Abstract
The assembly of the spliceosome involves dynamic rearrangements of interactions between snRNAs, protein components, and the pre-mRNA substrate. DExD/H-box ATPases are required to mediate structural changes of the spliceosome, utilizing the energy of ATP hydrolysis. Two DExD/H-box ATPases are required for the catalytic steps of the splicing pathway, Prp2 for the first step and Prp16 for the second step, both belonging to the DEAH subgroup of the protein family. The detailed mechanism of their action was not well understood until recently, when Prp2 was shown to be required for the release of U2 components SF3a and SF3b, presumably to allow the binding of Cwc25 to promote the first transesterification reaction. We show here that Cwc25 and Yju2 are released after the reaction in Prp16- and ATP-dependent manners, possibly to allow for the binding of Prp22, Prp18, and Slu7 to promote the second catalytic reaction. The binding of Cwc25 to the spliceosome is destabilized by mutations at the branchpoint sequence, suggesting that Cwc25 may bind to the branch site. We also show that Prp16 has an ATP-independent role in the first catalytic step, in addition to its known role in the second step. In the absence of ATP, Prp16 stabilizes the binding of Cwc25 to the spliceosome formed with branchpoint mutated pre-mRNAs to facilitate their splicing. Our results uncovered novel functions of Prp16 in both catalytic steps, and provide mechanistic insights into splicing catalysis.
Keywords: Prp16, spliceosome remodeling, catalytic steps
INTRODUCTION
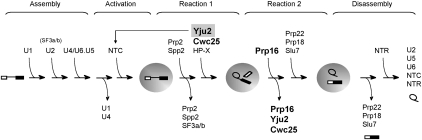
The spliceosome is assembled by sequential binding of five small nuclear RNAs (snRNAs)—U1, U2, U4/U6, and U5—in the form of ribonucleoprotein complexes and other protein components, to the pre-mRNA (Fig. 1) (for reviews, see Brow 2002; Wahl et al. 2009). After binding of all five snRNAs, a major structural rearrangement occurs on the spliceosome, which involves release of U1 and U4, and forming new base pairs between U2 and U6, and between U6 and the 5′ splice site. Moreover, a protein complex associated with Prp19, named NTC (NineTeen Complex), is added to the spliceosome to stabilize the interactions of U6 and U5 with the pre-mRNA in formation of the activated spliceosome (Tarn et al. 1994; Chan et al. 2003; Chan and Cheng 2005).
FIGURE 1.
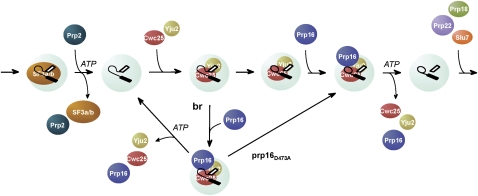
Scheme for the spliceosome pathway, divided into spliceosome assembly, spliceosome activation, catalysis (reactions 1 and 2), and spliceosome disassembly steps. Yju2 is shown joining the spliceosome with NTC or in the first catalytic step. Prp16, Yju2, and Cwc25 are highlighted for they are the focus of this work.
Each catalytic step of the splicing reaction requires a DEAH-box ATPase, Prp2 for the first step and Prp16 for the second step, and several other proteins. Each step proceeds through an ATP-dependent structural change in the spliceosome followed by an ATP-independent reaction that forms lariat intermediates and the mature message, respectively (Liu et al. 2007). Yju2, Cwc25, and a yet unidentified heat-resistant factor HP-X are required for the ATP-independent reaction in lariat formation (Chiu et al. 2009; Warkocki et al. 2009), and Prp22, Prp18, and Slu7 are required for exon ligation (Horowitz and Abelson 1993a, 1993b; Ansari and Schwer 1995; Jones et al. 1995; Schwer and Gross 1998). These proteins may facilitate or stabilize splice site alignment during catalytic reactions (James et al. 2002; Grotti et al. 2007; Chiu et al. 2009). Among them, Yju2, Cwc25, Prp22, and Slu7 are essential for cellular viability and for in vitro splicing. Yju2 and HP-X can be recruited to the spliceosome before the action of Prp2, despite the fact that their function is required in the first catalytic reaction in a post-Prp2 step (Liu et al. 2007; Warkocki et al. 2009). By contrast, Cwc25 only binds to the spliceosome after the action of Prp2 and in the presence of Yju2 (Chiu et al. 2009). After completion of splicing, the spliceosome first releases the mature mRNA, a step mediated by Prp22 (Company et al. 1991; Wagner et al. 1998; Schwer 2008). It is then disassembled by the concerted action of two DExD/H-box proteins, U5 component Brr2 (Small et al. 2006) and Prp43, in association with Ntr1 and Ntr2 in the form of a complex known as NTR complex (Arenas and Abelson 1997; Martin et al. 2002; Tsai et al. 2005; Boon et al. 2006; Pandit et al. 2006; Tsai et al. 2007). A scheme for the spliceosome pathway is shown in Figure 1.
The mechanisms underlying the structural change of the spliceosome mediated by Prp2 and Prp16 are not well understood. Recently, the function of Prp2 is shown to be associated with the release of U2 components SF3a and SF3b (Warkocki et al. 2009; Lardelli et al. 2010). Prp2 requires a cofactor Spp2, which was originally identified as a high-copy suppressor of prp2-1 mutation, for its function. Spp2 interacts with Prp2, and is required for the binding of Prp2 to the spliceosome (Last et al. 1987; Roy et al. 1995; Silverman et al. 2004). Prp16 was reported to promote a conformational change in the spliceosome, resulting in the protection of the 3′ splice site against oligonucleotide-directed RNase H cleavage (Schwer and Guthrie 1992). Although Prp16 has been shown to unwind RNA duplexes in vitro (Wang et al. 1998), it remains unclear whether such helicase activity is directly related to its function in the spliceosome pathway.
We show here that Prp16 functions in mediating the release of spliceosomal components Yju2 and Cwc25 in the second step. Yju2 and Cwc25 are required for the first catalytic reaction (Chiu et al. 2009), and are dissociated from the spliceosome in Prp16- and ATP-dependent manners, presumably to allow binding of Prp22, Prp18, and Slu7 to promote the second catalytic reaction. This result provides a possible explanation for how the first catalytic reaction could reverse on the Prp22-associated spliceosome independent of Prp16 and ATP (Tseng and Cheng 2008). Strikingly, Prp16 can bind the spliceosome prior to the first catalytic reaction to stabilize the binding of Cwc25 in the absence of ATP hydrolysis. This uncovers an ATP-independent role of Prp16 in the first catalytic step, and provides a biochemical explanation for how Prp16 might mediate proofreading of the branchpoint sequence.
RESULTS
Prp16 mediates the release of Yju2 and Cwc25 prior to the second catalytic reaction
It has recently been shown that U2 components SF3a/b are released from the spliceosome in the ATP-dependent Prp2 step (Warkocki et al. 2009; Lardelli et al. 2010). SF3b has been reported to bind to the branch site in the catalytic center of the spliceosome, and its component SAP155 (Hsh155 in yeast) directly contacts sequences across the branchpoint (Query and Sharp 1996; Gozani et al. 1998; Will et al. 2001). Conceivably, the removal of SF3a/b is necessary for binding of Cwc25, perhaps at the same site, to promote the transesterification reaction by fine-tuning the structure of the spliceosome for splice site alignment (Chiu et al. 2009; Warkocki et al. 2009). The second catalytic step also requires a DEAH-box protein Prp16 prior to the transesterification reaction, which requires Prp22, Prp18, and Slu7. Since Yju2 and Cwc25 have previously been found destabilized from the spliceosome prior to the second catalytic reaction (Liu et al. 2007; Chiu et al. 2009), we speculated that Prp16 might play an analogous role to Prp2 in exchanging components binding to the catalytic center. We therefore examined whether Yju2 and Cwc25 would accumulate on the spliceosome in the absence of Prp16 function.
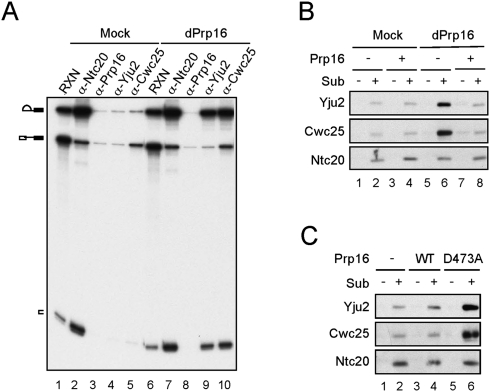
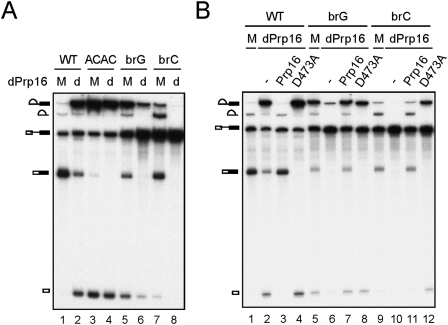
Spliceosome was formed in Prp16-depleted or mock-treated extracts and precipitated with antibodies against Ntc20, Prp16, Yju2, or Cwc25 (Fig. 2A). While only residual amounts of splicing intermediates were precipitated by anti-Yju2 and anti-Cwc25 antibodies in mock-treated extracts (lanes 4,5), large amounts were precipitated in Prp16-depleted extracts (lanes 9,10), indicating that Yju2 and Cwc25 accumulated on the spliceosome in the absence of Prp16. This was further confirmed by western blotting of affinity-purified spliceosome. Spliceosomes formed with biotinylated, 3′ splice site mutated ACAC pre-mRNA were precipitated with streptavidin Sepharose, and components of the spliceosome analyzed by western blotting (Fig. 2B). Yju2 and Cwc25 were found to accumulate on the spliceosome in large amounts in Prp16-depleted extracts (lane 6) as opposed to mock-treated extracts (lane 2), or mock-treated or Prp16-depleted extracts with the addition of recombinant Prp16 (lanes 4,8). A dominant-negative mutant of Prp16, D473A, which carries a mutation in the ATPase motif and blocks the second catalytic reaction (Hotz and Schwer 1998), also resulted in the accumulation of Yju2 and Cwc25 when added in the splicing reaction (Fig. 2C, lane 6). These results suggest that Prp16 might mediate the release of Yju2 and Cwc25, and the ATPase activity of Prp16 is required.
FIGURE 2.
Yju2 and Cwc25 were accumulated on the spliceosome in the absence of Prp16 function. (A) Splicing was carried out using ACAC pre-mRNA in mock-treated (lanes 1–5) or Prp16-depleted extract (lanes 6–10), and the reaction mixtures were precipitated with antibodies against Ntc20, Prp16, Yju2, or Cwc25. RXN: 1/10 of the reaction mixture. (B,C) Western blotting of the spliceosome formed with biotinylated ACAC pre-mRNA and pulled-down with streptavidin Sepharose. (B) Splicing was carried out in mock-treated (lanes 1–4) or Prp16-depleted (lanes 5–8) extracts, +/− recombinant Prp16. (C) Splicing without (lanes 1,2) or with the addition of wild-type Prp16 (lanes 3,4) or prp16D473A mutant protein (lanes 5,6). dPrp16: Prp16-depletion; Sub: substrate.
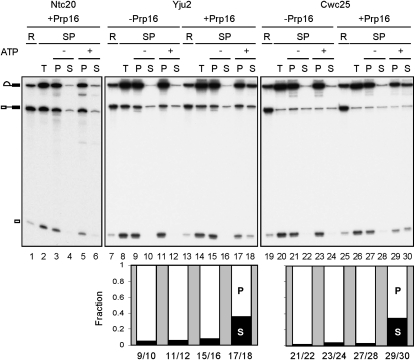
To confirm such a functional role of Prp16, splicing reactions were carried out in Prp16-depleted extracts followed by depletion of ATP. Recombinant Prp16 was added and allowed to bind to the spliceosome. The spliceosome was then precipitated with antibodies against Yju2, Cwc25, or Ntc20, and incubated in the presence or absence of ATP (Fig. 3). Indeed, while Ntc20 remained stably associated with the spliceosome (lanes 5,6), nearly 40% of Cwc25 and Yju2 were released from the lariat-containing spliceosome in the presence of ATP (lanes 17,18,29,30), indicating that the release of Yju2 and Cwc25 required the ATP-dependent function of Prp16. This result demonstrates that Yju2 and Cwc25 are removed from the spliceosome, in a step mediated by Prp16 in an ATP-dependent manner, after their action in the first catalytic step.
FIGURE 3.
Prp16 mediates the release of Yju2 and Cwc25. Splicing was carried out for 10 min with wild-type actin transcript in Prp16-depleted extracts, and ATP was depleted by incubation for 5 min upon the addition of 10 mM glucose. After addition or no addition of recombinant Prp16, the reaction mixtures were precipitated with antibody against Ntc20 (lanes 2–6), Yju2 (lanes 8–12,14–18) or Cwc25 (lanes 20–24,26–30). The precipitated spliceosome was reincubated in the presence or absence of ATP, and the pellet and supernatant fractions were separated. The amount of RNA was quantified by PhosphoImager and plotted in a bar graph with the sum of supernatant and pellet as 100%. The x-axis indicates lane numbers of the gel. R: 1/10 of the reaction mixture; SP: spliceosome; T: total precipitate; P: pellet; S: supernatant.
Branchpoint mutations destabilize the association of Cwc25 with the spliceosome
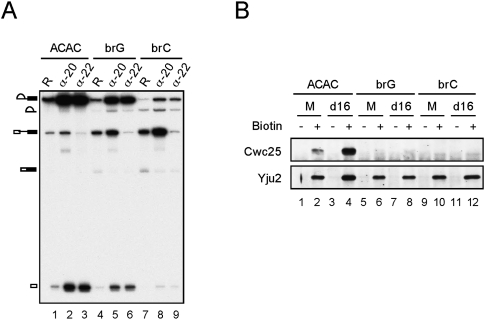
The occurrence of the first reaction immediately following Cwc25 binding suggests that Cwc25 may bind to the catalytic center of the spliceosome to promote the transesterification reaction. Consistent with this notion, the binding of Cwc25 to the spliceosome requires prior action of Prp2, presumably to remove SF3b from binding to the branch site. To see whether Cwc25 binds to the branch site, we examined whether the association of Cwc25 was affected by mutations at the branchpoint. Pre-mRNA containing an A to G (brG) or an A to C (brC) mutation at the branchpoint spliced less efficiently as compared to ACAC pre-mRNA in proceeding through the first reaction (Fig. 4A, lanes 1,4,7). Whereas the brG pre-mRNA was more strongly blocked for the second than the first reaction step (lane 4), the brC pre-mRNA was severely blocked for the first step (lane 7), consistent with previous results of in vivo analysis (Query and Konarska 2004). To see whether the block of the first step by brC mutation was due to a block in spliceosome assembly or inefficient catalytic reaction, we precipitated the post-activation spliceosome with antibody against Ntc20, which is deposited on the spliceosome as a component of the NTC during spliceosome activation (Chen et al. 2001a; Chan et al. 2003). The result shows that the amount of the activated spliceosome formed with brG and brC was 53% and 62%, respectively, of that with ACAC pre-mRNA, suggesting that the efficiency of spliceosome assembly decreased to half of its value with branchpoint mutations. The brC mutant was blocked primarily in the first catalytic reaction, as revealed by the accumulation of pre-mRNA on the activated spliceosome (lane 8). By contrast, less of the brG spliceosome stalled in the pre-catalytic state, and a larger amount of splicing intermediates than pre-mRNA was coprecipitated with Ntc20 (lane 5). The spliceosome containing splicing intermediates could be stalled either before the Prp16 step with Yju2 and Cwc25 accumulated (Fig. 2), or after the Prp16 step with step two factors accumulated as in the case of that formed with ACAC pre-mRNA (Chan et al. 2003). When the spliceosome was precipitated with anti-Prp22 antibody, splicing intermediates were precipitated at a comparable efficiency as with anti-Ntc20 in all three pre-mRNAs (lanes 2,3,5,6,8,9), suggesting that like the ACAC spliceosome, the catalytic brG and brC spliceosomes were mostly stalled in post-Prp16 steps after binding of Prp22.
FIGURE 4.
Mutations at the branchpoint destabilized the association of Cwc25 with the spliceosome. (A) Splicing was carried out with ACAC (lanes 1–3), brG (lanes 4–6), or brC (lanes 7–9) pre-mRNA and the reaction mixtures were precipitated with anti-Ntc20 or anti-Prp22 antibody. R: 1/10 of the reaction mixture; α-20: anti-Ntc20 antibody; α-22: anti-Prp22 antibody. (B) Spliceosome formed with biotinylated ACAC, brG, or brC pre-mRNA was precipitated with streptavidin Sepharose followed by Western blotting. M: mock; d16: Prp16 depletion.
To examine the association of Cwc25 and Yju2 with the spliceosome, the spliceosome formed with biotinylated pre-mRNA was isolated by precipitation with streptavidin Sepharose and the associated components were examined by western blotting. Figure 4B shows that while Cwc25 was seen to associate with the ACAC spliceosome, it was not detected to associate with the brG or brC spliceosome (lanes 6,10), indicating that mutations in the branchpoint prevented stable association of Cwc25 with the spliceosome. The association of Yju2 with the spliceosome was not affected by branchpoint mutation. Since Cwc25 has a very short life on the spliceosome, and is seen to accumulate on the spliceosome in large amounts only in the absence of a functional Prp16 (Fig. 2), we then depleted Prp16 from the extract to see whether Cwc25 could be detected on the brG or brC spliceosome. However, while Yju2 and Cwc25 accumulated in greater amounts on the ACAC spliceosome when Prp16 was depleted (lane 4), Cwc25 was still barely detected with brG and brC mutations, and the amount of Yju2 did not increase (lanes 8,12). The same results were obtained with A to U mutation in the branchpoint (brU) (data not shown). These results demonstrate that changes in the branchpoint sequence prevent stable association of Cwc25 with the spliceosome in the presence or absence of Prp16.
A functional link of Prp16 to the first catalytic reaction
Although Cwc25 was destabilized from the spliceosome by branchpoint mutations, splicing was not entirely blocked, suggesting that Cwc25 could still interact with the spliceosome, perhaps at a lower efficiency, to promote the reaction. Since depletion of Prp16 did not increase the amount of Yju2 accumulated on the brG or brC spliceosome (Fig. 4C), we speculated that depletion of Prp16 might affect splicing of brG or brC pre-mRNA. Splicing was assayed in mock-treated or Prp16-depleted extracts and compared between wild-type, ACAC, brG and brC pre-mRNA (Fig. 5A). In Prp16-depleted extracts, the splicing efficiency of brG pre-mRNA was greatly reduced (lanes 5,6), and splicing of brC pre-mRNA was completely inhibited (lanes 7,8), while splicing of wild-type or ACAC pre-mRNA was largely unaffected except for a block in the second step (lanes 1,4). Kinetic analysis also revealed no effect of Prp16 on the rate of the first reaction with wild-type or ACAC pre-mRNA (data not shown). This result suggests that Prp16 might stabilize or facilitate the binding of Cwc25 to the spliceosome formed on brG or brC pre-mRNA. To confirm that, the D473A mutant of Prp16 recombinant protein was added to the reaction to see whether it would facilitate splicing of brG or brC pre-mRNA (Fig. 5B). While the addition of recombinant wild-type Prp16 fully rescued the splicing activity of the extract (lanes 3,7,11), the D473A mutant of Prp16 also restored the splicing efficiency of brG and brC pre-mRNAs with the second step blocked (lanes 4,8,12). This indicates that, although Prp16 functions in mediating the release of Yju2 and Cwc25 after the first catalytic reaction, it may bind to the spliceosome prior to the reaction and stabilize the association of Cwc25 with brG and brC pre-mRNAs to promote the splicing reaction. Prp16 is known to be involved in the second catalytic step, and only until recently, while this manuscript was being prepared, shown to bind the pre-catalytic spliceosome formed with sulfur-substituted U80 residue of U6 (Koodathingal et al. 2010). We speculated that retention of Prp16 on the pre-catalytic spliceosome might be detectable with branchpoint mutants.
FIGURE 5.
Prp16 facilitated splicing of brG and brC pre-mRNA. (A) Splicing was carried out with wild-type (lanes 1,2), ACAC (lanes 3,4), brG (lanes 5,6), or brC (lanes 7,8) pre-mRNA in mock-treated (lanes 1,3,5,7) or Prp16-depleted (lanes 2,4,6,8) extracts. dPrp16: Prp16 depletion; M: mock; d: depletion. (B) Splicing was carried out with wild-type (lanes 1–4), brG (lanes 5–8), or brC (lanes 7,8) pre-mRNA in mock-treated (lanes 1,5,9) or Prp16-depleted (lanes 2–4,6–8,10–12) extracts without (lanes 2,6,10) or with the addition of wild-type (lanes 3,7,11) or D473A mutant (lanes 4,8,12) of Prp16.
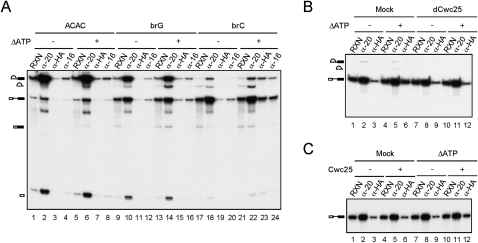
We then examined whether Prp16 and Cwc25 could be detected to associate with the pre-catalytic spliceosome by precipitation of splicing reaction mixtures with anti-Prp16 and anti-HA antibodies using Cwc25-HA extracts (Fig. 6A). To arrest Prp16 on the spliceosome, ATP was depleted by the addition of 10 mM glucose prior to precipitation. In the experiments without depleting ATP, a low level of pre-mRNA and lariat intron-exon 2 was found to coprecipitate with Prp16 (lanes 4,12,20), possibly due to nonspecific RNA binding of Prp16, or background from precipitation with anti-Prp16 antibody. Nevertheless, a significantly larger amount of brC pre-mRNA and splicing intermediates was coprecipitated with Prp16 and Cwc25 after ATP depletion (lanes 23,24) in comparison with the amount obtained without ATP depletion (lanes 19,20). Cwc25 was not significantly associated with ACAC pre-mRNA (lanes 3,7), but was associated with only a tiny amount of brG pre-mRNA after ATP depletion (lanes 11,15). This is consistent with the previous observation that brC was more severely impeded for the first catalytic step, while brG was more severely impeded for the second step at the post-Prp16 stage (Fig. 4A). The result demonstrates that Prp16 and Cwc25 can associate with the spliceosome before the first step with brC pre-mRNA. Together with the results shown in Figure 5, it suggests that Prp16 stabilizes the binding of Cwc25 to promote the first splicing reaction of brC pre-mRNA.
FIGURE 6.
Cwc25-dependent association of Prp16 with the pre-catalytic spliceosome. (A) Splicing was carried out for 30 min with ACAC, brG, or brC pre-mRNA in Cwc25-HA extracts, and the reaction mixtures were separated into two aliquots. Glucose was added to one aliquot at 10 mM and incubated for 5 min (lanes 5–8,13–16,21–24). All mixtures were precipitated with anti-Ntc20, anti-HA, or anti-Prp16 antibody. (B) Splicing was carried out with brC pre-mRNA in mock-treated (lanes 1–6) or Cwc25-depleted (lanes 7–12) Prp16-HA extracts, and the reaction mixtures were separated into two aliquots. Glucose was added to one aliquot at 10 mM and incubated for 5 min (lanes 4–6,10–12). All mixtures were precipitated with anti-Ntc20 or anti-HA antibody. (C) Splicing was carried out with brC pre-mRNA in Cwc25-depleted, Prp16-HA extracts, and the reaction mixtures were separated into two aliquots. Glucose was added (lanes 7–12) or not added (lanes 1–6) to each aliquot at 10 mM and incubated for 5 min, and each mixture was separated into two aliquots. Recombinant Cwc25 was added (lanes 4–6,10–12) or not added (lanes 1–3,7–9) to each aliquot, and further incubated for 10 min. All mixtures were precipitated with anti-Ntc20 or anti-HA antibody. ΔATP: ATP depletion; RXN: 1/50 of reaction mixture; α-20: 1/5 of precipitates from anti-Ntc20 antibody; dCwc25: Cwc25 depletion.
Since in normal splicing reactions, Prp16 is required only after the first catalytic reaction, which requires Cwc25, it is likely that the binding of Prp16 to the pre-catalytic spliceosome also depends on the presence of Cwc25. To find out whether Cwc25 was required for the association of Prp16 with the pre-catalytic brC spliceosome, the same experiment was performed with Cwc25-depleted Prp16-HA extracts (Fig. 6B). In this experiment, anti-HA antibody was used for precipitation of Prp16 to avoid possible background from precipitation with anti-Prp16 antibody. Although anti-HA antibody still precipitated a low level of pre-mRNA in the reaction without ATP depletion (lanes 3,9), a significantly higher level of pre-mRNA was coprecipitated in mock-treated extracts with ATP depletion (lane 6). Depletion of Cwc25 reduced the amount pre-mRNA coprecipitated with Prp16 to the level of no ATP depletion (lane 12), and adding back recombinant Cwc25 restored precipitation of pre-mRNA (Fig. 6C, lane 12), suggesting that binding of Prp16 to the pre-catalytic spliceosome requires the presence of Cwc25.
DISCUSSION
Remodeling of the spliceosome mediated by Prp2 and Prp16 during catalytic steps
The spliceosome undergoes repetitive remodeling during the successive catalytic steps to acquire components specifically required for each step. Prp2 was recently demonstrated to be functionally associated with the release of SF3a/b prior to the first catalytic reaction (Warkocki et al. 2009; Lardelli et al. 2010). Proteomic analysis of spliceosomes arrested at prp2-, prp16-, and prp22-arrested steps detected Yju2 and Cwc25 only in the prp16-arrested spliceosomes, suggesting removal of Yju2 and Cwc25 after the second step (Lardelli et al. 2010). We demonstrate here that Yju2 and Cwc25 are released from the spliceosome prior to the second catalytic reaction, and their release is associated with the function of Prp16. It is likely that SF3a/b leave the spliceosome to allow for the binding of Cwc25 to promote lariat formation. Cwc25 and Yju2 are then removed after their action to allow for the binding of Prp22, Prp18, and Slu7 to promote the second transesterification. Conceivably, SF3a/b, Cwc25/Yju2, and Prp22/Slu7/Prp18 may each bind to the catalytic center of the spliceosome at distinct stages in order to position the splice sites. Thus, the prior removal of factors occupying the position is necessary to afford their access. SF3b has been shown to bind to the branch site, presumably to stabilize U2-branch site base-pairing. We show here that mutations at the branchpoint destabilize the association of Cwc25 with the spliceosome, suggesting that Cwc25 may bind to the branch site during the first catalytic step.
The ATPase activity of Prp2 and Prp16 is associated with the release of SF3a/b, and the release of Yju2 and Cwc25, respectively. Coincidentally, other splicing DExD/H-box proteins are also functionally linked to the release of specific spliceosomal components. Prp28 has been implicated in the release of U1, and Brr2 in the release of U4 during spliceosome activation (Raghunathan and Guthrie 1998; Staley and Guthrie 1999; Chen et al. 2001b). Prp22 has a function in the second catalytic reaction; however, its ATPase activity is required only for the release of mRNA (Schwer and Gross 1998). Prp5 also has ATP-dependent and ATP-independent functions. Although the ATPase activity of Prp5 is not required for its role in formation of the pre-spliceosome, it is required for the dissociation of Cus2 from U2 snRNP to modulate the structure of U2 (Perriman et al. 2003). The function of Prp43 in disassembly of the spliceosome also involves separation of spliceosomal components from lariat-intron and from each other. Taken together, splicing DExD/H-box proteins appear to have a common role in remodeling the spliceosome that mediate the release of spliceosomal components no longer required for progression of the pathway at distinct stages.
How DExD/H-box proteins mediate the release of spliceosomal components is not completely clear. Multiple changes in the RNA-RNA and RNA-protein interactions take place at each step of the splicing pathway. It is conceivable that ATP hydrolysis by the DExD/H-box proteins may trigger conformational change of the spliceosome, leading to destabilization of specific components at specific stages. Alternatively, the DExD/H-box proteins may act as RNPases to remove proteins directly bound to RNA. Two DExD/H-box proteins have previously been implicated to function as RNPases in the splicing reaction (Schwer 2001). Sub2 has been proposed to remove Mud2 (yU2AF65) and BBP bound to the 3′ splice site and the branch site to allow U2 binding (Kistler and Guthrie 2001; Libri et al. 2001), and Prp28 to remove U1 from the 5′ splice site, possibly by destabilizing U1C (Chen et al. 2001b). Whether Prp2 and Prp16 function as RNPases in removing SF3a/b and Yju2/Cwc25 remains to be investigated.
Prp16 functions in the first step in proofreading the branchpoint sequence
The role of Prp16 in the catalytic steps is particularly interesting. Prp16 has long been known to be required for the second catalytic reaction, and depletion of Prp16 results in accumulation of splicing intermediates. Our results show that Prp16 has an additional role for the first catalytic step, which may serve to proofread the branchpoint sequence. With regard to the regular splicing reaction, Prp16 is not required for the binding of Yju2 or Cwc25 to the spliceosome, or for their function, and is only required for their release after the first catalytic reaction. With branchpoint mutants that weaken the binding of Cwc25, Prp16 could stabilize the association of Cwc25 to promote the reaction. Cwc25 is recruited as the last factor to the pre-catalytic spliceosome. Normally, the association of Cwc25 with pre-mRNA is not detected, presumably due to rapid conversion of the pre-mRNA to splicing intermediates upon Cwc25 binding. However, Cwc25 could be detected to associate with the pre-catalytic spliceosome when the brC mutant was employed, owing to a slower kinetics of the reaction, but only in the presence of Prp16 and in the absence of ATP. Prp16 was also detected to associate with the pre-catalytic spliceosome under such conditions, and its association requires the presence of Cwc25. The fact that Cwc25 and Prp16 could stall on the pre-catalytic spliceosome only in the absence of ATP suggests that hydrolysis of ATP by Prp16 results in the release of Cwc25. In this context, Prp16 could bind to the spliceosome after binding of Cwc25, either before or after the catalytic reaction, and mediate the release of Cwc25 upon ATP hydrolysis. It is conceivable that after binding of Cwc25, the catalytic reaction occurs faster than Prp16 binding or ATP hydrolysis-associated release of Cwc25, so that Cwc25 is dissociated only after the reaction. With the brC mutant, the catalytic reaction is impeded to allow binding of Prp16 before the reaction takes place; consequently, retention of Cwc25 and Prp16 on the spliceosome can be detected when ATP is depleted. Without depletion of ATP, Cwc25 may be rapidly dissociated upon ATP hydrolysis in a similar manner to post-catalytic dissociation of Cwc25 mediated by Prp16. A scheme illustrating the interplay between Prp16 and Cwc25, and how that might mediate proofreading of the branchpoint sequence, is shown in Figure 7.
FIGURE 7.
Schematic of the interplay between Prp16 and Cwc25 in the first catalytic step. After Prp2-mediated dissociation of SF3a/b, Yju2 and Cwc25 bind to the spliceosome to promote the first catalytic reaction. Prp16 binds to the spliceosome after the binding of Yju2 and Cwc25, and mediate their dissociation after the transesterification reaction if the reaction is fast, or before transesterification if the reaction is slow, as in the case of brC mutant. The D473A mutant of Prp16 can stabilize the association of Cwc25 with the spliceosome to promote the transesterification reaction.
PRP16 was initially identified as a suppressor of brC mutation (Burgess et al. 1990). Mutations in the helicase domain of Prp16 with reduced ATPase activity allow splicing of brC pre-mRNA. This has led to the hypothesis for a role of Prp16 in the fidelity control of branchpoint sequence by a kinetic proofreading mechanism (Burgess and Guthrie 1993a,b). Recently, Prp16 was shown to bind the pre-catalytic spliceosome and proofread the 5′ splice site cleavage of the spliceosome formed with sulfur-substituted U80 residue of U6 by distinguishing between different rates of the reaction (Koodathingal et al. 2010). In agreement with their results, our finding of the interplay between Prp16 and Cwc25 provides a biochemical explanation for how Prp16 may mediate proofreading of the branchpoint sequence. We propose that kinetic competition between the transesterification reaction and the Prp16- and ATP-dependent release of Cwc25 determines whether the first catalytic reaction can proceed. With wild-type pre-mRNA, the transesterification reaction occurs faster than the removal of Cwc25. With the brC mutant, Prp16 stabilizes the binding of Cwc25, but also hydrolyzes ATP to rapidly release Yju2 and Cwc25 before the reaction could occur. Consequently, the reaction is blocked. Reducing the rate of ATP hydrolysis allows prolonged retention of Cwc25 and the progression of the splicing reaction.
In contrast to brC mutant, Cwc25 or Prp16 was not as clearly seen to stably associate with the brG spliceosome at the pre-catalytic stage. Although binding of Cwc25 to the spliceosome was also weakened by the brG mutation, splicing of brG pre-mRNA was not completely abolished when Prp16 was depleted. Moreover, despite a similar efficiency in forming activated spliceosomes, the first catalytic reaction was less affected by brG mutation when compared to brC. We speculated that the reason Cwc25 was not detected to significantly associate with brG pre-mRNA was due to a lower amount of the pre-catalytic spliceosome present as the substrate for Cwc25 to bind after depletion of ATP, noting that performing splicing reactions for a shorter time only slightly increased the amount of the pre-catalytic spliceosome accumulated, and consequently the amount of pre-mRNA associated with Cwc25 (data not shown). Taken together, these results suggest that the first transesterification of brG pre-mRNA is faster than that of brC, so that Cwc25 is not detectably retained on the spliceosome before catalysis, and that the Prp16-mediated proofreading mechanism at the branchpoint sequence may not operate efficiently on the spliceosome carrying brG mutation. Another DExD/H-box ATPase, Prp22, has been shown to proofread exon ligation (Mayas et al. 2006). Splicing of pre-mRNA with mutations at the 3′ splice site is repressed for formation of mRNA by the ATP-dependent activity of Prp22, and such activity also represses aberrant exon ligation of the brG mutant (Mayas et al. 2006). All these results demonstrate that the branchpoint sequence is proofread at both catalytic steps by different DExD/H-box proteins, and that splicing can be repressed at either step, depending on the nature of the mutation.
The role of Prp16 in the second step
Prp16 has been hypothesized to mediate structural rearrangement of the spliceosome by destabilizing RNA-RNA base-pairings and/or RNA-protein interactions. Mutations that destabilize U2/U6 helix I or U2 stem IIc as well as mutations in PRP8 or ISY1 were found to suppress prp16 mutants (Madhani and Guthrie 1994; McPheeters 1996; Query and Konarska 2004; Villa and Guthrie 2005; Hilliker et al. 2007), suggesting that Prp16 may be required for disrupting structures that involve these components. As Cwc25 may bind to the catalytic center of the spliceosome, it is likely that mutations in any of these other components may also destabilize Cwc25, and consequently does not require full Prp16 activity. Supporting this notion, Cwc25 was found destabilized from the spliceosome formed in isy1Δ extracts, with or without depletion of Prp16 (data not shown).
A conserved mechanism for the two catalytic steps
The spliceosome utilizes a conserved mechanism in catalyzing two steps of transesterification. Each step requires a DExD/H-box ATPase for the exchange of components binding to the catalytic center to acquire specific proteins necessary for proper alignment of splice sites. Like Prp22, Prp16 also has both ATP-dependent and ATP-independent functions. Prp22 was originally identified as a factor mediating ATP-dependent release of mature mRNA (Company et al. 1991), and was later shown to have an ATP-independent function in the second catalytic step, in which it stabilizes the association of Slu7 with the spliceosome to promote exon ligation (James et al. 2002). Prp22 is needed for the second step only when the distance between the branchpoint and the 3′ splice site is greater than 21 nucleotides for actin pre-mRNA (Schwer and Gross 1998). Likewise, Prp16 stabilizes the binding of Cwc25 to the spliceosome on suboptimal substrates to promote the first transesterification reaction. This function does not require its ATPase activity. Prp16 could mediate the dissociation of Yju2 and Cwc25 prior to or after the first reaction, depending on the rate of transesterification, and its action prior to lariat formation plays a role in proofreading the branchpoint sequence. In analogy, Prp22 functions in proofreading the 3′ splice site and the branchpoint sequence prior to exon ligation (Mayas et al. 2006).
Implication for reverse splicing on the purified spliceosome
We have recently demonstrated that splicing reactions in the context of a fully assembled spliceosome are readily reversible without requiring ATP or DExD/H-box helicases (Tseng and Cheng 2008). Relatively subtle changes in the environment can drive the reaction in the forward or reverse direction. These observations support previous hypothesis of global conformational similarities of the spliceosome at various states (Smith et al. 2008). The finding that Yju2/Cwc25 are displaced by Prp22/Prp18/Slu7 during transition from the first to the second step suggests that Yju2/Cwc25 and Prp22/Prp18/Slu7 are likely positioned at the catalytic center of the spliceosome at each step to facilitate splice site alignment and to establish a conformation suitable for the transesterification reaction. We showed previously that the spliceosome, which is affinity-purified via its association with Prp22, could catalyze reverse splicing to convert splicing intermediates to pre-mRNA (Tseng and Cheng 2008). This indicates that switching between step one and step two conformation does not necessarily require exchange of components binding to the catalytic center given proper reaction conditions. Under normal splicing conditions, Prp16 is required to facilitate the conformational switch by displacing Yju2/Cwc25 from the catalytic center with Prp22/Prp18/Slu7, utilizing the energy from ATP hydrolysis. It will be interesting to see whether, under certain circumstances, the spliceosome can be switched from step one to step two conformation without having to displace Yju2/Cwc25 with Prp22/Prp18/Slu7.
MATERIALS AND METHODS
Yeast strains
The following yeast strains were used:
BJ2168 (MATa prc1 prb1 pep4 leu2 trp1 ura3)
YSCC163 (MATa prc1 prb1 pep4 leu2 trp1 ura3 PRP16-3HA)
YSCC2 (MATa prc1 prb1 pep4 leu2 trp1 ura3 YJU2-HA)
YSCC25 (MATa prc1 prb1 pep4 leu2 trp1 ura3 CWC25-HA)
Antibodies and reagents
Anti-V5 antibody was purchased from Serotech Inc. Anti-HA antibody 8G5F was produced by immunizing mice with the KLH-conjugated peptide of the HA-epitope (TY Tsao and SC Cheng, unpubl.). Anti-Ntc20, anti-Yju2, anti-Cwc25, anti-Prp8, anti-Prp16, and anti-Prp22 antibodies were produced by immunizing rabbits with the E. coli-expressed recombinant proteins (full-length protein for Ntc20, Yju2, and Cwc25, and amino acid residues 1–115 for Prp8, 1–298 for Prp16, and 1–484 for Prp22). Streptavidin Sepharose was purchased from Sigma Inc. and protein A-Sepharose from GE Healthcare Inc.
Splicing extracts, substrates, and splicing reactions
Splicing extracts were prepared according to Cheng et al. (1990). Wild-type, ACAC, brG and brC mutant (Vijayraghavan et al. 1986) pre-mRNAs were used in these experiments. Substrates were prepared by in vitro transcription with SP6 RNA polymerase of plasmid pSPact64-88 linearized with EcoRI. Splicing reactions were carried out according to Cheng et al. (1990) for 30 min at 25°C unless otherwise indicated. For reactions with Prp16- or Cwc25-depleted extracts, 50% of treated extracts were included.
Depletion of NTC, Yju2, Cwc25, and Prp16
Depletion of NTC, Yju2, and Prp16 was performed by incubation of 100 μl of splicing extracts with 100 μl of anti-Ntc20, 120 μl of anti-Yju2, 100 μl of anti-Cwc25, or 50 μl of anti-Prp16 anti-serum conjugated to 50 μl of protein A-Sepharose, on ice for 0.5–1 h, followed by removal of protein A-Sepharose beads.
Purification of Prp16 and prp16D473A
The PRP16 gene tagged with three copies of HA at the C-terminus was subcloned into pET28b for expression of the recombinant protein. An aspartate to alanine mutation at position 473 was introduced by site-directed mutagenesis. Both wild-type and prp16D473A mutant proteins were purified by consecutive chromatography on Ni-NTA column and anti-HA antibody conjugated protein A-Sepharose column.
Purification of the spliceosome
Precipitation of the spliceosome with antibodies and with streptavidin Sepharose, and assays for the release of the spliceosome were performed according to Chan et al. (2003). For immunoprecipitation, 20 μl each of splicing reaction mixture was precipitated by incubation with 1 μl of anti-V5, 5 μl of anti-Prp16, 10 μl of anti-Yju2, or 15 μl of anti-Cwc25 antiserum coupled to 10 μl of protein A-Sepharose. For precipitation with streptavidin Sepharose, 30 μl each of splicing reaction mixture was incubated with 10 μl of streptavidin Sepharose.
ACKNOWLEDGMENTS
We thank D.S. Horowitz and P. Lin for comments on the manuscript, H. Kuhn for English editing, and members of the Cheng lab for helpful discussions. This work was supported by Grant NSC 98-2745-B-001-ASP from the Academia Sinica and National Science Council of Taiwan.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2459611.
REFERENCES
- Ansari A, Schwer B 1995. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J 14: 4001–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas JE, Abelson JN 1997. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci 94: 11798–11802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K, Auchynnikava T, Edwalds-Gilbert G, Barrass JD, Droop AP, Dez C, Beggs JD 2006. Yeast Ntr1/Spp382 mediates Prp43 function in postspliceosomes. Mol Cell Biol 26: 6016–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA 2002. Allosteric cascade of spliceosome activation. Annu Rev Genet 36: 333–360 [DOI] [PubMed] [Google Scholar]
- Burgess SM, Guthrie C 1993a. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem Sci 18: 381–384 [DOI] [PubMed] [Google Scholar]
- Burgess SM, Guthrie C 1993b. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73: 1377–1392 [DOI] [PubMed] [Google Scholar]
- Burgess S, Couto JR, Guthrie C 1990. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell 60: 705–717 [DOI] [PubMed] [Google Scholar]
- Chan SP, Cheng SC 2005. The Prp19-associated complex is required for specifying interactions of U5 and U6 with Pre-mRNA during spliceosome activation. J Biol Chem 280: 31190–31199 [DOI] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC 2003. The Prp19p-associated complex in spliceosome activation. Science 302: 279–282 [DOI] [PubMed] [Google Scholar]
- Chen CH, Tsai WY, Chen HR, Wang CH, Cheng SC 2001a. Identification and characterization of two novel components of the Prp19p-associated complex, Ntc30p and Ntc20p. J Biol Chem 276: 488–494 [DOI] [PubMed] [Google Scholar]
- Chen JY, Stands L, Staley JP, Jackups RR Jr, Latus LJ, Chang TH 2001b. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol Cell 7: 227–232 [DOI] [PubMed] [Google Scholar]
- Cheng SC, Newman AN, Lin RJ, McFarland GD, Abelson JN 1990. Preparation and fractionation of yeast splicing extract. Methods Enzymol 181: 89–96 [DOI] [PubMed] [Google Scholar]
- Chiu YF, Liu YC, Chiang TW, Yeh TC, Tseng CK, Wu NY, Cheng SC 2009. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol Cell Biol 29: 5671–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M, Arenas J, Abelson J 1991. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349: 487–493 [DOI] [PubMed] [Google Scholar]
- Gozani O, Potashkin J, Reed R 1998. A potential role for U2AF SAP155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol 18: 4752–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotti LB, Bačíková D, Horowitz DS 2007. The Prp18 protein stabilizes interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes Dev 21: 1204–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker AK, Mefford MA, Staley JP 2007. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev 21: 821–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz DS, Abelson J 1993a. Stages in the second reaction of pre-mRNA splicing: the final step is ATP independent. Genes Dev 7: 320–329 [DOI] [PubMed] [Google Scholar]
- Horowitz DS, Abelson J 1993b. A U5 small nuclear ribonucleoprotein particle protein involved only in the second step of pre-mRNA splicing in Saccharomyces cerevisiae. Mol Cell Biol 13: 2959–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz HR, Schwer B 1998. Mutational analysis of the yeast DEAH-box splicing factor Prp16. Genetics 149: 807–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S, Turner W, Schwer B 2002. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA 8: 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, Frank DN, Guthrie C 1995. Characterization and functional ordering of Slu7p and Prp17p during the second step of pre-mRNA splicing in yeast. Proc Natl Acad Sci 92: 9687–9691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler AL, Guthrie C 2001. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for Sub2, an essential spliceosomal ATPase. Genes Dev 15: 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koodathingal P, Novak T, Piccirilli JA, Staley JP 2010. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol Cell 39: 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardelli RM, Thompson JX, Yates Jr III, Stevens SW 2010. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last RL, Maddock JR, Woolford JLJ 1987. Evidence for related functions of the RNA genes of Saccharomyces cerevisiae. Genetics 117: 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Graziani N, Saguez C, Boulay J 2001. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev 15: 36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Chen HC, Wu NY, Cheng SC 2007. A novel splicing factor Yju2 is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol Cell Biol 27: 5403–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C 1994. Genetic interactions between the yeast RNA helicase homolog Prp16 and spliceosomal snRNAs identify candidate ligands for the Prp16 RNA-dependent ATPase. Genetics 136: 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Schneider S, Schwer B 2002. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J Biol Chem 277: 17743–17750 [DOI] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Staley JP 2006. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol 13: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters DS 1996. Interactions of the yeast U6 RNA with the pre-mRNA branch site. RNA 2: 1110–1123 [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Lynn B, Rymond BC 2006. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc Natl Acad Sci 103: 13700–13705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R, Barta I, Voeltz GK, Abelson J, Ares JM 2003. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc Natl Acad Sci USA 100: 13857–13862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query CC, Konarska MM 2004. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell 14: 343–353 [DOI] [PubMed] [Google Scholar]
- Query CC, Sharp PA 1996. Three recognition events at the branch-site adenine. EMBO J 15: 1392–1402 [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C 1998. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol 8: 847–855 [DOI] [PubMed] [Google Scholar]
- Roy J, Kim K, Maddock JR, Anthony JG, Woolford JL Jr 1995. The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA 1: 375–390 [PMC free article] [PubMed] [Google Scholar]
- Schwer B 2001. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat Struct Biol 8: 113–116 [DOI] [PubMed] [Google Scholar]
- Schwer B 2008. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell 30: 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Gross CH 1998. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J 17: 2086–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Guthrie C 1992. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J 11: 5033–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman EJ, Maeda A, Wei J, Smith P, Beggs JD, Lin RJ 2004. Interaction between a G-patch protein and a spliceosome DEXD/H-box ATPase that is critical for splicing. Mol Cell Biol 24: 10101–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small EC, Leggett SR, Winans AA, Staley JP 2006. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H Box ATPase. Mol Cell 23: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM 2008. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol Cell 30: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell 3: 55–64 [DOI] [PubMed] [Google Scholar]
- Tarn WY, Hsu CH, Huang KT, Chen HR, Kao HY, Lee KR, Cheng SC 1994. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J 13: 2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC 2005. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 19: 2991–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RT, Tseng CK, Lee PJ, Chen HC, Fu RH, Chang KJ, Yeh FL, Cheng SC 2007. Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol Cell Biol 27: 8027–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CK, Cheng SC 2008. Both catalytic steps of nuclear pre-mRNA splicing are reversible. Science 320: 1782–1784 [DOI] [PubMed] [Google Scholar]
- Vijayraghavan U, Parker R, Tamm J, Iimura Y, Rossi J, Abelson J, Gurthrie C 1986. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J 5: 1683–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T, Guthrie C 2005. The Isy1p component of the NineTeen Complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev 19: 1894–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JD, Jankowsky E, Company M, Pyle AM, Abelson JN 1998. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J 17: 2926–2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann RL 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wagner JD, Guthrie C 1998. The DEAH-box splicing factor Prp16 unwinds RNA duplexes in vitro. Curr Biol 8: 441–451 [DOI] [PubMed] [Google Scholar]
- Warkocki Z, Odenwälder P, Schmitzová J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R 2009. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol 16: 1237–1243 [DOI] [PubMed] [Google Scholar]
- Will CL, Schneider C, MacMillan AM, Katopodis NF, Neubauer G, Wilm M, Lührmann R, Query CC 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J 20: 4536–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]