Abstract
The expression of p63 (TP63/p51) occurs in the basal cells of stratified epithelia and is strongly enhanced at the early stages of squamous cell carcinomas (SCCs) of the head and neck, skin, cervix, and others. We analyzed a promoter/enhancer region (2kΔN) that drives the predominant expression of ΔNp63 for sensitivity to Smad signaling pathways. Reporter assays in HepG2 cells showed a moderate activation of 2kΔN by Smad2 and IκB kinase α (IKKα), partners of the newly identified keratinocyte-specific transforming growth factor β (TGF-β) signaling, but not by other Smad molecules. In A431 cells, 2kΔN was activated by Smad2 and IKKα, for which a Smad binding element (SMD2) at -204 was essential. Binding of Smad2 to the chromosomal SMD2 site was detectable. The association of Smad2 with IKKα was evident in the nucleus of A431, accounting for the enhancement of ΔNp63 expression by TGF-β. Moreover, both ΔNp63 and IKKα were necessary to maintain the noninvasive phenotype of this cell line. FaDu, an invasive, Smad4-deficient SCC, also allowed 2kΔN transactivation by transfected Smad2 in the presence of endogenous IKKα. Reflecting the lack of chromosomal SMD2-Smad2 association and the absence of nuclear IKKα, however, endogenous ΔNp63 was not controlled by TGF-β or IKKα in FaDu. SCC tissue arrays showed nuclear accumulation of IKKα and p63 intensification in well-differentiated noninvasive lesions. This study indicates that p63 is a target gene of the proposed keratinocyte-specific TGF-β signal pathway for suppression of the malignant conversion of SCC.
Introduction
Unlike p53, which is ubiquitously expressed to exert the tumor-suppressing function, p63 (TP63, p51) [1,2] is required for the development of stratified epithelia including the skin and oral tissues [3,4]. A high level of expression of p63 occurs not only in keratinocyte stem cells of normal stratified epithelia [5,6] but also in squamous cell carcinomas (SCCs) of head and neck, skin, and cervix as well as in carcinomas of urothelia and others [7–9]. After the intensification at the lower-grade carcinomas, however, p63 expression diminishes during the malignant progression [10–13]. Although various genes induced by p63 have been reported [14–19], it remains obscure how p63 gene expression is enhanced at the limited stages of the specific lineages in tissue development and cancer progression.
The human p63 locus has two separate transcriptional initiation sites to produce transactivator protein TAp63 and N-terminally truncated protein ΔNp63. Because ΔNp63 isoform expression is significantly more predominant than TAp63 in normal keratinocytes and SCCs [2,8,15], the ΔNp63 promoter immediately upstream of exon 3′ is thought to control the level of ΔNp63 and overall functions of p63. An earlier report showed that ΔNp63 of zebrafish, the only p63 transcript of this species, is induced by bone morphogenetic protein 2 (BMP2) through Smad binding elements (SBEs) in the promoter/enhancer region [20].
In addition to the canonical Co-Smad/R-Smad signaling pathways of TGF-β and BMP, varied modes of cross-talk between the Smad systems and other cellular signaling mechanisms have been studied [21,22]. Interestingly, a keratinocyte-specific TGF-β signaling pathway has been recently identified, in which IκB kinase α (IKKα) instead of Smad4 acts like Co-Smad to interact with Smad2/3 (R-Smad) for transcriptional activation of the target genes [23,24]. Apart from the protein kinase activity required for the NF-κB pathway activation in the cytosol, IKKα needs to translocate to the nucleus for this function. As earlier studies showed, IKKα-deficient mice manifest severe defects in the skin and limbs because of the blockage of keratinocyte differentiation [25,26].
The Smad2/3-IKKα pathway is activated in noninvasive well-differentiated (grade 1) SCCs but seems switched off on the malignant conversion into invasive (grade 3) SCCs [27]. These processes are observed in conjunction with nuclear translocation of IKKα in grade 1 SCCs and its cytosolic sequestration in grade 3 SCC. Intriguingly, a line of evidence indicated that both TAp63 and ΔNp63 activate transcription of the IKKα gene (CHUK, also termed IKBKA, IKK-alpha, IKK1, etc) in humans [18,28,29].
Because the p63 expression pattern is well conserved over a wide range of species [20,30], we investigated how p63 is influenced by the human Smad signaling systems. Our results indicate that the ΔNp63 promoter is activated by the keratinocyte-specific TGF-β signaling mechanism with Smad2 and IKKα. Functional interactions between IKKα and p63 in SCC progression are discussed.
Materials and Methods
Cell Culture
HepG2 and A431 cells were obtained from the Japan Health Sciences Foundation. C2C12 and FaDu were from the American Type Culture Collection. HepG2, A431, C2C12 cells were propagated in Dulbecco modified minimum essential medium (D-MEM) with 10% fetal bovine serum (FBS). FaDu cells were maintained in MEM supplemented with sodiumpyruvate, nonessential amino acids, glutamine, and 10% FBS.
Luciferase Reporter Assay
Chromosomal DNA obtained from a healthy donor (Y.T.) was amplified by polymerase chain reaction (PCR) to obtain 2kΔN (-1848 to +152; Figure 1) and 2kTA (-1911 to +89, in which +1 was tentatively placed at the reported 5′ end of TAp63α complementary DNA [cDNA], NM_003722). The DNA segments were cloned into pGL3 basic (Promega, Madison, WI) at the Xho I site and sequenced. In vitro mutagenesis was carried out with the GeneTailor Site-directed Mutagenesis System (Invitrogen, Carlsbad, CA). The Smad1 expression vector pcDNA3-Flag(N)-Smad1 and the Id1 promoter linked to the luciferase gene (BRE-luc) were from Dr Kohei Miyazono, University of Tokyo. Smad2 was described [31]. The Smad3, Smad4, and Smad5 cDNA were isolated from a HeLa cell cDNA library (RIKEN, Tsukuba, Japan), cloned into pRc/CMV (Invitrogen), and sequenced. An IKKα expression vector (CHUK,NM_001278.3), pCMV6-XL5-CHUK was purchased from OriGene Technologies, Rockville, MD. Plasmid DNA transfection was carried out with Attractene (Qiagen, Hilden, Germany) for HepG2, C2C12, and FaDu and with Superfect (Qiagen) for A431. Eighteen hours after transfection, cells were starved in 0.1% serum for 6 hours and were further incubated for 24 hours with or without TGF-β1 (R&D Systems, Minneapolis, MN) at 10 ng/ml, a concentration relevant to experiments with epithelial cells [32]. We used the Luciferase Assay Kit (Promega) for FaDu and A431 and the Steady-Glo Luciferase Assay System (Promega) for HepG2 and C2C12 cells.
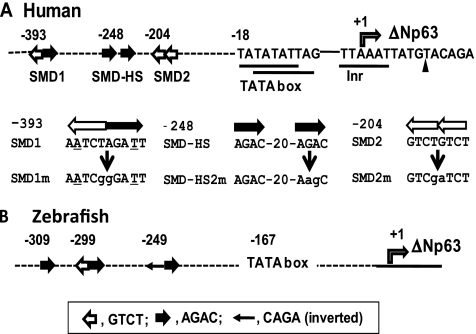
Figure 1.
SBE-like sequences found in the promoter/enhancer region of ΔNp63. (A) The proximal promoter region of human ΔNp63 is schematically shown. The 5′ end of the reported ΔNp63α cDNA data (NM_001114980) is marked by a filled triangle. Nucleotide stretches matching the consensus sequences of initiators, 5′-(C/T)(C/T)AN(A/T) (C/T)(C/T)-3′, and TATA box, 5′-TATA(T/A)A(T/A)(A/G)-3′, are underlined. Provisional initiation site (position +1) of ΔNp63 is marked by a rectangular arrow. Filled and open arrows represent the SBE motif 5′-AGAC-3′, and its complementary sequence 5′-GTCT-3′, respectively. Relative positions are shown for SMD1 (a complete palindrome including 5′-TCT-AGA-3′, SMD2 (a tandem repeat of 5′-GTCT-3′) and SMD-HS (a repeat of 5′-AGAC-3′ interrupted by 20 nucleotides). Mutated nucleotides in SMD1m, SMD2m and SMD-HS2m are in lower case. (B) The zebrafish (Danio rerio) ΔNp63 promoter/enhancer region with functional SBE sites are shown as described by Bakkers et al. [20].
Gene Silencing
SiLentfect (Bio-Rad, Hercules, CA) was used for small interfering RNA (siRNA) transfection. FaDu cells were transfected in MEM with 10% FBS. A431 cells were incubated with Opti-MEM serum-free medium (Invitrogen) during the siRNA transfection (18 hours) and refed with D-MEM with 10% FBS. A pair of siRNA duplex targeting p63 (p63si) was designed by us [15] and synthesized by Takara Bio (Otsu, Japan). Control siRNA (Csi) and IKKα-targeting siRNA (IKKαsi) were purchased from Ambion (Austin, TX; AM4611) and Dharmacon (Lafayette, CO; M-003473S), respectively.
Western Blot Analysis
Whole-cell lysates were subjected to Western blot analysis as described [14]. We purchased anti-p63 (4A4, mouse), anti-IKKα (sc-7218, rabbit), anti-Smad2 antibody (sc-101153, mouse), and anti-Smad4 (sc-7966, mouse) antibodies from Santa Cruz Biotechnology (Santa Cruz, CA). A rabbit anti-Flag antibody (PM020; MBL International, Woburn, MA) was used for Flag-Smad2 immunoprecipitation. Anti-HSP90 (ab1429, mouse) and anti-p84 (ab487, mouse) antibodies were from Abcam (Cambridge, UK). An anti-phospho-Smad2 (Ser465/467, #3101) antibody was from Cell Signaling Technology (Beverly, MA). Alkaline phosphatase (AP)-conjugated secondary antibodies (Cell Signaling Technology) were applied. Size markers (Biotinylated SDS-PAGE Standards; Bio-Rad) were detected with an AP-conjugated antibiotin immunoglobulin G (IgG; Cell Signaling Technology). Chemiluminescence (CDP-Star Reagent; New England Biolabs, Ipswich, MA) was captured with a camera luminometer (ECL Mini-camera; Amersham Pharmacia Biotech, Uppsala, Sweden), in which luminescence images appear white on the black background. Scanned digital images were processed by Photoshop (Adobe Systems, San Jose, CA) without a modification of the band intensity. Proteins were semiquantified with an imaging program, ImageJ (National Institutes of Health, Bethesda, MD).
Chromatin Immunoprecipitation
We used chromatin immunoprecipitation (ChIP) IT Expression Enzymatic, ChIP IT Control Kit (Active Motif, Carlsbad, CA), and an anti-Smad2 antibody (51-1300; Zymed Laboratories, South San Francisco, CA). A431 and FaDu cells required 100 strokes of homogenization in Dounce homogenizer (Wheaton, DuPage County, IL) for efficient nuclear release. A SMD2-containing segment was amplified by PCR with primers 5′-ATCATTCTTGAAACCCCAAATCTA-3′ (-411 to -388) and 5′-CCAGGAGACAGACAGGTAAAACTT-3′ (-215 to -192). A segment upstream of TAp63 was also amplified with 5′-TACATGTGCATGTGTTTGAGGTAG-3′ and 5′-TAGAGCTGCTTAGGGAAGTTTTGT-3′.
Reverse Transcription-Polymerase Chain Reaction
RNA isolated with RNAwiz (Ambion) was further cleaned up using the RNeasy MinElute Cleanup Kit (Qiagen). Reverse transcription-polymerase chain reaction (RT-PCR; 30 µl) contained the SuperScript One-Step RT-PCR reagent with Platinum Taq (Invitrogen), RNA (0.6 µg), and the primers described previously [14,15].
Immunofluorescence Staining
Cells were cultured on the Lab-Tek chamber slides (Nunc, Roskilde, Denmark). Cancer tissues were obtained from patients with informed consent at Kanagawa Dental College Hospital (Yokosuka, Japan). SCC tissue array slides, BC34011, and OR802 were from US Biomax (Rockville, MD). Formalin-fixed cells, tissue arrays, and sections were antigen-retrieved and probed with anti-p63 (4A4) and anti-IKKα (sc-7218) antibodies in combination with secondary antibodies conjugated with Alexa Fluor 594 or 488 (Molecular Probes, Eugene, OR). Fluorescence image was acquired using a Radiance 2000 laser scanning confocal microscope (Bio-Rad) with LaserSharp 2000 imaging software (Carl Zeiss, Oberkochen, Germany) or fluorescence microscope (BZ-9000; Keyence, Osaka, Japan).
Matrigel Invasion Assay
After rehydration, the Matrigel culture inserts (BD Biosciences, San Jose, CA) were placed in the wells (24-well culture plates) containing the medium (0.5 ml) with 10% FBS. Cell suspension (5 x 104) in 0.5 ml of the medium with 0.1% bovine serum albumin was added to the chamber and incubated for 18 hours.
Results
Lack of BMP Sensitivity in the TAp63 and ΔNp63 Promoter/Enhancer Regions
The TA and ΔN promoter/enhancer regions of 2 kbp (2kTA and 2kΔN, respectively) were linked 5′ to a firefly luciferase gene (luc) (Figure 1A). We first tested 2kTA and 2kΔN for sensitivity to BMP2 in C2C12 cells (Figure W1A). Activation of these promoters by the BMP-related Smad proteins was also assessed in HepG2 cells (Figure W1B). In comparison with the human Id1 promoter [33], neither one showed a positive response in either experiment.
We next tested to see whether 2kTA and 2kΔN can be activated by the canonical TGF-β signal by Smad2/3 paired with Smad4 in HepG2 cells (Figure W2A). Compared with the plasminogen activator inhibitor 1 (PAI-1) gene promoter [31], which was strongly activated by Smad3 transfection and the following TGF-β1 stimulation (to nearly 40-fold), 2kΔN showed a minimum detectable response to Smad2 (an approximately 2- to 3-fold activation), without an obvious response to TGF-β1. Smad4 was not a good partner of Smad2 in the 2kΔN activation.
Activation of the ΔNp63 Promoter by Smad2 and IKKα
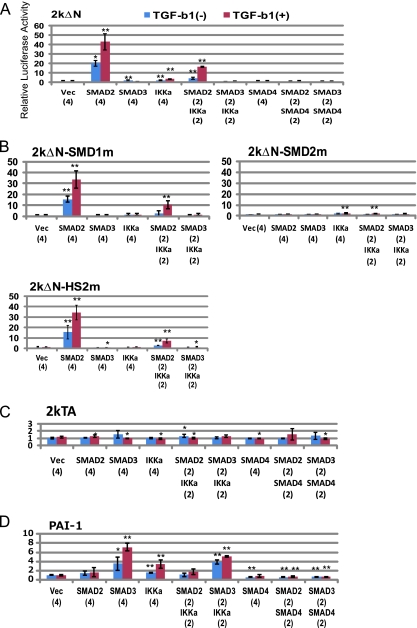
We considered the possibility of 2kΔN activation by the newly identified TGF-β signaling mechanism. A431 cells derived from an epidermoid SCC persistently expressed endogenous p63 at a high level [11]. In this cell line, the 2kΔN-driven luciferase expression was significantly (20-fold) increased by Smad2 transfection (Figure 2A). TGF-β1 (10 ng/ml) stimulation in the serum-starved (0.1% FBS) medium further enhanced the effect of Smad2 (to ∼40-fold). Plasmid-mediated IKKα expression elicited a minor response of 2kΔN (two-fold). Combined transfection of Smad2 and IKKα (2:2), followed by incubation with TGF-β1, also caused 2kΔN activation by a magnitude of ∼20. In contrast, no enhancement was detected by Smad3-only and Smad4-only transfection or by the combination of Smad2-Smad4 or Smad3-Smad4. Luciferase expression by 2kTA remained unchanged (Figure 2C). The PAI-1 promoter also responded to TGF-β1 (seven-fold activation) when Smad3 was transfected (Figure 2D). In contrast to the positive effects of Smad4 on PAI-1 in HepG2 cells (Figure W2A), Smad4 failed to activate this promoter in A431 cells, and even elicited a suppressive effect on Smad3.
Figure 2.
Activation of the ΔNp63 promoter by Smad2 and IKKα, and by stimulation with TGF-β in A431 cells. Results of the luciferase assay with 2kΔN (A),2kΔN mutants (B), 2kTA (C) and the PAI-1 promoter (D) are shown. Cells were incubated with (+) or without (-) TGF-β1 (TGF-b1) (10 ng/ml [32]) for 24 hours after serum starvation for 6 hours. Data (mean ± SD; n ≥ 3) are presented as relative luciferase activities in comparison with the control activity (1.0) produced by 2kΔN-luc transfected with empty expression vector pRcCMV (Vec). Statistic significance: **P < .01, *P < .05, versus control experiment with Vec. Indicated in parentheses are amounts of the cytomegalovirus promoter-driven gene expression vectors relative to 2kΔN-luc (= 1). IKKa indicates IKKα.
Within the 2kΔN region, we detected multiple SBE-related sequences referred to as SMD1 (starting at position -393), SMDHS-1/2 (-248), and SMD2 (-204) (Figure 1A). A double-nucleotide mutation was introduced to each site. The mutation of SMD2 (5′-GTCTGTCT-3′) to SMD2m (5′-GTCgaTCT-3′) was sufficient to cancel the activation of 2kΔN by Smad2 and IKKα, with or without stimulation with TGF-β1 (Figure 2B). The SMD1m and SMDHS2m mutations did not affect the luc expression significantly. Thus, SMD2 was determined as the common target site for Smad2 and IKKα. Furthermore, this result ruled out the possibility that IKKα activates 2kΔN through NF-κB because the Rel/NF-κB complex binds to 5′-GGGRNNYYCC-3′ (R, purine; Y, pyrimidine).
Requirement of IKKα for Smad2-Induced 2kΔN Activation
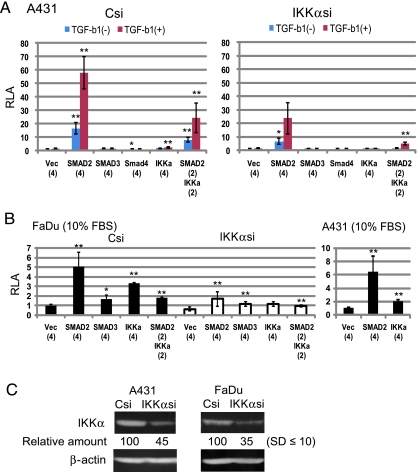
We performed IKKα silencing followed by luciferase assay with 2kΔN-luc (Figure 3A). In comparison with the control Csi-introduced A431 cells, luciferase activity produced by the IKKαsi-transfected cells was decreased by approximately 50% on Smad2 cotransfection with or without induction of TGF-β1. The levels of 2kΔN enhancement by IKKα and the Smad2/IKKα combination declined likewise (∼50%).
Figure 3.
Smad2 requires endogenously expressed IKKα for activation of the ΔNp63 promoter. (A) Luc reporter assay in A431 cells with IKKα silencing. Twenty-four hours after the siRNA (Csi, IKKαsi) introduction, cells were seeded onto 24-well plates to perform the luciferase assay with 2kΔN-luc and expression vectors of the Smad proteins and IKKα. Cells were incubated with (+) or without (-) TGF-β1 (TGF-b1) as described in the legend to Figure 2. Luciferase activity 48 hours after the 2kΔN-luc transfection was determined. Relative luciferase activity (RLA) is represented as mean ± SD; n ≥ 3). Statistic significance: **P <. 01, *P <. 05, versus control experiment with Vec. (B) IKKα silencing in FaDu cells. We determined luciferase activities produced by the Csi- and IKKαsi-transfected cells cultured with 10% FBS. Results with A431 under the same conditions are also shown. (C) Western blot analysis of A431 and FaDu cells with IKKα silencing. Day 3 cells after the transfection of Csi and IKKαsi were analyzed for IKKα (85 kDa) and β-actin (43 kDa). Relative amounts of IKKα were obtained by standardization with β-actin for each cell line.
FaDu cells derived from a hypopharyngeal SCC display an invasive phenotype [34–36], still expressing a level of p63 [8,15]. Without the entire Smad4 gene [37,38], FaDu allowed activation of 2kΔN by Smad2 and IKKα. When 10% FBS was contained in the culture medium throughout the reporter assay, Smad2 caused an approximately five-fold increase in the luciferase assay (Figure 3B, left), similar to the activation level in A431 (seven-fold) under the same conditions (right). We detected a blunted response of 2kΔN to both IKKα and Smad2 (∼70% reduction in the luciferase activity) in IKKαsi-transfected FaDu cells (Figure 3B, middle).
Western blot analysis indicated a significant (55%–65%) decrease in the IKKα protein 3 days after the IKKαsi transfection in the two cell lines (Figure 3C). Thus, endogenously expressed IKKα seemed essential for 2kΔN activation by transfected Smad2 in the reporter assay.
Endogenous ΔNp63 Induction by TGF-β in A431
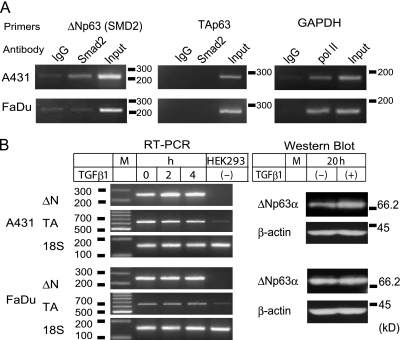
To assess whether Smad2 is physically associated with the identified sequences upstream of the ΔNp63-encoding sequences on chromosome 3, we performed ChIP experiments (Figure 4A). DNA fragments coprecipitated with an anti-Smad2 antibody, anti-RNA polymerase II antibody (anti-pol II), and a control nonimmune IgG were examined for the SMD2 target sequence and the glutaraldehyde phosphate dehydrogenase (GAPDH) gene promoter sequence by PCR. A corresponding segment in the TAp63 promoter/enhancer region was also tested as a control. In A431 cells, the SMD2 site was more enriched (four-fold or higher) in the anti-Smad2 precipitate than in the control IgG precipitate, and the GAPDH promoter was more enriched in the anti-pol II precipitate than in the control. The TA segment was not selectively coprecipitated by any antibody examined. Thus, association of Smad2 with the SMD2 site in A431 was strongly suggested.
Figure 4.
Endogenous ΔNp63 activation by TGF-β1 in A431. (A) ChIP. A431 and FaDu nuclear extracts were examined for association of Smad2 with the SMD2 site. Antibodies used and amplified chromosomal segments, SMD2 (220 bp), TA (282 bp), and GAPDH (166 bp) are indicated. (B) Induction of endogenous ΔNp63 by TGF-β1. Cells incubated with TGF-β1 (10 ng/ml) for indicated periods (h) were analyzed for TAp63 and ΔNp63 mRNA by RT-PCR. Total RNA from HEK293 was tested as a negative control of ΔNp63 expression. Ribosomal RNA (18S) was amplified as a quantitative control. A 100-bp DNA ladder was loaded in lane M. Cells incubated with (+) or without (-) TGF-β1 (for 20 hours) were also analyzed by Western blot analysis for ΔNp63α. The blots were reprobed with an anti-β-actin antibody. Sizes (kDa) and positions of the standard proteins are indicated.
In FaDu cells, however, the anti-Smad2 antibody failed to precipitate the segment containing SMD2. Results with the anti-pol II and control IgG were comparable to those in A431. Binding of Smad2 to the SMD2 site on the chromosomal DNA was less probable in FaDu.
We next examined whether TGF-β could influence the endogenous p63 expression. Two hours after the TGF-β1 (10 ng/ml) stimulation of serum-starved A431 cells, ΔNp63 messenger RNA (mRNA) was increased two-fold, which was retained to 4 hours (Figure 4B; RT-PCR). The TA-type mRNA was not altered. A 1.7-fold (1.7 ± 0.2) increase in ΔNp63α, the most predominant p63 protein, was obvious 20 hours after the TGF-β1 input (Figure 4B; Western blot). p63 expression in FaDu cells was not affected by TGF-β1 as observed in RT-PCR and Western blot analysis. Thus, TGF-β1 enhanced the chromosomal ΔNp63 transcription in A431 but not in FaDu.
Detection of Smad2-IKKα Association in the Nucleus of A431
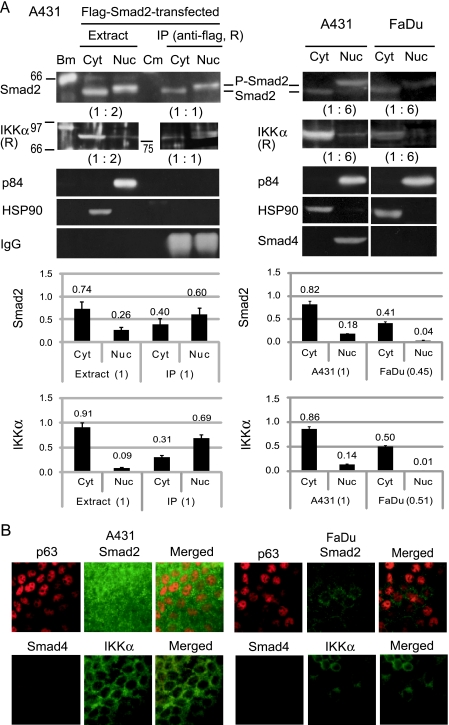
The status of Smad2 and IKKα in the two SCC lines was analyzed by Western blot analysis (Figure 5A). Under the normal culture condition of A431, Smad2 existed in the cytosol and nucleus at a ratio of 8:2, showing nuclear localization of the phosphorylated form, P-Smad, reactive with antibodies recognizing phosphorylation at Ser465/467 of this protein (Figure 5A, right). In addition to a large amount of IKKα (85 kDa) in the cytosol, a small population of this protein (∼14%) was detected in the nucleus. When Flag-tagged Smad2 was introduced into A431 by transfection, the cytoplasmic/nuclear distribution of the entire Smad2 molecules did not change significantly (Figure 5A, left). IKKα was efficiently coprecipitated with phosphorylated Flag-Smad2 from the nuclear extract by an anti-flag antibody. Although a large amount of IKKα was contained in the cytosol, it was poorly coprecipitated with Flag-Smad2 from the cytosol fraction.
Figure 5.
Status of the signaling molecules in A431 and FaDu. (A) Smad2 and IKKα were analyzed for the level of expression and subcellular localization by Western blot analysis. (Left panels) Cytosolic (Cyt) and nuclear (Nuc) fractions were obtained from A431 cells transfected with the Flag-Smad2 expression vector. Immunoprecipitation (IP) was carried out with a rabbit (R) anti-Flag antibody (PM020). Blot membrane was probed with anti-Smad2 (sc-101153), anti-IKKα (sc-7218), anti-p84 nuclear protein (ab487), and anti-HSP90 (ab1429) antibodies. Sizes (in kDa) and positions of the biotinylated standard proteins (Bm) and prestained color marker proteins (Cm) are indicated. Positions of Smad2 and phosphorylated Smad2 (P-Smad2) reactive with an anti-phospho-Smad2 (Ser465/467) antibody (#3101) are also indicated. Ratios of the sample amounts analyzed are indicated in parentheses. Graphs show relative amounts of Smad2 and IKKα in the fractions as determined by measurement of the band intensities (n = 3). (Right panels) Cytosol (Cyt) and nuclear (Nuc) fractions from untransfected A431 and FaDu cells were examined. Protein band intensities were measured and standardized with HSP90 and the ratio of sample amounts (1:6). Graphs show comparison of the total amount and subcellular distribution of Smad2 and IKKα between A431 and FaDu (n = 3). (B) Immunofluorescence analysis of formalin-fixed A431 and FaDu cells. Scale bars, 50 µm.
FaDu cells contained a lesser amount of Smad2, ∼45% in comparison with A431, showing its cytosolic and nuclear distributions at a ratio of 10:1 (Figure 5A, far right). Furthermore, IKKα was also decreased by ∼50% in the total amount and hardly detectable in the nuclear fraction.
These differences between the two cell lines were confirmed by immunofluorescence analyses (Figure 5B). A431 contained Smad2 inside the nucleus as well as in the cytosol, whereas FaDu had a decreased amount of Smad2 showing a brighter signal outside the nucleus. A significant decrease in IKKα and the lack of the Smad4 protein in FaDu were also observed.
Induction of an Invasion Activity Either by p63 or by IKKα Silencing
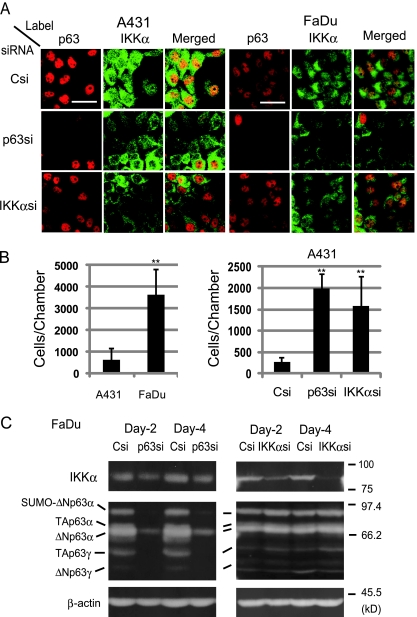
A431 cells displayed intense p63 immunofluorescence confined to the nucleus as observed by laser scanning microscopy (Figure 6A, left). Consistent with the Western blot analysis (Figure 5A), cells showing a stronger IKKα label in the cytosol than in the nucleus were seen predominantly. Nuclear accumulation of IKKα was rarely detected (≤5% cells). We carried out gene silencing by transfection of p63si and IKKαsi. As reported other studies [18,28,29], p63 knockdown cells showed a substantial decrease in IKKα. Conversely, IKKα knockdown cells also displayed an obvious decrease in p63. This result not only supported our results that IKKα facilitated p63 expression but also implied mutual activation between p63 and IKKα in A431.
Figure 6.
Induction of an invasion activity by p63 and IKKα knockdown in A431 cells. (A) Immunofluorescence analyses of siRNA-introduced A431 and FaDu cells. Two days after transfection with Csi, p63si, and IKKαsi, cells were formalin-fixed for immunostaining. Scale bars, 50 µm. (B) Invasion assay was carried out with Matrigel matrix cell culture inserts. A431 and FaDu cells without transfection (left) and A431 cells after siRNA (Csi, p63si, and IKKαsi) transfection (right) were examined. Statistic significance: **P < .01 versus A431 (left), **P < .01 versus Csi (right). (C) Western blot analysis for p63 and IKKα in FaDu cells transfected with siRNA. Positions of the p63 isoforms were determined as described [15,47]. IKKα, β-actin, and sizes of the standard proteins are also shown.
FaDu cells also showed a reduction of IKKα on the elimination of p63 (Figure 6A, right). However, IKKα knockdown cells maintained the p63 level as in the control Csi-transfected cells. In Western blot analysis, all of the detectable p63 isoforms including ΔNp63α and TAp63γ were greatly decreased by p63si transfection (Figure 6C, left), which was accompanied by an obvious reduction of IKKα. However, IKKα silencing did not affect the p63 isoforms (Figure 6C, right). Given the lack of Smad2-SMD2 association (Figure 4A) and the absence of IKKα in the nucleus (Figure 5), the endogenous ΔNp63 expression in FaDu cells may not depend on the Smad2-IKKα pathway of TGF-β.
In an invasion assay with Matrigel-coated membrane, FaDu cells penetrated through the matrix protein complex, whereas A431 failed to do so (Figure 6B). Interestingly, either by p63 or by IKKα gene silencing, A431 gained an invasive activity, implying that both IKKα and p63 are involved in the maintenance of the noninvasive phenotype of A431 cells.
Changes in p63 and IKKα in the Progression of SCC
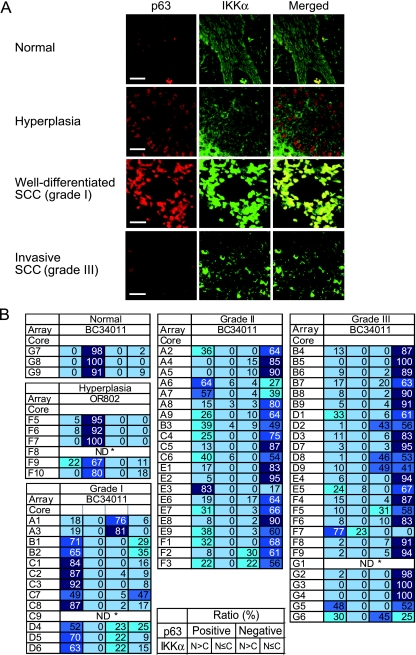
To assess alterations of p63 and IKKα during the SCC progression, we analyzed conventional oral carcinoma sections at different stages, hyperplasia (carcinoma in situ), well-differentiated (grade 1), and poorly differentiated (grade 3), for p63 and IKKα by immunostaining (Figure 7A). Four or six cases of each grade were examined. Normal gingival sections displayed nuclear p63 expression in the basal and suprabasal layers of the epithelium as described [3,4]. The IKKα label was spread throughout the cells, causing an enhanced signal in the cytoplasm. Hyperplasia lesions (4/4) displayed more enhanced p63 immunofluorescence than in the normal tissues. As observed in A431 (Figure 5), a majority of the IKKα molecules existed in the cytoplasm. Intriguingly, well-differentiated SCC lesions (6/6) displayed a significant enhancement of p63 as well as nuclear accumulation of IKKα. Furthermore, in poorly differentiated, invasive SCC sections (5/6), loss of p63 positive cells, and relocalization of IKKα to the cytoplasm were profound.
Figure 7.
Amplification of p63 with IKKα in well-differentiated SCC. (A) Oral carcinoma sections at different pathologic stages were analyzed by immunofluorescence staining for p63 and IKKα. Typical images representing four or six individual samples of the indicated stages are presented. The well-differentiated SCC images contain nests of keratin pearls in the dark areas. Scale bars, 50 µm. (B) Summary of the SCC tissue array analysis. Composition (%) of the p63(+)/IKKα (N > C), p63(+)/IKKα (N > C), p63(-)/IKKα (N > C), and p63(-)/IKKα (N ≤ C) populations was calculated for each core with predetermined cancer grade. Results are collectively shown for each category: normal, SCC grade 1, 2, 3 (indicated as I, II, III in array BC34011), and hyperplasia (OR802). ND* indicates not determined. (OR802-F8 and BC34011-G1 contained less than 10 labeled cells in each field of view. BC34011-C9 showed cells with cytoplasmically localized p63 and was therefore eliminated.)
To study more quantitatively the status of these proteins in SCCs, we analyzed SCC tissue arrays. The BC34011 slide contained 60 cores of different stages (grades 1–3) of SCC and 3 cores of normal tissues. Six cores of hyperplasia on OR802 were also examined. All of the tissue array immunofluorescence images are presented in Figures W3 and W4. Cells (100 < n ≤ 350) in a typical field of view were sorted by 1) the presence (+) or absence (-) of p63 and 2) nuclear accumulation (N > C) or cytoplasmic/whole-cell distribution (N ≤ C) of IKKα. We obtained, for each core, composition (%) of the four categories: p63(+)/IKKα (N > C), p63(+)/IKKα (N ≤ C), p63(-)/IKKα (N > C), and p63(-)/IKKα (N ≤ C) (Figure 7B).
In normal gingiva and hyperplasia tissues, a large population (≥90%) showed the p63(+)/IKKα (N ≤ C) phenotype in six of eight cores. Interestingly, p63(+)/IKKα (N > C) cells were predominant (>50%) in 9 of 12 cores of grade 1 SCC. Furthermore, the p63(+)/IKKα (N > C) population was diminished markedly in grade 2 (moderately well differentiated) tissues with a concomitant increase in the p63(-)/IKKα (N ≤ C) population. In grade 3 SCCs, p63(-)/IKKα (N ≤ C) cells were predominant (> 50%) in 22 of 25 cores. Only a small number (≤10%) of p63(+)/IKKα (N > C) cells remained in 16 cores of grade 3.
These results suggest that the maximum expression of p63 at the stage of well-differentiated SCC is associated with IKKα nuclear accumulation. Furthermore, the loss of p63 during the malignant conversion seemed to occur concurrently with the decline of IKKα.
Discussion
We have shown that the ΔNp63 promoter is activated by the recently identified keratinocyte-specific TGF-β signal pathway with Smad2 and IKKα in SCC. This mechanism may underlie the suppression of malignant conversion by p63 and IKKα.
The SMD2 (5′-GTCTGTCT-3′) site matching the SBE consensus was determined as the cis-acting sequence to respond to the Smad2/IKKα signal. The zebrafish ΔNp63 promoter also contains SBE-like sequences, with which epithelial cells responded to BMP2 in situ [20]. Our results do not necessarily dismiss the possibility that the human BMP signaling also contributes to p63 expression in other cell types. However, the functional SBE-like sequences in the ΔNp63 promoter are little conserved between human and zebrafish (Figure 1), implying that the disparity reflects diversification of the cis-regulatory sequences and the transcription factor usage in mammalian evolution.
By the ChIP, TGF-β1 stimulation and gene silencing experiments, we concluded that transcription of chromosomal ΔNp63 is activated by the Smad2/IKKα signaling in A431 cells. In contrast, the pathway seemed dispensable for ΔNp63 in FaDu cells, despite the observation of 2kΔN transactivation by Smad2 in an IKKα-dependent mode. This is not a contradiction between the assay results. Transfection of Smad2 in the reporter assay may have caused the pathway activation in FaDu. Furthermore, transcription of ΔNp63 on the chromosomal DNA can be influenced by the sequences upstream and downstream of the 2kΔN region and by proteins interacting with them.
Conversely, recent studies showed transcriptional activation of the IKKα gene by p63 [18,28,29], the reverse reaction of our finding, hypothesizing mutual activation between p63 and IKKα. This hypothesis seems relevant to A431 cells in which IKKα knockdown decreased p63 and vice versa. The same level of invasion activity was induced by p63 and IKKα silencing in A431, implying that the mutual activation mechanism is important to maintain the noninvasive phenotype.
As many other SCC lines, A431 expresses both keratinocyte stem cell markers and an early differentiation marker, involucrin [39]. When the IKKα distribution pattern and the result of invasion assay are taken into account, we consider A431 to be an “advanced” hyperplasia. FaDu cells produce proinvasive molecules including furin, matrix metalloproteinase 3 (MMP-3, also termed stromelysin-1), and other MMPs, and characterized as an invasive SCC line [34,36], which was consistent with our invasion assay. Earlier studies showed that cell lines derived from SCCs all fail, to a greater or lesser extent, to complete the process of keratinization [40,41], indicating a limitation in well-differentiated SCC isolation as a cell line.
By an extensive histologic study in conjunction with biochemical analyses, nuclear translocation of IKKα was found essential for the keratinocyte-specific TGF-β pathway activation and for suppression of the malignant conversion of SCC [23,27]. Our immunofluorescence analyses with tissue arrays are consistent with the previous studies as well as our finding that IKKα acts in the p63-inducing mechanism. The down-regulation of p63 during the malignant conversion [10,11,13] may be associated with the significant decrease of IKKα as demonstrated by the gene silencing and invasion assay with A431 (Figure 6).
Invasion and metastasis of SCC involve “epithelial-to-mesenchymal transition” controlled by varied intracellular signaling pathways [42], production of proteases [43], and functional interactions with fibroblasts [44] and endothelial cells [45]. The p63 and Smad2/IKKα systems independently control many target genes in keratinocytes [14,16–19,23,27,46], which may collectively facilitate the proposed invasion-suppressing function of the keratinocyte-specific TGF-β signaling pathway. Mechanisms of the nuclear accumulation of IKKα and its elimination in the course of SCC progression await extensive studies.
Supplementary Material
Acknowledgments
The authors thank K. Miyazono (University of Tokyo) for plasmids, M. Shibuya and colleagues (University of Tokyo) for discussion, and N. Shirato (Photography Department, Tokyo Medical and Dental University) for data file making.
Footnotes
This study was supported by Grant-in-Aid for Scientific Research (C) 20590380 to S.K. and (C) 21590434 to I.K., and by the High-Tech Research Center Project (to R.H.) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology.
This article refers to supplementary materials, which are designated by Figures W1 to W4 and are available online at www.neoplasia.com.
References
- 1.Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M, et al. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med. 1998;4:839–843. doi: 10.1038/nm0798-839. [DOI] [PubMed] [Google Scholar]
- 2.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 3.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 4.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 5.Pellegrini G, Dellambra E, Golisano O, Martinelli E, Fantozzi I, Bondanza S, Ponzin D, McKeon F, De Luca M. p63 identifies keratinocyte stem cells. Proc Natl Acad Sci USA. 2001;98:3156–3161. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senoo M, Pinto F, Crum CP, McKeon F. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 7.Barbareschi M, Pecciarini L, Cangi MG, Macri E, Rizzo A, Viale G, Doglioni C. p63, a p53 homologue, is a selective nuclear marker of myoepithelial cells of the human breast. Am J Surg Pathol. 2001;25:1054–1060. doi: 10.1097/00000478-200108000-00010. [DOI] [PubMed] [Google Scholar]
- 8.Hibi K, Trink B, Patturajan M, Westra WH, Caballero OL, Hill DE, Ratovitski EA, Jen J, Sidransky D. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci USA. 2000;97:5462–5467. doi: 10.1073/pnas.97.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quade BJ, Yang A, Wang Y, Sun D, Park J, Sheets EE, Cviko A, Federschneider JM, Peters R, McKeon FD, et al. Expression of the p53 homologue p63 in early cervical neoplasia. Gynecol Oncol. 2001;80:24–29. doi: 10.1006/gyno.2000.5953. [DOI] [PubMed] [Google Scholar]
- 10.Koga F, Kawakami S, Kumagai J, Takizawa T, Ando N, Arai G, Kageyama Y, Kihara K. Impaired ΔNp63 expression associates with reduced β-catenin and aggressive phenotypes of urothelial neoplasms. Br J Cancer. 2003;88:740–747. doi: 10.1038/sj.bjc.6600764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higashikawa K, Yoneda S, Tobiume K, Taki M, Shigeishi H, Kamata N. Snail-induced down-regulation of ΔNp63α acquires invasive phenotype of human squamous cell carcinoma. Cancer Res. 2007;67:9207–9213. doi: 10.1158/0008-5472.CAN-07-0932. [DOI] [PubMed] [Google Scholar]
- 12.Fukushima H, Koga F, Kawakami S, Fujii Y, Yoshida S, Ratovitski E, Trink B, Kihara K. Loss of ΔNp63α promotes invasion of urothelial carcinomas via N-cadherin/Src homology and collagen/extracellular signal-regulated kinase pathway. Cancer Res. 2009;69:9263–9270. doi: 10.1158/0008-5472.CAN-09-1188. [DOI] [PubMed] [Google Scholar]
- 13.Barbieri CE, Tang LJ, Brown KA, Pietenpol JA. Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res. 2006;66:7589–7597. doi: 10.1158/0008-5472.CAN-06-2020. [DOI] [PubMed] [Google Scholar]
- 14.Kurata S, Okuyama T, Osada M, Watanabe T, Tomimori Y, Sato S, Iwai A, Tsuji T, Ikawa Y, Katoh I. p51/p63 Controls subunit α3 of the 978 Induction of p63 by a TGF-β major epidermis integrin anchoring the stem cells to the niche. J Biol Chem. 2004;279:50069–50077. doi: 10.1074/jbc.M406322200. [DOI] [PubMed] [Google Scholar]
- 15.Okuyama T, Kurata S, Tomimori Y, Fukunishi N, Sato S, Osada M, Tsukinoki K, Jin HF, Yamashita A, Ito M, et al. p63(TP63) elicits strong transactivation of the MFG-E8/lactadherin/BA46 gene through interactions between the TA and ΔN isoforms. Oncogene. 2008;27:308–317. doi: 10.1038/sj.onc.1210646. [DOI] [PubMed] [Google Scholar]
- 16.Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA, Brugge JS, Ellisen LW. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- 17.Osada M, Park HL, Nagakawa Y, Yamashita K, Fomenkov A, Kim MS, Wu G, Nomoto S, Trink B, Sidransky D. Differential recognition of response elements determines target gene specificity for p53 and p63. Mol Cell Biol. 2005;25:6077–6089. doi: 10.1128/MCB.25.14.6077-6089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, Roop DR. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci USA. 2007;104:3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuyama R, Ogawa E, Nagoshi H, Yabuki M, Kurihara A, Terui T, Aiba S, Obinata M, Tagami H, Ikawa S. p53 homologue, p51/p63, maintains the immaturity of keratinocyte stem cells by inhibiting Notch1 activity. Oncogene. 2007;26:4478–4488. doi: 10.1038/sj.onc.1210235. [DOI] [PubMed] [Google Scholar]
- 20.Bakkers J, Hild M, Kramer C, Furutani-Seiki M, Hammerschmidt M. Zebrafish ΔNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev Cell. 2002;2:617–627. doi: 10.1016/s1534-5807(02)00163-6. [DOI] [PubMed] [Google Scholar]
- 21.Zhang YE. Non-Smad pathways in TGF-β signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranganathan P, Agrawal A, Bhushan R, Chavalmane A, Kalathur R, Takahashi T, Kondaiah P. Expression profiling of genes regulated by TGF-β: differential regulation in normal and tumour cells. BMC Genomics. 2007;8:98. doi: 10.1186/1471-2164-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Descargues P, Sil AK, Sano Y, Korchynskyi O, Han G, Owens P, Wang X-J, Karin M. IKKα is a critical coregulator of a Smad4-independent TGFβ-Smad2/3 signaling pathway that controls keratinocyte differentiation. Proc Natl Acad Sci USA. 2008;105:2487–2492. doi: 10.1073/pnas.0712044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Descargues P, Sil AK, Karin M. IKKα, a critical regulator of epidermal differentiation and a suppressor of skin cancer. EMBO J. 2008;27:2639–2647. doi: 10.1038/emboj.2008.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, Johnson R, Karin M. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKα subunit of IκB kinase. Science. 1999;284:316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- 26.Takeda K, Takeuchi O, Tsujimura T, Itami S, Adachi O, Kawai T, Sanjo H, Yoshikawa K, Terada N, Akira S. Limb and skin abnormalities in mice lacking IKK. Science. 1999;284:313–316. doi: 10.1126/science.284.5412.313. [DOI] [PubMed] [Google Scholar]
- 27.Marinari B, Moretti F, Botti E, Giustizieri ML, Descargues P, Giunta A, Stolfi C, Ballaro C, Papoutsaki M, Alema S, et al. The tumor suppressor activity of IKKα in stratified epithelia is exerted in part via the TGF-α antiproliferative pathway. Proc Natl Acad Sci USA. 2008;105:17091–17096. doi: 10.1073/pnas.0809288105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Candi E, Terrinoni A, Rufini A, Chikh A, Lena AM, Suzuki Y, Sayan BS, Knight RA, Melino G. p63 is upstream of IKKα in epidermal development. J Cell Sci. 2006;119:4617–4622. doi: 10.1242/jcs.03265. [DOI] [PubMed] [Google Scholar]
- 29.Marinari B, Ballaro C, Koster MI, Giustizieri ML, Moretti F, Crosti F, Papoutsaki M, Karin M, Alema S, Chimenti S, et al. IKKα is a p63 transcriptional target involved in the pathogenesis of ectodermal dysplasias. J Invest Dermatol. 2008;129:60–69. doi: 10.1038/jid.2008.202. [DOI] [PubMed] [Google Scholar]
- 30.Tomimori Y, Katoh I, Kurata S, Okuyama T, Kamiyama R, Ikawa Y. Evolutionarily conserved expression pattern and trans-regulating activity of Xenopus p51/p63. Biochem Biophys Res Commun. 2004;313:230–236. doi: 10.1016/j.bbrc.2003.11.113. [DOI] [PubMed] [Google Scholar]
- 31.Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin CH, Miyazono K, et al. TGF-β receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grande M, Franzen A, Karlsson J-O, Ericson LE, Heldin N-E, Nilsson M. Transforming growth factor-β and epidermal growth factor synergistically stimulate epithelial to mesenchymal transition (EMT) through a MEK-dependent mechanism in primary cultured pig thyrocytes. J Cell Sci. 2002;115:4227–4236. doi: 10.1242/jcs.00091. [DOI] [PubMed] [Google Scholar]
- 33.Lopez-Rovira T, Chalaux E, Massague J, Rosa JL, Ventura F. Direct binding of smad1 and smad4 to two distinctmotifsmediates bonemorphogenetic protein-specific transcriptional activation of Id1 gene. J Biol Chem. 2002;277:3176–3185. doi: 10.1074/jbc.M106826200. [DOI] [PubMed] [Google Scholar]
- 34.De Angelis T, Noe A, Chatterjee M, Mulholland J. Stromelysin-1 activation correlates with invasiveness in squamous cell carcinoma. J Invest Dermatol. 2002;118:759–766. doi: 10.1046/j.1523-1747.2002.01755.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W, Matrisian L, Holmbeck K, Vick C, Rosenthal E. Fibroblast-derived MT1-MMP promotes tumor progression in vitro and in vivo. BMC Cancer. 2006;6:52. doi: 10.1186/1471-2407-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daniel EB, Haleh M, Luma A-S, Ricardo Lopez De C, John AR, Andres JPK-S. Elevated furin expression in aggressive human head and neck tumors and tumor cell lines. Mol Carcinog. 2001;31:224–232. doi: 10.1002/mc.1057. [DOI] [PubMed] [Google Scholar]
- 37.Hummer BT, Bartlett C, Henry E, Weissman BE. Expression of Smad4 in the FaDu cell line partially restores TGF-β growth inhibition but is not sufficient to regulate fibronectin expression or suppress tumorigenicity. J Cell Physiol. 2003;194:289–302. doi: 10.1002/jcp.10202. [DOI] [PubMed] [Google Scholar]
- 38.Reiss M, Stash EB, Vellucci VF, Zhou ZL. Activation of the autocrine transforming growth factor α pathway in human squamous carcinoma cells. Cancer Res. 1991;51:6254–6262. [PubMed] [Google Scholar]
- 39.Jensen KB, Jones J, Watt FM. A stem cell gene expression profile of human squamous cell carcinomas. Cancer Lett. 2008;272:23–31. doi: 10.1016/j.canlet.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rheinwald JG, Beckett MA. Defective terminal differentiation in culture as a consistent and selectable character of malignant human keratinocytes. Cell. 1980;22:629–632. doi: 10.1016/0092-8674(80)90373-6. [DOI] [PubMed] [Google Scholar]
- 41.Fynan TM, Morgan D, Yuspa SH, Longley BJ, Jr, Zhou ZL, Reiss M. Restoration of differentiation and suppression of tumorigenicity in somatic cell hybrids of human squamous carcinoma cells and keratinocytes. Cell Growth Differ. 1994;5:1293–1300. [PubMed] [Google Scholar]
- 42.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 43.Bassi DE, Zhang J, Cenna J, Litwin S, Cukierman E, Klein-Szanto AJ. Proprotein convertase inhibition results in decreased skin cell proliferation, tumorigenesis, and metastasis. Neoplasia. 2010;12:516–526. doi: 10.1593/neo.92030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neiva KG, Zhang Z, Miyazawa M, Warner KA, Karl E, Nor JE. Cross talk initiated by endothelial cells enhances migration and inhibits anoikis of squamous cell carcinoma cells through STAT3/Akt/ERK signaling. Neoplasia. 2009;11:583–593. doi: 10.1593/neo.09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hembruff SL, Jokar I, Yang L, Cheng N. Loss transforming growth factor-β signaling in mammary fibroblasts enhances CCL2 secretion to promote mammary tumor progression through macrophage-dependent and -independent mechanisms. Neoplasia. 2009;12:425–433. doi: 10.1593/neo.10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu B, Xia X, Zhu F, Park E, Carbajal S, Kiguchi K, DiGiovanni J, Fischer SM, Hu Y. IKKα is required to maintain skin homeostasis and prevent skin cancer. Cancer Cell. 2008;14:212–225. doi: 10.1016/j.ccr.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghioni P, D'Alessandra Y, Mansueto G, Jaffray E, Hay RT, La Mantia G, Guerrini L. The protein stability and transcriptional activity of p63α are regulated by SUMO-1 conjugation. Cell Cycle. 2005;4:183–190. doi: 10.4161/cc.4.1.1359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.