Abstract
Severe stress causes plant and animal cells to form large cytoplasmic granules containing RNA and proteins. Here, we demonstrate the existence of stress-induced cytoplasmic RNA granules in Schizosaccharomyces pombe. Homologs to several known protein components of mammalian processing bodies and stress granules are found in fission yeast RNA granules. In contrast to mammalian cells, poly(A)-binding protein (Pabp) colocalizes in stress-induced granules with decapping protein. After glucose deprivation, protein kinase A (PKA) is required for accumulation of Pabp-positive granules and translational down-regulation. This is the first demonstration of a role for PKA in RNA granule formation. In mammals, the translation initiation protein eIF2α is a key regulator of formation of granules containing poly(A)-binding protein. In S. pombe, nonphosphorylatable eIF2α does not block but delays granule formation and subsequent clearance after exposure to hyperosmosis. At least two separate pathways in S. pombe appear to regulate stress-induced granules: pka1 mutants are fully proficient to form granules after hyperosmotic shock; conversely, eIF2α does not affect granule formation in glucose starvation. Further, we demonstrate a Pka1-dependent link between calcium perturbation and RNA granules, which has not been described earlier in any organism.
Keywords: post-transcriptional regulation, processing bodies, stress granules, Schizosaccharomyces pombe
BACKGROUND
In order to maintain viability and conserve energy resources, the cell has to adapt in a coordinated way on several levels when encountering stressful conditions. Protein synthesis, being one of the top consumers of cellular energy, is subject to strict regulation under conditions that limit cellular energy production, such as nutrient starvation or external chemical and physical stress. Thus, global translation is severely down-regulated during cellular stress and nutrient starvation (Shenton et al. 2006; Swaminathan et al. 2006; Asp et al. 2008; Melamed et al. 2008; Warringer et al. 2010). Regulation at the level of transcription initiation is not fast enough for these drastic changes, and so post-transcriptional regulation has to be used as a main regulatory device. Also, during stress and recovery, mRNA turnover is dynamically regulated according to an apparently complex program (Molina-Navarro et al. 2008; Molin et al. 2009; Romero-Santacreu et al. 2009). The cell enters a program where resources are directed from rapid growth to stress protection. Components of ribosomes and gene products required for ribosomal biogenesis are strictly down-regulated on the levels of transcriptional initiation, translation, and mRNA turnover (Tang et al. 2001; Grigull et al. 2004; Halbeisen and Gerber 2009). On the other hand, mRNAs encoding proteins required for survival in stress conditions are stabilized and preferentially translated (Lü et al. 2006; Halbeisen and Gerber 2009; Molin et al. 2009; Powley et al. 2009).
In addition to these general measures to cope with stress, cells of many eukaryotic species have been shown to form cytoplasmic granules containing mRNAs, proteins involved in translation and RNA metabolism, and ribosomal components from the light 40S subunit. Such stress-induced cytoplasmic RNA granules have been observed in hypoxic and arsenite-exposed plant cells (Weber et al. 2008). In mammalian cells, cytoplasmic RNA granules appear after, e.g., oxidative stress, heat shock, and UV irradiation (Kedersha et al. 1999, 2000). A dynamic relationship exists between RNA granules and actively translating polysomes, in that drugs that block polysome disassembly also prevent RNA granule formation (Kedersha et al. 1999, 2000; Cougot et al. 2004; Kedersha et al. 2005). RNA granules have been identified not only in the cytoplasm, but also in chloroplasts, in the single-celled green alga Chlamydomonas reinhardtii (Uniacke and Zerges 2008). A distinction is made between, on one hand, processing bodies (PB) containing proteins involved in 5′ to 3′ mRNA degradation, silencing, and nonsense-mediated decay, and, on the other hand, stress granules (SG) consisting of preinitiation complexes (40S ribosomal subunit and translation initiation factors) and several other RNA-binding proteins. PBs are considered to be focal points for mRNA degradation and exist also in unstressed cells albeit in lower numbers, whereas SGs appear only during severe stress. It should be noted, however, that pathways of transcript degradation independent of PBs also exist, and the extent of PB-dependent mRNA degradation is under debate. Thus, decapping and decay of mRNA has been shown to occur cotranslationally (Hu et al. 2009), and blocking of PB formation does not substantially affect basal mRNA degradation rates (Decker et al. 2007). SGs are larger and of more irregular shape than PBs (for review, see Anderson and Kedersha 2006, 2008). In S. pombe, the composition, appearance, and genetic control of RNA granules have not been defined; one observation has been reported, however, that eIF3i and eIF4A are seen in cytoplasmic foci after heat shock (Dunand-Sauthier et al. 2002). Being large, not bounded by a membrane, and presumably held together by weak interactions, PBs and SGs have proven refractory to biochemical purification efforts. They are mainly studied by light microscopy, using fluorescence tagged proteins known to localize to RNA granules. The absence of pure SG or PB preparations has so far precluded comprehensive analyses of the composition of the mRNA population in the RNA granules. It has been demonstrated for specific cases, however, that mRNAs encoding proteins required for stress survival, such as heat shock proteins, are excluded from SGs (Nover et al. 1989; Kedersha and Anderson 2002). While PBs have long been studied in S. cerevisiae (Sheth and Parker 2003; Teixeira et al. 2005; Sheth and Parker 2006), the existence of SGs in the budding yeast was demonstrated only recently (Buchan et al. 2008; Grousl et al. 2009). In S. cerevisiae, SG formation is dependent on the prior formation of PBs (Buchan et al. 2008).
Here, we demonstrate that upon heat shock, oxidative stress, hyperosmotic shock, and glucose deprivation, fission yeast cells form granules containing homologs of several protein components known from mammalian SGs and PBs, and that they colocalize with RNA. In mammalian cells, signaling through phosphorylation by Gcn2 or other stress-activated kinases of the translation initiation factor eIF2α is crucial for SG formation. The nonphosphorylatable allele eIF2α-S51A completely blocks SG formation (Kedersha et al. 1999). In fission yeast, the homologous nonphosphorylatable eIF2α-S52A allele does not block but confers a delay both in formation and subsequent dissolution of Pabp-positive RNA granules. We show that RNA granule formation induced by glucose deprivation is fully dependent on protein kinase A (PKA) signaling, since a pka1 mutant is unable to form Pabp granules under these conditions. Granules induced by other stress types appear unaffected by Pka1 activity however. In a novel development, we describe Ca2+ perturbations to have a major impact on RNA granule formation, through interactions with signaling from glucose deprivation. In summary, it appears that two separate pathways regulate Pabp- and RNA-positive granules upon stress. The first, responsive to hyperosmosis, arsenic and oxidative stress, involves eIF2α and its kinase Gcn2. The second, responsive to glucose deprivation and perturbations of calcium homeostasis, is dependent on Pka1. In contrast to Pabp, Dcp2 forms granules upon stress independently of either pathway.
MATERIALS AND METHODS
Fission yeast genetic techniques
PCR-based genomic epitope tagging was performed by standard methods (Bähler et al. 1998). pFA6a-GFP(S65T)-kanMX6 (Bähler et al. 1998) and pTY24-DsRed.T1.N1 (Yeast Genetic Resource Center) were used as templates for generating PCR fragments for transformation. In all cases, proteins were C-terminally tagged with GFP or RFP and expressed from the respective endogenous loci. Strains are listed in Table 1.
TABLE 1.
Fission yeast strains
Growth conditions and stress treatment
Unless otherwise stated, cells were cultivated at 30°C in YES rich medium (Moreno et al. 1991). Prior to stress treatment, cells were grown to mid-log phase (OD500 nm = 0.5). If cells were to be examined by microscopy, cells were concentrated about 10-fold by brief centrifugation before exposure to the respective stress condition. For hyperosmotic stress, the KCl concentration was raised to 1 M by adding an equal volume of YES containing 2 M of KCl to the culture. For glucose starvation, cells were washed once in YES without glucose and then resuspended and incubated in YES without glucose. Heat shock was imposed by transferring culture tubes to a water bath at 42°C for 20 min. EGTA was added as 1/10 volume of 375 mM EGTA stock solution. For CaCl2 experiments, appropriate media with working concentrations ranging from 50 to 100 mM CaCl2 were prepared from YES with or without glucose containing 1 M of CaCl2. Staurosporine (Sigma Aldrich) was prepared as a stock solution at 200 μg/mL in DMSO and used at a final concentration of 2 μg/mL. Stock solutions of dithiothreitol (DTT; 1 M in H2O), cycloheximide (10 mg/mL in H2O), 1-10-phenanthroline (100 mg/mL stock solution in ethanol) were stored at −20°C. To rule out any possible effects by dilution, controls were made by diluting cells in either water or 20 mM Tris, pH 8.0.
In situ RNA hybridization
Ten mL of culture were grown to mid-log phase and stressed with either 1 M KCl, 37.5 mM EGTA, or YES without glucose, and cells were prepared as previously described (Hannus et al. 2000) with minor modifications. Cells were harvested and resuspended in PBS containing 3% formaldehyde and 0.04% glutaraldehyde and incubated at 30°C for 30 min. Cells were pelleted and washed twice in 500 μL 0.1 M potassium phosphate buffer pH = 6.4 and once in 500 μL wash buffer (0.1 M potassium phosphate, 1.2 M sorbitol) and resuspended in 250 μL of wash buffer. Permeabilization was carried out by incubating at 30°C for 45 min with 0.8 mg/mL of Zymolyase 100T (Zymo Research). Dehydration was performed by incubations with 1 mL of −80°C methanol for 6 min and 1 mL of acetone followed by two sequential washing steps in 1 mL of 2 × SSC for 5 min. For poly(A) RNA staining, cells were preincubated in 400 μL of hybridization solution at 30°C. After 1 h, Alexa Fluor 594-labeled Oligo(dT) probe (InVitroGen) was added to a final concentration of 1.5 pmol/μL and the incubation continued overnight. The excess oligo d(T) probe was removed by washing the cells three times in 2 × SSC. Cells were finally resuspended in 2 × SSC, spotted on glass slides, and examined by fluorescence microscopy.
Fluorescence microscopy
Cells were spotted on poly-L-lysine coated glass slides. For wide-field microscopy, they were examined immediately at room temperature with a Zeiss Axioplan 2 microscope running Axiovision v4.6.3.0 software. Images were acquired with an Axiocam digital camera using FITC, GFP, and Cy3 filter sets. Confocal imaging was performed at the Center for Cellular Imaging, University of Gothenburg. Single plane images were acquired with an inverted Zeiss Axiovert 200 microscope equipped with a Plan-Apochromat 63x/1.4 oil DIC objective (Zeiss LSM 510 META system). Image adjustments were made in the Zeiss LSM image browser.
Image quantitation
Data from at least three individual experiments with a minimum of 50 cells counted were used for quantification. Since wide-field microscopy was used, the number of granules per cell reported is not necessarily absolute, but should be seen as a relative indicator. Image analysis was carried out by using Image J (http://rsbweb.nih.gov/ij/) and Adobe Photoshop as previously described (Nissan and Parker 2008).
Enrichment of granules
Protein lysates from 20 mL of cultures were prepared in buffer D (100 mM Hepes, pH 7.6; 15 mM KCl; 1.5 mM MgCl2; 0.25% NP-40; 0.2% Triton X-100; 4% glycerol). Lysates were cleared by centrifugation at 2500g and the supernatant was transferred to a fresh 2-mL tube. The supernatant was cleared once more by centrifugation at 3000g and transferred to a new 2-mL tube. A third centrifugation at 5700g resulted in a pellet that was washed twice in buffer D. The supernatant was removed, and the pellet was resuspended in 40 μL of SDS-PAGE loading buffer and solubilized by boiling for 5 min. Gel electrophoresis and Western blotting was carried out as described earlier (Asp and Sunnerhagen 2003).
Polysome separation and analysis
Cells (125 mL of culture) were grown to mid-log phase in rich medium. For glucose starvation experiments, cells were pelleted by centrifugation and washed with 50 mL of sterile H2O. Cells were then resuspended in 50 mL YES minus glucose and incubated for 15 min at 30°C. Polysomes were prepared and separated by sedimentation ultracentrifugation in sucrose gradients as previously described (Asp et al. 2008).
RESULTS
Stress-induced fission yeast granules contain core protein components of mammalian PBs and SGs
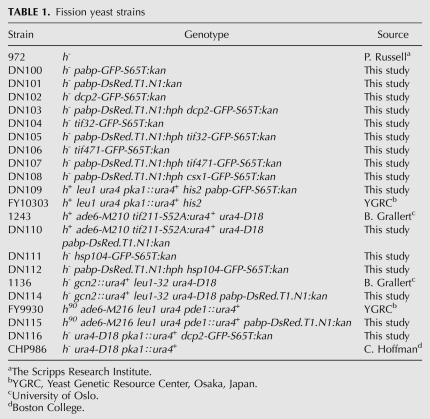
In mammalian cells, poly(A)-binding protein (PABP) is an abundant constituent of SGs (Kedersha et al. 1999) and decapping protein in PBs (Kedersha et al. 2005). We used RFP-tagged Pabp (a.k.a. Pab1, SPAC57A7.04c), the major poly(A)-binding protein of S. pombe, and a GFP-tagged version of the decapping protein Dcp2 as fluorescent markers to investigate possible formation of granules under different stress conditions. Indeed, both under hyperosmotic stress (1 M KCl) and glucose deprivation, cytoplasmic granules positive for Pabp and Dcp2 are visible (Fig. 1). Under hyperosmotic stress (1 M KCl), granules appear within 15 min of incubation, reaching a maximum at 30 min (Fig. 1), which gradually disappear within 1 h after osmotic shock. Glucose deprivation induces fewer, but more distinct, granules within 15 min, which rapidly disperse upon readdition of glucose (Fig. 1). By contrast, no granule formation was seen during nitrogen starvation (not shown). In cells exposed to heat shock (42°C), 1 mM H2O2 or sodium arsenite (5 mM), cytoplasmic granules containing Pabp are formed (Supplemental Fig. S1). Heat shock causes a rapid (10 min) emergence of granules, while during oxidative stress they appear gradually.
FIGURE 1.
Pabp, Dcp2, eIF4G, Csx1, and eIf3a localize to cytoplasmic foci after stress treatment. Wild-type cells expressing GFP or RFP tagged proteins were grown to mid-log-phase in normal medium (unstressed) and then subjected to 1 M KCl. Alternatively, cells were deprived of glucose for 15 min (−glucose) and thereafter given back glucose (+glucose). The localization of fluorescent epitope-tagged proteins was determined by fluorescence microscopy. First row, Pabp-RFP; second row, Dcp2-GFP; third row, eIF4G-GFP; fourth row, Csx1-GFP; fifth row, eIF3a-GFP.
Besides poly(A)-binding and decapping proteins, a number of RNA-binding proteins, translation initiation factors, ribosome 40S subunit components, and RNA silencing factors have been identified in mammalian PBs and SGs (Kedersha et al. 1999; Ingelfinger et al. 2002; Kedersha et al. 2002; Cougot et al. 2004; Stoecklin et al. 2004; Kedersha et al. 2005). To further characterize the composition of RNA granules in S. pombe, we investigated the translation initiation factor eIF4G and found it to form granules under the same conditions as Pabp and Dcp2 (Fig. 1, third row). The RNA-binding protein Csx1 is required for the stability of certain mRNAs during oxidative stress (Rodriguez-Gabriel et al. 2003). We observe Csx1 granules after osmotic stress and glucose starvation, albeit less distinct than seen for the previously mentioned proteins (Fig. 1, fourth row). The translation initiation factor eIF3b has not been found in budding yeast SGs upon glucose starvation (Buchan et al. 2008); eIF3a is found in granules after heat shock (Grousl et al. 2009), however. In fission yeast, we similarly did observe eIF3a (Tif32) in granules after KCl shock, but not in glucose-deprived cells (Fig. 1, bottom row).
Colocalization of protein components in stress-induced granules
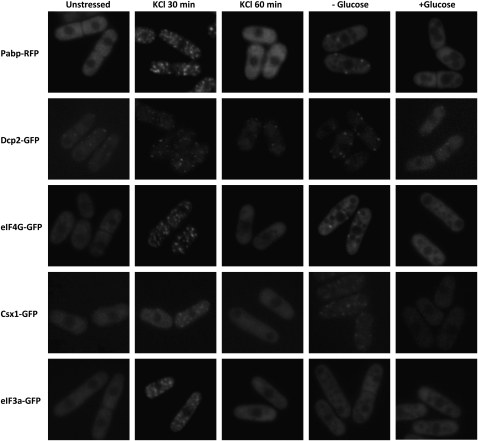
In mammalian cells, decapping protein is a marker for PBs, while poly(A)-binding protein is found in SGs. We wanted to investigate the relationship between Pabp and Dcp2, the major decapping protein in S. pombe, in RNA granules. When challenged with 1 M KCl, cells responded by forming both larger granules positive for Pabp, as well as smaller granules containing Dcp2 (Fig. 2A). With the higher spatial resolution of confocal images, it is clear that, although Dcp2 and Pabp granules generally are found in close vicinity, they do not always fully colocalize (Fig. 2B). Dcp2 persisted in KCl-induced granules up to 60 min, whereas Pabp started to disappear from these structures after 30 min (Fig. 1). Such a division in two categories also occurs early (within 10 min) after glucose deprivation (Fig. 2A, middle column), but not later (20 min) (Fig. 2A, right column). It should be noted that in contrast to other proteins investigated, a low number of Dcp2-positive granules are present also in unstressed cells (Fig. 1, first column, second row). We wanted to ascertain whether the different tagged proteins found in granular structures represented one or different types of granules. As seen in Figure 2A–D, Pabp also colocalizes with Csx1, eIF4G (Tif471), and eIF3a, under both stress conditions investigated (hyperosmosis and glucose deprivation), the only exception being eIF3a, which does not form granules in glucose starvation (Fig. 1).
FIGURE 2.
Pabp colocalizes with Csx1, eIF4G, and eIF3a, but only partially with Dcp2, in cytoplasmic foci after stress. Pabp-RFP was examined for colocalization with Dcp2-GFP, Csx1-GFP, eIF4G-GFP, and eIF3a-GFP, under stress (1 M KCl for 30 min or glucose deprivation for 20 min), using wide-field microscopy except where indicated. (A) Colocalization of Pabp-RFP with Dcp2-GFP. (B) Localization of Pabp-RFP and Dcp2-GFP using confocal microscopy (C) Colocalization of Pabp-RFP with Cxs1-GFP. (D) Colocalization of Pabp-RFP with eIF4G-GFP. (E) Colocalization of Pabp with eIF3a.
In mammalian cells, RNA granule formation by external stress can be blocked through simultaneous inhibition of protein synthesis by cycloheximide (Kedersha et al. 2000). We investigated if cycloheximide could similarly prevent RNA granule formation in S. pombe. Under conditions of glucose deprivation, heat, or hyperosmotic shock, cycloheximide at 100 μg/mL completely prevented the formation of both Pabp and Dcp2-containing RNA granules (not shown). Further, inhibition of RNA polymerase prevents formation of SGs and PBs in mammalian cells (Ohn et al. 2008). We found that in fission yeast, the RNA polymerase II inhibitor 1-10-phenanthroline (100 μg/mL), under the same conditions as tested for cycloheximide, likewise stopped both Pabp and Dcp2-positive RNA granules from forming (not shown). The fact that these inhibitors prevent granule formation demonstrates that this is not a passive, but a biologically regulated process.
Cytoplasmic Pabp granules contain RNA and are distinct from Hsp104 aggregates
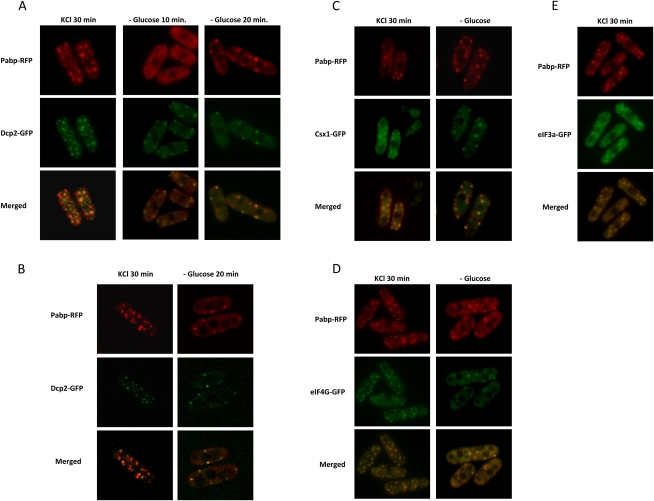
Cytoplasmic protein-containing granules of different types can be formed after environmental stress. For instance, misfolded and/or oxidatively damaged proteins will form aggregates, to which chaperones including Hsp104 will localize (Lum et al. 2004). It was therefore essential to show that the Pabp- and Dcp2-positive granules actually contain RNA. We visualized cellular poly(A)+ RNA by hybridization in vivo of Alexa Fluor 594-labeled Oligo(dT) in cells expressing GFP-tagged Pabp, demonstrating near-complete colocalization in the cytoplasm both after hyperosmotic shock and glucose deprivation (Fig. 3A). By contrast, RFP-tagged Pabp and GFP-tagged Hsp104 display a complete lack of overlap in glucose-starved cells (Fig. 3B) or after hyperosmotic shock (not shown), demonstrating that these represent distinct structures.
FIGURE 3.
Stress-induced Pabp granules colocalize with poly(A)+ RNA (A) but exclude Hsp104 (B). The distribution of poly(A)+ RNA after treatment with 1 M KCl for 30 min, or without glucose for 15 min, was examined in wt cells expressing Pabp-GFP or Hsp104-GFP. Poly(A)+ RNA was visualized by (dT)25-oligomers coupled to Alexa Fluor 568 (Invitrogen) (see Materials and Methods).
Pabp and Csx1 become part of an insoluble protein complex after hyperosmotic stress or glucose deprivation
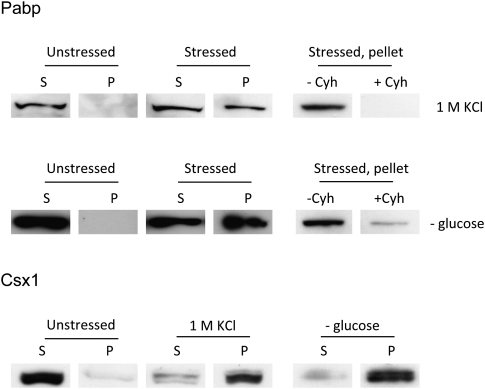
We wanted to demonstrate that our microscopic observations of cytoplasmic foci containing proteins and poly(A)+ RNA after stress correspond to physical aggregates. Mildly denatured protein lysates from unstressed cells or cells at 30 min after addition of 1 M KCl, or at 20 min after glucose deprivation, were centrifuged at low speed, and the supernatant and pellet fractions were analyzed by Western blot (see Materials and Methods). While all tagged Pabp remains in the supernatant in unstressed cells, a substantial fraction of Pabp is detected in the pellet from stressed cells (Fig. 4), confirming that Pabp indeed exists in an insoluble state under such conditions. Similar to what was seen by microscopy, the addition of cycloheximide is able to prevent Pabp from entering the insoluble fraction (Fig. 4, right-hand panels). For Csx1, the shift is even more pronounced, in that almost all Csx1 protein appears in the insoluble pellet after stress, and none in the supernatant.
FIGURE 4.
Pabp and Csx1 are enriched in the insoluble protein fraction after stress. Wild-type cells expressing Pabp-RFP or Csx1-GFP were grown to mid-log phase and exposed to 1 M KCl for 30 min or deprived of glucose for 20 min. Protein lysates from equal numbers of stressed and unstressed cells were prepared and the protein concentration determined by reading absorbance at 280 nm. Equal protein amounts were then subjected to sequential centrifugations as described in Materials and Methods. Pellets (P) formed by centrifugation and samples from supernatant (S) were prepared for Western blot analysis; 10% of each supernatant, without further concentration, and 100% of each pellet fraction, after solubilization, was applied to the gel. Pabp was detected with anti-RFP (Genescript) and Csx1 with anti-GFP (Santa Cruz Biotechnology). Cycloheximide (Cyh) was added to 100 μg/mL before application of stress in some samples to demonstrate prevention of the insoluble fraction (right panels).
Pathway control of Pabp-positive RNA granule formation under hyperosmosis
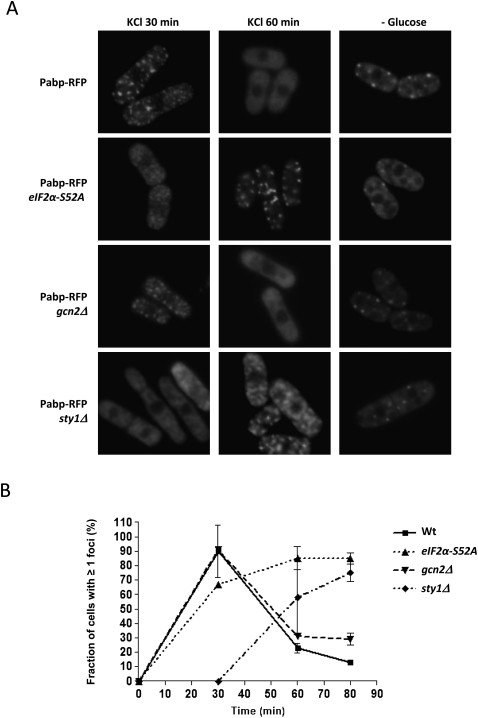
In mammalian cells, phosphorylation of the translation initiation factor eIF2α exerts strong control over SG formation. An allele expressing a phosphomimetic eIF2α version is sufficient to induce SGs on its own, whereas expression of a mutant, nonphosphorylatable, version of eIF2α blocks SG formation upon stress (Kedersha et al. 1999). We investigated RFP-Pabp in a strain expressing the corresponding fission yeast nonphosphorylatable eIF2α-S52A mutant protein. As shown in Figure 5, the kinetics of RNA granule appearance after KCl stress is somewhat slower in eIF2α-S52A (tif211-S52A) cells than in the wild type (wt). The ensuing decline is markedly slower in the mutant, however. Analysis of polysome profiles shows that in wt and eIF2α-S52A mutants alike, there is a complete elimination of polysomes, with little or no recovery during the first 80 min (Supplemental Fig. S2). Upstream kinases phosphorylate eIF2α upon stress; in S. cerevisiae expression of an activated Gcn2 allele leads to increased numbers and intensities of SGs upon stress (Buchan et al. 2008). However, the effect of the gcn2Δ mutation in S. pombe on granule formation was only slight if any (Fig. 5A, third row). In order to investigate the extent of the control exerted by eIF2α, we exposed eIF2α-S52A and gcn2Δ mutants to heat shock; in both cases, the response was indistinguishable from wild type (Supplemental Fig. S3).
FIGURE 5.
Effects of eIF2α, Sty1, and Gcn2 on granule formation and disintegration after hyperosmotic shock. (A) Pabp-RFP localization after stress in wt cells, or strains lacking wt eIF2α, Sty1, or Gcn2. First row, wt; second row, eIF2α-S52A; third row, gcn2Δ; fourth row, sty1Δ. (B) Diagram representing the percentage, from each strain, of cells with ≥1 granules plotted against time after exposure to 1 M KCl.
The MAP kinase Sty1 is activated in fission yeast cells by a wide range of stress conditions, and sty1Δ mutants display quite poor stress survival. In sty1Δ mutants, RNA granules appear with a marked delay during hyperosmosis. Thus, in wt cells, the maximal number of granules is reached after 30 min, the vast majority of which have disappeared after 60 min (Fig. 5, top). In sty1Δ mutants, by contrast, no granules are visible by 30 min, but instead they persist up to 80 min after hyperosmotic shock (Fig. 5B; data not shown). However, Sty1 did not affect RNA granule formation after glucose deprivation, as sty1 mutants formed granules with normal kinetics under such circumstances (Fig. 5, right lane).
Pka1 controls formation of Pabp-positive RNA granule under glucose deprivation and perturbation of calcium homeostasis
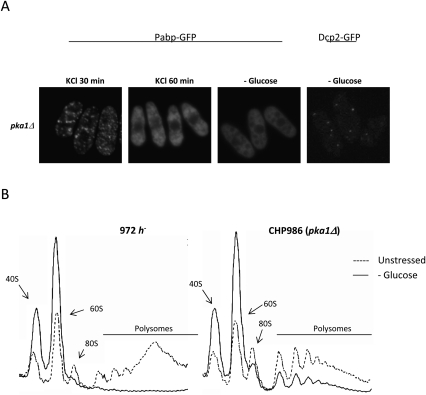
Given the rapid appearance of RNA granules after glucose deprivation, we wanted to investigate if glucose signaling through PKA would be involved in control of granule formation. In pka1Δ mutants, defective in PKA signaling, (Fig. 6A, lower row) Pabp granule formation is completely abolished after glucose starvation, whereas granules do form normally in hyperosmosis (Fig. 6) and heat shock (Supplemental Fig. S3). Dcp2-positive granules are still formed in pka1Δ mutants to the same extent as in wt, however (Fig. 6A). Glucose deprivation caused a rapid (15 min) and complete elimination of polysomes in wt, as expected (Fig. 6B). Notably, in pka1Δ mutants, by the same time a substantial fraction of polysomes remained (Fig. 6B). However, the global protein synthesis rates declined upon glucose deprivation to similar extents in wt and pka1Δ mutants (Supplemental Fig. S6). Thus, Pka1 has a profound effect on regulation of the translation process and RNA granule formation on glucose deprivation. However, neither an effect on Pabp-positive RNA granules was noticed by addition of external cyclic AMP (not shown), nor were any significant deviations from wt behavior seen in a pde1Δ mutant, lacking cyclic AMP phosphodiesterase (not shown).
FIGURE 6.
pka1Δ mutants are unable to form granules containing Pabp and down-regulate translation upon glucose deprivation, but do form granules in hyperosmotic stress. (A) The localization of Pabp-GFP or Dcp2-GFP in a pka1Δ background was examined after different types of stress treatment. (B) Polysomal profiles upon glucose starvation (15 min). Ribosomal material and polysomes were separated by sedimentation ultracentrifugation and detected online (see Materials and Methods). Left chromatogram, wt cells; right chromatogram, pka1Δ. Dashed line, no stress; solid line, −glucose.
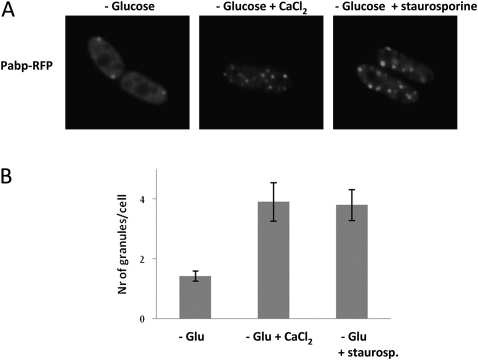
Communication seems to exist at several instances between glucose and Ca2+ signaling in yeast. We wanted to explore if perturbations of calcium homeostasis would impact RNA granule formation through glucose signaling, and examined the effect of increasing the external Ca2+ concentration to 100 mM at the same time as depriving cells of glucose. As seen in Figure 7 (cf. left and center panels), this markedly increased the number of RNA granules. By contrast, external Ca2+ did not affect the number of granules after hyperosmotic shock (not shown), and increased Ca2+ by itself did not induce granule formation (not shown).
FIGURE 7.
External CaCl2 and staurosporine enhance granule formation upon glucose starvation. Wild-type cells expressing Pabp-RFP were resuspended in glucose-free medium, or glucose-free medium supplemented with 100 mM of CaCl2 or 2 μg/mL staurosporine. (A) Left panel, glucose deprivation; middle panel, glucose deprivation + CaCl2; right panel, glucose deprivation + staurosporine. (B) Graph depicting the number of granules per cell formed after each condition. RNA granules were quantified using Image J and Adobe Photoshop.
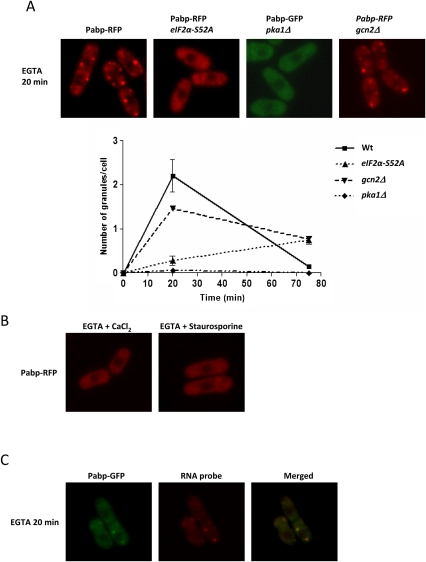
For pka1Δ mutants, addition of Ca2+ to glucose-starved cells did not produce any granules after 10 min (not shown); only at longer incubation (45 min) did a small number of granules appear. The chelator ethylene glycol tetraacetic acid (EGTA) depletes free Ca2+ with slow binding kinetics, and may cause local disturbances in intracellular Ca2+ gradients if added extracellularly (Stern 1992). Strikingly, EGTA triggered extensive production of Pabp and Dcp2 granules in wt cells within 20 min when added by itself to the medium (Fig. 8A; data not shown); these granules were negative for eIF3a, however (Fig. 8A; Supplemental Fig. S4). To ensure that this was not simply due to protein aggregation caused by, e.g., ER stress, we demonstrated colocalization of these EGTA-induced granules with poly(A)+ RNA (Fig. 8C). Further, EGTA-driven granule induction was blocked by cycloheximide addition (not shown). We added dithiothreitol (DTT), a known inducer of ER stress (Shen et al. 2002) in concentrations from 1 to 10 mM, but did not observe any effect on RNA granules (not shown), arguing against ER stress as a main contributor to granule formation. EGTA-induced RNA granules were rapidly dispersed by addition of Ca2+ (Fig. 8B), and prior addition of Ca2+ prevented granule formation by EGTA. This demonstrates that no other divalent cation is required for this process. To examine the relationship of EGTA-induced RNA granule production to glucose signaling, we added EGTA to cells at the same time as depriving them of glucose. This resulted in a strongly increased number of granules, compared to glucose deprivation only, at later times 20 min after medium shift (not shown). We also added EGTA to pka1Δ mutants. Here, no granules were formed (Fig. 8A). These observations strengthen the link between Ca2+ and glucose signaling in the control of RNA granule formation. In eIF2α-S52A mutants treated with EGTA, granules appear only after a long delay (Fig. 8A).
FIGURE 8.
EGTA-treated cells form RNA granules, and this can be blocked by CaCl2 or staurosporine. The cytoplasmic distribution of Pabp-RFP was examined after treatment with 37.5 mM EGTA, with or without the addition of 100 mM CaCl2 or 2 μg/mL staurosporine. (A) EGTA treatment. First row, wt; second row, eIF2α-S52A; third row, pka1Δ. Graph to the right shows the number of granules per cell as a function of time. (B) Wild-type cells treated with EGTA + CaCl2 or EGTA + staurosporine. (C) Colocalization of poly(A)+ RNA and Pabp-GFP after EGTA treatment.
Protein kinase C (PKC) is required for some biological processes involving Ca2+. To explore a possible role for PKC, we added the PKC inhibitor staurosporine to cells exposed to EGTA. As shown in Figure 8B, this completely blocked formation of RNA granules. Neither staurosporine by itself nor the solvent, DMSO, had any effect on granules (not shown). We further examined how staurosporine would affect RNA granule formation after glucose deprivation or hyperosmotic stress. As seen in Figure 7, glucose-deprived cells display an enhanced number of granules upon staurosporine addition. In line with other observations, no such enhancement by staurosporine was observed in glucose-starved pka1Δ mutants, where no granules were formed (not shown). No stimulatory or inhibitory effect by staurosporine was observed in wt cells stressed by 1 M KCl (not shown).
Finally, we wanted to examine if the complete suppression of RNA granule formation in pka1Δ mutants under the above-mentioned stress conditions significantly affects cell survival. As seen in Supplemental Fig. S7, there is only a minimal reduction of colony forming ability of pka1Δ mutants compared to wt, indicating that the effect on short-term survival is slight.
DISCUSSION
Cytoplasmic RNA granules appear to be a ubiquitous cellular stress survival mechanism among eukaryotes. Here, we show that these structures also exist in the fission yeast. Many of the characteristics of RNA granules of mammalian cells are shared by the fission yeast counterparts, including several structural components and circumstances where granules appear. In this work, we also demonstrate a novel pathway for RNA granule regulation. The novel pathway is responsive to glucose deprivation and altered calcium balance, and involves protein kinase A.
Appearance and protein composition of RNA granules
In mammalian cells, a clear distinction has been made between SGs and PBs, based on their composition and appearance. Notably, decapping protein is excluded from SGs, while poly(A)-binding protein is found in SGs but not PBs (Kedersha et al. 1999; Ingelfinger et al. 2002; Kedersha et al. 2005). We show that in fission yeast, by contrast, these proteins coexist in the same RNA-containing granules, at least for most of the duration of the stress response. However, it is also clear that distinct granules exist that contain only Pabp but not Dcp2 and vice versa; and these two types of granules tend to cluster in the vicinity of each other (Fig. 2). A similar situation is found in the budding yeast (Buchan et al. 2008). It is thus not clear to what extent the same criteria that distinguish mammalian PBs and SGs can be used to classify cytoplasmic RNA granules in yeast. In S. cerevisiae, Dcp2 granules persist longer than poly(A)-binding protein after glucose starvation (Buchan et al. 2008). Our data show, by contrast, that in S. pombe, separation of Dcp2 and Pabp is more apparent after KCl than after glucose starvation (Fig. 2A, first and third columns). A plausible interpretation is that, as little or no recovery of translation is seen in 1 M KCl within 60 min, mRNAs sequestered in granules dissociate from Pabp and get degraded. It is also clear that Dcp2-positive granules form independently of the genetic controls that govern Pabp-positive RNA granules, as the pka1 mutation did not interfere with the accumulation of Dcp2 granules. It is thus entirely possible that separate classes of RNA granules, PBs and SGs, exist in fission yeast. For the purposes of this discussion we will not use this terminology, however. We note that all the other RNA-binding proteins that we have tested do colocalize with Pabp; eIF4G, and Csx1 under all stress conditions examined, and eIF3a after KCl shock. Pabp, in turn, colocalizes with poly(A)+ RNA; whereas Hsp104, by contrast, is not found in the same structures.
The appearance of cytoplasmic RNA granules generally correlates with the disappearance of polysomes, indicative of translational arrest. However, in the recovery phase following hyperosmotic shock (60–80 min), RNA granules are cleared in wt cells even though polysomes are not yet restored (Fig. 5; Supplemental Figs. S2, S5). By contrast, in the eIF2α-S52A mutant, RNA granules persist in this phase (but translation is still not reactivated). This shows that in fission yeast, eIF2α is involved in the clearance of RNA granules, and that they are not maintained as an obligatory consequence of translational arrest.
Genetic control of granule formation
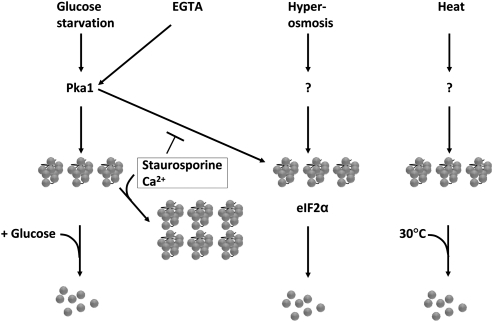
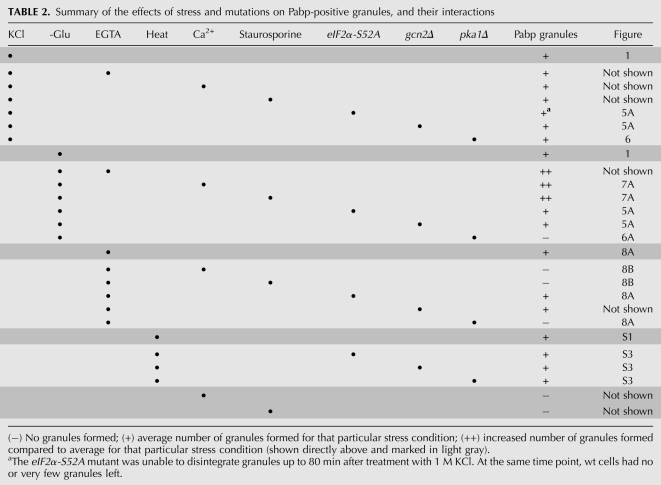
The genetic control of SG formation in mammalian cells has been shown to revolve around the phosphorylation status of eIF2α: a phosphomimetic allele of this protein forces SG formation even in the absence of stress, while expression of a nonphosphorylatable allele blocks SG formation entirely (Kedersha et al. 1999). Several upstream kinases including Gcn2, PKR, PERK, and HRIK, contribute to eIF2α phosphorylation and thus to SG formation. In budding yeast, assembly of stress granules is reported to be independent of eIF2α phosphorylation (Grousl et al. 2009). However, a constitutively active GCN2 allele does induce granules (Buchan et al. 2008). Both heat and arsenite stress-induced SG formation depends on eIF2α phosphorylation in mammalian cells, but in Drosophila cells, SG formation in arsenite is dependent on eIF2α phosphorylation, while heat induces SG independently of this pathway (Farny et al. 2009). Similarly, granules appear after heat shock in fission yeast independently of either eIF2α or Pka1 (Fig. 9; Table 2). It appears that other pathways contribute to RNA granule formation in several organisms. We find that in fission yeast, the dependence on eIF2α is only partial, in that stress-induced RNA granule formation is delayed, but not prevented, in cells expressing the nonphosphorylatable eIF2α mutant version. In particular, the recovery phase, where RNA granules are cleared, is delayed in such mutants (Table 2).
FIGURE 9.
Schematic model of control of RNA granule formation and disaggregation Pka1 is required for formation of RNA granules after glucose starvation or EGTA exposure, while it is unclear what controls formation after hyperosmotic or heat shock. Staurosporine and Ca2+ enhance granule formation after glucose starvation, but block granule formation after EGTA exposure. After hyperosmotic shock, eIF2α is needed to disaggregate granules.
TABLE 2.
Summary of the effects of stress and mutations on Pabp-positive granules, and their interactions
We have shown that PKA activity is required for RNA granule formation upon glucose deprivation, insofar as pka1 mutants, lacking the catalytic PKA subunit, are unable to form RNA granules under such circumstances (Table 2). It is noteworthy that in S. cerevisiae, Tpk2 and Tpk3 (catalytic subunits of PKA), but not Bcy1 (inhibitory regulatory subunit of PKA), are found in SGs and PBs (Tudisca et al. 2010). It is not straightforward to establish a possible genetic requirement of PKA for RNA granule formation in the budding yeast. This is because the S. cerevisiae equivalent of an S. pombe pka1Δ mutant, lacking the sole catalytic PKA subunit, would be a tpk1Δ tpk2Δ tpk3Δ triple mutant lacking all three catalytic PKA subunits; such a triple mutant is not viable however (Toda et al. 1987). Nevertheless, and relevant for this work, it has been shown that S. cerevisiae tpk1-w tpk2Δ tpk3Δ and tpk1Δ tpk2-w tpk3Δ mutants are unable to down-regulate translation when deprived of glucose (Ashe et al. 2000). It is also notable that despite the persistence of polysomes in pka1Δ mutants on glucose deprivation, such cells shut down protein synthesis to the same extent as wt cells. This behavior parallels that of S. cerevisiae cells expressing rck2-kd, which are likewise unable to dissociate polysomes under stress, but which nevertheless do diminish protein synthesis as much as the wt (Swaminathan et al. 2006), indicating that the attached ribosomes are trapped in unproductive complexes.
Relationship between Pka1 signaling and perturbation of calcium homeostasis
There appears to be a separation between signaling through Gcn2/eIF2α on one hand, and PKA on the other, with respect to stress-induced RNA granules in S. pombe (Fig. 9). Thus, while a pka1 mutant forms no Pabp granules upon glucose starvation, it is still fully proficient in granule formation upon hyperosmotic shock (Fig. 6). Conversely, an eIF2α-S52A mutant behaves like the wt when deprived of glucose (Fig. 5), but is deficient in its response to hyperosmosis (Fig. 5). This is further compounded by the observation that phenotypes elicited by glucose starvation are enhanced by the addition of external Ca2+ or EGTA, but abolished or strongly diminished in a pka1 mutant background (Figs. 6; 8A). These phenotypes are not affected by eIF2α or gcn2 mutations, with the sole exception of EGTA addition, which gives a weaker effect in eIF2α-S52A mutants than in the wt (Fig. 8A). Granules induced by both types of stresses contain Pab1 and Dcp2, however, eIF3a is absent from those induced by glucose starvation. Finally, staurosporine, a protein kinase inhibitor with a relative specificity for PKC, blocks the RNA granule accumulation caused by EGTA, but paradoxically enhances formation of RNA granules induced by glucose deprivation (Fig. 7). It is noteworthy that in mammalian cells, staurosporine causes an elevation of intracellular Ca2+ (Maurer et al. 1995). Indeed, the effects of staurosporine on RNA granules in fission yeast mimic those of added Ca2+ (Table 2). Collectively, this indicates that active biological processes are required for the granule induction by EGTA, or glucose starvation. At this point, it does not define which biological pathways are involved however, beyond cAMP/PKA. Numerous tentative interactions between glucose signaling and calcium homeostasis have been reported in the literature from mammalian cells and yeast, but the dominant mechanism remains obscure. Thus, in the budding yeast, glucose readdition to glucose-starved S. cerevisiae cells causes an increase of free Ca2+ in the cytoplasm in a phospholipase C-dependent way, through uptake from the external medium or release from the vacuole (Eilam et al. 1990; Tisi et al. 2002; Kellermayer et al. 2004). The S. pombe-specific protein Git1, with a role in cAMP signaling, possesses a C2 domain, which has also been in PKC, where it is required for association with phospholipids in a calcium-dependent way (Kao et al. 2006). Ssp1 is a putative calmodulin-dependent kinase in S. pombe, and ssp1 mutants are defective in the tolerance to limited glucose (Hanyu et al. 2009). We thus find it likely that the effects on RNA granules that we observe on calcium homeostasis perturbations are functionally linked to and funneled through the glucose-sensing system and PKA. Interestingly, while this work was in preparation, a functional link in S. cerevisiae between calcium homeostasis perturbations and RNA granule formation was also reported (Kilchert et al. 2010). There is a notable difference between the species in that the addition of Ca2+ alone does not induce RNA granule formation in S. pombe, whereas in the budding yeast, exposure to increased external Ca2+ robustly increases RNA granule formation. We have noted that the Ca2+ content of commonly used yeast growth media varies greatly. Thus, the Ca2+ concentration of rich YES medium is on the order of 10 mM, and that of the commonly used YPD rich medium formulation for growth of S. cerevisiae ∼50 mM (Farrell et al. 1993). A chemically defined S. pombe medium, such as EMM (Moreno et al. 1991), contains only 0.1 mM Ca2+, and SC for S. cerevisiae (Sherman 1991) 1 mM Ca2+. Such wide differences might have made uncovering of a consistent behavior in yeast cells more difficult, and might explain the contrasting behavior of fission vs. budding yeast cells.
CONCLUSION
Stress-induced RNA granules have so far been found in all eukaryotes examined, and appear like a universal and versatile mechanism to manage cellular stress. We show here that they are regulated by different signaling pathways depending on the stress condition. Recently, regulation of SGs has been proposed to indirectly affect the tendency of mammalian cells to undergo apoptosis under stressful conditions (Arimoto et al. 2008). Conditions that favor formation of SGs, such as hypoxia, lead to sequestration of Rack1. Since Rack1 is required for activation of the p38 and JNK MAP kinases, which promote apoptosis, SG formation favors cell survival functions. This shifted balance has implications for tumor treatment, and this example demonstrates the far-reaching biological consequences of regulation of cytoplasmic RNA granules.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Beáta Grallert and Erik Boye for useful discussions and the tif211-S52A mutant, and Charles Hoffman for mutants and helpful advice on the PKA pathway. This work was supported by the Swedish Research Council (2007-5460) and the Swedish Cancer Fund (09-0772).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2268111.
REFERENCES
- Anderson P, Kedersha N 2006. RNA granules. J Cell Biol 172: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N 2008. Stress granules: the Tao of RNA triage. Trends Biochem Sci 33: 141–150 [DOI] [PubMed] [Google Scholar]
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M 2008. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol 10: 1324–1332 [DOI] [PubMed] [Google Scholar]
- Ashe MP, De Long SK, Sachs AB 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell 11: 833–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp E, Sunnerhagen P 2003. Mkp1 and Mkp2, two MAPKAP-kinase homologues in Schizosaccharomyces pombe, interact with the MAP kinase Sty1. Mol Genet Genomics 268: 585–597 [DOI] [PubMed] [Google Scholar]
- Asp E, Nilsson D, Sunnerhagen P 2008. Fission yeast mitogen-activated protein kinase Sty1 interacts with translation factors. Eukaryot Cell 7: 328–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol 183: 441–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Séraphin B 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol 165: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179: 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunand-Sauthier I, Walker C, Wilkinson C, Gordon C, Crane R, Norbury C, Humphrey T 2002. Sum1, a component of the fission yeast eIF3 translation initiation complex, is rapidly relocalized during environmental stress and interacts with components of the 26S proteasome. Mol Biol Cell 13: 1626–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilam Y, Othman M, Halachmi D 1990. Transient increase in Ca2+ influx in Saccharomyces cerevisiae in response to glucose: effects of intracellular acidification and cAMP levels. J Gen Microbiol 136: 2537–2543 [DOI] [PubMed] [Google Scholar]
- Farny NG, Kedersha NL, Silver PA 2009. Metazoan stress granule assembly is mediated by P-eIF2α-dependent and -independent mechanisms. RNA 15: 1814–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell RE, Germida JJ, Huang PM 1993. Effects of chemical speciation in growth media on the toxicity of mercury(II). Appl Environ Microbiol 59: 1507–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol Cell Biol 24: 5534–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grousl T, Ivanov P, Frydlova I, Vasicova P, Janda F, Vojtova J, Malinska K, Malcova I, Novakova L, Janoskova D, et al. 2009. Robust heat shock induces eIF2α-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J Cell Sci 122: 2078–2088 [DOI] [PubMed] [Google Scholar]
- Halbeisen RE, Gerber AP 2009. Stress-dependent coordination of transcriptome and translatome in yeast. PLoS Biol 7: e105 doi: 10.1371/journal.pbio.1000105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannus S, Buhler D, Romano M, Séraphin B, Fischer U 2000. The Schizosaccharomyces pombe protein Yab8p and a novel factor, Yip1p, share structural and functional similarity with the spinal muscular atrophy-associated proteins SMN and SIP1. Hum Mol Genet 9: 663–674 [DOI] [PubMed] [Google Scholar]
- Hanyu Y, Imai KK, Kawasaki Y, Nakamura T, Nakaseko Y, Nagao K, Kokubu A, Ebe M, Fujisawa A, Hayashi T, et al. 2009. Schizosaccharomyces pombe cell division cycle under limited glucose requires Ssp1 kinase, the putative CaMKK, and Sds23, a PP2A-related phosphatase inhibitor. Genes Cells 14: 539–554 [DOI] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J 2009. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8: 1489–1501 [PMC free article] [PubMed] [Google Scholar]
- Kao RS, Morreale E, Wang L, Ivey FD, Hoffman CS 2006. Schizosaccharomyces pombe Git1 is a C2-domain protein required for glucose activation of adenylate cyclase. Genetics 173: 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans 30: 963–969 [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J Cell Biol 147: 1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell 13: 195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fitzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermayer R, Szigeti R, Kellermayer M, Miseta A 2004. The intracellular dissipation of cytosolic calcium following glucose re-addition to carbohydrate depleted Saccharomyces cerevisiae. FEBS Lett 571: 55–60 [DOI] [PubMed] [Google Scholar]
- Kilchert C, Weidner J, Prescianotto-Baschong C, Spang A 2010. Defects in the secretory pathway and high Ca2+ induce multiple P-bodies. Mol Biol Cell 21: 2624–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü X, de la Peña L, Barker C, Camphausen K, Tofilon PJ 2006. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res 66: 1052–1061 [DOI] [PubMed] [Google Scholar]
- Lum R, Tkach JM, Vierling E, Glover JR 2004. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem 279: 29139–29146 [DOI] [PubMed] [Google Scholar]
- Maurer JA, Wenger BW, Guan Z, Stokes BT, McKay DB 1995. Staurosporine affects calcium homeostasis in cultured bovine adrenal chromaffin cells. Eur J Pharmacol 288: 163–172 [DOI] [PubMed] [Google Scholar]
- Melamed D, Pnueli L, Arava Y 2008. Yeast translational response to high salinity: Global analysis reveals regulation at multiple levels. RNA 14: 1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin C, Jauhiainen A, Warringer J, Nerman O, Sunnerhagen P 2009. mRNA stability changes precede changes in steady-state mRNA amounts during hyperosmotic stress. RNA 15: 600–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Navarro MM, Castells-Roca L, Bellí G, García-Martínez J, Marín-Navarro J, Moreno J, Pérez-Ortín JE, Herrero E 2008. Comprehensive transcriptional analysis of the oxidative response in yeast. J Biol Chem 283: 17908–17918 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P 1991. Molecular genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Nissan T, Parker R 2008. Analyzing P-bodies in Saccharomyces cerevisiae. Methods Enzymol 448: 507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D 1989. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol 9: 1298–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P 2008. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol 10: 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powley IR, Kondrashov A, Young LA, Dobbyn HC, Hill K, Cannell IG, Stoneley M, Kong YW, Cotes JA, Smith GC, et al. 2009. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev 23: 1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Gabriel MA, Burns G, McDonald WH, Martin V, Yates JR 3rd, Bähler J, Russell P 2003. RNA-binding protein Csx1 mediates global control of gene expression in response to oxidative stress. EMBO J 22: 6256–6266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Santacreu L, Moreno J, Pérez-Ortín JE, Alepuz P 2009. Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA 15: 1110–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell 3: 99–111 [DOI] [PubMed] [Google Scholar]
- Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, Ashe MP, Grant CM 2006. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem 281: 29011–29021 [DOI] [PubMed] [Google Scholar]
- Sherman F 1991. Getting started with yeast. Methods Enzymol 194: 3–21 [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125: 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern MD 1992. Buffering of calcium in the vicinity of a channel pore. Cell Calcium 13: 183–192 [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P 2004. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J 23: 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Masek T, Molin C, Pospisek M, Sunnerhagen P 2006. Rck2 is required for reprogramming of ribosomes during oxidative stress. Mol Biol Cell 17: 1472–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Hornstein E, Stolovich M, Levy G, Livingstone M, Templeton D, Avruch J, Meyuhas O 2001. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol Cell Biol 21: 8671–8683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisi R, Baldassa S, Belotti F, Martegani E 2002. Phospholipase C is required for glucose-induced calcium influx in budding yeast. FEBS Lett 520: 133–138 [DOI] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Zoller M, Wigler M 1987. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50: 277–287 [DOI] [PubMed] [Google Scholar]
- Tudisca V, Recouvreux V, Moreno S, Boy-Marcotte E, Jacquet M, Portela P 2010. Differential localization to cytoplasm, nucleus or P-bodies of yeast PKA subunits under different growth conditions. Eur J Cell Biol 89: 339–348 [DOI] [PubMed] [Google Scholar]
- Uniacke J, Zerges W 2008. Stress induces the assembly of RNA granules in the chloroplast of Chlamydomonas reinhardtii. J Cell Biol 182: 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warringer J, Hult M, Regot S, Posas F, Sunnerhagen P 2010. The HOG pathway dictates the short-term translational response after hyperosmotic shock. Mol Biol Cell 21: 3080–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C, Nover L, Fauth M 2008. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J 56: 517–530 [DOI] [PubMed] [Google Scholar]