Abstract
HIV-1 RNA undergoes a complex splicing process whereby over 40 different mRNA species are produced by alternative splicing. In addition, approximately half of the RNA transcripts remain unspliced and either are used to encode Gag and Gag-Pol proteins or are packaged into virions as genomic RNA. It has previously been shown that HIV-1 splicing is regulated by cis elements that bind to cellular factors. These factors either enhance or repress definition of exons that are flanked by the HIV-1 3′ splice sites. Here we report that expression of modified U1 snRNPs with increased affinity to HIV-1 downstream 5′ splice sites and to sequences within the first tat coding exon act to selectively increase splicing at the upstream 3′ splice sites in cotransfected 293T cells. This results in a decrease of unspliced viral RNA levels and an approximately 10-fold decrease in virus production. In addition, excessive splicing of viral RNA is concomitant with a striking reduction in the relative amounts of Gag processing intermediates and products. We also show that T cell lines expressing modified U1 snRNAs exhibit reduced HIV-1 replication. Our results suggest that induction of excessive HIV-1 RNA splicing may be a novel strategy to inhibit virus replication in human patients.
The HIV-1 primary RNA transcript undergoes a complex splicing pathway leading to the production of over 40 different spliced mRNA species as well as unspliced (US) RNA (30). Unspliced HIV-1 mRNA, which is transported into the cytoplasm by a Rev-dependent process, encodes Gag and Gag-Pol structural proteins. Unspliced viral RNA is also packaged into the virus particles as genome RNA. Spliced HIV-1 mRNAs are either incompletely spliced (IS) and serve as mRNAs for viral Env, Vpu, Vpr, and Vif proteins or completely spliced (CS) to serve as mRNAs for Tat, Rev, or Nef protein. Splicing is a two-step catalytic reaction that occurs within a large complex called the spliceosome. One of the earliest steps in spliceosome assembly is the recognition of 5′ splice donor sites (5′ss) by cellular U1 snRNP (28, 39). The extent of base pairing between the 5′ss and the 5′-terminal sequence of U1 snRNA determines the strength of the 5′ss and thus contributes to the recognition and frequency of U1 snRNP usage (17, 32). In retroviruses, 3′ splice acceptor sites (3′ss) are weak, and recognition of these sites is assisted by the binding of cellular splicing regulatory proteins which regulate alternative splicing and gene expression. These regulatory proteins (SR proteins and hnRNPs) bind to exonic splicing enhancers (ESE) or exonic or intronic splicing silencers (ESS or ISS) within the viral genome (10, 38). U1 snRNP bound at the 5′ss also activates splicing at the upstream 3′ss by a mechanism referred to “exon definition” or “exon bridging” (18, 31).
Our laboratory has shown that splicing of the mRNAs encoding HIV-1 accessory proteins Vif and Vpr is regulated by suboptimal 5′ss D2 and D3 downstream of 3′ss A1 and A2, respectively (Fig. 1A) (13, 23, 24). Mutations of 5′ss D2 that increase its binding affinity for U1 snRNP result in increased splicing at 3′ss A1 and increased vif mRNA, whereas mutations which lower the affinity to U1 snRNP result in decreased splicing at 3′ss A1 and reduced vif mRNA (13, 25). Decreased splicing at 3′ss A1 reduces the level of Vif and inhibits HIV-1 replication in cells that are expressing elevated levels of the restriction factor APOBEC3G (25). Increased splicing at 3′ss A1, on the other hand, results in an increased level of Vif, a 10-fold inhibition of virus production, and also, surprisingly, a defect of Gag processing (24, 26). We showed in those studies that the strength of U1 snRNP binding at 5′ss D2 is important for regulation of splicing at 3′ss A1 but that splicing at 5′ss D2 is not required for this regulatory effect (25). Similarly, mutations of 5′ss D3 that change it to a consensus 5′ss result in increased splicing at 3′ss A2 (9). We also have shown that disruption of an ESE (ESSV) downstream of 3′ss A2 leads to increased splicing at 3′ss A2 and an approximately 10-fold decrease in virus production (23). Splicing at 3′ss A3 is regulated by two different ESS elements (ESS2p and ESS2) (3, 4, 35). Mutations of ESS2 result in increased splicing at 3′ss A3, reduced levels of unspliced RNA, and decreased virus production (Z. Feng and C. M. Stoltzfus, unpublished data).
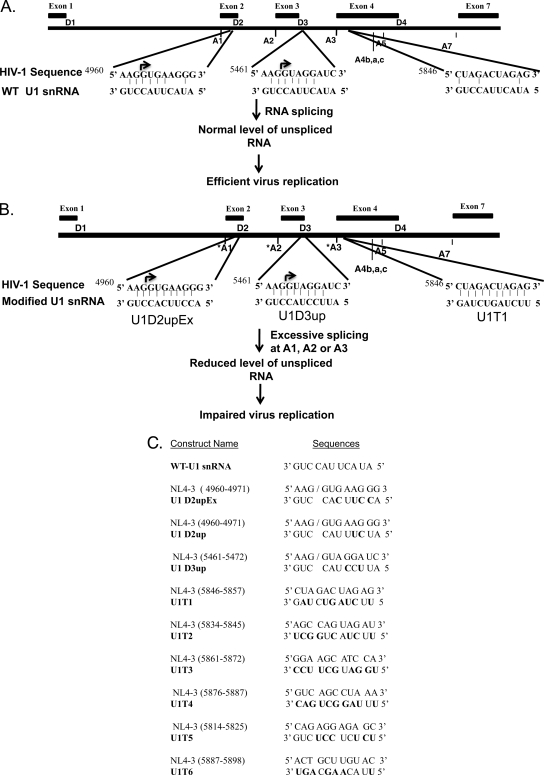
FIG. 1.
Proposed strategy to inhibit virus replication by inducing excessive splicing of HIV-1 RNA. (A) Organization of major splice donor (D1 to D4) and acceptor (A1 to A7) sites and locations of exons on the HIV-1 genome (30). (B) Expression of 5′-end-mutated U1 snRNAs that increase base pairing to 5′ss D2 or D3 compared to wild-type U1 snRNA increases splicing at upstream acceptor sites (U1D2upEx and U1D3up, respectively). Increased splicing at 3′ss A3 is induced by expression of modified U1 snRNAs that bind to sequences downstream of A3 and upstream of 3′ss A4a, 4b, 4c, and A5. Induction of excessive splicing will increase the level of splicing and inhibit virus replication. (C) Modified U1 snRNAs used in this study. Sequences of wild-type and 5′-end-mutated U1 snRNAs are shown. Target sequences of these modified U1 snRNAs within the HIV-1 genome (NL4-3) and their respective positions are indicated.
Based on these results, we reasoned that wild-type virus replication should be inhibited by promoting excessive splicing of HIV-1 RNA and reducing the amount of unspliced viral RNA available for synthesis of Gag and for use as genome RNA. To test this hypothesis, we evaluated whether modified U1 snRNAs can promote excessive splicing of HIV-1 RNA and inhibit virus replication. We show that excessive splicing of HIV-1 mRNA at 3′ss A1, A2, or A3 is induced by expressing U1 snRNA with exact complementarity to sequences downstream of these splice sites. We also show that this results in a drastic inhibition of virus replication in T cells. This strategy may be a novel approach to inhibit HIV-1 replication in infected cells of human patients.
MATERIALS AND METHODS
Plasmids.
Modified U1 snRNAs U1D2up, U1D2upEx, U1D3up, U1T2, U1T3, U1T4, U1T5, and U1T6 were generated by site-directed mutagenesis of the pUC13-U1 clone, which was obtained from Mark McNally at the Medical College of Wisconsin, Milwaukee, WI. Sequences of the primers used for site-directed mutagenesis are listed in Table 1. U1T1 was generated by PCR using mutagenic reverse primers with a BglII restriction site (UST1) and a forward primer (UAS) (Table 1) with a BspE1 restriction site. Amplified products were digested with BglII and BspE1 enzymes and cloned into the BglII- and BspE1-digested pUC13-U1. Clones were confirmed by sequencing. To clone the mutated U1 snRNA fragment into lentiviral vector pHIV7-SF-GFP (37), pUC13-U1-based U1 snRNA clones were digested with BamHI, and the fragments were inserted into the BamHI site of the pHIV7-SF-GFP vector. Infectious HIV-1 plasmid pNL4-3 (1) was obtained from the NIH AIDS Research and Reagent Program.
TABLE 1.
Oligonucleotides used to generate mutated U1 snRNAs
| Oligonucleotide | Sequence |
|---|---|
| U1D2upEx-F | 5′CTCCCCTGCCAGGTGAAGGTGAGATCTTCGG3′ |
| U1D2upEx-R | 5′CCGAAGATCTCACCTTCACCTGGCAGGGGAG3′ |
| U1D3up-F | 5′CTCCCCTGCCAGGTAGGAATGAGATCTTCGG3′ |
| U1D3Up-R | 5′CCGAAGATCTCATTCCTACCTGGCAGGGGAG3′ |
| U1D2up-F | 5′CTCCCCTGCCAGGTAAAGATGAGATCTTCGG3′ |
| U1D2up-R | 5′CCGAAGATCTCATCTTTACCTGGCAGGGGAG3′ |
| UST1 | 5′GGGCGAAGATCTCATTCTAGTCTAGCAGGGGAGATACCATGATCACGAACGG3′ |
| UAS | 5′GGGTCAGCACATCCGGAGTG3′ |
| U1T2-F | 5′CCACCTTCGTGATCATGGTATCTCCCCTGAGCCAGTAGAATGAGATCTTCGGGCTCTGCCCCGACACAGCC3′ |
| U1T2-R | 5′GGCTGTGTCGGGGCAGAGCCCGAAGATCTCATTCTACTGGCTCAGGGGAGATACCATGATCACGAAGGTGG3′ |
| U1T3-F | 5′CCACCTTCGTGATCATGGTATCTCCCCTGGGAAGCATCCATGAGATCTTCGGGCTCTGCCCCGACACAGCC3′ |
| U1T3-R | 5′GGCTGTGTCGGGGCAGAGCCCGAAGATCTCATGGATGCTTCCCAGGGGAGATACCATGATCACGAAGGTGG3′ |
| U1T4-F | 5′CCACCTTCGTGATCATGGTATCTCCCCTGGTCAGCCTAAATGAGATCTTCGGGCTCTGCCCCGACACAGCC3′ |
| U1T4-R | 5′GGCTGTGTCGGGGCAGAGCCCGAAGATCTCATTTAGGCTGACCAGGGGAGATACCATGATCACGAAGGTGG3′ |
| U1T5-F | 5′CCACCTTCGTGATCATGGTATCTCCCCTGCAGAGGAGAGATGAGATCTTCGGGCTCTGCCCCGACACAGCC3′ |
| U1T5-R | 5′GGCTGTGTCGGGCAGAGAGCCCGAAGATCTCATCTCTCCTCTGCAGGGGAGATACCATGATCACGAAGGTGG3′ |
| U1T6-F | 5′CCACCTTCGTGATCATGGTATCTCCCCTGACTGCTTGTAATGAGATCTTCGGGCTCTGCCCCGACACAGCC3′ |
| U1T6-R | 5′GGCTGTCGGGGCAGAGCCCGAAGATCTCATTACAAGCAGTCAGGGGAGATACCATGATCACGAAGGTGG3′ |
RNA isolation and analysis of viral RNA species.
Total RNA was isolated from transfected cells using the Tri-reagent (Molecular Research Center, Inc.) according to the protocol supplied by the manufacturer. Northern blotting analysis of total RNA was performed as previously described (13, 23). Reverse transcriptase PCR (RT-PCR) analysis of 1.8- and 4.0-kb classes of HIV-1 mRNA species was performed as previously described (13).
Quantitative real-time PCR to determine the level of unspliced RNA.
Total RNA from transfected 293T cells expressing modified U1 snRNAs was isolated, and cDNAs were prepared as described previously (13). The cDNAs were used as templates for real-time PCR. Unspliced RNA was measured using primers 5′GGG AAGTGACATAGCAGGAACTACT3′ and 5′TGGGATAGGTGGATTATGTGT CATC3′ (nucleotides [nt] 1485 to 1509 and 1536 to 1560, respectively), and total RNA was measured using 5′AAGGGCTAATTCACTCCCAA3′ and 5′TCTAACACTTCTCTCTCTCCGGG3′ (nucleotides 9079 to 9098 and 9311 to 9331, respectively). For internal controls, the amounts of β-actin mRNA were determined as previously described (13).
Reverse transcriptase assays and virus production.
Cell-free supernatants (10 μl) were assayed for RT activity by [α-32P]dTTP incorporation as previously described (36). Incorporation of radioactivity was quantitated using an Instant Imager (Packard). For transfection experiments, RT data were normalized to those for the wild type after correction for transfection efficiencies based on beta-galactosidase assays of extracts from transfected cells (34).
Detection of viral proteins by immunoblotting.
Cellular proteins (50 μg) were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, transferred to nitrocellulose membrane by the semidry transfer method, and immunoblotted according to previously described methods to detect Gag (13). Gag protein was detected using rabbit anti-p24 polyclonal antibody (NIH AIDS Research and Reagent Program). The amounts of Gag precursor and processed products were determined by chemiluminescence of the protein bands by using the LAS-4000 Fuji imager, and the ratios of p24 Gag and Pr-41 Gag to Pr-55 Gag were then calculated. Tubulin was detected as an internal control using a mouse antitubulin monoclonal antibody (Developmental Studies Hybridoma Bank, University of Iowa).
Lentiviral vector production, titration, and transduction.
Lentiviral vectors containing modified U1 snRNAs were generated by transfecting 293T cells with 5 μg pHIV7-SF-GFP construct with modified U1snRNAs, 5 μg pGag-Pol, 1 μg pRev, and 2.5 μg vesicular stomatitis virus G protein (VSV-G) plasmid. After 48 h of transfection supernatants from the transfected cells were collected and passed through 0.45-μm filters. In order to determine titers, 293T cells were infected with viral vectors and green fluorescent protein (GFP)-positive cells were counted. The virus vectors were then used to transduce CEMSS cells, GFP-positive cells were selected by fluorescence-activated cell sorting (FACS), and sorted cells were then propagated. A second round of selection was repeated to obtain pure GFP-expressing cells, and selected cells were then propagated.
Expression of modified U1 snRNAs.
The expression levels of 5′-end-modified U1 snRNAs in both transfected cells and stable T cells were determined by quantitative real-time PCR. RNA from both transfected 293T and stable CEMSS cells expressing the modified U1 snRNAs was isolated, cDNAs were prepared as described previously (13), and the cDNAs were then used as templates for real-time PCR. Quantitative real-time PCR was performed using 5′-GCAATGGATAAGCCTCGCCCTGG-3′ as the forward primer and 5′-CCGGAAATACTTACCTGG-3′, 5′-CCGGAAACCTTCACCTG-3′, and 5′-CCGGAAATTCTAGTCTAGCAG-3′ (R-WT, R-D2upEx, and R-T1, respectively) as reverse primers specific for endogenous U1snRNA, U1D2upEx, and U1T1, respectively. The amounts of cellular β-actin in the cDNA samples were determined by using specific primers (13), and the amounts of endogenous or mutant U1 snRNAs relative to β-actin mRNA were calculated. Different dilutions of plasmid DNA from pUC13U1, U1D2upEx, or U1T1 clones were used to generate standard curves with the same primers used for quantitative PCR. The levels of expression of the mutated U1 snRNAs relative to endogenous U1 snRNA were then determined.
Infection and multiday replication assay.
GFP-positive CEMSS cells expressing modified U1 snRNAs were infected with equal amounts of WT viruses as determined by RT assays for 4 h at 37°C in serum-free RPMI medium. Infected cells were then centrifuged, washed, and resuspended in RPMI medium containing 10% fetal bovine serum (FBS). Aliquots of cell-free medium were harvested at regular intervals, and virus production was measured by RT assays.
RESULTS
Expression of modified U1snRNAs promotes excessive splicing of HIV-1 RNA and results in reduced levels of unspliced RNA.
To promote excessive splicing at HIV-1 3′ss A1 and A2, we created mutations at the 5′ end of U1 snRNA that were designed to increase base paring at the relatively weak HIV-1 donor site 5′ss D2 or D3 (Fig. 1). In order to promote excessive splicing selectively at 3′ss A3, we used an alternative strategy. In contrast to donor sites D2 and D3, donor site 5′ss D4 is a consensus 5′ss which has been shown to be relatively strong (20, 29). Furthermore, there are multiple alternative 3′ss (3′ss A3, A4a, A4b, 4c, and A5) that act to define multiple exons flanked by 5′ss D4. Thus, in order to specifically promote excessive splicing at 3′ss A3, we targeted sequences in the first tat coding exon downstream of A3 (exon 4) and upstream of 3′ss A4a, A4b, A4c, and A5 (Fig. 1C). For this purpose, we created U1 snRNA mutants (U1T1 to U1T6) that were predicted to base pair to 10-nt HIV-1 sequences within the stem-loop structure SLS3-A3 in exon 4 (16, 19).
To investigate whether expression of the modified U1 snRNAs described above increases splicing of HIV-1 RNA, we cotransfected 293T cells with plasmids expressing modified U1 snRNAs together with the infectious HIV-1 plasmid pNL4-3. After 48 h of transfection, total RNA from the cells was isolated and HIV-1 RNA was analyzed by Northern blotting in order to determine the effect on splicing. Expression of U1D2upEx resulted in a drastic reduction in the level of unspliced (US) RNA compared to pNL4-3 alone or pNL4-3 cotransfected with a control U1 snRNA plasmid, pUC13U1. There was also a concomitant increase in the level of incompletely spliced (IS) mRNA (Fig. 2A). The bulk of the IS mRNA migrated more slowly than the IS band of the wild-type virus RNA. We have previously shown that this slower-migrating band corresponds to vif mRNA (13). There was less effect on the level of US RNA in the presence of U1D2up, although we also detected an increase in the band corresponding to vif mRNA. Quantitation of the labeled bands indicated that the ratios of unspliced to spliced RNA were approximately 2-fold higher in the presence of U1D2up compared to U1D2upEx. A reduction of US RNA and increased levels of IS and completely spliced (CS) mRNAs were also observed in the presence of U1D3up (Fig. 2A). Dramatic decreases in the level of US RNA were also observed in the presence of U1T1, U1T3, U1T4, or U1T6. Less effect on the level of US RNA was seen in the presence of U1T2. Expression of U1T5 had little or no effect on the relative amounts of spliced or unspliced RNA compared to those with the virus alone or in the presence of control U1 snRNA (Fig. 2A).
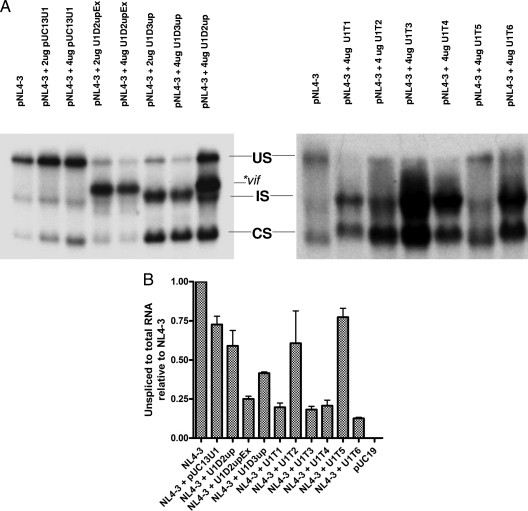
FIG. 2.
Analysis of HIV-1 RNA. (A)Total RNAs isolated from 293T cells transfected with pNL4-3 (6 μg) in the presence or absence of modified U1 snRNAs as shown in Fig. 1 were analyzed by Northern blotting as described in Materials and Methods. Unspliced (US), incompletely spliced (IS), and completely spliced (CS) RNA species are indicated. The location of vif mRNA produced in the presence of U1D2upEx and U1D2up is indicated. (B) Amounts of unspliced and total viral RNA from 293T cells were quantified by real-time PCR, and ratios of unspliced to total RNA relative to pNL4-3 in the absence of added U1 snRNA were determined and normalized to the β-actin levels. Error bars indicate standard deviations.
In order to more quantitatively determine the level of unspliced RNA produced in the presence of the modified U1 snRNAs, we analyzed RNA extracted from transfected cells by quantitative real-time PCR using primers specific for the Gag p24 sequence. We also determined the amounts of total viral RNA using primers specific for the HIV-1 U3 region of the long terminal repeat (LTR), which is common to all HIV-1 mRNAs. The ratios of unspliced to total viral RNA were then determined. For an internal control, we determined the amount of β-actin mRNA by quantitative real-time PCR. As shown in Fig. 2B, the ratios of unspliced to total RNA levels of NL4-3 in the presence of modified U1 snRNA plasmids U1D2upEx, U1T1, U1T3, U1T4, and U1T6 were reduced (≤25%) compared to that of pNL4-3 in the presence of control U1 snRNA plasmid (Fig. 2B). In the presence of U1D3up and U1D2up plasmids, there were moderate reductions in the level of unspliced RNA (unspliced/total RNA ratios of ∼40% and ∼60%, respectively, compared to that for pNL4-3 in the presence of control U1 snRNA). U1T2 and U1T5 had little or no effect on the unspliced RNA level and were similar to pNL4-3 in the presence of control U1 snRNA (Fig. 2B).
Excessive splicing of HIV-1 RNA induced by modified U1 snRNAs is selectively directed to upstream 3′ss.
To determine whether the observed increase in splicing promoted by the modified U1 snRNAs occurred at the upstream 3′ss flanking the exon, as expected based on the exon definition hypothesis, we analyzed the CS (1.8-kb) and IS (4-kb) mRNA species by RT-PCR. Expression of modified U1 snRNAs targeted to 5′ss D2 (U1D2up and U1D2upEx) resulted in greatly increased inclusion of noncoding exon 2 in CS RNA species, as evidenced by increased levels of 1.2.5.7 (nef) and 1.2.4.7 (tat) mRNA species accompanied by reduced levels of 1.5.7 (nef) and 1.4.7 (tat) mRNA species (Fig. 3A). In contrast, expression of control U1 snRNA (pUC13U1) had little or no effect on the HIV-1 splicing pattern. Expression of U1D2up also resulted in increased inclusion of exon 2, although there was somewhat less reduction in the level of the 1.5.7 species. Increased splicing at A1 promoted by U1D2up and U1D2upEx was also evident from the analysis of 4.0-kb IS mRNA species (Fig. 3A). This resulted in increased inclusion of exon 2, promoted by both U1D2up and U1D2upEx, as shown by the increases in mRNA species 1.2.5I (env/vpu). Expression of modified U1 snRNA with increased base pairing to the 5′ss D3 sequence (U1D3up) resulted in increased inclusion of noncoding exon 3 into the CS mRNA and IS mRNAs. This was evidenced by an increase in the 1.3.5.7 (nef) and 1.3.5I (env/vpu) mRNA species and the appearance of a labeled band corresponding to vpr mRNA (1.3I). This increase in inclusion of exon 3 was accompanied by decreases in 1.5.7 and 1.5I (env/vpu) (Fig. 3A).
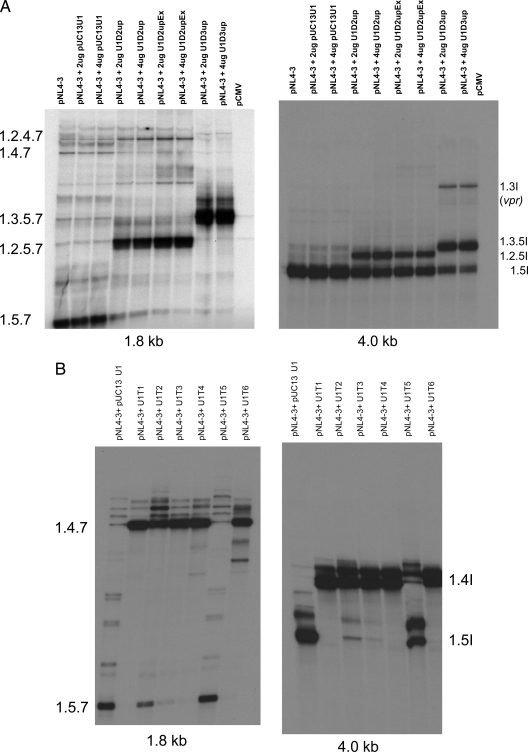
FIG. 3.
RT-PCR analysis of completely (1.8-kb) and incompletely (4.0-kb) spliced RNA species. Total RNA was isolated from 293T cells transfected with pNL4-3 proviral clone (6 μg) or in the presence of modified U1 snRNAs. RNAs were then subjected to RT-PCR, and 32P-labeled RT-PCR products were analyzed on 6% denaturing gels as described previously (13). The 1.8-kb and 4.0-kb spliced RNA species are indicated. (A) Analysis of 1.8-kb and 4.0-kb HIV-1 RNA species in the presence or absence of U1D2upEx, U2D2up, and U1D3up. (B) Analysis of 1.8-kb and 4.0-kb HIV-1 RNA species in the presence of U1T1, U1T2, U2T3, U1T4, U1T5, and U1T6.
We showed above (Fig. 2) that we could also promote excessive splicing of HIV-1 RNA by targeting sequences downstream of 3′ss A3 within the first tat coding exon (exon 4). Expression of all the modified U1 snRNAs tested except U1T5 resulted in increased inclusion of exon 4 into both CS and IS mRNAs, as shown by increases in the levels of two-exon tat (1.4.7) and single-exon tat (1.4I) mRNA species (Fig. 3B). There were also concomitant decreases in the levels of nef (1.5.7) and vpu/env (1.5I) mRNA species. The decrease in mRNA species 1.5.7 and 1.5I by expression of U1T2 appeared somewhat reduced compared to those with U1T1, U1T3, U1T4, and U1T6. There was little or no effect of U1T5 on the splicing patterns, consistent with the data shown in Fig. 2. Thus, our results indicated that in all cases, the increase in HIV-1 splicing promoted by the modified U1 snRNAs occurred at the corresponding upstream 3′ss and was consistent with the exon definition hypothesis.
Gag processing is defective in the presence of modified U1 snRNAs.
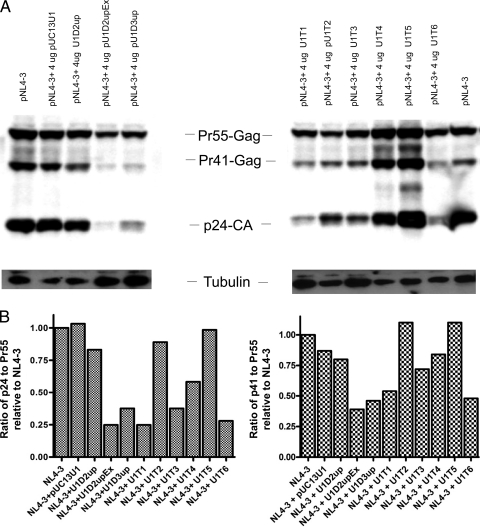
We have previously shown that mutants of 5′ss D2 that exhibit excessive splicing of HIV-1 RNA also demonstrate Gag protein processing defects (26). Thus, we next asked whether excessive splicing of HIV-1 RNA induced by modified U1 snRNAs also affected Gag processing. To examine this question, we analyzed total cellular protein by Western blotting using Gag CA-specific antibody from cells cotransfected with pNL4-3 and modified U1 snRNA expression clones (Fig. 4A). We quantitatively determined the amounts of both Pr55-Gag precursor and processed Gag (p24 and Pr-41) by directly measuring the chemiluminescence of the bands on the Western blot. The p24/Pr-55 and Pr-41/Pr-55 ratios were then determined (Fig. 4B). In the presence of U1D2upEx, whose expression was shown above to greatly increase splicing of HIV-1 RNA at 3′ss A1, aberrant Gag processing occurred, as indicated by reduced levels of Gag processing intermediate (Pr41) and product (p24-CA) relative to Gag precursor Pr55gag. This was confirmed by the reduced p24/Pr55 and Pr41/Pr55 ratios compared to those for virus alone (Fig. 4A and B). In contrast, expression of U1D2up, which we showed above also resulted in increased splicing at A1 at lower levels than for U1D2upEx, had less effect on Gag processing (Fig. 4A and B). Gag processing was also aberrant in the presence of U1D3up as well as U1T1, U1T3, and U1T6 (Fig. 4A and B), which we showed above to markedly affect HIV-1 RNA splicing (Fig. 2). Expression of U1T4 had a moderate effect on Gag processing, whereas U1T2 and U1T5 had little or no effect on Gag processing (Fig. 4A and B). These results were also consistent with the effects on HIV-1 splicing and the level of unspliced RNA as shown in Fig. 2A and B.
FIG. 4.
(A) Analysis of Gag protein isolated from 293T cells transfected with pNL4-3 (6 μg) alone or in the presence of modified U1 snRNAs. Total protein was isolated from transfected cells and analyzed on 10% SDS-polyacrylamide gels. Gag protein was detected by immunoblotting using anti p24-CA antibody. Gag precursor (Pr55-Gag), intermediate (Pr41-Gag), and processed product (p24-CA) are indicated. The β-tubulin loading control bands are also indicated. (B) Amounts of p24-CA, Pr-41 Gag, and Pr-55 Gag produced in the presence of different modified U1 snRNAs were measured directly from the chemiluminescence, and p24/Pr-55 and Pr41/Pr55 ratios relative to pNL4-3 in the absence of added U1 snRNA were calculated.
We have also analyzed virion proteins, but we did not observe any processing defect of Gag isolated from virion particles. These results suggested that Gag processing in the transfected cell was not due to a defect in viral protease function (data not shown).
Impaired virus production in the presence of modified U1 snRNAs.
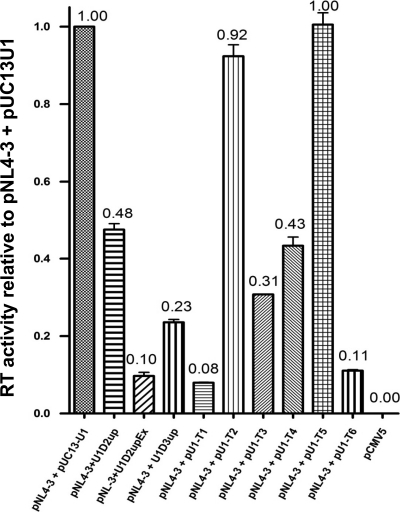
We next determined the effect of expressing the modified U1 snRNAs on HIV-1 virion production. Cells were cotransfected with pNL4-3 proviral plasmid and modified U1 snRNA constructs. At 48 h posttransfection the medium was harvested and analyzed for virus production. Expression of U1D2upEx resulted in a 10-fold decrease in virus production compared to that with pNL4-3 in the presence of pUC13U1 control U1 snRNA. Expression of U1D2up, on the other hand, resulted in only a 2-fold decrease in virus production. Expression of U1D3up resulted in an approximately 4-fold reduction in virus production compared to that with the wild-type control (Fig. 5). Expression of U1T1 and U1T6 caused approximately 10-fold reductions in virus production compared to that with expression of control U1 snRNA, whereas expression of U1T3 and U1T4 reduced virus production by 70% and 60%, respectively (Fig. 5). On the other hand, expression of U1T2 only slightly reduced virus production, and expression of U1T5 did not significantly affect virus production (Fig. 5). Thus, the effects of modified U1 snRNAs on splicing correlated with their effects on virus production.
FIG. 5.
Analysis of HIV-1 virus production. 293T cells were transfected with pNL4-3 alone (6 μg) or in the presence of modified U1 snRNAs (4 μg). Cell-free supernatants were collected at 48 h posttransfection, and virus production was determined by RT assays as described previously (36). Error bars indicate standard deviations.
Inhibition of virus replication in T cell lines expressing modified U1 snRNAs.
Our results above (Fig. 5) showed that in transfected 293T cells expression of several modified U1 snRNAs that promoted splicing of HIV-1 RNA inhibited virus production approximately 10-fold. To further examine whether modified U1 snRNAs inhibit virus replication, we generated stable T cell lines expressing several modified U1 snRNAs that were particularly effective in the transient assays. Stable CEMSS T cell lines were obtained by transducing lentiviral vectors expressing U1D2upEx, U1D3up, and U1T1, which were targeted to increase splicing at A1, A2, and A3, respectively. These cell lines were not different from the control cell lines in viability and did not show any significant cytopathic effect, suggesting that these modified U1 snRNAs do not have any significant off-target effects (data not shown).
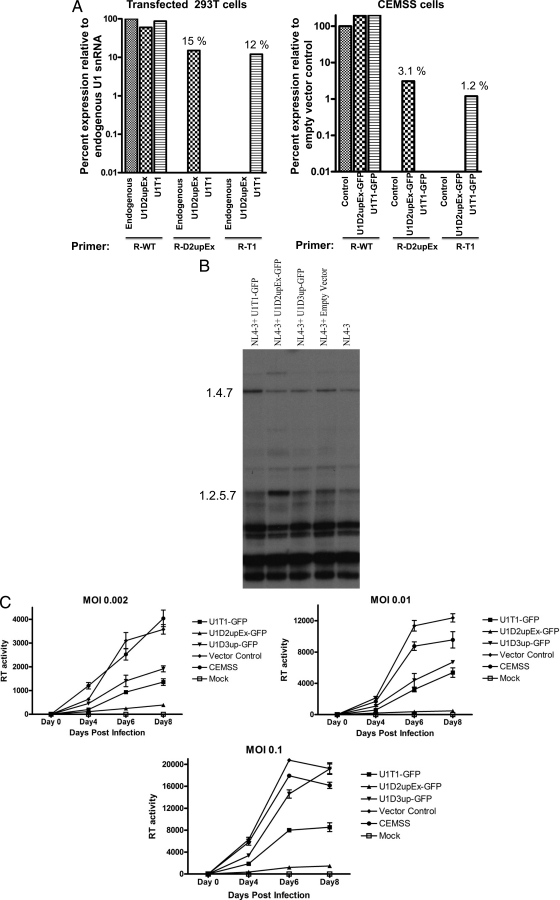
We first determined the relative expression levels of individual U1 snRNAs by quantitative real-time PCR assays as described in Materials and Methods. We used primers that specifically detected U1D2upEx and U1T1. We also attempted to measure U1D3up expression but were unable to find conditions and primers that were specific for U1D3up and that would allow us to distinguish its expression from wild-type expression. As shown in Fig. 6A, in transfected 293T cells, the expression of U1D2upEx and U1T1 was approximately 15% and 12%, respectively, of that of endogenous U1snRNA (Fig. 6A). In stable CEMSS cells, the expression of U1D2upEx and U1T1 was 3.1% and 1.2%, respectively, compared to that of the empty vector control (Fig. 6A).
FIG. 6.
Virus production in stable CEMSS T cells expressing modified U1 snRNA. (A) Determination of expression levels of modified U1 snRNAs by quantitative real-time PCR. Total RNA was isolated from transfected 293T cells or stable CEMSS cells transduced with lentiviral vectors expressing modified U1 snRNAs. cDNAs synthesized by RT from the RNA samples were subjected to quantitative real-time PCR. The relative amounts of WTU1, U1D2upEx, and U1T1 snRNAs, expressed as a percentage of wild-type U1 levels, were determined for each cell type using primers specific for wild-type (R-WT) and mutant (R-D2upEx and R-T1) U1 snRNAs as described in Materials and Methods. The values shown are the averages of three independent determinations. (B) Analysis of CS (1.8-kb) mRNA species from stable CEMSS T cells expressing modified U1 snRNAs infected with wild-type HIV-1. Total RNA was isolated from infected cells, and spliced RNA species were analyzed as described in the legend to Fig. 3. (C) HIV-1 replication in stable CEMSS T cells expressing modified U1 snRNAs. Control CEMSS T cells, cells expressing modified U1 snRNAs, and cells transduced with empty vector were infected with NL4-3 virus at different MOI as indicated. Virus production was determined by RT assays of cell-free supernatant collected at different time points shown. Error bars indicate standard deviations.
We then analyzed the HIV-1 RNA species by RT-PCR in stable cell lines infected with HIV-1. Figure 6B shows splicing pattern of CS RNA from HIV-1-infected U1T1-, U1D2upEx-, and U1D3up-expressing CEMSS cells. As expected, there was an increased level of 1.4.7 (tat) mRNA species in U1T1-expressing cells compared to that in control cell lines. In U1D2upEx-expressing cells, as expected, there was an increased level of the exon 2-containing mRNA species 1.2.5.7. In CEMSS cells expressing U1D3up, on the other hand, there were no detectable differences in the splicing pattern compared to that of the wild type.
We then tested whether expression of several modified U1 snRNAs that promoted excessive splicing also affected multiround replication of HIV-1. Stable CEMSS cell lines expressing U1D2upEx, U1T1, or U1D3up were challenged with NL4-3 virus at three different multiplicities of infection (MOI). Culture media were collected at 2-day intervals after infection, and RT activity was measured to determine virus production. At an MOI of 0.002, virus replication was significantly inhibited in U1D2upEx-, U1D3up-, and U1T1-expressing cells (Fig. 6C). Virus production was inhibited greater than 90% in cells expressing U1D2upEx, whereas in cells expressing U1D3up and U1T1, virus production was inhibited 70 to 80% compared to virus produced in empty vector control cells or CEMSS cells (Fig. 6C). Similar effects on virus production were observed at an MOI of 0.01 (Fig. 6C). At the relatively high MOI of 0.1, expression of U1D2upEx and U1T1 still resulted in significant inhibition of virus replication (90% and 60%, respectively). On the other hand, virus replication at an MOI of 0.1 was not significantly affected in U1D3up-expressing cells (Fig. 6C).
DISCUSSION
Modified snRNAs have previously been used to inhibit HIV-1 gene expression and virus replication. One approach was to inhibit the accumulation of HIV-1 RNA by targeting HIV-1 sequences in the 3′-terminal exon (33). This approach was adapted from prior studies indicating that expression of cellular genes could be inhibited by targeting 3′-terminal exons with modified U1 snRNAs (7, 21). This inhibition of RNA accumulation was proposed to be either a direct inhibition of 3′ polyadenylation machinery by U1 snRNPs or, alternatively, disruption of interactions between the 3′ss and the poly(A) signal as proposed by the exon definition hypothesis (31). In either case, binding of the modified U1 snRNPs would lead to incompletely processed RNA transcripts that are rapidly degraded (21). It has also been previously shown that HIV-1 replication was inhibited by inducing skipping of internal exons of tat, rev, and nef mRNAs by expression of modified U7 snRNA targeted to HIV-1 splice junctions and nearby regulatory elements (5, 6).
In this report we have shown for the first time that specifically designed U1 snRNAs targeting key splicing regulatory elements can be used to induce excessive HIV-1 splicing and, by this mechanism, inhibit virus replication. We expressed modified U1 snRNAs that are able to bind with high affinity to HIV-1 5′ss D2 and D3 and tat exon sequences downstream of 3′ss A3. Expression of some of these modified U1 snRNAs resulted in excessive splicing of HIV-1 RNA and a drastic inhibition of virus production. In addition we found that expression of modified U1 snRNPs that induced excessive splicing of HIV-1 also induced defective Gag processing. The effects of these modified U1 snRNAs on wild-type HIV-1 gene expression and replication are similar to the phenotype of some HIV-1 splicing element mutants (23, 24, 26).
Human cells express over one million copies of U1 snRNA per cell (22). To alter the splicing pattern by modified U1 snRNA, efficient expression of modified U1 snRNAs is necessary to compete with the unmodified cellular U1 snRNAs. In transfected 293T cells, expression of either of two 5′-mutated U1 snRNA molecules, U1D2upEx and U1T1, was sufficient to inhibit HIV-1 production by 90%. Assuming a transfection efficiency of greater than 90%, which is typical for 293T cells transfected under the conditions used here, our data imply that the expression levels of U1D2upEx and U1T1 in the transfected cells are approximately 15% and 12% of that of endogenous U1 snRNA. Strong inhibition of HIV-1 replication was also obtained in stable T cell lines in which U1D2upEx and U1T1 were expressed at levels of only 3.1% and 1.2% compared to the level of cellular U1 snRNA. These expression levels are comparable to those described in a previous study in which modified U1 snRNAs were used to inhibit expression of cellular genes (7). We believe that the modified U1 snRNAs are effective at these low levels because of their increased binding affinities to HIV-1 RNA compared to the wild-type U1 snRNA.
We showed that HIV-1 splicing at 3′ss A1 or A2 can be activated by increasing U1 base pairing to the relatively weak 5′ss D2 or D3, respectively. This activation occurs by increasing interactions between the downstream 5′ss and upstream 3′ss as predicted by the exon definition hypothesis. Interestingly, 3′ss A3 was activated by targeting modified U1 snRNAs to several different sequences within the first tat coding exon (exon 4). In this case, there was increased production of completely and incompletely spliced tat mRNAs. Thus, binding of U1 snRNPs at several locations within the tat exon is sufficient to activate splicing at 3′ss A3 and promote exon 4 inclusion by increased splicing at the 5′ss flanking exon 4, 5′ss D4. Some of the U1 snRNAs targeted to exon 4 were more effective than others in inducing excessive splicing. One of the reasons for this may be that the targets overlap binding sites for other cellular factors. For instance, U1T1, which was very effective in inducing splicing at 3′ss A3, overlaps the splicing regulatory element ESS2 which binds hnRNP A/B proteins and represses splicing at 3′ss A3 (4, 11, 35). However, overlap of a negative regulatory element cannot explain the results with U1T6, which we showed was equally effective in inducing excessive splicing. Another possibility is sequestration of target sequences in regions of base-paired secondary structures (16, 19). Insertion of target sequences into duplex structures has previously been shown to greatly reduce the inhibition induced by modified U1 snRNAs targeted to the 3′-terminal exons (14). However, target sequences for U1T1 and U1T6, both of which were effective, have been shown to be within base-paired regions. In contrast, U1T5, which was directed to an unpaired loop region, did not induce excessive splicing. Finally, it is possible that definition of exon 4 may require a minimum distance of the upstream 3′ss to the U1 snRNA binding site. This could explain the ineffectiveness of U1T5, which is only 26 to 35 nt downstream from 3′ss A3.
We showed that increasing the complementarity between U1 snRNP and 5′ss D2 and D3 resulted in increased splicing at 3′ss A1 and A2, respectively. In the case of U1D2upEx, the number of H bonds involved in U1 binding to the 5′ss, assuming Watson-Crick base pairing, increased from 14 to 23, and in the case of U1D3up, it increased from 17 to 23. These results are consistent with a model proposed by Freund et al. for the role of extended base pairing of U1 snRNA beyond the 5′ss consensus sequence −2AG/GU(A/G)AGU+6 to positions + 7 and + 8 (15). Increased binding of U1 snRNA would then be expected to lead to increased exon definition and increased splicing at the upstream 3′ss.
The effect of excessive splicing on HIV-1 Gag processing is not yet understood. Our results have indicated that several HIV-1 mutants demonstrating excessive splicing also show this same effect on Gag processing (24, 26). We have obtained preliminary data indicating that this defect in processing results from the failure of Gag to assemble and to associate with plasma membrane (D. Mandal and C. M. Stoltzfus, unpublished data). Thus, excessive splicing of wild-type HIV-1 RNA that is induced by modified U1 snRNPs may also cause defective Gag assembly, and this defect may contribute to the inhibition of virus replication. It is of interest that reduced levels of unspliced viral RNA, inefficient virus assembly, and low virus production have also been found for HIV-1 infections of murine and other rodent cell lines that were engineered to express the HIV-1 receptor and coreceptor as well as cyclin T1 required for Tat function (8, 27). In this case, there appears to be a reasonable correlation between levels of unspliced RNA and virus production in mouse and rat cell lines but not in hamster cell lines (8).
Using subgenomic HIV-1 env expression constructs, several studies have indicated that U1 snRNP binding to 5′ss D4 is important in HIV-1 transcription and stability (2, 12, 20). In our experiments we did not directly measure the effects of U1 snRNA on transcription or stability of HIV-1 mRNAs. The results shown in Fig. 2A indicated that the presence of control U1 plasmid resulted in an increase in the overall level of the Northern blot signal. This was not a consistent finding, however, in multiple independent experiments. We also could not demonstrate a significant difference in the levels of total HIV-1 RNA in the presence or absence of control U1 snRNA by quantitative real-time PCR assays (D. Mandal and C. M. Stoltzfus, unpublished observations).
We showed that U1D2upEx and U1T1 expressed by lentivirus-transduced CEMSS T cells result in excessive splicing and inhibition of HIV-1 replication. We believe that this strategy, if the challenges for delivery of the modified U1 snRNAs can be surmounted, may be an effective therapeutic approach and add to our arsenal of antiretrovirals. One reason is that the inhibition is mediated by RNA rather than protein and therefore would not be expected to generate an immune response. Second, the modified U1 snRNAs that we tested should be broadly effective, since they targeted to conserved HIV-1 sequences in 5′ss D2 and D3 and within the first tat coding exon. Third, the modified U1 snRNAs act to inhibit HIV-1 at a different step of gene expression, RNA splicing in the nucleus, compared to inhibitory small interfering RNAs (siRNAs), which act to reduce levels of HIV-1 mRNAs in the cytoplasm. It will be necessary to test these two RNA-based approaches together to investigate whether they can act synergistically to inhibit HIV-1 replication. It will also be necessary to assay for the frequency of resistant variants that arise when HIV-1 is passaged on cells expressing the modified U1 snRNAs. Finally, the approach described here may be useful for inhibiting the replication of other viruses that are dependent on the host cell splicing process.
Acknowledgments
We thank Wendy Maury, University of Iowa, for a critical reading of the manuscript. We thank Mark McNally, Medical College of Wisconsin, Milwaukee, WI, for the U1 snRNA expression vector pUC13-U1 and Tom Hope, Northwestern University School of Medicine, for the pCMV110 β-galactosidase expression plasmid. We also acknowledge the NIH AIDS Research and Reference Reagent Program for supplying HIV-1-related reagents.
This research was supported by PHS grant AI36073 from the National Institute of Allergy and Infectious Diseases and by a grant from the Levitt Center, University of Iowa.
Footnotes
Published ahead of print on 6 October 2010.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, M. R., A. K. Wheatley, R. J. Center, and D. F. Purcell. 2010. Efficient transcription through an intron requires the binding of an Sm-type U1 snRNP with intact stem loop II to the splice donor. Nucleic Acids Res. 38:3041-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amendt, B. A., D. Hesslein, L. J. Chang, and C. M. Stoltzfus. 1994. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 14:3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amendt, B. A., Z. H. Si, and C. M. Stoltzfus. 1995. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol. Cell. Biol. 15:4606-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asparuhova, M. B., I. Barde, D. Trono, K. Schranz, and D. Schumperli. 2008. Development and characterization of a triple combination gene therapy vector inhibiting HIV-1 multiplication. J. Gene Med. 10:1059-1070. [DOI] [PubMed] [Google Scholar]
- 6.Asparuhova, M. B., G. Marti, S. Liu, F. Serhan, D. Trono, and D. Schumperli. 2007. Inhibition of HIV-1 multiplication by a modified U7 snRNA inducing Tat and Rev exon skipping. J. Gene Med. 9:323-334. [DOI] [PubMed] [Google Scholar]
- 7.Beckley, S. A., P. Liu, M. L. Stover, S. I. Gunderson, A. C. Lichtler, and D. W. Rowe. 2001. Reduction of target gene expression by a modified U1 snRNA. Mol. Cell. Biol. 21:2815-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bieniasz, P. D., and B. R. Cullen. 2000. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J. Virol. 74:9868-9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bilodeau, P. S., J. K. Domsic, A. Mayeda, A. R. Krainer, and C. M. Stoltzfus. 2001. RNA splicing at human immunodeficiency virus type 1 3′ splice site A2 is regulated by binding of hnRNP A/B proteins to an exonic splicing silencer element. J. Virol. 75:8487-8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 11.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damgaard, C. K., S. Kahns, S. Lykke-Andersen, A. L. Nielsen, T. H. Jensen, and J. Kjems. 2008. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol. Cell 29:271-278. [DOI] [PubMed] [Google Scholar]
- 13.Exline, C. M., Z. Feng, and C. M. Stoltzfus. 2008. Negative and positive mRNA splicing elements act competitively to regulate human immunodeficiency virus type 1 vif gene expression. J. Virol. 82:3921-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortes, P., Y. Cuevas, F. Guan, P. Liu, S. Pentlicky, S. P. Jung, M. L. Martinez-Chantar, J. Prieto, D. Rowe, and S. I. Gunderson. 2003. Inhibiting expression of specific genes in mammalian cells with 5′ end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc. Natl. Acad. Sci. U. S. A. 100:8264-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freund, M., M. J. Hicks, C. Konermann, M. Otte, K. J. Hertel, and H. Schaal. 2005. Extended base pair complementarity between U1 snRNA and the 5′ splice site does not inhibit splicing in higher eukaryotes, but rather increases 5′ splice site recognition. Nucleic Acids Res. 33:5112-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallay, H., N. Locker, L. Ayadi, D. Ropers, E. Guittet, and C. Branlant. 2006. Biochemical and NMR study on the competition between proteins SC35, SRp40, and heterogeneous nuclear ribonucleoprotein A1 at the HIV-1 Tat exon 2 splicing site. J. Biol. Chem. 281:37159-37174. [DOI] [PubMed] [Google Scholar]
- 17.Hartmann, L., S. Theiss, D. Niederacher, and H. Schaal. 2008. Diagnostics of pathogenic splicing mutations: does bioinformatics cover all bases? Front. Biosci. 13:3252-3272. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman, B. E., and P. J. Grabowski. 1992. U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev. 6:2554-2568. [DOI] [PubMed] [Google Scholar]
- 19.Jacquenet, S., D. Ropers, P. S. Bilodeau, L. Damier, A. Mougin, C. M. Stoltzfus, and C. Branlant. 2001. Conserved stem-loop structures in the HIV-1 RNA region containing the A3 3′ splice site and its cis-regulatory element: possible involvement in RNA splicing. Nucleic Acids Res. 29:464-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kammler, S., C. Leurs, M. Freund, J. Krummheuer, K. Seidel, T. O. Tange, M. K. Lund, J. Kjems, A. Scheid, and H. Schaal. 2001. The sequence complementarity between HIV-1 5′ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA 7:421-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, P., M. Kronenberg, X. Jiang, and D. Rowe. 2004. Modified U1 snRNA suppresses expression of a targeted endogenous RNA by inhibiting polyadenylation of the transcript. Nucleic Acids Res. 32:1512-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lund, E., and J. E. Dahlberg. 1984. True genes for human U1 small nuclear RNA. Copy number, polymorphism, and methylation. J. Biol. Chem. 259:2013-2021. [PubMed] [Google Scholar]
- 23.Madsen, J. M., and C. M. Stoltzfus. 2005. An exonic splicing silencer downstream of the 3′ splice site A2 is required for efficient human immunodeficiency virus type 1 replication. J. Virol. 79:10478-10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madsen, J. M., and C. M. Stoltzfus. 2006. A suboptimal 5′ splice site downstream of HIV-1 splice site A1 is required for unspliced viral mRNA accumulation and efficient virus replication. Retrovirology 3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandal, D., C. M. Exline, Z. Feng, and C. M. Stoltzfus. 2009. Regulation of Vif mRNA splicing by human immunodeficiency virus type 1 requires 5′ splice site D2 and an exonic splicing enhancer to counteract cellular restriction factor APOBEC3G. J. Virol. 83:6067-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandal, D., Z. Feng, and C. M. Stoltzfus. 2008. Gag-processing defect of human immunodeficiency virus type 1 integrase E246 and G247 mutants is caused by activation of an overlapping 5′ splice site. J. Virol. 82:1600-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mariani, R., G. Rutter, M. E. Harris, T. J. Hope, H. G. Krausslich, and N. R. Landau. 2000. A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 74:3859-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mount, S. M., I. Pettersson, M. Hinterberger, A. Karmas, and J. A. Steitz. 1983. The U1 small nuclear RNA-protein complex selectively binds a 5′ splice site in vitro. Cell 33:509-518. [DOI] [PubMed] [Google Scholar]
- 29.O'Reilly, M. M., M. T. McNally, and K. L. Beemon. 1995. Two strong 5′ splice sites and competing, suboptimal 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology 213:373-385. [DOI] [PubMed] [Google Scholar]
- 30.Purcell, D. F., and M. A. Martin. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 67:6365-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robberson, B. L., G. J. Cote, and S. M. Berget. 1990. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 10:84-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roca, X., R. Sachidanandam, and A. R. Krainer. 2005. Determinants of the inherent strength of human 5′ splice sites. RNA 11:683-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sajic, R., K. Lee, K. Asai, D. Sakac, D. R. Branch, C. Upton, and A. Cochrane. 2007. Use of modified U1 snRNAs to inhibit HIV-1 replication. Nucleic Acids Res. 35:247-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Si, Z., B. A. Amendt, and C. M. Stoltzfus. 1997. Splicing efficiency of human immunodeficiency virus type 1 tat RNA is determined by both a suboptimal 3′ splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 25:861-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willey, R. L., D. H. Smith, L. A. Lasky, T. S. Theodore, P. L. Earl, B. Moss, D. J. Capon, and M. A. Martin. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 62:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yam, P. Y., S. Li, J. Wu, J. Hu, J. A. Zaia, and J. K. Yee. 2002. Design of HIV vectors for efficient gene delivery into human hematopoietic cells. Mol. Ther. 5:479-484. [DOI] [PubMed] [Google Scholar]
- 38.Zheng, Z. M. 2004. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 11:278-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhuang, Y., and A. M. Weiner. 1986. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell 46:827-835. [DOI] [PubMed] [Google Scholar]