A temperature-sensitive chimeric transmembrane protein reveals a mechanism for disposing misfolded proteins that make it to the plasma membrane.
Abstract
Cellular protein homeostasis profoundly depends on the disposal of terminally damaged polypeptides. To demonstrate the operation and elucidate the molecular basis of quality control of conformationally impaired plasma membrane (PM) proteins, we constructed CD4 chimeras containing the wild type or a temperature-sensitive bacteriophage λ domain in their cytoplasmic region. Using proteomic, biochemical, and genetic approaches, we showed that thermal unfolding of the λ domain at the PM provoked the recruitment of Hsp40/Hsc70/Hsp90 chaperones and the E2–E3 complex. Mixed-chain polyubiquitination, monitored by bioluminescence resonance energy transfer and immunoblotting, is responsible for the nonnative chimera–accelerated internalization, impaired recycling, and endosomal sorting complex required for transport–dependent lysosomal degradation. A similar paradigm prevails for mutant dopamine D4.4 and vasopressin V2 receptor removal from the PM. These results outline a peripheral proteostatic mechanism in higher eukaryotes and its potential contribution to the pathogenesis of a subset of conformational diseases.
Introduction
Proteostatic mechanisms are fundamental in preventing the accumulation of misfolded, aggregation-prone, and potentially cytotoxic polypeptides that are generated by mutations, transcriptional and translational errors, or cellular stresses (Sherman and Goldberg, 2001; Arvan et al., 2002; Ellgaard and Helenius, 2003; Powers et al., 2009). This is achieved in part by protein quality control (QC) mechanisms that assist folding as well as eliminate terminally misfolded polypeptides (Ellgaard and Helenius, 2001; Powers et al., 2009). Molecular chaperones, in conjunction with cochaperones, shield exposed hydrophobic residues to suppress aggregation and promote folding of membrane proteins in the ER lumen and cytoplasm (Young et al., 2004). In addition, chaperone machines are involved in triage decision by recruiting chaperone-dependent ubiquitination machinery to irreversibly misfolded polypeptides. Pioneering works have uncovered that this process culminates in the ER-associated degradation of nonnative membrane proteins mediated by the ubiquitin (Ub) proteasome system (UPS) after the retrotranslocation of client proteins into the cytoplasm (Brodsky and McCracken, 1999; Cyr et al., 2002; Hampton, 2002; Ellgaard and Helenius, 2003; Hirsch et al., 2009).
Ubiquitination, a covalent posttranslational modification, is mediated by the coordinated function of the E1 Ub-activating enzymes and the combination of several E2 Ub-conjugating and hundreds of E3 Ub-ligating enzymes that confer substrate specificity. Ubiquitination catalyzes the attachment of either mono-, multiple mono–, or poly-Ub chains to client proteins (Hicke, 2001; Piper and Luzio, 2007). Poly-Ub chains are linked to one of the seven Lys residues on the acceptor Ub, endowing distinct structural characteristics that are recognized by Ub-binding adaptors (Dunn and Hicke, 2001; Katzmann et al., 2001; Pickart, 2001; Dikic et al., 2009; Mittal and McMahon, 2009). Proteasome-dependent degradation of misfolded polypeptides is primarily mediated by K48- and K11-linked Ub chains (Xu et al., 2009). The signal-dependent down-regulation and lysosomal-associated degradation of native plasma membrane (PM) receptors, however, are preferentially catalyzed by K63-linked Ub chains (Duncan et al., 2006; Barriere et al., 2007; Varghese et al., 2008; Boname et al., 2010).
Integral membrane proteins with limited conformational defects may escape the ER and are either retrieved from the Golgi compartment back to the ER or targeted for vacuolar/lysosomal proteolysis (Cole et al., 1998; Tsigelny et al., 2005; Wang and Ng, 2010). The latter process can be initiated from the Golgi compartment or from the PM (Wolins et al., 1997; Reggiori and Pelham, 2002; Ehrlich et al., 2009). Although rapid elimination of mutant PM proteins is attributed to conformational defects, the underlying structural perturbations remain poorly defined (Ljunggren et al., 1990; Li et al., 1999; Zaliauskiene et al., 2000; Benharouga et al., 2001; Gong and Chang, 2001; Sharma et al., 2001; Wilson et al., 2001; Fayadat and Kopito, 2003; Schaheen et al., 2009). Ubiquitination of a subset of membrane proteins (Pma-1, bile salt export pump [BSEP], cystic fibrosis transmembrane conductance regulator [CFTR], and Na/H exchanger [NHE6]) was proposed to play a determinant role in their rapid turnover at the PM (Gong and Chang, 2001; Sharma et al., 2004; Hayashi and Sugiyama, 2009; Roxrud et al., 2009). These observations, along with the discovery of the endosomal sorting complex required for transport (ESCRT)–dependent lysosomal degradation of ubiquitinated native cargo molecules (Katzmann et al., 2001; Raiborg and Stenmark, 2009), led to the proposition that the peripheral QC system can recognize, ubiquitinate, and eliminate nonnative polypeptides from the PM or endosomes in both yeast and mammalian cells (Arvan et al., 2002; Sharma et al., 2004). The molecular machinery of peripheral QC systems, however, remains unknown.
To assess the molecular consequences of unfolding at the PM, we designed a chimeric membrane protein with a temperature-sensitive folding defect. Unfolding induced the chaperone- and E2–E3-dependent polyubiquitination, internalization, and ESCRT-dependent lysosomal destruction of the chimera from the PM. Similar cellular and biochemical processes are involved in the mutant vasopressin V2 receptor (V2R) and dopamine D4.4 receptor (DRD4) disposal from the PM, uncovering the potential contribution of the peripheral QC to the pathogenesis of conformational diseases. A preliminary study of this work has been published in abstract form (Apaja, P., and G. Lukacs. 2009. Membrane protein quality control in post-Golgi compartments. Experimental Biology. Abstr. 668.3).
Results
PM model protein with a thermosensitive conformational defect
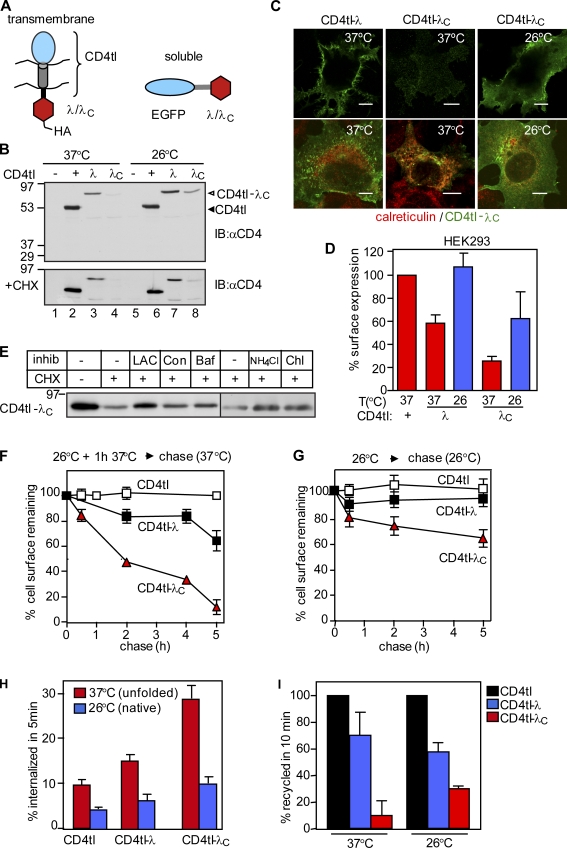
We designed a monotopic transmembrane model protein to study the cellular and biochemical consequences of misfolding at the PM. The C-terminally truncated CD4 incorporating a flexible cytoplasmic linker was used as a reporter molecule (CD4tl; Fig. 1 A) because it lacks a specific sorting signal and is constitutively targeted to the PM (Barriere et al., 2006). The N-terminal DNA-binding domain of the wild type (wt) or L57C mutant bacteriophage λ repressor was fused to the truncated CD4 (CD4tl-λ and CD4tl-λC, respectively; Fig. 1 A). The melting temperature (Tm) of the recombinant wt λ domain was ∼54°C. The Tm of the mutant containing the L57C substitution (λC) was decreased to ∼32°C (Parsell and Sauer, 1989). The two-state unfolding profile and the reduced Tm enabled us to shift the conformational equilibrium of the CD4tl-λC cytosolic domain from the largely native state at 26°C to a predominantly nonnative state at 37°C.
Figure 1.
Unfolding induced down-regulation of CD4tl-λC at the PM. (A) Schematic picture of membrane-tethered and soluble bacteriophage λ model proteins. (B) Immunoblot (IB) analysis of CD4tl and CD4tl-λ/λC expression before (top) or after CHX chase at 37°C for 1.5 h (bottom). Transiently transfected COS7 cells were cultured at 37°C or 26°C before the CHX chase. (C) Indirect immunostaining of thermally unfolded (37°C) or nativelike (26°C) CD4tl-λC in nonpermeabilized (top) or permeabilized (bottom) COS7 cells. Calreticulin was used as an ER marker. Bars, 10 µm. (D) The PM density of chimeras was determined by ELISA as described in Materials and methods. (E) Pharmacological characterization of rescued and then unfolded CD4tl-λC degradation as measured by CHX chase and immunoblotting. Lactacystine (LAC), concanamycin (Con), and bafilomycin A1 (Baf) were added simultaneously with CHX for 3.5 h. NH4Cl and chloroquin (Chl) were present during the last 2 h of the CHX chase. Equal amounts of proteins were loaded. (F and G) The PM stability of rescued and then unfolded model proteins at 37°C for 1.5 h was determined at 37°C (F) or in the absence of unfolding at 26°C (G) by ELISA. (H) Internalization of unfolded and rescued model proteins was monitored by Ab uptake at 37°C. Data are expressed as the percentage of initial Ab binding. (I) Recycling efficiency was determined by a biotin-streptavidin sandwich assay as described in Materials and methods and expressed as the percentage of CD4tl. Molecular mass is given in kilodaltons. The data shown represent means ± SEM.
The CD4tl as well as CD4tl-λ and CD4tl-λC were detected with the expected apparent molecular masses (∼55 and ∼75 kD, respectively) by immunoblotting with anti-CD4 antibody (Ab) in transiently transfected COS7 and stable Flp-In tetracycline-inducible HEK293 cells at 26°C (Fig. 1 B and Fig. S1 A). Attachment of the λ to the CD4tl decreased, whereas the fusion of the λC almost completely eliminated the expression of the respective chimera at 37°C (Fig. 1 B, top, lanes 2–4), suggesting that the mutant processing differs from the wt at the restrictive temperature. Indeed, the CD4tl-λC was predominantly confined to the ER at 37°C in COS7 and HEK293 cells as visualized by coimmunostaining and laser confocal fluorescence microscopy using calreticulin and calnexin as specific ER markers (Fig. 1 C and not depicted). The cell surface density of the CD4tl-λC was only ∼30% of the CD4tl when measured by the immunoperoxidase assay with anti-CD4 primary and HRP-conjugated secondary Ab (Fig. 1 D and Fig. S1 B). Rescuing the λC conformational defect at 26°C increased the CD4tl-λC cellular and PM expression (Fig. 1, B–D; and Fig. S1 B).
The temperature-rescued CD4tl-λC, but not the CD4tl-λ or the CD4tl expression, was eliminated upon inhibiting translation with cycloheximide (CHX) for 1.5 h at 37°C (Fig. 1 B, bottom, lanes 6–8). The CHX-induced disposal of CD4tl-λC was delayed by inhibitors of the proteasome (lactacystine) and lysosomal proteolysis (concanamycin, bafilomycin A, NH4Cl, and chloroquine), suggesting that the unfolded chimera is disposed by ER-associated and lysosomal degradation at 37°C (Fig. 1 E).
Unfolding of the λC accelerates endocytosis and impedes recycling of the CD4tl-λC
To assess the cellular consequence of the conformational destabilization of the λC domain, the CD4tl-λC turnover was monitored by the immunoperoxidase assay. After thermal unfolding at 37°C for 1 h, the rescued CD4tl-λC was eliminated three times faster than its wt counterpart from the PM (Fig. 1 F). Consistently, CD4tl-λC that constitutively reached the PM at 37°C exhibited a three- to fourfold accelerated turnover in HEK293 and COS7 cells (Fig. S1, C and D). In contrast, only ∼40% of the CD4tl-λC was eliminated after 5-h chase at 26°C, whereas the turnover of CD4tl and CD4tl-λ was negligible (Fig. 1 G).
Changes in internalization, recycling, and lysosomal delivery kinetics may account for the rapid disposal of CD4tl-λC from the PM at 37°C. Tethering the λ or the λC domain accelerated the internalization rate of the CD4tl (2% per minute) by ∼50% and ∼290%, respectively (Fig. 1 H and Fig. S1 E). The internalization of the CD4tl-λC, measured by anti-CD4 Ab uptake at 37°C (see Materials and methods), was attenuated by rescuing the λC conformational defect at 26°C (Fig. 1 H).
Recycling of unfolded CD4tl-λC was inhibited when compared with that of the CD4tl and CD4tl-λ (Fig. 1 I). However, rescuing the CD4tl-λC folding defect at 26°C partially restored the chimera recycling at 37°C (Fig. 1 I) as determined by using a modified biotin-neutravidin sandwich technique as described in Materials and methods. These results suggest that conformation-dependent cargo sorting occurs at endosomes.
CD4tl-λC unfolding provokes ubiquitination at the PM
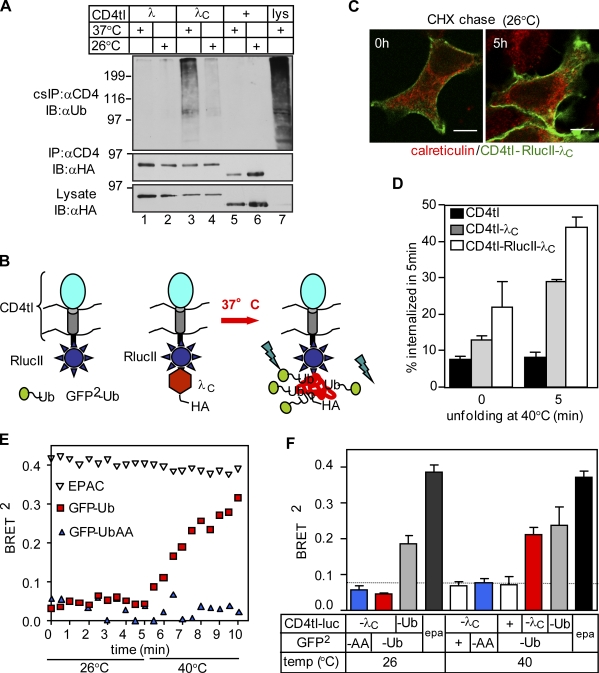
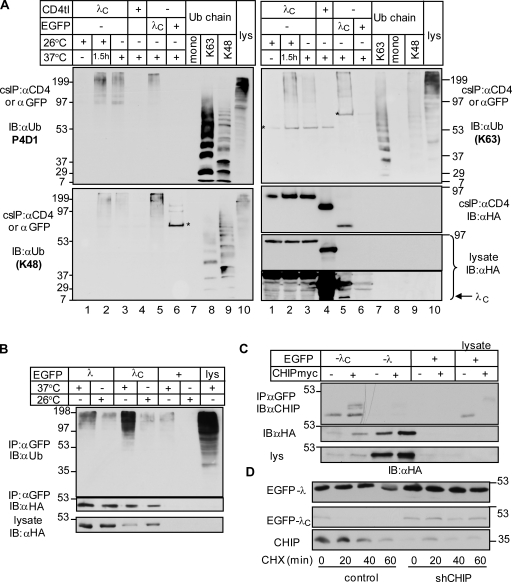
To assess whether Ub conjugation serves as a signal for the chimera down-regulation from the PM, the chimeras were selectively isolated using cell surface immunoprecipitation (IP [cs-IP]). Anti-CD4 Ab was bound to intact cells at 4°C. Immunocomplexes were isolated using a two-step denaturing precipitation protocol as described in Materials and methods. Precipitates were probed with the P4D1 anti-Ub Ab recognizing K63-, K48-, and K11-linked poly-Ub chains, but not the mono-Ub. Densitometry revealed that the ubiquitination of the unfolded CD4tl-λC was 8.9 ± 1.6 (means ± SEM; n = 4) times more augmented at 37°C than at 26°C when normalized for protein expression (Fig. 2 A, lanes 3 and 4). Both CD4tl and CD4tl-λ displayed marginal ubiquitination (Fig. 2 A). Isolation of ubiquitinated CD4tl-λC from intracellular compartments was ruled out because the dissociation of anti-CD4 Ab was negligible during the course of the precipitation (Fig. S2, A and B).
Figure 2.
Unfolding initiates ubiquitination of CD4tl-λC at the PM. (A) Ubiquitination of CD4 chimeras isolated by denaturing cell surface immunoprecipitation (cs-IP) was measured by immunoblotting using the HRP-conjugated P4D1 anti-Ub Ab. Three times more CD4tl-λC–expressing cells were harvested than CD4tl-λ–expressing cells. Molecular mass is given in kilodaltons. IB, immunoblot. (B) Approach to monitor the ubiquitination of CD4tl-RlucII-λC by BRET2 in vivo. (C) The temperature-rescued CD4tl-RlucII-λC (26°C) is largely confined to the PM and shows negligible colocalization with calreticulin. Indirect immunostaining was performed before and after CHX chase for 5 h at 26°C. Bars, 5 µm. (D) 26°C temperature-rescued cells were incubated for the indicated times at 40°C, and the internalization rates of CD4tl-λC and CD4tl-RlucII-λC were determined by Ab uptake at 37°C. (E) Unfolding stimulates of CD4tl-RlucII-λC ubiquitination detected by BRET2. HEK293 cells coexpressing rescued CD4tl-RlucII-λC and GFP2-Ub or GFP2-UbiAA were treated with CHX for 5 h at 26°C. BRET2 was measured in the presence of 5 µM coelenterazine 400a. EPAC expression served as a positive control. (F) Summary of BRET2 results of experiments shown in E. BRET2 was measured on cells cultured at 26°C before or after 5-min exposure to 40°C. The dotted line indicates the background BRET2 signal. epa, EPAC. The data shown represent means ± SEM.
To provide direct evidence for unfolding-provoked ubiquitination at the PM and in early endosomes, we implemented a bioluminescence resonance energy transfer (BRET2) assay based on principles that allowed the detection of the β-arrestin-2-Rluc ubiquitination in real time (Perroy et al., 2004). We rationalized that conjugation of GFP2-Ub to unfolded CD4tl-λC containing the Renilla luciferase II (CD4tl-RlucII-λC) would enhance the BRET2 efficiency (Fig. 2 B). To confine the native chimera to the PM, CD4tl-RlucII-λC were rescued at 26°C and chased in the presence of CHX for 5 h. Indirect immunostaining verified that the chimera was predominantly localized to the PM (Fig. 2 C). Thermal unfolding accelerated the endocytosis rate of rescued CD4tl-λC and CD4tl-RlucII-λC proportional to the incubation time at 40°C (Fig. 2 D). The progressively increasing BRET2 signal between the CD4tl-RlucII-λC and the GFP2-Ub during thermal unfolding suggests that ubiquitination of the chimera accounts for its accelerated internalization (Fig. 2 E). Neither the positive control exchange protein directly activated by cAMP (EPAC; Ponsioen et al., 2004), the constitutively polyubiquitinated CD4tl-Ub (Barriere et al., 2006), nor the GFP-UbAA (UbAA) exhibited temperature-dependent BRET2 (Fig. 2, E and F). GFP-UbAA lacks the terminal Gly residues of Ub that are essential for Ub conjugation (Coulon et al., 2008).
Endpoint measurements revealed a threefold increase in the BRET2 efficiency after unfolding the CD4tl-RlucII-λC, taking into consideration the nonspecific BRET2 signal in the presence of GFP2 (Fig. 2 F, fifth bar). Similar results were obtained using coelenterazine 400a or ViviRen as substrates (Fig. S2 C). BRET2 efficiency was reduced to background if GFP2-Ub or the CD4tl-RlucII-λC was substituted with GFP2-UbAA or CD4tl-RlucII, respectively (Fig. 2 F). These observations jointly suggest that thermal unfolding provokes the chimera ubiquitination at the PM and in early endosomes.
The activity of the cellular ubiquitination machinery is indispensable for the CD4tl-λC rapid disposal from the PM. The mutant chimera was stabilized in ts20, but not in E36, cells expressing a temperature-sensitive and wt E1 Ub-activating enzyme, respectively (Ciechanover et al., 1991). Down-regulation of the E1 enzyme at 40°C delayed the PM turnover of CD4tl-λC in ts20 cells and reduced the CD4tl-λC internalization rate to the CD4tl level (Fig. S2, D–F) but did not interfere with CD4-LAMP1 and transferrin (Tf) receptor (Tf-R) endocytosis signaled by linear endocytic motifs (Barriere et al., 2006). These results suggest a causal relationship between ubiquitination and the PM down-regulation of unfolded CD4tl-λC at 37°C.
Identification of the ubiquitination machinery of unfolded CD4tl-λC at the cell surface
The molecular machinery responsible for the CD4tl-λC ubiquitination at the PM was identified by mass spectrometry (MS). Unfolded CD4tl-λC protein complexes were isolated by cs-IP and trypsin digested, and peptide profiles were determined by liquid chromatography (LC)–MS/MS and database searches (Cloutier et al., 2009). C-terminal Hsp70-interacting protein (CHIP), a cytosolic QC Ub ligase, and molecular chaperones (Hsc70/Hsp70/Hsp90) were preferentially isolated in the CD4tl-λC complex when compared with that of the CD4tl (Fig. S3, B and C).
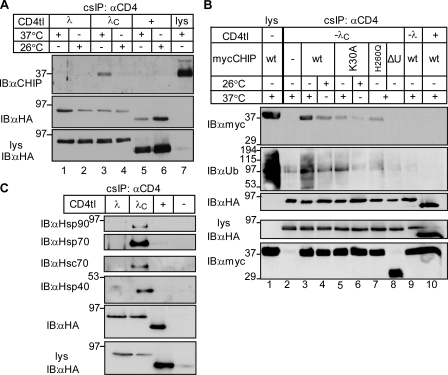
cs-IP confirmed that both endogenous and heterologously expressed myc-CHIP associates with the unfolded chimera (Fig. 3, A and B, lane 3). CHIP was unable to bind to the nativelike CD4tl-λC (26°C; Fig. 3, A and B, lane 4), CD4tl (Fig. 3, A [lanes 5 and 6] and B [lane 10]), or CD4tl-λ (Fig. 3, A [lanes 1 and 2] and B [lane 9]), indicating that the complex formation is conformation sensitive at the PM.
Figure 3.
CHIP and molecular chaperones are recruited to unfolded CD4tl-λC at the PM. (A and B) CHIP associates with unfolded CD4tl-λC at the PM. Native cs-IP was performed, and precipitates were probed with anti-CHIP and anti-HA Abs (A). Exogenous CHIP-myc association was probed in HEK293 cells cotransfected with the indicated construct and cultured at 26°C or 37°C (B). (C) Molecular chaperones associate with the CD4tl-λC but not with CD4tl-λ at the PM. Nondenaturing cs-IP was performed and probed as described in A. Molecular mass is given in kilodaltons. IB, immunoblot.
To probe the mechanism of CHIP association with unfolded CD4tl-λC, we used three CHIP variants in concert with cs-IP. Preventing chaperone binding to the CHIP N-terminal tetratricopeptide (TPR) domain by the K30A mutation (K30A-CHIP), inhibiting the E2 binding and the Ub ligase activity by the H260Q substitution, or deleting the U-box domain (CHIP-ΔU-box) significantly attenuated CHIP-myc interaction with the PM-resident CD4tl-λC at 37°C (Fig. 3 B, lanes 5–8). These results suggest that chaperone-dependent CHIP recruitment is, at least in part, responsible for the recognition and ubiquitination of the chimera (Connell et al., 2001; Rosser et al., 2007). This inference was confirmed by demonstrating the selective association of Hsp90/Hsp40/Hsc70/Hsp70 with unfolded CD4tl-λC, but not with CD4tl-λ and CD4tl at the PM (Fig. 3 C).
CHIP-dependent polyubiquitination down-regulates the unfolded CD4tl-λC from the PM
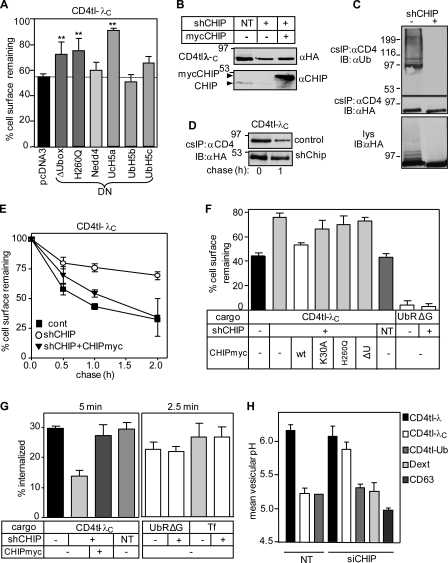
The function of CHIP in the peripheral fate of misfolded CD4tl-λC was examined next. Overexpression of the catalytically inactive H260Q-CHIP, ΔU-box CHIP, or UbcH5a delayed the CD4tl-λC disposal from the PM but had a marginal effect on the CD4tl-λ (Fig. 4 A and Fig. S3 D). In contrast, dominant-negative Nedd-4, AIP4, Casitas B-lineage lymphoma (Cbl)–b, UbcH5b, and UbcH5c had negligible effects on the chimera turnover (Fig. 4 A and not depicted).
Figure 4.
Cellular and biochemical phenotype of the CD4tl-λC upon inactivation of CHIP. (A) Effects of dominant-negative (DN) E3 and E2 enzymes on the PM stability of CD4tl-λC were measured in transiently cotransfected HEK293 cells and monitored by cell surface ELISA after 30-min chase at 37°C. Fig. S3 D depicts the CD4tl-λ PM stability (**, P < 0.01; n = 5). The dotted line indicates the chimera cell surface turnover in mock-transfected cells. (B) CHIP ablation was accomplished by lentiviral shCHIP in Flp-In T-Rex HEK293 cells expressing CD4tl-λC and verified by immunoblotting. CHIP-myc overexpression was detectable in the shCHIP cells. CHIP depletion had an adverse effect on tetracycline transactivation, reducing the induction level of CD4tl-λC in shCHIP cells. (C) Ubiquitination level of the PM CD4tl-λC was probed by denaturing cs-IP and immunoblotting. Three times more shCHIP cells were used. (D) Stability of internalized CD4tl-λC was measured by labeling the PM-resident chimera with anti-CD4 Ab at 4°C and chasing for 0–1 h at 37°C in shNT and shCHIP cells. Ab–chimera complexes were precipitated and immunoblotted. (E) The PM turnover of the CD4tl-λC was measured by cell surface ELISA in control, shCHIP, and CHIP-myc–transfected cells. (F) The CD4tl-λC and CD4cc-UbAllRΔG (UbRΔG) PM stability was determined after 1-h chase in shCHIP cells coexpressing the CHIP-myc variant. (G) The internalization rate of CD4tl-λC was monitored in the absence or presence of CHIP-myc at 37°C by Ab uptake in shCHIP cells for 5 min. Endocytosis of tetrameric CD4cc-UbAllRΔG (UbRΔG) and transferrin (Tf) was measured for 2.5 min. (H) Mean vesicular pH of internalized cargo was determined by FRIA in small interfering NT and small interfering CHIP (siCHIP) cells as described in Materials and methods. Anti-CD4 Ab and FITC-Fab were bound on ice, and FRIA was performed after 1-h chase at 37°C. NT, nontargeted siRNA. The lysosomal targeting of CD4tl-Ub, CD63, and dextran was not influenced by shCHIP. Molecular mass is given in kilodaltons. IB, immunoblot. The data shown represent means ± SEM.
As a second approach, CHIP was ablated by stably expressing a CHIP-specific short hairpin RNA (shRNA; shCHIP) in HEK293 cells. CHIP depletion (Fig. 4 B) resulted in attenuated ubiquitination and turnover of the CD4tl-λC at the PM (Fig. 4, C–E). Similar results were obtained with siRNA-mediated CHIP depletion, strongly suggesting that the CHIP activity accounts for the unfolded chimera turnover (Fig. S3, E–G). Notably, CHIP-myc overexpression destabilized CD4tl-λC in shCHIP cells at the PM (Fig. 4, E–G). The stability and internalization rates of tetrameric CD4Tcc-UbAllRΔG and the Tf-R signaled by tetrameric mono-Ub (Barriere et al., 2006) and Ub-independent endocytic motifs, respectively, were unaltered, ruling out nonspecific effects of CHIP ablation (Fig. 4, F and G).
Remarkably, the rapid PM turnover of the CD4tl-λC could not be restored by overexpressing K30A, H260Q, or ΔU-box CHIP-myc in shCHIP-expressing cells (Fig. 4, E and F; and see Fig. 3 B for CHIP expression), substantiating the notion that chaperone binding and ligase activity of CHIP are required for the chimera down-regulation from the PM.
Poly-Ub–chain configuration of unfolded CD4tl-λC and EGFP-λC
The high molecular mass smear of CD4tl-λC isolated by cs-IP and visualized by the P4D1 anti-Ub Ab suggested that the unfolded chimera is polyubiquitinated in post-Golgi compartments (Fig. 2 A and Fig. 5 A). Because the P4D1 Ab recognizes K63-, K11-, and K48-linked Ub chains, the chain configuration was probed by K48 (Apu2.07) and K63 (Apu3.A8) linkage–specific anti-Ub Abs (Newton et al., 2008). Purified K48-, K63-, and K11-linked oligo-Ub’s were used as standards (Fig. 5 A and Fig. S2, G and H). The native and unfolded chimera was isolated by cs-IP from the PM (Fig. 5 A, lanes 1, 3, and 4) using Ab capture at 4°C or at 37°C for 1.5 h (Fig. 5 A, lane 2). The latter protocol isolated PM and endosomal resident chimeras. Because K48 and K68 anti-Ub Abs do not recognize mono-Ub (Fig. 5 A, lanes 7 [left] and 8 [right]), a significant fraction of the λC was polyubiquitinated. The unfolded CD4tl-λC had a higher content of K63- than K48-linked Ub chains based on the chain specificities of the Abs (Fig. 5 A, lanes 2 and 3). The formation of other Ub-chain configurations, however, cannot be ruled out (Fig. S2, G and H).
Figure 5.
Unfolded CD4tl-λC undergoes mixed-chain polyubiquitination at the PM. (A) Ub-chain configuration of native and unfolded CD4tl-λC at the PM and endosomes. Denaturing cs-IP of the CD4tl-λC was performed after (a) temperature rescue at 26°C (native), (b) unfolding the rescued chimera at 37°C for 1.5 h, and (c) culturing the cells at 37°C. Anti-CD4 Ab was bound at 4°C, except in b, in which Ab labeling was performed at 37°C for 1.5 h. Precipitates were probed with anti-Ub Abs. Denaturing IP of EGFP-λC and EGFP was accomplished with anti-EGFP Ab. Ab specificity was probed by 2 µg mono-Ub and 0.2 µg K63- and K48-linked poly-Ub chains. Asterisks indicate nonspecific Ab reactivity. (B) Ubiquitination of soluble EGFP-λ and -λC was detected after denaturing IP and immunoblotting of the cells exposed to 26°C or 37°C. (C) Association of the EGFP-λ and -λC chimeras with endogenous and exogenous CHIP was examined by nondenaturing IP and immunoblotting. (D) Turnover rates of the EGFP-λ and -λC were measured in control and shCHIP cells by CHX chase and immunoblotting. Molecular mass is given in kilodaltons. IB, immunoblot.
Although all seven Ub-chain configurations can be synthesized by CHIP in vitro (Xu et al., 2006), CHIP activity is primarily responsible for the UPS-dependent degradation of K48- and K11-linked polyubiquitinated proteins at the ER and in the cytoplasm (Kim et al., 2007; Kundrat and Regan, 2010). Therefore, we asked whether the degradation pathway and the Ub-chain configuration were altered by attaching the λC domain to the soluble cytoplasmic EGFP (EGFP-λC; Fig. 1 A).
The temperature-sensitive expression, stability defect, and polyubiquitination as well as proteasome-dependent degradation of the EGFP-λC were documented (Fig. 5 B and Fig. S4, A–E). As expected, CHIP-dependent polyubiquitination is largely responsible for the proteasome-dependent disposal of the unfolded EGFP-λC because (a) both endogenous CHIP and CHIP-myc are coprecipitated with EGFP-λC, but not with EGFP-λ or EGFP (Fig. 5 C); and (b) CHIP depletion prevented both ubiquitination as well as the rapid degradation of the EGFP-λC (Fig. S4 F and Fig. 5 D). The EGFP-λC was enriched in K48-linked Ub chains relative to the CD4tl-λC, though K63-linked chains were also detected (Fig. 5 A, lanes 3 and 5). Likewise, the ER-retained CD4tl-λC contained predominantly K48-linked Ub chains (unpublished data). Thus, CHIP-mediated polyubiquitination of unfolded λC can lead to either proteasome- or lysosome-dependent degradation as a function of the chimera subcellular localization.
Multivesicular body (MVB)/lysosomal targeting of unfolded CD4tl-λC is ESCRT dependent
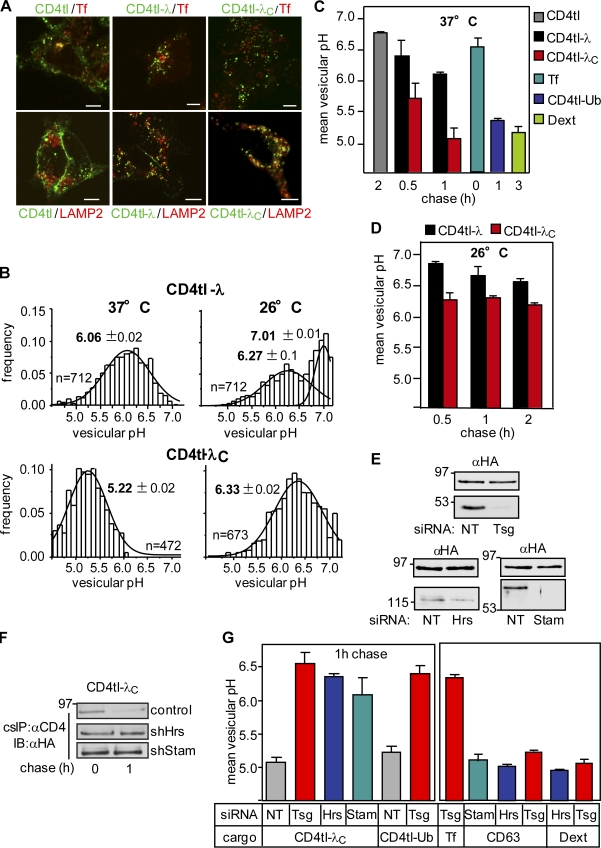
The recycling defect suggested that the chimera is subjected to conformation-dependent postendocytic sorting. This was assessed first by indirect immunostaining. Internalized chimeras were labeled by anti-CD4 Ab capture for 20 min and chased for 1 h at 37°C. The CD4tl predominantly, whereas the CD4tl-λ partially, colocalized with Tf- and EEA1-positive early endosomes and were precluded from dextran- and LAMP2-labeled MVB/lysosomes (Fig. 6 A and not depicted). In contrast, unfolded CD4tl-λC was largely delivered into MVB/lysosomes and precluded from early endosomes (Fig. 6 A).
Figure 6.
Lysosomal delivery of unfolded CD4tl-λC is ESCRT dependent. (A) Indirect immunolocalization of internalized CD4 chimeras with transferrin (Tf) and LAMP2 by laser confocal fluorescence microscopy. CD4 chimeras were labeled with anti-CD4 Ab capture for 20 min at 37°C and chased in Ab-free medium for 1 h. Bars, 5 µm. (B–D) Luminal pH of vesicles containing endocytosed CD4 chimeras and control cargoes was measured by FRIA. Internalized chimeras were labeled with anti-CD4 Ab and FITC-Fab on ice and chased for 1 h (B) or for the indicated time (C and D) at 37°C. In B, the Gaussian distribution (curved lines) and mean vesicular pH (bold numbers) are indicated. The chimera was rescued at 26°C before FRIA (D). (E) Verification of Tsg101 (Tsg), Hrs, and Stam1 depletion in siRNA-treated cells. (F) The stability of internalized CD4tl-λC, labeled by anti-CD4 Ab capture, was measured in shNT, shHrs, and shStam cells as described in Fig. 4 D. IB, immunoblot. (G) The mean vesicular pH of CD4tl-λC and other cargoes was determined after 1-h chase in cells depleted for Tsg101, Hrs, and Stam1. Molecular mass is given in kilodaltons. The data shown represent means ± SEM.
The lysosomal targeting of unfolded CD4tl-λC was confirmed by measuring the luminal pH of chimera-containing vesicles using fluorescence ratio image analysis (FRIA). FRIA has been used to determine the endocytic sorting of various receptors and channels (Barriere et al., 2006, 2007; Kumar et al., 2007; Barriere and Lukacs, 2008; Duarri et al., 2008; Varghese et al., 2008; Glozman et al., 2009). The chimera was labeled with anti-CD4 Ab and secondary FITC-conjugated Fab as a pH sensor in live cells and chased for 0–2 h. Considering that early and late endosomes have a luminal pH of ∼6.0–6.5 and 5.5–6.0, respectively, whereas the lysosomal pH is <5.5 (Mukherjee et al., 1997), localization of the chimera could be established based on the vesicular pH. The luminal pH of FITC-Tf–labeled endosomes as well as CD4tl-Ub and dextran-loaded lysosomes were used as references (Fig. 6 B; Barriere and Lukacs, 2008). Representative pH distribution patterns of CD4tl-λ– and CD4tl-λC–containing vesicles are shown in Fig. 6 B with a mean pH of 6.2 ± 0.19 and 5.2 ± 0.13, respectively. Thus, CD4tl-λC is efficiently targeted to MVB/lysosomes, whereas CD4tl-λ recycles back to the PM at 37°C, confirming the localization and recycling results (Fig. 1 I and Fig. 6 A). Rescuing the folding defect at 26°C led to a substantial delay in the CD4tl-λC lysosomal delivery at 37°C (Fig. 6, B and D).
ESCRT-0 (Hrs [hepatocyte growth factor–regulated tyrosine kinase substrate] and Stam1 [signal-transducing adaptor molecule]) and ESCRT-I (TSG101 [tumor susceptibility gene 101]) components are essential Ub-binding adaptors for ubiquitinated native cargo delivery into MVB/lysosomes (Katzmann et al., 2001). Down-regulation of Hrs, Stam1, and TSG101 by si/shRNA (Fig. 6 E) delayed the unfolded CD4tl-λC degradation and removal from the PM (Fig. 6 F and Fig. S3 A) and induced early endosomal retention in a compartment with a luminal pH of 6.0–6.5 (Fig. 6 G). As a consequence, the chimera post-Golgi stability was increased, and lysosomal delivery was impeded in the presence of Hrs and TSG101 siRNA (Fig. 6, F and G). The lysosomal delivery of CD63, CD4tl-Ub, and dextran as well as the Tf-R recycling were preserved in siRNA-treated cells (Fig. 6 G). Thus, Hrs, Stam1, and TSG101 have critical roles in the proteolytic disposal of nonnative chimeras from the cell surface. Accordingly, CHIP activity was indispensable for the lysosomal sorting of unfolded CD4tl-λC, but not dextran or CD63 (Fig. 4 H). Likewise, the Ub-dependent lysosomal targeting of the CD4tl-Ub chimera was unaltered, ruling out the possibility that CHIP knockdown interferes with the function of the Ub-dependent endosomal sorting machinery nonspecifically (Fig. 4 H).
Proteolytic QC of mutant G protein–coupled receptors (GPCRs) at the PM
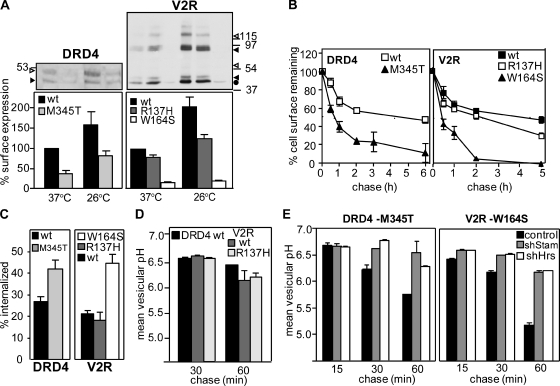
To test whether the peripheral QC system is involved in the degradation of other conformationally defective PM proteins, we selected the DRD4 and the V2R. The DRD4-M345T mutation is implicated in attention deficit hyperactivity disorder, and the V2R-W164S variant causes nephrogenic diabetes insipidus (Rosenthal et al., 1992; van den Ouweland et al., 1992; Swanson et al., 2000). These mutations are localized in the predicted transmembrane and cytosolic segments (Fig. S5 A) and impair the biosynthetic processing of the respective GPCR (Oksche et al., 1996; Van Craenenbroeck et al., 2005). Both DRD4-M345T–Flag and V2R-W164S–myc displayed temperature-sensitive cellular and cell surface expression defects relative to their wt counterparts in HEK293 cells as determined by immunoblotting and cell surface Ab binding using the extracellular Flag and myc tag of the DRD4 and V2R variants, respectively (Fig. 7 A). The V2R-R137H harbors a minor conformation defect and was expressed near the wt level (Fig. 7 A; Barak et al., 2001).
Figure 7.
The disposal of mutant V2R and DRD4 from the PM is CHIP and ESCRT dependent. (A) Cellular (top) and PM (bottom) expression of Flag-DRD4 and myc-V2R variants in transiently transfected HEK-293 cells. Immunoblotting and cell surface density were measured as described in Materials and methods. Molecular mass is given in kilodaltons. Open arrowhead, complex glycosylated forms. Closed arrowhead, core glycosylated forms. Circle, nonglycosylated form. (B) Turnover of GPCRs that accumulate constitutively at the PM was determined by using anti-Flag or anti-myc Ab and ELISA assay at 37°C. (C) The internalization of DRD4s and V2Rs was measured by the Ab uptake assay. (D and E) Postendocytic sorting of DRD4 and V2R variants was followed by FRIA after 15–60-min chase in control, shStam-, or shHrs-expressing cells as described in Fig. 6 B. The data shown represent means ± SEM.
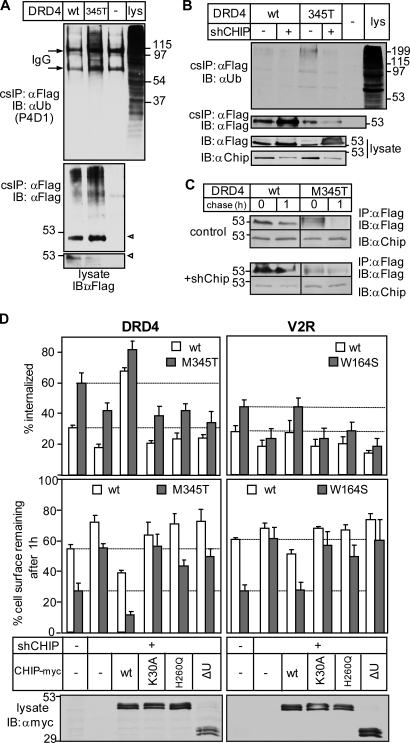
Remarkably, DRD4-M345T and V2R-W164S that constitutively escaped the ER QC exhibited a four- to ninefold faster turnover rate relative to their wt counterparts at the PM (Fig. 7 B). Both accelerated internalization and preferential MVB/lysosomal targeting account for the reduced PM residence of mutant GPCRs (Fig. 7, C–E). Several observations suggest that chaperone- and CHIP-dependent ubiquitination are responsible, at least in part, for the PM down-regulation of mutant GPCRs. (a) The DRD4-M345T exhibited increased ubiquitination and CHIP binding compared with wt DRD4 as measured by denaturating and native cs-IP, respectively (Fig. 8 A and Fig. S5 B). (b) Inactivation of the E1 enzyme stabilized the mutants at the PM and impeded internalization in ts20 cells (Fig. S5, C and D). (c) Although internalized wt GPCRs were recycled, DRD4-M345T and V2R-W164S were delivered into late endosomes/lysosomes in 1 h based on FRIA (Fig. 7, D and E; and Fig. S5 E). Ablation of Hrs and Stam1 by shRNA compromised the lysosomal delivery of mutant GPCRs as indicated by their retention in the early endosomes of HeLa cells (Fig. 7 E). (d) The reduced PM stability is unlikely linked to ligand-induced activation of GPCRs and prolonged β-arrestin-2 binding (Martin et al., 2003) because serum depletion failed to suppress the turnover and internalization rate (not depicted), and inhibiting ubiquitination by shCHIP (Fig. 8 B) was sufficient to attenuate internalization and stabilize the DRD4-M345T and V2R-W164S at the PM and in post-Golgi compartments (Fig. 8, C and D; and Fig. S5, F and G). Only the wt, but neither the catalytically inactive nor the TPR mutant, CHIP-myc overexpression restored the rapid internalization and PM turnover of the DRD4-M345T and V2R-W164S (Fig. 8 D). These results highlight the general role of chaperone-dependent CHIP activity in membrane protein QC at the PM.
Figure 8.
CHIP-dependent ubiquitination contributes to mutant V2R and DRD4 disposal from the PM. (A) Ubiquitination of wt and M345T-DRD4 was measured by denaturing cs-IP as described in Fig. 2 A using anti-Flag and P4D1 anti-Ub Abs. Open arrowhead, complex glycosylated form. (B) Effect of CHIP depletion (shCHIP) on the ubiquitination of wt and M345T-DRD4 at the PM as measured in A. (C) Metabolic stability of internalized DRD4 was determined after in vivo labeling with anti-Flag Ab in control and CHIP-ablated cells as in Fig. 4 D. (D) The impeded internalization and PM turnover of the M345T-DRD4 and W164S-V2R in shCHIP cells were reverted to the mutant phenotype by overexpressing the wt but not the catalytically inactive or TPR mutant CHIP-myc. The internalization and PM stability of GPCRs were monitored with cell surface ELISA. Western blots show the expression of CHIP-myc variants. Molecular mass is given in kilodaltons. IB, immunoblot. The data shown represent means ± SEM.
Discussion
Using a temperature-sensitive model protein and mutant GPCRs associated with conformational diseases, this work represents one of the first attempts to define some of the molecular constituents of the peripheral membrane protein QC that likely plays a fundamental role in the homeostasis of PM protein composition in mammalian cells.
Nonnative PM proteins are subjected to chaperone- and ubiquitination-dependent disposal
To establish a link between the conformational defect and ubiquitination susceptibility of a PM protein, we constructed a CD4 chimera containing the wt or a temperature-sensitive bacteriophage λ domain (Parsell and Sauer, 1989). Controlled thermal unfolding of the CD4tl-λC enabled us to isolate the ubiquitination machinery and compare the consequences of the chimera unfolding with that of mutant GPCRs at the cellular and biochemical levels.
In addition to the temperature-sensitive biosynthetic processing defect, we showed that the reduced PM residence time, accelerated internalization, and impaired recycling of unfolded CD4tl-λC are also hallmarks of mutant V2R and DRD4 membrane trafficking. cs-IP of CD4 chimeras in concert with LC-MS/MS identified Hsp40/Hsc70/Hsp90 and CHIP as major components of the peripheral QC machinery. The unfolded CD4tl-λC and the DRD4-M345T ubiquitination by the chaperone–CHIP complex likely represents a functionally relevant QC mechanism of nonnative PM proteins, an inference supported by several observations. (a) Ablating the E1 Ub-activating enzyme or CHIP inhibited both ubiquitination and the rapid turnover of the chimera and DRD4-M345T at the PM. Although cs-IP of the V2R was not feasible, depletion of the E1 and CHIP also stabilized the V2R-W164S. (b) A similar effect was observed by overexpressing the dominant-negative CHIP and UbH5a. (c) Preventing the Hsc70/Hsp90 association with the TPR domain of the K30A-CHIP compromised the ability of CHIP to destabilize the chimera and mutant GPCRs in CHIP knockdown cells. (d) The unfolded rescued ΔF508CFTR is also eliminated by a chaperone- and CHIP-dependent mechanism from the PM (Okiyoneda et al., 2010).
Although we cannot rule out that nonnative PM proteins are directly recognized by CHIP at post-Golgi compartments (Rosser et al., 2007), the results are consistent with the notion that Hsp40/Hsc70 is recruited to exposed hydrophobic patches of nonnative cytosolic regions followed by CHIP–UbcH5 binding. A similar sequence of events was documented for the mutant CFTR ubiquitination at the ER (Meacham et al., 2001). We envision that mutations in the predicted transmembrane segments of V2R and DRD4 as well as in monotopic membrane proteins (Fayadat and Kopito, 2003) provoke structural rearrangements that lead to the cytosolic ubiquitination of client proteins.
CHIP activity has been recognized as an essential component of the ER and cytosolic QC mechanisms (Murata et al., 2001; Cyr et al., 2002; McDonough and Patterson, 2003). Our results highlight the role of CHIP as a QC E3 ligase in the degradation of conformationally defective PM proteins that either have escaped the ER or are generated by environmental stress in situ. Down-regulation of the ErbB2 tyrosine kinase receptor from the PM may follow an overlapping mechanism with the difference that inhibition of Hsp90 activity is indispensable for the receptor disposal (Xu et al., 2001; Barr et al., 2008). It is assumed that geldanamycin-induced Hsp90 inhibition provokes partial unfolding of the ErbB2 kinase domain followed by the sequential recruitment of Hsc70/Hsp70 and Cullin5 or CHIP (Xu et al., 2002; Zhou et al., 2003; Jung et al., 2007; Ehrlich et al., 2009). Subsequent ubiquitination accounts for internalization, proteasome-dependent processing, and lysosomal targeting of the receptor (Lerdrup et al., 2006).
Although CHIP ablation prevented CD4 chimera and DRD4 ubiquitination and restored some of their metabolic and PM stability, the turnover of the mutants remained faster than their wt counterpart. This could be attributed to residual CHIP activity and/or up-regulation of alternative cellular proteostatic mechanisms in analogy to the redundant role of Doa10, Ubr1, and San1 in the UPS-dependent degradation of misfolded cytosolic proteins in the yeast cytoplasm (Lewis and Pelham, 2009; Heck et al., 2010). Likewise, cytosolic substrate recognition could be accomplished by a chaperone-dependent function of Parkin, Mdm2, and Cullin5 Ub ligases, implying that multiple QC E3 enzymes are probably involved in misfolded PM protein degradation in mammalian cells (Nagata et al., 1999; Imai et al., 2002; Ehrlich et al., 2009). In addition, autophagocytosis of endocytic vesicles enriched in ubiquitinated cargoes (Filimonenko et al., 2007; Lee et al., 2007; Raiborg and Stenmark, 2009), proteasome-dependent processing of misfolded polypeptides (Ciechanover et al., 1980; Waxman et al., 1987), and/or recruitment of arrestin domain–containing adaptors, the mammalian orthologues of arrestin-related adaptor proteins (ARTs), in concert with their cognate E3 ligase can be envisioned as auxiliary mechanisms of the peripheral protein QC in eukaryotes (Lin et al., 2008; Nikko et al., 2008).
Although direct comparison of the peripheral QC efficiency between mammalian and yeast cells is presently not possible, several studies suggest that proteostatic mechanisms in post-Golgi compartments are not restricted to higher eukaryotes. The conformationally impaired Pma-1 (H-ATPase) and general amino acid permease Gap1 are rapidly cleared from the cell surface via ubiquitination-dependent vacuolar degradation in yeast (Gong and Chang, 2001; Pizzirusso and Chang, 2004; Liu and Chang, 2006; Lauwers et al., 2007). In contrast, stabilization of the temperature-sensitive Ura3 or λ-L57A cytosolic domains was observed upon tethering them to the PM as compared with their rapid degradation at ERs (Lewis and Pelham, 2009). Ubiquitination was also indispensable for the vacuolar degradation of various transporters during stress-induced remodeling of the yeast PM, a process mediated by ART1–9 and Rsp5 recruitment (Lin et al., 2008; Nikko et al., 2008; Nikko and Pelham, 2009). Though phosphorylation of certain transporters serves as a recognition signal for ART binding (Nikko and Pelham, 2009), the appealing hypothesis that ARTs can recognize stress-induced nonspecific conformational changes remains to be demonstrated.
Cell surface polyubiquitination as a sorting signal for a nonnative PM protein
Accumulating evidence suggests that most Ub-chain configurations, including K48-, K11-, and K63-linked Ub chain, are involved in the proteasome-mediated degradation of client proteins (Xu et al., 2009). Conversely, besides the K63-linked chain, the contribution of K11-, K29-, and K48-linked Ub chains has been recognized in the internalization of PM proteins (Chastagner et al., 2006; Varghese et al., 2008; Boname et al., 2010). The K63-linked Ub chain was more abundant in the unfolded CD4tl-λC at the PM, whereas the ER-entrapped chimera and the EGFP-λC contained preferentially K48-linked Ub chains. Although we cannot rule out additional differences in the Ub-chain configuration between the soluble and membrane-bound λC, our results are consistent with the emerging model that both K48- and K63-linked Ub chains can participate in both proteasome- and lysosomal-dependent protein degradation with variable efficiency (Xu et al., 2009; Boname et al., 2010). The escape mechanism of nonnative PM proteins from extensive proteasome-dependent processing remains to be elucidated. Forked Ub-chain synthesis by the CHIP–UbcH5 complex (Kim et al., 2007) as well as complex formation with Ub-binding sorting adaptors may represent some of the protective mechanisms against premature proteolysis.
Consistent with the diverse configurations of Ub chains on PM proteins, the selectivity of Ub-binding adaptors in the internalization and endosomal sorting machinery appears to be limited (Shields et al., 2009; Ren and Hurley, 2010). Both epsin1 and eps15 clathrin adaptors recognize the K63-linked and K48-linked poly-Ub chain, though the K63 chain with higher avidity (Barriere et al., 2006; Hawryluk et al., 2006). Likewise, the heterodimeric ESCRT-0 complex has only a threefold higher affinity to K63- than to the K48-linked poly-Ub chain (Ren and Hurley, 2010). This modest Ub-chain selectivity, together with reconfiguration of Ub chains by deubiquitinating enzymes, likely ensures efficient lysosomal degradation of cargoes with various Ub-chain topologies.
The possible role of peripheral QC machinery as genetic modifiers in the loss-of-function phenotype associated with mutant PM proteins
Although the peripheral QC of the mutant V2R and DRD4 phenotypically is reminiscent of the ligand-induced desensitization of wt GPCRs, the molecular machinery accounting for ubiquitination is distinct. Instead of CHIP, ligand-induced down-regulation of GPCR is mediated by cellular Cbl (Jacob et al., 2005), Mdm2 (Shenoy et al., 2001), Nedd4 (Shenoy et al., 2008), and/or related E3 ligases, including AIP4 (Marchese et al., 2003). These E3 enzymes are recruited by posttranslational modification or by exposing a linear peptide-binding site for the E3 enzyme on either the GPCRs or their cognate adaptor β-arrestin-2 (Shenoy et al., 2001).
Among mutant membrane proteins that are subjected to rapid clearance from the PM, ubiquitination of CFTR, BSEP, and NHE6 has been established (Plass et al., 2004; Sharma et al., 2004; Roxrud et al., 2009). The phenotype of certain mutant BSEPs and CFTRs associated with progressive familial intrahepatic cholestasis type 2 disease and cystic fibrosis, respectively, suggests that metabolic destabilization correlates with their ubiquitination at the PM (Sharma et al., 2004; Hayashi and Sugiyama, 2009). The severity of cholestasis and cystic fibrosis is inversely proportional to the PM density of the BSEP and CFTR, respectively (Benharouga et al., 2001; Lam et al., 2007). In light of the limited fidelity of the ER QC (Wang and Ng, 2010) and delayed unfolding of mutant polypeptides, it is tempting to speculate that the peripheral QC may exacerbate the phenotype of conformational diseases by prematurely disposing partially functional mutants from post-Golgi compartments. Indeed, selected mutants of CFTR, MLC1 (megalencephalopathy leukodistrophy with subcortical cyst 1), V2R, and BSEP could escape the ER QC and be targeted for endolysosomal degradation from the PM in primary cells and heterologous expression systems (Morello et al., 2000; Sharma et al., 2004; Robben et al., 2005; Duarri et al., 2008; Hayashi and Sugiyama, 2009).
In summary, we have provided evidence for the role of chaperone–CHIP and ESCRT in the recognition, ubiquitination, and lysosomal degradation of a conformationally impaired model protein and two mutant GPCRs from the cell surface. Thus, the peripheral protein QC system complements the function of established protein surveillance mechanisms of the ER, cytoplasm, nucleus, and mitochondria by preserving proteostasis at the PM and in endocytic organelles in higher eukaryotes.
Materials and methods
Plasmids, cell culture, and transfection
The wt and L57C mutation (λC) of the N-terminal domain of λ bacteriophage (available from GenBank/EMBL/DDBJ under accession no. X00166; Parsell and Sauer, 1989) were fused in frame to the C terminus of the CD4tl or the EGFP-C1 (Fig. 1 A) by PCR mutagenesis. The cDNA of the λ domain was provided by B. Sauer (Massachusetts Institute of Technology, Cambridge, MA) and A. Davidson (University of Toronto, Toronto, Canada). Construction of the CD4tl and CD4tl-Ub has been previously described (Barriere et al., 2006). The following plasmids were gifts: Flag-DRD4.4 was obtained from H.H. van Tol (University of Toronto; Van Craenenbroeck et al., 2005); myc-V2R variants, GFP2-Ub (K48A and K63A), GFP2-UbAA (K48A, K63A, G75A, and G76A), and EPAC were given by M. Bouvier (University of Montréal, Montréal, Canada; Terrillon et al., 2003; Perroy et al., 2004; Kocan et al., 2008); Nedd4 was given by A. Fotia (Hanson Institute, Adelaide, Australia; Fotia et al., 2006); Cbl-b was obtained from S. Lipkowitz (National Cancer Institute, National Institutes of Health, Bethesda, MD); AIP4 was obtained from A. Marchese (Loyola University, Chicago, IL; Marchese et al., 2003); and UbcH5a, UbcH5b, and UbcH5c were given by K. Iwai (Kyoto University, Kyoto, Japan; Gonen et al., 1999). RlucII (Rluc T781A and F782A) was genetically fused to the C terminus of the CD4tl or inserted between the CD4tl and Ub or λC by PCR mutagenesis using EPAC cDNA as a template (Fig. 2 B). Stable inducible Flp-In T-Rex HEK293 cell lines expressing CD4 chimeras were generated by using the Flp-In T-Rex Core kit (Invitrogen) as previously described (Apaja et al., 2006) and used in most experiments if not indicated otherwise. Transgene expression was induced in the presence of 0.5 µg/ml tetracycline for 24 h at 37°C. Transient transfection of HEK293, COS7, or HeLa cells was performed using FuGENE 6 (Roche) or Lipofectamine 2000 (Invitrogen) 48 h before analysis. Cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and antibiotics.
RNA interference
siRNA SMARTpools to human CHIP (GenBank accession no. NM_005861), Hrs (GenBank accession no. NM_004712), Tsg101 (GenBank accession no. NM_006292), Stam1 (GenBank accession no. NM_003473), and nontarget siRNA (NT; 5′-UAGCGACUAAACACAUCAA-3′; GenBank accession no. D-001210-01) were purchased from Thermo Fisher Scientific. Flp-In T-Rex HEK293 cells were transfected two to three times with 50 nM siRNA using Oligofectamine (Invitrogen) in 2-d intervals. After transfection, the chimera expression was induced with 0.5 µg/ml tetracycline for 24 h. When indicated, the mutant chimera was temperature rescued at 26°C for 24 h after tetracycline induction at 37°C.
Constitutive (pGIPZ) and inducible (pTRIPZ) lentivirus vectors encoding microRNA-adapted shRNA specific for CHIP (GIPZ, V2LHS_208833), Stam1 (TRIPZ, V2THS_172428), and Hrs (TRIPZ, V2THS_36954) or NT (5′-ATCTCGCTTGGGCGAGAGTAAG-3′) were obtained from Thermo Fisher Scientific. Lentivirus was produced in HEK293 cells using the expression vector and packaging plasmids (psPAX2 and pMD2G) at a 4:3:1 ratio according to the instructions of the manufacturer in complex with FuGENE 6. The virus was concentrated by ultracentrifugation (Optima L-80 SW40 Ti; Beckman Coulter) at 27,000 rpm for 2 h at 4°C.
The parental or CD4tl-λC–expressing Flp-In T-Rex HEK293 cells were transduced with lentivirus (∼5 MOI) and selected in the presence of 5 µg/ml puromycin to generate shCHIP- and NT shRNA–expressing cell lines. The methodology of preparing HeLa cell lines expressing the inducible shStam1 or shHrs (∼10 MOI) was similar. Transcription of shRNA was verified by the expression of TurboGFP and TurboRFP (Thermo Fisher Scientific), respectively, using fluorescence microscopy. The knockdown efficiency of target proteins was determined by immunoblotting.
Cell surface density measurements of CD4 chimeras and GPCRs
The PM density of CD4 chimeras and GPCRs was determined by ELISA-based assay. Cells were seeded on 24-well plates and incubated at 37°C or rescued at 26°C. Primary and secondary Abs were bound on ice in Ham’s F12 media containing 5% bovine serum (Invitrogen). The following Abs were used to detect extracellular epitopes: CD4 chimeras (mouse monoclonal OKT4 ascites fluid, 1:500 dilution; American Type Culture Collection), myc-V2Rs (mouse monoclonal anti-myc Ab, 1:1,000 dilution; Covance), and Flag-DRD4 (M2 anti-Flag Ab, 1:2,000 dilution; Sigma-Aldrich). Excess Ab was washed away, and specific binding was determined by HRP-conjugated secondary Ab (GE Healthcare) with Amplex red (Invitrogen) as a substrate. The fluorescence intensity was measured from quadruplicate samples using a fluorescence plate reader (POLARstar OPTIMA; BMG Labtech Inc.) at 544-nm excitation and 590-nm emission wavelengths. Total Ab binding was corrected with nonspecific Ab binding as determined in mock-transfected cells.
IP and protein analyses
Selective isolation of CD4 chimeras and GPCRs from the PM was accomplished by cs-IP. CD4 chimeras and DRD4 variants were labeled with rat monoclonal anti-CD4 (1:2,000; AbD Serotec) and mouse monoclonal anti-Flag Ab (M2, 1:2,000; Sigma-Aldrich), respectively, in tissue culture medium containing 20 µM MG132, 10 µg/ml pepstatin, and 10 µg/ml leupeptin in living cells on ice for 1 h. Unbound Ab was washed away with PBS, and cells were chased for the indicated time at 37°C and solubilized in NP-40 lysis buffer (0.2% NP-40, 25 mM Tris-Cl, 150 mM NaCl, 10 µM MG132, 10 µg/ml pepstatin + leupeptin, 1 mM phenylmethylsulfonyl fluoride, and 5 mM N-ethylmaleimide, pH 7.4) on ice. Immunoprecipitates were bound to protein G agarose (20 µl of 50% slurry; Invitrogen), washed three times with lysis buffer, and eluted with Laemmli sample buffer for immunoblotting.
To monitor the ubiquitination of the PM-resident CD4 chimeras and DRD4s, membrane proteins were Ab labeled as described for cs-IP. Alternatively, for total IP, anti-CD4 Ab was added after lysis (1:2,000). Cells were lysed in radioimmunoprecipitation assay buffer containing 10 µM MG132, 10 µg/ml pepstatin + leupeptin, 1 mM phenylmethylsulfonyl fluoride, and 5 mM N-ethylmaleimide. CD4 chimeras and DRD4s were denatured in 1% SDS for 5 min at room temperature and immunoprecipitated after adjusting the SDS concentration to 0.1% (wt/vol) with radioimmunoprecipitation assay buffer. Polypeptides were separated by SDS-PAGE and probed by immunoblotting using the Enhanced Chemiluminescence Detection system (GE Healthcare) or Western Lightning (PerkinElmer). K48- and K63-specific tetra-Ub chains were obtained from Boston Biochem, and ([K11-only]Ub)n ubiquitinylated substrate was purchased from Enzo Life Sciences, Inc. K48- and K63-Ub chain–specific Apu2.07 and Apu3.A8 Abs (Newton et al., 2008), respectively, were provided by Genentech. P4D1 (Santa Cruz Biotechnology, Inc.) and K63 (Enzo Life Sciences, Inc.) anti-Ub Abs were also used. The following heat shock protein Abs were obtained from Stressgen: Hsp90 (SPA839), Hsp70 (SPA810), Hsp70 (SPA815), and Hsp40 (SPA900). Other Abs were purchased from the following sources: HA (cl11; Covance), HRP-Flag (M2, Sigma-Aldrich), Tsg101 (GeneTex Inc.), GFP (Takara Bio Inc.), GFP for IP (3E6; Invitrogen), and Stam1 and Hrs (gift from H. Stenmark, Oslo University Hospital, Oslo, Norway). We used the following inhibitors: 10 µM concanamycin A (Sigma-Aldrich), 1 µM bafilomycin A1 (LC Laboratories), 1 µM epoxomicin (EMD), 10 µM MG132 (Sigma-Aldrich or Cayman Chemical), and 5 µM clastolactacystin β-lactone (Sigma-Aldrich).
Metabolic stability of CD4 chimeras and GPCRs at the cell surface and endocytic pathway
The cell surface stability was monitored by the cargo–Ab complex disappearance from the PM. The Ab incubation, washing, and detection were performed as described in Cell surface density measurements of CD4 chimeras and GPCRs. To determine the PM stability, primary Ab–labeled cargo was chased for 0–5 h at 37°C or 26°C as indicated in Fig. 1 F, and the remaining PM signal was measured.
Metabolic stability of the CD4tl-λC and DRD4 mutant residing at the cell surface and in endocytic compartments was accomplished by incubating cells with primary Ab on ice for 1 h and chasing the labeled cargo for 0 or 1 h at 37°C. Cargo–Ab complexes were isolated by nondenaturating IP as described in the preceding paragraph and detected by immunoblotting.
LC-MS/MS
PM-resident protein complexes of CD4tl-λC and CD4tl were isolated by cs-IP under native conditions as described in the preceding paragraph. Immunoprecipitates were bound to Dynabeads (M280, sheep anti–mouse; Invitrogen), washed with 50 mM ammonium bicarbonate, pH 7.8, and eluted with 500 mM ammonium hydroxide, pH 11. The LC-MS/MS was performed on the Proteomics Discovery Platform (Institute de Recherches Cliniques de Montréal, Montréal, Canada). The LC-MS/MS procedure has been described in detail by Cloutier et al. (2009). In brief, after trypsin digestion of precipitates, peptides were isolated on a NanoLC-2D system (Eksigent) and injected into an LTQ Orbitrap (Thermo Fisher Scientific). Protein database searching was performed with Mascot 2.1 (Matrix Science) against the human NCBInr protein database. The mass tolerances for precursor and fragment ions were set to 10 parts per million and 0.6–0.8 D, respectively. Trypsin was used as the enzyme, allowing for up to two missed cleavages. Carbamidomethyl and oxidation of methionine were allowed as variable modifications. Protein identification data are derived from at least three separate experiments.
Internalization and recycling assay
The internalization and recycling assays were based on monitoring the cargo–Ab complex disappearance from the PM and exocytosis from the endosomal compartment, respectively. The Ab binding and detection were performed as described in the Cell surface density measurements of CD4 chimeras and GPCRs section. Cargo internalization was measured by determining the uptake of the primary Ab bound at 4°C and then was internalized for 5 min at 37°C as described in Barriere et al. (2006) if not specified otherwise. To follow recycling, the surface-bound anti-CD4 Ab (1:1,000; BD) was labeled with biotin-conjugated anti–mouse Fab (216–1,806; KPL, Inc.) for 1 h on ice. CD4 chimeras were internalized for 40 min at 37°C, and the remaining PM biotin-Fab was blocked by 10 µg/ml streptavidin at 0°C (Sigma-Aldrich). Recycling was activated for 10 min at 37°C. Exocytosed CD4 Ab–chimera complexes were measured by binding HRP-NeutrAvidin at 0°C (1:1,000; Thermo Fisher Scientific) using Amplex red as a substrate. The nonspecific Ab background as well as the residual signal derived after streptavidin blocking was taken into account when calculating the chimera recycling efficiency.
Immunostaining and confocal microscopy
Transiently transfected COS7 or Flp-In T-Rex HEK293 cells were cultured on 100 µg/ml poly-l-lysine (Sigma-Aldrich)–coated coverslips. Cells were fixed with 4% paraformaldehyde in PBS for 10 min and blocked with PBS supplemented with 0.5% BSA for 1 h at room temperature. Intracellular antigens were visualized in fixed, permeabilized (0.05% saponin or 0.1% Triton X-100) cells using the indicated primary Ab: mouse monoclonal CD4 (1:1,000; BD), rat monoclonal CD4 (1:1,000; AbD Serotec), rabbit polyclonal calreticulin (1:1,000, SPA-600; Stressgen), mouse monoclonal KDEL (1:1,000, SPA-827; Stressgen), polyclonal early endosomal antigen 1 (1:600; Thermo Fisher Scientific), or LAMP2 (1:100, H4B4-s; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA).
To examine the postendocytic distribution of CD4 chimeras and GPCRs, proteins were labeled by Ab capture for 20 and 60 min, respectively, in live cells at 37°C and chased for 1 h in the absence of extracellular Ab before indirect immunostaining. Lysosomes were labeled with 10 kD Oregon 488– or Texas red–dextran at 50 µg/ml (Invitrogen) by overnight fluid-phase endocytosis and chased for >3 h at 37°C. 10 µg/ml Alexa Fluor 594– or Cy3-labeled Tf uptake for 1 h at 37°C and mouse monoclonal CD4 cargo labeling for 90 min at 37°C were performed as previously described (Barriere et al., 2006). Cy2- and Cy3-labeled secondary Abs were purchased from Jackson ImmunoResearch Laboratories, Inc., and Alexa Fluor 488– and Alexa Fluor 555–labeled secondary Abs were obtained from Invitrogen. Fluorescence micrographs were obtained by a microscope (LSM510 or LSM710; Carl Zeiss, Inc.) and equipped with a Plan-Apochromat 63×/NA 1.4 objective in multitrack mode. Single optical sections are shown. Images were processed by Photoshop CS3 (Adobe). Colocalization of CD4tl-λC with ER markers was performed in permeabilized, fixed Flp-In T-Rex HEK293 cells. Thresholded Pearson’s correlation coefficients for CD4tl-λC colocalization with calreticulin and anti-KDEL Ab were 0.667 ± 0.03 and 0.75 ± 0.02 (n = 10–12 cells), respectively, using the Volocity suite 5.3.2 (PerkinElmer).
FRIA
The pH of endocytic vesicles containing CD4 chimeras, GPCR variants, Tf-R, CD63, or dextran was measured by FRIA as described previously (Sharma et al., 2004; Barriere et al., 2007). Cargo labeling was accomplished by allowing purified mouse monoclonal anti-CD4 (1:400; BD), anti-Flag (M2, 1:400), anti-myc (1:400, 9E10; Covance), or concentrated ascites fluid against CD63 (1:100, H5C6-c; Developmental Studies Hybridoma Bank) to form complexes with FITC-conjugated goat anti–mouse secondary Fab (Jackson ImmunoResearch Laboratories, Inc.) for 15 min at room temperature. The complex was internalized at 37°C. Alternatively, the primary and secondary Abs were incubated sequentially on ice before internalization, which was initiated at 37°C. To measure the lysosomal pH, 10 kD at 50 µg/ml FITC-dextran (Invitrogen) was endocytosed overnight and chased for >3 h at 37°C. Recycling endosomes were labeled with 5 µg/ml FITC-Tf (Jackson ImmunoResearch Laboratories, Inc.) for 1 h after 45 min of serum depletion at 37°C. CD63 loading was performed for 1 h and chased for 2 h at 37°C. FRIA was performed on an inverted fluorescence microscope (Axiovert 100; Carl Zeiss, Inc.) equipped with a cooled charge-coupled device camera (ORCA-ER 1394; Hamamatsu). The acquisition was performed at 490 ± 5– and 440 ± 10–nm excitation wavelengths using a 535 ± 25–nm emission filter and was analyzed with MetaFluor software (MDS Analytical Technologies).
BRET2 assay
BRET2 between CD4tl-RlucII chimeras and GFP2-Ub, GFP2-UbAA, or GFP2 was determined in the presence of coelenterazine 4a (Biotium) or ViviRen (Promega) in transiently transfected HEK293 cells (1:4 ratio, RlucII:GFP2) as described in Perroy et al. (2004). Cells were suspended in 0.1% glucose-containing PBS, and BRET2 signal was monitored by luminometer (POLARstar OPTIMA). The BRET2 signal was calculated by determining the ratio of the GFP2 fluorescence (at 500–530 nm) and the luminescence of the RlucII-containing proteins (at 370–450 nm). The values were corrected by subtracting the background signals of nontransfected cells. The nonspecific BRET2 signal between CD4tl-RlucII-λC and GFP2 was routinely determined at a GFP2 expression level comparable to that detected for GFP2-Ub. This was accomplished by transiently transfecting HEK293 cells with increasing amounts of the expression plasmid encoding the GFP2.
Statistical analysis
Experiments were repeated at least three times, and data were expressed as means ± SEM. Two-tailed p-values were calculated at a 95% confidence level with unpaired t test using Prism software (GraphPad Software, Inc.).
Online supplemental material
Fig. S1 shows the inducible expression, stability, and internalization of CD4 chimeras. Fig. S2 demonstrates the cellular and biochemical consequences of unfolded CD4tl-λC ubiquitination at the PM. Fig. S3 shows the proteomic identification of interacting proteins with unfolded PM-resident CD4tl-λC and the consequence of CHIP ablation on the chimera turnover. Fig. S4 illustrates the cellular and biochemical phenotype of the EGFP-λC and the role of CHIP in its ubiquitination and turnover. Fig. S5 depicts the effect of the E1 enzyme and CHIP inactivation on the stability of mutant GPCRs and CHIP association with the mutant DRD4. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201006012/DC1.
Acknowledgments
We thank F. Pampinella, M. Popov, and K. Du for conducting pilot studies; H. Stenmark for antibodies; B. Sauer, A. Davidson, M. Bouvier, H.H. van Tol, A. Fotia, S. Lipkowitz, and K. Iwai for expression plasmids; and M. Bouvier, T. Okiyoneda, D. Faubert, and S. Wisnovsky for helpful advice.
We thank the Canadian Institutes of Health Research, Canadian Foundation for Innovation, and the Research Group Focused on Protein Structure for grant support. P.M. Apaja has been partially funded by the Restracomp fellowship of the Hospital for Sick Children, and G.L. Lukacs is a holder of a Canada Research Chair.
Footnotes
Abbreviations used in this paper:
- Ab
- antibody
- ART
- arrestin-related adaptor protein
- BRET2
- bioluminescence resonance energy transfer
- BSEP
- bile salt export pump
- Cbl
- Casitas B-lineage lymphoma
- CFTR
- cystic fibrosis transmembrane conductance regulator
- CHIP
- C-terminal Hsp70-interacting protein
- CHX
- cycloheximide
- cs-IP
- cell surface IP
- DRD4
- dopamine D4.4 receptor
- EPAC
- exchange protein directly activated by cAMP
- ESCRT
- endosomal sorting complex required for transport
- FRIA
- fluorescence ratio image analysis
- GPCR
- G protein–coupled receptor
- IP
- immunoprecipitation
- LC
- liquid chromatography
- MS
- mass spectrometry
- MVB
- multivesicular body
- PM
- plasma membrane
- QC
- quality control
- shRNA
- short hairpin RNA
- Tf
- transferrin
- Tf-R
- Tf receptor
- Tm
- melting temperature
- TPR
- tetratricopeptide
- Ub
- ubiquitin
- UPS
- Ub proteasome system
- V2R
- vasopressin V2 receptor
- wt
- wild type
References
- Apaja P.M., Tuusa J.T., Pietilä E.M., Rajaniemi H.J., Petäjä-Repo U.E. 2006. Luteinizing hormone receptor ectodomain splice variant misroutes the full-length receptor into a subcompartment of the endoplasmic reticulum. Mol. Biol. Cell. 17:2243–2255 10.1091/mbc.E05-09-0875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvan P., Zhao X., Ramos-Castaneda J., Chang A. 2002. Secretory pathway quality control operating in Golgi, plasmalemmal, and endosomal systems. Traffic. 3:771–780 10.1034/j.1600-0854.2002.31102.x [DOI] [PubMed] [Google Scholar]
- Barak L.S., Oakley R.H., Laporte S.A., Caron M.G. 2001. Constitutive arrestin-mediated desensitization of a human vasopressin receptor mutant associated with nephrogenic diabetes insipidus. Proc. Natl. Acad. Sci. USA. 98:93–98 10.1073/pnas.011303698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr D.J., Ostermeyer-Fay A.G., Matundan R.A., Brown D.A. 2008. Clathrin-independent endocytosis of ErbB2 in geldanamycin-treated human breast cancer cells. J. Cell Sci. 121:3155–3166 10.1242/jcs.020404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriere H., Lukacs G.L. 2008. Analysis of endocytic trafficking by single-cell fluorescence ratio imaging. Curr. Protoc. Cell Biol. Chapter 15:Unit 15.13. [DOI] [PubMed] [Google Scholar]
- Barriere H., Nemes C., Lechardeur D., Khan-Mohammad M., Fruh K., Lukacs G.L. 2006. Molecular basis of oligoubiquitin-dependent internalization of membrane proteins in Mammalian cells. Traffic. 7:282–297 10.1111/j.1600-0854.2006.00384.x [DOI] [PubMed] [Google Scholar]
- Barriere H., Nemes C., Du K., Lukacs G.L. 2007. Plasticity of polyubiquitin recognition as lysosomal targeting signals by the endosomal sorting machinery. Mol. Biol. Cell. 18:3952–3965 10.1091/mbc.E07-07-0678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benharouga M., Haardt M., Kartner N., Lukacs G.L. 2001. COOH-terminal truncations promote proteasome-dependent degradation of mature cystic fibrosis transmembrane conductance regulator from post-Golgi compartments. J. Cell Biol. 153:957–970 10.1083/jcb.153.5.957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boname J.M., Thomas M., Stagg H.R., Xu P., Peng J., Lehner P.J. 2010. Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic. 11:210–220 10.1111/j.1600-0854.2009.01011.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J.L., McCracken A.A. 1999. ER protein quality control and proteasome-mediated protein degradation. Semin. Cell Dev. Biol. 10:507–513 10.1006/scdb.1999.0321 [DOI] [PubMed] [Google Scholar]
- Chastagner P., Israël A., Brou C. 2006. Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep. 7:1147–1153 10.1038/sj.embor.7400822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Heller H., Elias S., Haas A.L., Hershko A. 1980. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc. Natl. Acad. Sci. USA. 77:1365–1368 10.1073/pnas.77.3.1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Gropper R., Schwartz A.L. 1991. The ubiquitin-activating enzyme is required for lysosomal degradation of cellular proteins under stress. Biomed. Biochim. Acta. 50:321–332 [PubMed] [Google Scholar]
- Cloutier P., Al-Khoury R., Lavallée-Adam M., Faubert D., Jiang H., Poitras C., Bouchard A., Forget D., Blanchette M., Coulombe B. 2009. High-resolution mapping of the protein interaction network for the human transcription machinery and affinity purification of RNA polymerase II-associated complexes. Methods. 48:381–386 10.1016/j.ymeth.2009.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole N.B., Ellenberg J., Song J., DiEuliis D., Lippincott-Schwartz J. 1998. Retrograde transport of Golgi-localized proteins to the ER. J. Cell Biol. 140:1–15 10.1083/jcb.140.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell P., Ballinger C.A., Jiang J., Wu Y., Thompson L.J., Höhfeld J., Patterson C. 2001. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3:93–96 10.1038/35050618 [DOI] [PubMed] [Google Scholar]
- Coulon V., Audet M., Homburger V., Bockaert J., Fagni L., Bouvier M., Perroy J. 2008. Subcellular imaging of dynamic protein interactions by bioluminescence resonance energy transfer. Biophys. J. 94:1001–1009 10.1529/biophysj.107.117275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr D.M., Höhfeld J., Patterson C. 2002. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 27:368–375 10.1016/S0968-0004(02)02125-4 [DOI] [PubMed] [Google Scholar]
- Dikic I., Wakatsuki S., Walters K.J. 2009. Ubiquitin-binding domains - from structures to functions. Nat. Rev. Mol. Cell Biol. 10:659–671 10.1038/nrm2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarri A., Teijido O., López-Hernández T., Scheper G.C., Barriere H., Boor I., Aguado F., Zorzano A., Palacín M., Martínez A., et al. 2008. Molecular pathogenesis of megalencephalic leukoencephalopathy with subcortical cysts: mutations in MLC1 cause folding defects. Hum. Mol. Genet. 17:3728–3739 10.1093/hmg/ddn269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L.M., Piper S., Dodd R.B., Saville M.K., Sanderson C.M., Luzio J.P., Lehner P.J. 2006. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 25:1635–1645 10.1038/sj.emboj.7601056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn R., Hicke L. 2001. Multiple roles for Rsp5p-dependent ubiquitination at the internalization step of endocytosis. J. Biol. Chem. 276:25974–25981 10.1074/jbc.M104113200 [DOI] [PubMed] [Google Scholar]
- Ehrlich E.S., Wang T., Luo K., Xiao Z., Niewiadomska A.M., Martinez T., Xu W., Neckers L., Yu X.F. 2009. Regulation of Hsp90 client proteins by a Cullin5-RING E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA. 106:20330–20335 10.1073/pnas.0810571106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. 2001. ER quality control: towards an understanding at the molecular level. Curr. Opin. Cell Biol. 13:431–437 10.1016/S0955-0674(00)00233-7 [DOI] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4:181–191 10.1038/nrm1052 [DOI] [PubMed] [Google Scholar]
- Fayadat L., Kopito R.R. 2003. Recognition of a single transmembrane degron by sequential quality control checkpoints. Mol. Biol. Cell. 14:1268–1278 10.1091/mbc.E02-06-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E.M., Isaacs A., Brech A., Stenmark H., Simonsen A. 2007. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179:485–500 10.1083/jcb.200702115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotia A.B., Cook D.I., Kumar S. 2006. The ubiquitin-protein ligases Nedd4 and Nedd4-2 show similar ubiquitin-conjugating enzyme specificities. Int. J. Biochem. Cell Biol. 38:472–479 10.1016/j.biocel.2005.11.006 [DOI] [PubMed] [Google Scholar]
- Glozman R., Okiyoneda T., Mulvihill C.M., Rini J.M., Barriere H., Lukacs G.L. 2009. N-glycans are direct determinants of CFTR folding and stability in secretory and endocytic membrane traffic. J. Cell Biol. 184:847–862 10.1083/jcb.200808124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen H., Bercovich B., Orian A., Carrano A., Takizawa C., Yamanaka K., Pagano M., Iwai K., Ciechanover A. 1999. Identification of the ubiquitin carrier proteins, E2s, involved in signal-induced conjugation and subsequent degradation of IkappaBalpha. J. Biol. Chem. 274:14823–14830 10.1074/jbc.274.21.14823 [DOI] [PubMed] [Google Scholar]
- Gong X., Chang A. 2001. A mutant plasma membrane ATPase, Pma1-10, is defective in stability at the yeast cell surface. Proc. Natl. Acad. Sci. USA. 98:9104–9109 10.1073/pnas.161282998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R.Y. 2002. ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 14:476–482 10.1016/S0955-0674(02)00358-7 [DOI] [PubMed] [Google Scholar]
- Hawryluk M.J., Keyel P.A., Mishra S.K., Watkins S.C., Heuser J.E., Traub L.M. 2006. Epsin 1 is a polyubiquitin-selective clathrin-associated sorting protein. Traffic. 7:262–281 10.1111/j.1600-0854.2006.00383.x [DOI] [PubMed] [Google Scholar]
- Hayashi H., Sugiyama Y. 2009. Short-chain ubiquitination is associated with the degradation rate of a cell-surface-resident bile salt export pump (BSEP/ABCB11). Mol. Pharmacol. 75:143–150 10.1124/mol.108.049288 [DOI] [PubMed] [Google Scholar]
- Heck J.W., Cheung S.K., Hampton R.Y. 2010. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. USA. 107:1106–1111 10.1073/pnas.0910591107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L. 2001. A new ticket for entry into budding vesicles-ubiquitin. Cell. 106:527–530 10.1016/S0092-8674(01)00485-8 [DOI] [PubMed] [Google Scholar]
- Hirsch C., Gauss R., Horn S.C., Neuber O., Sommer T. 2009. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 458:453–460 10.1038/nature07962 [DOI] [PubMed] [Google Scholar]
- Imai Y., Soda M., Hatakeyama S., Akagi T., Hashikawa T., Nakayama K.I., Takahashi R. 2002. CHIP is associated with Parkin, a gene responsible for familial Parkinson’s disease, and enhances its ubiquitin ligase activity. Mol. Cell. 10:55–67 10.1016/S1097-2765(02)00583-X [DOI] [PubMed] [Google Scholar]
- Jacob C., Cottrell G.S., Gehringer D., Schmidlin F., Grady E.F., Bunnett N.W. 2005. c-Cbl mediates ubiquitination, degradation, and down-regulation of human protease-activated receptor 2. J. Biol. Chem. 280:16076–16087 10.1074/jbc.M500109200 [DOI] [PubMed] [Google Scholar]
- Jung Y., Xu W., Kim H., Ha N., Neckers L. 2007. Curcumin-induced degradation of ErbB2: A role for the E3 ubiquitin ligase CHIP and the Michael reaction acceptor activity of curcumin. Biochim. Biophys. Acta. 1773:383–390 10.1016/j.bbamcr.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Katzmann D.J., Babst M., Emr S.D. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 106:145–155 10.1016/S0092-8674(01)00434-2 [DOI] [PubMed] [Google Scholar]
- Kim H.T., Kim K.P., Lledias F., Kisselev A.F., Scaglione K.M., Skowyra D., Gygi S.P., Goldberg A.L. 2007. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282:17375–17386 10.1074/jbc.M609659200 [DOI] [PubMed] [Google Scholar]
- Kocan M., See H.B., Seeber R.M., Eidne K.A., Pfleger K.D. 2008. Demonstration of improvements to the bioluminescence resonance energy transfer (BRET) technology for the monitoring of G protein-coupled receptors in live cells. J. Biomol. Screen. 13:888–898 10.1177/1087057108324032 [DOI] [PubMed] [Google Scholar]
- Kumar K.G., Barriere H., Carbone C.J., Liu J., Swaminathan G., Xu P., Li Y., Baker D.P., Peng J., Lukacs G.L., Fuchs S.Y. 2007. Site-specific ubiquitination exposes a linear motif to promote interferon-α receptor endocytosis. J. Cell Biol. 179:935–950 10.1083/jcb.200706034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundrat L., Regan L. 2010. Identification of residues on Hsp70 and Hsp90 ubiquitinated by the cochaperone CHIP. J. Mol. Biol. 395:587–594 10.1016/j.jmb.2009.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P., Pearson C.L., Soroka C.J., Xu S., Mennone A., Boyer J.L. 2007. Levels of plasma membrane expression in progressive and benign mutations of the bile salt export pump (Bsep/Abcb11) correlate with severity of cholestatic diseases. Am. J. Physiol. Cell Physiol. 293:C1709–C1716 10.1152/ajpcell.00327.2007 [DOI] [PubMed] [Google Scholar]
- Lauwers E., Grossmann G., André B. 2007. Evidence for coupled biogenesis of yeast Gap1 permease and sphingolipids: essential role in transport activity and normal control by ubiquitination. Mol. Biol. Cell. 18:3068–3080 10.1091/mbc.E07-03-0196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.A., Beigneux A., Ahmad S.T., Young S.G., Gao F.B. 2007. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 17:1561–1567 10.1016/j.cub.2007.07.029 [DOI] [PubMed] [Google Scholar]
- Lerdrup M., Hommelgaard A.M., Grandal M., van Deurs B. 2006. Geldanamycin stimulates internalization of ErbB2 in a proteasome-dependent way. J. Cell Sci. 119:85–95 10.1242/jcs.02707 [DOI] [PubMed] [Google Scholar]
- Lewis M.J., Pelham H.R. 2009. Inefficient quality control of thermosensitive proteins on the plasma membrane. PLoS One. 4:e5038 10.1371/journal.pone.0005038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Kane T., Tipper C., Spatrick P., Jenness D.D. 1999. Yeast mutants affecting possible quality control of plasma membrane proteins. Mol. Cell. Biol. 19:3588–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.H., MacGurn J.A., Chu T., Stefan C.J., Emr S.D. 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 135:714–725 10.1016/j.cell.2008.09.025 [DOI] [PubMed] [Google Scholar]
- Liu Y., Chang A. 2006. Quality control of a mutant plasma membrane ATPase: ubiquitylation prevents cell-surface stability. J. Cell Sci. 119:360–369 10.1242/jcs.02749 [DOI] [PubMed] [Google Scholar]
- Ljunggren H.G., Stam N.J., Ohlén C., Neefjes J.J., Höglund P., Heemels M.T., Bastin J., Schumacher T.N., Townsend A., Kärre K., et al. 1990. Empty MHC class I molecules come out in the cold. Nature. 346:476–480 10.1038/346476a0 [DOI] [PubMed] [Google Scholar]
- Marchese A., Raiborg C., Santini F., Keen J.H., Stenmark H., Benovic J.L. 2003. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev. Cell. 5:709–722 10.1016/S1534-5807(03)00321-6 [DOI] [PubMed] [Google Scholar]
- Martin N.P., Lefkowitz R.J., Shenoy S.K. 2003. Regulation of V2 vasopressin receptor degradation by agonist-promoted ubiquitination. J. Biol. Chem. 278:45954–45959 10.1074/jbc.M308285200 [DOI] [PubMed] [Google Scholar]
- McDonough H., Patterson C. 2003. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 8:303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham G.C., Patterson C., Zhang W., Younger J.M., Cyr D.M. 2001. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3:100–105 10.1038/35050509 [DOI] [PubMed] [Google Scholar]
- Mittal R., McMahon H.T. 2009. Arrestins as adaptors for ubiquitination in endocytosis and sorting. EMBO Rep. 10:41–43 10.1038/embor.2008.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello J.P., Salahpour A., Laperrière A., Bernier V., Arthus M.F., Lonergan M., Petäjä-Repo U., Angers S., Morin D., Bichet D.G., Bouvier M. 2000. Pharmacological chaperones rescue cell-surface expression and function of misfolded V2 vasopressin receptor mutants. J. Clin. Invest. 105:887–895 10.1172/JCI8688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Ghosh R.N., Maxfield F.R. 1997. Endocytosis. Physiol. Rev. 77:759–803 [DOI] [PubMed] [Google Scholar]
- Murata S., Minami Y., Minami M., Chiba T., Tanaka K. 2001. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2:1133–1138 10.1093/embo-reports/kve246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Y., Anan T., Yoshida T., Mizukami T., Taya Y., Fujiwara T., Kato H., Saya H., Nakao M. 1999. The stabilization mechanism of mutant-type p53 by impaired ubiquitination: the loss of wild-type p53 function and the hsp90 association. Oncogene. 18:6037–6049 10.1038/sj.onc.1202978 [DOI] [PubMed] [Google Scholar]
- Newton K., Matsumoto M.L., Wertz I.E., Kirkpatrick D.S., Lill J.R., Tan J., Dugger D., Gordon N., Sidhu S.S., Fellouse F.A., et al. 2008. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 134:668–678 10.1016/j.cell.2008.07.039 [DOI] [PubMed] [Google Scholar]
- Nikko E., Pelham H.R. 2009. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic. 10:1856–1867 10.1111/j.1600-0854.2009.00990.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E., Sullivan J.A., Pelham H.R. 2008. Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep. 9:1216–1221 10.1038/embor.2008.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T., Barrière H., Bagdány M., Rabeh W.M., Du K., Höhfeld J., Young J.C., Lukacs G.L. 2010. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 329:805–810 10.1126/science.1191542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksche A., Schülein R., Rutz C., Liebenhoff U., Dickson J., Müller H., Birnbaumer M., Rosenthal W. 1996. Vasopressin V2 receptor mutants that cause X-linked nephrogenic diabetes insipidus: analysis of expression, processing, and function. Mol. Pharmacol. 50:820–828 [PubMed] [Google Scholar]
- Parsell D.A., Sauer R.T. 1989. The structural stability of a protein is an important determinant of its proteolytic susceptibility in Escherichia coli. J. Biol. Chem. 264:7590–7595 [PubMed] [Google Scholar]
- Perroy J., Pontier S., Charest P.G., Aubry M., Bouvier M. 2004. Real-time monitoring of ubiquitination in living cells by BRET. Nat. Methods. 1:203–208 10.1038/nmeth722 [DOI] [PubMed] [Google Scholar]
- Pickart C.M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503–533 10.1146/annurev.biochem.70.1.503 [DOI] [PubMed] [Google Scholar]
- Piper R.C., Luzio J.P. 2007. Ubiquitin-dependent sorting of integral membrane proteins for degradation in lysosomes. Curr. Opin. Cell Biol. 19:459–465 10.1016/j.ceb.2007.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzirusso M., Chang A. 2004. Ubiquitin-mediated targeting of a mutant plasma membrane ATPase, Pma1-7, to the endosomal/vacuolar system in yeast. Mol. Biol. Cell. 15:2401–2409 10.1091/mbc.E03-10-0727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass J.R., Mol O., Heegsma J., Geuken M., de Bruin J., Elling G., Müller M., Faber K.N., Jansen P.L. 2004. A progressive familial intrahepatic cholestasis type 2 mutation causes an unstable, temperature-sensitive bile salt export pump. J. Hepatol. 40:24–30 10.1016/S0168-8278(03)00483-5 [DOI] [PubMed] [Google Scholar]
- Ponsioen B., Zhao J., Riedl J., Zwartkruis F., van der Krogt G., Zaccolo M., Moolenaar W.H., Bos J.L., Jalink K. 2004. Detecting cAMP-induced Epac activation by fluorescence resonance energy transfer: Epac as a novel cAMP indicator. EMBO Rep. 5:1176–1180 10.1038/sj.embor.7400290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers E.T., Morimoto R.I., Dillin A., Kelly J.W., Balch W.E. 2009. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 78:959–991 10.1146/annurev.biochem.052308.114844 [DOI] [PubMed] [Google Scholar]
- Raiborg C., Stenmark H. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 458:445–452 10.1038/nature07961 [DOI] [PubMed] [Google Scholar]
- Reggiori F., Pelham H.R. 2002. A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat. Cell Biol. 4:117–123 10.1038/ncb743 [DOI] [PubMed] [Google Scholar]
- Ren X., Hurley J.H. 2010. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 29:1045–1054 10.1038/emboj.2010.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robben J.H., Knoers N.V., Deen P.M. 2005. Characterization of vasopressin V2 receptor mutants in nephrogenic diabetes insipidus in a polarized cell model. Am. J. Physiol. Renal Physiol. 289:F265–F272 10.1152/ajprenal.00404.2004 [DOI] [PubMed] [Google Scholar]
- Rosenthal W., Seibold A., Antaramian A., Lonergan M., Arthus M.F., Hendy G.N., Birnbaumer M., Bichet D.G. 1992. Molecular identification of the gene responsible for congenital nephrogenic diabetes insipidus. Nature. 359:233–235 10.1038/359233a0 [DOI] [PubMed] [Google Scholar]
- Rosser M.F., Washburn E., Muchowski P.J., Patterson C., Cyr D.M. 2007. Chaperone functions of the E3 ubiquitin ligase CHIP. J. Biol. Chem. 282:22267–22277 10.1074/jbc.M700513200 [DOI] [PubMed] [Google Scholar]
- Roxrud I., Raiborg C., Gilfillan G.D., Strømme P., Stenmark H. 2009. Dual degradation mechanisms ensure disposal of NHE6 mutant protein associated with neurological disease. Exp. Cell Res. 315:3014–3027 10.1016/j.yexcr.2009.07.012 [DOI] [PubMed] [Google Scholar]
- Schaheen B., Dang H., Fares H. 2009. Derlin-dependent accumulation of integral membrane proteins at cell surfaces. J. Cell Sci. 122:2228–2239 10.1242/jcs.048892 [DOI] [PubMed] [Google Scholar]
- Sharma M., Benharouga M., Hu W., Lukacs G.L. 2001. Conformational and temperature-sensitive stability defects of the delta F508 cystic fibrosis transmembrane conductance regulator in post-endoplasmic reticulum compartments. J. Biol. Chem. 276:8942–8950 10.1074/jbc.M009172200 [DOI] [PubMed] [Google Scholar]
- Sharma M., Pampinella F., Nemes C., Benharouga M., So J., Du K., Bache K.G., Papsin B., Zerangue N., Stenmark H., Lukacs G.L. 2004. Misfolding diverts CFTR from recycling to degradation: quality control at early endosomes. J. Cell Biol. 164:923–933 10.1083/jcb.200312018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy S.K., McDonald P.H., Kohout T.A., Lefkowitz R.J. 2001. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science. 294:1307–1313 10.1126/science.1063866 [DOI] [PubMed] [Google Scholar]
- Shenoy S.K., Xiao K., Venkataramanan V., Snyder P.M., Freedman N.J., Weissman A.M. 2008. Nedd4 mediates agonist-dependent ubiquitination, lysosomal targeting, and degradation of the beta2-adrenergic receptor. J. Biol. Chem. 283:22166–22176 10.1074/jbc.M709668200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M.Y., Goldberg A.L. 2001. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 29:15–32 10.1016/S0896-6273(01)00177-5 [DOI] [PubMed] [Google Scholar]
- Shields S.B., Oestreich A.J., Winistorfer S., Nguyen D., Payne J.A., Katzmann D.J., Piper R. 2009. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J. Cell Biol. 185:213–224 10.1083/jcb.200811130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J., Oosterlaan J., Murias M., Schuck S., Flodman P., Spence M.A., Wasdell M., Ding Y., Chi H.C., Smith M., et al. 2000. Attention deficit/hyperactivity disorder children with a 7-repeat allele of the dopamine receptor D4 gene have extreme behavior but normal performance on critical neuropsychological tests of attention. Proc. Natl. Acad. Sci. USA. 97:4754–4759 10.1073/pnas.080070897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrillon S., Durroux T., Mouillac B., Breit A., Ayoub M.A., Taulan M., Jockers R., Barberis C., Bouvier M. 2003. Oxytocin and vasopressin V1a and V2 receptors form constitutive homo- and heterodimers during biosynthesis. Mol. Endocrinol. 17:677–691 10.1210/me.2002-0222 [DOI] [PubMed] [Google Scholar]
- Tsigelny I., Hotchko M., Yuan J.X., Keller S.H. 2005. Identification of molecular determinants that modulate trafficking of DeltaF508 CFTR, the mutant ABC transporter associated with cystic fibrosis. Cell Biochem. Biophys. 42:41–53 10.1385/CBB:42:1:041 [DOI] [PubMed] [Google Scholar]
- Van Craenenbroeck K., Clark S.D., Cox M.J., Oak J.N., Liu F., Van Tol H.H. 2005. Folding efficiency is rate-limiting in dopamine D4 receptor biogenesis. J. Biol. Chem. 280:19350–19357 10.1074/jbc.M414043200 [DOI] [PubMed] [Google Scholar]
- van den Ouweland A.M., Dreesen J.C., Verdijk M., Knoers N.V., Monnens L.A., Rocchi M., van Oost B.A. 1992. Mutations in the vasopressin type 2 receptor gene (AVPR2) associated with nephrogenic diabetes insipidus. Nat. Genet. 2:99–102 10.1038/ng1092-99 [DOI] [PubMed] [Google Scholar]
- Varghese B., Barriere H., Carbone C.J., Banerjee A., Swaminathan G., Plotnikov A., Xu P., Peng J., Goffin V., Lukacs G.L., Fuchs S.Y. 2008. Polyubiquitination of prolactin receptor stimulates its internalization, postinternalization sorting, and degradation via the lysosomal pathway. Mol. Cell. Biol. 28:5275–5287 10.1128/MCB.00350-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Ng D.T. 2010. Evasion of endoplasmic reticulum surveillance makes Wsc1p an obligate substrate of Golgi quality control. Mol. Biol. Cell. 21:1153–1165 10.1091/mbc.E09-10-0910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman L., Fagan J.M., Goldberg A.L. 1987. Demonstration of two distinct high molecular weight proteases in rabbit reticulocytes, one of which degrades ubiquitin conjugates. J. Biol. Chem. 262:2451–2457 [PubMed] [Google Scholar]
- Wilson M.H., Highfield H.A., Limbird L.E. 2001. The role of a conserved inter-transmembrane domain interface in regulating alpha(2a)-adrenergic receptor conformational stability and cell-surface turnover. Mol. Pharmacol. 59:929–938 [DOI] [PubMed] [Google Scholar]
- Wolins N., Bosshart H., Küster H., Bonifacino J.S. 1997. Aggregation as a determinant of protein fate in post-Golgi compartments: role of the luminal domain of furin in lysosomal targeting. J. Cell Biol. 139:1735–1745 10.1083/jcb.139.7.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Duong D.M., Seyfried N.T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. 2009. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 137:133–145 10.1016/j.cell.2009.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Mimnaugh E., Rosser M.F., Nicchitta C., Marcu M., Yarden Y., Neckers L. 2001. Sensitivity of mature Erbb2 to geldanamycin is conferred by its kinase domain and is mediated by the chaperone protein Hsp90. J. Biol. Chem. 276:3702–3708 10.1074/jbc.M006864200 [DOI] [PubMed] [Google Scholar]
- Xu W., Marcu M., Yuan X., Mimnaugh E., Patterson C., Neckers L. 2002. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. USA. 99:12847–12852 10.1073/pnas.202365899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Devlin K.I., Ford M.G., Nix J.C., Qin J., Misra S. 2006. Structure and interactions of the helical and U-box domains of CHIP, the C terminus of HSP70 interacting protein. Biochemistry. 45:4749–4759 10.1021/bi0601508 [DOI] [PubMed] [Google Scholar]
- Young J.C., Agashe V.R., Siegers K., Hartl F.U. 2004. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5:781–791 10.1038/nrm1492 [DOI] [PubMed] [Google Scholar]
- Zaliauskiene L., Kang S., Brouillette C.G., Lebowitz J., Arani R.B., Collawn J.F. 2000. Down-regulation of cell surface receptors is modulated by polar residues within the transmembrane domain. Mol. Biol. Cell. 11:2643–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fernandes N., Dodge I.L., Reddi A.L., Rao N., Safran H., DiPetrillo T.A., Wazer D.E., Band V., Band H. 2003. ErbB2 degradation mediated by the co-chaperone protein CHIP. J. Biol. Chem. 278:13829–13837 10.1074/jbc.M209640200 [DOI] [PubMed] [Google Scholar]