Abstract
Enveloped virus particles are formed by budding from infected-cell membranes. For paramyxoviruses, viral matrix (M) proteins are key drivers of virus assembly and budding. However, other paramyxovirus proteins, including glycoproteins, nucleocapsid (NP or N) proteins, and C proteins, are also important for particle formation in some cases. To investigate the role of NP protein in parainfluenza virus 5 (PIV5) particle formation, NP protein truncation and substitution mutants were analyzed. Alterations near the C-terminal end of NP protein completely disrupted its virus-like particle (VLP) production function and significantly impaired M-NP protein interaction. Recombinant viruses with altered NP proteins were generated, and these viruses acquired second-site mutations. Recombinant viruses propagated in Vero cells acquired mutations that mainly affected components of the viral polymerase, while recombinant viruses propagated in MDBK cells acquired mutations that mainly affected the viral M protein. Two of the Vero-propagated viruses acquired the same mutation, V/P(S157F), found previously to be responsible for elevated viral gene expression induced by a well-characterized variant of PIV5, P/V-CPI−. Vero-propagated viruses caused elevated viral protein synthesis and spread rapidly through infected monolayers by direct cell-cell fusion, bypassing the need to bud infectious virions. Both Vero- and MDBK-propagated viruses exhibited infectivity defects and altered polypeptide composition, consistent with poor incorporation of viral ribonucleoprotein complexes (RNPs) into budding virions. Second-site mutations affecting M protein restored interaction with altered NP proteins in some cases and improved VLP production. These results suggest that multiple avenues are available to paramyxoviruses for overcoming defects in M-NP protein interaction.
Virus infections are usually transmitted from cell to cell and from host to host in the form of particles. For many viruses, the particles are enveloped, and the viral membranes are acquired from host cell membranes as the viruses bud. For negative-strand RNA viruses, the assembly and budding of virus particles is organized by viral matrix proteins. These proteins link together the major structural components of the viruses, interacting on the one hand with viral ribonucleoproteins (RNPs) and on the other hand with viral glycoproteins via their cytoplasmic tails (reviewed in reference 39). This results in a coordinated assembly process in which the different viral structural components accumulate together at specific sites on cellular membranes from which budding will occur.
Although viral matrix proteins are key drivers of virus particle formation and, in many cases, virus-like particles (VLPs) can be produced efficiently from cells transfected to produce viral matrix proteins in the absence of other viral proteins (17, 39), several negative-strand RNA viruses appear to require coordination among multiple viral components for efficient budding of particles to occur. For example, the presence of viral glycoproteins and, in particular, the glycoprotein cytoplasmic tails, is important for the efficient budding of influenza virus (21), vesicular stomatitis virus (VSV) (41), and several paramyxoviruses (5, 13, 38, 45). Recent evidence suggests that the hemagglutinin (HA) glycoprotein is the main driving force for the budding of influenza VLPs (7). The nonstructural C protein can function in certain contexts to facilitate the budding of Sendai virus (18, 43). For other viruses, including parainfluenza virus 5 (PIV5) (40), mumps virus (26), Ebola virus (27), and Tacaribe virus (16), a role for nucleocapsid structures in efficient virus budding has been suggested, as the expression of viral nucleocapsid proteins in transfected cells can enhance the budding of VLPs. The mechanism by which some nucleocapsid proteins can enhance particle budding remains to be explored.
Paramyxovirus nucleocapsid (NP or N) proteins function to bind and encapsidate viral genomic and antigenomic RNAs, forming helical structures which are the templates for viral RNA-dependent RNA polymerase complexes. Studies with numerous paramyxoviruses have defined distinct functional regions (core domains and tail domains) within the nucleocapsid proteins. The N-terminal core region generally comprises roughly three-quarters of the protein. This highly structured region is required for RNA binding and encapsidation, while the smaller C-terminal tail is dispensable for RNA binding and encapsidation (1, 3, 9, 22, 32, 34). The C-terminal tail domain appears to be intrinsically disordered (31) and functions to mediate interactions with various viral and host proteins. For example, in the cases of Sendai virus (2), measles virus (24, 30), human respiratory syncytial virus (42), and the henipaviruses (6), interactions with viral P proteins are mediated by the C-terminal tail domains of nucleocapsid proteins, thereby allowing the attachment of viral polymerases to their templates. However, other viruses, including mumps virus and Newcastle disease virus, direct interaction with P proteins via the N-terminal core domains of the nucleocapsid proteins (23, 24). For measles virus, the C-terminal tail region of N protein binds to host factors, including Hsp72, in addition to P protein (52, 53).
Paramyxovirus nucleocapsid proteins also mediate interactions with viral matrix (M) proteins, thereby allowing efficient incorporation of viral genomes into budding virions. For Sendai virus, M protein interaction has been mapped to the C-terminal tail portion of NP protein (8), and for measles virus, pairwise yeast two-hybrid tests and coimmunoprecipitation studies have defined amino acid residues very close to the C-terminal end of N protein that are critical for M protein binding (20).
Here, experiments aimed at defining regions of PIV5 NP protein that are important for its VLP production function were conducted. A region near the C-terminal end of the protein was identified that was critical for both the VLP production function and M protein binding. NP proteins with alterations in this region could still function to allow the formation of templates for the viral polymerase, however. Recombinant viruses with alterations affecting the C-terminal region of NP protein were generated, and these viruses were found to acquire second-site mutations. Characterization of these viruses and their second-site mutations revealed multiple avenues of adaptation which are available to paramyxoviruses for overcoming defective NP-M protein interactions.
MATERIALS AND METHODS
Plasmids.
Plasmids pCAGGS-PIV5 NP, pCAGGS-PIV5 M, and pCAGGS-PIV5 HN have been described before (40), as have pCAGGS-PIV5 L (47) and pCAGGS-PIV5 P (28). Altered PIV5 NP and M cDNAs were generated by PCR mutagenesis of the wild-type (wt) sequences. The resulting cDNAs were subcloned into the eukaryotic expression vector pCAGGS (33), and sequencing of the entire genes was carried out to verify their identities (Macrogen, Inc., South Korea). NP-altered derivatives of the PIV5 genomic clone (strain W3A) were constructed by subcloning altered cDNAs into plasmid pBH276 (19) using the unique AgeI and StuI restriction sites. Cloning details will be provided upon request. The PIV5 minigenome plasmid pSMG-RL and the firefly luciferase control plasmid pT7-F-Luc have been described before (46).
Measurements of VLP production.
293T cells in 6-cm-diameter dishes (70 to 80% confluent) grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) were transfected with pCAGGS plasmids encoding PIV5 proteins to generate VLPs. Transfections were carried out in Opti-MEM using Lipofectamine Plus reagents (Invitrogen, Carlsbad, CA). The quantities of plasmids per dish were as follows: pCAGGS-PIV5 M and derivatives, 400 ng; pCAGGS-PIV5 NP and derivatives, 100 ng; and pCAGGS-PIV5 HN, 1.5 μg.
At 24 h posttransfection (p.t.), the culture medium was replaced with DMEM containing 1/10 the normal amount of methionine and cysteine and 37 μCi of 35S-Promix/ml (Perkin Elmer, Waltham, MA). After an additional 16 h, cell and medium fractions were harvested. For VLP purification, the culture medium was centrifuged at 8,000 × g for 2 min to remove cell debris and then layered onto a 20% sucrose cushion (5 ml in NTE [0.1 M NaCl, 0.01 M Tris-HCl, pH 7.4, 0.001 M EDTA]). After centrifugation at 110,000 × g for 1.5 h, pellets were resuspended in 0.9 ml of NTE and mixed with 2.4 ml of 80% sucrose in NTE. Concentrations of 50% sucrose (3.6 ml) and 10% sucrose (0.6 ml) in NTE were layered on the tops of samples, which were then centrifuged at 110,000 × g for 3 h. An amount of 2 ml was collected from the top of each gradient, and the VLPs contained within this fraction were pelleted by centrifugation at 146,000 × g for 1.5 h. VLP pellets were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer containing 2.5% (wt/vol) dithiothreitol.
Cell lysate preparation and immunoprecipitation of proteins was performed as described previously (40). Monoclonal antibodies used for immunoprecipitation (M-f, HN1b, and NPa, specific to the PIV5 M, HN, and NP proteins, respectively [36]) were kind gifts of Richard Randall (St. Andrews University, St. Andrews, Scotland). The precipitated proteins and VLPs were analyzed by SDS-PAGE using 10% gels. Detection and quantification of protein bands was performed with a Fuji FLA-7000 laser scanner (FujiFilm Medical Systems, Stamford, CT). Budding efficiency was calculated as the quantity of M protein in purified VLPs divided by the quantity of M protein in the corresponding cell lysates and normalized relative to values obtained in control experiments.
Coflotation assays to measure M-NP protein interaction.
293T cells in 10-cm-diameter dishes were transfected with pCAGGS plasmids encoding M and/or NP proteins (1.5 μg and 150 ng of plasmid DNA, respectively). Transfections were carried out in Opti-MEM using Lipofectamine Plus reagents. At 24 h p.t., cells were harvested and resuspended in a hypotonic solution (25 mM NaCl, 25 mM HEPES, pH 7.3, 1 mM phenylmethylsulfonyl fluoride). The cells were subjected to 40 strokes of Dounce homogenization, and the resulting extracts were centrifuged at 1,500 × g for 5 min to remove nuclei and debris. Supernatants were mixed with 2.2 ml of 80% sucrose in NTE. Layers of 50% sucrose (1.6 ml) and 10% sucrose (0.6 ml) in NTE were placed on the tops of samples, which were then centrifuged at 160,000 × g for 4 h in a Beckman SW55Ti rotor. Six equal fractions were collected from the top of each gradient. Proteins from the gradient fractions were fractionated on 10% SDS gels and subjected to immunoblot analysis using the monoclonal antibodies M-f and NPb. Detection and quantification of protein bands were performed with a Fuji FLA-7000 laser scanner. Membrane-bound NP protein was calculated as the amount of NP protein detected in the top three fractions divided by the amount of NP protein detected in all six fractions.
Isolation of nucleocapsid-like structures.
293T cells in 10 cm-diameter dishes were transfected with pCAGGS plasmids to produce NP proteins (200 ng per dish) or M protein (500 ng per dish). Transfection and metabolic labeling of cells were performed as described above for VLP production, except that the labeling period was reduced to 3 h. Cell extracts were prepared by Dounce homogenization as described above. Nucleocapsid-like structures were isolated by centrifugation through CsCl density gradients as described previously (40). Viral proteins were immunoprecipitated from gradient fractions using monoclonal antibodies NPa and M-f. Precipitated proteins were fractionated on 10% SDS gels and visualized using a Fuji FLA-7000 laser scanner.
PIV5 minigenome replication.
Dual-luciferase assays to measure PIV5 minigenome reporter gene expression were performed essentially as described elsewhere (46). BSR-T7 cells, cultured in 24-well plates and at 70% confluence, were transfected using Lipofectamine and Plus reagents according to the manufacturer's instructions. The plasmid quantities used for transfection per well were as follows: pSMG-RL, 200 ng; pT7-F-Luc, 0.1 ng; pCAGGS-PIV5 L, 300 ng; pCAGGS-PIV5 P, 20 ng; and pCAGGS-PIV5 NP or derivatives, 40 ng. At 24 h p.t., cells were lysed in 100 μl of passive lysis buffer (Promega, Madison, WI). Twenty microliters of lysate from each well was subjected to dual luciferase assay, according to the manufacturer's protocol (Promega). A Veritas microplate luminometer (Turner BioSystems, Sunnyvale, CA) was used for luciferase activity measurements. Relative luciferase activity was calculated as Renilla luciferase activity divided by firefly luciferase activity.
Recombinant virus generation.
Infectious PIV5 was recovered from cloned DNA as described previously (19), using a modified procedure which avoids the use of vaccinia virus (47). BSR-T7 cells were transfected with variants of the infectious clone pBH276 having alterations to the NP gene, together with helper plasmids as described previously (47). At 4 days p.t., culture supernatants were harvested, clarified by low-speed centrifugation, and used to infect Vero or MDBK cells. Viruses were propagated by passaging 2 to 3 times in these cells, and titers were determined by plaque assay on BHK-21F cells as described previously (35). For sequencing of virus genomes, total RNA was isolated from infected Vero or MDBK cells and viral genomic RNA was used as template for reverse transcription and PCR amplification as described previously (19). DNA sequencing was performed by the Penn State University Genomics Core Facility. Sequence analysis spanned nucleotides (nt) 121 through 15,201 of the 15,246-nt full-length PIV5 genome.
Measurements of virus multiplication.
Growth curve analysis was performed essentially as described before (38). Vero or MDBK cells, cultured in 24-well plates, were infected with recombinant viruses at a multiplicity of infection (MOI) of 0.01 PFU per cell. After incubation of cells with viruses for 1.5 h at 37°C, the inocula were removed, the cells were washed twice with phosphate-buffered saline (PBS), and 0.5 ml of DMEM supplemented with 2% FBS was added to each well. The infected cell cultures were incubated at 37°C for various time periods (0, 24, 48, 72, 96, 120, or 144 h). Culture supernatants were collected, and 1/10 volume of 10× virus freezing solution (1 M MgSO4, 0.5 M HEPES, pH 7.5) was added. Samples were flash-frozen in a dry ice-ethanol bath. Virus titers were determined by plaque assay on BHK-21F cells as described previously (35).
Syncytia formation.
Vero cells, cultured in 10-cm-diameter dishes and grown to 90% confluence, were infected with recombinant viruses at an MOI of 0.05 PFU/cell. At 1.5 h postinfection (p.i.), the inocula were removed and replaced with 8 ml DMEM supplemented with 2% FBS. At various time intervals, monolayers were visualized using a Nikon Eclipse TS100-F microscope and photographed using a Nikon DS-Fi1 digital camera.
Measurements of virion production and virion infectivity.
MDBK cells in 6-cm-diameter dishes were infected with recombinant viruses at an MOI of 2.0 PFU/cell. Concentration of recombinant virus stocks, to allow infection at this MOI, was carried out via positive pressure filtration using an Amicon stirred-cell 8400 filtration device (Millipore, Bedford, MA). At 17 h p.i., the cells were starved for 30 min by incubation in DMEM lacking cysteine and methionine, after which the cells were incubated for another 30 min in DMEM lacking cysteine and methionine and supplemented with 37 μCi of 35S-Promix/ml. The labeling medium was then replaced with DMEM supplemented with 2% FBS, and the cells were incubated for an additional 5 h. Cell and medium fractions were then harvested. The medium fraction was clarified by low-speed centrifugation, and a 0.1-ml aliquot (5% of the total volume) was removed for titer determination by plaque assay. The remainder of the medium fraction was used for virion purification, which was performed according to the same procedure described above for VLP purification. Cell lysates were prepared and viral proteins were immunoprecipitated as described for VLP purification. PIV5 F protein was detected using Fsol polyclonal antibody (a kind gift of Robert Lamb, Northwestern University, Evanston, IL). The precipitated proteins and purified virions were fractionated on 10% SDS gels and detected using a Fuji FLA-7000 laser scanner.
Amino acid sequence comparisons.
Sequence data used for comparison of paramyxovirus NP/N protein C-terminal regions was derived from files with the following GenBank accession numbers: PIV5, AF052755; mumps virus, AF467767; human PIV2 (hPIV2), M55320; hPIV4, AB543337; Tioman virus, AF298895; Menangle virus, AF326114; Newcastle disease virus, AF064091; measles virus, AB016162; Nipah virus, AF212302; Sendai virus, M30202.
RESULTS
Alterations affecting the C-terminal end of PIV5 NP protein block VLP production function.
To define regions of PIV5 NP protein important for VLP production, truncated proteins were generated. One series of truncations targeted the C-terminal end of the 509-amino-acid protein, and another series targeted the N-terminal end. The expression of the various altered NP proteins in 293T cells was examined by transient transfection followed by immunoblotting (not shown). The truncated proteins illustrated in Fig. 1A were all expressed at levels comparable to the level of wt protein and were selected for further study. More severe truncations (140 or more amino acid residues removed from the C-terminal end or 40 or more amino acid residues removed from the N-terminal end) resulted in either complete or partial loss of protein accumulation, likely as a result of protein instability (data not shown). These polypeptides were not studied further. Assuming that the architecture of PIV5 NP protein is similar to that of other paramyxovirus nucleocapsid proteins, it is likely that C-terminal truncations of roughly 140 amino acid residues or more would begin to encroach into the core region of the protein.
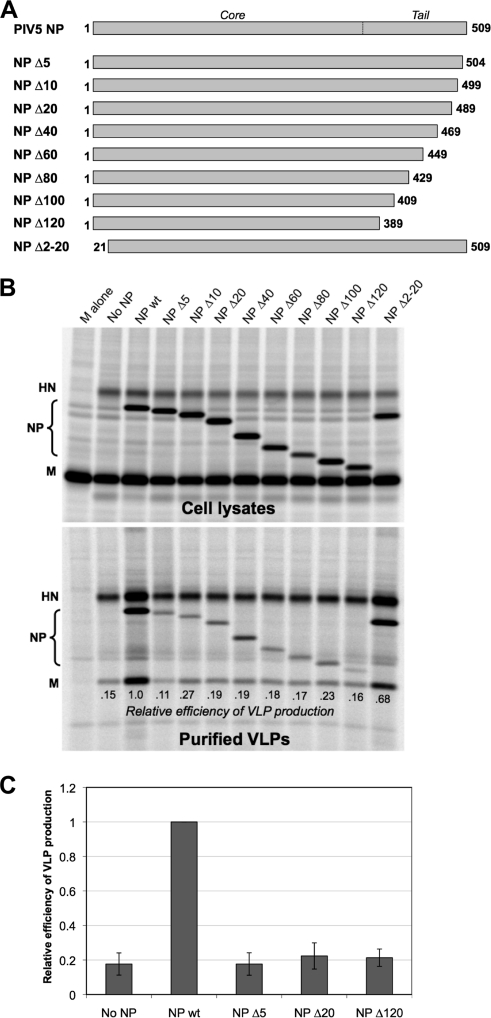
FIG. 1.
Minor truncations to the C-terminal end of PIV5 NP protein disrupt VLP production function. (A) Schematic illustration of truncated PIV5 NP proteins. Numbers indicate the span of amino acid residues that are present in each construct. An approximate boundary between the core and tail regions of NP protein, based on homology with other paramyxovirus nucleocapsid proteins, is indicated. (B) 293T cells were transfected to produce PIV5 M and HN proteins together with the indicated PIV5 NP protein variants. Viral proteins from cell lysates were collected by immunoprecipitation after metabolic labeling of cells. VLPs from culture supernatants were purified by centrifugation through sucrose cushions followed by flotation on sucrose gradients. Viral proteins were separated by SDS-PAGE and visualized using a phosphorimager. Efficiency of VLP production was calculated as the amount of M protein detected in VLPs divided by the amount of M protein detected in the corresponding cell lysate fraction, normalized to the value obtained with wt NP protein. (C) Three independent experiments were performed as described for panel B, and VLP production efficiencies were calculated, with error bars indicating standard deviations.
To test the effect of NP protein truncation on VLP production, the altered NP proteins were coexpressed together with PIV5 M and HN proteins in 293T cells by transient transfection. VLPs were harvested from the culture medium of the transfected cells and purified using sucrose gradients, allowing quantification of VLP release. Consistent with previous observations (40), no VLPs were produced when M protein was expressed by itself (Fig. 1B). Coexpression of HN glycoprotein together with M protein led to VLP production but at a level that was suboptimal. Optimal VLP production was obtained only upon coexpression of the M and HN proteins together with wt NP protein. In this case, VLP production was about 5.5-fold more efficient, on average, than that observed in the absence of NP protein expression (Fig. 1C). NP proteins with C-terminal truncations were completely defective in VLP production function, as VLP production in the presence of the truncated proteins was very similar to the level observed in the absence of NP protein expression (Fig. 1B and C). This was true even for NP Δ5, in which only 5 amino acid residues were removed from the C-terminal end. A different result was obtained with the N-terminal truncation variant, NP Δ2-20. Here, the VLP production function remained mostly intact, indicating that the N terminus of NP protein is not critical for this activity. From the experiments whose results are shown in Fig. 1, we conclude that even minor truncations of the C-terminal end of PIV5 NP protein completely eliminate the VLP production function of the protein.
We further explored the contribution to VLP production made by the 5 C-terminal amino acid residues of PIV5 NP protein. A sequence comparison of this region among several paramyxovirus NP/N proteins is shown in Fig. 2A. The 5 C-terminal amino acid residues are relatively well conserved among closely related paramyxoviruses within the Rubulavirus genus (Fig. 2A, upper portion). Paramyxoviruses outside the Rubulavirus genus (Fig. 2A, lower portion) have NP/N proteins that are more divergent from the PIV5 NP protein near the C-terminal end. A series of substitution mutants targeting the 5 C-terminal amino acid residues was generated, illustrated in Fig. 2B. The majority of these substitutions completely eliminated the VLP production function of NP protein (Fig. 2C and D). In particular, multiple substitutions that affected residues D506, L507, and/or D508 were detrimental to VLP production. One of these substitutions, D508A, resulted in a VLP production defect that was less severe than the others. Substitutions that did not affect residues D506, L507, or D508 did not impair VLP production at all. G505A substitution moderately increased VLP production, and I509A substitution had no significant effect on VLP production. Truncation of 4 amino acid residues from the C terminus (NP Δ4) completely abolished the VLP production function, similar to NP Δ5. Together, the results in Fig. 1 and 2 demonstrate that amino acid residues near the C-terminal end of PIV5 NP protein are important for the VLP production function of the protein.
FIG. 2.
Amino acid residues near the C-terminal end of PIV5 NP protein are important for VLP production. (A) Comparison of paramyxovirus NP/N protein C-terminal amino acid sequences. Sequence identities with PIV5 that are within 5 amino acid residues of the C-terminal end are shown in boldface. (B) Schematic illustrating NP protein amino acid substitutions. (C) 293T cells were transfected to produce PIV5 M and HN proteins together with the indicated NP protein variants, and VLP production was measured as described in the legend to Fig. 1. (D) Relative VLP production efficiency values were calculated from three independent experiments performed as indicated for panel C; error bars indicate standard deviations.
Alterations to the C-terminal end of NP protein impair interaction with M protein.
We tested whether NP protein alterations that affect VLP production also affect other NP protein functions, such as NP protein binding to M protein or the formation of NP-encapsidated RNA templates suitable for the viral polymerase. To test for NP-M protein interaction, we employed a membrane coflotation assay similar to one that has been described previously for measles virus (37). For this interaction, we found coflotation to be a more reliable assay than pulldown-type assays, likely because the NP and M proteins can each self-associate to form dense structures in transfected cells. Here, the basis for membrane coflotation is that M protein intrinsically binds to cellular membranes, whereas NP protein does not (Fig. 3). M-NP protein interaction indirectly brings NP protein to membranes via M protein, allowing coflotation of NP protein with membranes on sucrose gradients. We recovered approximately 28% of the wt NP protein in the membrane-bound (floated) fractions of the gradients when NP protein was coexpressed with M protein (Fig. 3). In contrast, most of the altered NP proteins were defective in membrane coflotation when expressed together with M protein. The only altered NP proteins which exhibited normal M protein binding were NP505A and NP509A, the same two NP protein variants that exhibited normal (or greater than normal) VLP production function (Fig. 2). From the results shown in Fig. 1 to 3, we conclude that alterations near the C-terminal end of NP protein block the VLP production function of the protein and impair interaction with M protein.
FIG. 3.
Alterations near the C-terminal end of PIV5 NP protein impair interaction with M protein. (A) 293T cells were transfected to produce the indicated viral proteins. Detergent-free cell extracts were prepared by Dounce homogenization and placed at the bottoms of sucrose flotation gradients. After centrifugation to allow flotation of membrane-bound components, fractions were collected from the tops of the gradients. Fractions were analyzed by immunoblotting, using antibodies specific to the PIV5 M and NP proteins. (B) NP protein was quantified from immunoblots using a laser scanner. The membrane-bound fraction was calculated as the amount of NP protein detected in the top three fractions divided by the total NP protein detected in all six fractions. Values represent averages from three independent experiments, with error bars indicating standard deviations.
The VLP production function of NP protein is distinct from its ability to form templates for the viral polymerase.
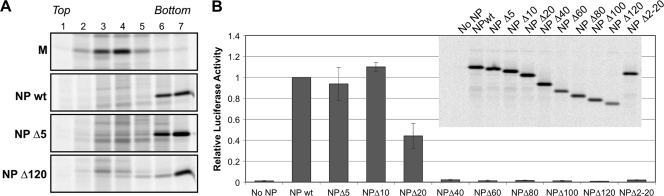
In infected cells, NP proteins function to bind and encapsidate viral genomic and antigenomic RNAs, forming templates for the viral polymerase. We tested whether C-terminal truncations to NP protein affect these functions. First, the ability of truncated NP proteins to form dense structures in transfected cells, consistent with the formation of nucleocapsid-like structures, was assessed. Lysates prepared from transfected cells were centrifuged through CsCl density gradients, a procedure which separates nucleocapsid-like structures from free NP protein (3). The wt PIV5 NP protein has previously been shown to form dense nucleocapsid-like structures in transfected cells (40), and it sedimented to the bottoms of CsCl density gradients as anticipated, while M protein in control experiments failed to sediment to the bottoms of the gradients (Fig. 4A). NP Δ5 and NP Δ120 proteins sedimented to the bottoms of CsCl gradients similarly to wt NP protein (Fig. 4A), as did other NP proteins with intermediate truncations (data not shown). Thus, even severe truncation to the C-terminal end of PIV5 NP protein does not impair its ability to form dense structures in transfected cells. This finding agrees with earlier observations made with other paramyxovirus nucleocapsid proteins, in which RNA encapsidation functions were mapped to the N-terminal core regions and the C-terminal tail regions were found to be dispensable for nucleocapsid-like structure formation (1, 3, 9, 22, 32, 34).
FIG. 4.
Minor truncations to the C-terminal end of PIV5 NP protein do not impair the formation of templates for minigenome reporter expression. (A) 293T cells were transfected to produce the indicated viral proteins. After metabolic labeling with 35S-amino acids, cells were collected and Dounce-homogenized cell extracts were prepared. Cell extracts were placed at the tops of CsCl density gradients and centrifuged, and seven equal gradient fractions were collected. PIV5 proteins were collected from the fractions by immunoprecipitation, fractionated on SDS gels, and detected using a phosphorimager. (B) BSR-T7 cells, stably expressing T7 RNA polymerase, were transfected with a PIV5 minigenome plasmid that contains a Renilla luciferase reporter. Transcription by T7 polymerase results in the production of a negative-sense PIV5 minigenome. In addition to the minigenome plasmid, cells were transfected with additional plasmids to produce PIV5 P and L proteins, together with wt or truncated NP proteins, as indicated. An additional plasmid, encoding firefly luciferase, was included as a transfection efficiency control. At 24 h posttransfection, cell lysates were prepared and luciferase activities were measured using a luminometer. Relative luciferase activity was calculated as Renilla luciferase activity divided by firefly luciferase activity, normalized to the value obtained with wt NP protein. Values represent averages from three independent experiments, with error bars indicating standard deviations. (Inset) NP protein expression levels were verified from replicate transfections of BSR-T7 cells. After metabolic labeling of cells with 35S-amino acids, lysates were prepared and subjected to immunoprecipitation using antibody specific to PIV5 NP protein, followed by SDS-PAGE and detection using a phosphorimager.
To determine if truncated NP proteins can contribute to the formation of templates useable by the viral polymerase, a PIV5 minigenome transcription/replication system was employed. Here, a viral minigenome RNA is produced in which PIV5 leader and trailer sequences flank a single transcriptional unit which encodes Renilla luciferase (29, 46). Reporter gene expression in the transfected cells occurs only in the presence of viral polymerase and only if the minigenome RNA has become encapsidated with NP protein to form a suitable template. Omission of NP protein expression results in no reporter gene expression, while inclusion of wt NP protein results in robust reporter gene expression (Fig. 4B). The substitution of NP Δ5 or NP Δ10 in place of wt NP protein had no significant effect on minigenome reporter gene expression (Fig. 4B). Hence, the VLP production and M protein binding functions of NP protein are separable from the template formation function. Further truncation of NP protein did eventually cause negative effects on template formation ability. NP Δ20 protein was partially defective, while truncation of 40 or more amino acid residues completely abolished the template formation function, judged by minigenome reporter gene expression. The N-terminal NP protein truncation, NP Δ2-20, was also completely defective for minigenome reporter gene expression. To confirm that the defects in minigenome reporter gene expression were not caused by differences in NP protein expression levels, replicate transfections were performed and NP proteins were detected by immunoprecipitation (Fig. 4B, inset). Only very minor fluctuations in NP protein expression levels were observed. NP Δ20 and NP Δ40 proteins were expressed to the same level as the wt NP protein yet failed to support normal minigenome reporter activity. We conclude that the C-terminal tail region of PIV5 NP protein does contain one or more regions important for proper minigenome template formation, with at least one such region located between 10 and 40 amino acid residues distant from the C-terminal end of the protein. However, the 10 amino acid residues at the very C-terminal end of NP protein are dispensable for minigenome template formation. This suggested to us that it may be feasible to recover recombinant viruses harboring certain NP protein variants, such as NP Δ4, that lack VLP production and M-binding function, as these alterations seemingly would not interfere with viral transcription and genome replication.
Recombinant viruses with alterations near the C-terminal end of NP protein acquire second-site mutations.
To probe the importance of the NP protein C-terminal region in the context of a live virus infection, we set out to generate recombinant viruses. A reverse genetics system for PIV5 has been established previously (19). Here, we modified the PIV5 genome plasmid, pBH276, to encode altered NP proteins in place of the wt NP protein. In initial experiments, we attempted to recover virus encoding NP Δ4 protein. The NP Δ4 modification changes the overall PIV5 genome length by a multiple of 6 nucleotides (whereas NP Δ5 modification would not) and thereby conforms to the “rule of six” for paramyxovirus genomes (4). We confirmed that NP Δ4 protein behaves similarly to wt NP protein in minigenome transcription/replication assays (data not shown). BSR-T7 cells were transfected with the modified PIV5 genome plasmid together with support plasmids necessary for the initiation of an infectious cycle (encoding wt NP, P, and L proteins). Approximately 3 days posttransfection, small syncytia were observed within the BSR-T7 monolayers, consistent with successful virus rescue. Supernatants from the transfected BSR-T7 cells were used to inoculate Vero cells for virus propagation and generation of virus stocks. After two to three passages in Vero cells, sufficient titers were achieved (between 104 and 106 PFU/ml) to allow subsequent experiments. Four viruses, designated v1, v2, v6, and v10, were recovered in this way (Table 1). Genome sequence analysis confirmed the presence of the NP Δ4 modification for each of these viruses. However, in each case, second-site mutations were discovered within the viral genomes (Table 1). Multiple viral proteins were affected by the second-site mutations (NP, P, V, M, F, and L).
TABLE 1.
Second-site mutations acquired by NP-altered recombinant viruses
| Virus | Engineered mutation | Second-site mutation(s) | Cell type used for propagation |
|---|---|---|---|
| rPIV5 NP Δ4 v1 | NP Δ4 | NP(S244R), P(K385N), F(R100I), L(G1199D) | Vero |
| rPIV5 NP Δ4 v2 | NP Δ4 | P(S308N), M(L376Q), L(A466T) | Vero |
| rPIV5 NP Δ4 v6 | NP Δ4 | L(L579F) | Vero |
| rPIV5 NP Δ4 v10 | NP Δ4 | NP(A252D), V/P(S157F) | Vero |
| rPIV5 NP L507A v11 | NP L507A | V/P(I159S), L(F2176L) | Vero |
| rPIV5 NP L507A v12 | NP L507A | M(G318D), L(T704A), L(S1562T) | Vero |
| rPIV5 NP D508A v13 | NP D508A | P(K370N) | Vero |
| rPIV5 NP D508A v14 | NP D508A | V/P(S157F) | Vero |
| rPIV5 NP 506,507AA v15 | NP 506,507AA | P(K370T), L(Y1948D) | Vero |
| rPIV5 NP L507A m1 | NP L507A | M(I278 M) | MDBK |
| rPIV5 NP D508A m2 | NP D508A | M(I278 M) | MDBK |
| rPIV5 NP 506,507AA m3 | NP 506,507AA | M(S250I), F(N524S) | MDBK |
We refined our strategy and attempted to recover alternative viruses in which NP protein would be affected by amino acid substitutions rather than the Δ4 truncation. Five additional viruses were recovered, two harboring the L507A substitution, two harboring the D508A substitution, and one harboring the 506,507AA double substitution. Viruses were recovered from transfected BSR-T7 cells and propagated in Vero cells to generate stocks as before. Once again, second-site mutations were observed within each of the viral genomes. Interestingly, all nine viruses which were recovered and propagated in this way were found to have second-site mutations affecting the viral L and/or P proteins, which together make up the viral polymerase complex. Two of these viruses, v10 and v14, acquired a specific mutation, V/P(S157F), that has been characterized before and that affects the shared region of the P and V proteins. Residue S157 of P protein plays a critical role in restraining viral polymerase activity by mediating an interaction with Polo-like kinase 1 (PLK1) (44, 46). Substitutions at position S157, including S157F, prevent PLK1 binding to the viral polymerase and thereby accelerate the replication of viral genomes, resulting in elevated viral gene expression (44, 46). P/V-CPI−, a PIV5 recombinant related to the CPI− natural canine isolate of the virus, harbors the S157F alteration (among others), and this virus causes elevated viral gene expression, induces the expression of host antiviral proteins, and causes rapid cell death (14, 15, 48, 51).
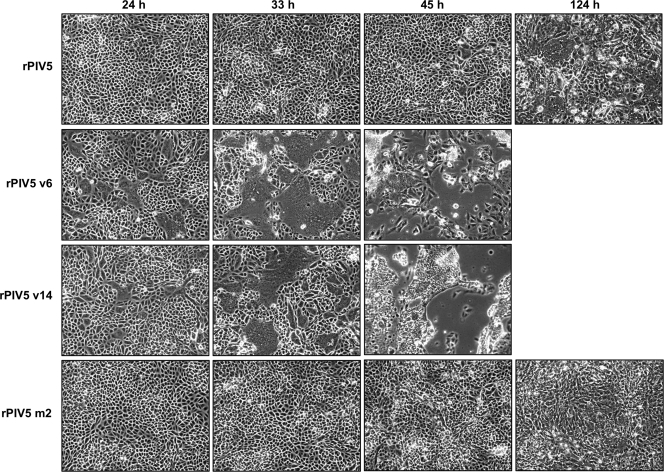
While propagating recombinant viruses in Vero cells, we noticed for all of these viruses that cell-cell fusion appeared to occur very rapidly and extensively compared with that in wt virus infection. To confirm this observation, Vero cells were infected with recombinant viruses, and at various times postinfection, syncytia within the cell monolayers were observed and photographed. We found that infections with Vero-propagated recombinant viruses caused more rapid and extensive syncytia formation than infection with wt virus, especially at early times postinfection (Fig. 5 and data not shown). At later times postinfection, these infected cell monolayers were destroyed, whereas cell monolayers infected with wt PIV5 remained mostly intact (Fig. 5). This observation raises the interesting possibility that defective NP-M protein interactions may have been overcome at least in part through a shift in the mode of virus spread toward one that favors direct cell-to-cell transmission of infection, bypassing the need to assemble and release infectious particles.
FIG. 5.
Rapid and extensive cell-cell fusion by Vero-propagated viruses but not by MDBK-propagated viruses. Vero cells were infected with the indicated viruses at an MOI of 0.05 PFU/cell. At the indicated times p.i., cell monolayers were visualized by phase-contrast microscopy and representative fields were photographed.
We wondered if the route of virus adaptation might be altered if virus propagation following transfection of BSR-T7 cells could be carried out in MDBK cells, which do not readily fuse to form syncytia, instead of Vero cells. Several attempts were made to recover virus harboring the NP Δ4 mutation in this way, but none were successful, even though supernatants from the same transfected BSR-T7 cells readily led to virus recovery when propagated in Vero cells. Additional attempts were made to generate viruses having NP proteins with amino acid substitutions instead of the NP Δ4 truncation, and in three cases, virus was successfully recovered after propagation in MDBK cells (Table 1). Second-site mutations were found in the genomes of all three viruses. However, none of these second-site mutations affected the viral polymerase complex, and none of these viruses caused accelerated syncytia formation in Vero cells (Fig. 5 and data not shown). Instead, all three viruses acquired second-site mutations that affected their M proteins (Table 1). Two of the viruses, m1 and m2, independently acquired the same mutation in M protein, I278M. This is despite the fact that the engineered mutations for the m1 and m2 viruses are different (NP L507A for m1 and NP D508A for m2). The third virus, m3, acquired two mutations. One mutation results in the S250I substitution within M protein, and the other mutation results in the N524S substitution within the F protein cytoplasmic tail (Table 1). In summary, we found that amplification of recombinant virus stocks in MDBK cells resulted in mutations affecting viral components involved in virus assembly, while amplification of stocks in Vero cells resulted in mutations which mostly affected the viral polymerase complex.
Recombinant viruses exhibit altered multiplication kinetics and impaired infectious virion production.
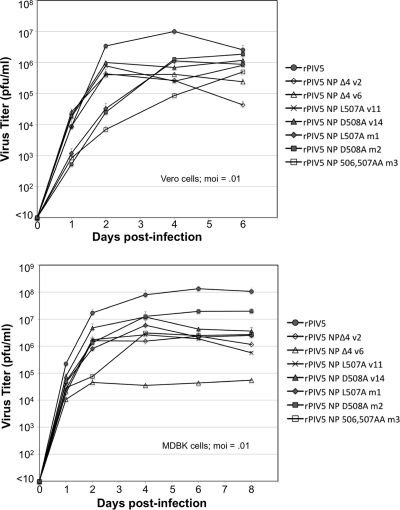
Multiple-step growth curve experiments were performed with mutant viruses both in Vero cells and MDBK cells (Fig. 6). Four of the nine Vero-amplified viruses were analyzed, together with all three MDBK-amplified viruses. In Vero cells, the Vero-amplified viruses spread very quickly compared with the spread of the MDBK-amplified viruses, resulting in higher titers in the culture medium on days 1 and 2 postinfection. At later time points, however, the MDBK-amplified virus titers continued to increase, while Vero-amplified virus titers declined. The most significant overall impairments in virus multiplication were observed for viruses v2, v6, and m3. The peak titers released into cell culture supernatants for these viruses were between 20-fold and 24-fold less than the titer of wt virus. The defects were less severe for viruses v11, v14, m1, and m2, with peak titers between 5-fold and 12-fold less than the wt virus.
FIG. 6.
Altered multiplication kinetics of recombinant viruses. Vero cells (upper panel) or MDBK cells (lower panel) were infected with recombinant viruses at an MOI of 0.01 PFU/cell. Culture medium was harvested at the indicated times, and virus titers were measured by plaque assay on BHK-21F cells. Values represent averages from three independent experiments, with error bars indicating standard deviations.
In many cases, the defects were more severe in MDBK cells than in Vero cells (Fig. 6). By far the most debilitated virus in MDBK cells was v6, with a peak titer of 4.5 × 104 PFU/ml, more than 2,000-fold less than the peak titer reached by wt virus (1.1 × 108 PFU/ml). This virus harbors the NP Δ4 truncation and acquired only a single second-site mutation, affecting its L protein. Other viruses (v2, v11, and m3) exhibited significant but less extreme defects in multiplication (peak titers between 35-fold and 50-fold less than the titer of wt virus). The viruses with the least severe defects in MDBK cells were v14 and m2, with peak titers reduced by less than 10-fold compared to the titer of wt virus. Both of these viruses harbor the NP 508A mutation. Overall, viruses which were engineered to carry the NP 508A mutation were among the least impaired in both cell types. Viruses which were engineered to carry the NP Δ4 mutation were among the most severely impaired in both cell types.
To assess the quantity and quality of virus particle formation, MDBK cells were infected with recombinant viruses at an MOI of 2 PFU/cell, and physical particle release, as well as infectious particle release, was measured (Fig. 7 and Table 2). In one case, that of virus v6, this analysis could not be carried out in a meaningful way because the majority of infected MDBK cells were killed within the timeframe of the experiment. In a replicate experiment, we observed that the viability at 26 h postinfection of MDBK cells infected with the v6 virus, judged by trypan blue staining, was 45% that of mock-infected cells, while the viability was at least 77% for cells infected with the other viruses analyzed in Fig. 7. Thus, a significant component of the severe defect for this virus in MDBK cells is likely related to rapid killing of infected cells.
FIG. 7.
Virions produced upon infection with recombinant viruses. MDBK cells were infected with recombinant viruses at an MOI of 2.0 PFU/cell. Infected cells were metabolically labeled with 35S-amino acids, and 5 h later, cell and medium fractions were harvested. Viral proteins from cell lysates were collected by immunoprecipitation. Virions from culture supernatants were purified by centrifugation through sucrose cushions followed by flotation on sucrose gradients and then loaded directly onto SDS gels. Viral proteins were separated by SDS-PAGE and visualized using a phosphorimager. The data shown are representative of two independent experiments.
TABLE 2.
Quantity and quality of virions produced upon infection of MDBK cells with recombinant viruses
| Virus | Budding efficiencya | Physical particles releasedb | Infectious particles releasedc | PFU/particle ratiod | Virion NP/M ratioe |
|---|---|---|---|---|---|
| rPIV5 | 1.0 | 1.0 | 1.5 × 107 | 1.0 | 1.0 |
| rPIV5 NP L507A v12 | 0.72 | 1.3 | 3.0 × 106 | 0.16 | 0.37 |
| rPIV5 NP D508A v14 | 0.80 | 3.0 | 1.4 × 107 | 0.31 | 0.69 |
| rPIV5 NP D508A m2 | 0.96 | 0.72 | 1.4 × 106 | 0.13 | 0.40 |
| rPIV5 NP 506,507AA m3 | 0.57 | 0.44 | 5.0 × 105 | 0.08 | 0.32 |
Calculated as the quantity of M protein detected in purified virions divided by the quantity of M protein detected in cell lysates, normalized to the value obtained with rPIV5.
Calculated as the quantity of M protein detected in purified virions, normalized to the value obtained with rPIV5.
Calculated as the total number of PFU detected in the culture medium following the 5-h labeling period.
Calculated as infectious particles released divided by physical particles released, normalized to the value obtained with rPIV5.
Calculated as the quantity of NP protein detected in purified virions divided by the quantity of M protein detected in purified virions, normalized to the value obtained with rPIV5.
Of the remaining viruses which were analyzed, the one with the most severe defects was m3. Although physical particle release and budding efficiency (particle release normalized to M protein expression) were reduced for this virus, these minor reductions did not account for the large decrease in infectious particle release. Rather, the main defect for this virus is the budding of particles which are noninfectious, judged by a reduction in PFU/particle ratio by more than 10-fold compared with that of wt virus (Table 2). Virions released after infection with the m3 virus had a 3-fold reduction in NP/M protein ratio compared with that of wt virions (Table 2). This suggests a defect in the packaging of RNPs into budding virions, consistent with poor NP-M protein interaction. Virus m2 displayed characteristics that were very similar to those of the m3 virus but with reduced severity. Here, the quantity of particle budding was not significantly different from that of the wt virus, but particle infectivity was reduced to 13% of the infectivity of the wt virus. This was coupled with a 2.5-fold reduction in virion NP/M protein ratio.
Viruses v12 and v14 caused elevated expression of viral proteins in the infected MDBK cells (Fig. 7). The S157F mutation found in the V/P gene of virus v14 has already been linked to elevated viral gene expression (46), and here, we observed M and HN protein expression levels that were 3.8-fold and 2.7-fold greater, respectively, than those found in wt virus-infected cells. Infection with virus v14 also resulted in an F protein expression level that was 1.6-fold greater than that found in wt virus-infected cells. It is possible that the elevated levels of F protein at least partially account for the rapid syncytia formation observed in cells infected with this virus (Fig. 5). Elevated viral protein expression was apparent but less pronounced after infection of MDBK cells with virus v12. M and HN protein expression levels were 1.8-fold and 1.9-fold greater, respectively, than those found in wt virus-infected cells, although in this case, no significant change in F protein expression could be observed (1.1-fold increase compared to the level in wt infection). The quantity of physical particle release was enhanced for virus v14 compared with that of wt virus (Fig. 7 and Table 2). However, the observed 3-fold increase in particle release was not due to increased budding efficiency and instead was likely a function of elevated M protein expression inside the infected cells. Furthermore, the virus buds the same amount of infectious particles as the wt virus, despite the elevated particle production. Hence, the extra particles released during infection with virus v14 are noninfectious, resulting in a PFU/particle ratio that is 3-fold lower than that of wt virus (Table 2). Despite this defect, virus v14 was the least impaired of the viruses subjected to this analysis. Virus v12 exhibited an intermediate level of impairment, similar to virus m2. The particle release quantity was similar to that of wt virus, but the PFU/particle ratio was reduced to 16% of the wt level. This was coupled with a 2.7-fold reduction in the virion NP/M protein ratio. Overall, we found that the main defect exhibited by these viruses was poor infectivity of virions, to various degrees. The severity of the infectivity defects correlated with reductions in virion NP/M protein ratios. The nature of this defect is suggestive of a problem with the incorporation of RNPs into budding virions. From this analysis, it was difficult to directly confirm the conclusion made from the data in Fig. 1 and 2, that alterations to NP protein reduce the quantity of particle release. This could be due to the nature of the second-site mutations. Viruses v12 and v14 affected M protein expression levels, which in turn influence particle production. Viruses v12, m2, and m3 harbor altered M proteins, and the alterations could potentially affect the quantity of particle release.
Second-site mutations to M protein restore NP protein interaction and VLP production.
Several of the second-site mutations acquired by recombinant viruses affected M protein. To test if M protein mutations could restore M-NP protein interactions, we generated cDNAs corresponding to the I278M and G318D mutant M proteins found in viruses m1, m2, and v12. Altered M proteins were expressed in 293T cells by transient transfection, together with NP proteins, and M-NP protein interactions were assessed by coflotation with membranes (Fig. 8). The results suggest that at least one of the viruses, m2, likely adapted through direct restoration of M-NP protein interaction, as the I278M M protein induced coflotation of the mutant NP protein 508A (Fig. 8A), whereas the NP 508A protein does not cofloat with wt M protein (Fig. 3). The gain in the ability to bind NP 508A protein did not appear to be the result of a specificity switch, as binding to wt NP protein remained intact. In fact, binding to wt NP protein appeared to be improved (Fig. 3 and 8A). The I278M M protein was not able to induce other NP protein variants, such as NP Δ4, to cofloat. Even the NP 507A protein tested negative for coflotation with the I278M M protein (Fig. 8A). This was somewhat surprising and raises the question of how acquisition of the I278M M protein provides any benefit to virus m1, which was engineered to contain the NP 507A protein.
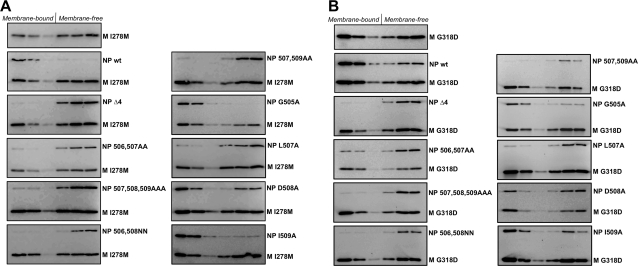
FIG. 8.
Second-site mutations to M protein restore interaction with NP 508A protein. 293T cells were transfected to produce the indicated NP protein variants together with M protein harboring the I278M second-site mutation (A) or M protein harboring the G318D second-site mutation (B). Detergent-free cell extracts were prepared by Dounce homogenization, and coflotation analysis was performed as described in the legend to Fig. 3. The data shown are representative of two independent experiments.
Similar results were obtained with the G318D M protein. This protein has gained the ability to induce coflotation of NP 508A but is not capable of inducing any of the other defective NP proteins to cofloat (Fig. 8B). Again, this was somewhat surprising, because virus v12 which acquired G318D M protein was not engineered to contain NP 508A. It was engineered to contain NP 507A (Table 1). One possible explanation is that the NP 508A substitution impairs M-NP protein interaction less severely than the other NP protein alterations but that this difference was not discernible using the coflotation assay, perhaps because the assay is only capable of detecting the most robust interactions that can survive gradient centrifugation. Consistent with this interpretation, NP 508A protein showed only a moderate defect for VLP production (Fig. 2C and D), whereas it appeared to be completely defective for M protein binding by coflotation (Fig. 3). Under this scenario, second-site mutations could generally enhance M-NP protein binding and improve virus infectivity, but only in the case of NP 508A protein would the interaction be restored to a level of robustness that would allow coflotation.
We tested whether second-site mutations to M protein allow efficient VLP production even in the presence of altered NP proteins. Altered M proteins (I278M or G318D) were expressed in 293T cells together with altered NP proteins (L507A or D508A) and wt HN protein for VLP production (Fig. 9). VLP production in the presence of altered NP proteins was substantially improved as a result of M protein second-site mutations (Fig. 9, compare lanes 7, 8, 11, and 12 to lanes 3 and 4). This is consistent with a partial restoration of M-NP protein interaction, even in the case of NP 507A. Some of the improvement in VLP production appeared to be independent of NP protein. Both of the altered M proteins direct VLP production that is reasonably efficient (but not optimal) even in the absence of NP protein (Fig. 9, lanes 5 and 9). VLP production directed by I278M M protein is further enhanced (by more than 5-fold) upon the inclusion of wt NP protein (lane 6). This results in VLP production that is hyperefficient compared to that which can be achieved with wt M protein (lane 2). In contrast, VLP production directed by G318D M protein is only enhanced by approximately 2-fold upon the inclusion of wt NP protein (lane 10), resulting in a VLP production efficiency that is only slightly elevated compared to that achieved with wt M protein. In summary, second-site mutations to M proteins improved VLP production, and the improvements consist of both NP-dependent and NP-independent components.
FIG. 9.
Second-site mutations to M protein improve VLP production. 293T cells were transfected to produce HN protein together with the indicated M and NP proteins. VLP production was measured as described in the legend to Fig. 1. The data shown are representative of two independent experiments.
DISCUSSION
In this study, we have defined a region of PIV5 NP protein, very close to the C-terminal end, that is critical for its virus assembly functions. NP protein truncations or substitutions that affected residues D506, L507, and/or D508 impaired both M protein interaction and VLP production but did not impair replication and transcription from a PIV5 minigenome. These findings are consistent with those described in a recent report on measles virus N protein. There, pairwise yeast two-hybrid tests together with coimmunoprecipitation experiments defined amino acid residues near the C-terminal end of N protein that are critical for M protein binding (20). The removal of a single amino acid residue from the C-terminal end of N protein did not impair M protein binding, but the removal of 2 or more amino acid residues resulted in failure to bind M protein (20). Hence, multiple paramyxoviruses appear to direct M protein binding via the extreme C-terminal regions of their NP/N proteins.
PIV5 recombinants harboring altered NP proteins acquired second-site mutations, and we were unable to generate NP-altered recombinant viruses lacking second-site mutations. We were also unable to propagate recombinant viruses in Vero or MDBK cells if the viruses were plaque purified after the initial recovery from BSR-T7 cells, suggesting that heterogeneity within the initially recovered virus population may have been necessary for subsequent propagation. Even after the acquisition of adaptive mutations, many recombinant viruses exhibited significant defects in growth curve experiments. It is likely, based on these findings, that impaired NP-M protein interaction is highly detrimental to PIV5 fitness. For measles virus, in contrast, a recombinant virus was recovered that harbors N protein with 3 amino acid residues removed from the C-terminal end, and there was no reported acquisition of second-site mutations (20). This altered virus replicated much less effectively than the parental virus in both Vero/hSLAM and CV-1/hSLAM cells (20). It should be noted that the mode of spread even for wt measles virus is shifted toward direct cell-to-cell transmission, compared to the mode of spread of many other paramyxoviruses, such as PIV5. It is possible that this could have partially alleviated any selective pressure and allowed the virus to more readily cope with NP-M protein interaction defects without the benefit of adaptive mutations.
Two of the viruses generated in this study, v10 and v14, independently acquired the same second-site mutation that is known to be involved in the regulation of viral polymerase activity. The mutation, S157F, affects the shared region of the viral P and V proteins and is one of six amino acid residues that is altered in the PIV5 variant, P/V-CPI− (48). This virus exhibits elevated expression of viral proteins, induces the expression of antiviral responses, and causes cell death more rapidly than wt PIV5 infection (14, 15, 48, 51). Interestingly, both the alterations to V protein and the alterations to P protein contribute to these phenotypes (11). The elevated gene expression phenotype has been attributed to the substitution at residue 157 of P protein, based on results obtained with minigenome reporter assays (46). P protein residue S157 was found to be phosphorylated, and this phosphorylation is required to allow binding of the host kinase PLK1 (44, 46). Upon binding to P protein at residue S157, PLK1 phosphorylates P protein at another residue (S308), which has the effect of downregulating viral gene expression (44). Consistent with these earlier findings, we observed that virus v14 caused increased expression of viral proteins compared with their expression in wt virus. We also observed elevated protein expression levels upon infection of cells with all other Vero-propagated recombinant viruses that were examined (v1, v2, and v12) (Fig. 7 and data not shown). Indeed, all of the recombinant viruses generated in this study which were propagated in Vero cells acquired second-site mutations affecting the viral polymerase subunits P and/or L (Table 1). We hypothesize that propagation of viruses defective in NP-M interaction in Vero cells results in a selective pressure that favors polymerase mutations that elevate viral gene expression. Viruses which were propagated in MDBK cells instead of Vero cells did not acquire polymerase mutations and did not induce elevated viral gene expression. One consequence of elevated viral gene expression is overproduction of the viral fusion (F) protein. The extent of PIV5 F protein-mediated membrane fusion has been shown to be dependent on the cell surface density of F protein (12), and indeed, we observed extensive and accelerated cell-cell fusion after infection with recombinant viruses harboring polymerase mutations. Thus, polymerase mutations may have benefited these viruses during their propagation in cell culture, by providing an alternative route for virus spread that does not rely on the formation of infectious virions. It is possible that the extent of cell-cell fusion induced by these viruses could have been even further accelerated as a direct consequence of M-NP protein interaction defects. Work with canine distemper virus has shown that M protein expression negatively affects fusion directed by the viral F and H proteins and that this negative effect is potentiated by the viral N protein (49). Hence, the mere absence of RNP structures at virus assembly sites could have contributed to the observed increase in cell-cell fusion to some extent.
Although polymerase mutations likely were beneficial to recombinant viruses through providing an alternative means of spread in cell cultures, it appears that these mutations must have provided other benefits as well. For example, virus v14 was among the least-defective viruses in terms of particle production, even in comparison with viruses m2 and m3, which harbor adaptive mutations in their M proteins (Table 2). Particle production was measured after infection of MDBK cells, which do not readily form syncytia, yet virus v14 exhibited greater particle release and improved particle infectivity compared with these other recombinant viruses. In this context, increased expression of the viral M protein may have been beneficial to virus v14. Paramyxovirus particle production is directly influenced by the level of M protein accumulation (25, 50), and in fact, virus v14 released a large quantity of particles even in comparison to the particle production of wt virus (Table 2). It is possible that elevated M protein expression levels counteracted any negative effects on particle release caused by the engineered NP protein mutations. In addition to the elevated viral protein production, infection with virus v14 may have caused increased accumulation of viral RNPs within the infected cells. Experiments with PIV5 minigenome reporter systems have demonstrated that elevated gene expression upon infection with P/V-CPI− is caused indirectly through an increase in genome replication and not by a direct effect on viral transcription (46). Hence, elevated particle production may be coupled with elevated RNP concentration in cells infected with virus v14, and this could favor the production of infectious particles in which the RNPs are incorporated by chance, without the benefit of a strong NP-M interaction. Although elevated viral gene expression is possibly beneficial for virion production, it may come at a cost, as the P/V alterations found in virus P/V-CPI− facilitate the triggering of host antiviral responses (14, 48, 51). This effect could have been mitigated during the generation of recombinant viruses here, however, as most were propagated in Vero cells, which do not produce interferon.
An important component of viral M protein function involves establishing protein-protein and protein-lipid interactions that organize virus assembly. These include M protein interactions with cell membranes, with viral glycoproteins, with viral nucleocapsids, and with other M proteins to allow self-association (39). However, it has been challenging to map the regions of M proteins that are important for each of these protein interactions. Truncation analysis frequently provides limited benefit, because the truncated M proteins oftentimes cannot be stably expressed in transfected cells. Even alanine scanning has been problematic, because a substantial fraction of substitution mutants are unstable and potentially misfolded (Ming Li and Anthony P. Schmitt, unpublished observations). Recently, Dancho et al. employed a NAAIRS scanning approach, designed to minimize the risk of protein misfolding, to fine-map sites within the M protein of VSV that are responsible for the membrane binding function, as well as interaction with viral nucleocapsids. Distinct but overlapping sequences within the N-terminal portion of M protein were identified, with sequences affecting plasma membrane association and VLP release being slightly more N terminal than those affecting nucleocapsid association (10). In the present study, second-site mutations, several affecting the viral M protein, were identified in PIV5 recombinants engineered to have defective NP-M protein interactions. The M protein substitutions targeted the C-terminal half of the protein (residues 250, 278, 318, and 376 of the 377-amino-acid M protein), and the substitutions at positions 278 and 318 were confirmed to have effects on NP protein interaction and VLP production. These residues may provide a useful starting point for efforts to fine-map regions within PIV5 M protein that are important for nucleocapsid association.
Acknowledgments
We thank Rick Randall and Bob Lamb for PIV5 antibody reagents. We are grateful to Khalid Timani and Biao He for assistance with minigenome reporter assays.
This work was supported by research grant AI-070925 from the National Institute of Allergy and Infectious Diseases to A.P.S.
Footnotes
Published ahead of print on 13 October 2010.
REFERENCES
- 1.Bankamp, B., S. M. Horikami, P. D. Thompson, M. Huber, M. Billeter, and S. A. Moyer. 1996. Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology 216:272-277. [DOI] [PubMed] [Google Scholar]
- 2.Buchholz, C. J., C. Retzler, H. E. Homann, and W. J. Neubert. 1994. The carboxy-terminal domain of Sendai virus nucleocapsid protein is involved in complex formation between phosphoprotein and nucleocapsid-like particles. Virology 204:770-776. [DOI] [PubMed] [Google Scholar]
- 3.Buchholz, C. J., D. Spehner, R. Drillien, W. J. Neubert, and H. E. Homann. 1993. The conserved N-terminal region of Sendai virus nucleocapsid protein NP is required for nucleocapsid assembly. J. Virol. 67:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calain, P., and L. Roux. 1993. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J. Virol. 67:4822-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, Y. P., C. L. Koh, S. K. Lam, and L. F. Wang. 2004. Mapping of domains responsible for nucleocapsid protein-phosphoprotein interaction of Henipaviruses. J. Gen. Virol. 85:1675-1684. [DOI] [PubMed] [Google Scholar]
- 7.Chen, B. J., G. P. Leser, E. Morita, and R. A. Lamb. 2007. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 81:7111-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coronel, E. C., T. Takimoto, K. G. Murti, N. Varich, and A. Portner. 2001. Nucleocapsid incorporation into parainfluenza virus is regulated by specific interaction with matrix protein. J. Virol. 75:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curran, J., H. Homann, C. Buchholz, S. Rochat, W. Neubert, and D. Kolakofsky. 1993. The hypervariable C-terminal tail of the Sendai paramyxovirus nucleocapsid protein is required for template function but not for RNA encapsidation. J. Virol. 67:4358-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dancho, B., M. O. McKenzie, J. H. Connor, and D. S. Lyles. 2009. Vesicular stomatitis virus matrix protein mutations that affect association with host membranes and viral nucleocapsids. J. Biol. Chem. 284:4500-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dillon, P. J., and G. D. Parks. 2007. Role for the phosphoprotein P subunit of the paramyxovirus polymerase in limiting induction of host cell antiviral responses. J. Virol. 81:11116-11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutch, R. E., S. B. Joshi, and R. A. Lamb. 1998. Membrane fusion promoted by increasing surface densities of the paramyxovirus F and HN proteins: comparison of fusion reactions mediated by simian virus 5 F, human parainfluenza virus type 3 F, and influenza virus HA. J. Virol. 72:7745-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouillot-Coriou, N., and L. Roux. 2000. Structure-function analysis of the Sendai virus F and HN cytoplasmic domain: different role for the two proteins in the production of virus particle. Virology 270:464-475. [DOI] [PubMed] [Google Scholar]
- 14.Gainey, M. D., P. J. Dillon, K. M. Clark, M. J. Manuse, and G. D. Parks. 2008. Paramyxovirus-induced shutoff of host and viral protein synthesis: role of the P and V proteins in limiting PKR activation. J. Virol. 82:828-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gainey, M. D., M. J. Manuse, and G. D. Parks. 2008. A hyperfusogenic F protein enhances the oncolytic potency of a paramyxovirus simian virus 5 P/V mutant without compromising sensitivity to type I interferon. J. Virol. 82:9369-9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groseth, A., S. Wolff, T. Strecker, T. Hoenen, and S. Becker. 2010. Efficient budding of the Tacaribe virus matrix protein Z requires the nucleoprotein. J. Virol. 84:3603-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison, M. S., T. Sakaguchi, and A. P. Schmitt. 2010. Paramyxovirus assembly and budding: building particles that transmit infections. Int. J. Biochem. Cell Biol. 42:1416-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasan, M. K., A. Kato, M. Muranaka, R. Yamaguchi, Y. Sakai, I. Hatano, M. Tashiro, and Y. Nagai. 2000. Versatility of the accessory C proteins of Sendai virus: contribution to virus assembly as an additional role. J. Virol. 74:5619-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, B., R. G. Paterson, C. D. Ward, and R. A. Lamb. 1997. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237:249-260. [DOI] [PubMed] [Google Scholar]
- 20.Iwasaki, M., M. Takeda, Y. Shirogane, Y. Nakatsu, T. Nakamura, and Y. Yanagi. 2009. The matrix protein of measles virus regulates viral RNA synthesis and assembly by interacting with the nucleocapsid protein. J. Virol. 83:10374-10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, H., G. P. Leser, J. Zhang, and R. A. Lamb. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 16:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kho, C. L., W. S. Tan, B. T. Tey, and K. Yusoff. 2003. Newcastle disease virus nucleocapsid protein: self-assembly and length-determination domains. J. Gen. Virol. 84:2163-2168. [DOI] [PubMed] [Google Scholar]
- 23.Kho, C. L., W. S. Tan, B. T. Tey, and K. Yusoff. 2004. Regions on nucleocapsid protein of Newcastle disease virus that interact with its phosphoprotein. Arch. Virol. 149:997-1005. [DOI] [PubMed] [Google Scholar]
- 24.Kingston, R. L., W. A. Baase, and L. S. Gay. 2004. Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins. J. Virol. 78:8630-8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo, T., T. Yoshida, N. Miura, and M. Nakanishi. 1993. Temperature-sensitive phenotype of a mutant Sendai virus strain is caused by its insufficient accumulation of the M protein. J. Biol. Chem. 268:21924-21930. [PubMed] [Google Scholar]
- 26.Li, M., P. T. Schmitt, Z. Li, T. S. McCrory, B. He, and A. P. Schmitt. 2009. Mumps virus matrix, fusion, and nucleocapsid proteins cooperate for efficient production of virus-like particles. J. Virol. 83:7261-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Licata, J. M., R. F. Johnson, Z. Han, and R. N. Harty. 2004. Contribution of Ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J. Virol. 78:7344-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, G. Y., and R. A. Lamb. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 74:9152-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, Y., F. Horvath, J. A. Aligo, R. Wilson, and B. He. 2005. The role of simian virus 5 V protein on viral RNA synthesis. Virology 338:270-280. [DOI] [PubMed] [Google Scholar]
- 30.Liston, P., R. Batal, C. DiFlumeri, and D. J. Briedis. 1997. Protein interaction domains of the measles virus nucleocapsid protein (NP). Arch. Virol. 142:305-321. [DOI] [PubMed] [Google Scholar]
- 31.Longhi, S., V. Receveur-Brechot, D. Karlin, K. Johansson, H. Darbon, D. Bhella, R. Yeo, S. Finet, and B. Canard. 2003. The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 278:18638-18648. [DOI] [PubMed] [Google Scholar]
- 32.Murphy, L. B., C. Loney, J. Murray, D. Bhella, P. Ashton, and R. P. Yeo. 2003. Investigations into the amino-terminal domain of the respiratory syncytial virus nucleocapsid protein reveal elements important for nucleocapsid formation and interaction with the phosphoprotein. Virology 307:143-153. [DOI] [PubMed] [Google Scholar]
- 33.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 34.Ong, S. T., K. Yusoff, C. L. Kho, J. O. Abdullah, and W. S. Tan. 2009. Mutagenesis of the nucleocapsid protein of Nipah virus involved in capsid assembly. J. Gen. Virol. 90:392-397. [DOI] [PubMed] [Google Scholar]
- 35.Paterson, R. G., and R. A. Lamb. 1993. The molecular biology of influenza viruses and paramyxoviruses, p. 35-73. In A. Davidson and R. M. Elliott (ed.), Molecular virology: a practical approach. IRL/Oxford University Press, Oxford, United Kingdom.
- 36.Randall, R. E., D. F. Young, K. K. Goswami, and W. C. Russell. 1987. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J. Gen. Virol. 68:2769-2780. [DOI] [PubMed] [Google Scholar]
- 37.Runkler, N., C. Pohl, S. Schneider-Schaulies, H.-D. Klenk, and A. Maisner. 2007. Measles virus nucleocapsid transport to the plasma membrane requires stable expression and surface accumulation of the viral matrix protein. Cell. Microbiol. 9:1203-1214. [DOI] [PubMed] [Google Scholar]
- 38.Schmitt, A. P., B. He, and R. A. Lamb. 1999. Involvement of the cytoplasmic domain of the hemagglutinin-neuraminidase protein in assembly of the paramyxovirus simian virus 5. J. Virol. 73:8703-8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt, A. P., and R. A. Lamb. 2004. Escaping from the cell: assembly and budding of negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:145-196. [DOI] [PubMed] [Google Scholar]
- 40.Schmitt, A. P., G. P. Leser, D. L. Waning, and R. A. Lamb. 2002. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J. Virol. 76:3952-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnell, M. J., L. Buonocore, E. Boritz, H. P. Ghosh, R. Chernish, and J. K. Rose. 1998. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 17:1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stokes, H. L., A. J. Easton, and A. C. Marriott. 2003. Chimeric pneumovirus nucleocapsid (N) proteins allow identification of amino acids essential for the function of the respiratory syncytial virus N protein. J. Gen. Virol. 84:2679-2683. [DOI] [PubMed] [Google Scholar]
- 43.Sugahara, F., T. Uchiyama, H. Watanabe, Y. Shimazu, M. Kuwayama, Y. Fujii, K. Kiyotani, A. Adachi, N. Kohno, T. Yoshida, and T. Sakaguchi. 2004. Paramyxovirus Sendai virus-like particle formation by expression of multiple viral proteins and acceleration of its release by C protein. Virology 325:1-10. [DOI] [PubMed] [Google Scholar]
- 44.Sun, D., P. Luthra, Z. Li, and B. He. 2009. PLK1 down-regulates parainfluenza virus 5 gene expression. PLoS Pathog. 5:e1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tahara, M., M. Takeda, and Y. Yanagi. 2007. Altered interaction of the matrix protein with the cytoplasmic tail of hemagglutinin modulates measles virus growth by affecting virus assembly and cell-cell fusion. J. Virol. 81:6827-6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Timani, K. A., D. Sun, M. Sun, C. Keim, Y. Lin, P. T. Schmitt, A. P. Schmitt, and B. He. 2008. A single amino acid residue change in the P protein of parainfluenza virus 5 elevates viral gene expression. J. Virol. 82:9123-9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waning, D. L., A. P. Schmitt, G. P. Leser, and R. A. Lamb. 2002. Roles for the cytoplasmic tails of the fusion and hemagglutinin-neuraminidase proteins in budding of the paramyxovirus simian virus 5. J. Virol. 76:9284-9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wansley, E. K., and G. D. Parks. 2002. Naturally occurring substitutions in the P/V gene convert the noncytopathic paramyxovirus simian virus 5 into a virus that induces alpha/beta interferon synthesis and cell death. J. Virol. 76:10109-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiener, D., P. Plattet, P. Cherpillod, L. Zipperle, M. G. Doherr, M. Vandevelde, and A. Zurbriggen. 2007. Synergistic inhibition in cell-cell fusion mediated by the matrix and nucleocapsid protein of canine distemper virus. Virus Res. 129:145-154. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida, T., Y. Nagai, K. Maeno, M. Linuma, M. Hamaguchi, T. Matsumoto, S. Nagayoshi, and M. Hoshino. 1979. Studies on the role of M protein in virus assembly using a ts mutant of HVJ (Sendai virus). Virology 92:139-154. [DOI] [PubMed] [Google Scholar]
- 51.Young, V. A., P. J. Dillon, and G. D. Parks. 2006. Variants of the paramyxovirus simian virus 5 with accelerated or delayed viral gene expression activate proinflammatory cytokine synthesis. Virology 350:90-102. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, X., J. M. Bourhis, S. Longhi, T. Carsillo, M. Buccellato, B. Morin, B. Canard, and M. Oglesbee. 2005. Hsp72 recognizes a P binding motif in the measles virus N protein C-terminus. Virology 337:162-174. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, X., C. Glendening, H. Linke, C. L. Parks, C. Brooks, S. A. Udem, and M. Oglesbee. 2002. Identification and characterization of a regulatory domain on the carboxyl terminus of the measles virus nucleocapsid protein. J. Virol. 76:8737-8746. [DOI] [PMC free article] [PubMed] [Google Scholar]