Abstract
Ubiquitination plays important roles in plant growth and development. Whereas ubiquitin-dependent protein degradation and modulation in the cytoplasm and nucleus are well established in plants, ubiquitination events mediated by E3 ubiquitin ligases at the plasma membrane are largely unknown. Here, it is demonstrated that the suppressor of premature senescence and cell death SENESCENCE-ASSOCIATED UBIQUITIN LIGASE 1 (SAUL1), a plant U-box armadillo repeat (PUB-ARM) E3 ubiquitin ligase, localizes at the plasma membrane. Among the members of the PUB-ARM protein family, this localization is unique to SAUL1 and its two closest homologues. A novel armadillo repeat domain was identified at the SAUL1 C-terminus that directs specific association with the plasma membrane and is crucial for SAUL1 function in vivo. The data suggest that a small subgroup of PUB-ARM proteins including SAUL1 have functions at the plasma membrane probably by modifying target proteins by ubiquitination.
Keywords: Armadillo repeat, ARM repeat, E3 ubiquitin ligase, plasma membrane, SAUL1, U-box
Introduction
The regulation of many cellular processes as important as transcription and cell cycle control, and a plethora of physiological responses require specific degradation of key regulatory proteins. In most cases, the ubiquitin–26S proteasome pathway is important for regulated proteolysis by mediating polyubiquitination and thus degradation of target proteins. This pathway comprises ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), ubiquitin ligases (E3s), the proteasome complex, and de-ubiquitinating enzymes (DUBs) (Kerscher et al., 2006; Vierstra, 2009). The pathway also governs monoubiquitination of target proteins to modulate protein functions in processes such as endocytosis and internalization of plasma membrane proteins (Sigismund et al., 2004). Binding of target proteins prior to ubiquitination occurs via E3 ubiquitin ligases, thus defining the specificity of the ubiquitin–26S proteasome pathway.
In various eukaryotes, large numbers of E3 ubiquitin ligases are found. In the model plant Arabidopsis thaliana probably >1300 E3s exist that can be grouped into four main types: HECT, RING, U-box, and cullin-RING ligases (including SCF E3s). Together, RING-containing and SCF E3s/F-box proteins account for the majority of E3 ligases and cover essential functions in plant biology such as organ morphogenesis and hormone signalling (Schwechheimer and Calderon Villalobos, 2004; Stone and Callis, 2007; Santner and Estelle, 2009; Vierstra, 2009). In contrast, the plant U-box (PUB) protein family representing the most recently identified type of E3 ligases contains only 64 predicted members (Azevedo et al., 2001; Wiborg et al., 2008). This protein family is characterized by the highly conserved U-box originally described for the yeast UFD2 protein (Koegl et al., 1999). The U-box motif consists of ∼70 amino acids and resembles a modified RING-domain that lacks crucial residues for metal binding (Aravind and Koonin, 2000). It has been demonstrated that the U-box is essential for activity of these ubiquitin ligases (Hatakeyama et al., 2001; Zeng et al., 2004; Gonzalez-Lamothe et al., 2006).
In addition to the U-box, most PUB proteins carry tandem armadillo (ARM) repeats and thus form the subgroup of PUB-ARM proteins (Mudgil et al., 2004). The ARM nomenclature originates from a motif of ∼42 amino acids discovered in the gene product of the Drosophila segment polarity gene armadillo (Nusslein-Volhard and Wieschaus, 1980; Riggleman et al., 1989). ARM repeats give rise to conserved protein structures. One such repeat consists of three α-helices, and multiple ARM repeats interact and form interfaces for protein–protein interactions (Huber et al., 1997; Conti and Kuriyan, 2000; Coates, 2003). ARM repeat proteins serve diverse functions in eukaryotic cells. The Saccharomyces cerevisiae protein Vac8 is a vacuolar membrane protein involved in vacuole inheritance and vacuolar membrane fusion (Pan and Goldfarb, 1998; Fleckenstein et al., 1998). Importin-α and its homologues function in the transport of proteins into the nucleus in conjunction with importin-β and in inhibition of mitotic spindle formation (Gorlich and Kutay, 1999; Gruss et al., 2001). Recently, it has been shown that a small fraction of importin-α was also detectable in membrane fractions (Hachet et al., 2004). The best-studied ARM protein, β-catenin, has essential roles in cell adhesion through maintenance of cell–cell adherens junctions and in Wnt signal transduction as a transcriptional coactivator. At the cell periphery, β-catenin interacts with the cytoplasmic end of cadherins. Wnt signalling blocks degradation of β-catenin in the cytoplasm via a protein complex including adenomatous polyposis coli (APC) and supports translocation of β-catenin into the nucleus where it interacts with transcription factors (Zhurinsky et al., 2000).
During a plant’s life cycle, PUB-ARM proteins play critical roles in diverse processes. The potato PHOR1 (PHOTOPERIOD RESPONSIVE 1) protein functions in light and gibberellin signalling (Amador et al., 2001). The related protein ARC1 (ARM repeat containing 1) from Brassica is involved in the self-incompatibility response (Stone et al., 2003). In Arabidopsis, PUB22 and PUB23 have been implicated in drought signalling (Cho et al., 2008). In addition, a number of PUB-ARM proteins from different plant species have functions in the pathogen response, including tobacco ACRE276 (Avr/Cf-9 Rapidly elicited 276) and CMPG1, as well as Arabidopsis PUB17, PUB22, PUB23, and PUB24 (Gonzalez-Lamothe et al., 2006; Yang et al., 2006; Trujillo et al., 2008). The rice SPL11 (spotted leaf 11) and the Arabidopsis SAUL1 (SENESCENCE-ASSOCIATED UBIQUITIN LIGASE 1) protein have regulatory functions in cell death/senescence control (Zeng et al., 2004; Raab et al., 2009). To this end, PUB-ARM proteins have been localized in the nucleus, in the cytosol, and in proteasome structures at the endoplasmic reticulum (ER) (Amador et al., 2001; Stone et al., 2003; Cho et al., 2008; Samuel et al., 2008).
In this study, properties of the PUB-ARM E3 ubiquitin ligase SAUL1 that has been described as a suppressor of premature senescence in Arabidopsis (Raab et al., 2009) were further investigated. Protoplast transformation and confocal laser scanning microscopy were applied to study the localization of fusion proteins consisting of different fluorescent proteins and full-length SAUL1 or mutant SAUL1 with different deletions. It could be shown that SAUL1 is localized at the plasma membrane, and five ARM repeats were identified at its C-terminal end that comprise a domain which is crucial for this localization. Furthermore, this protein domain was sufficient to drive plasma membrane association of PUB-ARM proteins that usually reside in the cytoplasm. Impaired plasma membrane association abolished SAUL1 function in vivo. The data suggest that SAUL1 is involved in ubiquitination of proteins at the plasma membrane.
Materials and methods
Cloning of DNA constructs
For generation of fusion proteins between full-length PUB-ARM proteins or SAUL1 carrying terminal truncations and green fluorescent protein (GFP), yellow fluorescent protein (YFP), or haemagglutinin (HA), the respective fragments were amplified from cDNA or genomic DNA using the primer pairs listed in Supplementary Table S1 available at JXB online. The Umsrt1 open reading frame was amplified using primers 5′-CACCATGGCGTCGTCTTCTCC-3′ and 5′-GGATCMTTGTGGACTCGGCTGC-3′ from yeast expression vectors carrying Umsrt1 (Wahl et al., 2010). For generation of N- or C-terminal deletions of the SAUL1 protein, fragments were generated by PCR using the primers depicted in Suppementary Table S2. For deletion of internal ARM repeats, a two-step PCR mutagenesis was performed generating two DNA fragments harbouring the up- and downstream sequences of the corresponding ARM repeat in a first PCR. The primers used in this PCR introduced overlapping DNA sequences over-spanning the corresponding ARM repeat. By using these fragments as templates for a second PCR, DNA fragments lacking the ARM repeats 7–8 or 9 were generated. The amplified fragments were cloned into pENTR/D-TOPO® (Invitrogen, Karlsruhe, Germany), sequenced, and recombined into destination vectors pEARLEYGATE104 and pEARLEYGATE201 (Earley et al., 2006) for the YFP and HA constructs, respectively, pMDC43 for fusion of GFP to the N-terminus (Curtis and Grossniklaus, 2003), pK7FWG2.0 for fusion of GFP to the C-terminus (Karimi et al., 2002), or pH7RWG2.0 for the Umsrt1-RFP construct (Karimi et al., 2002). For generation of the CaMV35S-promoter:PUB27-SAUL1ARM7–11-GFP chimeric constructs, a SAUL1ARM7–11 fragment carrying BamHI restriction sites at both ends was amplified using primers 5′-CAGGATCCATTAGCAACACAGGTCCAG-3′ and 5′-CTTGGATCCTGCGATGTTTGGGAATATAC-3′. The PCR fragment was subcloned into the vector pJET 1.2/blunt (Fermentas, St Leon-Rot, Germany), sequenced, and cloned into the expression vector CaMV35S-promoter:PUB27-GFP via the introduced BamHI sites. The generated plasmids were used for transformation of Arabidopsis mesophyll protoplasts, Nicotiana benthamiana leaves, and Arabidopsis plants, respectively.
Protoplast isolation and transformation
Protoplasts were isolated from fully expanded leaves of 3- to 4-week-old Arabidopsis plants grown on soil. Leaves were roughened using sandpaper, transferred to protoplasting buffer (500 mM sorbitol, 1 mM CaCl2, 0,03% pectolyase Y23, 0.75% cellulose YC, and 10 mM MES-KOH, pH 5.6–6.0), and incubated in the dark at 22 °C for 1.5 h with gentle agitation (60–75 rpm). Protoplasts were separated from undigested material by filtration through a 50 μm nylon mesh and sedimented by centrifugation for 8 min at 100 g. The pellet was resuspended in MaMg buffer (400 mM sorbitol, 15 mM MgCl2, 5 mM MES-KOH, pH 5.6). Protoplast transformation was performed essentially as previously described (Abel and Theologis, 1994). Transformed protoplasts were transferred into small Petri dishes and incubated for 24 h in the dark at 22 °C prior to analysis by confocal laser scanning microscopy.
Stable and transient plant transformation
The generated plasmids CaMV35S-promoter:YFP-SAUL1 and CaMV35S-promoter:HA-SAUL1 were used for stable transformation of Arabidopsis wild-type (Columbia) plants by floral dip (Clough and Bent, 1998; Raab et al., 2009). The plasmids CaMV35S-promoter:SAUL1ΔARM7–11-GFP and CaMV35S-promoter:SAUL1-GFP were used to transform saul1-1 mutants. Transformants were obtained by BASTA selection. For transient transformation of N. benthamiana leaves, Agrobacterium tumefaciens strain C58C1 (Deblaere et al., 1985) harbouring the P35S:GFP-SAUL1 construct was grown at 29 °C in LB supplemented with 50 μg ml−1 kanamycin to the stationary phase. Bacteria were sedimented by centrifugation at 5000 g for 15 min at room temperature and resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, KOH pH 5.7). Cells were infiltrated into the abaxial air spaces of 2- to 4-week-old N. benthamiana plants. GFP–SAUL1 fluorescence was monitored by confocal laser scanning microscopy 24 h post-infiltration.
Confocal laser scanning microscopy
Fluorescence of YFP fusions, GFP fusions, and UmSrt1-red fluorescent protein (RFP) was monitored by confocal laser scanning microscopy (Leica TCS SP II; Leica Microsystems, Wetzlar, Germany) using 488 nm (GFP), 514 nm (YFP), and 543 nm (RFP) laser light for excitation. Detection windows ranged from 497 nm to 527 nm for GFP, from 525 nm to 575 nm for YFP, and from 570 nm to 625 nm for RFP.
Generation of anti-SAUL1 antibodies and western blot analysis
Purified 6×His-SAUL1 protein (Raab et al., 2009) was used for immunization of rabbits, performed by Dr Pineda Antibody Service. For affinity purification of anti-SAUL1 antibodies, nitrocellulose filters soaked with 1 mg ml−1 MBP–SAUL1 fusion peptide (Raab et al., 2009) and blocked with skim milk-containing buffer (50 mM TRIS, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 1% skim milk powder) were incubated in raw antiserum for at least 60 min at 4 °C, washed, and bound antibodies were released as described (Sauer and Stadler, 1993).
For total membrane preparation Arabidopsis plant material was ground in liquid nitrogen and grinding buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol (DTT), and protease inhibitor mix (Complete Mini tablets, EDTA-free, Roche) was added. Samples were incubated on ice for 10 min and centrifuged twice for 10 min at 3500 g and 4 °C. The supernatant was centrifuged at 100 000 g and 4 °C for 1 h to separate soluble protein and total membrane fractions. Protein samples were resolved on 10% SDS–PAGE, followed by gel-blot analysis using purified anti-SAUL1 antibodies.
Results
The PUB-ARM E3 ubiquitin ligase SAUL1 is localized at the plasma membrane.
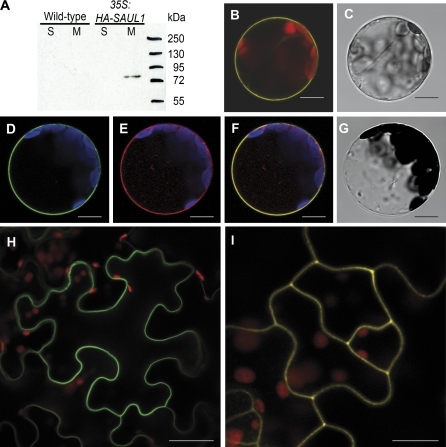
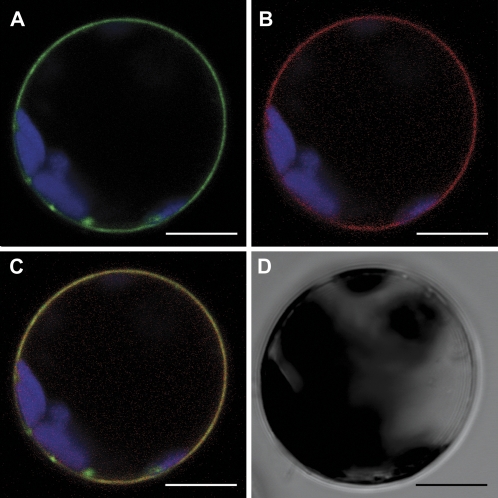
Recent results obtained by monitoring gene expression changes during premature senescence in saul1 mutants suggested that the E3 ubiquitin ligase SAUL1 acts upstream of these transcriptional changes (Raab et al., 2009). The aim of this study was therefore to identify the compartment harbouring SAUL1 activity. Soluble proteins and total membrane fractions were isolated, specific anti-SAUL1 antibodies were established, and western blot analyses of SAUL1 protein were performed in wild-type and CaMV35S:HA-SAUL1 plants. Whereas SAUL1 protein could not be detected in wild-type samples, HA-SAUL1 protein was present in membrane fractions of CaMV35S:HA-SAUL1 plants (Fig. 1A). To define the membrane with which SAUL1 is associated, Arabidopsis leaf protoplasts were transiently transformed with CaMV35S-promoter:YFP-SAUL1 or CaMV35S-promoter:GFP-SAUL1 fusion constructs. Consecutively, confocal laser scanning microscopy was used and fluorescence signals were detected at the plasma membrane (Fig. 1B–D). To prove plasma membrane association of SAUL1, the plasma membrane sugar transport protein UmSrt1 from the phytopathogenic fungus Ustilago maydis fused to RFP was used for co-localization experiments (Wahl et al., 2010). Co-transformation of protoplasts with CaMV35S-promoter:GFP-SAUL1 and CaMV35S-promoter:Umsrt1-RFP resulted in a match of green and red fluorescence, indicating that SAUL1 and the plasma membrane marker UmSrt1 were co-localized (Fig. 1D–G). Localization of SAUL1 at the plasma membrane was also confirmed in planta via transient expression of GFP–SAUL1 fusion proteins in N. benthamiana leaves and stable expression of YFP–SAUL1 fusion proteins in A. thaliana plants (Fig. 1H, I).
Fig. 1.
Plasma membrane localization of SAUL1. (A) SAUL1 localization in membrane fractions. Soluble (S) and total membrane fractions (M) were isolated from wild-type and CaMV35S:HA-SAUL1 plants. Western blot analysis using anti-SAUL1 antibodies did not result in detection of SAUL1 protein in wild-type plants, but revealed the presence of the HA-tagged SAUL1 in total membrane fractions at the expected size in CaMV35S:HA-SAUL1 plants. (B) Localization of YFP–SAUL1 fusion proteins at the plasma membrane. Yellow fluorescence signals from Arabidopsis protoplasts transformed with CaMV35S-promoter:YFP-SAUL1 DNA constructs recorded by confocal laser scanning microscopy showed plasma membrane localization of YFP–SAUL1 fusion proteins. (C) Transmission light picture of the protoplast from B. (D–G) Co-localization of SAUL1 and the plasma membrane marker UmSrt1. After co-transformation of Arabidopsis protoplasts with CaMV35S-promoter:GFP-SAUL1 and CaMV35S-promoter:Umsrt1-RFP DNA constructs, green and red fluorescence of GFP–SAUL1 (D) and UmSrt1–RFP (E) fusion proteins, respectively, was detected at the plasma membrane. Merging both pictures resulted in yellow colour, indicating co-localization of both proteins (F). (G) Transmission light picture of the protoplast from D and E. (H) Localization of GFP–SAUL1 fusion proteins in transiently transformed epidermal cells of Nicotiana benthamiana. Green fluorescence was observed at the plasma membrane. (I) Yellow fluorescence in transgenic CaMV35S-promoter:YFP-SAUL1 plants indicated localization of YFP–SAUL1 fusion proteins at the plasma membrane in planta. Autofluorescence of chlorophyll is shown in red in B, H, and I, or in blue in D–F. Scale bars represent 10 μm and 30 μm in H.
These results suggest that the plant U-box E3 ubiquitin ligase SAUL1 may have regulatory functions by modifying target proteins at the plasma membrane.
Association with the plasma membrane is specific to a small subgroup of PUB-ARM proteins
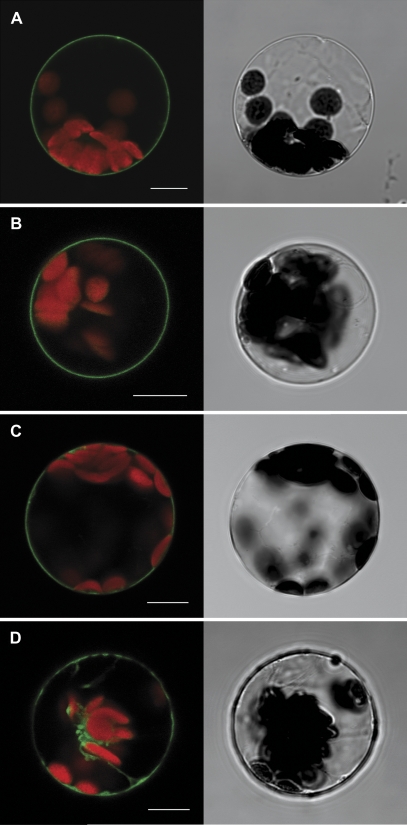
Localization of SAUL1 at the plasma membrane was independent of the position of the GFP tag (Figs 1B, D, 2A). To find out whether plasma membrane association is unique to SAUL1 among the Arabidopsis PUB-ARM proteins, the aim was to determine the subcellular localization of at least one member of each subgroup of the PUB-ARM protein family (Mudgil et al., 2004). The closest homologue carrying a similar arrangement of ARM repeats is PUB43. GFP–PUB43 fusions were generated, expressed in Arabidopsis leaf protoplasts under control of the CaMV35S-promoter, and the fluorescence signals were analysed using confocal laser scanning microscopy. GFP signals were found exclusively at the plasma membrane, indicating that not only SAUL1, but also its closest homologue PUB43, is associated with the plasma membrane (Fig. 2B). PUB42 is in the same subgroup as SAUL1 and PUB43, and turned out to be localized at the plasma membrane, too (Fig. 2C). In contrast, the next homologues PUB5 and PUB17, and other PUB-ARM proteins localized to the cytoplasm and/or to the nucleus (Fig 2D, Supplementary S1 at JXB online).
Fig. 2.
Unique localization of SAUL1 and its homologues at the plasma membrane. Transformation of Arabidopsis protoplasts followed by analyses of fluorescence signals by confocal laser scanning microscopy indicated the localization of SAUL1–GFP (A), GFP–PUB43 (B), and GFP–PUB42 (C) fusion proteins at the plasma membrane, and of GFP–PUB5 fusion proteins in the cytoplasm (D). Autofluorescence of chlorophyll is shown in red. Transmitted light pictures of the transformed protoplasts are shown next to the respective fluorescence picture. Scale bars represent 10 μm.
These localization studies indicated that association with the plasma membrane was specific to members of only one subgroup of the PUB-ARM protein family, namely SAUL1, PUB43, and PUB42.
Plasma membrane localization of SAUL1 does not require transmembrane helices or terminal lipid modifications
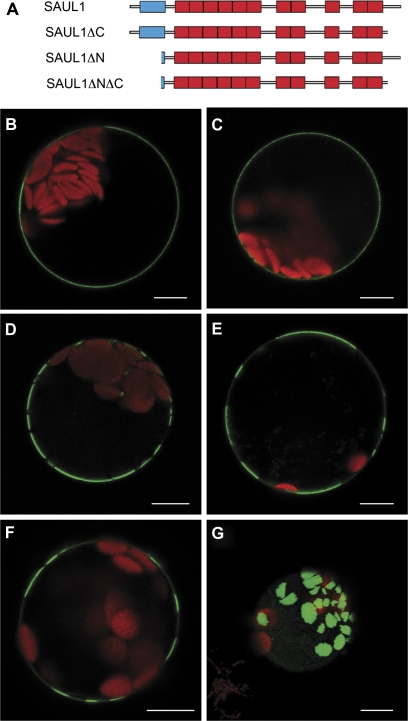
To investigate how SAUL1 is associated with the plasma membrane, DNA constructs were generated that lead to the expression of GFP fusion proteins containing different additions or deletions in Arabidopsis protoplasts (Fig. 3A). Transmembrane helices were not predicted to be present in SAUL1 (Schwacke et al., 2003). To exclude fully that SAUL1 is sequestered to the plasma membrane via the ER, ER retention signals were added, which would trap SAUL1 in the ER. After transient transformation of protoplasts with CaMV35S-promoter:SAUL1-GFP-HDEL constructs, GFP signals resembled the signals from GFP–SAUL1 fusion proteins, indicating that SAUL1 is not found in the ER at any time (Figs 1D, 3B).
Fig. 3.
Analysis of the contribution of the C- and N-terminus for localization of SAUL1 at the plasma membrane. (A) Schematic representation of SAUL1 deletion constructs. The U-box and ARM repeats are shown in blue and red, respectively. (B) Localization of SAUL1-GFP-HDEL in the plasma membrane indicated that SAUL1 does not require the ER pathway to reach the membrane. (C–E) Localization of SAUL1ΔC–GFP (C), SAUL1ΔNΔC–GFP (D), and SAUL1ΔN–GFP (E) at the plasma membrane. Note the distribution of N-terminally deleted fusion proteins in membrane patches. (F and G) Patchy distribution of GFP–SAUL1. In 25% of transformed protoplasts, GFP–SAUL1 was not equally distributed in the plasma membrane but instead was found in patches. (G) Top view of an Arabidopsis protoplast transformed with the CaMV35S:GFP-SAUL1 DNA construct. The GFP–SAUL1 fusion protein localized to large membrane patches. Autofluorescence of chlorophyll is shown in red. Scale bars represent 10 μm.
Lipid modifications can generally mediate membrane association of proteins (Nambara and McCourt, 1999; Yalovsky et al., 1999; Schwacke et al., 2003). Because sites for these modifications most frequently reside in the N- or C-terminus, SAUL1–GFP fusion proteins carrying N- or C-terminal deletions were generated (Fig. 3A). After transient transformation of protoplasts with CaMV35S-promoter:GFP-SAUL1ΔC constructs, fluorescence signals still indicated equal association of SAUL1ΔC–GFP with the plasma membrane (Fig. 3C). Additional deletion of the N-terminus in SAUL1ΔNΔC–GFP fusion proteins also did not abolish plasma membrane association (Fig. 3D). However, the association of SAUL1ΔNΔC–GFP fusion proteins did not appear to be equally distributed, but occurred in patches, suggesting that the N-terminus is important for equal distribution. This was supported through transformation of protoplasts with CaMV35S-promoter:GFP-SAUL1ΔN constructs. Deletion of only the N-terminus was sufficient to abolish equal distribution at the plasma membrane (Fig. 3E). In this context, it is important to mention that GFP–SAUL1 fusion proteins also featured patchy distribution in the plasma membrane in ∼25% of protoplasts, probably due to masking of the N-terminus. This indicated that this part of the protein including the U-box is important for equal distribution of SAUL1 at the plasma membrane (Fig. 3F, G).
These findings suggest that SAUL1 is not integrated in the plasma membrane through membrane helices or attached to the plasma membrane via lipid modifications at the N- or C-terminus, respectively. Lipid modification sites have also not been predicted in the rest of the SAUL1 protein. In addition, the N-terminus carrying the U-box is important for equal distribution of SAUL1 at the plasma membrane.
Plasma membrane localization of SAUL1 requires the C-terminal ARM repeat domain
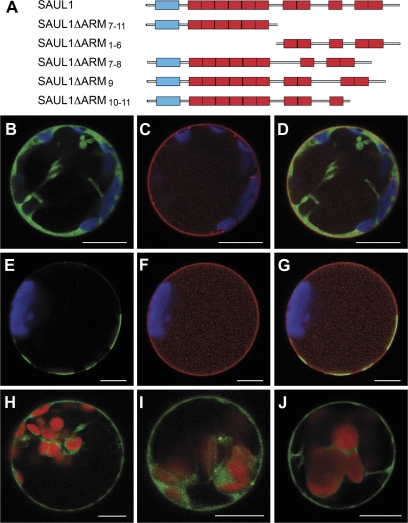
The SAUL1 protein contains 11 ARM repeats that may enable interactions with proteins in the plasma membrane. SAUL1 and PUB43 share a unique domain structure in contrast to all other PUB-ARM proteins in that they carry nearly twice the number of ARM repeats. Hence, they possess an elongated C-terminal half that is also found in PUB42 (Mudgil et al., 2004). Deletion constructs were generated to study the contribution of different ARM repeats for plasma membrane association (Fig. 4A). In a first step, the additional ARM repeats 7–11 in SAUL1 were deleted and SAUL1ΔARM7–11–GFP was expressed in Arabidopsis protoplasts. GFP signals indicated that the fusion proteins reside in intracellular structures other than the plasma membrane (Fig. 4B). To prove the loss of plasma membrane association, the plasma membrane sugar transport protein UmSrt1 from the phytopathogenic fungus U. maydis fused to RFP was used for co-localization experiments (Fig. 4C). Co-transformation of protoplasts with CaMV35S-promoter:SAUL1ΔARM7–11-GFP and CaMV35S-promoter:Umsrt1-RFP did not result in a match of green and red fluorescence signals, indicating that GFP–SAUL1ΔARM7–11 fusion proteins were not associated with the plasma membrane (Fig. 4D).
Fig. 4.
Contribution of ARM repeats to localization of SAUL1 at the plasma membrane. (A) Schematic representation of constructs deleting ARM repeats of SAUL1. (B–D) Co-localization of SAUL1ΔARM7–11–GFP and UmSrt1–RFP. (B) GFP signals of SAUL1ΔARM7–11–GFP were detected in the cytoplasm of transformed protoplasts. (C) Red fluorescence indicated localization of UmSrt1–RFP at the plasma membrane. (D) Merging pictures from B and C did not result in overlapping GFP and RFP signals. (E–G) Co-localization of SAUL1ΔARM1–6–GFP and UmSrt1–RFP. (E) SAUL1ΔARM1–6–GFP fluorescence was observed in plasma membrane patches. (F) Red fluorescence indicated localization of UmSrt1–RFP at the plasma membrane. (G) Merging pictures from E and F led to yellow signals by overlapping GFP and RFP fluorescence, indicating localization of SAUL1ΔARM1–6–GFP at the plasma membrane. Deletion of single C-terminal ARM repeats resulted in loss of plasma membrane association in SAUL1ΔARM7–8–GFP (H), SAUL1ΔARM9–GFP (I), and SAUL1ΔARM10–11–GFP (J) fusion proteins. Autofluorescence of chlorophyll is shown in blue in B–G or in red in H–J. Scale bars represent 10 μm.
Consecutively, ARM repeats 7–11 were fused to GFP, thus deleting ARM repeats 1–6, and GFP–SAUL1ΔARM1–6 was expressed in Arabidopsis leaf protoplasts. Fluorescence signals were detected at the plasma membrane but, due to the absence of the N-terminus, in patches rather than equally distributed (Fig. 4E, cf. Fig. 3). Co-transformation of protoplasts with CaMV35S-promoter:GFP-SAUL1ΔARM1–6 and CaMV35S-promoter:Umsrt1-RFP led to a match of green and red fluorescence, indicating that GFP–SAUL1ΔARM1–6 and the plasma membrane marker UmSrt1 were co-localized at the plasma membrane (Fig. 4F, G).
To narrow down the ARM repeats that mediate plasma membrane association, ARM repeats 7–8 in SAUL1ΔARM7–8–GFP, ARM repeat 9 in GFP–SAUL1ΔARM9, and the last two C-terminal ARM repeats in GFP–SAUL1ΔARM10–11 fusion proteins were deleted (Fig. 4A). All three of these fusion proteins showed fluorescence signals comparable with GFP–SAUL1ΔARM7–11 fusion proteins in intracellular structures. These results led to the conclusion that all ARM repeats in this ARM repeat domain consisting of ARM repeats 7–11 are crucial for plasma membrane association of SAUL1 (Fig. 4H–J).
The C-terminal ARM repeat domain of SAUL1 mediates membrane association of the cytosolic PUB27 protein
With regard to the specific localization of SAUL1, PUB43, and PUB42 at the plasma membrane in contrast to all other PUB-ARM proteins tested, it was tempting to speculate that the elongated C-terminus could drive plasma membrane association of PUB-ARM proteins, which were normally localized in intracellular structures (Figs 2D, Supplementary S1 at JXB online). Therefore, CaMV35S-promoter:PUB27-SAUL1ARM7–11-GFP constructs were generated to equip PUB27 with the additional ARM repeat domain of SAUL1. After transformation of Arabidopsis leaf protoplasts, PUB27-SAUL1ARM7–11–GFP fusion proteins indeed appeared to associate with the plasma membrane as indicated by green fluorescence (Fig. 5A). To validate this observation, co-transformation experiments were performed using CaMV35S-promoter:PUB27-SAUL1ARM7–11-GFP and CaMV35S-promoter:Umsrt1-RFP constructs (Fig. 5). Red fluorescence of UmSrt1–RFP fusion proteins was detected at the plasma membrane as expected (Fig. 5B), and red and green fluorescence matched in co-transformed protoplasts (Fig. 5C).
Fig. 5.
ARM repeats 7–11 mediate localization of PUB27 at the plasma membrane. (A) Green fluorescence of PUB27–SAUL1ARM7–11–GFP fusion proteins was predominantly detected at the plasma membrane. (B) Red fluorescence indicated localization of UmSrt1–RFP at the plasma membrane. (C) Merging pictures from A and B led to yellow signals by overlapping GFP and RFP fluorescence, indicating localization of PUB27–SAUL1ARM7–11–GFP at the plasma membrane. (D) Transmitting light picture of the protoplast recorded in A–C. Autofluorescence of chlorophyll is shown in blue. Scale bars represent 10 μm.
Taken together, the findings indicate that the C-terminal ARM repeat domain consisting of up to six ARM repeats in addition to the common core ARM repeats are essential and sufficient for association of PUB-ARM proteins with the plasma membrane.
Plasma membrane association is crucial for SAUL1 function
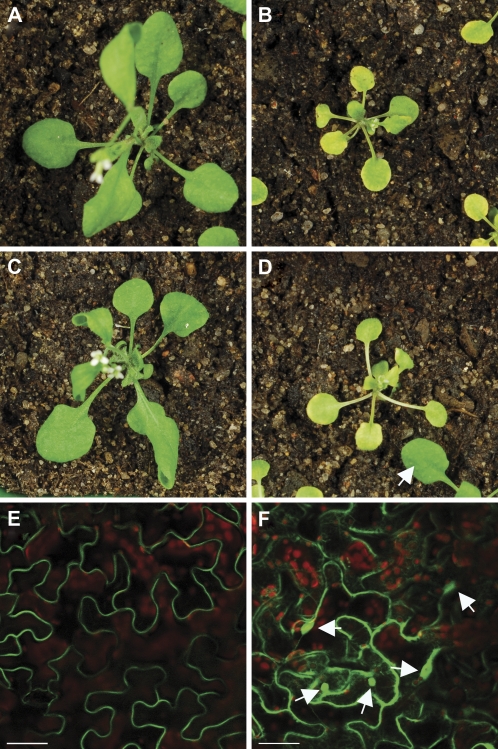
To investigate whether localization of SAUL1 at the plasma membrane is important for its function in vivo, complementation analyses with respect to the saul1 mutant phenotype were performed. Stable transformation of saul1-1 mutant plants with the CaMV35S-promoter:SAUL1-GFP construct resulted in complementation of the early senescence phenotype of saul1-1 mutant seedlings (Fig. 6A–C). In contrast, transformation of saul1-1 mutants with the CaMV35S-promoter:SAUL1ΔARM7–11-GFP construct did not complement the phenotype (Fig. 6D). In these transgenic lines, localization of SAUL1–GFP at the plasma membrane and of SAUL1ΔARM7–11–GFP in the cytosol/nucleus was confirmed by confocal laser scanning microscopy (Fig. 6E, F). These data indicated that loss of plasma membrane association through deletion of the C-terminal ARM repeats in the SAUL1ΔARM7–11–GFP protein abolished SAUL1 function in vivo.
Fig. 6.
Plasma membrane association of SAUL1 is important for its in vivo function. (A and B) In contrast to wild-type plants (A), saul1-1 mutants challenged with low light showed growth arrest and early senescence (B). (C) saul1-1 mutants stably transformed with the CaMV35S-promoter:SAUL1-GFP construct showed complementation of the senescence phenotype. (D) Stable transformation of saul1-1 mutants with the CaMV35S-promoter:SAUL1ΔARM7–11-GFP construct did not complement the early senescence phenotype. The arrow points to a leaf of a seedling that is not homozygous for the mutation in SAUL1 and thus does not show a senescence phenotype. (E) Plasma membrane association of SAUL–GFP was confirmed by monitoring GFP fluorescence in saul1-1 plants transformed with CaMV35S-promoter:SAUL1-GFP. (F) SAUL1ΔARM7–11–GFP localized to the cytosol and nucleus as indicated by GFP fluorescence in saul1-1 plants transformed with CaMV35S-promoter:SAUL1ΔARM7–11-GFP. Arrows point to nuclei in focus. Scale bars in E and F represent 50 μm.
Discussion
The aim of this study was to identify the subcellular localization of the PUB-ARM E3 ubiquitin ligase SAUL1 that is a suppressor of premature senescence and cell death in order to gain further insight into the possible mode of action of SAUL1. SAUL1 was detected in total membrane fractions through western blot analysis using anti-SAUL1 antibodies. Interestingly, expression of SAUL1 fused to different fluorescent proteins in Arabidopsis leaf protoplasts revealed fluorescence signals at the plasma membrane (Fig. 1). To date, PUB-ARM proteins have been localized in the nucleus (StPHOR1, BnARC1, AtPUB9, AtPUB13), in the cytoplasm (StPHOR1, BnARC1, AtPUB13, AtPUB22, AtPUB23), and in proteasome structures at the ER (BnARC1) (Amador et al., 2001; Stone et al., 2003; Cho et al., 2008; Samuel et al., 2008). It was demonstrated that the association with the plasma membrane is specific for SAUL1 and its two closest homologues AtPUB43 and AtPUB42 (Fig. 2). This was supported by analysing the subcellular localization of 18 members of the PUB-ARM protein family that all localized to the cytoplasm and/or nucleus and not to the plasma membrane (Supplementary Fig. S1 at JXB online). Together with published PUB-ARM proteins localized in the cytoplasm and nucleus, the analysis of this collection covered almost two-thirds of the protein family and all branches of the phylogenetic tree published previously (Mudgil et al., 2004).
What is the structural determinant for plasma membrane association of SAUL1, AtPUB42, and AtPUB43? These three proteins differ from all other PUB-ARM proteins in that they carry an extended C-terminus containing an additional domain of up to six ARM repeats (Mudgil et al., 2004). It could be demonstrated that this domain is essential and sufficient for plasma membrane association of SAUL1. The deletion of this domain or of single ARM repeats in this domain resulted in the loss of plasma membrane association (Fig. 4). Generally, ARM repeat domains are important for protein–protein interactions (Huber et al., 1997; Conti and Kuriyan, 2000; Coates, 2003). In addition to the PUB-ARM proteins, sequence analyses have detected >50 additional proteins containing tandem ARM repeats in Arabidopsis (Coates, 2003; Mudgil et al., 2004). However, clear plasma membrane association has not been reported for any of these proteins yet. Instead, the Arabidopsis ARM repeat proteins IMPA-1, IMPA-4, ARABIDILLO-1, ARABIDILLO-2, ARIA, LFR, ARO1, and ABAP1 have been shown to localize to the nucleus and/or the cytoplasm (Kim et al., 2004; Coates et al., 2006; Bhattacharjee et al., 2008; Gebert et al., 2008; Masuda et al., 2008; Wang et al., 2009). In a small fraction of analysed cells, plasma membrane association of Arabidopsis ARO1 in onion epidermis cells has been observed (Gebert et al., 2008). Taken together, these observations indicate a specific composition of ARM repeats in SAUL1 and its homologues, which mediates association with plasma membrane components.
The importance of tandem ARM repeat domains for protein targeting in plants has been studied in a few examples. The two F-box-containing proteins ARABIDILLO-1 and ARABIDILLO-2 that promote lateral root development carry a nuclear localization signal (NLS) and localize to the nucleus. In these proteins, the ARM repeat composition appears to allow for nuclear targeting in an NLS-independent manner (Coates et al., 2006). An ARM domain-containing fragment of the kinesin MRH2 that is involved in root hair tip growth was shown to bind to polymerized actin in vitro (Yang et al., 2007). Here, an ARM repeat domain was identified in plants that is sufficient to trigger plasma membrane association not only of SAUL1, but also of GFP and of the PUB-ARM protein PUB27 that normally resides in the cytoplasm (Figs 4, 5). Association of ARM repeat-containing proteins with membranes has been reported in other eukaryotes. The S. cerevisiae armadillo Vac8p, which functions in vacuole fusion and inheritance, is associated with the vacuolar membrane. In contrast to the situation in SAUL1, however, this association does not depend on the ARM repeats but on lipid modification (Fleckenstein et al., 1998; Pan and Goldfarb, 1998; Wang et al., 1998). The plasma membrane interaction domain in the Paramecium Nd9p involved in exocytotic membrane fusion is also located outside of its ARM repeat domain (Froissard et al., 2001). In animals, the cytosolic protein β-catenin, the mammalian homologue of the Drosophila armadillo gene product, can bind to interaction partners in different compartments (Yap et al., 1997). β-Catenin may serve as adaptor protein to anchor plasma membrane-localized cadherins, which regulate intercellular contact between epithelial cells at adherens junctions, to the actin cytoskeleton. Internal ARM repeats of β-catenin have been shown to be important for binding of β-catenin to the cytoplasmic domain of cadherins (Aberle et al., 1994; Hulsken et al., 1994; Pai et al., 1996). In addition to its cytoskeletal function, β-catenin has an important role in the regulation of gene expression through Wnt signalling during development (Behrens, 2000). This function requires the interaction with interaction partners in the nucleus and cytoplasm, namely TCF-type transcription factors, the tumour suppressor APC, and axin. Interestingly, binding to these proteins depends on the same ARM repeat domain as binding to cadherin (Graham et al., 2000; von Kries et al., 2000; Huber and Weis, 2001; Xing et al., 2003, 2004). In the SAUL1 protein, the composition and arrangement of the ARM repeats appear to be a case sui generis, thus mediating localization specifically at the plasma membrane.
Previously, it has been shown that the PUB-ARM E3 ubiquitin ligase SAUL1 is a suppressor of premature senescence and cell death (Raab et al., 2009). Together with the plasma membrane association and its relevance for SAUL1 function in vivo presented here, this suggests that SAUL1 modifies target proteins at the plasma membrane to suppress premature senescence and cell death in young Arabidopsis plants. Similarly, the Arabidopsis RING1 E3 ubiquitin ligase localizes to plasma membrane lipid rafts to modulate the fumonisin B1-induced programmed cell death pathway (Lin et al., 2008). In animals, extrinsic signalling leading to programmed cell death has recently also been shown to involve ubiquitin-dependent steps at the plasma membrane (Jin et al., 2009). Regarding dramatic changes in the transcriptome during senescence, it is tempting to speculate that SAUL1 may participate in processing membrane-bound transcription factors at the plasma membrane. Membrane-bound transcription factors have been described in Arabidopsis and, based on genome-scale analyses, many others have been predicted to localize to membranes (Iwata and Koizumi, 2005; Kim et al., 2006; Seo et al., 2008). Regulated ubiquitin/proteasome-dependent processing (RUP) at membranes has been suggested as a possible mechanism of transcription factor regulation and thus gene expression control (Hoppe et al., 2001). To address the actual function of SAUL1, future research is required to identify in vivo targets modified by SAUL1 at the plasma membrane.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Subcellular localization of PUB-ARM proteins.
Table S1. Primers used for amplification of PUB-ARM full-length fragments.
Table S2. Primers used for amplification of SAUL1 deletion fragments.
Supplementary Material
Acknowledgments
We thank Tanja Bender for technical assistance and Petra Dietrich for discussions. This work was supported by DFG grants HO 2234/4-1 (to SH).
References
- Abel S, Theologis A. Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. The Plant Journal. 1994;5:421–427. doi: 10.1111/j.1365-313x.1994.00421.x. [DOI] [PubMed] [Google Scholar]
- Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H. Assembly of the cadherin–catenin complex in vitro with recombinant proteins. Journal of Cell Science. 1994;107:3655–3663. doi: 10.1242/jcs.107.12.3655. [DOI] [PubMed] [Google Scholar]
- Amador V, Monte E, Garcia-Martinez JL, Prat S. Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell. 2001;106:343–354. doi: 10.1016/s0092-8674(01)00445-7. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The U box is a modified RING finger—a common domain in ubiquitination. Current Biology. 2000;10:R132–R134. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- Azevedo C, Santos-Rosa MJ, Shirasu K. The U-box protein family in plants. Trends in Plant Science. 2001;6:354–358. doi: 10.1016/s1360-1385(01)01960-4. [DOI] [PubMed] [Google Scholar]
- Behrens J. Control of beta-catenin signaling in tumor development. Annals of the New York Academy of Sciences. 2000;910:21–33. doi: 10.1111/j.1749-6632.2000.tb06698.x. discussion 33–25. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S, Lee LY, Oltmanns H, Cao H, Veena Cuperus J, Gelvin SB. IMPa-4, an Arabidopsis importin alpha isoform, is preferentially involved in agrobacterium-mediated plant transformation. The Plant Cell. 2008;20:2661–2680. doi: 10.1105/tpc.108.060467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Ryu MY, Song C, Kwak JM, Kim WT. Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. The Plant Cell. 2008;20:1899–1914. doi: 10.1105/tpc.108.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coates JC. Armadillo repeat proteins: beyond the animal kingdom. Trends in Cell Biology. 2003;13:463–471. doi: 10.1016/s0962-8924(03)00167-3. [DOI] [PubMed] [Google Scholar]
- Coates JC, Laplaze L, Haseloff J. Armadillo-related proteins promote lateral root development in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2006;103:1621–1626. doi: 10.1073/pnas.0507575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure. 2000;8:329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, De Greve H, Deboeck F, Schell J, Van Montagu M, Leemans J. Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Research. 1985;13:4777–4788. doi: 10.1093/nar/13.13.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. The Plant Journal. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Fleckenstein D, Rohde M, Klionsky DJ, Rudiger M. Yel013p (Vac8p), an armadillo repeat protein related to plakoglobin and importin alpha is associated with the yeast vacuole membrane. Journal of Cell Science. 1998;111:3109–3118. doi: 10.1242/jcs.111.20.3109. [DOI] [PubMed] [Google Scholar]
- Froissard M, Keller AM, Cohen J. ND9P, a novel protein with armadillo-like repeats involved in exocytosis: physiological studies using allelic mutants in paramecium. Genetics. 2001;157:611–620. doi: 10.1093/genetics/157.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert M, Dresselhaus T, Sprunck S. F-actin organization and pollen tube tip growth in Arabidopsis are dependent on the gametophyte-specific Armadillo repeat protein ARO1. The Plant Cell. 2008;20:2798–2814. doi: 10.1105/tpc.108.061028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Lamothe R, Tsitsigiannis DI, Ludwig AA, Panicot M, Shirasu K, Jones JD. The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. The Plant Cell. 2006;18:1067–1083. doi: 10.1105/tpc.106.040998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annual Review of Cell and Developmental Biology. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Graham TA, Weaver C, Mao F, Kimelman D, Xu W. Crystal structure of a beta-catenin/Tcf complex. Cell. 2000;103:885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Hachet V, Kocher T, Wilm M, Mattaj IW. Importin alpha associates with membranes and participates in nuclear envelope assembly in vitro. EMBO Journal. 2004;23:1526–1535. doi: 10.1038/sj.emboj.7600154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. Journal of Biological Chemistry. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- Hoppe T, Rape M, Jentsch S. Membrane-bound transcription factors: regulated release by RIP or RUP. Current Opinion in Cell Biology. 2001;13:344–348. doi: 10.1016/s0955-0674(00)00218-0. [DOI] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- Hulsken J, Birchmeier W, Behrens J. E-cadherin and APC compete for the interaction with beta-catenin and the cytoskeleton. Journal of Cell Biology. 1994;127:2061–2069. doi: 10.1083/jcb.127.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N. An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proceedings of the National Academy of Sciences, USA. 2005;102:5280–5285. doi: 10.1073/pnas.0408941102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annual Review of Cell and Developmental Biology. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Kim S, Choi HI, Ryu HJ, Park JH, Kim MD, Kim SY. ARIA, an Arabidopsis arm repeat protein interacting with a transcriptional regulator of abscisic acid-responsive gene expression, is a novel abscisic acid signaling component. Plant Physiology. 2004;136:3639–3648. doi: 10.1104/pp.104.049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Park JE, Park HY, Lim MH, Chua NH, Park CM. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. The Plant Cell. 2006;18:3132–3144. doi: 10.1105/tpc.106.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Lin SS, Martin R, Mongrand S, Vandenabeele S, Chen KC, Jang IC, Chua NH. RING1 E3 ligase localizes to plasma membrane lipid rafts to trigger FB1-induced programmed cell death in Arabidopsis. The Plant Journal. 2008;56:550–561. doi: 10.1111/j.1365-313X.2008.03625.x. [DOI] [PubMed] [Google Scholar]
- Masuda HP, Cabral LM, De Veylder L, Tanurdzic M, de Almeida Engler J, Geelen D, Inze D, Martienssen RA, Ferreira PC, Hemerly AS. ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO Journal. 2008;27:2746–2756. doi: 10.1038/emboj.2008.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil Y, Shiu SH, Stone SL, Salt JN, Goring DR. A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiology. 2004;134:59–66. doi: 10.1104/pp.103.029553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, McCourt P. Protein farnesylation in plants: a greasy tale. Current Opinion in Plant Biology. 1999;2:388–392. doi: 10.1016/s1369-5266(99)00010-2. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Pai LM, Kirkpatrick C, Blanton J, Oda H, Takeichi M, Peifer M. Drosophila alpha-catenin and E-cadherin bind to distinct regions of Drosophila Armadillo. Journal of Biological Chemistry. 1996;271:32411–32420. doi: 10.1074/jbc.271.50.32411. [DOI] [PubMed] [Google Scholar]
- Pan X, Goldfarb DS. YEB3/VAC8 encodes a myristylated armadillo protein of the Saccharomyces cerevisiae vacuolar membrane that functions in vacuole fusion and inheritance. Journal of Cell Science. 1998;111:2137–2147. doi: 10.1242/jcs.111.15.2137. [DOI] [PubMed] [Google Scholar]
- Raab S, Drechsel G, Zarepour M, Hartung W, Koshiba T, Bittner F, Hoth S. Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. The Plant Journal. 2009;59:39–51. doi: 10.1111/j.1365-313X.2009.03846.x. [DOI] [PubMed] [Google Scholar]
- Riggleman B, Wieschaus E, Schedl P. Molecular analysis of the armadillo locus: uniformly distributed transcripts and a protein with novel internal repeats are associated with a Drosophila segment polarity gene. Genes and Development. 1989;3:96–113. doi: 10.1101/gad.3.1.96. [DOI] [PubMed] [Google Scholar]
- Samuel MA, Mudgil Y, Salt JN, Delmas F, Ramachandran S, Chilelli A, Goring DR. Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiology. 2008;147:2084–2095. doi: 10.1104/pp.108.123380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- Sauer N, Stadler R. A sink-specific H+/monosaccharide co-transporter from Nicotiana tabacum: cloning and heterologous expression in baker’s yeast. The Plant Journal. 1993;4:601–610. doi: 10.1046/j.1365-313x.1993.04040601.x. [DOI] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flugge UI, Kunze R. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiology. 2003;131:16–26. doi: 10.1104/pp.011577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Calderon Villalobos LI. Cullin-containing E3 ubiquitin ligases in plant development. Current Opinion in Plant Biology. 2004;7:677–686. doi: 10.1016/j.pbi.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Kim SG, Park CM. Membrane-bound transcription factors in plants. Trends in Plant Science. 2008;13:550–556. doi: 10.1016/j.tplants.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Sigismund S, Polo S, Di Fiore PP. Signaling through monoubiquitination. Current Topics in Microbioogy andl Immunology. 2004;286:149–185. doi: 10.1007/978-3-540-69494-6_6. [DOI] [PubMed] [Google Scholar]
- Stone SL, Anderson EM, Mullen RT, Goring DR. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. The Plant Cell. 2003;15:885–898. doi: 10.1105/tpc.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Callis J. Ubiquitin ligases mediate growth and development by promoting protein death. Current Opinion in Plant Biology. 2007;10:624–632. doi: 10.1016/j.pbi.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Trujillo M, Ichimura K, Casais C, Shirasu K. Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Current Biology. 2008;18:1396–1401. doi: 10.1016/j.cub.2008.07.085. [DOI] [PubMed] [Google Scholar]
- Vierstra RD. The ubiquitin–26S proteasome system at the nexus of plant biology. Nature Reviews Molecular Cell Biology. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- von Kries JP, Winbeck G, Asbrand C, Schwarz-Romond T, Sochnikova N, Dell’Oro A, Behrens J, Birchmeier W. Hot spots in beta-catenin for interactions with LEF-1, conductin and APC. Nature Structural Biology. 2000;7:800–807. doi: 10.1038/79039. [DOI] [PubMed] [Google Scholar]
- Wahl R, Wippel K, Goos S, Kämper J, Sauer N. A novel high-affinity sucrose transporter is required for virulence of the plant pathogen Ustilago maydis. PLoS Biology. 2010;8 doi: 10.1371/journal.pbio.1000303. e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Catlett NL, Weisman LS. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. Journal of Cell Biology. 1998;140:1063–1074. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yuan T, Yuan C, Niu Y, Sun D, Cui S. LFR, which encodes a novel nuclear-localized Armadillo-repeat protein, affects multiple developmental processes in the aerial organs in Arabidopsis. Plant Molecular Biology. 2009;69:121–131. doi: 10.1007/s11103-008-9411-8. [DOI] [PubMed] [Google Scholar]
- Wiborg J, O’Shea C, Skriver K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochemical Journal. 2008;413:447–457. doi: 10.1042/BJ20071568. [DOI] [PubMed] [Google Scholar]
- Xing Y, Clements WK, Kimelman D, Xu W. Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex. Genes and Development. 2003;17:2753–2764. doi: 10.1101/gad.1142603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Clements WK, Le Trong I, Hinds TR, Stenkamp R, Kimelman D, Xu W. Crystal structure of a beta-catenin/APC complex reveals a critical role for APC phosphorylation in APC function. Molecular Cell. 2004;15:523–533. doi: 10.1016/j.molcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Yalovsky S, Rodríguez-Concepcion M, Gruissem W. Lipid modifications of proteins—slipping in and out of membranes. Trends in Plant Science. 1999;4:439–445. doi: 10.1016/s1360-1385(99)01492-2. [DOI] [PubMed] [Google Scholar]
- Yang CW, Gonzalez-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H, O’Donnell E, Jones JD, Sadanandom A. The E3 ubiquitin ligase activity of arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. The Plant Cell. 2006;18:1084–1098. doi: 10.1105/tpc.105.039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Gao P, Zhang H, Huang S, Zheng ZL. A mutation in MRH2 kinesin enhances the root hair tip growth defect caused by constitutively activated ROP2 small GTPase in Arabidopsis. PLoS One. 2007;2:e1074. doi: 10.1371/journal.pone.0001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap AS, Brieher WM, Gumbiner BM. Molecular and functional analysis of cadherin-based adherens junctions. Annual Reviews of Cell and Devopmental Biology. 1997;13:119–146. doi: 10.1146/annurev.cellbio.13.1.119. [DOI] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. The Plant Cell. 2004;16:2795–2808. doi: 10.1105/tpc.104.025171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurinsky J, Shtutman M, Ben-Ze'ev A. Plakoglobin and beta-catenin: protein interactions, regulation and biological roles. Journal of Cell Science. 2000;113:3127–3139. doi: 10.1242/jcs.113.18.3127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.