Abstract
Histone deacetylase inhibitors induce cell cycle arrest and apoptosis in tumor cells and are, therefore, promising anti-cancer drugs. The cyclin-dependent kinase inhibitor p21 is activated in histone deacetylase (HDAC) inhibitor-treated tumor cells, and its growth-inhibitory function contributes to the anti-tumorigenic effect of HDAC inhibitors. We show here that induction of p21 by trichostatin A involves MAP kinase signaling. Activation of the MAP kinase signaling pathway by growth factors or stress signals results in histone H3 serine 10 phosphorylation at the p21 promoter and is crucial for acetylation of the neighboring lysine 14 and recruitment of activated RNA polymerase II in response to trichostatin A treatment. In non-induced cells, the protein phosphatase PP2A is associated with the p21 gene and counteracts its activation. Induction of p21 is linked to simultaneous acetylation and phosphorylation of histone H3. The dual modification mark H3S10phK14ac at the activated p21 promoter is recognized by the phospho-binding protein 14-3-3ζ, which protects the phosphoacetylation mark from being processed by PP2A. Taken together we have revealed a cross-talk of reversible phosphorylation and acetylation signals that controls the activation of p21 by HDAC inhibitors and identify the phosphatase PP2A as chromatin-associated transcriptional repressor in mammalian cells.
Keywords: Chromatin Histone Modification, RNA Polymerase II, Signal Transduction, Transcription Regulation, Tumor Therapy, Histone Acetylation, Histone Phosphorylation
Introduction
Post-translational modifications of histones, such as acetylation, methylation, phosphorylation, and sumoylation play important roles in the regulation of chromatin accessibility and gene expression. The transformation of a differentiated resting cell into a highly proliferative tumor cell is accompanied by a severe change of the gene expression profile, which in turn is the direct consequence of acquired/accumulated tumor suppressor and oncogene mutations. In addition to these genetic alterations, epigenetic alterations are also involved in the development of cancer. These include covalent post-translational modifications of histones and changes in the methylation status of cytosines within CpG islands (1, 2). Therefore, compounds that target these epigenetic changes are of potential interest for anti-tumor therapies. Indeed, histone deacetylase (HDAC)5 inhibitors have been shown to induce cell cycle arrest, differentiation, or apoptosis in tumor cells and are, therefore, promising anti-tumor drugs (3–5).
One of the genes that is activated by different HDAC inhibitors encodes the cyclin-dependent kinase (CDK) inhibitor p21cip1/waf1 (referred to as p21 thereafter) (3, 4). The p21 protein is a member of the CIP/KIP family of cyclin-dependent kinase inhibitors consisting of p21, p27, and p57. The different members of the CIP/KIP family share a conserved region at the N terminus that is required and sufficient for the inhibition of cyclin-Cdk complexes (6, 7). The p21 protein preferentially inactivates cyclin E/Cdk2 complexes, thereby blocking the inactivating phosphorylation of pRb, which represses genes important for S phase entry. In addition, p21 can bind to proliferating cell nuclear antigen, thus, blocking DNA synthesis (8, 9). Paradoxically, p21 also promotes in a concentration-dependent manner the formation of active complexes by stabilizing the interaction between Cdk4/Cdk6 and D-cyclins (10). In agreement with this finding, expression of the p21 gene is activated by growth factors (11). Interestingly, p21 can also directly affect transcription by association with specific promoter regions or by binding to specific transcription factors or co-activators (12–15).
The p21 gene is a transcriptional target of p53 and plays a crucial role in mediating growth arrest when cells are exposed to DNA-damaging agents such as doxorubicin and γ-irradiation (16, 17). In addition to its role in DNA damage response, p21 is also implicated in terminal differentiation, replicative senescence, and protection from p53-dependent and -independent apoptosis (18, 19). The p21 gene is regulated by a plethora of transcription factors and transcriptional regulators, and its activation is accompanied by histone hyperacetylation, suggesting a negative regulatory role for HDACs (20–22). Indeed, several members of the HDAC family have been shown to repress p21 expression (3, 23, 24). Importantly, HDAC inhibitors can activate p21 transcription in the absence of p53, allowing for p21 induction in tumor cells lacking functional p53 (25, 26).
The activity of p21 contributes to the growth inhibitory effect of HDAC inhibitors, but it is evident that also other factors are involved in this process (27–29). For instance, several other cell cycle regulators including gelsolin, p16, p27, and different cyclins are deregulated upon HDAC inhibition (5). In addition, p21, which is usually considered to act as a tumor suppressor, might also act as an oncogene (30). Along this line, a tumor promoting activity of p21 was recently shown in different mouse tumor models (31, 32). Given the opposite functions of p21 in growth control and tumorigenesis, it is crucial to understand the mechanism underlying the activation of the p21 gene by HDAC inhibitors.
In this study we analyze in detail the mechanisms regulating p21 expression in response to HDAC inhibition. We show that histone H3 phosphorylation at the p21 promoter is indispensable for the activation of the p21 gene by HDAC inhibitors. The activation of stress or mitogen-activated signaling cascades in concert with HDAC inhibition leads to simultaneous Ser-10 phosphorylation and Lys-14 acetylation (phosphoacetylation) of histone H3 at the p21 promoter. We identify mitogen- and stress-activated kinase 1 (MSK1) to be one of the kinases responsible for histone H3 phosphorylation and demonstrate its importance for p21 gene activation. Furthermore, we show for the first time that chromatin-associated protein phosphatase PP2A can act as transcriptional repressor in mammalian cells. Finally, we demonstrate that recognition of the phosphoacetylation mark by the adaptor protein 14-3-3ζ is important for p21 activation, and this might be in part due to protection of the mark from dephosphorylation by PP2A.
EXPERIMENTAL PROCEDURES
Cell Culture and Drug Treatment
Swiss 3T3 mouse fibroblasts were grown in DMEM medium supplemented with 10% (v/v) fetal calf serum (FCS) and antibiotics. For cell cycle arrest in G0, cells were serum-deprived for 72 h using DMEM medium containing 0.2% (v/v) FCS. Re-entry into the cell cycle was induced with 20% (v/v) FCS. The human glioblastoma cell line T98G was cultured in DMEM containing 10% (v/v) FCS and was arrested in G0 by reducing FCS to 0.2% (v/v) for 48 h.
Proliferating and resting cells were treated with trichostatin A (TSA, 165.5 nm, Wako Pure Chemical Industries), a subinhibitory concentration of anisomycin (188.5 nm, Sigma) as previously described (33), and okadaic acid (50 nm, Sigma), H89 (10 μm, Alexis Biochemicals) for different periods of time.
RNA Extraction and RT-Real Time PCR
Total RNA was prepared from Swiss 3T3, and T98G cells using the TRIzol reagent (Invitrogen) according to the manufacture's protocol. For RT-real time PCR, 1 μg of total RNA was reverse-transcribed in cDNA using the iScriptTMcDNA synthesis kit (Bio-Rad). 5 μl of a 1:10 dilution of cDNA was analyzed by SYBER Green real time PCR using the Bio-Rad iCycler iQ system. Expression of p21 was normalized to the housekeeping genes mouse hypoxanthine-guanine phosphoribosyltransferase (HPRT) or human glycerinaldehyd-3-phosphate dehydrogenase (GAPDH), whose expression is not affected by the nucleosomal response. Primer sequences are available on request.
Histone extraction, dot blot, and Western blot analysis. Histone preparation and Western blot analysis were performed as previously described (35). The following antibodies were used in this study: modification-specific antibodies against acetylated histone H3 (06-599) and H4 (06-866), H3K14ac (07-353) from Millipore Upstate Biotechnology; H3S10ph (Santa Cruz 8656-R); H3S10phK14ac (Seiser and co-workers (34)); C-terminal histone H3 antibody (Abcam 1791); phospho-MSK1 (Thr-581) (Cell Signaling #959); 14-3-3ζ (Santa Cruz Biotechnology); Ser-5-phosphorylated RNA polymerase II (Bethyl Laboratories A300-655A); PP2A catalytic subunit (the E. Ogris laboratory); β-tubulin (Sigma T4026; normal rabbit IgG (Invitrogen 10500C)).
Chromatin Immunoprecipitation Assays
ChIP assays were performed as described previously (35). Antibodies used in this study are listed above. Quantification of precipitated material was performed via quantitative real time (qRT) PCR using an iCycler iQ system from Bio-Rad and KAPA SYBR FAST qPCR MasterMix (Peqlab). Primer sequences are available on request.
RNA Interference Experiments
T98G cells were transfected with siRNAs at a final concentration of 24 nm using Lipofectamine 2000 (Invitrogen). The next day cells were either serum-deprived (0.2% (v/v) FCS) for 48 h or kept in proliferation medium containing 10% (v/v) FCS. The following siRNA oligonucleotides were used in this study: hMSK1 (Santa Cruz, sc-35977); h14-3-3ζ (Santa Cruz, sc-29583); hPP2A-Cα (Dharmacon, L-003598-00); hPP2A-Cβ (Dharmacon, L-003599-00); unspecific hRNA-A (Santa Cruz, sc-37007).
In Vitro Phosphatase Assay and Protection Assay
Histones were isolated from resting 3T3 fibroblasts, which were treated with anisomycin (188.5 nm) for 1 h by acid extraction as described (35). Extracted histones were diluted to 1 μm in phosphatase assay buffer (50 mm Tris-HCl, pH 7.5, 75 mm NaCl, 0.1% β-mercaptoethanol, 0.1 mm EDTA) and either preincubated with an equimolar 2 or 4× molar excess of recombinant 14-3-3ζ-GST for 2h at 4 °C. In addition, control samples without 14-3-3ζ were performed. Phosphatase reactions were carried out by adding 0.5 units of PP2A A/C dimer (Millipore Upstate Biotechnology) for 2 h at 30 °C. Reactions were lyophilized resuspended in SDS sample buffer and analyzed by immunoblotting.
Statistical Analysis
p values were calculated using the right-tailed Fisher exact test.
RESULTS
Activation of p21 Expression by HDAC Inhibition Requires a Second Signal
In this study we intended to analyze the molecular mechanisms regulating the activation of the p21 gene in response to HDAC inhibitor treatment. Induced p21 transcription in response to HDAC inhibitor treatment of human tumor cells was previously shown to be accompanied by increased histone acetylation at the proximal p21 promoter region but not at the 3′ end of the gene (36) (Fig. 1A). Similar acetylation patterns were also obtained for the mouse p21 gene in HDAC inhibitor treated embryonic stem cells (data not shown). The proximal promoter region contains multiple binding sites for the transcription factors Sp1, Sp3, and AP2. Sp1/Sp3 was shown to be involved in the recruitment of HDACs and histone acetyltransferases and to mediate the effect of the phosphatase inhibitor okadaic acid (20, 23). In agreement with previous reports (23, 37), HDAC inhibition by TSA led to strong induction of p21 expression in proliferating untransformed Swiss 3T3 mouse fibroblasts and transformed fibroblast lines COP8 and Tau2 (Fig. 1B and data not shown). To test whether TSA-mediated activation of p21 in untransformed fibroblasts is also linked to hyperacetylation of core histones, we performed ChIP assays. As shown in Fig. 1C, ChIP experiments with antibodies specific for either acetylated histone H3 or H4 demonstrated that TSA treatment of proliferating fibroblasts results in increased H3 and H4 acetylation at the p21 promoter region. In agreement with previously published results (36), histone acetylation at the 3′ end of the p21 gene was not affected by TSA treatment.
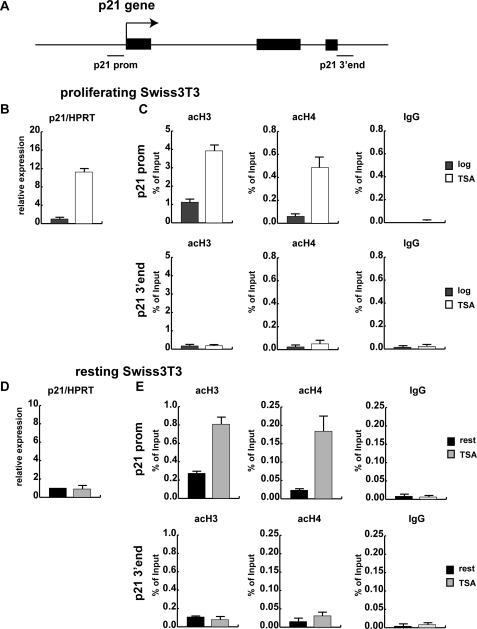
FIGURE 1.
Proliferating and resting mouse fibroblasts respond differently to the HDAC inhibitor TSA. A, shown is a schematic view of the murine p21 gene. The three exons are depicted as black boxes, and the regions analyzed by qRT-ChIP are indicated. B, proliferating Swiss 3T3 mouse fibroblasts were kept either untreated or were treated with 165.5 nm TSA for 3 h. Total RNA was extracted, and reverse-transcribed cDNA was used to quantify p21 mRNA levels by quantitative real time qRT PCR using hypoxanthine-guanine phosphoribosyltransferase (HPRT) as a housekeeping gene for normalization. C, formaldehyde cross-linked chromatin was prepared from proliferating cells treated with 165.5 nm TSA for 3 h. ChIP experiments were performed with an unspecific rabbit IgG antibody as control, two antibodies specific for acetylated histones H3 (acH3) and H4 (acH4), and an antibody recognizing the C terminus of histone H3 to correct for potential changes in nucleosomal density. DNA from the antibody-bound fraction and a dilution of total input DNA (Input) isolated from the respective chromatin sample was analyzed by qRT PCR using primers specific for the p21 promoter (p21 prom) and the 3′ end of the p21 gene (p21 3′ end) as control. ChIP results are shown as the percentage of input corrected for changes in nucleosomal density. D, Swiss 3T3 cells were arrested by serum deprivation for 72 h, treated with 165.5 nm TSA for 3 h, and analyzed by qRT PCR for p21 expression. E, qRT-ChIP with unspecific antibody (IgG) and two antibodies specific for acetylated histones H3 (acH3) and H4 (acH4) was performed as described in panel C.
The majority of cells in an adult mammalian organism is in the G0/G1 phase of the cell cycle. Therefore, we asked next whether the p21 gene is also activated by TSA in non-proliferating cells. To this end we analyzed p21 mRNA levels in serum-deprived resting Swiss 3T3 fibroblasts. In comparison to proliferating Swiss 3T3 cells, expression of p21 was about 8-fold lower in resting fibroblasts. Interestingly, in the absence of growth factors the HDAC inhibitor TSA failed to activate p21 transcription (Fig. 1D). Even longer periods of TSA treatment up to 9 h did not induce p21 mRNA expression (data not shown). Strikingly, ChIP assays with acetyl H3 and acetyl H4 antibodies revealed a strong TSA-mediated increase in H3 and H4 acetylation at the p21 promoter in resting cells (Fig. 1E). However, despite the observed histone hyperacetylation, p21 transcription was not activated in resting cells, suggesting the requirement for an additional signal.
Expression of p21 Is Activated by Inducers of the Mitogen-activated Protein (MAP) Kinase Pathway and Correlates with H3 Phosphoacetylation
An obvious difference between proliferating and resting cells is the activation status of MAP kinase cascades. Serum-deprived cells lack stimulation of growth factor-activated signal transduction pathways and, thus, arrest in the G0/G1 phase. In addition to growth factors, cellular stress also activates MAP kinase cascades in resting fibroblasts but without inducing cell cycle entry. We, therefore, asked whether activation of the p38 MAP kinase pathway would affect p21 expression in TSA-treated cells.
The stress inducer anisomycin activates the p38 MAP kinase pathway at low concentrations that have no effect on protein synthesis (38). To test the effect of stress and growth factor signals on p21 expression, serum-deprived mouse fibroblasts were stimulated for 1 h with subinhibitory concentrations of anisomycin (188.5 nm) or with 20% FCS alone or in combination with TSA. Analysis of p21 mRNA expression by real time PCR revealed a 2-fold increase by both serum stimulation and anisomycin treatment, whereas TSA had hardly any effect on p21 levels (Fig. 2A). Importantly, when TSA was used together with either of the MAPK activators anisomycin or FCS, we observed an enhanced transcriptional activation, resulting in a 3.5- and a 3-fold induction of p21 expression, respectively.
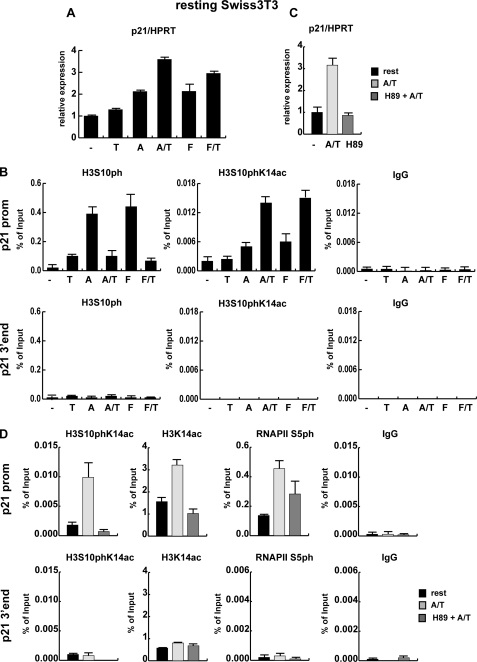
FIGURE 2.
Activated MAP kinase cascades are required for p21 expression. A, resting Swiss 3T3 mouse fibroblasts were left untreated (-) or were treated with 165.5 nm TSA alone (T), 188.5 nm anisomycin alone (A), or both drugs simultaneously (A/T) or were stimulated with 20% serum (F) either alone or in combination with 165.5 nm TSA (F/T) for 1 h. Total RNA was extracted, and reverse-transcribed cDNA was used to quantify p21 mRNA levels by qRT PCR using hypoxanthine-guanine phosphoribosyltransferase (HPRT) as a housekeeping gene for normalization. The difference in p21 expression between untreated resting cells and TSA-treated cells is not significant (p value > 0.05), whereas all other treatments resulted in significant changes in p21 expression. B, chromatin was prepared from cells treated as described in panel A and precipitated using an unspecific antibody (IgG), an histone H3S10ph antibody, and an antibody specifically recognizing histone H3 phosphoacetylated on serine 10 and lysine 14 (H3S10phK14ac). Precipitated DNA was analyzed by qRT PCR using primers specific for the promoter (upper panel) and the 3′ end (lower panel) of the p21 gene. Panels C and D, resting Swiss 3T3 mouse fibroblasts were treated for 3 h with 188.5 nm anisomycin and 165.5 nm TSA with (H89 + A/T) or without (A/T) pretreatment for 15 min with 10 μm H89. C, total RNA was extracted from stimulated cells, and reverse-transcribed cDNA was used for p21 expression analysis by qRT PCR. D, ChIP assays of resting and anisomycin/TSA stimulated 3T3 cells in the presence and absence of H89 are shown. Chromatin was precipitated with an unspecific control antibody (IgG) or antibodies specific for H3S10phK14ac, H3K14ac, H3 C terminus, and initiation-competent RNA polymerase II (RNAP II phS5), and qRT PCR was performed with primers for the p21 promoter (p21 prom) and the 3′end of the p21 gene (p21 3′ end) as control. ChIP results are shown as percentage of input; for histone modifications the values were corrected for changes in nucleosomal density.
Activation of mitogen- and stress-activated kinase cascades leads not only to the activation of a variety of transcription factors but also to phosphorylation of HMG-14 (HMGN-1) and histone H3 known as the nucleosomal response (39). Phosphorylation of histone H3 at serine 10 was previously shown to be associated with transcriptional activation of a variety of mammalian genes (40–45). Phosphorylation is targeted to a minute fraction of histone H3, which is also especially susceptible to hyperacetylation, resulting in simultaneous phosphorylation and acetylation of histone H3 referred to as phosphoacetylation (46). It is important to note that in proliferating cells a considerable part of the Ser-10-phosphorylated histone H3 (H3S10ph) and phosphoacetylated histone H3 (H3S10phK14ac) stems from the mitotic fraction. Indeed, compared with resting cells, the abundance of histone isoforms H3S10ph and H3S10phK14ac, but also H3K14ac, was enhanced in proliferating Swiss 3T3 cells and anisomycin/TSA-treated resting cells (supplemental Fig. 1A). To monitor local changes in histone H3 phosphorylation, we performed quantitative ChIP assays with an H3S10ph-specific antibody. As shown in Fig. 2B, local H3 phosphorylation at the p21 promoter increased in response to activation of MAP kinase cascades in resting cells by anisomycin or FCS. Simultaneous TSA treatment led to reduction in the H3S10ph signal. However, it is known that acetylation of neighboring lysine residues can negatively affect the recognition of the H3S10ph mark by this antibody due to epitope occlusion (supplemental Fig. 1B and Refs. 35, 46). Therefore, we performed ChIP assays with an antibody specifically recognizing dually modified S10phK14ac histone H3 (Ref. 35 and supplemental Fig. 1B). Using the H3S10phK14ac antibody, we detected H3 phosphoacetylation in response to anisomycin or FCS in resting fibroblasts (Fig. 2B). In both cases the phosphoacetylation signal was increased upon additional HDAC inhibition by TSA. Thus, the histone H3 phosphoacetylation patterns at the proximal p21 promoter closely parallel the transcriptional activation the p21 gene. In contrast, histone H3 phosphorylation and phosphoacetylation were not induced at the 3′ end of the p21 gene (Fig. 2B, lower panel). Taken together, active MAP kinase cascades are important for the induction of the p21 gene by TSA.
Histone Phosphorylation by MSK1 Kinase Is Crucial for Full p21 Activation
Studies with MSK1 and MSK2 single and double knock-out cells identified MSK1 and MSK2 as the major histone H3 serine 10 effector kinases downstream of the ERK and p38 pathways (47). As a first step to analyze the contribution of histone phosphorylation to the activation of the p21 gene by HDAC inhibition, we used the protein kinase inhibitor H89. This compound was shown to inhibit rather specifically MSK1/MSK2 and the nucleosomal response without affecting phosphorylation of relevant transcription factors (48). As shown in Fig. 2C, pretreatment of resting mouse fibroblasts with H89 efficiently blocked the induction of p21 expression by combinatorial treatment with TSA and anisomycin. Similarly, H89 treatment significantly reduced the activation of the p21 gene by anisomycin, TSA, and anisomycin/TSA in proliferating Swiss 3T3 cells (supplemental Fig. 2). Thus, the nucleosomal response is important for activation of the p21 gene by these drugs. Accordingly, ChIP analysis revealed that H89 pretreatment of anisomycin/TSA-treated resting cells completely inhibited H3 phosphoacetylation at the p21 promoter (Fig. 2D).
Previously, a possible interrelationship between H3S10 phosphorylation and the acetylation of neighboring lysine residues was discussed (49, 50). Therefore, we examined next whether H3S10 phosphorylation at the p21 promoter had an impact on acetylation of H3K14 in response to TSA/anisomycin treatment. ChIP assays using the H3K14ac antibody revealed that acetylation of histone H3 at Lys-14 was induced by combinatorial treatment with TSA and anisomycin (Fig. 2D). Blocking of the nucleosomal response by H89 significantly reduced H3K14 acetylation at the p21 promoter in anisomycin/TSA-treated cells, suggesting a link between MAP kinase signaling and recruitment or activity of a H3K14 histone acetyltransferase.
Next, we analyzed the effect of the nucleosomal response on transcriptional initiation of the p21 gene. To this end we monitored by quantitative real time (qRT)-ChIP the recruitment of initiation-competent RNA polymerase II (RNAPII S5ph) (51) to the p21 promoter. As shown in Fig. 2D, RNA polymerase II (RNAPII) S5ph was recruited to the p21 promoter in response to combinatorial treatment with TSA and anisomycin, and this recruitment was sensitive to the inhibitor of the nucleosomal response H89. Taken together, these data strongly suggest that H3 phosphorylation via the nucleosomal response is required for local H3K14 acetylation, recruitment of activated RNA polymerase II, and induction of the p21 gene in response to anisomycin/TSA.
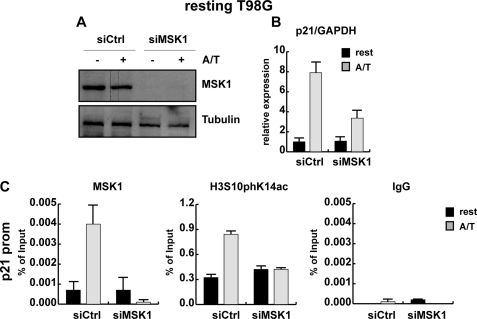
However, it is important to note that H89 not only affects the enzymatic activity of MSK1 and MSK2 but can also inhibit other protein kinases including PKA, S6K1, and ROCK-II (52). Therefore, we performed siRNA knockdown experiments to ask whether MSK1 is important for p21 gene activation. To this end, we abolished MSK1 expression in T98G human glioblastoma cells. Pilot experiments indicated significantly higher knockdown efficiencies in this cell line compared with mouse fibroblasts. T98G cells have a fibroblast like morphology and, similar to mouse fibroblasts, can be arrested in G0 by serum deprivation (53). This is of advantage to study interphase phosphorylation of histone H3, as cells can be synchronized in G0 to exclude interference with mitotic phosphorylation. T98G cells were transfected with MSK1 siRNA or control siRNA, arrested for 48 h, and either left untreated or stimulated with anisomycin and TSA for 3 h. Both mRNA and protein levels of MSK1 were significantly reduced in siRNA-treated cells as shown by real time PCR and Western blot analysis (Fig. 3A and supplemental Fig. 3A). Resting T98G glioblastoma cells showed an 8-fold increase in p21 expression upon combinatorial anisomycin and TSA treatment (Fig. 3B). Importantly, p21 activation by anisomycin alone or anisomycin/TSA was reduced in MSK1-depleted cells compared with resting control cells (Fig. 3B and supplemental Fig. 3B). Activation of the p21 gene in resting T98G cells was accompanied by enhanced recruitment of MSK1 and induced histone H3 phosphoacetylation at the proximal p21 promoter (Fig. 3C). Importantly, the presence of the H3S10phK14ac mark at the p21 promoter was significantly reduced upon loss of MSK1, suggesting that this enzyme is one of the crucial mediators of the nucleosomal response in T98G cells (Fig. 3C). These data demonstrate an important role of MSK1 in p21 expression in response to stress signaling.
FIGURE 3.
The effector kinase MSK1 is important for p21 gene induction by TSA/anisomycin. Proliferating T98G cells were transfected with control siRNA oligonucleotides and siRNA oligonucleotides specifically targeting MSK1 mRNA for 5 h. Next day-transfected T98G cells were arrested by serum deprivation for 48 h. Whole cells extracts, total RNA, and chromatin were gained from control (siCtrl) and knockdown cells (siMSK1) that were left untreated or were stimulated with 188.5 nm anisomycin and 165.5 nm TSA (+ or A/T) for 1 h. A, knockdown efficiency was analyzed by Western blot using an antibody against MSK1. Antibody against β-tubulin ensured equal loading. B, p21 expression levels in control and MSK1 knockdown cells were monitored by qRT PCR using GAPDH for normalization. C, qRT-ChIP analysis was performed with antibodies specific for MSK1, H3S10phK14ac, or an unspecific control antibody (IgG) and primers specific for the proximal p21 promoter. ChIP results are shown as percentage of input; for histone modifications, the values were corrected for changes in nucleosomal density.
The Phosphatase Inhibitor Okadaic Acid Induces p21 Expression and Histone H3 Phosphoacetylation in Proliferating Cells
As histone H3 phosphorylation is a reversible mark, specific phosphatases counteract kinase-mediated phosphorylation and its biological read-out. At the end of mitosis, the phosphatase PP1 removes H3S10ph marks that are placed at the G2/M transition by Aurora B (54, 55). However, little is known about the phosphatases regulating Ser-10 dephosphorylation during interphase. Genetic studies in Drosophila suggest a role for PP2A in the regulation of heat shock-regulated genes, which is linked to histone H3 phosphorylation (56, 57). We, therefore, asked whether PP2A would play a role in the control of dynamic histone H3 phosphorylation and p21 gene expression in mammalian cells.
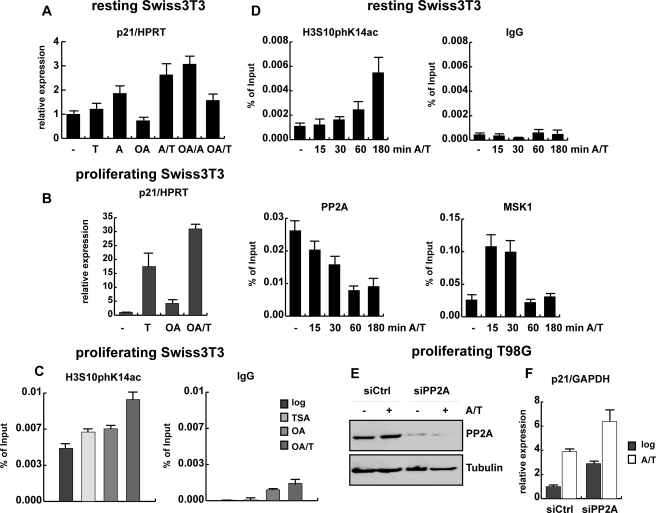
Fibroblasts were treated with the phosphatase inhibitor okadaic acid, which at low concentrations (50 nm) specifically inhibits PP2A (and PP2A-related enzymes) without affecting PP1 activity (58, 59). Okadaic acid treatment has been previously shown to activate p21 expression in human cancer cells (60). In resting mouse fibroblasts, okadaic acid treatment in the absence of MAP kinase signaling had no major impact on global phosphoacetylation levels and on p21 expression (Fig. 4A and data not shown). Only in combination with the MAP kinase inducer anisomycin was p21 expression elevated by okadaic acid to levels comparable with anisomycin/TSA-treated cells. Therefore, we analyzed a putative effect of okadaic acid on phosphoacetylation and p21 expression in proliferating Swiss 3T3 fibroblasts. Cells were stimulated for 5 h either with okadaic acid or TSA alone or in combination. Treatment with TSA or okadaic acid alone induced phosphoacetylation of bulk histones and combination of okadaic acid with TSA gave even higher H3S10phK14ac signals (data not shown). Treatments with TSA or okadaic acid led to a 17- or 4-fold increase, respectively, in p21 expression levels (Fig. 4B). Combinations of okadaic acid with anisomycin or TSA had a synergistic effect on the activation of the p21 promoter in proliferating fibroblasts. In accordance with higher p21 expression in proliferating cells (Fig. 1), ChIP assays with the H3S10phK14ac antibody showed higher levels of H3 phosphoacetylation at the p21 promoter in untreated proliferating cells compared with resting cells (compare Figs. 4C with 2D). This is in part due to the presence of mitotic cells with high H3S10 phosphorylation levels. Combined treatment with TSA and okadaic acid further enhanced H3 phosphoacetylation at the p21 promoter (Fig. 4C). These findings suggest that in the presence of growth factors (i.e. in proliferating cells) HDAC inhibitors and phosphatase inhibitors cooperate in the induction of phosphoacetylation and p21 expression, as the turnover of phosphate and acetyl moieties is blocked. These data also suggest PP2A (or a PP2A like phosphatase) to be involved in the regulation of p21 transcription.
FIGURE 4.
PP2A acts a as negative regulator of p21 expression. A, resting Swiss 3T3 cells obtained by serum deprivation for 72 h were left untreated or treated for 5 h as indicated with the following concentrations: 165.5 nm TSA (T), 188.5 nm anisomycin (A), or 50 nm okadaic acid (OA) alone or in combination. Expression of p21 mRNA was examined by qRT PCR normalized to hypoxanthine-guanine phosphoribosyltransferase (HPRT) and is shown relative to untreated cells. The difference in p21 expression between untreated and TSA-treated cells is not significant (p value: 0.051). B, proliferating Swiss 3T3 cells were induced with the same stimuli and for the same time period as described in panel A. Total RNA was extracted, and reverse-transcribed cDNA was used for p21 expression analysis by qRT PCR as described for panel A. C, qRT-ChIP analysis of proliferating Swiss 3T3 fibroblasts either left untreated or stimulated with 50 nm okadaic acid (OA), and 165.5 nm TSA alone or simultaneously (OA/T) using a phosphoacetylhistone H3 antibody (H3S10phK14ac) or an unspecific antibody (IgG). D, a ChIP assay of a time course is shown in which resting cells were stimulated with 188.5 nm anisomycin and 165.5 nm TSA for short time periods (indicated in minutes) using an unspecific antibody (IgG) or antibodies specific for H3S10phK14ac, the catalytic subunit of PP2A, and MSK1. ChIP results are shown as percentage of input; for histone modifications the values were corrected for changes in nucleosomal density. E and F, proliferating T98G cells were transfected with control siRNA oligonucleotides and siRNA oligonucleotides specifically targeting the two isoforms of the catalytic subunit of PP2A. Protein extracts and total RNA were gained from control (siCtrl) and knockdown cells (siPP2A) that were left untreated or were stimulated with 188.5 nm anisomycin and 165.5 nm TSA (A/T) for 1 h. E, knockdown efficiency was analyzed by Western blot using an antibody against the catalytic subunit of PP2A. An antibody specific for β-tubulin was used to ensure equal loading. F, p21 mRNA expression levels in control and PP2A knockdown cells were monitored by qRT PCR using GAPDH for normalization.
Chromatin-associated PP2A Is a Negative Regulator of p21 Expression
Next, we examined whether PP2A is directly associated with the p21 promoter. We performed ChIP assays with resting 3T3 cells after short-term treatment with anisomycin and TSA using the phosphoacetyl-H3 antibody and an antibody specific for catalytic C-subunit of PP2A. To assess a potential kinase recruitment, short term kinetics of MSK1 association and phosphoacetylation turnover were determined by ChIP assays. Resting fibroblasts stimulated with anisomycin and TSA for 15, 30, 60, and 180 min were subjected to ChIP analysis using antibodies specific for phosphoacetylated histone H3 (H3S10phK14ac), the catalytic subunit of PP2A and MSK1 (Fig. 4D). Signaling to the p21 gene was fast and coincided with a rapid recruitment of MSK1 to the p21 promoter only after 15 min of induction followed by increasing phosphoacetylation of p21-associated chromatin. MSK1 recruitment seemed to be a dynamic and transient event, which was reduced after 30 min of stimulation. PP2A was present at the p21 promoter in resting, non-stimulated cells but was gradually reduced upon treatment with TSA and anisomycin, whereas local H3 phosphoacetylation increased at the same time (Fig. 4D). In agreement with the H3S10phK14ac ChIP data shown in Fig. 2D, both MSK1 and PP2A were absent at the 3′ end of the p21 gene (data not shown).
To test whether PP2A is indeed a transcriptional repressor of the p21 gene, we ablated the expression of the catalytic C-subunit of PP2A in T98G cells. As mentioned above, the phosphatase inhibitor okadaic acid induced p21 expression only in proliferating cells. Therefore, we depleted proliferating T98G cells simultaneously of both C-subunit isoforms (α and β) by siRNA-mediated knockdown. PP2A C-subunit mRNA and protein levels were efficiently reduced in knockdown cells (Fig. 4E and supplemental Fig. 3C). Notably, loss of PP2A resulted in a more than 2-fold increase in p21 expression in untreated proliferating T98G cells (Fig. 4F). The levels of p21 mRNA were also induced upon loss of PP2A in anisomycin/TSA-treated cells, suggesting a modulating function of the phosphatase in MAP kinase stimulated cells. In summary, these data indicate a novel role of PP2A as negative regulator of p21 expression.
The H3S10ph Binding Module 14-3-3 Is Required for p21 Activation by the Nucleosomal Response
14-3-3 proteins are well established phosphoserine-binding proteins, and recently, members of the 14-3-3 family were shown to bind histone H3 phosphorylated serine 10 (61). Importantly, the binding affinities of 14-3-3 to phosphorylated histone H3 are increased by acetylation of neighboring lysine residues, especially lysine 14 (62, 63). Moreover, 14-3-3 epsilon and, in particular, 14-3-3ζ, are required for transcriptional activation of the HDAC1 gene by anisomycin and TSA (64).
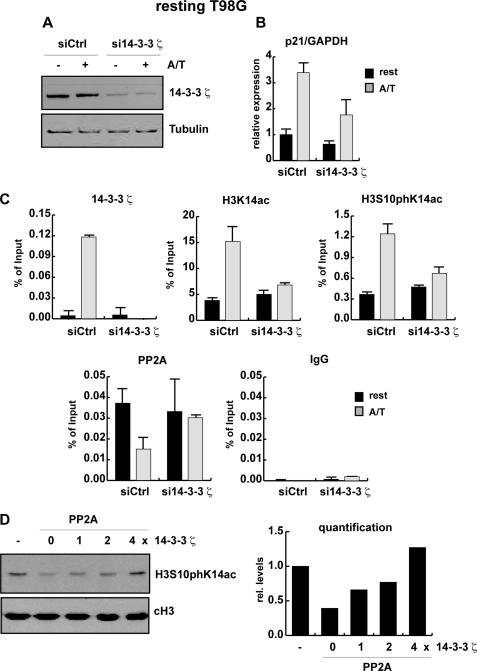
We, therefore, investigated whether 14-3-3ζ is required for p21 gene activation. To this end proliferating T98G cells were transfected with either 14-3-3ζ specific siRNA or control siRNA, resulting in efficient reduction of both mRNA and protein expression (Fig. 5A and supplemental Fig. 3D). Expression of p21 in untreated and anisomycin/TSA-stimulated 14-3-3 knockdown and control cells was monitored by real time PCR analysis. In resting T98G control cells, p21 mRNA levels were 3.5-fold increased by combined anisomycin and TSA treatment (Fig. 5B). In 14-3-3ζ knockdown cells, activation by anisomycin and TSA was significantly reduced, indicating a crucial role of 14-3-3ζ for p21 expression. Of note, in the absence of TSA/anisomycin treatment, 14-3-3ζ-deficient proliferating T98G cells show a 50% reduction of p21 expression. In ChIP assays with a 14-3-3ζ antibody, we observed efficient recruitment of 14-3-3ζ to the proximal p21 promoter (Fig. 5C). In a similar manner, 14-3-3ζ was detected at the p21 promoter in anisomycin/TSA-treated resting Swiss 3T3 fibroblasts (data not shown). MSK1 knockdown experiments revealed that 14-3-3ζ recruitment to the p21 promoter was reduced in the absence of MSK1 (supplemental Fig. 4). To test whether loss of 14-3-3ζ has effects on H3 acetylation and phosphoacetylation at the p21 gene, we performed ChIP assays with H3K14ac and H3S10phK14ac antibody in control and 14-3-3ζ knockdown cells. As shown in Fig. 5C depletion of 14-3-3ζ in resting T98G cells interfered with the TSA/anisomycin-mediated increase in H3K14 acetylation and H3 phosphoacetylation at the p21 promoter. Interestingly, dissociation of PP2A from the p21 promoter in response to anisomycin/TSA is abolished in the absence of 14-3-3ζ (Fig. 5C). Similarly, H89 pretreatment blocked the dissociation of PP2A in anisomycin/TSA-treated resting Swiss 3T3 fibroblasts, and the presence of PP2A at the p21 promoter was enhanced upon loss of MSK1 (data not shown). These data support the idea that the recruitment of 14-3-3 in response to MAP kinase signaling has a positive impact on histone H3 phosphoacetylation at the p21 promoter.
FIGURE 5.
14-3-3ζ is important for p21 activation. A–C, proliferating T98G cells were transfected with control siRNA oligonucleotides and siRNA oligonucleotides specifically targeting 14-3-3ζ mRNA. Transfected T98G cells were arrested by serum deprivation. Whole cells extracts, total RNA, and chromatin were gained from control (siCtrl) and knockdown cells (si14-3-3ζ) that were left untreated (rest) or were stimulated with 188.5 nm anisomycin and 165.5 nm TSA (A/T) for 1 h. A, knockdown efficiency was analyzed by Western blot using an antibody against 14-3-3ζ. Equal loading was controlled with an antibody specific for β-tubulin. B, p21 expression levels in control and 14-3-3ζ knockdown cells were monitored by real time PCR using GAPDH for normalization. C, shown are qRT-ChIP assays using antibodies specific for 14-3-3ζ, H3K14ac, H3S10phK14ac, the catalytic subunit of PP2A, and an unspecific antibody (IgG) as control. ChIP results are shown as percentage of input; for histone modifications, the values were corrected for changes in nucleosomal density. D, shown are an in vitro phosphorylation assay and 14-3-3 protection. Phosphoacetylated histone H3 was incubated without (-) or with recombinant PP2A in the absence (PP2A) or presence of increasing amounts of recombinant 14-3-3ζ. Phosphoacetylated histone H3 was detected with an H3S10phK14ac antibody, and equal loading was confirmed with an antibody specific for the C terminus of histone H3 (cH3). Intensities of histone H3S10phK14ac signals were quantified by densitometric scanning and normalized to the loading control (cH3).
It was previously shown that 14-3-3 proteins are able to protect phosphorylated substrates against dephosphorylation by PP2A (65). To test for a potential protective role of 14-3-3 in the context of histone H3 phosphorylation, we performed in vitro H3 dephosphorylation assays with recombinant PP2A and 14-3-3ζ. Phosphoacetylated histone H3 was isolated from serum-starved fibroblasts stimulated with anisomycin and TSA and used as a substrate for dephosphorylation by PP2A in the presence and absence of 14-3-3ζ. Recombinant PP2A dephosphorylated serine 10 of histone H3 in vitro. Strikingly, the addition of increasing amounts of recombinant 14-3-3ζ prevented H3S10 dephosphorylation by PP2A (Fig. 5D). In conclusion, our data suggest that one important function of 14-3-3 proteins in the context of p21 gene regulation might be to protect the of phosphoacetylation mark at histone H3 from being processed by PP2A.
DISCUSSION
MAP Kinase Activation Sensitizes Resting Cells to HDAC Inhibitor Treatment
In this report we analyzed in details the mechanisms responsible for the activation of the CDK inhibitor gene p21 by HDAC inhibitor treatment. Activation of p21 by HDAC inhibitors can occur in the absence of functional p53 (25, 26) and is, therefore, of particular interest for cancer treatment, as mutations of the tumor suppressor p53 often contribute to tumor development. Here we report that MAP kinase activation by growth factors or stress has an important role in the transcriptional regulation of the p21 gene by HDAC inhibitors. In transformed cells and proliferating fibroblasts the HDAC inhibitor TSA induces histone hyperacetylation at the p21 regulatory region and p21 expression. In contrast, TSA-induced histone hyperacetylation is not sufficient to activate p21 expression in resting cells, i.e. in the absence of growth factors. The inability of HDAC inhibitors to activate the p21 gene in resting cells is not limited to TSA but was also observed with other HDAC inhibitors such as valproic acid and MS-275.6 Our data suggest that stimulation of the MAP kinase pathway with growth factors or stress inducers is required to sensitize resting fibroblasts to HDAC inhibitor treatment.
Regulation of p21 by the Nucleosomal Response
MAP kinase signaling to the nucleosome, the so-called nucleosomal response, is involved in the regulation of several important biological processes such as stress response, carcinogenesis, control of circadian rhythms, and neuronal signaling circuits (66–68). We show here that the nucleosomal response is required for activation of the p21 gene by HDAC inhibitor treatment. Blocking kinase signaling to the nucleosome with the inhibitor H89 prevents H3 phosphoacetylation at the p21 promoter, recruitment of activated RNA polymerase II, and expression of p21. Simultaneous treatment with the stress inducer anisomycin and TSA had a synergistic effect on p21 expression and H3 phosphoacetylation levels. Similarly, we observed induced H3 phosphoacetylation at the p21 promoter along with increased expression upon TSA treatment of cells with activated MAP kinase pathways such as proliferating fibroblasts and transformed cells.6 These data suggest that in both resting and proliferating cells the active p21 gene is marked by H3S10phK14ac.
A variety of genes have been shown to be induced by mitogen- or stress-dependent signaling to the nucleosome (40). However, the detailed mechanism regulating the appearance of acetylation and phosphorylation marks is different for individual target genes. For instance, similar to p21, c-fos, and c-jun promoter-associated chromatin becomes hyperacetylated in response to HDAC inhibitor treatment without concomitant induced gene expression (33). However, p21 induction by anisomycin is enhanced by simultaneous TSA treatment, whereas transcriptional activation of c-jun in response to stress signaling was shown to be abolished by HDAC inhibitors (69). In contrast to p21, c-fos, and c-jun, the HDAC1 gene can be activated in resting fibroblasts by TSA alone (35). It is conceivable that parameters such as promoter architecture, presence of individual transcription factor binding sites, and additional epigenetic marks affect the recruitment of histone kinases and histone acetyltransferases in response to MAP kinase signaling.
Phosphorylated histone H3 is highly susceptible to HDAC inhibitor-induced hyperacetylation of neighboring lysine residues, resulting in phosphoacetylated histone H3 (46). Therefore, a direct link between histone acetylation and H3S10 phosphorylation was previously discussed. For instance, in vitro assays showed that Ser-10 phosphorylation increased the affinity of the H3K14-acetylating histone acetyltransferase Gcn5 for histone H3, suggesting a direct link between H3S10 phosphorylation and Lys-14 acetylation (50, 70). On the other hand, blocking the nucleosomal response by kinase inhibitors did not diminish histone H3 acetylation at the c-jun promoter in response to anisomycin (33). We show here that the nucleosomal response is not only required for RNA polymerase II recruitment to the p21 promoter but also important for efficient local H3K14 acetylation. Recruitment or activation of the H3K14 specific histone acetyltransferase activity seems to be largely dependent on H3S10 phosphorylation, whereas inhibitors of the nucleosomal response did not affect the association of the H3K9-specific activity.
MSK1 and PP2A as Antagonistic Regulators of p21 Expression
MSK1 and MSK2 have been shown to be the major histone H3 kinases mediating the nucleosomal response (47). In agreement with this report, we find a prominent role of MSK1 in p21 activation. The presence of the kinase at the p21 promoter was rapidly enhanced upon anisomycin/TSA stimulation, leading to local histone H3 phosphorylation. As antagonist of MSK1, protein phosphatase PP2A, is associated with chromatin at the silent p21 promoter in resting cells. A first link between PP2A activity and transcriptional regulation came from genetic studies in Drosophila. In response to heat shock, transcriptionally active heat shock loci show a dramatic accumulation of H3S10ph marks, whereas global histone H3 phosphorylation decreases (71). Interestingly, Drosophila PP2A mutants display reduced H3 dephosphorylation at non-heat-shock genes during heat shock (56).
In our study treatment of proliferating cells with concentrations of okadaic acid known to specifically inhibit PP2A resulted in enhanced histone phosphoacetylation and a significant increase in p21 expression, indicating that phosphatase activity is involved in p21 gene repression. Interestingly, the highest p21 expression was observed upon simultaneous treatment with okadaic acid and TSA, suggesting that the presence and stability of the phosphoacetyl-H3 mark is crucial for transcriptional induction of p21.
Remarkably, the reduced presence of PP2A at the p21 gene upon MAP kinase activation coincided with the onset of histone H3 phosphorylation, suggesting a direct link between the PP2A activity and dephosphorylation of histone H3. The potential of PP2A to dephosphorylate histone H3 was also shown in in vitro experiments (Ref. 56 and this study). Direct evidence that PP2A acts as a transcriptional repressor of the p21 gene was finally demonstrated by specifically depleting the catalytic subunit of PP2A. PP2A is a multisubunit holoenzyme that achieves substrate specificity via interaction with regulatory subunits. At the moment it is unclear which of the known regulatory subunits mediate the specificity for phosphorylated histone H3. In summary our study reveals a novel function of PP2A as chromatin-modifier and transcriptional repressor in mammalian cells.
14-3-3ζ Is important for p21 Gene Activation by the Nucleosomal Response
We and others have previously shown that 14-3-3 proteins recognize and bind phospho-acetylated histone H3 (61–63). In the present study, we demonstrate that 14-3-3ζ plays an important role in the activation of the p21 gene by the nucleosomal response. The recruitment of 14-3-3ζ to the p21 promoter strongly correlates with the presence of the H3S10phK14ac mark and activation of p21 expression. Depletion of 14-3-3ζ results in significant reduction of p21 expression. Recently, 14-3-3 has been shown to act as a scaffold protein for the recruitment of chromatin-remodeling complexes to target genes of the nucleosomal response (72). In addition, 14-3-3 has been implicated in the regulation of transcriptional elongation upon kinase signaling. In mammalian cells, 14-3-3 was shown to recruit the H4K16 acetyltransferase males absent on the first (MOF), resulting in acetylation-dependent recruitment of the elongation factor P-TEFb via the bromodomain protein BRD4 (73). In Drosophila, 14-3-3 recruits the acetyltransferase Elongator protein 3, which acetylates histone H3 at Lys-9 (74).
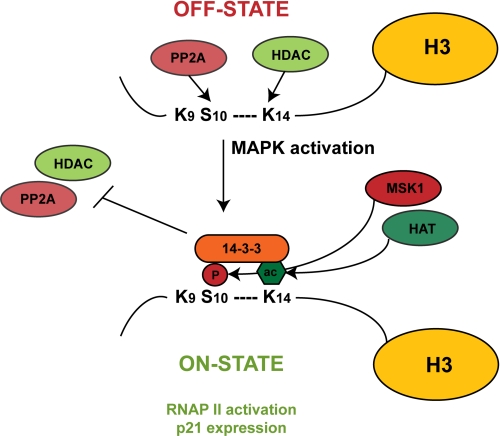
Our experiments establish a crucial role for 14-3-3 proteins in the regulation of p21 promoter activity and suggest a potential novel role of 14-3-3 in the context of chromatin modifications. The data presented in this study are compatible with the idea that 14-3-3ζ stabilizes the phosphoacetylation mark at the p21 promoter by protecting against PP2A activity. According to our model shown in Fig. 6, triggering the nucleosomal response by anisomycin alone results in transient H3 phosphoacetylation and p21 induction. Combined treatment with TSA leads to enhanced phosphoacetylation at the p21 promoter and thereby promotes stable recruitment of 14-3-3ζ. Indeed, additional acetylation of neighboring lysine residues was shown to stabilize the interaction between 14-3-3 and the phosphorylated histone H3 tail (62, 63). Stable binding of 14-3-3ζ to phosphoacetylated histone H3 prevents the dual mark from being processed by PP2A and might thereby stabilize the H3S10phK14ac mark at the p21 promoter, resulting in prolonged p21 expression in response to combinatorial TSA/anisomycin treatment. This study adds an additional dimension to the concept of gene regulation by the nucleosomal response by introducing PP2A as chromatin-associated dephosphorylating enzyme and 14-3-3 as potential protector of the H3S10 mark.
FIGURE 6.
Model for the regulation of the p21 gene by reversible phosphorylation and acetylation. In the absence of growth factor or stress signals, HDACs and PP2A are present at the p21 promoter, and the gene is silent. TSA-induced hyperacetylation is not sufficient for p21 activation. MAP kinase signaling leads to MSK1-mediated H3S10 phosphorylation and subsequent acetylation of H3K14. Inhibition of HDACs by TSA stabilizes the phosphoacetylation mark. The H3S10phK14ac mark is recognized and bound by 14-3-3 and thereby protected against PP2A activity. The presence of the H3S10phK14ac mark and 14-3-3 is required for RNA polymerase II recruitment and p21 expression. HAT, histone acetyltransferase.
Based on our findings that histone acetylation and phosphorylation act in concert in the regulation of the cell cycle regulator p21 (and maybe other proliferation regulators), it is tempting to speculate that treatment of tumor cells with combinations of HDAC inhibitors and drugs with impact on H3 phosphorylation might be more effective and/or specific as anti-proliferative agents. Experiments testing this hypothesis are ongoing in our laboratory.
Supplementary Material
Acknowledgments
We thank Reinhard Brunmeir for helpful support, Claudia Miccolo for performing part of the siRNA experiments, Hande Nayman and Benjamin Fuernsinn for their contribution to the dot blot analysis, and Ingrid Mudrak for technical assistance.
This work was supported by the Austrian Science Fund (FWF P18746 and P22340), the GEN-AU project “Epigenetic Plasticity of the Mammalian Genome” (Austrian Ministry of Science and Research), the Herzfelder Family Foundation (to C. S.), and by the Italian Association for Cancer Research (to S. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
E. Simboeck and C. Seiser, unpublished data.
- HDAC
- histone deacetylase
- MSK1
- mitogen- and stress-activated protein kinase 1
- TSA
- trichostatin A
- CDK
- cyclin-dependent kinase
- qRT
- quantitative real time.
REFERENCES
- 1.Jones P. A., Baylin S. B. (2007) Cell 128, 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballestar E., Esteller M. (2008) Adv. Genet. 61, 247–267 [DOI] [PubMed] [Google Scholar]
- 3.Glozak M. A., Seto E. (2007) Oncogene 26, 5420–5432 [DOI] [PubMed] [Google Scholar]
- 4.Xu W. S., Parmigiani R. B., Marks P. A. (2007) Oncogene 26, 5541–5552 [DOI] [PubMed] [Google Scholar]
- 5.Mottet D., Castronovo V. (2008) Clin. Exp. Metastasis 25, 183–189 [DOI] [PubMed] [Google Scholar]
- 6.Polyak K., Lee M. H., Erdjument-Bromage H., Koff A., Roberts J. M., Tempst P., Massagué J. (1994) Cell 78, 59–66 [DOI] [PubMed] [Google Scholar]
- 7.Chen J., Jackson P. K., Kirschner M. W., Dutta A. (1995) Nature 374, 386–388 [DOI] [PubMed] [Google Scholar]
- 8.Waga S., Hannon G. J., Beach D., Stillman B. (1994) Nature 369, 574–578 [DOI] [PubMed] [Google Scholar]
- 9.Luo Y., Hurwitz J., Massagué J. (1995) Nature 375, 159–161 [DOI] [PubMed] [Google Scholar]
- 10.LaBaer J., Garrett M. D., Stevenson L. F., Slingerland J. M., Sandhu C., Chou H. S., Fattaey A., Harlow E. (1997) Genes Dev. 11, 847–862 [DOI] [PubMed] [Google Scholar]
- 11.Michieli P., Chedid M., Lin D., Pierce J. H., Mercer W. E., Givol D. (1994) Cancer Res. 54, 3391–3395 [PubMed] [Google Scholar]
- 12.Dotto G. P. (2000) Biochim. Biophys. Acta 1471, M43–M56 [DOI] [PubMed] [Google Scholar]
- 13.Chang B. D., Watanabe K., Broude E. V., Fang J., Poole J. C., Kalinichenko T. V., Roninson I. B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4291–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coqueret O., Gascan H. (2000) J. Biol. Chem. 275, 18794–18800 [DOI] [PubMed] [Google Scholar]
- 15.Wang J., Devgan V., Corrado M., Prabhu N. S., El-Deiry W. S., Riccardi C., Pandolfi P. P., Missero C., Dotto G. P. (2005) J. Biol. Chem. 280, 37725–37731 [DOI] [PubMed] [Google Scholar]
- 16.el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 17.el-Deiry W. S., Tokino T., Waldman T., Oliner J. D., Velculescu V. E., Burrell M., Hill D. E., Healy E., Rees J. L., Hamilton S. R. (1995) Cancer Res. 55, 2910–2919 [PubMed] [Google Scholar]
- 18.Gartel A. L., Serfas M. S., Tyner A. L. (1996) Proc. Soc. Exp. Biol. Med. 213, 138–149 [DOI] [PubMed] [Google Scholar]
- 19.Gartel A. L., Najmabadi F., Goufman E., Tyner A. L. (2000) Oncogene 19, 961–964 [DOI] [PubMed] [Google Scholar]
- 20.Gartel A. L., Tyner A. L. (1999) Exp. Cell Res. 246, 280–289 [DOI] [PubMed] [Google Scholar]
- 21.Sambucetti L. C., Fischer D. D., Zabludoff S., Kwon P. O., Chamberlin H., Trogani N., Xu H., Cohen D. (1999) J. Biol. Chem. 274, 34940–34947 [DOI] [PubMed] [Google Scholar]
- 22.Richon V. M., Sandhoff T. W., Rifkind R. A., Marks P. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10014–10019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagger G., Doetzlhofer A., Schuettengruber B., Haidweger E., Simboeck E., Tischler J., Chiocca S., Suske G., Rotheneder H., Wintersberger E., Seiser C. (2003) Mol. Cell. Biol. 23, 2669–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mottet D., Pirotte S., Lamour V., Hagedorn M., Javerzat S., Bikfalvi A., Bellahcène A., Verdin E., Castronovo V. (2009) Oncogene 28, 243–256 [DOI] [PubMed] [Google Scholar]
- 25.Nakano K., Mizuno T., Sowa Y., Orita T., Yoshino T., Okuyama Y., Fujita T., Ohtani-Fujita N., Matsukawa Y., Tokino T., Yamagishi H., Oka T., Nomura H., Sakai T. (1997) J. Biol. Chem. 272, 22199–22206 [DOI] [PubMed] [Google Scholar]
- 26.Huang L., Sowa Y., Sakai T., Pardee A. B. (2000) Oncogene 19, 5712–5719 [DOI] [PubMed] [Google Scholar]
- 27.Archer S. Y., Meng S., Shei A., Hodin R. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6791–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wharton W., Savell J., Cress W. D., Seto E., Pledger W. J. (2000) J. Biol. Chem. 275, 33981–33987 [DOI] [PubMed] [Google Scholar]
- 29.Vaziri C., Stice L., Faller D. V. (1998) Cell Growth Differ. 9, 465–474 [PubMed] [Google Scholar]
- 30.Gartel A. L., Kandel E. S. (2006) Biomol. Eng. 23, 17–34 [DOI] [PubMed] [Google Scholar]
- 31.Liu Y., Yeh N., Zhu X. H., Leversha M., Cordon-Cardo C., Ghossein R., Singh B., Holland E., Koff A. (2007) EMBO J. 26, 4683–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viale A., De Franco F., Orleth A., Cambiaghi V., Giuliani V., Bossi D., Ronchini C., Ronzoni S., Muradore I., Monestiroli S., Gobbi A., Alcalay M., Minucci S., Pelicci P. G. (2009) Nature 457, 51–56 [DOI] [PubMed] [Google Scholar]
- 33.Thomson S., Clayton A. L., Mahadevan L. C. (2001) Mol. Cell 8, 1231–1241 [DOI] [PubMed] [Google Scholar]
- 34.Brunmeir R., Lagger S., Simboeck E., Sawicka A., Egger G., Hagelkruys A., Zhang Y., Matthias P., Miller W. J., Seiser C. (2010) PLoS Genet. 6, e1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hauser C., Schuettengruber B., Bartl S., Lagger G., Seiser C. (2002) Mol. Cell. Biol. 22, 7820–7830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gui C. Y., Ngo L., Xu W. S., Richon V. M., Marks P. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sowa Y., Orita T., Minamikawa S., Nakano K., Mizuno T., Nomura H., Sakai T. (1997) Biochem. Biophys. Res. Commun. 241, 142–150 [DOI] [PubMed] [Google Scholar]
- 38.Mahadevan L. C., Edwards D. R. (1991) Nature 349, 747–748 [DOI] [PubMed] [Google Scholar]
- 39.Mahadevan L. C., Willis A. C., Barratt M. J. (1991) Cell 65, 775–783 [DOI] [PubMed] [Google Scholar]
- 40.Clayton A. L., Mahadevan L. C. (2003) FEBS Lett. 546, 51–58 [DOI] [PubMed] [Google Scholar]
- 41.Martens J. H., Verlaan M., Kalkhoven E., Zantema A. (2003) Mol. Cell. Biol. 23, 1808–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto Y., Verma U. N., Prajapati S., Kwak Y. T., Gaynor R. B. (2003) Nature 423, 655–659 [DOI] [PubMed] [Google Scholar]
- 43.Vicent G. P., Ballaré C., Nacht A. S., Clausell J., Subtil-Rodríguez A., Quiles I., Jordan A., Beato M. (2006) Mol. Cell 24, 367–381 [DOI] [PubMed] [Google Scholar]
- 44.Espino P. S., Li L., He S., Yu J., Davie J. R. (2006) Cancer Res. 66, 4610–4616 [DOI] [PubMed] [Google Scholar]
- 45.Lefevre P., Witham J., Lacroix C. E., Cockerill P. N., Bonifer C. (2008) Mol. Cell 32, 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clayton A. L., Rose S., Barratt M. J., Mahadevan L. C. (2000) EMBO J. 19, 3714–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soloaga A., Thomson S., Wiggin G. R., Rampersaud N., Dyson M. H., Hazzalin C. A., Mahadevan L. C., Arthur J. S. (2003) EMBO J. 22, 2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomson S., Clayton A. L., Hazzalin C. A., Rose S., Barratt M. J., Mahadevan L. C. (1999) EMBO J. 18, 4779–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheung P., Tanner K. G., Cheung W. L., Sassone-Corsi P., Denu J. M., Allis C. D. (2000) Mol. Cell 5, 905–915 [DOI] [PubMed] [Google Scholar]
- 50.Lo W. S., Trievel R. C., Rojas J. R., Duggan L., Hsu J. Y., Allis C. D., Marmorstein R., Berger S. L. (2000) Mol. Cell 5, 917–926 [DOI] [PubMed] [Google Scholar]
- 51.Oelgeschläger T. (2002) J. Cell. Physiol. 190, 160–169 [DOI] [PubMed] [Google Scholar]
- 52.Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stein G. H. (1979) J. Cell. Physiol. 99, 43–54 [DOI] [PubMed] [Google Scholar]
- 54.Hsu J. Y., Sun Z. W., Li X., Reuben M., Tatchell K., Bishop D. K., Grushcow J. M., Brame C. J., Caldwell J. A., Hunt D. F., Lin R., Smith M. M., Allis C. D. (2000) Cell 102, 279–291 [DOI] [PubMed] [Google Scholar]
- 55.Murnion M. E., Adams R. R., Callister D. M., Allis C. D., Earnshaw W. C., Swedlow J. R. (2001) J. Biol. Chem. 276, 26656–26665 [DOI] [PubMed] [Google Scholar]
- 56.Nowak S. J., Pai C. Y., Corces V. G. (2003) Mol. Cell. Biol. 23, 6129–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivaldi M. S., Karam C. S., Corces V. G. (2007) Genes Dev. 21, 2818–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Millward T. A., Zolnierowicz S., Hemmings B. A. (1999) Trends Biochem. Sci. 24, 186–191 [DOI] [PubMed] [Google Scholar]
- 59.Chowdhury D., Keogh M. C., Ishii H., Peterson C. L., Buratowski S., Lieberman J. (2005) Mol. Cell 20, 801–809 [DOI] [PubMed] [Google Scholar]
- 60.Kobayashi H., Tan E. M., Fleming S. E. (2004) Int. J. Cancer 109, 207–213 [DOI] [PubMed] [Google Scholar]
- 61.Macdonald N., Welburn J. P., Noble M. E., Nguyen A., Yaffe M. B., Clynes D., Moggs J. G., Orphanides G., Thomson S., Edmunds J. W., Clayton A. L., Endicott J. A., Mahadevan L. C. (2005) Mol. Cell 20, 199–211 [DOI] [PubMed] [Google Scholar]
- 62.Winter S., Fischle W., Seiser C. (2008) Cell Cycle 7, 1336–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walter W., Clynes D., Tang Y., Marmorstein R., Mellor J., Berger S. L. (2008) Mol. Cell. Biol. 28, 2840–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winter S., Simboeck E., Fischle W., Zupkovitz G., Dohnal I., Mechtler K., Ammerer G., Seiser C. (2008) EMBO J. 27, 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin M., Potente M., Janssens V., Vertommen D., Twizere J. C., Rider M. H., Goris J., Dimmeler S., Kettmann R., Dequiedt F. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4727–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dunn K. L., Espino P. S., Drobic B., He S., Davie J. R. (2005) Biochem. Cell Biol. 83, 1–14 [DOI] [PubMed] [Google Scholar]
- 67.Crosio C., Cermakian N., Allis C. D., Sassone-Corsi P. (2000) Nat. Neurosci. 3, 1241–1247 [DOI] [PubMed] [Google Scholar]
- 68.Stipanovich A., Valjent E., Matamales M., Nishi A., Ahn J. H., Maroteaux M., Bertran-Gonzalez J., Brami-Cherrier K., Enslen H., Corbillé A. G., Filhol O., Nairn A. C., Greengard P., Hervé D., Girault J. A. (2008) Nature 453, 879–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hazzalin C. A., Mahadevan L. C. (2005) PLoS Biol. 3, e393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheung P., Allis C. D., Sassone-Corsi P. (2000) Cell 103, 263–271 [DOI] [PubMed] [Google Scholar]
- 71.Nowak S. J., Corces V. G. (2000) Genes Dev. 14, 3003–3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Drobic B., Pérez-Cadahía B., Yu J., Kung S. K., Davie J. R. (2010) Nucleic Acids Res. 38, 3196–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zippo A., Serafini R., Rocchigiani M., Pennacchini S., Krepelova A., Oliviero S. (2009) Cell 138, 1122–1136 [DOI] [PubMed] [Google Scholar]
- 74.Karam C. S., Kellner W. A., Takenaka N., Clemmons A. W., Corces V. G. (2010) PLoS Genet. 6, e1000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.