Abstract
Amyotrophic lateral sclerosis (ALS) is a disorder characterized by the death of both upper and lower motor neurons and by 3- to 5-yr median survival postdiagnosis. The only US Food and Drug Administration-approved drug for the treatment of ALS, Riluzole, has at best, moderate effect on patient survival and quality of life; therefore innovative approaches are needed to combat neurodegenerative disease. Some familial forms of ALS (fALS) have been linked to mutations in the Cu/Zn superoxide dismutase (SOD1). The dominant inheritance of mutant SOD1 and lack of symptoms in knockout mice suggest a “gain of toxic function” as opposed to a loss of function. A prevailing hypothesis for the mechanism of the toxicity of fALS-SOD1 variants, or the gain of toxic function, involves dimer destabilization and dissociation as an early step in SOD1 aggregation. Therefore, stabilizing the SOD1 dimer, thus preventing aggregation, is a potential therapeutic strategy. Here, we report a strategy in which we chemically cross-link the SOD1 dimer using two adjacent cysteine residues on each respective monomer (Cys111). Stabilization, measured as an increase in melting temperature, of ∼20 °C and ∼45 °C was observed for two mutants, G93A and G85R, respectively. This stabilization is the largest for SOD1, and to the best of our knowledge, for any disease-related protein. In addition, chemical cross-linking conferred activity upon G85R, an otherwise inactive mutant. These results demonstrate that targeting these cysteine residues is an important new strategy for development of ALS therapies.
Keywords: mass spectrometry, thiol-disulfide
Innovative approaches are needed to combat neurodegenerative disease, among the most serious of which is amyotrophic lateral sclerosis (ALS), a disorder characterized by the death of both upper and lower motor neurons and by 3- to –5-yr median survival postdiagnosis. The only US Food and Drug Administration-approved drug for the treatment of ALS, Riluzole, has at best, moderate effect on patient survival and quality of life (1–3). Although the causes of sporadic neurodegenerative diseases remain a mystery, mutations causing familial forms of many of these diseases (e.g., Alzheimer’s, Parkinson, and ALS) are known. For example, mutations in the gene encoding Cu/Zn superoxide dismutase (SOD1) are responsible for ∼20% of the familial ALS cases (fALS) and 2% of all ALS (4, 5). Two such mutations are G93A, which maintains wild-type-like enzymatic activity, and the metal-deficient G85R, which is essentially inactive. Posttranslational modifications of proteins involved in familial diseases have been invoked in the etiology of the corresponding sporadic diseases, for example, alpha-synuclein (6) and Parkin (7) modification in Parkinson, Abeta (8) and tau (9) modification in Alzheimer’s, and TDP43 (10) and SOD1 (11–14) modification in ALS. The hope, therefore, is that strategies for treating familial diseases may translate to at least a subset of sporadic diseases.
Both dominant inheritance of mutant SOD1 (15) and lack of symptoms in knockout mice (16) suggest a “gain of toxic function” as opposed to a loss of function (16–22). Aggregation propensity and loss of stability of SOD1 are synergistic risk factors for fALS patient disease severity (23), and it has been suggested that a common property of fALS variants in vitro and in vivo is their propensity to aggregate (24, 25). A prevailing hypothesis for the mechanism of the toxicity of fALS-SOD1 variants involves dimer destabilization and dissociation into monomers, which then nucleate the formation of higher-order aggregates (13, 26–28). Indeed, variant proteins, such as the G85R SOD1 used in this study, are found as monomers in vivo (29–31). A number of modifications, including loss of Cu or Zn (32), cleavage of the native, intramolecular disulfide (33), oxidation (13, 34), and fALS-associated mutation (35), predispose the SOD1 dimer to dissociate. X-ray crystal structures of both A4V, and to a lesser extent I113T (35); yeast two hybrid analysis of H46R, A4V, and H48Q (36); dissociation of G85R, G93R, E100G, and I113T by chaotrophs (37); and molecular dynamics simulations (38, 39) are all consistent with this hypothesis. Consistent with this, stabilization of the SOD1 dimer interface either by tethering subunits with a genetically engineered intersubunit disulfide (40) or through the use of small molecules (41) prevents protein aggregation and is being pursued as a therapeutic strategy (40).
Here we present unique chemical approaches for stabilizing SOD1, which has the distinction of being the only protein where loss of stability and propensity to aggregate are known to correlate with increased disease severity (23, 33). Increasing SOD1 stability, therefore, has the potential to directly improve prognosis. Chemical cross-linking of both the G93A and G85R variants using maleimide and thiol-disulfide exchange chemistries resulted in stabilization of ∼20 °C and ∼45 °C, respectively. These are the largest increases in SOD1 stability to date, and to the best of our knowledge, the largest for any disease protein. In addition to stabilizing G85R, cross-linking the dimer conferred activity to an otherwise inactive mutant. Therefore, these results suggest a unique target and mechanism for developing potential therapeutic compounds.
Results
Cross-Linking Using Maleimide Functional Groups Stabilizes Dimeric SOD1.
To stabilize the human SOD1 dimer while minimizing the potential for toxicity (off-target binding to other proteins), we took advantage of the three following SOD1 characteristics: (i) the presence of two symmetrically arranged Cys111 residues on opposite sides of the dimer interface that are separated by ∼9 or 13 Å, depending upon Cys rotomer (4) (Fig. 1); (ii) the high nucleophilicity (unique reactivity) of cysteine residues in general; and (iii) the entropic benefits of cross-linking. Compound binding strength was increased through the use of covalent bonding, as opposed to van der Waals forces, such that binding is essentially irreversible in vitro. All of the maleimide cross-linkers tested that had chain lengths in the range of 8–14 Å resulted in the stabilization of SOD1 dimer, including dithio-bismaleimidoethane (DTME, spacer arm 13.3 Å), 1,4-bismaleimidyl-2,3-dihydroxybutane (BMDB, spacer arm 10.2 Å), 1,8-bis-maleimidodiethyleneglycol (BM(PEG)2, spacer arm 14.7 Å), 1,4-bismaleimidobutane (BMB, spacer arm 10.9 Å), and Tris[2-maleimidoethyl]amine (TMEA, 10.3 Å) (Figs. S1 and S2). Alternatively, the one vinylsulfone tested, 1,6-hexane-bis-vinylsulfone (HBVS, spacer arm 14.7 Å), did not result in formation of dimeric SOD1 (Fig. S2).
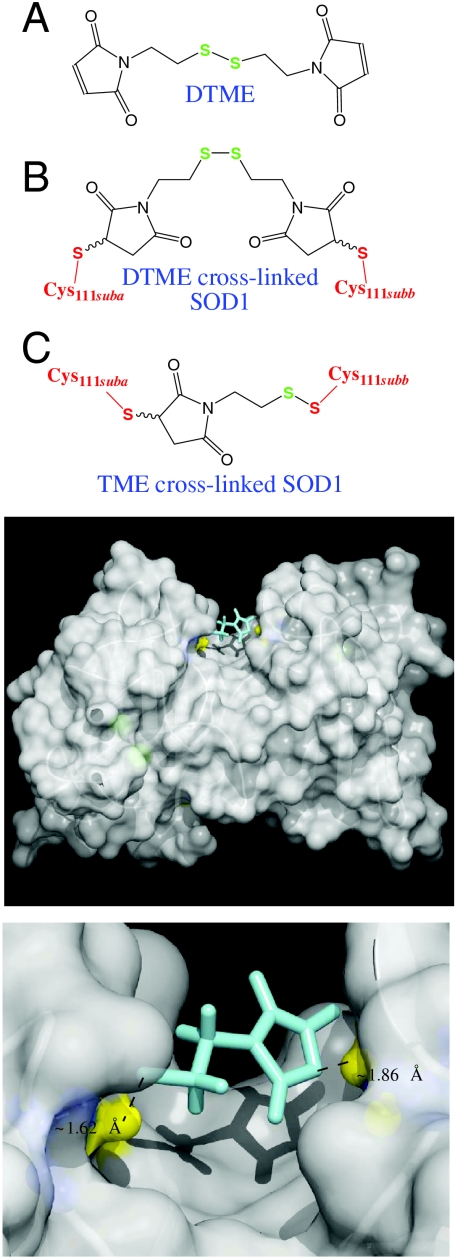
Fig. 1.
Model of SOD1 after cross-linking DTME with adjacent C111 residues at the dimer interface. (Top, A) Free (unreacted) DTME. (B) Maleimide-chemistry mediated, “cross-linked” reaction product of SOD1 and DTME, with DTME bridging individual SOD1 monomers. SOD1 Cys111 constituents are shown in red, and the two SOD1 monomer/subunits are labeled “suba” and “subb.” Cysteine111 rotomers are oriented such that the cysteinyl sulfur spacing is 13 Å (the minor rotomer observed in crystal structures). (C) Thiol-disulfide exchange- and maleimide-chemistry mediated cross-linked reaction product of SOD1 and DTME. Cysteine111 rotomers are oriented such that the cysteinyl sulfur spacing is 9 Å (the major rotomer observed in crystal structures). Note that thiomaleimidoethane (TME) is comprised of half of the DTME structure. TME results from both binding a maleimide moiety to one subunit (Left), and the reaction of the Cysteine111 thiol of the second subunit with the disulfide of DTME (Right). Note that the Cys111 thiol (red) exchanges with a disulfide sulfur of DTME (green) to form a new disulfide bond. (Bottom) Cysteine 111 sulfur (PDB ID code 2C9V) (44) is denoted by yellow balls. TME, which results from thiol-disulfide exchange (Fig. S3) and maleimide cross-linking as seen in the bottom panel, is shown in cyan. This model, including site of cross-linking, was confirmed via LC-FTMS analysis (Fig. S4).
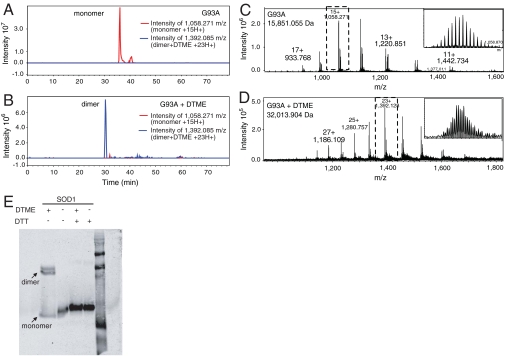
To investigate the stoichiometry of cross-linker binding, we compared the molecular weights of cross-linked [DTME or bis(maleimido)ethane (BMOE), spacer arm 8.0 Å] versus non-cross-linked G93A variants using liquid chromatography (LC)-Fourier transform mass spectrometry (FTMS). The experimental monoisotopic mass for the monomeric, non-cross-linked species was determined to be 15,851.1 Da (theoretical 15,850.9 Da), and the experimental monoisotopic mass for the dimeric, DTME cross-linked species was determined to be 32,013.9 Da [0.2 Da difference from the theoretical value of the molecular weight of two G93A monomers plus one DTME (312.4 Da)]. The experimental molecular weight of dimeric DTME cross-linked G93A, 32,013.9 Da, suggests that one equivalent of cross-linker produced one equivalent of dimer (Fig. 2), consistent with the binding of a single cross-linker to two monomers. Similar results were obtained for G93A and BMOE cross-linking.
Fig. 2.
Cross-linker dependent dimerization of SOD1. (A) Extracted ion chromatogram for the 15 H+ charge state of the SOD1 monomer (red) and (B) 23 H+ charge state of the DTME-cross-linked dimer (blue) demonstrate the stoichiometric conversation of monomeric as-isolated SOD1 to dimeric cross-linked SOD1. (C) Mass spectrum of as-isolated (non-cross-linked) G93A SOD1 and (D) DTME cross-linked G93A SOD1 confirm cross-linker dependent dimerization. (E) Chemical cross-linking of WT SOD1 with a reductively labile molecule. One of the maleimide cross-linkers tested, DTME, has a disulfide bond in its spacer arm that can be cleaved by reducing agents such as DTT. WT SOD1 was cross-linked with a 1∶1 molar ratio of DTME and analyzed by SDS-PAGE gel in both the presence and in the absence of reducing agent, DTT. The SOD1 cross-linked dimer became a monomer in the presence of reducing agent (Fig. 2B, lane 2), suggesting that the dimer being formed is specific to the cross-linkers being used, and was not the result of cross-linker catalyzed dimer formation.
To rule out the occurrence of cross-linker-catalyzed reactions, a reductively labile cross-linker, DTME, was used, and cross-linking was monitored using Western blotting. Cross-linking using DTME resulted in dimeric SOD1; however, in the presence of reducing agent only monomer was observed (Fig. 2E), suggesting that the creation of SOD1 dimer was specifically due to covalently bound chemical cross-linkers.
Thiol-Disulfide Exchange Mediated SOD1 Dimer Stabilization.
The mass of intact SOD1 and MS/MS fragmentation data using funnel-skimmer dissociation (FSD) (42, 43) of the DTME cross-linked G93A variant (precursor ions) revealed a unique reaction mechanism, thiol-disulfide exchange, for cross-linking SOD1 (Fig. S3). The species modified by half DTME suggests that exchange occurred between the disulfide bond within DTME’s spacer and the thiol moiety of a cysteine residue on G93A. The possibility of thiol-disulfide exchange was further confirmed by modeling half of DTME at the dimer interface, specifically Cys111, of the SOD1 structure (2C9V) (44) (Fig. 1). Thus, we were able to cross-link SOD1 monomers efficiently using numerous molecules (Fig. S2) and two distinct reaction mechanisms, namely maleimide (Fig. 2 and Figs. S1 and S2) and thiol-disulfide exchange (Fig. S3), with the latter being suitable for peptide-based therapeutic strategies.
Cys111 Is the Site of Chemical Cross-Linking.
In order to identify the site of cross-linking, we compared proteolysis and LC-FTMS/MS data, which provided structural data of the digested peptides, for non-cross-linked and cross-linked samples (Fig. S4). Because of the protease resistance of WT and G93A SOD1, likely conferred via metal binding (45) and an intramolecular disulfide bond (32, 44, 46, 47), the G85R variant, which is less protease resistant, was used. The m/z 487.8 (Mr 973.6; residues 1–9, acetylated N terminus) is observed in both samples and is presented as a positive control, highlighting similar intensities of the peptide in the cross-linked and non-cross-linked samples. We observed two Mr’s that were in the cross-linked but not in the native sample: 5,232. 7 (m/z 873.1) and 5,347.7 (m/z 892.3). MS-Bridge (http://prospector.ucsf.edu) identified both 5,232. 7 and 5,347.7 as being involved in a cross-link (residues 103–125 of one monomer cross-linked via BMOE to residues 103–126 of a second monomer) and (residues 103–126 of one monomer cross-linked by BMOE to residues 103–126 of a second monomer), respectively. The predicted cross-link for 5,232.7 had a 6.2-ppm accuracy, whereas 5,347.7 had a 5.9-ppm accuracy. Therefore, mass spectrometry confirmed that cross-linking was stoichiometric and occurred mainly through Cys111, and that cross-reactivity with other SOD1 cysteine residues Cys6, Cys57, and Cys146 was relatively low (Fig. S4). Because of the complexity of cross-linking, there was a lack of precursor ion fragmention of the cross-linked peptides.
Chemical Cross-Linking Stabilizes the G93A and G85R Dimers.
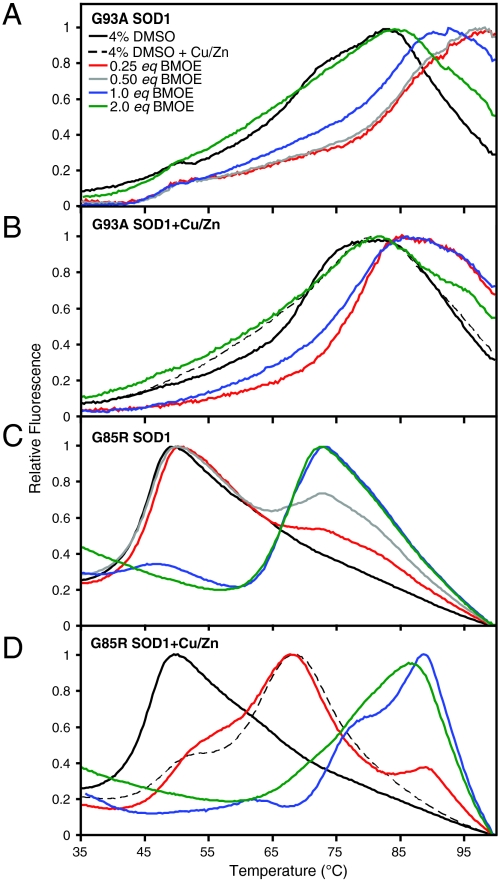
Sypro orange binds preferentially to hydrophobic patches that become exposed as a protein unfolds. A fluorescence-based assay monitoring Sypro orange binding as a function of temperature was used to observe trends in the unfolding temperatures of both G93A and G85R. This assay is not reversible, probably as the result of aggregation of SOD1, and although the unfolding temperatures resemble the previously observed melting temperatures of SOD1 (37, 48–51), they are not proper thermodynamic stabilities. We observed unprecedented increases in the stability of the two mutants analyzed (Fig. 3 and Fig. S5). G93A SOD1, for example, was stabilized by ∼20 °C; incubation with excess copper and zinc had no effect on G93A stability; and the lower concentrations of cross-linker had a greater effect on G93A stability. G85R SOD1, on the other hand, was stabilized ∼20 °C in the presence of excess copper and zinc (absence of cross-linker); was further increased ∼45 °C (from ∼40 °C to 85 °C) in the presence of copper, zinc, and cross-linker; and the higher concentrations of cross-linker had a greater effect on G85R stability. The degree of stabilization achieved here by what are essentially (covalent) pharmacological chaperones is the highest ever achieved for SOD1.
Fig. 3.
Stabilization of fALS-associated SOD1 variants by chemical cross-linking. Stability of mutant SOD1 measured by thermofluorescence assay (see Fig. S5). Ten micromolar G93A (A) and G85R SOD1 (C) were incubated with concentrations of BMOE ranging from 2.5–20 μM. Ten micromolar G93A (B) and G85R (D) were incubated with 20 μM copper/zinc and concentrations of BMOE ranging from 2.5–20 μM. The cross-linkers used here were resuspended in DMSO; therefore, SOD1 in 4% DMSO controls were used. WT SOD1 was also investigated; however, due to its near-boiling point melting temperature, the assay used here is not capable of detecting whether or not stabilization occurred. G85R and G93A were stabilized in a cross-linker concentration-dependent manner by approximately 45 °C and 20 °C, respectively. The addition of copper and zinc had little effect on the wild-type like (metallated) G93A variant; however, the addition of copper and zinc to G85R, a metal-deficient variant, along with cross-linkers stabilizes the protein to almost wild-type levels. Fig. 4 illustrates that this highly stable form of G85R has regained wild-type-like levels of SOD1 activity. The above graphs represent the average of three replicates of each respective concentration; these experiments were repeated in triplicate.
Activity of the fALS Variant G85R SOD1 Is Restored by Stabilizing the SOD1 Dimer.
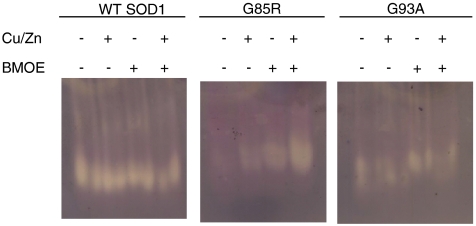
In addition to stability we investigated the effect chemical cross-linking had on SOD1 activity using a gel-based assay. WT and G93A SOD1 activity were unaffected by the addition of copper, zinc, or chemical cross-linking. However, in addition to increasing the thermostability of G85R SOD1, chemical cross-linking increased its qualitative metal-binding affinity, resulting in the transformation of G85R SOD1 from a protein that is catalytically inactive in vitro and in vivo (52), to a protein that is active in vitro and potentially in vivo (Fig. 4). The increase in G85R activity observed during DTME cross-linking was reversed upon cleavage of DTME’s internal disulfide bond using tris(2-carboxyethyl)phosphine (TCEP) (Fig. S6). Thus, in addition to the improvement in the thermostability of G85R SOD1, these compounds have the potential to increase the resistance of the organism to oxidative stress, and our approach may be considered for other loss-of-function diseases.
Fig. 4.
Rescue of fALS-variant G85R SOD1 activity by chemical cross-linking (see Fig. S6). G85R is a metal-deficient and inactive variant. In the presence of excess copper, zinc, or BMOE G85R activity increases. Most notably, G85R in the presence of copper, zinc, and DTME confers WT-like levels of activity. Thus, the increase in stability of G85R also corresponds to an increase in activity (Fig. S6). As expected WT SOD1 and G93A SOD1, a fully active mutant, appear to be unaffected.
SOD1 Metal Content Plays a Role in Stabilization of SOD1.
The metal contents of as-isolated WT, G93A, and G85R SOD1 were determined using inductively coupled plasma mass spectrometry (ICP-MS), which are listed in Table S1: as-isolated WT SOD1 contained approximately two molecules of copper and zinc per monomer, as-isolated G93A contained one molecule of copper and one and a half molecules of zinc per monomer, whereas G85R contained less than one copper and one and a half zinc per monomer. From these data it appears that WT and G93A SOD1 were metal replete, and that in addition some adventitious (nonactive site) metal binding occurred. We therefore also analyzed metal content using FTMS; we have observed that the desolvation process tends to remove most adventitious metals: as-isolated WT SOD1 appeared to be fully metallated, as-isolated G93A SOD1 appeared to be ∼95% metallated, and as-isolated G85R SOD1 appeared to be partially metallated, with the majority either unmetallated or singly metallated. These data may explain the increase in stability observed when exogenous metals were added to the G85R sample versus the G93A sample, because the population of G85R consists of a larger percentage of partially metallated and unmetallated forms.
Discussion
We developed strategies for increasing the thermostability of fALS-associated SOD1 variants that took advantage of the unique nucleophilicity of cysteine residues in general, and the proximity of the Cys111 of adjacent SOD1 monomers in particular. We present both maleimide and thiol-disulfide exchange-mediated stabilization of SOD1 using two adjacent cysteines (Cys111) on each respective SOD1 monomer. Mass spectrometry data are consistent with one equivalent of cross-linker producing one equivalent of dimer, and a reductively labile cross-linker (DTME) ruled out the occurrence of cross-linker-catalyzed reactions. Although the cross-linking agents described here are not likely valid therapeutic agents, they are suited for use as scaffolds for the design of high affinity and/or specificity compounds that may serve as potential therapeutics that can be validated in ALS cell culture and mouse models. Thiol-disulfide exchange is well-suited for further investigation as a therapeutic strategy and is known to occur in vivo on SOD1 Cys111, resulting in SOD1 binding the tripeptide glutathione (GSH) (53–55). Moreover, thiol-disulfide exchange is a strategy used in tethering experiments, which are a fragment-based drug discovery approach (56, 57). These suggest that the Cys111 residues at the dimer interface of SOD1 are a potential target for peptide-based therapeutics. However, in addition to the hypothesis that dimer destabilization causes aggregation, another hypothesis for aggregation is that newly translated fALS SOD1 causing variants never dimerize due to lack of intrasubunit disulfide formation, metal deficiency, etc., resulting in unstable monomers (58, 59). Even if fALS variants never dimerize and exist as monomers in vivo (29–31), and thus C111 residues are rarely in close proximity to each other, our data with G85R suggest that this strategy may be an effective one.
Chemical cross-linking stabilized G93A by ∼20 °C and excess copper, zinc, and chemical cross-linking stabilized G85R by ∼45 °C, which is the highest ever achieved for SOD1, and as far as we know, for any disease-associated protein. In addition to stabilizing G85R, excess copper, zinc, and chemical cross-linking increased its enzymatic activity. In addition to increasing stability and activity while decreasing aggregation, the strategy presented here is expected to decrease Cys111-dependent intersubunit disulfide bond formation, which has been observed in some animal models of ALS (60, 61), as well as Cys111 oxidization to sulfonic acid, which has been observed in Parkinson and Alzheimer’s brains (62), in Lewy body-like hyaline inclusions, and vacuole rims of fALS-SOD1 mice (63). Moreover, human SOD1 Cys111 is one of two residues [the other being SOD1 W32 (64)] that modulate the toxicity of fALS mutations.
Notably, the increased activity achieved here by targeting residues distal to the active site is in contrast to the most popular strategy for designing pharmacological chaperones, which involves binding reversible inhibitors to the active site (65). For such active site inhibitors a primary source of toxicity is the enzyme inhibition that is fundamental to the approach, which results in compromising on a dose that may be suboptimal for stabilization. In contrast, our molecules, interacting at the dimer interface, both stabilize SOD1 and increase SOD1 activity for at least some inactive mutants.
Materials and Methods
Protein Expression and Purification.
WT SOD1 was purchased from Sigma-Aldrich. Constructs for expression of G93A and G85R in Saccharomyces cerevisiae have been obtained through the generous gift of P. John Hart (University of Texas Health Science Center, San Antonio, TX). The expression and purification of G93A and G85R were carried out as previously published (26, 66). Briefly, each construct in the yeast expression vector YEp-351 was transformed into EGy118ΔSOD1 yeast and grown at 30 °C for 36–48 h. Cultures were pelleted, lysed using 0.5-mm glass beads and a blender, and subjected to a 60% ammonium sulfate cut. After ammonium sulfate precipitation, the sample was pelleted and the supernatant was diluted with 0.19 volumes to a final concentration of 2.0 M ammonium sulfate. This sample was then purified using a phenyl-sepherose 6 fast flow (high sub) hydrophobic interaction chromatography column (GE Life Sciences) using a 300-mL linearly decreasing salt gradient from a high salt buffer (2.0 M ammonium sulfate, 50 mM sodium phosphate, 150 mM sodium chloride, 0.1 M EDTA, 0.25 mM DTT, pH 7.0) to a low salt buffer (50 mM sodium phosphate, 150 mM sodium chloride, 0.1 M EDTA, 0.25 mM DTT, pH 7.0). Samples containing SOD1 were eluted between 1.6 and 1.1 M ammonium sulfate and identified using SDS-PAGE analysis, pooled, and exchanged to a 10-mM Tris, pH 8.0 buffer. The protein was then loaded onto a Mono Q 10/100 anion exchange chromatography column (GE Life Sciences) and eluted using a 200-mL linearly increasing salt gradient from a low salt buffer (10 mM Tris, pH 8.0) to a high salt buffer (10 mM Tris, pH 8.0, 1 M sodium chloride). The gradient was run from 0–30% 10 mM Tris, pH 8.0, 1 M sodium chloride and SOD1 eluted between 5 and 12% 10 mM Tris, pH 8.0, 1 M sodium chloride. SOD1 protein was confirmed via SDS PAGE, Western blot, MALDI-TOF, and FTMS.
Cross-Linking and Western Blots.
WT, G93A, or G85R SOD1 were incubated with 5–25-mM DTT for approximately 20 min and either buffer exchanged using Amicon Ultra-4 centrifugal spin concentrators (MWCo 10K) or using reversed phase chromatography (ZIPTIP, Millipore, Inc). Samples cleaned by a C18 tip were also subjected to incubation with 5 mM EDTA. SOD1 samples that were buffer exchanged using Amicon concentrators were exchanged into in HPLC water, whereas C18 purified samples were further exchanged into PBS, pH 7.4 or HPLC water. DTT reduced SOD1 was incubated at a 1∶1 (20 μM∶20 μM or 10 μM∶10 μM) or 1∶3 (20 μM∶60 μM or 10 μM∶30 μM) ratio of protein to cross-linker. A variety of cross-linkers (Thermo-Fisher Scientific) were used: DTME, spacer arm 13.3 Å; BMDB, spacer arm 10.2 Å; BM(PEG)2, spacer arm 14.7 Å; BMB, spacer arm 10.9 Å; TMEA, 10.3 Å; and HBVS, spacer arm 14.7 Å. Cross-linking was achieved by incubating the reaction in either PBS pH 7.4 or water at room temperature for 1 h. After an hour the reactions were analyzed on a 15% SDS-PAGE gel with a non-cross-linked control, transferred to nitrocellulose membrane and Western blotted using a polyclonal antibody to SOD1. These experiments were repeated in triplicate.
In addition, DTME is a cleavable sulfhydryl-sulfhydryl cross-linking agent. Therefore, a cross-linking reaction containing 1∶1 molar ratio of WT SOD1 to DTME was performed at room temperature for 1 h. After cross-linking, the reaction was split in half, and half of the sample was run in a sample buffer containing DTT (reducing) and the other half in one containing no DTT (nonreducing). These samples along with non-cross-linked controls were then analyzed on a 15% SDS-PAGE gel and Western blotted as above.
MALDI-TOF.
WT SOD1 and G93A SOD1 were DTT treated and cross-linked at a 1∶1 molar ratio as described above. WT SOD1 was cross-linked with all the cross-linkers mentioned previously, whereas G93A was cross-linked with DTME and bis(maleimido)ethane (BMOE, spacer arm 8.0 Å). BMOE was used because of its shorter spacer arm length. After cross-linking, 1 μL of sample was spotted on a MALDI target containing 1 μL of matrix, 20 mg/mL sinipic acid, and analyzed on a Bruker Daltonics Microflex. The MALDI was calibrated each time using a high molecular weight protein calibration standard, Protein Calibration Standard 1 (Bruker Daltonics). The MALDI-TOF was operated in linear mode using a laser power of between 72–90%. MALDI-TOF spectra of cross-linked and non-cross-linked samples were analyzed using FlexAnalysis software (Bruker Daltonics). These experiments were repeated in triplicate.
LC-FTMS and FSD.
G93A was cross-linked using DTME or BMOE as previously described at a 1∶1 molar ratio (5 μM∶5 μM). After 1 h, 3% acetonitrile and 1% formic acid were added to the sample and spun at 14,000 rpm for 10 min to pellet any precipitated protein. The cross-linked sample was placed in an autosampler and 1 μL of cross-linked sample was aspirated into either a Proxeon 1D HPLC or Eksigent 2D ultraperformance liquid chromatography (UPLC) with the following gradient: 3–50% B for 30 min, 50–95% B for 7 min, 95% B for 5 min, 95–3% B for 1 min, and 3% B for 15 min. Buffer A is HPLC water with 0.1% formic acid and buffer B is 100% acetonitrile, 0.1% formic acid. After liquid chromatography, the sample was ionized using nanospray ionization and analyzed using a 9.4-Tesla Bruker Daltonics FTMS. Mass spectrometry parameters have been described previously (42, 43). Briefly, the FTMS was controlled using Apex control software and source parameters were controlled using the Apollo II software. Spectra (monomeric and dimeric G93A) were collected using a skimmer 1 voltage of 35–40 V, whereas funnel skimmer dissociation was used to fragment the cross-linked G93A by increasing skimmer one voltage to 80–120 V. LC-FTMS data of cross-linked and non-cross-linked G93A were analyzed using Data Analysis software (Bruker Daltonics). These experiments were repeated in triplicate.
Peptide Sequencing Using LC-FTMS.
G85R was cross-linked using BMOE as previously described at a 1∶1 molar ratio (5 μM∶5 μM). After 1 h, the cross-linked protein was heated at 99 °C for 30 min and then incubated with 10 mM TCEP for 10 min. The heated and reduced cross-linked G85R was then incubated with 1.5 μL 0.5 mg/mL Glu-C at 30 °C overnight. The digested sample was spun at 14,000 rpm for 10 min and then injected into the Eksigent UPLC using the gradient above. After liquid chromatography, the samples were introduced using nanospray ionization and MS/MS data were collected using collision-induced dissociation. Compounds were identified using Bruker Daltonics Data Analysis software, deconvoluted, and exported to a generic mascot file. Cross-linked and non-cross-linked analysis was performed by uploading the generic mascot files into the MASCOT search engine selecting none as the enzyme, using the NCBIr database, with a 1.2 Da MS error tolerance and 0.6 Da MS/MS error tolerance. MASCOT searches for the non-cross-linked and cross-linked samples were compared and m/z’s identified in the cross-linked sample and not in the non-cross-linked sample were submitted to an MS Bridge (protein prospector, UCSF, http://prospector.ucsf.edu) search using BMOE as the cross-linker (220.05 Da). MS-Bridge searches all the potential molecular weights of cross-linked peptides plus the molecular weight of the cross-linker. Peptides identified from the MS-Bridge search as being involved in cross-linked were further characterized by extracted ion chromatograms in the Data Analysis software.
Metal Analysis.
Metal analysis was preformed using ICP-MS at the University of Georgia Chemical Analysis Lab. Briefly, buffer alone was sent for analysis as a blank along with 1 μM of each variant; analysis was repeated in triplicate. In addition, 5 μM of each variant was analyzed using the FTMS in electrospray ionization mode using the direct infusion method.
Thermofluorescence Stability Assay.
The melting curves of G93A and G85R SOD1 were monitored in the presence or absence of DTME or BMOE and an excess of both copper and zinc where an increase in melting temperature suggests binding and an increase in protein stability. Therefore, protein samples in the absence of cross-linker (which are resuspended in DMSO) or copper and zinc were analyzed in the presence of 2–4% DMSO to determine the effects of DMSO on protein stability. In the first sequence of reactions, 10 μM of mutant SOD1 was incubated with increasing concentrations of 0–20 μM DTME or BMOE, incubated with 20X SYPRO™ Orange, and added to a 96-well plate. Alternatively, in the second sequence of reactions, 10 μM of mutant SOD1 was incubated with 20 μM copper, 20 μM zinc, and 0–20 μM BMOE or DTME. The melting temperature of the protein was monitored using an RT-PCR machine (Applied Biosystems) with a 0.3-°C increase in temperature every minute from 25–100 °C. Data were analyzed by subtracting a dye alone blank from each respective well, normalized to 1, and temperature versus fraction of unfolded protein was graphed (67). Subtracting a DTME or BMOE, copper, zinc, dye blank gave similar results as subtracting a dye alone blank. These experiments were repeated in triplicate.
SOD1 Activity Assay.
SOD1 activity was monitored using a nitroblue tetrazoleum (NBT) gel-based assay (68–71). Ten micrograms (∼42 μM) of WT or mutant SOD1 was incubated in the presence or absence of 80 μM copper and zinc and/or 42 μM BMOE, then analyzed on a 12.5% polyacrylamide gel. Alternatively, 10 μg (∼42 μM) of WT or mutant SOD1 was incubated in the presence or absence of 80 μM copper and zinc and/or 42 μM DTME. The DTME cross-linked samples were divided in half, where one-half was incubated with 10 mM TCEP and one was not, then analyzed on a 12.5% polyacrylamide gel. The gel was stained using a solution containing 50 mM potassium phosphate, pH 7.8, 1 tablet NBT (10 mg/tablet), and 0.1 mg/mL riboflavin for 45 min in the dark. After the 45-min incubation 1 μl/mL N,N,N′,N′-tetramethylethylenediamine was added and the gel was exposed to light for 2 min. Superoxide radicals create insoluble blue formazon from NBT. SOD1 activity is seen as a colorless band because SOD1 scavenges the superoxide, thus inhibiting the blue color formation. These experiments were repeated in triplicate.
Supplementary Material
Acknowledgments.
We thank Dr. P. John Hart for the generous gift of the YEP351-SOD1 plasmids (WT, G85R, and G93A) and EGy118-(ΔSOD1) yeast cells used to express SOD1 and its variants in this study. We also thank members of the Agar and Petsko/Ringe Labs for thoughtful discussions, insights, and critically reviewing this manuscript. This work was supported in part by a National Institutes of Health R21 (to J.N.A.) (1R21NS071256) and Fidelity Biosciences Reserach Initiative (to G.A.P. and D.R.).
Footnotes
Conflict of interest statement: A patent has been filed by the authors relating to the strategy of SOD1 stabilization described here.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015463107/-/DCSupplemental.
References
- 1.Bensimon G, Lacomblez L, Meininger V. A controlled trial of riluzole in amyotrophic lateral sclerosis ALS/Riluzole Study Group. N Engl J Med. 1994;330:585–591. doi: 10.1056/NEJM199403033300901. [DOI] [PubMed] [Google Scholar]
- 2.Lacomblez L, Bensimon G, Leigh PN, Guillet P, Meininger V. Dose-ranging study of riluzole in amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis/Riluzole Study Group II. Lancet. 1996;347:1425–1431. doi: 10.1016/s0140-6736(96)91680-3. [DOI] [PubMed] [Google Scholar]
- 3.Wagner ML, Landis BE. Riluzole: A new agent for amyotrophic lateral sclerosis. Ann Pharmacother. 1997;31:738–744. doi: 10.1177/106002809703100614. [DOI] [PubMed] [Google Scholar]
- 4.Deng HX, et al. Amyotrophic lateral sclerosis and structural defects in Cu, Zn superoxide dismutase. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 5.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 6.Giasson BI, et al. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science. 2000;290:985–989. doi: 10.1126/science.290.5493.985. [DOI] [PubMed] [Google Scholar]
- 7.LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, Selkoe DJ. Dopamine covalently modifies and functionally inactivates parkin. Nat Med. 2005;11:1214–1221. doi: 10.1038/nm1314. [DOI] [PubMed] [Google Scholar]
- 8.Shimizu T, Fukuda H, Murayama S, Izumiyama N, Shirasawa T. Isoaspartate formation at position 23 of amyloid beta peptide enhanced fibril formation and deposited onto senile plaques and vascular amyloids in Alzheimer’s disease. J Neurosci Res. 2002;70:451–461. doi: 10.1002/jnr.10350. [DOI] [PubMed] [Google Scholar]
- 9.Yan SD, et al. Glycated tau protein in Alzheimer disease: A mechanism for induction of oxidant stress. Proc Natl Acad Sci USA. 1994;91:7787–7791. doi: 10.1073/pnas.91.16.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackenzie IR, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol. 2007;61:427–434. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 11.Bredesen DE, Ellerby LM, Hart PJ, Wiedau-Pazos M, Valentine JS. Do posttranslational modifications of CuZnSOD lead to sporadic amyotrophic lateral sclerosis? Ann Neurol. 42:135–137. doi: 10.1002/ana.410420202. [DOI] [PubMed] [Google Scholar]
- 12.Kabashi E, Valdmanis PN, Dion P, Rouleau GA. Oxidized/misfolded superoxide dismutase-1: The cause of all amyotrophic lateral sclerosis? Ann Neurol. 2007;62:553–559. doi: 10.1002/ana.21319. [DOI] [PubMed] [Google Scholar]
- 13.Rakhit R, et al. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J Biol Chem. 2004;279:15499–15504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 14.Shibata N, et al. Cu/Zn superoxide dismutase-like immunoreactivity in Lewy body-like inclusions of sporadic amyotrophic lateral sclerosis. Neurosci Lett. 1994;179:149–152. doi: 10.1016/0304-3940(94)90956-3. [DOI] [PubMed] [Google Scholar]
- 15.Andersen PM. Amyotrophic lateral sclerosis associated with mutations in the CuZn superoxide dismutase gene. Curr Neurol Neurosci. 2006;6:37–46. doi: 10.1007/s11910-996-0008-9. [DOI] [PubMed] [Google Scholar]
- 16.Reaume AG, et al. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 17.Durham HD, Kabashi E, Taylor DM, Agar JN. The proteasome in neurodegeneration. In: Stefanis L, Keller JN, editors. Motor Neuron Disease. New York: Springer; 2007. pp. 247–264. [Google Scholar]
- 18.Gurney ME, et al. Motor neuron degeneration in mice that express a human Cu, Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 19.Ripps ME, Huntley GW, Hof PR, Morrison JH, Gordon JW. Transgenic mice expressing an altered murine superoxide dismutase gene provide an animal model of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 1995;92:689–693. doi: 10.1073/pnas.92.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiwari A, Hayward LJ. Mutant SOD1 instability: Implications for toxicity in amyotrophic lateral sclerosis. Neurodegener Dis. 2005;2:115–127. doi: 10.1159/000089616. [DOI] [PubMed] [Google Scholar]
- 21.Valentine JS, Doucette PA, Zittin Potter S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 22.Wong PC, et al. An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q, Johnson JL, Agar NY, Agar JN. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS Biol. 2008;6:e170. doi: 10.1371/journal.pbio.0060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaw BF, et al. Detergent-insoluble aggregates associated with amyotrophic lateral sclerosis in transgenic mice contain primarily full-length, unmodified superoxide dismutase-1. J Biol Chem. 2008;283:8340–8350. doi: 10.1074/jbc.M707751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, et al. Coincident thresholds of mutant protein for paralytic disease and protein aggregation caused by restrictively expressed superoxide dismutase cDNA. Neurobiol Dis. 2005;20:943–952. doi: 10.1016/j.nbd.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Doucette PA, et al. Dissociation of human copper-zinc superoxide dismutase dimers using chaotrope and reductant. Insights into the molecular basis for dimer stability. J Biol Chem. 2004;279:54558–54566. doi: 10.1074/jbc.M409744200. [DOI] [PubMed] [Google Scholar]
- 27.Hornberg A, Logan DT, Marklund SL, Oliveberg M. The coupling between disulphide status, metallation and dimer interface strength in Cu/Zn superoxide dismutase. J Mol Biol. 2007;365:333–342. doi: 10.1016/j.jmb.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 28.Rakhit R, et al. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat Med. 2007;13:754–759. doi: 10.1038/nm1559. [DOI] [PubMed] [Google Scholar]
- 29.Jonsson PA, et al. Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain. 2004;127:73–88. doi: 10.1093/brain/awh005. [DOI] [PubMed] [Google Scholar]
- 30.Jonsson PA, et al. Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models. Brain. 2006;129:451–464. doi: 10.1093/brain/awh704. [DOI] [PubMed] [Google Scholar]
- 31.Zetterstrom P, et al. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc Natl Acad Sci USA. 2007;104:14157–14162. doi: 10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnesano F, et al. The unusually stable quaternary structure of human Cu,Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J Biol Chem. 2004;279:47998–48003. doi: 10.1074/jbc.M406021200. [DOI] [PubMed] [Google Scholar]
- 33.Lindberg MJ, Bystrom R, Boknas N, Andersen PM, Oliveberg M. Systematically perturbed folding patterns of amyotrophic lateral sclerosis (ALS)-associated SOD1 mutants. Proc Natl Acad Sci USA. 2005;102:9754–9759. doi: 10.1073/pnas.0501957102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rakhit R, et al. Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J Biol Chem. 2002;277:47551–47556. doi: 10.1074/jbc.M207356200. [DOI] [PubMed] [Google Scholar]
- 35.Hough MA, et al. Dimer destabilization in superoxide dismutase may result in disease-causing properties: structures of motor neuron disease mutants. Proc Natl Acad Sci USA. 2004;101:5976–5981. doi: 10.1073/pnas.0305143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, et al. Disease-associated mutations at copper ligand histidine residues of superoxide dismutase 1 diminish the binding of copper and compromise dimer stability. J Biol Chem. 2007;282:345–352. doi: 10.1074/jbc.M604503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vassall KA, Stathopulos PB, Rumfeldt JA, Lepock JR, Meiering EM. Equilibrium thermodynamic analysis of amyotrophic lateral sclerosis-associated mutant apo Cu,Zn superoxide dismutases. Biochemistry. 2006;45:7366–7379. doi: 10.1021/bi0600953. [DOI] [PubMed] [Google Scholar]
- 38.Khare SD, Caplow M, Dokholyan NV. FALS mutations in Cu, Zn superoxide dismutase destabilize the dimer and increase dimer dissociation propensity: A large-scale thermodynamic analysis. Amyloid. 2006;13:226–235. doi: 10.1080/13506120600960486. [DOI] [PubMed] [Google Scholar]
- 39.Schmidlin T, Kennedy BK, Daggett V. Structural changes to monomeric CuZn superoxide dismutase caused by the familial amyotrophic lateral sclerosis-associated mutation A4V. Biophys J. 2009;97:1709–1718. doi: 10.1016/j.bpj.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray SS, Lansbury PT., Jr A possible therapeutic target for Lou Gehrig’s disease. Proc Natl Acad Sci USA. 2004;101:5701–5702. doi: 10.1073/pnas.0401934101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ray SS, Nowak RJ, Brown RH, Jr, Lansbury PT., Jr Small-molecule-mediated stabilization of familial amyotrophic lateral sclerosis-linked superoxide dismutase mutants against unfolding and aggregation. Proc Natl Acad Sci USA. 2005;102:3639–3644. doi: 10.1073/pnas.0408277102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cobb JS, Easterling ML, Agar JN. Structural characterization of intact proteins is enhanced by prevalent fragmentation pathways rarely observed for peptides. J Am Soc Mass Spectr. 2010;21:949–959. doi: 10.1016/j.jasms.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karabacak NM, et al. Sensitive and specific identification of wild-type and variant proteins from 8 to 669 kDa using top-down mass spectrometry. Mol Cell Proteomics. 2009;8:846–856. doi: 10.1074/mcp.M800099-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strange RW, et al. Variable metallation of human superoxide dismutase: Atomic resolution crystal structures of Cu-Zn, Zn-Zn and as-isolated wild-type enzymes. J Mol Biol. 2006;356:1152–1162. doi: 10.1016/j.jmb.2005.11.081. [DOI] [PubMed] [Google Scholar]
- 45.Roberts BR, et al. Structural characterization of zinc-deficient human superoxide dismutase and implications for ALS. J Mol Biol. 2007;373:877–890. doi: 10.1016/j.jmb.2007.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiDonato M, et al. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J Mol Biol. 2003;332:601–615. doi: 10.1016/s0022-2836(03)00889-1. [DOI] [PubMed] [Google Scholar]
- 47.Parge HE, Hallewell RA, Tainer JA. Atomic structures of wild-type and thermostable mutant recombinant human Cu, Zn superoxide dismutase. Proc Natl Acad Sci USA. 1992;89:6109–6113. doi: 10.1073/pnas.89.13.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furukawa Y, O’Halloran TV. Amyotrophic lateral sclerosis mutations have the greatest destabilizing effect on the apo- and reduced form of SOD1, leading to unfolding and oxidative aggregation. J Biol Chem. 2005;280:17266–17274. doi: 10.1074/jbc.M500482200. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez JA, et al. Destabilization of apoprotein is insufficient to explain Cu, Zn-superoxide dismutase-linked ALS pathogenesis. Proc Natl Acad Sci USA. 2005;102:10516–10521. doi: 10.1073/pnas.0502515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stathopulos PB, et al. Calorimetric analysis of thermodynamic stability and aggregation for apo and holo amyotrophic lateral sclerosis-associated Gly-93 mutants of superoxide dismutase. J Biol Chem. 2006;281:6184–6193. doi: 10.1074/jbc.M509496200. [DOI] [PubMed] [Google Scholar]
- 51.Stathopulos PB, et al. Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis show enhanced formation of aggregates in vitro. Proc Natl Acad Sci USA. 2003;100:7021–7026. doi: 10.1073/pnas.1237797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao X, et al. Structures of the G85R variant of SOD1 in familial amyotrophic lateral sclerosis. J Biol Chem. 2008;283:16169–16177. doi: 10.1074/jbc.M801522200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Briggs RG, Fee JA. Sulfhydryl reactivity of human erythrocyte superoxide dismutase. On the origin of the unusual spectral properties of the protein when prepared by a procedure utilizing chloroform and ethanol for the precipitation of hemoglobin. Biochim Biophys Acta. 1978;537:100–109. doi: 10.1016/0005-2795(78)90606-2. [DOI] [PubMed] [Google Scholar]
- 54.Schinina ME, et al. Amino acid sequence of chicken Cu, Zn-containing superoxide dismutase and identification of glutathionyl adducts at exposed cysteine residues. Eur J Biochem. 1996;237:433–439. doi: 10.1111/j.1432-1033.1996.0433k.x. [DOI] [PubMed] [Google Scholar]
- 55.Wilcox KC, et al. Modifications of superoxide dismutase (SOD1) in human erythrocytes: A possible role in amyotrophic lateral sclerosis. J Biol Chem. 2009;284:13940–13947. doi: 10.1074/jbc.M809687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buck E, Wells JA. Disulfide trapping to localize small-molecule agonists and antagonists for a G protein-coupled receptor. Proc Natl Acad Sci USA. 2005;102:2719–2724. doi: 10.1073/pnas.0500016102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erlanson DA, Wells JA, Braisted AC. Tethering: Fragment-based drug discovery. Annu Rev Biophys Biomol Struct. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- 58.Seetharaman SV, et al. Immature copper-zinc superoxide dismutase and familial amyotrophic lateral sclerosis. Exp Biol Med. 2009;234:1140–1154. doi: 10.3181/0903-MR-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winkler DD, et al. Structural and biophysical properties of the pathogenic SOD1 variant H46R/H48Q. Biochemistry. 2009;48:3436–3447. doi: 10.1021/bi8021735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng HX, et al. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc Natl Acad Sci USA. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Furukawa Y, Fu R, Deng HX, Siddique T, O’Halloran TV. Disulfide cross-linked protein represents a significant fraction of ALS-associated Cu, Zn-superoxide dismutase aggregates in spinal cords of model mice. Proc Natl Acad Sci USA. 2006;103:7148–7153. doi: 10.1073/pnas.0602048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi J, et al. Oxidative modifications and aggregation of Cu, Zn-superoxide dismutase associated with Alzheimer and Parkinson diseases. J Biol Chem. 2005;280:11648–11655. doi: 10.1074/jbc.M414327200. [DOI] [PubMed] [Google Scholar]
- 63.Fujiwara N, et al. Oxidative modification to cysteine sulfonic acid of Cys111 in human copper-zinc superoxide dismutase. J Biol Chem. 2007;282:35933–35944. doi: 10.1074/jbc.M702941200. [DOI] [PubMed] [Google Scholar]
- 64.Taylor DM, et al. Tryptophan 32 potentiates aggregation and cytotoxicity of a copper/zinc superoxide dismutase mutant associated with familial amyotrophic lateral sclerosis. J Biol Chem. 2007;282:16329–16335. doi: 10.1074/jbc.M610119200. [DOI] [PubMed] [Google Scholar]
- 65.Lieberman RL, et al. Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease. Nat Chem Biol. 2007;3:101–107. doi: 10.1038/nchembio850. [DOI] [PubMed] [Google Scholar]
- 66.Hayward LJ, et al. Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. J Biol Chem. 2002;277:15923–15931. doi: 10.1074/jbc.M112087200. [DOI] [PubMed] [Google Scholar]
- 67.Vedadi M, et al. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci USA. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Culotta VC, et al. The copper chaperone for superoxide dismutase. J Biol Chem. 1997;272:23469–23472. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- 69.Flohe L, Otting F. Superoxide dismutase assays. Methods Enzymol. 1984;105:93–104. doi: 10.1016/s0076-6879(84)05013-8. [DOI] [PubMed] [Google Scholar]
- 70.Schmidt PJ, Ramos-Gomez M, Culotta VC. A gain of superoxide dismutase (SOD) activity obtained with CCS, the copper metallochaperone for SOD1. J Biol Chem. 1999;274:36952–36956. doi: 10.1074/jbc.274.52.36952. [DOI] [PubMed] [Google Scholar]
- 71.Zhang F, Zhu H. Intracellular conformational alterations of mutant SOD1 and the implications for fALS-associated SOD1 mutant induced motor neuron cell death. Biochim Biophys Acta. 2006;1760:404–414. doi: 10.1016/j.bbagen.2005.11.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.