Abstract
Production of heat via nonshivering thermogenesis (NST) is critical for temperature homeostasis in mammals. Uncoupling protein UCP1 plays a central role in NST by uncoupling the proton gradients produced in the inner membranes of mitochondria to produce heat; however, the extent to which UCP1 homologues, UCP2 and UCP3, are involved in NST is the subject of an ongoing debate. We used an evolutionary approach to test the hypotheses that variants that are associated with increased expression of these genes (UCP1 −3826A, UCP2 −866A, and UCP3 −55T) show evidence of adaptation with winter climate. To that end, we calculated correlations between allele frequencies and winter climate variables for these single-nucleotide polymorphisms (SNPs), which we genotyped in a panel of 52 worldwide populations. We found significant correlations with winter climate for UCP1 −3826G/A and UCP3 −55C/T. Further, by analyzing previously published genotype data for these SNPs, we found that the peak of the correlation for the UCP1 region occurred at the disease-associated −3826A/G variant and that the UCP3 region has a striking signal overall, with several individual SNPs showing interesting patterns, including the −55C/T variant. Resequencing of the regions in a set of three diverse population samples helped to clarify the signals that we found with the genotype data. At UCP1, the resequencing data revealed modest evidence that the haplotype carrying the −3826A variant was driven to high frequency by selection. In the UCP3 region, combining results from the climate analysis and resequencing survey suggest a more complex model in which variants on multiple haplotypes may independently be correlated with temperature. This is further supported by an excess of intermediate frequency variants in the UCP3 region in the Han Chinese population. Taken together, our results suggest that adaptation to climate influenced the global distribution of allele frequencies in UCP1 and UCP3 and provide an independent source of evidence for a role in cold resistance for UCP3.
Keywords: climate, obesity, nonshivering thermogenesis, natural selection, evolution
Introduction
Nonshivering thermogenesis (NST) plays an important role in cold-induced heat production across many eutherian mammal species (Cannon and Nedergaard 2004), and this pathway has been studied extensively in humans because of its potential importance in obesity risk (Wijers et al. 2009). UCP1 contributes to NST by uncoupling the proton gradient in the inner membrane of mitochondria from the formation of ATP, resulting in heat production. UCP1 is expressed highly and specifically in brown adipose tissue (BAT), a tissue that is characterized by a high density of mitochondria and that is the main site of NST. In mice, UCP1 expression in BAT is over 3,000-fold higher than its median expression across tissues, and it is the only protein found to have such a high level of relative expression in this tissue (Su et al. 2004). Until recently, BAT was thought to exist only in newborns where it contributed to the maintenance of normal body temperature. However, recent studies based on positron emission tomographic and computed tomographic scans showed that active BAT is present in adult humans, that UCP1 is expressed adult BAT, and that the amount of active BAT in adult humans exhibits substantial seasonal variation (Cypess et al. 2009; Saito et al. 2009; Virtanen et al. 2009). Therefore, BAT and UCP1 are likely to influence body temperature regulation throughout human life, with potential effects on infant as well as adult survival and on evolutionary fitness. In addition, UCP1 knockout mice exhibit increased sensitivity to cold and increased weight gain over time (Enerback et al. 1997; Kontani et al. 2005), and overexpressing UCP1 leads to an increase in metabolic rate (Li et al. 2000). An increase in UCP1 expression is expected to result in an increase in the total capacity for heat production and a concomitant decrease in the amount of energy stored as fat. Therefore, variants that increase UCP1 function are expected to be protective against obesity. Consistent with this expectation, the A allele of the −3826A/G variant upstream of UCP1, which is associated with increased expression of UCP1 (Esterbauer et al. 1998), is also associated with higher postprandial thermogenesis (Nagai et al. 2003) and resting metabolic rate and a slower rate of weight gain (Clement et al. 1996; Matsushita et al. 2003) relative to the G allele.
Two uncoupling protein homologues, UCP2 and UCP3, located about 10 kb apart on chromosome 11, were discovered in 1997 (Boss et al. 1997; Fleury et al. 1997) and were hypothesized to have thermogenic activity based on homology to UCP1; however, the functions of these two proteins are still the subject of substantial debate. Although it is generally accepted that neither of these proteins play as central a role in NST as UCP1 (Krauss et al. 2005), there is evidence that both proteins have uncoupling activity (Gong et al. 1997; Cline et al. 2001; Krauss et al. 2002), that heat production increases in response to overexpression of UCP3 in yeast (Paulik et al. 1998), and that UCP3 expression increases in response to stimulation by thyroid hormone (Gong et al. 1997; Larkin et al. 1997). Further, loss of function of UCP3 specific to BAT in Djungarian hamsters results in impaired NST (Nau et al. 2008). However, there are strong arguments against a role for UCP2 and UCP3 in NST. First, UCP2 and UCP3 are only expressed at low levels in BAT. Second, although uncoupling activity was observed in yeast cells containing mouse uncoupling proteins, subsequent research shows that this could be an artifact of the experimental procedure (Stuart et al. 2001; Harper et al. 2002). Third, although both starvation and a high fat diet lead to increased expression of UCP2 and UCP3, no change in mitochondrial uncoupling activity has been detected in response to the increased expression of the proteins (Cadenas et al. 1999; Hesselink et al. 2003). Fourth, although an UCP3 knockout rat exhibited lower uncoupling function in skeletal muscle (Lanni et al. 1999), no change in uncoupling activity was found for an UCP3 knockout mouse model (Bezaire et al. 2001). Alternative functions to NST that have been suggested for UCP2 and UCP3 include transporting fatty acids across the mitochondrial membrane (Himms-Hagen and Harper 2001; Dulloo et al. 2002; Schrauwen et al. 2006), regulating insulin signaling through changes in reactive oxygen species levels (Krauss et al. 2005; Choi et al. 2007) and regulating reactive oxygen species levels themselves (Krauss et al. 2005).
Although the UCP2 and UCP3 genes are adjacent to one another in the human genome and the similarity between the proteins is high, it is likely that the two proteins have quite different physiological functions. UCP2 is widely expressed among cell types, including white adipose tissue and pancreatic beta cells, whereas UCP3 is expressed mainly in skeletal muscle and to a lesser extent in BAT (Harper et al. 2001). In addition, evidence from association studies indicates that variation upstream of UCP2 and UCP3 may influence expression of the proteins and obesity risk, although no genome-wide significant association signal was found at these genes. Specifically, the A allele of the UCP2 −866G/A variant and the T allele of the UCP3 −55C/T variant are each associated with increased expression of the respective proteins (Schrauwen et al. 1999a, 1999b; Esterbauer et al. 2001) and with reduced risk of obesity (Esterbauer et al. 2001; Halsall et al. 2001; Kimm et al. 2002; Krempler et al. 2002; Liu et al. 2005; Hamada et al. 2008).
Human populations inhabit a wide range of environments with respect to temperature. Although cultural adaptations and acclimation are clearly important for survival in extremely cold environments, several lines of evidence indicate that genetic adaptations likely played an important role as well. Body shape and size varies with winter temperature among human populations, with populations in colder regions tending to be more robust, and populations in hot, equatorial regions tending to be slimmer, with longer arms and legs (Roberts 1978; Leonard et al. 2005). Furthermore, resting energy expenditure differs among populations; it tends to be high in arctic populations (Snodgrass et al. 2007), intermediate in whites of European ancestry, and lower in African Americans (Kushner et al. 1995; Kaplan et al. 1996; Morrison et al. 1996; Albu et al. 1997; Foster et al. 1997; Yanovski et al. 1997; Weyer et al. 1999; Wong et al. 1999), differences which may be explained, at least in part, by differences in NST.
We hypothesized that, as some human populations left Africa and migrated to colder regions at higher latitudes, alleles that increased thermogenic function increased in frequency as a result of natural selection. Because no previous studies provided sufficient data to test this hypothesis, we genotyped variants previously associated with thermogenesis and obesity phenotypes in UCP1, UCP2, and UCP3 in a worldwide population sample and calculated correlations with climate using both a linear model method that takes differences in population size into account and controls for population history and a nonparametric method that does not make assumptions about the relationship between allele frequencies and climate variables. In addition, we resequenced ∼10-kb regions for each of these genes in three populations (Italians, Han Chinese, and the Hausa of Cameroon) and examined evidence for the signature of positive natural selection on patterns of linked variation using haplotype- and frequency spectrum–based tests. Taken together, our results suggest evidence of positive selection at UCP1 primarily based on haplotype structure but also on the distribution of allele frequencies. At UCP3, there is strong evidence of selection due to cold climates from population allele frequency distributions, but the overall picture is more complex. The genetic variants that are correlated with climate variables belong to multiple distinct haplotypes, and there is no strong signal of selection from the haplotype structure. These observations suggest that the pattern at UCP3 may result from selection on standing variation. Moreover, the results for UCP3 contribute to the debate concerning its function by providing independent evidence for a role in cold resistance.
Materials and Methods
Climate Data
We obtained climate data collected for the past 50 years and averaged over the three winter months (December–February) from the NCEP/NCAR Climate Reanalysis Project, a joint project from the National Centers for Environmental Prediction and the National Center for Atmospheric Research (Kistler et al. 2001), for several variables related to winter climate: minimum temperature, maximum temperature, mean temperature, solar radiation flux, precipitation rate, and relative humidity. Although recent and ancient climates are unlikely to be the same, many human phenotypes and polymorphisms are correlated with measures of recent climate; this suggests that the recent climate is a reasonable proxy for the long-term climate experienced by human populations. In addition, we included in our analysis the absolute latitude for each population because latitude does not change over time and thus may be a better measure of long-term climate. Supplementary table 1 (Supplementary Material online) shows the climate data for each population included in the analysis.
Population Samples and Genotype Data
Initially, we selected three single-nucleotide polymorphisms (SNPs) that had previously been associated with thermogenesis or obesity for genotyping in 952 unrelated members of the Centre d'Etude du Polymorphisme Humain (CEPH) Human Genome Diversity Project (HGDP) panel (Cann et al. 2002; Rosenberg 2006) using Taqman assays. The probes used for UCP1 −3826G/A were GTGCAGCGATTTCTGATTGAC (forward) and CTGAATGTAACAAATTCTCCTTTCCTT (reverse), for UCP2 −866A/G were CCAGAGGGCCCAATTGTTG (forward) and ATGGACCGGGCCTGGTT (reverse), and for UCP3 −55C/T were TGTCAACCAACTTCTCTAGGATAAGG (forward) and CACTGTTGTCTCTGCTGCTTCTG (reverse) (nucleotide positions for these SNPs refers to distance from the start codon of the corresponding gene). To learn more about the evidence of selection over each region and to generate a null distribution against which we could assess significance for the UCP data, we obtained genotype data for the same individuals from two published studies. The first study genotyped tagging SNPs across the UCP2 and UCP3 regions (Hancock et al. 2008), and the second one generated genome-wide SNP genotype data using the Illumina 650Y platform (Li et al. 2008); both studies used the HGDP panel, thus providing the opportunity to integrate the allele frequency data for different SNPs in the same set of populations.
Associations between SNP Allele Frequencies and Climate Variables
We assessed the strength of the relationships between SNP allele frequencies and each climate variable using two complementary methods. The first is a Bayesian linear model (BLM) method that accounts for differences in sample sizes among populations and controls for population history by means of a covariance matrix that is incorporated into the model (Coop et al. 2010). This method yields a Bayes factor that is a measure of the support for a model in which an SNP allele frequency distribution is linearly dependent on both an environmental variable and population structure, relative to a model in which the allele frequency distribution is dependent on population structure alone. The BLM method has the greatest power when the relationship between the SNP frequencies and climate variable is linear. The second method, the Spearman rank correlation (SRC), makes no specific assumptions about the relationship between allele frequencies and climate and therefore may perform better when linearity assumptions are seriously violated. However, by controlling for populations structure, the BLM method should, in general, have lower false-positive and false-negative rates.
Because population history alone can result in strong correlations between allele frequencies and climate variables, using P values based on the theoretical distribution of correlations would often cause us to overestimate the significance for these SNPs. Therefore, for each SNP and for each climate variable, we assigned a transformed rank statistics (i.e., an empirical P value) by finding the rank of the SNP based on its correlation with the climate variable compared with other SNPs with similar characteristics. Assessing significance by comparing our results to the genome-wide distribution of correlations also allowed us to control for differences in power for SNPs with different global allele frequencies. Before computing the transformed rank statistic, we binned SNPs based on their derived allele frequencies (0–10%, 10–20%, 20–30%, 30–40%, 40–50%, 50–60%, 60–70%, 70–80%, 80–90%, 90–100%), and we calculated the rank compared with all other SNPs in the same bin. We inferred the ancestral state for each SNP using chimp and macaque sequence data obtained from the panTro2 and the rheMac2 genome assemblies, respectively. In addition, SNPs that were genotyped as part of the Illumina 650Y genotyping chip were further separated into one of three ascertainment panels for the determination of rank. For simplicity, the resulting rank-based P values will be referred to simply as P values below.
Gene-Based Scores
In addition to the signals of selection on individual SNPs, we wanted to assess the evidence for selection acting on each UCP gene as a whole. To this end, we calculated gene-based scores in two different ways. The first combines the evidence for selection across each SNP within 10 kb of the gene, by taking the average of the negative log-transformed P values across all SNPs in the region (eq. 1).
| (1) |
For the second method, we simply identified the minimum P value across the SNPs within 10 kb of the gene. For each UCP region and for each method, we created a null distribution for both statistics by calculating scores for 10,000 randomly selected contiguous genic regions (defined to be within 10 kb of a gene) that contained the same number of SNPs as the UCP gene itself. Then, we compared the observed score for each UCP gene region with the distribution of scores from the randomly selected regions to arrive at a gene-based empirical P value.
Collection of Resequencing Data
We conducted a survey of sequence variation in the UCP regions by sequencing a total of about 34 kb across UCP1, UCP2, and UCP3 genic and conserved 5′ regions in 16 individuals from each of 3 ethnically diverse populations (Hausa of Cameroon, Han Chinese, Italians). DNA was polymerase chain reaction (PCR) amplified, and the PCR products, after Exo-SAP purification, were sequenced with the ABI BigDye Terminator v. 3.1 Cycle Sequencing kit. The products were analyzed on an ABI 3730 automated sequencer (Applied Biosystems), and the resulting sequences were scored using the software Polyphred (version 6.11) (Bhangale et al. 2006).
Next, we inferred haplotypes and imputed missing data using PHASE v.2.028 (settings: 100 burn-in, 100 iterations, thinning interval = 1) (Stephens et al. 2001). Percentages of missing data were 4.3% for UCP1, 0.68% for UCP2, and 2.7% for UCP3. Subsequent analyses use the most likely haplotypes from the PHASE output. We calculated summary statistics for each region and population using SLIDER (http://genapps.uchicago.edu/labweb/index.html). The population recombination rate parameter (ρ = 4Nc, where N is the effective population size and c is the crossover rate between adjacent nucleotides) was estimated with Maxdip (Hudson 2001) under a scenario in which gene conversion is 10 times as likely as crossover and the mean conversion tract length is 55 bp.
Testing for Departures from Neutrality Based on the Allele Frequency Spectrum and Haplotype Structure
For each gene, we assessed the evidence for a deviation from neutrality based on Tajima's D statistic and on haplotype structure in both non-African populations. We focused on non-African populations because we hypothesized that variation in these genes was adaptive in populations that moved away from warm, equatorial regions to temperate regions at higher latitudes.
We calculated the Tajima's D statistic, the normalized difference between two measures of nucleotide diversity: the number of segregating sites (θW) and the mean number of pairwise differences between individuals (Tajima 1989). The values of these statistics reflect both the demographic history of a population and the history of selective pressures on a genomic region. To control for the effects of population history, we assessed significance for the observed statistics by comparing them to the distributions of statistics from 1,000 replicates of sequence data simulated under a neutral model of evolution as well as to the empirical distributions from actual sequence data previously collected in the same populations for 50 randomly selected genomic regions (Pluzhnikov et al. 2002). Neutral coalescent simulations were conducted using the computer program, ms (Hudson 2002). For these simulations, we matched simulated data to the true genomic regions by setting polymorphism levels and population recombination rate parameter values to those estimated for the regions. Two different bottleneck scenarios (representing the extremes based on previous modeling of the neutral variation in the same populations [Voight et al. 2005]) were used for each population. For both bottleneck models and both populations, the starting population size was assumed to be 10,000 individuals and the bottlenecks were assumed to have started 1,600 generations ago. For both populations, simulated bottlenecks ended 1,200 (model 1) and 200 (model 2) generations ago, with 25 years per generation. For Italians, bottleneck population sizes were 10% (model 1) and 30% (model 2) of the original population sizes, and for the Han, bottleneck population sizes were 5% (model 1) and 25% (model 2) of the original population sizes.
We also used a haplotype-based test to ask whether haplotypes defined by phenotype-associated variants appear to have been driven quickly to high frequency, a pattern that is consistent with a model of an incomplete selective sweep (Hudson et al. 1994). This test asks whether the subset of sequences defined by a given variant contains less variation than expected given the number of chromosomes and a neutral model of evolution. To assess significance for each haplotype, we ran 1,000 neutral coalescent simulations as described above. Then, for each simulation, we asked whether a subset of chromosomes existed that contained the number of chromosomes in the haplotype of interest with as few or fewer segregating sites.
Results
Correlations with Winter Climate for Disease-Associated Variants
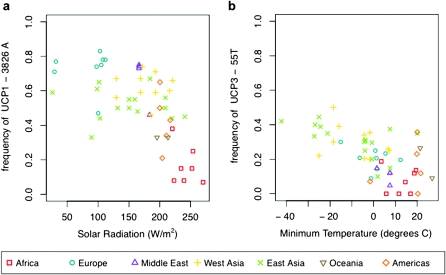
As shown in table 1, two of the three obesity-associated SNPs were marginally significantly correlated with at least one climate variable using both the SRC and BLM methods. The UCP1 −3826A variant that is associated with increased expression tends to have higher frequencies in populations at high latitude and where solar radiation is low. The UCP3 allele associated with increased expression (−55T) increases in frequency with decreasing minimum temperature, suggesting that it is linked to variation that plays a role in cold resistance. The UCP2 −866 variant was not suggestive or significant with either method (table 1). Figure 1 shows the distributions of allele frequencies across populations relative to the climate variables with the strongest signals for UCP1 −3826A and UCP3 −55T. A particularly compelling feature of the geographic pattern for UCP3 −55T is that the Native American populations (allele frequency = 0.16) living in equatorial environments have allele frequencies more similar to those of the African populations (mean allele frequency = 0.073) than to those of the more closely related East Asian Yakut (allele frequency = 0.42). This observation is potentially informative with regard to the strength of selection, in that selection was strong enough to increase the allele frequency within the 12–15 kya that elapsed since the peopling of the Americas.
Table 1.
Climate Correlations for SNPs Previously Associated with Phenotypes.
| Climate Correlationa (P valueb) |
|||||||||||||||
| Gene/SNP | Method | Absolute Latitude |
Minimum Temperature |
Maximum Temperature |
Mean Temperature |
Precipitation Rate |
Solar Radiation |
Relative Humidity |
|||||||
| UCP1 | SRC | 0.62 | (0.01) | −0.36 | (0.11) | −0.48 | (0.04) | −0.41 | (0.08) | −0.25 | (0.14) | −0.61 | (0.01) | 0.17 | (0.25) |
| −3826A | BLM | 1.37 | (0.15) | 0.21 | (0.74) | 0.27 | (0.55) | 0.23 | (0.66) | 0.20 | (0.73) | 5.16 | (0.04) | 0.15 | (0.79) |
| UCP2 | SRC | 0.23 | (0.56) | −0.21 | (0.50) | −0.22 | (0.56) | −0.21 | (0.53) | −0.10 | (0.53) | −0.20 | (0.67) | 0.16 | (0.43) |
| −866A | BLM | 1.30 | (0.15) | 0.73 | (0.20) | 1.10 | (0.15) | 0.84 | (0.18) | 0.26 | (0.46) | 0.31 | (0.41) | 0.16 | (0.71) |
| UCP3 | SRC | 0.46 | (0.13) | −0.58 | (0.01) | −0.56 | (0.03) | −0.57 | (0.02) | −0.16 | (0.34) | −0.36 | (0.26) | 0.13 | (0.50) |
| −55T | BLM | 2.00 | (0.11) | 5.58 | (0.05) | 2.17 | (0.09) | 4.04 | (0.06) | 0.22 | (0.55) | 0.62 | (0.21) | 0.15 | (0.78) |
For the SRC method, the correlation coefficient is reported.
For the BLM, the Bayes factor, which represents the strength of the evidence, is reported.
FIG. 1.
Plots showing the variation in population frequencies of UCP1 −3826A with solar radiation and UCP3 −55T with minimum temperature. Plots are displayed for the two alleles that were previously associated with thermogenesis or obesity phenotypes and that have significant correlations with climate; these SNPs are (a) UCP1 −3826A and (b) UCP3 −55T. The plotted allele is the one hypothesized to increase cold resistance (the allele that is protective against obesity). The color and symbol for each point represent the major geographic region to which the population is associated.
Correlations with Winter Climate across the UCP Regions
To ask whether there was evidence for climate adaptations in the UCP regions overall, we combined SNP data for the associated variants with additional genotype data for SNPs from two previously published reports (Hancock et al. 2008; Li et al. 2008). Then, we assessed the evidence for correlations with climate for each UCP gene region using two gene-based approaches. The first method combines the evidence for a correlation at each individual SNP in the region, and the second method considers the evidence of correlation only for the most strongly correlated SNP. For both approaches, we assessed significance for each gene region by comparing the statistic for the focal region to the distribution of the corresponding statistic calculated for 10,000 randomly sampled contiguous genic regions that had the same number of SNPs as the region of interest. The results from the two methods were highly concordant (table 2 and supplementary table 2, Supplementary Material online). This analysis revealed a very strong signal with temperature variables in the UCP3 region from both the BLM and SRC methods. Further, the strongest signal for this region was with minimum temperature, which is the variable most directly linked to cold resistance. We found weaker evidence for selection across the UCP1 and UCP2 regions. UCP1 was significant with temperature variables, latitude, and solar radiation with the SRC method but was not with the BLM method. Because the BLM assumes a linear relationship between a variant and climate variable, this discrepancy could be due to a geographic pattern of adaptive allele frequencies that violates the linearity assumption. For the UCP2 region, we observe the opposite pattern—the region is significant with the BLM method but not with the nonparametric SRC method. This could be because the power of the BLM method is higher than the power of the nonparametric method if the relationship between the climate variable and SNP allele frequencies is linear (Coop et al. 2010). Alternatively, the signal at UCP2 may be due to the partial overlap between SNPs in the UCP2 and UCP3 regions because SNPs within 10 kb of each gene were included in this analysis and UCP2 and UCP3 are located only about 11 kb apart.
Table 2.
Gene-Based Scores that Summarize the Combined Evidence for Selection across Each UCP Gene.
| Gene-Based Scorea (P valueb) |
|||||||||||||||
| Gene | Method | Absolute Latitude |
Minimum Temperature |
Maximum Temperature |
Mean Temperature |
Precipitation Rate |
Solar Radiation |
Relative Humidity |
|||||||
| UCP1 | SRC | 2.8 | (0.010) | 2.1 | (0.036) | 2.4 | (0.017) | 2.4 | (0.023) | 0.6 | (0.720) | 2.8 | (0.008) | 0.6 | (0.760) |
| BLM | 1.3 | (0.228) | 0.6 | (0.766) | 0.8 | (0.562) | 0.7 | (0.649) | 0.8 | (0.583) | 1.7 | (0.105) | 0.7 | (0.698) | |
| UCP2 | SRC | 0.8 | (0.636) | 0.6 | (0.791) | 0.7 | (0.724) | 0.6 | (0.782) | 0.7 | (0.646) | 0.8 | (0.601) | 0.9 | (0.455) |
| BLM | 2.5 | (0.026) | 2.5 | (0.023) | 3.0 | (0.011) | 2.7 | (0.014) | 1.0 | (0.383) | 1.8 | (0.097) | 1.2 | (0.290) | |
| UCP3 | SRC | 1.1 | (0.371) | 2.2 | (0.015) | 1.7 | (0.050) | 2.0 | (0.026) | 1.0 | (0.432) | 0.8 | (0.616) | 0.7 | (0.718) |
| BLM | 3.0 | (0.003) | 3.3 | (0.001) | 3.1 | (0.002) | 3.3 | (0.002) | 1.5 | (0.115) | 2.4 | (0.017) | 1.0 | (0.460) | |
The gene-based score is the average of the negative log-transformed P values across all SNPs in a gene.
P values are calculated by comparing the score for an UCP region to the scores for 10,000 randomly chosen genic regions with the same number of SNPs.
Motivated by the evidence of selection on the UCP1 and UCP2–UCP3 regions, we assessed evidence for correlation for all genotyped SNPs within 50 kb of each gene for the variables with the strongest gene-based evidence (i.e., solar radiation for UCP1 and minimum temperature for the UCP2–UCP3 region). Figures 2 and 3 show the variation in P values across the regions for both methods, and supplementary tables 3–6 (Supplementary Material online) show statistics and P values from the BLM and SRC methods for all SNPs with all climate variables. Although the evidence overall for the UCP1 region is weaker than for the UCP2–UCP3 region, it is interesting that the peak of signal with solar radiation occurs at the −3826A/G SNP previously associated with obesity and thermogenesis-related phenotypes. Conversely, for the UCP2-UCP3 region, the evidence of selection overall is striking, but the phenotype-associated SNPs (UCP2 −866A/G and UCP3−55C/T) do not have the strongest signals in the region; rather, several other SNPs (rs11235965, rs590336, rs7930460, rs647126, rs3741135, rs1726764, rs12417424) have P values that are as low or lower for at least one of the two methods used to assess correlation. Supplementary figures 1 and 2 (Supplementary Material online) show how the population allele frequencies vary with climate for SNPs that were significant in either the BLM or SRC analysis.
FIG. 2.
Correlations with solar radiation across the region within 50 kb of UCP1. The figure shows P values for the SRC method in black and for the BLM method in gray. A horizontal black line denotes a P value of 0.05. The position and orientation of UCP1 are indicated by the arrow at the bottom and by vertical hashed lines at the start and stop positions of the gene. Significant correlations (P < 0.05) were observed using both methods for the −3826A/G SNP, and these values are circled in the plot. The position of this SNP is indicated by a black line at the bottom of the plot, and positions of other SNPs for which climate correlations were calculated are indicated by gray lines.
FIG. 3.
Correlations with solar radiation across the region within 50 kb of UCP2 or UCP3. The figure shows P values for the SRC method in black and for the BLM method in gray. A horizontal black line denotes a P value of 0.05. The positions and orientations of UCP2 and UCP3 are indicated by the arrows at the bottom and by vertical hashed lines at the start and stop positions of the gene. Significant correlations (P < 0.05) were observed using both methods for the −55C/T SNP, and these values are circled in the plot. The position of this SNP is indicated by a black line at the bottom of the plot, and positions of other SNPs for which climate correlations were calculated are indicated by gray lines.
Sequence Variation in the UCP Regions
To learn more about sequence variation in the UCP genes and to test for departures from neutrality, we resequenced coding and conserved upstream regions of the UCP genes in three geographically distinct human populations (Hausa of Cameroon, Han Chinese and Italians). In total, we resequenced 11,781 bp in the UCP1 region and 22,093 bp over a region spanning 36,014 bp in the UCP2–UCP3 region for a total of nearly 34 kb. In UCP1, we identified 60 polymorphic sites, two of which were nonsynonymous (NS); in UCP2, there were 67 polymorphic sites, one of which was NS; and in UCP3, we identified 64 polymorphic sites, of which three were NS and one occurred at a splice site junction, which is known to result in truncation of the protein (Chung et al. 1999).
The three NS polymorphisms and the splice site variant detected in UCP3 were present only in the African population where one NS polymorphism was present on six chromosomes (Val102Ile), one was present on three chromosomes (Val9Met) and the Arg143X site, and the splice site were present on one chromosome each. Although this pattern could be consistent with a relaxation of purifying selection for the region, it may also be due to chance.
Summary statistics for each region and population are shown in table 3. Consistent with previous findings (Ramachandran et al. 2005; Liu et al. 2006; Jakobsson et al. 2008; Li et al. 2008) and expectations from models of human population history (Schaffner et al. 2005; Voight et al. 2005), we find that nucleotide diversity is highest in Africa and lower in non-African populations.
Table 3.
Summary Statistics of Polymorphism Data for the UCP Genes.
| Gene | Population | Length | Sa | πb | θWc | TDd | ρe |
| UCP1 | Hausa | 11781 | 45 | 1.05 | 0.95 | 0.40 | 1.48 |
| Italian | 43 | 0.90 | 0.91 | −0.04 | 0.75 | ||
| Han | 28 | 0.78 | 0.59 | 1.17 | 0.74 | ||
| UCP2 | Hausa | 10981 | 70 | 1.05 | 1.58 | −1.26 | 0.51 |
| Italian | 34 | 0.68 | 0.77 | 1.48 | 0.11 | ||
| Han | 12 | 0.40 | 0.27 | −0.40 | 0.66 | ||
| UCP3 | Hausa | 11112 | 56 | 1.16 | 1.25 | −0.27 | 1.13 |
| Italian | 37 | 0.96 | 0.83 | 0.59 | 0.36 | ||
| Han | 30 | 1.08 | 0.67 | 2.19 | 0.21 |
Number of segregating sites.
Nucleotide diversity per base pair (×10−3).
Watterson's estimator of the population mutation rate parameter θ (=4Nm) per base pair (×10−3) (Watterson 1975).
Tajima's D (Tajima 1989).
Composite likelihood estimator of the population recombination rate parameter (4Nr) per base pair (×10−3) under a model with gene conversion.
Using the Resequencing Data to Clarify the Signals with Climate
Figures 4 and 5 show the haplotype structure of the UCP1 and UCP2–UCP3 regions, with reference to SNPs previously associated with phenotypes, NS SNPs, and SNPs with significant climate correlations; only the climate variables with the strongest gene-based evidence were considered in this analysis (i.e., solar radiation for UCP1 and minimum temperature for the UCP2–UCP3 region). The UCP1 −3826A, UCP2 −866A, and UCP3 −55T alleles have been associated with increased mRNA levels of the downstream genes and reduced risk of obesity; however, these SNPs have not been tested functionally and therefore may merely be in strong LD with the variants that underlie the observed associations. Thus, our resequencing data provide the opportunity to uncover these causative variants.
FIG. 4.
Inferred haplotypes for the UCP1 region. To aid visualization, singleton sites were removed and identical chromosomes were combined (counts are shown in the far right column). Positions of the SNPs relative to the National Center for Biotechnology Information (NCBI) reference sequence (build 36) and reference sequence ID numbers for SNPs genotyped in the HGDP panel or of special interest are shown across the top of the figure. SNPs genotyped in the HGDP are shown in bold. Asterisks denote the SNPs that have significant correlations with minimum temperature using either the SRC or BLM method. In addition, positions for features of special interest are shown in italics. These include the UCP1−3826A/G SNP that was previously associated with obesity-related phenotypes and two NS SNPs. For all SNPs, the derived allele is shown in black and the ancestral in white.
FIG. 5.
Inferred haplotypes for the UCP2–UCP3 region. To aid visualization, singleton sites were removed and identical chromosomes were combined (counts are shown in the far right column). Positions of the SNPs relative to the NCBI reference sequence (build 36) and reference sequence id numbers for SNPs genotyped in the HGDP panel or of special interest are shown across the top of the figure. SNPs genotyped in the HGDP are shown in bold. Asterisks denote the SNPs that have significant correlations with minimum temperature using either the SRC or BLM method. In addition, positions for features of special interest are shown in italics. These include the UCP −866G/A and UCP3 −55C/T SNPs that were previously associated with phenotypes, three NS SNPs, and a 45-bp indel that was previously associated with type 2 diabetes. For all SNPs, the derived allele is shown in black and the ancestral in white.
At UCP1, the ancestral A allele at the −3826 variant, which has the strongest correlation with climate and is associated with increased postprandial thermogenesis and protection against other obesity-related phenotypes, is at low frequency in Africa and increases in frequency with decreasing solar radiation. Strong linkage disequilibrium extends across the sequenced region for the haplotype carrying this allele in both the Italian and Han Chinese populations, suggesting that, although UCP1 −3826A is associated with obesity-related traits, the possibility that the observed associations are due to other linked variation cannot be ruled out. Further, a subset of chromosomes carrying the −3826A allele is at fairly high frequency in the Italian population and contains only modest variation, suggesting that this allele may represent a haplotype driven quickly to high frequency due to positive selection. This hypothesis is tested below.
The haplotype structure is more complex for the UCP2–UCP3 region; here, several SNPs are strongly correlated with minimum winter temperature. An examination of figure 5 shows that in the Han population, there are three well-defined groups of UCP3 haplotypes, each occurring at moderate frequency, whereas in the European population, only one UCP3 haplotype occurs at intermediate frequency and two UCP3 haplotypes at lower frequency. To clarify the relationships among the SNPs that are correlated with temperature and to put these signals into context relative to the sequence data, we examined the correlations (r2) between the significant SNPs in the HapMap Project samples (supplementary figure 3, Supplementary Material online). Combined information from these different sources shows that there are multiple distinct haplotypes containing SNPs that are strongly correlated with minimum winter temperature. One of these haplotypes, which is at intermediate frequency in the Han, contains the −55C/T SNP (rs1800849) as well as two other SNPs: rs3741135 and rs11235965. Of the other five SNPs with strong correlations with climate, one (rs7960460) is in strong LD with rs11235965, two are in low LD with −55C/T (r2 < 0.4), but in perfect LD with one another in Asians and Europeans (rs647126 and rs590336), and the third is not in strong LD with any other SNPs with significant climate correlations (rs3781907). Notably, the SNPs rs647126 and rs590336 show extremely strong signals with temperature using the BLM method but show no such signal using the SRC method. The plots in supplementary figure 2 (Supplementary Material online) help to clarify this discrepancy. Both these SNPs have strong signals with temperature within major geographic regions, but on a global scale, the pattern is not striking. These signals exemplify how the BLM approach, which controls for the effects of population structure, can increase power to detect SNPs with strong correlations with climate, especially when the patterns vary across regions.
To compare the geographic structure of the UCP genes and also to ask whether a particular region of UCP3 appeared to show the strongest differentiation, we calculated the KST statistic, which is a measure of genetic differentiation between populations based on sequence data, and assessed its significance by permutations using the permtest software (http://home.uchicago.edu/∼rhudson1/source/permtest.html) (Hudson et al. 1992). We observed significant differentiation overall for UCP1, but not UCP2 or UCP3 (supplementary table 7, Supplementary Material online). However, when we ran the analysis over 20 SNP windows (with 10 SNP overlap) for UCP3, we observed significant evidence of differentiation (P = 0.013) only for the first window, which contains only one SNP strongly correlated with winter temperature: UCP3 −55C/T. Although this analysis suggests that the −55C/T variant shows the strongest evidence of driving the signal we observe for this gene, it remains possible that either variation outside the sequenced region or multiple variants within the UCP3 region contribute to the signal and, thus, to function.
Neutrality Tests Based on Resequencing Data
We observed a strongly positive Tajima's D statistic (2.19) for UCP3 in the Han population, suggesting an excess of intermediate frequency variants in the region. This is consistent with findings from the climate analysis that multiple distinct alleles show evidence of adaptation to climate. To assess the significance for this value of Tajima's D, we compared the observed statistic with results from coalescent simulations. The results were moderately significant when compared with simulations under two demographic scenarios inferred from a previous modeling study (Voight et al. 2005) and to the distribution of Tajima's D over a set of 50 randomly chosen loci sequenced in the same individuals (Voight et al. 2005) (P ≤ 0.043 and P = 0.04, respectively, using one-tailed tests of significance).
Next, we asked whether there was evidence that a haplotype defined by an SNP previously associated with obesity-related phenotypes (UCP1 −3826A, UCP2 −866A, or UCP3 −55T) was driven quickly to high frequency by selection (Hudson et al. 1994). Although we did not find a significant signal for any of these haplotypes overall, we examined the haplotype carrying the UCP1 −3826A allele in Italians more carefully because a subset of chromosomes carrying this variant appeared to have a deficit of variation relative to its frequency (supplementary table 8, Supplementary Material online). Although the haplotype strictly defined by the −3826A variant in this population includes a small number of chromosomes with fairly high variation (4 of the 32 chromosomes contain 29 polymorphic sites), a subset of 17 chromosomes contain very little variation (two polymorphic sites). A haplotype test for this subset of chromosomes was marginally significant (P ≤ 0.031) under both demographic models.
There are at least two reasons why a subset of chromosomes might exist with a stronger signature of selection than the total set defined by a selected variant. First, results of coalescent simulations under a model with selection showed that the haplotype strictly defined by the selected allele is not necessarily the one associated with the strongest evidence of selection (Kudaravalli 2008). Alternatively, if selection acted on standing variation, the selected variant may be present on multiple haplotypes. This can occur when the time between the emergence of the allele and the onset of positive selection is sufficiently long for recombination to break down the class of haplotypes carrying the allele (Hermisson and Pennings 2005; Przeworski et al. 2005). This may be a particularly likely scenario given that the −3826A variant that increases in frequency with latitude is the ancestral allele.
Discussion
We used genotype data from a worldwide panel of populations and resequencing data for three distinct populations to test the hypothesis that the distributions of variants associated with increased expression of candidate genes for thermogenesis (UCP1, UCP2, and UCP3) have been shaped by adaptations to climate. Although UCP1 has a well-established role in NST, the functions of UCP2 and UCP3 are the subjects of considerable debate (Krauss et al. 2005). Our results suggest that positive selection related to climate shaped the global distribution of the allele carrying the −3826A/G variant in UCP1, a gene known to have a critical role in cold resistance through the NST pathway. Further, our results provide a new line of evidence that variation in the UCP3 gene is implicated in cold resistance, possibly via NST.
We found that multiple variants within the UCP3 region have strong correlations with temperature, and these variants appear to represent multiple independent signals. Given the excess of intermediate frequency variants that we observed for the UCP3 region in the Han population, our results are consistent with a scenario in which multiple alleles that each enhance the function of UCP3 (via increased expression or some other mechanism) increased in frequency due to selection for cold resistance. This proposed scenario is also supported by the results of association studies. Two variants in UCP3 have been associated with variation in obesity-related phenotypes. The most commonly studied variant is the −55C/T variant, which is associated with increased mRNA levels of the UCP3 protein, resting metabolic rate, decreased body mass index, and higher resting energy expenditure (Jia et al. 2009); the other variant is a Tyr210Tyr polymorphism, which is associated with resting energy expenditure (Kimm et al. 2002) and with baseline body mass index, fat mass, and leptin levels (Lanouette et al. 2002). Although we do not have genotype data for the Tyr210Tyr polymorphism in the HGDP panel, in all three HapMap populations, this SNP is in strong LD with rs647126 (r2 ≥ 0.84) (supplementary figure 4, Supplementary Material online), an SNP with a strong correlation with temperature and that is on the haplotype not linked to the −55 variant in our analysis.
Another noteworthy feature of the UCP3 region is that there are three NS variants and one splice site variant that are polymorphic in the Hausa sample, but not in the Italian or Han Chinese samples. This pattern may be consistent with relaxed purifying selection within the Hausa population, which lives in sub-Saharan Africa where the selection pressure for high levels of NST is likely to be low compared with that expected for the Italian or Han Chinese populations, but it may also reflect the greater overall diversity in Africa. Previous authors have noted the difference in allele frequencies between populations of African descent and non-African populations for three of these variants (Val102Ile, Val9Met, and the splice site variant) (Kimm et al. 2002), and a further review of the literature showed that all these variants are consistently more common in samples of African ancestry compared with non-African samples (Argyropoulos et al. 1998; Chung et al. 1999; Kimm et al. 2002; Lanouette et al. 2002). However, it is unclear whether these variants influence human traits. For the most common of the variants (Val102Ile), no significant association with obesity-related traits has been observed and functional assays using yeast heterologous expression assays found no evidence for functional effects for either the Val102Ile or Val9Met variant. The two singleton variants, Arg143X and the splice site variant, are more likely to have functional effects. Both these variants result in premature truncation of the protein, and the splice site variant is associated with large effects on respiratory quotient and fat oxidation in carriers (Argyropoulos et al. 1998). Furthermore, effects on uncoupling were observed for Arg143X (Brown et al. 1999) and for the splice site variant (Hagen et al. 1999) in yeast heterologous expression assays. Therefore, even though the evidence for lower selective constraints in populations of African origin is inconclusive, at least some of these less common variants may play an important role in obesity risk, consistent with the notion that rare variants may be important in common disease etiology (Cohen et al. 2004; Romeo et al. 2007; Slatter et al. 2008; Lakoski et al. 2009; Romeo et al. 2009).
Our results are consistent with recent empirical and theoretical work arguing that selection on standing variation may play an important role in adaptation (Orr and Betancourt 2001; Hermisson and Pennings 2005; Przeworski et al. 2005; Pennings and Hermisson 2006a, 2006b; Hancock et al. 2010; Pritchard et al. 2010). Although we observe strong correlations with climate for UCP1 and UCP3, there are not strong signatures of an ongoing sweep either from frequency spectrum-based tests or from haplotype-based tests. Moreover, at UCP3, several SNPs from distinct haplotypes are strongly correlated with climate, suggesting that either there are multiple variants driving the signal, each on a different haplotype, or one undetected variant that is in weak LD with both these haplotypes. Either of these scenarios is consistent with selection on standing variation acting on the region. Standard models of selection on newly arisen variants tend to have low power to detect evidence of selection on standing variation; however, methods that assess evidence for selection based on spatial patterns of allele frequencies may have greater power in these cases (Novembre and Di Rienzo 2009). Furthermore, methods to assess evidence for positive selection based on correlations between phenotypes or genetic variants and environmental variables have historically been important for identifying phenotypes and loci under selection (e.g., loci involved in malaria resistance and lactose tolerance [Haldane 1949; Allison 1954; Livingstone 1964; Simoons 1970; Tishkoff et al. 2007]) and can be used to address particular biological hypotheses.
In conclusion, our study corroborates the idea that population genetic approaches can provide useful insights into the functional role of genes and of genetic variation (Nielsen et al. 2007). In the specific case of the uncoupling proteins, our population genetic data are relevant to both the function of the UCP3 gene and to the role of specific genetic variants that are inferred to have fitness and, therefore, functional effects. Ultimately, these evolutionary inferences will have to be validated through functional and phenotypic studies. Because variants associated with signatures of selection are noncoding, it is likely that these SNPs influence UCP gene function by modulating their expression levels, which in turn determine the total capacity for heat production. A prominent role for regulatory variants is consistent with the findings of the recent genome-wide association studies, which showed that most variation associated with common diseases and traits is noncoding (Nicolae et al.). Whether the noncoding regulatory SNPs in the UCP genes influence baseline transcript levels or expression in response to environmental stimuli is an open question. The growing repertoire of tools for functional genomic analyses will undoubtedly help in elucidating the role of variants associated with signatures of natural selection.
Supplementary Material
Supplementary tables 1–8 and supplementary figures 1–4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We are grateful to members of the Di Rienzo laboratory for helpful discussions about this research, to Molly Przeworski for comments on an earlier version of the manuscript, to Richard Hudson for advice on the geographic structure analysis, and to two anonymous reviewers for helpful comments. This work was supported by National Institutes of Health (NIH) Grants DK56670 and GM79558. A.M.H. was supported in part by American Heart Association Graduate Fellowship 0710189Z and by NIH Genetics and Regulation Training Grant GM07197.
References
- Albu J, Shur M, Curi M, Murphy L, Heymsfield SB, Pi-Sunyer FX. Resting metabolic rate in obese, premenopausal black women. Am J Clin Nutr. 1997;66:531–538. doi: 10.1093/ajcn/66.3.531. [DOI] [PubMed] [Google Scholar]
- Allison AC. Protection afforded by sickle-cell trait against subtertian malarial infection. Br Med J. 1954;1:290–294. doi: 10.1136/bmj.1.4857.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyropoulos G, Brown AM, Willi SM, Zhu J, He Y, Reitman M, Gevao SM, Spruill I, Garvey WT. Effects of mutations in the human uncoupling protein 3 gene on the respiratory quotient and fat oxidation in severe obesity and type 2 diabetes. J Clin Invest. 1998;102:1345–1351. doi: 10.1172/JCI4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezaire V, Hofmann W, Kramer JK, Kozak LP, Harper ME. Effects of fasting on muscle mitochondrial energetics and fatty acid metabolism in Ucp3(−/−) and wild-type mice. Am J Physiol Endocrinol Metab. 2001;281:E975–E982. doi: 10.1152/ajpendo.2001.281.5.E975. [DOI] [PubMed] [Google Scholar]
- Bhangale TR, Stephens M, Nickerson DA. Automating resequencing-based detection of insertion-deletion polymorphisms. Nat Genet. 2006;38:1457–1462. doi: 10.1038/ng1925. [DOI] [PubMed] [Google Scholar]
- Boss O, Samec S, Paoloni-Giacobino A, Rossier C, Dulloo A, Seydoux J, Muzzin P, Giacobino JP. Uncoupling protein-3: a new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett. 1997;408:39–42. doi: 10.1016/s0014-5793(97)00384-0. [DOI] [PubMed] [Google Scholar]
- Brown AM, Dolan JW, Willi SM, Garvey WT, Argyropoulos G. Endogenous mutations in human uncoupling protein 3 alter its functional properties. FEBS Lett. 1999;464:189–193. doi: 10.1016/s0014-5793(99)01708-1. [DOI] [PubMed] [Google Scholar]
- Cadenas S, Buckingham JA, Samec S, Seydoux J, Din N, Dulloo AG, Brand MD. UCP2 and UCP3 rise in starved rat skeletal muscle but mitochondrial proton conductance is unchanged. FEBS Lett. 1999;462:257–260. doi: 10.1016/s0014-5793(99)01540-9. [DOI] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, et al. (41 co-authors) A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Choi CS, Fillmore JJ, Kim JK, et al. (16 co-authors) Overexpression of uncoupling protein 3 in skeletal muscle protects against fat-induced insulin resistance. J Clin Invest. 2007;117:1995–2003. doi: 10.1172/JCI13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WK, Luke A, Cooper RS, et al. (26 co-authors) Genetic and physiologic analysis of the role of uncoupling protein 3 in human energy homeostasis. Diabetes. 1999;48:1890–1895. doi: 10.2337/diabetes.48.9.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement K, Ruiz J, Cassard-Doulcier AM, Bouillaud F, Ricquier D, Basdevant A, Guy-Grand B, Froguel P. Additive effect of A→G (-3826) variant of the uncoupling protein gene and the Trp64Arg mutation of the beta 3-adrenergic receptor gene on weight gain in morbid obesity. Int J Obes Relat Metab Disord. 1996;20:1062–1066. [PubMed] [Google Scholar]
- Cline GW, Vidal-Puig AJ, Dufour S, Cadman KS, Lowell BB, Shulman GI. In vivo effects of uncoupling protein-3 gene disruption on mitochondrial energy metabolism. J Biol Chem. 2001;276:20240–20244. doi: 10.1074/jbc.M102540200. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Coop G, Witonsky D, Di Rienzo A, Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:1411–1423. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, et al. (12 co-authors) Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulloo AG, Stock MJ, Solinas G, Boss O, Montani JP, Seydoux J. Leptin directly stimulates thermogenesis in skeletal muscle. FEBS Lett. 2002;515:109–113. doi: 10.1016/s0014-5793(02)02449-3. [DOI] [PubMed] [Google Scholar]
- Enerback S, Jacobsson A, Simpson EM, Guerra C, Yamashita H, Harper ME, Kozak LP. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature. 1997;387:90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- Esterbauer H, Oberkofler H, Liu YM, Breban D, Hell E, Krempler F, Patsch W. Uncoupling protein-1 mRNA expression in obese human subjects: the role of sequence variations at the uncoupling protein-1 gene locus. J Lipid Res. 1998;39:834–844. [PubMed] [Google Scholar]
- Esterbauer H, Schneitler C, Oberkofler H, et al. (12 co-authors) A common polymorphism in the promoter of UCP2 is associated with decreased risk of obesity in middle-aged humans. Nat Genet. 2001;28:178–183. doi: 10.1038/88911. [DOI] [PubMed] [Google Scholar]
- Fleury C, Neverova M, Collins S, et al. (11 co-authors) Uncoupling protein-2: a novel gene linked to obesity and hyperinsulinemia. Nat Genet. 1997;15:269–272. doi: 10.1038/ng0397-269. [DOI] [PubMed] [Google Scholar]
- Foster GD, Wadden TA, Vogt RA. Resting energy expenditure in obese African American and Caucasian women. Obes Res. 1997;5:1–8. doi: 10.1002/j.1550-8528.1997.tb00276.x. [DOI] [PubMed] [Google Scholar]
- Gong DW, He Y, Karas M, Reitman M. Uncoupling protein-3 is a mediator of thermogenesis regulated by thyroid hormone, beta3-adrenergic agonists, and leptin. J Biol Chem. 1997;272:24129–24132. doi: 10.1074/jbc.272.39.24129. [DOI] [PubMed] [Google Scholar]
- Hagen T, Zhang CY, Slieker LJ, Chung WK, Leibel RL, Lowell BB. Assessment of uncoupling activity of the human uncoupling protein 3 short form and three mutants of the uncoupling protein gene using a yeast heterologous expression system. FEBS Lett. 1999;454:201–206. doi: 10.1016/s0014-5793(99)00811-x. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. Disease and evolution. Ric Sci Suppl A. 1949;19:68–76. [Google Scholar]
- Halsall DJ, Luan J, Saker P, Huxtable S, Farooqi IS, Keogh J, Wareham NJ, O'Rahilly S. Uncoupling protein 3 genetic variants in human obesity: the c-55t promoter polymorphism is negatively correlated with body mass index in a UK Caucasian population. Int J Obes Relat Metab Disord. 2001;25:472–477. doi: 10.1038/sj.ijo.0801584. [DOI] [PubMed] [Google Scholar]
- Hamada T, Kotani K, Fujiwara S, Sano Y, Domichi M, Tsuzaki K, Sakane N. The common -55 C/T polymorphism in the promoter region of the uncoupling protein 3 gene reduces prevalence of obesity and elevates serum high-density lipoprotein cholesterol levels in the general Japanese population. Metabolism. 2008;57:410–415. doi: 10.1016/j.metabol.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Ehler E, et al. (11 co-authors) Human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proc Natl Acad Sci U S A. 2010;107(Suppl 2):8924–8930. doi: 10.1073/pnas.0914625107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Gordon AS, Eshel G, Pritchard JK, Coop G, Di Rienzo A. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 2008;4:e32. doi: 10.1371/journal.pgen.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JA, Stuart JA, Jekabsons MB, Roussel D, Brindle KM, Dickinson K, Jones RB, Brand MD. Artifactual uncoupling by uncoupling protein 3 in yeast mitochondria at the concentrations found in mouse and rat skeletal-muscle mitochondria. Biochem J. 2002;361:49–56. doi: 10.1042/0264-6021:3610049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper ME, Dent RM, Bezaire V, Antoniou A, Gauthier A, Monemdjou S, McPherson R. UCP3 and its putative function: consistencies and controversies. Biochem Soc Trans. 2001;29:768–773. doi: 10.1042/bst0290768. [DOI] [PubMed] [Google Scholar]
- Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselink MK, Greenhaff PL, Constantin-Teodosiu D, Hultman E, Saris WH, Nieuwlaat R, Schaart G, Kornips E, Schrauwen P. Increased uncoupling protein 3 content does not affect mitochondrial function in human skeletal muscle in vivo. J Clin Invest. 2003;111:479–486. doi: 10.1172/JCI16653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himms-Hagen J, Harper ME. Physiological role of UCP3 may be export of fatty acids from mitochondria when fatty acid oxidation predominates: an hypothesis. Exp Biol Med (Maywood) 2001;226:78–84. doi: 10.1177/153537020122600204. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Two-locus sampling distributions and their application. Genetics. 2001;159:1805–1817. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Bailey K, Skarecky D, Kwiatowski J, Ayala FJ. Evidence for positive selection in the superoxide dismutase (Sod) region of Drosophila melanogaster. Genetics. 1994;136:1329–1340. doi: 10.1093/genetics/136.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographic subdivision. Mol Biol Evol. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Scholz SW, Scheet P, et al. (24 co-authors) Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Jia JJ, Zhang X, Ge CR, Jois M. The polymorphisms of UCP2 and UCP3 genes associated with fat metabolism, obesity and diabetes. Obes Rev. 2009;10:519–526. doi: 10.1111/j.1467-789X.2009.00569.x. [DOI] [PubMed] [Google Scholar]
- Kaplan AS, Zemel BS, Stallings VA. Differences in resting energy expenditure in prepubertal black children and white children. J Pediatr. 1996;129:643–647. doi: 10.1016/s0022-3476(96)70143-9. [DOI] [PubMed] [Google Scholar]
- Kimm SY, Glynn NW, Aston CE, Damcott CM, Poehlman ET, Daniels SR, Ferrell RE. Racial differences in the relation between uncoupling protein genes and resting energy expenditure. Am J Clin Nutr. 2002;75:714–719. doi: 10.1093/ajcn/75.4.714. [DOI] [PubMed] [Google Scholar]
- Kistler R, Kalnay E, Collins W, et al. (13 co-authors) The NCEP-NCAR 50-year reanalysis: monthly means CD-ROM and documentation. Bull Am Meteor Soc. 2001;82:247–267. [Google Scholar]
- Kontani Y, Wang Y, Kimura K, Inokuma KI, Saito M, Suzuki-Miura T, Wang Z, Sato Y, Mori N, Yamashita H. UCP1 deficiency increases susceptibility to diet-induced obesity with age. Aging Cell. 2005;4:147–155. doi: 10.1111/j.1474-9726.2005.00157.x. [DOI] [PubMed] [Google Scholar]
- Krauss S, Zhang CY, Lowell BB. A significant portion of mitochondrial proton leak in intact thymocytes depends on expression of UCP2. Proc Natl Acad Sci U S A. 2002;99:118–122. doi: 10.1073/pnas.012410699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S, Zhang CY, Lowell BB. The mitochondrial uncoupling-protein homologues. Nat Rev Mol Cell Biol. 2005;6:248–261. doi: 10.1038/nrm1592. [DOI] [PubMed] [Google Scholar]
- Krempler F, Esterbauer H, Weitgasser R, Ebenbichler C, Patsch JR, Miller K, Xie M, Linnemayr V, Oberkofler H, Patsch W. A functional polymorphism in the promoter of UCP2 enhances obesity risk but reduces type 2 diabetes risk in obese middle-aged humans. Diabetes. 2002;51:3331–3335. doi: 10.2337/diabetes.51.11.3331. [DOI] [PubMed] [Google Scholar]
- Kudaravalli S. Recent positive selection in the human genome [doctoral dissertation] [Chicago (IL)]: University of Chicago; 2008. [Google Scholar]
- Kushner RF, Racette SB, Neil K, Schoeller DA. Measurement of physical activity among black and white obese women. Obes Res. 1995;3(Suppl 2):261s–265s. doi: 10.1002/j.1550-8528.1995.tb00472.x. [DOI] [PubMed] [Google Scholar]
- Lakoski SG, Lagace TA, Cohen JC, Horton JD, Hobbs HH. Genetic and metabolic determinants of plasma PCSK9 levels. J Clin Endocrinol Metab. 2009;94:2537–2543. doi: 10.1210/jc.2009-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanni A, Beneduce L, Lombardi A, Moreno M, Boss O, Muzzin P, Giacobino JP, Goglia F. Expression of uncoupling protein-3 and mitochondrial activity in the transition from hypothyroid to hyperthyroid state in rat skeletal muscle. FEBS Lett. 1999;444:250–254. doi: 10.1016/s0014-5793(99)00061-7. [DOI] [PubMed] [Google Scholar]
- Lanouette CM, Chagnon YC, Rice T, et al. (12 co-authors) Uncoupling protein 3 gene is associated with body composition changes with training in HERITAGE study. J Appl Physiol. 2002;92:1111–1118. doi: 10.1152/japplphysiol.00726.2001. [DOI] [PubMed] [Google Scholar]
- Larkin S, Mull E, Miao W, Pittner R, Albrandt K, Moore C, Young A, Denaro M, Beaumont K. Regulation of the third member of the uncoupling protein family, UCP3, by cold and thyroid hormone. Biochem Biophys Res Commun. 1997;240:222–227. doi: 10.1006/bbrc.1997.7636. [DOI] [PubMed] [Google Scholar]
- Leonard WR, Snodgrass JJ, Sorensen MV. Metabolic adaptation in indigenous Siberian populations. Annu Rev Anthropol. 2005;34:451–471. [Google Scholar]
- Li B, Nolte LA, Ju JS, Han DH, Coleman T, Holloszy JO, Semenkovich CF. Skeletal muscle respiratory uncoupling prevents diet-induced obesity and insulin resistance in mice. Nat Med. 2000;6:1115–1120. doi: 10.1038/80450. [DOI] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, et al. (11 co-authors) Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Liu H, Prugnolle F, Manica A, Balloux F. A geographically explicit genetic model of worldwide human-settlement history. Am J Hum Genet. 2006;79:230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Liu PY, Long J, Lu Y, Elze L, Recker RR, Deng HW. Linkage and association analyses of the UCP3 gene with obesity phenotypes in Caucasian families. Physiol Genomics. 2005;22:197–203. doi: 10.1152/physiolgenomics.00031.2005. [DOI] [PubMed] [Google Scholar]
- Livingstone FB. Aspects of the population dynamics of the abnormal hemoglobin and glucose-6-phosphate dehydrogenase deficiency genes. Am J Hum Genet. 1964;16:435–450. [PMC free article] [PubMed] [Google Scholar]
- Matsushita H, Kurabayashi T, Tomita M, Kato N, Tanaka K. Effects of uncoupling protein 1 and beta3-adrenergic receptor gene polymorphisms on body size and serum lipid concentrations in Japanese women. Maturitas. 2003;45:39–45. doi: 10.1016/s0378-5122(03)00088-4. [DOI] [PubMed] [Google Scholar]
- Morrison JA, Alfaro MP, Khoury P, Thornton BB, Daniels SR. Determinants of resting energy expenditure in young black girls and young white girls. J Pediatr. 1996;129:637–642. doi: 10.1016/s0022-3476(96)70142-7. [DOI] [PubMed] [Google Scholar]
- Nagai N, Sakane N, Ueno LM, Hamada T, Moritani T. The -3826 A→G variant of the uncoupling protein-1 gene diminishes postprandial thermogenesis after a high fat meal in healthy boys. J Clin Endocrinol Metab. 2003;88:5661–5667. doi: 10.1210/jc.2003-030672. [DOI] [PubMed] [Google Scholar]
- Nau K, Fromme T, Meyer CW, von Praun C, Heldmaier G, Klingenspor M. Brown adipose tissue specific lack of uncoupling protein 3 is associated with impaired cold tolerance and reduced transcript levels of metabolic genes. J Comp Physiol B. 2008;178:269–277. doi: 10.1007/s00360-007-0219-7. [DOI] [PubMed] [Google Scholar]
- doi: 10.1371/journal.pgen.1000888. Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. 2010. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6:e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. Recent and ongoing selection in the human genome. Nat Rev Genet. 2007;8:857–868. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Di Rienzo A. Spatial patterns of variation due to natural selection in humans. Nat Rev Genet. 2009;10:745–755. doi: 10.1038/nrg2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, Betancourt AJ. Haldane's sieve and adaptation from the standing genetic variation. Genetics. 2001;157:875–884. doi: 10.1093/genetics/157.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulik MA, Buckholz RG, Lancaster ME, Dallas WS, Hull-Ryde EA, Weiel JE, Lenhard JM. Development of infrared imaging to measure thermogenesis in cell culture: thermogenic effects of uncoupling protein-2, troglitazone, and beta-adrenoceptor agonists. Pharm Res. 1998;15:944–949. doi: 10.1023/a:1011993019385. [DOI] [PubMed] [Google Scholar]
- Pennings PS, Hermisson J. Soft sweeps II—molecular population genetics of adaptation from recurrent mutation or migration. Mol Biol Evol. 2006a;23:1076–1084. doi: 10.1093/molbev/msj117. [DOI] [PubMed] [Google Scholar]
- Pennings PS, Hermisson J. Soft sweeps III: the signature of positive selection from recurrent mutation. PLoS Genet. 2006b;2:e186. doi: 10.1371/journal.pgen.0020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluzhnikov A, Di Rienzo A, Hudson RR. Inferences about human demography based on multilocus analyses of noncoding sequences. Genetics. 2002;161:1209–1218. doi: 10.1093/genetics/161.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M, Coop G, Wall JD. The signature of positive selection on standing genetic variation. Evolution. 2005;59:2312–2323. [PubMed] [Google Scholar]
- Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci U S A. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DF. Climate and human variability. 2nd ed. Menlo Park (CA): Cummings; 1978. [Google Scholar]
- Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, Hobbs HH, Cohen JC. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo S, Yin W, Kozlitina J, Pennacchio LA, Boerwinkle E, Hobbs HH, Cohen JC. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest. 2009;119:70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. Standardized subsets of the HGDP-CEPH human genome diversity cell line panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Saito M, Okamatsu-Ogura Y, Matsushita M, et al. (12 co-authors) High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, Altshuler D. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen P, Hoeks J, Hesselink MK. Putative function and physiological relevance of the mitochondrial uncoupling protein-3: involvement in fatty acid metabolism? Prog Lipid Res. 2006;45:17–41. doi: 10.1016/j.plipres.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Schrauwen P, Xia J, Bogardus C, Pratley RE, Ravussin E. Skeletal muscle uncoupling protein 3 expression is a determinant of energy expenditure in Pima Indians. Diabetes. 1999a;48:146–149. doi: 10.2337/diabetes.48.1.146. [DOI] [PubMed] [Google Scholar]
- Schrauwen P, Xia J, Walder K, Snitker S, Ravussin E. A novel polymorphism in the proximal UCP3 promoter region: effect on skeletal muscle UCP3 mRNA expression and obesity in male non-diabetic Pima Indians. Int J Obes Relat Metab Disord. 1999b;23:1242–1245. doi: 10.1038/sj.ijo.0801057. [DOI] [PubMed] [Google Scholar]
- Simoons FJ. Primary adult lactose intolerance and the milking habit: a problem in biologic and cultural interrelations. II. A culture historical hypothesis. Am J Dig Dis. 1970;15:695–710. doi: 10.1007/BF02235991. [DOI] [PubMed] [Google Scholar]
- Slatter TL, Jones GT, Williams MJ, van Rij AM, McCormick SP. Novel rare mutations and promoter haplotypes in ABCA1 contribute to low-HDL-C levels. Clin Genet. 2008;73:179–184. doi: 10.1111/j.1399-0004.2007.00940.x. [DOI] [PubMed] [Google Scholar]
- Snodgrass JJ, Sorensen MV, Tarskaia LA, Leonard WR. Adaptive dimensions of health research among indigenous Siberians. Am J Hum Biol. 2007;19:165–180. doi: 10.1002/ajhb.20624. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JA, Harper JA, Brindle KM, Jekabsons MB, Brand MD. Physiological levels of mammalian uncoupling protein 2 do not uncouple yeast mitochondria. J Biol Chem. 2001;276:18633–18639. doi: 10.1074/jbc.M011566200. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, et al. (13 co-authors) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Ranciaro A, et al. (19 co-authors) Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, et al. (11 co-authors) Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Voight BF, Adams AM, Frisse LA, Qian Y, Hudson RR, Di Rienzo A. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci U S A. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975;2:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Weyer C, Snitker S, Bogardus C, Ravussin E. Energy metabolism in African Americans: potential risk factors for obesity. Am J Clin Nutr. 1999;70:13–20. doi: 10.1093/ajcn/70.1.13. [DOI] [PubMed] [Google Scholar]
- Wijers SL, Saris WH, van Marken Lichtenbelt WD. Recent advances in adaptive thermogenesis: potential implications for the treatment of obesity. Obes Rev. 2009;10:218–226. doi: 10.1111/j.1467-789X.2008.00538.x. [DOI] [PubMed] [Google Scholar]
- Wong WW, Butte NF, Ellis KJ, Hergenroeder AC, Hill RB, Stuff JE, Smith EO. Pubertal African-American girls expend less energy at rest and during physical activity than Caucasian girls. J Clin Endocrinol Metab. 1999;84:906–911. doi: 10.1210/jcem.84.3.5517. [DOI] [PubMed] [Google Scholar]
- Yanovski SZ, Reynolds JC, Boyle AJ, Yanovski JA. Resting metabolic rate in African-American and Caucasian girls. Obes Res. 1997;5:321–325. doi: 10.1002/j.1550-8528.1997.tb00558.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.