Abstract
The eyes absent 1 protein (Eya1) plays an essential role in the development of various organs in both invertebrates and vertebrates. Mutations in the human EYA1 gene are linked to BOR (branchio-oto-renal) syndrome, characterized by kidney defects, hearing loss, and branchial arch anomalies. For a better understanding of Eya1's function, we have set out to identify new Eya1-interacting proteins. Here we report the identification of the related proteins Sipl1 (Shank-interacting protein-like 1) and Rbck1 (RBCC protein interacting with PKC1) as novel interaction partners of Eya1. We confirmed the interactions by glutathione S-transferase (GST) pulldown analysis and coimmunoprecipitation. A first mechanistic insight is provided by the demonstration that Sipl1 and Rbck1 enhance the function of Eya proteins to act as coactivators for the Six transcription factors. Using reverse transcriptase PCR (RT-PCR) and in situ hybridization, we show that Sipl1 and Rbck1 are coexpressed with Eya1 in several organs during embryogenesis of both the mouse and zebrafish. By morpholino-mediated knockdown, we demonstrate that the Sipl1 and Rbck1 orthologs are involved in different aspects of zebrafish development. In particular, knockdown of one Sipl1 ortholog as well as one Rbck1 ortholog led to a BOR syndrome-like phenotype, with characteristic defects in ear and branchial arch formation.
The human EYA1 gene is an ortholog of the fruit fly eyes absent gene (eya), which was identified as a regulator of compound eye development. In contrast to the single eya gene found in Drosophila melanogaster, mammals possess four Eya paralogs, designated Eya1-4. The respective gene products are characterized by a C-terminal domain called the Eya domain, which is conserved in both length (271 to 274 amino acids [aa]) and sequence. The Eya domain has an intrinsic phosphatase activity (24, 39, 53) and is required for protein-protein interactions (38). Recent studies provided evidence that the phosphatase function of Eya is involved in the innate immune system and the regulation of DNA damage response (8, 35). Interestingly, all Eya interaction partners that have been identified so far, for example, the Six and Dach proteins or inhibitory G protein α (Gα) subunits, bind to the Eya domain (5, 10, 34). It has been demonstrated that the cooperative action of Eya and Six is essential for the development of several tissues and organs in a variety of species throughout evolution (reviewed in reference 6). In vertebrates, Six induces nuclear translocation of Eya and recruitment to the DNA, and Eya enhances Six-mediated activation of target gene expression (34).
The N termini of Eya proteins, which are highly divergent between Eya family members, harbor a proline-serine-threonine (PST)-rich transactivation domain, which is indispensable for their function as coactivators of transcription (31, 45, 50, 58). Natural target genes of the vertebrate Eya-Six complex are, for example, Six2, Sall1, and Myogenin. Activation of Six2 and Sall1 expression has been shown to be essential for proper kidney development in the mouse, whereas activation of Myogenin is required for muscle development (3, 4, 46).
Mutations in the human EYA1 gene are associated with several congenital disorders, like BOR (branchio-oto-renal) and BO (branchio-oto) syndrome, as well as ocular defects. BOR syndrome patients suffer from severe malformations of the ears, the branchial arch derivatives, and the kidneys, while in BO syndrome patients, the kidneys are not affected (11, 12, 30). The physiological importance of Eya1 was underlined by knockout studies in mice. It was shown that the phenotype of Eya1-heterozygous mice resembles the characteristic symptoms of human patients suffering from BOR syndrome with renal abnormalities, including hypoplasia and unilateral agenesis, and conductive hearing loss. In contrast, Eya1 homozygote mice die at birth, showing severe craniofacial and skeletal defects and the complete absence of the thymus, parathyroid glands, ears, and kidneys due to defective inductive tissue interactions (57, 59). Interestingly, similar to EYA1, the paralogous EYA4 gene has also been demonstrated to be associated with human disease. EYA4 is most closely related to EYA1 rather than EYA2 and EYA3. Mutations in EYA4 have been linked to cardiomyopathy and hearing loss (37, 42, 55). In contrast, there is no evidence for the existence of disease-associated mutations in EYA2 and EYA3 up to now.
The identification and characterization of an Eya1 ortholog in zebrafish revealed a remarkable conservation in both the structure and expression pattern between Eya1 genes in fish and humans. In zebrafish, eya1 expression was detected in several organs during embryogenesis, such as the ears, the lateral line organ, the branchial arches, and the somites (41). As in mammals, eya1 in zebrafish is essential for the development of several tissues and organs. A quite well-studied example is the zebrafish ear, whose proper formation depends on eya1 expression, as shown by morpholino-mediated knockdown analysis (23).
Since Eya1 is involved in the development of many different organs, we proposed that additional factors bind to it and, thereby, modulate its function. Using a yeast two-hybrid-based approach, we identified the related proteins Sipl1 (Shank-interacting protein-like 1) and Rbck1 (RBCC protein interacting with PKC1) as so-far-unknown Eya1 interaction partners. We characterized the interactions and demonstrated that both Sipl1 and Rbck1 enhance the function of Eya proteins to act as coactivators for the Six transcription factors. Furthermore, we showed that Sipl1 and Rbck1 are coexpressed with Eya1 in several organs during embryogenesis of both the mouse and zebrafish. Importantly, the morpholino-mediated knockdown of Sipl1 and Rbck1 orthologs in zebrafish led to a BOR syndrome-like phenotype, indicating the relevance of the Eya1-Sipl1/Rbck1 interaction in vivo.
MATERIALS AND METHODS
Expression constructs and antibodies.
The full-length cDNAs of Sipl1 and Rbck1 were purchased from the ATCC (GenBank accession numbers BC016203 for Sipl1 and BC034555 for Rbck1). Mammalian expression constructs for mouse Flag-Sipl1 and Flag-Rbck1 as well as bacterial expression constructs for glutathione S-transferase (GST)-Sipl1 and GST-Rbck1 were generated by cloning of the full-length cDNAs into pcDNA3.1-Flag and pGEX-KG (15), respectively. Mammalian expression constructs for mouse Eya1, Eya2, and Eya3 (pHM6-Eya1, -Eya2, -Eya3) were generous gifts from Kyoshi Kawakami. Yeast expression constructs for Eya1, Eya2, and Eya3 were obtained by PCR amplification of the respective cDNA fragments and subsequent cloning into pGBT9 for interaction assays or pGBKT7 for expression analysis. Similarly, Sipl1 and its deletion fragments were introduced into the yeast expression vector pGADT7. Yeast expression constructs for mouse Eya4 were kindly provided by Richard J. H. Smith, and the plasmids pGL3-MEF3/TATA, pGL3-TATA, and pCR3-Six4 used for luciferase reporter assays were provided by Pascal Maire.
Mouse monoclonal anti-Flag M2 (Sigma), anti-HA 6E2 and anti-c-Myc 9B11 (Cell Signaling), and anti-β-actin (ab8224; Abcam) antibodies were purchased from the indicated manufacturers. The hybridoma cell line for production of the mouse monoclonal antihemagglutinin (anti-HA) 12CA5 antibody was obtained from Ed Harlow.
Yeast two-hybrid analysis.
Yeast two-hybrid screens were performed according to the manufacturer's instructions (Clontech). Briefly, the DNA fragment encoding the C terminus of murine Eya1 (aa 291 to 591) was PCR amplified and ligated into the bait vector pGBT9. The resultant plasmid was used to transform the yeast strain KFY1 (generated by Thomas Munder, Jena, Germany), which was then mated to the yeast strain Y187 pretransformed with a cDNA library of an 11-day-old mouse embryo (Clontech). About 2 × 106 clones were screened for activity of the reporter genes HIS3 and lacZ by growth on selection medium lacking histidine and colony-lift filter assay, respectively. Positive clones were subjected to plasmid rescue to isolate the prey plasmid (18) and confirmed by retransformation into S. cerevisiae KFY1 together with the bait plasmid.
To assess the strength of a protein-protein interaction, the yeast strain KFY1 was cotransformed with the indicated bait and prey plasmids. Colonies were analyzed by quantitative β-galactosidase (β-Gal) liquid assay, according to the Clontech protocol. Expression of the constructs used for the β-Gal liquid assay was confirmed by yeast protein extraction as described previously (27).
In vitro transcription/translation and GST pulldown assays.
Full-length HA-Eya1 or HA-Eya2 were synthesized in vitro using pHM6-Eya1 or pHM6-Eya2 as a template in a coupled transcription/translation system (Promega). Recombinant GST-Sipl1 and GST-Rbck1 fusion proteins or GST alone was purified and coupled to glutathione-agarose beads as described previously (56).
For GST pulldown assays, the GST fusion proteins precoupled to agarose beads were resuspended in 100 μl HB buffer (20 mM HEPES at pH 7.8, 100 mM KCl, 5 mM MgCl2, 0.5% NP-40) and 30 μl of in vitro-translated HA-Eya1 or HA-Eya2. After incubation overnight at 4°C, the beads were washed in HB buffer. Proteins were eluted in 2× Laemmli loading buffer, separated on a 10% sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel, and analyzed by immunoblotting.
Coimmunoprecipitation.
Cos-7 cells (1 × 106) were seeded into 10-cm plates and transfected with the indicated expression constructs using SuperFect (Qiagen). To increase protein input, cells were treated with the proteasome inhibitor MG132 (1 μM; Sigma) at 24 h posttransfection. The following day, cells were scraped in phosphate-buffered saline (PBS), collected by centrifugation (3,000 rpm, 5 min, 4°C), and lysed in 500 μl HEPES lysis buffer (25 mM HEPES at pH 7.9, 0.5 mM EDTA, 150 mM NaCl, 0.5% NP-40). The lysate was cleared by centrifugation (10,000 rpm, 5 min, 4°C) and added to anti-HA 12CA5 antibody, which had been coupled to protein G-agarose (Calbiochem) previously. After incubation overnight at 4°C and being washed in lysis buffer, immunocomplexes were eluted by being boiled in 2× Laemmli loading buffer. Samples were analyzed by 10% SDS-PAGE and immunoblotting.
RNA isolation and RT-PCR analysis.
Total RNA obtained from mouse tissues or zebrafish embryos was isolated using the RNeasy minikit (Qiagen). An aliquot of the RNA (500 ng) was reverse transcribed into cDNA using SuperScript II RNase H− reverse transcriptase (RT) (Invitrogen) and random primers (Promega). The following primers were used for RT-PCR analysis: for mouse Eya1, for-TGGCCCTACCCCTTCCCCAC and rev-TGACAATCCACTTTCCGTCTT; for mouse Sipl1, for-CCTGTGTATGCCTGAACGAA and rev-AGAGGATCCCAAGCACAGG; for mouse Rbck1, for-AACACGTCACTCAACCCACA and rev-CTGTTTCCGCTGCTGGTACT; for mouse Lim1, for-TCACCTCAACTGCTTCACCT and rev-CATCCTGCGATGGATCTTG; for mouse Tbp, for-GGCCTCTCAGAAGCATCACTA and rev-GCCAAGCCCTGAGCATAA.
In order to quantify the expression levels of Eya1, Sipl1, and Rbck1 in different mouse tissues, quantitative real-time RT-PCR (qRT-PCR) analysis was performed using SYBR green I nucleic acid gel stain (BioWhittaker Molecular Applications) and 1 μM fluorescein calibration dye (Bio-Rad). The factor difference in expression of each of the analyzed genes was calculated according to the ΔΔCT (threshold cycle) method (26), including normalization to expression of the housekeeping gene Tbp.
Luciferase reporter assay.
Transfection of Cos-7 cells for the luciferase reporter assay was performed using Lipofectamine 2000 (Invitrogen). Cos-7 cells (1.5 × 105) were seeded into 6-well dishes and transfected on the following day with 1 μg pGL3-MEF3/TATA reporter construct, 1.6 μg pHM6-Eya2, 0.4 μg pCR3-Six4, and 1 μg Flag-Sipl1 or Flag-Rbck1. To normalize for transfection efficiency, 0.05 μg phRL-TK control plasmid was added to each sample. At 48 h posttransfection, cells were harvested, and luciferase activities were determined using the dual-luciferase reporter assay (Promega).
In situ hybridization analysis.
Zebrafish embryos were obtained from matings of wild-type fish of the TüAB strain, which has been kept in laboratory stocks in Jena, Germany, for many generations. Embryos were raised at 28°C and staged according to Kimmel et al. (22).
In situ hybridizations on 10-μm paraffin sections of mouse or zebrafish embryos were performed essentially as described previously (32). For double in situ hybridization, sections of the zebrafish head were incubated with a fluorescein-labeled probe for eya1 and a digoxigenin-labeled probe for sipl1-rbck1. Fluorescein-labeled eya1 was detected using a sheep antifluorescein antibody coupled to alkaline phosphatase. Staining was developed using Fast Red (Roche). After staining, slides were washed twice in 0.1 M Tris-0.1% Tween, and antibody was stripped by incubation in 0.1 M glycine, pH 2.2, containing 0.1% Tween for 20 min. Subsequently, hybridization of the digoxigenin-labeled sipl1-rbck1 probe was detected as described previously (32).
Whole-mount in situ hybridizations were performed as described by Hauptmann and Gerster (16). The cDNA templates for the synthesis of digoxigenin-labeled riboprobes were amplified with the following primers: for mouse Sipl1, for-ACGTGAATTCATGTCGCCGCCCGCCGG and rev-ACGTCTCGAGCTAGTCGAGGAAGTGCACGCTG; for mouse Rbck1, for-CCCTCAGGGTGCAAGTAAAA and rev-CTCAAGGTGCTTCGGTTCTC; for zebrafish eya1, for-GGACTATCCTTCCTACCCGACG and rev-GTGGCAGCAGCGTGGAATCCG (23); for zebrafish sipl1, for-GTGGGCTCCGACTCTCTG and rev-GCACAAACACTGAGAGATGATCC; for zebrafish sipl1-rbck1, for-AGTTTGGCAACACCTCCACA and rev-CAATTGTGGAGTGTGGGAAG; for zebrafish rbck1, for-TATGGCTTCCATCCGTCTCT and rev-TCCAGCATCTCTGTGGTCTG. The zebrafish probes are located within the coding regions of the respective mRNAs and correspond to aa 229 to 431 for Eya1, aa 23 to 239 for Sipl1, aa 113 to 248 for Sipl1-Rbck1, and aa 98 to 440 for Rbck1. Each cDNA fragment was cloned into pCRII-TOPO (Invitrogen), which was used as a template for in vitro transcription to generate probes in antisense and sense orientations. The riboprobe for mouse Eya1 was generated by HindIII digestion of pHM6-Eya1. The HindIII fragment was cloned into pBSK, which was used as a template for in vitro transcription.
Injection of zebrafish embryos with morpholinos and Alcian Blue staining.
Injections into zebrafish embryos were performed as described previously (36). Briefly, morpholino antisense oligonucleotides (MO) (Gene Tools) were diluted to a concentration of 1 or 2 mM in water with 0.1% phenol red (Sigma) and injected into the yolks of 1- to 2-cell embryos. Morpholinos were directed against splice donor sites of the pre-mRNA. The morpholino sequences are as follows: for eya1, 5′-AAACAAAGATGATAGACCTACTTCC-3′ (exon 10-intron 10) (23); for sipl1, 5′-AGGCCCTATGATATACCTGATGTCT-3′ (exon 4-intron 4) and 5′-TATTTTGATCCTCCTACCTGCTGCT-3′ (exon 5-intron 5); for sipl1-rbck1, 5′-CAAGTTGGACATTTACTCACCACAC-3′ (exon 1-intron 1) and 5′-GCAGAAGAAATGCAAACCTCTGTGT-3′ (exon 2-intron 2); and for rbck1, 5′-AATGCATTACCATCGATCTGCCTCT-3′ (exon 5-intron 5) and 5′-AGAGGCATCAAGAGCAGCCATACTT-3′ (translational start site).
In each case, knockdown efficiency was confirmed by RT-PCR analysis. For this, total RNA from 10 injected zebrafish embryos at 24 hours postfertilization (hpf) was isolated and reverse transcribed into cDNA as described above. The following primers were used: for zebrafish eya1, for-GGACTATCCTTCCTACCCGACG and rev-GTGGCAGCAGCGTGGAATCCG (23); for zebrafish sipl1, for-ATGAGCTGCGTCTCCTCAAG and rev-GCACAAACACTGAGAGATGATCC; for zebrafish sipl1-rbck1, for-TCGTTCAAATTTCGGTGTGA (exon 2-intron 2 MO) or for-TATACGGCCGCTATGTCTCC (exon 1-intron 1 MO) and rev-GGAGGTAGTCGCCCTTCTTC; and for zebrafish rbck1, for-TGAATAAACCGACACGTCCA and rev-CATGATGGTGCCAAAACAAA. The identity of the resulting product was verified by cloning and sequencing in each case.
Alcian Blue staining of morpholino-injected zebrafish embryos was performed at 5 days postfertilization, according to the protocol by Walker and Kimmel (54).
RESULTS
Sipl1 is a novel Eya1-binding protein.
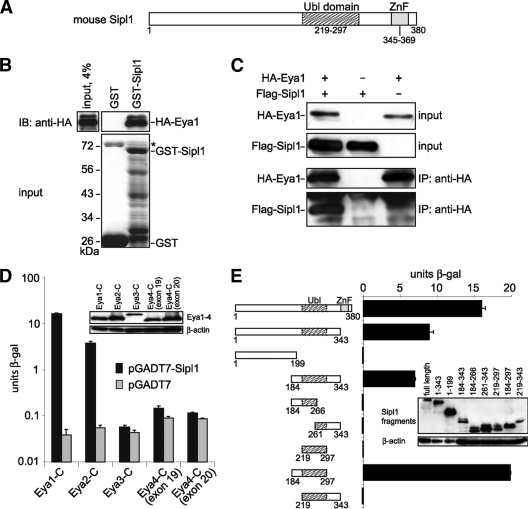
In order to identify novel interaction partners of Eya1, we performed a yeast two-hybrid screen, using the highly conserved C terminus of Eya1 as bait against a mouse embryonic cDNA library. Within this screen, we isolated 35 positives from approximately 2 × 106 clones. Upon retransformation, 10 clones turned out to be true positives. One clone encoded a part of Six2, a well-established Eya1-interacting protein, thus confirming the validity of the screen. Another positive clone coded for a fragment of Sipl1 (aa 1 to 343). The Sipl1 protein consists of 380 aa and was previously described as an interaction partner of the Shank1 scaffolding protein in rat brain (25). The C-terminal part of Sipl1 includes the following two conserved regions: a Ubl (ubiquitin-like) domain and a Ran-BP2 (Ran binding protein 2)-type zinc finger (ZnF) (Fig. 1A). In order to confirm the interaction between Eya1 and Sipl1, we performed GST pulldown experiments using GST-Sipl1 as bait and in vitro-synthesized HA-tagged Eya1 as prey. We could detect binding of Eya1 to GST-Sipl1 but not to GST alone (Fig. 1B), indicating a physical association between the two proteins in vitro. Furthermore, we verified the interaction in mammalian cells by coimmunoprecipitation. To this end, we transfected Cos-7 cells with expression constructs for HA-tagged Eya1 and Flag-tagged Sipl1. As shown in Fig. 1C after precipitation of Eya1, we were able to detect coprecipitation of Sipl1 (Fig. 1C).
FIG. 1.
Sipl1 is a novel interaction partner of Eya1. (A) Sipl1 protein domain structure. Ubl, ubiquitin-like; ZnF, zinc finger. (B) Eya1 and Sipl1 interact directly with each other. GST pulldown assay with in vitro-synthesized HA-Eya1 and recombinant GST-Sipl1 or GST as a control. HA-Eya1 was visualized by immunoblotting (IB) with anti-HA 6E2 antibody (top) and GST fusion proteins by SDS-PAGE and Coomassie blue staining (bottom). The asterisk indicates bacterial protein copurifying with GST. (C) Sipl1 and Eya1 interact in mammalian cells. Cos-7 cells were transfected with expression constructs for HA-Eya1 and Flag-Sipl1. At 48 h posttransfection, cells were lysed, and HA-Eya1 was precipitated with anti-HA 12CA5 antibody. Cell lysates before immunoprecipitation (IP) and precipitated complexes were analyzed by immunoblotting using anti-HA 6E2 antibody for the detection of HA-Eya1 and anti-Flag M2 antibody for the detection of Flag-Sipl1. (D) Sipl1 interacts with Eya1 and Eya2 but not with Eya3 and Eya4. Cotransformation of S. cerevisiae KFY1 with constructs encoding the C-terminal fragments of Eya1-4 and pGADT7-Sipl1 or empty vector. In each case, the interaction strength was determined by β-Gal liquid assay from three pooled colonies in triplicate. (E) Eya1 binds to the conserved Ubl domain of Sipl1. S. cerevisiae KFY1 was transformed with a pGBT9 vector encoding the C terminus of Eya1 and pGADT7 encoding the indicated Sipl1 deletion fragments. For each sample, a β-Gal liquid assay was performed with 3 pooled colonies and measured in triplicate. The data represent the means and standard deviations from the results of one representative experiment. Expression of the yeast constructs was confirmed by protein extraction and immunoblot analysis (insets).
Sipl1 interacts with Eya1 and Eya2 via its Ubl domain.
The C terminus of Eya1, which served as bait for the yeast two-hybrid screen, contains the Eya domain. This domain is highly conserved in the Eya paralogs Eya2, Eya3, and Eya4. To address the specificity of the interaction, we tested whether Sipl1 can also interact with these paralogs using a yeast two-hybrid-based approach. We were able to detect an interaction of Sipl1 with Eya1 and Eya2 but not with Eya3 and two alternative splice forms of Eya4 (Fig. 1D).
To further characterize the Eya1-Sipl1 interaction, we narrowed down the binding sites using a yeast two-hybrid assay and different deletion fragments for each of the interaction partners. The strongest interaction with Eya1 could be observed with a Sipl1 fragment containing the whole Ubl domain and an additional 35 aa located N terminally of it (Fig. 1E). Binding in this case was even stronger than with the full-length Sipl1 protein, which might be explained by different folding of the respective protein fragments. In the case of Eya1, we were not able to identify a subregion within the Eya1 C terminus, which mediates binding to Sipl1. Any deletion led to complete loss of interaction with Sipl1 (data not shown), suggesting that the whole Eya domain of Eya1 is required for the Sipl1 interaction or that subfragments of the Eya domain may not fold correctly.
Eya1 interacts with the Sipl1-related protein Rbck1.
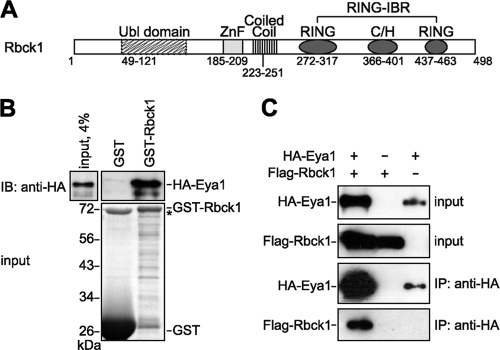
The Ubl domain of Sipl1, which has been shown to mediate the interaction with Eya1, is a conserved domain present in other proteins as well. One of those is Rbck1 (RBCC protein interacting with PKC1). In fact, the N terminus of Rbck1 is highly similar to the C terminus of Sipl1, including both conserved regions, namely, the Ubl and ZnF domains. The C terminus of Rbck1 contains a coiled-coil region and a RING-IBR (RING-in between RING) domain (Fig. 2A). Recently, it has been shown that Rbck1 acts as an E3 ubiquitin ligase (49, 60). In addition to that, Rbck1 possesses a transactivation activity and functions as a transcriptional coactivator (7, 47). Based on the fact that the Ubl domain shows a high degree of conservation between Sipl1 and Rbck1 (42% identity), we analyzed whether Rbck1 also interacts with Eya1. Therefore, we performed GST pulldown experiments and could show that Eya1 binds to Rbck1 in vitro (Fig. 2B). Furthermore, by coimmunoprecipitation analysis, we were able to detect complex formation of Eya1 and Rbck1 in Cos-7 cells (Fig. 2C).
FIG. 2.
Eya1 interacts with the Sipl1-related protein Rbck1. (A) Rbck1 protein domain structure. RING, really interesting new gene; IBR, in-between RING; C/H, cysteine/histidine rich. (B) Eya1 and Rbck1 bind directly to each other. A GST pulldown assay was performed by incubating in vitro-synthesized HA-Eya1 with GST-Rbck1 or GST alone as a control. HA-Eya1 was detected by immunoblotting using anti-HA 6E2 antibody (top). The input levels of GST fusion proteins were determined by SDS-PAGE and Coomassie blue staining (bottom). The asterisk indicates bacterial protein copurifying with GST. (C) Eya1 interacts with Rbck1 in mammalian cells. For coimmunoprecipitation analysis, Cos-7 cells were transfected with expression constructs for HA-Eya1 and Flag-Rbck1. After lysis of the cells, HA-Eya1 was precipitated using anti-HA 12CA5 antibody. Whole-cell extracts and immunocomplexes were analyzed by SDS-PAGE and immunoblotting. HA-Eya1 was detected with anti-HA 6E2 antibody, and Flag-Sipl1 was detected with anti-Flag M2 antibody.
Sipl1 and Rbck1 are coexpressed with Eya1 in several tissues of a mouse embryo.
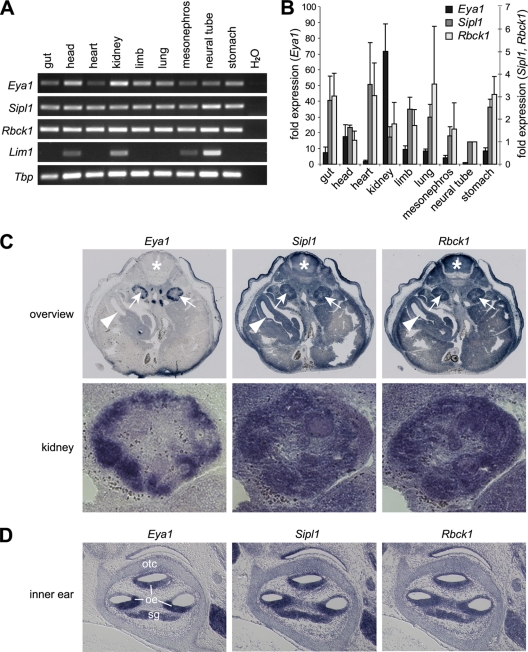
As a first step to address the functional relevance of the interactions of Eya1 with its novel interaction partners Sipl1 and Rbck1 in vivo, we performed expression studies. For this, we analyzed RNA from several tissues of a 13.5-day-old mouse embryo for expression of Sipl1, Rbck1, and Eya1 using RT-PCR and quantitative real-time RT-PCR (Fig. 3A and B). In line with already published data, we could detect Eya1 mRNA in multiple tissues during this stage of embryonic development, with the lowest expression level in the neural tube and the highest expression level in the kidney (approximately 70-fold compared to that in the neural tube) (Fig. 3B) (58). Furthermore, our analysis shows that at this stage of development, both Sipl1 and Rbck1 are ubiquitously expressed (Fig. 3A and B). As an additional control for PCR specificity and cDNA quality, we have used Lim1, a gene that is expressed specifically in the brain and the kidney (13). In situ hybridization analysis on transverse sections of the kidney region of a 13.5-day-old mouse embryo confirmed the results of the RT-PCR analyses, with ubiquitous expression of Sipl1 and Rbck1 as well as prominent Eya1 expression in the embryonic kidney (Fig. 3C). In the latter, coexpression of Eya1, Sipl1, and Rbck1 could be observed in developing nephrons in the outer region of the kidney. The apparent differences in Eya1 expression when comparing the RT-PCR data with the in situ hybridization results can be attributed to the different sensitivities of the methods, as well as the fact that in situ hybridization is performed on a section that represents only a very specific area of the embryo. An additional site of Eya1 expression is the inner ear (62). We analyzed gene expression of Eya1, Sipl1, and Rbck1 on adjacent sections of the inner ear of a 14.5-day-old mouse embryo and found all three genes coexpressed in the otic epithelium and the spiral ganglion (Fig. 3D).
FIG. 3.
Sipl1 and Rbck1 are expressed in several tissues of a developing mouse embryo. (A) Eya1, Sipl1, and Rbck1 expression in the tissue of a 13.5-day-old mouse embryo was analyzed by RT-PCR using gene-specific primers. As an additional control, expression analysis of Lim1 was included. For control of the cDNA input, Tbp expression was determined in parallel in the same samples. (B) Quantification of Eya1, Sipl1, and Rbck1 expression by quantitative real-time RT-PCR. Relative expression levels normalized to Tbp were calculated using the ΔΔCT method. For comparison, expression in the neural tube was set to 1. The left y axis corresponds to Eya1 expression, and the right y axis corresponds to Sipl1 and Rbck1 expression. The data represent means and standard deviations from the results of three independent experiments. (C) Analysis of Eya1, Sipl1, and Rbck1 expression by in situ hybridization on adjacent sections of a 13.5-day-old mouse embryo. (Top) Overview. Arrows mark the kidneys, arrowheads mark the guts, and asterisks mark the neural tubes of the mouse embryo. Please note that staining of the tissue surrounding the embryos is unspecific. (Bottom) Higher magnification of the right embryonic kidney. (D) Analysis of Eya1, Sipl1, and Rbck1 expression in the inner ear by in situ hybridization on adjacent sections of a 14.5-day-old mouse embryo. oe, otic epithelium; otc, otic capsule; sg, spiral ganglion.
Both Sipl1 and Rbck1 enhance the transactivation potential of the Eya-Six complex.
The best understood function of Eya proteins is their role as coactivators for the Six-transcription factors. In fact, complex formation of Eya and Six proteins and subsequent activation of organ-specific genes are regarded to be essential for the development of various organs, such as the kidney, thymus, and muscle (17, 21, 57, 59). In order to assess the effect of the two interaction partners Sipl1 and Rbck1 on the function of Eya proteins as coactivators of transcription, we established a transactivation assay. Based on already reported data, we used a luciferase reporter construct containing 6 MEF3 (myogenic enhancing factor 3) sites in the background of a minimal TATA promoter, which can be efficiently activated by the complex of Eya2 and Six4 (10). As shown previously, Sipl1 also interacts with Eya2 in the yeast two-hybrid assay, an activity that can be assumed for Rbck1 as well, since the binding regions of the two proteins are conserved. We indeed confirmed direct binding of both Sipl1 and Rbck1 to Eya2 by GST pulldown analysis (Fig. 4A). In line with published data, we observed that Six4 alone activated the reporter to about 4-fold, and this activation was further enhanced to about 12-fold in the presence of Eya2. The presence of Sipl1 or Rbck1 did not influence transactivation by Six4 alone but enhanced transactivation by Eya2 and Six4 to about 16-fold, indicating that this effect is mediated via the interaction of Sipl1 and Rbck1 with Eya2 (Fig. 4B).
FIG. 4.
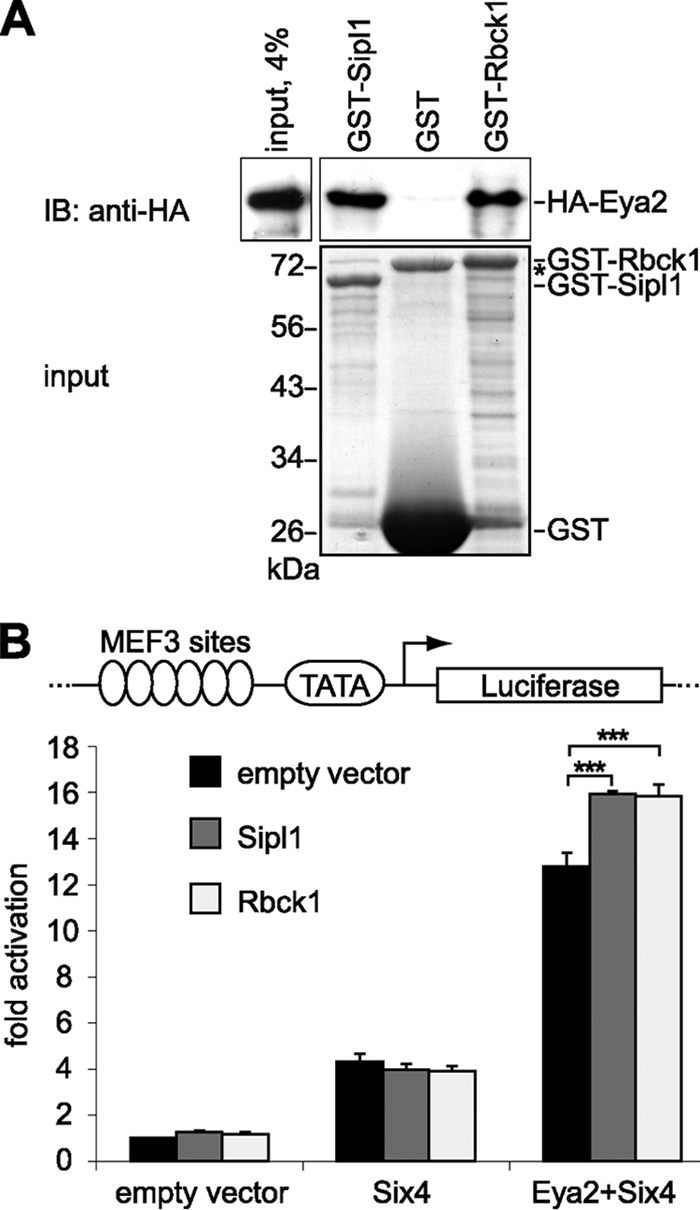
Sipl1 and Rbck1 interact with Eya2 and enhance Eya2-Six4-mediated transactivation. (A) Sipl1 and Rbck1 bind directly to Eya2. A GST pulldown assay was performed using in vitro-synthesized HA-Eya2 and recombinant GST-Sipl1, GST-Rbck1, or GST as a control. HA-Eya2 was visualized by immunoblotting with anti-HA 6E2 antibody (top) and GST fusion proteins by SDS-PAGE and Coomassie blue staining (bottom). The asterisk indicates bacterial protein copurifying with GST. (B) Sipl1 and Rbck1 enhance Eya2-mediated transactivation. Scheme of the luciferase reporter construct containing six MEF3 sites in the background of a minimal TATA promoter (top). For luciferase reporter assays, Cos-7 cells were cotransfected with the pGL3-MEF3/TATA or pGL3-TATA reporter construct, indicated expression constructs, and Renilla control plasmid. Each transfection was performed in triplicate. After normalization to Renilla activity, the fold reporter gene activation of samples containing pGL3-MEF3/TATA was calculated relative to respective samples containing pGL3-TATA. Activity of the reporter alone was set to 1. The graph represents one of three experiments showing similar results. Error bars indicate standard deviations, and asterisks indicate statistically significant differences (P value < 0.001; Student's t test).
Identification of orthologs of Sipl1 and Rbck1 in zebrafish.
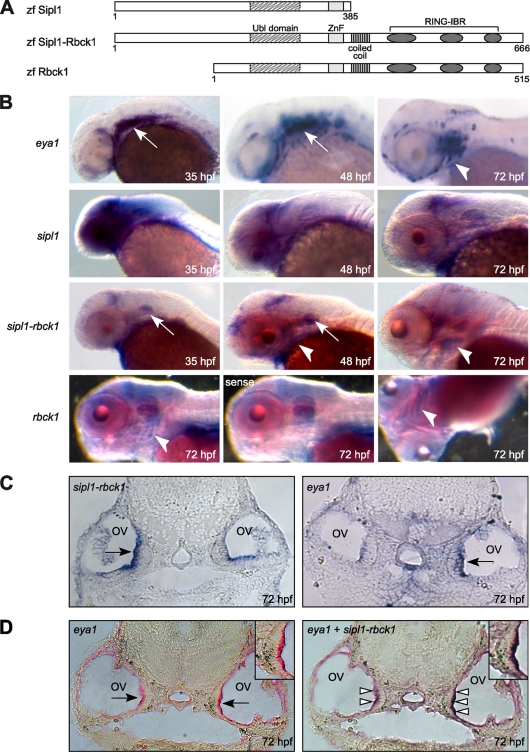
Since our analysis had shown that Sipl1 and Rbck1 are expressed in multiple tissues in the embryo, we wanted to investigate a potential role of both genes during embryonic development. We therefore turned to zebrafish as a model system. By bioinformatic analysis, we were able to identify putative orthologs of Sipl1 and Rbck1 in zebrafish (Fig. 5A). One of them, zebrafish sipl1, is located on chromosome 2 and is an ortholog of mouse Sipl1. This gene encodes a protein, which contains the conserved regions of the Ubl domain and of the Ran-BP-type ZnF in its C-terminal part. Furthermore, our analysis revealed zebrafish rbck1 on chromosome 22 to be an ortholog of mouse Rbck1, with the respective proteins sharing all the conserved domains, as follows: the Ubl domain, the Ran-BP-type ZnF, the coiled-coil region, and the RING-IBR domain. Interestingly, we identified a third ortholog on zebrafish chromosome 7, which we have named sipl1-rbck1, because it seems to be a fusion of both sipl1 and rbck1. According to our analysis, a sipl1-rbck1 ortholog is present in the genomes of other teleost fish species, such as stickleback and fugu, but not in higher vertebrates like mice or humans. An alignment of the minimal Eya1-binding region in mouse Sipl1 or Rbck1 to each of the identified zebrafish proteins revealed that zebrafish Sipl1 and Sipl1-Rbck1 are more closely related to mouse Sipl1 (50% identity in the case of Sipl1 and 49% identity in the case of Sipl1-Rbck1), whereas Rbck1 is more closely related to mouse Rbck1 (41% identity) within this region.
FIG. 5.
The zebrafish orthologs of Sipl1 and Rbck1 are expressed during embryonic development. (A) Protein domain structures of the zebrafish orthologs of Sipl1 and Rbck1. zf, zebrafish. (B) Expression of eya1, sipl1, sipl1-rbck1, and rbck1 at different embryonic stages was analyzed with the help of whole-mount in situ hybridization using gene-specific probes in antisense or sense orientation. White arrows point at expression in the otic vesicle, and white arrowheads point at expression in the pharyngeal arches. (C) Expression of sipl1-rbck1 and eya1 in otic vesicles was analyzed at 72 hpf using in situ hybridization on a transverse section of the zebrafish head. Black arrows point at the sensory epithelium of the otic vesicle (ov). The differences in signal intensities between the respective left and right otic vesicles are most likely due to the fact that the sections are slightly askew. (D) Double in situ hybridization on a transverse section of the zebrafish head (72 hpf) using a fluorescein-labeled eya1 probe and a digoxigenin sipl1-rbck1 probe. (Left) Staining for eya1 alone; (right) double staining for eya1 and sipl1-rbck1 on the same section. Insets show higher magnifications of the sensory epithelium. Black arrows point at the sensory epithelium of the otic vesicle, and white arrowheads indicate areas of coexpression of eya1 and sipl1-rbck1.
Coexpression of eya1 and the orthologs of Sipl1 and Rbck1 in developing zebrafish embryos.
In zebrafish, eya1 expression has been described in several organs during embryogenesis, for example, the otic vesicle and the pharyngeal arches (41). In order to investigate the expression pattern of each of the newly identified zebrafish orthologs of Sipl1 and Rbck1 during embryonic development, we performed whole-mount in situ hybridization using gene-specific probes at different stages of development.
In agreement with published data, we could detect eya1 expression in the otic vesicle, the region of the pharyngeal arches, and the lateral line organ (Fig. 5B). In the case of sipl1, we observed a high level of expression in the zebrafish head at 35 hpf (hours postfertilization), with prominent expression in the brain, the eyes, the otic vesicle, and the pharyngeal arches. At later stages (48 and 72 hpf), sipl1 expression was still detectable in the same organs although decreasing in intensity.
In contrast to sipl1, expression of sipl1-rbck1 was more restricted during all developmental stages analyzed. At 35 hpf and 48 hpf, sipl1-rbck1 mRNA was detected in the otic vesicle and in a region in the zebrafish brain probably representing the midbrain-hindbrain boundary. In addition to that, at 48 hpf and 72 hpf, sipl1-rbck1 expression was detectable in the pharyngeal arches. In order to have a closer look at a potential coexpression of sipl1-rbck1 and eya1 in the otic vesicle, we performed in situ hybridization on transverse sections of the zebrafish head at 72 hpf (Fig. 5C). We detected expression of sipl1-rbck1 and eya1 in an overlapping region corresponding to the sensory epithelium. Coexpression was confirmed by double in situ hybridization of the same section using probes against both genes simultaneously (Fig. 5D).
Regarding rbck1, we were not able to detect expression at earlier stages of zebrafish development. However, at 72 hpf, we observed a specific signal for rbck1 mRNA in the pharyngeal arches, which was not present when we used the corresponding probe in sense orientation.
In summary, we have shown by whole-mount in situ hybridization that the orthologs of Sipl1 and Rbck1 are expressed in distinct but overlapping domains during zebrafish embryonic development. While sipl1 is expressed ubiquitously in the head of the zebrafish embryo, both sipl1-rbck1 and rbck1 show coexpression with eya1 in the otic vesicle and/or the region of the pharyngeal arches.
The zebrafish orthologs of Sipl1 and Rbck1 are essential for craniofacial development.
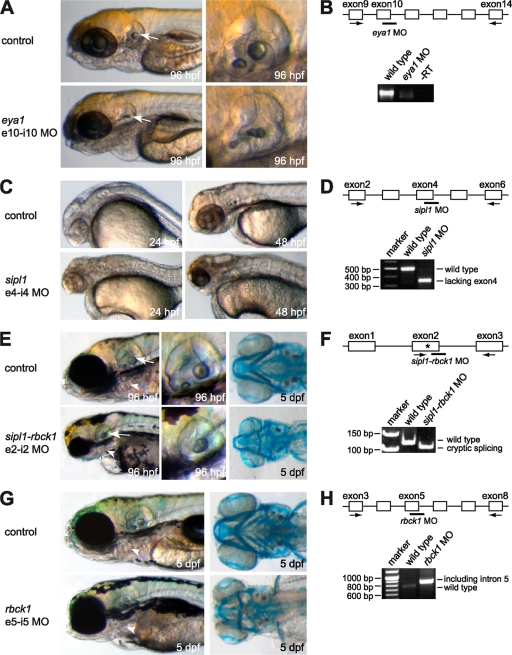
Mutations in the zebrafish eya1 gene have been shown to result in defects in the formation of the inner ear and the lateral line sensory systems during embryogenesis (23). Furthermore, it has been demonstrated that microinjection of an eya1 morpholino complementary to the exon 10-intron 10 splice site in the eya1 primary mRNA phenocopies this mutant phenotype (23). We reproduced the previously published results using the same eya1-specific morpholino and observed an abnormal morphology of the otic vesicle (Fig. 6A and B).
FIG. 6.
The zebrafish orthologs of Sipl1 and Rbck1 are essential for embryonic development. Expression of each of the orthologs was targeted by injection of splice donor site morpholinos into 1- to 2-cell embryos. Noninjected control embryos compared to embryos injected with morpholino directed against eya1 (A), sipl1 (C), sipl1-rbck1 (E), and rbck1 (G) are shown at the indicated stages (left). Arrows point to the region of the otic vesicle, and arrowheads point to the region of the pharyngeal arches. In the cases of eya1 and sipl1-rbck1, higher magnifications of the otic vesicles of control embryos and morpholino-injected embryos at 96 hpf are shown. Sipl1-rbck1- and rbck1-morphant embryos were subjected to Alcian Blue staining at 5 day postfertilization to visualize the pharyngeal arches. In each case, knockdown was confirmed by RT-PCR at 24 hpf (B, D, F, and H) using gene-specific primers, as indicated by black arrows. The bars in the diagrams mark the target sites for the respective morpholinos. The asterisk indicates a cryptic splice site.
To address the question of whether the zebrafish orthologs of Sipl1 and Rbck1 are involved in developmental processes, we employed a morpholino-mediated knockdown approach. Morpholinos are chemically modified antisense oligonucleotides that prevent gene expression on two different levels. When targeting the translational initiation codon AUG, translation of the corresponding mRNA is inhibited. We have used an alternative strategy, namely, splice morpholinos that are directed against specific splice donor or acceptor sites. Possible consequences are the inclusion of an intron, the exclusion of an exon, or usage of an alternative splice site. In each case, generation of the normal mRNA is prevented. We designed two different morpholinos for each gene and tested them independently. In each case, the two morpholinos directed against the same gene showed identical and consistent phenotypes (Fig. 6 and data not shown). In the following, results for only one of the gene-specific morpholinos are presented. Injection of a sipl1 morpholino directed against the splice donor site of intron 4 led to efficient knockdown of sipl1 expression via exclusion of exon 4 from the mRNA, leading to a frameshift and an early stop codon, as confirmed by sequencing (Fig. 6D). In line with the above-described expression pattern of sipl1, knockdown of sipl1 gene expression resulted in a severe phenotype affecting the cranial structures of the embryo (Fig. 6C). The head itself as well as individual organs, such as the eyes and the ears, were significantly smaller in size compared to those of noninjected control embryos. Furthermore, the brain appeared to be severely malformed. Thus, the observed sipl1 knockdown phenotype affects specifically those organs, which display high levels of sipl1 expression in the zebrafish head.
In contrast to sipl1, knockdown of sipl1-rbck1 using a morpholino targeting the splice donor site of intron 2 led to a milder phenotype affecting mainly the region of the pharyngeal arches and the developing ears of the zebrafish embryo (Fig. 6E). We could show by RT-PCR analysis of injected embryos at 24 hpf that in the presence of the morpholino, cryptic splicing occurred 15 bp upstream of the natural splice site (Fig. 6F). Interestingly, the observed sipl1-rbck1 knockdown phenotype resembled the morphology of BOR syndrome in human patients (29). In sipl1-rbck1 morphants, the pharyngeal arches and the lower jaw, which is derived from the pharyngeal arches, were shortened. Staining of the cartilage using Alcian Blue at 5 days postfertilization revealed that the pharyngeal arches were reduced in number and disorganized compared to those of control embryos at the same age. Moreover, knockdown of sipl1-rbck1 inhibited proper ear development, in that the otic vesicle of sipl1-rbck1 morphants was smaller in size and that structures were not properly developed compared to those of control embryos. The sipl1-rbck1 knockdown phenotype resembles the expression pattern of the respective gene, with major expression in the otic vesicle and the pharyngeal arches, and is partly similar to the phenotype of eya1 knockdown embryos, affecting the developing ear (23). Interestingly, rbck1 also seems to be involved in the development of the pharyngeal arches (Fig. 6G). Knockdown of rbck1 by injection of a morpholino directed against the splice donor site of intron 5 resulted in a mild phenotype affecting only the development of the pharyngeal arches, including the lower jaw of the developing zebrafish embryo. Alcian Blue staining confirmed a high degree of disorganization in the pharyngeal arches, particularly in the first two, and an almost complete lack of arches 3 to 7 after knockdown of rbck1, similar to the results shown previously for sipl1-rbck1. We confirmed knockdown efficiency by RT-PCR analysis and showed that injection of the rbck1 morpholino inhibited splicing of intron 5, resulting in a premature stop codon (Fig. 6H).
DISCUSSION
Eya1 is required for the development of several organs in vertebrates, including the kidney, ear, thymus, and muscle (14, 57, 59). In addition to its activity as a phosphatase, Eya1 can also act as a transcriptional cofactor, together with interacting proteins, e.g., of the Six and Dach families. In this work, we have identified Sipl1 and Rbck1 as two novel interaction partners of Eya1.
Sipl1 was first described in the rat as an interaction partner of Shank1, a protein that functions as a scaffold factor in the formation and maintenance of postsynaptic densities (44). However, Sipl1 is expressed not only in the brain but also in many other adult tissues, for example, in the heart, muscle, kidney, and spleen (25). This is in line with our data that show Sipl1 expression in multiple tissues in the developing mouse embryo. Homologs of Sipl1 have been identified in the human and mouse, for which they have been implicated in enteric nervous system function (9). Furthermore, recent studies indicated that mutations in the mouse Sipl1 gene result in multiorgan inflammation, immune system dysregulation, and dermatitis (43).
We have identified the Ubl domain in the C terminus of Sipl1 as the region that mediates binding to Eya1. Since the Ubl domain is a conserved domain, we proposed that other Ubl domain-containing proteins might also interact with Eya1. The N terminus of Rbck1, including the Ubl domain and a Ran-BP2-type zinc finger, is highly similar to the C terminus of Sipl1 and was previously considered to be an independent domain, termed the Rbck1 homology domain (25). Based on the homology within this region, we have speculated that Rbck1 might also interact with Eya1 and could indeed demonstrate interaction of the two proteins.
Rbck1 belongs to the family of RING-IBR-containing proteins, which have been shown to possess E3 ubiquitin ligase activity (28). In fact, Rbck1 has been described to act as an E3 ubiquitin ligase, mediating the degradation of several proteins, like IRP2 (iron regulatory protein 2), PKC (protein kinase C), Bach1, and TAB2/3 (TAK1-binding protein 2/3) (33, 51, 60, 61). Based on our observation that Eya1 and Rbck1 can associate, one could therefore speculate that Eya1 itself or other Eya1-associated proteins might be subject to regulation by Rbck1-mediated ubiquitination.
Rbck1 also possesses transactivation activity (7) and has been shown to shuttle between the cytoplasm and the nucleus (48). Eya proteins have been shown to act as coactivators in several transcriptional activation complexes, too. One of the best-studied examples is the Eya-Six complex, which has been demonstrated to mediate activation of gene expression during the development of a variety of organs in both invertebrates and vertebrates (2, 3, 17, 21, 46). Therefore, we tested whether Rbck1 and Sipl1 can also act as cofactors for the Eya-Six complex and could indeed show that both proteins could increase Eya-Six-mediated transactivation. This result provides a potential mechanistic basis for the role of Eya1 and its associated proteins in organ development. Moreover, Eya1 and Rbck1 share an interaction partner, CBP (CREB binding protein). CBP is a well-characterized coactivator that functions as a key integrator in various transcription-activating complexes (1). CBP has been described to act as a linker for the interaction between mammalian Eya and Dach proteins, thereby mediating target gene activation (20). Strikingly, interaction with CBP has also been implicated in the regulation of Rbck1 transactivation function (48). It is tempting to speculate that CBP is also part of the Eya1-Rbck1 complex and presumably involved in target gene activation.
Sipl1 and Rbck1 have been shown to be expressed ubiquitously in adult mammalian tissues (9, 25, 52). However, expression and function of Sipl1 and Rbck1 during embryonic development has not been described yet. We show here that both Sipl1 and Rbck1 are expressed together with Eya1 in many tissues of a mouse embryo, which is in line with the hypothesis that the proteins act together during embryonic development. We underlined this assumption by the identification and functional characterization of zebrafish orthologs of Sipl1 and Rbck1. We identified not only one Sipl1 ortholog and one Rbck1 ortholog but also a gene which seems to represent a fusion of both, sipl1-rbck1. Using morpholino-mediated knockdown analysis, we could show that each of the zebrafish orthologs is involved in organogenesis. This is in line with the expression of each respective ortholog in the affected tissues. For example, zebrafish sipl1 is expressed in the brain region and essential for its proper development. Moreover, deficiency for both zebrafish sipl1-rbck1 and rbck1 leads to anomalies in the regions of the lower jaw and the pharyngeal arches. Also, knockdown of sipl1-rbck1 leads to anomalies in formation of the zebrafish ear. Interestingly, the development of the ears and the pharyngeal arches also depends on proper function of Eya1. It has been shown that eya1 directs ear development in zebrafish (23). Moreover, in Eya1 knockout mice, several organs which are derived from the pharyngeal arches are malformed or even completely absent, such as the thymus, cranial skeleton, and middle ear. Furthermore, mutations in human EYA1 lead to BOR/BO syndrome, affecting mainly the cranial skeleton and the ear, including the inner, middle, and outer ear.
As described above, there is a remarkable overlap between the phenotypes of sipl1-, rbck1-, and sipl-rbck1-deficient zebrafish embryos and the characteristic symptoms of BOR and BO syndrome. The latter is caused by mutations in EYA1 but also in SIX1 and SIX5, as recently demonstrated (19, 40). Since SIX1 and SIX5 encode interaction partners of Eya1, it is tempting to speculate that mutations in human SIPL1 or RBCK1 could also be associated with BOR syndrome.
In conclusion, we identified Sipl1 and Rbck1 as novel Eya1-interacting proteins and provided a first insight into the physiological importance of the respective interactions. Using the zebrafish model system, we showed that orthologs of Sipl1 and Rbck1 direct the development of several organs, for example, the ears and the pharyngeal arches, which also depend on expression of Eya1. On the molecular level, we showed that both Sipl1 and Rbck1 act as cofactors for the Eya-Six complex. Further experiments regarding the functional consequences of the interaction of Sipl1 or Rbck1 with Eya1 should clarify the importance of the respective interaction for vertebrate organogenesis.
Acknowledgments
We thank Eric Rivera-Milla for critically reading and improving the manuscript, as well as Frank-Dietmar Böhmer and Amna Musharraf for stimulating discussions throughout the project. We also thank Pascal Maire for providing the myogenin reporter, Kiyoshi Kawakami for providing Eya1-3 expression constructs, and Richard J. Smith for providing the Eya4 expression construct. We are grateful to Ulrike Gausmann for help with bioinformatic analysis, Helmut Pospiech for establishing the cartilage staining, and Claudia Franke, Uta Schmidt, and Katrin Sulik for technical support. We appreciate the help of the rotating students Doreen Köhler, Andreas Boland, Daniela Endt, and Andrea Wetzel.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB604, project C7) to C.E.
Footnotes
Published ahead of print on 18 October 2010.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Bonini, N. M., Q. T. Bui, G. L. Gray-Board, and J. M. Warrick. 1997. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development 124:4819-4826. [DOI] [PubMed] [Google Scholar]
- 3.Brodbeck, S., B. Besenbeck, and C. Englert. 2004. The transcription factor Six2 activates expression of the Gdnf gene as well as its own promoter. Mech. Dev. 121:1211-1222. [DOI] [PubMed] [Google Scholar]
- 4.Chai, L., J. Yang, C. Di, W. Cui, K. Kawakami, R. Lai, and Y. Ma. 2006. Transcriptional activation of the SALL1 by the human SIX1 homeodomain during kidney development. J. Biol. Chem. 281:18918-18926. [DOI] [PubMed] [Google Scholar]
- 5.Chen, R., M. Amoui, Z. Zhang, and G. Mardon. 1997. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell 91:893-903. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, K. L., A. N. Patrick, E. L. McCoy, and H. L. Ford. 2008. The six family of homeobox genes in development and cancer. Adv. Cancer Res. 101:93-126. [DOI] [PubMed] [Google Scholar]
- 7.Cong, Y. S., Y. L. Yao, W. M. Yang, N. Kuzhandaivelu, and E. Seto. 1997. The hepatitis B virus X-associated protein, XAP3, is a protein kinase C-binding protein. J. Biol. Chem. 272:16482-16489. [DOI] [PubMed] [Google Scholar]
- 8.Cook, P. J., B. G. Ju, F. Telese, X. Wang, C. K. Glass, and M. G. Rosenfeld. 2009. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature 458:591-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daigo, Y., I. Takayama, S. M. Ward, K. M. Sanders, and M. A. Fujino. 2003. Novel human and mouse genes encoding a shank-interacting protein and its upregulation in gastric fundus of W/WV mouse. J. Gastroenterol. Hepatol. 18:712-718. [DOI] [PubMed] [Google Scholar]
- 10.Fan, X., L. F. Brass, M. Poncz, F. Spitz, P. Maire, and D. R. Manning. 2000. The alpha subunits of Gz and Gi interact with the eyes absent transcription cofactor Eya2, preventing its interaction with the six class of homeodomain-containing proteins. J. Biol. Chem. 275:32129-32134. [DOI] [PubMed] [Google Scholar]
- 11.Fraser, F. C., D. Ling, D. Clogg, and B. Nogrady. 1978. Genetic aspects of the BOR syndrome—branchial fistulas, ear pits, hearing loss, and renal anomalies. Am. J. Med. Genet. 2:241-252. [DOI] [PubMed] [Google Scholar]
- 12.Fraser, F. C., J. R. Sproule, and F. Halal. 1980. Frequency of the branchio-oto-renal (BOR) syndrome in children with profound hearing loss. Am. J. Med. Genet. 7:341-349. [DOI] [PubMed] [Google Scholar]
- 13.Fujii, T., J. G. Pichel, M. Taira, R. Toyama, I. B. Dawid, and H. Westphal. 1994. Expression patterns of the murine LIM class homeobox gene lim1 in the developing brain and excretory system. Dev. Dyn. 199:73-83. [DOI] [PubMed] [Google Scholar]
- 14.Grifone, R., J. Demignon, J. Giordani, C. Niro, E. Souil, F. Bertin, C. Laclef, P. X. Xu, and P. Maire. 2007. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev. Biol. 302:602-616. [DOI] [PubMed] [Google Scholar]
- 15.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 16.Hauptmann, G., and T. Gerster. 1994. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 10:266. [DOI] [PubMed] [Google Scholar]
- 17.Heanue, T. A., R. Reshef, R. J. Davis, G. Mardon, G. Oliver, S. Tomarev, A. B. Lassar, and C. J. Tabin. 1999. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 13:3231-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 19.Hoskins, B. E., C. H. Cramer, D. Silvius, D. Zou, R. M. Raymond, D. J. Orten, W. J. Kimberling, R. J. Smith, D. Weil, C. Petit, E. A. Otto, P. X. Xu, and F. Hildebrandt. 2007. Transcription factor SIX5 is mutated in patients with branchio-oto-renal syndrome. Am. J. Hum. Genet. 80:800-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda, K., Y. Watanabe, H. Ohto, and K. Kawakami. 2002. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol. Cell. Biol. 22:6759-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawakami, K., S. Sato, H. Ozaki, and K. Ikeda. 2000. Six family genes—structure and function as transcription factors and their roles in development. Bioessays 22:616-626. [DOI] [PubMed] [Google Scholar]
- 22.Kimmel, C. B., W. W. Ballard, S. R. Kimmel, B. Ullmann, and T. F. Schilling. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203:253-310. [DOI] [PubMed] [Google Scholar]
- 23.Kozlowski, D. J., T. T. Whitfield, N. A. Hukriede, W. K. Lam, and E. S. Weinberg. 2005. The zebrafish dog-eared mutation disrupts eya1, a gene required for cell survival and differentiation in the inner ear and lateral line. Dev. Biol. 277:27-41. [DOI] [PubMed] [Google Scholar]
- 24.Li, X., K. A. Oghi, J. Zhang, A. Krones, K. T. Bush, C. K. Glass, S. K. Nigam, A. K. Aggarwal, R. Maas, D. W. Rose, and M. G. Rosenfeld. 2003. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 426:247-254. [DOI] [PubMed] [Google Scholar]
- 25.Lim, S., C. Sala, J. Yoon, S. Park, S. Kuroda, M. Sheng, and E. Kim. 2001. Sharpin, a novel postsynaptic density protein that directly interacts with the shank family of proteins. Mol. Cell. Neurosci. 17:385-397. [DOI] [PubMed] [Google Scholar]
- 26.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 27.Lussier, M. P., S. Cayouette, P. K. Lepage, C. L. Bernier, N. Francoeur, M. St-Hilaire, M. Pinard, and G. Boulay. 2005. MxA, a member of the dynamin superfamily, interacts with the ankyrin-like repeat domain of TRPC. J. Biol. Chem. 280:19393-19400. [DOI] [PubMed] [Google Scholar]
- 28.Marin, I., and A. Ferrus. 2002. Comparative genomics of the RBR family, including the Parkinson's disease-related gene parkin and the genes of the ariadne subfamily. Mol. Biol. Evol. 19:2039-2050. [DOI] [PubMed] [Google Scholar]
- 29.Melnick, M., D. Bixler, W. E. Nance, K. Silk, and H. Yune. 1976. Familial branchio-oto-renal dysplasia: a new addition to the branchial arch syndromes. Clin. Genet. 9:25-34. [DOI] [PubMed] [Google Scholar]
- 30.Melnick, M., D. Bixler, K. Silk, H. Yune, and W. E. Nance. 1975. Autosomal dominant branchiootorenal dysplasia. Birth Defects Orig. Artic. Ser. 11:121-128. [PubMed] [Google Scholar]
- 31.Mermod, N., E. A. O'Neill, T. J. Kelly, and R. Tjian. 1989. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell 58:741-753. [DOI] [PubMed] [Google Scholar]
- 32.Moorman, A. F., A. C. Houweling, P. A. de Boer, and V. M. Christoffels. 2001. Sensitive nonradioactive detection of mRNA in tissue sections: novel application of the whole-mount in situ hybridization protocol. J. Histochem. Cytochem. 49:1-8. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura, M., F. Tokunaga, S. Sakata, and K. Iwai. 2006. Mutual regulation of conventional protein kinase C and a ubiquitin ligase complex. Biochem. Biophys. Res. Commun. 351:340-347. [DOI] [PubMed] [Google Scholar]
- 34.Ohto, H., S. Kamada, K. Tago, S. I. Tominaga, H. Ozaki, S. Sato, and K. Kawakami. 1999. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol. Cell. Biol. 19:6815-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okabe, Y., T. Sano, and S. Nagata. 2009. Regulation of the innate immune response by threonine-phosphatase of Eyes absent. Nature 460:520-524. [DOI] [PubMed] [Google Scholar]
- 36.Perner, B., C. Englert, and F. Bollig. 2007. The Wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros. Dev. Biol. 309:87-96. [DOI] [PubMed] [Google Scholar]
- 37.Pfister, M., T. Toth, H. Thiele, B. Haack, N. Blin, H. P. Zenner, I. Sziklai, P. Nurnberg, and S. Kupka. 2002. A 4-bp insertion in the eya-homologous region (eyaHR) of EYA4 causes hearing impairment in a Hungarian family linked to DFNA10. Mol. Med. 8:607-611. [PMC free article] [PubMed] [Google Scholar]
- 38.Pignoni, F., B. Hu, and S. L. Zipursky. 1997. Identification of genes required for Drosophila eye development using a phenotypic enhancer-trap. Proc. Natl. Acad. Sci. U. S. A. 94:9220-9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rayapureddi, J. P., C. Kattamuri, B. D. Steinmetz, B. J. Frankfort, E. J. Ostrin, G. Mardon, and R. S. Hegde. 2003. Eyes absent represents a class of protein tyrosine phosphatases. Nature 426:295-298. [DOI] [PubMed] [Google Scholar]
- 40.Ruf, R. G., P. X. Xu, D. Silvius, E. A. Otto, F. Beekmann, U. T. Muerb, S. Kumar, T. J. Neuhaus, M. J. Kemper, R. M. Raymond, Jr., P. D. Brophy, J. Berkman, M. Gattas, V. Hyland, E. M. Ruf, C. Schwartz, E. H. Chang, R. J. Smith, C. A. Stratakis, D. Weil, C. Petit, and F. Hildebrandt. 2004. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc. Natl. Acad. Sci. U. S. A. 101:8090-8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sahly, I., P. Andermann, and C. Petit. 1999. The zebrafish eya1 gene and its expression pattern during embryogenesis. Dev. Genes Evol. 209:399-410. [DOI] [PubMed] [Google Scholar]
- 42.Schonberger, J., L. Wang, J. T. Shin, S. D. Kim, F. F. Depreux, H. Zhu, L. Zon, A. Pizard, J. B. Kim, C. A. Macrae, A. J. Mungall, J. G. Seidman, and C. E. Seidman. 2005. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat. Genet. 37:418-422. [DOI] [PubMed] [Google Scholar]
- 43.Seymour, R. E., M. G. Hasham, G. A. Cox, L. D. Shultz, H. Hogenesch, D. C. Roopenian, and J. P. Sundberg. 2007. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun. 8:416-421. [DOI] [PubMed] [Google Scholar]
- 44.Sheng, M., and E. Kim. 2000. The Shank family of scaffold proteins. J. Cell Sci. 113(Pt. 11):1851-1856. [DOI] [PubMed] [Google Scholar]
- 45.Silver, S. J., E. L. Davies, L. Doyon, and I. Rebay. 2003. Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol. Cell. Biol. 23:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spitz, F., J. Demignon, A. Porteu, A. Kahn, J. P. Concordet, D. Daegelen, and P. Maire. 1998. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc. Natl. Acad. Sci. U. S. A. 95:14220-14225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tatematsu, K., C. Tokunaga, N. Nakagawa, K. Tanizawa, S. Kuroda, and U. Kikkawa. 1998. Transcriptional activity of RBCK1 protein (RBCC protein interacting with PKC 1): requirement of RING-finger and B-Box motifs and regulation by protein kinases. Biochem. Biophys. Res. Commun. 247:392-396. [DOI] [PubMed] [Google Scholar]
- 48.Tatematsu, K., N. Yoshimoto, T. Koyanagi, C. Tokunaga, T. Tachibana, Y. Yoneda, M. Yoshida, T. Okajima, K. Tanizawa, and S. Kuroda. 2005. Nuclear-cytoplasmic shuttling of a RING-IBR protein RBCK1 and its functional interaction with nuclear body proteins. J. Biol. Chem. 280:22937-22944. [DOI] [PubMed] [Google Scholar]
- 49.Tatematsu, K., N. Yoshimoto, T. Okajima, K. Tanizawa, and S. Kuroda. 2008. Identification of ubiquitin ligase activity of RBCK1 and its inhibition by splice variant RBCK2 and protein kinase C{beta}. J. Biol. Chem. 283:11575-11585. [DOI] [PubMed] [Google Scholar]
- 50.Theill, L. E., J. L. Castrillo, D. Wu, and M. Karin. 1989. Dissection of functional domains of the pituitary-specific transcription factor GHF-1. Nature 342:945-948. [DOI] [PubMed] [Google Scholar]
- 51.Tian, Y., Y. Zhang, B. Zhong, Y. Y. Wang, F. C. Diao, R. P. Wang, M. Zhang, D. Y. Chen, Z. H. Zhai, and H. B. Shu. 2007. RBCK1 negatively regulates tumor necrosis factor- and interleukin-1-triggered NF-kappaB activation by targeting TAB2/3 for degradation. J. Biol. Chem. 282:16776-16782. [DOI] [PubMed] [Google Scholar]
- 52.Tokunaga, C., S. Kuroda, K. Tatematsu, N. Nakagawa, Y. Ono, and U. Kikkawa. 1998. Molecular cloning and characterization of a novel protein kinase C-interacting protein with structural motifs related to RBCC family proteins. Biochem. Biophys. Res. Commun. 244:353-359. [DOI] [PubMed] [Google Scholar]
- 53.Tootle, T. L., S. J. Silver, E. L. Davies, V. Newman, R. R. Latek, I. A. Mills, J. D. Selengut, B. E. Parlikar, and I. Rebay. 2003. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature 426:299-302. [DOI] [PubMed] [Google Scholar]
- 54.Walker, M. B., and C. B. Kimmel. 2007. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 82:23-28. [DOI] [PubMed] [Google Scholar]
- 55.Wayne, S., N. G. Robertson, F. DeClau, N. Chen, K. Verhoeven, S. Prasad, L. Tranebjarg, C. C. Morton, A. F. Ryan, G. Van Camp, and R. J. Smith. 2001. Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum. Mol. Genet. 10:195-200. [DOI] [PubMed] [Google Scholar]
- 56.Wollmann, Y., U. Schmidt, G. D. Wieland, P. F. Zipfel, H. P. Saluz, and F. Hanel. 2007. The DNA topoisomerase IIbeta binding protein 1 (TopBP1) interacts with poly (ADP-ribose) polymerase (PARP-1). J. Cell. Biochem. 102:171-182. [DOI] [PubMed] [Google Scholar]
- 57.Xu, P. X., J. Adams, H. Peters, M. C. Brown, S. Heaney, and R. Maas. 1999. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet. 23:113-117. [DOI] [PubMed] [Google Scholar]
- 58.Xu, P. X., J. Cheng, J. A. Epstein, and R. L. Maas. 1997. Mouse Eya genes are expressed during limb tendon development and encode a transcriptional activation function. Proc. Natl. Acad. Sci. U. S. A. 94:11974-11979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu, P. X., W. Zheng, C. Laclef, P. Maire, R. L. Maas, H. Peters, and X. Xu. 2002. Eya1 is required for the morphogenesis of mammalian thymus, parathyroid and thyroid. Development 129:3033-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamanaka, K., H. Ishikawa, Y. Megumi, F. Tokunaga, M. Kanie, T. A. Rouault, I. Morishima, N. Minato, K. Ishimori, and K. Iwai. 2003. Identification of the ubiquitin-protein ligase that recognizes oxidized IRP2. Nat. Cell Biol. 5:336-340. [DOI] [PubMed] [Google Scholar]
- 61.Zenke-Kawasaki, Y., Y. Dohi, Y. Katoh, T. Ikura, M. Ikura, T. Asahara, F. Tokunaga, K. Iwai, and K. Igarashi. 2007. Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol. Cell. Biol. 27:6962-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zou, D., C. Erickson, E. H. Kim, D. Jin, B. Fritzsch, and P. X. Xu. 2008. Eya1 gene dosage critically affects the development of sensory epithelia in the mammalian inner ear. Hum. Mol. Genet. 17:3340-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]