Visualization of VASP tetramers interacting with static and growing actin filaments in vitro by TIRF microscopy leads to a new model for VASP-mediated actin filament assembly.
Abstract
Ena/VASP proteins regulate the actin cytoskeleton during cell migration and morphogenesis and promote assembly of both filopodial and lamellipodial actin networks. To understand the molecular mechanisms underlying their cellular functions we used total internal reflection fluorescence microscopy to visualize VASP tetramers interacting with static and growing actin filaments in vitro. We observed multiple filament binding modes: (1) static side binding, (2) side binding with one-dimensional diffusion, and (3) processive barbed end tracking. Actin monomers antagonize side binding but promote high affinity (Kd = 9 nM) barbed end attachment. In low ionic strength buffers, VASP tetramers are weakly processive (Koff = 0.69 s−1) polymerases that deliver multiple actin monomers per barbed end–binding event and effectively antagonize filament capping. In higher ionic strength buffers, VASP requires profilin for effective polymerase and anti-capping activity. Based on our observations, we propose a mechanism that accounts for all three binding modes and provides a model for how VASP promotes actin filament assembly.
Introduction
Assembly of actin filament networks drives many fundamental cellular processes, including cell polarization and migration, endocytosis, intracellular trafficking, and maintenance of membrane-bound compartments. The functions of these different actin networks are determined in part by differences in their localization, composition, and architecture (Fletcher and Mullins, 2010). These basic properties are, in turn, specified by proteins that regulate the assembly and cross-linking of actin filaments (Pollard et al., 2001; Welch and Mullins, 2002). Two important and widely studied actin-based structures—lamellipodia and filopodia—are assembled at the leading edge of motile cells and play important roles in directed cell migration. Lamellipodia are three-dimensional, space-filling networks that resist deformation and generate forces that advance the cell membrane (Pollard and Borisy, 2003), whereas filopodia are parallel bundles of actin filaments required for sensing chemical gradients and interacting with appropriate cellular targets (Adler et al., 2006). The filaments in a lamellipodial actin network are kept short because their growth is rapidly terminated by capping protein (Iwasa and Mullins, 2007; Akin and Mullins, 2008). The molecular dynamics of filopodia are not well understood but they appear, at least in some cases, to arise from a subset of lamellipodial filaments that first increase in length and then become aligned and cross-linked (Svitkina et al., 2003; Mejillano et al., 2004; Mogilner and Rubinstein, 2005). The increase in filament length required for lamellipod to filopod transitions could be caused by faster elongation or by delayed capping. In vivo, these transitions are important for many processes, including neurite dilation and outgrowth, tumor metastasis, and chemotaxis.
One set of factors that modulates the architecture of lamellipodial networks and promotes their reorganization into filopodia is the Enabled/vasodilator-stimulated phosphoprotein (Ena/VASP) family of proteins. The first member of this family to be described was VASP, a human protein originally identified as a target of cAMP/cGMP-dependent kinases during platelet activation (Halbrügge and Walter, 1989; Halbrügge et al., 1990). Subsequent work identified additional family members, including the Drosophila protein Ena (Gertler et al., 1990) and its mammalian orthologues, Mena and EVL, which are highly expressed in neurons and the spleen/thymus, respectively (Gertler et al., 1996; Lanier et al., 1999).
The Ena/VASP proteins are modular and consist of several functional domains separated by unstructured linkers. The N-terminal EVH1 (Ena/VASP homology 1) domain binds specific targeting sequences in upstream regulators (Prehoda et al., 1999; Krause et al., 2004; Boëda et al., 2007). The C-terminal region contains a coiled-coil motif that mediates self-assembly of Ena/VASP proteins into stable tetramers (Bachmann et al., 1999; Kühnel et al., 2004) and that appears to be required for proper function in vivo (Applewhite et al., 2007). Between the EVH1 and the coiled-coil is a set of sequence motifs that mediate the interaction of Ena/VASP proteins with both monomeric and filamentous actin. The so-called GAB (globular actin binding) and FAB (filamentous actin binding) domains have been shown to bind actin monomers and filaments (Bachmann et al., 1999; Hüttelmaier et al., 1999). Adjacent to these domains is a central proline-rich region, which interacts with the actin monomer-binding protein profilin to recruit profilin–actin complexes (Mahoney et al., 1997; Ferron et al., 2007).
Since their discovery, Ena/VASP proteins have emerged as important regulators of actin network architecture. Deletion of all three Ena/VASP proteins in mice is embryonic lethal (Dent et al., 2007; Kwiatkowski et al., 2007). In migrating cells, VASP localizes across the lamellipodial network and at the tips of filopodial protrusions (Lanier et al., 1999; Rottner et al., 1999). VASP is thought to play a central role in formation of these filopodial protrusions because, in fibroblasts, removal of VASP dampens filopodium formation and favors assembly of lamellipodia (Bear et al., 2002). Loss of VASP, however, also decreases the coherence of lamellipodial networks and impairs migration of fish keratocytes (Lacayo et al., 2007) and protrusion of fibroblast lamellipodia (Bear et al., 2002).
Although the effects of perturbing localization and expression of Ena/VASP proteins have been well characterized in vivo, the mechanism underlying their effect on actin assembly remains controversial. Previous studies demonstrated that purified VASP can bind and bundle actin filaments in vitro (Hüttelmaier et al., 1999; Barzik et al., 2005). In vivo, however, VASP localizes to the distal tips rather than the sides of filopodia (Lanier et al., 1999; Svitkina et al., 2003; Applewhite et al., 2007), indicating that filament bundling is not its major function in the filopodium. Interestingly, purified VASP has been shown to capture actin filament barbed ends and prevent termination of filament elongation by capping protein in solution (Bear et al., 2002; Barzik et al., 2005; Pasic et al., 2008). Recently, however, Breitsprecher et al. (2008) reported that VASP tetramers cannot antagonize capping protein in solution, but must be densely clustered to sterically hinder capping protein barbed end association. Previous studies have also reached different conclusions regarding whether VASP accelerates actin filament elongation. One report detected marginal effects of VASP on the elongation rates of single actin filaments (Pasic et al., 2008), whereas another study reported a significant acceleration of barbed end filament elongation (Breitsprecher et al., 2008). In addition, two studies concluded that VASP accelerates barbed end filament elongation in the presence of profilin–actin (Barzik et al., 2005; Pasic et al., 2008), whereas another study reached the opposite conclusion (Breitsprecher et al., 2008). One significant difference between these studies turns out to be the ionic strength of the buffers used in the experiments.
To better understand the mechanisms underlying the cellular function of VASP and to address questions raised by previous studies, we used total internal reflection fluorescence (TIRF) microscopy to visualize single, fluorescently labeled VASP tetramers bound to static and growing actin filaments. In low ionic strength buffers we find that VASP is a processive actin polymerase that localizes to growing barbed ends where it accelerates polymerization in both the absence and presence of profilin. VASP barbed end association requires both the GAB and FAB domains as well as monomeric actin. In higher ionic strength buffers the combination of VASP plus profilin–actin retains effective polymerase activity while the polymerase activity of VASP, plus actin alone decreases significantly. Our observations help reconcile discrepancies between previous studies of VASP activity and enable us to construct a molecular model of how VASP functions to promote actin filament assembly.
Results
VASP accelerates actin assembly from the barbed end
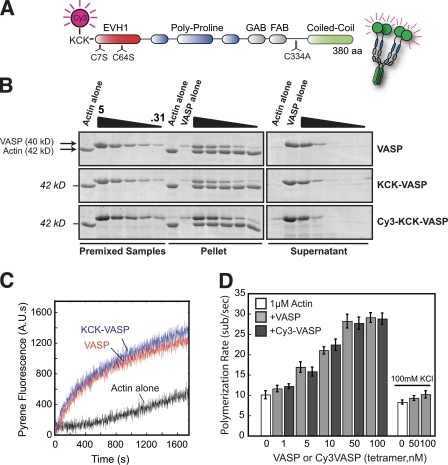
We began by constructing a fluorescent derivative of human VASP and visualizing its interaction with F-actin using time-lapse TIRF microscopy. This required mutating all the endogenous cysteines of human VASP (C7S, C64S, C334A) and introducing an N-terminal Lys-Cys-Lys peptide sequence, which we labeled with Cy3-maleimide (Cy3-VASP; Fig. 1 A). Consistent with results from Hüttelmaier et al. (1999) and Barzik et al. (2005), our recombinant VASP cosedimented with F-actin and accelerated actin assembly in bulk assays (Fig. 1, B and C). We also found that, in low ionic strength buffers (50 mM KCl) similar to those used by Breitsprecher et al. (2008), both unlabeled and Cy3-VASP accelerated barbed end growth up to threefold (Fig. 1 D). Acceleration was dose dependent, with a maximum rate of ∼30 subunits/second in the presence of 50 nM VASP (Fig. 1 D, Video 1). Interestingly, in the higher ionic strength buffers (100 mM KCl) used by Pasic et al. (2008) we observed only a modest acceleration (20%) of actin assembly in the presence of VASP (Fig. 1 D). Because the behavior of Cy3-VASP was indistinguishable from unlabeled, wild-type VASP in all our assays, we considered the two proteins functionally equivalent.
Figure 1.
Cy3-KCK-hVASPCCC-SSA and unlabeled hVASP are functionally equivalent. (A) Cartoon of Cy3-KCK-hVASP1-380aa (C7S, C64S, C334A) domain architecture. (B) Cosedimentation of 2 µM F-actin with VASP, KCK-VASP, or Cy3-KCK-VASP (0–5 µM monomeric concentrations). VASP migrates slower than predicted. Note that Cy3-VASP is slightly brighter due to spectral overlap with SYPRO Red. (C) Pyrene actin polymerization assay. VASP and KCK-VASP (63 nM tetrameric/250 nM monomeric) equally enhance the polymerization of 2 µM actin (5% pyrene labeled). (D) Concentration-dependent increase in the barbed end polymerization rate of single actin filaments in the presence of 1 µM Mg-ATP-actin (30% Alexa Fluor 488, 50–100 mM KCl) in the presence of VASP or Cy3-VASP. Error bars indicate SD (n ≥ 30 filaments from ≥2 slides).
Multiple modes of association between VASP and actin filaments
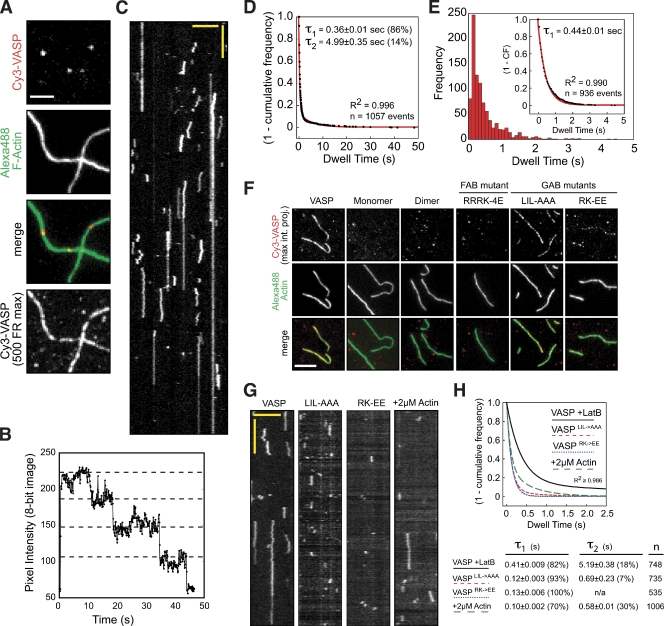
Multi-step photobleaching verified that the fluorescent Cy3-VASP foci observed in our TIRF assays were single tetramers (Fig. 2, A and B). These molecules displayed two distinct modes of interaction with actin filaments: (1) static side binding and (2) side binding, followed by one-dimensional diffusion along the filament (Fig. 2 C, Video 2). The most common interaction was diffusive binding (68%, n = 719), whereas a minority of these tetramers (11%, n = 116) stopped moving and became statically attached. We also observed a large number of transient interactions (21%, n = 222) whose lifetimes were too short to judge whether they were static or diffusive. A histogram of the association lifetimes of all interactions was best fit by a sum of two exponentials (Fig. 2 D) with characteristic dwell times corresponding to: (1) short-lived, 1D diffusion (τ1 = 0.36 ± 0.01 s, n = 909) and (2) static binding (τ2 = 4.99 ± 0.35 s, n = 148). Supporting our conclusion that the shorter dwell time represents diffusive binding, our data fit well to a single exponential when we removed the lifetimes for the static VASP molecules (τ1 = 0.44 ± 0.01 s, n = 936; Fig. 2 E).
Figure 2.
Mechanisms regulating the interaction between VASP and F-actin. (A) Localization of single Cy3-VASP tetramers binding to phalloidin-stabilized Alexa Fluor 488 actin filaments. Intensity differences are due to photobleaching of Cy3-VASP. Bottom panel shows the maximum intensity projection of 0.25 nM Cy3-VASP (500 frames) bound to F-actin. Bar, 5 µm. (B) Multi-step photobleaching of a Cy3-VASP tetramer (τ1 = 11.5 ± 0.07 s for a single Cy3 fluorophore, n = 229 molecules; not depicted). (C) Kymograph of 0.25 nM tetrameric Cy3-VASP bound to an actin filament in TIRF buffer plus oxygen scavenger (see Materials and methods). Vertical bar, 2 s; horizontal bar, 5 µm. (D and E) Dwell times for Cy3-VASP binding to F-actin. (D) Double exponential fit of 1-cumulative frequency (CF) for all binding modes. (E) Single exponential fit of molecules exhibiting diffusive binding mode. (F) Localization of 0.25 nM Cy3-VASP, wild-type, and mutants, binding to phalloidin-stabilized Alexa Fluor 488 actin filaments. Bar, 5 µm. (G) Kymographs of 0.25 nM Cy3-VASP, GAB mutants (Cy3-VASPL226A,I230A,L235A and Cy3-VASPR236E,K237E), and Cy3-VASP plus 2 µM Mg-ATP-actin (+10 µM Latrunculin B) binding to actin filaments. Vertical bar, 2 s; horizontal bar, 5 µm. (H) Double exponential fits of 1-cumulative frequency for GAB mutants and Cy3-VASP plus 2 µM Mg-ATP-actin (+10 µM LatB). Percentage of molecules with each characteristic dwell time are printed in parentheses.
To determine whether tetramerization is required for side binding, we visualized the localization of monomeric Cy3-VASP1-343aa or dimeric Cy3-VASP1-343aa-LZ (leucine zipper) on F-actin (Fig. 2 F). The oligomerization state of each construct was verified by equilibrium ultracentrifugation and gel filtration (not depicted). Surprisingly, we detected no interactions between monomeric or dimeric VASP and F-actin under conditions in which tetrameric VASP strongly localized to actin filaments (Fig. 2 F).
Previous studies identified two regions of VASP that bind actin, called the GAB and FAB domains. These sequences have generally been thought to act independently and to carry out distinct functions (Barzik et al., 2005; Ferron et al., 2007). The FAB domain has been proposed to bind the sides of actin filaments and promote bundling, whereas the GAB domain is thought to recruit actin monomers for barbed end polymerization. We explored the function of these domains in more detail using point mutants (Fig. S1 A). Consistent with a role in binding F-actin, we found that mutation of basic residues in the FAB domain abolished actin filament binding (Fig. 2 F and Fig. S1, C and D). We also observed a reduced affinity of Cy3-VASP for actin filaments in buffers of increasing ionic strength (Fig. S2, A–C). Based on the crystal structure of an actin monomer bound to the VASP GAB domain (Ferron et al., 2007) and on previously reported actin binding mutants (Loureiro et al., 2002), we also generated the following GAB mutants: VASPL226A,I230A,L235A and VASPR236E,K237E. As expected, both mutations had reduced affinities for monomeric actin (unpublished data). Surprisingly, however, both GAB mutations also compromised filament binding, albeit less severely than the FAB mutations (Fig. 2, F–H; Video 3). The majority of interactions between GAB mutants and actin filaments were too short-lived to be characterized as either diffusive or static (Fig. 2 G). This result indicates that in the absence of soluble actin monomers the GAB domain can contribute to the interaction with the sides of actin filaments.
How does the presence of monomeric actin affect the side-binding activity of VASP? If the GAB domain interacts with both monomers and filaments using the same residues, then the presence of monomeric actin should produce an effect similar to mutating the GAB domain, namely a decrease in the dwell time on actin filaments. To test this prediction we compared the dwell times of wild-type VASP in the absence of monomeric actin to those measured in the presence of 2 µM monomeric actin. For these experiments we used Latrunculin B to keep actin in the monomeric state and to avoid confounding effects due to filament elongation. Similar to the effect observed when the GAB domain was mutated, addition of 2 µM monomeric actin dramatically reduced the lifetime of VASP on actin filaments (τ1 = 0.10 ± 0.002 s and τ2 = 0.58 ± 0.01 s, n = 1,006 molecules) (Fig. 2, G and H; Video 4). Together, these data indicate that binding of monomeric actin to the GAB domain antagonizes lateral association of VASP with actin filaments.
VASP barbed end association requires actin monomer binding
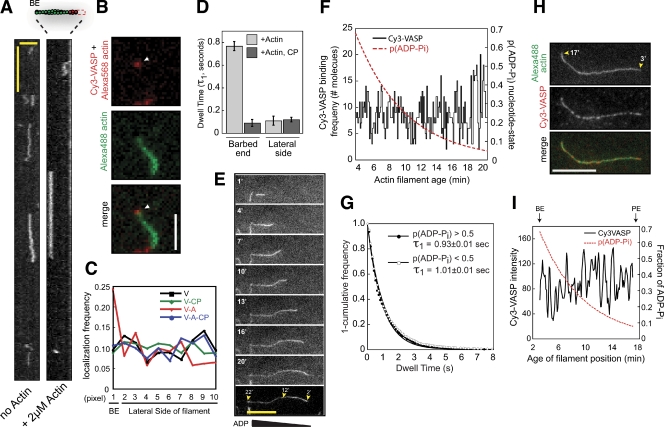
Previous work by Bear et al. (2002) and Pasic et al. (2008) suggested that VASP has an intrinsic affinity for actin filament barbed ends. In addition, the C-terminal fragment, VASP280-380aa, has been proposed to contain a “cryptic” barbed end capture motif (Pasic et al., 2008). To test these proposals we compared the localization of Cy3-VASP and Cy3-VASP280-380aa (C-terminal fragment) on the sides and barbed ends of polarity-marked actin filaments (Alexa Fluor 488, barbed end; Alexa Fluor 568, pointed end; Fig. S3 B). First we measured the relative frequency and dwell time of Cy3-VASP binding to the apparent barbed end and the sides of filaments in the presence or absence of monomeric actin and/or capping protein. We found that, in the absence of monomeric actin, single VASP tetramers bound uniformly along the filament with no bias toward the barbed end (Fig. 3, A–C). In addition, the dwell time of single VASP tetramers near the barbed end was indistinguishable from molecules bound elsewhere on the filament (not depicted). The presence of capping protein did not change the distribution or the binding probability of VASP alone along the filament (Fig. 3 C). This suggests that the molecules we observe near the barbed end under these conditions are actually bound to the sides of the filaments.
Figure 3.
Mechanism for VASP barbed end capture. (A) Kymograph showing Cy3-VASP barbed end association in the absence (left) or presence (right) of 2 µM Mg-ATP-actin (+10 µM Latrunculin B). Vertical bar, 1 s; horizontal bar, 2.5 µm. (B) Barbed end localization of single Cy3-VASP tetramer (arrowhead) in the presence of 2 µM Mg-ATP-actin/LatB. Bar, 2.5 µm. (C) Binding probability for Cy3-VASP at different filament positions in the presence or absence of 2 µM Mg-ATP-actin/LatB and/or 5 nM MmCP. V, VASP; A, actin; CP, capping protein. Pixel position 1 is the apparent barbed end; positions 2–10 are actin filament sides. For experiments including MmCP, filaments were capped for 5 min before adding Cy3-VASP and 5 nM MmCP (≥200 events scored per condition). (D) Dwell times for 0.25 nM Cy3-VASP binding to the apparent barbed end versus the sides of actin filaments in the presence of 2 µM actin/LatB ± 5 nM MmCP. (E) Life history of a single actin filament elongating in the presence of 0.75 µM actin (30% Alexa Fluor 488). Bar, 10 µm. (F) The binding frequency of 0.25 nM Cy3-VASP (n = 1,045 molecules) is independent of the probability of the ADP-Pi nucleotide state at each position. (G) Single molecule dwell times for 0.25 nM Cy3-VASP binding to regions of F-actin with p(ADP-Pi) ≥ 0.5 or p(ADP-Pi) ≤ 0.5 were fit to a single exponential (p(ADP-Pi) ≥ 0.5, τ1 = 0.93 ± 0.01 s, n = 235; p(ADP-Pi) ≤ 0.5, τ1 = 1.0 ± 0.01 s, n = 1,135). (H) Localization of 2.5 nM Cy3-VASP bound to an ADP-Pi/ADP actin filament (3–17 min old). Bar, 10 µm. (I) Intensity profile of Cy3-VASP bound to actin filament in H, overlaid with probability distribution of the ADP-Pi nucleotide state. The probability distribution of each nucleotide state was solved numerically in MatLab (Fig. S3 E).
Next we tested whether the C-terminal fragment, VASP280-380aa, had an intrinsic barbed end capture activity. No binding of Cy3-VASP280-380aa on the sides or barbed ends of F-actin could be detected (Fig. S3, A and C). Furthermore, VASP280-380aa did not change the actin filament elongation rate, as would be expected for a protein that binds to the barbed end (Fig. S3 D). We also tested whether a barbed end capture motif could be unmasked by mutating both the GAB and FAB domains. We failed, however, to detect an interaction between Cy3-VASP(L226A,I230A,L235A)/(RRRK-4A) and F-actin (Fig. S3, A and D). Together, these results suggest that VASP lacks an intrinsic or cryptic barbed end capture motif.
Because VASP alone does not preferentially localize to barbed ends, we hypothesized that monomeric actin might be required to target VASP to barbed ends. When we visualized the localization the Cy3-VASP in the presence of 2 µM actin (+LatB), we observed a shift from uniform localization to preferential barbed end binding (9% vs. 24% of molecules barbed end localized; Fig. 3, A and C). This shift correlated with an increase in the dwell time at the apparent barbed end (τ1 = 0.77 ± 0.04 s, n = 205 molecules) and a decrease in the dwell time elsewhere on the filament (τ1 = 0.11 ± 0.01 s, n = 181), due to actin monomer binding antagonizing lateral filament interactions (Fig. 3 D). Whereas capping protein had no effect on the uniform localization of VASP alone, in the presence of monomeric actin it blocked barbed end association of VASP (Fig. 3, C and D). When we mutated either the GAB or FAB domain, actin-dependent localization to the barbed end was abolished (not depicted). Together, these data indicate that actin monomer binding is required for VASP barbed end association.
Several actin regulators have been shown to bind preferentially to either ATP or ADP actin filaments (Blanchoin and Pollard, 1999; Mahaffy and Pollard, 2006), so we next asked whether the nucleotide state of the actin filament biases the binding of VASP toward the barbed end. To address this question, we correlated the localization and dwell time of Cy3-VASP to the age of F-actin (Fig. S3, E–G; nucleotide state: ADP-Pi vs. ADP). We followed the life history of elongating actin filaments, terminated barbed end growth with capping protein, and then imaged the localization of Cy3-VASP on these filaments in the absence of free actin monomers (Fig. 3 E; Fig. S3). First, we found that the binding frequency of Cy3-VASP molecules was independent of the ADP-Pi/ADP nucleotide state of the filament (Fig. 3 F). Second, the dwell times for single molecules binding to regions of filaments with a high or low probability of containing ADP-Pi were nearly identical (Fig. 3 G; τ1 = 0.93 ± 0.01 s, n = 235, p(ADP-Pi) > 0.5; τ1 = 1.0 ± 0.01 s, n = 1,135, p(ADP-Pi) < 0.5). Third, the steady-state localization of a subsaturating concentration of Cy3-VASP was independent of the probability of the ADP-Pi nucleotide state along the filament length (Fig. 3, H and I). Together, these data suggest that VASP does not recognize the ADP-Pi/ADP nucleotide state of the filament to which it binds.
Processivity of single VASP tetramers
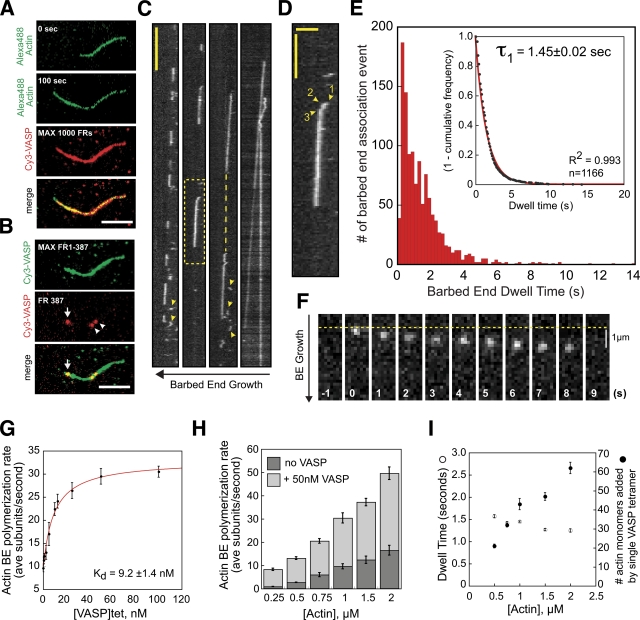
We next wondered whether, similar to formins, VASP associates processively with growing filament ends and delivers multiple actin monomers before dissociating. When we imaged Cy3-VASP barbed end association events in the presence of polymerizable actin (Fig. 4 A), we found that single VASP tetramers persistently associated with and surfed along the growing barbed ends (Fig. 4, B–F; Video 5). Interestingly, we occasionally observed molecules that initially laterally associated with actin filaments and then diffused along the filament before capturing the growing end (Fig. 4 D). Once attached to the barbed end, VASP tetramers surfed along for 1.45 ± 0.02 s (n = 1,166 molecules) in the presence of 1 µM actin before dissociating (Fig. 4 E). Strikingly, we found that the average velocity of single VASP tetramers bound to growing filament ends (33 ± 6.5 sub/sec, n = 7 highly processive VASP tetramers; Fig. 4 F and Video 6) closely matched the maximum rate of filament growth measured in the presence of 50 nM VASP (29.1 ± 1.2 sub/sec, n ≥ 30 filaments; Fig. 4, G and H), while unoccupied barbed ends elongated at the basal polymerization rate (∼10 s/sec; dashed line in Fig. 4 C). By fitting the rate of actin filament growth versus the concentration of VASP, we calculated a Kd of 9.2 nM for the VASP barbed end interaction (Fig. 4 G). Using the barbed end dwell time (τ1 =1.45 s in the presence of 1 µM actin) and maximum polymerization rate (+50 nM VASP), we estimate that, under these conditions, a VASP tetramer delivers, on average, 42 monomers before dissociating from the barbed end. We were unable to determine the effect of ionic strength on processivity because of rapid filament motion in higher ionic strength buffers.
Figure 4.
Processivity of barbed end–associated VASP tetramers. (A) Actin filament lengths before and after burst-phase imaging of 0.25 nM Cy3-VASP in the presence of 1 µM Mg-ATP-actin (30% Alexa Fluor 488). Maximum intensity projections of 0.25 nM Cy3-VASP (1,000 frames) superimposed on the Alexa Fluor 488 actin filament (t = 100 s). Bar, 5 µm. (B). Barbed end localization of Cy3-VASP (middle image; arrow) merged with maximum intensity projection of Cy3-VASP actin filament binding (frames 1–387; top image). Two molecules are also laterally bound to the actin filament (arrowheads). Bar, 5 µm. (C) Representative kymographs showing single Cy3-VASP tetramers processively tracking on actin filament barbed ends. Kymographs from left to right show molecules with increasing processivity. The dashed line connecting two processive VASP tetramers has a slope equivalent to an ∼10 sub/sec barbed end elongation rate in the absence of VASP. Arrowheads point to molecules transiently binding to ATP/ADP-Pi actin, near barbed ends (τ1 = 0.22 ± 0.01 s, n = 280). Vertical bar, 5 s. (D) Magnification of dashed line box in C showing: (1) side binding, (2) diffusion toward barbed end, and (3) barbed end attachment and surfing of Cy3-VASP. Horizontal bar, 2 µm; vertical bar, 2.5 s. (E) Histogram of barbed end dwell times measured in the presence of 0.25 nM Cy3-VASP and 1 µM actin (30% Alexa Fluor 488). Inset plot is an exponential fit of 1-cumulative frequency (τ1 = 1.45 ± 0.02 s, n = 1,166). (F) Time series for a highly processive Cy3-VASP tetramer tracking an actin filament barbed end. (G) Barbed end growth rates measured in the presence of 1 µM Mg-ATP-actin (30% Alexa Fluor 488), plus varying concentrations of VASP. Data fit to Michaelis-Menten equation with a y-axis offset of 10 (Kd = 9.2 ± 1.4 nM). From the Kd and Koff (0.69 s−1) we calculated a barbed end association rate constant of 75 µM−1 sec−1 for VASP. (H) Barbed end polymerization rates in the presence of 0–50 nM VASP, plus 0.25–2 µM Mg-ATP-actin (30% Alexa Fluor 488). (I) Barbed end dwell times for 0.25 nM Cy3-VASP in the presence of 0.5–2 µM Mg-ATP-actin. The number of actin monomers delivered to the barbed end by single VASP tetramers is the product of the barbed end dwell time and the polymerization rate from H.
Consistent with monomeric actin being required for barbed end association, we did not observe an interaction between VASP GAB mutants (VASPL226A,I230A,L235A and VASPR236E,K237E) and growing actin filament barbed ends (unpublished data). Furthermore, neither of these mutants could enhance barbed end filament elongation (Fig. S1 B). Similar to the phenotype observed for the GAB mutants, we found that mutations in the FAB domain (VASPRRRK-EEEE) abolished processive barbed end filament tracking (unpublished data).
Contrary to the model proposed by Breitsprecher et al. (2008), VASP barbed end association rarely transitioned to a lateral side-binding interaction (0.5%; n = 1,166 molecules). Instead, VASP tetramers dissociated directly from the barbed end with an off rate of 0.69 s−1 (1/τ1) in the presence of 1 µM actin. Although VASP tetramers rarely transition from barbed end to a static lateral interaction, molecules from solution frequently bound the sides of growing filaments, including the region adjacent to the barbed end. The lifetimes of these molecules were significantly shorter (τ1 = 0.22 ± 0.01 s, n = 280; Fig. 4 C, arrowheads) than the processive barbed end–tracking tetramers.
To better understand the mode of interaction between VASP and growing barbed ends, we determined the effect of saturating VASP concentrations on filaments elongating in varying concentrations of Mg-ATP-actin (0.25–2 µM; Fig. 4 H). Across all actin concentrations tested, 50 nM tetrameric VASP increased the barbed end elongation rate by 2.7-fold (9 µM−1sec−1 in actin alone vs. 24 µM−1sec−1 plus 50 nM VASP; Fig. 4 H, curve fit not shown). Next, we measured the lifetimes of single Cy3-VASP tetramers surfing on growing barbed ends (Fig. 4 I). At all actin concentrations tested, the lifetimes of barbed end–associated VASP tetramers were exponentially distributed. Strikingly, although we observed a fourfold increase in the maximum rate of barbed end polymerization between 0.5 µM and 2 µM actin, we observed only a 19% reduction in the barbed end dwell time for VASP tetramers (Fig. 4 I). This demonstrates that VASP tetramers deliver more actin monomers per binding event at higher elongation rates. Based on actin monomer binding data obtained by sedimentation equilibrium, it appears that human VASP tetramers are not completely saturated in the presence of 2 µM monomeric actin (Fig. S5, A–C). We hypothesize that human VASP will promote faster barbed end elongation and deliver more subunits at higher actin concentrations.
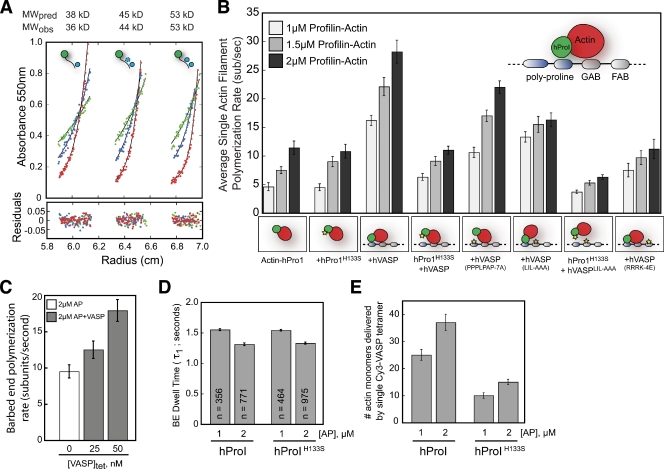
Because the majority of actin monomers are bound to profilin in vivo (Kaiser et al., 1999), we next investigated the effect of human profilin I on VASP-mediated actin polymerization. From sedimentation equilibrium experiments, we found that monomeric VASP interacts with two molecules of human profilin I (Fig. 5 A). This interaction requires the poly-proline domain located in the central domain of VASP. Attempts to measure the stoichiometry of profilin–actin binding to full-length tetrameric VASP were foiled by the complexity of interactions between the three proteins. Compared with barbed end polymerization in the presence of 2 µM profilin–actin alone (11.4 ± 1.2 sub/sec), we observed a 2.5-fold rate enhancement in the presence of 50 nM VASP (28.2 ± 2.0 sub/sec; Fig. 5 B; Video 7). Enhancement was dependent on the interaction between profilin and the poly-proline domain (2 µM Actin-hPro1H133S + VASP; 11 ± 0.7 sub/sec; Fig. 5 B) and was marginally affected by increasing the ionic strength of the buffer (Fig. 5 C). Mutations in the high affinity poly-proline domain (VASPPPPLPPAP-AAAAAAA) also reduced the maximum barbed end polymerization rate (Fig. 5 B). In the presence of a profilin mutant that cannot bind actin (hPro1Y59A), filaments assembled at a rate indistinguishable from that of VASP plus actin alone (not depicted). In conclusion, these results show that VASP can promote barbed end growth in the presence of profilin–actin and, consistent with the results of Barzik et al. (2005), this acceleration is dependent on the interaction between profilin–actin and the poly-proline domain. Interestingly, the interaction between profilin–actin and the poly-proline domain can partially rescue the inability of VASPL226A,I230A,L235A to accelerate barbed end filament elongation in the presence of actin alone (2 µM Actin-hPro1 + VASPL226A,I230A,L235A, 16.3 ± 1.2 sub/sec; Fig. 5 B).
Figure 5.
VASP enhances barbed end polymerization in the presence of profilin–actin. (A) Interaction between Cy3-VASP1-240aa and human profilin I measured by sedimentation equilibrium. Equilibrium traces from left to right: 5 µM Cy3-VASP1-240aa plus 30, 60, or 90 µM hProI. A predicted size of 24.8 kD for Cy3-VASP1-240aa alone closely matched the observed molecular weight of 24.5 kD (not depicted). Based on a global fit of all traces, Cy3-VASP1-240aa interacts with two hProI proteins (15 kD per hPro1). (B) Barbed end growth rates measured in the presence of 1, 1.5, 2 µM profilin–actin (10% Alexa Fluor 488), plus 50 nM VASP. Mutations made to profilin or VASP are indicated with yellow stars. From left to right: (1) hPro1-Actin alone, (2) hPro1H133S-Actin, (3) hPro1-Actin-VASP, (4) hPro1H133S-Actin-VASP, (5) hPro1-Actin-VASPPPPLPAP-8A, (6) hPro1-Actin-VASPGAB*(LIL-AAA), (7) hPro1H133S-Actin-VASPGAB*(LIL-AAA), and (8) hPro1-Actin-VASPFAB*(RRRK-4E). (C) Barbed end filament growth rates in the presence of 2 µM profilin–actin, 100 mM KCl, and 0–50 nM VASP. (D) Barbed end dwell times for Cy3-VASP measured in the presence hPro1-actin and hPro1H133S-actin. (E) The number of actin monomers delivered to the barbed end equals the product of the polymerization rate (B) and dwell time (D).
Because the interaction between profilin and the poly-proline domain is essential for rapid VASP-mediated polymerization of profilin–actin in vitro, we investigated what effect slower polymerization rates have on VASP barbed end processivity. When we measured the barbed end association lifetime for VASP tetramers in the presence of hPro1 or hPro1H133S, we observed no significant differences (Fig. 5 D). Because the dwell time was insensitive to slower barbed end polymerization kinetics, VASP tetramers simply delivered fewer actin monomers per association event in the presence of hPro1H133S (Fig. 5 E). In the presence of 1 µM hPro1-actin, VASP tetramers deliver ∼30 actin monomers within 1.45 s (Fig. 5 E), whereas only 10 actin monomers are delivered by VASP tetramers over the same period of time in the presence of 1 µM hPro1H133S-actin.
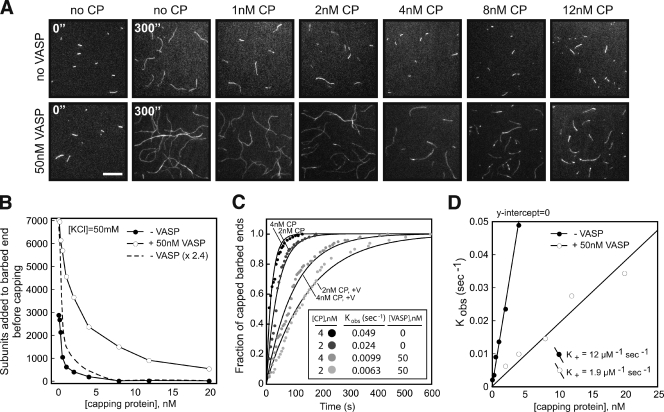
VASP antagonizes capping protein barbed end association in the presence of profilin–actin
To test whether VASP antagonizes capping protein, we polymerized profilin–actin in the presence or absence of VASP and/or capping protein (Fig. 6 A). Comparing the average filament length distribution after 5 min of barbed end growth, we found that filaments grown in the presence of 0–20 nM CP plus 50 nM VASP were significantly longer (Fig. 6, A and B). Interestingly, the difference in filament length was not solely due to a VASP-dependent rate enhancement of barbed end growth (Fig. 6 B, dashed line). Rather, VASP barbed end polymerase activity significantly delayed association of capping protein. Based on the rate at which barbed end growth was terminated by capping protein in the presence of VASP (Fig. 6 C), we calculated a 6.3-fold reduction in the barbed end association rate constant for capping protein (12 µM−1 sec−1, no VASP; 1.9 µM−1 sec−1, plus 50 nM VASP; Fig. 6 D). We also performed these experiments in the presence of 100 mM KCl and observed a 2.7-fold reduction in the association rate constant for capping protein (Fig. S2 D).
Figure 6.
VASP antagonizes capping protein barbed end association in the presence of profilin–actin. (A) Actin filament length distribution after 5 min of barbed end growth in the presence of 2 µM Mg-ATP-actin (10% Alexa Fluor 488), 2 µM hPro1, 0–20 nM MmCP, and 0–50 nM VASP. Bar, 10 µm. (B) Average number of actin subunits delivered to barbed end before termination of growth by MmCP. The dash line represents the predicted actin filament length if the filaments had been elongating 2.4 times faster than 2 µM profilin–actin alone (n ≥ 100 filaments). (C) Rate of actin filament capping in the presence of 2 µM Mg-ATP-actin (10% Alexa Fluor 488), 2 µM hPro1, 2–4 nM MmCP, and 0–50 nM VASP. Calculation of Kobs was achieved by fitting data to the [BE]free * exp^(−-k/τ1) equation in KaleidaGraph (n ≥ 150 filaments tracked). (D) Plot of Kobs (sec−1) versus MmCP concentration. The slope gave a capping protein association rate constant of 12 µM−1 sec−1 and 1.9 µM−1 sec−1, in the absence or presence of 50 nM VASP, respectively.
Discussion
The polymerase activity of VASP: polymerization, processivity, and profilin
Two previous TIRF microscopy studies reported different effects of VASP on actin filament elongation. Pasic et al. (2008) observed minor effects, whereas Breitsprecher et al. (2008) observed a significant concentration-dependent acceleration of barbed end growth. One difference between the previous studies is the ionic strength of the buffers used in the assays of filament elongation. In the present study we find that, in the low ionic strength buffers (50 mM KCl) used by Breitsprecher et al. (2008), human VASP accelerates barbed end elongation of actin filaments by 300%, whereas, in the higher ionic strength buffers (100 mM KCl) used by Pasic et al. (2008), the effect on elongation is much more modest (∼20%). In our hands, the polymerase activity of VASP requires the presence of both the GAB and FAB domains. Interestingly, we find that VASP also accelerates filament elongation by ∼250% in the presence of human profilin 1 and that this effect is insensitive to the ionic strength. In the presence of profilin, the acceleration of filament assembly requires an interaction between profilin–actin complexes and the poly-proline domain of VASP. Remarkably, when both profilin and an intact poly-proline domain are present, the polymerase activity of VASPL226A,I230A,L235A (GAB mutant) was partially rescued.
The simplest interpretation of these results is that VASP can add actin monomers to the barbed end from either the GAB domain or from a profilin–actin complex bound to the poly-proline domain. The difference in sensitivity to ionic strength is likely due to the fact that binding of actin monomers to the GAB domain is primarily an electrostatic interaction, whereas binding of profilin to proline-rich domains is primarily a hydrophobic interaction (Petrella et al., 1996). Similarly, the affinity of profilin for monomeric actin is relatively insensitive to ionic strength (Vinson et al., 1998). In ameboid cells most unpolymerized actin is bound to profilin (Kaiser et al., 1999; Pollard and Borisy, 2003), and so we suggest that the salt-independent polymerase activity of VASP in the presence of profilin–actin complexes is a more physiologically relevant phenomenon than the polymerase activity observed in the absence of profilin.
Because the filament elongation rate increases with VASP concentration, Breitsprecher et al. (2008) proposed that VASP binds barbed ends distributively, delivering a maximum of four actin monomers before either dissociating or remaining statically attached to the side of the filament. We find, however, that in the low ionic strength buffers used for these experiments, individual human VASP tetramers interact processively with growing filament ends and, in the presence of 2 µM actin, can deliver more than 60 monomers before dissociating. The average lifetime of a VASP tetramer on the end of a growing actin filament is 1.45 s, which makes its activity much less processive than some formins (Kovar et al., 2006). Interestingly, however, the lifetime of barbed end association is only weakly sensitive to the rate of filament elongation and this is true both in the presence and absence of profilin. For technical reasons we could not explore the effects of actin concentrations above 2 µM but, at physiological actin concentrations, we estimate a single VASP tetramer could deliver several hundred monomers per barbed end association event. In summary, we propose that actin, or profilin–actin, targets VASP to the barbed end of actin filaments where it remains weakly tethered by the FAB domain and delivers monomers for a dwell time that is independent of the rate of filament elongation.
Multiple modes of actin interaction
In the absence of monomeric actin, VASP tetramers undergo both static and diffusive binding to the sides of actin filaments. Based on mobility and lifetime, diffusive binding is the weaker of the two modes and mutagenesis indicates that the majority of binding energy in this mode is contributed by the FAB domain. The simplest model to account for the difference between the two modes is that diffusive binding involves engagement of a smaller number of FAB domains than the longer-lived, static mode. The occasional transitions from diffusive to static binding probably represent engagement of more FAB domains. The low frequency of transitions suggests that steric constraints limit the ability of all four FAB domains in a tetramer to contact one filament. This interpretation also fits with the fact that VASP tetramers bind more strongly to closely apposed pairs of filaments (Fig. S4, A–D; Video 8) because a second filament provides additional FAB-binding sites. Also, the flexibility of VASP means that the orientation of the overlapping filaments (parallel vs. anti-parallel) makes little difference (Fig. S5 D).
The GAB domain also contributes to diffusive binding and the diffusion along a filament likely reflects binding and unbinding of FAB domains under conditions where the weak interaction of the GAB domains is sufficient to keep the VASP tetramer from diffusing away before another FAB domain binds. The flexibility of VASP and the four FAB domains in a tetramer means that rebinding will likely occur on a different subunit of the filament, producing a diffusive, random walk.
The mechanism of processive interaction with growing barbed ends
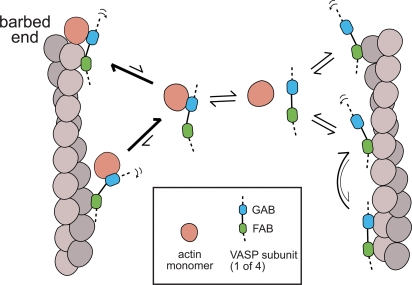
Monomeric actin destabilizes side-binding modes and stabilizes the interaction of VASP with filament ends. This result suggests that the GAB domain uses the same residues to bind monomers and filaments and that it does not interfere with the ability of a bound monomer to interact with a filament barbed end. We propose that, similar to diffusive side binding, processive barbed end binding is driven by a combination of GAB and FAB domains. When the GAB domains are bound to monomeric actin they cannot contribute to filament tethering unless the VASP tetramer is located near the barbed end of the filament. At the barbed end, the GAB domains can contribute their bound monomers to the growing filament and maintain the VASP tetramer in the vicinity of the filament end. The affinity of the GAB domain for actin monomers appears to be higher than for filaments so (similar to WH2 domains and profilin) incorporation of a bound monomer into the filament promotes dissociation of the GAB domain and frees it up to bind another monomer. The GAB domain is related to WH2 domains found in other actin regulatory proteins (Chereau and Dominguez, 2006; Ferron et al., 2007) and, consistent with our results, Co et al. (2007) found that WH2 domains from N-WASP can interact with barbed ends of actin filaments and recruit the molecule to sites with a high density of free barbed ends. In a VASP tetramer, the multiple GAB domains tethered near the end of the filament likely increase the rate of filament elongation by capturing actin monomers at a higher rate than a barbed end by itself (Fig. 7). In vivo the FAB domain might also help bundle tips of actin filaments in the distal regions of filopodia, and this activity might be enhanced by higher-order oligomerization of VASP, perhaps mediated by proteins such as Lamellipodin (Krause et al., 2004).
Figure 7.
Model for VASP barbed end association. In the absence of monomeric actin (right side), the GAB and FAB domains of VASP contribute to actin filament binding. VASP binds with equal affinity along the sides and near the ends of the filament. Apparent barbed end association of VASP alone is indistinguishable from lateral interaction with F-actin. When the GAB domain is bound to monomeric actin (left side), binding to the sides of filaments is weakened and the bound actin monomer allows VASP to capture an actin filament barbed end. Note: although the mechanism described occurs in the context of a VASP tetramer, a single subunit is drawn for simplicity.
In vivo relevance
Previous in vivo studies indicate that recruitment of VASP to the plasma membrane significantly increases the average length of the membrane-proximal actin filaments (Bear et al., 2002). This has been interpreted as evidence that VASP prevents or delays capping of actin filaments and enables them to grow for longer periods of time (Bear and Gertler, 2009). Such results are, however, also consistent with VASP-dependent acceleration of actin assembly. Our data indicate that, even though the lifetime of VASP on the end of an actin filament (1.5 s) is much shorter than the lifetime of capping protein (∼30 min in vitro; Kuhn and Pollard, 2007), VASP can delay the binding of capping protein to filament ends, producing a 6.3-fold decrease in the association rate constant for capping protein. The effectiveness of this competition relies on the very fast association rate of VASP (75 µM−1 sec−1), which gives it nanomolar affinity for barbed ends. We believe that 1D diffusion along the sides of the actin filaments facilitates barbed end capture and increases the effective association rate above the diffusion limit (Fig. 4 D). Observations made by Breitsprecher et al. (2008) suggest that clustering of VASP molecules into dense foci further enhances this effect. More work is required to measure the densities of VASP molecules at various locations in various cell types and to quantify the relationship between VASP density and anti-capping activity.
Previous experiments using cytochalasin D and RNAi knockdown demonstrated that VASP is recruited to the leading edge by both free barbed ends and by membrane-associated proteins such as Mig-10/RIAM/Lamellipodin or CXCR2 (Krause et al., 2004; Lacayo et al., 2007; Neel et al., 2009). Based on our results, the contribution of barbed ends to localization is analogous to the WH2-dependent recruitment of N-WASP to free barbed ends (Co et al., 2007). Svitkina et al. (2003) reported that the intensity of GFP-VASP increases locally at sites on the plasma membrane that subsequently give rise to filopodia. At present it is unclear whether convergence of actin filament barbed ends and/or clustering of VASP tetramers is sufficient to drive filopodia formation. Other activities, such as nucleation by formin family proteins or bundling by fascin are likely required (Vignjevic et al., 2006). Reconstituting the interplay between these factors will lead to a better understanding of how cells dynamically regulate the architecture of functional actin networks.
Single molecule imaging of GFP-VASP in vivo suggests that the majority of the proteins are rapidly binding and dissociating from actin (Miyoshi et al., 2006). In light of our results, we believe this population of protein represents transient interaction with the sides of actin filament in the lamellipod. However, a smaller fraction of the molecules have lifetimes that are greater than 5 s. Interestingly, the proteins with the long lifetimes are localized to the leading edge or membrane of a polarized cell.
Materials and methods
Molecular biology
Human VASP1-380aa and derivates were cloned into a modified pET vector containing an N-terminal his6 tag with a TEV protease cleavage site (HHHHHHDYDIPTTENLYFQ–GS…). A Gly–Ser remains N-terminal post-TEV-cleavage. QuikChange site-directed mutagenesis was used to mutate cysteines, GAB, FAB, and poly-proline domain residues of human VASP. The following point mutations in hVASP were made to generate the “cysteine light” version: C7S, C64S, C334A. Mutation C334S caused VASP to be proteolyzed during expression in Escherichia coli, whereas C334A did not. We made the constitutive VASP dimer by replacing the C-terminal coiled-coil with the leucine zipper derived from Saccharomyces cerevisiae, Gcn4 (KQLEDKVEELASKNYHLENEVARLKKLV; Tomishige et al., 2002). Primer sequences are available upon request.
Protein purification and labeling
Expression of his6-TEV-hVASP and his6-TEV-KCK-hVASPCCC-SSA constructs were performed in BL21 Rosetta E. coli, induced with 50 µM IPTG at 18°C for 16 h. Cultures were harvested, spun down, and stored at −80°C before lysis. Using a microfluidizer, bacteria were lysed into 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 10 mM β-mercaptoethanol, and 1 mM PMSF, pH 8. High speed supernatant was then batch bound to Ni-NTA resin (QIAGEN). Resin was washed with lysis buffer containing 20 mM imidazole and eluted with lysis buffer containing 500 mM imidazole. TEV protease was then added and the protein was dialyzed overnight in 50 mM NaH2PO4, pH 8, 300 mM NaCl, and 10 mM β-ME. Cleaved VASP was then dialyzed into 10 mM Hepes, pH 7.5, 50 mM KCl, and 0.5 mM TCEP to precipitate uncleaved his6-VASP. We find that his6-VASP is mostly insoluble in buffers containing 50 mM KCl. Precipitant was removed by syringe filtration or ultracentrifugation. Protein was then flowed over DEAE resin to remove high molecular weight contaminants and Ni-NTA resin (QIAGEN) to remove uncleaved his6-VASP and TEV protease. VASP tetramers were further purified using a Superdex200 gel filtration column, followed by a cation exchange (MonoS) column. Based on sedimentation equilibrium, VASP was a soluble tetramer (162 kD measured molecular weight compared with 161 kD expected for a VASP tetramer). Pure untagged hVASP1-380aa was dialyzed into storage buffer (10 mM Hepes, pH 7.5, 1 mM TCEP, 50 mM KCl, and 25% glycerol) and frozen with liquid nitrogen before −80°C storage. As judged by pyrene actin polymerization assay, single actin filament TIRF microscopy, and cosedimentation with F-actin, freezing did not reduce the activity of VASP. The extinction coefficient for VASP was determined from a SYPRO Red (Invitrogen)–stained quantitative gel (€280 = 36565 M−1 cm−1).
Labeling was achieved by combining reduced GS-KCK-hVASPCCC-SSA with 5 molar excess Cy3-maleimide (GE Healthcare) on ice for 15 min before quenching with 10 mM DTT. Insoluble material was then removed by high speed ultracentrifugation, whereas soluble free dye was removed with a G25 Sephadex column. Labeling efficiency was assessed with a spectrophotometer. The contribution of Cy3 to the A280 signal (8.6%) was subtracted to accurately calculate the final protein concentration. A labeling efficiency of 85–95% was typically achieved.
Cytoplasmic actin was purified from Acanthamoeba castellani based on the methods described in Gordon et al. (1976). Gel-filtered monomeric actin was stored in buffer containing 2 mM Tris, pH 8.0, 0.5 mM TCEP, 0.1 mM CaCl2, and 0.2 mM ATP. Actin was labeled on Cys-374 with Alexa Fluor 488 maleimide (Invitrogen) using the same method used for labeling KCK-VASP. Human profilin I, hProIH133S, and hProIY59A were purified using established protocols (Kaiser et al., 1989; Lu and Pollard, 2001). Recombinant mouse capping protein was purified using a protocol adapted from Palmgren et al. (2001). Heavy meromyosin (HMM) was a gift from Roger Cooke (University of California, San Francisco, San Francisco, CA). We biotinylated HMM with EZ-link maleimide-PEO11-biotin (Thermo Fisher Scientific).
Actin cosedimentation assay
4 µM (2x final concentration) A. castellani G-actin was polymerized for 60 min in KMEI (50 mM KCl, 1 mM MgCl2, 1 mM EDTA, and 10 mM imidazole, pH 7) plus an equal molar concentration of dark phalloidin. A twofold serial dilution of 10 µM VASP (diluted in KMEI) was combined with an equal volume of F-actin to obtain the final concentrations of 2 µM F-actin, plus 0.31–5 µM VASP (twofold dilutions; monomeric concentrations). F-actin and VASP were allowed to equilibrate for 60 min at room temperature in 7 × 20-mm PC tubes (Beckman Coulter) before ultracentrifugation in a rotor (model TLA100; Beckman Coulter) for 30 min at 48,000 rpm (102,611 g), at 25°C. Supernatants were removed and pellets were resuspended in 1x sample buffer. To normalize for gel loading differences, a no-spin load of known actin and VASP concentrations was used to quantify the amount VASP that cosedimented with F-actin. SDS-PAGE gels were stained with SyproRed and scanned with a Typhoon gel imaging system (532 nm/580 nm; EX/EM).
Pyrene actin polymerization assay
Actin purified from A. castellani was labeled with pyrene iodoacetamide as described previously (Cooper et al., 1983). Polymerization reactions were performed in 1x KMEI (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7.0) and initiated by converting Ca2+-ATP-actin to Mg2+-ATP-actin with 10x ME (0.5 mM MgCl2, 2 mM EGTA). A final concentration of 2 µM Mg-ATP-actin (5% pyrene labeled) was polymerized in the presence or absence of 63 nM tetrameric VASP (250 nM monomeric concentration). Pyrene fluorescence was measured with an ISS PCI/K2 fluorimeter.
Preparation of glass
Coverslips were sonicated in Coplin jars for 20 min in ethanol, 0.5 M KOH, and ethanol with Milli-Q washes in between each step. Glass was then washed with isopropanol before a 30-min sonication in silane solution (95% isopropanol, 1 mM glacial acetic acid, 3% 3-aminopropyltriethoxysilane [Sigma-Aldrich], and 2% Milli-Q water). Slides were washed with isopropanol and then baked at 90°C for ≥8 h. Baked slides were then sonicated twice in ethanol for 20 min before being stored in 100% ethanol. Before PEGylation, glass was washed with Milli-Q water and oven dried for 10 min. PEGylation of silanized slides required assembly of flow cells with double-stick tape to create a 10-µl volume flow space. Flow cells were washed with 100 mM Hepes, pH 8, before adding a 10-mg/ml solution containing 90% PEG-NHS and 10% Biotin-PEG-NHS (JenKem Technology USA). Three rounds of PEGylation were performed at room temperature in a humidity chamber over the course of 90 min. Flow cells were then washed with Milli-Q water and stored hydrated at 4°C for up to 1 wk.
Single actin filament TIRF assays
PEGylated TIRF chambers were incubated with 1 µM streptavidin (Rockland), followed by an incubation with 1 µM biotin (0.8 µM free biotin, plus 0.2 µM biotinylated heavy meromyosin). For barbed end polymerization experiments we followed procedures outlined in Kuhn and Pollard (2005) with modifications. In brief, we combined 9 µl of 4.44 µM actin (30% Alexa Fluor 488 labeled) with 1 µl of 10x ME (0.5 mM MgCl2, 2 mM EGTA) for 2 min; followed by the addition of a solution containing 10 µl Cy3-VASP (1x KMEI, 1 mg/ml BSA), plus 20 µl of 2x TIRF buffer (75 mM KCl, 1.5 mM MgCl2, 1.5 mM EGTA, 15 mM imidazole, pH 7.0, 2 mM DTT, 0.2 mM ATP, 0.4% methycellulose cP400, 2 mg/ml BSA, 200 µM n-propyl gallate, 4 mM Trolox, 2 mM 3,4-dihydroxybenzoic acid [PCA], and 100 nM protocatechuate 3,4-dioxygenase). For polymerization off actin filament barbed ends, the polymerization was performed in a TIRF chamber containing phalloidin-stabilized Alexa Fluor 488 actin filaments. For experiments involving profilin–actin, we used 10% Alexa Fluor 488 actin because Cys-374–labeled actin has a reduced affinity for profilin (Vinson et al., 1998). Profilin–actin was always combined in equal stoichiometry for TIRF experiments. Unless noted in the figure legends, the final KMEI buffer composition was 10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, and 1 mM EGTA for all single actin filament TIRF assays. All concentrations of VASP mentioned in the manuscript represent the tetramer, unless otherwise noted. The details of the oxygen scavenging system are described in Aitken et al. (2008) and Rasnik et al. (2006).
Dual-colored actin filaments were generated by polymerizing 0.5 µM monomeric actin (40% Alexa Fluor 488 labeled; green) in the presence of 20 nM Alexa Fluor 568 phalloidin–stabilized actin filaments (red) for 4 min. Dual-colored seeds were then flowed into the TIRF chamber, captured by biotin-HMM, and chased with 1 µM dark phalloidin to prevent barbed end depolymerization.
Capping protein–VASP barbed end competition
Small phalloidin-stabilized actin filament seeds (50% Alexa Fluor 488 labeled, ∼2–3 µm length) were immobilized in a TIRF chamber. Separately, 2 µM monomeric actin (10% Alexa Fluor 488), 2 µM profilin, 0–20 nM MmCP α1β2, 50 nM VASP, and buffers were combined and flowed into a chamber. Elapsed time was ∼60 s between mixing and imaging. Only filaments that appeared in the first frame (t = 60 s) were followed throughout the experiment. This approach ensured that only the free barbed ends at the start of the experiment were scored for filament length (t = 5 min) and the time at which growth was terminated by capping protein. If filaments were capped at the start of image acquisition, the time at which growth was terminated was calculated from the length of new growth and the average barbed end polymerization rate for a given condition. Filaments falling from solution onto the coverslip (polymerization history unknown) or new filaments appearing due to spontaneous nucleation (results in an underestimate of average filament length) imposed artifacts. These filaments were not scored in our analysis of VASP anti-capping. Lengths of ≥150 filaments were measured from ≥3 slides.
Analytical ultracentrifugation
For sedimentation equilibrium experiments, full-length Cy3-VASP or Cy3VASP1-240aa were buffer exchanged into 1x KMEI containing 1 mM TCEP and then spun at 70,000 rpm in a rotor (model TLA100.4; Beckman Coulter) to remove protein aggregates. A final concentration of 3–7 µM Cy3-VASP was then combined with an increasing concentration of human profilin I (30, 60, 90 µM). Samples were loaded into a 6-well chamber, placed in a rotor (model An-60; Beckman Coulter), and spun at three different speeds (e.g., 10, 14, and 20 K rpm) in an ultracentrifuge (16–20 h per speed; model XL-I; Beckman Coulter). Continuous scans were acquired every 2 h at 527/550 nm to monitor the sedimentation of Cy3-VASP constructs. Extinction coefficient of 150,000 M−1 cm−1 was used to determine the Cy3-VASP protein concentration from the absorbance at 550 nm. Global fitting of three equilibrium traces (e.g., 10, 14, 20 K rpm) for each condition was performed using NIH Sedphit and Sedphat software. More complex data analysis of actin monomer binding to VASP was performed using MatLab (refer to Fig. S5).
Microscopy, software, and data analysis
All microscopy was performed on a TIRF microscope (Eclipse TE2000-E; Nikon) with Nikon Perfect Focus at 23°C. All images were acquired using a cooled EM CCD camera (iXon; Andor Technology) and Micromanager 3.0 software (Stuurman et al., 2007). Fluorophores (e.g., Cy3 and Alexa Fluor 488) were excited through a 100x Apo TIRF objective (NA 1.49; Nikon) using either a 40-mW 488/514 argon ion laser or 40-mW 542 crystal laser. For rapid dual-color imaging of Alexa Fluor 488 actin and Cy3-VASP we used a hybrid Chroma/Shemrock dual bandpass filter. For image analysis of microscopy data, we used ImageJ. An ImageJ Kymograph plugin, designed by J. Rietdorf (FMI Basel, Basel, Switzerland) and A. Seitz (EMBL Heidelberg, Heidelberg, Germany), was used to measure barbed end polymerization rates of single actin filaments. Because VASP is a potent actin bundler, we limited our measurements of barbed end growth to single actin filaments. Error bars for all actin filament growth rates are SDs (n ≥ 30 filaments from ≥2 slides for each condition). Error bars for dwell times are SEM from curve fitting. Burst-phase image sequences of Cy3-VASP were collected for 50–100 s at a frame rate of 50–100 ms. Processivity of single Cy3-VASP barbed end association events were visualized by superimposing the path of actin filament growth onto the Cy3-VASP burst-phase image sequence. Cy3-VASP molecules falling on the diagonal of the kymograph were scored as barbed end association events. This method of analysis was previously used by Bieling et al. (2007) to measure the lifetime of microtubule plus end–tracking proteins. Dwell times for single Cy3-VASP tetramers were determined by fitting 1-cumulative frequency to a single exponential equation, f(x) = x0 * exp^(−x/τ1), or a double exponential equation, f(x) = x0 * exp^(−x/τ1) + (1 − x0) * exp^(−x/τ2). All curve fitting and data plots were made in KaleidaGraph. Final figures were generated with Adobe Photoshop CS3 and Illustrator CS3.
Online supplemental material
Fig. S1 provides additional characterization of the Cy3-VASP GAB and FAB mutants (e.g., barbed polymerization rates, binding to F-actin, and bundling of actin filaments). Fig. S2 shows salt dependence of Cy3-VASP actin filament binding and anti-capping. Fig. S3 investigates whether a cryptic barbed capture motif and/or recognition of the actin filament nucleotide state contributes to VASP barbed end capture. Fig. S4 shows that localization of Cy3-VASP during filament bundling occurs independent of the actin filament orientation. Fig. S5 contains mathematical equations used for determining the affinity and binding capacity of full-length tetrameric Cy3-VASP for monomeric actin. Video 1 shows VASP-dependent acceleration of barbed end polymerization using single actin filament TIRF assay. Video 2 shows dynamic localization of Cy3-VASP on static phalloidin-stabilized actin filaments. Video 3 shows that mutations in the GAB or FAB domain reduce the dwell time on F-actin. Video 4 demonstrates that actin monomer binding to VASP antagonizes lateral interactions with F-actin. Video 5 shows examples of processive Cy3-VASP barbed end tracking. Video 6 provides an example of a highly processive Cy3-VASP tetramer tracking on the barbed end. Video 7 demonstrates that VASP enhances barbed end filament elongation in the presence of profilin–actin. Video 8 show that Cy3-VASP localizes to sites of filament bundling independent of the filament orientations. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201003014/DC1.
Acknowledgments
We thank Orkun Akin for technical support, reagents, and demonstrating how to be a meticulous scientist; Brad Zuchero and Peter Bieling for critical reading of the manuscript; Roger Cooke and Kathy Franks-Skiba for heavy meromyosin; Bethany Simmons for assistance with MatLab; and members of the Mullins, Vale, and Weiner laboratories for reagents and feedback. Scott Hansen dedicates this work to Dr. Galen George, Gary Decicco, and Dennis Fujita for teaching him chemistry at Santa Rosa Junior College.
This work was supported by grants from the National Institutes of Health (ROI #GM61010 to R.D. Mullins), UCSF/UC Berkeley Nanomedicine Development Center (NDC), and the National Science Foundation (to S.D. Hansen).
Footnotes
Abbreviations used in this paper:
- Ena/VASP
- Enabled/vasodilator-stimulated phosphoprotein
- FAB
- filamentous actin binding
- GAB
- globular actin binding
- TIRF
- total internal reflection fluorescence
References
- Adler C.E., Fetter R.D., Bargmann C.I. 2006. UNC-6/Netrin induces neuronal asymmetry and defines the site of axon formation. Nat. Neurosci. 9:511–518 10.1038/nn1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken C.E., Marshall R.A., Puglisi J.D. 2008. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 94:1826–1835 10.1529/biophysj.107.117689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akin O., Mullins R.D. 2008. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 133:841–851 10.1016/j.cell.2008.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applewhite D.A., Barzik M., Kojima S., Svitkina T.M., Gertler F.B., Borisy G.G. 2007. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol. Biol. Cell. 18:2579–2591 10.1091/mbc.E06-11-0990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann C., Fischer L., Walter U., Reinhard M. 1999. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J. Biol. Chem. 274:23549–23557 10.1074/jbc.274.33.23549 [DOI] [PubMed] [Google Scholar]
- Barzik M., Kotova T.I., Higgs H.N., Hazelwood L., Hanein D., Gertler F.B., Schafer D.A. 2005. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J. Biol. Chem. 280:28653–28662 10.1074/jbc.M503957200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J.E., Gertler F.B. 2009. Ena/VASP: towards resolving a pointed controversy at the barbed end. J. Cell Sci. 122:1947–1953 10.1242/jcs.038125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J.E., Svitkina T.M., Krause M., Schafer D.A., Loureiro J.J., Strasser G.A., Maly I.V., Chaga O.Y., Cooper J.A., Borisy G.G., Gertler F.B. 2002. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 109:509–521 10.1016/S0092-8674(02)00731-6 [DOI] [PubMed] [Google Scholar]
- Bieling P., Laan L., Schek H., Munteanu E.L., Sandblad L., Dogterom M., Brunner D., Surrey T. 2007. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 450:1100–1105 10.1038/nature06386 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Pollard T.D. 1999. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J. Biol. Chem. 274:15538–15546 10.1074/jbc.274.22.15538 [DOI] [PubMed] [Google Scholar]
- Boëda B., Briggs D.C., Higgins T., Garvalov B.K., Fadden A.J., McDonald N.Q., Way M. 2007. Tes, a specific Mena interacting partner, breaks the rules for EVH1 binding. Mol. Cell. 28:1071–1082 10.1016/j.molcel.2007.10.033 [DOI] [PubMed] [Google Scholar]
- Breitsprecher D., Kiesewetter A.K., Linkner J., Urbanke C., Resch G.P., Small J.V., Faix J. 2008. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 27:2943–2954 10.1038/emboj.2008.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D., Dominguez R. 2006. Understanding the role of the G-actin-binding domain of Ena/VASP in actin assembly. J. Struct. Biol. 155:195–201 10.1016/j.jsb.2006.01.012 [DOI] [PubMed] [Google Scholar]
- Co C., Wong D.T., Gierke S., Chang V., Taunton J. 2007. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 128:901–913 10.1016/j.cell.2006.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.A., Walker S.B., Pollard T.D. 1983. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil. 4:253–262 10.1007/BF00712034 [DOI] [PubMed] [Google Scholar]
- Dent E.W., Kwiatkowski A.V., Mebane L.M., Philippar U., Barzik M., Rubinson D.A., Gupton S., Van Veen J.E., Furman C., Zhang J., et al. 2007. Filopodia are required for cortical neurite initiation. Nat. Cell Biol. 9:1347–1359 10.1038/ncb1654 [DOI] [PubMed] [Google Scholar]
- Ferron F., Rebowski G., Lee S.H., Dominguez R. 2007. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 26:4597–4606 10.1038/sj.emboj.7601874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher D.A., Mullins R.D. 2010. Cell mechanics and the cytoskeleton. Nature. 463:485–492 10.1038/nature08908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler F.B., Doctor J.S., Hoffmann F.M. 1990. Genetic suppression of mutations in the Drosophila abl proto-oncogene homolog. Science. 248:857–860 10.1126/science.2188361 [DOI] [PubMed] [Google Scholar]
- Gertler F.B., Niebuhr K., Reinhard M., Wehland J., Soriano P. 1996. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 87:227–239 10.1016/S0092-8674(00)81341-0 [DOI] [PubMed] [Google Scholar]
- Gordon D.J., Eisenberg E., Korn E.D. 1976. Characterization of cytoplasmic actin isolated from Acanthamoeba castellanii by a new method. J. Biol. Chem. 251:4778–4786 [PubMed] [Google Scholar]
- Halbrügge M., Walter U. 1989. Purification of a vasodilator-regulated phosphoprotein from human platelets. Eur. J. Biochem. 185:41–50 10.1111/j.1432-1033.1989.tb15079.x [DOI] [PubMed] [Google Scholar]
- Halbrügge M., Friedrich C., Eigenthaler M., Schanzenbächer P., Walter U. 1990. Stoichiometric and reversible phosphorylation of a 46-kDa protein in human platelets in response to cGMP- and cAMP-elevating vasodilators. J. Biol. Chem. 265:3088–3093 [PubMed] [Google Scholar]
- Hüttelmaier S., Harbeck B., Steffens O., Messerschmidt T., Illenberger S., Jockusch B.M. 1999. Characterization of the actin binding properties of the vasodilator-stimulated phosphoprotein VASP. FEBS Lett. 451:68–74 10.1016/S0014-5793(99)00546-3 [DOI] [PubMed] [Google Scholar]
- Iwasa J.H., Mullins R.D. 2007. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr. Biol. 17:395–406 10.1016/j.cub.2007.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D.A., Goldschmidt-Clermont P.J., Levine B.A., Pollard T.D. 1989. Characterization of renatured profilin purified by urea elution from poly-l-proline agarose columns. Cell Motil. Cytoskeleton. 14:251–262 10.1002/cm.970140211 [DOI] [PubMed] [Google Scholar]
- Kaiser D.A., Vinson V.K., Murphy D.B., Pollard T.D. 1999. Profilin is predominantly associated with monomeric actin in Acanthamoeba. J. Cell Sci. 112:3779–3790 [DOI] [PubMed] [Google Scholar]
- Kovar D.R., Harris E.S., Mahaffy R., Higgs H.N., Pollard T.D. 2006. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 124:423–435 10.1016/j.cell.2005.11.038 [DOI] [PubMed] [Google Scholar]
- Krause M., Leslie J.D., Stewart M., Lafuente E.M., Valderrama F., Jagannathan R., Strasser G.A., Rubinson D.A., Liu H., Way M., et al. 2004. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev. Cell. 7:571–583 10.1016/j.devcel.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Kuhn J.R., Pollard T.D. 2005. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88:1387–1402 10.1529/biophysj.104.047399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.R., Pollard T.D. 2007. Single molecule kinetic analysis of actin filament capping. Polyphosphoinositides do not dissociate capping proteins. J. Biol. Chem. 282:28014–28024 10.1074/jbc.M705287200 [DOI] [PubMed] [Google Scholar]
- Kühnel K., Jarchau T., Wolf E., Schlichting I., Walter U., Wittinghofer A., Strelkov S.V. 2004. The VASP tetramerization domain is a right-handed coiled coil based on a 15-residue repeat. Proc. Natl. Acad. Sci. USA. 101:17027–17032 10.1073/pnas.0403069101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski A.V., Rubinson D.A., Dent E.W., Edward van Veen J., Leslie J.D., Zhang J., Mebane L.M., Philippar U., Pinheiro E.M., Burds A.A., et al. 2007. Ena/VASP Is Required for neuritogenesis in the developing cortex. Neuron. 56:441–455 10.1016/j.neuron.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Lacayo C.I., Pincus Z., VanDuijn M.M., Wilson C.A., Fletcher D.A., Gertler F.B., Mogilner A., Theriot J.A. 2007. Emergence of large-scale cell morphology and movement from local actin filament growth dynamics. PLoS Biol. 5:e233 10.1371/journal.pbio.0050233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier L.M., Gates M.A., Witke W., Menzies A.S., Wehman A.M., Macklis J.D., Kwiatkowski D., Soriano P., Gertler F.B. 1999. Mena is required for neurulation and commissure formation. Neuron. 22:313–325 10.1016/S0896-6273(00)81092-2 [DOI] [PubMed] [Google Scholar]
- Loureiro J.J., Rubinson D.A., Bear J.E., Baltus G.A., Kwiatkowski A.V., Gertler F.B. 2002. Critical roles of phosphorylation and actin binding motifs, but not the central proline-rich region, for Ena/vasodilator-stimulated phosphoprotein (VASP) function during cell migration. Mol. Biol. Cell. 13:2533–2546 10.1091/mbc.E01-10-0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Pollard T.D. 2001. Profilin binding to poly-l-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol. Biol. Cell. 12:1161–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffy R.E., Pollard T.D. 2006. Kinetics of the formation and dissociation of actin filament branches mediated by Arp2/3 complex. Biophys. J. 91:3519–3528 10.1529/biophysj.106.080937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney N.M., Janmey P.A., Almo S.C. 1997. Structure of the profilin-poly-l-proline complex involved in morphogenesis and cytoskeletal regulation. Nat. Struct. Biol. 4:953–960 10.1038/nsb1197-953 [DOI] [PubMed] [Google Scholar]
- Mejillano M.R., Kojima S., Applewhite D.A., Gertler F.B., Svitkina T.M., Borisy G.G. 2004. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 118:363–373 10.1016/j.cell.2004.07.019 [DOI] [PubMed] [Google Scholar]
- Melki R., Fievez S., Carlier M.F. 1996. Continuous monitoring of Pi release following nucleotide hydrolysis in actin or tubulin assembly using 2-amino-6-mercapto-7-methylpurine ribonucleoside and purine-nucleoside phosphorylase as an enzyme-linked assay. Biochemistry. 35:12038–12045 10.1021/bi961325o [DOI] [PubMed] [Google Scholar]
- Miyoshi T., Tsuji T., Higashida C., Hertzog M., Fujita A., Narumiya S., Scita G., Watanabe N. 2006. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J. Cell Biol. 175:947–955 10.1083/jcb.200604176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A., Rubinstein B. 2005. The physics of filopodial protrusion. Biophys. J. 89:782–795 10.1529/biophysj.104.056515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel N.F., Barzik M., Raman D., Sobolik-Delmaire T., Sai J., Ham A.J., Mernaugh R.L., Gertler F.B., Richmond A. 2009. VASP is a CXCR2-interacting protein that regulates CXCR2-mediated polarization and chemotaxis. J. Cell Sci. 122:1882–1894 10.1242/jcs.039057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren S., Ojala P.J., Wear M.A., Cooper J.A., Lappalainen P. 2001. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J. Cell Biol. 155:251–260 10.1083/jcb.200106157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasic L., Kotova T., Schafer D.A. 2008. Ena/VASP proteins capture actin filament barbed ends. J. Biol. Chem. 283:9814–9819 10.1074/jbc.M710475200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella E.C., Machesky L.M., Kaiser D.A., Pollard T.D. 1996. Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry. 35:16535–16543 10.1021/bi961498d [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Borisy G.G. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Blanchoin L., Mullins R.D. 2001. Actin dynamics. J. Cell Sci. 114:3–4 [DOI] [PubMed] [Google Scholar]
- Prehoda K.E., Lee D.J., Lim W.A. 1999. Structure of the enabled/VASP homology 1 domain-peptide complex: a key component in the spatial control of actin assembly. Cell. 97:471–480 10.1016/S0092-8674(00)80757-6 [DOI] [PubMed] [Google Scholar]
- Rasnik I., McKinney S.A., Ha T. 2006. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods. 3:891–893 10.1038/nmeth934 [DOI] [PubMed] [Google Scholar]
- Rottner K., Behrendt B., Small J.V., Wehland J. 1999. VASP dynamics during lamellipodia protrusion. Nat. Cell Biol. 1:321–322 10.1038/13040 [DOI] [PubMed] [Google Scholar]
- Stuurman N., Amodaj N., Vale R.D. 2007. µManager: Open Source software for light microscope imaging. Microscopy Today. 15:42–43 [Google Scholar]
- Svitkina T.M., Bulanova E.A., Chaga O.Y., Vignjevic D.M., Kojima S., Vasiliev J.M., Borisy G.G. 2003. Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160:409–421 10.1083/jcb.200210174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige M., Klopfenstein D.R., Vale R.D. 2002. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 297:2263–2267 10.1126/science.1073386 [DOI] [PubMed] [Google Scholar]
- Vignjevic D., Kojima S.I., Aratyn Y., Danciu O., Svitkina T.M., Borisy G.G. 2006. Role of fascin in filopodial protrusion. J. Cell Biol. 174:863–875 10.1083/jcb.200603013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson V.K., De La Cruz E.M., Higgs H.N., Pollard T.D. 1998. Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry. 37:10871–10880 10.1021/bi980093l [DOI] [PubMed] [Google Scholar]
- Welch M.D., Mullins R.D. 2002. Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 18:247–288 10.1146/annurev.cellbio.18.040202.112133 [DOI] [PubMed] [Google Scholar]