Abstract
Hepadnaviruses are DNA viruses that are found in several mammalian and avian species. These viruses replicate their genome through reverse transcription of an RNA intermediate termed pregenomic RNA (pgRNA). pgRNA is reverse transcribed by the viral polymerase into a minus-strand DNA, followed by synthesis of the plus-strand DNA. There are multiple cis-acting sequences that contribute to the synthesis of minus-strand DNA for human hepatitis B virus (HBV). Less is known about the cis-acting sequences of avian hepadnaviruses that contribute to synthesis of minus-strand DNA. To identify cis-acting sequences of duck hepatitis B virus (DHBV) and heron hepatitis B virus (HHBV), we analyzed variants containing 200-nucleotide (nt) deletions. Most variants of DHBV synthesized minus-strand DNA to 50 to 100% of the wild-type (WT) level, while two variants synthesized less than 50%. For HHBV, most variants synthesized minus-strand DNA to less than 50% the WT level. These results differ from those for HBV, where most of the genome can be removed with little consequence. HBV contains a sequence, φ, that contributes to the synthesis of minus-strand DNA. It has been proposed that DHBV has an analogous sequence. We determined that the proposed φ sequence of DHBV does not contribute to the synthesis of minus-strand DNA. Finally, we found that the DR2 sequence present in all hepadnaviruses is important for synthesis of minus-strand DNA in both DHBV and HHBV but not in HBV. These differences in cis-acting sequences suggest that the individual hepadnaviruses have evolved differences in their mechanisms for synthesizing minus-strand DNA, more so than for other steps in replication.
Hepadnaviruses are a family of DNA viruses that primarily infect the liver (4). Members can be placed into one of two subfamiles, either mammalian or avian hepadnaviruses. Hepatitis B virus (HBV), the prototype member of the family, is a worldwide health concern. It is estimated that nearly two billion people have been exposed to HBV through either acute or chronic infections (23). Approximately 360 million people are chronically infected. These individuals have an increased risk for developing liver disease such as hepatocellular carcinoma.
The infectious particle consists of a double-stranded DNA genome in a relaxed circular conformation (RC DNA) within a nucleocapsid. A lipid bilayer envelope surrounds the nucleocapsid. A distinct feature of hepadnaviruses is that genome replication involves reverse transcription of an RNA intermediate, the pregenomic RNA (pgRNA) (for a review, see reference 3). After entry into a host cell, the genome is converted into covalently closed circular DNA (cccDNA) in the nucleus. All viral RNAs are transcribed from cccDNA by RNA polymerase II. Central to genome replication is pgRNA, which is the mRNA for both the nucleocapsid protein (C protein) and the multifunctional polymerase (P protein) as well as the template for reverse transcription. pgRNA is terminally redundant and contains three copies of a short (∼11- to 12-nucleotide [nt]) direct repeat sequence (DR) (Fig. 1A). Two copies of DR (DR1) are found in the terminal redundancies (R) of pgRNA. A third copy (DR2) is upstream of the 3′ copy of DR1.
FIG. 1.
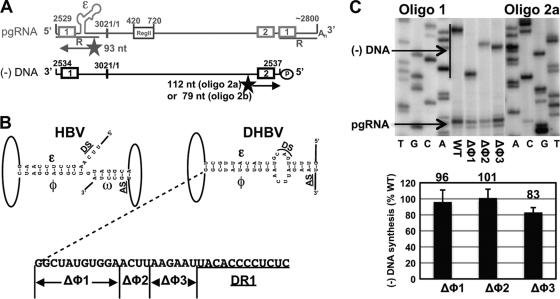
A φ-like sequence does not contribute to synthesis of minus-strand DNA in DHBV. (A) Representation of DHBV pgRNA and minus-strand DNA and relative locations of oligonucleotides used in primer extension. Oligonucleotide 1 anneals near the 5′ end of the pgRNA and produces an extension product of approximately 93 nt. Oligonucleotide 2a anneals near the 5′ end of minus-strand DNA, and produces an extension product of 112 nt, while oligonucleotide 2b produces an extension product of 79 nt. Gray and black boxes 1 and 2, direct repeats 1 and 2 (nt 2535 to 2546 and 2477 to 2488, respectively). ɛ, epsilon (nt 2560 to 2616) (3). Region II, nt 450 to 720. R, terminal redundancy (nt 2529 to ca. 2800). 5′ and 3′ end coordinates are depicted, as well as the unique EcoRI site (at nt 3021 and nt 1). (B) Base pairing between ɛ, φ, and ω on pgRNA for HBV (2), model for DHBV as proposed by Tang and McLachlan (20), and deletions analyzed in DHBV. DS, donor site for minus-strand template switch. AS, acceptor site for minus-strand template switch. φ, phi (nt 1771 to 1789). ω, omega (nt 1830 to 1835). (C) Levels of minus-strand DNA as determined by primer extension. Sequencing ladders produced from linearized plasmid DNA and the individual oligonucleotides were markers. The histogram shows levels of minus-strand DNA for initial deletion mutants (see text for calculation). n = 4 for Δφ3. Standard deviations are shown.
Genome replication begins in the cytoplasm with binding of the P protein to the encapsidation signal (ɛ) on pgRNA. This complex then becomes encapsidated into a nascent nucleocapsid. Synthesis of the first-strand (or minus-strand) DNA begins, using a tyrosine residue within P protein as a primer and a bulge within ɛ as the template. After synthesis of several nucleotides, there is a template switch from ɛ to an acceptor site located near the 3′ end of pgRNA. After this template switch, synthesis of minus-strand DNA [(−)DNA] resumes and proceeds to the 5′ end of pgRNA. As minus-strand DNA is synthesized, the P protein (which has RNase H activity) digests the pgRNA. The final cleavage leaves a short segment (∼17 nt) of pgRNA annealed to the 3′ end of the minus-strand DNA. This RNA segment contains DR1 at its 3′ end and is the primer for initiation of synthesis of the second-strand (or plus-strand) DNA. Synthesis of plus-strand DNA proceeds via one of two pathways. In the minor pathway, the RNA primer remains annealed to the 3′ end of the minus-strand DNA and is extended from the site of the final cleavage. This pathway gives rise to a duplex linear (DL) form of the genome. The major pathway results in the formation of RC DNA. The RNA primer is translocated to the complementary DR2 sequence, which is near the 5′ end of the minus-strand template. Synthesis of plus-strand DNA initiates from DR2 and proceeds to the 5′ end of the template. At this time the final template switch occurs, from the 5′ end to the 3′ end of the template. Elongation results in RC DNA. Nucleocapsids containing both forms of DNA are enveloped and secreted, but only virions containing RC DNA can sustain an infection (24).
For many viruses, the replication templates contain cis-acting sequences that participate in replication (for examples, see references 19, 21, 22, 25, and 26). Previous studies have identified cis-acting sequences within pgRNA that are important for its encapsidation for both mammalian and avian hepadnaviruses (9, 16). Additionally, sequences within the minus-strand DNA that contribute to the synthesis of plus-strand DNA have been identified (8, 12). Recent studies have characterized cis-acting sequences within pgRNA of HBV that contribute to synthesis of minus-strand DNA (1, 2). These studies also found that most of the pgRNA could be deleted without affecting levels of minus-strand DNA. Much less is known about sequences within pgRNA of avian hepadnaviruses that participate in synthesis of minus-strand DNA. We examined the genomes of two avian hepadnaviruses, duck HBV (DHBV) and heron HBV (HHBV), for the presence of cis-acting sequences that contribute to synthesis of minus-strand DNA. We analyzed variants of the two viruses that contained deletions in the genomic sequence. Collectively these deletions represent the majority of the sequence of pgRNA. We found that DHBV and HHBV differ in the sequences necessary for synthesis of minus-strand DNA. Deletions in pgRNA of DHBV sometimes decreased levels of minus-strand DNA, while deletions in the HHBV pgRNA almost always led to decreased levels. We found one similarity between the two avian hepadnaviruses: the sequence within DR2 contributes to synthesis of minus-strand DNA. However, we found that the sequence within DR2 does not play a role in the synthesis of minus-strand DNA for HBV. Finally, we found that a proposed secondary structure (ɛ:φ [20]) whose analog in HBV contributes to synthesis of minus-strand DNA does not play a role for DHBV. Our studies indicate that HBV, DHBV, and HHBV have different cis-acting sequences for the synthesis of minus-strand DNA. The differences between HBV, DHBV, and HHBV are likely a reflection of significant differences in the mechanisms they use to synthesize minus-strand DNA.
MATERIALS AND METHODS
Molecular clones.
All DHBV plasmids were derived from subtype DHBV3 (GenBank accession no. DQ195079) (18). All HHBV plasmids were derived from subtype HHBV4 (GenBank accession no. NC_001486) (17). All HBV plasmids were derived from subtype ayw (GenBank accession no. V01460 J02203) (5). LacZ sequence (GenBank accession no. V00296) (10) was amplified from pON249 (6). Nucleotide position 1 in DHBV3 and ayw is the C of the unique EcoRI site present in the genome, while position 1 in HHBV is the C of the analogous EcoRI site of DHBV. The wild type (WT) reference clone of DHBV, MM166, expresses pgRNA under the control of the cytomegalovirus (CMV) immediate-early promoter. The WT reference clone of HBV, NL84 (6), also expresses pgRNA using the CMV promoter. The WT HHBV reference clone, HHBV P−C− (11), expresses pgRNA from the endogenous HHBV promoter. The DHBV and HHBV reference plasmids do not express C or P proteins due to premature stop codons in the respective genes; the HBV reference does not express any viral proteins. To engender replication in our cell cultures, the C and P proteins were expressed from donor plasmid 538-10 (DHBV) (8), KM99 (HHBV) (14), or LJ96 (HBV) (6), all of which lack a functional encapsidation signal, ɛ. Details of construction and sequence of any plasmid will be provided upon request. All deletion variants described are lacking the specific sequence denoted. All variants were constructed either by removing restriction fragments or by PCR-mediated, site-specific mutagenesis. All deletion junctions were determined by DNA sequencing. All fragments generated by PCR were sequenced in their entirety to ensure that fortuitous mutations were absent.
Cell cultures and transfections.
The chicken hepatoma cell line LMH was used in the analyses of DHBV and HHBV. The human hepatoma cell line Huh7 was used to study HBV. Cells were grown on 60-mm plates at 37°C with 5% CO2, in Dulbecco modified Eagle medium (DMEM)/F12 medium supplemented with 5% fetal bovine serum (FBS). Cells were approximately 75% confluent or greater at the time of transfection. DNA transfections were performed using calcium phosphate precipitation. In each transfection, 5 μg of the expression plasmid for pgRNA and 4.5 μg of the expression plasmid for C and P proteins (DHBV and HHBV) or C, P, and X proteins (HBV) were used; 0.5 μg of a green fluorescent protein (GFP) expression plasmid was also included to monitor the efficiency of the transfection. Typically, medium containing the calcium phosphate precipitate was removed after 16 h, fresh medium was added, and the cultures were grown for additional 72 h.
Isolation of viral nucleic acid.
Nucleic acid (both RNA and DNA) from cytoplasmic capsids was isolated as described previously (11). Briefly, cells were lysed in a solution containing 50 mM Tris, 1 mM EDTA, and 0.2% NP-40 (pH 8.0). Nuclei were pelleted via centrifugation, and supernatant was collected. This cytoplasmic lysate was treated with 45 units of micrococcal nuclease in the presence of 2 mM CaCl2 to digest plasmid DNA and unencapsidated pgRNA, followed by treatment with 0.4% SDS and 0.4 mg/ml pronase to digest nucleocapsids and P protein. Encapsidated nucleic acids were extracted with phenol-chloroform, precipitated with ethanol and sodium chloride, and resuspended in 30 μl of a solution of 10 mM Tris and 0.1 mM EDTA (pH 8.0).
Primer extension analysis.
Primer extension was performed with two DNA oligonucleotides that were end labeled with [γ-32P]ATP (Fig. 1A). For analysis of DHBV and HHBV, oligonucleotide 1 (5′-GGTACATACCAAAGGTACAGTCAC-3′) anneals near the 5′ end of pgRNA of both DHBV (nt 2622) and HHBV (nt 2628) and generates a product of 93 nt. Oligonucleotide 2a (5′-TCCTGCTGACGGCCCATCCAGGC-3′) anneals near the 5′ end of minus-strand DNA of DHBV (nt 2425) and generates a product of 112 nt. Oligonucleotide 2b (5′-TGATTGGACGGCTTTTCCA-3′) anneals near the 5′ end of minus-strand DNA of DHBV (nt 2458) and generates a product of 79 nt. Oligonucleotide 3 (5′-TCCTGCTGATGGCCCAACCAGGC-3′) anneals near the 5′ end of minus-strand DNA of HHBV (nt 2431) and generates a product of 111 nt. For HBV analysis, oligonucleotide 1 (5′-GAGAGTAACTCCACAGTAGCTCC-3′) anneals near the 5′ end of pgRNA (nt 1948) and generates a product of 132 nt. Oligonucleotide 2 (5′-CTCTTGGACTCTCAGCAATGTCAAC-3′) anneals near the 5′ end of minus-strand DNA beginning at nt 1661 and generates a product of 165 nt. A single reaction, with a mixture containing two oligonucleotides, was performed using nucleic acid isolated from cytoplasmic capsids corresponding to 1/10 of the cells from a transfected plate. Samples were heat denatured prior to the primer extension reactions. The primer extension reactions were performed using a mixture of Vent exo− DNA polymerase (NEB; 1 U per reaction) and Transcriptor reverse transcriptase (Roche; 1 U per reaction), 1pmol of each oligonucleotide, and 200 μM deoxynucleoside triphosphates (dNTPs). Reactions were performed in 1× Vent reaction buffer [20 mM Tris-HCl (pH 8.8 at 25°C), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100]. Oligonucleotide annealing and extension were performed at 55°C for 45 min, and samples were heat denatured in a formamide loading dye prior to electrophoresis. These DNA products were electrophoresed through a 6% polyacrylamide sequencing gel containing 7.6 M urea and 1× Tris-borate EDTA (TBE), using 1× TBE as an electrophoresis buffer. Gels were dried under vacuum, and autoradiography was performed using either a Storm 860 or Typhoon 8600 PhosphorImager (Molecular Dynamics). Levels of DNA were measured using the software ImageQuant 5.2 (Molecular Dynamics).
Statistical analysis.
Statistical analysis was performed using the software Mstat 5.2 (Norman Drinkwater, McArdle Laboratory for Cancer Research, University of Wisconsin [http://www.mcardle.wisc.edu/mstat]). All data were analyzed using the Wilcoxon rank sum test (two sided). A P value of <0.05 was considered significant. Unless indicated otherwise, n was ≥5 for all comparisons between variants and WT references.
RESULTS
DHBV does not have a φ element.
HBV has a cis-acting sequence, φ, that makes an important contribution to synthesis of minus-strand DNA (20). φ is located on pgRNA ∼35 nt upstream of the acceptor site for the minus-strand template switch. φ contributes to synthesis of minus-strand DNA mainly through base pairing with two other sequences on HBV pgRNA; the 5′ half of ɛ and ω, which is immediately downstream of the acceptor site for the minus-strand template switch (2) (Fig. 1B). It has been proposed that DHBV pgRNA contains a sequence in an analogous position to φ that could potentially base pair with the 5′ half of ɛ (20). To date a functional analysis of this sequence has not been reported. To determine whether the proposed φ sequence in DHBV contributes to the synthesis of minus-strand DNA, we analyzed three mutants that contained deletions within this sequence (Fig. 1B). Plasmids expressing these variants, as well as a plasmid expressing a WT reference, were transfected into LMH cells. The levels of encapsidated pgRNA and minus-strand DNA in each sample were measured by primer extension (Fig. 1C). The ability of these variants to synthesize minus-strand DNA was calculated as the level of minus-strand DNA as a function of total pgRNA encapsidation events. The total number of pgRNA encapsidation events is the sum of the levels of pgRNA and minus-strand DNA in each sample. The ability of a variant to synthesize minus-strand DNA relative to a WT reference is described by the following formula:
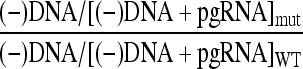 |
Using this formula, we will detect defects in both the early steps of minus-strand synthesis, such as template switching, and the later steps, such as elongation of minus-strand DNA.
We found that all variants were indistinguishable from the WT reference (Fig. 1C). Similar types of mutations introduced into HBV φ lead to significant decreases in the levels of minus-strand DNA (1). We conclude that the φ:ɛ base-pairing mechanism, as proposed by Tang and McLachlan (20), is not used by DHBV.
Identification of pgRNA sequences that contribute to synthesis of minus-strand DNA in DHBV.
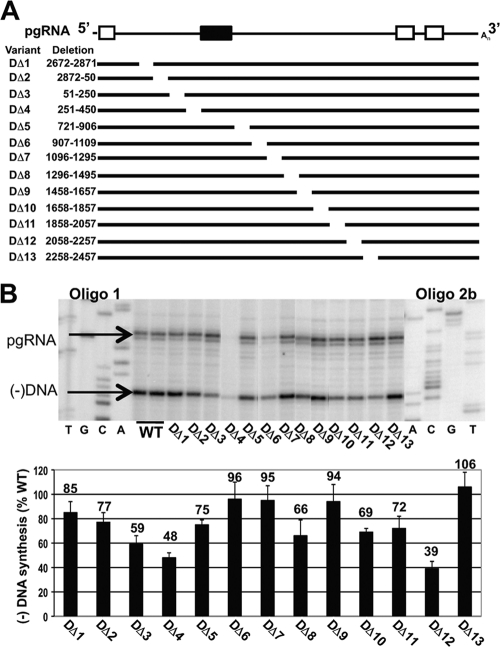
We hypothesized that DHBV contained one or more discrete cis-acting sequences that are important for the synthesis of minus-strand DNA. To identify these sequences, we analyzed a set of 13 variants that each contained deletions of 200 nt and collectively represent most of the DHBV genome (Fig. 2A). It was necessary to avoid deleting regions known to be critical in steps in replication such as pgRNA encapsidation (e.g., ɛ or region II). We analyzed these variants by Southern blotting (not shown) and primer extension (Fig. 2B). Using primer extension, we found that five variants accumulated minus-strand DNA to levels ranging from 85% to 106% of the WT reference level and that six other variants were within the range from 59% to 77% (Fig. 2B). Of the remaining two variants, DΔ12 (nt 2058 to 2257 removed) supported synthesis of minus-strand DNA at 39% of the WT reference level. This decrease in intracellular DNA levels was not due to increased release of virions and naked capsids into the cell culture medium (as analyzed by Southern blotting [data not shown]). The remaining variant, DΔ4 (nt 251 to 450 removed), accumulated low levels of both encapsidated pgRNA and minus-strand DNA. This mutant accumulated low levels of total pgRNA in the cytoplasm as well (as analyzed by Northern blotting [data not shown]). Nevertheless, we determined that DΔ4 synthesized minus-strand DNA at 48% of the WT reference level. In summary, most DHBV variants synthesized minus-strand DNA at between 48 to 106% of the WT reference level, with one variant, DΔ12, displaying a larger decrease. Levels of minus-strand DNA detected by Southern blotting (not shown) yielded results similar to those of the primer extension analysis.
FIG. 2.
Analysis of 200-nt deletions in DHBV. (A) Representation of 200-nt deletion variants. Nucleotide coordinates refer to the specific sequence deleted or substituted in the WT pgRNA. Oligonucleotide 2b anneals near the 5′ end of minus-strand DNA and produces an extension product of approximately 79 nt. cis sequences necessary for encapsidation (i.e., ɛ and region II) were not analyzed. White boxes, DR sequences. Black box, region II. (B) Primer extension analysis of variants. Products representing the 5′ ends of pgRNA and minus-strand DNA are indicated. Relative levels of minus-strand DNA were calculated as described in text.
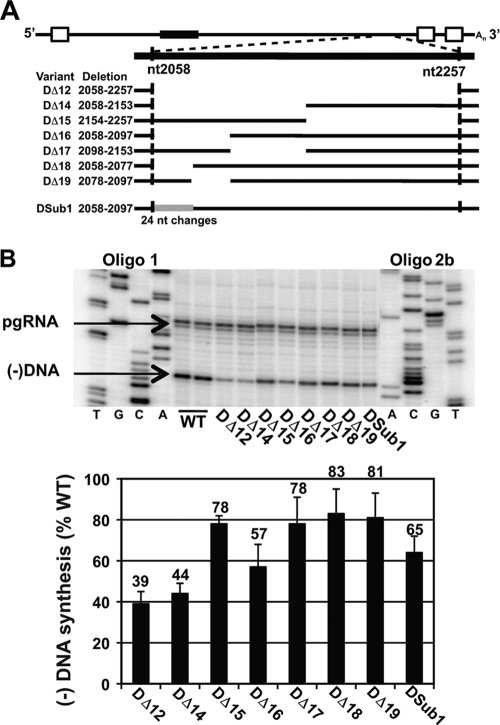
We hypothesized that a discrete cis-acting element was removed in DΔ12 (nt 2058 to 2257). We made smaller deletions within this sequence (Fig. 3A). Analysis of deletions of approximately 100 nt (DΔ14 and DΔ15) indicated that the putative element was located within nt 2058 to 2153 (Fig. 3B). When we analyzed deletions within nt 2058 to 2153 (variants DΔ16 and DΔ17), we found decreases in levels of minus-strand DNA for both, albeit less severe than for DΔ14 (57% and 78% of the WT reference level). The magnitudes of the decreases were different from that for DΔ14 (P = 0.03 and 0.008, respectively). However, subsequent dissection of the region from nt 2058 to 2097 (DΔ18 and DΔ19) led to even less severe decreases. One interpretation of these results is that there are multiple discrete elements contained within nt 2058 to 2153, with most within nt 2058 to 2097. When we compare the levels of minus-strand DNA of two subdeletions to that of the larger deletion, we find an additive effect. For example, when the relative levels of minus-strand DNA for DΔ16 and DΔ17 (57% and 78%, respectively) are multiplied, the product is 44%, which is the level of minus-strand DNA for DΔ14. This trend is true for every comparison of a deletion and its subdeletions in this region.
FIG. 3.
Localization of cis-acting sequences within nt 2058 to 2257 of DHBV. (A) Deletion variants within nt 2058 to 2257 and substitution variant DSub1. Nucleotide coordinates refer to the specific sequence deleted or substituted in the WT pgRNA. See text for description of the Dsub1 sequence. (B) Primer extension analysis of deletion variants and DSub1. Products representing the 5′ ends of pgRNA and minus-strand DNA are indicated. Sequencing ladders are as indicated in Fig. 2. Relative levels of minus-strand DNA were calculated as described in the text.
Lastly, we analyzed a variant (DSub1) in which HHBV4 sequence (nt 2064 to 2103) was substituted for nt 2058 to 2097 in our WT reference (Fig. 3A). This substitution resulted in 24 nucleotide changes. DSub1 made minus-strand DNA at a level comparable to that for its deletion counterpart (DΔ14) (Fig. 3B). This result supports the interpretation that cis-acting sequences for synthesis of minus-strand DNA of DHBV are contained within this region.
For HHBV the majority of pgRNA is indispensable for efficient synthesis of minus-strand DNA.
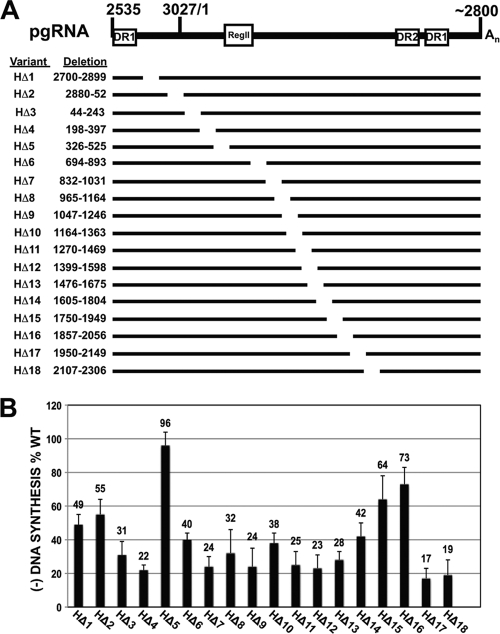
To determine whether the distribution of cis-acting sequences for synthesis of minus-strand DNA was conserved among avian hepadnaviruses, we carried out a similar analysis with HHBV. Relative to DHBV, HHBV is the most distantly related avian hepadnavirus based on nucleotide sequence phylogeny. We analyzed a previously described set of 18 variants of HHBV that each contained a 200-nt deletion (15) (Fig. 4A). Unlike for DHBV, most (14 of 18) of the variants synthesized minus-strand DNA at <50% of the WT reference level, with many being at less than 25% (Fig. 4B). Only one variant (HΔ5, nt 326 to 525) supported synthesis of minus-strand DNA at a level similar to the WT reference level. This difference between DHBV and HHBV indicates that these two avian hepadnaviruses have a significant difference in their mechanisms of synthesis of minus-strand DNA.
FIG. 4.
Deletion analysis of HHBV pgRNA. (A) Representation of HHBV pgRNA and deletion variants analyzed. Nucleotide coordinates refer to the specific sequence deleted in the WT pgRNA. Similar to the DHBV analysis, cis sequences necessary for encapsidation (i.e., ɛ and region II) were not analyzed. (B) Relative levels of minus-strand DNA as determined by primer extension analysis. For these and subsequent HHBV mutants, oligonucleotide 3 was used to measure levels of minus-strand DNA. This oligonucleotide produces a DNA product of 111 nt. Measurements were calculated as described in text.
DR2 contributes to minus-strand DNA synthesis in both DHBV and HHBV but not in HBV.
For all hepadnaviruses, synthesis of the plus strand of RC DNA initiates at DR2 and, as such, plays a critical role in synthesis of plus-strand DNA. Previously it was reported that mutations in DR2 of DHBV had the unexpected consequence of decreasing the level of minus-strand DNA (13). The mechanism by which this sequence contributes to synthesis of minus-strand DNA in DHBV is not understood. To determine whether sequence within DR2 plays a role in the synthesis of minus-strand DNA for other hepadnaviruses, we analyzed mutations of DR2 in HHBV and HBV. We found that removing DR2 from the HHBV genome resulted in the synthesis of less minus-strand DNA (Fig. 5A). The magnitude of the decrease for HHBV (24% of the WT reference level) was comparable to the same mutation in DHBV (21% of the WT reference level). Thus, despite clear differences in the sequence requirements of DHBV and HHBV in minus-strand DNA synthesis (see, e.g., Fig. 2 and 4), both viruses (and possibly all avian hepadnaviruses) use some aspect of DR2 in the synthesis of minus-strand DNA. This finding illustrates a shared mechanism in the synthesis of minus-strand DNA between HHBV and DHBV.
FIG. 5.
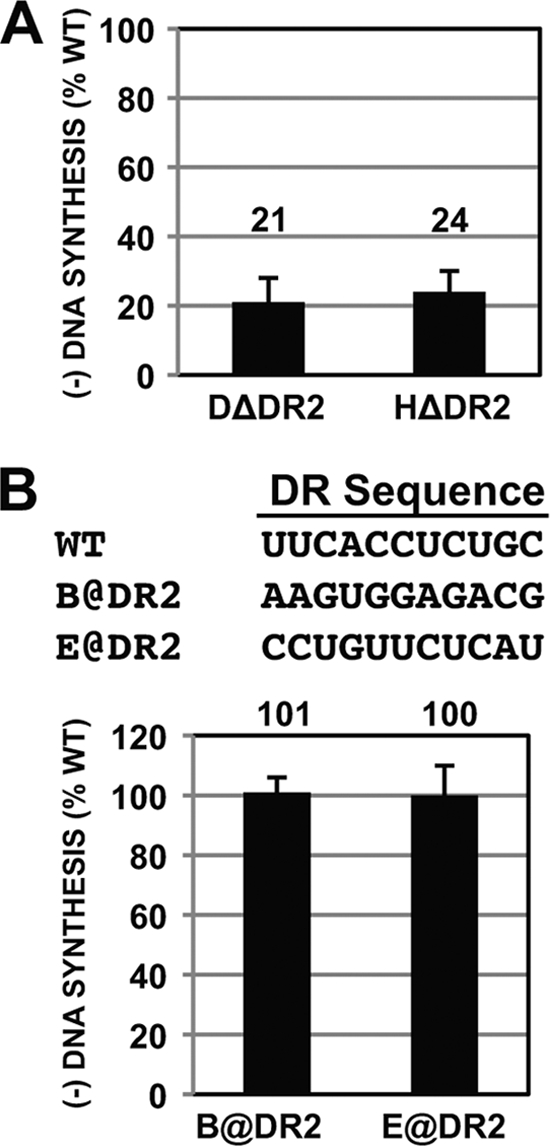
DR2 contributes to synthesis of minus-strand DNA for both DHBV and HHBV but not for HBV. (A) For both DHBV and HHBV, variants containing deletions of DR2 were analyzed for their ability to synthesize minus-strand DNA. Relative levels of minus-strand DNA were determined by primer extension analysis. (B) Sequence of HBV DR2, nucleotide changes in DR2 mutants, and primer extension analysis of HBV DR2 variants. The histogram shows relative levels of minus-strand DNA for DR2 variants. Measurements were calculated as described in the text.
To determine the role of DR2 in synthesis of minus-strand DNA in HBV, we analyzed two variants that have been characterized previously for their ability to support synthesis of plus-strand DNA (7) (Fig. 5B). These variants have the entire 11 nt of DR2 substituted (they are called variants A@DR2 and E@DR2 in Table 1 of reference 7). We found that these mutants synthesized minus-strand DNA at a level similar to that for the WT reference. Thus, HHBV and DHBV share a requirement for DR2 (or some sequence within it) in the synthesis of minus-strand DNA, while HBV does not.
Unlike for HBV, pgRNAs of DHBV with larger deletions are not reverse transcribed as efficiently as the WT.
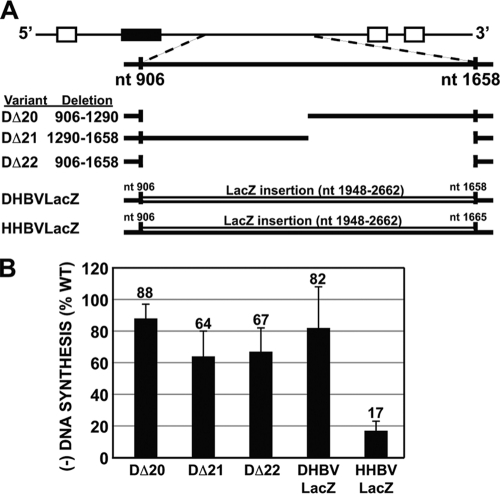
It was shown that derivatives of HBV pgRNA that were shorter than the WT (as small as ∼900 nt) could be reverse transcribed efficiently, indicating that HBV pgRNA does not need to be a specific size or within a size range (2). We asked whether DHBV pgRNAs with deletions larger than 200 nt would serve as templates for synthesis of normal levels of minus-strand DNA. Clearly, having a pgRNA template 200 nt smaller than normal did not, per se, affect the synthesis of minus-strand DNA (see, e.g., DΔ6 and DΔ13 in Fig. 2). We chose to remove sequence from the central region (nt 906 to 1658) of DHBV pgRNA because it was relatively insensitive to mutation (Fig. 2). We analyzed two variants containing deletions of 385 nt (DΔ20) and 368 nt (DΔ21) and a variant that combined these two deletions (DΔ22; 753 nt removed) (Fig. 6A). DΔ20 and DΔ21 showed levels of minus-strand DNA that were 88% and 64% of the WT reference level, respectively (Fig. 6B). The variant with the combined deletion, DΔ22, supported minus-strand DNA synthesis at a level similar to that for DΔ21 (67% versus 64%, respectively). All three variants were significantly different than the WT reference (P = 0.02, 0.0005, and 0.0004, respectively).
FIG. 6.
Larger deletions in DHBV lead to lower levels of minus-strand DNA; sequence substitutions have different effects in DHBV and HHBV. (A) Mutants analyzed to examine the contribution of deletion size and sequence specificity in DHBV and HHBV. For deletions, nucleotide coordinates refer to the specific sequence deleted in the WT pgRNA. For LacZ substitutions, nucleotide coordinates refer to the sequence of LacZ inserted into DHBV or HHBV. (B) Relative levels of minus-strand DNA as determined by primer extension analysis. Measurements were calculated as described in the text.
LacZ substitutions have different effects on synthesis of minus-strand DNA for DHBV and HHBV.
Building upon the previous analysis, we asked whether synthesis of minus-strand DNA could be restored to wild-type levels when we restored the DHBV pgRNA to wild-type size, albeit with a different sequence. We replaced the sequence removed from DΔ22 with a fragment of LacZ sequence (DHBVLacZ) and found that levels of minus-strand DNA were at 82% of the WT reference level. When a similar substitution (HHBVLacZ) in HHBV was examined, it made decreased levels of minus-strand DNA (17% of the WT reference level) compared to DHBV (Fig. 6B). While both substitutions are different than their WT references (P = 0.005 and 0.02, respectively; n = 4 for HHBV samples), clearly these substitutions had dramatically different effects on the synthesis of minus-strand DNA. These results are consistent with the earlier interpretation (Fig. 4) that HHBV contains multiple sequences that make contributions to synthesis of minus-strand DNA.
DISCUSSION
This study allows us to compare and contrast the cis-acting requirements for synthesis of minus-strand DNA for DHBV, HHBV, and HBV. We found more differences than similarities, not only between HBV and the two avian hepadnaviruses but also between DHBV and HHBV. We take our results and interpretations to indicate that the process of synthesis of minus-strand DNA has multiple mechanistic components and that it is not uncommon for a given family member to evolve and use distinct mechanisms to accomplish the same goal.
Consideration of DHBV.
In HBV, base pairing between specific sequences on the pgRNA (ɛ, φ, and ω) is critical for efficient synthesis of minus-strand DNA (2). For DHBV, a secondary structure similar to the base pairing between ɛ and φ of HBV was proposed but not tested for its role in synthesis of minus-strand DNA (20). When we deleted portions of the proposed φ sequence of DHBV, we saw no reduction in the synthesis of minus-strand DNA (Fig. 1). Thus, we conclude that the ɛ/φ base pairing, as proposed by Tang and McLachlan (20), does not take place with DHBV.
We searched ∼2,500 nt of the DHBV genome for the presence of cis-acting sequences involved in synthesis of minus-strand DNA. Many segments of the genome could be removed with no impact or, at most, a 50% reduction in the level of minus-strand DNA. We found two regions, one between nt 251 and 450 and the other between nt 2058 and 2257, that when removed resulted in 48% and 39%, respectively, of the normal level of minus-strand DNA. We attempted to map the location of the cis-acting sequence within nt 2058 to 2257 and were able to further localize this sequence to be within a 96-nt region (nt 2058 to 2153). Attempts to further localize the element were ambiguous, possibly indicating the presence of multiple discrete elements that act independently. How the sequence within nt 2058 to 2153 contributes mechanistically to synthesis of minus-strand DNA is not known at this time. Because the conformation of pgRNA plays an important role in synthesis of minus-strand DNA for HBV, as well as in synthesis of plus-strand DNA for both HBV and DHBV, it is tempting to postulate that DHBV also uses a similar strategy in synthesis of minus-strand DNA. Computer-aided searches for potential base pairing between this region (nt 2058 to 2153) and other sequences (ɛ, acceptor site, DR2, and DΔ4) known to contribute to minus-strand DNA synthesis did not suggest any testable leads (data not shown).
Consideration of HHBV.
The analysis of HHBV yielded a picture quite different from that for DHBV; most deletions led to large decreases in the levels of minus-strand DNA synthesized (Fig. 4). Only one deletion mutant (HΔ5, nt 326 to 525) synthesized minus-strand DNA at a level similar to the WT reference level. Only three deletion mutants synthesized minus-strand DNA at greater than 60% of the WT reference level. This sensitivity to perturbation suggests that HHBV has many cis-acting requirements for synthesis of minus-strand DNA.
Comparison of DHBV and HHBV.
In the 200-nt deletion analyses (Fig. 2 and 4), we scanned 2,536 nt of DHBV pgRNA and 2,466 nt of HHBV pgRNA. Comparison of the histograms in Fig. 2B and 4B indicates a significant difference between DHBV and HHBV. For HHBV, removal of sequences over a cumulative span of 1,822 nt (HΔ3 to HΔ4, 354 nt; HΔ6 to HΔ14, 1,111 nt; HΔ17 to HΔ18, 357 nt) resulted in a reduction in minus-strand DNA to less than or equal to 42% of the WT reference level. This sensitivity of HHBV is not a complete intolerance for a smaller-than-WT-size pgRNA, because the variant HΔ5 (Fig. 4), which has 200 nt removed, synthesizes minus-strand DNA as well as the WT reference. In contrast, only one DHBV deletion (DΔ12) was below 48%. These results suggest that HHBV has more cis-acting requirements for synthesis of minus-strand DNA than DHBV. This interpretation is reinforced upon consideration of the variants DHBVLacZ and HHBVLacZ (Fig. 6), in which the analogous ∼750-nt segments of the two genomes were replaced with the same segment of LacZ. The DHBV derivative made minus-strand DNA at 82% of the WT reference level, while the analogous HHBV derivative was at 17%. The underlying mechanistic reason for the sensitivity of HHBV to mutation is not clear. It seems to indicate that HHBV has more cis-acting requirements than DHBV for synthesis of minus-strand DNA.
Mutations within the DR2 sequences of DHBV and HHBV, but not HBV, lead to less synthesis of minus-strand DNA. We think that this finding is not influenced by the nature of the mutation (deletions within DHBV and HHBV versus substitutions within HBV). An earlier study (13) showed that for DHBV, both deletions and substitutions of DR2 resulted in less synthesis of minus-strand DNA. DR2 is the acceptor site during primer translocation, which occurs early in the synthesis of plus-strand DNA for all hepadnaviruses. A role for this sequence in synthesis of minus-strand DNA is surprising. It is possible that DR2 plays a role in the earliest steps of minus-strand DNA synthesis by contributing a conformation of pgRNA favorable for synthesis of the first nucleotides of minus-strand DNA or the minus-strand DNA template switch. It is also possible that the role of DR2 in synthesis of minus-strand DNA does not develop until after the template switch, during elongation of minus-strand DNA. This role would need to manifest during the early stages of elongation, as our assay detects a minus-strand DNA with a minimum size of 79 nt. As this requirement for DR2 is shared by two phylogenetically divergent avian hepadnaviruses, it is a mechanism that warrants further study.
Taken as a whole, our analysis allows us to highlight a central concept: hepadnaviruses may share an overall strategy for synthesis of minus-strand DNA (i.e., priming from ɛ, a template switch after initiation of 4 nt, and elongation to the 5′ end of pgRNA) but individual members appear to have evolved significant differences in their mechanisms for synthesis of minus-strand DNA. At this time we cannot discern the exact nature of these differences, due in part to the lack of a suitable in vitro system to study the early steps of minus-strand synthesis. These differences between DHBV, HHBV, and HBV could be a reflection of the use of different, unidentified host factors in the synthesis of minus-strand DNA. Alternatively, there may be structural differences in the viral components involved in synthesis of minus-strand DNA (e.g., pgRNA, the viral polymerase, or even the nature of the capsid) that lead to the different sequence requirements. These differences could influence interactions between the different viral components directly, or they may contribute to an optimal environment for efficient synthesis of minus-strand DNA.
Acknowledgments
We thank Kristin Ostrow for constructing the HHBV deletion plasmids used in this study. We thank Natalie Greco, Thomas Lentz, Eric Lewellyn, and Dan Yuan for thoughtful and constructive criticism of the manuscript.
This work was supported by National Institutes of Health grant R01-AI060018.
Footnotes
Published ahead of print on 6 October 2010.
REFERENCES
- 1.Abraham, T. M., and D. D. Loeb. 2006. Base pairing between the 5′ half of epsilon and a cis-acting sequence, phi, makes a contribution to the synthesis of minus-strand DNA for human hepatitis B virus. J. Virol. 80:4380-4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham, T. M., and D. D. Loeb. 2007. The topology of hepatitis B virus pregenomic RNA promotes its replication. J. Virol. 81:11577-11584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck J., and M. Nassad. 2007. Hepatitis B replication. World J. Gastroenterol. 13:48-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fields, B. N., D. M. Knipe, P. M. Howley, and D. E. Griffin. 2001. Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 5.Galibert, F., E. Mandart, F. Fitoussi, P. Tiollais, and P. Charnay. 1979. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature 281:646-650. [DOI] [PubMed] [Google Scholar]
- 6.Geballe, A. P., R. R. Spaete, and E. S. Mocarski. 1986. A cis-acting element within the 5′ leader of a cytomegalovirus beta transcript determines kinetic class. Cell 46:865-872. [DOI] [PubMed] [Google Scholar]
- 7.Haines, K. M., and D. D. Loeb. 2007. The sequence of the RNA primer and the DNA template influence the initiation of plus-strand DNA synthesis in hepatitis B virus. J. Mol. Biol. 370:471-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Havert, M. B., and D. D. Loeb. 1997. cis-acting sequences in addition to donor and acceptor sites are required for template switching during synthesis of plus-strand DNA for duck hepatitis B virus. J. Virol. 71:5336-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch, R. C., D. D. Loeb, J. R. Pollack, and D. Ganem. 1991. cis-acting sequences required for encapsidation of duck hepatitis B virus pregenomic RNA. J. Virol. 65:3309-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalnins, A., K. Otto, U. Ruther, and B. Muller-Hill. 1983. Sequence of the lacZ gene of Escherichia coli. EMBO J. 2:593-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewellyn, E. B., and D. D. Loeb. 2007. Base pairing between cis-acting sequences contributes to template switching during plus-strand DNA synthesis in human hepatitis B virus. J. Virol. 81:6207-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, N., L. Ji, M. L. Maguire, and D. D. Loeb. 2004. cis-acting sequences that contribute to the synthesis of relaxed-circular DNA of human hepatitis B virus. J. Virol. 78:642-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loeb, D. D., R. Tian, and K. J. Gulya. 1996. Mutations within DR2 independently reduce the amount of both minus- and plus-strand DNA synthesized during duck hepatitis B virus replication. J. Virol. 70:8684-8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller-Hill, K., and D. D. Loeb. 1996. Previously unsuspected cis-acting sequences for DNA replication revealed by characterization of a chimeric heron/duck hepatitis B virus. J. Virol. 70:8310-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostrow, K. M., and D. D. Loeb. 2002. Characterization of the cis-acting contributions to avian hepadnavirus RNA encapsidation. J. Virol. 76:9087-9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollack, J. R., and D. Ganem. 1993. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J. Virol. 67:3254-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sprengel, R., E. F. Kaleta, and H. Will. 1988. Isolation and characterization of a hepatitis B virus endemic in herons. J. Virol. 62:3832-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sprengel, R., C. Kuhn, H. Will, and H. Schaller. 1985. Comparative sequence analysis of duck and human hepatitis B virus genomes. J. Med. Virol. 15:323-333. [DOI] [PubMed] [Google Scholar]
- 19.Suo, Z., and K. A. Johnson. 1998. DNA secondary structure effects on DNA synthesis catalyzed by HIV-1 reverse transcriptase. J. Biol. Chem. 273:27259-27267. [DOI] [PubMed] [Google Scholar]
- 20.Tang, H., and A. McLachlan. 2002. A pregenomic RNA sequence adjacent to DR1 and complementary to epsilon influences hepatitis B virus replication efficiency. Virology 303:199-210. [DOI] [PubMed] [Google Scholar]
- 21.Topping, R., M. A. Demoitie, N. H. Shin, and A. Telesnitsky. 1998. cis-acting elements required for strong stop acceptor template selection during Moloney murine leukemia virus reverse transcription. J. Mol. Biol. 281:1-15. [DOI] [PubMed] [Google Scholar]
- 22.Wang, J. B., D. E. Nixon, and M. A. McVoy. 2008. Definition of the minimal cis-acting sequences necessary for genome maturation of the herpesvirus murine cytomegalovirus. J. Virol. 82:2394-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organization. 2010, posting date. Epidemic and pandemic alert and response (EPR). http://www.who.int/mediacentre/factsheets/fs204/en/.
- 24.Yang, W., and J. Summers. 1998. Infection of ducklings with virus particles containing linear double-stranded duck hepatitis B virus DNA: illegitimate replication and reversion. J. Virol. 72:8710-8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu, L., M. Nomaguchi, R. Padmanabhan, and L. Markoff. 2008. Specific requirements for elements of the 5′ and 3′ terminal regions in flavivirus RNA synthesis and viral replication. Virology 374:170-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, B., H. Dong, D. A. Stein, P. L. Iversen, and P. Y. Shi. 2008. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology 373:1-13. [DOI] [PubMed] [Google Scholar]