Abstract
Since their discovery in the early 1990s, microRNAs (miRs) have gone from initially being considered an oddity to being recognized as a level of gene expression regulation that is integral to the normal function of cells and organisms. They are implicated in many if not all biological processes in animals, from apoptosis and cell signaling to organogenesis and development. Our understanding of cell regulatory states, as determined primarily by transcription factor (TF) profiles, is incomplete without consideration of the corresponding miR profile. The miR complement of a cell provides robust and redundant control over the output of hundreds of possible targets for each miR. miRs are common components of regulatory pathways, and in some cases can constitute on–off switches that regulate crucial fate decisions. In this review, we summarize our current knowledge about the biogenesis and regulation of miRs and describe their involvement in the pathways that regulate cell division, pluripotency, and reprogramming to the pluripotent state.
Keywords: MicroRNAs, embryonic stem cells, cell cycle
Embryonic stem cells (ESCs) are usually derived from the inner cell mass of a blastocyst stage embryo. Their defining trait is self-renewal: the ability to proliferate indefinitely in vitro (absence of a Hayflick limit) while maintaining their main in vivo characteristic, which is the ability to give rise to all differentiated cell types of the adult organism, termed pluripotency. As with any specialized cell type, the phenotype of ESCs is the result of complex regulatory interactions between transcription factors (TFs), chromatin remodeling proteins, signaling molecules, and noncoding RNAs. While capable of continuous cell division in the undifferentiated state, ESCs are permanently “poised” to differentiate as soon as the proper cues arise. This massive transformation of cell phenotype poses a major regulatory challenge to the cell, as the entire makeup of the network must be changed within a short developmental window. What mechanisms does an ESC use to bring about such rapid changes in its proteome? A main regulatory component are the microRNAs (miRs), the subgroup of noncoding RNAs that is best characterized and for which, unlike other noncoding RNAs, we have a general mechanistic model. miRs were initially considered a species-specific peculiarity (Lee et al. 1993), but today it is widely recognized that they constitute a level of post-transcriptional regulation that is integral to normal cell and organism function in metazoans, and their ability for post-transcriptional coregulation of hundreds of potential targets makes them well suited to bring about rapid transformations in cell phenotype. In this review, we summarize our current knowledge of the involvement of miRs in multiple aspects of ESC function, and argue that to say that miRs have a role in ESC biology is an understatement: Quite simply, the network responsible for ESC behavior cannot function without its miR component.
Walking down the miR path
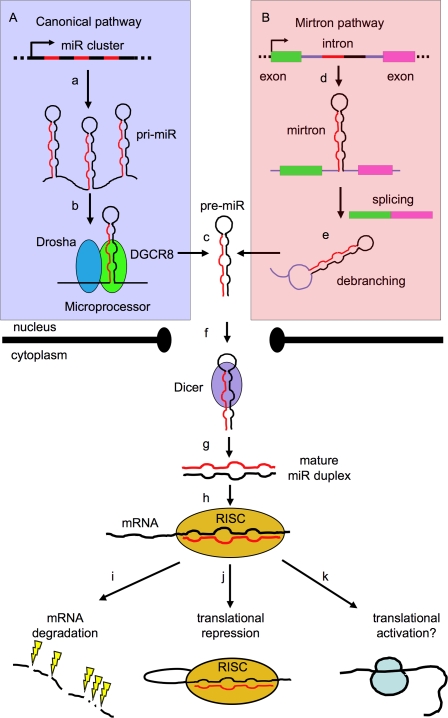
We assume the reader is familiar with the basic events of miR biogenesis and summarize them only briefly in Figure 1. Essentially, they are small 21- to 23-nucleotide (nt) noncoding RNAs capable of modulating mRNA expression by base pair interaction with mRNAs (mostly within their 3′ untranslated regions [UTRs]) in the context of a miR-containing ribonucleoprotein (miRNP) complex. In many instances, their expression is tissue-specific and developmentally regulated. miRs number >700 in the human genome, and >60% of human coding genes are predicted to have miR target sites in their 3′UTRs. Bioinformatic and experimental approaches suggest that any given miR can have hundreds of targets (Friedman et al. 2009). The simplest view of the miR pathway is that, directed by the tissue-specific profile of TFs, it constitutively produces a cellular “miR-ome,” which fine-tunes the protein output of the transcriptome (Selbach et al. 2008). In this view, the regulatory power of the system is seen mainly in terms of the combinatorial flexibility afforded by the potential of any given miR to target multiple mRNAs, and that a 3′UTR can contain several miR target sites, allowing for extensive coregulation of transcript sets. However, recent research is highlighting the less widely recognized fact that the miR pathway itself is subject to post-transcriptional regulation on multiple levels. For example, Drosha has been found in two complexes: a smaller one with DGCR8, and a larger one with other proteins, including RNA-binding proteins, RNA helicases, and the Ewing's sarcoma family of proteins (Gregory et al. 2004). In the cytoplasm, Dicer, the core ribonuclease responsible for generating miR duplexes from pre-miRs, interacts with, among others, the TAR RNA-binding protein (TRBP) (Chendrimada et al. 2005), originally characterized in relation to the HIV life cycle (Gatignol et al. 1991), and the PKR-activating protein (PACT) (Y Lee et al. 2006). A number of miRNP effector complexes (of which the RNA-induced silencing complex [RISC] found in Drosophila is the best characterized) have been identified, but their exact structure is still unknown. Proteomic analyses have identified a considerable number of Argonaute-interacting proteins (Hock et al. 2007; Landthaler et al. 2008). Therefore, each level of miR biogenesis can undergo protein–protein interactions that offer multiple regulatory options, and RISCs constitute a versatile mRNA-regulating platform that uses miRs (or other small RNAs) to direct several different types of regulatory responses.
Figure 1.
Biogenesis of miRs. (A) The canonical pathway is shown boxed in violet. Most miR promoters characterized to date are typical of RNA polymerase II transcribed genes (Lee and Dutta 2009), and give rise to a 5′-capped, spliced, and polyadenylated primary precursor (pri-miR) (a), formed by one or several concatenated hairpin structures (Lee et al. 2002, 2004; Altuvia et al. 2005) consisting of a stem (containing a miR) and a terminal loop. The pri-miR is recognized cotranscriptionally in the nucleus by the Microprocessor complex (b), whose catalytic core (formed by the Drosha and DGCR8 proteins) processes the base of the stem of the pri-miR and trims away the flanking sequences to release an intermediate stem–loop structure (known as pre-miR) of ∼70 nt (c). (B) A second, noncanonical pathway is shown boxed in pink. (d) Some animal miRs (“mirtrons”) are embedded in mRNA introns and completely bypass processing by Microprocessor. (e) Instead, they are processed by splicing and debranching (Okamura et al. 2007). In both cases, the resulting pre-miR is transported to the cytoplasm (via the exportin–RanGTP pathway) (f) (Yi et al. 2003), where the loop is further processed by the Dicer complex to release a mature miR duplex (g), which is finally incorporated into the RISC (h) (Hammond et al. 2000). The RISC recognizes the miR duplex, unwinds it, selects the miR strand while degrading the passenger strand, and mediates recognition of the target mRNA. The downstream regulatory effect is degradation of the mRNA (the prevalent mechanism in mammalian cells (i) (Guo et al. 2010) or its translational repression (j) (Mourelatos et al. 2002; Pratt and MacRae 2009), although instances of up-regulation have also been reported (k) (Vasudevan et al. 2007).
Adequate levels of both components of Microprocessor (Drosha and DGCR8) are ensured by a positive–negative feedback loop in which DGCR8 stabilizes Drosha and Drosha down-regulates the DGCR8 mRNA by targeting two hairpin structures in the 5′UTR and early coding sequence of DGCR8 that resemble those found in pri-miRs (Han et al. 2009). Many pri-miRs accumulate without being efficiently processed, until specific developmental or environmental cues arise (Thomson et al. 2006). Such control of Microprocessor activity is determined by interaction with several Drosha-binding partners. For instance, down-regulation of p68 or p72, two DEAD-box RNA helicases that act as cofactors of Drosha, results in reduction of a specific set of miRs (Fukuda et al. 2007). Mediated through p68 and/or p72, several signaling pathways regulate Microprocessor activity with effects on the miR-ome that can be global or more limited to few or even single miRs. In ovariectomized mice, exogenous estradiol binds to the nuclear estrogen receptor ERα and leads to a global p68/p72-dependent down-regulation of Microprocessor activity in the uterus (Yamagata et al. 2009). Cellular stress leads to interaction of the tumor suppressor p53 with Drosha and p68, resulting in enhanced processing of miR-16-1 and miR-143 (Suzuki et al. 2009). Similarly, in smooth muscle cells, BMP4 or TGF-β signaling results in interaction of SMADs with p68 and increased processing of pri-miR21 and pri-miR199a (Davis et al. 2008).
In addition, processing along the miR pathway is dependent on the miR precursor substrates being available to the processing complexes; binding of other RNA-binding proteins to these substrates could therefore modulate maturation. For example, the RNA-binding protein Lin28 has been suggested to inhibit processing of pri-let7 by binding to a conserved sequence in the precursor loop (Viswanathan and Daley 2010). Another RNA-binding protein, KSRP, binds to a different sequence on the loop of pri-let7 (and other miR precursors), resulting in enhanced processing. A regulatory mechanism has been proposed in which both proteins regulate pri-let7 processing by competing for binding, as both binding sites are close enough to cause steric hindrance (Trabucchi et al. 2009). While in principle such a mechanism seems plausible, it still awaits direct experimental confirmation. The exportin pathway is known to be rate-limiting, but no major regulatory event has yet been detected (Yi et al. 2005).
In the cytoplasm, the MAPK/ERK pathway can promote miR maturation by stabilizing Dicer through phosphorylation of TRBP (Paroo et al. 2009). Mature let7 is highly expressed in differentiated cells, and let7 target sites have been found in Dicer mRNA, suggesting that Dicer activity might be dampened to a certain extent in differentiated cells (Forman et al. 2008). In summary, increasing evidence points to the fact that the miR pathway itself is inherently flexible and subject to regulation at multiple stages.
The stem cell clockworks
The ESC phenotype is supported by a molecular program formed by a specific collection of TFs, signaling pathways, chromatin modifiers, and noncoding RNAs. The ongoing study of ESCs has revealed certain general characteristics, including a hierarchical transcriptional network and a particular cell cycle profile.
The Oct4–Sox2–Nanog triumvirate rules the ESC transcriptional hierarchy
A substantial body of literature accumulated since the discovery of the TF Oct4 in 1989 (Scholer et al. 1989) has clearly determined that the TFs Oct4, Sox2, and Nanog act coordinately and are central to the establishment and maintenance of the ESC regulatory program, and has been demonstrated dramatically in the context of direct reprogramming of somatic cells to induced pluripotent stem cells (iPSCs). Reprogramming was first achieved using a group of four TFs: Oct4, Sox2, Klf4, and c-Myc (Takahashi and Yamanaka 2006). Oct4, Sox2, Nanog, and Lin28 (Yu et al. 2007) formed a second group of four TFs also used to reprogram fibroblasts to iPSCs. Research in the last 3 years has shown that certain cell types can be reprogrammed with fewer factors: Human cord blood progenitors can be reprogrammed with only Oct4 and Sox2 (Giorgetti et al. 2009), and neural stem cells can be reprogrammed with only Oct4 (Kim et al. 2009). The current thinking is that the number and identity of the reprogramming proteins can vary depending on which factors are initially expressed—and at what level—in the starting cell type. However, while the central role of the Oct4–Sox2–Nanog triumvirate is widely accepted, the regulatory network of pluripotency involves other players whose level of expression also influence the self-renewal state, such as Esrrb, Zfx, Klf4, c-Myc, STAT3, and Ronin, among others (Niwa et al. 1998; Cartwright et al. 2005; Ivanova et al. 2006; Galan-Caridad et al. 2007; Chen et al. 2008; Dejosez et al. 2008; Jiang et al. 2008).
The “modus operandi” of the triumvirate involves three levels of action (Fig. 3, below). First, the trio forms a positive autoregulatory circuit in which each member binds to (and activates) its own promoter as well as to the promoters of the two other members of the group (Boyer et al. 2005), resulting in maintenance of high levels of expression of all three TFs (Bosnali et al. 2009). Second, the interaction of specific members of the trio with a small number of other TFs regulates crucial early development fate decisions. For example, the interaction of Oct4 and the caudal-type homeodomain TF Cdx2 determines the choice between inner cell mass fate and trophoectoderm fate (Niwa et al. 2005), and, similarly, the interplay between Nanog and the TFs GATA4 and GATA6 regulates the switch to the primitive endoderm fate (Fujikura et al. 2002; Capo-Chichi et al. 2005). Third, mapping of genomic binding sites (by chromatin immunoprecipitation followed by sequencing) of the three factors in mouse and human ESCs indicates that, despite some species-specific differences, they co-occupy the regulatory regions of hundreds of genes (Boyer et al. 2005; Loh et al. 2006) divided in two sets. The first set is transcriptionally active in ESCs and includes ESC-specific TFs, chromatin modifiers, and components of stem cell-specific signaling pathways. The second set is silenced in ESCs and comprises a number of TFs involved in differentiation and lineage commitment. Their silent state is explained by the fact that many of the genes of this set are also co-occupied by members of the polycomb group (PcG) proteins (Bernstein et al. 2006; TI Lee et al. 2006); PcG proteins are involved in the formation of at least two distinct polycomb repressor complexes (PRC1 and PRC2–PRC3), which ultimately result in chromatin condensation and epigenetic silencing (Schuettengruber et al. 2007). While this setup (up-regulation of stem cell functions alongside repression of differentiation functions) for maintaining the ESC state makes sense, how the system determines what genes belong to which set, how PcG proteins are recruited to the proper sites, and the molecular details of how the regulatory network changes when differentiation cues arise are currently far from clear. It must also be kept in mind that this relatively straightforward view of the transcriptional regulation of ESCs is a consequence of limiting the analysis to just three TFs. When mapping of genomic binding sites is extended to other ESC-specific TFs, the complexity increases significantly. As an example, one such analysis done on 10 TFs in addition to Oct4, Sox2, and Nanog has described that the number of genomic binding sites for the 13 TFs varied roughly between 1000 and 40,000; a total of 3586 genomic sites were co-occupied by four or more TFs, with an extreme example being provided by the Oct4 distal enhancer, which was bound by 11 TFs. When analyzing the collection of genomic sites binding multiple TFs, four major combinations of TFs that tended to be found together could be discerned. Furthermore, an analysis of the distribution of each of the TFs among these four major combinations revealed that, while each of them showed clear “preferences,” all of them could be found in all groups with significant frequencies (Chen et al. 2008). Therefore, while certain general correlations are indeed evident, the highly combinatorial nature of TF binding to regulatory sequences across the genome underscores the fact that our understanding of the regulatory network governing ESC transcriptional regulation is only beginning.
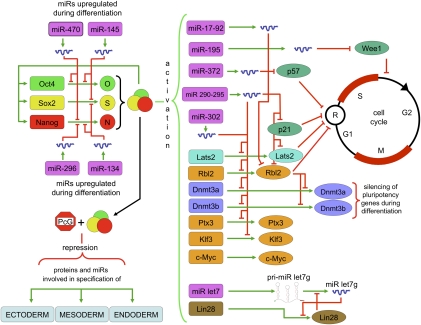
Figure 3.
Regulatory network of ESCs integrating miRs and proteins controlled by the Oct4–Sox2–Nanog trio of TFs. Genes are represented by boxes and proteins are represented by ovals or circles. Green arrows represent activation. Red lines represent repression. Proteins and genes are color-coded according to class/function: miRs are in purple, cell cycle regulators are in dark green, transcriptional regulators are in orange, DNA methytransferases are in violet, miR regulators are in brown, and signaling molecules are in light blue. Oct4, Sox2, and Nanog are depicted in green, yellow, and red, respectively.
Undifferentiated ESCs display a particular cell cycle profile
In striking contrast to differentiated somatic cells, in which the regulators of the cell cycle fluctuate periodically, mouse ESCs (mESCs) show stable and high levels of activators of the cell cycle (high activity of cdk2–cyclin E/A and cdk6–cyclin D3) and an absence of cell cycle inhibitors (cdk inhibitors p21cip and p27Kip1, and the INK4a family member p16INK4a) (Faast et al. 2004; Becker et al. 2010). In somatic cells, transition from G1 to S phase requires overcoming the G1-to-S restriction (R) checkpoint, where the cell cycle becomes independent of external growth factors and is determined by the inactivation of the pRb protein followed by the subsequent release of E2F factors (Blagosklonny and Pardee 2002). In mESCs, pRb is permanently inactivated by hyperphosphorylation, leading to constitutive activity of E2F TFs, which in turn allows an R-point-independent short transition through G1 phase (Savatier et al. 1994; Stead et al. 2002). Although differences were observed when compared with mESCs (Conklin and Sage 2009), similar mechanisms are presumed to regulate human ESCs (hESCs). Indeed, the ability to support at least two independent rounds of cell division in the absence of external growth factors suggests the existence of autocrine mechanisms supporting R-point-independent cell cycling in hESCs (Becker et al. 2010). Hence, in contrast to somatic cells, which depend on mitogenic signaling to proceed through the R point, ESCs proliferate in a mitogen-independent manner, leading to a short G1 phase. It is thought that ESCs initiate differentiation during G1 phase, which constitutes a “window of opportunity” in which, for example, a developmental signal can accumulate until it surpasses the threshold level that triggers a differentiation pathway. Therefore, control of the G1-phase length becomes a way to control the gateway to differentiation (Savatier et al. 1996; Burdon et al. 2002; Jirmanova et al. 2002). This concept is re-enforced by the fact that pRb, depending on the specific cellular context, can also act as a transcriptional activator or repressor of genes that function as master inducers of differentiation. pRb has been shown recently to have a clear role in mesenchymal stem cell choice between the adipocyte versus osteogenic fate; in this instance, lack of pRb biases the choice toward the adipocyte fate and, furthermore, can restore committed preosteoblasts to a progenitor multipotent state (Calo et al. 2010). However, a direct mechanistic link between cell cycle and differentiation is still lacking.
The role of miRs in ESC cell cycle control
Insight into the role of these ESC-specific miRs has been gained through the analysis of Dicer-null and DGCR8-null ESC lines. In both cases, the lines are viable, completely lack their repertoire of mature canonical miRs, and show similar phenotypes. Both cell lines maintain expression of pluripotency markers and proliferate slowly compared with wild-type ESCs. They also fail to efficiently down-regulate pluripotency markers and up-regulate differentiation markers when induced to differentiate. However, the phenotypes are not identical. Upon deletion of both Dicer alleles, mESCs experience a complete proliferation block (Kanellopoulou et al. 2005; Murchison et al. 2005) and resemble the phenotype resulting from knockout of all four Argonaut family members (Su et al. 2009). Continued culture of the Dicer-null cells eventually gives rise to clones that proliferate at rates comparable with that of DGCR8-null ESCs (Murchison et al. 2005). In contrast, DGCR8-null cells do not show this complete initial block of proliferation (Wang et al. 2007). Lack of Dicer, but not DGCR8, has been reported to result in heterochromatin instability (Kanellopoulou et al. 2005), but this result failed to be reproduced by a different research group (Murchison et al. 2005). Dicer has also been implicated in telomere maintenance and DNA methylation (Benetti et al. 2008), while DGCR8 has not yet been studied in this regard. Some of these differences might be explained by the involvement of Dicer in the maturation of non-Microprocessor-dependent endogenous siRNAs and other small RNAs (Babiarz et al. 2008). Therefore, the role of miRs in ESCs is better illustrated in the DGCR8-null lines. Cell cycle analysis of DGCR8−/− cells revealed that they accumulated in the G1 phase of the cell cycle, indicating a defect in the G1-to-S-phase transition. A screen testing for the effect of individually reintroducing 461 miRs on proliferation of DGCR8-null cells showed that the defect could be rescued by 14 different miRs. These rescuing miRs belong to several different families of miRs (mainly the miR-290, miR-302, and miR-17–92 clusters) that are highly expressed in ESCs and down-regulated upon differentiation. They were collectively called ESCC (for ESC-specific cell cycle-regulating) miRs, and, significantly, they shared the same or a very similar seed sequence, suggesting that they were redundantly directed against the same targets. A search for these targets uncovered p21cip, the Retinoblastoma-like 2 protein (Rbl2), and Lats2, all previously known inhibitors of the cyclinE/cdk2 pathway, which regulates the G1/S transition. ESCC miRs ensure rapid progression through the R point by down-regulating these inhibitors and therefore increasing cyclinE/cdk2 activity (Wang et al. 2008; Smith et al. 2010). At least one ESCC family member, miR-106, has been confirmed to induce cell cycle progression by inhibition of p21cip independently (Ivanovska et al. 2008). These results were consistent with a previous report showing that p21cip protein levels (but not mRNA levels) increased upon differentiation of ESCs (Sabapathy et al. 1997). Significantly, other miRs have been identified in similar roles. miR-372 and miR-92b (abundantly expressed in hESCs) target p21cip and p57 (another inhibitor of G1/S progression), respectively, and miR-195 has been shown to down-regulate the G2–M checkpoint inhibitory kinase WEE1, an inhibitor of the G2 cyclin B–Cdk complex (Qi et al. 2009; Sengupta et al. 2009). These results clearly show that, in ESCs, miRs redundantly counteract inhibitors of the cell cycle (Fig. 3, below), effectively removing its brakes and playing a crucial role in the establishment and maintenance of the peculiar ESC cell cycle profile.
The role of miRs in ESC differentiation
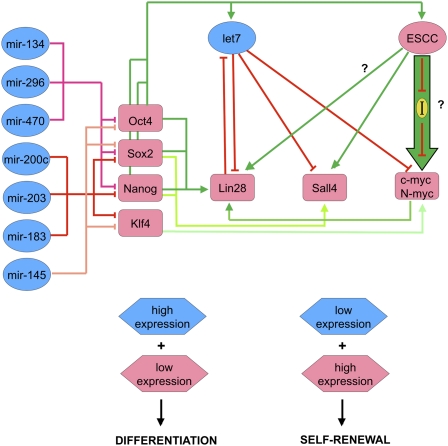
The molecular mechanism underlying the inability of ESCs lacking miRs to efficiently silence pluripotency markers upon differentiation has been investigated in differentiating Dicer-null ESCs (Benetti et al. 2008; Sinkkonen et al. 2008). Two groups identified Rbl2, a transcriptional repressor, as a target of the miR-290 miR family, and found these cells to have decreased levels of DNA methyltransferases Dnmt3a and Dnmt3b, involved in de novo DNA methylation. The de novo DNA methylation activity could be restored by exogenous expression of de novo methyltransferases or reintroduction of miR-290 family members. Considering a previous study (Feldman et al. 2006) had established that silencing of pluripotency markers requires de novo methylation, Benetti et al. (2008) and Sinkkonen et al. (2008) suggest that absence of the miR-290 family leads to up-regulation of Rbl2, which transcriptionally represses de novo DNA methyltransferases and results in the observed inability of Dicer−/− cells to silence the pluripotency markers and differentiate. However, other miRs are also required to turn over key pluripotency proteins for differentiation to proceed (Fig. 2; Wang et al. 2008). Loss-of-function and gain-of-function experiments have shown that, during differentiation, mESCs up-regulate miR-134, miR-296, and miR-470, which target and down-regulate Nanog, Oct4, and Sox2 (Tay et al. 2008); similarly, miR-200c, miR-203, and miR-183 repress Sox2 and Klf4 (Wellner et al. 2009). In differentiating hESCs, miR-145 represses Oct4, Sox2, and Klf4 (Xu et al. 2009).
Figure 2.
Regulatory network illustrating the role of miRs in ESC differentiation. Proteins are in boxes, and miRs are in ovals. Pro-self-renewal elements are in pink, and prodifferentiation elements are in blue. Green-shaded arrows represent activating interactions, and red-shaded lines represent repressive interactions. See the text.
Another example involves the let7 family and illustrates how the transition from the undifferentiated to differentiated state can be influenced by an intricate interplay between different miR families. pri-let7 is transcribed in ESCs (Thomson et al. 2006; Wulczyn et al. 2007), processed to pre-let7, and exported to the cytoplasm (Rybak et al. 2008). However, mature let7 family members are essentially absent in ESCs and accumulate only upon ESC differentiation, eventually ending up broadly expressed in differentiated cell types (Viswanathan et al. 2008). This observation prompted the hypothesis that the let7 family could be involved in shutting down the pluripotency program upon differentiation (Melton et al. 2010). Transfection of let7 into DGCR8−/− cells rescued the differentiation defect, allowing the cells to shut down the self-renewal program more efficiently. However, transfection of let7 into DGCR8 wild-type cells (or into DGCR8-null cells that received ESCC miR family members along with let7) had no effect on expression of pluripotency genes. This led to a model in which the ESCC and let7 family members oppose each other, with ESCC favoring the pluripotency state and the let7 family opposing it (Fig. 2; Melton et al. 2010). Microarray analysis revealed that the mechanism underlying this effect was mediated not only by the effect of each miR family on mRNAs bearing their target binding sites, but also by opposing effects on both families of miRs on the TFs c-Myc and n-Myc. ESCC miR family members up-regulate c-Myc through a still unknown mechanism, presumably by down-regulating a c-Myc inhibitor. In certain cellular contexts, members of the let-7 family directly target and down-regulate c-Myc (Kumar et al. 2007). In ESCs, the effect of let7 on c-Myc is smaller, but it does strongly down-regulate n-Myc, and consequently the let7 family down-regulates the set of mRNA transcripts that are under positive transcriptional regulatory control by c-Myc and n-Myc. Other transcripts are also found to be subject to this “tug of war,” and, among them, two stand out significantly. One is Sall4, a TF involved in pluripotency. The second is Lin28, which is highly expressed in ESCs and is down-regulated upon differentiation (Newman et al. 2008; Viswanathan et al. 2008). Lin 28 has been found to bind to pre-let7 and promote its polyuridylation, resulting in both an inhibition of Dicer activity and targeting of the pre-let7 for degradation (Heo et al. 2008). Lin28 also regulates other miRs (miR-107, miR-143, and miR-200c) similarly (Heo et al. 2009). The Lin28 promoter is co-occupied by the Oct4–Sox2–Nanog trio as well as by Tcf3 (Marson et al. 2008), suggesting that it is under their transcriptional control. Lin28 depletion does not, in isolation, cause cells to differentiate. However, as differentiation starts, the levels the Oct4–Sox2–Nanog trio begin to fall, possibly leading to down-regulation of Lin28. This would allow accumulation of mature let7 and further down-regulation of the expression of Lin28 by binding directly to its mRNA (Reinhart et al. 2000; Rybak et al. 2008). Once a critical threshold is surpassed, the let7 family dominates the ESCC family and the transition is stabilized. Hence, the Lin28/let7 interaction provides a degree of robustness to the differentiation switch (Melton et al. 2010). These results highlight how miRs can be used at several different levels to quickly and coordinately turn over the key regulatory proteins of a given cell phenotype to facilitate the establishment of a new one (Fig. 2).
Integration of miRs into the basic molecular circuit of pluripotency
Integration of miR expression into the network governing pluripotency has required mapping of miR promoters and analysis of their occupancy by a tetrad of key pluripotency TFs (Oct4, Sox2, Nanog, and Tcf3) (Marson et al. 2008). Approximately 20% of all known miRs are under the control of this tetrad of TFs, and these miRs can be divided into two sets: one comprised of miRs active in ESCs and whose promoters are occupied by the tetrad (many of which have already been implicated in pluripotency maintenance, such as the ESCC miR group and the let7 family), and a second set of miRs inactive in ESCs (but up-regulated in differentiated cells) whose promoters are occupied by the tetrad and PcG-repressive complexes. Other reports have established direct connections between expression of ESC-specific miRs and the Myc family of TFs. Both c-Myc and N-Myc have been shown to bind to the promoter of the ESC-specific miR-290 cluster (Chen et al. 2008); c-Myc has also been demonstrated to activate the expression of several ESC-specific miRs (Card et al. 2008; Lin et al. 2009; Smith et al. 2010), and, in turn, miR-294 can indirectly activate the expression of c-Myc (Melton et al. 2010). Therefore, the general strategy of transcriptional control used by the ESC regulatory state applies equally to protein and miR-encoding genes (Fig. 3).
The role of miRs in reprogramming
The two basic results on which the field of direct reprogramming is founded was the discovery that fibroblasts can be reprogrammed to iPSCs by retroviral-mediated delivery of two groups of ESC regulators: Oct4, Sox2, Klf4, and c-Myc on one hand (Takahashi and Yamanaka 2006), and Oct4, Sox2, Nanog, and Lin28 on the other (Yu et al. 2007). Recently, a number of reports have highlighted a role for miRs in reprogramming. Several of the members of the reprogramming cocktails have “miR connections.” As mentioned above, Oct4, Sox2, Nanog, and c-Myc have been found to control the expression of ESC-specific miR families, with Oct4, Sox2, and Nanog inducing the expression of the miR-290 and miR-302 clusters (Barroso-delJesus et al. 2008; Card et al. 2008; Marson et al. 2008) and c-Myc inducing the miR-17–92 cluster (O'Donnell et al. 2005). c-Myc can also repress let7 family members indirectly through up-regulation of Lin28B (Chang et al. 2009), while Lin28 controls and is controlled by the let7 family (John et al. 2004; Heo et al. 2008; Newman et al. 2008; Rybak et al. 2008). Interestingly, transient overexpression of ESC-specific miRs could replace c-Myc when reprogramming fibroblasts with Oct4, Sox2, and Klf4, with miR-294 increasing iPSC derivation efficiencies by ∼20 fold (Judson et al. 2009), but adding both c-Myc and miR-294 at the same time had no effect, suggesting that one reason behind the enhancing effect of c-Myc on reprogramming is the induction of ESC-specific miRs. There has also been one report showing that overexpression of miR-302 alone can reprogram human cancer cells to the iPSC state (Lin et al. 2008), and, recently, this observation has been extended to human hair follicle cells (Lin et al. 2010). The opposing effect of the ESCC and let7 miR families prompted testing of the hypothesis that down-regulating the let7 family could increase reprogramming efficiency of fibroblasts, and it was found that, indeed, inhibition of let7 by means of an antisense inhibitor could enhance reprogramming efficiency when using Oct4, Sox2, and Klf4, regardless of whether c-Myc was added to the mix (Melton et al. 2010).
Another instance in which miRs have been implicated with reprogramming relates to the question of whether iPSCs are identical (i.e., completely reprogrammed) to ESCs. This question is still being debated, but a recent finding has been that, when mESCs and iPSCs from identical genetic backgrounds are compared, the transcriptional profile is extremely similar, with the exception of one locus: Dlk1–Dio3 (Stadtfeld et al. 2010). This locus is paternally imprinted, and therefore the genes it encodes are expressed from the maternal allele in ESCs. Interestingly, a majority of iPSCs failed to reactivate the maternal allele, and the reactivation status of the Dlk1–Dio3 locus in iPSCs was shown to be correlated with the ability of these clones to give rise to live mice by tetraploid complementation; lack of reactivation resulted in embryonic lethality. Of note, the Dlk1–Dio3 locus encodes ∼50 miRs, and 18 of them were expressed in ESCs but not iPSCs. However, the evidence suggests that the miRs of the Dlk1–Dio3 locus are involved in embryonic development and are not part of the self-renewal network of pluripotency.
Transitions between cell states
Transitions between cell states can involve generation of intermediate states. Two recent studies (Li et al. 2010; Samavarchi-Tehrani et al. 2010) suggest that the earliest state of reprogramming is remarkably similar to a mesenchymal-to-epithelial transition (MET), showing that initiation of reprogramming is marked by down-regulation of mesenchymal markers such as Snail and N-Cadherin and up-regulation of epithelial markers such as E-cadherin and Epcam. Overexpression of Snail- or RNAi-mediated down-regulation of E-cadherin—both events known to inhibit MET—substantially reduces iPSC formation. Overexpression of miR-200 and miR-205 (which had been shown previously to down-regulate mesenchymal genes in the context of MET) in fibroblasts accelerated the up-regulation of MET-related genes compared with the control (Li et al. 2010; Samavarchi-Tehrani et al. 2010). These results underscore the concept that transitions between cell states can be driven by down-regulating the molecular support for the initial state while up-regulating the molecular support for the final state, regardless of whether the support itself is TF-based, miR-based, or both.
Conclusions
miRs are important components of the regulatory network that governs ESCs. The study of gene regulation in many different systems has led to the observation that certain regulatory motifs are found repeatedly. The study of their structure has allowed certain basic types to be defined: Examples of these motifs are positive or negative feedback loops and coherent or incoherent feed-forward loops, where a small number of molecules (proteins or RNAs) are functionally related to each other in a way that forms an operational unit with predictable behavior (Alon 2007). A gene regulatory network can be seen as a complex structure formed by the association of many such operational units. In many cases, miRs are structural components of these regulatory motifs, and their absence can have major consequences on the behavior of the regulatory network they are a part of. The concept that miRs are central to the ESC phenotype is highlighted by the simple observation that ESCs that lack miRs cease, in fact, to be stem cells. Indeed, the definition of an ESC is an operational one: Above all, a stem cell must self-renew; i.e., it must be capable of ongoing cell division in vitro while retaining the ability to differentiate to all cell types of the adult organism. The analysis of the phenotype of DGCR8- or Dicer-null ESCs clearly shows that they no longer fulfill this requirement, as lack of miRs results in the cells remaining trapped in a state of ongoing cell division. When induced to differentiate, they fail to turn off the pluripotency regulatory program. Turning off pluripotency and switching to differentiation are aspects of a single molecular network, which cannot function normally in the absence of miRs. It would not be surprising to find novel ways in which miRs are integrated into (and necessary for) the normal function of the self-renewal regulatory program.
Acknowledgments
Work in the laboratory of J.C.I.B. was supported by grants from the Cellex and G. Harold and Leila Y. Mathers Charitable Foundations, CIBER, MICINN, and Sanofi-Aventis.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1982910.
References
- Alon U 2007. Network motifs: Theory and experimental approaches. Natl Rev 8: 450–461 [DOI] [PubMed] [Google Scholar]
- Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H 2005. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res 33: 2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-delJesus A, Romero-Lopez C, Lucena-Aguilar G, Melen GJ, Sanchez L, Ligero G, Berzal-Herranz A, Menendez P 2008. Embryonic stem cell-specific miR302-367 cluster: Human gene structure and functional characterization of its core promoter. Mol Cell Biol 28: 6609–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS 2010. Human embryonic stem cells are pre-mitotically committed to self-renewal and acquire a lengthened G1 phase upon lineage programming. J Cell Physiol 222: 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, et al. 2008. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol 15: 268–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV, Pardee AB 2002. The restriction point of the cell cycle. Cell Cycle 1: 103–110 [PubMed] [Google Scholar]
- Bosnali M, Munst B, Thier M, Edenhofer F 2009. Deciphering the stem cell machinery as a basis for understanding the molecular mechanism underlying reprogramming. Cell Mol Life Sci 66: 3403–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T, Smith A, Savatier P 2002. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol 12: 432–438 [DOI] [PubMed] [Google Scholar]
- Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, Lees JA 2010. Rb regulates fate choice and lineage commitment in vivo. Nature 466: 1110–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-Chichi CD, Rula ME, Smedberg JL, Vanderveer L, Parmacek MS, Morrisey EE, Godwin AK, Xu XX 2005. Perception of differentiation cues by GATA factors in primitive endoderm lineage determination of mouse embryonic stem cells. Dev Biol 286: 574–586 [DOI] [PubMed] [Google Scholar]
- Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK 2008. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol 28: 6426–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S 2005. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 132: 885–896 [DOI] [PubMed] [Google Scholar]
- Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, et al. 2009. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci 106: 3384–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin JF, Sage J 2009. Keeping an eye on retinoblastoma control of human embryonic stem cells. J Cell Biochem 108: 1023–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A 2008. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454: 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, Songyang Z, Thomson JA, Zwaka TP 2008. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell 133: 1162–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faast R, White J, Cartwright P, Crocker L, Sarcevic B, Dalton S 2004. Cdk6-cyclin D3 activity in murine ES cells is resistant to inhibition by p16(INK4a). Oncogene 23: 491–502 [DOI] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y 2006. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol 8: 188–194 [DOI] [PubMed] [Google Scholar]
- Forman JJ, Legesse-Miller A, Coller HA 2008. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci 105: 14879–14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura J, Yamato E, Yonemura S, Hosoda K, Masui S, Nakao K, Miyazaki Ji J, Niwa H 2002. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev 16: 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, et al. 2007. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol 9: 604–611 [DOI] [PubMed] [Google Scholar]
- Galan-Caridad JM, Harel S, Arenzana TL, Hou ZE, Doetsch FK, Mirny LA, Reizis B 2007. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell 129: 345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatignol A, Buckler-White A, Berkhout B, Jeang KT 1991. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science 251: 1597–1600 [DOI] [PubMed] [Google Scholar]
- Giorgetti A, Montserrat N, Aasen T, Gonzalez F, Rodriguez-Piza I, Vassena R, Raya A, Boue S, Barrero MJ, Corbella BA, et al. 2009. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell 5: 353–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN 2009. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN 2008. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 32: 276–284 [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708 [DOI] [PubMed] [Google Scholar]
- Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G 2007. Proteomic and functional analysis of Argonaute-containing mRNA–protein complexes in human cells. EMBO Rep 8: 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR 2006. Dissecting self-renewal in stem cells with RNA interference. Nature 442: 533–538 [DOI] [PubMed] [Google Scholar]
- Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, et al. 2008. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol 28: 2167–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH 2008. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol 10: 353–360 [DOI] [PubMed] [Google Scholar]
- Jirmanova L, Afanassieff M, Gobert-Gosse S, Markossian S, Savatier P 2002. Differential contributions of ERK and PI3-kinase to the regulation of cyclin D1 expression and to the control of the G1/S transition in mouse embryonic stem cells. Oncogene 21: 5515–5528 [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS 2004. Human MicroRNA targets. PLoS Biol 2: e363 doi: 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R 2009. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 27: 459–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Greber B, Arauzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Scholer HR 2009. Direct reprogramming of human neural stem cells by OCT4. Nature 461: 649–653 [DOI] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T 2007. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 39: 673–677 [DOI] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T 2008. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 14: 2580–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Dutta A 2009. MicroRNAs in cancer. Annu Rev Pathol 4: 199–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN 2002. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J 21: 4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN 2006. The role of PACT in the RNA silencing pathway. EMBO J 25: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. 2010. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7: 51–63 [DOI] [PubMed] [Google Scholar]
- Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, Ying SY 2008. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 14: 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN 2009. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J 28: 3157–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Chang DC, Lin CH, Ying SY, Leu D, Wu DTS 2010. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res doi: 10.1093/nar/gkq850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440 [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R 2010. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G 2002. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev 16: 720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ 2005. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci 102: 12135–12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM 2008. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev 12: 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J 2005. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123: 917–929 [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843 [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroo Z, Ye X, Chen S, Liu Q 2009. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell 139: 112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt AJ, MacRae IJ 2009. The RNA-induced silencing complex: A versatile gene-silencing machine. J Biol Chem 284: 17897–17901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Yu JY, Shcherbata HR, Mathieu J, Wang AJ, Seal S, Zhou W, Stadler BM, Bourgin D, Wang L, et al. 2009. microRNAs regulate human embryonic stem cell division. Cell Cycle 8: 3729–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906 [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG 2008. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 10: 987–993 [DOI] [PubMed] [Google Scholar]
- Sabapathy K, Klemm M, Jaenisch R, Wagner EF 1997. Regulation of ES cell differentiation by functional and conformational modulation of p53. EMBO J 16: 6217–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL 2010. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7: 64–77 [DOI] [PubMed] [Google Scholar]
- Savatier P, Huang S, Szekely L, Wiman KG, Samarut J 1994. Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene 9: 809–818 [PubMed] [Google Scholar]
- Savatier P, Lapillonne H, van Grunsven LA, Rudkin BB, Samarut J 1996. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene 12: 309–322 [PubMed] [Google Scholar]
- Scholer HR, Hatzopoulos AK, Balling R, Suzuki N, Gruss P 1989. A family of octamer-specific proteins present during mouse embryogenesis: Evidence for germline-specific expression of an Oct factor. EMBO J 8: 2543–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G 2007. Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Sengupta S, Nie J, Wagner RJ, Yang C, Stewart R, Thomson JA 2009. MicroRNA 92b controls the G1/S checkpoint gene p57 in human embryonic stem cells. Stem Cells 27: 1524–1528 [DOI] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W 2008. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol 15: 259–267 [DOI] [PubMed] [Google Scholar]
- Smith KN, Singh AM, Dalton S 2010. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell 7: 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K 2010. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465: 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead E, White J, Faast R, Conn S, Goldstone S, Rathjen J, Dhingra U, Rathjen P, Walker D, Dalton S 2002. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene 21: 8320–8333 [DOI] [PubMed] [Google Scholar]
- Su H, Trombly MI, Chen J, Wang X 2009. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev 23: 304–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K 2009. Modulation of microRNA processing by p53. Nature 460: 529–533 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I 2008. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455: 1124–1128 [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM 2006. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20: 2202–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG 2009. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459: 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA 2007. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 318: 1931–1934 [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ 2010. Lin28: A microRNA regulator with a macro role. Cell 140: 445–449 [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI 2008. Selective blockade of microRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R 2007. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39: 380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R 2008. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 40: 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. 2009. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 11: 1487–1495 [DOI] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R 2007. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J 21: 415–426 [DOI] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS 2009. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137: 647–658 [DOI] [PubMed] [Google Scholar]
- Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, Takeyama K, Minami Y, O'Malley BW, Kato S 2009. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell 36: 340–347 [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR 2005. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA 11: 220–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]