hdeB in E. coli appears to be regulated by those transcription factors mentioned above in the context of AR-2 [36]. Acid stress specifically induced by growth in acetate media in E. coli caused widespread protein abundance changes, including the enzymes Pta and YfiD and the stress response protein WrbA [28]. YfiD (see section 3.2) and WrbA were increased in abundance in SD1 cells in vivo (Table 1). This data further supported the key role of YfiD in the pyruvate-formate lyase branch of fermentation under acid stress conditions. Whether WrbA, annotated as a NADH:quinone oxidoreductase, is functionally linked to acid stress remains to be shown. In summary, we gained insight into protein expression changes likely required for the survival of S. dysenteriae during acid stress in the host’s intestine. Proteins such as GadB, AdiA, HdeA, HdeB and WrbA are interesting targets for functional and structural studies, and possibly inhibitor design.
3.4. S. dysenteriae type III secretion and invasion plasmid antigens
The type III secretion system (T3SS) encoded by the 30 kb spa-mxi region of large Shigella plasmids forms a cell envelope-spanning machinery responsible for the secretion of virulence factors that facilitate invasion of the intestinal mucosa [6, 8]. The invasive process via tight junctions between epithelial cells, colonocytes and polymorphonuclear cells is not entirely understood. Invasion is followed by entry of Shigella into colonocytes via the basolateral membrane and requires invasion plasmid antigens (Ipa proteins) translocated by the T3SS. Intra- and intercellular spreading of Shigella, which do not produce flagella, is mediated by host cell actin polymerization [6]. There is also increasing evidence that Shigella manipulates the innate and adaptive host immune system via activities of proteins belonging to the Osp family [10, 40]. The fact that SD1 cells isolated from the piglets’ intestines likely represented various stages of adhesion to the luminal and basolateral side of colonic and cecal epithelia and invasion of epithelial cells, polymorphonuclear leukocytes and macrophages prevented us from associating abundance changes of the T3SS proteins and effectors with specific stages of the infection. Most membrane-associated Mxi and Spa proteins were of low abundance or not resolved well in 2D spots, resulting in difficulties to differentially quantitate them. Spa15 is a soluble chaperone for T3SS proteins with broad specificity and was profiled as a highly acidic, low Mr protein (Figure 3). Spa15 is involved in folding Osp family effectors, IpaA, IpgB1 and IpgB2 [9]. IpgC was also detected as a highly acidic, low Mr protein (Figure 3) and facilitates the folding of the invasion plasmid antigens IpaB and IpaC. Expression of Shigella mxi-spa genes was reported to be induced upon contact with host epithelial cells [8], which we could not confirm in the SD1 proteome analysis. Post-translational processes mediated by proteases are known to influence the integrity and the activity of the T3SS. Dominant proteases observed in 2D spot profiles of SD1 lysates were ClpB, HtrA and HslU (Figure 3) and deserve further functional analysis regarding the modification of T3SS proteins.
Following secretion by the T3SS, formation of the S. flexneri T3SS translocator pore in various mammalian cell types requires IpaB, IpaC and IpaD [8, 41]. IpaB and IpaC disrupt the host cell membrane integrity, which results in bacterial uptake via macropinocytosis [42]. The SD1 orthologs were detected as streaky spots in 2D gels (Figure 3). IpaB contains several TMDs, accounting for its poor resolution and detection as a minor component of a GroEL spot. IpaC and IpaD were increased in abundance in vivo (Table 1). Other T3SS effectors characterized in S. flexneri were either more abundant or solely detected in vivo (piglet #1 data; Table 1). IpgB1, OspF and IcsB are displayed in Figure 3, IpaA in Figure 4. With the exception of ospF, the virulence factor genes are clustered in the mxi-spa-ipa-ipg region, ranging from the ORFs P151 to P194 of the plasmid Sd197 (Figure 2). The expression of the respective S. flexneri genes is controlled by the transcription factor VirB [8]. Our data support the induction of the sub-cluster ipa-ipg via VirB which was detected as a weak antigen (Figure 6). OspF is a phosphothreonine lyase inactivating mitogen-activated protein kinase (MAPK) in macrophages [43]. MAPK is an essential kinase for the human innate immune response via macrophages. IpgB1 activates Rho GTPases. This interaction promotes membrane ruffling in epithelial cells, a process that promotes Shigella invasion of host cells [44]. IpaA binds to vinculin, a focal adhesion protein implicated in the depolymerization of actin filaments [45, 46]. IcsB was reported to prevent IcsA binding to the host cell autophagy protein Atg5 [47]. None of the effectors have been characterized to date in S. dysenteriae. Many of them displayed basic pI values in 2D gels, such as IpaC, IpgB1, IcsB and OspF. It is not known whether such high pI values are physiologically relevant. Additional virulence plasmid-encoded proteins, PhoN1 (Figure 3), VirA (Figure 4) and PhoN2, were decreased in abundance in vivo or detected only in vitro. PhoN2 was reported to hydrolyze dNTPs and modulate the localization of IcsA at the cell surface of S. flexneri [48]. VirA has been implicated in membrane ruffling in HeLa cells [49].
Fig. 6.
S. dysenteriae serotype 1 antigens detected with a piglet antiserum in a 2D western blot. Left: cell lysate; right: usb-MBR fraction. Matching CBB-stained 2D gels were used to identify the spots by MS. Thorough validation was required to designate a protein confidently as an antigen: (1) highest LC-MS/MS score from at least two gel spots matching the western blot signal; (2) same spot shape in 2D gel and western blot; (3) availability of MALDI-TOF data (IpaB and VirB were the exceptions); (4) cross-validation of data via SDS-PAGE western blotting after fractionation of a cell lysate by anion exchange chromatography (data not shown here).
The uncharacterized OspC2 protein was markedly increased in abundance in vivo (Table 1). The ospC2 gene locus on plasmid Sd197 is P070 (Figure 2). The streaky spot appearance of OspC2 in gels (Figure 3) and western blots (Figure 6) was reminiscent of Ipa proteins and suggested membrane or cell surface association. The predicted Mr and pI values are 55.6 kDa and 6.6, respectively. Conserved motifs for SpI- or SpII-type signal sequences or TMDs were not found. S. flexneri OspC2 was reported to share the chaperone Spa15 with other Osp family proteins [9]. A mutation in the ospC2 gene did not interfere with the motility of S. flexneri in the polymorphonuclear leukocyte trans-epithelial migration assay [50]. Other S. flexneri Osp family proteins were recently characterized as prominent effectors subverting the host innate immune response, including OspB [51], OspG [52] and OspZ [53].
3.5. Other proteins implicated in the pathogenicity of S. dysenteriae
SD1 is the only Shigella serotype liberating Shiga toxin 1 (Stx1), considered a serious virulence factor characteristic of the genus and thought to contribute significantly to the manifestation of pathology and severity of disease [54]. The stx genes were acquired via lateral gene transfer from a phage [55]. Both subunits (Stx1-A and Stx1-B) were only detected in vitro (Table 1). This was an unexpected observation since the ability to liberate Stx1 is thought to exacerbate shigellosis. Yet, it may explain why other serotypes, such as S. flexneri 2a which cause dysenteric illness of equal severity, lack this gene. The lack of in vivo expression of Stx1 and its pathological and clinical implications require further investigation. O-antigens, outer components of LPS, are highly diverse in Shigella spp. (at least 46 serotypes) and have evolved to promote evasion from the human immune system [12]. The small SD1 plasmid-encoded galactosyltransferase (Rfp), responsible for the addition of a galactosyl residue into the tetrasaccharide repeat unit of the SD1 O-antigen [56], the O-antigen polymerase RfaC and ligase RfaL were not detected. In contrast, complete sets of enzymes involved in the biosynthesis of the O-antigen sugars galactose and rhamnose were identified (GalE, GalT, GalK, RfbA, RfbB, RfbC, Figures S1-S6, Suppl. Information). Differential display of these enzymes did not suggest increased O-antigen biosynthesis rates in vivo.
Protein profiles of hs-MBR and usb-MBR fractions from one of the in vivo samples (Figure 4) revealed high abundance of soluble heat shock proteins (DnaK, GroEL and HtpG) and other stress response proteins such as Dps, which sequesters iron and protects DNA from damage, and the protease ClpB. There is increasing evidence for the participation of heat shock proteins in protein-membrane phospholipid microdomains upon exposure to thermal stress [57]. It is plausible that membrane stress in S. dysenteriae is enhanced in vivo, although we need to confirm this assumption with a more rigorous differential 2D display analysis. ClpB is a peptidase involved in the elimination of protein aggregates in cooperation with DnaK under stress conditions [58]. Interestingly, membrane protein disaggregation functions were attributed to ClpB derived from the intracellular pathogen Francisella tularensis [59]. This study also described the inability of a clpB mutant to multiply in target organs of F. tularensis-infected mice. Moreover, Campylobacter jejuni was reported to induce expression of clpB under acid shock conditions [60]. We hypothesize that ClpB functions as a repair chaperone in SD1 cells and preserves the membrane proteome under hostile conditions in the host environment, which includes exposure to strong acids and toxic compounds. ClpB was not only increased in abundance in vivo in the cell lysate (Table 1), but also quite abundant in the hs-MBR and usb-MBR fractions (Figure 5D). Finally, abundance of the protein YgaG (Figure 3) was markedly decreased in vivo. YgaG is orthologous to E. coli LuxS. This enzyme plays a role in the activated methyl cycle and in quorum sensing [61], as well as in transcriptional activation of the T3SS gene locus in enterohemorrhagic E. coli [62]. Consequences of reduced YgaG expression in vivo remain to be elucidated.
3.6. Identification of S. dysenteriae antigens in infected piglets
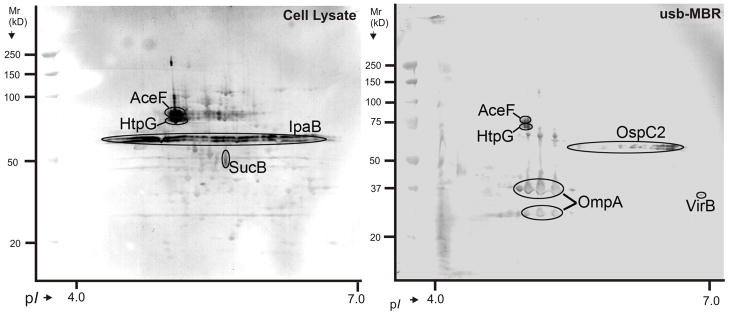
Immunoblots measuring the reactivity of serum IgG antibodies towards SD1 proteins were performed. The antisera were from four S. dysenteriae-infected convalescent piglets, two from strain Sd1617 infections, two from Sd1617 ΔstxAB infections (the latter strain did not produce the toxin Stx1). The disease symptoms were significantly milder in the piglets challenged with Sd1617 ΔstxAB, which was consistent with a diminished immune reaction based on western blot data. Only faint signals were observed for OmpA and IpaB (not shown here). Immunoblot data for the SD1-infected piglets varied markedly. Using one antiserum, strong signals were observed for IpaB, HtpG, AceF, OmpA and OspC2, weaker signals for SucB and VirB (Figure 6). Using the other antiserum, all but the three plasmid-encoded proteins (IpaB, OspC2 and VirB) were also identified as antigens. The significance of such differences is unknown. Both AceF and SucB are highly conserved dihydrolipoamide acetyltransferases of large intracellular protein complexes. The translocator pore component IpaB, currently studied as a S. flexneri subunit vaccine target, and the β-barrel protein OmpA form oligomeric complexes at the OM cell surface of SD1. Bu et al. constructed a ΔhtpG mutant of S. flexneri 2a strain 2457T and found that HtpG evoked an inflammatory response in mice [63]. This is in accordance with our preliminary data (personal communications, Q. Zhang). Heat shock proteins such as HtpG have been implicated in immune responses to cancer and microbial pathogens, particularly as carriers of immunogenic peptides and targets of the cellular arm of immunity, e.g. NK cells [64]. OmpA has antigen characteristics in other pathogenic bacteria including Shigella flexneri 2a [17, 19]. A prominent Shigella antigen is the extracellular LPS whose O-antigen variants modulate the mammalian immune response. O-antigens have been useful in diagnostic tests of shigellosis and as targets for vaccine development [2]. LPS was not targeted with our protein-specific antigen identification methods. Although swine and human immune systems are not identical and few sera were tested here, HtpG, AceF, OmpA, OspC2 and IpaB are potential subunit vaccine candidates for the prevention of shigellosis.
4. Concluding Remarks
The first extensive proteomic survey of S. dysenteriae and of any Shigella spp. isolated from infected animals was performed. Abundance increases of translocator pore proteins and a few other T3SS effectors were detected in cells isolated from piglets’ guts compared to cells grown in vitro to stationary phase. The response of bacterial cells to acid stress seemed to require high expression levels of cytoplasmic amino acid decarboxylases (GadB and AdiA), periplasmic acid stress chaperones (HdeA and HdeB), and a protein disaggregation protease (ClpB). Immunoproteomic analysis led to the identification of strong antigens (HtpG, AceF, OmpX, OspC2 and IpaB). The design of functional inhibitors to proteins such as OspC2, IpaB, GadB and HdeA could be a useful strategy for the development of therapeutic drugs. In vivo proteome profiles seemed to reflect the pathogen’s response to the hostile mammalian gut environment, including the lack of oxygen, acid fluctuation, toxic agents and heat (fever). This proteomic survey will help us understand host-pathogen interactions and select novel vaccine candidates with the right balance of immunogenicity and reactogenicity.
Supplementary Material
Acknowledgments
At Tufts, thanks to D. Girouard, who performed the C-sections, and the animal technicians who cared for the piglets. This part of the work was supported by NIAID, FWD IRN contract N01-AI-30050. At the JCVI, we thank S. Kuntumalla and L. Papazisi for advice and assistance using tools for proteomic and genomic analysis. This part of the work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under contract No. N01-AI15447.
Abbreviations
- AR-1, AR-2, AR-3
acid resistance systems 1, 2 and 3, respectively
- hs-MBR
high salt-extracted membrane fraction
- IM
inner membrane
- LPS
lipopolysaccharide
- OM
outer membrane
- SD1
S. dysenteriae serotype 1
- T3SS
type III secretion system
- TCA
tricarboxylic acid
- TMD
transmembrane domain
- usb-MBR
urea/thiourea/ASB-14-extracted membrane fraction
Footnotes
Conflict of interests. None of the authors has conflicts of interest.
References
- 1.World Health Organization. Diarrhoeal diseases (update February 2009) 2009 http://www.who.int/vaccine_research/diseases/diarrhoeal/en/print.html.
- 2.Niyogi SK. Shigellosis. J Microbiol. 2005;43:133–143. [PubMed] [Google Scholar]
- 3.Shapiro RL, Kumar L, Phillips-Howard P, Wells JG, et al. Antimicrobial-resistant bacterial diarrhea in rural western Kenya. J Infect Dis. 2001;183:1701–1704. doi: 10.1086/320710. [DOI] [PubMed] [Google Scholar]
- 4.Herold S, Karch H, Schmidt H. Shiga toxin-encoding bacteriophages--genomes in motion. Int J Med Microbiol. 2004;294:115–121. doi: 10.1016/j.ijmm.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 5.Buchrieser C, Glaser P, Rusniok C, Nedjari H, et al. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol. 2000;38:760–771. doi: 10.1046/j.1365-2958.2000.02179.x. [DOI] [PubMed] [Google Scholar]
- 6.Morita-Ishihara T. Supramolecular structure of the Shigella type III secretion machinery. Bioscience Microflora. 2007;26:29–36. [Google Scholar]
- 7.Microbial Genome Center of Chinese Ministry of Public Health. Shigella dysenteriae Sd197 genome sequencing project. 2007 http://gib.genes.nig.ac.jp/single/main.php?spid=Sdys_SD197.
- 8.Parsot C. Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiol Lett. 2005;252:11–18. doi: 10.1016/j.femsle.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 9.Parsot C. Shigella type III secretion effectors: how, where, when, for what purposes? Curr Opin Microbiol. 2009 doi: 10.1016/j.mib.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Phalipon A, Sansonetti PJ. Shigella’s ways of manipulating the host intestinal innate and adaptive immune system: a tool box for survival? Immunol Cell Biol. 2007;85:119–129. doi: 10.1038/sj.icb7100025. [DOI] [PubMed] [Google Scholar]
- 11.Sperandio B, Regnault B, Guo J, Zhang Z, et al. Virulent Shigella flexneri subverts the host innate immune response through manipulation of antimicrobial peptide gene expression. J Exp Med. 2008;205:1121–1132. doi: 10.1084/jem.20071698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B, Knirel YA, Feng L, Perepelov AV, et al. Structure and genetics of Shigella O antigens. FEMS Microbiol Rev. 2008;32:627–653. doi: 10.1111/j.1574-6976.2008.00114.x. [DOI] [PubMed] [Google Scholar]
- 13.Dmitriev BA, Knirel YA, Kochetkov NK. Somatic antigens of shigella. Structural investigation on the O-specific polysaccharide chain of Shigella dysenteriae type 1 lipopolysaccharide. Eur J Biochem. 1976;66:559–566. doi: 10.1111/j.1432-1033.1976.tb10582.x. [DOI] [PubMed] [Google Scholar]
- 14.Yao Z, Valvano MA. Genetic analysis of the O-specific lipopolysaccharide biosynthesis region (rfb) of Escherichia coli K-12 W3110: identification of genes that confer group 6 specificity to Shigella flexneri serotypes Y and 4a. J Bacteriol. 1994;176:4133–4143. doi: 10.1128/jb.176.13.4133-4143.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao X, Ying T, Wang H, Wang J, et al. A two-dimensional proteome map of Shigella flexneri. Electrophoresis. 2003;24:2864–2882. doi: 10.1002/elps.200305519. [DOI] [PubMed] [Google Scholar]
- 16.Zhu L, Liu XK, Zhao G, Zhi YD, et al. Dynamic proteome changes of Shigella flexneri 2a during transition from exponential growth to stationary phase. Genomics Proteomics Bioinformatics. 2007;5:111–120. doi: 10.1016/S1672-0229(07)60021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jennison AV, Raqib R, Verma NK. Immunoproteome analysis of soluble and membrane proteins of Shigella flexneri 2457T. World J Gastroenterol. 2006;12:6683–6688. doi: 10.3748/wjg.v12.i41.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng X, Ye X, Wang S. Identification of novel immunogenic proteins of Shigella flexneri 2a by proteomic methodologies. Vaccine. 2004;22:2750–2756. doi: 10.1016/j.vaccine.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 19.Ying T, Wang H, Li M, Wang J, et al. Immunoproteomics of outer membrane proteins and extracellular proteins of Shigella flexneri 2a 2457T. Proteomics. 2005;5:4777–4793. doi: 10.1002/pmic.200401326. [DOI] [PubMed] [Google Scholar]
- 20.Pieper R, Huang ST, Clark DJ, Robinson JM, et al. Characterizing the dynamic nature of the Yersinia pestis periplasmic proteome in response to nutrient exhaustion and temperature change. Proteomics. 2008;8:1442–1458. doi: 10.1002/pmic.200700923. [DOI] [PubMed] [Google Scholar]
- 21.Mendizabal-Morris CA, Mata LJ, Gangarosa EJ, Guzman G. Epidemic Shiga-bacillus dysentery in Central America. Derivation of the epidemic and its progression in Guatemala, 1968–69. Am J Trop Med Hyg. 1971;20:927–933. doi: 10.4269/ajtmh.1971.20.927. [DOI] [PubMed] [Google Scholar]
- 22.Gatlin CL, Pieper R, Huang ST, Mongodin E, et al. Proteomic profiling of cell envelope-associated proteins from Staphylococcus aureus. Proteomics. 2006;6:1530–1549. doi: 10.1002/pmic.200500253. [DOI] [PubMed] [Google Scholar]
- 23.Yang F, Yang J, Zhang X, Chen L, et al. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33:6445–6458. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei C, Yang J, Zhu J, Zhang X, et al. Comprehensive proteomic analysis of Shigella flexneri 2a membrane proteins. J Proteome Res. 2006;5:1860–1865. doi: 10.1021/pr0601741. [DOI] [PubMed] [Google Scholar]
- 25.Sargent F. Constructing the wonders of the bacterial world: biosynthesis of complex enzymes. Microbiology. 2007;153:633–651. doi: 10.1099/mic.0.2006/004762-0. [DOI] [PubMed] [Google Scholar]
- 26.McNicholas PM, Gunsalus RP. The molybdate-responsive Escherichia coli ModE transcriptional regulator coordinates periplasmic nitrate reductase (napFDAGHBC) operon expression with nitrate and molybdate availability. J Bacteriol. 2002;184:3253–3259. doi: 10.1128/JB.184.12.3253-3259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Encyclopedia of Escherichia coli K-12 Genes and Metabolism. 2009 http://www.ecocyc.org/
- 28.Kirkpatrick C, Maurer LM, Oyelakin NE, Yoncheva YN, et al. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J Bacteriol. 2001;183:6466–6477. doi: 10.1128/JB.183.21.6466-6477.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner AF, Schultz S, Bomke J, Pils T, et al. YfiD of Escherichia coli and Y06I of bacteriophage T4 as autonomous glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvate formate-lyase. Biochem Biophys Res Commun. 2001;285:456–462. doi: 10.1006/bbrc.2001.5186. [DOI] [PubMed] [Google Scholar]
- 30.Fee JA. Regulation of sod genes in Escherichia coli: relevance to superoxide dismutase function. Mol Microbiol. 1991;5:2599–2610. doi: 10.1111/j.1365-2958.1991.tb01968.x. [DOI] [PubMed] [Google Scholar]
- 31.Bearson S, Bearson B, Foster JW. Acid stress responses in enterobacteria. FEMS Microbiol Lett. 1997;147:173–180. doi: 10.1111/j.1574-6968.1997.tb10238.x. [DOI] [PubMed] [Google Scholar]
- 32.Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 33.Iyer R, Iverson TM, Accardi A, Miller C. A biological role for prokaryotic ClC chloride channels. Nature. 2002;419:715–718. doi: 10.1038/nature01000. [DOI] [PubMed] [Google Scholar]
- 34.Jennison AV, Verma NK. The acid-resistance pathways of Shigella flexneri 2457T. Microbiology. 2007;153:2593–2602. doi: 10.1099/mic.0.2007/006718-0. [DOI] [PubMed] [Google Scholar]
- 35.Capitani G, De Biase D, Aurizi C, Gut H, et al. Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. Embo J. 2003;22:4027–4037. doi: 10.1093/emboj/cdg403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuda N, Church GM. Regulatory network of acid resistance genes in Escherichia coli. Mol Microbiol. 2003;48:699–712. doi: 10.1046/j.1365-2958.2003.03477.x. [DOI] [PubMed] [Google Scholar]
- 37.Lin J, Smith MP, Chapin KC, Baik HS, et al. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kern R, Malki A, Abdallah J, Tagourti J, Richarme G. Escherichia coli HdeB is an acid stress chaperone. J Bacteriol. 2007;189:603–610. doi: 10.1128/JB.01522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malki A, Le HT, Milles S, Kern R, et al. Solubilization of protein aggregates by the acid stress chaperones HdeA and HdeB. J Biol Chem. 2008;283:13679–13687. doi: 10.1074/jbc.M800869200. [DOI] [PubMed] [Google Scholar]
- 40.Zurawski DV, Mumy KL, Faherty CS, McCormick BA, Maurelli AT. Shigella flexneri type III secretion system effectors OspB and OspF target the nucleus to downregulate the host inflammatory response via interactions with retinoblastoma protein. Mol Microbiol. 2008 doi: 10.1111/j.1365-2958.2008.06524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Picking WL, Nishioka H, Hearn PD, Baxter MA, et al. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect Immun. 2005;73:1432–1440. doi: 10.1128/IAI.73.3.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menard R, Dehio C, Sansonetti PJ. Bacterial entry into epithelial cells: the paradigm of Shigella. Trends Microbiol. 1996;4:220–226. doi: 10.1016/0966-842X(96)10039-1. [DOI] [PubMed] [Google Scholar]
- 43.Li H, Xu H, Zhou Y, Zhang J, et al. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 2007;315:1000–1003. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- 44.Ohya K, Handa Y, Ogawa M, Suzuki M, Sasakawa C. IpgB1 is a novel Shigella effector protein involved in bacterial invasion of host cells. Its activity to promote membrane ruffling via Rac1 and Cdc42 activation. J Biol Chem. 2005;280:24022–24034. doi: 10.1074/jbc.M502509200. [DOI] [PubMed] [Google Scholar]
- 45.Bourdet-Sicard R, Rudiger M, Jockusch BM, Gounon P, et al. Binding of the Shigella protein IpaA to vinculin induces F-actin depolymerization. Embo J. 1999;18:5853–5862. doi: 10.1093/emboj/18.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramarao N, Le Clainche C, Izard T, Bourdet-Sicard R, et al. Capping of actin filaments by vinculin activated by the Shigella IpaA carboxyl-terminal domain. FEBS Lett. 2007;581:853–857. doi: 10.1016/j.febslet.2007.01.057. [DOI] [PubMed] [Google Scholar]
- 47.Ogawa M, Yoshimori T, Suzuki T, Sagara H, et al. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 48.Santapaola D, Del Chierico F, Petrucca A, Uzzau S, et al. Apyrase, the product of the virulence plasmid-encoded phoN2 (apy) gene of Shigella flexneri, is necessary for proper unipolar IcsA localization and for efficient intercellular spread. J Bacteriol. 2006;188:1620–1627. doi: 10.1128/JB.188.4.1620-1627.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida S, Katayama E, Kuwae A, Mimuro H, et al. Shigella deliver an effector protein to trigger host microtubule destabilization, which promotes Rac1 activity and efficient bacterial internalization. Embo J. 2002;21:2923–2935. doi: 10.1093/emboj/cdf319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zurawski DV, Mitsuhata C, Mumy KL, McCormick BA, Maurelli AT. OspF and OspC1 are Shigella flexneri type III secretion system effectors that are required for postinvasion aspects of virulence. Infect Immun. 2006;74:5964–5976. doi: 10.1128/IAI.00594-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zurawski DV, Mumy KL, Faherty CS, McCormick BA, Maurelli AT. Shigella flexneri type III secretion system effectors OspB and OspF target the nucleus to downregulate the host inflammatory response via interactions with retinoblastoma protein. Mol Microbiol. 2009;71:350–368. doi: 10.1111/j.1365-2958.2008.06524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim DW, Lenzen G, Page AL, Legrain P, et al. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci U S A. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zurawski DV, Mumy KL, Badea L, Prentice JA, et al. The NleE/OspZ family of effector proteins is required for polymorphonuclear transepithelial migration, a characteristic shared by enteropathogenic Escherichia coli and Shigella flexneri infections. Infect Immun. 2008;76:369–379. doi: 10.1128/IAI.00684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tzipori S, Sheoran A, Akiyoshi D, Donohue-Rolfe A, Trachtman H. Antibody therapy in the management of shiga toxin-induced hemolytic uremic syndrome. Clin Microbiol Rev. 2004;17:926–941. doi: 10.1128/CMR.17.4.926-941.2004. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Unkmeir A, Schmidt H. Structural analysis of phage-borne stx genes and their flanking sequences in shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect Immun. 2000;68:4856–4864. doi: 10.1128/iai.68.9.4856-4864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe H, Timmis KN. A small plasmid in Shigella dysenteriae 1 specifies one or more functions essential for O antigen production and bacterial virulence. Infect Immun. 1984;43:391–396. doi: 10.1128/iai.43.1.391-396.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Horvath I, Multhoff G, Sonnleitner A, Vigh L. Membrane-associated stress proteins: more than simply chaperones. Biochim Biophys Acta. 2008;1778:1653–1664. doi: 10.1016/j.bbamem.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 58.Doyle SM, Wickner S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 59.Meibom KL, Dubail I, Dupuis M, Barel M, et al. The heat-shock protein ClpB of Francisella tularensis is involved in stress tolerance and is required for multiplication in target organs of infected mice. Mol Microbiol. 2008;67:1384–1401. doi: 10.1111/j.1365-2958.2008.06139.x. [DOI] [PubMed] [Google Scholar]
- 60.Reid AN, Pandey R, Palyada K, Naikare H, Stintzi A. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Appl Environ Microbiol. 2008;74:1583–1597. doi: 10.1128/AEM.01507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol. 2005;3:383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- 62.Sperandio V, Mellies JL, Nguyen W, Shin S, Kaper JB. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:15196–15201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bu X, Zhu L, Liu X, Zhao G, et al. Wei Sheng Wu Xue Bao. 2008;48:905–910. [PubMed] [Google Scholar]
- 64.Multhoff G. Heat shock proteins in immunity. Handb Exp Pharmacol. 2006:279–304. doi: 10.1007/3-540-29717-0_12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.