Abstract
The Mediator complex forms the bridge between transcriptional activators and the RNA polymerase II. Med1 (also known as PBP or TRAP220) is a key component of Mediator that interacts with nuclear hormone receptors and GATA transcription factors. Here, we show dynamic recruitment of GATA-1, TFIIB, Mediator, and RNA polymerase II to the β-globin locus in induced mouse erythoid leukemia cells and in an erythropoietin-inducible hematopoietic progenitor cell line. Using Med1 conditional knockout mice, we demonstrate a specific block in erythroid development but not in myeloid or lymphoid development, highlighted by the complete absence of β-globin gene expression. Thus, Mediator subunit Med1 plays a pivotal role in erythroid development and in β-globin gene activation.
Keywords: blood, hematopoiesis, erythropoiesis, transcriptional regulation, globin
Studies of globin gene regulation have served as a useful paradigm to dissect the molecular mechanism of gene expression in higher eukaryotes (as reviewed in ref. 1). Several transcription factors like GATA-1, EKLF, and NF-E2 have been found to be essential for adult globin gene expression (2). Mice deficient in GATA-1 succumb to fatal anemia and do not produce any β-globin transcript (3). Efforts to better understand the molecular mechanism, how this master regulator GATA-1 acts, have identified a number of additional transcription factors such as SCL/TAL-1, EKLF, PU.1, Sp1, Rb (reviewed in refs. 2 and 4), and recently p53 (5). In addition, several GATA-1 cofactors, FOG-1, CBP/p300, PIASy the SWI/SNF (subunit Brg1), and the Mediator complex (subunit Med1/TRAP220/PBP), have been shown to play crucial roles in GATA-1–mediated transcriptional control (refs. 6–10 and as reviewed in refs. 2 and 4).
The Mediator complex was first discovered in yeast and is required for RNA polymerase II (RNAP II)-dependent gene transcription. In the past decade, several studies on mammalian Mediator demonstrated the central role of Mediator as a bridging factor between transcriptional regulators and the RNA polymerase II itself (reviewed in refs. 11–13). Mediator is recruited to the promoter independently of RNAP II and resides at enhancer elements rather than at core promoters (14, 15).
The mammalian Mediator complex consists of 31 subunits (11, 13). The best-studied subunit is Med1/PBP/TRAP220 (also termed ARC/DRIP205 or Med220). Med1 was first discovered as a coactivator for nuclear receptors (16, 17 and as reviewed in ref. 18). Consistent with an important role in mouse development, ablation of Med1 leads to lethality at midgestation, day 11.5 postcoitum (19, 20). Using a conditional null mutation of Med1 (PBP), it was demonstrated that Med1 is an essential cofactor for PPARα in the liver (21).
Here, we demonstrate the central role of the Mediator complex in β-globin gene activation. Using time-course chromatin immunoprecipitation (ChIP) experiments, we identified the sequence of events controlling β-globin gene activation: Following the initial binding of the GATA-1 transcription factor and the presence of histone acetylation, dynamic recruitment of TFIIB, Mediator, and subsequently RNAP II was observed. In addition, the use of Med1-conditional knockout mice allowed us to address the physiological relevance of Med1/TRAP220 during adult hematopoiesis as these mice showed a specific block in adult erythropoiesis whereas lymphopoiesis and myelopoeisis remained unaffected. Although the Mediator subunit Med1 is not required for global gene transcription per se, it is an essential cofactor for the activation of β-globin gene expression.
Results
Mediator Complex Plays a Central Role in β-Globin Gene Activation.
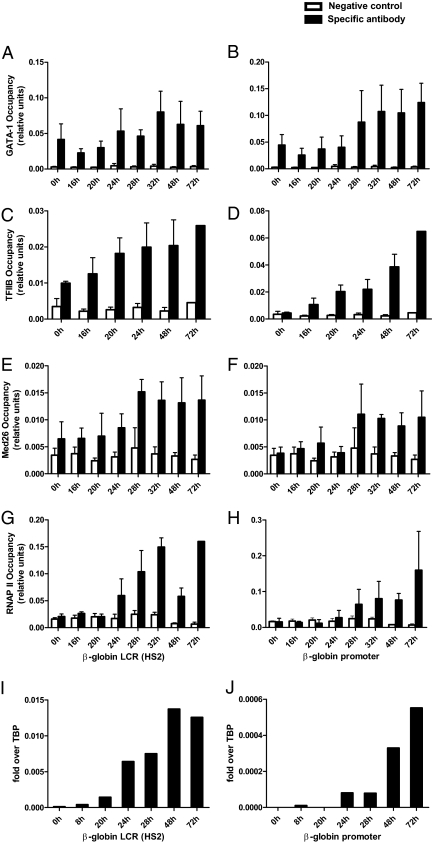
To dissect the spatial and temporal cofactor recruitment to the β-globin locus, we took advantage of the mouse erythroid leukemia (MEL) cell line as previously done by other investigators (22, 23). MEL cells are arrested at the proerythroblast stage and express high levels of GATA-1 but not β-globin mRNA unless differentiation is further induced by DMSO treatment. ChIP experiments using anti–GATA-1 antibody show that GATA-1 is already found at both the locus control region (LCR) and 50 kb away at the β-major promoter (Fig. 1 A and B) even before DMSO induction. The H3K9ac histone mark was also found at both of these regulatory elements before induction (Fig. S1 A and B). Most importantly, Mediator (Fig. 1 E and F and Fig. S2) and the general transcription factor TFIIB (Fig. 1 C and D) were detected only at the LCR and not at the β-major promoter in noninduced MEL cells whereas RNAP II (Fig. 1 G and H) recruitment could not be detected at any of these sites. RNAP II was first detected at the LCR 24 h after induction and 4 h later at the promoter. The dynamic appearance of RNAP II at the β-globin gene locus was further confirmed by the detection of noncoding RNA transcripts first at the LCR (Fig. 1I) and then at the β-major promoter (Fig. 1J). The observation that Mediator binds first to the LCR and then at the β-globin promoter before the recruitment of RNAP II is reminiscent of what we previously reported in yeast (15). Thus, Mediator is found at enhancer elements independently of RNAP II.
Fig. 1.
Dynamic recruitment of GATA-1, Mediator, TFIIB, and RNAP II at the β-globin locus in differentiating MEL cells. ChIP was performed at the β-globin LCR and β-major promoter at several time points after DMSO induction. GATA-1 is already bound at the hypersensitive site 2 (HS2) of the LCR (A) and at the β-major promoter (B) in noninduced MEL cells. Association of TFIIB occurs before induction only at the LCR (C) and after induction at the promoter (D). Mediator behaves similarly to TFIIB and is first found at the LCR (E) and only upon induction at the core promoter (F). However, RNAP II is not present at the LCR or at the β-major promoter before induction (G and H). The ChIP data for RNAP II are supported by measuring noncoding transcripts by RT-PCR at the LCR (I) and at the β-globin promoter (J). The y axis represents the absolute enrichment (mean ± SD, four to seven independent experiments) normalized to input. Background is defined as the unspecific binding of each individual antibody to an unrelated control sequence (−106 kb upstream in the GATA-1 locus).
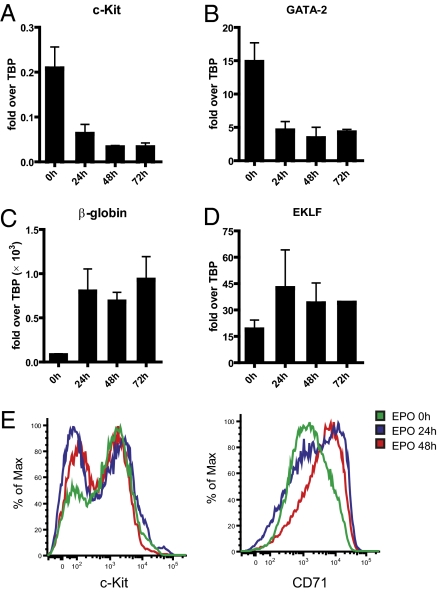
To investigate the recruitment of transcription factors in β-globin gene activation in a more physiological setting, we established a hematopoietic progenitor cell line in which differentiation along the erythroid lineage is controlled by erythropoietin (EPO). On the basis of the work of independent groups on bone marrow and embryonic stem cells (24, 25), transduction of the homeobox factor HoxB4 led to the isolation of progenitor cells that could be differentiated into erythroid cells ex vivo. We observed that the erythroid potential of HoxB4-transduced progenitor cells was lost when the culture was maintained over an extended period. Therefore, we transduced HoxB4 together with AML1-ETO fusion protein. As expected, addition of EPO to HoxB4/AML1-ETO–expressing progenitor cells down-regulated the genes typically expressed in stem/progenitor cells such as c-Kit and GATA-2 (Fig. 2 A and B). The expression of erythroid-specific transcription factors such as EKLF (Fig. 2D) and globin β-major (Fig. 2C) was induced. c-Kit down-regulation was further confirmed at the protein level by a decrease in cell surface expression as seen by FACS analysis (Fig. 2E, Left). In contrast, the erythroid surface marker CD71 (transferrin receptor) was up-regulated upon EPO induction (Fig. 2E, right).
Fig. 2.
HoxB4/AML1-ETO–transduced hematopoietic progenitor cells as a model system to study erythroid differentiation. RT-PCR of c-Kit (A), GATA-2 (B), β-globin (C), and EKLF (D) expression before and 24, 48, or 72 h after EPO induction of HoxB4/AML1-ETO–transduced hematopoietic progenitors. (E) FACS analysis of c-Kit and CD71 protein expression before and 24 or 48 h after EPO induction.
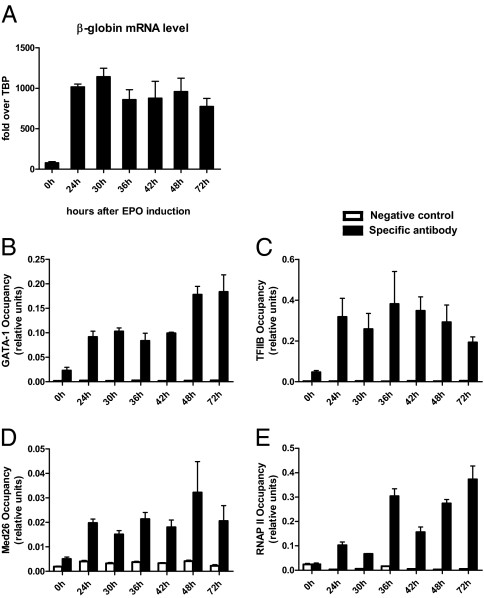
In ChIP experiments, transcription factor GATA-1 was detected at the β-major promoter at low levels even before induction (Fig. 3B). TFIIB and Mediator were also detected at a low level in noninduced HoxB4/AML1-ETO cells (Fig. 3 C and D) but not RNAP II (Fig. 3E). After EPO induction, occupancy of TFIIB, Mediator, and RNAP II at the globin-promoter region increased significantly. We could not detect Mediator or RNA Pol II above background at the LCR in HoxB4/AML1-ETO cells; occupancy or looping between LCR and β-major promoter is possibly significantly lower in HoxB4/AML1-ETO cells compared with MEL cells.
Fig. 3.
ChIP analysis at the β-globin locus using EPO-inducible hematopoietic progenitor cells. (A) RT-PCR of globin β-major at different time points after EPO induction. (B–E) ChIP analysis was performed at the β-globin promoter at different time points after EPO induction. GATA-1 (B), TFIIB (C), and Mediator (D) are found before induction and occupancy increases upon EPO induction. RNAP II (E) is not present before induction but is found at the promoter upon EPO induction. The y axis represents the absolute enrichment (mean ± SD, two independent experiments) normalized to input. Background is defined as the unspecific binding of each individual antibody to an unrelated control sequence (−106 kb upstream in the GATA-1 locus).
Med1 Conditional Knockout Mice.
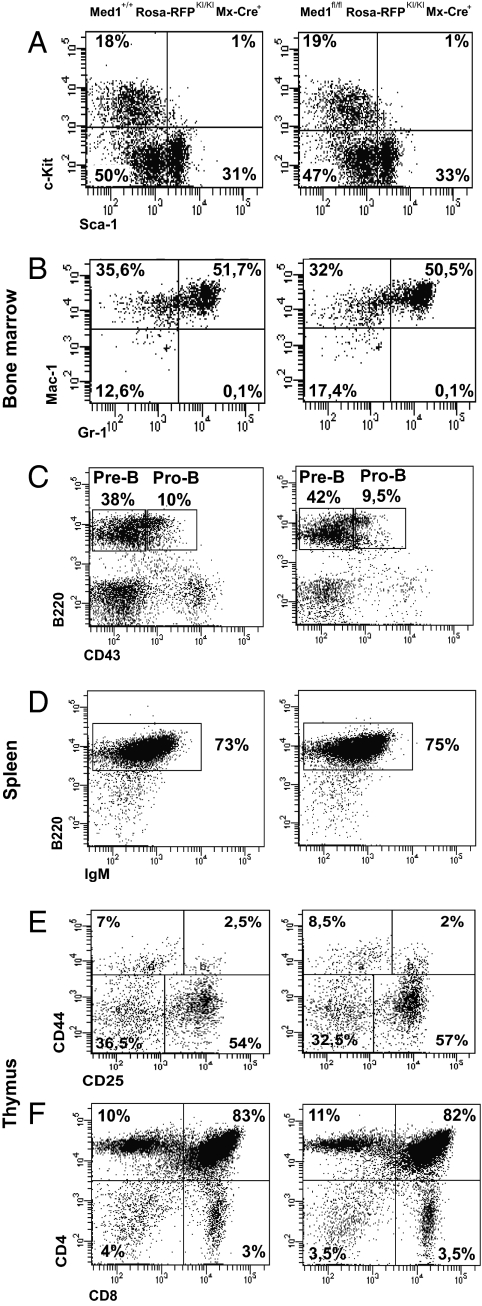
It was previously shown that Med1 knockout mice die during embryogenesis before the fetal liver stage. To directly investigate the role of Med1 in hematopoiesis, we crossed mice with a conditional knockout allele (Med1fl/fl) to a strain containing a Cre-recombinase gene controlled by the IFN-inducible Mx1 promoter (Mx-Cre) (26), leading to inducible expression of the Cre recombinase and excision of the floxed DNA elements after injection of poly-I:C. To trace the cells in which Cre is expressed, we crossed this compound mouse with the ROSA26-tdRFP Cre-reporter mouse (27). Red fluorescent protein (RFP) is expressed only upon Cre-mediated deletion of a floxed neomycin gene, and progeny of these cells are permanently RFP+. Five weeks after poly-IC injection, total bone marrow cells and splenocytes were isolated to examine the effect of Med1 deletion in different hematopoietic subsets by flow cytometry (Fig. 4). Overall, RFP expression in splenocytes corresponded well with efficient deletion of Med1 as measured by RT-PCR with primers for the Cre-excised Med1 exons (Fig. 5B, Left, and Fig. S3A) and by RT-PCR detecting Med1 mRNA levels (Fig. 5B, Right, and Fig. S3B). Efficient Cre recombination, indicated by RFP expression, could be detected in 30% of total splenocytes in Med1+/+/RFPKI/KI/Mx-Cre+ mice. Cell surface expression of defined sets of markers was analyzed on stem/progenitor cells (Fig. 4A), myeloid cells (Fig. 4B), B lymphocytes (Fig. 4 C and D), and T lymphocytes (Fig. 4 E and F) from either Med1fl/fl/RFPKI/KI/Mx-Cre+ mice (Med1fl/fl, Fig. 4, Right panels) or Med1 WT/RFPKI/KI/Mx-Cre+ littermates (Med1WT, Fig. 4, Left panels).
Fig. 4.
Development of stem/progenitor cells, lymphocytes, and myeloid cells is normal in the absence of Med1. FACS analysis reveals equal numbers of stem/progenitor cells (A), monocytes and granylocytes (B), B lymphocytes in the bone marrow (C) and spleen (D), and T lymphocytes in the thymus (E and F) in Med1 conditional knockout (Med1 fl/fl: floxed Med1 alleles; ROSA-RFP: Cre reporter; Mx-Cre: inducible Cre-transgene) and wild-type mice (littermates but not floxed).
Fig. 5.
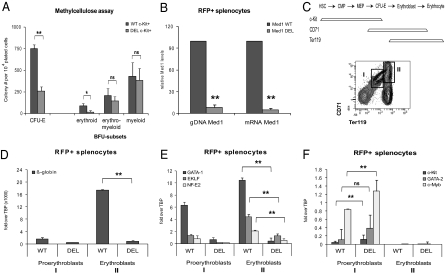
Specific defect in erythroid development in the absence of Med1. (A) FACS-sorted Med1-deficient c-Kit+ bone marrow progenitors fail to form erythroid CFUs and BFUs but not myeloid colonies in vitro. (B) Expression of Cre recombinase results in efficient deletion of the floxed Med1 genomic DNA sequence accompanied by the loss of Med1 mRNA transcripts. (C) (Upper) Schematic representation of c-Kit, CD71, and Ter119 expression throughout different stages of erythroid development (HSC: hematopoietic stem cell; CMP: common myeloid progenitor; MEP: megakaryocyte-erythrocyte progenitor; CFU-E: colony-forming unit erythrocyte). (Lower) FACS scheme referring to the proerythroblast (I) and erythroblast (II) populations sorted for the expression profile analysis in D–F. (D) RT-PCR analysis demonstrated a severe abrogation of globin β-major expression in Med1-deficient erythroblasts and (E) an overall decrease of GATA-1, EKLF, and NF-E2 transcript levels in Med1-deficient blast populations. (F) In contrast, expression of the immature markers c-Kit, c-Myb, and GATA-2 is elevated in the absence of Med1. RNA expression levels were examined by RT-PCR and values were normalized to the expression level of TATA binding protein (TBP). Data points are mean ± SD, n ≥ 3, *P < 0.05, **P < 0.01, Student's t test.
Lymphopoiesis Remains Unaffected in Med1 Conditional Knockout Mice.
Deletion of Med1 did not affect thymocyte development as seen in immature CD4/CD8 double-negative pre–T-cell subsets (Fig. 4E) and CD4/CD8 double- and single-positive thymocytes (Fig. 4F). B-lymphoid lineage differentiation was also unaffected as reflected by normal numbers and frequencies of mature splenic B cells (Fig. 4D) and immature pre- and pro-B cells found in the bone marrow (Fig. 4C). In addition, by analyzing the myeloid compartment using flow cytometry, we detected equal numbers of Gr-1+/Mac-1+–positive cells in either Med1 wild type or Med1-deficient bone marrow cells (Fig. 4B). Thus, we show that Med1 deficiency does not affect T-cell, B-cell, or myeloid cell development.
Impairment of Erythroid Development in Med1 Conditional Knockout Mice.
Hematopoietic stem/progenitor cells were found in equal numbers in Med1-deficient or wild-type littermates as stained by surface markers c-Kit and Sca-1 (Fig. 4A). In both Med1+/+ and Med1fl/fl mice, we found comparable numbers of Lin−/tdRFP+ stem/progenitor cells. To investigate the functional consequences of Med1 deletion, we performed methylcellulose colony assays (Fig. 5A). When assayed for erythroid colony-forming ability, we identified a threefold decrease in erythroid CFUs and an eightfold decrease in erythroid burst-forming units (BFUs) in Med1-deficient cells compared with Med1+/+ cells. Erythro-myeloid colonies were only slightly reduced compared with Med1+/+, and myeloid colony formation was completely unaffected (Fig. 5A), further supporting our previous flow cytometry analysis.
As previously published (27), expression of the tdRFP reporter could not be detected in mature erythrocytes, possibly due to enucleation (Fig. S4 A and B). Thus, we cannot investigate whether Med1-deficient cells are able to progress to the final reticulocyte stage. However, the Cre reporter expression in the proerythroblast and erythroblast stage allowed us to compare these populations in Med1-deficient mice and littermate controls (Fig. 5 and Fig. S4). We identified that Med1 deficiency resulted in a 1.4-fold increase of proerythroblasts and in an almost 2-fold decrease in erythroblasts compared with Med1+/+ littermates (Fig. S4E).
When measuring gene expression of β-globin at the proerythroblast or erythroblast stage, we observed a complete abrogation in Med1-deficient cells (Fig. 5D). In addition, the expression of the key transcription factors GATA-1, EKLF, and NF-E2 was also severely impaired (Fig. 5E). Interestingly, the progenitor-specific genes GATA-2, c-Kit, and c-Myb are slightly up-regulated in Med1-deficient cells as measured by RT-PCR (Fig. 5F).
To circumvent the essential positive feedback loop of GATA-1 itself, we performed Med1 knockdown experiments in G1E cells reconstituted with GATA1-ER (Fig. S5). This GATA-1 knockout erythroid cell line was reconstituted with GATA-1 fused to the estrogen receptor ligand-binding domain. Although Med1 can be efficiently knocked down as seen in Western blotting experiments (Fig. S5 A and B), Med1 is not essential for induction of β-globin in the GATA1-ER cell line (Fig. S5C, Left) and for GATA-1 target gene Band3 (Fig. S5C, Right). One possible scenario is that immediate high levels of nuclear GATA-1 make these cells Med1-independent.
In summary, conditional Med1-deficient mice display a surprisingly specific block in erythroid development but not in other hematopoietic lineages. This indicates that certain Mediator subunits are specialized to interact with specific transcription factors.
Discussion
Med1 as a Cofactor for GATA Factors.
In the present study, we showed that Mediator subunit Med1 is essential for erythroid development and correct induction of β-globin gene expression. As shown previously by others and us, Med1 is an important coactivator for the master regulator GATA-1 (9, 10, 28). Whereas other GATA factors have been shown to be essential for T-cell development (GATA-3) or hematopoietic stem cells (GATA-2) (as reviewed in ref. 2), conditional deletion of the Med1 subunit did not substantially affect either of these cell populations. It is likely that GATA protein levels play an important role in cofactor requirements. Other known cofactors such as p300/CBP or FOG may substitute for Med1 in T cells, but Med1 is essential in the erythroid context. Alternatively, it is possible that GATA-3 not only interacts with Med1 but also can complex with other Mediator subunits.
Recruitment of GATA1, Mediator, and RNA Polymerase II.
As shown by ChIP experiments, binding of transcription factors increases upon induction first at the LCR and subsequently at the β-major promoter in line with the generation of intergenic transcripts at these sites as shown by others (29) and us. This is consistent with previous studies showing that acetylated lysine-9 on histone H3, which is associated with active transcription, is first detected at the LCR, and to a lesser extent at the promoter of the silent β-globin locus, and then increases upon transcriptional activation. This is mainly due to the enhanced GATA-1 binding, CBP/p300 recruitment, and the SWI/SNF chromatin-remodeling complex at these sites (30–32). By varying the temperature of the culture of the G1E cells, Bresnick and colleagues have also described different GATA-1–triggered events and have determined that the initial events were mainly found at the LCR and not at the β-major promoter (22). In addition, mice with transgenes lacking the LCR are capable only of recruiting the inactive form of RNA Pol II, which lacks Ser-5 phosphorylation (33). Our data together with these reports imply that Mediator is required for the initial events of transcription initiation at the LCR, acting after CBP/p300 and the SWI/SNF complex. Using a system where GATA-1 knockout cells were functionally complemented with a GATA-1/estrogen receptor fusion transgene, Pope and Bresnick (34) came to a different conclusion. In this context, Med1 is required for maximal GATA-1 activity but not for GATA-ER–induced erythroid maturation. We do not know the cause of the discrepancy observed between Med1-knockdown experiments using the GATA1-ER cell line and the phenotype observed in Med1 conditional knockout mice. One possibility is that the remaining Med1 levels in the Med1 knockdown are sufficient for GATA-1 function. Alternatively, it could be that nuclear GATA-1 levels are so high in the GATA1-ER cell line upon induction with tamoxifen that Med1 is dispensable.
Mediator Recruitment at Enhancer Elements.
Biochemical data from several laboratories (12, 13, 35) show that Mediator is found in a complex with RNAP II as confirmed by electron microscopic studies. This would suggest that Mediator is found at the core promoter similarly to RNAP II. However, in the present study at the β-globin locus as well as in previous work performed in yeast (14, 15), we demonstrate that Mediator is clearly found at enhancer elements in vivo. It is plausible that Mediator acts as a scaffold factor or molecular bridge that initiates the recruitment of canonical transcription factors and RNAP II. In addition, Mediator could be involved in the looping process between LCR and a core promoter, as shown for FOG-1 (36). However, this question should be addressed using another system rather than Med1 conditional knockout mice because we have shown that GATA-1 levels are already down-regulated in Med1-deficient erythroid precursors.
Med1 Presence in Different Mediator Complexes.
Several laboratories provided evidence for mammalian Mediator complexes that differ in composition (37–39). Med1 deficiency clearly does not lead to the disintegration of the Mediator complex (38). One can speculate that Med1-containing Mediator is crucial for the recruitment of other Mediator subunits and RNAP II. Alternatively, Med1-containing Mediator complexes could have a special conformation that would act as a scaffold for the enhanceosome at the β-globin gene locus.
Using conditional knockout mice, we could show that Mediator subunit Med1 is essential for erythroid development. Thus, Med1 is a unique, essential cofactor for the activation of the gene expression of the well-studied β-globin locus. Mediator subunit Med1 recruitment to β-globin promoter elements is temporally and spatially regulated: Med1 is recruited after GATA-1 but before RNAP II, first at the LCR and then at the β-major globin promoter. In the future, genome-wide mapping of Mediator, RNAP II, and active RNAP II phosphorylated at Ser-5 could be used to validate whether Mediator is found primarily at enhancers and/or at core promoters and how it globally correlates with gene activity.
Materials and Methods
Animals and Tissue Preparation.
C57BL/6 Med1 conditional knockout mice (21), C57BL/6 Rosa26-tdRFP Cre-reporter mice (27), and Mx-Cre conditional deleter mice (26) were generated previously. In the floxed-Med1 mice carrying the Mx-Cre transgene, deletion of the Med1 allele was performed by i.p. injection of 250 μg poly I:C every other day over a period of 6 d. After 5 wk, mice were killed and organs of interest were taken. Spleen and thymus were mashed between glass slides, and bone marrow cells were obtained by flushing femurs with PBS/5% FCS. Cells were filtered through a cell strainer to obtain single-cell suspensions and kept under sterile conditions on ice until further use.
Flow Cytometry and Cell Sorting Analysis.
Cells were blocked with 10 μg of rat IgG antibody in PBS/5% FCS (Jackson ImmunoResearch) for 15 min on ice. Cells were washed once with PBS/FCS and stained with fluorescently labeled antibodies [CD4, CD8, CD11b, CD25, CD44, and IgM (Beckton Dickinson); CD43, CD45R, CD117, Gr-1, and Ter119 (eBioscience/Natutec); and CD71 and Sca-1 (Caltag)] to identify different hematopoietic lineages and progenitors. Flow cytometry analysis and cell sorting were performed using the BD FACS LSRII and Aria (Beckton Dickinson). Data were analyzed using FlowJo software (Treestar).
ChIP Assays.
Chromatin of 8 × 106 MEL or HoxB4/AML1-ETO cells was cross-linked using a final concentration of 1% formaldehyde for 10 min at room temperature. Ice-cold PBS (10 mL) was added to quench the reaction. Cells were centrifuged for 10 min at 1,500 × g at 4 °C, and the resulting cell pellet was washed three times in 10 mL cold PBS for 10 min on ice. Cell pellets were resuspended in 600 μL of lysis buffer (1% SDS; 5 mM EDTA, pH 8.0; 50 mM Tris·HCl, pH 8.0). Samples were then sonicated [seven times for 10 s, 50% amplitude with a 45-s pause between pulses (Vibracell; Sonics)]. Resulting chromatin fragments were ∼600 bp on average. A total of 150–200 μL of this preparation was diluted 1:10 with dilution buffer (0.5% Nonidet P-40; 200 mM NaCl; 5 mM EDTA; 50 mM Tris·HCl, pH 8.0) and precleared for 1–2 h with proteinA Sepharose beads presaturated with salmon sperm DNA (Amersham Biosience, Invitrogen). Precleared chromatin solution (60 μL) was used as “input” to normalize primer-binding efficiency. Immunoprecipitation was done overnight at 4 °C using 2 μg antibody. Immunocomplexes were collected with proteinA Sepharose beads over 45 min at 4 °C. The resin was washed four times with NaCl wash buffer (0.1% SDS; 1% Nonidet P-40; 2 mM EDTA, pH 8.0; 500 mM NaCl; 50 mM Tris·HCl, pH 8.0) and three times with TE buffer (10 mM Tris·HCl; 1 mM EDTA, pH 8.0). Elution was performed using 2× 50-μL elution buffer (1× TE, pH 8.0; 2% SDS) for 5 min at 1,200 × g at room temperature. Reverse cross-linking was done over night at 65 °C. The following day DNA was isolated using a PCR purification kit (Qiagen) according to the manufacturer's procedure. In all buffers, except elution buffer, 20 μM PMSF in isopropanol was added to inhibit protease activity. For the GATA-1 ChIP, preclearing of the chromatin was done with Sepharose-G beads overnight at 4 °C, and the GATA-1 protein–DNA complexes were immunoprecipitated for 3 h in an additional step using an AffiniPure rabbit anti-rat antibody (Jackson ImmunoResearch). Additional materials and methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. J. Fehling (University of Ulm, Ulm, Germany) for providing us with Cre-reporter mice and Dr. S. Saccani and Dr. D. van Essen for help with the HoxB4 cell system. This work was supported by Deutsche Forschungsgemeinschaft Grants BOR-1639/4-1, the Emmy-Noether Fellowship BO1639/3, Collaborative Research Center Grant SFB592/C3, and funds from the Max Planck Society (to T.B.), and National Institutes of Health Grants GM23750 and DK083163 (to J.K.R). X.Y. is part of the International Max-Planck-Research-School (IMPRS) of Molecular and Cellular Biology.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005794107/-/DCSupplemental.
References
- 1.Dean A. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 2006;22:38–45. doi: 10.1016/j.tig.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol. 2005;25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pevny L, et al. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 4.Kim SI, Bresnick EH. Transcriptional control of erythropoiesis: Emerging mechanisms and principles. Oncogene. 2007;26:6777–6794. doi: 10.1038/sj.onc.1210761. [DOI] [PubMed] [Google Scholar]
- 5.Trainor CD, Mas C, Archambault P, Di Lello P, Omichinski JG. GATA-1 associates with and inhibits p53. Blood. 2009;114:165–173. doi: 10.1182/blood-2008-10-180489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsang AP, et al. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 7.Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SI, Bultman SJ, Kiefer CM, Dean A, Bresnick EH. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc Natl Acad Sci USA. 2009;106:2259–2264. doi: 10.1073/pnas.0806420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford SE, et al. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J Biol Chem. 2002;277:3585–3592. doi: 10.1074/jbc.M107995200. [DOI] [PubMed] [Google Scholar]
- 10.Stumpf M, et al. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc Natl Acad Sci USA. 2006;103:18504–18509. doi: 10.1073/pnas.0604494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Blazek E, Mittler G, Meisterernst M. The mediator of RNA polymerase II. Chromosoma. 2005;113:399–408. doi: 10.1007/s00412-005-0329-5. [DOI] [PubMed] [Google Scholar]
- 13.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Cosma MP, Panizza S, Nasmyth K. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol Cell. 2001;7:1213–1220. doi: 10.1016/s1097-2765(01)00266-0. [DOI] [PubMed] [Google Scholar]
- 15.Kuras L, Borggrefe T, Kornberg RD. Association of the Mediator complex with enhancers of active genes. Proc Natl Acad Sci USA. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- 17.Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belakavadi M, Fondell JD. Role of the mediator complex in nuclear hormone receptor signaling. Rev Physiol Biochem Pharmacol. 2006;156:23–43. doi: 10.1007/s10254-005-0002-0. [DOI] [PubMed] [Google Scholar]
- 19.Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell. 2000;5:683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 20.Zhu Y, et al. Deletion of PBP/PPARBP, the gene for nuclear receptor coactivator peroxisome proliferator-activated receptor-binding protein, results in embryonic lethality. J Biol Chem. 2000;275:14779–14782. doi: 10.1074/jbc.C000121200. [DOI] [PubMed] [Google Scholar]
- 21.Jia Y, et al. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARalpha-regulated gene expression in liver. J Biol Chem. 2004;279:24427–24434. doi: 10.1074/jbc.M402391200. [DOI] [PubMed] [Google Scholar]
- 22.Kim SI, Bultman SJ, Jing H, Blobel GA, Bresnick EH. Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation. Mol Cell Biol. 2007;27:4551–4565. doi: 10.1128/MCB.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilat S, et al. HOXB4 enforces equivalent fates of ES-cell-derived and adult hematopoietic cells. Proc Natl Acad Sci USA. 2005;102:12101–12106. doi: 10.1073/pnas.0505624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carotta S, et al. Directed differentiation and mass cultivation of pure erythroid progenitors from mouse embryonic stem cells. Blood. 2004;104:1873–1880. doi: 10.1182/blood-2004-02-0570. [DOI] [PubMed] [Google Scholar]
- 26.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 27.Luche H, Weber O, Nageswara Rao T, Blum C, Fehling HJ. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur J Immunol. 2007;37:43–53. doi: 10.1002/eji.200636745. [DOI] [PubMed] [Google Scholar]
- 28.Gordon DF, et al. MED220/TRAP220 functions as a transcriptional coactivator with Pit-1 and GATA2 on the TSH{beta} promoter in thyrotropes. Mol Endocrinol. 2006;20:1073–1079. doi: 10.1210/me.2005-0115. [DOI] [PubMed] [Google Scholar]
- 29.Ling J, Baibakov B, Pi W, Emerson BM, Tuan D. The HS2 enhancer of the beta-globin locus control region initiates synthesis of non-coding, polyadenylated RNAs independent of a cis-linked globin promoter. J Mol Biol. 2005;350:883–896. doi: 10.1016/j.jmb.2005.05.039. [DOI] [PubMed] [Google Scholar]
- 30.Kiekhaefer CM, Grass JA, Johnson KD, Boyer ME, Bresnick EH. Hematopoietic-specific activators establish an overlapping pattern of histone acetylation and methylation within a mammalian chromatin domain. Proc Natl Acad Sci USA. 2002;99:14309–14314. doi: 10.1073/pnas.212389499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letting DL, Rakowski C, Weiss MJ, Blobel GA. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol Cell Biol. 2003;23:1334–1340. doi: 10.1128/MCB.23.4.1334-1340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Im H, et al. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc Natl Acad Sci USA. 2005;102:17065–17070. doi: 10.1073/pnas.0506164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawado T, Halow J, Bender MA, Groudine M. The beta-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev. 2003;17:1009–1018. doi: 10.1101/gad.1072303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pope NJ, Bresnick EH. Differential coregulator requirements for function of the hematopoietic transcription factor GATA-1 at endogenous loci. Nucleic Acids Res. 2010;38:2190–2200. doi: 10.1093/nar/gkp1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 36.Vakoc CR, et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 37.Mo X, Kowenz-Leutz E, Xu H, Leutz A. Ras induces mediator complex exchange on C/EBP beta. Mol Cell. 2004;13:241–250. doi: 10.1016/s1097-2765(03)00521-5. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, et al. MED1/TRAP220 exists predominantly in a TRAP/Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol Cell. 2005;19:89–100. doi: 10.1016/j.molcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 39.Taatjes DJ, Tjian R. Structure and function of CRSP/Med2: A promoter-selective transcriptional coactivator complex. Mol Cell. 2004;14:675–683. doi: 10.1016/j.molcel.2004.05.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.