Abstract
DNA methylation is an epigenetic event involved in a variety array of processes that may be the foundation of genetic phenomena and diseases. DNA methyltransferase is a key enzyme for cytosine methylation in DNA, and can be divided into two functional families (Dnmt1 and Dnmt3) in mammals. All mammalian DNA methyltransferases are encoded by their own single gene, and consisted of catalytic and regulatory regions (except Dnmt2). Via interactions between functional domains in the regulatory or catalytic regions and other adaptors or cofactors, DNA methyltransferases can be localized at selective areas (specific DNA/nucleotide sequence) and linked to specific chromosome status (euchromatin/heterochromatin, various histone modification status). With assistance from UHRF1 and Dnmt3L or other factors in Dnmt1 and Dnmt3a/Dnmt3b, mammalian DNA methyltransferases can be recruited, and then specifically bind to hemimethylated and unmethylated double-stranded DNA sequence to maintain and de novo setup patterns for DNA methylation. Complicated enzymatic steps catalyzed by DNA methyltransferases include methyl group transferred from cofactor Ado-Met to C5 position of the flipped-out cytosine in targeted DNA duplex. In the light of the fact that different DNA methyltransferases are divergent in both structures and functions, and use unique reprogrammed or distorted routines in development of diseases, design of new drugs targeting specific mammalian DNA methyltransferases or their adaptors in the control of key steps in either maintenance or de novo DNA methylation processes will contribute to individually treating diseases related to DNA methyltransferases.
Keywords: DNA methyltransferase, epigenetics, enzyme catalysis, protein-DNA interactions
INTRODUCTION
The discovery of C5-methylated cytosine in DNA was about half a century ago [1]. Extensive researches in last decade have led scientists to realize that self-controlled DNA methylation can be a new inheritable route affecting genetic process, including X chromosome inactivation and uniparental disomy [2]. A number of experimental and clinical studies demonstrated that disturbance (even subtle deflexion) of DNA methylation may induce various disorders, including different types of cancer, genomic imprinting-associated diseases (e.g. Beckwith-Wiedemann syndrome, transient neonatal diabetes, etc.), repeat-instability diseases (e.g. Huntington’s disease, spinocerebellar ataxia, etc.), and DNA methylation machinery-defected diseases [e.g. systemic lupus erythematosus, immunodeficiency, centromeric instability, and facial anomalies syndrome (ICF)] [3]. In addition, dynamic and programmed DNA methylation is critical during development [4], and recent studies strongly suggest that reprogramming of DNA methylation is an important mechanism of “fetal origin” diseases [5].
DNA methyltransferase (DNA MTase) is a key enzyme in the process of DNA methylation concerning reactions of methyl group transferred to the C5 position in the cytosine base. In mammals, DNA MTases are divided into three families encoded by the respective genes, including Dnmt1 (DNA nucleotide methyltransferase 1), Dnmt2, and Dnmt3. Each family possesses several transcriptional variants [6]. All mammalian DNA MTases can be incorporated in synergistically determining and regulating DNA methylation patterns involved in various cellular events, including cell proliferation, differentiation, and transformation [7]. Such enzymatic methyl transfer reaction is suggested almost identical among different mammalian DNA MTases. Nevertheless, selectivity and specificity of substrates (i.e. DNA sequence), and spatial and temporal activation or regulation may be unique for each of mammalian DNA MTases. As a result, their functional and pathological roles may be remarkably different [8].
In this review, we briefly summarized molecular and biochemical properties of mammalian DNA MTases. Focusing on enzymatic and structural basis, we assessed each step of C5 position methylation of cytosine in DNA sequence, including substrate sequence targeting, base flipping, cofactor binding, methyl group transferring, and reaction processivity, as well as miscellaneous adaptors in regulation of those processes. Drugs targeting certain DNA methylation processes and possible targets for new drug designs also will be briefly discussed.
1. MOLECULAR AND STRUCTURAL CHARACTERISTICS OF MAMMALIAN DNA MTases
More than three decades ago, the first mammalian DNA MTases—Dnmt1, from Friend murine erythroleukemia (MEL), was isolated and purified [9, 10] with biochemical and molecular assays. The studies at that time indicated that mammalian DNA MTases, as putative DNA methylation machineries, were considerably conserved throughout the eukaryotes [10]. Later, attempts to identify novel DNA MTases using expressed sequence tag (EST) databases screening resulted in the discovery of three candidates of Dnmt1: Dnmt2, Dnmt3a, and Dnmt3b [11, 12]. Another protein Dnmt3L (Dnmt3-like), with its sequence similarity to that of Dnmt3a and Dnmt3b, was identified without enzymatic activity in vitro in DNA methylation [13]. By that time, there emerged a flood of scientific efforts at biochemical, molecular, and structural levels to elucidate properties of the four members of active mammalian DNA MTases, including Dnmt1, Dnmt3a, Dnmt3b, and Dnmt2 encoded by four independent genes [13]. In simplicity, those mammalian DNA MTases can be categorized into three families: Dnmt1, Dnmt2, and Dnmt3 family (Table 1). In general, they are composed of the regulatory N-terminal region and catalytic C-terminal region, although the catalytic target is not cytosine nucleotide and with no regulatory domain in Dnmt2, and the catalytic domain in Dnmt3L lacks enzymatic capability for DNA methylation [6]. Attributing to the established gene and protein models of DNA MTases, some antisense oligodeoxynucleotide (e.g. MG98) and peptide derivative (e.g. RG108) were successfully annotated for inhibiting expression and functions of Dnmt1 [14, 15].
Table 1.
| 1a. Molecular Characteristics of Mammalian DNA Methyltransferases | ||||
|---|---|---|---|---|
| Species | Dnmt1 | Dnmt3a | Dnmt3b | |
| Chromosome Location | human | 19p13.2 | 2p23 | 20q11.2 |
| mouse | chromosome 9 | chromosome 12 A2–A3 | chromosome 2 H1 | |
| Gene | human | 60 kb, 40exon/39intron 45 kb, | 110 kb, 26exon/25intron 79 kb, | 47 kb, 24exon/23intron 38 kb, |
| mouse | 40exon/39intron | 23exton/22intron | 24exon/23intron | |
| Transcript | human | DNMT1 variant: DNMT1B | DNMT3A variant: DNMT3A2 | DNMT3B variants: DNMT3B2~ DNMT3B6 |
| mouse | Dnmt1 variants: Dnmt1o and Dnmt1p | Dnmt3a variant: Dnmt3a2 | Dnmt3b variants: Dnmt3b2~Dnmt3b9 | |
| Protein (Predominant) | human | 1616aa | 912aa | 908aa |
| mouse | 1619aa | 853aa | 859aa | |
| 1b. Molecular Characteristics of Mammalian DNA Methyltransferases | |||
|---|---|---|---|
| Species | Dnmt3L | Dnmt2 | |
| Chromosome | human | 21q22.3 | 10p15.1 |
| Location | mouse | chromosome 10 | chromosome 2 A1 |
| Gene | human mouse |
16 kb, 12exon/11intron 190 kb, 15exon/14intron |
58 kb, 11exon/10intron 34 kb, 11exon/10intron |
| Transcript | human | DNMT3L variant: DNMT3L2 Dnmt3L | TRDMT1 variants: TRDMT1B and TRDMT1C Trdmt1 |
| mouse | variant: Dnmt3L2 | variants: Trdmt1b and Trdmt1c | |
| Protein | human | 387aa | 391aa |
| (predominant) | mouse | 421aa | 415aa |
Following sections will discuss some research data at molecular, biochemical, and structural levels regarding mammalian DNA MTases, and provide information for drug development targeting at the enzymes that play important roles in DNA methylation mediated diseases.
1.1. Dnmt1
Dnmt1 has been generally believed to be constitutively expressed in proliferating cells and somatic tissues, which is closely related to replication foci in S phase of cell cycle. A large body of findings has showed that Dnmt1 mainly functions in the maintenance of DNA methylation, which is crucial for genomic integrity by preserving DNA methylation patterns throughout development and during whole life periods [6].
1.1.1. Molecular Architecture of Dnmt1
1.1.1.1. Genomic Mapping of Dnmt1 Gene
The gene encoded for Dnmt1 is localized at 19p13.2 in human and at proximity (5.0 cM) of chromosome 9 in mouse [10, 16]. The DNMT1 gene in human spans more than 60kb (kilobases) in the genome, composing at least 40 exons and 39 introns, and its canonical single transcript spreads about 5.2 kb long [16, 17]. Whereas, the Dnmt1 gene in mouse occupies about 45kb in the genome (containing 40 exons and 39 introns), producing a 5.3kb transcript. The protein Dnmt1 is predominantly expressed in somatic tissues and proliferating cells, and contains 1616 amino acid residues with molecular mass of about 190 kDa [18].
1.1.1.2. Dnmt1 Variants
Several isoforms have been detected and they could be the consequence of intrinsic RNA splicing in vivo [19]. During the gametogenesis in mouse, research evidence showed that an alternative splicing event related to the sex-specific 5’ exons produced at least two Dnmt1 mRNA variants—Dnmt1o and Dnmt1p.
Dnmt1o (for oocyte specific), an oocyte-specific short protein isoform encoded by Dnmt1o variant, was demonstrated to be due to translational initiation codon shifted to axon 4 in RNA editing, with molecular weight of about 170kDa and lacking 118 amino acid residues at the N-terminus. It is expressed in oocytes and pre-implantation embryo and fully functional in vivo to participate in maintaining methylation patterns of imprinted loci [20].
The other larger isoform of Dnmt1 mRNA variant in mouse, Dnmt1p (for pachytene spermatocyte specific), was primarily detected in pachytene spermatocytes. Dnmt1p mRNA was initially suggested without translation function attributed to inhibitory effects induced by multiple upstream out-of-frame initiation codons [20]. Later, Leonhardt et al. showed that an alternative Dnmt1 transcript isoform identical to Dnmt1p mRNA was present in skeletal muscle and differentiated myotubes [21]. Dnmt1p mRNA could be translated in the testis and skeletal muscle, and the protein isoform Dnmt1p could be active in the regulation of DNA methylation patterns during myogenesis and gametogenesis; however, the details for molecular enzymolgy of Dnmt1p, and whether Dnmt1p plays critical roles in development, still need to be elucidated.
In human and other primates, owing to in-frame insertion of an Alu element, there is an additional fragment extending 48nt (nucleotides) between exons 4 and 5. Consequently, a longer splice variant—DNMT1b derived from the primary DNMT1 gene is produced, and distributed throughout somatic tissues and all human cell lines tested. However, DNMT1b abundance is less than DNMT1 [22]. Furthermore, the evidence showed that DNMT1b, the product of DNMT1b variant, with additional 16 amino acid residues incorporated in the very N-terminus of Dnmt1, possesses some similar enzymatic features to that of DNMT1 in vitro [23]. Shen and colleagues indicated that during RNA editing in mouse, an acceptor site at the 3’ end of intron 4 in Dnmt1 was also functional. As a result, another protein isoform Dnmt1b in somatic tissues of mouse lacks 2 amino acid residues compared to that of Dnmt1. Dnmt1b was a translational product of the mRNA variant Dnmt1b [24]. Nevertheless, whether methyltransferase activity of Dnmt1b (also DNMT1b) is intact in vivo remains unclear, and how and to which extent Dnmt1b and DNMT1b are involved in epigenetic modulation processes also should be further investigated.
1.1.2. Biochemical and Structural Organization of Dnmt1
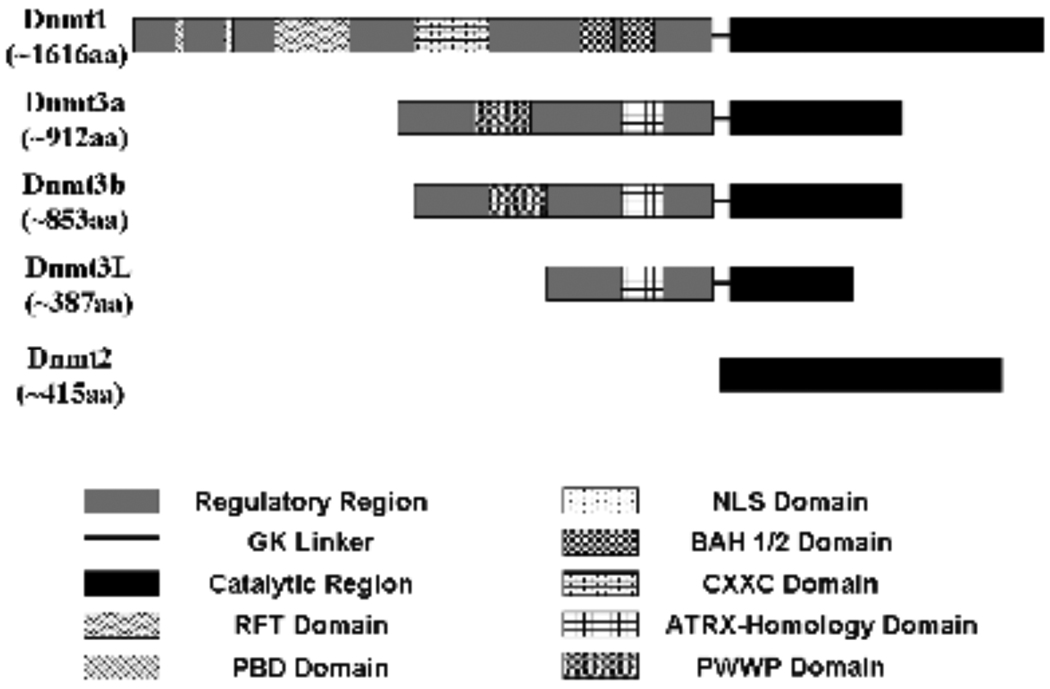
Overall, Dnmt1 is composed of two parts that could be separated by V8 protease treatment: a fairly long diversified N-terminal extensions containing about 1,100 amino acid residues (the largest in all mammalian DNA MTases), and a relatively conserved C-terminal domain which occupies the remaining residues (Fig. 1). These two regions are joined by a linker of glycine-lysine (GK) repeats [13].
Fig. 1.
Various functional protein domains in mammalian DNA MTases. Except Dnmt2, all mammalian DNA MTases are divided into catalytic and regulatory regions. From N-terminal to C-terminal, domains in the regulatory region of Dnmt1 are: PBD domain [PCNA (proliferating cell nuclear antigen) binding domain], NLS (nuclear localization sequence) domain, RFT (replication-foci targeting) domain, CXXC (cysteine-rich, cysteine-x-x-cysteine type) domain, and randomly arranged BAH (bromo/Brahma-adjacent homology) domain. The ATRX (alpha-thalassemia and mental retardation on X chromosome)-homology domain is located in the C-terminal part of the regulatory region of Dnmt3a, Dnmt3b, and Dnmt3L. Another essential functional domain—PWWP (proline-tryptophan-tryptophan-proline motif) domain locates at C-terminal to the ATRX-homology domain of the regulatory region in Dnmt3a and Dnmt3b.
1.1.2.1. Catalytic Region of Dnmt1
The C-terminal catalytic region in Dnmt1 contains ten characteristic sequence motifs (i.e. conserved motifs I–X), and the spacing sequences between each conserved motifs is referred as variable regions. Several lines of evidence further indicate that about six of the conserved motifs, that is motifs I, IV, VI, VIII, IX, and X, might be highly conserved in mammalian DNA MTases. In fact, the C-terminal part of Dnmt1 seems to be more closely related to that in prokaryotic DNA MTases (especially for bacterial restriction methyltransferases serving in the restriction-modification system) than to other mammalian DNA MTases (i.e. Dnmt2, Dnmt3a, Dnmt3b, and Dnmt3L) [10, 13]. This indicates that three families of mammalian DNA MTases may be generated in relatively independent ways during evolution. To date, no structural information for any part (even domain) of the Dnmt1 is acquired. However, due to the characteristic of evolutionary conservation of the conserved motifs between Dnmt1 and prokaryotic DNA MTases, the X-ray crystallography data on prokaryotic DNA MTases (especially the M.HhaI referred as a member of C-MTases) provided important hints concerning the structural features of the C-terminal catalytic region of Dnmt1 [25]. Grossly, it is suggested that the C-terminal catalytic region of Dnmt1 could be oriented and folded into two domains. The large and small domain were separated by a large cleft [26].
1.1.2.1.1. Large Domain
The distribution of motifs arranged in the two domains is extremely asymmetric. The large domain encompasses the most conserved motifs, including motifs I–VIII and the most C-terminal part of motif X, which could be participated in Ado-Met [S-adenosyl-L-methionine (SAM)] cofactor binding, substrate (cytosine) targeting, and essential catalysis events. The “core” structure in the large domain is composed of the highly conserved motifs I, IV, VI, and VIII. Most of the constant amino acid residues, in the “core” structure such as PC dipeptidyl residues (proline-cysteine) in motif IV constituting the catalytic loop, are indicated to be situated facing the cleft and to be clustered around the active site in the C-terminal region of the Dnmt1 molecule [26].
The large domain could have a characteristic structural appearance which is composed of six-stranded parallel β sheet accompanying the seventh anti-parallel strand between the fifth and sixth strands, throughout which almost every β strand could be flanked by 1–2 α helices on either side to form an α/β/α sandwich-like structure [25].
Furthermore, there was evidence that the first strand initiated in middle of the β sheet to separate the large domain into two subdomains, (β1–β3) and (β4–β7) subdomain. The former (involved in conserved motifs I–III and X) could create the Ado-Met binding site, and the latter (involved in conserved motifs IV–VIII) could provide extrahelical cytosine targeting site. Both the biding sites are part of respective hydrophobic binding pockets embedded in equivalent areas within the two subdomains. Thus, the functional and structural similarities between the two subdomains in the large domain indicated that the C-terminal catalytic region can be arisen by gene duplication during evolution, although further investigation is needed [27].
1.1.2.1.2. Small Domain
The small domain comprises an extremely long variable region between conserved motif VIII and IX, conserved motif IX and partial N-terminal region of conserved motif X. The long variable sequence emanates as a stalk from the large domain, transverses almost full length of the C-terminal region, and links motif IX and small initial segment of motif X to be folded as a bulk mass in the small domain [26]. A variety array of evidence showed that the small domain is partially responsible for the DNA sequence targeting process. Consequently, the secondary and more detailed structural features of Dnmt1 could be very different with other DNA MTases. Thus, corresponding structural features of the small domain may not be extrapolated directly from that of prokaryotic DNA MTases [25]. As a result, to gain further refined structural information of the domains, we may have to wait for the direct and high-resolution conformational data on this macromolecule with methods of X-ray crystallography and NMR spectroscopy.
1.1.2.2. Regulatory Domain of Dnmt1
In addition to the C-terminal catalytic region, Dnmt1 also possesses a long N-terminal region (about 1,100 amino acid residues) that is accreted many domains over the course of evolution and is the basis for differing from other mammalian DNA MTases (i.e. Dnmt2 and Dnmt3 families) at the biochemical level [13]. Using molecular and biochemical assays, as well as detailed analysis of the biological functions of Dnmt1, it is now well accepted that there exist multiple functional domains within the N-terminal regulatory extensions of Dnmt1 (Fig. 1).
In 1992, Leonhardt et al. initially showed that replication-foci targeting (RFT) domain, also known as targeting sequence (TS), was located in the N-terminus region (256–629 amino acid residues) of Dnmt1 by mutagenesis analysis. This domain was considered to target Dnmt1 toward DNA in S phase during cell cycle [28]. Another PBD [proliferating cell nuclear antigen (PCNA) binding domain] sequence with 158–171 amino acid residues located proximal to TS domain also was demonstrated to be associated with replication during S phase [29]. Between the PBD and TS domain, a short sequence termed as nuclear localization sequence (NLS) domain was suggested to induce Dmnt1 importing into nucleus [30]. In the C-terminus of N-terminal region adjacent to the GK linker, there extends a region homologous to the polybromo-1 protein that was indicated to build two tandemly arranged bromo/Brahma-adjacent homology (BAH) domains. The BAH1 and BAH2 domains were proposed to act as protein-protein interaction modular motifs [31, 32]. In the centre of the N-terminal region, there is a cysteine-rich region present in almost all regulatory regions of mammalian DNA MTases (except Dnmt2 that lacks the N-terminal regulatory domain). The cysteine-rich region referred to as a zinc-binding domain containing eight conserved cysteine residues seems to function as CXXC-type zinc-finger protein in Dnmt1 [33].
Subsequent researches indicated that there were additional functional binding domains: Dnmt1-associated protein (DMAP1) binding domain was located near the very N-terminus [32], retinoblastoma tumor suppressor gene product (Rb) binding domain also was resided in the proximity of N-terminal region where could be overlapped with that in DMAP1 binding domain [35, 36]. Both the DMAP1 and Rb binding domains are related to transcriptional repression roles of Dnmt1. However, specific protein-protein interaction modules and definitive interfaces for physical interactions may need to be resolved further in depth.
In addition, the regulatory region of Dnmt1 also could directly interact with methyl-CpG binding proteins, including methyl-CpG binding protein-2 (MeCP2), methyl-CpG binding domain protein (MBD2), and MBD3 [37, 38]. Moreover, lines of evidence showed that the regulatory region could recruit several chromatin associated elements such as HDAC1 (histone deacetylase-1), HDAC2, H3-K9 methyltransferase, Suv39h1 (suppressor of variegation 3–9 homologue 1), and HP1β (heterochromatin protein 1β) [34, 39, 40]. However, data concerning direct interactions between the regulatory region and methyl-CpG binding proteins as well as chromatin associated elements is limited. Therefore, exact sequences of those binding domains and detailed direct interactions between the functional domains and their targeting proteins should be further investigated.
1.2. Dnmt3 Family
In 1998, Okano et al. reported the cloning and initial characterization of two enzymatically functional Dnmt3 members, Dnmt3a and Dnmt3b [11]. Later, researchers identified a novel DNA methylation regulator, Dnmt3-like (Dnmt3L), which lacks active methyltransferase activity with its sequence similar to Dnmt3a and Dnmt3b [41]. The Dnmt3 family members, especially Dnmt3L, are relatively specifically expressed in embryo and germ cells. Unlike Dnmt3L that is generally considered to be a regulatory factor in DNA machinery, both Dnmt3a and Dnmt3b are widely accepted as de novo DNA MTases for setting DNA methylation patterns [6].
1.2.1. Dnmt3a and Dnmt3b
1.2.1.1. Genomic Mapping of Dnmt3a and Dnmt3b Genes
In human, the coding genes DNMT3A and DNMT3B are localized at 2p23 and 20q11.2; and in mouse, genes encoding Dnmt3a and Dnmt3b are believed to be localized in chromosome 12A2–A3 and chromosome 2H1. The human genes DNMT3A and DNMT3B are composed of 26exon/25intron and 24exon/23intron, spanning nearly 110kb and 47kb in the genome, respectively; whereas in mouse, Dnmt3a and Dnmt3b genes are composed of 23exton/22intron and 24exon/23intron, extending approximately 79kb and 38kb in the genome, respectively [42, 43]. In human, the main mRNAs of DNMT3A and DNMT3B are approximately 4.3kb and 4.4kb, whose protein products are DNMT3A and DNMT3B that are composed of 912 and 853 amino acid residues, respectively; in mouse, the dominate mRNAs of Dnmt3a and Dnmt3b are about 4.2kb and 4.3kb, encoding protein Dnmt3a and Dnmt3b containing 908 and 859 amino acid residues, respectively (Table 1) [11, 42]. Moreover, preliminary data showed that there appeared 98% and 94% identity between the human DNMT3 (i.e. DNMT3A and DNMT3B) and the corresponding murine homologues [42].
1.2.1.2. Variants of Dnmt3a and Dnmt3b
Multiple alternative transcripts have been detected involved in both Dnmt3a (also DNMT3A) and Dnmt3b (also DNMT3B) in human and mouse [44].
1.2.1.2.1. Variants in Dnmt3a
As for Dnmt3a, by northern blot analysis, researchers reported three distinct tissue-specific splicing variants with 4.0kb-, 4.4kb-, 9.5kb-length in human, and 4.0kb-, 4.2kb-, 9.5kb-length in mouse. Recently, another variant as a candidate similar to the 9.5kb-length transcript was isolated, which was believed to be important for DNA methylation machinery during development rather than DNA methylation patterning in somatic cells generated by 9.5kb-length transcript. The newly determined splicing variant, named β transcript, was suggested to be the result of replacement of the 1α exon in 9.5 kb-length transcript by the 1β exon related to alternative splicing in process of Dnmt3a (also DNMT3A) RNA editing [45].
However, whether the splicing variants could possess their own corresponding translational isoforms (proteins) is still an unresolved issue. In 2002, Chen et al. demonstrated that a novel short isoform—Dnmt3a2, about 100kDa, in which the variable region possessing 219 amino acid residues in the N-terminus of Dnmt3a (termed as a long isoform, approximately 130 kDa) was lacked. Dnmt3a2 could be the translational product of the 4.0kb or 4.2 kb transcript variants in mouse [46]. Later, evidence showed that the isoform Dnmt3a2 was translated underlying the mechanism by which, with a promoter sequence rich in GC nucleotides within the intron 6, the transcriptional initiation could be started at the sequences residing in the specific exon 7. Unlike Dnmt3a that is expressed ubiquitously in both embryo and most adult tissues and concentrated on heterochromatin, Dnmt3a2 is thought to act as a major isoform in embryo, and restricted in adult tissues (might only in the testis, ovary, thymus, and spleen) [46]. In addition, DNMT3A2, a homologue to Dnmt3a2 in human, also had been detected as a short isoform lacking 223 amino acid residues in the N-terminus of DNMT3A; nonetheless, its relationship to the splicing variants have to be further determined [46].
1.2.1.2.1. Variants in Dnmt3b
In contrast to Dnmt3a, the situation concerning splicing polymorphism and the corresponding isoforms is less controversial in Dnmt3b. At beginning, EST database screening showed that DNMT3B (also Dnmt3b) could generate two different transcripts by alternative splicing [11]. Several years later, researchers identified tissue-specific splicing variants of DNMT3B (also Dnmt3b), including DNMT3B1, DNMT3B2, DNMT3B3, DNMT3B4, DNMT3B5, and DNMT3B6 in human; as well as Dnmt3b1, Dnmt3b2, Dnmt3b3, Dnmt3b4, Dnmt3b5, Dnmt3b6, Dnmt3b7, and Dnmt3b8 [45, 46].
For underlying mechanism of in-frame alternative splicing during RNA editing, transcript variants generate their corresponding protein isoforms, in which isoform products of the variants Dnmt3b7 and Dnmt3b8 have not been fully determined [44, 46, 47]. Several lines of evidence showed that some peptide sequences localized at functional parts of Dnmt3b (DNMT3B) (for instance: conserved motif VII, VIII, target recognition domain between conserved motif VIII and IX, and etc.) could be deteriorated in isoforms of Dnmt3b (also DNMT3B) [47]. Thus, unlike Dnmt1 and Dnmt3a, the alternative splicing could affect enzymatic integrity in Dnmt3b (also in DNMT3B). Consequently, isoforms of DNMT3B (Dnmt3b) can be divided into two categories: those do not alter catalytic activity of DNA MTase; and others may be inactive in catalysis at least in vitro. In human, DNMT3B6 (854aa, full-length), DNMT3B1 (853aa, nearly full-length), and DNMT3B2 (833aa, its transcript disrupted in exon 10 and 11 by in-frame alternative splicing) can be categorized into the former class; whereas, DNMT3B3 (770aa), DNMT3B4 (745aa), and DNMT3B5 (812aa), those can be affected in the C-terminal region by inframe alternative splicing involved in 3’ exons (e.g. exon 22 and/or exon 23), are categorized into the later class. For those in the later class, they also may disrupt target recognition domain as a result of alternative splicing related to 10 and 11 exons. In mouse, Dnmt3b1 (859aa, full-length), Dnmt3b2 (839aa, the 11 exon is disrupted during in-frame alternative splicing), Dnmt3b5 (860aa, the 2 exon can be alternatively spliced), and Dnmt3b6 (similar to that in Dnmt3b3) can be included in the former class; whereas Dnmt3b3 (776aa, 3 exons are spliced involved in sequences of C-terminal region and target recognition domain) and Dnmt3b4 (796aa, 2 exons in 3’ region expressing C-terminal catalytic domain are disrupted by in-frame alternative splicing) are in the later class. Moreover, the isoform products of Dnmt3b7 and Dnmt3b8 are resembled to that of DNMT3B4 and DNMT3B5, and categorized into the later class [44, 46, 47].
Although increasing transcript variants and the corresponding protein isoforms of Dnmt3a and Dnmt3b were discovered in dominating specific-tissue expression patterns in embryo and postnatal life, corresponding functions of mRNA and proteins of the variants are unclear [46, 47]. Specific genetic manipulation (i.e. isoform-specific gene knockout, mutagenesis, and etc.) towards the specific splicing variant and protein isoforms may be a clue to unwind such a “Gordian knot”.
1.2.1.3. Biochemical and Structural Organization of Dnmt3a and Dnmt3b
Similar to that of Dnmt1, the architecture of Dnmt3a and Dnmt3b also can be viewed via N-terminal regulatory region and C-terminal catalytic region. The research evidence showed that Dnmt3a and Dnmt3b were closely related proteins in structures, especially in their C-terminal regions with about 84% identity (Fig. 1) [48].
1.2.1.3.1. Catalytic Region of Dnmt3a and Dnmt3b
Unlike Dnmt1, the crystal structure of the C-terminal domain of Dnmt3a2 had been resolved by Cheng and his colleagues in 2007 [49]. Molecular modeling data indicated that the C-terminal region of Dnmt3a superimposed well with the representative prokaryotic DNA MTases—M.HhaI [26, 49]. The C-terminal catalytic part of Dnmt3a and Dnmt3b could also be packed into two-domain structure pattern (i.e. large domain and small domain separated by a large cleft). Following structure-based sequence alignment, the researchers found seven β strands and six α helices forming the typical α/β/α sandwich-like super-secondary structure in the large domain, and three β strands and two α helices in the small domain [49].
In the large domain, loops in the conserved motif IV flanked by N-terminal β strand and C-terminal α helix is the active site for enzymatic activities of Dnmt3a and Dnmt3b [49]. There also appears substrate binding pocket in the large domain. Crystallography data even showed that residues located in the conserved motif I, II, and III were essential for Ado-Met substrate binding activity of the catalytic domain. They are resided in those loops flanked by N-terminal β strand and C-terminal α helix. Meanwhile, Phe residue within α helix in motif V and Arg residue between α helices in motif X are also important for Ado-Met binding [49].
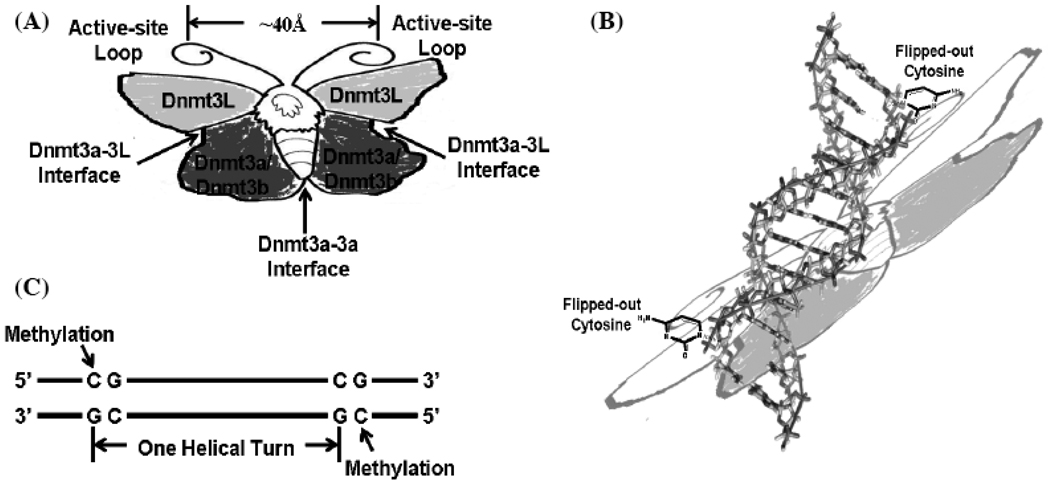
Moreover, size-exclusive chromatography and co-crystal structure evidence also indicated that Dnmt3a/Dnmt3b could be formed as a homodimer, and Dnmt3a/Dnmt3b homodimer could link to two Dnmt3L to form a tetramer [with the form of Dnmt3L-(Dnmt3a/Dnmt3b)2-Dnmt3L] [49] (Fig. 4). Structure-based sequence alignment indicated that the variable sequence linker between conserved motif VIII and IX in the small domain mediated the interface between the two Dnmt3a/Dnmt3b proteins, acting as DNA sequence recognition domain [49]. The interface between Dnmt3a/Dnmt3b and Dnmt3L is mediated by three α helices in the conserved motif III, V, and VII, respectively, in the large domain [50].
Fig. 4.
CG spacing pattern in Dnmt3-mediated de novo DNA methylation. (A): Two Dnmt3a/3b and two Dnmt3L can form a tetramer complex, in which there may be three interfaces: one central Dnmt3a-Dnmt3a homodimer interface and two lateral Dnmt3a-Dnmt3L heterodimer interfaces. The overall Dnmt3L-(Dnmt3a)2-Dnmt3L complex can be proposed as an elongated, butterfly-like structure, in which the distance between two active-site loops in each Dnmt3a/3b is approximately 40 Å apart. (B), (C): Dnmt3L-(Dnmt3a)2-Dnmt3L complex can target the unmethylated DNA duplex to simultaneously de novo methylate the two flipped-out cytosines in CG sites of both the strands that is one-helical-turn apart.
1.2.1.3.2. Regulatory Region of Dnmt3a and Dnmt3b
Compared to that of Dnmt1, the N-terminal regulatory part of Dnmt3a and Dnmt3b is only about half in length (Fig. 1). Unlike the C-terminal region, similarity of N-terminal region between Dnmt3a and Dnmt3b is limited. Two conserved domains exist in Dnmt3a and Dnmt3b. However, the most N-terminal region in the N-terminal regulatory domain of Dnmt3a/Dnmt3b is variable.
A cysteine-rich zinc-binding domain shares homology with a region in ATRX (alpha-thalassemia and mental retardation on X chromosome), and is belong to the Swi2/Snf2 family (mating type swithching-2/sucrose non-fermenting factor-2) of ATP-dependent chromatin remodeling factors. It is located in the C-terminal area near the GK linker of the N-terminal regulatory region of Dnmt3a/Dnmt3b to function as a transcriptional repressor domain [51]. The cysteine-rich domain (also named ATRX-homology domain) in Dnmt3a and Dnmt3b possesses a C2-C2 zinc finger, and contains a plant homology domain (PHD)-like sequence that is involved in protein-protein interactions [51]. Further studies have demonstrated that Dnmt3a/Dnmt3b can be interacted directly with several essential transcriptional and epigenetic regulators mediated by the cysteine-rich domain, including RP58 (repression protein of 58kDa), Myc, HDAC1, Suv39h1, and HP1 [40, 52–55].
Another conserved sequence located at N-terminal to the cysteine-rich domain is the PWWP domain (rich in proline and tryptophan residues as a highly conserved proline-tryptophan-tryptophan-proline motif), and spanned 100–150 amino acid residues in both Dnmt3a and Dnmt3b [56]. A variety body of evidence demonstrated that the PWWP domain is a widely distributed modular protein domain found in more than 60 eukaryotic proteins, especially chromatinassociated proteins [57]. Preliminary clinical findings showed that the ICF syndrome also could be associated with mutation in the N-terminal region of DNMT3B related to a missense defection in the PWWP domain [58]. It is generally accepted that the PWWP domain could be an essential functional module to play roles in targeting Dnmt3a/Dnmt3b to pericentric heterochromatin and metaphase chromosomes [59, 60]. According to the structural information resolved in Dnmt3b PWWP domain, the PWWP domain in Dnmt3a/Dnmt3b can be plausibly divided into two subdomains [56]. The N-terminal half is folded as a barrel-like structure composing five β strands, which is believed to be fairly conserved throughout almost all PWWP domains; and the C-terminal half would be packed into five-α helix bundle, which could be variable between different PWWP domains so as to be inferred to participate protein-protein interactions [56, 61]. Recently, Li et al. proposed that the PWWP domain may mediate direct interactions between Dnmt3a and a polycomb group protein, chromobox 4 (Cbx4), and function as a sumoylation E3 ligase to promote Dnmt3a via SUMO (small ubiquitin-related modifier) modification [62]. Furthermore, two subdomains are packed into an integrated structure entity, in which the overall module exhibits an apparently positive potential as a basic surface for DNA binding [56].
As mentioned above, the most its variable region in the N-terminal regulatory part does not show much homology between Dnmt3a and Dnmt3b: about 280aa-length in Dnmt3a and approximately 220aa-length in Dnmt3b. It was noted that the short isoform of Dnmt3a and Dnmt3a2 lacks the most part of the variable region (219aa), and represents a diffuse localizing pattern in nuclei [46]. These data could provide hints that the variable region should be involved in guiding subnuclear localization of Dnmt3a and Dnmt3b. Several studies later further proposed that the variable region may hold some unspecified domain for protein-protein interactions. Dnmt3a is bound to small ubiquitin-like modifier peptide-1 (SUMO-1) and ubiquitin conjugating enzyme-9 (Ubc9) as sumoylational E2 conjugating enzyme and protein inhibitor of activated STAT (PIAS). Dnmt3b is also proposed to interact with SUMO-1 and Ubc9 [63, 64]. This provides information that the variable region could regulate Dnmt3a/Dnmt3b functions mediated by protein sumoylation.
The resolved structures of C-terminal domain of Dnmt3a and PWWP domain of Dnmt3b have been taken into explaining several key events of enzymatic processes of DNA MTase [50, 56]. Nevertheless, it should be noted that the truncated form of the protein can not be represented as a native structure of DNA MTase; and thus, to elucidate the crystal structure and even the in situ structure of the full-length DNA MTases should be seriously put on the agenda so as to unmask the sobering reality of DNA cytosine methylation in mammalian kingdom.
1.2.2. Dnmt3L
1.2.2.1. Genomic Mapping the Dnmt3L Gene
In human, encoding gene DNMT3L is located at 21q22.3, spanning nearly 16kb in the genome, existing 12exon/II intron. Its dominate mRNA is about 1.7kb, coding the protein DNMT3L of 387 amino acid residues in length [41]. Whereas in mouse, the Dnmt3L gene is located at the distal locus in chromosome 10 (41.6cM), extending approximately 190kb in length holding 15exon/14intron in the genome. Its main transcript is nearly 1.7kb, translating the protein Dnmt3L with 421 amino acid residues. The genes of Dnmt3L in human and mouse share about 60% identity (Table 1) [65].
1.2.2.2. Variants in Dnmt3L
In contrast to Dnmt1 and Dnmt3a/Dnmt3b, to date, variants in Dnmt3L are not so complicated. As a result of inframe splicing during RNA editing, in both human and mouse, there is only one additional transcriptional variant: 1.7kb variant in human and 1.8kb variant in mouse. In human, DNMT3L2 mRNA variant could be resulted from replacing of the exon 12 in DNMT3L mRNA, whose translational isoform DNMT3L2 lacks 1 amino acid residue in C-terminus of DNMT3L. Meanwhile, in mouse, Dnmt3L2 mRNA variant is the transcript disrupting in exon 3; whereas in the dominant Dnmt3L transcrip, exon 1–2 is spliced out. Although Dnmt3L2 isoform translated from the mRNA variant Dnmt3L2 also holds 421 amino acid residues, its N-terminal part is different from that of Dnmt3L [41, 65, 66]. However, it is largely unknown of those transcript variants and protein isoforms in tissue-specific expression, enzymatic properties, and definitive functions in development and disease pathogenesis.
1.2.2.3. Biochemical and Structural Organization of Dnmt3L
The overall structure of Dnmt3L is tightly related to Dnmt3a/Dnmt3b, and is believed to be relatively restricted during gametogenesis as a regulator in DNA methylation machinery, involved in genomic imprinting and DNA methylation setting processes [67]. However, either the N-terminal regulatory region or the C-terminal catalytic part in Dnmt3L seems to be a truncated form compared to that of Dnmt3a/Dnmt3b (Fig. 1).
1.2.2.3.1. C-terminal Region of Dnmt3L
The C-terminal region in Dnmt3L is much shorter compared to that of Dnmt3a/Dnmt3b (~200aa in Dnmt3L vs. ~300aa in Dnmt3a/Dnmt3b). Furthermore, structure-based sequence alignment indicated that several critical functional motifs commonly seen in other DNA MTases (e.g. Dnmt1 and Dnmt3a/Dnmt3b) are subject to certain unconservative substitutions. For instance, PC dipeptidyl motif in the conserved motif IV for the function of active loop could be replaced by PP dipeptide; several residues for Ado-Met cofactor binding are mutated or absent; variable region (DNA recognition domain) responsible for DNA targeting is almost disappeared [49, 68]. This may be one of reasons why Dnmt3L has not been detected to possess DNA methyltransferase activity to date.
Crystal structures of the human DNMT3L showed that the C-terminal region of Dnmt3L can be folded into a classical methyltransferase-like fold that is canonical in the prokaryotic DNA MTase (e.g. M.HhaI), and is conserved in Dnmt3a/Dnmt3b [26, 68]. The large domain is composed of seven β strands and four α helices packed into so-called α/β/α super-secondary fold; and the small domain only contains two α helices separated from the large domain by a small cleft [49, 68].
Although the C-terminal region of Dnmt3L is inactive concerning the DNA methyltransferase activity, Chedin et al demonstrated that Dnmt3L could functionally interact with Dnmt3a/Dnmt3b via the C-terminal domain, and to be involved in the de novo DNA methylation processes [69–71]. Co-crystallization research indicated that the intermolecular interactions found in the interface between the C-terminal region of Dnmt3L and Dnmt3a/Dnmt3b could stabilize conformation of the catalytic loop, and facilitate the Ado-Met cofactor binding potency of Dnmt3a/Dnmt3b [49]. In addition, the structure-based sequence alignment data showed that the interface between Dnmt3L and Dnmt3a/Dnmt3b might be associated with two α helices in each large domains of the catalytic region, which should be localized in the conserved motif V and VII [49].
1.2.2.3.2. N-Terminal Region of Dnmt3L
The N-terminal regulatory region in Dnmt3L seems to be truncated at the N-terminus compared with that of Dnmt3a/Dnmt3b (~200aa in Dnmt3L vs.~600aa in Dnmt3a/Dnmt3b) (Fig. 1). The cysteine-rich domain also exists, however, the PWWP domain in the regulatory region of Dnmt3a/Dnmt3b lacks in Dnmt3L [48].
The PHD sequence of cysteine-rich domain in Dnmt3L has been demonstrated to be directly interacted with HDAC1 [72, 73]. In addition, crystallography studies showed that the cysteine-rich domain, accompanying with the zinc ions in its C2-C2 zinc finger domain, could bind with the N-terminal tail of H3 histone [68]. Ooi et al. also indicated that the methylation level in the lysine 4 residue of H3 histone was inversely related to the binding capacity between Dnmt3L and H3 histone [68]. Furthermore, it was noted that the N-terminal region possessed an active nuclear localization sequence occupying 156–159 amino acid residues in Dnmt3L [72], suggesting that Dnmt3L may convert the H3 histone methylation signal into de novo DNA methylation execution commands. Taken together, the data indicated that Dnmt3L is a functional modulator assisting Dnmt3a/Dnmt3b in targeting the specific loci of nucleosome for regulation of de novo DNA methylation, and functions as a molecular linker tangling DNA methylation and histone modifications.
1.3. Dnmt2
Like Dnmt3a/Dnmt3b, Dnmt2 also was discovered by molecular informational manipulation in 1998 [11]. It was strikingly exciting that Dnmt2 was extremely conserved from yeast to mammals, and was the most widely distributed DNA MTases homologue [12]. Dnmt2 only contains the C-terminal catalytic region just like a prokaryotic DNA MTase with ten conserved motifs that are fairly conserved. There was evidence that Dnmt2 was functionally located in the cytoplasm. However, its DNA methyltransferase activity and definitive roles in vivo and in vitro might still be an enigma to date [74, 75]. Recently, emerging information indicated that Dnmt2 was a DNA MTase-like RNA methyltransferase that could specifically methylate cytosine 38 in the anticodon loop of aspartic acid transfer RNA (tRNAAsp) [76, 77]. Moreover, a glycolytic enzyme—enolase was shown to be an interacting partner of Dnmt2 in inhibiting tRNAAsp methylation in Entamoeba histolytica. This could encourage a hypothesis that Dnmt2 may link the metabolic status and DNA methylation programming/reprogramming during evolution [78].
Although functions of Dnmt2 in mammals are still a mystery, its roles in other metazoan model organisms (e.g. drosophila, zebrafish, and ceanorhabditis) have been extensively studied [79–81]. Recently, researches found that Dnmt2 also as a nuclear protein associated with nuclear matrix in fly, involved in mitotic divisions, retrotransposon silencing, and telomere integrity in somatic cells [82, 83].
1.3.1. Genomic Mapping of Dnmt2 Gene
The gene coding DNMT2 (391 amino acid residues in length) in human has been designated as TRDMT1 (tRNA aspartic acid methyltransferase 1) that is located at 10p15.1 (initially reported to be localized at 10p12–14), spanning about 58kb holding 11exon/10intron in the genome. In mouse, the locus for the Dnmt2 (415 amino acid residues in length) gene—Trdmt1, is believed to occupy chromosome 2 A1 area, expending approximately 34kb with 11exon/10intron in the genome (Table 1) [11, 12].
1.3.2. Variants in Dnmt2
To date, none of distinct transcript variant and protein isoform of Dnmt2 in mouse was found. In human, it is proposed that the gene TRDMT1 could generate additional two short transcriptional variants—TRDMT1b and TRDMT1c, which lacks exon4, exon4 and 5, respectively, resulting from the in-frame alternative splicing. The corresponding protein isoforms are 367 amino acid residues and 345 amino acid residues in length. It was believed that Dnmt2 is ubiquitously expressed in prenatal and adult tissues at a low level, and not prerequisite to mammalian development [13, 76, 84]. However, compared to that in Dnmt1 and Dnmt3, whether there is differential expression and localization of the transcript variants and protein isoforms of Dnmt2 is not clear, and corresponsive enzymatic activity and functions of these variants have not been elucidated.
1.3.3. Biochemical and Structural Organization of Dnmt2
As described above, Dnmt2 is the most conserved member in eukaryotic DNA MTases, even present in two species of the bacterial genus Geobacter [48]. In addition, Dnmt2 is a unique mammalian DNA MTase that lacks the entire N-terminal regulatory region. Similar to that in Dnmt1 and Dnmt3, the motifs I–X are conserved in Dnmt2 (Fig. 1). Sequence alignment researches showed that the variable sequences locating between conserved motif VIII and IX also were well conserved in Dnmt2, and acted as a DNA/RNA recognition domain. The central CFTXXYXXY motif (CFT motif) in DNA recognition domain of Dnmt2 is absent in other mammalian DNA MTases, although its roles need further investigation [48]. The crystal structure resolved in human DNMT2 showed that the structure was extremely superimposed well with the prokaryotic DNA MTase M.HhaI. Overall, Dnmt2 is composed of a large domain and a small domain. They are separated by the CFT motif as a cleft with the corresponding functional motifs and certain domains in well-organized and correct orientations [85].
The large domain contains conserved motif I–VIII and most of the motif X, in which α/β/α sandwich-like structure (eight-stranded β sheet with three α helices on one side and four α helices on the opposite side) are well conserved. Unlike that of M.HhaI, additional β strand is recently demonstrated in Dnmt2. In the small domain of Dnmt2, another three α helices evolutionarily emerges and they are absent in M.HhaI. The conformational structure of small domain of Dnmt2 is a propeller-like short four-stranded β sheet with five surrounding α helices (two α helices on one side and three on the opposite) [85]. GRASP representation of the electrostatic surface of Dnmt2 showed that the acidic pocket for Ado-Met binding is adjacent to the basic pocket for targeting DNA [85]. Also, limited data indicated that the CFT motif as a cleft in the interface between the large and small domain could make Dnmt2 contact DNA with the sequence-independent manner. Structural analysis indicated that some key residues (e.g. tyrosine) in CFT motif could make hydrogen bond with clashed unpaired base in order to increase target DNA binding affinity and to retain the target base in the “flipped out” state [48, 86].
In contrast to other mammalian DNA MTases, Dnmt2 has been recently shown to be involved in cytosine methylation of tRNA [76, 77]. In the light of these findings, Dnmt2 may be the functional dichotomy of either DNA or RNA cytosine methylation. However, the detailed dynamic expression, distribution, and definitive catalytic activity, as well as mechanisms of Dnmt2 are still a mystery. Although this protein is widely distributed and well conserved throughout the eukaryotes, the biological roles of Dnmt1 in mammals are largely unknown [87]. Thus, to further explore the fascinating paradox of Dnmt2 may enrich our knowledge on mammalian DNA MTases and DNA/RNA methylation network.
2. MOLECULAR ENZYMOLOGY OF MAMMALIAN DNA MTases
It has been generally believed that all active mammalian DNA MTases, including Dnmt1, Dnmt3a/Dnmt3b, and Dnmt2, utilize Ado-Met cofactor as a methyl group donor. Variety arrays of evidence showed that DNA MTases could be induced to dock in the nucleus, and then be actively or passively targeted to the relatively specific DNA sequences (might be bound with one or more “chaperones”). Following rotating the target cytosine completely out of double-stranded DNA helix (referred to as base flipping), DNA MTases can provide a platform for interaction between the substrate—cytosine and the cofactor—Ado-Met, in the catalytic active-site loop and hydrophobic binding pocket, respectively. Their enzymatic efficacy can be modulated by self regulating domain as an allosteric effect or by other regulatory partners. During the process involved in the C5 carbon atom in the cytosine making nucleophilic bound with the methyl group from Ado-Met, the covalent intermediate could be transiently formed during dearomatic and rearomatic reactions on cytosine by DNA MTases. Subsequently, DNA MTases could encode certain epigenetic marks with differential processivity genuine to regulate DNA methylation patterns involved in development and diseases pathogenesis. In the context below, we will further discuss these issues in fascinating events in molecular enzymatic processes of mammalian DNA MTases.
2.1. Catalytic Mechanism of Mammalian DNA MTases
Information derived from the classical prokaryotic cytosine methyltransferases (e.g. M.HhaI) showed that Ado-Met is buried in the hydrophobic pocket, and stabilized via hydrogen bonds and hydrophobic interactions with amino acid residues in the conserved motif I–III and X. Following the base flipping of cytosine, the covalent intermediate could endow the cytosine C5 carbon to activate a nucleophilic attack towards the methyl group in a sulfonium centre of Ado-Met. Then, the proton in C5 position of cytosine is eliminated followed by release of cysteine moiety from the covalent intermediate to generate the C5 methylated cytosine [25]. The sophisticated DNA cytosine methylation processes involved in the sequential covalent catalysis and acid-base catalysis previously demonstrated in the prokaryotic kingdom could be well suitable for mammalian DNA MTases. These were supported, at least in part, by the data from comparative sequence alignment between prokaryotic and mammalian DNA MTases, and also by the biochemical and structural analysis of certain mammalian DNA MTases [49, 88].
2.1.1. Base Flipping
In general, one base buried in the double-stranded DNA is stabilized by Watson/Crick hydrogen bonds between the corresponding paired base regardless of conformation (A-, B-, Z-, or other type) and style (free or bound) of DNA. As a result, the targeted base (cytosine) is unable to get access to the hydrophobic binding pocket or to expose their active N3, C5, and C6 positions for interactions within DNA MTases. Preliminary thermodynamic studies on prokaryotic DNA MTases (even not restricted in DNA cytosine methyltransferases) indicate that binding affinity between enzymes and substrates is inversely related to stability of the target base pair [89, 90]. This indicated that disruption of both the Watson/Crick hydrogen bonds of the target base and the stacking π interactions between adjacent base pairs could make DNA MTases bind substrate more favorable [89].
X-ray crystallography of the complex of M.HhaI and target sequence showed that the target base (cytosine) as well as the corresponding deoxy-ribose moiety could be flipped out of the canonical position together, and be localized within the concave active-site pocket out of the double-stranded DNA [91, 92]. Thus, “nucleotide flipping” is the reasonable nomenclature. In addition, there was evidence that some interactions between phosphodiester groups and DNA MTases could alter either distance or angle of the flanking sugar-phosphate ester bonds on either side of the targeted cytosine, and simultaneously induce conformational changes of DNA MTases [93, 94]. Meanwhile, researchers also found that the DNA MTase could move the sugarphosphate backbone to be everted, even when the target site in DNA substrate was abasic (apurinic/apyrimidinic) pair [93]. The “base flipping” event also is manifested by other various DNA-interacting enzymes (e.g. DNA glycosylase) [95, 96]. These suggested that the sole target base (cytosine) in DNA methylation may not induce conformational changes of DNA per se. Moreover, mutagenesis investigation indicated that the glutamine residue in the variable region (i.e. DNA recognition domain spanning between conserved motif VIII and IX) could be inserted into the target DNA for filling the place of cytosine for contacting with guanine [97, 98]. Later, studies further indicated that valine and glutamate residues in the conserved motif VI and arginine residue in the conserved motif VIII immediately adjacent to the target cytosine were essential for both stabilization of the flipped-out target base and maintenance of DNA MTases enzymatic activity [99, 100]. In addition, interactions between target base and DNA MTase could dislocate adjacent DNA backbone atoms. For instance, the theonine residue in the DNA recognition domain of DNA MTases could contact with the 5’phosphate of the target nucleotide [101]. In the light of previous kinetic results, a hypothesis has been proposed that changes of free energy (binding energy) induced by interactions between DNA MTase and DNA might be the resource for base pair destruction and conformational changes of sugar-phosphate bonds. Furthermore, researchers discovered that the induced-fit involved in the active loop rearrangement in DNA MTases, which should be the basis for enzymatic reactions in DNA methylation, was directly coupled to the “base flipping” process [102]. Therefore, the specific cytosine methylation in DNA duplex could be essentially contributed by substrate selectivity of DNA MTases, especially by selective interactions between the active-catalytic loop and the target cytosine.
As described above, the C-terminal catalytic region of all mammalian DNA MTases is homologous to the prokaryotic DNA MTases (especially M.HhaI). Consequently, to flip the cytosine out of its native DNA double helix should also be the key determinant for DNA methylation in mammals. This is supported at least by various enzyme kinetic experiments and numerical molecular simulations concerning mammalian DNA MTases. It is indicated that the “base flipping” may be the very initial fast-equilibrium kinetic process modulating catalytic reaction rate of DNA cytosine methylation [103]. Recently, Jia et al. reported that the cocrystal complex of the C-terminal catalytic domain of Dnmt3a and Dnmt3L fitted well to the DNA fragment cocrystallized with M.HhaI where the target cytosine was flipped out [49]. Thus, the data further supports the reality of “base flipping” event in the enzymatic processes of mammalian DNA MTases. Interestingly, emerging findings indicated that UHRF1 [ubiquitin-like, containing PHD (plant homology domain) and RING (really interesting new gene) finger domains 1] could recruit Dnmt1 to the replication fork, and act as a “base flipping” executor [104, 105]. However, the target base is the C5 methylated cytosine in hemimethylated double-stranded DNA [106–108]. Thus, cytosine in DNA double helix in mammals might not be the specific target for “base flipping” per se, as presented in the prokaryotic DNA MTases. Protein-DNA interactions could be a plausible reality for the reconstruction of intermolecular interactions involved in the target cytosine eversion executed by mammalian DNA MTases. To date, there still lacks direct evidence supporting whether the target base (cytosine) can undergo the “base flipping” event in the process of DNA methylation by mammalian DNA MTases. Compared to that of prokaryotic DNA MTases, mammalian DNA MTases possess the functional N-terminal regulatory region (except for Dnmt2), and also interact with various regulatory elements, so as to synergistically function as an enzymatic entity. Whether and to which extent the self-regulatory domain or other modulators are involved in the “base flipping” event is unclear, and needs further studies.
2.1.2. Cofactor (Ado-Met) Binding
Ado-Met is a methyl group donor involved in most of methyl transfer reactions, as well as in DNA methylation. In prokaryotes, Ado-Met was demonstrated to be buried in the hydrophobic binding pocket related to conserved motifs I–III and X [26]. The high binding affinity between DNA MTases and Ado-Met was supported by the chromatographic experiments showing that Ado-Met could be co-purified together with M.HhaI [25]. Relevant evidence further indicated that almost all hydrogen bond donors and acceptors in Ado-Met were interacted with the corresponding atoms of the certain conserved amino acid residues in DNA MTases [109]. In addition, some hydrophobic interactions also exist between Ado-Met and DNA MTases [110].
In addition, the binding site of Ado-Met can be changed in association with DNA binding state of DNA MTase. O'Gara et al. provided evidence that the primed orientation of Ado-Met in the hydrophobic binding pocket of the DNA—M.HhaI—Ado-Met tertiary complex was changed when DNA was absent in the M.HhaI—Ado-Met binary complex [111]. The S-adenosyl-L-homocysteine, a product of Ado-Met in DNA methylation, can be buried in a differential conformational orientation, so as to be interacted with the active-site loop in DNA MTases in inhibiting catalytic potency of DNA methylation [112]. These data led us to consider that cofactor binding in DNA MTases could manifest differential modes in a sequential order regulated by DNA targeting and enzymatic reaction processes per se [103]. In prokaryotes, more than one Ado-Met binding site was detected in DNA MTases. Ado-Met binding was suggested to be able to allosterically modulate the enzymatic activity through the pathway influencing the active site in DNA MTases [25]. Thus, we would like to postulate that DNA, Ado-Met, and DNA MTase in an enzymatic tertiary complex could regulate reciprocally each other.
The structure-based sequence comparison and the resolved crystal structure of Dnmt3a-Dnmt3L complex indicated that Ado-Met in mammals also were embedded in the hydrophobic binding pocket where the corresponding amino acid residues were resided in the conserved motifs I–III, V, and X of mammalian DNA MTases [49]. Whether the native Ado-Met binding site also resided in the resemble orientation should be further investigated.
2.1.3. Cytosine C5 Methylation Reaction Mechanism
To fulfill methylation reaction in the C5 position of the target cytosine, the nucleophilic potency of cytosine is the foundation for transferring methyl group from the sulfonium centre in Ado-Met to the C5 position in cytosine. Nevertheless, distribution of reactive electrons in the cytosine ring is delocalized. Thus, cytosine alone could not provide an effective nucleophilic attack in the C5 position to methyl group in Ado-Met. In this context, destruction of the aromaticity of cytosine ring could be prerequisite to DNA cytosine methylation. DNA MTases facilitate these processes by lowering the energy barrier under the mechanism that a transient covalent intermediate could be created followed by a series of acid-base catalysis. This is critically attributed to sophisticated interactions among target base, cofactor, and enzyme.
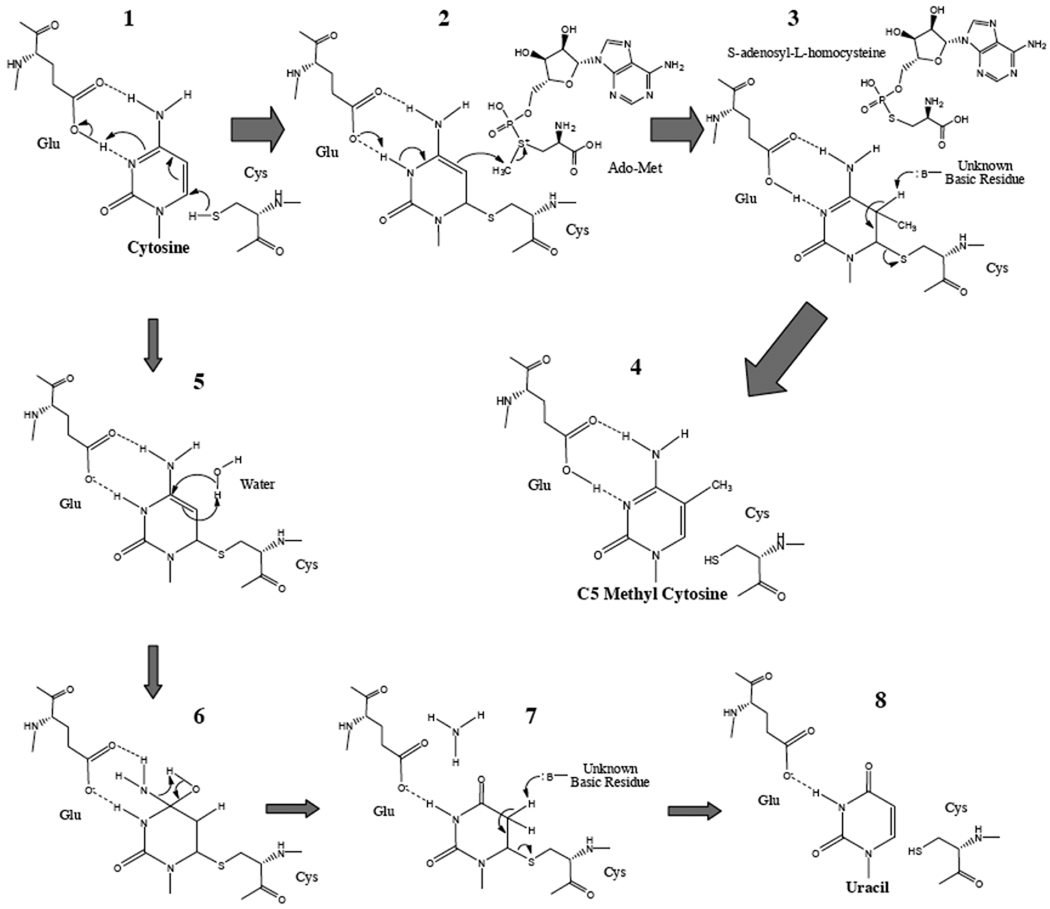
The reaction mechanism of methyl group transferring from Ado-Met to the cytosine C5 position had been extensively investigated in the prokaryotic DNA MTases [25]. The essential step is the formation of covalent intermediate resulted from a nucleophilic attack of the key cysteine in the active-site loop of DNA MTase on the C6 position in the target cytosine [113, 114]. Owing to the enzyme, this reaction might be facilitated by transient protonation of the N3 position with assistance of glutamate residue in the conserved motif VI of DNA MTase [115]. Consequently, the aromaticity of cytosine could be broken; and the activity of nucleophilic attack of the C5 position in cytosine can be potentially strengthened. As a result, C5 carbon could be potently activated to attack methyl group in the sulfonium centre of Ado-Met, followed by deprotonation of the N3 position in cytosine. Finally, the covalent enzyme—cytosine complex could be resolved by eliminating the proton on the C5 position in cytosine, so as to release thiol group of the cysteine moiety (Fig. 2). However, none of amino acid residue(s) has been shown to act as a Lewis base (basic residue) responsible for the deprotonation of the C5 position in cytosine. Theoretical calculation showed that hydroxide anion rather than basic residue(s) in the active site, achieved through an essential water channel in DNA MTases, may be actually involved in the deprotonation step immediately prior to release of the cytosine moiety [116]. In concert, the nature concerning the proton abstracting involved in the β-elimination reaction is still delusive and has to be further dissected. Currently, frequently used clinic drugs against cancers like cytosine analogues [e.g. 5-azacytidine, 2′-deoxy-5-azacytidine (decitabine), arabinosyl-5-azacytidine (fazarabine), and dihydro-5-azacytidine (DHAC)], are considered to incorporate into genome to mimic cytidine binding with DNA MTases. Owing to the nitrogen atom at position 5 in the bases of cytosine analogues, the β-elimination reaction could not occur between drugs and enzymes. Consequently, DNA MTases may be degraded and the DNA methylation process is inhibited [117].
Fig. 2.
The chemical mechanism of cytosine C5 methylation (1–4) and its possible side-effect reactions (5–8). The sulfhydryl group at the side chain of the key cysteine residue, in the active-site loop of DNA MTase, could nucleophilically attack to the C6 position of cytosine followed by transient protonation of the N3 nitrogen atom via the glutamate residue in conserved motif VI of DNA MTase (1–2). Consequently, aromaticity of cytosine is broken so as to strengthen activity of nucleophilic attack of the C5 position in cytosine to the methyl group in the sulfonium centre of Ado-Met (2). Following the methyl group transferred to the C5 position of cytosine, the N3 position in cytosine is re-deprotonated (b). To release enzyme moiety covalently bound in the C6 position of cytosine, the C5 position could be deprotonated with assistance of unknown residues in the enzyme (3). Following the β-elimination reaction involved in C5 and C6 positions in cytosine, C5-methyl cytosine could finally be formed (4). Under certain conditions (e.g. Ado-Met inadequate), water can penetrate into the active site of DNA MTase towards the relatively localized π electron in the activated cytosine (5). Then the C4 position in cytosine is hydroxylated so that deamination reaction is triggered (6). The amino group in the C4 position of cytosine then is replaced to form a carbonyl oxygen atom (7). The conversion of cytosine to uracil is accomplished (7–8).
Furthermore, it is notable that to generate the transient covalent intermediate described above might be very complicated. So far, the mechanism underlying cytosine methylation mentioned above is almost derived from structural information of the tertiary complex of DNA—enzyme—cofactor in postchemistry state and from mutagenesis combined with enzyme kinetic studies [25, 103]. Recently, a prechemical crystal structure of the DNA—enzyme—cofactor tertiary complex has been described. Following the refined comparison between prechemical and postchemical tertiary structures, Youngblood et al. suggested that the target cytosine could be tilted up to the cysteine residue in the active-site loop prior to the formation of covalent intermediate [118]. The compressive arrangement was critically important for facilitating the nucleophilic attach on C6 carbon in cytosine ring derived from the thiol group in the cysteine moiety. In addition, functional and simulating data showed that, besides glutamate residue, another two arginine residues in the conserved motif VIII of DNA MTases could also play critical roles in protonation of N3 position in the target cytosine [119–121]. Quantum mechanical/molecular mechanics analysis further indicated that the two arginine residues in the conserved motif VIII may be essential for stabilizing the delocalized electrons in cytosine moiety of the transient covalent intermediate through their intrinsic electrostatic interactions [116]. It was also indicated that the free water molecule, buried between the two arginine residues in the conserved motif VIII of DNA MTases stabilized by hydrogen bonds, was directly detected in the prechemical crystal structure of the tertiary complex and was firmly coupled to the prime orientation of Ado-Met [118]. Therefore, it should be inferred that the free water molecule coupled with two arginine residues might modulate active-site loop in DNA MTases in stretching to and maintaining the closed conformation status, and that the compressive arrangement of cytosine—catalytic loop—Ado-Met could be transiently stated in order to effectively promote methyl transferring reaction.
Unfortunately, DNA cytosine base might be deaminated as a mutagenic reaction catalyzed by DNA MTase in prokaryotes [122]. Solvent such as water could penetrate into active site of DNA MTase towards the relatively localized π electrons in the activated cytosine induced by covalent linkage between C6 position in cytosine and thiol group in cysteine moiety. Therefore, the amino group in the C4 position of cytosine could be replaced to form a carbonyl oxygen atom as such the conversion of cytosine to uracil is accomplished [123] (Fig. 2). The transient covalent intermediate is a prerequisite for the mutagenic cytosine deamination catalyzed by DNA MTases in prokaryotes [124]. In bacteria, the mutagenic cytosine deamination is triggered in a case of undernutrition (e.g. limiting Ado-Met). This is an evolved physiological advantage for evading lethal deterioration digested by phage restrictive endonucleases [123, 124]. So far, the mutation of cytosine deamination has not been considered to be benefit for mammalian cells. In mammals, the steric blocks derived from the binding of Ado-Met or S-adenosyl-L-homocysteine in mammalian DNA MTases could prevent mutagenic cytosine deamination triggered by harmful water accession [103]. Furthermore, mammalian DNA MTases can not effectively attack the target base without Ado-Met binding [125, 126]. These studies represent that susceptibility of cytosine mutation induced by DNA MTases in eukaryotes is less likely than that in prokaryotes. Thus, in eukaryotes, at least in mammals, epigenetic events may not interfere with intrinsic genetic states, and such the vital biological stability could be well maintained.
Globally, according to the structure-based sequence alignment data, DNA methylation reaction mechanism in mammalian DNA MTases is similar to that of prokaryotes [49]. Interestingly, evidence showed that covalent catalysis might not be the crucial step in mammalian DNA cytosine methylation, at least in the case of Dnmt3A [127]. These findings suggested that conformational changes of the catalytic loop in mammalian DNA MTases can be regulated by unknown modulators, and the enzyme could then directly catalyze the methyl transferring reaction under the assistance of glutamate residue in the conserved motif VI. Later, geometrical orientations among cytosine, cysteine, and S-adenosyl-L-homocysteine in the crystal structure of Dnmt3A showed that the covalent intermediate might still be important for DNA methylation [49]. However, the active site of mammalian DNA MTases could also change its conformation postchemically. Consequently, the actual arrangement of the tertiary complex cytosine—catalytic loop—Ado-Met might not simply extrapolated from the postchemical crystal structure described above. In addition, mammalian DNA MTases still possess the N-terminal regulatory domain and bind to various functional partners; and the active site loop could be effectively modulated by interactions in the interfaces between Dnmt3A and Dnmt3L [49]. Thus, DNA methylation reaction is surely more complicated. The roles of conserved residues in the active-site loop of mammalian DNA MTases might be evolutionarily changed. Whether there may emerge additional functional residues or small molecules (e.g. water, free ions) attributing to this enzymatic process should be a good topic for detailed mechanisms in DNA methylation in mammals.
2.2. Substrate Targeting of Mammalian DNA MTases
All mammalian DNA MTases may preferentially select CG sites in DNA [128]. As a consequence, some small molecule compounds, for instance: procaine and procainamide, have been designed to bind to CG-rich sequences so as to interfere with the binding efficacy of DNA MTases. Interestingly, procainamide specifically inhibits Dnmt1 but not Dnmt3a/3b; however, the underlying reasons should be further elucidated [129, 130].
Subsequent researches on binding or kinetic preferences showed that substrate selectivity of mammalian DNA MTases may depend on the status of DNA sequence (CG sites and the flanking sequence), DNA structures (single vs. double strand), DNA methylation patterns (unmethylated vs. hemimethylated, fully methylated), and DNA length [131–133]. Furthermore, a great array of evidence demonstrated that degree of target sequence selectivity and magnitude of preference for different methylation states of DNA represented remarkable divergence among different mammalian DNA MTases [13, 48]. As described above, Dnmt2 is the RNA MTase whose DNA methylation activity is not well ascertained and the DNA sequence selectivity is obscured [87]. Certain tRNA sequence could be the effective target of Dnmt2.
2.2.1. Dnmt1 Targeting Hemimethylated DNA as a Maintenance DNA MTase
It has been widely accepted that Dnmt1 methylated target symmetric CG sites in hemimethylated DNA sequence with a high specificity [103]. This property is essential for imparting existing DNA methylation patterns after DNA replication, as Dnmt1 serves as maintenance DNA MTase. Recently, it has been shown that UHRF1 could, through PHD or SRA [SET (Su(var), E(z), Trithorax) and RING module associated] domain, physically bind to regions in the regulatory domain of Dnmt1 [104, 105, 134]. Moreover, as mentioned above, detailed crystallographic researches showed that UHRF1 could specifically project the C5-methyl cytosine out of the hemimethylated DNA double helix via its SRA domain [106–108], suggesting that UHRF1 could be a new target interfering with DNA MTases. Small molecules such as 5-methylcytosine-like compounds may inhibit SRA domain's functions is block DNA methylation processes. However, it is still far beyond its clinical uses [135].
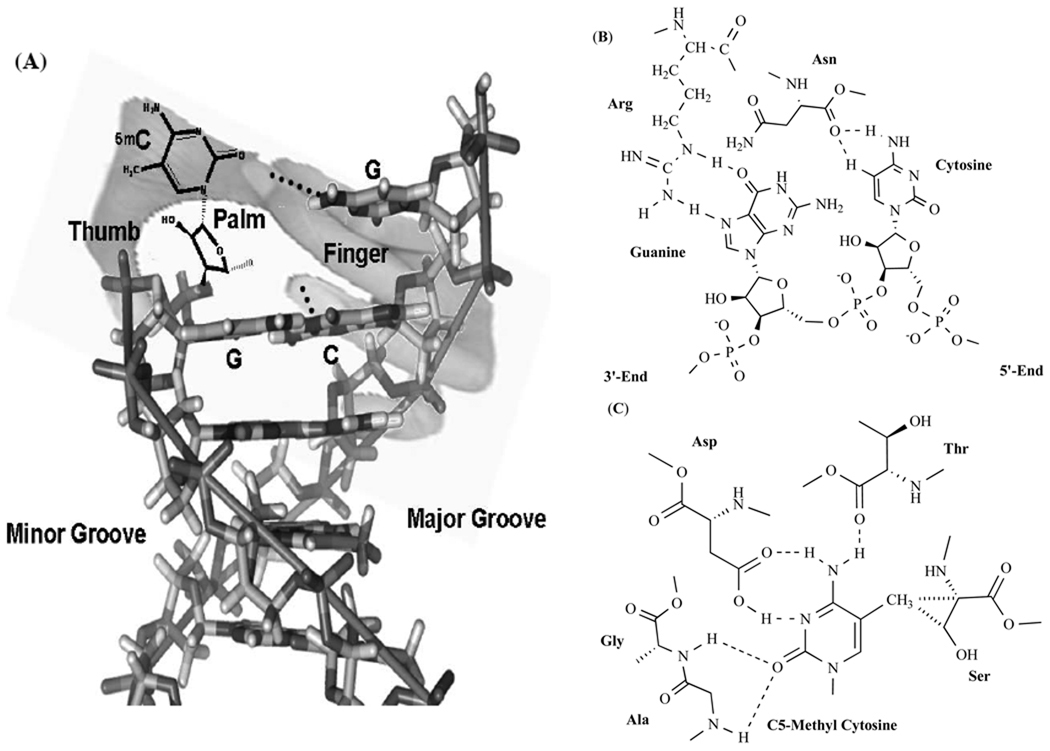
The SRA domain, of about 210 residues in UHRF1, a single globular structure contains two twisted β sheets (composed of mixed five β strands and anti-parallel three β strands, respectively) and scattered four α helices and one 310 helix surrounding the shell-like β sandwich. This domain is characterized as a crescent moon-like conformation revealed by X-ray crystallography [108]. The concave surface of the SRA domain interacts with the target DNA sequence to be responsible for recognition of hemimethylated CG site and promotion of C5-methyl cytosine eversion [107]. The overall structural mode of the SRA domain-DNA duplex complex was imagined as a hand clamping the double-stranded DNA, with the Arg-associated loop (between β5 strand and α2 helix) and Val-associated loop (between α1 helix and β2 strand) corresponding to a finger and a thumb projecting into the major and minor groove of DNA, respectively, and the positively charged inner patch as a palm holding a deep pocket for binding C5-methyl cytosine [107] (Fig. 3). Evidence indicated that several amino acid residues in both the finger and of the thumb could interact extensively with nucleotides in the target DNA sequence. The Arg residue in the finger is important for binding of the SRA domain and targeting DNA sequence, whose guanidinium side chain could form hydrogen bonds with O6 and N7 positions in the intrahelical orphaned guanine ring, so as to simulate the Watson-Crick hydrogen bonds. It was also represented that the Arg residue might even stack with adjacent bases in the methylated strand to compensate for eversion of the C5-methyl cytosine. Another amino acid residue—Asn, located near the Arg residue in the finger, which is prerequisite for specificity of binding to hemimethylated CG sites, could be collaboratively organized with the Arg residue to recognize the opposite adjacent unmethylated cytosine and effectively interact with the phosphor-diester in the complementary strand. Lacking steric clashes derived from methyl group in the C5 position, the main-chain carbonyl group of the Asn residue in the finger could be effectively form essential hydrogen bonds with the cytosine ring in the unmethylated strand (Fig. 3). Moreover, the Val residue in the thumb may occupy the vacancy left by the extrusion of C5-methyl cytosine so as to approach a proper distance from the minor groove side, leading a van der Waals interaction between its methyl group and the aliphatic portion of Arg residue in the finger located in the opposite major groove [106–108]. In the light of these, the finger and the thumb in the SRA domain is termed as hemimethylated CG recognition loop and base-flipping promotion loop, respectively [108]. Furthermore, researchers found that, following a dramatic conformational change of the SRA domain induced by its binding with the target sequence DNA, the space of the target base binding pocket was approximately 6×10×6 Å in the palm that was not enough for binding purines [107]. In addition, crystallography data showed that the C5 methylated cytosine was well stabilized by the π stacking force via the two benzene rings of two Tyr residues in the palm located in the two sides of the pyrimidine ring. In the palm, the amide groups in the main chain of the Gly and Ala residues, the carbonyl groups in the side chain of the Asp residue, and the main chain of the Thr residue, are well accepted to form hydrogen bonds with O2 and N4 positions in the target cytosine, respectively, so as to discriminate the thymine base [106–108]. To explain specificity for C5-methylated cytosine binding, researches showed that, deep in the pocket, there existed a hemisphere spacing, of about 2 Å in radius, explicitly fitted the methyl group. Additionally, crystallographic data demonstrated that the C5 methyl group in the target cytosine was tightly packed in a cavity strengthened by various hydrogen bonds and van de Waals interactions with the Tyr, Thr, and Gly residues in the palm [106]. Especially, van de Waals interactions are tightly stabilized between the C5 methyl group of the target cytosine and the Cα and Cβ atoms of the Ser residue in the palm, and the hydroxyl group of that Ser residue interacts with the phospho-diester in the methylate strand [108] (Fig. 3).
Fig. 3.
Interactions between hemimethylated DNA duplex and SRA domain of UHRF1. (A): The interface toward DNA in the SRA [SET (Su(var), E(z), Trithorax) and RING module associated] domain of UHRF1 [ubiquitin-like, containing PHD (plant homology domain) and RING (really interesting new gene) finger domains 1] is imagined as a grasping hand, with its finger and thumb projecting into the major and minor groove of DNA duplex, respectively. Its palm holding a deep pocket for binding C5-methyl cytosine. (B): The guanidinium side chain of Arg residue in the finger can form hydrogen bonds with O6 and N7 positions in the intrahelical orphaned guanine ring in order to simulate the Watson-Crick hydrogen bonds, which is important for binding of the SRA domain and target DNA duplex. The main-chain carbonyl group of another amino acid residue—Asn, located near the Arg residue in the finger, can effectively form essential hydrogen bonds with the cytosine ring in the unmethylated strand. The collaborative organization of the Arg and Asn residues in the finger is prerequisite for specificity of the SRA domain binding to hemimethylated CG site. (C): In the palm, amide groups in the main chain of the Gly and Ala residues, and carbonyl groups in the side chain of the Asp residue and the main chain of the Thr residue, are toward to O2 and N4 positions of C5-methyl cytosine to form hydrogen bonds so as to discriminate cytosine from thymine base. Moreover, the van de Waals interaction is formed by interaction of C5 methyl group of the target methylated cytosine and the Cα and Cβ atoms of the Ser residue of the deep pocket in the palm. In addition, π stacking force given by the two benzene rings of two Tyr residues in the palm located in the two sides of the C5-methyl cytosine ring.
The crucial Gly residue at the entrance to the deep binding pocket in the palm is a key determinant for the high affinity of the SRA domain to the hemimethylated DNA sequence [107]. Detailed analysis of the X-ray crystallography data indicated that the contact area of the SRA domain to one everted C5-methylated cytosine in hemimethylated CG site was comparable to that of methyl-DNA-binding domain (MBD) to the two C5-methylated cytosines in fully methylated CG site, and the contact area in the interface of SRA-DNA complex was also much larger than that of the MBD-DNA interface [106, 108]. Consequently, interactions with the target base flipped out could be essential and economic for binding of UHRF1 to the target DNA sequence. Based on those findings, we could propose that the mechanism underlying “base flipping” concerning the methylated cytosine executed by UHRF1 exclusively in the hemimethylated CG site may be the determinant for maintenance of methylation by Dnmt1 recruited by UHRF1. The initiation of methylation reaction by Dnmt1 might follow the release of UHRF1 from the hemimethylated DNA sequence [106]. Thus, whether UHRF1 could determine or elevate the binding specificity and affinity should be further determined [104]. However, it was noted that conformation of the unbound state of the SRA domain is largely different from that of bounded form, leading to an imagination that the initial selection or primary binding of UHRF1 and DNA may be nucleotide sequence-independent [107]. Given this fact, what should be the so-called “a priori” for the specific binding of UHRF1 to the hemimethylated DNA sequence? This basic issue has yet not been illuminated.
In addition, in vitro enzyme kinetic studies indicated that Dnmt1 not only binds to hemimethylated double-stranded DNA sequence, but also may interact directly with CG-rich single-stranded oligonucleotide in which one CG site was methylated [136]. Furthermore, binding studies showed that Dnmt1 could bind to both unmethylated and fully methylated double-stranded DNA, and could weakly methylate cytosine in the unmethylated DNA duplex [137]. After binding with that type of single-stranded oligonucleotide as mentioned above, or with unmethylated double-stranded DNA, catalytic activity of classical maintenance of DNA methylation by Dnmt1 could be inhibited [136]. Nonetheless, fully methylated double-stranded DNA can “buffer” the inhibitory effect of unmethylated double-stranded DNA to Dnmt1 that catalyze its own binding to unmethylated DNA duplex [138]. Recent researches further demonstrated that non-coding RNA could also bind to the regulatory domain of Dnmt1 in the control of DNA methylation [139]. Consequently, an explanation for the phenomenon was introduced from those findings. It states that several nucleotide sequences (either single-stranded or double-stranded, unmethylated or fully methylated DNA duplex, DNA or RNA) adjacent or distant (in the same or other DNA strands) from the DNA region bound to the catalytic domain of Dnmt1, might allosterically regulate activity of methylation reaction through interactions with the regulatory domain of Dnmt1 [103]. However, the “allosteric regulatory model” applied in enzymatic reactions by Dnmt1 is only imagined from in vitro kinetic experiments, it still lacks real 3D structural information on the full-length Dnmt1 complexed with its interacted nucleotide sequences. Those issues should be solved in future.
In general, Dnmt1 selectively targets hemimethylated double-stranded DNA to apply its intrinsic roles in molecular memory of DNA methylation status after cellular proliferation. However, as described above, DNA methylation in the unmethylated DNA duplex also can be catalyzed by Dnmt1. This may be strengthened under allosteric activation by presence of fully methylated DNA [103]. In addition, forthcoming experimental data showed that overexpression of Dnmt1 could hypermethylate the global genome, and Dnmt1 may function as de novo methylation fashion interacted with Dnmt3a/Dnmt3b [140, 141]. These results further indicated that the selective binding with the hemimethylated DNA duplex by Dnmt1 for maintenance of DNA methylation may be relative. The de novo-like DNA methylation reaction by Dnmt1 could be induced by certain conditions during development or diseases. The bimodal characteristics of Dnmt1 in canonical maintenance and inducible de novo-like property in vivo are essential for stabilization of epigenetic inheritance and efficiency of gene regulation switched on/off by the epigenetic network.
2.2.2. The Destination of Dnmt3: Combination and Uncertainty
As discussed above, although Dnmt3a/Dnmt3b is the key enzyme for the de novo DNA methylation. Without assistance of Dnmt3L, DNA methylation patterns of imprinted loci germ in cells could be potentially affected [142, 143]. Cocrystallographic information showed that hydrophobic interactions on the interface of Dnmt3a and Dnmt3L may be induced by the Phe residues in both Dnmt3a and Dnmt3L. Moreover, the Asp residue and Arg residue in Dnmt3L may form polar interactions with the Arg residue and Glu residue in Dnmt3a, respectively, at the neighborhood of the activesite loop of Dnmt3a. Furthermore, the polar interaction between Dnmt3a and Dnmt3L is stabilized by additional intermolecular hydrogen bond and intramolecular polar interaction. Thus, native open conformation of the active-site loop in Dnmt3a can be clashed to the closed state induced by formation of the Dnmt3a-Dnmt3L interface, and increase catalytic activity of Dnmt3a [50]. Meanwhile, Dnmt3a also can self contact to generate the Dnmt3a-Dnmt3a interface formed by polar interaction between Arg and Asp residue in one Dnmt3a and the corresponsive Asp and Arg residue in another Dnmt3a [49]. Subsequently, two Dnmt3a and two Dnmt3L may form a tetramer complex possessing three interfaces: one central Dnmt3a-Dnmt3a homodimer interface and two lateral Dnmt3a-Dnmt3L heterodimer interfaces. The overall Dnmt3L-(Dnmt3a)2-Dnmt3L complex is an elongated (approximately 160×60×50Å), butterfly-like structure [49] (Fig. 4). The length of the Dnmt3L-(Dnmt3a)2-Dnmt3L tetramer (about 16nm) is apparently greater than the diameter of the nucleosome (about 11nm). This is the structural foundation for directly physical interactions between Dnmt3L of Dnmt3L-(Dnmt3a)2-Dnmt3L tetramer and H3 histone tail of nucleosome. Consequently, Dnmt3a could be targeted to DNA duplex assisted by Dnmt3L [68].
Unlike Dnmt1, Dnmt3a/Dnmt3b did not discriminate between hemimethylated and unmethylated DNA duplex so that the nomenclature of de novo DNA MTase is assigned for Dnmt3a/Dnmt3b [144]. Interestingly, in vitro experiments showed that cytosine methylation positions, in a long DNA duplex catalyzed by Dnmt3a, represented a periodical property with its periodicity of approximately 10 base pairs (about one DNA helical turn) [49]. Analysis of the structural data showed that the successive two active-site loops, facing roughly from the opposite direction in the Dnmt3L-(Dnmt3a)2-Dnmt3L tetramer, were about 40 Å apart which was equal to the distance of one helical turn [49] (Fig. 4). This may explain specific DNA methylation in a spacing fashion catalyzed by Dnmt3a in the target genes [49].
Surprisingly, besides the classical symmetric CG site, Dnmt3a/Dnmt3b could methylate non-CG sites, especially the CA site, although the enzymatic activity is apparently lower than that in the CG site [145, 146]. In light of these, Dnmt3L-(Dnmt3a)2-Dnmt3L complex and histone modification status (methylation or acetylation) could affect subsequent regulations of various genes via direct interactions between Dnmt3L and histone tails, especially during development [147]. A possibility might exist that the DNA region where de novo DNA methylation or histone modification occurs may lack symmetrical CG sites. Dnmt3a/Dnmt3b could be forced to use non-CG site as methylation target, together with other epigenetic machinery, to synergistically control expression of the gene. Another speculation postulated that non-CG DNA methylation might be a rapid onset repression of gene expression in early embryonic periods. After emergence and maturation of other epigenetic mechanisms, non-CG site methylation catalyzed by Dnmt3a/Dnmt3b may be no longer required. Obviously, this issue should be further investigated. To dynamically analyze the functional development of epigenetic mechanisms and compare them in details during embryogenesis may provide clues for the explanation.
Whether mammalian DNA MTases only bind to CG site in the nucleotide sequence is another interesting question. According to typical protein-DNA interaction, mammalian DNA MTases (i.e. Dnmt1 and Dnmt3a/Dnmt3b) could not merely exclusively bind to CG dinucleotides [148]. Evidence showed that changes in other nucleotides adjacent to the central CG site, about 6–12 nucleotides, termed as the “flanking sequence”, may modify catalytic activity of mammalian DNA MTases. It is suggested that Dnmt3a and Dnmt3b preferentially select the flanking sequence with the purine base at the 5’end and pyrimidine base at the 3’end in the central CG site [149]. However, structural basis and refined modulation by the flanking sequence need additional studies.
2.3. Reaction Processivity of Mammalian DNA MTases
DNA MTases are enzymes acting on a long substrate. This could methylate more than one target cytosine without release from DNA. In vitro kinetic findings showed that the turnover rate of mammalian DNA MTases is obviously faster than its dissociation rate. This behavior of the enzyme is termed as the processivity. Dnmt1 is highly processive in the DNA methylation on a long hemimethylated DNA duplex, especially during DNA replication with its binding to PCNA [150]. Recent findings demonstrated that methylation process was clearly slower than DNA replication in the heterochromatic region, in which high density of CG sites were present [151]. This indicated that in the heterochromatin region DNA methylation may lag behind DNA replication that can be impeded. To overcome complications, Dnmt1 is first released from the replication fork and then rebind to the newly replicated DNA duplex to catalyze cytosine methylation in unmethylated CG sites. Further studies showed that active lowering of enzyme efficacy might even applied by Dnmt3a/Dnmt3b [152]. These results could suggest that processivity of Dnmt1 is relative, and it may be spatially or temporarily regulated.
Dnmt3a and Dnmt3b represent distributive and processive enzymes, respectively; although there is nearly 85% identity in amino acid sequences between their catalytic domains [48]. Comparative amino acid sequence alignment analysis showed more basic amino acid residues in the catalytic domain of Dnmt3b than that in Dnmt3a. Thus, more positive charges in the catalytic domain of Dnmt3b may be the reason for its higher efficiency grasping negatively charged nucleotides [144]. As a result, Dnmt3a and Dnmt3b can methylate cytosine in certain defined CG sites and be functional in an extensive large domain in some genes. Via those mechanisms, the de novo DNA methylation could be set up effectively, and regulated precisely and separately.
CONCLUSION REMARKS
In mammalian cells, maintenance and de novo methylation events can be carried by Dnmt1 and Dnmt3a/Dnmt3b, respectively, while there may exist some functional overlap. Although the primary protein structures among them are fairly similar, each mammalian DNA MTase is the product of different genes located in different chromosomes. The catalytic domain of mammalian DNA MTases is almost identical. The organized enzymatic reaction of methyl group transferring, concerning target nucleotide flipping out of the DNA duplex, Ado-Met cofactor binding, and multiple covalent and acid-base catalysis for the methyl group transferring from Ado-Met to the C5 position of cytosine, could be constantly conserved from bacteria to mammals. A variety array of evidence indicated that UHRF1 and Dnmt3L may serve as the regulatory adaptor to recruit and target the Dnmt1 and Dnmt3a/Dnmt3b binding to selective and specific regions and status of methylation in the DNA sequence, however, the exact interaction between them is unclear. In particular, primary motivation of the key step—base flipping, involved in the sequence recognition by either DNA MTases or adaptors (i.e. UHRF1), has yet not been elucidated. In addition, several regions in the regulatory domain also may be related to other epigenetic events, such as histone acetylation, histone methylation, and RNA interference. To data, there have emerged various inhibitors of mammalian DNA MTases, applying mainly in cancer therapy. Only cytosine analogues and some certain small molecule compounds get approval for clinical treatment. The anti-cancer DNA MTase inhibitors could merely target or interfere with a general event in the enzymatic process with no specificity. As a consequence, only overall DNA methylation at genome level may be adjusted. Furthermore, different DNA MTase can play a different role in pathogenesis of either cancer or other diseases. Therefore, drugs with more specificity targeting unique DNA MTase have to be designed and annotated.
ACKNOWLEDGEMENTS
Partially supported by NIH HL090920, National Natural Science Foundation (30973211) and NSCF key grant, Jiangsu Grant (BK2009122, 08KJB32001), and Suzhou Grant (No: 90134602, SZS0602).
REFERENCES
- 1.Hochkiss RD. The quantitative separation of purine, pyrimidines and nucleoside by paper chromatography. J. Biol. Chem. 1948;168:315–332. [PubMed] [Google Scholar]
- 2.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2001;2(1):21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 3.Robertson KD. DNA methylation and human disease. Nat. Rev. Genet. 2005;6(8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 4.Latham KE. Epigenetic modification and imprinting of the mammalian genome during development. Curr. Top. Dev. Biol. 1999;43:1–49. doi: 10.1016/s0070-2153(08)60377-4. [DOI] [PubMed] [Google Scholar]
- 5.Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod. Toxicol. 2007;23(3):297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell Mol. Life Sci. 2004;61(19–20):2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PubMed] [Google Scholar]
- 7.Turek-Plewa J, Jagodziński PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol. Biol. Lett. 2005;10(4):631–647. [PubMed] [Google Scholar]
- 8.Bogdanović O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118(5):549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bestor TH, Ingram VM. Growth-dependent expression of multiple species of DNA methyltransferase in murine erythroleukemia cells. Proc. Natl. Acad. Sci. 1985;82(9):2674–2678. doi: 10.1073/pnas.82.9.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 1988;203(4):971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 11.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998;19(3):219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 12.Yoder JA, Bestor TH. A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum. Mol. Genet. 1998;7(2):279–284. doi: 10.1093/hmg/7.2.279. [DOI] [PubMed] [Google Scholar]
- 13.Bestor TH. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9(16):2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 14.Amato RJ. Inhibition of DNA methylation by antisense oligonucleotide MG98 as cancer therapy. Clin. Genitourin. Cancer. 2007;5(7):422–426. doi: 10.3816/CGC.2007.n.029. [DOI] [PubMed] [Google Scholar]
- 15.Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65(14):6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- 16.Yen RW, Vertino PM, Nelkin BD, Yu JJ, el-Deiry W, Cumaraswamy A, Lennon GG, Trask BJ, Celano P, Baylin SB. Isolation and characterization of the cDNA encoding human DNA methyltransferase. Nucleic Acids Res. 1992;20(9):2287–2291. doi: 10.1093/nar/20.9.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramchandani S, Bigey P, Szyf M. Genomic structure of the human DNA methyltransferase gene. Biol. Chem. 1998;379(4–5):535–540. doi: 10.1515/bchm.1998.379.4-5.535. [DOI] [PubMed] [Google Scholar]
- 18.Leonhardt H, Bestor TH. Structure, function and regulation of mammalian DNA methyltransferase. EXS. 1993;64:109–119. doi: 10.1007/978-3-0348-9118-9_5. [DOI] [PubMed] [Google Scholar]
- 19.Deng J, Szyf M. Multiple isoforms of DNA methyltransferase are encoded by the vertebrate cytosine DNA methyltransferase gene. J. Biol. Chem. 1998;273(36):22869–22872. doi: 10.1074/jbc.273.36.22869. [DOI] [PubMed] [Google Scholar]
- 20.Mertineit C, Yoder JA, Taketo T, Laird DW, Trasler JM, Bestor TH. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development. 1998;125(5):889–897. doi: 10.1242/dev.125.5.889. [DOI] [PubMed] [Google Scholar]
- 21.Aguirre-Arteta AM, Grunewald I, Cardoso MC, Leonhardt H. Expression of an alternative Dnmt1 isoform during muscle differentiation. Cell Growth Differ. 2000;11(10):551–559. [PubMed] [Google Scholar]
- 22.Hsu DW, Lin MJ, Lee TL, Wen SC, Chen X, Shen CK. Two major forms of DNA (cytosine-5) methyltransferase in human somatic tissues. Proc. Natl. Acad. Sci. 1999;96(17):9751–9756. doi: 10.1073/pnas.96.17.9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonfils C, Beaulieu N, Chan E, Cotton-Montpetit J, MacLeod AR. Characterization of the human DNA methyltransferase splice variant Dnmt1b. J. Biol. Chem. 2000;275(15):10754–10760. doi: 10.1074/jbc.275.15.10754. [DOI] [PubMed] [Google Scholar]
- 24.Lin MJ, Lee TL, Hsu DW, Shen CK. One-codon alternative splicing of the CpG MTase Dnmt1 transcript in mouse somatic cells. FEBS Lett. 2000;469(1):101–104. doi: 10.1016/s0014-5793(00)01254-0. [DOI] [PubMed] [Google Scholar]
- 25.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2000;3(4):274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 26.Kumar S, Cheng X, Klimasauskas S, Mi S, Posfai J, Roberts RJ, Wilson GG. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994;22(1):1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 1995;253(4):618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 28.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71(5):865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 29.Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277(5334):1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 30.Bestor TH, Verdine GL. DNA methyltransferases. Curr. Opin. Cell. Biol. 1994;6:3803–3809. doi: 10.1016/0955-0674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Oakeley EJ, Sun L, Jost JP. Multiple domains are involved in the targeting of the mouse DNA methyltransferase to the DNA replication foci. Nucleic Acids Res. 1998;26(4):1038–1045. doi: 10.1093/nar/26.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Callebaut I, Courvalin JC, Mornon JP. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999;446(1):189–1893. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- 33.Chuang LS, Ng HH, Chia JN, Li BF. Characterisation of independent DNA and multiple Zn-binding domains at the N terminus of human DNA-(cytosine-5) methyltransferase: modulating the property of a DNA-binding domain by contiguous Zn-binding motifs. J. Mol. Biol. 1996;257(5):935–948. doi: 10.1006/jmbi.1996.0213. [DOI] [PubMed] [Google Scholar]
- 34.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 2000;25(3):269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 35.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000;25(3):338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 36.Pradhan S, Kim GD. The retinoblastoma gene product interacts with maintenance human DNA (cytosine-5) methyltransferase and modulates its activity. EMBO J. 2002;21(4):779–788. doi: 10.1093/emboj/21.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tatematsu KI, Yamazaki T, Ishikawa F. MBD2-MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication foci in late S phase. Genes Cells. 2000;5(8):677–688. doi: 10.1046/j.1365-2443.2000.00359.x. [DOI] [PubMed] [Google Scholar]
- 38.Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J. Biol. Chem. 2003;278(7):4806–48012. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- 39.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 2000;24(1):88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 40.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31(9):2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, Shintani A, Kawasaki K, Minoshima S, Krohn K, Antonarakis SE, Shimizu N, Kudoh J, Peterson P. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65(3):293–298. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- 42.Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, Okumura K, Li E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236(1):87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- 43.Okano M, Takebayashi S, Okumura K, Li E. Assignment of cytosine-5 DNA methyltransferases Dnmt3a and Dnmt3b to mouse chromosome bands 12A2-A3 and 2H1 by in situ hybridization. Cytogenet. Cell Genet. 1999;86(3–4):333–334. doi: 10.1159/000015331. [DOI] [PubMed] [Google Scholar]
- 44.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27(11):2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weisenberger DJ, Velicescu M, Preciado-Lopez MA, Gonzales FA, Tsai YC, Liang G, Jones PA. Identification and characterization of alternatively spliced variants of DNA methyltransferase 3a in mammalian cells. Gene. 2002;298(1):91–99. doi: 10.1016/s0378-1119(02)00976-9. [DOI] [PubMed] [Google Scholar]
- 46.Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 2002;277(41):38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- 47.Weisenberger DJ, Velicescu M, Cheng JC, Gonzales FA, Liang G, Jones PA. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol. Cancer Res. 2004;2(1):62–72. [PubMed] [Google Scholar]
- 48.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 49.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449(7159):248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng X. Blumenthal, R.M. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16(3):341–350. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burgers WA, Fuks F, Kouzarides T. DNA methyltransferases get connected to chromatin. Trends Genet. 2002;18(6):275–277. doi: 10.1016/S0168-9525(02)02667-7. [DOI] [PubMed] [Google Scholar]
- 52.Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20(10):2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 2001;276(34):32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 54.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13(14):1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 55.Brenner C, Deplus R, Didelot C, Loriot A, Viré E, De Smet C, Gutierrez A, Danovi D, Bernard D, Boon T, Pelicci PG, Amati B, Kouzarides T, de Launoit Y, Di Croce L, Fuks F. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005;24(2):336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiu C, Sawada K, Zhang X, Cheng X. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat. Struct. Biol. 2002;9(3):217–224. doi: 10.1038/nsb759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stec I, Nagl SB, van Ommen GJ, den Dunnen JT. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS. Lett. 2000;473(1):1–5. doi: 10.1016/s0014-5793(00)01449-6. [DOI] [PubMed] [Google Scholar]
- 58.Shirohzu H, Kubota T, Kumazawa A, Sado T, Chijiwa T, Inagaki K, Suetake I, Tajima S, Wakui K, Miki Y, Hayashi M, Fukushima Y, Sasaki H. Three novel DNMT3B mutations in Japanese patients with ICF syndrome. Am. J. Med. Genet. 2002;112(1):31–37. doi: 10.1002/ajmg.10658. [DOI] [PubMed] [Google Scholar]
- 59.Chen T, Tsujimoto N, Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell. Biol. 2004;24(20):9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ge YZ, Pu MT, Gowher H, Wu HP, Ding JP, Jeltsch A, Xu GL. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 2004;279(24):25447–25454. doi: 10.1074/jbc.M312296200. [DOI] [PubMed] [Google Scholar]
- 61.Lukasik SM, Cierpicki T, Borloz M, Grembecka J, Everett A, Bushweller JH. High resolution structure of the HDGF PWWP domain: a potential DNA binding domain. Protein Sci. 2006;15(2):314–323. doi: 10.1110/ps.051751706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li B, Zhou J, Liu P, Hu J, Jin H, Shimono Y, Takahashi M, Xu G. Polycomb protein Cbx4 promotes SUMO modification of de novo DNA methyltransferase Dnmt3a. Biochem. J. 2007;405(2):369–378. doi: 10.1042/BJ20061873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang ES, Park CW, Chung JH. Dnmt3b, de novo DNA methyltransferase, interacts with SUMO-1 and Ubc9 through its N-terminal region and is subject to modification by SUMO-1. Biochem. Biophys. Res. Commun. 2001;289(4):862–868. doi: 10.1006/bbrc.2001.6057. [DOI] [PubMed] [Google Scholar]
- 64.Ling Y, Sankpal UT, Robertson AK, McNally JG, Karpova T, Robertson KD. Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription. Nucleic Acids Res. 2004;32(2):598–610. doi: 10.1093/nar/gkh195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aapola U, Lyle R, Krohn K, Antonarakis SE, Peterson P. Isolation and initial characterization of the mouse Dnmt3l gene. Cytogenet. Cell. Genet. 2001;92(1–2):122–126. doi: 10.1159/000056881. [DOI] [PubMed] [Google Scholar]
- 66.El-Maarri O, Kareta MS, Mikeska T, Becker T, Diaz-Lacava A, Junen J, Nüsgen N, Behne F, Wienker T, Waha A, Oldenburg J, Chédin F. A systematic search for DNA methyltransferase polymorphisms reveals a rare DNMT3L variant associated with subtelomeric hypomethylation. Hum. Mol. Genet. 2009;18(10):1755–1768. doi: 10.1093/hmg/ddp088. [DOI] [PubMed] [Google Scholar]
- 67.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294(5551):2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 68.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448(7154):714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. 2007;99(26):16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suetake I, Shinozaki F, Miyagawa J, Takeshima H. Tajima, S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 2004;279(26):27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 71.Chen ZX, Mann JR, Hsieh CL, Riggs AD. Chédin, F. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J. Cell Biochem. 2005;95(5):902–917. doi: 10.1002/jcb.20447. [DOI] [PubMed] [Google Scholar]
- 72.Aapola U, Liiv I, Peterson P. Imprinting regulator DNMT3L is a transcriptional repressor associated with histone deacetylase activity. Nucleic Acids Res. 2002;30(16):3602–3608. doi: 10.1093/nar/gkf474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deplus R, Brenner C, Burgers WA, Putmans P, Kouzarides T, de Launoit Y, Fuks F. Dnmt3L is a transcriptional repressor that recruits histone deacetylase. Nucleic Acids Res. 2002;30(17):3831–3838. doi: 10.1093/nar/gkf509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hermann A, Schmitt S, Jeltsch A. The human Dnmt2 has residual DNA-(cytosine-C5) methyltransferase activity. J. Biol. Chem. 2003;278(34):31717–31721. doi: 10.1074/jbc.M305448200. [DOI] [PubMed] [Google Scholar]
- 75.Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebra fish. Genes Dev. 2007;21(3):261–266. doi: 10.1101/gad.1472907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311(5759):395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 77.Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G, Jeltsch A. Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism. RNA. 2008;14(8):1663–1670. doi: 10.1261/rna.970408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ayala T, Rama ST, Ricarda G, Mark H, Serge A. A New Nuclear Function of the Entamoeba histolytica Glycolytic Enzyme Enolase: The Metabolic Regulation of Cytosine-5 Methyltransferase 2 (Dnmt2) Activity. PLoS. Pathog. 2010;6(2):e1000775. doi: 10.1371/journal.ppat.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kunert N, Marhold J, Stanke J, Stach D, Lyko F. A Dnmt2-like protein mediates DNA methylation in Drosophila. Development. 2003;130(21):5083–5090. doi: 10.1242/dev.00716. [DOI] [PubMed] [Google Scholar]
- 80.Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007;21(3):261–266. doi: 10.1101/gad.1472907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arturo G, Ralf JS. Evolution of dnmt-2 and mbd-2-like genes in the free-living nematodes Pristionchus pacificus, Caenorhabditis elegans and Caenorhabditis briggsae. Nucleic Acids Res. 2004;32(21):6388–6396. doi: 10.1093/nar/gkh982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schaefer M, Steringer JP, Lyko F. The Drosophila Cytosine-5 Methyltransferase Dnmt2 Is Associated with the Nuclear Matrix and Can Access DNA during Mitosis. PLoS ONE. 2008;3(1):e1414. doi: 10.1371/journal.pone.0001414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Phalke S, Nickel O, Walluscheck D, Hortig F, Onorati MC, Reuter G. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat. Genet. 2009;41(6):696–702. doi: 10.1038/ng.360. [DOI] [PubMed] [Google Scholar]
- 84.Okano M, Xie S, Li E. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 1998;26(11):2536–2540. doi: 10.1093/nar/26.11.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dong A, Yoder JA, Zhang X, Zhou L, Bestor TH, Cheng X. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 2001;29(2):439–448. doi: 10.1093/nar/29.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee YF, Tawfik DS, Griffiths AD. Investigating the target recognition of DNA cytosine-5 methyltransferase HhaI by library selection using in vitro compartmentalisation. Nucleic. Acids. Res. 2002;30(22):4937–4944. doi: 10.1093/nar/gkf617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schaefer M, Lyko F. Solving the Dnmt2 enigma. Chromosoma. 2010;119:35–40. doi: 10.1007/s00412-009-0240-6. [DOI] [PubMed] [Google Scholar]
- 88.Reither S, Li F, Gowher H, Jeltsch A. Catalytic mechanism of DNA-(cytosine-C5)-methyltransferases revisited: covalent intermediate formation is not essential for methyl group transfer by the murine Dnmt3a enzyme. J. Mol. Biol. 2003;329(4):675–684. doi: 10.1016/s0022-2836(03)00509-6. [DOI] [PubMed] [Google Scholar]
- 89.Klimasauskas S, Roberts RJ. M.HhaI binds tightly to substrates containing mismatches at the target base. Nucleic Acids Res. 1995;23(8):1388–1395. doi: 10.1093/nar/23.8.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jeltsch A. The cytosine N4-methyltransferase M.PvuII also modifies adenine residues. Biol. Chem. 2001;382(4):707–710. doi: 10.1515/BC.2001.084. [DOI] [PubMed] [Google Scholar]
- 91.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76(2):357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 92.Reinisch KM, Chen L, Verdine GL, Lipscomb WN. The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell. 1995;82(1):143–153. doi: 10.1016/0092-8674(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 93.O'Gara M, Horton JR, Roberts RJ, Cheng X. Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base. Nat. Struct. Biol. 1998;5(10):872–877. doi: 10.1038/2312. [DOI] [PubMed] [Google Scholar]
- 94.Roberts RJ, Cheng X. Base flipping. Annu. Rev. Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 95.Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barrett TE, Scharer OD, Savva R, Brown T, Jiricny J, Verdine GL, Pearl LH. Crystal structure of a thwarted mismatch glycosylase DNA repair complex. EMBO. J. 1999;18:6599–6609. doi: 10.1093/emboj/18.23.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng X, Blumenthal RM. Finding a basis for flipping bases. Structure. 1996;4(6):639–645. doi: 10.1016/s0969-2126(96)00068-8. [DOI] [PubMed] [Google Scholar]
- 98.Mi S, Alonso D, Roberts RJ. Functional analysis of Gln-237 mutants of HhaI methyltransferase. Nucleic Acids Res. 1995;23(4):620–627. doi: 10.1093/nar/23.4.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Estabrook RA, Lipson R, Hopkins B, Reich N. The coupling of tight DNA binding and base flipping: identification of a conserved structural motif in base flipping enzymes. J. Biol. Chem. 2004;279(30):31419–31428. doi: 10.1074/jbc.M402950200. [DOI] [PubMed] [Google Scholar]
- 100.Shieh FK, Youngblood B, Reich NO. The role of Arg165 towards base flipping, base stabilization and catalysis in M.HhaI. J. Mol. Biol. 2006;362(3):516–527. doi: 10.1016/j.jmb.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 101.Vilkaitis G, Dong A, Weinhold E, Cheng X, Klimasauskas S. Functional roles of the conserved threonine 250 in the target recognition domain of HhaI DNA methyltransferase. J. Biol. Chem. 2000;275(49):38722–38730. doi: 10.1074/jbc.m005278200. [DOI] [PubMed] [Google Scholar]
- 102.Estabrook RA, Reich N. Observing an induced-fit mechanism during sequence-specific DNA methylation. J. Biol. Chem. 2006;281(48):37205–37214. doi: 10.1074/jbc.M607538200. [DOI] [PubMed] [Google Scholar]
- 103.Svedruzić ZM. Mammalian cytosine DNA methyltransferase Dnmt1: enzymatic mechanism, novel mechanism-based inhibitors, and RNA-directed DNA methylation. Curr. Med. Chem. 2008;15(1):92–106. doi: 10.2174/092986708783330700. [DOI] [PubMed] [Google Scholar]
- 104.Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317(5845):1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 105.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450(7171):908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 106.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455(7214):818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 107.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455(7214):822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 108.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455(7214):826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Roth M, Helm-Kruse S, Friedrich T, Jeltsch A. Functional roles of conserved amino acid residues in DNA methyltransferases investigated by site-directed mutagenesis of the EcoRV adenine-N6-methyltransferase. J. Biol. Chem. 1998;273(28):17333–17342. doi: 10.1074/jbc.273.28.17333. [DOI] [PubMed] [Google Scholar]
- 110.Tran PH, Korszun ZR, Cerritelli S, Springhorn SS, Lacks SA. Crystal structure of the DpnM DNA adenine methyltransferase from the DpnII restriction system of streptococcus pneumoniae bound to S-adenosylmethionine. Structure. 1998;6(12):1563–1575. doi: 10.1016/s0969-2126(98)00154-3. [DOI] [PubMed] [Google Scholar]
- 111.O'Gara M, Zhang X, Roberts RJ, Cheng X. Structure of a binary complex of HhaI methyltransferase with S-adenosyl-L-methionine formed in the presence of a short non-specific DNA oligonucleotide. J. Mol. Biol. 1999;287(2):201–209. doi: 10.1006/jmbi.1999.2608. [DOI] [PubMed] [Google Scholar]
- 112.Schluckebier G, Kozak M, Bleimling N, Weinhold E, Saenger W. Differential binding of S-adenosylmethionine S-adenosylhomocysteine and Sinefungin to the adenine-specific DNA methyltransferase M.TaqI. J. Mol. Biol. 1997;265(1):56–67. doi: 10.1006/jmbi.1996.0711. [DOI] [PubMed] [Google Scholar]
- 113.Chen L, MacMillan AM, Chang W, Ezaz-Nikpay K, Lane WS, Verdine GL. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991;30(46):11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- 114.Wyszynski MW, Gabbara S, Kubareva EA, Romanova EA, Oretskaya TS, Gromova ES, Shabarova ZA, Bhagwat AS. The cysteine conserved among DNA cytosine methylases is required for methyl transfer, but not for specific DNA binding. Nucleic. Acids. Res. 1993;21(2):295–301. doi: 10.1093/nar/21.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.O'Gara M, Klimasauskas S, Roberts RJ, Cheng X. Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaL methyltransferase-DNA-AdoHcy complexes. J. Mol. Biol. 1996;261(5):634–645. doi: 10.1006/jmbi.1996.0489. [DOI] [PubMed] [Google Scholar]
- 116.Zhang X, Bruice TC. The mechanism of M.HhaI DNA C5 cytosine methyltransferase enzyme: a quantum mechanics/molecular mechanics approach. Proc. Natl. Acad. Sci. 2006;103(16):6148–6153. doi: 10.1073/pnas.0601587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer. 2008;123(1):8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 118.Youngblood B, Shieh FK, Buller F, Bullock T, Reich NO. S-adenosyl-L-methionine-dependent methyl transfer: observable precatalytic intermediates during DNA cytosine methylation. Biochemistry. 2007;46(30):8766–8775. doi: 10.1021/bi7005948. [DOI] [PubMed] [Google Scholar]
- 119.Shieh FK, Youngblood B, Reich NO. The role of Arg165 towards base flipping, base stabilization and catalysis in M.HhaI. J. Mol. Biol. 2006;362(3):516–527. doi: 10.1016/j.jmb.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 120.Gabbara S, Sheluho D, Bhagwat AS. Cytosine methyltransferase from Escherichia coli in which active site cysteine is replaced with serine is partially active. Biochemistry. 1995;34(27):8914–8923. doi: 10.1021/bi00027a044. [DOI] [PubMed] [Google Scholar]
- 121.Lau EY, Bruice TC. Active site dynamics of the HhaI methyltransferase: insights from computer simulation. J. Mol. Biol. 1999;293(1):9–18. doi: 10.1006/jmbi.1999.3120. [DOI] [PubMed] [Google Scholar]
- 122.Shen JC, Rideout WM, Jones PA. High frequency mutagenesis by a DNA methyltransferase. Cell. 1992;71(7):1073–1080. doi: 10.1016/s0092-8674(05)80057-1. [DOI] [PubMed] [Google Scholar]
- 123.Zingg JM, Shen JC, Jones PA. Enzyme-mediated cytosine deamination by the bacterial methyltransferase M.MspI. Biochem. J. 1998;332:223–230. doi: 10.1042/bj3320223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sharath AN, Weinhold E, Bhagwat AS. Reviving a dead enzyme: cytosine deaminations promoted by an inactive DNA methyltransferase and an S-adenosylmethionine analogue. Biochemistry. 2000;39(47):14611–14616. doi: 10.1021/bi001610e. [DOI] [PubMed] [Google Scholar]
- 125.Svedruzić ZM, Reich NO. The mechanism of target base attack in DNA cytosine carbon 5 methylation. Biochemistry. 2004;43(36):11460–11473. doi: 10.1021/bi0496743. [DOI] [PubMed] [Google Scholar]
- 126.Svedruzić ZM, Reich NO. DNA cytosine C5 methyltransferase Dnmt1: catalysis-dependent release of allosteric inhibition. Biochemistry. 2005;44(27):9472–9485. doi: 10.1021/bi050295z. [DOI] [PubMed] [Google Scholar]
- 127.Reither S, Li F, Gowher H, Jeltsch A. Catalytic mechanism of DNA-(cytosine-C5)-methyltransferases revisited: covalent intermediate formation is not essential for methyl group transfer by the murine Dnmt3a enzyme. J. Mol. Biol. 2003;329(4):675–684. doi: 10.1016/s0022-2836(03)00509-6. [DOI] [PubMed] [Google Scholar]
- 128.Gruenbaum Y, Cedar H, Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982;295(5850):620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- 129.Lin X, Asgari K, Putzi MJ, Gage WR, Yu X, Cornblatt BS, Kumar A, Piantadosi S, DeWeese TL, De Marzo AM, Nelson WG. Reversal of GSTP1 CpG island hypermethylation and reactivation of pi-class glutathione S-transferase (GSTP1) expression in human prostate cancer cells by treatment with procainamide. Cancer Res. 2001;61(24):8611–8616. [PubMed] [Google Scholar]
- 130.Lee BH, Yegnasubramanian S, Lin X, Nelson WG. Procainamide is a specific inhibitor of DNA methyltransferase 1. J. Biol. Chem. 2005;280(49):40749–40756. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bolden A, Ward C, Siedlecki JA, Weissbach A. DNA methylation. Inhibition of de novo and maintenance methylation in vitro by RNA and synthetic polynucleotides. J. Biol. Chem. 1984;259(20):12437–12443. [PubMed] [Google Scholar]
- 132.Flynn J, Glickman JF, Reich NO. Murine DNA cytosine-C5 methyltransferase: pre-steady- and steady-state kinetic analysis with regulatory DNA sequences. Biochemistry. 1996;35(23):7308–7315. doi: 10.1021/bi9600512. [DOI] [PubMed] [Google Scholar]
- 133.Flynn J, Azzam R, Reich N. DNA binding discrimination of the murine DNA cytosine-C5 methyltransferase. J. Mol. Biol. 1998;279(1):101–116. doi: 10.1006/jmbi.1998.1761. [DOI] [PubMed] [Google Scholar]
- 134.Achour M, Jacq X, Rondé P, Alhosin M, Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A, Hughes AD, Schini-Kerth VB, Bronner C. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene. 2008;27:2187–2197. doi: 10.1038/sj.onc.1210855. [DOI] [PubMed] [Google Scholar]
- 135.Unoki M, Brunet J, Mousli M. Drug discovery targeting epigenetic codes: the great potential of UHRF1, which links DNA methylation and histone modifications, as a drug target in cancers and toxoplasmosis. Biochem. Pharmacol. 2009;78(10):1279–1288. doi: 10.1016/j.bcp.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 136.Flynn J, Fang JY, Mikovits JA, Reich NO. A potent cell-active allosteric inhibitor of murine DNA cytosine C5 methyltransferase. J. Biol. Chem. 2003;278(10):8238–8243. doi: 10.1074/jbc.M209839200. [DOI] [PubMed] [Google Scholar]
- 137.Bacolla A, Pradhan S, Roberts RJ, Wells RD. Recombinant human DNA (cytosine-5) methyltransferase. II. Steady-state kinetics reveal allosteric activation by methylated dna. J. Biol. Chem. 1999;274(46):33011–33019. doi: 10.1074/jbc.274.46.33011. [DOI] [PubMed] [Google Scholar]
- 138.Goyal R, Reinhardt R, Jeltsch A. Accuracy of DNA methylation pattern preservation by the Dnmt1 methyltransferase. Nucleic. Acids. Res. 2006;34(4):1182–1188. doi: 10.1093/nar/gkl002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128(4):763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 140.Biniszkiewicz D, Gribnau J, Ramsahoye B, Gaudet F, Eggan K, Humpherys D, Mastrangelo MA, Jun Z, Walter J, Jaenisch R. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol. Cell. Biol. 2002;22(7):2124–2135. doi: 10.1128/MCB.22.7.2124-2135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Datta J, Ghoshal K, Sharma SM, Tajima S, Jacob ST. Biochemical fractionation reveals association of DNA methyltransferase (Dnmt) 3b with Dnmt1 and that of Dnmt 3a with a histone H3 methyltransferase and Hdac1. J. Cell. Biochem. 2003;88(5):855–864. doi: 10.1002/jcb.10457. [DOI] [PubMed] [Google Scholar]
- 142.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294(5551):2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 143.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429(6994):900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 144.Gowher H, Jeltsch A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J. Biol. Chem. 2002;277(23):20409–20414. doi: 10.1074/jbc.M202148200. [DOI] [PubMed] [Google Scholar]
- 145.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. 2000;97(10):5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E. De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation. Gene. 2002;289(1–2):41–48. doi: 10.1016/s0378-1119(02)00469-9. [DOI] [PubMed] [Google Scholar]
- 147.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 148.Lin IG, Han L, Taghva A, O'Brien LE, Hsieh CL. Murine de novo methyltransferase Dnmt3a demonstrates strand asymmetry and site preference in the methylation of DNA in vitro. Mol. Cell. Biol. 2002;22(3):704–723. doi: 10.1128/MCB.22.3.704-723.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Handa V, Jeltsch A. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J. Mol. Biol. 2005;348(5):1103–1012. doi: 10.1016/j.jmb.2005.02.044. [DOI] [PubMed] [Google Scholar]
- 150.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004;279(46):48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 151.Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 2002;22(2):480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 2003;23(16):5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]