Abstract
Staphylococcus aureus (S. aureus) is a Gram-positive bacterial pathogen that has emerged as a major public health threat. Here we report that the cell wall of S. aureus can be covalently re-engineered to contain non-native small molecules. This process makes use of endogenous levels of the bacterial enzyme sortase A (SrtA), which ordinarily functions to incorporate proteins into the bacterial cell wall. Thus, incubation of wild-type bacteria with rationally designed SrtA substrates results in covalent incorporation of functional molecular handles (fluorescein, biotin, and azide) into cell wall peptidoglycan. These conclusions are supported by data obtained through a variety of experimental techniques (epifluorescence and electron microscopy, biochemical extraction, and mass spectrometry), and azide incorporation was exploited as a chemical handle to perform an azide-alkyne cycloaddition reaction on the bacterial cell surface. This report represents the first example of cell wall engineering of S. aureus or any other pathogenic Gram-positive bacteria, and has the potential for widespread utility.
Staphylococcus aureus (S. aureus) is a Gram-positive bacterial pathogen that has become a major public health threat. Most hospital isolates of S. aureus are resistant to many if not all available treatments,(1) and recent reports suggest that more Americans die every year as a result of S. aureus infection than due to HIV/AIDS, Parkinson’s Disease, or emphysema.(2, 3) The virulence of this organism is mediated in part by proteins in its cell wall, which enable it to interact with animal cells and tissues, and to evade the human immune system.(4, 5) Flexible strategies for modulating the molecular composition of the S. aureus cell wall would therefore be highly desirable, and could enable both fundamental and therapeutic applications.
Here we demonstrate for the first time that the cell surface of wild-type S. aureus can be re-engineered biosynthetically to incorporate non-native small molecules. Exposure of wild-type bacteria to rationally designed, low molecular weight substrates for the enzyme sortase A (SrtA)(6) leads to covalent incorporation of functional small molecules (fluorescein, biotin, or azide) into the S. aureus cell wall (Figure 1). Diverse experimental techniques are employed to support these conclusions including epifluorescence and electron microscopy, flow cytometry, mass spectrometry, and biochemical cell wall extraction. Furthermore, azide incorporation is exploited as a chemical handle to perform an azide-alkyne cycloaddition reaction on the bacterial cell surface. This report represents the first example of cell wall engineering of S. aureus or any other pathogenic Gram-positive bacteria, and represents a significant advance with the potential to enable wide-ranging applications.
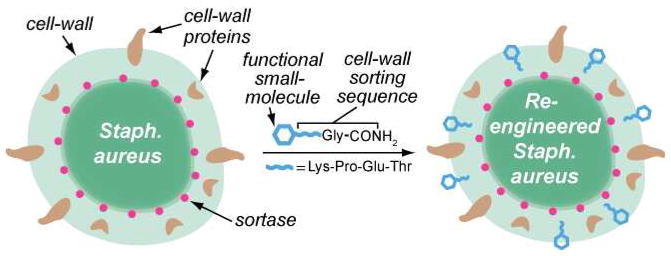
Figure 1.
Schematic depiction of the reported method for sortase-mediated tagging of the S. aureus cell wall. Bacteria are fed peptides containing small molecule tags. Upon penetrating the cell wall, these compounds are recognized by the periplasmic enzyme Sortase A (red dots), which cleaves between the C-terminal Thr and Gly residues and covalently attaches the remaining N-terminal portion to lipid II in the forming cell wall, as shown.
RESULTS AND DISCUSSION
Substrate Design and Incorporation into S. aureus
Our design of synthetic SrtA substrates was based upon the enzyme’s mechanism of action, which involves recognition of a conserved pentapeptide motif, typically LPETG in S. aureus, near the C-terminus of various secreted proteins.(6) Upon substrate recognition, the enzyme cleaves the threonine-glycine bond, forming an acyl-enzyme intermediate. This complex is attacked by the pentaglycine motif of lipid II, a peptidoglycan precursor, and the resulting adduct is covalently attached to the bacterial cell wall.(6) We speculated that since natural SrtA substrates are structurally diverse, the native enzyme present in the bacterial periplasm might be able to integrate LPXTG-tagged small molecules into the cell walls of living bacteria in an identical manner to protein substrates. Indeed, previous in vitro studies have demonstrated that SrtA is capable of catalyzing transpeptidation reactions with non-natural LPXTG-modified substrates.(7–11) Also, purified, recombinant SrtA has been shown to catalyze transpeptidation reactions on the surfaces of eukaryotic cells that have been genetically manipulated to express LPXTG-modified surface proteins.(12–14) To our knowledge, however, no one has metabolically incorporated small molecule functionality into the S. aureus cell wall.
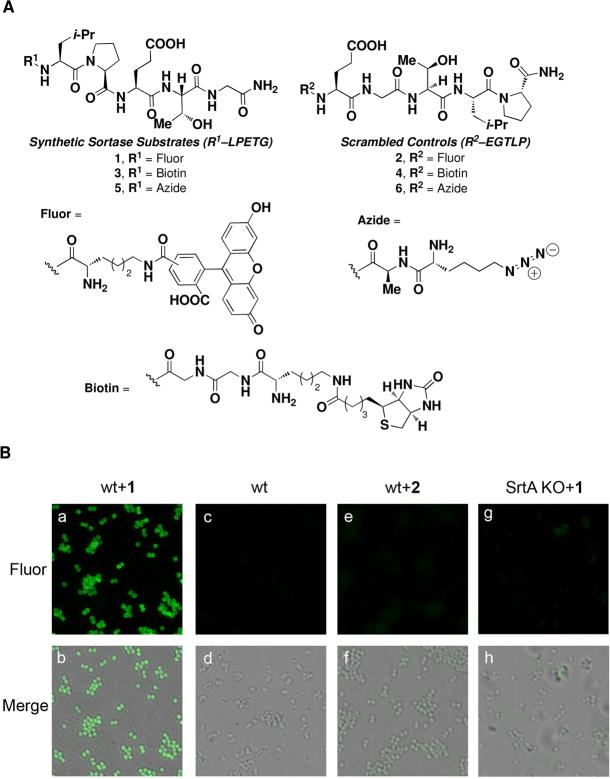
As a convenient means of assessing integration of non-native small molecules into wild-type S. aureus, we designed compounds 1–6, which were prepared through Fmoc solid-phase peptide synthesis (Figure 2A). Notably, since the presence of a carboxylic acid at the LPETG C-terminus has been suggested to obviate SrtA activity,(15) we chose to incorporate an amide at this position. Incubating wild-type S. aureus cultures with compound 1, which contains a fluorescein-derivatized lysine residue at the N-terminus of the LPETG pentapeptide, led to highly fluorescent bacteria as measured by epifluorescence microscopy (Figure 2B, panels a and b). Neither control experiments with vehicle alone (Figure 2B, panels c and d), nor with 2, a fluorescein-containing peptide isomeric to 1 containing a scrambled sorting sequence (Figure 2B, panels e and f), demonstrated detectable cellular fluorescence. This latter control argues against non-specific protein binding as the cause of bacterial fluorescence, and is consistent with the proposed mechanism for small molecule integration. Furthermore, an isogenic S. aureus strain lacking the SrtA gene(16) exhibited a greatly reduced ability to incorporate 1 into the cell wall, consistent with a critical involvement of SrtA in this process (Figure 2B, panels g and h).
Figure 2.
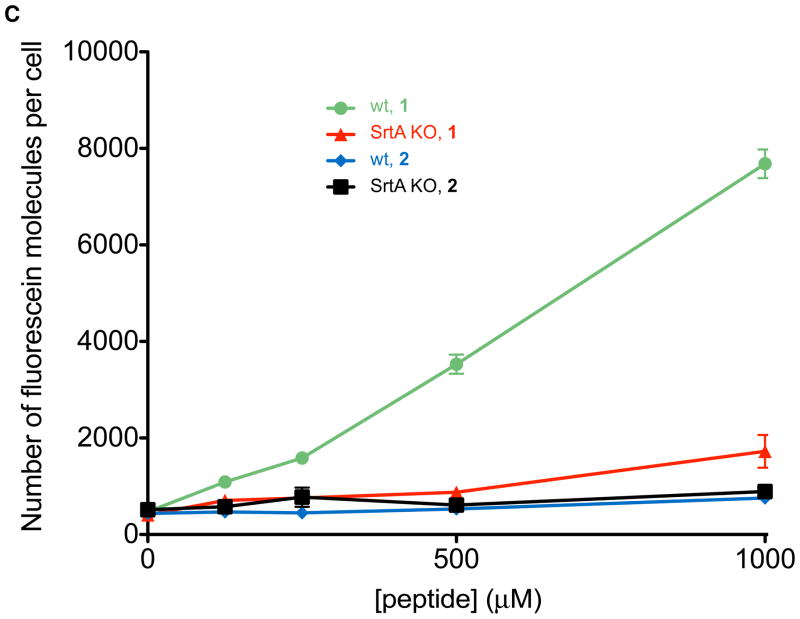
Observation and measurement of incorporated fluorescence into wild-type or SrtA-knockout S. aureus following small molecule treatment. (A) Synthetic small molecules employed in these studies. (B) Epifluorescence microscopy. Abbreviations: “wt” indicates wild-type S. aureus, “SrtA KO” indicates a Sortase A knockout strain of S. aureus,(16) “Fluor” indicates epifluorescence images, and “Merge” corresponds to fluorescent images superimposed atop brightfield images. All micrographs were acquired under identical exposure settings, and processed in an identical fashion. The depicted trends were reproduced on at least three separate occasions. (C) Quantitation of fluorescein-incorporation via flow cytometry.
We then quantified the levels of small molecule incorporation as a function of synthetic small molecule concentration using flow cytometry (Figure 2C). Detectable levels of fluorescein (approximately 1000 molecules/cell) were observed at concentrations of 1 as low as 125 μM, and the levels of incorporated small molecules were observed to be roughly proportional to the concentration of 1 in treatment media within a range up to 6500 molecules/cell. Notably, the native S. aureus collagen adhesin protein is surface exposed at a level of approximately 5000 molecules per cell in wild-type organisms.(17) Thus, the amount of incorporated small molecule observed in our studies is comparable to expression levels of this native protein. Importantly, treatment of bacteria with compound 1 neither affects cell viability as measured by re-plating colonies after peptide treatment at concentrations up to 1 mM (Supplementary Figure 1), nor abrogates endogenous levels of protein A, a cell surface protein and SrtA substrate (Supplementary Figure 2). These results indicate the potential of this technology for the non-disruptive labeling of living bacteria.
Subcellular Localization of SrtA Substrates
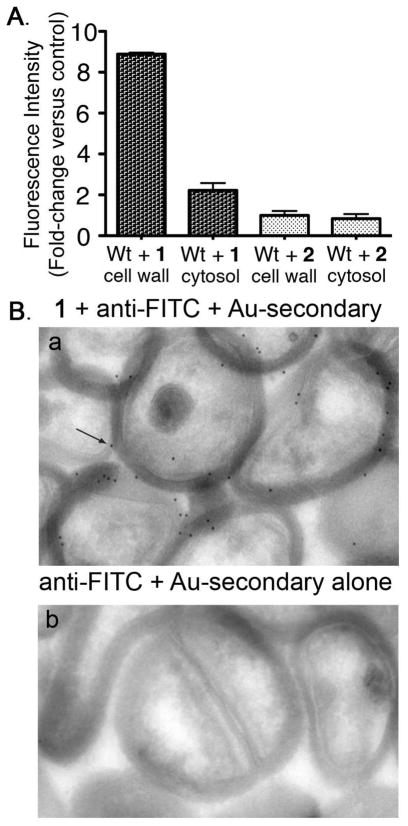
To evaluate further the subcellular localization of incorporated small molecules, cell walls were extracted from wild-type S. aureus that had been pre-treated with either 1 or 2 (Figure 3A). Cell wall extracts from bacteria incubated with 1 exhibited approximately 8-fold greater fluorescence versus those isolated from organisms treated with scrambled peptide 2. Further, a 3.5-fold increase in the fluorescence of cell wall versus cytosolic fractions following treatment with 1 was observed. Interestingly, 1-treated bacteria exhibited a small increase in fluorescence compared to the control, indicating that fluorescein-LPETG constructs – but not scrambled sequences – partition into cytosolic extracts. This may result from contamination of cytosolic extracts with membrane vesicles containing peptide bound to SrtA but not yet incorporated into cell wall, interactions with other membrane-associated enzymes (such as S. aureus LPXTGase),(18, 19) or recycling of peptidoglycan components into the cytosol for re-use.(20)
Figure 3.
SrtA-targeted small molecules are localized to the cell wall. (A) Fractionation of wild-type S. aureus cells into cell wall and cytosolic extracts following treatment with compounds 1 or 2. Data represent the average of triplicate experiments ± SD. (B) Immunocryoelectron microscopy studies of a protein A-knockout S. aureus strain in the Newman genetic background. Cells were incubated in the presence (panel a) and absence (panel b) of 1. Arrow points to an example of black dots, which correspond to gold-labeled antibody binding. Abbreviations: anti-FITC = rabbit anti-fluorescein antibody; Au-secondary = gold-labeled anti-rabbit secondary antibody.
Localization of 1 to the peptidoglycan layer was confirmed via immunocryoelectron microscopy (Figure 3B, panels a and b). Here a S. aureus mutant deficient in cell wall protein A(21) was incubated with peptide 1, then subjected to thin sectioning and immunostaining with an anti-fluorescein antibody followed by a gold-conjugated secondary antibody. Use of a protein A-knockout was necessary here to prevent background labeling of cell wall through interactions between protein A and the antibody’s Fc region. As shown, greater than 90% of gold labeling was observed in the vicinity of the cell wall. A small amount of apparent intracellular labeling was also detected, perhaps due to antibody background or to the presence of overlapping cells in thin sections; some degree of intracellular uptake or recycling of compound 1 cannot be ruled out.
Measuring Covalency of SrtA-targeted small molecules
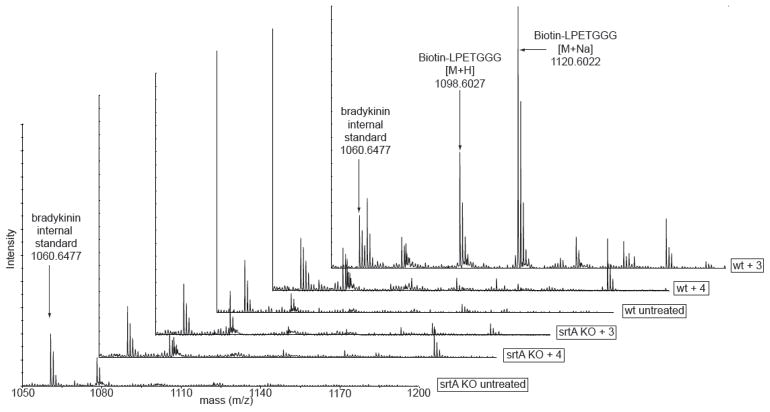
To demonstrate unambiguously that small molecules were covalently incorporated into the cell wall, we utilized a MALDI-TOF MS-based strategy.(22) These experiments provide direct evidence for covalent attachment of synthetic small molecules into the Gram-positive bacterial cell wall. S. aureus cultures treated with and without constructs 3 and 4 were first digested with the enzymes mutanolysin and lysostaphin to degrade the cell wall and release cell wall associated proteins. Digests were then subjected to streptavidin affinity chromatography followed by mass spectrometric analysis. Because lysostaphin is a Gly-Gly endopeptidase and S. aureus cell wall proteins are tethered to the peptidoglycan through a pentaglycine cross-link, we anticipated that processing of 3-treated bacteria in our mass spectrometry protocol would provide a biotin-LPET fragment attached to multiple glycine residues (between 1 and 4).
Indeed, extracts from cells treated with 3 and analyzed through the above sequence exhibited strong peak enrichments at 1098.60 and 1120.60 a.m.u., which were absent from all control conditions (Figure 4). These masses correspond to [M+H] and [M+Na] ions for the biotin-LPETGGG sequence, whose identity we were able to confirm via MALDI MS/MS sequencing (Supplementary Figure 3). Masses corresponding to biotin-LPETGG and biotin-LPETGGGG were not well resolved in these spectra, which is consistent with observations from previous mass spectrometry studies of cell wall proteins released from S. aureus following lysostaphin digestion.(23) Taken together, these data provide strong evidence supporting covalent attachment of our synthetic materials into the S. aureus cell wall through a mechanism involving SrtA.
Figure 4.
MALDI-TOF mass spectra of streptavidin affinity-purified S. aureus cell wall extracts after treatment of both wild-type and SrtA-knockout strains with and without biotin-peptide hybrids 3 and 4.
A Bioorthogonal [3+2] Cycloaddition on the S. aureus Surface
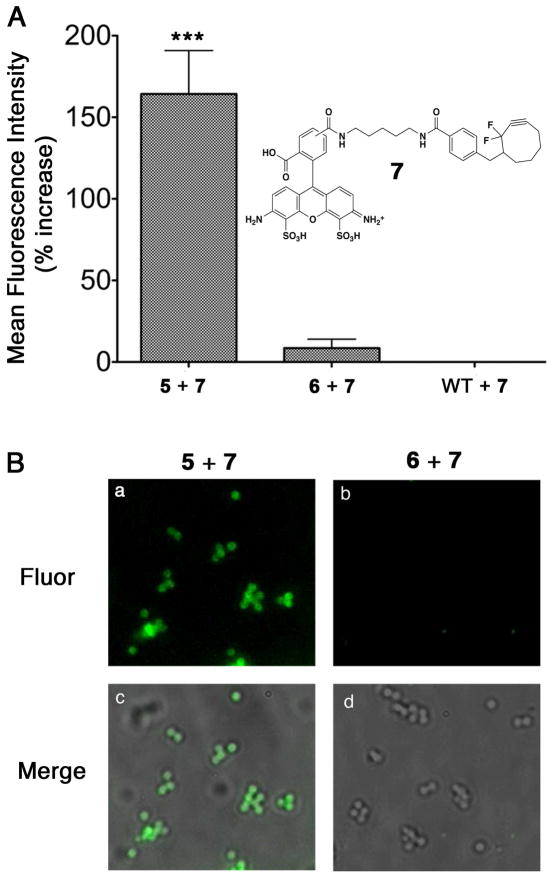
Bioorthogonal chemical reactions have proven useful in the study of a wide array of biological processes.(24, 25) Only a few studies, however, have employed bioorthogonal reactions in bacteria.(26–32) Furthermore, none of these to our knowledge have ever been executed in S. aureus. Accordingly, we synthesized molecules 5 and 6, which both contain Nε-azido-L-lysine residues at their N-termini (Figure 2A). The azide functionality in 5 and 6 was introduced because of its ability to undergo a [3+2] cycloaddition (“click”) reaction with alkyne-containing probes.(33, 34) To this end, we prepared Alexa Fluor 488-conjugated cyclooctyne probe 7. Structural analogues of compound 7 have been shown to participate efficiently in bioorthogonal copper-free click chemistry.(24, 35–38) Wild-type S. aureus cultured in the presence of putative SrtA substrate 5, then treated with compound 7, demonstrated a marked increase in fluorescence versus both background and scrambled control 6, as determined by flow cytometry and epifluorescence microscopy (Figure 5). These results demonstrate the ability of non-natural SrtA substrates to decorate the staphylococcal cell wall with reactive groups, and represent the first bioorthogonal chemical reaction performed on the surface of S. aureus.
Figure 5.
Conjugation of a fluorescent probe to S. aureus via copper-free click chemistry. (A) Flow Cytometry-based analysis of conjugation of 7 to wild-type S. aureus labeled with 5 or 6 and reported as percent increase over control (WT in the absence of compounds 5 or 6). Data is presented as the mean of three replicate experiments ± standard deviation. Data in the left-hand column was found to exhibit a highly statistically significant increase in fluorescence (***, P < 0.0001) compared to that in the middle or right-hand columns following a one-way analysis of variance (ANOVA) with Tukey’s multiple comparison post-hoc test. (B) Epifluorescence microscopy. “Fluor” indicates epifluorescence images, and “Merge” corresponds to fluorescent images superimposed atop brightfield images. All micrographs were acquired under identical exposure settings, and processed in an identical fashion. The depicted trends were reproduced on at least two separate occasions.
Conclusions
Strategies for re-engineering cell-surfaces with small molecules have emerged as powerful tools to perturb and study biological processes both in vitro and in vivo.(39, 40) Despite this, relatively few such approaches involving bacteria have appeared.(26–32, 41, 42) Here we demonstrate for the first time that the S. aureus surface can be modified to incorporate exogenous, non-native small molecules by way of its own biosynthetic machinery. Epifluorescence microscopy and flow cytometry experiments demonstrate that these molecules are incorporated into bacteria at high levels in a SrtA- and LPETG-dependent fashion. Subsequent cell wall extraction and electron microscopy data indicate that these non-native materials are localized to the cell wall, and mass spectrometry data confirms that this labeling phenomenon involves covalent bond formation. Finally, we exploit this biosynthetic labeling technology to perform a bioorthogonal chemical reaction on the S. aureus surface.
A diverse range of applications of this strategy can be envisioned. For example, SrtA-mediated biosynthetic incorporation of fluorescent materials into bacterial peptidoglycan could enable the development of novel in situ imaging protocols. Because labeling is SrtA-dependent, highly selective, and does not affect bacterial viability, it should prove advantageous in comparison to conventional techniques for labeling bacteria with small molecules. Such strategies often involve promiscuous electrophiles (e.g., FITC), and can perturb bacterial viability and/or behavior.(43) The reported strategy might also be useful in developing engineered organisms with entirely novel functions. Indeed, by enabling the display of chemical functionality that cannot yet be genetically encoded, SrtA-mediated biosynthetic incorporation processes could complement ongoing efforts in synthetic biology, which rely predominantly on genetic manipulations.(44) Given that SrtA is widely distributed among Gram-positive bacteria, this strategy could be applied in diverse species including Streptococcus pneumoniae, Bacillus anthracis, Corynebacterium diphtheriae, and many others. Also, because sortase enzymes in species other than S. aureus often possess unique recognition and anchoring elements (other than LPXTG),(45) this strategy may prove highly selective, even in complex settings. Finally, biosynthetic incorporation of small molecules into bacteria could lead to novel therapeutic approaches. Strategies that employ small molecules to enhance immunological recognition of pathogens have proven quite promising,(46–51) however, none of these have utilized biosynthetic mechanisms to incorporate immunotargeting moieties. Such technologies could possess unique advantages over available antibiotic modalities, and lead to novel paradigms in the treatment of patients suffering from bacterial illness.
METHODS
Cell Culture and Reagents
Wild-type S. aureus Newmann and a matching srtA− strain were kindly donated by O. Schneewind (University of Chicago).(16) S. aureus strain spa::kan,(21) a protein A knockout strain, was obtained from T. J. Foster (Trinity College, Dublin, Ireland).
Peptide Synthesis Procedure
Rink amide resin preloaded with either Nα-Fmoc-Gly or Nα-Fmoc-Pro was mixed with four column volumes (CV) of dimethylformamide (DMF) for one hour prior to the commencement of each peptide synthesis. Deprotection of the resin prior to coupling was accomplished on shaking in 4 CV 20% piperidine/DMF for thirty minutes. For all subsequent coupling reactions, Fmoc-protected amino acid (5 equiv), O-benzotriazole-N, N, N′, N′-tetramethyl-uronium-hexafluoro-phosphate (HBTU, 5 equiv), and i-Pr2NEt (10 equiv) were suspended with the resin in DMF (4 CV) and shaken at room temperature for 4 hours. In all syntheses, Fmoc-protecting groups were removed by shaking for five minutes in 50% piperidine/DMF (4 CV).
For peptide cleavage reactions, resin was first treated with a mixture of trifluoroacetic acid (TFA), triisopropylsilane (TIPS), and water (94:3:3, 10 mL) at room temperature for 90 minutes. Cleavage solutions were then filtered through cotton into cold ether, pelleted, resuspended twice in ether, and dried in vacuo. Peptides were purified via High Pressure Liquid Chromatography (HPLC), lyophilized, and their identities were confirmed via high-resolution mass spectrometry.
Synthesis of Alexa Fluor 488-DIFO conjugates (7)
Perfluorophenyl 4-((2,2-difluorocyclooct-3-ynyl)methyl)benzoate (3.1 mg, 7 μmol, 2 equiv) and i-Pr2NEt (2.4 μL, 14 μmol, 4 equiv) were dissolved in DMF (0.5 mL) followed by Alexa Fluor 488 cadaverine (2 mg, 3.1 μmol, 1 equiv), whose transfer was aided by the use of DMF (0.5 mL). The reaction mixture stirred for 48 hours, concentrated, and purified by HPLC to give 7. Suspension in 2 mL of PBS led to a 200 μM solution (0.4 μmol, 13% yield) as determined by UV absorbance at 495 nm.
Generic Bacterial Labeling Procedure
Bacteria were grown for twenty-four hours in Luria Broth at 37°C (supplemented with 10 μg mL−1 erythromycin for sortase-deficient mutants) containing compounds at the desired concentration (1 mM unless otherwise noted, final volume 500 μL). Subsequently, 100 μL aliquots of culture were taken, pelleted at 12000g for one minute, and washed three times with PBS buffer. Cells were then used in the following experiments.
Microscopy/Flow Cytometry
Labeled cells were resuspended in 4% paraformaldehyde in PBS and incubated at room temperature on a nutating mixer for 45 minutes. Following three additional PBS washes, bacteria were resuspended in 1 mL of PBS and analyzed on an Accuri C6 Flow Cytomer. Levels of fluorescein incorporation we quantitated by comparison to Quantum FITC MESF beads (Bangs Laboratories). 100 μL aliquots of the above were removed (prior to cytometric analysis), pelleted, and resuspended in 20 μL anti-fade media (Vectashield Mounting Media), mounted onto Esco Superfrost® Microscope slides, and allowed to incubate overnight at 4 °C before images were obtained.
Quantitation of Fluorescence via Cell Wall Extraction
Following labeling, bacteria were diluted with PBS to normalize total bacterial count and subjected to ultrasonication. The sonicated cells were filtered via centrifugal filtration using Amicon YM-10 filters at 4 °C for thirty minutes. The filtrate was taken and used to determine the fluorescence intensity of the cytosol fraction, while the high-molecular weight pellet was collected and treated for 30 minutes at 37°C with 500 μL of SMM buffer containing 50 μg mL−1 lysostaphin prewarmed to 37°C. The mixture was then centrifuged at 14000 rpm for one minute, and the supernatant was removed and placed on ice. 500 μL of 10% sodium dodecyl sulfate (SDS) was added to the cold supernatant mixtures. 100 μL of each sample was taken, and fluorescence intensities were determined in triplicate using a Tecan Platereader (em 485 nm, ex 535 nm). Gain was set to the intensity of the wild type bacterial cell wall that had been incubated with peptide. Cell wall fractionation data were derived by first subtracting the appropriate blank (cell wall or cytosol) from fluorescence intensity values, then measuring the difference between this value and the fluorescence reading obtained for control conditions (i.e., wild-type bacteria not treated with peptide). Entries represent means from triplicate experiments and error bars indicate standard deviations.
Electron Microscopy
Following the generic protocol for bacterial labeling outlined above, bacteria were resuspended in 4% paraformaldehyde in PBS and incubated at room temperature on a nutating mixer for 45 minutes. Samples were then pelleted, washed three additional times with PBS, and then suspended in 5% gelatin. Suspensions were trimmed to small blocks and placed in 2.3M sucrose solution (cryoprotectant) overnight on a rotor at 40 °C. They were transferred to aluminum pins and frozen rapidly in liquid nitrogen. The frozen block was trimmed on a Leica CryoUltraCut and 75 nm thick sections were collected using the Tokoyasu method.(52) The frozen sections were collected on a drop of sucrose, thawed, placed on a nickel formvar/carbon coated grid, and floated in a dish of PBS ready for immunolabeling.
For immunolabeling, grids were placed on drops of 0.1 M ammonium chloride for 10 minutes to quench untreated aldehyde groups, then blocked for nonspecific binding on 1% fish skin gelatin for 20 mins. Grids were incubated on primary antibody (rabbit anti-FITC IgG, Invitrogen) at 1:10 and 1:50 dilutions for 30 min. Controls were also performed using Rabbit IgG (Jackson) at the same dilution. Samples were then rinsed 6 times on drops of PBS, and labeled using 12 nm anti-rabbit colloidal gold (Jackson) for 30 minutes. Grids were rinsed in PBS and fixed using 1% gluteraldehyde for 5 min, and a final rinse step with distilled water was performed before transfering grids to a UA/methylcellulose drop for 10 min. Grids were dried and viewed in a FEI Tencai Biotwin TEM at 80Kv. Images were acquired using Morada CCD and iTEM (Olympus) software.
MALDI-ToF Mass Spectrometry
Following the generic protocol for labeling outlined above, bacteria were diluted with 3.5 mL of PBS and subjected to ultrasonication. The pellet was then subjected to enzymatic digest with mutanolysin (150 μg mL−l) and lysostaphin (100 μg mL−1) in 500 μL of Tris-HCl buffer (0.5 mM, pH 6.5). After gentle mixing, the solution was incubated at 37°C for 4 hours. Samples were then pelleted and the supernatant heated at 90°C for 5 minutes before isolation of biotinylated adducts.
Biotinylated peptides were isolated using an Avidin cartridge (ABI) with the ABI syringe setup and ABI protocols. The avidin eluted biotinylated peptides were dried in a Savant SVC100 SpeedVac, prior to dissolving in 4 μL of matrix solvent (0.1% trifluoroacetic acid, 50 % acetonitrile/water). Matrix was prepared by dissolving alpha-cyano-4-hydroxycinnamic acid (CHCA) in the matrix solvent to a concentration of 3.0 mg mL−1. Bradykinin was added to the matrix as an internal calibrant at a final concentration of 1.67 fmol μL−1. Spotting mixtures were prepared by mixing 1 μL of sample with 4 μL of matrix in an Eppendorf tube. 0.8 μL of each mixture was spotted on a 384 well sample target plate and allowed to air dry. Matrix assisted laser desorption ionization mass spectrometry (MALDI-MS) was acquired on an Applied Biosystems/MDS SCIEX 4800 MALDI TOF/TOF Analyzer in reflector positive mode. The mass range from 700–4000 m/z was screened, and the spectra were internally calibrated to the bradykinin standard (protonated, monoisotopic mass of 1060.569 Da).
Cell-Surface Labeling with DIFO Reagents
Following the generic protocol for labeling outlined above, bacteria were resuspended in ice-cold PBS (500 μL). Click-reagent 7 was then added to the suspended bacteria to a final concentration of 10 μM, and cultures were then incubated for 48 hours at 4ºC on a nutating mixer. Cells were washed three times with PBS and then fixed with 4% PFA for 30 minutes. Following three more washes, the bacteria were resuspended in 1 mL of PBS and analyzed on an Accuri C6 Flow Cytometer.
For microscopy, 100 μL aliquots of the fixed samples from above were removed prior to cytometric analysis. The aliquots were pelleted by centrifugation, resuspended in 5 μL of Fluoro-Gel, mounted on Esco Superfrost® Microscope slides, and allowed to incubate overnight at 4 °C before images were obtained.
Supplementary Material
Acknowledgments
We are grateful to S. Campbell (Yale University School of Medicine), O. Schneewind (University of Chicago), and T. Foster (Trinity College, Dublin, Microbiology Department) for providing us with Wild-type (Newman), SrtA knockout, and protein A knockout S. aureus strains, respectively. We also thank Z. Gartner (UCSF) for a gift of the carboxylic acid precursor to cyclooctyne 7. Finally, we acknowledge J. Kanyo, K. Stone, T. Lam and T. Voss of the Keck Biotechnology Resource for assistance with mass spectrometry, and M. Graham and C. Rahner for assistance with electron microscopy. This work was funded by the National Institutes of Health through the NIH Director’s New Innovator Award Program (DP22OD002913) and through a Bill and Melinda Gates Grand Challenges Explorations Award.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet.
References
- 1.Taubes G. The bacteria fight back. Science. 2008;321:356–361. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- 2.Klevens R, Morrison M, Nadle J, Petit S, Gershman K, Ray S, Harrison L, Lynfield R, Dumyati G, Townes J, Craig A, Zell E, Fosheim G, Mcdougal L, Carey R, Fridkin S. Invasive methicillin-resistant Staphylococcus aureus Infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 3.Center for Disease Control. HIV/AIDS Surveillance Report: HIV Infection and AIDS in the United States and Dependent Areas. 2006 Can be found at http://www.cdc.gov/hiv/topics/surveillance/resources/reports/2006report/table7.htm.
- 4.Rooijakkers SH, van Kessel KP, van Strijp JA. Staphylococcal innate immune evasion. Trends Microbiol. 2005;13:596–601. doi: 10.1016/j.tim.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Serruto D, Rappuoli R, Scarselli M, Gros P, van Strijp JA. Molecular mechanisms of complement evasion: learning from staphylococci and meningococci. Nat Rev Microbiol. 2010;8:393–399. doi: 10.1038/nrmicro2366. [DOI] [PubMed] [Google Scholar]
- 6.Maresso A, Schneewind O. Sortase as a target of anti-infective therapy. Pharmacol Rev. 2008;60:128–141. doi: 10.1124/pr.107.07110. [DOI] [PubMed] [Google Scholar]
- 7.Parthasarathy R, Subramanian S, Boder ET. Sortase A as a novel molecular “stapler” for sequence-specific protein conjugation. Bioconjug Chem. 2007;18:469–476. doi: 10.1021/bc060339w. [DOI] [PubMed] [Google Scholar]
- 8.Antos JM, Miller GM, Grotenbreg GM, Ploegh HL. Lipid modification of proteins through sortase-catalyzed transpeptidation. J Am Chem Soc. 2008;130:16338–16343. doi: 10.1021/ja806779e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo X, Wang Q, Swarts BM, Guo Z. Sortase-catalyzed peptide-glycosylphosphatidylinositol analogue ligation. J Am Chem Soc. 2009;131:9878–9879. doi: 10.1021/ja903231v. [DOI] [PubMed] [Google Scholar]
- 10.Pritz S, Wolf Y, Kraetke O, Klose J, Bienert M, Beyermann M. Synthesis of biologically active peptide nucleic acid-peptide conjugates by sortase-mediated ligation. J Org Chem. 2007;72:3909–3912. doi: 10.1021/jo062331l. [DOI] [PubMed] [Google Scholar]
- 11.Wu Z, Guo X, Wang Q, Swarts BM, Guo Z. Sortase A-catalyzed transpeptidation of glycosylphosphatidylinositol derivatives for chemoenzymatic synthesis of GPI-anchored proteins. J Am Chem Soc. 132:1567–1571. doi: 10.1021/ja906611x. [DOI] [PubMed] [Google Scholar]
- 12.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: a versatile method for protein labeling. Nat Chem Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto T, Nagamune T. Expansion of the sortase-mediated labeling method for site-specific N-terminal labeling of cell surface proteins on living cells. Chem Commun. 2009:1022–1024. doi: 10.1039/b818792d. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka T, Yamamoto T, Tsukiji S, Nagamune T. Site-specific protein modification on living cells catalyzed by Sortase. Chembiochem. 2008;9:802–807. doi: 10.1002/cbic.200700614. [DOI] [PubMed] [Google Scholar]
- 15.Popp MW, Antos JM, Ploegh HL. Site-specific protein labeling via sortase-mediated transpeptidation. Curr Protoc Protein Sci Supplement. 2009;56(Unit 15.13) doi: 10.1002/0471140864.ps1503s56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci. 2000;97:5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohamed N, Visai L, Speziale P, Ross JM. Quantification of Staphylococcus aureus cell surface adhesins using flow cytometry. Microb Pathog. 2000;29:357–361. doi: 10.1006/mpat.2000.0399. [DOI] [PubMed] [Google Scholar]
- 18.Lee SG, Fischetti VA. Purification and characterization of LPXTGase from Staphylococcus aureus: the amino acid composition mirrors that found in the peptidoglycan. J Bacteriol. 2006;188:389–398. doi: 10.1128/JB.188.2.389-398.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SG, Pancholi V, Fischetti VA. Characterization of a unique glycosylated anchor endopeptidase that cleaves the LPXTG sequence motif of cell surface proteins of Gram-positive bacteria. J Biol Chem. 2002;277:46912–46922. doi: 10.1074/jbc.M208660200. [DOI] [PubMed] [Google Scholar]
- 20.Litzinger S, Duckworth A, Nitzsche K, Risinger C, Wittmann V, Mayer C. Muropeptide rescue in Bacillus subtilis involves sequential hydrolysis by beta-N-acetylglucosaminidase and N-acetylmuramyl-L-alanine amidase. J Bacteriol. 2010;192:3132–3143. doi: 10.1128/JB.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel AH, Foster TJ, Pattee PA. Physical and genetic mapping of the Protein A gene in the chromosome of Staphylococcus aureus 8325–4. J Gen Microbiol. 1989;135:1799–1807. doi: 10.1099/00221287-135-7-1799. [DOI] [PubMed] [Google Scholar]
- 22.Ton-That H, Faull KF, Schneewind O. Anchor structure of staphylococcal surface proteins. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J Biol Chem. 1997;272:22285–22292. doi: 10.1074/jbc.272.35.22285. [DOI] [PubMed] [Google Scholar]
- 23.Schneewind O, Fowler A, Faull KF. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 24.Sletten EM, Bertozzi CR. Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew Chem Int Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Best MD. Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry. 2009;48:6571–6584. doi: 10.1021/bi9007726. [DOI] [PubMed] [Google Scholar]
- 26.Yi W, Liu X, Li Y, Li J, Xia C, Zhou G, Zhang W, Zhao W, Chen X, Wang PG. Remodeling bacterial polysaccharides by metabolic pathway engineering. Proc Natl Acad Sci. 2009;106:4207–4212. doi: 10.1073/pnas.0812432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadamoto R, Niikura K, Ueda T, Monde K, Fukuhara N, Nishimura SI. Control of bacteria adhesion by cell-wall engineering. J Am Chem Soc. 2004;126:3755–3761. doi: 10.1021/ja039391i. [DOI] [PubMed] [Google Scholar]
- 28.Sadamoto R, Niikura K, Sears PS, Liu HT, Wong CH, Suksomcheep A, Tomita F, Monde K, Nishimura SI. Cell-wall engineering of living bacteria. J Am Chem Soc. 2002;124:9018–9019. doi: 10.1021/ja026133x. [DOI] [PubMed] [Google Scholar]
- 29.Sadamoto R, Niikura K, Monde K, Nishimura SI. Cell wall engineering of living bacteria through biosynthesis. Methods Enzymol. 2003;362:273–286. doi: 10.1016/s0076-6879(03)01019-x. [DOI] [PubMed] [Google Scholar]
- 30.Sadamoto R. Artificial modification of bacterial surface with cell-wall engineering. Trends Glycosci Glycotechnol. 2005;17:97–105. [Google Scholar]
- 31.Link AJ, Vink MK, Tirrell DA. Presentation and detection of azide functionality in bacterial cell surface proteins. J Am Chem Soc. 2004;126:10598–10602. doi: 10.1021/ja047629c. [DOI] [PubMed] [Google Scholar]
- 32.Beatty KE, Xie F, Wang Q, Tirrell DA. Selective dye-labeling of newly synthesized proteins in bacterial cells. J Am Chem Soc. 2005;127:14150–14151. doi: 10.1021/ja054643w. [DOI] [PubMed] [Google Scholar]
- 33.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise Huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Tornøe C, Christensen C, Meldal M. Peptidotriazoles on Solid Phase: [1,2,3]-Triazoles by Regiospecific Copper(I)-Catalyzed 1,3-Dipolar Cycloadditions of Terminal Alkynes to Azides. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 35.Codelli JA, Baskin JM, Agard NJ, Bertozzi CR. Second-generation difluorinated cyclooctynes for copper-free click chemistry. J Am Chem Soc. 2008;130:11486–11493. doi: 10.1021/ja803086r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang PV, Prescher JA, Hangauer MJ, Bertozzi C. Imaging Cell Surface Glycans with Bioorthogonal Chemical Reporters. J Am Chem Soc. 2007;129:8400–8401. doi: 10.1021/ja070238o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laughlin ST, Baskin JM, Amacher SL, Bertozzi C. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prescher JA, Bertozzi CR. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 39.Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR. Copper-free click chemistry in living animals. Proceedings of the National Academy of Sciences. 2010;107:1821–1826. doi: 10.1073/pnas.0911116107. and references contained therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agard NJ, Bertozzi CR. Chemical Approaches To Perturb, Profile, and Perceive Glycans. Acc Chem Res. 2009;42:788–797. doi: 10.1021/ar800267j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schilling B, Goon S, Samuels NM, Gaucher SP, Leary JA, Bertozzi CR, Gibson BW. Biosynthesis of sialylated lipooligosaccharides in Haemophilis ducreyi is dependent on exogenous sialic acid and not mannosamine. Incorporation studies using N-acylmannosamine analogues, N-glycolylneuraminic acid, and C-13-labeled N-acetylneuraminic acid. Biochemistry. 2001;40:12666–12677. doi: 10.1021/bi0107849. [DOI] [PubMed] [Google Scholar]
- 42.Goon S, Schilling B, Tullius MV, Gibson BW, Bertozzi CR. Metabolic incorporation of unnatural sialic acids into Haemophilus ducreyi lipooligosaccharides. Proc Natl Acad Sci. 2003;100:3089–3094. doi: 10.1073/pnas.0437851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weingart CL, Broitman-Maduro G, Dean G, Newman S, Peppler M, Weiss AA. Fluorescent labels influence phagocytosis of Bordetella pertussis by human neutrophils. Infect Immun. 1999;67:4264–4267. doi: 10.1128/iai.67.8.4264-4267.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agapakis CM, Silver PA. Synthetic biology: exploring and exploiting genetic modularity through the design of novel biological networks. Mol Biosyst. 2009;5:704–713. doi: 10.1039/b901484e. [DOI] [PubMed] [Google Scholar]
- 45.Boekhorst J, de Been MWHJ, Kleerebezem M, Siezen RJ. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-Like sorting motifs. J Bacteriol. 2005;187:4928–4934. doi: 10.1128/JB.187.14.4928-4934.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carlson CB, Mowery P, Owen RM, Dykhuizen EC, Kiessling LL. Selective tumor cell targeting using low-affinity, multivalent interactions. ACS Chem Biol. 2007;2:119–127. doi: 10.1021/cb6003788. and references contained therein. [DOI] [PubMed] [Google Scholar]
- 47.Low P, Henne W, Doorneweerd D. Discovery and development of folic-acid-based receptor targeting for imaging and therapy of cancer and inflammatory diseases. Acc Chem Res. 2008;41:120–129. doi: 10.1021/ar7000815. and references contained therein. [DOI] [PubMed] [Google Scholar]
- 48.Murelli RP, Zhang AX, Michel J, Jorgensen WL, Spiegel DA. Chemical Control over Immune Recognition: A Class of Antibody-Recruiting Small Molecules That Target Prostate Cancer. J Am Chem Soc. 2009:17090–17092. doi: 10.1021/ja906844e. and references contained therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker CG, Domaoal RA, Anderson KS, Spiegel DA. An antibody-recruiting small molecule that targets HIV gp120. J Am Chem Soc. 2009;131:16392–16394. doi: 10.1021/ja9057647. and references contained therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Popkov M, Gonzalez B, Sinha SV, Barbas CF., III Instant immunity through chemically programmable vaccination and covalent self-assembly. Proc Natl Acad Sci. 2009;106:4378–4383. doi: 10.1073/pnas.0900147106. and references contained therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krishnamurthy VM, Quinton LJ, Estroff LA, Metallo SJ, Isaacs JM, Mizgerd JP, Whitesides GM. Promotion of opsonization by antibodies and phagocytosis of Gram-positive bacteria by a bifunctional polyacrylamide. Biomaterials. 2006;27:3663–3674. doi: 10.1016/j.biomaterials.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 52.Tokuyasu KT. A technique for ultracryotomy of cell suspensions and tissues. J Cell Biol. 1973;57:551–565. doi: 10.1083/jcb.57.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.