Changes in protein tyrosine phosphatase 1B expression affect duration and amplitude of EphA3 phosphorylation and cell surface concentration.
Abstract
Eph receptors orchestrate cell positioning during normal and oncogenic development. Their function is spatially and temporally controlled by protein tyrosine phosphatases (PTPs), but the underlying mechanisms are unclear and the identity of most regulatory PTPs are unknown. We demonstrate here that PTP1B governs signaling and biological activity of EphA3. Changes in PTP1B expression significantly affect duration and amplitude of EphA3 phosphorylation and biological function, whereas confocal fluorescence lifetime imaging microscopy (FLIM) reveals direct interactions between PTP1B and EphA3 before ligand-stimulated receptor internalization and, subsequently, on endosomes. Moreover, overexpression of wild-type (w/t) PTP1B and the [D-A] substrate–trapping mutant decelerate ephrin-induced EphA3 trafficking in a dose-dependent manner, which reveals its role in controlling EphA3 cell surface concentration. Furthermore, we provide evidence that in areas of Eph/ephrin-mediated cell–cell contacts, the EphA3–PTP1B interaction can occur directly at the plasma membrane. Our studies for the first time provide molecular, mechanistic, and functional insights into the role of PTP1B controlling Eph/ephrin-facilitated cellular interactions.
Introduction
Eph receptor tyrosine kinases (Eph RTKs) and cognate cell surface ephrin ligands orchestrate patterns of cellular organization by arranging cell–cell contacts through control of adhesive and contractile cell functions (Lackmann and Boyd, 2008; Pasquale, 2008). They assemble multivalent (Himanen et al., 2001) signaling clusters, which initiate Eph receptor forward signaling via conserved juxtamembrane and activation loop phosphotyrosines (PYs; Wybenga-Groot et al., 2001), and reverse signaling by clustered ephrins (Pasquale, 2008). The overall signal strength largely determines if cells respond to ephrin contact by repulsion or by adhesion (Holmberg and Frisén, 2002; Wimmer-Kleikamp et al., 2008). Similar to other RTKs, specific protein tyrosine phosphatases (PTPs) are thought to control Eph activation and shape cellular responses ensuing from contacts between Eph- and ephrin-expressing cells (Lackmann and Boyd, 2008). Consistent with this notion, PTPRO controls EphA4 phosphorylation in retinal ganglion cells and modulates their sensitivity to ephrin contact (Shintani et al., 2006), and EphB2 activation is controlled by the leukocyte common antigen-related tyrosine phosphatase receptor (LAR-1; Poliakov et al., 2008), whereas elevated PTP activity in EphA3-overexpressing leukemia cells shifts the response to ephrinA5 from cell–cell repulsion to adhesion (Wimmer-Kleikamp et al., 2008). Moreover, insulin secretion from pancreatic β cell granules, triggered by glucose-induced elevation of PTP activity, attenuates EphA5 forward and promotes ephrinA reverse signaling (Konstantinova et al., 2007).
PTP1B is a prototypic nonreceptor tyrosine phosphatase, with established roles as a negative regulator of several RTKs, including the receptors for insulin, epidermal growth factor, and platelet-derived growth factor β (Bourdeau et al., 2005; Tonks, 2006), and as a positive modulator of integrin and cadherin signaling (Burridge et al., 2006; Sallee et al., 2006). Within cells, PTP1B is anchored to the cytoplasmic face of the ER (Frangioni et al., 1992) so that its interaction with transmembrane or membrane-proximal substrates, as well as the timing and site of their dephosphorylation, poses a conceptual dilemma. Recent findings provide strong evidence for dynamic, spatially and temporally controlled interactions between PTP1B and its transmembrane or membrane-associated substrates, whereby dephosphorylation by PTP1B occurs when endocytosed RTKs transit past the ER (Haj et al., 2002; Boute et al., 2003). Other studies, however, suggest that PTP1B contacts transmembrane receptors and cell–matrix adhesion sites directly (Hernández et al., 2006; Anderie et al., 2007), and a recent study suggested the existence of microtubule-dependent positioning of ER-bound PTP1B to the periphery of growth cones that is stabilized by cell–cell contacts (Fuentes and Arregui, 2009).
We now demonstrate that rapid recruitment of PTP1B to the cell surface controls activity, trafficking, and function of EphA3 in cell contact with ephrinA5-expressing cells. We show that EphA3 phosphorylation and endocytosis is tightly controlled by PTP1B in normal and cancer cell lines, consequently regulating downstream cell morphological responses. Our study provides the first comprehensive evidence for a central role of PTP1B in controlling Eph receptor function by modulating the amplitude and biological consequences of Eph/ephrin signaling.
Results
PTP1B negatively regulates ephrinA5-induced EphA3 phosphorylation
We reported previously that EphA3 kinase activity and biological functions are tightly controlled by tyrosine phosphatase activity, although PTPs implicated in Eph signaling, including low molecular weight PTP (LMW-PTP) and Src homology 2 domain-containing PTP 2 (SHP2), appeared not to affect EphA3 phosphorylation directly (Wimmer-Kleikamp et al., 2008). However, biotin-iodoacetamide labeling of reactive oxygen-sensitive cysteine residues (Kim et al., 2000) in whole cell lysates from ephrinA5-stimulated cells identified a Mr 45–50-kD protein as a potential PTP that is transiently inactivated by reactive oxygen species (ROS; Tonks, 2005) during EphA3 signaling (unpublished data). A matching molecular weight and circumstantial evidence suggesting that the EphA3 activation loop tyrosine was a potential substrate (Mertins et al., 2008) prompted us to explore PTP1B for its potential role in controlling EphA3 signaling.
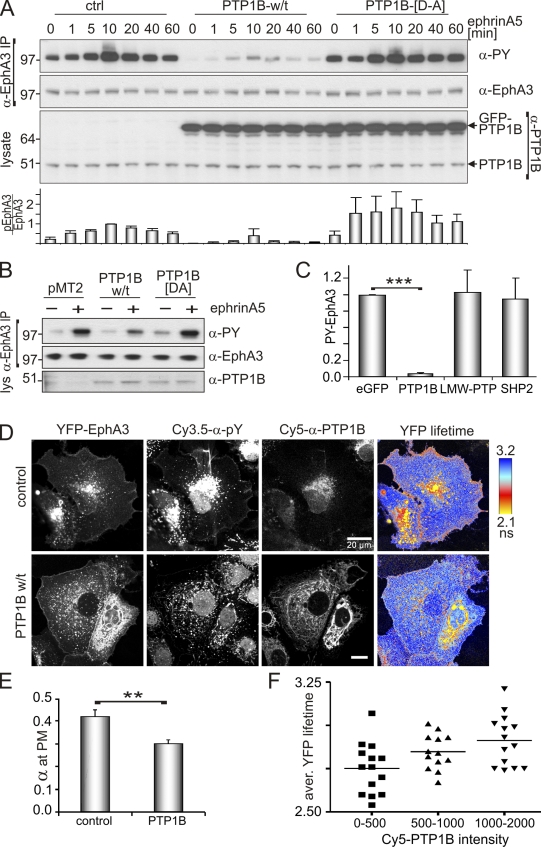
We first tested the impact of PTP1B overexpression on EphA3 activation in EphA3-positive HEK293T (EphA3/HEK293T) cells (Fig. 1 A) and in 22Rv1 prostate carcinoma cells with high endogenous EphA3 expression (Fig. 1 B). In both cell lines, expression of wild-type (w/t) PTP1B significantly reduced ephrinA5-induced EphA3 phosphorylation (Fig. 1, A–C; and Fig. S1 B), whereas overexpression of the catalytically impaired substrate trapping mutant PTP1B-[D-A] (Flint et al., 1997) enhanced the duration (Fig. 1 A) and amplitude (Fig. 1, A and B) of EphA3 phosphorylation in stimulated and nonstimulated cells. By comparison, overexpression of T cell PTP (TC-PTP), a phosphatase with high structural and functional homology to PTP1B, had only a moderate effect on EphA3 phosphorylation in stimulated 22Rv1 cells (Fig. S1, A and B). Similarly, LMW-PTP and SHP2, which had been implicated in Eph signaling (Stein et al., 1998; Miao et al., 2000; Parri et al., 2005), did not affect ephrin-induced EphA3 phosphorylation in EphA3/HEK293T cells (Fig. 1 C).
Figure 1.
PTP1B controls ephrin-induced phosphorylation of EphA3. (A) α-EphA3 immunoprecipitates and whole cell lysates from GFP-PTP1B, GFP-PTP1B-[D-A], or control vector-transfected EphA3/HEK293T cells, treated with ephrinA5-Fc, were analyzed by Western blotting with antibodies as indicated. The ratio of phosphorylated EphA3/total EphA3 (n = 3) was estimated (densitometry); mean ± SE are shown (error bars). (B) α-EphA3 immunoprecipitates and cell lysates of PTP1B, PTP1B-[D-A], or control vector (pMT2)-transfected 22Rv1 prostate carcinoma cells, with or without 10 min of ephrinA5 stimulation, were immunoblotted with the appropriate antibodies. Molecular mass standards are indicated next to the gel blots in kilodaltons. (C) EphrinA5-induced EphA3 phosphorylation in α-EphA3 immunoprecipitates from EphA3/HEK293T cells transfected with EGFP (control) vector, GFP-PTP1B, GFP-LMW-PTP, or GFP-SHP2, was quantified by densitometry analysis of Western blots (n = 3); mean PY-EphA3 levels, normalized to the EGFP-transfected control, ±SE are shown (error bars; ***, P < 0.001). (D) EphrinA5-induced EphA3 phosphorylation was monitored in PTP1B w/t and in control vector-transfected COS7 cells by confocal FLIM using YFP-EphA3 as fluorescence donor and Cy3.5-labeled PY72 as an acceptor. The YFP fluorescence lifetime maps are illustrated together with confocal micrographs revealing YFP-EphA3, PY, and PTP1B, the latter detected with labeled α-PY (Cy3.5PY72) and α-PTP1B (Cy5FG6) antibodies. Endogenous PTP1B (top) is shown at a higher display setting compared with recombinant w/t PTP1B (bottom). (E) The relative fraction (α) of membrane-proximal activated EphA3 receptors in PTP1B-overexpressing and control cells was calculated pixel-by-pixel from the ratio of the amplitude of the short lifetime component and the sum of the two amplitudes (n = 40 cells); mean ± SE are shown (error bars; **, P < 0.005). (F) The correlation between EphA3-YFP lifetimes (averaged across the whole cell) and the PTP1B expression level (determined from the fluorescence intensity of Cy5-α-PTP1B staining) is illustrated, where increased lifetime indicates decreased EphA3 phosphorylation. Each data point represents measurements from an individual cell.
We next examined PTP1B-controlled EphA3 phosphorylation in whole cells using confocal fluorescence lifetime imaging microscopy (FLIM) to map the energy transfer between YFP-tagged EphA3 as the fluorescence energy donor and Cy3.5-tagged anti-PY (α-PY) antibody (Reynolds et al., 2003) as the fluorescence acceptor. As anticipated, ephrinA5 stimulation yielded notable Förster resonance energy transfer (FRET) at the plasma membrane (PM) and in membrane-proximal vesicles (Fig. 1 D). PY-EphA3 was also present in perinuclear compartments of stimulated and nonstimulated cells (Fig. S1 C), which likely reflects cytosolic accumulation of activated receptors present because of transient overexpression. Transfection with w/t PTP1B markedly reduced FRET (i.e., increased the YFP fluorescence lifetime) in all cellular compartments (Fig. 1 D and Fig. S1 D), which indicates a consistent reduction in levels of ephrinA5-activated EphA3 receptors (Fig. 1 E). A positive correlation between increasing PTP1B expression (measured as Cy5-α-PTP1B intensity) and increasing YFP fluorescence lifetimes (which is indicative of reduced FRET; Fig. 1 F) implies that FRET is a genuine measure of PTP1B-catalyzed EphA3 dephosphorylation, which we observed consistently across whole cells or within relevant PM compartments (Fig. S1 D).
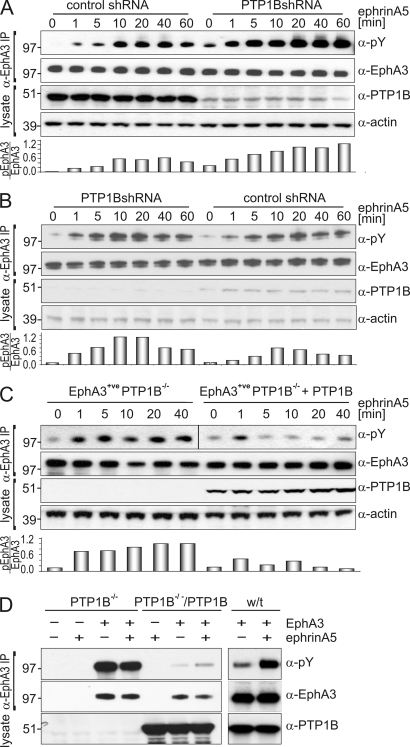
We further assessed if PTP1B counteracts EphA3 signaling by silencing PTP1B expression in EphA3/HEK293T and in 22Rv1 prostate carcinoma cells using small-hairpin RNA (shRNA). In clones from both cell lines, notably reduced PTP1B expression coincided with elevated ephrinA5-induced EphA3 phosphorylation (Fig. 2, A and B). Although basal EphA3 phosphorylation was only moderately elevated in EphA3/HEK293T cells, we noted a prominent increase in duration and amplitude of phosphorylation in both cell lines (Fig. 2, A and B). Likewise, in PTP1B−/− mouse embryonic fibroblasts (MEFs) engineered to stably express EphA3, ephrin-induced EphA3 phosphorylation was notably enhanced and prolonged compared with their PTP1B-reconstituted counterparts (Fig. 2 C). EphA3-transfected w/t MEFs with comparable levels of endogenous PTP1B responded to ephrinA5 stimulation with markedly increased phosphorylation, whereas transient EphA3 expression in PTP1B−/− MEFs, in addition, triggered EphA3 hyperphosphorylation even without ephrinA5 activation (Fig. 2 D). This deregulated EphA3 activation likely reflects an acute lack of PTP control over EphA3 phosphorylation in transiently transfected cells, which lack compensatory mechanisms that would be active in stably transfected cells (Fig. 2 C).
Figure 2.
Abrogated PTP1B expression causes enhanced and prolonged EphA3 activation. (A and B) EphA3/HEK293Ts (A) and 22Rv1 cells (B), stably transfected with PTP1B-shRNA– or nontarget control vector–containing lentiviral transduction particles, were subjected to ephrinA5 stimulation. α-EphA3 immunoprecipitates and whole cell lysates were examined by immunoblotting with the appropriate antibodies. Densitometry quantifies EphA3 phosphorylation relative to total EphA3 expression. Data are representative of at least five (A) and three (B) independent experiments, respectively. (C) The time course of ephrinA5-induced EphA3 phosphorylation in PTP1B-reconstituted or parental PTP1B−/− MEFs, both stably transfected with EphA3, was assessed by immunoblotting of anti-EphA3 immunoprecipitates using antibodies against EphA3, PY, and PTP1B, as indicated. Densitometry quantifies relative EphA3 phosphorylation, which is representative of two experimental repeats. The black line indicates that intervening lanes have been spliced out. (D) PTP1B−/− MEFs with or without reconstitution with w/t PTP1B and w/t MEFs were transiently transfected with EphA3. α-EphA3 immunoprecipitates from ephrinA5-stimulated or nonstimulated cells were analyzed by Western blot analysis using antibodies against PY-EphA3, EphA3, and PTP1B as indicated. Molecular mass standards are indicated next to the gel blots in kilodaltons.
Association of PTP1B-[D-A] with EphA3 relies on intact tyrosine phosphorylation sites
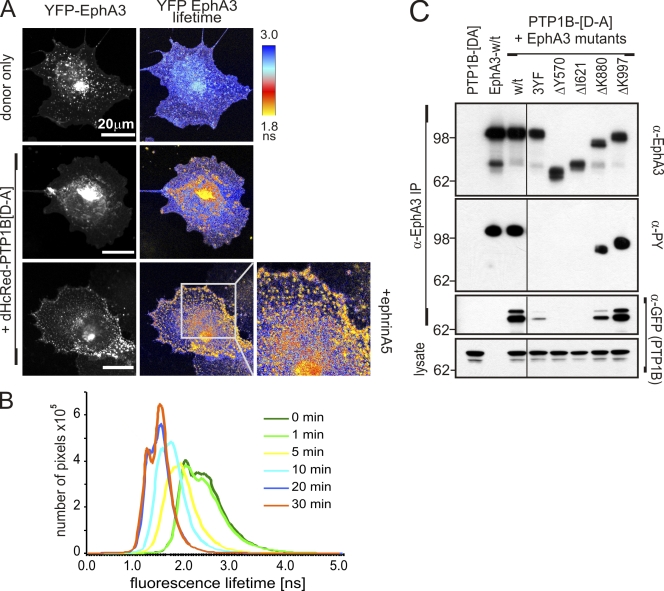
Our findings prompted us to study the interaction between YFP-EphA3 and dHcRed-PTP1B-[D-A] and map the subcellular location of PTP1B recruitment in intact cells using confocal FLIM. FRET-induced low YFP-EphA3 donor lifetimes in intracellular and in some PM-proximal compartments of nonstimulated cells (Fig. 3 A, middle, yellow-orange) imply that EphA3 interacts directly with PTP1B-[D-A], even in the absence of ephrin. Stimulation with ephrinA5 resulted in a dramatic increase in PM-proximal FRET, particularly in areas of cell–cell contact (Fig. 3 A, bottom), which confirmed prominent PTP1B-[D-A] recruitment to ligand-activated EphA3. Also, live-cell FLIM during ephrinA5-triggered EphA3 activation revealed a gradual decrease in YFP lifetimes (increased FRET) over 30 min, which confirmed the continued recruitment of PTP1B-[D-A] over and above the basal association observed in nonstimulated cells (Fig. 3 B and Fig. S2 A). Interestingly, we also detected FRET in cells expressing dHcRed-PTP1B-[D-A] and GFP-tagged EphB2, which suggests that PTP1B may also control phosphorylation of EphB receptors, whereas a [C-S] substrate–trapping mutant of LMW-PTP used in a control experiment appeared not to associate with EphA3 directly (Fig. S2 B).
Figure 3.
The interaction between PTP1B and EphA3 is kinase dependent. (A) The association between YFP-EphA3 and dHcRed-PTP1B-[D-A] in untreated or ephrinA5-stimulated COS7 cells was analyzed by confocal FLIM. Confocal micrographs of YFP-EphA3 (donor) fluorescence and the YFP fluorescence lifetime maps are shown. The boxed area in the bottom right is shown at higher magnification. (B) COS7 cells transfected with YFP-EphA3 and dHcRed-PTP1B-[D-A] were stimulated with ephrinA5, and live cells were analyzed by FLIM. The distribution of YFP fluorescence lifetimes at various time points during stimulation is plotted. The corresponding confocal images and lifetime maps are presented in Fig. S4 A. (C) NG108 cells, lacking endogenous Ephs (Elowe et al., 2001), cotransfected with cDNAs encoding GFP-PTP1B-[D-A] and w/t or mutant EphA3 (containing tyrosine-replacement or truncation mutants as indicated) were stimulated with ephrinA5 before lysis, α-EphA3 immunoprecipitation, and Western blot analysis with antibodies as indicated. The black line indicates that intervening lanes have been spliced out. Molecular mass standards are indicated next to the gel blots in kilodaltons.
To assess the protein domains and/or interaction motifs responsible for the interaction between EphA3 and PTP1B-[D-A], we performed coimmunoprecipitation analysis of various mutant EphA3 proteins: mutants lacking the essential juxtamembrane and activation loop tyrosines (3Y-F), the cytoplasmic domain (Δ570), or kinase domain (Δ621) showed no detectable tyrosine phosphorylation and marginal ([3YF] mutant) or no capacity to associate with PTP1B-[D-A] (Fig. 3 C). In agreement, we did not observe FRET in PTP1B−/− MEFs coexpressing GFP-EphA3-[3Y-F] and dHcRed-PTP1B-[D-A] (Fig. S2 C), which together suggest that the presence of the EphA3 consensus tyrosine phosphorylation sites (Wybenga-Groot et al., 2001) is a prerequisite for the PTP1B-[D-A]/EphA3 association. Although truncation of the PDZ binding motif (ΔK997) did not affect phosphorylation or PTP1B-[D-A] association, by comparison both were reduced in the EphA3 sterile alpha motif (SAM) domain truncation mutant (ΔK880; Fig. 3 C), which is in line with a potential role of the SAM domain in stabilizing EphA3 signaling complexes during activation and downstream signaling (Stapleton et al., 1999).
PTP1B directly interacts with activated EphA3 at contacts to ephrinA5-expressing cells
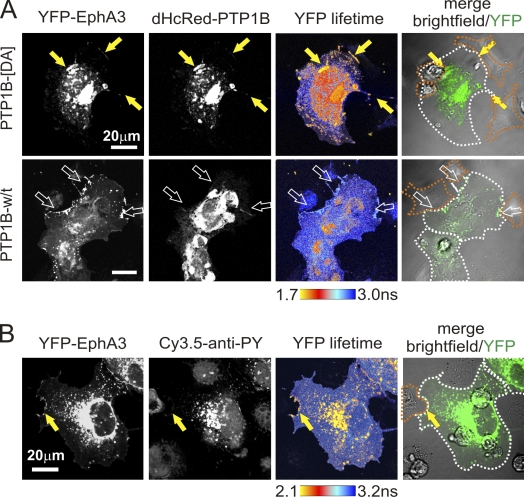
Considering that physiological Eph-ephrin signaling is only triggered at sites of contact between Eph and ephrin-expressing cells, the question arises if the ER-resident PTP1B can contact EphA3 that is already at the cell surface. To address this notion, we cotransfected dHcRed-PTP1B-[D-A] and YFP-EphA3 into COS7 cells and analyzed their interaction with ephrinA5-expressing HEK293T cells. FLIM revealed, in addition to obvious pools of the colocalized proteins in the cytosol, conspicuous clusters of EphA3-associated PTP1B-[D-A] clearly marking PM interfaces with ephrinA5-expressing cells (Fig. 4 A, yellow arrows). Using Cy3.5-labeled α-PY antibody as a fluorescence acceptor, FLIM suggested that these prominent sites of EphA3/PTP1B-[D-A] association corresponded with sites of marked EphA3 phosphorylation (Fig. 4 B). Of note, although FRET was not apparent in cells expressing w/t dHcRed-PTP1B as a fluorescence acceptor, their contacts with ephrinA5 cells were still lined with pronounced EphA3 accumulation (Fig. 4 A, bottom, open arrows). Together, these findings suggest that PTP1B is indeed recruited to ephrinA5–EphA3 complexes, which tether interacting cells; however, this analysis does not discern if the contact between PTP1B-[D-A] and activated EphA3 occurs at the PM or on PM-proximal, early endosomal vesicles.
Figure 4.
PTP1B is recruited to sites of Eph/ephrin contact. (A) COS7 cells overexpressing YFP-EphA3 and dHcRed-PTP1B-[D-A] or w/t dHcRed-PTP1B were co-cultured (20 min) with ephrinA5/HEK293T cells and analyzed by FLIM to monitor direct interactions between EphA3 and PTP1B. Confocal images and YFP-EphA3 lifetime maps are shown. White broken lines mark the boundaries of EphA3-expressing cells; orange dotted lines mark those of ephrinA5 cells. Yellow arrows indicate EphA3-associated PTP1B-[D-A] and FRET at PM interfaces with ephrinA5-expressing cells. Open arrows indicate accumulated EphA3 in w/t dHcRed-PTP1B–expressing cells at sites of contact with ephrinA5 cells. (B) Co-cultures (20 min) between YFP-EphA3 expressing COS7 cells and ephrinA5-expressing HEK/293T cells were fixed, permeabilized, and stained with Cy3.5-tagged α-PY antibody to monitor EphA3 phosphorylation by using FRET between YFP-EphA3 and the Cy3.5-α-PY antibody. Dotted lines are as described in A. Yellow arrows indicate EphA3-associated Cy3.5-α-PY antibody and FRET at sites of marked EphA3 phosphorylation.
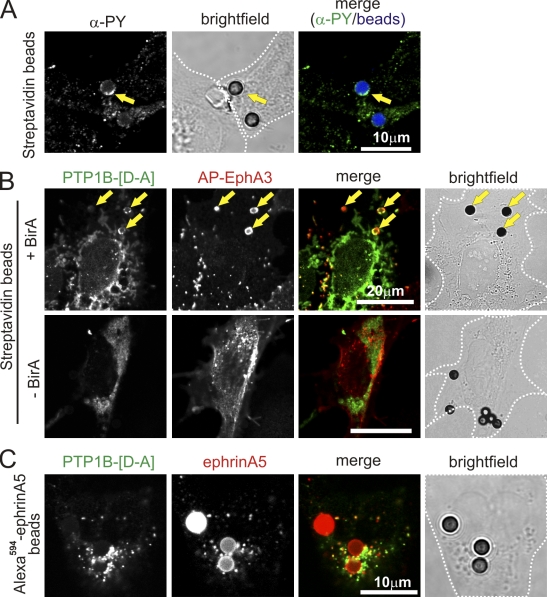
To address this question, we imaged PTP1B-[D-A] substrate trapping in COS 7 cells coexpressing EphA3 that is tagged on its N terminus with a biotin acceptor peptide (APN), and Escherichia coli biotin ligase (BirA; Howarth and Ting, 2008). EphA3 in these APN-EphA3/BirA COS 7 cells is constitutively biotinylated so that streptavidin (SA) conjugated to beads can be used as surface-tethered agonist that clusters (Janes et al., 2009) and activates the biotinylated APN-EphA3 (Fig. 5 A), but cannot be shed by EphA3-associated ADAM10 (Janes et al., 2005). This trapping of activated EphA3 at the cell surface effectively prevents ephrin-induced endocytosis and allows monitoring protein–protein interactions that normally may occur shortly before or during EphA3 internalization. Staining of intact (nonpermeabilized) cells with α-EphA3 monoclonal antibodies (mAb) and their analysis by confocal microscopy revealed coinciding EphA3 and GFP-PTP1B-[D-A]–associated fluorescence accumulating around many of the SA beads, a pattern that was absent in control cells not expressing biotin ligase (−BirA) and thus lacking the capacity to tether SA beads (Fig. 5 B). By comparison, incubation of APN-EphA3/HEK293T cells with Alexa Fluor 594–ephrinA5–Fc–coated beads yielded notable, but considerably weaker GFP-PTP1B–associated fluorescence that is directly associated with the beads (Fig. 5 C), which confirmed that rapid and continuous ephrin cleavage (Janes et al., 2005) and endocytosis of activated EphA3 (as indicated by fluorescent cytosolic vesicles) prevents its accumulation on the beads. Collectively, these data unambiguously demonstrate the capacity of PTP1B to associate with activated EphA3 at regions of the cell surface that are in contact with a (cell) surface-bound agonist.
Figure 5.
PTP1B interacts with EphA3 at the cell surface. (A) EphA3 clustering by SA beads provokes tyrosine phosphorylation. COS7 cells, cotransfected with APN-EphA3 and BirA, were stimulated with SA Dynabeads. Tyrosine phosphorylation was evaluated by confocal microscopy of fixed and permeabilized cells stained with Cy3.5α-PY (PY72) antibodies. SA Dynabeads appear blue in the merged image; yellow arrows mark beads with α-PY association. (B) COS7 cells, cotransfected with GFP-PTP1B-[D-A], APN-EphA3, and TM-BirA (as indicated), were stimulated with SA Dynabeads. Cell surface EphA3 was labeled on intact cells with IIIA4 α-EphA3 mAb and detected with Alexa Fluor 546–labeled secondary antibodies by confocal microscopy. Yellow arrows mark SA Dynabeads where PTP1B/EphA3 colocalization is apparent. (C) GFP-PTP1B-[D-A] and EphA3-transfected COS7 cells were stimulated with Alexa Fluor 594–ephrinA5–Fc–coated Protein A Dynabeads, then fixed and analyzed by confocal microscopy. White dotted lines mark cell boundaries from bright-field images.
Eph/ephrin-mediated cell contraction and cell sorting is modulated by PTP1B
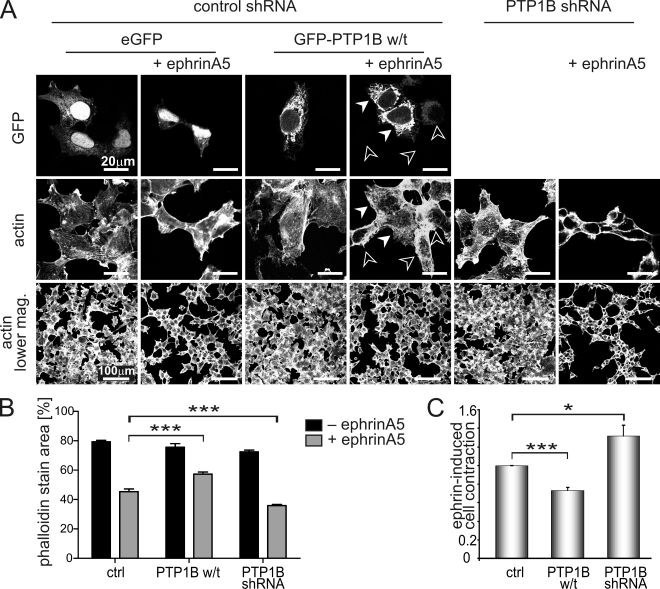
We next tested if modulating PTP1B expression by cDNA transfection or stable shRNA-mediated silencing would impact on cell biological responses to EphA3 signaling. As described previously (Lawrenson et al., 2002), stimulation of EphA3/HEK293T cells with preclustered ephrinA5-Fc caused pronounced contraction of the actin cytoskeleton (Fig. 6 A, EGFP ctrl), which is considered to be the prerequisite for cell–cell repulsion. Estimation of the cellular footprint from the actin-associated fluorescence before and after ephrinA5 treatment revealed that PTP1B overexpression significantly attenuates cytoskeletal contraction in ephrinA5-stimulated cells (Fig. 6, A–C, PTP1B w/t). Notably, cells with high exogenous PTP1B remained stretched out (Fig. 6 A, closed arrowheads), whereas neighboring cells with low/undetectable GFP-PTP1B were contracted. However, silencing of PTP1B significantly enhanced ephrin-induced cell contraction (Fig. 6, A–C, PTP1B shRNA).
Figure 6.
Modulation of PTP1B expression alters ephrin-induced cell contraction. (A) EphA3/HEK293T cells stably transfected with PTP1B-shRNA and nontarget control shRNA EphA3/HEK293T cells transiently expressing GFP alone (control) or w/t GFP-PTP1B were stimulated with ephrinA5 (40 min) or left untreated. Cell morphology, observed by Alexa Fluor 647–phalloidin staining and GFP-(PTP1B) expression, was analyzed by confocal microscopy. Closed and open arrowheads mark cells with high and low PTP1B levels, respectively. (B) Thresholded fluorescence area of Alexa Fluor 647–phalloidin staining was determined in 10 low-magnification images per condition of the experiment shown in A and plotted as mean ± SE (error bars; ***, P < 0.001). (C) Relative ephrinA5-induced cell contraction was estimated from the ratio of the actin-associated fluorescence footprint in micrographs taken after and before ephrinA5 treatment: whole 18-mm-diameter coverslips (equivalent to 100 low-magnification images; A) of phalloidin-stained cells were imaged by “tile scanning” on a fluorescence microscope. Mean ± SE from five separate experiments are shown (error bars; ***, P < 0.001; *, P < 0.05).
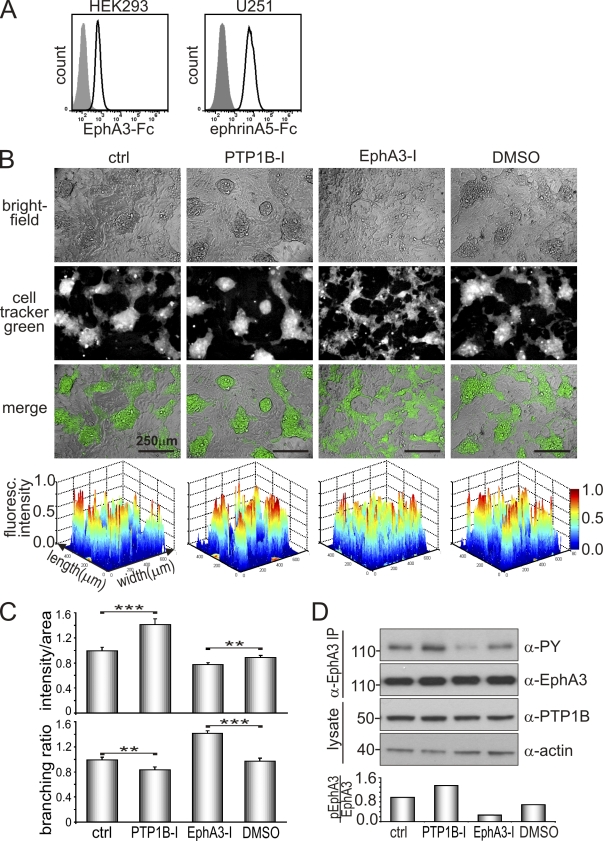
We assessed the role of PTP1B in regulating EphA3 function to facilitate cell–cell repulsion from ephrin-expressing cells in a more physiological setting by adopting a cell co-culture system that is regarded as a model of Eph/ephrin-mediated cell sorting (Poliakov et al., 2008; Jørgensen et al., 2009) during tissue boundary formation (Mellitzer et al., 1999). To avoid uncontrolled effects due to overexpression of exogenous proteins, we selected for this assay cell lines with endogenous Eph and ephrin expression. To this end, we analyzed co-cultures of U251 glioblastoma cells and HEK293 cells with demonstrable endogenous expression of Eph and ephrin, as shown by binding of ephrinA5-Fc and EphA3-Fc, respectively (Fig. 7 A), and of PTP1B (Fig. 7 D). As expected, fluorescent-labeled HEK293 cells were segregated into distinct, partially interconnected cell clusters that are bordered by U251 cells (Fig. 7 B, ctrl). Treatment with PTP1B inhibitor tightened these clusters, as indicated by reduced dispersion of fluorescence intensity peaks in corresponding 2D and 3D contour maps (Fig. 7 B, bottom; and Fig. S3, A and B), a significantly higher fluorescence intensity per area of cell cluster footprints, and reduced contacts (decreased branching) between cell clusters (Fig. 7, B and C). In agreement, silencing of PTP1B in Eph+ U251 cells also increased the segregation of the two cell populations and significantly reduced cell dispersion (Fig. S4, A and B), as revealed by significantly reduced numbers of fluorescence intensity peaks (Fig. S4 C). In contrast, treatment with an EphA3 kinase inhibitor significantly increased the intermingling and cell dispersion between the two cell populations (Fig. 7 B and Fig. S3), as judged from decreased overall fluorescence intensity per area and an increased branching ratio (Fig. 7 C). As expected, treatment with PTP1B inhibitor increased EphA3 phosphorylation, whereas EphA3 inhibitor treatment decreased it, which implies that their opposite effects on cell–cell segregation are likely caused by altered EphA3 activation (Fig. 7 D).
Figure 7.
PTP1B activity affects Eph/ephrin-mediated cell segregation. (A) HEK293 and U251 cells were allowed to bind EphA3-Fc and ephrinA5-Fc, respectively, before labeling with Alexa Fluor 647–α-human secondary antibodies and flow cytometry analysis. (B) Cell segregation was monitored in co-cultures of U251 cells and cell tracker green–labeled HEK293 cells in the presence or absence of PTP1B inhibitor (10 µM PTP1B-I), EphA3 inhibitor (10 µM EphA3-I), or DMSO (as control for EphA3-I) for 72 h. Bright-field, fluorescence, and merged images of PFA fixed cell cultures are shown. Contour plots of cell tracker green fluorescent images were generated (MatLab) to illustrate the accumulation of HEK293 cells and the effects of the inhibitor on sorting of mixed HEK293 and U251 cell populations (bottom); their quantification is shown in Fig. S3. (C) Additional quantification of the segregation assay was performed by analyzing ratios of overall fluorescence intensity/area, and the branching ratio of fluorescent areas using ImageJ software. For each setting, cell tracker green images equivalent to 60 fields of view (as shown in B) were analyzed for n = 3 independent experiments. Mean ± SE are shown (error bars; **, P < 0.01; ***, P < 0.001). (D) Co-cultures of HEK293 and U251 cells were treated with PTP1B or EphA3 inhibitor as indicated for 48 h. Lysates and α-EphA3 immunoprecipitates were subjected to Western blot analysis with the appropriate antibodies. Levels of phosphorylated EphA3 relative to total expression were determined by densitometry. Data are representative of at least two independent experiments. Molecular mass standards are indicated next to the gel blots in kilodaltons.
Collectively, these findings indicate that PTP1B, by controlling EphA3 phosphorylation, functions to modulate Eph-induced changes in cell morphology and cell–cell boundary formation.
PTP1B colocalizes with EphA3 and controls EphA3 trafficking
We next thought to examine cellular compartments and timing of the EphA3–PTP1B interplay during ephrinA5-induced signaling and EphA3 trafficking. Previous studies demonstrated rapid internalization of Eph/ephrin signaling complexes (Marston et al., 2003; Wimmer-Kleikamp et al., 2004; Janes et al., 2009), and costaining of ephrinA5-stimulated cells with antibodies against EphA3, endosomal (early endosomal antigen I [EEAI]), and lysosomal (LampI) markers revealed trafficking of activated EphA3 into the endosomal but not into the lysosomal compartment (Fig. S5, A and B). Interestingly in stimulated, GFP-PTP1B-[D-A]–expressing cells, PTP1B also was found colocalized with early endosomal vesicles, including those containing endocytosed EphA3 (Fig. S5 C).
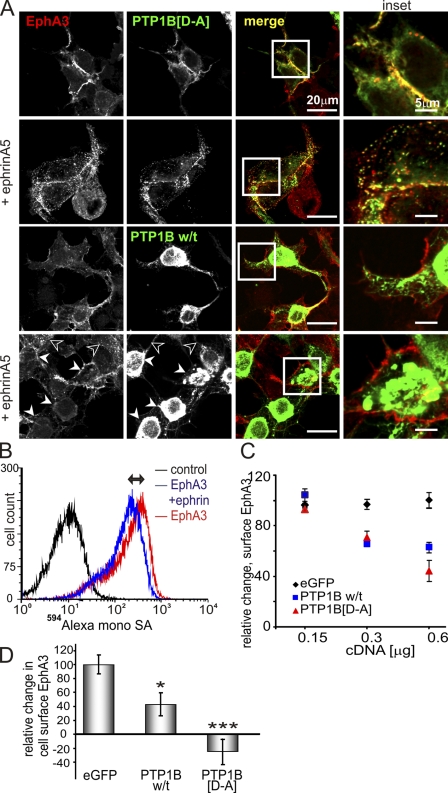
Further analysis of GFP-PTP1B-[D-A]–transfected EphA3/HEK293T cells (Fig. 8 A, top) and PTP1B−/− MEFs (Fig. S5 D) by confocal microscopy revealed both proteins at sites of cell–cell contact. After ephrinA5 stimulation, GFP-PTP1B-[D-A]– and EphA3-associated fluorescence also notably colocalized with endosomal vesicles (Fig. 8 A, top; and Fig. S5, A, C, and D). Live-cell imaging confirmed that internalization of Alexa Fluor 594–ephrinA5–bound EphA3 and GFP-PTP1B-[D-A] concurred with cell contraction and detachment (Video 1). In accord with the perception that interactions between w/t PTPs and RTKs are transient, confocal analysis of cells transfected with w/t GFP-PTP1B did not allow detecting its colocalization with EphA3 (Fig. 8 A, bottom). However, closer inspection suggested that in cells with high w/t GFP-PTP1B, the levels of internalized EphA3 were markedly reduced compared with cells with low PTP1B expression (Fig. 8 A, closed and open arrowheads, respectively).
Figure 8.
Elevated expression of w/t PTP1B or PTP1B-[D-A] inhibits EphA3 endocytosis. (A) EphrinA5-Fc stimulated (10 min) EphA3/HEK293T cells expressing exogenous GFP-PTP1B-[D-A] or w/t GFP-PTP1B were fixed, permeabilized, and labeled with α-EphA3 and Alexa Fluor 546–labeled secondary antibodies for confocal microscopy. Individual fluorescent channels and merged images are shown. Closed and open arrowheads denote cells with highly elevated and low levels of PTP1B, respectively. Areas of interest are boxed and shown at increased magnification (insets). (B) The level of biotinylated EphA3 on the surface of APN-EphA3/TM-BirA/HEK293T cells was determined by flow cytometry using Alexa Fluor 594–monoSA. The Alexa Fluor 594–monoSA intensity of ephrin-stimulated (20 min) cells is reduced compared with untreated cells, which reflects EphA3 internalization (↔). (C) The relative ephrinA5-induced change in cell surface EphA3 was determined as in B in cells transfected with pEGFP (control), w/t GFP-PTP1B, or GFP-PTP1B–[D-A] at cDNA concentrations of 0.15, 0.3, or 0.6 µg/well as indicated. Mean ± SE from three individual samples from each condition are shown (error bars). (D) 22Rv1 cells transfected with EGFP, GFP-PTP1B-[D-A], or w/t GFP-PTP1B were stimulated with ephrinA5-Fc for 20 min, and cell surface EphA3 was labeled using sheep α-EphA3 and Alexa Fluor 647 secondary antibodies, analyzed by flow cytometry, and evaluated as described in B. Mean ± SE from six individual samples of two separate experiments are shown (error bars; *, P < 0.05; ***, P < 0.001).
To evaluate this potential effect of PTP1B on ephrin-induced EphA3 trafficking quantitatively, we analyzed with flow cytometry APN-EphA3/BirA/HEK293T cells stably expressing constitutively biotinylated EphA3, allowing its rapid and explicit cell surface labeling with Alexa Fluor 594–tagged monovalent SA (Alexa Fluor 594–monoSA; Howarth and Ting, 2008). As expected, preclustered ephrinA5-Fc–induced EphA3 internalization caused a notable decrease in Alexa Fluor 594–monoSA fluorescence, providing a measure for the change in cell surface EphA3 levels (Fig. 8 B, ↔). Overexpression of either w/t PTP1B or PTP1B-[D-A] in these cells dose-dependently decreased this change, and thus the amount of internalizing EphA3 (Fig. 8 C). Likewise, overexpression of w/t PTP1B also decreased the trafficking of endogenous EphA3 in 22Rv1 prostate carcinoma cells. In this case, the exogenous PTP1B-[D-A] inhibited ephrin-induced EphA3 endocytosis completely so that the EphA3 cell surface levels were increased after ephrinA5 stimulation (Fig. 8 D). In control experiments, cell surface levels of the constitutively recycling Transferrin transmembrane receptor were not influenced by exogenous PTP1B expression (Fig. S4 E), which argues against a global effect of PTP1B on receptor recycling. In agreement with confocal microscopic analysis (Fig. 8 A), these experiments suggest that both EphA3 dephosphorylation by elevated w/t PTP1B, as well as binding of the PTP1B-[D-A]–trapping mutant likely blocking access to EphA3-phospho-tyrosines, markedly attenuate ephrinA5-induced EphA3 trafficking.
Discussion
Compelling evidence attests a crucial role of Ephs and ephrins in controlling cell–cell interactions during developmental cell positioning. Eph kinase signaling translates the ephrin cell surface abundance into precisely graded changes in adhesion and position of the cells: high kinase activity triggers cell–cell repulsion, whereas lacking or low activity leads to tight cell adhesion (Lackmann and Boyd, 2008; Pasquale, 2008). Not surprisingly, a critical role of PTPs in controlling initiation and transduction of the underlying signals has been anticipated, but the spatial and temporal aspects of their recruitment into Eph signaling clusters have remained largely unexplored.
We now demonstrate that PTP1B, a ubiquitous ER-resident phosphatase controlling the signaling of several growth factor receptors and cell adhesion proteins (Bourdeau et al., 2005; Burridge et al., 2006; Sallee et al., 2006; Tonks, 2006), negatively regulates agonist-induced EphA3 phosphorylation. Unlike other RTKs, Ephs are activated in vivo only by surface-tethered ephrins (Lackmann and Boyd, 2008; Pasquale, 2008), whereby, as we demonstrate here, PTP1B already interacts with activated cell-surface EphA3 before endocytosis, providing an example for a mechanism that allows the ER-tethered phosphatase to access its RTK substrates at sites of cell–cell contact (unpublished data). PTP1B activity has direct consequences on EphA3 receptor trafficking and downstream biological responses: PTP1B overexpression reduces EphA3 activation at the cell surface and in cytosolic compartments, thereby decreasing ephrinA5-induced cell contraction, whereas PTP1B silencing enhances EphA3 activation and ephrin-induced cell contraction. Accordingly, pharmacological inhibition of EphA3 kinase activation/signaling attenuates formation of boundaries with ephrin-expressing cells, whereas inhibition of PTP1B activity and PTP1B silencing increases cell–cell repulsion and boundary formation.
Eph receptors are activated in vivo within signaling clusters at the PM interface (Wimmer-Kleikamp et al., 2004) between Eph- and ephrin-expressing cells, and phosphorylation of the activation loop tyrosine, releasing the kinase from the Eph juxtamembrane segment, is required for full kinase activity (Wybenga-Groot et al., 2001; Wiesner et al., 2006). This conformational change relieves a steric hindrance, leading to functional association of the ADAM10 metalloprotease. This then facilitates ephrin shedding and endocytosis to proceed, which indicates that control of kinase activity, ephrin shedding, and endocytosis are linked (Janes et al., 2009). In agreement with this notion, our findings here reveal that overexpression of PTP1B results in EphA3 dephosphorylation and dose-dependent reduction in trafficking of cell surface EphA3, which suggests direct control of phosphorylation-dependent endocytosis.
Notionally, it is also possible that the observed effect of PTP1B overexpression on cell surface EphA3 trafficking is caused by very rapid receptor recycling, as suggested for other transmembrane receptors (Grant and Donaldson, 2009); thus, PTP1B inhibition was shown to extend the presence of phosphorylated insulin receptor in the endocytic recycling compartment (Cromlish et al., 2006). To date, the kinetics and mechanics of Eph endocytosis have remained largely unexplored, and a potential effect of PTP1B overexpression on EphA3 recycling clearly warrants further analysis. Nevertheless, the finding that phosphorylation of the activation loop tyrosine (EphA3-Y779), a recently identified PTP1B substrate (Mertins et al., 2008), is essential for ligand-induced endocytosis (Janes et al., 2009), together with our observation that PTP1B can interact with activated EphA3 at the PM, would suggest that elevated phosphatase levels and lack of EphA3 phosphorylation affect ephrin-induced endocytosis directly. Overexpression of PTP1B-[D-A] has a very similar effect to w/t PTP1B and at high concentrations inhibits EphA3 internalization completely, though we argue for a different mechanism: its effective binding to phosphorylated EphA3 competitively blocks access to the phosphorylated activation loop tyrosine that is required for endocytosis.
The interaction with EphA3 and apparent phosphatase activity of PTP1B at sites of cell–cell contact and on endosomal vesicles raises important questions about the control of PTP1B activity in different cellular compartments. It is likely that, similar to other RTKs, EphA3 activation triggers ROS production and local inhibition of PTP activity, which is gradually reversed once the endosomes have traveled into the cytosol. In the case of EGF and insulin receptors, where dephosphorylation by PTP1B occurs en route past the ER (Haj et al., 2002; Boute et al., 2003), an increase in PTP1B activity toward the nucleus explains why RTK phosphorylation declines with increasing PM distance (Yudushkin et al., 2007) and why extended residence at the cell surface prolongs EGF receptor phosphorylation (Offterdinger et al., 2004). For EphA3, our current evidence suggests that signal termination on endocytic vesicles is also facilitated through dephosphorylation by PTP1B.
ER-bound PTP1B (Frangioni et al., 1992) is thought to interact with transmembrane receptors mainly in the cytosol proximal to the ER (Haj et al., 2002; Boute et al., 2003) or after proteolytic release of the catalytic domain (Frangioni et al., 1993; Cortesio et al., 2008), although it was reported recently that PTP1B can contact some transmembrane receptors and cell–matrix adhesion sites directly (Hernández et al., 2006; Anderie et al., 2007). In the case of EphA3, an RTK which generally requires contact with an ephrin-expressing cell for activation, cell surface interactions with PTP1B occur prominently at sites of cell–cell contact. However, stimulation with soluble ephrinA5 also enhanced EphA3/PTP1B interactions elsewhere on the cell surface and in endocytic vesicles, presumably because this nonphysiological activation triggers more immediate endocytosis. In line with this argument, global phospho-proteomic analysis of Eph/ephrin signaling networks revealed significant qualitative and quantitative differences when cells were stimulated with soluble, preclustered ephrins or by ephrin–cell contact (Jørgensen et al., 2009). It is likely that assembly of signaling clusters with interacting cell-bound ephrins results in a local accumulation of EphA3 that promotes interaction with PTP1B already at the cell surface. Indeed, our in vitro model of this scenario, where tethered SA activates biotinylated EphA3 but cannot be cleaved and thus prevents EphA3 from leaving the cell surface, confirms this notion. In this case, distinct PTP1B-[D-A] recruitment to biotinylated EphA3 is focused to the surface of SA-conjugated beads, which indicates that the PTP1B/EphA3 interaction occurs before endocytosis. Thus, in agreement with its role in controlling EphA3 activation, trafficking, and signaling, we demonstrate that ER-bound PTP1B-[D-A] can target phosphorylated cell surface EphA3 in areas of cell–cell contact before endocytosis, thereby directing the overall outcome of Eph/ephrin-facilitated cell–cell interactions.
Both EphA3 and PTP1B have documented roles in tumor development: PTP1B is implicated in growth and progression of solid tumors (Lessard et al., 2010), whereas unscheduled expression of Ephs and ephrins correlates with tumor progression in a range of highly invasive and metastatic tumors (Janes et al., 2008) where EphA3 was identified as one of the genes most frequently mutated in colon (Sjöblom et al., 2006) and lung cancer (Ding et al., 2008). In view of our findings here, the balance between Eph kinase and counteracting PTP1B activity is likely to be a critical regulatory mechanism for EphA3-mediated cell positioning during tumor development and progression. In line with emerging evidence for a role of PTP1B in cell spreading, migration, and cancer cell invasion (Arregui et al., 1998; Cortesio et al., 2008), we propose that, similar to PTPRO in neuronal cells (Shintani et al., 2006), PTP1B acts as cell-autonomous control of Eph/ephrin-facilitated cell positioning by modulating the balance between phosphorylated and dephosphorylated Ephs during embryonic as well as oncogenic development. In support of the underlying notion that PTP1B may also regulate activity of other Ephs, our FRET analysis also suggested a direct interaction of the PTP1B substrate-trapping mutant with activated EphB2.
In summary, our study provides detailed molecular and mechanistic insights into phosphatase-controlled Eph receptor activation, trafficking, and cellular responses. We demonstrate for the first time that PTP1B is a principal regulator of EphA3 activity and function, and controls the steady-state concentration of cell surface EphA3. In view of emerging evidence for crosstalk between Ephs and other RTKs and adhesion receptors (Lackmann and Boyd, 2008; Pasquale, 2008), our finding that ER-tethered PTP1B interacts with activated EphA3 at the cell surface may suggest that activated Ephs are among the PTP1B substrates that polarize the ER toward the PM and provide PTP1B access to areas of cell–cell contacts (unpublished data). Importantly, we confirm that the crucial Eph-mediated switch between cell–cell repulsion and adhesion is controlled by the activity of Eph-specific PTPs such as PTP1B. Further elucidation of the mechanisms that shift the balance between dominating PTP and dominating Eph activity will impact significantly on our understanding of Eph function in normal and oncogenic development.
Materials and methods
Expression constructs
Mammalian pEFBos expression vectors for full-length EphA3, EphA3-GFP, and APN-EphA3 have been described previously (Wimmer-Kleikamp et al., 2004), and deletions and point mutations were made using site-directed mutagenesis (QuikChange XL; Agilent Technologies). EphA3-YFP was constructed by replacing the GFP sequence with YFP cDNA. Membrane-targeted BirA (TM-BirA) was generated from YFP-tagged TM-BirA (from A. Ting, Massachusetts Institute of Technology, Cambridge, MA). Fluorescent protein-tagged PTP1B expression constructs have been described previously (Haj et al., 2002). PTP1B and TC-PTP w/t and [D-A]-trapping mutants in pMT2 were provided by T. Tiganis (Monash University, Melbourne, Australia) and described previously (Flint et al., 1997; Tiganis et al., 1998).
Reagents and antibodies
Production of ephrinA5-Fc, Alexa Fluor 546–ephrinA5–Fc, EphA3-Fc, and the α-EphA3 mAb (IIIA4) have been described previously (Lawrenson et al., 2002; Wimmer-Kleikamp et al., 2008). Antigen affinity-purified sheep α-EphA3 was used for immunoblots. PTP1B mAbs were AE4-2J (EMD) for immunoblots and FG6 (Haj et al., 2002) for immunocytochemistry. Cy5-FG6 and Cy3.5-PY72 (Reynolds et al., 2003) were labeled using a α-Cy5 or Cy3.5 mono-Reactive Dye Pack (GE Healthcare). TC-PTP mAb (CF4) was provided by T. Tiganis. Other antibodies were obtained from Invitrogen (rabbit α-PY), Thermo Fisher Scientific (α-actin), Abcam (α-EEAI), BD (α-LampI; α-Transferrin receptor), Roche (α-GFP), and Millipore (α-phospho-EphA3). All HRP-labeled secondary antibodies for Western blotting and anti-human Fc were obtained from Jackson ImmunoResearch Laboratories, except α-mouse-HRP (GE Healthcare) and α-sheep-HRP (Millipore). Alexa Fluor–labeled secondary antibodies and rhodamine- or Alexa Fluor 647–phalloidin were obtained from Invitrogen. MonoSA (provided by A. Ting) was labeled with Alexa Fluor 594 (Invitrogen). Difluoromethylphosphonate PTP1B inhibitor (Han et al., 2008) was provided by B. Kennedy and K. McCusker (Merck Frosst Canada, Kirkland, Quebec, Canada), and EphA3 kinase inhibitor (Choi et al., 2009) was a gift of N. Gray (Harvard Medical School, Boston, MA).
Cell culture
Human kidney epithelial 293T (HEK293T), COS7 African green monkey kidney fibroblast cell line, U251 human glioblastoma cells (provided by A. Scott, Ludwig Institute For Cancer Research, Melbourne Centre for Clinical Sciences, Melbourne, Australia), PTP1B-deficient MEFs, and corresponding w/t PTP1B reconstituted cell lines (Haj et al., 2003) were maintained in DME supplemented with 10% FCS. Stable HEK293T and MEF cell lines expressing APN-EphA3 alone or with TM-BirA were generated through clonal selection (0.2 mg/ml zeocin and 400 µg/ml G418). Stable ephrinA5 (ephrinA5/HEK293T)-expressing cell clones have been described previously (Janes et al., 2005). NG108 cells (mouse neuroblastoma × rat glioma hybrid) were provided by T. Pawson (Mount Sinai Hospital, Toronto, Canada) and maintained in DME, 10% FCS, and 1× HAT supplement (Invitrogen). Human prostate carcinoma 22Rv1 cells (American Type Culture Collection) were maintained in RPMI 1640/10% FCS.
Transfections and gene knockdown by RNA interference
Transfections of cDNA were performed with Lipofectamine 2000 (Invitrogen) or FugeneHD (Roche) according to the manufacturer’s instructions. PTP1B silencing was achieved using the Mission Lentiviral Transduction Particles system (Sigma-Aldrich) according to the manufacturer’s instructions. Four different hairpin constructs in individual lentivirus clones for PTP1B silencing as well as nontarget control lentivirus particles were used to create stable cell lines by puromycin-induced killing (1 µg/ml for 22Rv1, 2 µg/ml for EphA3/HEK293T) of nontransduced cells.
Ephrin stimulation, immunoprecipitation, and Western blotting
For ligand stimulation, ephrinA5-Fc (1.5 µg/ml final) was preclustered using goat anti–human IgG (Fcγ-specific) and applied to the cells for 10 min or as indicated. Whole cell lysis was performed in 50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1 mM NaV04, 10 mM NaF, and protease inhibitors (Complete; Roche). EphA3 was immunoprecipitated from cell lysates with mAb IIIA4 conjugated to Mini-Leak beads (Kem-En-Tec) as described previously (Janes et al., 2005). Immunoprecipitates or whole cell lysates were analyzed by Western blotting with appropriate antibodies, and blots were visualized using an ECL substrate (Supersignal; Thermo Fisher Scientific).
Flow cytometry and internalization assay
HEK293 and U251 cells were detached using 0.5 mM EDTA/PBS and incubated with EphA3-Fc or ephrinA5-Fc in FACS buffer (1% FCS, 0.5 mM EDTA in PBS) for 30 min on ice before washing and labeling with Alexa Fluor 647–α-human secondary antibodies. Flow cytometry was performed on an LSRII (BD), and data were analyzed using FlowJo (Tree Star, Inc.).
To assess EphA3 internalization, HEK293T cell clones expressing biotinylated EphA3 (APN-EphA3/TM-BirA/HEK293T) were transfected with PTP1B cDNA as indicated, then treated with preclustered ephrinA5-Fc for 20 min followed by labeling with Alexa Fluor 594–monoSA in FACS buffer for 30 min at 4°C. 22Rv1 cells were transfected and stimulated in the same manner, and cell surface levels of endogenous EphA3 were labeled with sheep α-EphA3 pAb followed by an Alexa Fluor 647 secondary antibody.
Cell sorting assay
HEK293 cells were mixed with U251 cells at a ratio of 2:1. Cells were differentiated by labeling one population with CellTracker green according to the manufacturer’s instructions (Invitrogen) and cultured at a density of 55,000 cells/cm2 (100,000 cells/ml media) to reach confluency within 3 d. Cells fixed in 4% paraformaldehyde were imaged in PBS at 37°C on an inverted microscope (AF6000LX; Leica) equipped with a camera (DFC 350FX; Leica) and running LAS AF software using a 10× dry objective (NA 0.3) and a monochromator (Leica).
ImageJ was used to threshold and process the fluorescent images for subsequent analysis with a MATLAB (MathWorks Inc.) script (see supplemental material) to graph the fluorescence intensity as 3D contour maps and to quantitate the area (in pixels) and fluorescence intensity of individual peaks (representing cell aggregates) above a baseline intensity as a measure of cell sorting. In this context, cell sorting was regarded as loss of cell dispersion and is quantitated as a reduction in the number of small fluorescence intensity peaks in microscopic sections (0.4–0.8 mm2) randomly selected from a minimum of n = 4 independent experiments (a representative image from each group is shown). In addition, lower magnification images were captured using a 4× objective (NA 0.16) on a wide-field fluorescence microscope (Cell-R; Olympus) equipped with a charge-coupled device camera (XM10) and Cell-R image acquisition software (Olympus). These images were analyzed using ImageJ and AnalyzeSkeleton (Ignacio Arganda-Carreras, Massachusetts Institute of Technology, Cambridge, MA) plug-ins to determine the overall intensity/area ratio and degree of branching of fluorescent areas, respectively.
Confocal and wide-field fluorescence microscopy
Cells on coverslips, transfected and treated as indicated, were fixed in 4% paraformaldehyde (or methanol for FG6 mAb labeling), permeabilized in 0.1% Triton X-100/PBS, labeled with antibody or phalloidin (rhodamine- or Alexa Fluor 647–labeled) as indicated, and mounted in Mowiol (EMD). Stimulation with M280-Streptavin– and ephrinA5-coated Protein A Dynabeads (Invitrogen) was for 2 h at 37°C; IIIA4 mAb was applied to the cells 5 min before fixation. Slides were examined at room temperature on a microscope (FluoView 1000; Olympus) equipped with Ar (488 nm) and HeNe (543 nm and 633 nm) lasers and running FV10-ASW software using an oil immersion 60× lens (NA 1.4; Olympus). Images were analyzed with analySIS (Soft Imaging System; Olympus) and ImageJ (National Institutes of Health) software. Quantification of rhodamine-phalloidin fluorescence area as a measure of cell spreading in images representing a whole coverslip (acquired using the motorized XY stage and the tile scan function on the inverted microscope (AF6000LX) and a 10× dry objective lens, NA 0.3) was performed using Metamorph (MDS Analytical Technologies).
For live cell imaging, cells growing on coverslips or glass-bottom dishes (MatTek Corp.) were transfected as indicated and imaged in phenol red–free DME (Invitrogen) supplemented with 1% FCS. Stimulation with Alexa Fluor 594–ephrinA5 was performed at 37°C in a FCS2 perfusion chamber (Bioptecs) on an inverted microscope (AF6000LX) equipped with a monochromator (using a BGR emission filter and a 63× glycerol objective lens, NA 1.3) and a CO2 and temperature-controlled atmospheric chamber.
FLIM
Confocal FLIM based on time-correlated single photon counting (TCSPC) was performed in transiently transfected COS7 cells grown on coverslips or glass-bottom dishes and treated as described in the corresponding figure legends. FLIM data were obtained using a microscope (FluoView 1000; Olympus) equipped with a Picoharp 300 photon counting setup (PicoQuant) and a CO2 and temperature-controlled atmospheric chamber for live cell imaging. GFP and YFP were excited with a 470-nm diode (Sepia II; PicoQuant). Images of 512 × 512 pixels were acquired detecting ∼10 million photons. Images of the donor fluorescence were processed using the SymPhoTime software package (v4.2; PicoQuant), and fluorescence lifetime images are presented in pseudo-color.
Statistical analysis
Graphpad Prism (GraphPad Software) and Excel software (Microsoft) were used for graphs and statistical analysis. Data are presented as mean ± SE, and unpaired, two-tailed t tests were used to compare two variables and two-way analysis of variance (ANOVA) for multiple parameters.
Online supplemental material
Fig. S1 shows that TC-PTP1B overexpression affects EphA3 phosphorylation only moderately, whereas phosphorylated EphA3 in PM-proximal and perinuclear compartments of nonstimulated and stimulated cells is efficiently dephosphorylated by PTP1B. Fig. S2 demonstrates that PTP1B also directly interacts with EphB2, whereas LMW-PTP does not interact with EphA3. Fig. S3 illustrates contour plots of integrated fluorescence intensity and quantitative analysis of images displayed in Fig. 7 B. Fig. S4 reveals that shRNA-facilitated PTP1B silencing enhances Eph/ephrin-mediated cell–cell segregation by reducing the dispersion of EphA3-expressing cells. Fig. S5 shows that activated EphA3 traffics into the endosomal compartment, that PTP1B associates with endosomal vesicles, and that Transferrin receptor trafficking is not affected by PTP1B overexpression. Video 1 follows the co-internalization of EphA3 and PTP1B during ephrinA5 stimulation and the resulting cell contraction and detachment. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201005035/DC1.
Acknowledgments
We thank I. Harper and the team at Monash Micro Imaging for imaging advice, A. Fryga and his team at Monash Flowcore for help with flow cytometry, and Mark Hink for help with confocal FLIM. We are grateful to T. Tiganis for reagents and helpful discussions. We also thank B. Kennedy and K. McCusker for providing the PTP1B inhibitor, and N. Gray for the EphA3 kinase inhibitor.
This work was supported by National Health and Medical Research Council grants (334085 and 436774), an Australian Research Council Linkage grant (LP0882735), a Human Frontier Science Program grant (RGP0039/2009-C), a National Institutes of Health grant (R37 CA49152), a Juvenile Diabetes Research Foundation grant (1-2009-337) to F.G. Haj, and a Monash Postgraduate Scholarship to E. Nievergall. C. Stegmayer is a postdoctoral fellow of the Deutsche Forschungsgemeinschaft, P.W. James is an RD Wright Fellow, and M. Lackmann is a National Health and Medical Research Council Senior Research Fellow.
Footnotes
Abbreviations used in this paper:
- EEAI
- early endosomal antigen I
- FLIM
- fluorescence lifetime imaging microscopy
- FRET
- Förster resonance energy transfer
- LMW-PTP
- low molecular weight PTP
- MEF
- mouse embryonic fibroblast
- monoSA
- monovalent streptavidin
- PM
- plasma membrane
- PTP
- protein tyrosine phosphatase
- PY
- phosphotyrosine
- RTK
- receptor tyrosine kinase
- SA
- streptavidin
- SHP2
- src homology 2 domain-containing PTP
- shRNA
- small-hairpin RNA
- TC-PTP
- T cell PTP
- w/t
- wild type
References
- Anderie I., Schulz I., Schmid A. 2007. Direct interaction between ER membrane-bound PTP1B and its plasma membrane-anchored targets. Cell. Signal. 19:582–592 10.1016/j.cellsig.2006.08.007 [DOI] [PubMed] [Google Scholar]
- Arregui C.O., Balsamo J., Lilien J. 1998. Impaired integrin-mediated adhesion and signaling in fibroblasts expressing a dominant-negative mutant PTP1B. J. Cell Biol. 143:861–873 10.1083/jcb.143.3.861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdeau A., Dubé N., Tremblay M.L. 2005. Cytoplasmic protein tyrosine phosphatases, regulation and function: the roles of PTP1B and TC-PTP. Curr. Opin. Cell Biol. 17:203–209 10.1016/j.ceb.2005.02.001 [DOI] [PubMed] [Google Scholar]
- Boute N., Boubekeur S., Lacasa D., Issad T. 2003. Dynamics of the interaction between the insulin receptor and protein tyrosine-phosphatase 1B in living cells. EMBO Rep. 4:313–319 10.1038/sj.embor.embor767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K., Sastry S.K., Sallee J.L. 2006. Regulation of cell adhesion by protein-tyrosine phosphatases. I. Cell-matrix adhesion. J. Biol. Chem. 281:15593–15596 10.1074/jbc.R500030200 [DOI] [PubMed] [Google Scholar]
- Choi Y., Syeda F., Walker J.R., Finerty P.J., Jr., Cuerrier D., Wojciechowski A., Liu Q., Dhe-Paganon S., Gray N.S. 2009. Discovery and structural analysis of Eph receptor tyrosine kinase inhibitors. Bioorg. Med. Chem. Lett. 19:4467–4470 10.1016/j.bmcl.2009.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortesio C.L., Chan K.T., Perrin B.J., Burton N.O., Zhang S., Zhang Z.Y., Huttenlocher A. 2008. Calpain 2 and PTP1B function in a novel pathway with Src to regulate invadopodia dynamics and breast cancer cell invasion. J. Cell Biol. 180:957–971 10.1083/jcb.200708048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromlish W.A., Tang M., Kyskan R., Tran L., Kennedy B.P. 2006. PTP1B-dependent insulin receptor phosphorylation/residency in the endocytic recycling compartment of CHO-IR cells. Biochem. Pharmacol. 72:1279–1292 10.1016/j.bcp.2006.07.038 [DOI] [PubMed] [Google Scholar]
- Ding L., Getz G., Wheeler D.A., Mardis E.R., McLellan M.D., Cibulskis K., Sougnez C., Greulich H., Muzny D.M., Morgan M.B., et al. 2008. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 455:1069–1075 10.1038/nature07423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowe S., Holland S.J., Kulkarni S., Pawson T. 2001. Downregulation of the Ras-mitogen-activated protein kinase pathway by the EphB2 receptor tyrosine kinase is required for ephrin-induced neurite retraction. Mol. Cell. Biol. 21:7429–7441 10.1128/MCB.21.21.7429-7441.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint A.J., Tiganis T., Barford D., Tonks N.K. 1997. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc. Natl. Acad. Sci. USA. 94:1680–1685 10.1073/pnas.94.5.1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangioni J.V., Beahm P.H., Shifrin V., Jost C.A., Neel B.G. 1992. The nontransmembrane tyrosine phosphatase PTP-1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell. 68:545–560 10.1016/0092-8674(92)90190-N [DOI] [PubMed] [Google Scholar]
- Frangioni J.V., Oda A., Smith M., Salzman E.W., Neel B.G. 1993. Calpain-catalyzed cleavage and subcellular relocation of protein phosphotyrosine phosphatase 1B (PTP-1B) in human platelets. EMBO J. 12:4843–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes F., Arregui C.O. 2009. Microtubule and cell contact dependency of ER-bound PTP1B localization in growth cones. Mol. Biol. Cell. 20:1878–1889 10.1091/mbc.E08-07-0675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B.D., Donaldson J.G. 2009. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10:597–608 10.1038/nrm2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haj F.G., Verveer P.J., Squire A., Neel B.G., Bastiaens P.I. 2002. Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science. 295:1708–1711 10.1126/science.1067566 [DOI] [PubMed] [Google Scholar]
- Haj F.G., Markova B., Klaman L.D., Bohmer F.D., Neel B.G. 2003. Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatase-1B. J. Biol. Chem. 278:739–744 10.1074/jbc.M210194200 [DOI] [PubMed] [Google Scholar]
- Han Y., Belley M., Bayly C.I., Colucci J., Dufresne C., Giroux A., Lau C.K., Leblanc Y., McKay D., Therien M., et al. 2008. Discovery of [(3-bromo-7-cyano-2-naphthyl)(difluoro)methyl]phosphonic acid, a potent and orally active small molecule PTP1B inhibitor. Bioorg. Med. Chem. Lett. 18:3200–3205 10.1016/j.bmcl.2008.04.064 [DOI] [PubMed] [Google Scholar]
- Hernández M.V., Sala M.G., Balsamo J., Lilien J., Arregui C.O. 2006. ER-bound PTP1B is targeted to newly forming cell-matrix adhesions. J. Cell Sci. 119:1233–1243 10.1242/jcs.02846 [DOI] [PubMed] [Google Scholar]
- Himanen J.P., Rajashankar K.R., Lackmann M., Cowan C.A., Henkemeyer M., Nikolov D.B. 2001. Crystal structure of an Eph receptor-ephrin complex. Nature. 414:933–938 10.1038/414933a [DOI] [PubMed] [Google Scholar]
- Holmberg J., Frisén J. 2002. Ephrins are not only unattractive. Trends Neurosci. 25:239–243 10.1016/S0166-2236(02)02149-5 [DOI] [PubMed] [Google Scholar]
- Howarth M., Ting A.Y. 2008. Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat. Protoc. 3:534–545 10.1038/nprot.2008.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes P.W., Saha N., Barton W.A., Kolev M.V., Wimmer-Kleikamp S.H., Nievergall E., Blobel C.P., Himanen J.P., Lackmann M., Nikolov D.B. 2005. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell. 123:291–304 10.1016/j.cell.2005.08.014 [DOI] [PubMed] [Google Scholar]
- Janes P.W., Adikari S., Lackmann M. 2008. Eph/ephrin signalling and function in oncogenesis: lessons from embryonic development. Curr. Cancer Drug Targets. 8:473–479 10.2174/156800908785699315 [DOI] [PubMed] [Google Scholar]
- Janes P.W., Wimmer-Kleikamp S.H., Frangakis A.S., Treble K., Griesshaber B., Sabet O., Grabenbauer M., Ting A.Y., Saftig P., Bastiaens P.I., Lackmann M. 2009. Cytoplasmic relaxation of active Eph controls ephrin shedding by ADAM10. PLoS Biol. 7:e1000215 10.1371/journal.pbio.1000215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen C., Sherman A., Chen G.I., Pasculescu A., Poliakov A., Hsiung M., Larsen B., Wilkinson D.G., Linding R., Pawson T. 2009. Cell-specific information processing in segregating populations of Eph receptor ephrin-expressing cells. Science. 326:1502–1509 10.1126/science.1176615 [DOI] [PubMed] [Google Scholar]
- Kim J.-R., Yoon H.W., Kwon K.-S., Lee S.-R., Rhee S.G. 2000. Identification of proteins containing cysteine residues that are sensitive to oxidation by hydrogen peroxide at neutral pH. Anal. Biochem. 283:214–221 10.1006/abio.2000.4623 [DOI] [PubMed] [Google Scholar]
- Konstantinova I., Nikolova G., Ohara-Imaizumi M., Meda P., Kucera T., Zarbalis K., Wurst W., Nagamatsu S., Lammert E. 2007. EphA-Ephrin-A-mediated beta cell communication regulates insulin secretion from pancreatic islets. Cell. 129:359–370 10.1016/j.cell.2007.02.044 [DOI] [PubMed] [Google Scholar]
- Lackmann M., Boyd A.W. 2008. Eph, a protein family coming of age: more confusion, insight, or complexity? Sci. Signal. 1:re2 10.1126/stke.115re2 [DOI] [PubMed] [Google Scholar]
- Lawrenson I.D., Wimmer-Kleikamp S.H., Lock P., Schoenwaelder S.M., Down M., Boyd A.W., Alewood P.F., Lackmann M. 2002. Ephrin-A5 induces rounding, blebbing and de-adhesion of EphA3-expressing 293T and melanoma cells by CrkII and Rho-mediated signalling. J. Cell Sci. 115:1059–1072 [DOI] [PubMed] [Google Scholar]
- Lessard L., Stuible M., Tremblay M.L. 2010. The two faces of PTP1B in cancer. Biochim. Biophys. Acta. 1804:613–619 [DOI] [PubMed] [Google Scholar]
- Marston D.J., Dickinson S., Nobes C.D. 2003. Rac-dependent trans-endocytosis of ephrinBs regulates Eph-ephrin contact repulsion. Nat. Cell Biol. 5:879–888 10.1038/ncb1044 [DOI] [PubMed] [Google Scholar]
- Mellitzer G., Xu Q., Wilkinson D.G. 1999. Eph receptors and ephrins restrict cell intermingling and communication. Nature. 400:77–81 10.1038/21907 [DOI] [PubMed] [Google Scholar]
- Mertins P., Eberl H.C., Renkawitz J., Olsen J.V., Tremblay M.L., Mann M., Ullrich A., Daub H. 2008. Investigation of protein-tyrosine phosphatase 1B function by quantitative proteomics. Mol. Cell. Proteomics. 7:1763–1777 10.1074/mcp.M800196-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H., Burnett E., Kinch M., Simon E., Wang B. 2000. Activation of EphA2 kinase suppresses integrin function and causes focal-adhesion-kinase dephosphorylation. Nat. Cell Biol. 2:62–69 10.1038/35000008 [DOI] [PubMed] [Google Scholar]
- Offterdinger M., Georget V., Girod A., Bastiaens P.I.H. 2004. Imaging phosphorylation dynamics of the epidermal growth factor receptor. J. Biol. Chem. 279:36972–36981 10.1074/jbc.M405830200 [DOI] [PubMed] [Google Scholar]
- Parri M., Buricchi F., Taddei M.L., Giannoni E., Raugei G., Ramponi G., Chiarugi P. 2005. EphrinA1 repulsive response is regulated by an EphA2 tyrosine phosphatase. J. Biol. Chem. 280:34008–34018 10.1074/jbc.M502879200 [DOI] [PubMed] [Google Scholar]
- Pasquale E.B. 2008. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 133:38–52 10.1016/j.cell.2008.03.011 [DOI] [PubMed] [Google Scholar]
- Poliakov A., Cotrina M.L., Pasini A., Wilkinson D.G. 2008. Regulation of EphB2 activation and cell repulsion by feedback control of the MAPK pathway. J. Cell Biol. 183:933–947 10.1083/jcb.200807151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A.R., Tischer C., Verveer P.J., Rocks O., Bastiaens P.I. 2003. EGFR activation coupled to inhibition of tyrosine phosphatases causes lateral signal propagation. Nat. Cell Biol. 5:447–453 10.1038/ncb981 [DOI] [PubMed] [Google Scholar]
- Sallee J.L., Wittchen E.S., Burridge K. 2006. Regulation of cell adhesion by protein-tyrosine phosphatases: II. Cell-cell adhesion. J. Biol. Chem. 281:16189–16192 10.1074/jbc.R600003200 [DOI] [PubMed] [Google Scholar]
- Shintani T., Ihara M., Sakuta H., Takahashi H., Watakabe I., Noda M. 2006. Eph receptors are negatively controlled by protein tyrosine phosphatase receptor type O. Nat. Neurosci. 9:761–769 10.1038/nn1697 [DOI] [PubMed] [Google Scholar]
- Sjöblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science. 314:268–274 10.1126/science.1133427 [DOI] [PubMed] [Google Scholar]
- Stapleton D., Balan I., Pawson T., Sicheri F. 1999. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat. Struct. Biol. 6:44–49 10.1038/4917 [DOI] [PubMed] [Google Scholar]
- Stein E., Lane A.A., Cerretti D.P., Schoecklmann H.O., Schroff A.D., Van Etten R.L., Daniel T.O. 1998. Eph receptors discriminate specific ligand oligomers to determine alternative signaling complexes, attachment, and assembly responses. Genes Dev. 12:667–678 10.1101/gad.12.5.667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiganis T., Bennett A.M., Ravichandran K.S., Tonks N.K. 1998. Epidermal growth factor receptor and the adaptor protein p52Shc are specific substrates of T-cell protein tyrosine phosphatase. Mol. Cell. Biol. 18:1622–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks N.K. 2005. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 121:667–670 10.1016/j.cell.2005.05.016 [DOI] [PubMed] [Google Scholar]
- Tonks N.K. 2006. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 7:833–846 10.1038/nrm2039 [DOI] [PubMed] [Google Scholar]
- Wiesner S., Wybenga-Groot L.E., Warner N., Lin H., Pawson T., Forman-Kay J.D., Sicheri F. 2006. A change in conformational dynamics underlies the activation of Eph receptor tyrosine kinases. EMBO J. 25:4686–4696 10.1038/sj.emboj.7601315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer-Kleikamp S.H., Janes P.W., Squire A., Bastiaens P.I., Lackmann M. 2004. Recruitment of Eph receptors into signaling clusters does not require ephrin contact. J. Cell Biol. 164:661–666 10.1083/jcb.200312001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer-Kleikamp S.H., Nievergall E., Gegenbauer K., Adikari S., Mansour M., Yeadon T., Boyd A.W., Patani N.R., Lackmann M. 2008. Elevated protein tyrosine phosphatase activity provokes Eph/ephrin-facilitated adhesion of pre-B leukemia cells. Blood. 112:721–732 10.1182/blood-2007-11-121681 [DOI] [PubMed] [Google Scholar]
- Wybenga-Groot L.E., Baskin B., Ong S.H., Tong J., Pawson T., Sicheri F. 2001. Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region. Cell. 106:745–757 10.1016/S0092-8674(01)00496-2 [DOI] [PubMed] [Google Scholar]
- Yudushkin I.A., Schleifenbaum A., Kinkhabwala A., Neel B.G., Schultz C., Bastiaens P.I. 2007. Live-cell imaging of enzyme-substrate interaction reveals spatial regulation of PTP1B. Science. 315:115–119 10.1126/science.1134966 [DOI] [PubMed] [Google Scholar]