Abstract
In many eukaryotes, spliceosomal introns are able to influence the level and site of gene expression. The mechanism of this Intron Mediated Enhancement (IME) has not yet been elucidated, but regulation of gene expression is likely to occur at several steps during and after transcription. Different introns have different intrinsic enhancing properties, but the determinants of these differences remain unknown. Recently, an algorithm called IMEter, which is able to predict the IME potential of introns without direct testing, has been proposed. A computer program was developed for Arabidopsis thaliana and rice (Oryza sativa L.), but was only tested experimentally in Arabidopsis by measuring the enhancement effect on GUS expression of different introns inserted within otherwise identical plasmids. To test the IMEter potential in rice, a vector bearing the upstream regulatory sequence of a rice β-tubulin gene (OsTub6) fused to the GUS reporter gene was used. The enhancing intron interrupting the OsTub6 5′-UTR was precisely replaced by seven other introns carrying different features. GUS expression level in transiently transformed rice calli does not significantly correlate with the calculated IMEter score. It was also found that enhanced GUS expression was mainly due to a strong increase in the mRNA steady-state level and that mutations at the splice recognition sites almost completely abolished the enhancing effect. Splicing also appeared to be required for IME in Arabidopsis cell cultures, where failure of the OsTub6 5′ region to drive high level gene expression could be rescued by replacing the poorly spliced rice intron with one from Arabidopsis.
Keywords: IME, IMEter, intron, Oryza sativa, splicing, tubulin
Introduction
While promoters regulate transcription at the DNA level, many eukaryotic introns seem to play a similar role at the RNA level, by finely tuning either the amount of the mature mRNA or its translatability.
Spliceosomal introns capable of influencing the level of reporter gene expression have been reported in most eukaryotes (Le Hir et al., 2003). In plants, Intron Mediated Enhancement (IME) of gene expression has progressively been found to be a much more common event than originally suspected. The list of enhancing introns that have been experimentally tested includes 40 different members at least, yet the proportion of plant introns endowed with enhancing capacity cannot be predicted without experimental data.
More recently, several examples of introns capable of influencing the site of gene expression have been reported in plants: Intron-Dependent Spatial Expression (IDSE) has been shown in, for example, the potato Sus3 gene (Fu et al., 1995), the Arabidopsis histone replacement H3 gene (Chaubet-Gigot et al., 2001), the Petunia ADF1 intron (Jeong et al., 2007), the Arabidopsis PRF2 intron (Jeong et al., 2006), the tomato sucrose transporter LeSUT1 (Weise et al., 2008), and for different rice α- and β-tubulin genes (Jeon et al., 2001; Gianì et al., 2009). Despite several efforts to understand it, the mechanism by which introns affect gene expression is still elusive. However, a few points seem to be clearly defined.
(i) Intron position is important: introns must be located within the transcription unit (Callis et al., 1987; Mascarenhas et al., 1990; Clancy et al., 1994) and in the correct orientation (Mascarenhas et al., 1990; Mun et al., 2002; Curi et al., 2005) in order to enhance expression. Moreover, the same intron gradually weakens its effect as distance from the first ATG codon increases (Rose, 2004) and introns are generally inactive if located in the 3′-UTR sequence (Snowden et al., 1996; Rose, 2002). In some cases, promoter-proximal introns may also contain cis-acting elements (Giedeckel et al., 1996; Deyholos and Sieburth, 2000; Vitale et al., 2003): any enhancing effect would then occur at the DNA level and this mechanism is likely to be unrelated to IME. The two effects can, in fact, be separated: the Arabidopsis ACT1 intron does not sustain expression in root, shoot, and floral organ primordia when inserted in both forward and reverse orientations upstream of its own promoter, as it does when present in the leader region, but can still enhance expression in pollen (Vitale et al., 2003).
(ii) IME in plants is mainly a co- and a post-transcriptional event. The increase in reporter gene expression, measured as the protein amount or enzyme activity, correlates with an increased mRNA steady-state level. In some cases, run-on experiments have demonstrated a weak (about 2-ofold) increase in the rate of transcription (Rose and Last, 1997; Samadder et al., 2008), not enough to account for the observed increase in mRNA amount. An effect on mRNA stability has instead been excluded for the maize SalT intron (Rethmeier et al., 1997). Furthermore, some effect on translation has been reported in several cases (Tanaka et al., 1990; Bourdon et al., 2001; Rose, 2004; Curi et al., 2005; Lu et al., 2008; Samadder et al., 2008), while it has been excluded in others (Rethmeier et al., 1997). Studies performed in other eukaryotes have defined many steps where introns can affect mRNA metabolism: most of these effects are linked to mRNA splicing and are mediated by interactions between the spliceosome or the exon junction complex (EJC), the protein complex deposed after splicing upstream of the exon–exon junction, and other regulatory proteins of the ‘mRNA factory’ (Le Hir et al., 2003). The need for plant introns to be transcribed in order to enhance gene expression also suggests an involvement of splicing, but this is apparently one of the most controversial points. While mutations blocking the splicing of enhancing introns completely impaired IME in some systems (Mascarenhas et al., 1990; Sinibaldi and Mettler, 1992; Clancy and Hannah, 2002), they still allowed a sustained increase of GUS mRNA level in others (Rose and Beliakoff, 2000; Rose, 2002).
(iii) Enhancing properties are not common to all introns: while some introns are known to be required to drive high level and/or the correct expression pattern of the host gene, others have no, or even negative, effects on gene expression (Bourdon et al., 2001). Their enhancement effect is also quantitatively different. Intron deletion experiments favour the hypothesis that redundant and dispersed sequences are required for IME, a future target would be clearly to identify such determinants. In this regard, an algorithm named IMEter and based on the assumption that promoter-proximal introns have a high chance of being endowed with enhancement activity, has recently been developed (Rose et al., 2008). Literature data cannot be used to test the capacity of the IMEter for predicting intron enhancing properties, because the results obtained from different work are often not comparable. Too many factors can, in fact, contribute to the expression level of the tested gene: the promoter, the reporter gene, the intron position, the flanking exons, and the transformation and expression system.
In this work, new data, obtained by testing a set of plasmids derived from the previously described pProTub6-GUS vector (Gianì et al., 2009), are presented. This vector bears the rice OsTub6 upstream regulatory sequence (promoter and leader sequence, containing a strongly enhancing intron) followed by the first four β-tubulin codons fused in-frame with the GUS reporter gene. pProTub6-GUS drives higher expression levels in rice calli, as well as in transgenic rice plants, compared with a corresponding intronless vector (pProTub6ΔLI). Seven different plasmids were derived by precisely replacing the OsTub6 intron with introns from different sources, so that each produced identical transcripts after splicing.
To test GUS expression, a transient transformation approach was preferred, in order to avoid the unpredictable effects of flanking genomic sequences, gene copy number, silencing or co-suppression, all of which are events responsible for highly variable reporter gene expression among lines of stably transformed plants.
Materials and methods
Plasmid construction
All plasmids were constructed starting from pProTub6-GUS (Gianì et al., 2009), a pBI221-based plasmid bearing the 1.9 kb upstream region and the first eight nucleotides of the Ostub6 gene, fused in-frame with the GUS coding sequence through a 34 bp linker. All the replacing introns were PCR-amplified from genomic or plasmid DNA with specific primers bearing EcoRI and PstI restriction sites at their ends (Table 1; see Supplementary Table S1 at JXB online) and inserted in the corresponding sites of the plasmid, after excision of the endogenous 5′ intron. Mutation at the 3′ splice site was produced by digestion with PstI followed by filling-in of the protruding ends and re-ligation of the plasmid. The 5′ splice site mutation was introduced through PCR-mediated mutagenesis. Intron insertions and sequence mutations were checked by sequencing. Plasmids were amplified in E. coli DH5α and extracted with QIAprep spin extraction kits (www.qiagen.com). pUbi-GFP was described by Morello et al. (2002).
Table 1.
Sequence features and IME effect of the tested plasmid set
| Introna | Plant | Position | Length | %GC | %T | T-repeats (at least six Ts) | IME motifs | Reported IME effectb | Tested IME effect (this work) |
| OsTub6LI | Rice | Leader | 446 | 47 | 39 | 3 | 0 | + | 9 |
| OsTub4LLI | Rice | Leader | 863 | 41 | 37 | 1 | 2 | ++ | 13 |
| OsTub4SLI | Rice | Leader | 110 | 46 | 36 | – | 0 | + | 4 |
| OsTua2FI | Rice | Coding | 892 | 42 | 36 | 1 | 5 | + | 12 |
| OsTua3FI | Rice | Coding | 907 | 45 | 35 | 3 | 6 | + | 6 |
| AtTub6LI | At | Leader | 416 | 34 | 46 | 1 | 4 | nt | 10 |
| OsCpk2LLI | Rice | Leader | 969 | 44 | 40 | 3 | 1 | nt | 17 |
| OsCpk2SLI | Rice | Leader | 290 | 38 | 47 | 1 | 1 | ++ | 11 |
+/–, Low; +, high; ++, very high; nt, not tested. In bold, the only Arabidopsis intron.
LLI, long leader intron; SLI, short leader intron; FI, first intron.
Previously published works.
In vitro plant and cell cultures
Rice calli (Oryza sativa L., japonica type, cv. Arborio) generated from rice endosperm were cultivated in solid MS medium supplemented with 2 mg l−1 2,4-D. Arabidopsis suspension culture cells (ecotype Landsberg 310-14-2) from germinated seeds were a kind gift of Dr C Bonza, University of Milan, and were grown in liquid medium as described by Curti et al. (1993).
Young tobacco leaves (cv. SR1) were cut from plantlets at the third or fourth leaf stage, axenically grown in MS medium with 30 g l−1 sucrose.
Transient transformation
Rice calli were co-bombarded with the plasmid of interest and pUBI-GFP, using the Bio-Rad PDS-1000/He biolistic device (www.bio-rad.com) as described by Gianì et al. (2009). Arabidopsis suspension cells and tobacco leaves were bombarded in the same way, except that preincubation in osmotic medium was omitted. GUS expression was evaluated 24 h after transformation.
Expression analysis
For GFP and GUS activity measurement, transformed cell samples were frozen in liquid nitrogen, ground with a pestle and mortar, and solubilized with 500 μl GUS extraction buffer (Jefferson et al., 1987). Cell debris were pelleted by two subsequent centrifugation steps (15 min, 13 000 rpm, 4 °C) and GFP epifluorescence of 100 μl of the total cell extract was directly measured with a VersaFluor fluorometer (www.bio-rad.com) using a dedicated filter set (Ex 490/10; Em 510/10). Relative Fluorescent Units (RFU) were zeroed against an untransformed sample. For the GUS fluorometric assay, 100 μl of the same extract were incubated at 37 °C in 0.5 ml of GUS assay buffer. At different times, 50 μl aliquots of the reaction were stopped with 0.2 M Na2CO3 and fluorescence was measured with an opportune filter set (Ex 360/40; Em 460/10).
GUS activity, calculated as pmoliMU per 100 μl sample h−1, was normalized for the GFP activity of the same sample, measured as RFU per 100 μl. The mean of three different samples was calculated.
The fold enhancement was calculated as the mean amount of GUS activity induced by each tested plasmid, relative to the activity measured, in the same experiment, in samples transformed with the intronless plasmid.
Histochemical GUS staining was performed as described by Jefferson (1987).
RT-PCR
Total RNA was extracted from 100 μg of transformed or control cells (bombarded with uncoated microcarriers) with the plant RNeasy kit (www.qiagen.com). About 1 μg of RNA was treated with DNAse I, before retrotranscription with SuperScript II (www.invitrogen.com) following the protocol of the supplier, using an oligo(dT)12–18 for priming. 1 μl of each reaction was used as a template for specific PCR primer couples (Table 1; see Supplementary Table S1 at JXB online). PCR reactions were conducted in a total volume of 30 μl, under standard procedures, with the number of cycles indicated in the specific result section. Control reactions without any cDNA were always run in parallel. The whole reaction was loaded on agarose gel for analysis.
Accession numbers
OsTub6, AM400988; OsTub4, AJ306592; OsTua2, AJ488063; OsTua3, AJ488064; AtTub6, NC_003076; and OsCDPK2, Y13658.
Results
Different introns enhance gene expression to different extents in rice cells
Previous results obtained with plasmid pProTub6-GUS have shown that the leader intron of rice OsTub6 can increase GUS expression in rice calli by 10-fold (Gianì et al., 2009). Two simple features of this intron sequence suggested it may be used for exploring the mechanism of IME through a non-disrupting mutational approach. First, the intron is located within the 5′ untranslated sequence, which avoids any possible frameshift effect on the protein product. Second, the intron is naturally delimited by two restriction sites (5′-EcoRI/3′-PstI). This feature was exploited to generate a set of recombinant expression plasmids in which the intron of pProTub6-GUS was eliminated or precisely replaced by other plant introns, all but two of which are reported to have an IME effect (Table 1). To gain insights into the intron features possibly correlated to the IME effect, seven plant introns that were different with respect to their position within the original gene and largely variable in length and nucleotide composition were tested (Table 1). They were: two first introns from the coding regions of rice α2 and α3 tubulin genes (Fiume et al., 2004), two introns nested within one another in the 5′-UTR of the OsTub4 β-tubulin gene (Morello et al., 2002), two nested introns from the 5′-UTR of the calcium-dependent protein kinase gene OsCDPK2 (Morello et al., 2006), and one intron coming from the leader region of AtTub6, the putative Arabidopsis orthologue of OsTub6 (Snustad et al., 1992).
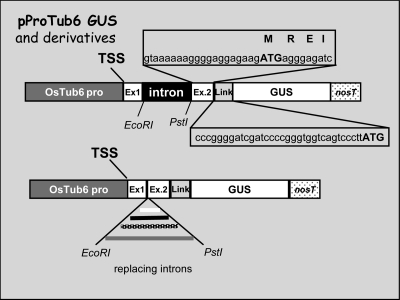
Intron sequences were PCR-amplified from genomic or plasmid DNA with primers complementary to their borders, all bearing the EcoRI and PstI sites as tails. After digestion, the PCR products were inserted into the corresponding sites of the GUS expression vector pProTub6-GUS, as shown in Fig. 1. All derived plasmids have the same splice sites and flanking exons, to minimize any possible difference in splicing efficiency or any other sequence effect, and all are expected to give rise to identical mature transcripts when correctly spliced. The context of the translation initiation site, only 20 bp away from the 3′ end of the intron, was also preserved so that all GUS fusion products were identical, with the first four amino acids of β-tubulin fused in-frame, through a short linker, to the GUS open reading frame. These four amino acids (MREI) are the same in all plant and animal β-tubulins and are known to be involved in the regulation of tubulin production (Theodorakis and Cleveland, 1992). For transient transformation experiments, rice calli were co-bombarded with the plasmid of interest and equimolar amounts of a GFP-expressing plasmid, pUBI-GFP, on triplicate samples. As a control for background activity, callus samples were bombarded with uncoated gold microcarriers. For quantitative analysis of gene expression, total fluorogenic GUS activity, measured on extracts from each bombarded sample, was normalized to the GFP fluorescence value of the same extract, after subtraction of the background value from untransformed cell extracts. In a typical experiment, CV (coefficient of variation) values among triplicate samples ranged from 7–8% to 20%. To compare results from different experiments, each value was then expressed as a percentage of the GUS activity from the pProTub6-GUS plasmid, taken as a reference.
Fig. 1.
Diagram of the pProTub6-GUS vector and derived plasmids.
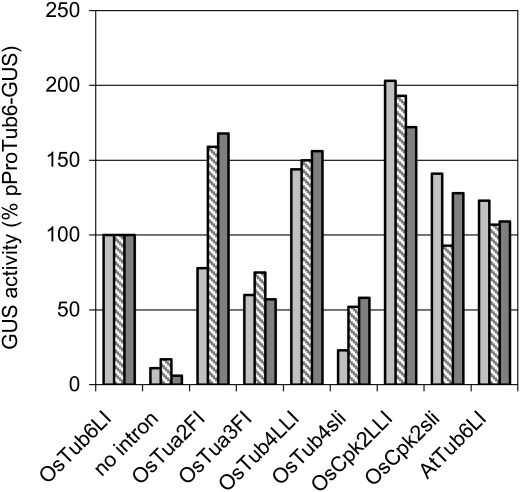
As shown in Fig. 2, that reports the results of three independent transformation experiments, all tested introns enhanced GUS expression over the level of the intronless plasmid, but the observed expression varied greatly among plasmids, with some introns having stronger effects than others. Since plasmid sequences were all identical except for the intron sequence, this latter should be held responsible for the observed differences.
Fig. 2.
Effect of different introns on GUS activity in transformed rice calli. GUS activity is expressed as a percentage of that induced by pProTub6-GUS. Each column represents the mean of triplicate samples within a single experiment. Data from three independent transformations are reported. Histograms are named from the name of the tested intron.
This result is in agreement with similar experiments conducted on the Arabidopsis TRP1 gene fused to GUS as a reporter (Rose, 2004; Rose et al., 2008). Replacement of the first TRP1 intron with other introns (all from Arabidopsis) led to different increases in steady-state mRNA levels in transgenic Arabidopsis plants (3–15-fold). Therefore, a model dicot plant (Arabidopsis) and a model monocot one (rice) display, in this respect, a similar behaviour.
However, from the data reported in Table 1, it is very hard to find any correlation between intron features and enhancement activity. Introns from both coding and leader regions show enhancing effects when placed in the 5′-UTR. None of the general features reported to affect splicing efficiency and/or enhancing ability (length, GC content, T content, presence of T-stretches) correlate with the enhancing effect measured, i.e. OsCDPK2sLI, OsTub6 LI, and OsTua2 FI show a similar degree of enhancement (10–12-fold) but have quite different lengths (290, 446, and 892 base pairs, respectively), GC content (38, 47, and 42%), and T content (47, 39, and 36%). Conversely, OsTua2 and OsTua3 introns, which are very similar in length and base composition, induce 12-fold and 6-fold increases in GUS expression, respectively. Therefore, the determinants of the observed differences in enhancing activity are not evident.
IME effect does not linearly correlate to the IMEter score in rice
Deletion experiments have often led to the conclusion that dispersed signals within introns are required for enhancement, though none have so far been identified. Very recently, a word-based algorithm predictive of the ability of an intron to stimulate gene expression was developed and trained with Arabidopsis and rice introns (Rose et al., 2008). The algorithm provides a measure of the abundance of unknown promoter-proximal signals in any intron, assigning a score that can be either negative or positive: a positive score indicates a good candidate for IME. The program was run on six experimentally tested introns inserted in the same position within TRP1::GUS fusion constructs. Data collected on transgenic Arabidopsis plants gave a good correlation (R2=0.89) between the algorithm prediction and the level of GUS mRNA level attributable to the presence of the intron (Rose et al., 2008).
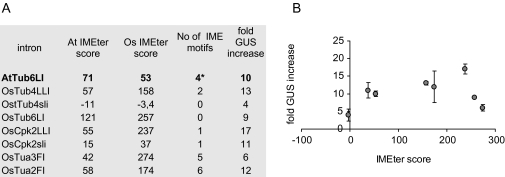
All our introns were therefore analysed with the on-line version of the IMEter program (http://korflab.ucdavis.edu/cgi-bin/web-imeter.pl) trained with rice introns, with the purpose of comparing the calculated predictive scores with our experimental data. All but one of the enhancing introns tested, scored positive with the IMEter (Fig 3a) and the slightly negative scoring intron (OsTub4 sli) was the less effective one. However, the enhancing effect does not significantly correlate to the IMEter score, calculated with the version of the IMEter available for rice (Fig. 3b). We also searched our introns for the sequence TC/AGATC/G which has been proposed o be the most common motif found in high-scoring rice introns (Rose et al., 2008). Sequences compatible with this consensus were found more than once only in OsTua2 and OsTua3, which are ORF introns, as reported in the table of Fig. 3A.
Fig. 3.
Calculated rice IMEter scores and measured enhancing activity of tested introns (A) features of the tested introns with respect to IME. *indicates the IME motif identified in Arabidopsis introns. (B) Distribution of the experimental data. GUS activity is reported as fold-increase with respect to that induced by the intronless plasmid.
Splicing is an absolute prerequisite for enhancement in rice cells
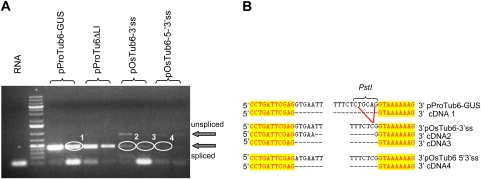
One controversial point related to IME is its relationship with splicing. Mutagenesis at splice sites resulted in the abolition of reporter gene expression enhancement for the maize Hsp82 and the Adh1 introns (Mascarenhas et al., 1990; Sinibaldi and Mettler, 1992), in electroporated maize protoplasts, and for the Sh1 intron in bombarded maize cell suspensions (Clancy and Hannah, 2002). Conversely, two unspliceable mutants of TRP1::GUS fusions were reported to induce mRNA accumulation by up to 75% of that inducible by the wild-type intron in transgenic Arabidopsis plants (Rose and Beliakoff, 2000; Rose, 2002). Therefore, the effect of mutations in the splice recognition sites of the OsTub6LI were tested in our transient rice expression system. Two mutants were produced: the first, pOsTub6-3′ss, with a short deletion affecting the PstI site, leading to an AG to CG transition at the 3′ splice site and the second, pOsTub6-5′-3′ss, with an additional single nucleotide substitution at the 5′ splice site (GT to AT). Since the intron is located upstream of the ATG, intron retention cannot affect the protein frame. Furthermore, no ATG triplets are present within the OsTub6 intron that can lead to aberrant, inactive protein production.
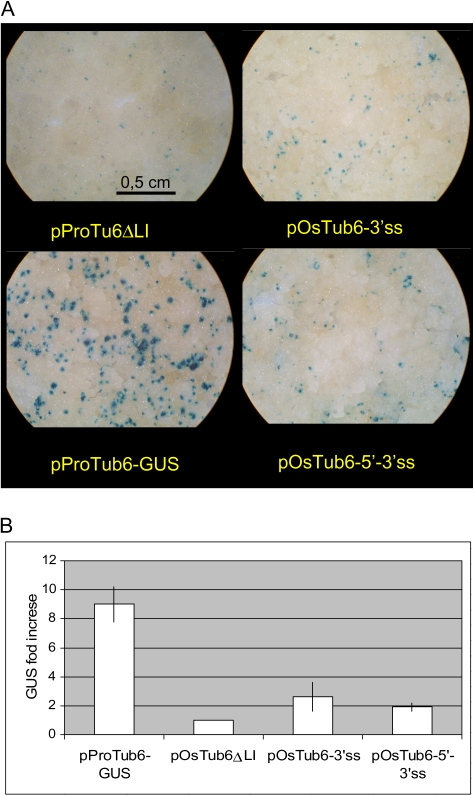
GUS activity in rice calli transformed with either splice site mutated plasmid was equal or only slightly higher than that observed with the intronless plasmid, indicating that impairing splicing significantly alters the intron-mediated enhancing effect (Fig. 4).
Fig. 4.
Effect of the OsTub6 intron splice sites mutations on GUS activity. (A) GUS histochemical assay of representative rice callus samples transformed with the indicated plasmids. (B) GUS activity enhancement with respect to the intronless plasmid, measured in three independent experiments with triplicate samples. Mean values ±SD.
Our results are thus in agreement with those obtained with the maize Sh1 gene, suggesting that a difference may exist between monocots (cereals, at least) and dicots.
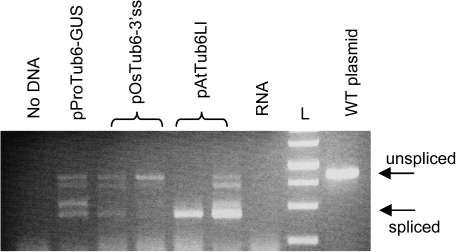
Effect of splice site mutations on splicing
Effective splicing inhibition in rice calli independently transformed with the two plasmids carrying the intron splice site mutations was actually tested by RT-PCR amplification with the use of primers flanking the intron. Compared with the wild-type plasmid, the amount of the spliced mRNA form was dramatically reduced when plasmid pOsTub6-3′ss was used for transformation and became barely detectable in cells transformed with pOsTub6-5′-3′ss, even after 40 PCR cycles (Fig. 5A), suggesting a strong, although not complete, inhibition of splicing. No bands co-migrating with the spliced RNA form were amplified from a cDNA sample from mock-treated cells or in the PCR mix with no added DNA (not shown). The unspliced transcript, absent from either the control RNA sample or the cDNA from callus transformed with the wild-type plasmid, was instead amplified from samples transformed with the two mutagenized plasmids, although at a very low level. The experiment was repeated on a different sample set with similar results. Thus, besides inefficient splicing, a strong reduction in the total level of mRNA was observed as a consequence of the introduced mutations.
Fig. 5.
Effect of mutations at the splice sites of the Ostub6 intron on splicing efficiency in rice cells. (A) RT-PCR amplification of cDNA derived from duplicate rice callus samples transformed with the indicated plasmids, with primers flanking the intron. (B) Sequence of the intron borders of the PCR-amplified spliced products shown by circles in panel A, compared to the DNA sequence of the corresponding plasmids. (This figure is available in colour at JXB online.)
The faint bands in Fig. 5A (numbers 2, 3, and 4), probably corresponding to the spliced transcript, were then analysed to verify if proper splicing could actually have occurred despite the presence of splice-site mutations. They were sequenced and it was found that both mutants were still able to produce small amounts of correctly spliced transcripts (cDNA 3 and 4; Fig. 5B). This suggests that the correct splicing position could still be selected, irrespective of the presence of the canonical GT/AG dinucleotides. It is likely that other strong signals, like the recognition of a strong transition point between AT-rich and GC-rich regions (Lou et al., 1993), were sufficient to drive the correct recognition of splice sites even in the absence of the original consensus dinucleotides. This is similar to what was reported for a natural Shrunken2 maize mutant, where the correct though inefficient, recognition of a 3′ splice site bearing a G to A transition was also observed (Lal et al., 1999a).
Fig. 6.
Effect of the different introns on GUS mRNA level. RT-PCR analysis of duplicated samples, transformed with the indicated plasmids. (A) GUS primers; (B) GFP primers.
In one case (cDNA2, Fig. 5B), we also found that the mutation in the 3′ splice site was enough to cause an incorrect recognition of the wild-type 5′ splice site, causing a sliding of five nucleotides at the intron 5′ splice site. The nucleotide sequence of this specific, aberrant cDNA was different from the DNA sequence of any plasmids used in these experiments. This rules out any possibility that the amplified bands were the result of an artefact due to plasmid contamination.
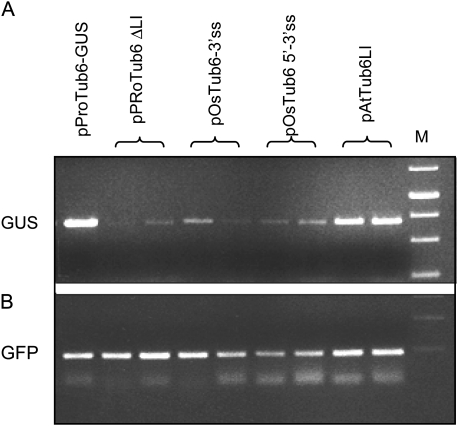
GUS activity correlates with transcript amount
To confirm that the low level of GUS activity, observed upon transformation with the splicing defective plasmids or with the intronless plasmid, was related to a reduction of the transcript amount, as already reported by others and suggested by our RT-PCR experiments, the steady-state mRNA level was determined through semi-quantitative end-point RT-PCR on cDNA produced from transiently transformed rice cells. As a reference, transiently transformed callus samples expressing high GUS activity, sustained by pProTub6-GUS and pAtTub6-GUS plasmids, were used. Comparison of the mRNA amounts was done by amplifying, by RT-PCR, a fragment of the GUS gene that was then analysed by agarose gel electrophoresis. As a control for transformation efficiency, a short GFP fragment was also amplified from the same cDNA samples Fig. 6B. The number of cycles, 27 and 28, respectively, was kept in the range of linearity (not shown).
As shown in Fig. 6A, mRNA accumulation, induced by the intronless plasmid and by plasmids bearing mutations at the splice sites (all associated with low GUS activity; see Fig. 4), was considerably reduced with respect to that induced by pProTub6-GUS and pAtTub6-GUS.
Thus, a high level of GUS activity sustained by plasmids bearing a spliceable intron was paralleled by a high accumulation of mRNA, while a low level of GUS activity, sustained by the unspliceable plasmids or by the intronless construct, was related to a low steady-state mRNA level. This evidence suggests that, in our system, IME acts mainly by increasing the mRNA steady-state level, influencing either transcript stability or transcription rate. However, in the case of the plasmids bearing the unspliceable intron, a feedback mechanism, mediated by a reduced translational efficiency due to the long, unprocessed 5′-UTR may also have contributed to reduce the steady-state level of mRNA further.
Efficient splicing is also required for IME in Arabidopsis cells
Intron splice site recognition differs between monocot and dicot plants. While monocot cells can efficiently splice most dicot plant introns as well as animal introns, dicot cells often fail to recognize and splice monocot introns correctly (Goodall and Filipowicz, 1991). Since intron recognition is related to their higher AT content with respect to exons, this occurrence is generally ascribed to the difference in the AT/GC ratio between the genomes of these two systematic groups. Therefore, some of our plasmids were tested in a dicot system by transient transformation of Arabidopsis cell cultures. The pProTub6-GUS vector sustained very low GUS activity in Arabidopsis cell (very tiny spots, activity not measurable with GUS fluorogenic assay; not shown), indicating that the rice promoter is not as strong as in rice cells. Furthermore, GUS expression was not significantly different from that sustained by the intronless vector, suggesting that the intron did not exert any positive or negative effect. However, the replacement of the rice OsTub6 intron with the leader intron of its putative orthologue, the AtTub6 gene of Arabidopsis, boosted GUS gene expression in cells, resulting in a significantly higher number of blue foci, all of bigger size and of higher colour intensity (Fig. 7). This is an important observation since it demonstrates that an endogenous intron can compensate for the low level of expression resulting from a weak promoter.
Fig. 7.
Effect of two different tubulin introns on GUS expression in tobacco, Arabidopsis, and rice. Arabidopsis suspension cells, rice calli, and tobacco leaves were transiently transformed with the three indicated plasmids and GUS activity was tested by histochemical analysis. Representative samples are shown.
Very similar results were obtained upon transient transformation of tobacco (Nicotiana tabacum) leaves, where stronger GUS expression was again observed when the rice OsTub6 promoter was coupled to the AtTub6 intron (Fig. 7). Results by simple spot counting are summarized in Table 2. Similar to rice, enhanced GUS activity in Arabidopsis cells was related to increased mRNA level, as demonstrated by RT-PCR analysis (not shown).
Table 2.
IME effect in different plant systems
| Target tissue | Transforming plasmid | ||
| pProTub6GUS | pAtTub6LI | pOsTub65′-3′ss | |
| Rice cells | 9 | 102 | |
| Arabidopsis cells | 1–1.5 | 2–3 | 1 |
| Tobacco leaves | 1 | 5-6 | 1 |
GUS activity is expressed as fold increase with respect to the activity induced by the intronless plasmid.
When looking at splicing efficiency, it was observed that splicing of the rice intron in Arabidopsis was inefficient, even when the wild-type rice intron was used (Fig. 8), since the unspliced and two mis-spliced forms were amplified by RT-PCR, together with a low amount of the spliced mRNA. Conversely, splicing of the Arabidopsis intron was far more effective, although not complete, producing a more intense band corresponding to the spliced transcript and accounting for the higher level of GUS expression.
Fig. 8.
Splicing efficiency of different introns in Arabidopsis suspension cells. RT-PCR analysis was performed as in Fig. 5, from duplicated cell samples transformed with the indicated plasmids.
This observation indicates that the enhancing role of introns also depends on splicing in dicot cells. Furthermore, it demonstrates that the presence of a more efficiently spliced intron can rescue a poor promoter activity by increasing the steady-state level of the mRNA.
Since splice sites context and exon sequences were the same in both plasmids, it can be concluded that the intron sequence itself is very important in determining splicing efficiency in Arabidopsis.
Discussion
Modulation of expression and intron-specificity
Many plant introns have been found to enhance gene expression when inserted within a transcription unit, in transgenic cells or plants, with effects varying between 5–6-fold to 1000-fold among different introns. However, there many factors that can determine intron effect on transgene expression level: the promoter, the reporter gene, the intron position, and the length and sequence of the flanking exons, so that it is often difficult to extrapolate the real contribution from the intron.
Intron specificity was clearly demonstrated in a dicot expression system, in a detailed comparative work in which the native Arabidopsis TRP1 intron within a TRP1::GUS1 fusion construct (promoter, leader, first exon and intron, fused in-frame with GUS), was precisely replaced by six different Arabidopsis introns, inserted in exactly the same position, resulting in different levels (3–15-fold) of enhancement of GUS steady-state mRNA level in transgenic Arabidopsis plants (Rose, 2002).
Comparable results were obtained by testing the effect on GUS expression of seven different rice introns and one from Arabidopsis, in a monocot expression system. Introns inserted in an identical context in the Ostub6 leader sequence, with identical flanking exons and splice sites, resulted in reproducibly different enhancement of GUS activity (4–17-fold) measured in rice cells. Such a difference was actually dependent only on the intron sequence.
Our results confirm the idea that specific determinants within introns are able to modulate downstream gene expression through a more complex mechanism than a simple on/off switch and that what was observed in Arabidopsis is also true for rice. Evidence from deletion experiments suggests that such signals must be redundant since great parts of an intron can be deleted with no or slight effect on IME (Luehrsen and Walbot, 1991; Clancy and Hannah, 2002). Portions of the intron1 sequence of Arabidopsis UBQ1 can be deleted individually and replaced by sequences of a non-enhancing intron without a significant reduction of enhancement (Rose et al., 2008). The presence of an informational content within introns is also evident from recent observations relating to tissue-specific effects of introns on gene expression (Jeon et al., 2001; Jeong et al., 2006, Kim et al., 2006).
Such specificity may rely on sequence or conformational determinants interacting with unknown tissue-specific elements. Extensive sequence homology is hardly found among enhancing introns from putative orthologue genes in distantly related species, although a higher level of sequence conservation among first introns with respect to downstream ones has been observed, (Bradnam and Korf, 2008). This suggests that sequence determinants, necessary for intron function, must be short or highly degenerated.
IMEter testing
Despite evidence for their existence, IME determinants have so far remained elusive. In one case, a specific T-rich region required for IME (but not for splicing) has been detected through deletion experiments (Clancy and Hannah, 2002) but no general roles or conclusions have been defined. Although in Arabidopsis an increase in the IME effect was obtained by increasing the intron T-content (Rose, 2002), no correlation was found between the T content or number of T-stretches in our intron set and the degree of enhancement. Complete splicing of all tested introns (not shown) in rice cells suggests that all of them fulfil the minimal sequence requirements for optimal splicing and the wide range of these parameters (Table 1) is in accordance with the great tolerance of monocots in this respect.
IMEter testing of our intron set, although not indicating a significant linear correlation between intron scoring and induced GUS activity, confirmed that this algorithm, previously experimentally tested only in Arabidopsis, may also have some predictive value for rice introns. This in view of the fact that the IMEter identifies as potentially enhancing (positively scoring, all with values over 30) seven introns out of eight in our set, while the vast majority of rice introns were reported to have a negative score (Rose et al., 2008). We believe that our experimentally uniform dataset, still missing for rice, could be useful to modify the parameters of the IMEter algorithm in view of its optimization for the rice model, as it is the case for Arabidopsis. Further research is however required to identify, inside rice promoter-proximal introns, those specific signatures that differentiate them from all other introns and that may figure as quantitative indicator of their enhancing capabilities. A linear correlation like that observed in Arabidopsis was even surprising, when considering two aspects: first, the degree of enhancement is not an absolute feature for one specific intron but strongly depends on the flanking exons (Luehrsen and Walbot, 1991). Second, being an intron or an exon is not always an intrinsic feature of a sequence but may even be a contingency: switches between the two conditions is the basis of alternative splicing.
The IMEter score was also calculated for the first intron within the ORF of all rice tubulin genes (eight β and three α), all in the same conserved position (Table 2; see Supplementary Table S2 at JXB online) It was found that five of them are negative-scoring introns: however, of these, one is moderately negative (–0.69) and two are not the first intron of the gene since an upstream intron is present in the leader sequence. In future, it would be interesting to test the IME effect of these negative scoring introns in our experimental system, to further evaluate the qualitative predictivity of the IMEter.
A search was made for the motif most commonly found in high-scoring rice introns, suggested by Rose et al. (2008) as a putative enhancing signal, and found it present at least once in all but two of our introns (Fig. 3A). Its presence was more relevant (five–six times) in the OsTua2 and Ostua3 introns, which are the only two ORF introns of our set. We speculate that this may reflect different features of IME motifs between leader introns and ORF introns, in accordance with previous work reporting distinct properties for the two classes (Chung et al., 2006; Hong et al., 2006; Bradnam and Korf, 2008). Since a minority of genes have introns in their leader rather then in their coding region (about 19% versus 72% in Arabidopsis), and since each 5′-UTRs rarely hosts more then one intron, leader introns are likely to be underrepresented in the dataset used to identify putative IME motifs. It is thus possible that IME signals may differ between the two intron classes and that those identified by Rose et al. (2008) are the most represented in introns of the coding sequence class. A further analysis that distinguishes these two classes of introns may help in identifying new or more specific sequences.
Need for splicing
Much circumstantial evidence has generally favoured the hypothesis that IME is strictly related to splicing. Correlation between intron splicing efficiency and enhanced IME activity was found through deletion experiments of the maize Adh1 intron in maize protoplasts (Luehrsen and Walbot, 1991). Furthermore, expression level of reporter genes were different when the same intron was flanked by different exons (Luehrsen and Walbot, 1991) which fits with the observation that splicing efficiency is the result of a combinatorial effect of exon, intron, and splice site sequences (Carle-Urioste et al., 1997; Latijnhouwers et al., 1999). Point mutations at the splice sites resulted in the abolition of reporter gene expression enhancement in different monocot systems (Mascarenhas et al., 1990; Sinibaldi and Mettler, 1992; Clancy and Hannah, 2002). Although splicing efficiency was not always documented, the early interpretation was that the mechanism of stimulation was dependent only on splicing per se (Mascarenas, 1990; Luehrsen and Walbot, 1991). Our data are in agreement with these reports, showing that plasmids carrying mutations at the intron splice sites could not significantly enhance GUS expression, measured at either protein or mRNA level, similar to plasmids lacking the intron. Conversely, other authors suggested that splicing is not required for IME. By testing deletion mutants of the PhADF1 intron (Jeong et al., 2007), no correlation was found between IME effect and splicing efficiency in transgenic Arabidopsis plants. However, splicing efficiency was inferred from the pre-RNA/mRNA ratio (measured by Real-Time experiments) which does not take into account different stability of the two forms. Other works, neglecting the contribution of splicing to IME, were based on the analysis of GUS transcriptional expression from two unspliceable mutants of a TRP1::GUS fusion plasmid, in transgenic Arabidopsis plants (Rose and Beliakoff, 2000; Rose, 2002). Such mutants were shown to increase mRNA accumulation relative to similar constructs lacking introns, leading to the conclusion that splicing was not required for IME. However no data on the protein level or activity were reported, so it is not known to what extent the accumulated transcript, spliced or unspliced, was translated. Splicing could actually affect translation as reported in the case of three introns of the maize RpoT gene, inducing highly differential levels of LUC activity that did not correspond to a similar difference in mRNA accumulation (Bourdon et al., 2001). Intron retention itself may also have affected normal RNA turnover, since the intron sequence was intentionally modified in order to place its premature termination codons out of frame, but the whole sequence in-frame with GUS. This genetic engineering work may have erased some signatures that are normally required for targeting unspliced mRNA for degradation through the Nonsense Mediated Decay pathway. This may have caused a non-physiological increase in the stability of the unspliced mRNA, yet unrelated to IME.
In conclusion, our results favour the idea that splicing, although not sufficient per se, is, nevertheless, required for IME. Whatever the mechanism, splicing of the OsTub6 intron occurring in its native location is needed for enhanced GUS expression. Additional alternative mechanisms, specific to the different experimental systems, may also contribute.
Intron effect in different plant systems
Splicing mechanisms show slightly different specificities between the two groups of angiosperms. Dicot introns have an higher mean AT content compared to monocot introns (71% and 61%, respectively) a feature that is important for efficient splicing (Goodall and Filipowicz, 1991; Gniadkowski et al., 1996), while monocots have less strict requirements: in fact animal and dicot introns are generally efficiently spliced in monocots, but not vice versa (Goodall and Filipowicz, 1991; Lou et al., 1993).
Since evidence that IME relies on splicing comes exclusively from monocot plants, while the reverse has only been proposed from data on dicots, some of our plasmids were exploited to test the relationship between IME and splicing, in different plant systems. GUS expression driven by the OsTub6 promoter-leader sequence was shown to be lower in Arabidopsis cell cultures than in rice cells, as expected for a monocot promoter in dicot cells. Furthermore, the presence of the corresponding rice intron had no effect on both GUS activity or GUS mRNA level. This evidence correlated with a partial and inaccurate splicing of the rice intron in Arabidopsis cells suggesting that this was the cause of the lack of enhancement. Splicing inefficiency is probably due to the requirement of an higher intron AT content of dicots, not fulfilled by the OsTtub6 intron (53% AT). The poorly spliceable rice intron in Arabidopsis thus mimicked the situation of the splice-site mutated intron in rice, giving exactly the same result. The fact that splicing of the AtTub6 intron, although much more efficient, was nevertheless incomplete, despite its elevated AT content, confirmed the importance of the flanking exon sequences (coming from the rice tubulin gene) for splicing.
The evidence that enhancing monocot introns fail to increase reporter gene expression in a dicot plant is not totally new (Maas et al., 1991; Last et al., 1991, Xu et al., 1994), but splicing efficiency was tested and demonstrated to be an associated process in only one previous case (Tanaka et al., 1990).
It is, therefore, concluded that similar mechanisms are controlling IME in the two different plant systems and that splicing inefficiency is the most probable cause of IME failure by otherwise enhancing introns. If an enhancing intron, is efficiently processed, it can have a strong IME effect both in rice and in Arabidopsis cells. Conversely, in the absence of introns, expression is kept at a low level. A similar result was also observed by testing the same plasmids in tobacco leaves: the plasmid pAtTub6LI sustained a much higher level of gene expression then pProTub6-GUS, confirming that similar rules are valid for two distantly related dicot plants.
In our experiments, expression analysis was mainly performed at the protein level, by measuring GUS activity. However, RT-PCR data collected from intronless plasmids or plasmids carrying mutations in the splicing sites, show a decrease in GUS mRNA that parallels that of GUS activity thus confirming the general evidence that most of the observed effect occurs at the transcript level. However, the small enhancement of GUS expression observed might somehow be underestimated, considering that a partial inhibition of translation may be caused by the increased length of the 5′-UTR.
The strong enhancing effect of the AtTub6 intron suggests that its function has been preserved throughout evolution. Both AtTtub6 and OsTub6 belong to a subfamily of plant β tubulin genes (class I; Oakley et al., 2007) that is characterized by the presence of an intron in the 5′-UTR. The putative Arabidopsis orthologue is endowed with IME activity like two of the three members of the rice subfamily (Morello et al., 2002; Gianì et al., 2009), despite no evident sequence homology being found. Such a functional conservation among introns has also been observed for the Petunia and Arabidopsis ADF1 genes (Jeong et al., 2007), both endowed with IME activity as well as for introns of the ubiquitin gene family (Christensen et al., 1992; Sivamani and Qu, 2006).
Conclusions
The experimental system set up here allowed the systematic analysis of the IME effect of different introns, within an identical and native context, through transient assays in different model plant systems.
Our results strongly support the conclusion that splicing, although required, is not sufficient to sustain IME. Additional intron sequence specificities must be present to modulate the enhancing properties exhibited by different introns placed in the same location. Such IME signals should influence the magnitude of the effect and their abundance or strength can reasonably be predicted by the IMEter program in Arabidopsis but not in rice. However, experimental evidence for their existence is still lacking. Sequence comparison among enhancing introns may help to identify putative IME signals whose effectiveness needs to be experimentally tested. We believe that our experimental system is suitable for testing such candidate sequences. This should allow the identification of specific trans-acting regulators, proteins or regulatory RNAs capable of recognizing the IME signals. Due to the difficulty associated with dissecting the complex interplaying mechanisms supporting IME, we believe that starting from intron specificities could provide a new approach to step further into the understanding of intron-mediated enhancement of gene expression.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. Sequence of the primers used for cloning and RT-PCR.
Supplementary Table S2. IMEter score of first introns of all rice tubulin genes.
Supplementary Material
Acknowledgments
This work was fully supported by funding from Consiglio Nazionale delle Ricerche.
References
- Bourdon V, Harvey A, Lonsdale DM. Introns and their positions affect the translational activity of mRNA in plant cells. EMBO Reports. 2001;2:394–398. doi: 10.1093/embo-reports/kve090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam KB, Korf I. Longer first introns are a general property of eukaryotic gene structure. PLoS ONE. 2008;3:e3093. doi: 10.1371/journal.pone.0003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J, Fromm M, Walbot V. Introns increase gene expression in cultured maize cells. Genes and Development. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- Carle-Urioste JC, Brendel V, Walbot V. A combinatorial role for exon, intron and splice site sequences in splicing in maize. The Plant Journal. 1997;11:1253–1263. doi: 10.1046/j.1365-313x.1997.11061253.x. [DOI] [PubMed] [Google Scholar]
- Chaubet-Gigot N, Kapros T, Flenet M, Kahn K, Gigot C, Waterborg JH. Tissue-dependent enhancement of transgene expression by introns of replacement histone H3 genes of Arabidopsis. Plant Molecular Biology. 2001;45:17–30. doi: 10.1023/a:1006487023926. [DOI] [PubMed] [Google Scholar]
- Christensen AH, Sharrock RA, Quail PH. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Molecular Biology. 1992;18:675–689. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- Chung BYW, Simons C, Firth AE, Brown CM, Hellens RP. Effect of 5′-UTR introns on gene expression in Arabidopsis thaliana. BMC Genomics. 2006;7:120. doi: 10.1186/1471-2164-7-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy M, Hannah LC. Splicing of the maize Sh1 first intron is essential for enhancement of gene expression, and a T-rich motif increases expression without affecting splicing. Plant Physiology. 2002;130:918–929. doi: 10.1104/pp.008235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy M, Vasil V, Hannah LC, Vasil IK. Maize Shrunken-1 intron and exon regions increase gene expression in maize protoplasts. Plant Science. 1994;98:151–161. [Google Scholar]
- Curi G, Chan RL, Gonzales DH. The leader intron of Arabidopsis thaliana genes encoding cytochrome c oxidase subunit 5c promotes high level expression by increasing transcript abundance and translation efficiency. Journal of Experimental Botany. 2005;56:2563–2571. doi: 10.1093/jxb/eri250. [DOI] [PubMed] [Google Scholar]
- Curti G, Massardi F, Lado P. Synergistic activation of the membrane H+-ATPase in Arabidopsis thaliana cells by turgor decrease by fusicoccin. Physiologia Plantarum. 1993;87:592–600. [Google Scholar]
- Deyholos MK, Sieburth LE. Tissue-dependent enhancement of transgene expression by introns of replacement histone H3 genes of Arabidopsis. The Plant Cell. 2000;12:1799–1810. [Google Scholar]
- Fiume E, Christou P, Gianì S, Breviario D. Introns are key regulatory elements of rice tubulin expression. Planta. 2004;218:693–703. doi: 10.1007/s00425-003-1150-0. [DOI] [PubMed] [Google Scholar]
- Fu H, Kim SY, Park W. A potato sucrose synthase gene contains a context-dependent 3′ element and a leader intron with both positive and negative tissue-specific effects. The Plant Cell. 1995;7:1395–1403. doi: 10.1105/tpc.7.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianì S, Altana A, Campanoni P, Morello L, Breviario D. In transgenic rice, α- and β-tubulin regulatory sequences control GUS expression amount and distribution through intron mediated enhancement and intron dependent spatial expression. Transgenic Research. 2009;18:151–162. doi: 10.1007/s11248-008-9202-7. [DOI] [PubMed] [Google Scholar]
- Giedeckel M, Jimenez B, Herrera-Estrella L. The first intron of the Arabidopsis thaliana gene coding for elongation factor 1b contains an enhancer-like element. Gene. 1996;170:201–206. doi: 10.1016/0378-1119(95)00837-3. [DOI] [PubMed] [Google Scholar]
- Gniadkowski M, Hemmings-Mieszczak M, Klahre U, Liu HX, Filipowicz W. Characterisation of intronic uridine-rich sequence elements acting as possible targets for nuclear proteins during pre-mRNA splicing in Nicotiana plumbaginifolia. Nucleic Acids Research. 1996;24:619–627. doi: 10.1093/nar/24.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall GJ, Filipowicz W. Different effects of intron nucleotide composition and secondary structure on pre-mRNA splicing in monocot and dicot plants. EMBO Journal. 1991;10:2635–2644. doi: 10.1002/j.1460-2075.1991.tb07806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K, Scofield G, Lynch M. Intron size, abundance and distribution within untranslated regions of genes. Molecular Biology Evolution. 2006;23:2392–2404. doi: 10.1093/molbev/msl111. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SJ, Kim C, An G. Tissue-preferential expression of a rice α-tubulin gene, OsTUA1, mediated by the first intron. Plant Physiology. 2001;123:1005–1014. doi: 10.1104/pp.123.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y-M, Mun JH, Kim H, Lee SY, Kim SG. An upstream region in the first intron of petunia actin-depolymerizing factor 1 affects tissue-specific expression in transgenic Arabidopsis. The Plant Journal. 2007;50:230–239. doi: 10.1111/j.1365-313X.2007.03053.x. [DOI] [PubMed] [Google Scholar]
- Jeong Y-M, Mun J-H, Lee I, Woo JC, Hong CB, Kim S- G. Distinct roles of the first introns on the expression of Arabidopsis profilin gene family members. Plant Physiology. 2006;140:196–209. doi: 10.1104/pp.105.071316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Kim H, Shin JS, Chung C-H, Ohlrogge JB, Suh MC. Seed-specific expression of sesame microsomal oleic acid desaturase is controlled by combinatorial properties between negative cis-regulatory elements in the SeFAD2 promoter and enhancers in the 5′-UTR intron. Molecular Genetics and Genomics. 2006;276:351–368. doi: 10.1007/s00438-006-0148-2. [DOI] [PubMed] [Google Scholar]
- Lal S, Choi J-H, Hannah LC. the AG dinucleotide terminating introns is important but not always required for pre-mRNA splicing in the maize endosperm. Plant Physiology. 1999;120:65–72. doi: 10.1104/pp.120.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last DL, Brettell RIS, Chamberlain DA, Chaudhury AM, Larkin PJ, Marsh EL, Peacock WJ, Dennis ES. pEmu: an improved promoter for gene expression in cereal cells. Theoretical and Applied Genetics. 1991;81:581–588. doi: 10.1007/BF00226722. [DOI] [PubMed] [Google Scholar]
- Latijnhouwers MJ, Pairoba CF, Brendel V, Walbot V, Carle-Urioste JC. Test of the combinatorial model of intron recognition in a native maize gene. Plant Molecular Biology. 1999;41:637–644. doi: 10.1023/a:1006329517740. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. TRENDS in Biochemical Science. 2003;28:215–221. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Lou H, McCullough AJ, Schuler MA. 3′ Splice site selection in dicot plant nuclei is position dependent. Molecular and Cellular Biology. 1993;13:4485–4493. doi: 10.1128/mcb.13.8.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Sivamani E, Azhakanandam K, Samadder P, Li X, Qu R. Gene expression enhancement mediated by the 5′-UTR intron of the rice rubi3 gene varied remarkably among tissues in transgneic rice plants. Molecular Genetics and Genomics. 2008;279:563–572. doi: 10.1007/s00438-008-0333-6. [DOI] [PubMed] [Google Scholar]
- Luehrsen KR, Walbot V. Intron enhancement of gene expression and the splicing efficiency of introns in maize cells. Molecular Genetics and Genomics. 1991;225:81–93. doi: 10.1007/BF00282645. [DOI] [PubMed] [Google Scholar]
- Maas C, Laufs J, Grant S, Korfhage C, Werr W. The combination of a novel stimulatory element in the first exon of the maize Shrunken-1 gene with the following intron 1 enhances reporter gene expression up to 1000-fold. Plant Molecular Biology. 1991;16:199–207. doi: 10.1007/BF00020552. [DOI] [PubMed] [Google Scholar]
- Mascarenhas D, Mettler IJ, Pierce DA, Lowe HW. Intron-mediated enhancement of heterologous gene expression in maize. Plant Molecular Biology. 1990;15:913–920. doi: 10.1007/BF00039430. [DOI] [PubMed] [Google Scholar]
- Morello L, Bardini M, Cricrì M, Sala F, Breviario D. Functional analysis of DNA sequences controlling the expression of the rice OsCDPK2 gene. Planta. 2006;223:479–491. doi: 10.1007/s00425-005-0105-z. [DOI] [PubMed] [Google Scholar]
- Morello L, Bardini M, Sala F, Breviario D. A long leader intron of the OsTUB16 rice β-tubulin gene is required for high-level gene expression and can autonomously promote transcription both in vivo and in vitro. The Plant Journal. 2002;29:33–44. doi: 10.1046/j.0960-7412.2001.01192.x. [DOI] [PubMed] [Google Scholar]
- Mun JH, Lee SY, Jeong YM, Shin MY, Kim H, Lee I, Kim SG. Petunia actin depolymerizing factor is mainly accumulated in vascular tissue and its gene expression is enhanced by the first intron. Gene. 2002;292:233–243. doi: 10.1016/s0378-1119(02)00646-7. [DOI] [PubMed] [Google Scholar]
- Oakley RV, Wang Y, Ramakrishna W, Harding S, Tsai C. Differential expansion and expression of α- and β-tubulin gene families in Populus. Plant Physiology. 2007;145:961–973. doi: 10.1104/pp.107.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rethmeier N, Seurinck J, Van Montangu M, Cornelissen M. Intron-mediated enhancement of transgene expression in maize is a nuclear, gene-dependent process. The Plant Journal. 1997;12:895–899. doi: 10.1046/j.1365-313x.1997.12040895.x. [DOI] [PubMed] [Google Scholar]
- Rose AB. Requirements for intron-mediated enhancement of gene expression in Arabidopsis. RNA. 2002;8:1444–1453. doi: 10.1017/s1355838202020551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AB. The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. The Plant Journal. 2004;40:744–751. doi: 10.1111/j.1365-313X.2004.02247.x. [DOI] [PubMed] [Google Scholar]
- Rose AB, Beliakoff JA. Intron-mediated enhancement of gene expression independent of unique intron sequences and splicing. Plant Physiology. 2000;122:535–542. doi: 10.1104/pp.122.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AB, Elfersi T, Parra G, Korf I. Promoter-proximal introns in Arabidopsis thaliana are enriched in dispersed signal and elevate gene expression. The Plant Cell. 2008;20:543–551. doi: 10.1105/tpc.107.057190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AB, Last RL. Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene Pat1. The Plant Journal. 1997;11:455–464. doi: 10.1046/j.1365-313x.1997.11030455.x. [DOI] [PubMed] [Google Scholar]
- Samadder P, Sivamani E, Lu J, Li X, Qu R. transcriptional and post-transcriptional enhancement of gene expression by the 5′-UTR intron of rice rubi3 gene in transgenic rice cells. Molecular Genetics and Genomics. 2008;279:429–439. doi: 10.1007/s00438-008-0323-8. [DOI] [PubMed] [Google Scholar]
- Sinibaldi RM, Mettler IJ. Intron splicing and intron-mediated enhanced expression in monocots. Progress in Nucleic Acid Research, Molecular Biology. 1992;42:229–255. doi: 10.1016/s0079-6603(08)60577-2. [DOI] [PubMed] [Google Scholar]
- Sivamani E, Qu R. Expression enhancement of a rice polyubiquitin gene promoter. Plant Molecular Biology. 2006;60:225–239. doi: 10.1007/s11103-005-3853-z. [DOI] [PubMed] [Google Scholar]
- Snowden KC, Bucholz WG, Hall T. Intron position affects expression of the tpi promoter in rice. Plant Molecular Biology. 1996;31:689–692. doi: 10.1007/BF00042241. [DOI] [PubMed] [Google Scholar]
- Snustad DP, Haas NA, Kopczak SD, Silflow CD. The small genome of Arabidopsis contains at least nine expressed beta-tubulin genes. The Plant Cell. 1992;4:549–556. doi: 10.1105/tpc.4.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Mita S, Ohta S, Kyozuka J, Shimamoto K, Nakamura K. Enhancement of foreign gene expression by a dicot intron in rice but not in tobacco is correlated with an increased level of mRNA and an efficient splicing of the intron. Nucleic Acids Research. 1990;18:6767–6770. doi: 10.1093/nar/18.23.6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorakis NG, Cleveland DW. Physical evidence for cotranslational regulation of beta-tubulin mRNA degradation. Molecular and Cellular Biology. 1992;12:791–799. doi: 10.1128/mcb.12.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A, Wu RJ, Cheng Z, Meagher RB. Multiple conserved 5′ elements are required for high-level pollen expression of the Arabidopsis reproductive actin ACT1. Plant Molecular Biology. 2003;52:1135–1151. doi: 10.1023/b:plan.0000004309.06973.16. [DOI] [PubMed] [Google Scholar]
- Weise A, Lalonde C, Kuhn C, Frommer WB, Ward JM. Introns control expression of sucrose transporter LeSUT1 in trichomes, companion cells and in guard cells. Plant Molecular Biology. 2008;68:251–262. doi: 10.1007/s11103-008-9366-9. [DOI] [PubMed] [Google Scholar]
- Xu Y, Yu H, Hall TC. Rice triosephosphate isomerase gene 5′ sequence directs β-glucuronidase activity in transgenic tobacco but requires an intron for expression in rice. Plant Physiology. 1994;106:459–467. doi: 10.1104/pp.106.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.