Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs that post-transcriptionally regulate expression of their target messenger RNAs. We recently demonstrated that primary miRNA transcripts (pri-miRNAs) retained at transcription sites are processed with enhanced efficiency, suggesting that pri-miRNA processing is coupled to transcription in mammalian cells. We also observed that transiently expressed pri-miRNAs accumulate in nuclear foci with splicing factor SC35 and Microprocessor components, Drosha and DGCR8. Here, we show that pri-miRNAs containing a self-cleaving hepatitis delta ribozyme accumulate in the nucleoplasm after release from their transcription sites, but are not efficiently processed. Pri-miRNAs with ribozyme-generated 3′ ends do not localize to SC35-containing foci, whereas cleaved and polyadenylated pri-miRNA transcripts with or without the pre-miRNA hairpin do. Pri-miRNA/SC35 foci contain a number of proteins normally associated with SC35 domains, including ASF/SF2, PABII, and the prolyl isomerase, Pin1. In contrast, RNA polymerase II and PM/Scl-100 do not strongly colocalize with pri-miRNAs in SC35-containing foci. These data argue that pri-miRNA/SC35-containing foci are not major sites of pri-miRNA processing and that pri-miRNA processing is coupled to transcription. We discuss the implications of our findings relative to recent insights into miRNA biogenesis, mRNA metabolism, and the nuclear organization of gene expression.
Keywords: microRNA, pri-miRNA, SC35, Drosha, cotranscriptional
Introduction
MicroRNAs (miRNAs) are short noncoding RNAs that imperfectly base pair with target messenger RNAs (mRNAs) to control their translational output.1 MiRNAs have critical roles in cell growth, development and disease,2–6 thus understanding the mechanisms involved in miRNA expression is essential. Most miRNAs are transcribed as long primary miRNA transcripts (pri-miRNAs) by RNA polymerase II (Pol II)7,8 and undergo two sequential processing reactions to produce the mature miRNA.9 First, the pri-miRNA is recognized by the Microprocessor complex, which minimally consists of the RNase III-like enzyme, Drosha and its essential cofactor, DGCR8, but likely includes additional components.10–13 Drosha cleavage releases a hairpin precursor miRNA (pre-miRNA) of ~70 nucleotides (nts) that is exported to the cytoplasm by Exportin-5,14–16 and further processed by a second RNase III-like enzyme, Dicer.17,18 The mature ~22 nt miRNA strand is then incorporated into an RNA-induced silencing complex (RISC) and guides this complex to target mRNAs.19 MiRNA expression can be regulated at multiple levels, including transcription20,21 and several posttranscriptional steps, such as Drosha22 and Dicer processing.23 Despite some progress,24–27 the mechanisms involved in regulated miRNA expression remain poorly understood.
A now well-established concept in gene expression is that many mRNA processing events, including capping, splicing and 3′-end formation, are tightly coupled to transcription by Pol II.28–33 This coupling is important for the efficiency as well as the regulation of mRNA maturation.30,33 We recently reported that pri-miRNA processing efficiency is enhanced when pri-miRNAs remain tethered to the DNA template.34 Furthermore, we observed that pri-miRNAs containing a viral RNA element, the ENE, accumulate to high levels after release from the site of transcription, but are not efficiently processed.34 Consistently, an independent study by Morlando et al.35 directly demonstrated that Drosha is recruited to endogenous pri-miRNA chromosomal loci and that pri-miRNAs are processed cotranscriptionally. Together, these data suggest that pri-miRNA processing might be yet another event that is coupled to transcription by Pol II.
In the course of our studies,34 we also observed that overexpressed, unprocessed pri-miRNAs accumulate in nuclear foci containing the splicing factor SC35, as well as Microprocessor components Drosha and DGCR8. SC35 is a marker for subnuclear organelles known as SC35 domains, or splicing “speckles”, which are sites of high concentration of several proteins involved in mRNA metabolism, and are visible by electron microscopy as interchromatin granule clusters (IGCs).36,37 SC35 domains/IGCs are unlikely to be major sites of transcription or mRNA processing.38 Rather, proteins involved in mRNA maturation are usually recruited from SC35 domains to sites of active transcription, which are distributed throughout the nucleoplasm.39,40 Recently, a number of studies have highlighted the importance of SC35 domains in the coordination of gene expression.37,41,42 The data obtained in these studies are consistent with the proposed model of SC35 domains as dynamic “hubs”, which facilitate the expression of highly active Pol II-transcribed genes by spatially connecting their synthesis with the recycling of numerous protein complexes involved in mRNA metabolism.37
Here, we further explore the link between pri-miRNA processing and retention at the site of transcription. We also characterize in greater detail the pri-miRNA/SC35-containing nuclear foci observed in transfected cells. Our data support a model in which pri-miRNAs that escape processing at transcription sites accumulate in SC35-containing foci, perhaps to allow both recycling of bound proteins and surveillance of pri-miRNAs for quality control. These findings, in combination with recent insights into mRNA metabolism and its spatial organization within the nucleus have important implications for miRNA biogenesis and its regulation.
Results
Pri-miRNAs released from transcription sites by ribozyme cleavage are not efficiently processed
To further investigate the relationship between pri-miRNA processing efficiency, retention at transcription sites, and localization to SC35-containing foci, we investigated the processing and localization of pri-miRNAs that are released from the DNA template upon cleavage by a hepatitis delta (HD) ribozyme. The HD ribozyme is an RNA sequence that undergoes rapid self-cleavage after transcription.43 Pri-miRNA constructs were generated that contain the HD ribozyme sequence inserted at the 3′ end of pri-lin-4 (pri-lin-4HD; Fig. 1A). Control pri-miRNA constructs contained a mutant HD ribozyme (pri-lin-4HDm) with a single inactivating alteration of the catalytic cysteine to uracil (C76-U).44,45 Consequently, pri-lin-4HDm produces only transcripts that are cleaved and polyadenylated at the downstream bovine growth hormone (BGH) cleavage and polyadenylation (CPA) signal. A hairpin stem loop derived from the 3′ end of histone mRNA was inserted one base upstream of the ribozyme cleavage site to stabilize the 5′ RNA cleavage product (based on refs. 44, 46 and 47).
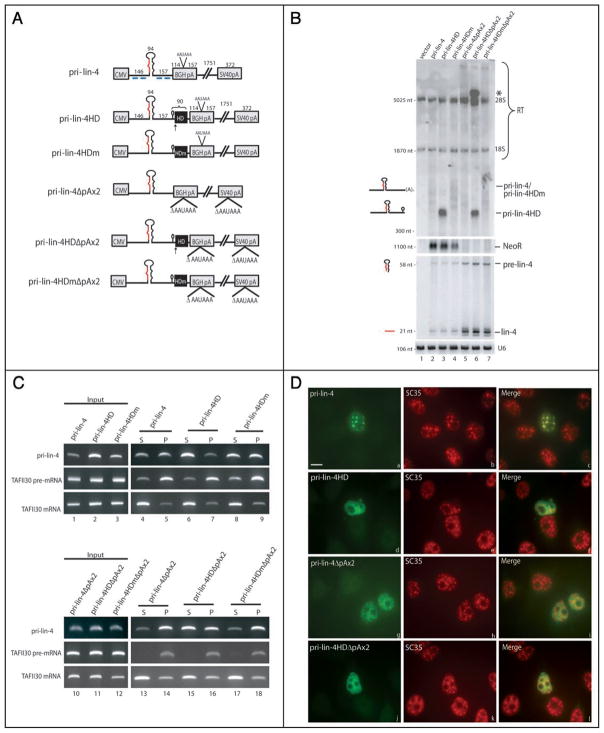
Figure 1.
Pri-miRNAs with ribozyme-generated 3′ ends are not processed efficiently and do not localize to SC35-containing nuclear foci. (A) Schematic of pri-miRNA constructs. CMV (cytomegalovirus), BGH (bovine growth hormone), and SV40 (simian virus 40) denote the vector-derived CMV promoter and the BGH or SV40 cleavage and polyadenylation signal. The pri-miRNA insert is shown in black, with the pre-miRNA represented as a large hairpin and the mature miRNA sequence indicated in red. Black boxes show the location of either the wild type (HD) or the mutant (HDm) ribozyme. Arrows indicate the site of ribozyme cleavage, with the small histone mRNA 3′ hairpin immediately upstream. The lengths in nts of the pri-miRNA insert and surrounding sequences are indicated above the pri-miRNAs. (B) Northern blot analysis of pri-miRNA processing. Constructs diagramed in A were transfected into HeLa cells, and total RNA was analyzed by Northern blotting using either a denaturing formaldehyde/agarose gel to detect pri-miRNAs (top) or a 15% denaturing urea/polyacrylamide gel to visualize the precursor and mature miRNAs (bottom). 28S and 18S rRNA are indicated and serve as loading controls. The neomycin resistance gene (NeoR) was probed as a transfection efficiency control (note that pri-lin-4ΔpAx2 constructs do not produce a NeoR transcript because the downstream SV40 CPA signal is deleted). Mature miRNA Northern blots were probed for the U6 small nuclear RNA as a loading control. RT indicates readthrough transcripts. The asterisk denotes the pri-lin-4HDΔpAx2 transcript that has transcribed around the plasmid and undergone ribozyme cleavage after the second encounter. (C) Nuclear fractionation of pri-miRNAs. Cells transfected as in (B) were fractionated into released, nucleoplasmic RNAs (supernatant [S]) and chromatin-associated RNAs (pellet [P]). Fractionated RNA was reverse transcribed and amplified by PCR. TAFII30 pre-mRNA and fully spliced mRNA were amplified as controls for loading and fractionation efficiency. (D) ISH to pri-lin-4 transcripts, followed by IF to SC35 on cells transfected as in (B), using DIG-labeled probes complementary to regions diagramed in (A) (blue lines). Scale bar, 10 μm.
To confirm that the inserted HD ribozyme is active, constructs diagramed in Figure 1A were transfected into HeLa cells, and the levels of pri-, pre- and mature miRNA were analyzed by Northern blotting. As seen in Figure 1B, pri-lin-4HD migrates at ~400 nts, the size predicted for transcripts that have undergone ribozyme cleavage just downstream of the pri-lin-4 insert (lane 3, upper). Pri-miRNAs with a ribozyme generated 3′ end were ~2.5-fold more abundant than the heterogeneous cleaved and polyadenylated pri-miRNAs that lack the HD ribozyme or contain its mutant form (lanes 2–4, upper; see also Fig. 1C, lanes 1–3). This increase in stability of pri-lin-4HD transcripts could be due to the presence of the histone stem loop or other variables (see Discussion). Importantly, despite the ~2.5-fold increase in the level of pri-miRNAs with a ribozyme generated 3′ end, no increase in pre- or mature miRNAs was observed (Fig. 1B, lanes 2–4, lower).
We hypothesized that pri-miRNAs containing ribozyme-generated 3′ ends may be inefficiently processed to pre-miRNAs because they are released from transcription sites. To test this hypothesis, we employed a nuclear fractionation procedure that separates chro- matin-associated transcripts from transcripts that have been released into the nucleoplasm.48,49 Constructs in Figure 1A were transfected into HeLa cells and nuclear fractionation was performed. TAFII30 mRNA and pre-mRNA served as controls for loading and fractionation efficiency. As shown in Figure 1C, pri-lin-4 transcripts are found approximately equally in the nucleoplasmic supernatant and in the chromatin-associated pellet (lanes 4 and 5). In contrast, pri-lin-4HD constructs are enriched in the nucleoplasmic supernatant, confirming that ribozyme-cleaved transcripts are indeed released from transcription sites (lanes 6 and 7). The presence of the mutant ribozyme did not alter the nuclear distribution of pri-lin-4 (lanes 8 and 9). These data demonstrate that ribozyme-cleaved pri-lin-4 transcripts accumulate after release from the DNA template, which correlates with inefficient processing (Fig. 1B).
We also investigated the processing of a pri-miRNA that contains the HD ribozyme but lacks a CPA signal (pri-lin-4HDΔpAx2 and pri-lin-4HDmΔpAx2; Fig. 1A). Consistent with our previous results, Figure 1B demonstrates that pri-miRNAs lacking a CPA signal are processed ~4-fold more efficiently than cleaved and polyadenylated pri-miRNAs (lanes 5 and 7). Nuclear fractionation confirmed that pri-miRNAs lacking a CPA signal are retained in the chromatin-associated pellet (Fig. 1C, lanes 13 and 14). For the ribozyme-cleaved pri-lin4HDΔpAx2 transcripts, we may have expected fewer mature miRNAs to be produced since they should be released from transcription sites. However, Figure 1B (lane 6) demonstrates that pri-lin-4HDΔpAx2 is processed with an efficiency comparable to that of pri-lin-4ΔpAx2 (lane 5) and pri-lin-4HDmΔpAx2 (lane 7), most likely because cleavage by the HD ribozyme is not 100% efficient; this is demonstrated by the clear accumulation of a pri-miRNA species above 5,100 nts (Fig. 1B, lane 6, asterisk), which is the size predicted for a transcript that escapes ribozyme cleavage initially, transcribes around the entire plasmid DNA template, and is cleaved when the HD ribozyme site is encountered the second time. Indeed, nuclear fractionation revealed that while pri-lin-4HDΔpAx2 yields a high level of transcripts released into the nucleoplasmic supernatant by ribozyme cleavage (Fig. 1C, lane 15), transcripts are also abundant in the chromatin-associated pellet (lane 16), confirming that some transcripts escape ribozyme cleavage. These data are consistent with our previous results and indicate that pri-miRNAs released into the nucleoplasm are not efficiently processed whereas pri-miRNAs retained at transcription sites are processed with enhanced efficiency.
Pri-miRNAs released from transcription sites by ribozyme cleavage do not localize to SC35-containing nuclear foci
We next investigated the localization of pri-miRNAs diagramed in Figure 1A by performing in situ hybridization (ISH) using DIG-tailed oligonucleotide probes complementary to regions of the pri-miRNA flanking the pre-miRNA hairpin (Fig. 1A, blue lines). Consistent with previous results,34 cleaved and polyadenylated pri-lin-4 transcripts accumulate in nuclear foci that colocalize with SC35 (Fig. 1D, parts a–c). In contrast, pri-lin-4HD transcripts do not strongly localize to SC35-containing foci (parts d–f). Presence of the mutant HD ribozyme sequence had no effect on localization, and pri-lin-4HDm accumulated in SC35-containing foci (data not shown). As demonstrated previously,34 efficiently processed pri-lin-4ΔpAx2 transcripts do not colocalize with SC35 domains, but rather localize diffusely in the nucleus (Fig. 1D, parts g–i), which is where the plasmid DNA templates also localize.34 Similarly, Figure 1D (parts j–l) reveals that pri-lin-4HDΔpAx2 transcripts distribute diffusely in the nucleus. This pattern likely arises from both the ribozyme-cleaved transcripts that are released into the nucleoplasm and transcripts that escape ribozyme cleavage and remain tethered to their DNA templates. These results, together with our previous observations,34 lead us to hypothesize that pri-miRNAs containing ribozyme-generated 3′ ends fail to localize to SC35-containing foci because they lack a polyadenylate (polyA) tail (see Discussion).
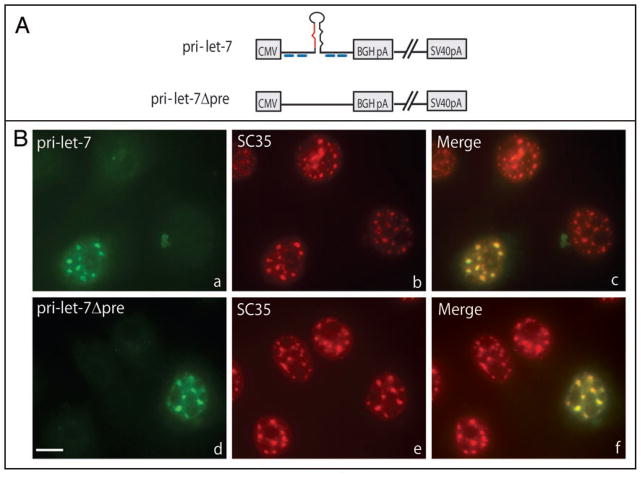
To determine if a miRNA sequence or a pre-miRNA hairpin is necessary for localization of transiently expressed pri-miRNAs to SC35-containing foci, we next investigated the localization of a pri-miRNA construct that undergoes CPA, but lacks the entire pre-miRNA sequence (Fig. 2A; pri-let-7Δpre). Figure 2B demonstrates that pri-let-7Δpre transcripts accumulate in nuclear foci (part d) that contain SC35 (part e), as do pri-let-7 transcripts (parts a–c). This result indicates that localization to SC35-containing nuclear foci is not dependent on the presence of a miRNA sequence or a pre-miRNA hairpin, but may be a more general feature of transcripts that are transcribed by Pol II and polyadenylated.
Figure 2.
Transcript localization to SC35-containing foci does not require a pre-miRNA hairpin. (A) Schematic of pri-miRNA constructs. (B) Cells transfected with pri-let-7 (a–c) or pri-let-7Δpre (d–f) were subjected to ISH (a and d) using DIG-labeled probes diagramed in (A) (blue lines), followed by IF for SC35 (b and e). Merged images are shown in (c and f). Scale bar, 10 μm.
Components of pri-miRNA/SC35-containing nuclear foci
Our previous results,34 as well as data presented above, demonstrate that pri-miRNA/SC35-containing nuclear foci are unlikely to be sites of transcription or RNA processing. To investigate the functional significance of pri-miRNA accumulation in SC35-containing foci, we further characterized the protein composition of these foci.
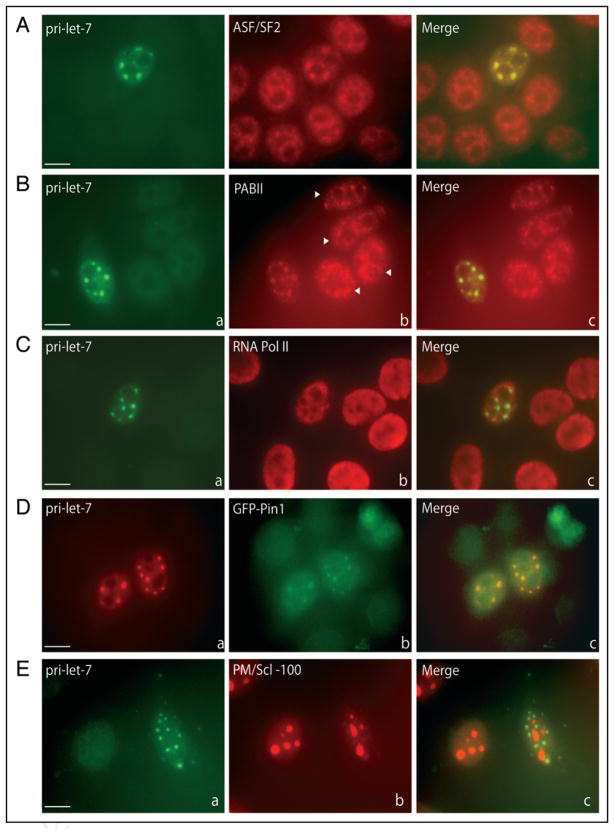
Several splicing factors in addition to SC35 have been shown to concentrate to varying extents in SC35 domains in untransfected cells.37 For example, the essential SR splicing factor ASF/SF2 distributes throughout the nucleoplasm, but with a fraction concentrating in SC35 domains.37,50 We investigated the relative localization of ASF/SF2 and pri-let-7 by performing immunofluorescence (IF) to ASF/SF2 followed by ISH to pri-let-7. Figure 3A demonstrates that ASF/SF2 and transiently expressed pri-let-7 colocalize in SC35-containing nuclear foci, but that ASF/SF2 also appears more diffusely in the nucleoplasm.
Figure 3.
Components of pri-miRNA/SC35-containing nuclear foci. Cells transfected with pri-let-7 were subjected to IF using antibodies directed against (A) ASF/SF2, (B) PABII, (C) Pol II or (E) PM/Scl-100, followed by in situ hybridization to pri-let-7. In (D), cells were cotransfected with pri-let-7 and GFP-Pin1, and ISH to pri-let-7 was performed. Arrowheads in (B) (part b) indicate untransfected cells. Scale bars, 10 μm.
Nuclear polyA binding protein (PABII) plays an important role in the addition of the polyA tail to newly synthesized Pol II transcripts.51 In untransfected cells, PABII localizes diffusely in the nucleoplasm as well as in SC35 foci52 (Fig. 3B, part b; see untransfected cells indicated by arrowheads), likely resulting from its binding to polyA RNA, which concentrates in these domains.53 We observe that transiently expressed pri-let-7 colocalizes with PABII in nuclear foci (Fig. 3B). These data are consistent with the interpretation that cleaved and polyadenylated pri-miRNAs released from their sites of transcription bind PABII and then accumulate together in SC35 domains.
SC35 domains also contain a small subpopulation of Pol II,54 which has been reported to represent an inactive form.55,56 The localization of Pol II in cells transfected with pri-let-7 was explored by performing IF using an antibody that recognizes both phosphorylated and unphosphoryated Pol II. Subsequent ISH for pri-let-7 (Fig. 3C) reveals Pol II does not strongly colocalize with pri-let-7 in nuclear foci. This result is consistent with the interpretation that pri-miRNA/SC35-containing nuclear foci are unlikely to represent major sites of pri-let-7 transcription.
The petidyl-prolyl cis-trans isomerase, Pin1, also associates with SC35 domains.57 Pin1 specifically isomerizes the phosphoserine/threonine-proline bond of its substrates, thus regulating their biological activity.58 Pin1 is critical for entry into mitosis and for tumor cell survival, and has been shown to play roles in the regulation of subcellular trafficking, transcriptional activity and RNA processing.59–61 The WW domain of Pin1 is essential for both substrate binding and localization to SC35 domains.57 To determine if Pin1 localizes to nuclear foci that correspond to pri-miRNA/SC35-containing foci, a construct expressing GFP-tagged Pin1 was cotransfected into HeLa cells with pri-let-7, and 24 hours later ISH was performed. Figure 3D demonstrates that GFP-Pin1 colocalizes with pri-let-7 transcripts in nuclear foci, arguing that the foci where pri-miRNAs and Pin1 accumulate are the same nuclear structures. A construct expressing a GFP-tagged version of the WW domain of Pin1 also colocalized in nuclear foci with pri-let-7, whereas the GFP-tagged petidyl-prolyl isomerase domain alone did not, but rather localized diffusely throughout the nucleoplasm (data not shown).
Finally, we investigated the localization of PM/Scl-100 in cells overexpressing pri-let-7. PM/Scl-100 is a component of the nuclear exosome, which mediates 3′ to 5′ mRNA decay.62 Recent studies using ChIP analysis demonstrated that PM/Scl-100 is recruited to nascent pri-miRNA transcripts after cleavage by Drosha, where it cotranscriptionally degrades the free 3′ end of the 5′ cleavage product.35 For intronic miRNAs, this cotranscriptional degradation of intronic sequences following Drosha cleavage was hypothesized to enhance the splicing efficiency of flanking exons.35 To determine if PM/Scl-100 is recruited to pri-miRNA/SC35-containing foci in cells transfected with pri-let-7, IF with anti-PM/Scl-100 was performed followed by ISH to pri-let-7. Consistent with previous results, PM/Scl-100 exhibits predominantly nucleolar localization (Fig. 3E), which was attributed to its roles in ribosomal RNA biogenesis.63 Importantly, no colocalization of PM/Scl-100 with exogenously expressed pri-let-7 in SC35-containing foci is observed, consistent with the idea that unprocessed pri-miRNAs are the predominant species that accumulate in SC35-containing foci. Instead, PM/Scl-100 is likely to be recruited only to pri-miRNAs that are efficiently processed at transcription sites, which are localized diffusely in the nucleoplasm34 and are probably not observed here because the levels are below the limit of detection.
Discussion
Here, we have presented data that corroborate our previous observations of enhanced pri-miRNA processing at the site of transcription. We also further characterized the components of pri-miRNA/SC35-containing nuclear foci and examined the requirements for transcript localization to these foci. We observe that pri-miRNAs released from transcription sites by a self-cleaving HD ribozyme accumulate in the nucleoplasm but are not efficiently processed (Fig. 1). The majority of ribozyme-cleaved pri-miRNAs do not localize to SC35-containing nuclear foci, but rather distribute diffusely in the nucleus. In contrast, cleaved and polyadenylated pri-miRNAs either with or without the pre-miRNA hairpin colocalize with SC35 in nuclear foci (Fig. 2). Components of pri-miRNA/SC35-containing nuclear foci include proteins known to concentrate to various extents in SC35 domains, such as ASF/SF2, PABII, and the prolyl isomerase, Pin1 (Fig. 3). Interestingly, Pol II and PM/Scl-100, a component of the nuclear exosome with a role in the cotranscriptional turnover of pri-miRNA flanking sequences,35 do not localize to pri-miRNA/SC35-containing foci (Fig. 3). These findings, along with those reported in other recent studies,35,42,64–67 have important implications for the mechanisms of pri-miRNA processing, as well as for the significance of pri-miRNA accumulation in SC35-containing nuclear foci.
Inefficient processing of pri-miRNAs released from sites of transcription
We found that ribozyme-cleaved pri-miRNAs accumulate in the nucleoplasm to steady-state levels that are ~2- to 3-fold higher than those of cleaved and polyadenylated pri-miRNAs (Fig. 1). Despite these elevated levels, the level of mature miRNAs produced from ribozyme-cleaved pri-miRNAs did not increase. Similarly, we previously observed that ENE-containing pri-miRNAs accumulate to high levels after release from sites of transcription, but are not efficiently processed.34
The increased stability of ribozyme-cleaved pri-lin-4 transcripts may be due to insertion of the histone mRNA stem loop upstream of the ribozyme cleavage site, to the 2′,3′-cyclic phosphate resulting from ribozyme cleavage, or to impairment of deadenylation-dependent decapping, as previously suggested.68 Alternatively, since the ENE, which dramatically stabilizes pri-miRNAs, inhibits entry into a deadenylation-dependent decay pathway,69 the lack of a polyA tail may prevent entry of ribozyme-cleaved pri-miRNAs into this normal decay pathway and result in enhanced stability. We presume that the mature miRNAs produced from pri-miRNAs containing the ENE or the HD ribozyme result from processing at the site of transcription before release into the nucleoplasm. Because the HD ribozyme is situated upstream of the BGH CPA signal, we may have expected fewer mature miRNAs to be produced from ribozyme-cleaved pri-miRNAs in comparison to those generated by CPA. However, no difference was observed (Fig. 1B), perhaps because HD ribozyme cleavage is not 100% efficient. Nonetheless, the observation (Fig. 1B–D) that ribozyme-cleaved pri-miRNAs accumulate in the nucleoplasm and are not efficiently processed supports the idea that pri-miRNA processing is less efficient when uncoupled from the transcription site (see Fig. 4).
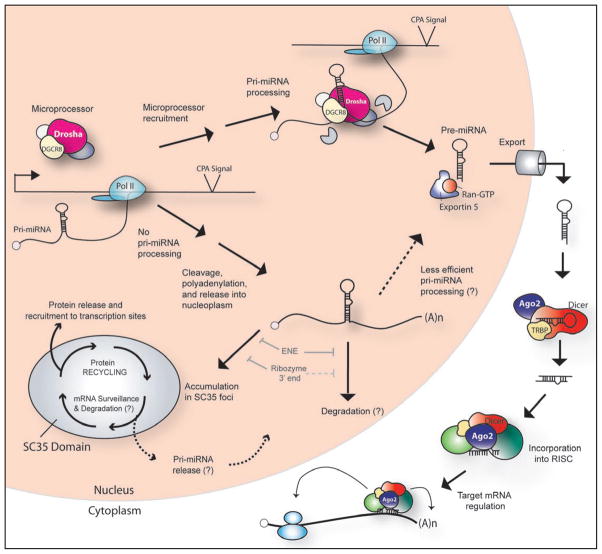
Figure 4.
Model of pri-miRNA processing and localization. In the cotranscriptional pri-miRNA processing pathway, the Microprocessor complex, consisting of Drosha, DGCR8, and other factors (represented as unlabeled circles, see text for examples), is recruited to sites of pri-miRNA synthesis and binds the nascent pri-miRNA. Drosha cleavage releases α ~70-nt pre-miRNA hairpin. Grey partial circles represent exonucleases that are recruited cotranscriptionally to degrade the 5′ and 3′ sequences flanking the former site of the pre-miRNA hairpin.35 The pre-miRNA is exported to the cytoplasm by the Exportin 5-Ran-GTP complex. In the cytoplasm, Dicer, its cofactor TRBP, and Ago2 recognize the pre-miRNA,118,119 and Dicer cleavage results in a miRNA:miRNA* duplex. The mature miRNA strand is then incorporated into the RISC effector complex, which targets an mRNA to regulate its translational output19,120 or stimulates miRNA-mediated deadenylation and decay.19 In the alternate pathway, where either Microprocessor binding or action is lacking, nascent pri-miRNAs undergo CPA and are released into the nucleoplasm. Unprocessed polyadenylated pri-miRNAs may then be transported to SC35 foci, degraded in the nucleoplasm, or processed to pre-miRNAs, but with lower efficiency than those processed at transcription sites. In SC35 foci, protein complexes are recycled and then released and recruited to sites of active transcription. 37 Pri-miRNAs that accumulate in SC35 foci undergo surveillance and may be retained or degraded within the domain, or they may be released into the nucleoplasm, followed by degradation or less efficient processing to pre-miRNAs. The inhibitory effects of the ENE and of ribozyme cleavage are indicated.
Recently, Morlando et al.35 presented data complementary to ours,34 demonstrating that Drosha is present at endogenous pri-miRNA transcription sites and that pri-miRNA processing occurs on the nascent pri-miRNA. These observations raise the question of how the Microprocessor complex is recruited to sites of transcription. It is possible that Drosha and DGCR8 accumulate via mechanisms similar to those involved in recruitment of pre-mRNA processing factors to nascent pre-mRNAs. Notably, the presence of Drosha at endogenous transcription sites correlates with transcriptional activity and with the presence of Pol II at these locations.35 Since the carboxy-terminal domain (CTD) of Pol II plays a major role in cotranscriptional recruitment of pre-mRNA processing factors,32,70–75 the CTD may likewise either directly or indirectly recruit Drosha and DGCR8. Indeed, several proteins that interact with Drosha and DGCR8 in the large Microprocessor complex10,11,13 play important roles in the regulation and coordination of transcription and pre-mRNA processing. For instance, the DEAD-box helicase Microprocessor components p68 and p72, which increase the efficiency of Drosha processing for a subset of pri-miRNAs,13 are recruited to gene-specific promoters,76,77 can interact with transcriptional coactivators and Pol II,78 and play a role in regulating alternative splicing.79–81 Similarly, all three members of the TET family of RNA binding proteins, including TLS/FUS, EWS and TAF15, interact with Drosha11 and are essential for coordinating transcription with mRNA maturation;82 they could serve as adaptors between Pol II and Drosha to link transcription and pri-miRNA processing. Finally, the FHA-domain-containing protein SNIP1 has recently been shown to interact with Drosha and play a role miRNA biogenesis.83 SNIP1 has documented roles in transcription regulation84,85 and is a component of a large SNIP1/SkIP-associated complex that is involved in cotranscriptional pre-mRNA processing.86 The ability of SNIP1 to recruit several proteins to active chromosomal loci86 suggests it could also recruit Drosha to sites of pri-miRNA synthesis.
Alternative mechanisms of recruitment of Microprocessor components to sites of transcription could also exist. Specific chromatin modifications are known to be important for recruitment of spliceosomal proteins to nascent pre-mRNAs.87 For example, the chromodomain-containing protein CHD1 binds the active chromatin mark of trimethylated histone H3 lysine 4,88,89 and interacts with the SF3a subunit of U2 snRNP, serving to enhance the efficiency of splicing by tethering core spliceosomal components to active DNA.87 The double-stranded RNA binding protein NFAR/NF90 was also found to affinity purify with histone H3 trimethylated at lysine 4,87 and has documented roles in regulating transcriptional elongation and mRNA processing.90 Its interaction with Drosha11 suggests that NF90 could likewise play a role in the recruitment of Drosha and DGCR8 to active pri-miRNA loci.
Other proteins involved in transcription or pre-mRNA maturation may also affect pri-miRNA processing. For example, the core transcription factor, TFIID recruits the 3′-end processing protein CPSF to mRNA promoters.91 Similarly, the cap-binding complex (CBC), which is composed of CBP20 and CBP80, plays a critical role in cotranscriptional recruitment of the splicing commitment complex to nascent pre-mRNAs,28,92 and stimulates both splicing93 and 3′-end formation in mammalian cells.94 Interestingly, in Arabidopsis thaliana, CBP20 and CBP80 contribute to pri-miRNA processing,95–97 and may do so by facilitating the cotranscriptional loading of DCL1, the RNase III that processes pri-miRNAs in Arabidopsis, onto nascent pri-miRNAs.96 In mammalian cells, the CBC interacts with hnRNPH,98 whose close homolog hnRNPH1 is a component of the larger form of the Microprocessor complex.11 Additional studies will be required to determine whether the CBC plays a role in cotranscriptional pri-miRNA processing in mammalian cells.
Regulation of pri-miRNA processing plays a critical role in development and disease.22 The evidence that pri-miRNA processing is cotranscriptional34,35 suggests that this regulation also occurs cotranscriptionally, as is the case for the regulation of many pre-mRNA processing events.30,33 For splicing in both yeast and mammals, not all Pol II transcripts are able to interact to the same degree with the spliceosome and therefore the levels of pre-mRNAs transcribed do not always correlate with the levels of mature mRNA;67,99 such disparities are often attributed to differences in the gene context.65,100–102 Similarly, promoter identity, chromatin structure, and the presence of pause sequences within pri-miRNA chromosomal loci may influence Drosha recruitment and processing. Since regulation of alternative splicing can be dramatically influenced by the rate of transcription elongation,30 the Pol II elongation rate could also affect the folding of nascent pri-miRNAs or the recruitment of processing factors.
These mechanisms of cotranscriptional recruitment of splicing factors are not mutually exclusive,31,87 so it is possible that multiple mechanisms contribute to cotranscriptional processing by Drosha and DGCR8. Finally, because pre-mRNA splicing can begin at transcription sites with the cotranscriptional recruitment of processing factors, but can continue posttranscriptionally for some pre-mRNAs,28 a combination of cotranscriptional and posttranscriptional regulatory mechanisms may exist for the processing of pri-miRNAs.
Pri-miRNA localization to SC35-containing nuclear foci
Our previous data suggested that pri-miRNA/SC35-containing nuclear foci are neither sites of transcription nor obligate sites of pri-miRNA processing.34 Here, we observed that pri-miRNAs released from transcription sites by ribozyme cleavage do not localize to SC35-containing foci, while cleaved and polyadenylated pri-miRNAs with or without a pre-miRNA hairpin do. Pri-miRNAs transcribed by RNA polymerase III, which lack a polyA tail, and pri-miRNAs in which the ENE sequesters the polyA tail also fail to localize to SC35-containing foci.34 This argues that a polyA tail may be necessary for transcript localization to SC35-containing foci and that proteins recruited to nascent Pol II transcripts during the CPA reaction or proteins interacting with the polyA tail could be instrumental in their localization or retention in SC35 domains. Notably, we observed an enhanced concentration of PABII in pri-miRNA/SC35-containing nuclear foci (Fig. 3B), suggesting that it binds the polyadenylated pri-miRNAs that accumulate there.
The amount of ASF/SF2 localized in SC35-containing foci in cells expressing pri-let-7 transcripts was slightly enhanced over that observed in untransfected cells (Fig. 3A), perhaps because of nonspecific binding of ASF/SF2 to the large quantity of pri-miRNAs expressed in transfected cells. In contrast, Pol II localization was not strongly affected by pri-miRNA overexpression (Fig. 3C). This observation is consistent with the hypothesis that pri-miRNA/SC35-containing nuclear foci are not major sites of transcription, in accord with recent studies indicating that the minor subpopulation of Pol II that accumulates in SC35 domains is inactive.56
A GFP-tagged form of the prolyl isomerase, Pin1, was found to accumulate with pri-miRNAs in SC35-containing nuclear foci (Fig. 3D). The localization of Pin1 to SC35-containing foci is hypothesized to result from its role in facilitating phosphorylation-dependent interactions of proteins involved in mRNA metabolism.58,59 Pin1 also interacts with elongating Pol II,103 suggesting a role for Pin1 in the integration of mRNA transcription and processing.
Together, our observations indicate that many proteins found in pri-miRNA/SC35-containing nuclear foci in transfected cells are also present in SC35 domains in untransfected cells. Therefore, although it remains unclear whether these foci represent bona fide pre-existing SC35 domains or domains that form de novo upon overexpression of a polyadenylated transcript, they are clearly similar to naturally occurring SC35 domains.
Interestingly, in Arabidopsis thaliana, a number of proteins involved in miRNA biogenesis colocalize in nuclear foci that have been referred to as nuclear Dicing bodies (D-bodies).104–107 Exogenously expressed plant pri-miRNAs localize to D-bodies, suggesting that these foci may represent sites of miRNA biogenesis.104,106 Alternatively, D-bodies could be sites of storage and assembly of protein complexes involved in pri-miRNA processing in plants.104,107 However, in contrast to the pri-miRNA/SC35-containing nuclear foci observed in mammalian cells,34 plant D-bodies do not contain SC35. Rather, plant D-bodies represent a subset of Cajal body-like foci.108 Furthermore, several differences between plant and mammalian miRNA biogenesis exist, including (1) a single RNase III protein, DCL1, performs the processing of both pri-miRNAs to pre-miRNAs and pre-miRNAs to mature miRNAs in the nucleus in plants,109 and (2) many proteins involved in biogenesis of other classes of small RNAs localize to D-bodies.108 Thus, current evidence favors the idea that plant D-bodies are unrelated to the pri-miRNA/SC35-containing nuclear foci observed in transfected mammalian cells.
A major question that arises concerns the significance of pri-miRNA localization to SC35-containing nuclear foci. While this localization could result from the process of transfection or the expression of large quantities of RNA, some sorting mechanism must exist since only cleaved and polyadenylated Pol II transcripts accumulate there. One possible explanation is that the recruitment of cleaved and polyadenylated pri-miRNAs to SC35-containing foci occurs because SC35 domains are critical for coordinating and integrating steps in the expression of Pol II-transcribed genes.37 Indeed, a vast amount of data has demonstrated that SC35 domains are preferentially associated with highly active genes.110–112 Moreover, the nonrandom gene pairing that occurs during the transient “kissing” of alleles during X inactivation or the colocalization of coregulated genes during differentiation,113,114 may result from their mutual interaction with a third party, a common SC35 speckle, rather than from actual gene-gene interactions.42,115 Localization of highly active genes near SC35 domains may occur because these domains are rich in metabolic complexes that can be rapidly supplied to facilitate efficient RNA processing.41 Once transcription and processing are complete, mature mRNAs often enter SC35 domains, perhaps to allow recycling of protein complexes before export.37 Transcripts that have not completed splicing are retained in SC35 domains,116,117 suggesting that these domains also serve as checkpoints in the mRNA processing pathway by preventing the export of defective transcripts to the cytoplasm.36,37 These observations indicate that SC35 domains are critical for the efficiency, regulation and quality control of gene expression. The high levels of polyadenylated pri-miRNAs produced in transfected cells may therefore accumulate in SC35 domains to allow rapid recycling of bound proteins, including Drosha and DGCR8, and enhance the efficiency of miRNA expression (see Fig. 4). Unprocessed pri-miRNAs may also be retained in SC35-containing nuclear foci as a result of the role of these domains in mRNA surveillance and quality control.
The results presented here, in combination with recent insights into transcription, mRNA maturation and SC35 domain function, converge on the common theme that integration of transcription and RNA processing are important for the efficiency and regulation of mammalian gene expression. The idea that there is tight coupling between transcription and pre-mRNA processing is underscored by recent findings that splicing factors, such as SC35, play essential roles in the transcription of specific genes64 and that transcription factors, such as c-myb, are critical for the regulation of alternative splicing.66 It will therefore be interesting to determine the influence of transcriptional activities on pri-miRNA processing, as well as the converse effects of pri-miRNA processing on transcription and pre-mRNA maturation.
Materials and Methods
Plasmids
The plasmids pri-lin-4, pri-let-7 and pri-lin-4ΔpAx2 have been described previously. 34 To construct pri-lin-4HD and pri-lin-4HDm, a histone stem loop followed by a variant HD ribozyme or mutant ribozyme with a C76-U substitution (based on 44) was inserted into pri-lin-4 digested with ApaI and blunted with T4 DNA polymerase. The sequence inserted was GGC CCT TAT CAG GGC CA*G GGC GGC ATG GTC CCA GCC TCC TCG CTG GCG CCG CCT GGG CAA CAT GCT TCG GCA TGG CGA ATG GGA CCA AAT, where the histone stem loop is in italics (stem underlined) followed by the ribozyme sequence. Ribozyme cleavage occurs one base after the histone stem loop (cleavage site indicated with an asterisk). The position of the C-76U substitution is underlined.
Pri-lin-4HDΔpAx2 and pri-lin-4HDmΔpAx2 were constructed by inserting the sequence above into pri-lin-4ΔpAx2 digested with ApaI and blunted with T4 DNA polymerase.
To generate pri-let-7Δpre, the hairpin precursor sequence was deleted from pri-let-7 using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) with the oligonucleotides (forward): CCT TTT CAC CAT TCA CCC TGG TAG AAA AGT CTG CAT CCA GGC G and (reverse) CGC CTG GAG CAG ACT TTT CTA CCA GGG TGA ATG GTG AAA AGG.
The plasmids encoding GFP-tagged Pin1 (GFP-Pin1) or truncation mutants GFP-WW and GFP-PPI (which contain only the WW domain or the peptidyl-prolyl isomerase domain of Pin1, respectively) have been previously described.57
Antibodies
Mouse monoclonal anti-SC35 antibody was from Sigma-Aldrich and mouse monoclonal anti-Pol II (4H8) was from Abcam. Anti-ASF/SF2, anti-PABII and anti-Pm/Scl-100 were gifts from Adrian Krainer (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), David Bear (University of New Mexico, Albuquerque, NM), and Ger Pruijn (Radboud University of Nijmegen, Nijmegen, the Netherlands), respectively.
Cell lines and transfection
HeLa cells were cultured in Dulbecco’s Modified Eagles’s medium (Invitrogen) supplemented with 10% Fetal Bovine Serum (BD Biosciences), 2 mM L-glutamine, and 1X penicillin streptomycin solution (Sigma-Aldrich) at 37°C in 5% carbon dioxide. Transfections were performed with HeLa Monster reagent (Mirus Bio Corporation) according to the manufacturer’s protocol.
RNA isolation and northern blot analysis
Total RNA was isolated using Trizol Reagent (Invitrogen), and analyzed by Northern blotting as previously described.34 Briefly, 5 μg of total RNA from each sample was run on two gels to detect (1) pri-miRNAs or (2) precursor and mature miRNAs. To detect pri-miRNAs, 5 μg of total RNA was resolved on a 1.2% agarose/6.5% formaldehyde gel and transferred to a Zeta probe membrane (Bio-Rad Laboratories). To detect precursor and mature miRNAs, 5 μg of total RNA was separated on a 15% polyacrylamide/8 M urea/1X TBE gel, and then transferred to a Hybond N+ membrane (GE Healthcare) using a semidry electroblotter. Pri-miRNAs were detected using an in vitro transcribed riboprobe complementary to pri-lin-4 uniformly labeled with [α-32P]UTP. Precursor and mature lin-4 were detected using an oligonucleotide probe complementary to mature lin-4 labeled with [γ-32P]ATP using T4 polynucleotide kinase. Results were visualized using a Storm PhosphorImager (Molecular Dynamics).
Fractionation of nucleoplasmic and chromatin-associated transcripts and RT-PCR
Fractionation of HeLa cell nuclei and RNA extraction from the nucleoplasmic supernatant and chromatin-associated pellet were performed as previously described.34,48,49 After nuclear fractionation, cDNA was generated using SuperScript II Reverse Transcriptase (Invitrogen) with random primers according to the manufacturer’s instructions. Primers used to amplify pri-lin-4 and TAFII30 mRNA have been previously described.34 Primers used to amplify TAFII30 pre-mRNA were (forward): GGG TGA GGG CAG AGG GTA TAG and (reverse): TTT GTC AGC AGG CTA GGT GG.
IF and ISH
ISH to pri-let-7 or pri-lin-4, followed by IF to SC35 was performed as described.34 In Figure 3, IF to ASF/SF2, Pol II, PABII or PM/Scl-100 was performed prior to ISH to pri-let-7 as described.34 For detection of pri-let-7 in Figure 3D, cells cotransfected with GFP-Pin1 and pri-let-7 were subjected to ISH as described,34 with the exception that DIG-tailed probes (complementary to regions diagramed in Fig. 2A, blue lines) were detected using a 1:200 dilution of anti-DIG antibody conjugated to Rhodamine (Roche).
Acknowledgments
We thank Adrian Krainer, David Bear and Ger Pruijn for antibodies, and Kun Ping Lu (Beth Israel Deaconess Medical Center, Boston, MA) for GFP-Pin1 constructs. We are grateful to Shobha Vasudevan, Kasandra Riley, Robin Lytle and Kristina Herbert for critical comments on this manuscript, and A. Miccinello for editorial assistance. We also thank all members of the Steitz lab for stimulating discussions. This work was supported by grant R01GM026154 from the NIGMS to J.A.S. and a National Science Foundation Graduate Research Fellowship to J.M.P. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the NIH. J.A.S. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- BGH
bovine growth hormone
- CBC
cap-binding complex
- CPA
cleavage and polyadenylation
- CTD
carboxy-terminal domain
- D-bodies
dicing bodies
- HD
hepatitis delta
- IF
immunofluoresence
- IGCs
interchromatin granule clusters
- ISH
in situ hybridization
- miRNA
microRNA
- mRNA
messenger RNA
- PABII
nuclear polyA binding protein
- Pol II
RNA polymerase II
- polyA
polyadenylate
- pre-miRNA
precursor miRNA
- pri-miRNA
primary miRNA
- RISC
RNA-induced silencing complex
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Bueno MJ, de Castro IP, Malumbres M. Control of cell proliferation pathways by microR-NAs. Cell Cycle. 2008;7:3143–8. doi: 10.4161/cc.7.20.6833. [DOI] [PubMed] [Google Scholar]
- 3.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–30. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 4.Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7:2643–6. doi: 10.4161/cc.7.17.6597. [DOI] [PubMed] [Google Scholar]
- 5.Lotterman CD, Kent OA, Mendell JT. Functional integration of microRNAs into oncogenic and tumor suppressor pathways. Cell Cycle. 2008;7:2493–9. doi: 10.4161/cc.7.16.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medina PP, Slack FJ. microRNAs and cancer: an overview. Cell Cycle. 2008;7:2485–92. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- 7.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, poly-adenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 10.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microR-NAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 11.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 12.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–11. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 14.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–91. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 16.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 18.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 19.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–26. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 20.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 21.Fukao T, Fukuda Y, Kiga K, Sharif J, Hino K, Enomoto Y, et al. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129:617–31. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- 22.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–7. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–7. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–4. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 27.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–49. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neugebauer KM. On the importance of being co-transcriptional. J Cell Sci. 2002;115:3865–71. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- 29.Bentley D. The mRNA assembly line: transcription and processing machines in the same factory. Curr Opin Cell Biol. 2002;14:336–42. doi: 10.1016/s0955-0674(02)00333-2. [DOI] [PubMed] [Google Scholar]
- 30.Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–98. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allemand E, Batsche E, Muchardt C. Splicing, transcription and chromatin: a menage a trois. Curr Opin Genet Dev. 2008;18:145–51. doi: 10.1016/j.gde.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–5. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kornblihtt AR. Coupling transcription and alternative splicing. Adv Exp Med Biol. 2007;623:175–89. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- 34.Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–9. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–12. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 37.Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:664–75. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cmarko D, Verschure PJ, Martin TE, Dahmus ME, Krause S, Fu XD, et al. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol Biol Cell. 1999;10:211–23. doi: 10.1091/mbc.10.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melcak I, Cermanova S, Jirsova K, Koberna K, Malinsky J, Raska I. Nuclear pre-mRNA compartmentalization: trafficking of released transcripts to splicing factor reservoirs. Mol Biol Cell. 2000;11:497–510. doi: 10.1091/mbc.11.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang G, Taneja KL, Singer RH, Green MR. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–12. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- 41.Moen PT, Jr, Johnson CV, Byron M, Shopland LS, de la Serna IL, Imbalzano AN, et al. Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis. Mol Biol Cell. 2004;15:197–206. doi: 10.1091/mbc.E03-06-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown JM, Green J, das Neves RP, Wallace HA, Smith AJ, Hughes J, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–97. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Been MD. HDV ribozymes. Curr Top Microbiol Immunol. 2006;307:47–65. doi: 10.1007/3-540-29802-9_3. [DOI] [PubMed] [Google Scholar]
- 44.Bird G, Fong N, Gatlin JC, Farabaugh S, Bentley DL. Ribozyme cleavage reveals connections between mRNA release from the site of transcription and pre-mRNA processing. Mol Cell. 2005;20:747–58. doi: 10.1016/j.molcel.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 45.Perrotta AT, Shih I, Been MD. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–6. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- 46.Eckner R, Ellmeier W, Birnstiel ML. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 1991;10:3513–22. doi: 10.1002/j.1460-2075.1991.tb04915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y, Carmichael GG. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–42. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14:7219–25. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol Cell. 2006;21:849–59. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 50.Caceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–26. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edmonds M. A history of poly A sequences: from formation to factors to function. Prog Nucleic Acid Res Mol Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 52.Krause S, Fakan S, Weis K, Wahle E. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp Cell Res. 1994;214:75–82. doi: 10.1006/excr.1994.1235. [DOI] [PubMed] [Google Scholar]
- 53.Carter KC, Taneja KL, Lawrence JB. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J Cell Biol. 1991;115:1191–202. doi: 10.1083/jcb.115.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mintz PJ, Patterson SD, Neuwald AF, Spahr CS, Spector DL. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 1999;18:4308–20. doi: 10.1093/emboj/18.15.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bregman DB, Du L, van der Zee S, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–98. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie SQ, Martin S, Guillot PV, Bentley DL, Pombo A. Splicing speckles are not reservoirs of RNA polymerase II, but contain an inactive form, phosphorylated on serine2 residues of the C-terminal domain. Mol Biol Cell. 2006;17:1723–33. doi: 10.1091/mbc.E05-08-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J Biol Chem. 2002;277:2381–4. doi: 10.1074/jbc.C100228200. [DOI] [PubMed] [Google Scholar]
- 58.Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–16. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 59.Rippmann JF, Hobbie S, Daiber C, Guilliard B, Bauer M, Birk J, et al. Phosphorylation-dependent proline isomerization catalyzed by Pin1 is essential for tumor cell survival and entry into mitosis. Cell Growth Differ. 2000;11:409–16. [PubMed] [Google Scholar]
- 60.Komuro A, Saeki M, Kato S. Npw38, a novel nuclear protein possessing a WW domain capable of activating basal transcription. Nucleic Acids Res. 1999;27:1957–65. doi: 10.1093/nar/27.9.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morris DP, Phatnani HP, Greenleaf AL. Phospho-carboxyl-terminal domain binding and the role of a prolyl isomerase in pre-mRNA 3′-End formation. J Biol Chem. 1999;274:31583–7. doi: 10.1074/jbc.274.44.31583. [DOI] [PubMed] [Google Scholar]
- 62.Vanacova S, Stefl R. The exosome and RNA quality control in the nucleus. EMBO Rep. 2007;8:651–7. doi: 10.1038/sj.embor.7401005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schilders G, van Dijk E, Pruijn GJ. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 2007;35:2564–72. doi: 10.1093/nar/gkm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–26. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bittencourt D, Dutertre M, Sanchez G, Barbier J, Gratadou L, Auboeuf D. Cotranscriptional splicing potentiates the mRNA production from a subset of estradiol-stimulated genes. Mol Cell Biol. 2008;28:5811–24. doi: 10.1128/MCB.02231-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Orvain C, Matre V, Gabrielsen OS. The transcription factor c-Myb affects pre-mRNA splicing. Biochem Biophys Res Commun. 2008;372:309–13. doi: 10.1016/j.bbrc.2008.05.054. [DOI] [PubMed] [Google Scholar]
- 67.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell. 2007;27:928–37. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607–17. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol Cell. 2006;24:943–53. doi: 10.1016/j.molcel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 70.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–8. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 71.Chapman RD, Heidemann M, Hintermair C, Eick D. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008;24:289–96. doi: 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 72.de Almeida SF, Carmo-Fonseca M. The CTD role in cotranscriptional RNA processing and surveillance. FEBS Lett. 2008;582:1971–6. doi: 10.1016/j.febslet.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 73.Hirose Y, Ohkuma Y. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J Biochem. 2007;141:601–8. doi: 10.1093/jb/mvm090. [DOI] [PubMed] [Google Scholar]
- 74.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–36. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 75.Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes Dev. 2005;19:1401–15. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- 76.Bates GJ, Nicol SM, Wilson BJ, Jacobs AM, Bourdon JC, Wardrop J, et al. The DEAD box protein p68: a novel transcriptional coactivator of the p53 tumour suppressor. EMBO J. 2005;24:543–53. doi: 10.1038/sj.emboj.7600550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watanabe M, Yanagisawa J, Kitagawa H, Takeyama K, Ogawa S, Arao Y, et al. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 2001;20:1341–52. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Rossow KL, Janknecht R. Synergism between p68 RNA helicase and the transcriptional coactivators CBP and p300. Oncogene. 2003;22:151–6. doi: 10.1038/sj.onc.1206067. [DOI] [PubMed] [Google Scholar]
- 79.Auboeuf D, Honig A, Berget SM, O’Malley BW. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–9. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- 80.Guil S, Gattoni R, Carrascal M, Abian J, Stevenin J, Bach-Elias M. Roles of hnRNP A1, SR proteins and p68 helicase in c-H-ras alternative splicing regulation. Mol Cell Biol. 2003;23:2927–41. doi: 10.1128/MCB.23.8.2927-2941.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Honig A, Auboeuf D, Parker MM, O’Malley BW, Berget SM. Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol Cell Biol. 2002;22:5698–707. doi: 10.1128/MCB.22.16.5698-5707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Law WJ, Cann KL, Hicks GG. TLS, EWS and TAF15: a model for transcriptional integration of gene expression. Brief Funct Genomic Proteomic. 2006;5:8–14. doi: 10.1093/bfgp/ell015. [DOI] [PubMed] [Google Scholar]
- 83.Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci USA. 2008;105:10073–8. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fujii M, Lyakh LA, Bracken CP, Fukuoka J, Hayakawa M, Tsukiyama T, et al. SNIP1 is a candidate modifier of the transcriptional activity of c-Myc on E box-dependent target genes. Mol Cell. 2006;24:771–83. doi: 10.1016/j.molcel.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 85.Roche KC, Rocha S, Bracken CP, Perkins ND. Regulation of ATR-dependent pathways by the FHA domain containing protein SNIP1. Oncogene. 2007;26:4523–30. doi: 10.1038/sj.onc.1210233. [DOI] [PubMed] [Google Scholar]
- 86.Bracken CP, Wall SJ, Barre B, Panov KI, Ajuh PM, Perkins ND. Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res. 2008;68:7621–8. doi: 10.1158/0008-5472.CAN-08-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–76. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–8. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 89.Daniel JA, Pray-Grant MG, Grant PA. Effector proteins for methylated histones: an expanding family. Cell Cycle. 2005;4:919–26. doi: 10.4161/cc.4.7.1824. [DOI] [PubMed] [Google Scholar]
- 90.Saunders LR, Perkins DJ, Balachandran S, Michaels R, Ford R, Mayeda A, et al. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem. 2001;276:32300–12. doi: 10.1074/jbc.M104207200. [DOI] [PubMed] [Google Scholar]
- 91.Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 92.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–68. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 93.Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 94.Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc Natl Acad Sci USA. 1997;94:11893–8. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gregory BD, O’Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14:854–66. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 96.Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Ratsch G, et al. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:8795–800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim S, Yang JY, Xu J, Jang IC, Prigge MJ, Chua NH. Two CAP BINDING PROTEINS CBP20 and CBP80 are involved in processing primary microRNAs. Plant Cell Physiol. 2008 doi: 10.1093/pcp/pcn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gamberi C, Izaurralde E, Beisel C, Mattaj IW. Interaction between the human nuclear cap-binding protein complex and hnRNP F. Mol Cell Biol. 1997;17:2587–97. doi: 10.1128/mcb.17.5.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moore MJ, Schwartzfarb EM, Silver PA, Yu MC. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol Cell. 2006;24:903–15. doi: 10.1016/j.molcel.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 100.Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci USA. 1997;94:11456–60. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roberts GC, Gooding C, Mak HY, Proudfoot NJ, Smith CW. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 1998;26:5568–72. doi: 10.1093/nar/26.24.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–22. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 103.Xu YX, Manley JL. Pin1 modulates RNA polymerase II activity during the transcription cycle. Genes Dev. 2007;21:2950–62. doi: 10.1101/gad.1592807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for microR-NA biogenesis in living Arabidopsis plants. Curr Biol. 2007;17:818–23. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ramachandran V, Chen X. Small RNA metabolism in Arabidopsis. Trends Plant Sci. 2008;13:368–74. doi: 10.1016/j.tplants.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fujioka Y, Utsumi M, Ohba Y, Watanabe Y. Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 2007;48:1243–53. doi: 10.1093/pcp/pcm099. [DOI] [PubMed] [Google Scholar]
- 107.Song L, Han MH, Lesicka J, Fedoroff N. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci USA. 2007;104:5437–42. doi: 10.1073/pnas.0701061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pontes O, Pikaard CS. siRNA and miRNA processing: new functions for Cajal bodies. Curr Opin Genet Dev. 2008;18:197–203. doi: 10.1016/j.gde.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen X. MicroRNA biogenesis and function in plants. FEBS Lett. 2005;579:5923–31. doi: 10.1016/j.febslet.2005.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xing Y, Johnson CV, Moen PT, Jr, McNeil JA, Lawrence J. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J Cell Biol. 1995;131:1635–47. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol. 1999;144:617–29. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shopland LS, Johnson CV, Byron M, McNeil J, Lawrence JB. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J Cell Biol. 2003;162:981–90. doi: 10.1083/jcb.200303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.de Laat W. Long-range DNA contacts: romance in the nucleus? Curr Opin Cell Biol. 2007;19:317–20. doi: 10.1016/j.ceb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 114.de Laat W, Grosveld F. Inter-chromosomal gene regulation in the mammalian cell nucleus. Curr Opin Genet Dev. 2007;17:456–64. doi: 10.1016/j.gde.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 115.Lawrence JB, Clemson CM. Gene associations: true romance or chance meeting in a nuclear neighborhood? J Cell Biol. 2008;182:1035–8. doi: 10.1083/jcb.200808121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Johnson C, Primorac D, McKinstry M, McNeil J, Rowe D, Lawrence JB. Tracking COL1A1 RNA in osteogenesis imperfecta. splice-defective transcripts initiate transport from the gene but are retained within the SC35 domain. J Cell Biol. 2000;150:417–32. doi: 10.1083/jcb.150.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shopland LS, Johnson CV, Lawrence JB. Evidence that all SC-35 domains contain mRNAs and that transcripts can be structurally constrained within these domains. J Struct Biol. 2002;140:131–9. doi: 10.1016/s1047-8477(02)00507-5. [DOI] [PubMed] [Google Scholar]
- 118.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 119.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can upregulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]