Abstract
MicroRNAs (miRNAs) have recently emerged as a new class of modulators of gene expression. miRNAs control protein synthesis by targeting mRNAs for translational repression or degradation at the posttranscriptional level. These noncoding RNAs are endogenous, single-stranded molecules approximately 22 nucleotides in length and have roles in multiple facets of immunity, from regulation of development of key cellular players to activation and function in immune responses. Recent studies have shown that dysregulation of miRNAs involved in immune responses leads to autoimmunity. Multiple sclerosis (MS) serves as an example of a chronic and organ-specific autoimmune disease in which miRNAs modulate immune responses in the peripheral immune compartment and the neuroinflammatory process in the brain. For MS, miRNAs have the potential to serve as modifying drugs. In this review, we summarize current knowledge of miRNA biogenesis and mode of action and the diverse roles of miRNAs in modulating the immune and inflammatory responses. We also review the role of miRNAs in autoimmunity, focusing on emerging data regarding miRNA expression patterns in MS. Finally, we discuss the potential of miRNAs as a disease marker and a novel therapeutic target in MS. Better understanding of the role of miRNAs in MS will improve our knowledge of the pathogenesis of this disease.
1. Introduction
MicroRNAs (miRNAs) represent a class of noncoding RNA molecules that play pivotal roles in cellular and developmental processes by regulating gene expression at the posttranscriptional level. miRNAs are endogenous, evolutionarily conserved, single-stranded RNAs approximately 22 nucleotides in length that suppress the expression of protein-coding genes by directing translational repression through base-pairing with complementary messenger RNA (mRNA) and/or by promoting degradation of target mRNA degradation [1, 2]. Since the identification of the miRNA lin-4 as a regulator of developmental timing in the nematode Caenorhabditis elegans (C. elegans) in 1993 [3, 4], more than 17000 miRNAs have been recognized in 142 species. Currently, 1048 human miRNAs are registered in the miRNA registry (miRBase) which is the most commonly used database for miRNA (September 2010, release 16, http://www.mirbase.org/) [5]. miRBase reports 672 miRNAs in mouse and 408 miRNAs in rat, with new miRNAs constantly being identified, though the biologic function of only a fraction of miRNAs has been elucidated. miRNAs are predicted to regulate up to one-third of all human protein-coding genes. Unraveling the miRNA translational silencing network remains a challenge in part because individual miRNAs typically target several transcripts rather just one specific gene and a single mRNA can be regulated by several distinct miRNAs that act cooperatively [2]. Ribosome profiling experiments showed that miRNAs mediate destabilization of target mRNAs resulting in reduced protein levels [6]. miRNAs play an important role in diverse biologic processes such as development, cell proliferation and differentiation, apoptosis, oncogenesis, metabolism, angiogenesis, and inflammation. The expression of miRNAs is initially controlled at the level of transcriptionby transcription factors that regulate the production of miRNA-containing primary transcripts in specific cell types during development or in response to different environmental signals. Dysregulation of miRNA expression and function is associated with a variety of human diseases including cancer, neurodegeneration and autoimmunity [7, 8].
The regulation of mammalian immune responses by miRNAs is a concept currently evidenced by rapidly accumulating data [9, 10]. miRNAs have unique expression profiles in cells of the innate and adaptive immune systems and have pivotal roles in the regulation of both cell development and function. Recent studies focused on the networkwide role of miRNA or the functions of individual miRNAs have revealed that these small noncoding RNAs are involved in T and B cell differentiation in the thymus and bone marrow, respectively. During the effector phases of adaptive immunity, miRNAs contribute to the differentiation of T cells into functional lineages, class switching and germinal centre formation in B cells and activation of antigen-presenting cells (APCs) through pattern recognition pathways [11]. miRNAs are also directly involved in innate immunity and transduction signalling by Toll-like receptors (TLRs) and the ensuing cytokine response [12]. Up to one half of innate immune genes are predicted to be under the direct regulation of miRNAs. With the capacity of miRNAs to regulate the survival and death of T and B cells, control over miRNA expression is essential to prevent adaptive immune cells from unregulated proliferation leading to cancer or autoimmunity [13, 14]. miRNAs are differentially expressed in autoimmune diseases and miRNA regulation may have an impact on the development or prevention of autoimmunity. miRNA dysregulation is linked to autoimmune diseases such as rheumatoid arthritis, systemic lupus erythematosus, Sjögren's syndrome, psoriasis, and MS [15–17]. MS is the most common autoimmune disease of the central nervous system (CNS). It is a chronic, neuroinflammatory, and demyelinating disease in which myelin specific autoreactive CD4+ T cells become activated in the peripheral immune compartment, cross the blood-brain barrier (BBB), and promote neurological disability [18, 19]. Both genetic (HLA type) and environmental causes for MS have been suggested. Recently, genomewide association studies have identified additional potential MS susceptibility loci [20, 21]. Recent studies suggest that miRNA dysregulation may contribute to the pathogenesis of MS. Thus, better understanding of miRNA mechanisms might shed light, not only on the pathogenesis of MS but also on potential approaches for managing or even suppressing the disease. In this review, we briefly overview the biogenesis and action mechanisms of miRNAs and summarize recent advances in our understanding of both the intended functions of miRNAs in managing immune responses. We then review evolving knowledge on the role of miRNA in autoimmunity and emerging data regarding miRNA expression patterns in MS. Finally, we also discuss the potential of miRNAs as a diagnostic and prognostic indicators of disease type and status and as a novel therapeutic target in MS.
2. MicroRNAs
2.1. MicroRNA Biogenesis
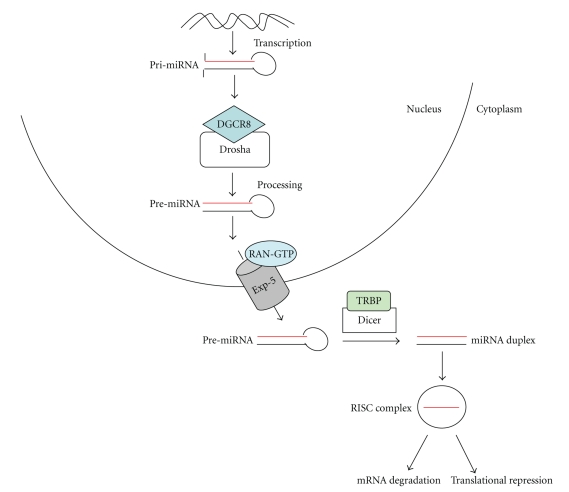
All miRNAs are processed and maturated through a complex biogenesis process involving multiple protein catalysts, accessory proteins, and macromolecular complexes following a coordinated series of events. The reader is referred to excellent recent reviews for detailed discussions of miRNA biogenesis and its regulation [22–25]. MicroRNAs can be encoded by independent genes but may also be processed from a variety of different RNA species, including introns, 3′-UTR of mRNAs, long noncoding RNAs, transposable elements, and genomic repeats [26–32]. miRNAs are expressed as 21–23 nucleotide RNA molecules initially transcribed by RNA polymerase II as long primary miRNAs (pri-microRNAs). Although most of the miRNA genes are transcribed by RNA Polymerase II, a cluster of human miRNAs have recently been shown to utilize RNA Polymerase III for their transcription [33]. Pri-miRNAs are typically 3 to 4 kilobases long single-stranded RNAs with 5′ cap, 3′ poly(A) tail and complicated secondary structure [34–37]. Pri-miRNAs are processed in the nucleus into one or more precursor-miRNAs (pre-miRNAs) with an approximately 70-nt loop structure. Processing is performed by a protein complex named microprocessor complex consisting of the nuclease Drosha (nuclear RNase III) and the stranded RNA-binding protein, human DiGeorge syndrome critical region gene 8 (DGCR8) (also known as Pasha in flies) [35, 37–42]. Drosha functions as the catalytic subunit while DGCR8 recognizes the RNA substrate. Pre-miRNAs are exported from the nucleus to the cytoplasm by exportin-5 which specifically recognizes the characteristic end structure of pre-miRNAs [43–46]. In the cytoplasm, another RNase III, known as Dicer, further processes the pre-miR into mature miRNA, which is double stranded (miRNA duplex) [47, 48]. After Dicer processing, the miRNA duplex is unwound and a strand (known as miRNA strand or guide strand) binds to an Argonaute 2 (Ago 2) protein (eIF2C2 in human) in a process that is referred to as miRNA loading or assembly, while the complementary strand (known as miRNA* strand, star strand or passenger strand) is degraded. The effector complex that mediates catalytic mRNA cleavage is known as RNA-induced silencing complex (RISC), and the effector complex that mediates translational repression directed by miRNAs is known as micro-ribonucleoprotein complex (miRNP) [49–51]. The single stranded mature microRNA must associate with the RISC. Mature microRNAs are incorporated into a miRNP (Figure 1). In this complex, which includes the Dicer-transactivation-responsive RNA-binding protein (TRBP)-PACT-Ago 2, microRNAs can direct downregulation by two mechanisms: translational inhibition and target mRNA cleavage [52–55]. Perfect match with the target results in mRNA degradation whereas partial match leads to translational inhibition.
Figure 1.
miRNA genes are transcribed in the form of Pri-miRNA. The DGCR8-Drosha complex processes in the form Pre-miRNA followed by transport into cytoplasm by Exportin-5. In cytoplasm, Pre-miRNA is processed by Dicer into miRNA duplex. Of miRNA duplex, one strand is loaded into RISC complex, which functions for either mRNA degradation or translational repression.
Inflammation has been reported to regulate miRNA biogenesis; TLR ligands, antigens, or cytokines can modulate miRNA expression level through regulation of specific transcription factors [2, 9, 56]. Cytokines have been shown to regulate Dicer expression resulting in alteration of pre-miRNA processing. Interferon-beta (IFN-β) has been shown to inhibit Dicer expression, which results in decrease of pre-miRNA processing, whereas IFN-γ induces pre-miRNA processing [57].
2.2. Detection of MicroRNAs
Information about miRNA and target expression patterns can help to assess the likelihood that a predicted miRNA-target relationship is relevant in vivo [58]. Expression of a miRNA can be measured by molecular biology techniques, such as Northern blotting, RNase protection assay, polymerase chain reaction- (PCR-) based techniques, and high throughput assays [59–61]. miRNA expression profiles were first generated by small RNA cloning and Northern blotting [4, 62–67]. The small size of miRNAs initially hampered PCR-based methods [61]. However, since the development of quantitative real-time PCR, PCR-based techniques have become very popular due to their high sensitivity [62, 68, 69]. In situ hybridization has provided further insight into the tissue-specific expression of pri- and mature miRNAs [62, 70–74]. Microarray techniques are widely used to comprehensively assay the entire miRNome (the global miRNA expression profile) in tissues or in cell lines [62, 68, 75–83]. In addition, serial analyses of gene expression (SAGE) adapted for small RNAs have been used to obtain miRNomes [84]. Interest in the SAGE approach was stimulated by recent innovations in next generation (deep) sequencing methods that provide a powerful tool for various genomics studies [85–87]. Overall, these technical improvements are expected to greatly widen the repertoire of known miRNAs in a variety of biological systems [61]. Emerging techniques for miRNA detection and quantification, including luminescence-based, fluorescence-based, electrochemical, colorimetric, and enzyme-based, and nanotechnology-based methods have recently been reviewed [88]. Whereas expression analyses are required to identify miRNAs with altered expression patterns in diseased tissues, functional analyses of the ability of these miRNAs to regulate expression of target mRNAs are essential to understand their impact on pathogenic pathways and processes.
3. MicroRNAs and Immunity
Clearly, both innate and adaptive immune responses are extremely highly regulated. Recent work from a number of laboratories has revealed that miRNAs play an important role in this intricate system (Table 1). miRNAs have unique expression patterns in immune cells and play a pivotal role in their development, maturation, and function.
Table 1.
miRNA in immune functions.
| miRNA | Expressing cells | Functions | Targets |
|---|---|---|---|
| Let-7e | macrophages | Innate immune response | TLR4 |
| miR-9 | myeloid cells | Immune response | NFK B1 |
| miR-17-5p | myeloid cells | monocyte proliferation and differentiation | RUNX1 |
| miR-17-92 | B and T cells | B and T cell development | BIM, PTEN |
| miR-21 | myeloid cells | macrophage activation | IL12a, PTEN, PDCD4 |
| miR-34 | DC and B cells | Myeloid DC differentiation | FOXP1, JAG1, WNT1 |
| miR-125b | monocyte | Innate immune response, TLR signaling | TNF-α |
| miR-126 | HSC | expansion of progenitor cells | HOXA9, PLK2 |
| miR-132 | monocyte | Innate immune response | not determined |
| miR-142 | Treg cell | Suppresor function of Treg cells | AC9 |
| miR-146a | monocyte | Innate immune response, TLR signaling | IRAK-1, IRAK-2, TRAF6 |
| miR-150 | B and T cells | mature B-cell production, T-cell activation | Myb |
| miR-155 | B and T cells, DC | Innate and adaptive immune response | AID, BACH1, CEBPB, CSFR |
| macrophages germinal center response | c-MAF, FADD, IKK, JARID2, | ||
| Ig G class-switch | PU.1, Ripk1, SOCS, TAB2 | ||
| Peripheral T cell development | |||
| miR-181a | T cells | T cell receptor signaling | AID, BCL2, CD69, DUSP5 |
| B cell development | DUSP6, PTPN22, SHP2 | ||
| miR-181b | macrophages, B cells | B cell class switch | AID |
| miR-196b | HSC | Hematopoietic stem-cell homeostasis | HOX |
| miR-223 | myeloid cells | Granulopoiesis | MEF2C |
| miR-326 | T cells | TH-17 cells development | ETS1 |
| miR-424 | myeloid cells | monocyte differentiation and maturation | NFIA, PU.1 |
AC9: adenylate cyclase 9; AID: Activation-Induced Cytidine Deaminase; BACH1: BTB and CNC homology 1, basic leucine zipper transcription factor 1; BCL2: B-cell lymphoma 2; BIM: BCL2-like 11; CEBPB: CCAAT/enhancer-binding protein beta; CSFR: Colony stimulating factor receptor; c-MAF: musculoaponeurotic fibrosarcoma oncogene homolog; DC: dendritic cell; DUSP5: Dual specificity protein phosphatase 5; DUSP6: Dual specificity protein phosphatase 6; ETS1: v-ets erythroblastosis virus E26 oncogene homolog 1; FADD: Fas-Associated protein with Death Domain; HSC: haematopoetic stem cell; HOX: Homeobox protein; HOXA9: Homeobox protein Hox-A9; FOXP1: Forkhead box P1; IKK: inhibitor of NF-kappaB kinase; IL12a: Interleukin-12 subunit alpha; IRAK-1: Interleukin-1 receptor-associated kinase 1; IRAK-2: Interleukin-1 receptor-associated kinase 2; JAG1: jagged 1; JARID2: Jumonji; Myb: Myb oncogene-like; MEF2C: Myocyte-specific enhancer factor 2C; NFIA: Nuclear factor 1 A-type; PDCD4: Programmed cell death protein 4; PTEN: phosphatase and tensin homolog; PLK2: pololike kinase 2; PTPN22: Tyrosine-protein phosphatase non-receptor type 22; PU.1: spleen focus forming virus (SFFV) proviral integration oncogene spi1; Ripk1: Receptor-interacting serine/threonine-protein kinase 1; RUNX1: Runt-related transcription factor; SHP2: SH2 domain containing protein thyrosine phosphatase; SOCS: Suppressor of cytokine signaling; TAB2: TAK1-associated binding protein 2 TRAF6: TNF receptor associated factor-6; TLR: Toll-like receptor; WNT1: wingless-related MMTV integration site 1.
3.1. Role of MicroRNAs in Immune Cell Development
miRNAs have an important role in regulating stem cell self-renewal and differentiation by repressing the translation of selected mRNAs in stem cells and differentiating into daughter cells. Such a role has been shown in embryonic stem cells, germline stem cells and various somatic tissue stem cells [89]. The first studies implicating miRNAs in immunological processes were originated from expression profiling of haematopoietic cells during their development. Haematopoietic stem cells reside mainly in the bone marrow and give rise to all blood cell lineages, including cells that constitute the immune system [9]. These cells must maintain a precise balance between self-renewal and differentiation into multipotent progenitors, which subsequently give rise to both the common lymphoid and common myeloid progenitors of the haematopoietic system [9, 90]. Systematic investigation of miRNA levels in hematopoietic cell lineages has identified miRNAs that are now considered as markers of these lineages [91–93]. Peculiar miRNA profiles in different haematopoietic organs and cell types suggest that miRNAs are dynamically regulated during early haematopoiesis, lineage commitment, and the development of immune cells and are involved in the regulation of these processes.
One of the first miRNAs described to have a role in immune cell development was miR-181a which is highly expressed in thymus cells and expressed at lower levels in the heart, lymph nodes, and bone marrow [91, 94]. In bone marrow-derived B cells, miR-181a expression has been shown to decrease during B cell development from the pro-B to the pre-B cell stage [91]. miR-181a inhibits the transition of pro-B to the pre-B cell stage. Moreover, miR-181a was identified as a positive regulator of B lymphocyte differentiation based on evidence that expression of miR-181a in hematopoietic stem and progenitor cells resulted in an increase in CD19+ B cells and a decrease in CD8+ T cells [91]. Interestingly, miR-181a is also involved in thymic T cell differentiation, by defining the activation threshold of T cell receptor (TCR) [94]. This miRNA modulates TCR signaling, thus affecting the sensitivity of T cells to antigens [94]. Other examples of miRNA-mediated regulation of immune cell development include miR-223 which was identified as an essential modulator of granulocytic differentiation [95] and miR-150 which has been shown to be critical for B cell differentiation [96, 97]. Collectively, these studies demonstrate that miRNAs play critical roles at distinct stages of immune cell development.
3.2. MicroRNAs in Adaptive Immune Responses
The adaptive or acquired immune system involves the selective recognition and removal of nonself by the TCRs on T cells and antibodies produced by B cells. The maturation, proliferation, differentiation, and activation of T and B cells are complex processes tightly controlled at different levels including miRNA-mediated posttranscriptional gene regulation [11]. Adaptive immunity refers to immune responses to antigens that undergo learning processes and provide specific memory. Once APCs capture a pathogen, they display foreign antigens complexed with major histocompatibility complexes (MHCs) on their surface to enable recognition of the antigen by naïve T cells to induce the adaptive immune response [19]. The combination of this interaction further drives the upregulation of both CD80 and CD86 on the surface of APCs. CD80 and CD86 identify two additional receptors, CD28 and Cytotoxic T-lymphocyte antigen 4 (CTLA4), on the surface of the T cells to provide a second signal to APCs [19]. CD28 is associated with activation of the T cell whereas CTLA4 is more regulatory. After this second signal, the T cells become activated, and APCs begin to secrete important cytokines, including IL-12 and IL-23, which bind to specific receptors on T cells and drive them to secrete different cytokines, such as IFN-γ or IL-17, depending on the cytokine milieu. T cells also begin to secrete IL-2, which then activates its own IL-2 receptor [19]. Upon activation of their TCR in the presence of costimulatory molecules, naïve T cells differentiate into various subsets of effector T cells with distinct effector functions (e.g., Th1, Th2, Th17, Th9). This differentiation is directed by a specific cytokine milieu leading to the expression of transcription factors specific for the respective lineages. The expression levels of all molecules involved in adaptive immune responses (transcription factors, cell surface receptors, cytokines, and their receptors) may be regulated by miRNAs as discussed below.
3.2.1. T Cells
The development of T cells in the thymus and their activation in the periphery are controlled by complex protein signalling networks that are subject to regulation by miRNAs [9, 98]. miRNA expression profiles vary between T cell subsets and different developmental stages [92, 99]. Specific deletion of Dicer in the T cell lineage resulted in impaired T cell development and aberrant T helper cell differentiation and cytokine production [100]. A severe block in peripheral CD8+ T cell development was observed upon Dicer deletion in the thymus. However, Dicer-deficient CD4+ T cells, although reduced in numbers, were viable and could be analyzed further. These cells were defective in microRNA processing, and upon stimulation, they proliferated poorly and underwent increased apoptosis [100]. Deletion of Dicer at an early stage of T cell development compromised the survival of alpha-beta lineage cells whereas the numbers of gamma-delta-expressing thymocytes were not affected in developing thymocytes [101]. Mice with higher expression of miR-17–92 in lymphocytes developed lymphoproliferative disease and autoimmunity and died prematurely. Lymphocytes from these mice showed more proliferation and less activation-induced cell death. The miR-17–92 miRNA suppressed expression of the tumor suppressor PTEN and the proapoptotic protein Bim [102]. T cell sensitivity to antigen is intrinsically regulated during maturation to ensure proper development of immunity and tolerance. Increasing miR-181a expression in mature T cells augments the sensitivity to peptide antigens while inhibiting miR-181a expression in the immature T cells reduces sensitivity and impairs both positive selection and negative selection [94]. These effects are in part achieved by the downregulation of multiple phosphatases, which leads to elevated steady-state levels of phosphorylated intermediates and a reduction of the TCR signaling threshold. T cell activation requires signaling through the TCR and costimulatory molecules, such as CD28. Costimulation-dependent upregulation of miR-214 promotes T cell activation by targeting the negative regulator Pten. Thus, the requirement for T cell costimulation is, in part, related to its ability to regulate expression of miRNAs that control T cell activation [103].
Recent data have also indicated a role for miRNAs in the differentiation of T cells into distinct effector T helper cell subsets. miR-155 has an important role in the mammalian immune system, specifically in regulating T helper cell differentiation and the germinal center reaction to produce an optimal T cell-dependent antibody response [104]. miR-155 exerts this control, at least in part, by regulating cytokine production. Many types of specialized Th cells, including Th1, Th2, Th17, Th9, follicular helper T, and Treg, have been identified. Different Th cells are committed to their paths but recent emerging evidence suggests that under certain conditions, seemingly committed Th cells possess plasticity and may convert into other types of effector cells [105]. There is growing evidence that clinically similar forms of autoimmune demyelinating disease can be driven by myelin-specific T cells of distinct lineages with different degrees of dependence on IL-17 production to achieve their pathological effects [106]. miRNAs play an important role in the development of Th17 cells [107]. Bcl-6, a transcriptional repressor, binds to the promoters of the Th1 and Th17 cell transcriptional regulators T-bet and RORgammat and represses IFN-γ and IL-17 production. Bcl-6 also represses expression of many miRNAs predicted to control the T follicular cell signature, including miR-17-92, which represses CXCR5 expression. Thus, Bcl-6 positively directs T follicular cell differentiation, through combined repression of miRNAs and transcription factors [108]. miRNAs are also essential in the development, differentiation, and function of Treg cells which are potent immune regulators [109]. Recent studies showed a crucial role for miRNAs in Treg cell biology and the prevention of spontaneous autoimmunity [110–112].
miR-155 deficiency in Treg cells results in increased suppressor of cytokine signaling 1 (SOCS1) expression accompanied by impaired activation of signal transducer and activator of transcription 5 (STAT5) transcription factor in response to limiting amounts of IL-2. Forkhead box P3- (Foxp3-) dependent regulation of miR155 maintains competitive fitness of Treg cell subsets by targeting SOCS1 [113]. miR-155-deficient mice have reduced numbers of Tregs, both in the thymus and periphery, due to impaired development. However, no evidence for defective suppressor activity of miR-155-deficient Tregs was found, either in vitro or in vivo, suggesting that miR-155 contributes to Treg development, but that additional miRNAs control Treg function [114]. The expression of miR-142-3p was recently shown to be repressed by Foxp3, leading to increased production of cyclic AMP and suppressor function of Treg cells [115]. Depleting miRNAs by eliminating Dicer reduces Treg cell numbers and results in immune pathology [116]. Dicer facilitates, in a cell-autonomous fashion, the development of Treg cells in the thymus and the efficient induction of Foxp3 by transforming growth factor-beta (TGF-β). These results suggest that Treg cell development involves Dicer-generated RNAs awaiting functional assessment. miR-31 negatively regulates Foxp3 expression by binding directly to its potential target site in the 3′-UTR of Foxp3 mRNA whereas miR-21 acts as a positive, though indirect, regulator of Foxp3 expression [117]. Finally, miR-155 inhibition sensitizes CD4+ Th cells for Treg-mediated suppression [118].
3.2.2. B Cells
The generation of B cells that express high affinity antigen receptors involves two main stages: antigen-independent development in the bone marrow and antigen-dependent selection in the secondary lymphoid organs, both of which are associated with dynamic regulation by miRNAs [9, 119]. Antigen receptors on the surface of B cells trigger adaptive immune responses after encountering their cognate antigens but also control a series of antigen-independent checkpoints during B cell development. These physiological processes are regulated by the expression and function of cell surface receptors, intracellular signaling molecules, transcription factors, and miRNAs [119]. Temporal regulation of several different miRNAs was observed and putative new cell type-specific miRNAs were identified in the development of B cells, suggesting the involvement of many, but undefined, regulatory pathways in B cell development and maturation [9]. The role of miRNAs in controlling the early development of B cells is now thought to involve the modulation of key protein factors that control these aspects of B cell development [97]. miR-181 is preferentially expressed in the B-lymphoid cells of mouse bone marrow and its ectopic expression in hematopoietic stem/progenitor cells leads to an increased fraction of B-lineage cells, without increase of T cells or myeloid cells in both tissue-culture differentiation assays and adult mice [91].
In contrast, mice with a conditional deletion of Dicer in B cells had a complete block in B cell development [120]. This block is related to dysregulated expression of the proapoptotic protein Bim, probably during the selection of effective antigen receptors. These results suggest a defect in the regulation of B cell selection. Regulation of apoptosis and cell cycle progression plays an essential role in the maintenance of B-cell homeostasis, because a fine balance of survival and expansion is critical for preventing lymphocytic disorders. Interestingly, the changes observed by gene expression profiling of Dicer-deficient B cell precursors are generally similar to those observed in B cells lacking the miR-17–92 family. Absence of miR-17–92 leads to increased levels of the proapoptotic protein Bim and inhibits B cell development at the pro-B to pre-B transition [121]. In addition to effects on antigen receptor selection, miRNAs also regulate the transcription factors involved in early B cell development [9]. Constitutive expression of miR-150, which is highly upregulated at the immature B cell stage, leads to a block at a proximal stage of B cell development, the pro-B to pre-B cell transition, indicating that miR-150 most likely downregulates mRNAs that are important for pre- and pro-B cell formation or function [96]. miR-150 controls B cell differentiation by targeting the transcription factor c-Myb [97]. miR-125b also promotes B cell diversification in the germinal center by inhibiting premature utilization of essential transcription factors for plasma cell differentiation [122].
The contribution of miRNAs in the antigen-driven stages of the humoral response in secondary lymphoid organs has also been described [9]. miR-155 is required in B cell responses to thymus-dependent and -independent antigens [123]. B cells lacking miR-155 generated reduced extrafollicular and germinal center responses and failed to produce high-affinity IgG1 antibodies. When transcription factor Pu.1 is overexpressed in wild-type B cells, fewer IgG1 cells are produced, suggesting that loss of Pu.1 regulation is a contributing factor to the miR-155-deficient phenotype [123]. The miR-23a cluster is a downstream target of PU.1 involved in antagonizing lymphoid cell fate determination [124]. miR-155 represses activation-induced cytidine deaminase, which is required for immunoglobulin gene diversification in B lymphocytes [125, 126]. A recent study showed that numerous miRNAs were expressed in a stage- or transformation-specific fashion in B cells, suggesting specific functional or pathological roles [127].
3.3. MicroRNAs in Innate Immune Responses
The innate immune response provides the initial defense against infection by external pathogens and is predominantly mediated via myeloid cells such as macrophages, DCs, monocytes, neutrophils, as well as natural killer (NK) cells. The presence of pathogens is commonly detected by tissue APCs such as macrophages and DCs via families of pattern recognition receptors that bind nonself-antigens such as microbial products. Many families of pattern recognition receptors have been identified, although the best characterised are the TLR which are composed of 11 members and the interleukin IL-1 receptors which have 10 members. On ligation, the APC is activated by the Nuclear factor kappa B (NF-κB) pathway that leads to the production of type 1 IFNs, including IFN-β. These processes are stereotypical and do not generate immunological memory. The distinction between the body's cells and unwanted foreign invaders becomes obscured in autoimmune diseases. Thus, the innate immune system plays an important role in autoimmunity. Emerging data have identified an important contribution of miRNAs to the development and function of innate immune cells. Furthermore, studies investigating myeloid cell development and function have identified a common theme of a dynamic interplay between lineage-specific transcription factors and miRNAs. miRNAs involved in the regulation of granulocytes, monocytes, macrophages, DCs, NK, and natural killer T cells have been identified [9, 98].
Several studies have shown that transcription factors involved in monocytopoiesis are regulated by, and/or regulate, specific miRNAs, which indicates a connection between these molecular species during development [9, 98]. Studies in human umbilical cord blood CD34+ haematopoietic progenitor cells induced to differentiate into monocytes upon exposure to macrophage-colony stimulating factor (M-CSF) showed that monocytopoiesis is controlled by a circuitry involving sequentially three miRNAs (i.e., miR-17-5p, miR-20a, and miR-106a, members of the miR-17–92 and related miR-106a–92 families) and the transcription factor acute myeloid leukaemia-1 (AML1) [128]. During monocytic differentiation, the expression of these miRNAs is downregulated, whereas the transcription factor AML1 is upregulated at the protein but not mRNA level. Accordingly, this process promotes M-CSF receptor (M-CSFR) transcription, which therefore enhances the differentiation and maturation of monocytes. While these miRNAs target AML1, this transcription factor binds and transcriptionally inhibits expression of miR-17-5p, miR-20a, and miR-106a in a mutual negative feedback loop [128]. PU.1 is another transcription factor that is crucial for monocyte and macrophage differentiation [129]. PU.1 activates the transcription of miR-424, and this upregulation is involved in stimulating monocyte differentiation through miR-424-dependent translational repression of nuclear factor I/A (NFI-A). In turn, the decrease in NFI-A levels is important for the activation of differentiation-specific genes such as M-CSFR [129]. Translational repression of NFI-A by miR-233 is also involved in myeloid cell differentiation [95].
Neutrophils arise from granulocyte-monocyte progenitors under the influence of the transcription factor growth factor independent 1 (GfI1) [9]. GfI1 was recently shown to bind to the promoter regions of pri-miR-21 and pri-miR-196b and repress their expression [130]. The sustained expression of miR-155 can increase immature granulocyte numbers in vivo, and several of its targets, including SH2-domain-containing inositol-5-phosphatase 1 (SHIP1), are probably involved in this process [131, 132]. In addition to regulating neutrophil development, miRNAs also regulate granulocyte function. Genetic deletion of miR-223 can positively influence myeloid cell development and function in vivo [133]. miR-223 is induced by the myeloid transcription factors PU.1 and CCAAT/enhancer-binding protein-β (C/EbPβ), and it negatively regulates both the proliferation and activation of neutrophils. Myeloid Elf1-like factor 2C (MEF2C) has been shown to be a direct target of miR-223. TLR4-activated NF-κB rapidly increases the expression of miR-9 that operates a feedback control of the NF-κB-dependent responses by fine tuning the expression of a key member of the NF-κB family [134]. Brief exercise alters the miRNA profile in circulating neutrophils in humans [135].
miRNAs regulate distinct aspects of DC biology and so are involved in the crucial connection between innate and adaptive immune responses. miR-34 and miR-21 have been shown to be important for human myeloid-derived DC differentiation by targeting the mRNAs encoding Jagged1 and WNT1 [136]. Myeloid-derived DCs from Bic−/− (miR-155-deficient) mice showed defects in antigen presentation to T cells [137]. In addition, miR-155 downregulated expression of DC-specific ICAM3-grabbing nonintegrin (DC-SIGN; also known as CD209) by human monocyte-derived DCs through suppression of PU.1 expression [138]. DC-SIGN is a cell surface C-type lectin that binds pathogens, implicating miRNAs in the regulation of pathogen uptake by DCs. In human myeloid-derived DCs, knockdown of miR-155 expression significantly increased protein expression of the proinflammatory cytokine interleukin-1β (IL-1β) [139]. miR-146a acts as a regulator of monocyte and DC activation but not myeloid/DC subset differentiation [140].
miRNAs have been implicated in the development and function of NK cells which are important components of immune surveillance against cancer and viral infection [9]. NK cells express the receptor natural killer group 2, member D (NKG2D), which recognizes ligands, MHC class I polypeptide-related sequence A (MICA), and MICB, expressed by cells undergoing stress triggered by events such as viral infection or cell transformation [9, 141]. Engagement of NKG2D on NK cells leads to direct killing of the target cell. A recent study showed that a set of miRNAs, many of which are overexpressed by various cancer cells, binds to MICA and MICB 3′-UTR sequences and maintains expression of MICA and MICB protein under a certain threshold and facilitates acute upregulation of MICA and MICB during cellular stres [142]. Certain herpesvirus family members, namely, cytomegalovirus, Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus, produce miRNAs that target MICB mRNA, suggesting a miRNA-based immunoevasion mechanism that appears to be exploited by human viruses [143]. Lipopolysaccharide (LPS) stimulation decreased expression of miRNAs, miR-17-5, miR-20a, and miR-93, which target MICA, implicating a novel role for miRNAs in NKG2D ligand expression. These results suggest that TLR stimulation allows expression of NKG2D ligands through multiple pathways, including downmodulation of specific miRNAs [144]. Invariant NKT (iNKT) cells are a class of innate-like T cells that express an invariant TCR that recognizes lipids presented by the MHC class I-like CD1d molecule and regulate diverse immune responses [9, 145]. Two recent studies showed that differentiation and homeostasis of iNKT cells require Dicer in a cell-autonomous fashion [146, 147]. Dicer deletion results in a substantial reduction of iNKT cells in thymus and their disappearance from the periphery. Without Dicer, iNKT cells do not complete their innate effector differentiation and display a defective homeostasis due to increased cell death.
Numerous studies clearly demonstrate that miRNAs play an essential role in the regulation of various aspects of innate immunity, including the regulation of direct microbial killing, the production of cytokines, and antigen presentation by MHC molecules. All of these mechanisms are important for host defense and are instrumental in initiating antigen-specific responses by cells of the adaptive immune system [9].
Both the induction and repression of miRNA expression in response to inflammatory stimuli can influence several biological processes and exert pro- or antinflammatory effects [16]. Microbial products are important proinflammatory stimuli and activation by TLR ligands has been shown to modulate several miRNAs including miR-9, miR-125b, miR-146a, and miR-155 [98]. Of these miRNAs, only miR-146a and miR-155 appear to be induced in multiple cell types. In macrophages and DCs, stimulation by TLRs ligands results in miR-155 induction via NFκB pathway and signalling through the c-jun-N-terminal kinase (JNK) pathway [148, 149]. The induction of miR-155 by LPS has also been demonstrated in vivo and is accompanied by a decrease in miR-125b expression [149].
The downregulation of miR-125b appears to be necessary in macrophages to prevent suppression of Tumor necrosis factor-alpha (TNF-α) during the inflammatory response. miR-155 is upregulated in murine macrophages by the synthetic triacylated lipopeptide Pam3CSK4, the synthetic double stranded RNA analog poly(I:C), LPS, and CpG oligonucleotides, suggesting that several TLR ligands can induce miR-155 expression and that miR-155 is involved in the regulation of both bacterial and viral innate immune responses [148]. Fas-associated death domain protein, IkB kinase ε and receptor interacting serine-threonine kinase 1 were experimentally validated as targets of miR-155 [139, 148]. Involvement of miR-155 in the TLR-induced antigen presentation pathway was confirmed by a study showing that miR-155-deficient DCs are unable to induce efficient T-cell activation, with impaired antigen presentation and costimulation [137]. miR-9 is upregulated in both polymorphonuclear neutrophils and monocytes after TLR4 activation. This miRNA is also induced by TLR2 and TLR7/8 agonists and by the proinflammatory cytokines TNF-α and IL-1β [134]. MiR-146, miR-147, and miR-21 are also upregulated after the activation of TLR4 upon stimulation via LPS [150–152]. However, in contrast to miR-155, these miRNAs are negative regulators of pattern-recognition response. miR-146a reduces the translation of tumour necrosis factor receptor-associated factor-6 (TRAF6) and IL-1 receptor-activated kinase-1 (IRAK1), which are two key components of the TLR signalling pathway [150]. These studies indicate an essential role of miRNAs as important regulators of inflammation.
4. MicroRNAs and Autoimmunity
The roles of miRNAs are only beginning to be explored in the context of autoimmunity, in which they may be involved in regulating immune responses against self-tissues [9]. Immune responses are normally targeted against microbial pathogens and not self-antigens by mechanisms that are only partially understood. Over the past few decades, multiple mechanisms have emerged that operate to prune the lymphocyte repertoire of self-reactive specificities and maintain immunological tolerance. miRNAs target immune transcripts to fine-tune gene expression and turn on negative feedback loops. Both of these actions are crucial to limit costimulation, set precise cellular activation thresholds, curtail inflammation, control lymphocyte growth, and maintain regulatory T cell homeostasis and suppressive function [153]. miRNA expression is tightly regulated during hematopoiesis and lymphoid cell differentiation and disruption of the entire miRNA network or selected miRNAs may lead to dysregulated immune responses. Dysregulation of single or a few miRNAs or miRNA clusters can result from genetic variation, hormonal influences, or environmental triggers including infections. In the light of this vast and promiscuous miRNA-mediated regulation of autoimmune genes, it is anticipated that changes in miRNA levels or their target sequences may help explain susceptibility to complex autoimmune diseases. Abnormalities in miRNA expression related to inflammatory cytokines, Th17 and Treg cells, as well as B cells have been described in several autoimmune diseases [9, 13, 14, 154].
In 2007, the involvement of miRNA in a new pathway regulating autoimmunity was discovered in T lymphocytes in the sanroque mouse [155]. The sanroque mouse was originally selected from screening mutant mice derived from the chemical mutagen N-ethyl-N-nitrosourea and has been shown to result from a mutation in the gene Roquin that encodes a RING-type ubiquitin ligase [14]. In normal T cells, Roquin normally limits the expression of inducible T-cell costimulator (ICOS) by promoting the degradation of ICOS mRNA. In sanroque mice, however, the absence of this regulation leads to an accumulation of lymphocytes that is associated with a lupus-like autoimmune syndrome. Yu et al. reported that miR-101 is required for the Roquin-mediated degradation of ICOS mRNA [155]. Introducing mutations into the miR-101 binding sites in the 3′-UTR of ICOS mRNA disrupted the repressive activity of Roquin. These results revealed a critical miRNA-mediated regulatory pathway that prevents lymphocyte accumulation and autoimmunity. More recently, deletion studies showed that targeted deletion of miRNAs in hematopoietic stem cells or in thymus disrupts T cell homeostasis and results in autoimmunity and abnormal cytokine production. Recent studies revealed the importance of miRNA regulation in safeguarding Treg function in the prevention of autoimmunity. miRNA biogenesis is indispensable for the function of Treg cells. Specific deletion of either Drosha or Dicer phenocopies mice lacking a functional Treg cell-specific transcription factor Foxp3 gene or Foxp3(+) cells whereas deletion throughout the T cell compartment also results in spontaneous inflammatory disease, but later in life [112]. Treg cell-mediated immune tolerance is critically dependent on the Dicer-controlled miRNA pathway. Mice with conditional Dicer knockout within the Treg cell lineage rapidly developed fatal systemic autoimmune disease resembling the Foxp3 knockout phenotype [110, 111]. Although thymic Treg cells developed normally in Dicer-deficient mice, the cells exhibited altered differentiation and dysfunction in the periphery. Interestingly, Dicer-deficient Treg cells retained some suppressive activity, albeit reduced compared to wild-type mice [111]. However, under inflammatory conditions Dicer-deficient Treg cells were completely devoid of any suppressor activity and instead showed a robust in vitro proliferative response leading to autoimmunity suggesting that miRNAs preserve the Treg cell functional program under inflammatory conditions. These findings support a central role for miRNAs in maintaining the stability of differentiated Treg cell function in vivo and homeostasis of the adaptive immune system.
Further support for a causal relationship between specific miRNAs and the onset of autoimmunity has come from studies involving miR-17–92 overexpression in mice [102]. Mice with higher expression of miR-17–92 in lymphocytes developed lymphoproliferative disease and autoimmunity and died prematurely. miR-17–92 overexpression promoted marked lymphoproliferation, the presence of serum autoantibodies, and tissue changes such as lymphoid infiltrates and antibody deposition. T cells seem to develop normally in these mice, but the number of mature CD4+ T cells was markedly increased and they had a highly activated profile, suggesting a failure of peripheral tolerance. Lymphocytes from these mice showed more proliferation and less activation-induced cell death. The miR-17–92 miRNA suppressed expression of the tumor suppressor Phosphatase and tensin homolog (Pten) and the proapoptotic protein Bim [102]. This mechanism probably contributed to the lymphoproliferative disease and autoimmunity of miR-17–92-transgenic mice. Dysregulation of miRNAs involved in immune cell development may cause autoimmunity. A recent study has shown that inhibition of miR-181a in T cells during thymic development converts endogenous positively selecting peptides into autoantigens [156].
Emerging evidence has demonstrated that miRNAs are differentially expressed in autoimmune diseases and miRNA regulation may impact in the development or prevention of autoimmunity. miRNA dysregulation is linked to autoimmune diseases that include rheumatoid arthritis, systemic lupus erythematosus, primary biliary cirrhosis, ulcerative colitis, psoriasis, Idiopathic thrombocytopenic purpura, primary Sjögren's syndrome, and MS.
These molecules have also been shown to be useful as diagnostic and prognostic indicators of disease type and severity [15–17]. Many autoimmunity and disease susceptibility genes are targeted by several miRNAs [153, 157]. The precise mechanisms miRNAs use to promote or hinder autoimmunity have yet to be elucidated. However, several potential mechanisms deserve consideration, including loss or downregulation of miRNA expression due to mutation, epigenetic activation, aberrant processing, or transcriptional downregulation; overexpression of particular miRNA consequent to gene amplification or mutation, especially miRNA promoter regions, or due to transcriptional upregulation that may result in the suppressed production of its target proteins; and mutation at the 3′-UTR of the target mRNA or its gene [14]. In most cases the role of specific miRNAs in autoimmune diseases has been established in vitro by association, and that causal roles in vivo remain a matter of investigation [14].
Clinical characteristics along with pathological heterogeneity make MS appealing to study many aspects of miRNAs in an organ-specific autoimmune disease, such as their potential as diagnostic or prognostic biomarkers and their role in pathogenesis of autoimmunity, neuroinflammation, and organ dysfunction. Thus, we will focus on the involvement of specific miRNAs in MS pathogenesis following the general overview of the immunopathobiology of the disease.
5. MicroRNAs and Multiple Sclerosis
5.1. Genetic and Epigenetic Factors in Multiple Sclerosis
MS is a chronic inflammatory demyelinating disease of the CNS that primarily affects young adults. Prevalence rates for MS vary between 2 and 160 per 100,000 in different countries, and more than 2 million individuals are affected by this disease worldwide [158]. Autoreactive T cell-mediated autoimmune response to myelin antigens results in both inflammation and axonal degeneration accounting for the disability of patients with MS [19]. The exact factors that initiate inflammation are unknown, but it is generally believed that MS is caused by environmental factors in a genetically susceptible host that trigger a T-cell autoimmune response against the CNS [18]. In the literature, several genetic factors have been described to influence the development and severity of MS and are responsible for disease susceptibility [20]. The major genetic factor in MS is the major histocompatibility complex [159], however recent genomewide association studies revealed new susceptibility alleles for MS that are all related with immune functions (e.g., CASP8, CD58, STAT3, interleukin 7 receptor (IL7RA; CD127), interleukin 2 receptor A (IL2RA)) [20, 160, 161]. Nevertheless, no locus has been detected of constant form in all the studies, suggesting the existence of genetic heterogeneity. MS is likely to be the result of interactions between environmental stimuli (e.g., infection), susceptibility genes (which predispose individuals to the development of neuroinflammation), and modifier genes (which affect disease phenotype in susceptible subjects). Although viruses may trigger MS relapses, there is no definitive evidence that there is an MS virus or an ongoing chronic infection of the nervous system. It is possible, however, that a self-limited CNS infection in childhood could trigger MS, and epidemiological evidence suggests that Epstein-Barr virus (EBV) may play a key role in MS [18]. Other nongenetic but nevertheless gene-regulation factors including epigenetic mechanisms such as DNA methylation and histone modification and miRNA-mediated posttranscriptional gene regulation might individually influence both susceptibility and severity of the disease [162].
Experimental allergic encephalomyelitis (EAE) serves as the primary and most widely used animal model for MS and can be induced in susceptible rodent strains by active immunization of myelin antigens [163, 164]. Different types of the model have been developed that mimic virtually all the clinical features of MS including relapsing, relapsing remitting, progressive, and opticospinal forms. The majority of treatments for MS have stemmed from studies in the EAE model, further supporting the concept that autoimmune processes in the EAE model are relevant to MS [18]. However, there are also examples of mechanisms that have worked in EAE but have failed in the clinic, such as the TNF-α antagonists and anti-p40 (a subunit of IL-12 and IL-23) [163, 164].
5.2. Immunopathobiology of Multiple Sclerosis
The clinical course of MS varies, with 80% of patients presenting with episodes of disability followed by a period of recovery classified as relapsing-remitting while 10%–15% exhibit a more progressive disease without remission, namely, primary progressive [165, 166]. The patient has a yearly risk of about 3% for a transition from the relapsing-remitting phase to the chronic, progressive form of MS. Over a period of 10 years, roughly half of relapsing-remitting patients enter a secondary progressive stage of disease characterized by accumulating disability while recovery between episodes diminishes.
There is consensus that a dysregulated immune system plays a critical role in the pathogenesis of MS. Relapses are driven by the adaptive immune system and involve waves of Th1, Th17, and CD8+ cells that infiltrate the CNS and provoke an attack. These cells are modulated by Treg and B cells. MS is initiated and maintained by continuous migration of inflammatory immune cells from the periphery into the target organ. The three ways that lymphocytes can enter the CNS include entry from the bloodstream across the choroid plexus into the Cerebrospinal fluid (CSF), from the blood in the subanachroid space into the CSF, or directly into the parenchyma under permissible conditions, such as inflammation, controlled by cell adhesion molecules and cytokines [167]. Subpopulations of T cells may employ different trafficking mechanisms [163]. Infiltration of T cells into the CNS initiates a complex immunological cascade consisting of epitope spreading, which triggers new attacks, and activation of the innate immune system composed of microglia, dendritic cells, and astrocytes [18]. The secondary progressive phase is due to neurodegeneration triggered by neuroinflammation and is driven by the innate immune system. The loss of axons and their neurons in the course of chronic neuroinflammation is a major factor determining long-term disability in patients and neurodegeneration as the major cause of irreversible neurological disability in MS patients. Thus, in the relapsing stage, a proinflammatory milieu that combines both the innate and adaptive immune system is present whereas in the progressive stage abnormalities of the innate immune system predominate [18].
5.2.1. Adaptive Immune Responses in Multiple Sclerosis
Pathogenic T Cells —
Among cells isolated from the inflammatory infiltrate in actively demyelinating MS lesions, approximately 10% are T cells [168, 169]. There, multiple T-cell subsets have been implicated: CD4+ Th1 and Th17, γ/δ T cells, CD8+, and Treg cells [163]. CD4+ T cells are the most prominent cells in active MS lesions but are not present in chronic MS lesions [170]. It is generally believed that the acute MS lesion is initiated by a myelin-reactive CD4+ T cell that is stimulated in the periphery and enters the brain and spinal cord [18]. Recent research has focused on the different roles of subsets of CD4+ T cells in MS and other autoimmune diseases. Th1 cells classically express IFN-γ, TNF-β, IL-2, and nitric oxide [171] and activate macrophages to stimulate cell-mediated immunity [168]. Th2 cells release IL-4, IL-5, IL-6, IL-10, IL-13, and TGF-β [168]. These cytokines may be associated with disease recovery in MS. Th17 cells are CD4+ T cells subtype that are associated with autoimmune diseases. Th17 cells are dependent on IL-23, TGF-β, IL-6, and IL-1 [163]. Th17 cells produce IL-17A and IL-17F, which are upregulated in chronic lesions [172, 173], and IL-22 which is also involved in MS pathogenesis. It is now recognized that Th17 cells play a crucial role in autoimmunity in the EAE model [174]. However, recent work by Haak et al. [175] has demonstrated that overexpression of IL17 in T cells did not exacerbate EAE. If Th17 cells are given with Th1 cells, then full disease induction occurs [176]. These results suggested that pure Th17 cells are not pathogenic. Both types of cells (Th1 and Th17) may play a role in MS and could account for the immunological and clinical heterogeneity of the disease [18, 177].
Most TCRs are composed of two linked polypeptides, α and β, which participate in the recognition of foreign antigen plus self-MHC [168, 178]. However, a small subtype of circulating lymphocytes expresses γ/δ TCR polypeptides which function in both innate and adaptive immunity [168, 179]. Clonal expansion of activated lymphocytes bearing the γ/δ TCR has been demonstrated in samples isolated from the CSF of patients with recent-onset MS but not from patients with chronic MS [180]. Recently, investigators demonstrated that γ/δ T cell-deficient mice were unable to recover from EAE [168, 179, 181]. Histopathologically, there was a prolonged presence of monocytes and lymphocytes in the CNS [179, 181]. CD8+ T cells are also implicated in MS pathology. Within MS plaques, clonal and oligoclonal expansion of CD8+ T cells reactive to myelin antigens has been observed [182]. A new effector T cell subset, Th9 cells, has been identified. Jager et al. showed that Th9 effector cells participate in induction of EAE [183]. These results suggested that Th9 cells may participate in MS pathogenesis.
Regulatory T Cells —
Defects in Treg-cell function have been described in MS, and a major goal of MS immunotherapy is to induce regulatory cells in a physiological fashion [184–186]. Clinical studies in MS patients showed that Treg cell dysfunction occurred in the initial stages of the disease [168]. In addition, experimental data suggest that regulatory cells may not be effective if there is ongoing CNS inflammation [187].
B Cells and Antibodies —
MS is generally thought to be a T cell-mediated immune disease although there is an important role of humoral immunity in pathogenesis of MS. Intrathecal antibody synthesis is a hallmark of the disease process and, in most of cases, consists of oligoclonal IgG production [18, 188]. A direct correlation has been reported between levels of immunoglobulin production and MS disease severity [189, 190]. Antibodies to self-antigen such as Myelin Basic Protein (MBP) and myelin oligodendrocyte glycoprotein (MOG) have been identified in the serum of patients with MS and clinically isolated syndromes (CISs) [191, 192]. B cells and plasma cells have been detected in brains and CSF of patients with MS [189]. Characterization of the B-cell compartment within the CSF of MS patients shows that short-lived plasmablasts, not plasma cells, are the predominant antibody-secreting cell in MS CSF [189, 193] and the B cell to monocyte ratio correlates with the rate of disease progression. B cells are also potent APCs and may play a prominent role in T-cell antigenic stimulation. Thus, B cells may well be active participants in initiating and maintaining disease [168].
5.2.2. Innate Immune Responses in Multiple Sclerosis
The innate immune system consists of monocytes, dendritic cells, and microglia. The innate immune system plays an important role in the immunopathogenesis of MS. The secondary progressive phase of MS has been believed to be related to neurodegenerative changes in the CNS [18]. Furthermore, chronic microglial activation occurs in MS [194]. The peripheral innate immune system changes cause the transition from the relapsing-remitting to the progressive stage. This raises important questions regarding the pathogenesis and treatment of different stages of MS. A major question is whether aggressive and early anti-inflammatory treatment will prevent the secondary progressive form of the disease. There are no specific therapies designed to affect the innate immune system in MS. Furthermore, like the adaptive immune system, there are different classes of innate immune responses, for example, protective and tolerogenic versus pathogenic and proinflammatory [18]. This fact should be kept in mind for new drug development studies that target innate immunity.
Antigen Presenting Cells —
Macrophages are the major MHC Class II positive cell in the CSF. Macrophages in EAE have an integral role in initiating disease, and depletion of macrophages significantly inhibits disease [195]. Macrophages are not the only class II positive cells that can present myelin antigens. Monocytes, DCs, microglia, and astrocytes have all been implicated in presenting antigen and involved in MS pathogenesis [163]. Greter et al. demonstrated that mice with MHC class II expression limited to DC can still develop disease [196]. DCs can be further subdivided into myeloid (mDCs) and plasmacytoid DCs (pDCs) depending on their lineage, and they also differ in function [163]. pDCs are the major CNS-infiltrating DC population during EAE and pDCs have both stimulatory and regulatory effects on T cells [197]. pDCs negatively regulate CD4+ inflammatory responses in the CNS [197]. Depletion of pDCs during either the acute or relapse phase of EAE resulted in exacerbation of disease severity [163]. In MS patients, pDC from peripheral blood showed an immature phenotype. The pDC had a lower capacity to secrete IFN-α upon TLR-9 stimulation. This may indicate why common infectious agents trigger MS attacks [198]. mDCs within the CNS activate myelin specific T cells that are recruited to the inflamed tissue and facilitate differentiation into Th1 and Th17 cells [199, 200]. However, Deshpande et al. reported that mDC isolated from the peak of disease are less efficient APCs than those isolated at disease onset, suggesting that changes in DC phenotype may contribute to remissions [200].
Microglial cells seem to be crucial for maintaining autoimmune responses in the CNS. It has been demonstrated that both a microglial cell-specific deficiency of CD40 expression and a transient inactivation of microglial cells reduce disease severity [201].
Astrocytes also express MHC Class II after IFNγ exposure and it has been reported that astrocytes can present antigen [202]. Astrocytes from mice deficient in Class II transactivator (CIITA) failed to activate MOG-specific CD4+ T cells due to a lack of MHC Class II expression [163, 203]. However, CIITA-deficient mice still were susceptible to EAE [203]. However, human astrocytes do not effectively activate encephalitogenic T cells in vitro [204]. They may also influence the disease by secretion of cytokines and chemokines.
5.3. Neurodegeneration in Multiple Sclerosis
The identification of MS susceptibility loci, of which at least 15 have a primary function in immunological systems, favors early immune dysregulation followed by secondary neurodegenerative processes [163]. Indeed, MS is not exclusively a white matter disease. Specific cognitive deficits such as memory impairment, attention deficit, and reduced mental reasoning are increasingly being explained by damage to neurons in the gray matter, which affects 45%–65% of MS patients [205]. Although the precise trigger for MS remains elusive, it is understood that autoimmune mechanisms underlie the pathology, and furthermore that activated T cells migrate through the BBB where they accumulate and proliferate because of antigen restimulation. These cells release a host of proinflammatory molecules, which, in turn, further activate microglia or infiltrated macrophages and B cells. Axonal and neuronal injury occurs as an early event in the disease and is strongly correlated with the degree of inflammation in the brain [206–208]. In MS, neurons in the cortex and spinal cord are also affected, albeit to varying extents [209, 210]. The latest events in the chain of neuronal damage processes following focal axonal lesions include axon degeneration and atrophy of neuronal cell bodies and dendrites [165]. The loss of neurons and their processes is the leading cause of atrophy and is the primary determinant of long-term disability in MS patients. This chain of events produces a marked inflammatory response, which causes axonal injury through various antigen specific and bystander mechanisms.
In MS, both soluble factors and surface molecules could participate in neurodegeneration. Besides injurious proinflammatory molecules, proapoptotic factors produced by T cells, including FasL, granzyme B, soluble TNF-related apoptosis-inducing ligand (TRAIL), glutamate, nitric oxide, and free radicals, are possible mediators of injury [208, 211–214]. Accumulating evidence suggests that the increased energy demand of impulse conduction along excitable demyelinated axons and reduced axonal ATP production induce a chronic state of virtual hypoxia in chronically demyelinated axons, ultimately leading to excessive stimulation of Ca2+-dependent degradative pathways [215]. Glutamate and nitric oxide can lead to enhanced expression of chemokine (C-C-motif) ligand 2 (CCL2) on astrocytes, which, in turn, leads to infiltration of CD11b cells and additional tissue damage [216]. Antiexcitotoxic compounds have an ameliorating effect in EAE model [18]. Another important component of neurodegeneration relates to changes in Na+ channels, and these are targets of therapy [217].
Axonal injury can be directly caused by immune cells. CD4+ and CD8+ T-cell subsets, once activated, are highly neurotoxic. These effects are mediated through a variety of contact-dependent mechanisms involving cell surface molecules such as FasL, LFA-1, and CD40. Th1 and Th17 proinflammatory classes of CD4+ T cells are neurotoxic whereas the anti-inflammatory Th2 subset is not [218]. Although activated T cells can clearly harm neurons, the converse has also been observed. Activated T cells underwent apoptosis that was mediated through neurons via a FasL-dependent mechanism [219]. In another context, neurons may induce encephalitogenic T cells to convert to T-regulatory cells that inhibit encephalitogenic T-cell action and suppress EAE [220]. It is likely that the adaptive immune system orchestrates the attack against CNS cells and drives microglia and macrophages to attack oligodendrocytes and neurons. Activated microglia and peripherally derived macrophages are shifted towards a strongly proinflammatory phenotype and produce apoptosis-inducing molecules such as the TRAIL and the proinflammatory cytokines TNF-α and IL-1β as well as potentially neurotoxic substances including nitric oxide, oxygen radicals and proteolytic enzymes [221, 222].
5.3.1. Neurodegeneration and MicroRNAs
Many recent studies provide a link between miRNA function and neurodegeneration [223–225]. Complete loss of miRNA expression in the brain leads to neurodegeneration in several animal models. Evidence from patient material is emerging that miRNA dysregulation could, indeed, contribute to neurodegenerative disorders. The translation of proteins previously implicated in familial forms of disease seems to be under control of miRNAs, and changes in miRNAs might explain how these proteins become affected in sporadic neurodegeneration. Thus, miRNAs are rapidly moving to center stage as key regulators of neuronal development and function as well as important contributors to neurodegeneration. The link between miRNAs and axonal neurodegeneration in the context of MS has not been focused on to date.
Endogenous tissue repair mechanisms such as myelin repair, gliogenesis, and neurogenesis in MS may also be modulated by specific miRNAs. Enhancing such repair mechanisms is an important, and increasingly realistic, therapeutic goal in MS [226]. Neurogenesis is defined as a process that includes the proliferation of neural stem/progenitor cells (NPCs) and the differentiation of these cells into new neurons that integrate into the existing neuronal circuitry. Recent studies point to the importance of miRNAs in regulating lineage-specific gene expression and determining neuronal identity during neurogenesis [227, 228]. These new observations suggest that miRNAs could function at many levels to regulate self-renewal of neural stem cells and neuronal fate specification, implicating miRNAs in the complexity of neurogenesis. miRNAs are also involved in adult neurogenesis which may imply the possible role of some miRNAs in endogenous repair mechanisms in MS [229, 230]. In addition, cross talk between miRNA and epigenetic regulation contributes to the modulation of adult neurogenesis [231]. The modulation of miRNAs involved in adult neurogenesis may stimulate the differentiation of NPCs into mature neurons that can replace neurons lost through the disease process in MS. Patient studies also suggest the presence of neuronal precursor cells in MS lesions [232].
Within the CNS, myelin is produced by oligodendrocytes. Developmentally, the oligodendrocyte lineage arises from subventricular zone progenitors that give rise to oligodendrocyte progenitor cells (OPCs), which divide and migrate throughout the CNS before terminally differentiating to generate mature oligodendrocytes which myelinate receptive axons [233]. Each step of progression along the lineage is under tight transcriptional control; elucidation of this control is vital for understanding developmental myelination and for developing strategies to promote repair in demyelinating diseases.
Remyelination following CNS demyelination restores rapid saltatory conduction of action potentials and contributes to the maintenance of axonal integrity [234]. Chronic demyelination predisposes axons to atrophy, an irreversible event that is a major pathological correlate of progressive functional decline. Remyelination in MS is in most cases insufficient, leading to irreversible disability. Different and nonexclusive factors account for this repair deficit [235]. Local inhibitors of the differentiation of OPCs might play a role as well as axonal factors impairing the wrapping process. Alternatively, a defect in the recruitment of OPCs toward the demyelinated area may be involved in lesions with oligodendroglial depopulation. Deciphering the mechanisms underlying myelin repair success or failure should open new avenues for designing strategies aimed at favoring endogenous remyelination [235]. The few treatments that are available for combating myelin damage in MS, which largely comprise anti-inflammatory drugs, only show limited efficacy in subsets of patients. More effective treatment of myelin disorders will probably be accomplished by early intervention with combinatorial therapies that target inflammation and other processes—for example, signaling pathways that promote remyelination [236]. However, the integration of these pathways with transcriptional and posttranscriptional regulatory networks is not fully understood. The interplay of transcription factors and epigenetic modifiers including histone modifications, DNA methylation, and miRNAs during development is essential for the acquisition of specific cell fates [237]. Recent studies have identified a number of new transcriptional regulators and miRNAs as having key roles in oligodendrocyte (OL) differentiation and CNS myelination, providing new targets for myelin repair [233].
Selective deletion of miRNA-processing enzyme, Dicer, in oligodendrocyte lineage cells results in severe myelinating deficits despite an expansion of the oligodendrocyte progenitor pool [238, 239]. Dugas et al. identified the miRNA pathways responsible for myelination using Dicer1-deleted transgenic mouse model [238]. In this study, they found the inhibition of OPC-OL miRNA processing resulting in defects in mature miRNA processing. They also identified three miRNAs: miR-219, miR-138, and miR-338. Of these miRNAs, miR-219 is important for OL differentiation, directly repressing PDGFRalpha, Sox6, FoxJ3, and ZFP238 which promote OPC differentiation [238]. Postnatal Dicer ablation in mature OLs results in inflammatory neuronal degeneration through increased demyelination, lipid accumulation, and peroxisomal and oxidative damage and therefore indicates that miRNAs play an essential role in the maintenance of lipids and redox homeostasis in mature OLs [240]. A small subset of miRNAs (e.g., miR-9, miR-23, miR-206, miR-219, miR-338, and miR-17-92 cluster), is important to orchestrate the switch from OPCs to myelin-forming oligodendrocytes [238–244]. Transcription factors, myelin proteins, signaling molecules, and cytoskletal proteins were identified as validated targets of these miRNAs. Interestingly, the highest differentially expressed miRNAs demonstrated a similar pattern of expression throughout all stages of differentiation, suggesting that they potentially regulate a common target or set of targets in this process [245].
Dysfunction of the BBB is a major hallmark of MS and may impair tissue homoeostasis, which may have effects on disease progression, repair mechanisms, and drug delivery [246–248]. Thus, restoration of BBB permeability may help endogenous tissue repair. Although the pivotal role of miRNAs in angiogenesis is well established [249–251], these molecules have not been focused on in the context of MS, BBB integrity, and cerebral angiogenesis. Only one study showed that a proapoptotic miRNA, miR-15a, was downregulated by peroxisome proliferator-activated receptor delta in brain endothelial cells [252]. Peroxisome proliferator-activated receptor delta is a nuclear receptor whose agonists have been shown to inhibit EAE [253–255]. However, the contribution of vascular protection by peroxisome proliferator-activated receptor delta through miRNA regulation in the recovery process is not known.
5.4. MicroRNA Studies in Multiple Sclerosis
Little is known about what drives the differential control of the immune system in MS patients compared to unaffected individuals. Thus, it is important to reveal the aberrant miRNA expression profiling in MS patients. To our knowledge there have been only seven publications investigating the role of miRNAs in MS, six of which focus on the immune system in MS and the other on active and inactive MS lesions (Table 2). Differences in miRNA expression patterns have been documented in MS compared to healthy controls and in relapse versus remission of the disease.
Table 2.
Differential miRNA expression in Multiple Sclerosis.
| Sampe type | Number of patients and disease status | Specificity of patients and treatment | Number of tested miRNA | Results | Target genes | Reference |
|---|---|---|---|---|---|---|
| Whole bood | 59 MS (18 PP, 17 SP, 24 RR) and 37 controls | Causian No IMT | 733 | miR-17 and miR-20a downregulated | ND | Cox |
| CD4+CD25+ | 12 MS (RR) and 14 controls | No IMT | 723 | miR-106b, MiR-19a, MiR-19b and miR-25 upregulated | TGF β signaling | De Santis |
| CD4+, CD8+, B | 8 MS (RR) and 10 controls (microarray) | No IMT | 365 | miR-17-5p upregulated in CD4+ cells | ND | Lindberg |
| 15 MS (RR) | ||||||
| and 10 controls (qPCR) | ||||||
| Peripheral blood leukocytes | 43 MS (RR) | Chinese | ND | miR 326 upreguated in CD4+ cells | Ets-1 | Du |
| 40 control | miR-326 promotes Th-17 differentiation | |||||
| 11 NMO | ||||||
| Whole bood | 20 MS (RR) | glatiramer acetate (9) | 866 | miR-145 upregulated in MS | ND | Keller |
| 19 controls | interferon-b (10) | |||||
| Whole bood | 21 MS (9 remission, 4 relaps) | ND | 364 | miR-18b and miR-599 upregulated in relapse | interleukine signaling | Otaegui |
| 8 control | miR-96 upregulated in remission | Wnt, glutamate | ||||
| Brain tissue | 20 MS (16 active, 5 inactive) | ND | 365 | miR-34a, miR-155 and miR-326 upregulated in active lesions | CD47 | Junker |
| 9 controls | ||||||
Ets-1: v-ets erythroblastosis virus E26 oncogene homolog 1; IMT: Immunmodulatory treatment; MS: Multiple sclerosis; ND: not determined; NMO: neuromyeliis optica; PP: primary progressive; RR: Relapsing remitting, Secondary progressive.
Studies in peripheral blood mononuclear cells (PBMCs) of patients with MS revealed different expression patterns compared to control individuals. Using qPCR, a pilot study of the expression of 346 miRNAs in PBMCs obtained from a small number of MS patients during relapse and remission, versus healthy controls, demonstrated differences in gene expression patterns not only between the MS patients and healthy controls but also between patients with and without active disease [256]. Two miRNAs (miR-18b and miR-599) have been shown to be associated with relapse whereas another miRNA (miR-96) was found to be involved in the remission of the disease. The genes targeted by miR-96 are involved in immunological pathways such as interleukin signaling and other pathways as wnt signaling [256]. In another recent study, Keller et al. [257] investigated the expression profiles of 866 human miRNAs; in whole blood cells of MS patients 165 miRNAs were identified that were significantly up- or downregulated in patients with RRMS as compared to healthy controls. The best single miRNA marker, miR-145, allowed discriminating MS patients from controls with a specificity of 89.5%, a sensitivity of 90.0%, and an accuracy of 89.7%. The authors concluded that single miRNAs, and even more so miRNA expression profiles, may have the potential to serve as diagnostic biomarkers for RRMS. However, MS patients in that study were treated with either glatiramer acetate or interferon-beta while one patient was not treated with anything. One of the difficulties of studying MS is the acquisition of samples unaffected by the influence of immunomodulatory treatment. These studies do not provide information about miRNA expression in various cell subpopulations and their importance during the differentiation and activation of lymphocytes in MS.
The recent study by Du et al. [258] identified a Th17 cell-associated miRNA, miR-326, as a major determinant of MS in a Chinese population but not of neuromyelitis optica. Its expression was highly correlated with disease severity in patients with MS and mice with EAE. In vivo silencing of miR-326 resulted in fewer Th17 cells and mild EAE, and its overexpression led to more Th17 cells and severe EAE. Du et al. also found that miR-326 promoted Th17 differentiation by targeting Ets-1, a negative regulator of Th17 differentiation [258, 259]. These results suggest a critical role for miR-326 in the regulation of Th17 differentiation and the pathogenesis of MS. Although a more recent study did not identify any statistically significant change in whole blood miR-326 expression between MS patients and controls [260], one of the three most upregulated miRNA detected in active MS lesions is miR-326 lending further support to the relevance of this miRNA for MS pathogenesis [261]. The discrepancies between the results of clinical studies may be caused by differences observed in MS patients from Asian or Caucasian origin [260]. In a group of MS patients in relapse, glucocorticoid treatment downregulates miR-326 expression indicating that this miRNA is under control of disease-modifying drugs and thus may be used in the monitoring of therapy responses [258]. Further exploration of the function of miR-326 in other cell types may be of great importance for understanding the immunopathogenesis of MS.
Although it is known that specific miRNAs are involved in each step of the maturation of pluripotent hematopoietic stem cells into the various blood cell lineages including B and T cells [262], little is known about miRNA involvement in the differentiation during T-cell activation under disease conditions such as MS. A recent study has analyzed the expression of 365 miRNA and revealed different miRNA expression profiles in CD4+, CD8+, and B cells of peripheral blood from eight RRMS patients compared with ten healthy volunteers and they have also validated miRNA in CD4+ cells with qPCR [263]. Importantly, all the patients had no immunomodulatory or other MS specific treatments in the six months before or during the study. Ten miRNAs in CD4+, four miRNAs in CD8+, and six miRNAs in B cells were differentially expressed in MS patients. Lindberg et al. found distinct and cell-specific expression patterns of miRNA in all cell subpopulations, which is well in line with reports about diverse miRNA expression in immune cells. Furthermore, the expression of potential target genes of these miRNA was altered. miR-17-5p, which is known to be involved in the development of autoimmunity and in numerous lymphoproliferative diseases [264], was detected in CD4+ lymphocytes of MS patients [263]. Functional experiments with a synthetic inhibitor of miR-17 also supported the link between miRNA expression and the altered target gene expression. Moreover, authors have found that miRNAs were also differentially expressed in the two study groups following in vitro stimulation of CD4+ T cells with anti-CD3/CD28. miR-17-5p and miR-193a were strongly upregulated, in contrast to the downregulation of miR-497, miR-1, and miR-126. This was correlated with alterations in the expression of potential target genes of miR-17-5p, that is, phosphatase and tensin homology and phosphatidyl-inositol-3-kinase regulatory subunit 1, which were downregulated upon stimulation of CD4+ cells in vitro. Other deregulated miRNAs did not respond to the stimulation probably due to other, non-T-cell activation related, mechanisms in their mode of action. These results support the role of miRNA-dependent regulatory mechanisms in the immunopathogenesis of MS. However, in a larger and more recent study, Cox et al. showed that miR-17 is underexpressed in MS whole blood [260]. This discrepancy between the studies may be due to methodological differences. Another cause of the discrepancy may be the material analyzed in those two studies, such as cell types. Patient number and disease activity status may also change the outcome of the analyses. In the study by Cox et al., the transcriptome of currently known miRNAs was investigated using miRNA microarray analysis in peripheral blood samples of 59 MS patients that were free of disease modifying therapy for at least 3 months before the study and 37 healthy age-matched controls. Of the patients, 18 had a primary progressive, 17 a secondary progressive, and 24 a relapsing remitting disease course. In all MS subtypes miR-17 and miR-20a were significantly underexpressed in MS, confirmed by qPCR. It was demonstrated that these miRNAs modulate T cell activation genes in a knock-in and knock-down T cell model. The same T cell activation genes are also upregulated in MS whole blood mRNA, suggesting that miR-17 and miR-20a are implicated in the development of MS [260].
It is known that Tregs play a key role in the autoimmune balance and their improper function may facilitate the expansion of autoreactive T cell clones. CD4+CD25+Foxp3+ Treg cells play a pivotal role in the maintenance of self-tolerance and controlling autoimmunity [109].
Recent evidence has been provided for a potential functional defect of CD4+CD25+Foxp3+ Treg cells in patients with RRMS [265]. The fact that ablation of miRNAs in Treg cells completely phenocopies the loss of Foxp3 cells clearly indicated that multiple immunosuppressive mechanisms used by Treg cells are ultimately controlled by miRNAs [109]. The miRNA expression profile in Treg cells from treatment naive RRMS patients has recently been analyzed by De Santis et al. [266]. The suppressive capacity of isolated CD4+CD25+ has been verified by in vitro suppression assays. When the expression levels of 723 human miRNAs were compared in CD4+CD25+ T cells obtained from 12 MS patients and 14 healthy donors using microarray assay, 23 human miRNAs were differentially expressed between study groups. Among the deregulated miRNAs, members of miR-106b-25 were found to be downregulated in the Treg cells of MS patients when compared to healthy donors as confirmed by qPCR. Unexpectedly, in a preliminary experiment performed in a very small number of subjects, the ratio between Treg cells (CD4+CD25+CD127DIM)/T effector cells (CD4+CD25+CD127high) showed an enrichment of these miRNA in Treg cells derived from patients as compared to healthy controls [266]. miR-106b and miR-25 modulate the TGF-beta signaling pathway through their action on cell cycle inhibitor CDKN1A/p21 and the proapoptotic gene BCL2L11/Bim. TGF-β is involved in Treg cell differentiation and maturation [267]. Therefore, the deregulation of this miRNA cluster may alter Treg cell activity during the course of MS, by altering TGF-β biological functions.
A recent in situ and in vitro study extends the current concepts of MS lesion activity to the level of miRNA-regulated gene expression and may have broad implications for the regulation of macrophage activation in autoimmune and inflammatory diseases in general [261]. In the study which used laser capture microdissection from active and inactive MS lesions to pool single cells and in vitro cultures, differentially expressed miRNAs were assigned to specific cell types by qPCR. Tissue specimens were obtained from the brains of 20 MS pateints and nine control subjects without any known neurological disease. In active MS lesions 20 miRNAs, including those that are associated with immune responses such as miR-155 and miR-146a, were at least twofold more abundant and eight miRNAs were at least twofold less abundant than in normal white matter. Eight of the active MS brain specimens are derived from MS cases with a very fulminant disease course called Marburg's variant. Some miRNAs were more prominently regulated in Marburg's variant than those in the other active MS lesions, probably reflecting the more intense tissue destruction in Marburg's variant. In inactive MS lesions, 22 miRNAs were at least twofold more abundant and 13 miRNAs at least twofold less abundant than in normal white matter. Among the significantly altered miRNAs, some showed differential regulation in active versus inactive lesions whereas others were modified in the same direction. Junker et al. found that three of the most upregulated miRNAs in active MS lesions, namely, miR-155, miR-326, and miR-34a, target CD47, which was one of the downregulated transcripts in the active lesions in comparison to normal brain white matter. CD47 is a membrane glycoprotein and mediator of macrophage inhibition via its receptor signal-regulatory protein alpha on myeloid cells. Using laser dissection microscopy combined with qPCR, CD47 gene expression was found to be downregulated in the center of chronic active and inactive MS lesions [268]. CD47 has been considered as a “don't eat me signal” and its reduction in brain cells of MS could promote phagocytosis of myelin by macrophage activation. Active MS lesions are defined by the presence of myelin degradation products in macrophages, and phagocytosis of myelin by activated macrophages/microglia is a crucial step in tissue destruction in MS. The results of the study by Junker et al. suggest that miRNAs dysregulated in MS lesions reduce CD47 in brain resident cells, releasing macrophages from inhibitory control, thereby promoting phagocytosis of myelin [261]. This mechanism may have broad implications for miRNA-regulated macrophage activation in inflammatory diseases.
Altered miRNA profiles detected in MS active lesions may reflect the presence of infiltrating immune cells, changes in brain resident cells such as glial cells, or both. MiRNA profiling in isolated cells by laser capture microdissection from active and inactive MS lesions showed that the most prominently upregulated miRNAs in active MS lesions, miR-155, miR-650, miR-34a, and miR-326 were detected in both microdissected astrocytes and infiltrating immune cells [261]. Novel techniques that allow detection of miRNAs and their targets at the same tissue sections may be used to confirm these results [269, 270]. Under in vitro culture conditions, human astrocytes contained all 10 miRNA that were most strongly upregulated in active MS lesions, including miR-155, which is known to modulate immune responses in different ways but so far had not been assigned to CNS resident cells [104, 137, 271, 272]. When cultured astrocytes were stimulated with various cytokines (i.e., IL-1β, TNF-α, IFNγ, and TGF-β), miR-23a, miR-146a, and miR-155 were strongly induced in vitro [261]. These results were also confirmed with cultured astroglial cells from miRNA-155−/−lacZ mice expressing lacZ reporter instead of miR-155 [104, 261].
Although Junker et al. have focused on only three upregulated miRNAs in active MS lesions (i.e., miR-155, miR-326, and miR-34a) and their common target CD47, other upregulated miRNAs, especially miR-146a and miR-34a deserve further mention. These miRNAs are known to modulate immune responses in different ways. They are also implicated in other CNS disorders accompanied by chronic neuroinflammatory conditions such as epilepsy, Alzheimer's disease (AD), and schizophrenia. miR-34a upregulation was determined in peripheral blood cells of sporadic AD patients, cerebral cortex of APPswe/PSDeltaE9 mice, and prefrontal cortex of schizophrenic patients [273–275]. Cortical expression of miR-34a was inversely correlated with the protein level of Bcl2 in a double transgenic mouse model of AD. In vitro experiments in SH-SY5Y neuronal cells verified anti-apoptotic gene bcl2 as a target of miR-34a [274]. The meaning of these results in the context of MS is currently unknown. Interestingly, mood stabilizators such as lithium and valproate modulate the expression level of miR-34a both in vitro and in vivo [262, 276, 277]. Metabotropic glutamate receptor 7, which is an important regulator of glutamatergic function and of fear, aversion, and cognition, was identified as a target of miR-34a [277, 278]. Although changes in miR-34a expression levels are in opposite directions (downregulation in rat hippocampus and upregulation in cultured lymphoblastoid cells) [262, 277], these results suggest that miRNAs and their predicted effectors may be targets for the action of psychotherapeutic drugs.
miR-146a has been recently identified as a potentially endogenous regulator of TLR and cytokine receptor signalling, suggesting a link between miRNAs and human inflammatory diseases [279]. In contrast to the emerging role of miR-146a in innate immunity, a role of this miRNA in the adaptive immune response has recently been identified. MiR-146a is among the most highly expressed miRNAs in murine Tregs, thus suggesting a possible role for miR-146a in maintaining differentiated T-cell lineages [116]. miR-146a modulates activation-induced cell death (AICD), acting as an antiapoptotic factor, and it is known that Fas-associated death domain (FADD) is a target of miR-146a [280]. Furthermore, miR-146a-enforced expression impairs both activator protein 1 (AP-1) activity and IL-2 production induced by TCR engagement, suggesting a role of this miRNA in the modulation of adaptive immunity. NF-κB and c-ETS binding sites were identified as required for the modulation of miR-146a transcription upon TCR engagement.
Recently, specific miRNAs have been shown to be significantly upregulated in response to cytokine stress and in affected regions of AD brain. The brain-enriched miRNA-146a is currently thought to be a key regulator of the immune and inflammatory signaling systems in both health and disease [150, 279, 281]. Inflammatory processes contribute to the onset, progression, and propagation of this common disorder and amyloid beta peptides, key pathological lesions of AD, are important inflammatory mediators, as are upregulated IL-1β and increased oxidative stress.
The established role of miR-146a in innate immunity responses may also contribute to the pathogenesis of neuroinflammatory CNS diseases such as MS and AD. miRNA-146a controls TLR and cytokine signaling through a negative feedback regulation loop involving downregulation of TRAF6 and IRAK1 levels [150]. In neuroinflammation, TLRs provide a critical link between immune stimulants and the initiation of host defense, and TLR activation modulates the release of inflammatory cytokines [282, 283]. TLRs are expressed on cells of the CNS and can influence local CNS immune responses. There is a marked increase in expression of TLRs in MS brain lesions and CSF mononuclear cells as well as in EAE brain lesions [284, 285]. The secondary progressive phase is characterized by progressive accumulation of disability in the absence of clinical attacks and is driven by the innate immune system [18]. However, the exact role of specific miRNAs in these processes is unknown.
The miRNA profiling of microglial cells in both unstimulated and stimulated conditions has not been reported. Our preliminary study using microarray and qPCR revealed that the expression levels of a set of miRNA were deregulated upon LPS stimulation in N9 murine microglial cell line (Table 3). Predicted target genes of upregulated miRNAs detected in this preliminary study were also found to be downregulated in a microarray study with N9 cells [286]. Validation experiments for the predicted target genes are ongoing. Interestingly, deregulated microglial miRNAs are somewhat different from those detected in murine primary macrophages or mouse Raw 264.7 macrophages upon LPS stimulation [149, 151, 287]. Further studies for the profiling of microglial miRNA expression in MS and EAE are still warranted.
Table 3.
Significantly altered miRNAs upon stimulation LPS in N9 microglial cells.
| MicroRNA | Microarray | qPCR | Targets | ||
|---|---|---|---|---|---|
| Fold change | P | Fold change | P | ||
| mmu-miR-105 | 0.35 | .02 | ns | not determined | |
| mmu-miR-125b-3p | 2.81 | ns | 0.16 | .047 | IL-1β, IL-13, TNF-α |
| mmu-miR-191 | 3.12 | ns | 0.21 | .032 | CCL9, CRP, IL-6, TLR-3 |
| mmu-miR-193* | 0.26 | .03 | 0.28 | .047 | CCL6, IL-10, IL-12Rγ |
| mmu-miR-208a | 3.01 | ns | 0.12 | .015 | CD8, IL-18BP, IL-24 |
| mmu-miR-224 | 3.73 | ns | 0.09 | .033 | CD53, CXCL-14, IL-11 |
| mmu-miR-297c* | 0.31 | ns | 0.12 | .033 | not determined |
| mmu-miR-324-3p | 0.33 | ns | 0.18 | .049 | not determined |
| mmu-miR-376c | 2.99 | .01 | ns | not determined | |
| mmu-miR-421 | 0.35 | ns | 0.03 | .033 | not determined |
| mmu-miR-431* | 4.62 | ns | 0.22 | .034 | CD5, CD81, DICER, IRAK1, TRAP 1 |
| mmu-miR-669g | 3.48 | ns | 0.15 | .015 | not determined |
| mmu-miR-1190 | 0.28 | .01 | 0.12 | .016 | not determined |
| mmu-miR-1894-5p | 0.34 | ns | 0.08 | .017 | not determined |
CCL9: chemokine (C-C) motif ligand 9; CXCL-14: chemokine (C-X-C) motif ligand 14; CRP: c reactive protein; IL-1β: interleukin 1-β; IL-6: interleukin-6; IL-10: interleukine-10; IL-11: interleukin-11; IL-12Rγ: interleukin 12 receptor γ; IL-13: interleukin-13; IL-18BP: interleukin-18 binding protein; IL-24: interleukin-24; IRAK1: interleukin-1 receptor associated kinase-1; TLR-3: Toll-like receptor-3; TRAP 1: TNF receptor associated protein 1; TNF-α: tumor necrosis factor-α.
6. Diagnostics and Therapeutic Perspectives
In patients with MS, intensive efforts are directed at identifying biomarkers in body fluids related to underlying disease mechanisms, disease activity and progression, and therapeutic response [288]. Without biomarkers, the clinical and pathological heterogeneity of MS makes treatment difficult. Thus, identification of biomarkers appears desirable for an improved diagnosis of MS as well as for monitoring of disease activity and treatment response. Biomarkers are defined as parameters that are objectively measurable biological characteristics, which can be used as indicators of physiologic or pathologic processes. A valuable biomarker requires robust and reproducible assays that work in clinically available samples as well as archived material. As miRNAs are more stable than mRNAs, they are good candidates for use as disease biomarkers and their use as biomarkers has gained growing interest in the last few years [289]. Blood serum and plasma are important sample types for investigating miRNAs as biomarkers. Blood biomarkers are attractive because blood samples are easy to collect, cheaper, and noninvasive. However, miRNA profiles in all body fluids such as CSF and their content including hematopoietic cells, exosomes, microvesicles, and microparticles can be used as diagnostic, prognostic, or monitoring marker [290, 291]. miRNA profiling has been established only in an AD study [292] and has not been evaluated in the context of MS. Besides magnetic resonance imaging parameters [166], CSF biomarkers provide important and specific information since changes in the CSF composition may reflect disease mechanisms inherent to MS [288, 293]. miRNA profiling in CSF may provide valuable information about key pathological processes of MS such as inflammation, demyelination, neuroaxonal injury, gliosis, and regeneration. In recent years, the field of biomarker discovery has gradually shifted from the aim of finding a single perfect surrogate marker to the construction of composite markers with higher performance. miRNA profiles may be coupled with diagnostic evaluation of miRNA targets, mRNAs, and protein output; therefore comparison of miRNA analysis with transcriptomic and proteomic studies represents one of the major challenges for clinical application. Another major challenge is represented by technological aspects of miRNA detection aimed at high throughput, sensitivity, and accurate analysis. The level of miRNAs in body fluid samples is very low and efficient and reproducible recovery of miRNA may be problematic. Due to their short length and high sequence similarity within miRNA families, reliable and accurate quantification is still a challenge. In addition, RNA-purified plasma can also contain inhibitors that affect qPCR efficiency [294]. Analyses of miRNA-associated SNPs (e.g., SNP in miRNA genes, in miRNA binding sites in the target mRNA, or in miRNA biogenesis pathway genes) are also potential biomarkers of the diseases associated with miRNA dysregulation [295–298]. While still not fully validated, profiling of blood cells, exosomes, or body fluid miRNAs would represent a tremendous and promising advance in noninvasive diagnostics of CNS disorders. Identification of suitable miRNA-based biomarker sets for MS based on parameters in peripheral blood is only in its infancy.
While we have only just begun to gain insights into miRNA biology, their apparent association with the onset and progression of human diseases such as MS has produced great interest in assessing the feasibility of therapeutic regulation of miRNAs [299]. miRNA-based therapies could involve administration of a specific miRNA mimic to downregulate target genes or antisense oligonucleotide for the blocking of certain miRNA to increase expression of target genes. Importantly, anti-miRNA strategies may be preferred over antisense mRNA strategies in complex human diseases because of the potential of miRNA to affect the regulation of multiple disease-related genes. However, because manipulation of one miRNA may have impact on multiple mRNAs and because one mRNA may be regulated by multiple miRNAs, it is important to guard against off-target effects. Also, miRNAs, instead of causing translational repression or mRNA degradation, may relieve translational repression and promote transcription [300, 301]. Another challenge is the risk of triggering a cellular immune response with RNA therapy. A very promising approach may be the use of LNAs (locked nucleic acids). These molecules comprise a class of bicyclic conformational analogues of RNA, which exhibit high binding affinity to complementary RNA molecules and high stability in blood and tissues in vivo [302]. Recent reports on LNA-mediated miRNA silencing in primates support the potential of LNA-modified oligonucleotides in studying miRNA functions in vivo and in the future development of miRNA-based therapeutics [303, 304]. LNA-modified miR-122 inhibitor has entered the clinic and it is in phase I trials with the goal of treating hepatitis C infection [299]. miRNAs could also be promising potential targets or tools for new therapeutic strategies in the treatment/prevention of autoimmunity. However, to date, no miRNA therapies have been tested in vivo for the treatment of autoimmune diseases.
7. Conclusion
The field of study of miRNAs is a very rapidly evolving new field in molecular biology. miRNAs are important regulators of gene expression, and they function by repressing specific target genes at the posttranscriptional level. miRNA-mediated regulation is essential for immune homeostasis and the prevention of autoimmune diseases. miRNA expression is tightly regulated during hematopoiesis and lymphoid cell differentiation and disruption of the entire miRNA network or specific miRNAs may lead to dysregulated immune responses. Abnormalities in miRNA expression related to inflammatory cytokines, Th17 and Treg cells, as well as B cells have been described in several autoimmune diseases. Emerging evidence suggests that miRNA dysregulation may contribute to the pathogenesis of MS. In the near future, further understanding of the role of miRNAs in intracellular signaling, the expression of proteins involved in immune responses, modulation of cytokines and chemokines, adhesion and costimulatory molecules and the interplay between the immune system and CNS should help to define the role of miRNAs in autoimmunity, and provide an exciting framework for developing new biomarkers and new therapeutic interventions in MS. It is reasonable to assume that future studies concerning the function of miRNAs involved in immune responses will extend our understanding about the complex regulatory networks in autoimmune diseases and MS. These efforts might allow the invention of novel strategies for the treatment of MS. miRNAs are promising reliable biomarkers of human diseases due to their stability being less susceptible to chemical modification and RNase degradation. Although there is much to be learned in the field, the role of miRNAs in regulating a great variety of targets and, as a consequence, multiple pathways makes their use in diagnostics a powerful tool to be exploited for early detection of MS, assessment for risk disease, and monitoring both disease progression and therapeutic responses to disease-modifying drugs.
8. Take-Home Messages
miRNAs have recently emerged as a new class of modulators of gene expression at the posttranscriptional level and are thought to play a critical role in many biological processes.
miRNAs are involved in the development, maturation, and the functions of immune cells, which suggest that they are implicated in the development of autoimmune diseases.
Changes of expression of some miRNAs have been reported in autoimmune pathologies such as rheumatoid arthritis, systemic lupus erythematosus, and MS.
MS serves an example of a chronic and organ-specific autoimmune disease in which miRNAs modulate immune responses in the peripheral immune compartment and the neuroinflammatory process in the brain.
The differential expression of miRNAs and their role in MS have been investigated by several studies.
miRNAs have the potential to serve as biomarkers for the assessment of disease activity and therapeutic response to disease-modifying drugs in MS.
Conflict of Interest Disclosure
The authors declare no competing financial interests.
Acknowledgment
The authors thank Professor Anne Frary for critical reading of the manuscript for English.
References
- 1.Bartel DP. microRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. microRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S. Current Protocols Bioinformatics. Chapter 12, Unit 12.9.1-10. 2010. miRBase: microRNA sequences and annotation. [DOI] [PubMed] [Google Scholar]
- 6.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanwar JR, Mahidhara G, Kanwar RK. MicroRNA in human cancer and chronic inflammatory diseases. Frontiers in Bioscience. 2010;2:1113–1126. doi: 10.2741/s121. [DOI] [PubMed] [Google Scholar]
- 8.Sonntag K-C. microRNAs and deregulated gene expression networks in neurodegeneration. Brain Research. 2010;1338:48–57. doi: 10.1016/j.brainres.2010.03.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nature Reviews Immunology. 2010;10(2):111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 10.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136(1):26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 11.Liston A, Linterman M, Lu L-F. MicroRNA in the adaptive immune system, in sickness and in health. Journal of Clinical Immunology. 2010;30(3):339–346. doi: 10.1007/s10875-010-9378-5. [DOI] [PubMed] [Google Scholar]
- 12.Gantier MP. New perspectives in MicroRNA regulation of innate immunity. Journal of Interferon and Cytokine Research. 2010;30(5):283–289. doi: 10.1089/jir.2010.0037. [DOI] [PubMed] [Google Scholar]
- 13.Jeker LT, Bluestone JA. Small RNA regulators of T cell-mediated autoimmunity. Journal of Clinical Immunology. 2010;30(3):347–357. doi: 10.1007/s10875-010-9392-7. [DOI] [PubMed] [Google Scholar]
- 14.Pauley KM, Cha S, Chan EKL. MicroRNA in autoimmunity and autoimmune diseases. Journal of Autoimmunity. 2009;32(3-4):189–194. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furer V, Greenberg JD, Attur M, Abramson SB, Pillinger MH. The role of microRNA in rheumatoid arthritis and other autoimmune diseases. Clinical Immunology. 2010;136(1):1–15. doi: 10.1016/j.clim.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Iborra M, Bernuzzi F, Invernizzi P, Danese S. microRNAs in autoimmunity and inflammatory bowel disease: crucial regulators in immune response. doi: 10.1016/j.autrev.2010.07.002. Autoimmunity Reviews. In press. [DOI] [PubMed] [Google Scholar]
- 17.Alevizos I, Illei GG. microRNAs in Sjögren's syndrome as a prototypic autoimmune disease. Autoimmunity Reviews. 2010;9(9):618–621. doi: 10.1016/j.autrev.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiner HL. The challenge of multiple sclerosis: how do we cure a chronic heterogeneous disease? Annals of Neurology. 2009;65(3):239–248. doi: 10.1002/ana.21640. [DOI] [PubMed] [Google Scholar]
- 19.Kasper LH, Shoemaker J. Multiple sclerosis immunology: the healthy immune system vs the MS immune system. Neurology. 2010;74(supplement 1):S2–S8. doi: 10.1212/WNL.0b013e3181c97c8f. [DOI] [PubMed] [Google Scholar]
- 20.Hoffjan S, Akkad DA. The genetics of multiple sclerosis: an update 2010. Molecular and Cellular Probes. 2010;24(5):237–243. doi: 10.1016/j.mcp.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Oksenberg JR, Baranzini SE. Multiple sclerosis genetics—is the glass half full, or half empty? Nature Reviews Neurology. 2010;6(8):429–439. doi: 10.1038/nrneurol.2010.91. [DOI] [PubMed] [Google Scholar]
- 22.Davis BN, Hata A. Regulation of MicroRNA Biogenesis: a miRiad of mechanisms. Cell Communication and Signalling. 2009;7:p. 18. doi: 10.1186/1478-811X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature Reviews Molecular Cell Biology. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 24.Lau P-W, MacRae IJ. The molecular machines that mediate microRNA maturation. Journal of Cellular and Molecular Medicine. 2009;13(1):54–60. doi: 10.1111/j.1582-4934.2008.00520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nature Cell Biology. 2009;11(3):228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 26.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13(3):313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He S, Su H, Liu C, et al. MicroRNA-encoding long non-coding RNAs. BMC Genomics. 2008;9:p. 236. doi: 10.1186/1471-2164-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piriyapongsa J, Mariño-Ramírez L, Jordan IK. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007;176(2):1323–1337. doi: 10.1534/genetics.107.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PloS ONE. 2007;2(2, article e203) doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sevignani C, Calin GA, Siracusa LD, Croce CM. Mammalian microRNAs: a small world for fine-tuning gene expression. Mammalian Genome. 2006;17(3):189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends in Genetics. 2005;21(6):322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Yoshimizu T, Miroglio A, Ripoche M-A, et al. The H19 locus acts in vivo as a tumor suppressor. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(34):12417–12422. doi: 10.1073/pnas.0801540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nature Structural and Molecular Biology. 2006;13(12):1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 34.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 36.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. The EMBO Journal. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(45):17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denli AM, Tops BBJ, Plasterk RHA, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 39.Gregory RI, Yan K-P, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 40.Han J, Lee Y, Yeom K-H, Kim Y-K, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes and Development. 2004;18(24):3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Current Biology. 2004;14(23):2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. The EMBO Journal. 2005;24(1):138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bohnsack MT, Czaplinski K, Görlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brownawell AM, Macara IG. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. Journal of Cell Biology. 2002;156(1):53–64. doi: 10.1083/jcb.200110082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of MicroRNA precursors. Science. 2004;303(5654):95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 46.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes and Development. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint É, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 48.Ketting RF, Fischer SEJ, Bernstein E, Sijen T, Hannon GJ, Plasterk RHA. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes and Development. 2001;15(20):2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135(7):1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 50.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes and Development. 2005;19(24):2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mourelatos Z, Dostie J, Paushkin S, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes and Development. 2002;16(6):720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;114(2):p. 269. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 53.Wienholds E, Plasterk RHA. MicroRNA function in animal development. FEBS Letters. 2005;579(26):5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 54.Harfe BD. microRNAs in vertebrate development. Current Opinion in Genetics and Development. 2005;15(4):410–415. doi: 10.1016/j.gde.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 55.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 56.Anglicheau D, Muthukumar T, Suthanthiran M. microRNAs: small RNAs with big effects. Transplantation. 2010;90(2):105–112. doi: 10.1097/TP.0b013e3181e913c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiesen JL, Tomasi TB. Dicer is regulated by cellular stresses and interferons. Molecular Immunology. 2009;46(6):1222–1228. doi: 10.1016/j.molimm.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bushati N, Cohen SM. MicroRNA functions. Annual Review of Cell and Developmental Biology. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 59.Berezikov E, Cuppen E, Plasterk RHA. Approaches to microRNA discovery. Nature Genetics. 2006;38(1):S2–S7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- 60.Cissell KA, Deo SK. Trends in microRNA detection. Analytical and Bioanalytical Chemistry. 2009;394(4):1109–1116. doi: 10.1007/s00216-009-2744-6. [DOI] [PubMed] [Google Scholar]
- 61.Yong SL, Dutta A. microRNAs in cancer. Annual Review of Pathology. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aravin AA, Lagos-Quintana M, Yalcin A, et al. The small RNA profile during Drosophila melanogaster development. Developmental Cell. 2003;5(2):337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 63.Berezikov E, Van Tetering G, Verheul M, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Research. 2006;16(10):1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen PY, Manninga H, Slanchev K, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes and Development. 2005;19(11):1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 66.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 67.Ruby JG, Jan C, Player C, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127(6):1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 68.Krichevsky AM. MicroRNA profiling: from dark matter to white matter, or identifying new players in neurobiology. The Scientific World Journal. 2007;7(2):155–166. doi: 10.1100/tsw.2007.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmittgen TD, Lee EJ, Jiang J, et al. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44(1):31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RHA. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nature Methods. 2006;3(1):27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 71.Nuovo GJ. In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations. Methods. 2008;44(1):39–46. doi: 10.1016/j.ymeth.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 72.Várallyay E, Burgyán J, Havelda Z. Detection of microRNAs by Northern blot analyses using LNA probes. Methods. 2007;43(2):140–145. doi: 10.1016/j.ymeth.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 73.Várallyay E, Burgyán J, Havelda Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nature Protocols. 2008;3(2):190–196. doi: 10.1038/nprot.2007.528. [DOI] [PubMed] [Google Scholar]
- 74.Wienholds E, Kloosterman WP, Miska E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 75.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11(3):241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li W, Ruan K. MicroRNA detection by microarray. Analytical and Bioanalytical Chemistry. 2009;394(4):1117–1124. doi: 10.1007/s00216-008-2570-2. [DOI] [PubMed] [Google Scholar]
- 77.Liu C-G, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nature Protocols. 2008;3(4):563–578. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 78.Liu C-G, Spizzo R, Calin GA, Croce CM. Expression profiling of microRNA using oligo DNA arrays. Methods. 2008;44(1):22–30. doi: 10.1016/j.ymeth.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miska EA, Alvarez-Saavedra E, Townsend M, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome biology. 2004;5(9):p. R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat Methods. 2004;1(2):155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- 81.Saba R, Booth SA. Target labelling for the detection and profiling of microRNAs expressed in CNS tissue using microarrays. BMC Biotechnology. 2006;6, article 47 doi: 10.1186/1472-6750-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1(1):47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 83.Yin JQ, Zhao RC, Morris KV. Profiling microRNA expression with microarrays. Trends in Biotechnology. 2008;26(2):70–76. doi: 10.1016/j.tibtech.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 84.Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(10):3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anisimov SV. Serial Analysis of Gene Expression (SAGE): 13 years of application in research. Current Pharmaceutical Biotechnology. 2008;9(5):338–350. doi: 10.2174/138920108785915148. [DOI] [PubMed] [Google Scholar]
- 86.Friedländer MR, Chen W, Adamidi C, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nature Biotechnology. 2008;26(4):407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 87.Mardis ER. Next-generation DNA sequencing methods. Annual Review of Genomics and Human Genetics. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 88.Kong W, Zhao J-J, Lili HE, Cheng JQ. Strategies for profiling MicroRNA expression. Journal of Cellular Physiology. 2009;218(1):22–25. doi: 10.1002/jcp.21577. [DOI] [PubMed] [Google Scholar]
- 89.Gangaraju VK, Lin H. microRNAs: key regulators of stem cells. Nature Reviews Molecular Cell Biology. 2009;10(2):116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Navarro F, Lieberman J. Small RNAs guide hematopoietic cell differentiation and function. Journal of Immunology. 2010;184(11):5939–5947. doi: 10.4049/jimmunol.0902567. [DOI] [PubMed] [Google Scholar]
- 91.Chen C-Z, Li L, Lodish HF, Bartel DP. microRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 92.Monticelli S, Ansel KM, Xiao C, et al. MicroRNA profiling of the murine hematopoietic system. Genome biology. 2005;6(8, article R71) doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neilson JR, Zheng GXY, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes and Development. 2007;21(5):578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li Q-J, Chau J, Ebert PJR, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129(1):147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 95.Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPα regulates human granulopoiesis. Cell. 2005;123(5):819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 96.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(17):7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xiao C, Calado DP, Galler G, et al. miR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131(1):146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 98.Tsitsiou E, Lindsay MA. microRNAs and the immune response. Current Opinion in Pharmacology. 2009;9(4):514–520. doi: 10.1016/j.coph.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu H, Neilson JR, Kumar P, et al. miRNA profiling of naïve, effector and memory CD8 T cells. PloS ONE. 2007;2(10) doi: 10.1371/journal.pone.0001020. Article ID e1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Muljo SA, Mark Ansel K, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. Journal of Experimental Medicine. 2005;202(2):261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cobb BS, Nesterova TB, Thompson E, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. Journal of Experimental Medicine. 2005;201(9):1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nature Immunology. 2008;9(4):405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jindra PT, Bagley J, Godwin JG, Iacomini J. Costimulation-dependent expression of microRNA-214 increases the ability of T cells to proliferate by targeting Pten. Journal of Immunology. 2010;185(2):990–997. doi: 10.4049/jimmunol.1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thai T-H, Calado DP, Casola S, et al. Regulation of the germinal center response by MicroRNA-155. Science. 2007;316(5824):604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 105.Wan YY. Multi-tasking of helper T cells. Immunology. 2010;130(2):166–171. doi: 10.1111/j.1365-2567.2010.03289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Segal BM. Th17 cells in autoimmune demyelinating disease. Seminars in Immunopathology. 2010;32(1):71–77. doi: 10.1007/s00281-009-0186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wei B, Pei G. microRNAs: critical regulators in Th17 cells and players in diseases. Cellular & Molecular Immunology. 2010;7(3):175–181. doi: 10.1038/cmi.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu D, Rao S, Tsai LM, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31(3):457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 109.Zhou L, Seo KH, Wong HK, Mi Q-S. microRNAs and immune regulatory T cells. International Immunopharmacology. 2009;9(5):524–527. doi: 10.1016/j.intimp.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 110.Zhou X, Jeker LT, Fife BT, et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. Journal of Experimental Medicine. 2008;205(9):1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liston A, Lu L-F, O’Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. Journal of Experimental Medicine. 2008;205(9):1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chong MMW, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. Journal of Experimental Medicine. 2008;205(9):2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lu L-F, Thai T-H, Calado DP, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30(1):80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. Journal of Immunology. 2009;182(5):2578–2582. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- 115.Huang B, Zhao J, Lei Z, et al. miR-142-3p restricts cAMP production in CD4+CD25- T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. The EMBO Reports. 2009;10(2):180–185. doi: 10.1038/embor.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cobb BS, Hertweck A, Smith J, et al. A role for Dicer in immune regulation. Journal of Experimental Medicine. 2006;203(11):2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rouas R, Fayyad-Kazan H, El Zien N, et al. Human natural Treg microRNA signature: role of microRNA-31 and microRNA-21 in FOXP3 expression. European Journal of Immunology. 2009;39(6):1608–1618. doi: 10.1002/eji.200838509. [DOI] [PubMed] [Google Scholar]
- 118.Stahl HF, Fauti T, Ullrich N, et al. miR-155 inhibition sensitizes CD4+ Th cells for TREG mediated suppression. PloS ONE. 2009;4(9, article e7158) doi: 10.1371/journal.pone.0007158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kurosaki T, Shinohara H, Baba Y. B cell signaling and fate decision. Annual Review of Immunology. 2010;28:21–55. doi: 10.1146/annurev.immunol.021908.132541. [DOI] [PubMed] [Google Scholar]
- 120.Koralov SB, Muljo SA, Galler GR, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132(5):860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 121.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17∼92 family of miRNA clusters. Cell. 2008;132(5):875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gururajan M, Haga CL, Das S, et al. MicroRNA 125b inhibition of B cell differentiation in germinal centers. International Immunology. 2010;22(7):583–592. doi: 10.1093/intimm/dxq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vigorito E, Perks KL, Abreu-Goodger C, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27(6):847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kong KY, Owens KS, Rogers JH, et al. MIR-23A microRNA cluster inhibits B-cell development. Experimental Hematology. 2010;38(8):629–640. doi: 10.1016/j.exphem.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dorsett Y, McBride KM, Jankovic M, et al. microRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28(5):630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Teng G, Hakimpour P, Landgraf P, et al. microRNA-155 is a negative regulator of activation induced cytidine deaminase. Immunity. 2008;28(5):621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Basso K, Sumazin P, Morozov P, et al. Identification of the human mature B cell miRNome. Immunity. 2009;30(5):744–752. doi: 10.1016/j.immuni.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fontana L, Pelosi E, Greco P, et al. microRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nature Cell Biology. 2007;9(7):775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 129.Rosa A, Ballarino M, Sorrentino A, et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(50):19849–19854. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Velu CS, Baktula AM, Grimes HL. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. 2009;113(19):4720–4728. doi: 10.1182/blood-2008-11-190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.O’Connell RM, Rao DS, Chaudhuri AA, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. Journal of Experimental Medicine. 2008;205(3):585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(17):7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 134.Bazzoni F, Rossato M, Fabbri M, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(13):5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Radom-Aizik S, Zaldivar Jr. F, Oliver S, Galassetti P, Cooper DM. Evidence for microRNA involvement in exercise-associated neutrophil gene expression changes. Journal of Applied Physiology. 2010;109(1):252–261. doi: 10.1152/japplphysiol.01291.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hashimi ST, Fulcher JA, Chang MH, Gov L, Wang S, Lee B. microRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the coordinate regulation of dendritic cell differentiation. Blood. 2009;114(2):404–414. doi: 10.1182/blood-2008-09-179150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Martinez-Nunez RT, Louafi F, Friedmann PS, Sanchez-Eisner T. microRNA-155 modulates the pathogen binding ability of dendritic cells (DCs) by down-regulation of DC-specific intercellular adhesion molecule-3 grabbing non-integrin (DC-SIGN) Journal of Biological Chemistry. 2009;284(24):16334–16342. doi: 10.1074/jbc.M109.011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ceppi M, Pereira AM, Dunand-Sauthier I, et al. microRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(8):2735–2740. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jurkin J, Schichl YM, Koeffel R, et al. miR-146a is differentially expressed by myeloid dendritic cell subsets and desensitizes cells to TLR2-dependent activation. Journal of Immunology. 2010;184(9):4955–4965. doi: 10.4049/jimmunol.0903021. [DOI] [PubMed] [Google Scholar]
- 141.Stern-Ginossar N, Mandelboim O. An integrated view of the regulation of NKG2D ligands. Immunology. 2009;128(1):1–6. doi: 10.1111/j.1365-2567.2009.03147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stern-Ginossar N, Gur C, Biton M, et al. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nature Immunology. 2008;9(9):1065–1073. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- 143.Stern-Ginossar N, Elefant N, Zimmermann A, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317(5836):376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Eissmann P, Evans JH, Mehrabi M, Rose EL, Nedvetzki S, Davis DM. Multiple mechanisms downstream of TLR-4 stimulation allow expression of NKG2D ligands to facilitate macrophage/NK cell crosstalk. Journal of Immunology. 2010;184(12):6901–6909. doi: 10.4049/jimmunol.0903985. [DOI] [PubMed] [Google Scholar]
- 145.Sakuishi K, Miyake S, Yamamura T. Role of NK cells and invariant NKT cells in multiple sclerosis. Results and Problems in Cell Differentiation. 2010;51:127–147. doi: 10.1007/400_2009_11. [DOI] [PubMed] [Google Scholar]
- 146.Fedeli M, Napolitano A, Wong MP, et al. Dicer-dependent microRNA pathway controls invariant NKT cell development. Journal of Immunology. 2009;183(4):2506–2512. doi: 10.4049/jimmunol.0901361. [DOI] [PubMed] [Google Scholar]
- 147.Zhou L, Seo K-H, He H-Z, et al. Tie2cre-induced inactivation of the miRNA-processing enzyme Dicer disrupts invariant NKT cell development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(25):10266–10271. doi: 10.1073/pnas.0811119106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. microRNA-155 is induced during the macrophage inflammatory response. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(5):1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tili E, Michaille J-J, Cimino A, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF- α stimulation and their possible roles in regulating the response to endotoxin shock. Journal of Immunology. 2007;179(8):5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 150.Taganov KD, Boldin MP, Chang K-J, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu G, Friggeri A, Yang Y, Park Y-J, Tsuruta Y, Abraham E. miR-147, a microRNA that is induced upon toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(37):15819–15824. doi: 10.1073/pnas.0901216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nature Immunology. 2010;11(2):141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 153.Vinuesa CG, Rigby RJ, Yu D. Logic and extent of miRNA-mediated control of autoimmune gene expression. International Reviews of Immunology. 2009;28(3-4):112–138. doi: 10.1080/08830180902934909. [DOI] [PubMed] [Google Scholar]
- 154.Carissimi C, Fulci V, Macino G. microRNAs: novel regulators of immunity. Autoimmunity Reviews. 2009;8(6):520–524. doi: 10.1016/j.autrev.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 155.Yu DI, Tan AH-M, Hu X, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450(7167):299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- 156.Ebert PJ, Jiang S, Xie J, Li QJ, Davis MM. An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nature immunology. 2009;10(11):1162–1169. doi: 10.1038/ni.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tomasi TB, Magner WJ, Wiesen JL, et al. MHC class II regulation by epigenetic agents and microRNAs. Immunologic Research. 2010;46(1–3):45–58. doi: 10.1007/s12026-009-8128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pugliatti M, Sotgiu S, Rosati G. The worldwide prevalence of multiple sclerosis. Clinical Neurology and Neurosurgery. 2002;104(3):182–191. doi: 10.1016/s0303-8467(02)00036-7. [DOI] [PubMed] [Google Scholar]
- 159.Ramagopalan SV, Knight JC, Ebers GC. Multiple sclerosis and the major histocompatibility complex. Current Opinion in Neurology. 2009;22(3):219–225. doi: 10.1097/WCO.0b013e32832b5417. [DOI] [PubMed] [Google Scholar]
- 160.Camiña-Tato M, Fernández M, Morcillo-Suárez C, et al. Genetic association of CASP8 polymorphisms with primary progressive multiple sclerosis. Journal of Neuroimmunology. 2010;222(1-2):70–75. doi: 10.1016/j.jneuroim.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 161.Jakkula E, Leppä V, Sulonen A-M, et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. American Journal of Human Genetics. 2010;86(2):285–291. doi: 10.1016/j.ajhg.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurology. 2009;8(11):1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- 163.Slavin A, Kelly-Modis L, Labadia M, et al. Pathogenic mechanisms and experimental models of multiple sclerosis. doi: 10.3109/08916931003674733. Autoimmunity. In press. [DOI] [PubMed] [Google Scholar]
- 164.Mix E, Meyer-Rienecker H, Hartung H-P, Zettl UK. Animal models of multiple sclerosis-Potentials and limitations. Progress in Neurobiology. 92(3):386–404. doi: 10.1016/j.pneurobio.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Siffrin V, Vogt J, Radbruch H, Nitsch R, Zipp F. Multiple sclerosis—candidate mechanisms underlying CNS atrophy. Trends in Neurosciences. 2010;33(4):202–210. doi: 10.1016/j.tins.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 166.Rudick RA, Polman CH. Current approaches to the identification and management of breakthrough disease in patients with multiple sclerosis. The Lancet Neurology. 2009;8(6):545–559. doi: 10.1016/S1474-4422(09)70082-1. [DOI] [PubMed] [Google Scholar]
- 167.Hamann I, Zipp F, Infante-Duarte C. Therapeutic targeting of chemokine signaling in Multiple Sclerosis. Journal of the Neurological Sciences. 2008;274(1-2):31–38. doi: 10.1016/j.jns.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 168.Bennett JL, Stüve O. Update on inflammation, neurodegeneration, and immunoregulation in multiple sclerosis: therapeutic implications. Clinical Neuropharmacology. 2009;32(3):121–132. doi: 10.1097/WNF.0b013e3181880359. [DOI] [PubMed] [Google Scholar]
- 169.Charo IF, Ransohoff RM. Mechanisms of disease: the many roles of chemokines and chemokine receptors in inflammation. New England Journal of Medicine. 2006;354(6):610–621. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- 170.Chitnis T. The role of CD4 T cells in the pathogenesis of multiple sclerosis. International Review of Neurobiology. 2007;79:43–72. doi: 10.1016/S0074-7742(07)79003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mosmann TR, Cherwinski H, Bond MW. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. Journal of Immunology. 1986;136(7):2348–2357. [PubMed] [Google Scholar]
- 172.Kebir H, Kreymborg K, Ifergan I, et al. Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nature Medicine. 2007;13(10):1173–1175. doi: 10.1038/nm1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lock C, Hermans G, Pedotti R, et al. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nature Medicine. 2002;8(5):500–508. doi: 10.1038/nm0502-500. [DOI] [PubMed] [Google Scholar]
- 174.Bettelli E, Korn T, Oukka M, Kuchroo VK. Induction and effector functions of TH17 cells. Nature. 2008;453(7198):1051–1057. doi: 10.1038/nature07036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Haak S, Croxford AL, Kreymborg K, et al. IL-17A and IL-17F do not contribute vitally to autoimmune neuro-inflammation in mice. Journal of Clinical Investigation. 2009;119(1):61–69. doi: 10.1172/JCI35997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.O’Connor RA, Prendergast CT, Sabatos CA, et al. Cutting edge: Th1 cells facilitate the entry of Th17 cells to the central nervous system during experimental autoimmune encephalomyelitis. Journal of Immunology. 2008;181(6):3750–3754. doi: 10.4049/jimmunol.181.6.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Stromnes IM, Cerretti LM, Liggitt D, Harris RA, Goverman JM. Differential regulation of central nervous system autoimmunity by T H1 and TH17 cells. Nature Medicine. 2008;14(3):337–342. doi: 10.1038/nm1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Marrack P, Kappler J. The T cell receptor. Science. 1987;238(4830):1073–1079. doi: 10.1126/science.3317824. [DOI] [PubMed] [Google Scholar]
- 179.Ponomarev ED, Dittel BN. γδ T cells regulate the extent and duration of inflammation in the central nervous system by a fas ligand-dependent mechanism. Journal of Immunology. 2005;174(8):4678–4687. doi: 10.4049/jimmunol.174.8.4678. [DOI] [PubMed] [Google Scholar]
- 180.Shimonkevitz R, Colburn C, Burnham JA, Murray RS, Kotzin BL. Clonal expansions of activated γ/δ T cells in recent-onset multiple sclerosis. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(3):923–927. doi: 10.1073/pnas.90.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ponomarev ED, Novikova M, Yassais M, Szczepanik M, Gorski J, Dittel BN. γδ T cell regulation of IFN-γ production by central nervous system-infiltrating encephalitogenic T cells: correlation with recovery from experimental autoimmune encephalomyelitis. Journal of Immunology. 2004;173(3):1587–1595. doi: 10.4049/jimmunol.173.3.1587. [DOI] [PubMed] [Google Scholar]
- 182.Babbe H, Roers A, Waisman A, et al. Clonal expansions of CD8+ T cells dominate the T cell infiltrate in active multiple sclerosis lesions as shown by micromanipulation and single cell polymerase chain reaction. Journal of Experimental Medicine. 2000;192(3):393–404. doi: 10.1084/jem.192.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jäger A, Dardalhon V, Sobel RA, Bettelli E, Kuchroo VK. Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. Journal of Immunology. 2009;183(11):7169–7177. doi: 10.4049/jimmunol.0901906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Viglietta V, Baecher-Allan C, Weiner HL, Hafler DA. Loss of functional suppression by CD4+CD25+ regulatory T cells in patients with multiple sclerosis. Journal of Experimental Medicine. 2004;199(7):971–979. doi: 10.1084/jem.20031579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Martinez-Forero I, Garcia-Munoz R, Martinez-Pasamar S, et al. IL-10 suppressor activity and ex vivo Tr1 cell function are impaired in multiple sclerosis. European Journal of Immunology. 2008;38(2):576–586. doi: 10.1002/eji.200737271. [DOI] [PubMed] [Google Scholar]
- 186.Astier AL, Meiffren G, Freeman S, Hafler DA. Alterations in CD46-mediated Tr1 regulatory T cells in patients with multiple sclerosis. Journal of Clinical Investigation. 2006;116(12):3252–3257. doi: 10.1172/JCI29251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Korn T, Reddy J, Gao W, et al. Myelin-specific regulatory T cells accumulate in the CNS but fail to control autoimmune inflammation. Nature Medicine. 2007;13(4):423–431. doi: 10.1038/nm1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ganes T, Brautaset NJ, Nyberg-Hansen R, Vandvik B. Multimodal evoked responses and cerebrospinal fluid oligoclonal immunoglobulins in patients with multiple sclerosis. Acta Neurologica Scandinavica. 1986;73(5):472–476. doi: 10.1111/j.1600-0404.1986.tb04587.x. [DOI] [PubMed] [Google Scholar]
- 189.Cepok S, Rosche B, Grummel V, et al. Short-lived plasma blasts are the main B cell effector subset during the course of multiple sclerosis. Brain. 2005;128(7):1667–1676. doi: 10.1093/brain/awh486. [DOI] [PubMed] [Google Scholar]
- 190.Villar LM, Masjuan J, González-Porqué P, et al. Intrathecal IgM synthesis in neurologic diseases: relationship with disability in MS. Neurology. 2002;58(5):824–826. doi: 10.1212/wnl.58.5.824. [DOI] [PubMed] [Google Scholar]
- 191.Berger T, Rubner P, Schautzer F, et al. Antimyelin antibodies as a predictor of clinically definite multiple sclerosis after a first demyelinating event. The New England Journal of Medicine. 2003;349(2):139–145. doi: 10.1056/NEJMoa022328. [DOI] [PubMed] [Google Scholar]
- 192.Gaertner S, de Graaf KL, Greve B, Weissert R. Antibodies against glycosylated native MOG are elevated in patients with multiple sclerosis. Neurology. 2004;63(12):2381–2383. doi: 10.1212/01.wnl.0000147259.34163.33. [DOI] [PubMed] [Google Scholar]
- 193.Winges KM, Gilden DH, Bennett JL, Yu X, Ritchie AM, Owens GP. Analysis of multiple sclerosis cerebrospinal fluid reveals a continuum of clonally related antibody-secreting cells that are predominantly plasma blasts. Journal of Neuroimmunology. 2007;192(1-2):226–234. doi: 10.1016/j.jneuroim.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 194.Kutzelnigg A, Lucchinetti CF, Stadelmann C, et al. Cortical demyelination and diffuse white matter injury in multiple sclerosis. Brain. 2005;128(11):2705–2712. doi: 10.1093/brain/awh641. [DOI] [PubMed] [Google Scholar]
- 195.Heppner FL, Greter M, Marino D, et al. Experimental autoimmune encephalomyelitis repressed by microglial paralysis. Nature Medicine. 2005;11(2):146–152. doi: 10.1038/nm1177. [DOI] [PubMed] [Google Scholar]
- 196.Greter M, Heppner FL, Lemos MP, et al. Dendritic cells permit immune invasion of the CNS in an animal model of multiple sclerosis. Nature Medicine. 2005;11(3):328–334. doi: 10.1038/nm1197. [DOI] [PubMed] [Google Scholar]
- 197.Bailey-Bucktrout SL, Caulkins SC, Goings G, Fischer JAA, Dzionek A, Miller SD. Cutting edge: central nervous system plasmacytoid dendritic cells regulate the severity of relapsing experimental autoimmune Encephalomyelitis. Journal of Immunology. 2008;180(10):6457–6461. doi: 10.4049/jimmunol.180.10.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bayas A, Stasiolek M, Kruse N, Toyka KV, Selmaj K, Gold R. Altered innate immune response of plasmacytoid dendritic cells in multiple sclerosis. Clinical and Experimental Immunology. 2009;157(3):332–342. doi: 10.1111/j.1365-2249.2009.03964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bailey SL, Schreiner B, McMahon EJ, Miller SD. CNS myeloid DCs presenting endogenous myelin peptides ’preferentially’ polarize CD4+ TH-17 cells in relapsing EAE. Nature Immunology. 2007;8(2):172–180. doi: 10.1038/ni1430. [DOI] [PubMed] [Google Scholar]
- 200.Deshpande P, King IL, Segal BM. Cutting edge: CNS CD11c+ cells, from mice with encephalomyelitis polarize Th17 cells, and support CD25+CD4+ T cell-mediated immunosuppression, suggesting dual roles in the disease process. Journal of Immunology. 2007;178(11):6695–6699. doi: 10.4049/jimmunol.178.11.6695. [DOI] [PubMed] [Google Scholar]
- 201.Becher B, Durell BG, Miga AV, Hickey WF, Noelle RJ. The clinical course of experimental autoimmune encephalomyelitis and inflammation is controlled by the expression of CD40 within the central nervous system. Journal of Experimental Medicine. 2001;193(8):967–974. doi: 10.1084/jem.193.8.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Weber F, Meinl E, Aloisi F, et al. Human astrocytes are only partially competent antigen presenting cells: possible implications for lesion development in multiple sclerosis. Brain. 1994;117(1):59–69. doi: 10.1093/brain/117.1.59. [DOI] [PubMed] [Google Scholar]
- 203.Stüve O, Youssef S, Slavin AJ, et al. The role of the MHC class II transactivator in class II expression and antigen presentation by astrocytes and in susceptibility to central nervous system autoimmune disease. Journal of Immunology. 2002;169(12):6720–6732. doi: 10.4049/jimmunol.169.12.6720. [DOI] [PubMed] [Google Scholar]
- 204.Xiao B-G, Diab A, Zhu J, Van Der Meide P, Link H. Astrocytes induce hyporesponses of myelin basic protein-reactive T and B cell function. Journal of Neuroimmunology. 1998;89(1-2):113–121. doi: 10.1016/s0165-5728(98)00123-4. [DOI] [PubMed] [Google Scholar]
- 205.Geurts JJ, Barkhof F. Grey matter pathology in multiple sclerosis. The Lancet Neurology. 2008;7(9):841–851. doi: 10.1016/S1474-4422(08)70191-1. [DOI] [PubMed] [Google Scholar]
- 206.Kuhlmann T, Lingfeld G, Bitsch A, Schuchardt J, Brück W. Acute axonal damage in multiple sclerosis is most extensive in early disease stages and decreases over time. Brain. 2002;125(10):2202–2212. doi: 10.1093/brain/awf235. [DOI] [PubMed] [Google Scholar]
- 207.Frischer JM, Bramow S, Dal-Bianco A, et al. The relation between inflammation and neurodegeneration in multiple sclerosis brains. Brain. 2009;132(5):1175–1189. doi: 10.1093/brain/awp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yong VW, Marks S. The interplay between the immune and central nervous systems in neuronal injury. Neurology. 2010;74(supplement 1):S9–S16. doi: 10.1212/WNL.0b013e3181c97d04. [DOI] [PubMed] [Google Scholar]
- 209.Gilmore CP, Deluca GC, Bö L, et al. Spinal cord neuronal pathology in multiple sclerosis. Brain Pathology. 2009;19(4):642–649. doi: 10.1111/j.1750-3639.2008.00228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Papadopoulos D, Dukes S, Patel R, Nicholas R, Vora A, Reynolds R. Substantial archaeocortical atrophy and neuronal loss in multiple sclerosis. Brain Pathology. 2009;19(2):238–253. doi: 10.1111/j.1750-3639.2008.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Medana I, Li Z, Flügel A, Tschopp J, Wekerle H, Neumann H. Fas ligand (CD95L) protects neurons against perforin-mediated T lymphocyte cytotoxicity. Journal of Immunology. 2001;167(2):674–681. doi: 10.4049/jimmunol.167.2.674. [DOI] [PubMed] [Google Scholar]
- 212.Aktas O, Smorodchenko A, Brocke S, et al. Neuronal damage in autoimmune neuroinflammation mediated by the death ligand TRAIL. Neuron. 2005;46(3):421–432. doi: 10.1016/j.neuron.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 213.Giuliani F, Goodyer CG, Antel JP, Wee Yong V. Vulnerability of human neurons to T cell-mediated cytotoxicity. Journal of Immunology. 2003;171(1):368–379. doi: 10.4049/jimmunol.171.1.368. [DOI] [PubMed] [Google Scholar]
- 214.Vercellino M, Merola A, Piacentino C, et al. Altered glutamate reuptake in relapsing-remitting and secondary progressive multiple sclerosis cortex: correlation with microglia infiltration, demyelination, and neuronal and synaptic damage. Journal of Neuropathology and Experimental Neurology. 2007;66(8):732–739. doi: 10.1097/nen.0b013e31812571b0. [DOI] [PubMed] [Google Scholar]
- 215.Trapp BD, Stys PK. Virtual hypoxia and chronic necrosis of demyelinated axons in multiple sclerosis. The Lancet Neurology. 2009;8(3):280–291. doi: 10.1016/S1474-4422(09)70043-2. [DOI] [PubMed] [Google Scholar]
- 216.Basso AS, Frenkel D, Quintana FJ, et al. Reversal of axonal loss and disability in a mouse model of progressive multiple sclerosis. Journal of Clinical Investigation. 2008;118(4):1532–1543. doi: 10.1172/JCI33464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Waxman SG. Mechanisms of disease: sodium channels and neuroprotection in multiple sclerosis—current status. Nature Clinical Practice Neurology. 2008;4(3):159–169. doi: 10.1038/ncpneuro0735. [DOI] [PubMed] [Google Scholar]
- 218.Hendrix S, Nitsch R. The role of T helper cells in neuroprotection and regeneration. Journal of Neuroimmunology. 2007;184(1-2):100–112. doi: 10.1016/j.jneuroim.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 219.Flügel A, Schwaiger FW, Neumann H, et al. Neuronal FasL induces cell death of encephalitogenic T lymphocytes. Brain Pathology. 2000;10(3):353–364. doi: 10.1111/j.1750-3639.2000.tb00267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Liu Y, Teige I, Birnir B, Issazadeh-Navikas S. Neuron-mediated generation of regulatory T cells from encephalitogenic T cells suppresses EAE. Nature Medicine. 2006;12(5):518–525. doi: 10.1038/nm1402. [DOI] [PubMed] [Google Scholar]
- 221.Ransohoff RM, Perry VH. Microglial physiology: unique stimuli, specialized responses. Annual Review of Immunology. 2009;27:119–145. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- 222.Genc S, Kizildag S, Genc K, Ates H, Atabey N. Erratum: Interferon gamma and lipopolysaccharide upregulate TNF-related apoptosis-inducing ligand expression in murine microglia. Immunology Letters. 2003;85(3):271–274. doi: 10.1016/s0165-2478(02)00245-6. [DOI] [PubMed] [Google Scholar]
- 223.Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nature Reviews Neuroscience. 2009;10(12):837–841. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Yelamanchili SV, Fox HS. Defining larger roles for “tiny” RNA molecules: role of miRNAs in neurodegeneration research. Journal of Neuroimmune Pharmacology. 2009;5(1):63–69. doi: 10.1007/s11481-009-9172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Bushati N, Cohen SM. microRNAs in neurodegeneration. Current Opinion in Neurobiology. 2008;18(3):292–296. doi: 10.1016/j.conb.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 226.Yong VW. Prospects of repair in multiple sclerosis. Journal of the Neurological Sciences. 2009;277(supplement 1):S16–S18. doi: 10.1016/S0022-510X(09)70006-1. [DOI] [PubMed] [Google Scholar]
- 227.Liu C, Zhao X. microRNAs in Adult and Embryonic Neurogenesis. NeuroMolecular Medicine. 2010;11(3):141–152. doi: 10.1007/s12017-009-8077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Li X, Jin P. Roles of small regulatory RNAs in determining neuronal identity. Nature Reviews Neuroscience. 2010;11(5):329–338. doi: 10.1038/nrn2739. [DOI] [PubMed] [Google Scholar]
- 229.Cheng L-C, Pastrana E, Tavazoie M, Doetsch F. MiR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nature Neuroscience. 2009;12(4):399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Shen Q, Temple S. Fine control: microRNA regulation of adult neurogenesis. Nature Neuroscience. 2009;12(4):369–370. doi: 10.1038/nn0409-369. [DOI] [PubMed] [Google Scholar]
- 231.Szulwach KE, Li X, Smrt RD, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. Journal of Cell Biology. 2010;189(1):127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Chang A, Smith MC, Yin X, Fox RJ, Staugaitis SM, Trapp BD. Neurogenesis in the chronic lesions of multiple sclerosis. Brain. 2008;131(9):2366–2375. doi: 10.1093/brain/awn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Emery B. Transcriptional and post-transcriptional control of CNS myelination. Current Opinion in Neurobiology. 2010;20(5):601–607. doi: 10.1016/j.conb.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 234.Bruce CC, Zhao C, Franklin RJM. Remyelination—an effective means of neuroprotection. Hormones and Behavior. 2010;57(1):56–62. doi: 10.1016/j.yhbeh.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 235.Piaton G, Williams A, Seilhean D, Lubetzki C. Remyelination in multiple sclerosis. Progress in Brain Research. 2009;175:453–464. doi: 10.1016/S0079-6123(09)17530-1. [DOI] [PubMed] [Google Scholar]
- 236.Taveggia C, Feltri ML, Wrabetz L. Signals to promote myelin formation and repair. Nature Reviews Neurology. 2010;6(5):276–287. doi: 10.1038/nrneurol.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Liu J, Casaccia P. Epigenetic regulation of oligodendrocyte identity. Trends in Neurosciences. 2010;33(4):193–201. doi: 10.1016/j.tins.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Dugas JC, Cuellar TL, Scholze A, et al. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65(5):597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhao X, He X, Han X, et al. microRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65(5):612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Shin D, Shin J-Y, McManus MT, Ptáček LJ, Fu Y-H. Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Annals of Neurology. 2009;66(6):843–857. doi: 10.1002/ana.21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lehotzky A, Lau P, Tokési N, Muja N, Hudson LD, Ovádi J. Tubulin polymerization-promoting protein (TPPP/p25) is critical for oligodendrocyte differentiation. GLIA. 2010;58(2):157–168. doi: 10.1002/glia.20909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. Journal of Neuroscience. 2008;28(45):11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Budde H, Schmitt S, Fitzner D, Opitz L, Salinas-Riester G, Simons M. Control of oligodendroglial cell number by the miR-17-92 cluster. Development. 2010;137(13):2127–2132. doi: 10.1242/dev.050633. [DOI] [PubMed] [Google Scholar]
- 244.Lin S-T, Fu Y-H. miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. DMM Disease Models and Mechanisms. 2009;2(3-4):178–188. doi: 10.1242/dmm.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Letzen BS, Liu C, Thakor NV, Gearhart JD, All AH, Kerr CL. microRNA expression profiling of oligodendrocyte differentiation from human embryonic stem cells. PloS ONE. 2010;5(5) doi: 10.1371/journal.pone.0010480. Article ID e10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.McQuaid S, Cunnea P, McMahon J, Fitzgerald U. The effects of blood-brain barrier disruption on glial cell function in multiple sclerosis. Biochemical Society Transactions. 2009;37(1):329–331. doi: 10.1042/BST0370329. [DOI] [PubMed] [Google Scholar]
- 247.Weiss N, Miller F, Cazaubon S, Couraud P-O. The blood-brain barrier in brain homeostasis and neurological diseases. Biochimica et Biophysica Acta. 2009;1788(4):842–857. doi: 10.1016/j.bbamem.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 248.Watzlawik J, Warrington AE, Rodriguez M. Importance of oligodendrocyte protection, BBB breakdown and inflammation for remyelination. Expert Review of Neurotherapeutics. 2010;10(3):441–457. doi: 10.1586/ern.10.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wu F, Yang Z, Li G. Role of specific microRNAs for endothelial function and angiogenesis. Biochemical and Biophysical Research Communications. 2009;386(4):549–553. doi: 10.1016/j.bbrc.2009.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wang S, Olson EN. AngiomiRs-Key regulators of angiogenesis. Current Opinion in Genetics and Development. 2009;19(3):205–211. doi: 10.1016/j.gde.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Suárez Y, Sessa WC. microRNAs as novel regulators of angiogenesis. Circulation Research. 2009;104(4):442–454. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Yin K-J, Deng Z, Hamblin M, et al. Peroxisome proliferator-activated receptor δ regulation of miR-15a in ischemia-induced cerebral vascular endothelial injury. Journal of Neuroscience. 2010;30(18):6398–6408. doi: 10.1523/JNEUROSCI.0780-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Dunn SE, Bhat R, Straus DS, et al. Peroxisome proliferator-activated receptor δ limits the expansion of pathogenic Th cells during central nervous system autoimmunity. Journal of Experimental Medicine. 2010;207(8):1599–1608. doi: 10.1084/jem.20091663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Kanakasabai S, Chearwae W, Walline CC, Iams W, Adams SM, Bright JJ. Peroxisome proliferator-activated receptor δ agonists inhibit T helper type 1 (Th1) and Th17 responses in experimental allergic encephalomyelitis. Immunology. 2010;130(4):572–588. doi: 10.1111/j.1365-2567.2010.03261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Polak PE, Kalinin S, Dello Russo C, et al. Protective effects of a peroxisome proliferator-activated receptor-β/δ agonist in experimental autoimmune encephalomyelitis. Journal of Neuroimmunology. 2005;168(1-2):65–75. doi: 10.1016/j.jneuroim.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 256.Otaegui D, Baranzini SE, Armañanzas R, et al. Differential micro RNA expression in PBMC from multiple sclerosis patients. PloS ONE. 2009;4(7) doi: 10.1371/journal.pone.0006309. Article ID e6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Keller A, Leidinger P, Lange J, et al. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PloS ONE. 2009;4(10) doi: 10.1371/journal.pone.0007440. Article ID e7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Du C, Liu C, Kang J, et al. microRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nature Immunology. 2009;10(12):1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 259.Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho I-C. Ets-1 is a negative regulator of Th17 differentiation. Journal of Experimental Medicine. 2007;204(12):2825–2835. doi: 10.1084/jem.20070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Cox MB, Cairns MJ, Gandhi KS, et al. microRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PloS ONE. 2010;5(8) doi: 10.1371/journal.pone.0012132. Article ID e12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Junker A, Krumbholz M, Eisele S, et al. microRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132(12):3342–3352. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- 262.Chen H, Wang N, Burmeister M, McInnis MG. microRNA expression changes in lymphoblastoid cell lines in response to lithium treatment. International Journal of Neuropsychopharmacology. 2009;12(7):975–981. doi: 10.1017/S1461145709000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lindberg RLP, Hoffmann F, Mehling M, Kuhle J, Kappos L. Altered expression of miR-17-5p in CD4+ lymphocytes of relapsing-remitting multiple sclerosis patients. European Journal of Immunology. 2010;40(3):888–898. doi: 10.1002/eji.200940032. [DOI] [PubMed] [Google Scholar]
- 264.Bonauer A, Dimmeler S. The microRNA-17∼92 cluster: still a miRacle? Cell Cycle. 2009;8(23):3866–3873. doi: 10.4161/cc.8.23.9994. [DOI] [PubMed] [Google Scholar]
- 265.Venken K, Hellings N, Liblau R, Stinissen P. Disturbed regulatory T cell homeostasis in multiple sclerosis. Trends in Molecular Medicine. 2010;16(2):58–68. doi: 10.1016/j.molmed.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 266.De Santis G, Ferracin M, Biondani A, et al. Altered miRNA expression in T regulatory cells in course of multiple sclerosis. Journal of Neuroimmunology. 2010;226(1-2):165–171. doi: 10.1016/j.jneuroim.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 267.Yamagiwa S, Gray JD, Hashimoto S, Horwitz DA. A role for TGF-β in the generation and expansion of CD4+CD25+ regulatory t cells from human peripheral blood. Journal of Immunology. 2001;166(12):7282–7289. doi: 10.4049/jimmunol.166.12.7282. [DOI] [PubMed] [Google Scholar]
- 268.Koning N, Bö L, Hoek RM, Huitinga I. Downregulation of macrophage inhibitory molecules in multiple sclerosis lesions. Annals of Neurology. 2007;62(5):504–514. doi: 10.1002/ana.21220. [DOI] [PubMed] [Google Scholar]
- 269.Nuovo GJ, Elton TS, Nana-Sinkam P, Volinia S, Croce CM, Schmittgen TD. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nature Protocols. 2009;4(1):107–115. doi: 10.1038/nprot.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Nuovo GJ. In situ detection of microRNAs in paraffin embedded, formalin fixed tissues and the co-localization of their putative targets. doi: 10.1016/j.ymeth.2010.08.009. Methods. In press. [DOI] [PubMed] [Google Scholar]
- 271.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochimica et Biophysica Acta. 2009;1792(6):497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 272.Tili E, Croce CM, Michaille JJ. miR-155: on the crosstalk between inflammation and cancer. International reviews of immunology. 2009;28(5):264–284. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- 273.Schipper HM, Maes OC, Chertkow HM, Wang E. microRNA expression in Alzheimer blood mononuclear cells. Gene Regulation and Systems Biology. 2007;1:263–274. doi: 10.4137/grsb.s361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Wang X, Liu P, Zhu H, et al. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer’s disease, inhibits bcl2 translation. Brain Research Bulletin. 2009;80(4-5):268–273. doi: 10.1016/j.brainresbull.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 275.Kim AH, Reimers M, Maher B, et al. microRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. doi: 10.1016/j.schres.2010.07.002. Schizophrenia Research. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Dinan TG. microRNAs as a target for novel antipsychotics: a systematic review of an emerging field. International Journal of Neuropsychopharmacology. 2010;13(3):395–404. doi: 10.1017/S1461145709990800. [DOI] [PubMed] [Google Scholar]
- 277.Zhou R, Yuan P, Wang Y, et al. Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology. 2009;34(6):1395–1405. doi: 10.1038/npp.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.O'Connor RM, Finger BC, Flor PJ, Cryan JF. Metabotropic glutamate receptor 7: at the interface of cognition and emotion. European Journal of Pharmacology. 2010;639(1–3):123–131. doi: 10.1016/j.ejphar.2010.02.059. [DOI] [PubMed] [Google Scholar]
- 279.Li L, Chen X-P, Li Y-J. microRNA-146a and human disease. Scandinavian Journal of Immunology. 2010;71(4):227–231. doi: 10.1111/j.1365-3083.2010.02383.x. [DOI] [PubMed] [Google Scholar]
- 280.Curtale G, Citarella F, Carissimi C, et al. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood. 2010;115(2):265–273. doi: 10.1182/blood-2009-06-225987. [DOI] [PubMed] [Google Scholar]
- 281.Lukiw WJ, Zhao Y, Jian GC. An NF-κB-sensitive micro RNA-146a-mediated inflammatory circuit in alzheimer disease and in stressed human brain cells. Journal of Biological Chemistry. 2008;283(46):31315–31322. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Buchanan MM, Hutchinson M, Watkins LR, Yin H. Toll-like receptor 4 in CNS pathologies. Journal of Neurochemistry. 2010;114(1):13–27. doi: 10.1111/j.1471-4159.2010.06736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Marta M, Meier UC, Lobell A. Regulation of autoimmune encephalomyelitis by toll-like receptors. Autoimmunity Reviews. 2009;8(6):506–509. doi: 10.1016/j.autrev.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 284.Fernández M, Montalban X, Comabella M. Orchestrating innate immune responses in multiple sclerosis: molecular players. Journal of Neuroimmunology. 2010;225(1-2):5–12. doi: 10.1016/j.jneuroim.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 285.Gandhi R, Laroni A, Weiner HL. Role of the innate immune system in the pathogenesis of multiple sclerosis. Journal of Neuroimmunology. 2010;221(1-2):7–14. doi: 10.1016/j.jneuroim.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.McKimmie CS, Roy D, Forster T, Fazakerley JK. Innate immune response gene expression profiles of N9 microglia are pathogen-type specific. Journal of Neuroimmunology. 2006;175(1-2):128–141. doi: 10.1016/j.jneuroim.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 287.Worm J, Stenvang J, Petri A, et al. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of c/ebp Beta and down-regulation of G-CSF. Nucleic Acids Research. 2009;37(17):5784–5792. doi: 10.1093/nar/gkp577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Tumani H, Hartung H-P, Hemmer B, et al. Cerebrospinal fluid biomarkers in multiple sclerosis. Neurobiology of Disease. 2009;35(2):117–127. doi: 10.1016/j.nbd.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 289.de Smaele E, Ferretti E, Gulino A. microRNAs as biomarkers for CNS cancer and other disorders. Brain Research. 2010;1338:100–111. doi: 10.1016/j.brainres.2010.03.103. [DOI] [PubMed] [Google Scholar]
- 290.Harrington MG, Fonteh AN, Oborina E, et al. The morphology and biochemistry of nanostructures provide evidence for synthesis and signaling functions in human cerebrospinal fluid. Cerebrospinal Fluid Research. 2009;6, article 10 doi: 10.1186/1743-8454-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Hunter MP, Ismail N, Zhang X, et al. Detection of microRNA expression in human peripheral blood microvesicles. PloS ONE. 2008;3(11) doi: 10.1371/journal.pone.0003694. Article ID e3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Cogswell JP, Ward J, Taylor IA, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. Journal of Alzheimer's Disease. 2008;14(1):27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 293.Awad A, Hemmer B, Hartung H-P, Kieseier B, Bennett JL, Stuve O. Analyses of cerebrospinal fluid in the diagnosis and monitoring of multiple sclerosis. Journal of Neuroimmunology. 2010;219(1-2):1–7. doi: 10.1016/j.jneuroim.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 294.Andreasen D, Fog JU, Biggs W, Salomon J, Dahslveen IK, Baker A. Improved microRNA quantification in total RNA from clinical samples. Methods. 2010;50(4):S6–S9. doi: 10.1016/j.ymeth.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 295.Glinsky GV. SNP-guided microRNA maps (MirMaps) of 16 common human disorders identify a clinically accessible therapy reversing transcriptional aberrations of nuclear import and inflammasome pathways. Cell Cycle. 2008;7(22):3564–3576. doi: 10.4161/cc.7.22.7073. [DOI] [PubMed] [Google Scholar]
- 296.Glinsky GV. An SNP-guided microRNA map of fifteen common human disorders identifies a consensus disease phenocode aiming at principal components of the nuclear import pathway. Cell Cycle. 2008;7(16):2570–2583. doi: 10.4161/cc.7.16.6524. [DOI] [PubMed] [Google Scholar]
- 297.Nicoloso MS, Sun H, Spizzo R, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Research. 2010;70(7):2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Saunders MA, Liang H, Li W-H. Human polymorphism at microRNAs and microRNA target sites. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(9):3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Seto AG. The road toward microRNA therapeutics. International Journal of Biochemistry and Cell Biology. 2010;42(8):1298–1305. doi: 10.1016/j.biocel.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 300.Beitzinger M, Meister G. microRNAs: from decay to decoy. Cell. 2010;140(5):612–614. doi: 10.1016/j.cell.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 301.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annual review of biochemistry. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 302.Stenvang J, Lindow M, Kauppinen S. Targeting of microRNAs for therapeutics. Biochemical Society Transactions. 2008;36(6):1197–1200. doi: 10.1042/BST0361197. [DOI] [PubMed] [Google Scholar]
- 303.Elmén J, Lindow M, Schütz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 304.Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]