Abstract
Quinone reductases are flavin-containing enzymes that have been implicated in protecting organisms from redox stress and, more recently, as redox switches controlling the action of the proteasome. The reactions of the catalytic cycle of the dimeric quinone reductase Lot6p from Saccharomyces cerevisiae were studied in anaerobic stopped-flow experiments at 4° C. Both NADH and NADPH reacted similarly, reducing the FMN prosthetic group rapidly at saturation, but binding with very low affinity. The enzyme stereospecifically transferred the proS-hydride of NADPH with an isotope effect of 3.6, indicating that hydride transfer, and not an enzyme conformational change, is rate-determining in the reductive half-reaction. No intermediates such as charge-transfer complexes were detected. In the oxidative half-reaction, reduced enzyme reacted in a single phase with the six quinone substrates tested. The observed rate constants increased linearly with quinone concentration up to the limits allowed by solubility, indicating either a bimolecular reaction or very weak binding. The logarithm of the bimolecular rate constant increase linearly with the reduction potential of the quinone, consistent with the notion that quinone reductases strongly disfavor radical intermediates. Interestingly, both half-reactions of the catalytic cycle strongly resemble bioorganic model reactions; the reduction of Lot6p by NAD(P)H is moderately faster than non-enzymatic models, while the oxidation of Lot6p by quinones is actually slower than non-enzymatic reactions. This curious situation is consistent with the structure of Lot6p, which has a crease we propose to be the binding site for pyridine nucleotides, and space – but no obvious catalytic residues – near the flavin allowing the quinone to react. The decidedly sub-optimized catalytic cycle suggests that selective pressures other than maximizing quinone consumption shaped the evolution of Lot6p. This may reflect the importance of suppressing other potentially deleterious side-reactions, such as oxygen reduction, or it may indicate that the role Lot6p plays as a redox sensor in controlling the proteasome is more important than its role as a detoxifying enzyme.
Several cellular oxidoreductases are involved in the metabolism of quinone compounds by catalyzing their one- or two-electron reduction. The cytotoxic and neoplastic effects of quinones are generally thought to arise from their one-electron reduction, for instance by cytochrome P450 reductases, thus resulting in oxidative cycling of harmful oxygen species (1). In contrast, flavin-dependent NAD(P)H:quinone oxidoreductases (QRs) catalyze the strict two-electron reduction yielding the fully reduced hydroquinone species, thereby affording protection against cytotoxic effects of quinones (2). This property has served as the starting point for a large number of studies to examine the possible relationship of QRs to oxygen toxicity. By 1-electron reduction of quinones, semiquinones are generated which react spontaneously with molecular oxygen to give the parent quinone and superoxide. They are also able to react with nucleophiles, such as reduced glutathione, thereby depleting the reduced thiol and dihydronicotinamide pools. Furthermore, quinone metabolites contribute considerably to the toxic and carcinogenic effects of aromatic hydrocarbons. By reducing quinones directly to hydroquinones, QRs are proposed to be a cellular defense against quinone toxicity (3;4).
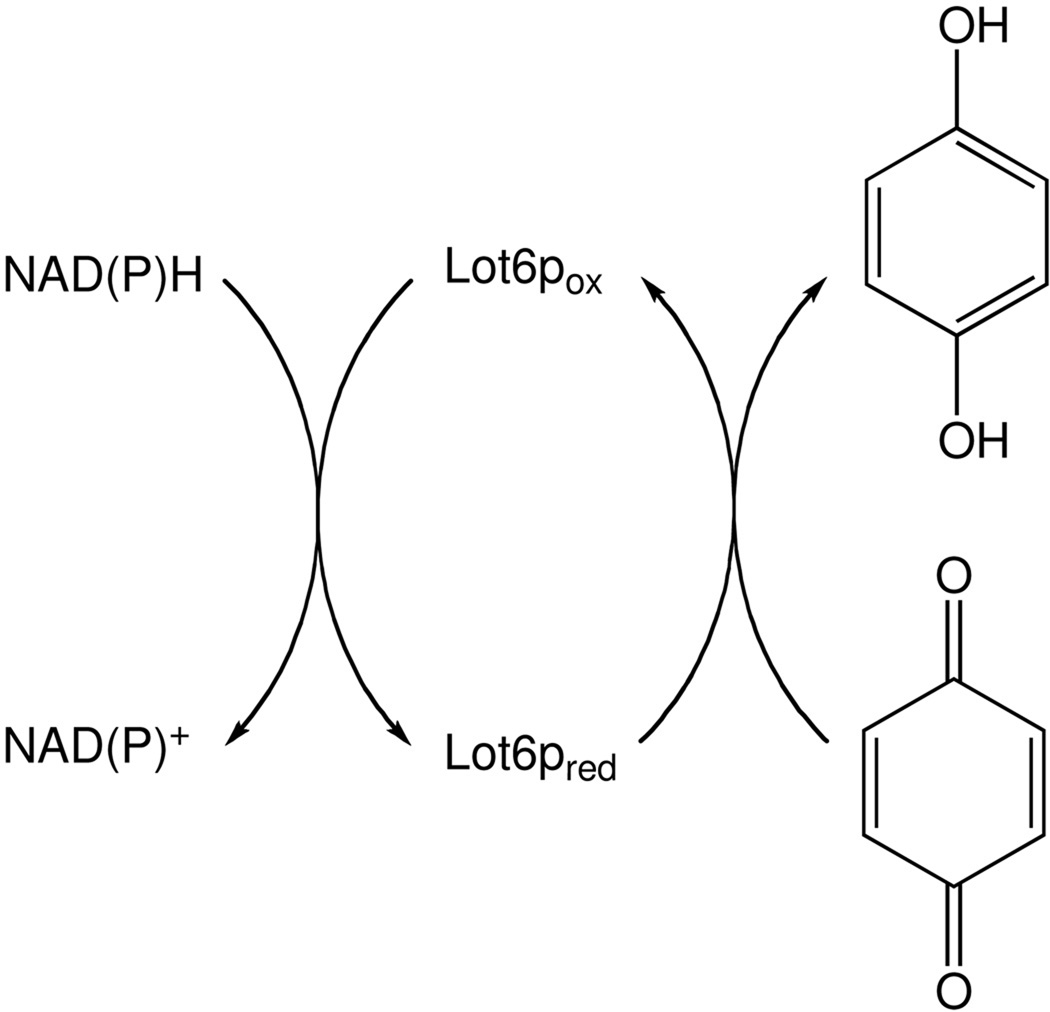
Flavin-dependent QRs from a number of species have been reported, e.g. human, mouse and rat, and from plant species. Activity was also found in the unicellular model organism Saccharomyces cerevisiae, where the FMN-dependent QR Lot6p was shown to possess similar biochemical properties as mammalian QRs (5). The similarity becomes evident on a structural level as well since all cytosolic QRs reported so far, including the yeast enzyme, adopt a flavodoxin-like fold and self-assemble as homodimers (1;6). The flavin cofactor (FAD for mammalian and plant enzymes, FMN in the case of Lot6p) is bound to the enzyme in the same manner, with the isoalloxazine ring system of the molecule accessible to the solvent and the ribityl phosphate and AMP (for FAD) moiety buried in the protein. The flavin prosthetic group is reduced by NAD(P)H, followed by the reaction of reduced flavin and the quinone (Figure 1) (5;7). X-ray structures of QRs with either electron donor or acceptor bound to the active center show that both substrates occupy the same site near the flavin prosthetic group (7). Binding of the electron acceptor cannot occur before the electron donor is released from the active site, thus providing a structure-based explanation for the observed ping-pong bi-bi mechanism which seems to be in operation in all QRs (1).
Figure 1.
Catalytic cycle of Lot6p. In the reductive half reaction, NADH or NADPH reduces the FMN prosthetic group. Subsequently, a quinone reoxidizes the reduced flavin and accepts two electrons to form a fully reduced hydroquinone in the oxidative half-reaction.
We recently reported the association of yeast QR Lot6p with the 20S proteasome via its FMN binding site (8). Interestingly, reduction of the flavin by either NADH or photoreduction triggers binding of a second interaction partner, namely the yeast transcription factor Yap4p, a member of the yeast activator protein (Yap) family which was previously shown to be involved in the oxidative stress response (9;10). Binding of Yap4p to the preformed QR-proteasome complex thus does not require the presence of a reduced pyridine nucleotide, but instead a reduced flavin in the active site. Association of Yap4p with the QR and the 20S proteasome was shown to affect both localization and ubiquitin-independent degradation of the transcription factor. Hence we proposed that the FMN prosthetic group in the Lot6p active site is a redox-regulated switch which controls stability and localization of Yap4p. Under reducing conditions, i.e. in the presence of excess NAD(P)H, Yap4p is associated with the QR-proteasome complex and thus is retained in the cytosol. In the presence of quinones, the flavin is rapidly reoxidized and the bound transcription factor is released from the complex and subsequently relocalizes to the nucleus, avoiding proteasomal degradation (8). A similar redox controlled mechanism might regulate the stabilization and localization of p53 and related transcription factors in mammalian cells (11).
Here, we report kinetic analysis of the half-reactions of Lot6p, using both NADH and NADPH as electron donors and a series of quinones as electron acceptors. Furthermore, by measuring kinetic isotope effects (KIEs), we show that breakage of the H-C bond of the reduced pyridine nucleotide is the rate-limiting step within the reductive half-reaction. Finally, we elucidated the stereochemistry of hydride transfer from NADPH to the active site flavin and correspondingly suggest a binding mode of NADPH to the QR.
Experimental procedures
Reagents
All chemicals were of highest grade commercially available and purchased from Sigma-Aldrich, Fluka, Merck or BioCatalytics.
Expression of Lot6p
Lot6p from Saccharomyces cerevisiae was overexpressed and purified according to published procedures (5). The fractions containing the enzyme were pooled, concentrated and stored at −70 °C after being diluted with glycerol to 20% (v/v). For use in experiments, the enzyme was exchanged into the appropriate buffer using Econo-Pac 10DG disposable desalting columns (Bio-Rad).
Synthesis of NADPD
The stereospecifically labelled coenzymes [4R-2H]-NADPH was synthesized enzymatically using [2-2H]-isopropanol/alcohol dehydrogenase (12). [4S-2H]-NADPH was synthesized from [1-2H]glucose/hexokinase/glucose-6-phosphate dehydrogenase (13).
Determination of the Stereospecificity of Lot6p
Lot6p was exchanged into appropriate buffer (30 mM Tris-HCl, pH 8.0, in D2O) using Econo-Pac 10DG desalting columns (Bio-Rad). A solution (1 mL) containing the buffer mentioned above, 10 µM Lot6p, and 3 mg of either [4R-2H]-NADPH or [4S-2H]-NADPH was left to react for 2 hours at 37 °C. Enzyme was removed using size-exclusion chromatography, the remaining solution was lyophilized and the product analyzed by 1H-NMR (12). All signals were referenced to the HOD peak and are given relative to TMS.
Instrumentation
Absorbance spectra were obtained using a Shimadzu UV-2501PC scanning spectrophotometer. Stopped-flow experiments were performed at 4 °C using a Hi-Tech Scientific KinetAsyst SF-61 DX2 stopped-flow spectrophotometer. NMR spectra were collected on a 600 MHz instrument (Brucker).
Preparation of Anaerobic Solutions
Enzyme solutions for rapid reaction studies were made anaerobic in glass tonometers by repeated cycles of evacuation and equilibration over an atmosphere of purified argon (14). Substrate solutions were made anaerobic within the syringes that were to be loaded onto the stopped-flow instrument by bubbling solutions with purified argon.
Reductive Half-Reaction
Anaerobic enzyme equilibrated in buffer (20 mM Tris-HCl, pH 8.0) was mixed with various concentrations of either NADH (0.5 to 5 mM after mixing) or NADPH (0.2 to 16 mM after mixing) in the same buffer. Reaction traces were collected at 456 nm. Kinetic traces were fit to two exponentials using KinetAsyst 3 (HiTech Scientific). The reduction rate constant (kred) was determined from the limiting value of the observed rate constant extrapolated to infinite NAD(P)H concentration obtained by fitting kobs versus NAD(P)H concentration to a hyperbola in SigmaPlot (Systat Software, Inc.). Deuterium isotope effects on flavin reduction by [4S-2H]-NADPH were determined by comparing the reductive half-reaction at pH 9.0 using labelled and unlabelled substrate. A diode-array detector was used in experiments with 4 mM NADPH in attempts to detect intermediates in the reductive half-reaction. Absorbance spectra were collected from 350 to 700 nm for 0.4 s with an integration time of 1.5 ms.
Oxidative Half-Reaction
The anaerobic enzyme was equilibrated in buffer (50 mM TAPS, pH 7.4) and reduced by titrating with one equivalent of dithionite. Quinone substrates were prepared as 10 mM stock solutions in EtOH and subsequently diluted into buffer. The reduced enzyme was then mixed with various concentrations of anaerobic quinone (1,4-benzoquinone, 25 to 100 µM; menadione, 25 to 300 µM; coenzyme Q0, 25 to 200 µM; 2-hydroxy-1,4-naphthoquinone, 25 to 200 µM; 1,2-naphthoquinone-4-sulfonic acid, 50 to 200 µM; Na-anthraquinone, α-sulfonate, 50 to 200 µM; all concentrations are those after mixing) at pH 7.4, 4 °C, and flavin oxidation was monitored at 456 nm. Kinetic traces were fit to two exponentials using KinetAsyst 3 (HiTech Scientific). The bimolecular rate constants were determined by fitting kobs versus quinone concentration to a line.
Results
The catalytic cycle of Lot6p follows a ping-pong bi-bi mechanism and consists of a reductive half-reaction, in which NADH or NADPH reduces the FMN prosthetic group, and an oxidative half-reaction, in which a quinone or other substrate oxidizes the reduced flavin (Figure 1) (5). Each of the half-reactions of the catalytic cycle of Lot6p was studied in detail in anaerobic stopped-flow experiments. Additional information was obtained by determining the stereochemistry of hydride transfer and the kinetic isotope effect.
Reductive Half-Reaction
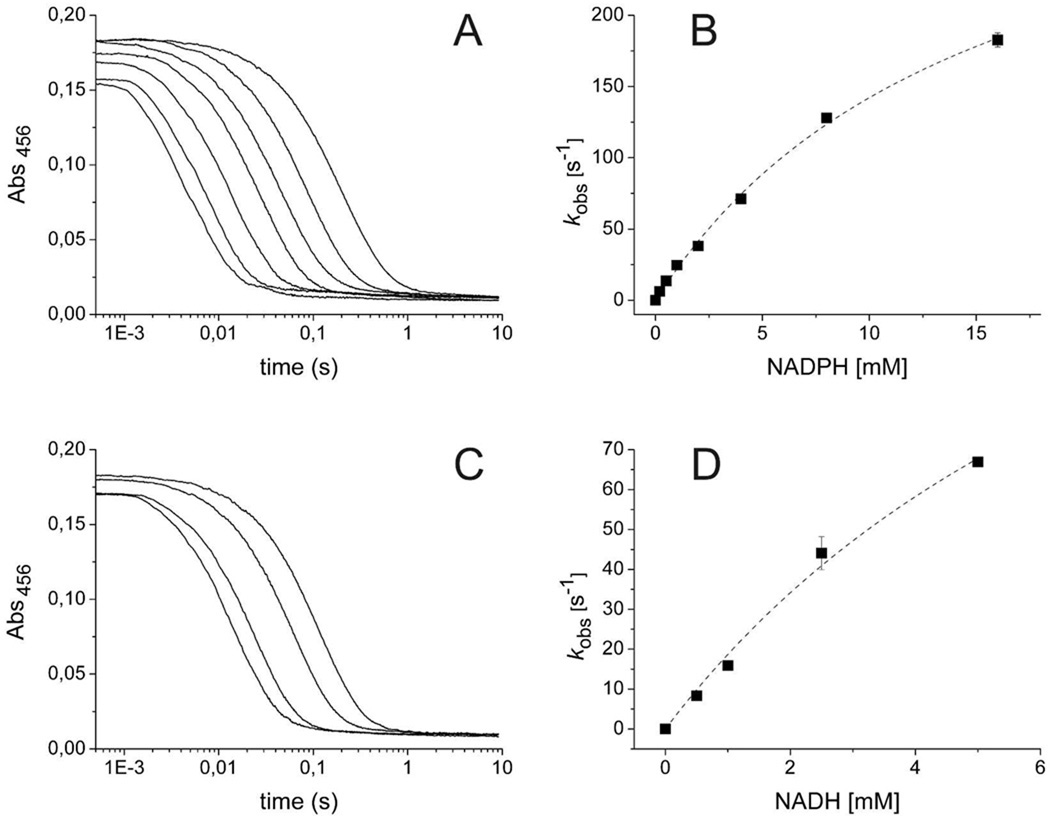
Anaerobic enzyme was mixed in a stopped-flow instrument at pH 8.0, 4 °C, with various concentrations of NADPH or NADH. FMN reduction was monitored by the decrease in absorbance at 456 nm. The decrease in flavin absorbance versus time occurred in a single phase, and the total change was always equal to the expected change in absorbance for the complete reduction of Lot6p (Figures 2A and 2C). Traces were fitted to single exponentials. The rate constant for reduction, obtained by extrapolating the observed rate constant to infinite pyridine nucleotide, was 311 ± 16 s−1 for NADPH and 198 ± 36 s−1 for NADH. The dissociation constant, obtained from the half-saturating concentration, was 12.5 ± 1.1 mM for NADPH and 9.6 ± 2.4 mM for NADH (Figures 2B and 2D). These results indicate that, at saturation, NADPH is a marginally better reductant compared to NADH. Often, flavin-pyridine nucleotide reactions produce charge-transfer complexes with characteristic absorbances at long wavelengths. However, no charge-transfer absorbance could be observed for either of the two pyridine nucleotides in traces collected at 550, 600, or 700 nm, nor were any observed in spectra collected with a diode-array detector.
Figure 2.
Reduction of Lot6p with NADH and NADPH. (A) Anaerobic Lot6p (final concentration of 15 µM) was mixed with anaerobic NADPH (concentrations after mixing: 0.2, 0.5, 1, 2, 4, 8 and 16 mM) at pH 8.0. Note that the reaction traces are shown on a logarithmic time scale. (B) Observed rate constants of flavin reduction as a function of NADPH concentration. The dashed line represents the fit to the data points using a hyperbolic function giving a Kd of 12.5 ± 1.1 mM. (C) Anaerobic Lot6p (final concentration of 15 µM) was mixed with anaerobic NADH (concentrations after mixing: 0.5, 1, 2.5 and 5 mM) at pH 8.0. Again, the reaction traces are shown on a logarithmic time scale. (D) Observed rate constants of flavin reduction as a function of NADH concentration. The dashed line represents the fit to the data points using a hyperbolic function giving a Kd of 9.6 ± 2.4 mM.
Stereochemistry
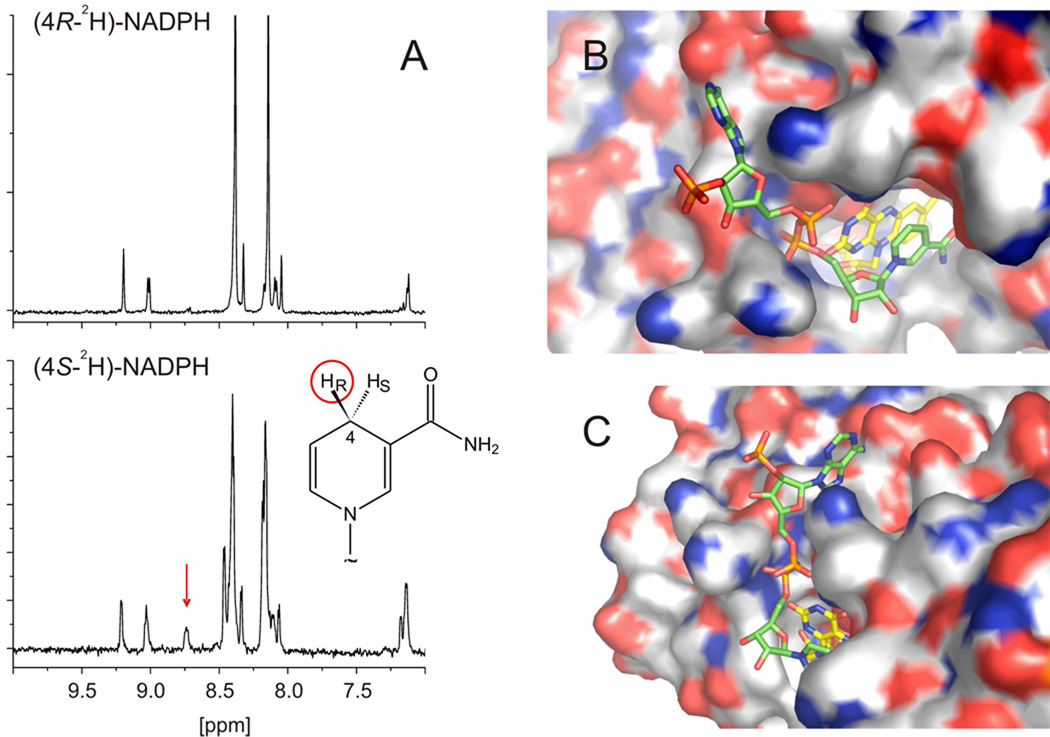
Almost all enzymes that use pyridine nucleotides as substrates have an absolute preference for transferring one of the two diastereotopic hydrides at the 4-position. We synthesized [4S-2H]-NADPH and [4R-2H]-NADPH and oxidized them (in separate reactions) using O2 as the oxidizing substrate. After complete oxidation, the NADP+ produced was analyzed by 1H-NMR spectroscopy. A proton signal for the 4-position of the nicotinamide ring was observed at 8.74 ppm when [4S-2H]-NADPH was used as substrate, and this signal was absent when [4R-2H]-NADPH was tested (Figure 3A), indicating that Lot6p specifically transfers the pro-S hydride of NADPH.
Figure 3.
Stereochemistry of hydride transfer from NADPH to Lot6p. [4S-2H]-NADPH and [4R-2H]-NADPH was oxidized using Lot6p and O2 as the oxidizing substrate. After complete oxidation, the NADP+ produced was analyzed by 1H-NMR spectroscopy. (A) No proton signal for the 4-position of the nicotinamide ring was observed when [4R-2H]-NADPH was used (upper panel), whereas the signal at 8.74 ppm was present in the case of [4S-2H]-NADPH (lower panel, red arrow). The inset shows the stereochemistry of protons in position 4 of the nicotinamide moiety of NADPH. The proR proton of NADPH that is transformed into the aromatic proton visible in the NMR spectra of [4S-2H]-NADPH is indicated by a red circle. (B) and (C) Possible binding mode of NADPH to the Lot6p active site, as suggested by stereochemistry. Lot6p is shown in a surface representation, color-coded according to the elements (carbon, grey; nitrogen, blue; oxygen, red; sulfur, orange). FMN is shown as a color-coded stick model (carbon, yellow), the NADP+ is shown in the same manner (carbon, green). The model was prepared by manually docking NADP+ taken from 1d1g.pdb to the structure of Lot6p (1t0i.pdb) using PyMOL (35).
Kinetic Isotope Effects
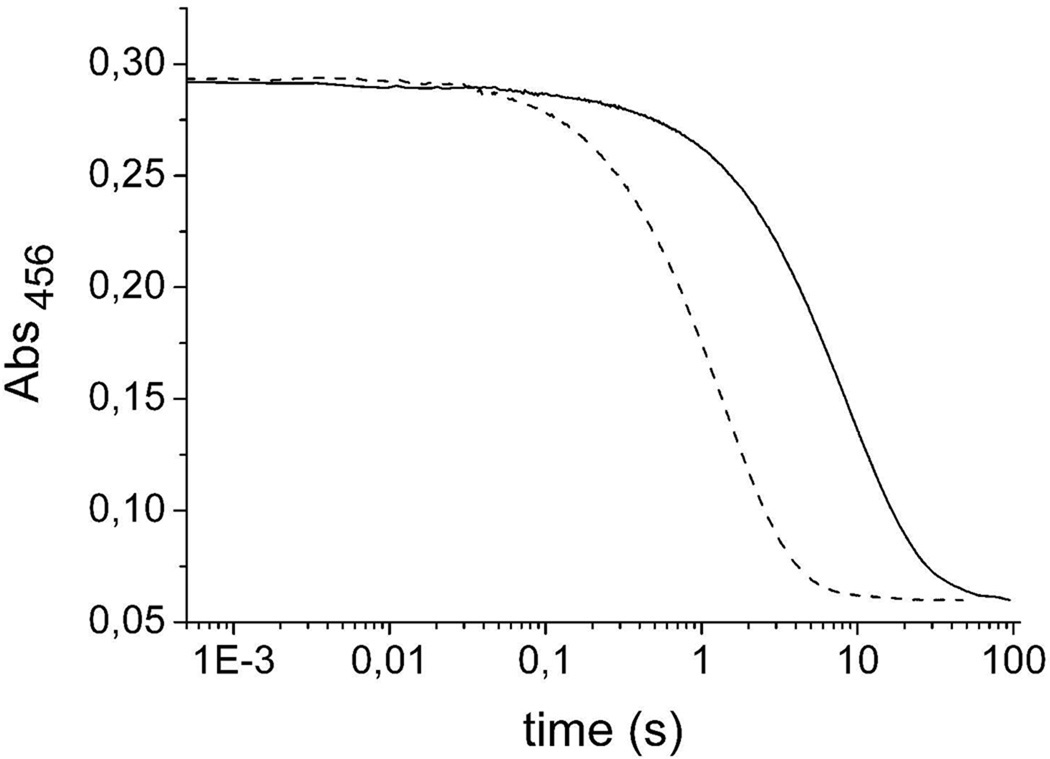
The kinetic isotope effect on flavin reduction by [4S-2H]-NADPH was determined at pH 9.0 in stopped-flow experiments using 200 µM – 400 µM pyridine nucleotide. An average KIE on kred/Kd of 3.6 ± 0.3 was obtained (Figure 4).
Figure 4.
The kinetic isotope effect on flavin reduction by [4S-2H]-NADPH was determined in the reductive half-reaction using the labelled substrate in stopped-flow experiments. Anaerobic Lot6p (final concentration of 15 µM) was mixed with 200 µM anaerobic NADPH (dashed curve) or 200 µM [4S-2H]-NADPH (solid curve) in 100 mM CHES, pH 9.7 at 4 °C. Reaction traces (456 nm) are shown on a logarithmic time scale.
Oxidative Half-Reactions
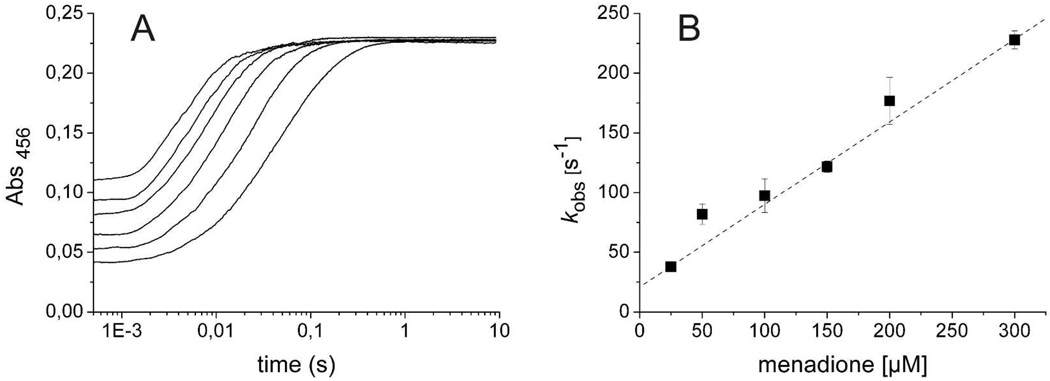
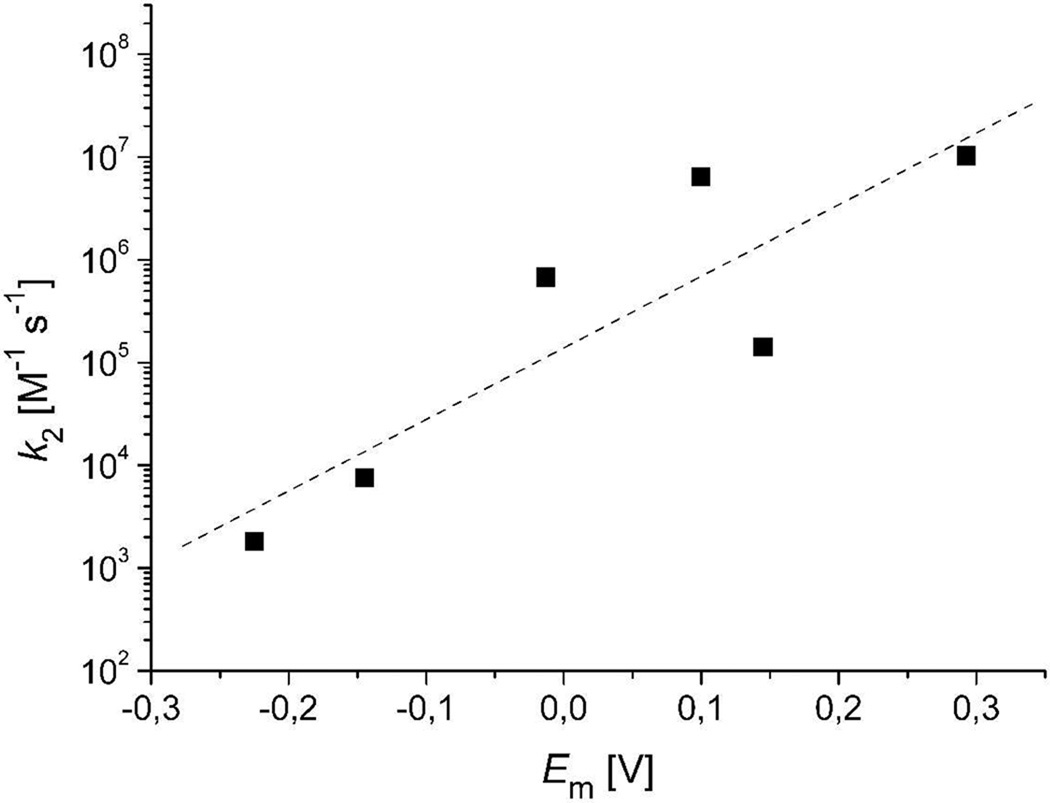
Reduced flavoenzymes often react with many oxidants. Quinone reductases usually have a broad substrate tolerance. Therefore, we investigated the ability of several quinones to oxidize the reduced enzyme. Anaerobic enzyme was reduced by titrating with one equivalent of dithionite. The reduced enzyme was mixed with various concentrations of anaerobic quinone at pH 7.4, 4 °C, and flavin oxidation was monitored at 456 nm (Figure 5A for menadione). The observed rate constants varied linearly with quinone concentration up to the highest values tested, suggesting a bimolecular reaction, i.e., there was no evidence for quinone binding prior to reaction (Figure 5B for menadione). However, it should be noted that many of the quinones used have low solubilities in water, preventing the use of high quinone concentrations and, therefore, the detection of weak binding. With some quinones (e.g., menadione, Figure 5B), the linear dependence of the observed rate constant extrapolated to a finite value as the quinone concentration approached zero. If the quinone actually did react in a complex with the reduced enzyme, then the non-zero intercept would be the value of the rate constant for the reverse reaction. Traces obtained at longer wavelengths showed no evidence of transient charge-transfer complexes. Bimolecular rate constants for enzyme oxidation ranged from 1.81 × 103 M−1 s−1 for anthraquinone α-sulfonate, the quinone tested with the lowest reduction potential, to 1.03 × 107 M−1 s−1 for 1,4-benzoquinone, the quinone with the highest reduction potential tested (Table 1). Interestingly, there was a linear increase of the logarithm of the flavin oxidation rate constant with increasing quinone reduction potential (Figure 6).
Figure 5.
Oxidation of reduced Lot6p with menadione. Anaerobic enzyme (20 µM after mixing) was reduced by titrating with one equivalent dithionite. Subsequently, reduced enzyme was mixed with various concentrations of menadione (25, 50, 100, 150, 200 and 300 µM) at pH 7.4, and flavin oxidation was monitored at 456 nm. (B) The observed rate constants varied linearly with quinone concentration, suggesting a bimolecular reaction with a rate constant of 6.77 × 105 M−1 s−1.
Table 1.
Comparison of rate constants and midpoint redox potentials for the oxidative half reaction.
| Electron acceptor | k2 [M−1 s−1] | Em (V) |
|---|---|---|
| 1,4-Benzoquinone | 1.03 × 107 | +0.293 (31) |
| Coenzyme Q0 | 6.45 × 106 | +0.100 (32) |
| Menadione | 6.77 × 105 | −0.013 (33) |
| 1,2-Napthoquinone-4-sulfonic acid, Na salt | 1.42 × 105 | +0.145 (34) |
| 2-Hydroxy-1,4-naphthoquinone | 7.58 × 103 | −0.145 (33) |
| Na-anthraquinone, α-sulfonate | 1.81 × 103 | −0.225 (33) |
Figure 6.
Comparison of bimolecular rate constants of the oxidative half-reaction and the reduction potentials of substrates. A linear increase of the logarithm of the flavin oxidation rate with increasing quinone reduction potential is found. The dashed line represents a linear fit to the data points.
Discussion
NAD(P)H:quinone oxidoreductases catalyze strict two-electron reductions of a variety of quinones at the expense of both NADH and NADPH. Thus, they are thought to protect against the cytotoxic and neoplastic effects of electrophilic quinones. For example, QR-null mice were shown to be more susceptible to quinone toxicity than their wild-type counterparts. Furthermore, QRs are found at increased levels in many solid tumours and therefore can be used to target tumour cells through bioreductive activation of quinone-based prodrugs (1).
However, recent findings suggest an additional role of QRs beyond detoxification. They were shown to be able to act as redox switches of the proteasome thereby regulating proteasomal degradation of certain transcription factors and possibly intrinsically unstructured proteins. To this end QR binds to the 20S core particle of the proteasome and recruits transcription factors to the complex. Binding of transcription factors such as p53 appears to be governed by the redox state of the flavin and therefore links stability of transcription factors to cellular events such as oxidative stress (11). Interestingly, recruitment of a transcription factor to the reduced QR:proteasome complex seems to be conserved from mammals to yeast. We recently reported that reduction of Lot6p, the yeast ortholog of mammalian NQO1, triggers binding of the b-Zip transcription factor Yap4p to the preformed QR:proteasome complex. Binding of Yap4p to the complex not only protects it from ubiquitin-independent proteasomal degradation, but also regulates its cellular localization. Thus, the Lot6p:proteasome complex can be regarded as redox switch in which the QR acts as a sensor for oxidative stress (8).
The possible biological roles for QR – either as a quinone detoxifier or as redox sensor regulating proteasomal action – are facilitated by the curiously atypical catalytic behavior of Lot6p. The enzyme was reduced by saturating concentrations of NADPH and NADH nearly equally rapidly (~300 s−1 and ~200 s−1, respectively), but binding was quite weak, with Kd values estimated near ~10 mM. Typical cellular pyridine nucleotide levels are much lower (~1 mM for NADH and an order of magnitude lower for NADPH (15)), so that reduction of the enzyme will be controlled by the second-order rate constants kred/Kd which are nearly equal for both reductants – 2.5 × 104 M−1s−1 for NADPH and 2.1 × 104 M−1s−1 for NADH. Therefore, under second-order conditions, Lot6p shows essentially no intrinsic preference for a reducing substrate. Reduction of mammalian NAD(P)H:quinone oxidoreductase by NAD(P)H also followed second-order kinetics, although it was much faster than with Lot6p (3.13 × 108 M−1 s−1 for NADPH at 15 °C (16) and 2.2 × 107 and 1.5 × 107 M−1 s−1 for NADPH and NADH, respectively, at 4 °C) (4). However, in those studies the electron donor concentration was tested only up to 50 µM NAD(P)H.
Interestingly, the enzyme-free reactions of model dihydronicotinamides with flavins (17;18) are about as fast at 25 °C as the reactions of Lot6p with NAD(P)H are at 4 °C, and dihydronicotinamides form complexes with isoalloxazines in aqueous solution with an affinity only about ten-fold weaker than the binding of pyridine nucleotides to Lot6p (17). Thus the enzyme enhances the reaction rate only moderately over the non-enzymatic rate. Unlike the reactions of model compounds, no charge-transfer absorbance was observed in the enzyme system. Charge-transfer complexes are frequently observed in the reduction of flavoenzymes by NAD(P)H, but not always. In some instances this may be because the complex never accumulates to an appreciable amount for kinetic reasons, or that the extinction coefficient is low in the particular orientation of dihydronicotinamide and flavin. It has been shown recently that rapid hydride transfer to flavin is possible even without a charge-transfer intermediate (19). NAD(P)H reduces Lot6p with an isotope effect of 3.6. The corresponding primary isotope effect on the reduction of model flavins by model dihydronicotinamides is 2.7 (17). Therefore, hydride transfer determines the rate of flavin reduction in Lot6p, and the enzymatic transition state for reduction is more symmetrical than in unstructured aqueous solvent.
The X-ray structure of Lot6p suggests that its binding site for NAD(P)H will be significantly different from that of mammalian NQO1. In NQO1 from rat, NAD and competitive analogs bind over the re-face of the flavin, with the nucleotide stretched almost perpendicularly to the plane of the isoalloxazine and the adenosine moiety making contacts with residues from an adjoining protein subunit. Analogous residues are not provided by the dimer of Lot6p (6), indicating that the nucleotide must bind differently. We found that Lot6p specifically transfers the proS hydride of NADPH and thus hydride transfer occurs with B-side stereospecificity (Figure 3A). A plausible model of a pyridine nucleotide complex was built by manually docking NADP taken from the structure of dihydrofolate reductase from Thermotoga maritima (1d1g.pdb). NADP was positioned so that the si-face of the nicotinamide ring was over the re-face of the isoalloxazine, C4 of the nicotinamide was ~3.4 Å from N5 of the flavin (a position ideal for hydride transfer), and the pyrophosphate moiety of the nucleotide was next to the loop connecting α4 and β4 (6), in contact with a histidine, and was also at the positive end of a helix dipole (Figures 3B and 3C). The adenine and ribose phosphate moieties run along the α4 helix, with the phosphate moiety pointing away from the protein and into solvent, thus not participating in any interactions with the protein. Hence, our model structure explains why Lot6p has no intrinsic preference for NADPH over NADH.
The lack of substrate specificity is even more prominent in the oxidative half-reaction. QRs are remarkable in their ability to reduce a variety of oxidants including quinones (4;20), azo-compounds (1;21), chromium VI compounds (22), benzotriazine di-N-oxides (23), and nitro-compounds (24). In our studies, six different quinone electron acceptors were used as substrates. Each reacted in a single phase whose observed rate constant varied linearly with quinone concentration – there was no evidence for the formation of a reduced enzyme-substrate complex. However, it should be noted that many of the quinones tested have low solubilities in water, preventing the use of high concentrations and therefore the detection of weak binding. Interestingly, quinone binding was observed in titrations of the oxidized enzyme (5), suggesting a redox-linked change to the active site is responsible for the weak (or non-existent) quinone binding in the oxidative half-reaction.
Quinones could either be reduced by hydride transfer or by sequential single-electron transfers. No evidence was obtained for single-electron transfer reactions – no semiquinones were observed. This is in line with the fact that the enzyme does not thermodynamically stabilize the flavin semiquinone in redox titrations (5). However, these observations alone do not preclude radical intermediates; if quinone substrates are reduced in sequential single-electron transfers, the intermediate radical pair will not be observed kinetically if the first electron transfer from reduced flavin to quinone is rate-determining, and radical products will not escape the enzyme to be detected by analytical methods if the rate constant for dissociation is significantly lower than the rate constant for the second electron transfer. Ferricyanide, an obligate one-electron oxidant, reacts with reduced Lot6p, demonstrating that the flavin semiquinone is accessible to this powerful oxidizing reagent. In contrast, the reaction with molecular oxygen was very slow, with a bimolecular rate constant of 1.1 M−1s−1 (8), indicating that the reduced QR is unable to activate oxygen with an efficiency similar to many other flavoproteins (25). The structural basis for the low oxygen reactivity of reduced QRs is not understood. In human NQO1, which was also reported to have very low oxidase activity (16), Tyr-128 gates the catalytic site and was thought to protect the reduced flavin from reoxidation through reaction with molecular dioxygen. However, the loop containing Tyr-128 in the human enzyme is entirely missing in Lot6p. Since oxygen activation by flavoproteins is not yet understood clearly (25), the relative importance of possible gating residues and the stabilization of the flavin semiquinone and superoxide (obligate intermediates in the reaction of flavin hydroquinone with O2) cannot be assessed. Nonetheless, an aromatic gate does not appear to explain the low oxygen reactivity in QRs.
The reactions of Lot6p with quinones closely resembles the non-enzymatic reactions of model flavins with quinones. For those quinones that react slowly enough to be studied, the bimolecular rate constant increases with the reduction potential of the quinone, and there is no evidence for radical intermediates. The reaction of benzoquinone with reduced flavin was too rapid to be observed, placing a limit of >108 M−1s−1 on the bimolecular rate constant (26;27). The reduction of benzoquinone by Lot6p was slower, with a rate constant of 107 M−1s−1. Thus, the apoprotein of Lot6p actually slows the reduction of this quinone substrate, strongly suggesting that selective pressures other than quinone consumption rates were paramount in the evolution of the enzyme. As with non-enzymatic quinone reduction, reduction by Lot6p was faster with higher-potential quinones; the logarithms of second-order rate constants increased linearly with the reported 2-electron reduction potentials of the substrates (Figure 6). The correlation of the two-electron potential with rate constant again suggests that semiquinones are not intermediates. It is worth noting, however, that most of the single-electron reduction potentials of the quinones used in this work are not known, so a possible correlation with the rate constant of oxidation could not be assessed.
The lack of enzymatic acceleration of quinone reduction may not be general to all quinone reductases. The structure of the human NQO1-duraquinone complex shows that duraquinone is held over the face of the flavin by hydrogen bonds to both of its carbonyls – a tyrosine on one and a water molecule held by two tyrosines and a histidine on the other (28). These hydrogen-bond donors, especially the presumably positively charged network, are ideally positioned to protonate the nascent hydroquinone formed by reduction. Thus the structure suggests that the human enzyme uses general acid catalysis in quinone reduction, although this has not been checked directly in half-reactions. In contrast, Lot6p lacks these potential catalytic residues (6). This may explain why the reactivity of Lot6p is lower than that of free flavin – the protein offers no catalytic machinery, but partially limits access to the flavin. These results are in accordance with the lack of specificity shown by a number of QRs with respect to the electron accepting substrate, suggesting that the active site of this enzyme can accommodate molecules of varying size and structure (29).
Biological Implications
Lot6p is a curiously sub-optimal catalyst. The rate of the reductive half-reaction is only moderately higher than the rates of non-enzymatic model reactions between dihydronicotinamides and flavins; the oxidative half-reaction is actually slower than model reactions. This calls into question the necessity of an enzyme for quinone reduction, when free reduced flavin would apparently be as competent kinetically and could be produced by several enzymes with flavin reductase activity. Two hypotheses are suggested, depending on the true physiological importance of Lot6p. If the most important role for Lot6p is the detoxification of quinones, then our results suggest that maximizing catalytic flux is not as important as some other property, such as minimizing the rate of reaction with the wrong oxidants. When bound to the apoprotein of Lot6p, the reaction of reduced FMN with O2 is significantly inhibited. In contrast, free reduced FMN reacts in a chain-reaction to rapidly convert O2 to superoxide and hydrogen peroxide (after a lag). Thus, free reduced FMN, while capable of detoxifying quinones to the hydroquinones non-enzymatically, is also an effective generator of reactive oxygen species; Lot6p, on the other hand, avoids this.
Alternatively, we have recently shown that yeast QR Lot6p not only plays a role in intracellular quinone detoxification (5), but also acts as a redox switch for the 20S proteasome when it shifts between its two redox states (8). Under reducing conditions, i.e. when an excess of NADH and/or NADPH is present in the cytosol, Lot6p is reduced and uses one of its reduced flavin cofactors to associate with the transcription factor Yap4p. Importantly, Lot6p is hardly oxidized by dioxygen (k = 1.1 M−1s−1), thereby ensuring that reoxidation of the active site flavin (and thus release of the transcription factor) only occurs in the presence of quinones (or other as yet unidentified oxidizing agents) (8). This signalling role for Lot6p might not require extremely rapid reaction rates with quinones, shielding the kinetics of the oxidative half-reaction from extreme selective pressure. This scenario presumes that the kinetics of the reactions of free Lot6p, determined in this work, would be unchanged when the enzyme is bound to the proteasome. This remains to be tested; it is possible that reaction rates are different in theLot6p-proteasome complex.
A third scenario is also imaginable – that Lot6p is actually not a non-specific quinone reductase – that it is actually optimized for an as-yet unidentified specific substrate. However, the identity of this hypothetical quinone is not apparent. During apoptosis, mitochondrial membrane permeability increases and release of mitochondrial proteins and compounds into the cytosol leads to the apoptotic phenotype (30). Consequently, ubiquinones originating from mitochondria could be responsible for reoxidation of Lot6p and subsequently relocalization of the transcription factor. However, the insolubility of ubiquinone and the fact that ubiquinone is not the best electron acceptor for Lot6p argue against this scenario.
Acknowledgements
The authors are grateful to Lance W. Rider, Rebecca L. Fagan and Mheran Ebadi-Tehrani for their excellent technical support.
Abbreviations
- QR
quinone reductase
- NADH
β-nicotinamide adenine dinucleotide reduced
- NADPH
β-nicotinamide adenine dinucleotide phosphate, reduced
- FMN
flavin mononucleotide
- FAD
flavin adenine dinucleotide
- KIE
kinetic isotope effect
- NADPD
[4-2H]-NADPH
- TMS
trimethylsilane
Footnotes
This work was supported in part by the Fonds zur Förderung der wissenschaftlichen Forschung (FWF); the Doktoratskolleg “Molecular Enzymology” Grant W901-B05 from the Austrian Science Fund FWF to P. M; and by NIH Grant GM61087 to B.A.P.
References
- 1.Deller S, Macheroux P, Sollner S. Flavin-dependent quinone reductases. Cell Mol. Life Sci. 2008;65:141–160. doi: 10.1007/s00018-007-7300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lind C, Cadenas E, Hochstein P, Ernster L. DT-diaphorase: purification, properties, and function. Methods Enzymol. 1990;186:287–301. doi: 10.1016/0076-6879(90)86122-c. [DOI] [PubMed] [Google Scholar]
- 3.Prochaska HJ, Talalay P. In: Oxidative Stress: Oxidants and Antioxidants. Sies H, editor. New York: Academic Press, Ltd.; 1991. pp. 195–211. [Google Scholar]
- 4.Tedeschi G, Chen S, Massey V. DT-diaphorase. Redox potential, steady-state, and rapid reaction studies. J. Biol. Chem. 1995;270:1198–1204. doi: 10.1074/jbc.270.3.1198. [DOI] [PubMed] [Google Scholar]
- 5.Sollner S, Nebauer R, Ehammer H, Prem A, Deller S, Palfey BA, Daum G, Macheroux P. Lot6p from Saccharomyces cerevisiae is a FMN-dependent reductase with a potential role in quinone detoxification. FEBS J. 2007;274:1328–1339. doi: 10.1111/j.1742-4658.2007.05682.x. [DOI] [PubMed] [Google Scholar]
- 6.Liger D, Graille M, Zhou CZ, Leulliot N, Quevillon-Cheruel S, Blondeau K, Janin J, van TH. Crystal structure and functional characterization of yeast YLR011wp, an enzyme with NAD(P)H-FMN and ferric iron reductase activities. J. Biol. Chem. 2004;279:34890–34897. doi: 10.1074/jbc.M405404200. [DOI] [PubMed] [Google Scholar]
- 7.Li R, Bianchet MA, Talalay P, Amzel LM. The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: mechanism of the two-electron reduction. Proc. Natl. Acad. Sci. U. S. A. 1995;92:8846–8850. doi: 10.1073/pnas.92.19.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sollner S, Schober M, Wagner A, Prem A, Lorkova L, Palfey BA, Groll M, Macheroux P. Quinone reductase acts as a redox switch of the 20S yeast proteasome. EMBO Rep. 2009;10:65–70. doi: 10.1038/embor.2008.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nevitt T, Pereira J, Azevedo D, Guerreiro P, Rodrigues-Pousada C. Expression of YAP4 in Saccharomyces cerevisiae under osmotic stress. Biochem. J. 2004;379:367–374. doi: 10.1042/BJ20031127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nevitt T, Pereira J, Rodrigues-Pousada C. YAP4 gene expression is induced in response to several forms of stress in Saccharomyces cerevisiae. Yeast. 2004;21:1365–1374. doi: 10.1002/yea.1188. [DOI] [PubMed] [Google Scholar]
- 11.Sollner S, Macheroux P. Hiding near the lion's den: emerging role of quinone reductases as regulators of proteasomal degradation. FEBS J. 2009 doi: 10.1111/j.1742-4658.2009.07143.x. [DOI] [PubMed] [Google Scholar]
- 12.Ottolina G, Riva S, Carrea G, Danieli B, Buckmann AF. Enzymatic synthesis of [4R-2H]NAD (P)H and [4S-2H]NAD(P)H and determination of the stereospecificity of 7 alpha- and 12 alpha hydroxysteroid dehydrogenase. Biochim. Biophys. Acta. 1989;998:173–178. doi: 10.1016/0167-4838(89)90270-7. [DOI] [PubMed] [Google Scholar]
- 13.Rider LW, Ottosen MB, Gattis SG, Palfey BA. Mechanism of Dihydrouridine Synthase 2 from Yeast and the Importance of Modifications for Efficient tRNA Reduction. J. Biol. Chem. 2009;284:10324–10333. doi: 10.1074/jbc.M806137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palfey BA. Time resolved spectral analysis. In: Johnson KA, editor. Kinetik analysis of macromolecules. New York: Oxford University Press; 2003. pp. 203–227. [Google Scholar]
- 15.Saez MJ, Lagunas R. Determination of intermediary metabolites in yeast. Critical examination of the effect of sampling conditions and recommendations for obtaining true levels. Mol. Cell Biochem. 1976;13:73–78. doi: 10.1007/BF01837056. [DOI] [PubMed] [Google Scholar]
- 16.Hosoda S, Nakamura W, Hayashi K. Properties and reaction mechanism of DT diaphorase from rat liver. J. Biol. Chem. 1974;249:6416–6423. [PubMed] [Google Scholar]
- 17.Porter DJ, Blankenhorn G, Ingraham LL. The kinetics of lumiflavin reduction by N-methyl-1,4-dihydronicotinamide: direct evidence for a preequilibrium complex between oxidized lumiflavin and N-methyl-1,4-dihydronicotinamide. Biochem. Biophys. Res. Commun. 1973;52:447–452. doi: 10.1016/0006-291x(73)90732-8. [DOI] [PubMed] [Google Scholar]
- 18.Blankenhorn G. Intermolecular complexes between N-methyl-1,4-dihydronicotinamide and flavines. The influence of steric and electronic factors on complex formation and the rate of flavine-dependent dihydronicotinamide dehydrogenation. Biochemistry. 1975;14:3172–3176. doi: 10.1021/bi00685a021. [DOI] [PubMed] [Google Scholar]
- 19.Pennati A, Zanetti G, Aliverti A, Gadda G. Effect of salt and pH on the reductive half-reaction of Mycobacterium tuberculosis FprA with NADPH. Biochemistry. 2008;47:3418–3425. doi: 10.1021/bi702250h. [DOI] [PubMed] [Google Scholar]
- 20.Buffinton GD, Ollinger K, Brunmark A, Cadenas E. DT-diaphorase-catalysed reduction of 1,4-naphthoquinone derivatives and glutathionyl-quinone conjugates. Effect of substituents on autoxidation rates. Biochem. J. 1989;257:561–571. doi: 10.1042/bj2570561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang MT, Miwa GT, Lu AY. Rat liver cytosolic azoreductase. Purification and characterization. J. Biol. Chem. 1979;254:3930–3934. [PubMed] [Google Scholar]
- 22.De FS, Morelli A, Basso C, Romano M, Serra D, De FA. Prominent role of DT-diaphorase as a cellular mechanism reducing chromium(VI) and reverting its mutagenicity. Cancer Res. 1985;45:3188–3196. [PubMed] [Google Scholar]
- 23.Riley RJ, Workman P. Enzymology of the reduction of the potent benzotriazine-di-N-oxide hypoxic cell cytotoxin SR 4233 (WIN 59075) by NAD(P)H:(quinone acceptor) oxidoreductase (EC 1.6.99.2) purified from Walker 256 rat tumour cells. Biochem. Pharmacol. 1992;43:167–174. doi: 10.1016/0006-2952(92)90274-m. [DOI] [PubMed] [Google Scholar]
- 24.Sugimura T, Okabe K, Nagao M. The metabolism of 4-nitroquinoline-1-oxide, a carcinogen. 3. An enzyme catalyzing the conversion of 4-nitroquinoline-1-oxide to 4-hydroxyaminoquinoline-1-oxide in rat liver and hepatomas. Cancer Res. 1966;26:1717–1721. [PubMed] [Google Scholar]
- 25.Mattevi A. To be or not to be an oxidase: challenging the oxygen reactivity of flavoenzymes. Trends Biochem. Sci. 2006;31:276–283. doi: 10.1016/j.tibs.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Gibian MJ, Rynd JA. Oxidation of reduced flavins by quinones. Biochem. Biophys. Res. Commun. 1969;34:594–599. doi: 10.1016/0006-291x(69)90779-7. [DOI] [PubMed] [Google Scholar]
- 27.Bruice TC, Yano Y. Radical mechanisms for 1,5-dihydro-5-methylflavine reduction of carbonyl compounds. J. Am. Chem. Soc. 1975;97:5263–5271. doi: 10.1021/ja00851a041. [DOI] [PubMed] [Google Scholar]
- 28.Faig M, Bianchet MA, Talalay P, Chen S, Winski S, Ross D, Amzel LM. Structures of recombinant human and mouse NAD(P)H:quinone oxidoreductases: species comparison and structural changes with substrate binding and release. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3177–3182. doi: 10.1073/pnas.050585797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Z, Fisher D, Spidel J, Greenfield J, Patson B, Fazal A, Wigal C, Moe OA, Madura JD. Kinetic and docking studies of the interaction of quinones with the quinone reductase active site. Biochemistry. 2003;42:1985–1994. doi: 10.1021/bi026518s. [DOI] [PubMed] [Google Scholar]
- 30.Fleury C, Mignotte B, Vayssiere JL. Mitochondrial reactive oxygen species in cell death signaling. Biochimie. 2002;84:131–141. doi: 10.1016/s0300-9084(02)01369-x. [DOI] [PubMed] [Google Scholar]
- 31.Clark WM. Oxidation-Reduction Potentials of Organic Systems. Baltimore, Md: Williams and Wilkens; 1960. [Google Scholar]
- 32.Crane FL. Biochemical functions of coenzyme Q10. J. Am. Coll. Nutr. 2001;20:591–598. doi: 10.1080/07315724.2001.10719063. [DOI] [PubMed] [Google Scholar]
- 33.Denke E, Merbitz-Zahradnik T, Hatzfeld OM, Snyder CH, Link TA, Trumpower BL. Alteration of the midpoint potential and catalytic activity of the rieske iron-sulfur protein by changes of amino acids forming hydrogen bonds to the iron-sulfur cluster. J. Biol. Chem. 1998;273:9085–9093. doi: 10.1074/jbc.273.15.9085. [DOI] [PubMed] [Google Scholar]
- 34.Brugna M, Albouy D, Nitschke W. Diversity of cytochrome bc complexes: example of the Rieske protein in green sulfur bacteria. J. Bacteriol. 1998;180:3719–3723. doi: 10.1128/jb.180.14.3719-3723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]