Abstract
L-amino acid oxidases (LAOs), because they produce hydrogen peroxide as a by-product, function in innate immune defenses of both vertebrates and mollusks. Phylogenetic analysis revealed two major subfamilies of LAOs: (1) a subfamily including LAOs from vertebrates and mainly from Terrabacteria and (2) a subfamily including LAOs from mollusks and Hydrobacteria. These subfamilies thus originated early in the history of life, implying that their innate immune functions in vertebrates and mollusks have evolved separately. Mammalian LAOs were found to belong to three separate clades: (1) LAO1, (2) LAO2, and (3) IL4I1. Phylogenetic analysis supported the hypothesis that LAO1 and LAO2 arose by a gene duplication prior to the divergence of marsupials from placental mammals, while IL4I1 duplicated from the ancestor of the LAO1 and LAO2 prior to the divergence of tetrapods from bony fishes. Mammalian IL4I1 clustered with LAOs from bony fishes, and these molecules shared a number of unique sequence features, including both amino acid replacements and a unique two-codon deletion. It is certain such unique features may be functionally important, especially three unique amino acid replacements in close proximity to the putative active site.
Keywords: Interleukin 4-induced 1, Innate immune defense, L-amino acid oxidase
Introduction
The L-amino acid oxidases (LAO) are homodimeric enzymes that, with noncovalently bound flavin adenine dinucleotide (FAD) as a cofactor, catalyze the oxidative deamination of L-amino acids and the concomitant reduction of FAD; re-oxidation of FAD by molecular O2 results in turn in the production of hydrogen peroxide (H2O2; Moustafa et al. 2006). Because of the cytotoxic effects of H2O2, LAO family members play a variety of roles in the innate immune defenses of animals. In the rockfish, Sebastes schlegeli, an LAO expressed in skin mucus has an antibacterial function (Kitani et al. 2007), while in mammals, LAO has an antibacterial activity in milk and has been shown to be important in defense against bacterial infections of the mammary glands (Nagaoka et al. 2009). A related molecule, encoded by a gene known in human as IL-4-induced gene-1 (IL4I1), is expressed in immune system cells and B cell lymphomas (Boulland et al. 2007; Carbonelle-Puscian et al. 2009; Mason et al. 2004). The L-amino acid oxidase activity of the IL4I1 protein is mainly directed toward phenylalanine, and its effects include the suppression of T lymphocyte proliferation (Boulland et al. 2007). LAOs are also prominent components of snake venom (Du and Clemetson 2002) and of the ink of the sea hare Aplysia californica and related gastropod mollusks (Butzke et al. 2005; Yang et al. 2005). The latter also are known to have antimicrobial properties (Yang et al. 2005). Flavin-containing amine oxidases homologous to animal LAOs have been found in certain bacteria, including a LAO of Streptococcus oligofermentans used to outcompete the related oral pathogen Streptococcus mutans (Tong et al. 2008).
No phylogenetic analysis has addressed the relationship between mammalian LAO and IL4I1 or attempted to time the gene duplication that gave rise to the genes encoding these related proteins. In addition, the phylogenetic relationships of the LAO family members found in vertebrates and mollusks have not been investigated. The latter relationships are of interest because vertebrate innate immune defenses of vertebrates and invertebrates share relatively few homologous proteins (Hughes 1998). Here, I present a phylogenetic analysis of the LAO protein family in animals, along with sequences of related flavin-containing amine oxidases of bacteria. In several gene families whose members have immune system functions in both vertebrates and invertebrates, phylogenetic analyses have supported the hypothesis that these functions have arisen independently (convergently) in deuterostomes (including vertebrates) and protostomes (including insects and mollusks; Hughes 1998; Friedman and Hughes 2002). The present phylogenetic analysis provides a test of whether the function of LAO in innate immunity has arisen independently in vertebrate and mollusk lineages.
There has been a great interest recently in computational identification of amino acid sites important for adaptive differences between proteins, but unfortunately, several commonly used methods are based on faulty logic, generally misidentifying as functionally important residues that are in fact functionally unconstrained (Hughes 2007, 2008; Hughes and Friedman 2008). Here, I apply to the LAO family a biologically sound approach for identifying residues of potential functional importance, based on the principle that functionally important genomic regions tend to be conserved by purifying selection (Kimura and Ohta 1974). This approach assumes that an amino acid residue that is important for distinctive functional properties shared by members of a protein subfamily is likely to have the following properties: (1) The residue is unique to that protein subfamily, having arisen by mutation in the ancestor of that subfamily, and is not found in other subfamilies of the same protein family that differ functionally from the subfamily of interest, and (2) the residue is conserved in the subfamily of interest (Hughes et al. 2003). In the language of phylogenetic analysis, the goal of this method was to identify unique, conserved apomorphies that characterize the amino acid sequence of a clade of proteins sharing a common function.
Materials and methods
Sequences used in the phylogenetic analysis (N=54, Table 1) were found by BLAST homology search (Altschul et al. 1997) of the Genbank protein sequence database. Representative sequences from mammals and birds (with emphasis on completely sequenced genomes) were included along with selected snake and fish LAOs (Table 1). All available LAO sequences from mollusks were also included, but no other invertebrate LAOs were discovered by homology search. Selected bacterial flavin-containing amine oxidases of bacteria, homologous to animal LAO, representing five different bacterial phyla were included in order to root the tree of vertebrate sequences, as well as one sequence from Marseillevirus, a large dsDNA virus isolated from an amoeba (Boyer et al. 2009).
Table 1.
Sequences used in analyses
| Taxonomic group |
Species and sequences |
|---|---|
| dsDNA viruses | Marseillevirus XP_003406846 |
| Actinobacteria | Streptosporangium roseum ACZ88355 |
| Bacteroidetes | Bacillus cereus ACK96881; Chitinophaga pinensis YP_003122191; Dyadobacter fermentans YP_003085902; Flavobacterium johnsoniae YP_001196816 |
| Chloroflexi | Roseoflexus castenholzii YP_001431975 |
| Firmicutes | Clostridium botulinum YP_001886528; Staphylococcus aureus ADA81071 |
| Proteobacteria | Burkholderia ubonensis ZP_02381245; Chromobacterium violaceum NP_902772; Hahella chejuensis XP_433240; Pseudomonas entomophila XP_607749 |
| Mollusca | Achatina fulica acacin X64584; Aplysia californica aplysianin A AY161041, escapin AY615888; Aplysia punctata cyplasin L AJ304802, cyplasin S AJ304801, ink toxin 1 AY442281, ink toxin 2 AY442282, ink toxin 3 AY442283 |
| Actinopterygii | Flounder Platichthys stellatus LA01 AB495360, LAO2 AB495361, LAO3 AB495362; Green pufferfish Tetraodon nigroviridis LAO CAF89975; Rockfish Sebastes schlegeli LAO AB218876; Zebrafish Danio rerio LAO XM_679381 |
| Reptilia | Elapidae: Many-banded krait Bungarus multicinctus EF080832; Black whip snake Demancia vestigiata DQ917521; King cobra Ophiophagus hannah EF080831; Coastal taipan Oxyuranus scutellatus DQ088990; Mulga snake Pseudechis australis DQ08892 |
| Viperidae: Western diamondback rattlesnake Crotalus atrox AF093248; Jararacussu Bothrops jararacussu AY398691; Ocellated carpet viper Echis ocellatus FM177950; Massasauga Sistrurus catenatus ABG26996; Stejneger's pit viper Trimesurus stejnegeri AY338966; Halys viper Gloydeus halys AY450403 | |
| Aves | Chicken Gallus gallus LAO NM_001099351; Turkey Meleagris gallopavo ACA64754; Zebrafinch Taeniopygia guttata XM_002189400 |
| Mammalia | Opossum Monodelphis domestica LAO1 XM_001916389, LAO2 XM_001362797; Mouse Mus musculus LAO1 NM_133892, IL4I1 NM_010215; Horse Equus caballus LAO1 XM_001916389, LAO2 XM_ 001489515; Bovine Bos taurus LAO1 XM_001251177; Dog Canis familiaris LAO1 XM_539553, IL4I1 XM_541486; Panda Ailuropoda melanoleuca LAO1 EFB24797, IL4I1 EFB23924; Human Homo sapiens IL4I1 NM_152899 |
Amino acid sequences were aligned by the CLUSTALX program (Electronic supplementary material (ESM) Fig. S1; Thompson et al. 1997). Each site at which the alignment postulated a gap in any one of a given set of aligned sequences was excluded from phylogenetic analysis involving that set of sequences so that a comparable set of sites was used for each pairwise distance estimation. Phylogenetic trees were constructed by the neighbor-joining (NJ) method (Saitou and Nei 1987) on the basis of the JTT amino acid sequence distance (Jones et al. 1992), assuming that rate variation among sites followed a gamma distribution, using the MEGA 4 program (Tamura et al. 2007). The shape parameter (a) of the gamma distribution was estimated by the TREE-PUZZLE program (Schmidt et al. 2002). A strategy of successive phylogenetic analyses was used. First, an overall phylogenetic analysis was used to establish major groupings; then, a subset of sequences was analyzed in order to better understand relationships within that subset (Hughes 1994). The reliability of branching patterns was assessed by bootstrapping (Felsenstein 1985); 1,000 bootstrap samples were used.
Information regarding LAO structure was inferred by analogy with published structures of snake venom LAOs (Pawelek et al. 2000; Moustafa et al. 2006) and previous comparative analysis of the IL4I1 primary structure (Chavan et al. 2002).
Results
Phylogenetic analysis
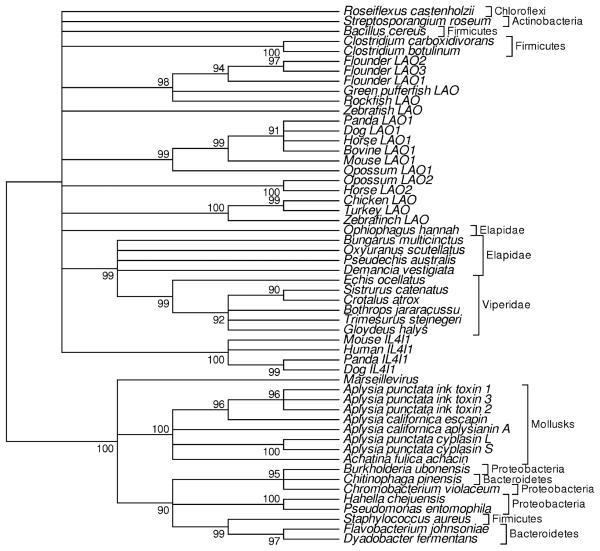
A phylogenetic tree of animal LAO sequences and related flavin-containing amine oxidases of bacteria showed two major clusters, each including both animal and bacterial sequences, which were separated by a branch receiving 100% bootstrap support (Fig. 1). I refer to these clusters as the mollusk-related subfamily and the vertebrate-related subfamily, respectively. The former subfamily included all of the sequences from mollusks, flavin-containing amine oxidase sequences from three bacterial phyla (Bacteroidetes, Firmicutes, and Proteobacteria), and the sequence from Marseillevirus (Fig. 1). Within this cluster, the mollusk sequences formed a monophyletic group, which received 100% bootstrap support (Fig. 1). The second cluster included all the sequences from vertebrates and sequences from three bacterial phyla (Actinobacteria, Chloroflexi, and Firmicutes; Fig. 1). Within the latter cluster, sequences from vertebrates did not form a monophyletic group, although bootstrap support for this hypothesis was not strong (Fig. 1). In general, deep branches within the vertebrate-related group were not well resolved (Fig. 1).
Fig. 1.
Condensed NJ tree (showing topology only) of L-amino acid oxidases based on the JTT distance with gamma correction (parameter a=2.25) at 218 aligned sites. Numbers on the branches are percentages of 1000 bootstrap samples supporting the branch; branches with bootstrap support <90% are collapsed
The phylogenetic relationships of LAO-related oxidases in bacteria (Fig. 1) were not generally consistent with bacterial phylogeny. Firmicutes was the only bacterial phylum having representatives of both subfamilies (Fig. 1). The phyla Actinobactetia and Chloroflexi were found to possess only members of the vertebrate-related subfamily, whereas Bacteroidetes and Proteobactetria possessed only members of the mollusk-related subfamily (Fig. 1). Even within the latter subfamily, the clustering patterns did not reflect phylogeny. Sequences from Burkholderia and Chromobacterium (Proteobacteria) clustered with a sequence from Chitinophaga (Bacteroidetes), and this pattern received 95% bootstrap support (Fig. 1). But other sequences from Proteobacteria and Bacteroidetes fell outside this cluster (Fig. 1).
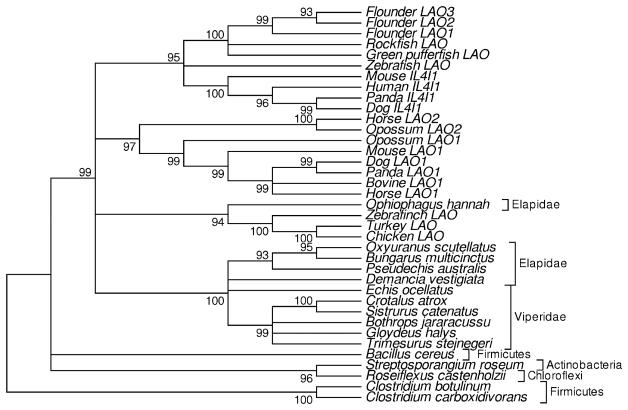
A total of 218 aligned amino acid residues were available for the phylogenetic tree of all 54 sequences (Fig. 1). In contrast, 373 aligned residues were available for the phylogenetic analysis of the vertebrate-related group alone; therefore, the vertebrate-related group was analyzed separately in order to obtain better resolution of relationships within that group (Fig. 2). In the phylogenetic tree of the vertebrate-related group, all vertebrate sequences clustered together, apart from bacterial sequences, and this topology received 99% bootstrap support (Fig. 2).
Fig. 2.
Condensed NJ tree (showing topology only) of the vertebrate-related subfamily of L-amino acid oxidases based on the JTT distance with gamma correction (parameter a=1.37) at 373 aligned sites. Numbers on the branches are percentages of 1,000 bootstrap samples supporting the branch; branches with bootstrap support <90% are collapsed
In the phylogenetic tree of the vertebrate-related group, mammalian sequences were found in three distinct clades: (1) LAO1, which was found in the orders Marsupialia (opossum), Rodentia (mouse), Perissodactyla (horse), Artiodactyla, bovine, and Carnivora (dog and panda); (2) LAO2, which was found in the orders Marsupialia and Perissodactyla; and (3) a clade including LAOs from bony fishes along with mammalian IL4I1, found in Rodentia, Carnivora, and Primates (Fig. 2). Each of these clades received strong bootstrap support (Fig. 2). The support was 95% for the clustering of vertebrate IL4I1 with LAOs of bony fishes, and the mammalian LAO1 and LAO2 clades received 99% and 100% bootstrap support, respectively (Fig. 2). In addition, mammalian LAO1 and LAO2 clades clustered together with 97% bootstrap support (Fig. 2).
All but one of the LAOs from snakes clustered together in a clade receiving 100% bootstrap support (Fig. 2), a pattern that was also seen in the tree based on all 54 sequences (Fig. 1). The one exception to this pattern was a sequence from the king cobra Ophiophagus hannah, which clustered with LAOs from birds; the latter clustering pattern received 94% bootstrap support (Fig. 2). Within the cluster containing most snake LAOs, the sequences from the families Elapidae and Viperidae formed separate clusters, but neither received strong bootstrap support (Fig. 2).
Conserved and divergent sequence features
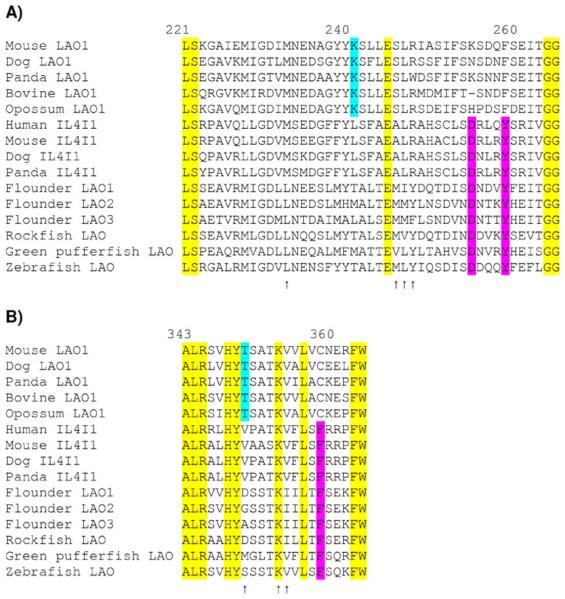
In the alignment of 54 amino acid sequences from bacteria, mollusks, and vertebrates, the following seven amino acid residues were conserved in all sequences (numbered here as in human IL4I1; Chavan et al. 2002): G66, G68, G71, G95, G96, G113, G265, and W364 (ESM Fig. S1). The first five of these residues (all glycines) are located in folds involved in FAD binding (Chavan et al. 2002). G265 and W364 are located in proximity to active site residues (Moustafa et al. 2006; Fig. 3).
Fig. 3.
Alignment of mammalian LAO1, mammalian IL4I1, and fish LAO sequences for the regions corresponding to residues 221–265 (a) and residues 343–364 (b) of human IL4I1. Residues conserved in all sequences illustrated are highlighted in yellow; sites where there is a unique amino acid residue found in all mammalian LAO1 are highlighted in turquoise, and sites where there is a unique residue conserved in all mammalian IL4I1 and fish LAO are highlighted in magenta. Arrows indicate active sire residues (by analogy with snake venom LAO; Moustafa et al. 2006)
Amino acid sequence features unique to the clade of mammalian IL4I1 and bony fish LAO (the IL4I1+Fish clade) are listed in Table 2. These include several unique residues that are in the vicinity of active site residues, namely, D255, Y259, and F359 (Fig. 3). Also of interest is a unique deletion of two residues (following G428 of human IL4I1) found in all members of the IL4I1+Fish clade and only in that clade (Table 2).
Table 2.
Unique conserved features of amino acid sequences of two clades of vertebrate LAOs
| Clade | Residues |
|---|---|
| IL4I1+Fish | D255a (H, K, N, Q, Y)b |
| Y259 (F) | |
| F359 (C) | |
| G428 (D,Q,N,R) | |
| Deletion of 2 amino acid residues after position 428 | |
| Mammalian LAO1 | S56 (I,P,T,V) I187 (L,F) |
| A213 (D,E,R) | |
| K241 (E,L,M,N,T,V) | |
| T350 (A,D,G,K,M,S,N,R,V) |
Sites are numbered according to the human IL4I1 as in Chavan et al. (2002)
Residues in parentheses are those found in other vertebrate LAOs outside the clade in question.
Discussion
Phylogenetic analysis of LAO family members showed that vertebrate and mollusk LAOs belong to two distinct subfamilies, consistent with the hypothesis that the functions of these proteins in innate immune defense have evolved independently in the deuterostome and protostome lineages. This conclusion is consistent with other analyses showing independent origin of deutersostome and protostome immune mechanisms (Hughes 1998). The vertebrate-related and mollusk-related groups of LAOs each included flavin-containing amine oxidases from bacteria. One explanation for this pattern is that these two subfamilies if LAOs are very ancient, resulting from a gene duplication that occurred prior to the most recent common ancestor of bacteria and eukaryotes. On this hypothesis, both bacterial and eukaryotic lineages would have ancestrally possessed both subfamilies of LAO, and one of them would have been lost in the vertebrate lineage while the other was lost in the mollusk lineage. If both subfamilies were present in the ancestor of animals, then it must be assumed that both lost in certain lineages of animals. For example, despite the availability of several complete insect genomes, BLAST homology searches revealed no insect LAOs, suggesting loss of all LAO genes in insects. In sea hares, LAOs are components of a secretion released into the aquatic environment (Butzke et al. 2005; Yang et al. 2005), raising the possibility that the adaptation to terrestrial life may have played a role in the loss of these genes in insects.
While much remains unresolved regarding the evolutionary relationships among the bacterial phyla, it was interesting that the vertebrate-related subfamily of LAOs was found mainly in phyla belonging to bacterial group I or Terrabacteria, whereas the mollusk-related subfamily was found exclusively in phyla belonging to group II or Hydrobacteria (Battistuzzi and Hedges 2009). This pattern suggests that the separation of the two subfamilies may have occurred when these two major clades of bacteria diverged about three billion years ago (Battistuzzi and Hedges 2009). The reported gene of the mollusk-related subfamily from Staphylococcus (Firmicutes), if it is not due to a contaminated sample, would plausibly be explained by horizontal gene transfer (HGT).
In addition to the Staphylococcus gene, there were other aspects of the phylogeny of the mollusk-related subfamily that suggest the possibility of HGT. Sequences from two species of Proteobacteria formed a well-supported cluster with a sequence from Bacteroidetes, while other sequences from Proteobacteria and Bacteroidetes fell outside this cluster. In addition, a sequence from this subfamily was found in Marseillevirus, a virus for which there is evidence of numerous HGT events (Boyer et al. 2009). The evidence for HGT in LAOs of microorganisms suggests the possibility that LAOs of animals resulted from two independent HGT events: (1) from Terrabacteria to an ancestor of vertebrates and (2) from Hydrobacteria to an ancestor of mollusks. However, it is at present impossible to decide definitively between the HGT hypothesis and the hypothesis of differential gene loss in these two lineages of animals.
Phylogenetic analysis of the vertebrate-related subfamily of LAOs showed a number of distinct clades. Most of the LAOs expressed in snake venom clustered together, except for one from king cobra which clustered with LAOs from birds (Fig. 2). The latter topology suggests that the snake venom LAOs may have resulted from a gene duplication unique to the snake lineage, although this hypothesis needs to be tested by further sequencing of LAOs from reptiles. In mammals, there were three distinct well-supported clades of LAOs: LAO1, LAO2, and IL4I1. LAO2 was found both in a marsupial (the opossum) and in one placental mammal (the horse), while LAO1 was found in the opossum and in several placental mammals (Fig. 2). This topology supports the hypothesis that LAO1 and LAO2 arose from a gene duplication that occurred before marsupials diverged from placental mammals.
In contrast, IL4I1 of mammals clustered with LAOs from bony fishes, supporting the hypothesis that IL4I1 arose by gene duplication before tetrapods diverged from bony fishes. The close relationship between mammalian IL4I1 and fish LAOs was supported by a number of unique shared sequence features, including both amino acid replacements and a unique two-codon deletion (Table 2) and involving unique amino acid residues in the vicinity of the active site (Fig. 3). Particularly striking among the latter is F359, at a position where all vertebrate LAOs except IL4I1 and fish LAOs have C (Fig. 3). It is possible that the presence of these shared residues reflects shared functions in IL4I1 and fish LAOs. However, at least one fish LAO appears not to share the preference for phenylalanine seen in mammalian IL4I1 (Boulland et al. 2007) since rockfish LAO shows a preference for l-lysine (Kitani et al. 2007). Experiments involving techniques such as site-directed mutagenesis may be able to shed light on the functional effects of the residues shared between IL4I1 and fish LAOs. However, in order to understand the functional diversification of this family, it will be necessary to know the substrate preferences of a wide variety of LAOs. By identifying major clades of LAOs, the phylogenetic analyses presented here provide useful background information to guide such functional studies.
The hypothesis that IL4I1 and the ancestor of LAO1 and LAO2 arose from an ancient duplication prior to the origin of tetrapods implies that one or other of these lineages has been lost repeatedly in the evolution of vertebrates. For example, the LAO1/LAO2 lineage appears to have been lost in bony fishes, while the IL4I1 lineage has apparently been lost in many non-mammalian tetrapods. Patterns of differential loss of members of this family have apparently continued within mammalian evolution. Although LAO2 arose prior to the divergence of marsupials and placentals, it is not found in most placentals with sequenced genomes, including human and mouse. In the human, LAO1 also has been lost, leaving IL4I1 as the apparent sole representative of the family. Thus, the overall pattern of evolution of the LAO family in vertebrates seems to have been one of “birth-and-death,” involving repeated duplications and deletions, a pattern characteristic of numerous gene families involved in both specific and innate immune processes of vertebrates (Hughes and Nei 1989; Nei and Rooney 2005).
Supplementary Material
Acknowledgments
This research was supported by grant GM43940 from the National Institutes of Health.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00251-010-0482-8) contains supplementary material, which is available to authorized users.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistuzzi FU, Hedges SB. A major clade of prokaryotes with ancient adaptations to life on land. Mol Biol Evol. 2009;26:335–343. doi: 10.1093/molbev/msn247. [DOI] [PubMed] [Google Scholar]
- Boulland M-L, Marquet J, Molinier-Frenkel MP, Guiter C, Lasoudris F, Copie-Bergman C, Baia M, Gaulard P, Leroy K, Castellano F. Human IL4I1 is a secreted l-phenylalanine oxidase expressed by mature dendritic cells that inhibits T-lymphocyte proliferation. Blood. 2007;110:220–227. doi: 10.1182/blood-2006-07-036210. [DOI] [PubMed] [Google Scholar]
- Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, Suzan-Monti M, La Scola B, Koonin EV, Raoult D. Giant Marseillevirus highlights the role of amoebae as a melting pot in the emergence of chimeric microorganisms. Proc Natl Acad Sci USA. 2009;106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butzke D, Hurwitz R, Thiede B, Godert S, Rudel T. Cloning and biochemical characterization of APIT, a new L-amino acid oxidase from Aplysia punctata. Toxicon. 2005;46:479–489. doi: 10.1016/j.toxicon.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Carbonelle-Puscian A, Copie-Bergaman C, Baia M, Martin-Garcia N, Allory Y, Haioun C, Crémades A, Abd-Alsamad I, Farcet J-P, Gaulard P, Castellano F, Molinier-Frenkel V. The novel immunosuppressive enzyme IL4I1 is expressed by neoplastic cells of several B-cell lymphomas and by tumor-associated macrophages. Leukemia. 2009;23:952–960. doi: 10.1038/leu.2008.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavan SS, Tian W, Hsueh K, Jahaweer D, Gregersen PK, Chu CC. Characterization of the human homolog of the IL-4 induced gene (Fig1) Biochem Biophys Acta. 2002;1576:70–80. doi: 10.1016/s0167-4781(02)00295-6. [DOI] [PubMed] [Google Scholar]
- Du X-Y, Clemetson KJ. Snake venom L-amino acid oxidases. Toxicon. 2002;40:659–665. doi: 10.1016/s0041-0101(02)00102-2. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Friedman R, Hughes AL. Molecular evolution of the NF-κB signaling system. Immunogenetics. 2002;53:964–974. doi: 10.1007/s00251-001-0399-3. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Evolution of cysteine proteinases in eukaryotes. Mol Phyl Evol. 1994;3:310–321. doi: 10.1006/mpev.1994.1038. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Protein phylogenies provide evidence of a radical discontinuity between arthropod and vertebrate immune systems. Immunogenetics. 1998;47:283–296. doi: 10.1007/s002510050360. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99:364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- Hughes AL. The origin of adaptive phenotypes. Proc Natl Acad Sci USA. 2008;105:13193–19194. doi: 10.1073/pnas.0807440105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Evolution of the major histocompatibility complex: independent origin of nonclassical class I genes in different groups of mammals. Mol Biol Evol. 1989;6:559–579. doi: 10.1093/oxfordjournals.molbev.a040573. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Codon-based tests of positive selection, branch lengths, and the evolution of mammalian immune system genes. Immunogenetics. 2008;60:495–506. doi: 10.1007/s00251-008-0304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Green JA, Piontkivska H, Roberts RM. Aspartic proteinase phylogeny and the origin of pregnancy-associated glycoproteins. Mol Biol Evol. 2003;20:1940–1945. doi: 10.1093/molbev/msg217. [DOI] [PubMed] [Google Scholar]
- Jones D, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kimura M, Ohta T. On some principles governing molecular evolution. Proc Natl Acad Sci USA. 1974;71:2848–2852. doi: 10.1073/pnas.71.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitani Y, Tsukamoto C, Zhang G, Nagai H, Ishida M, Ishazaki S, Shimakura K, Shiomi K, Nagashima Y. Identification of an antibacterial protein as L-amino acid oxidase in the skin mucus of rockfish Sebastes schlegeli. FEBS J. 2007;275:125–136. doi: 10.1111/j.1742-4658.2006.05570.x. [DOI] [PubMed] [Google Scholar]
- Mason JM, Naidu MD, Barcia M, Porti D, Chavan SS, Chu CC. IL-4-induced gene-1 is a leukocyte L-amino acid oxidase with an unusual acidic pH preference and lysosomal localization. J Immunol. 2004;173:4561–4567. doi: 10.4049/jimmunol.173.7.4561. [DOI] [PubMed] [Google Scholar]
- Moustafa IM, Foster S, Lyubimov AY, Vrielink A. Crystal structure of LAAO from Calloselasma rhodostoma with an l-phenylalanine substrate: insights intro structure and mechanism. J Mol Biol. 2006;364:991–1002. doi: 10.1016/j.jmb.2006.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka K, Aoki F, Hayashi M, Muroi Y, Sakurai T, Itoh K, Ikawa M, Okabe M, Imakawa K, Sakai S. L-amino acid oxidase plays a crucial role in host defense in the mammary glands. FASEB J. 2009;23:2514–2520. doi: 10.1096/fj.08-126466. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney A. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelek PD, Cheah J, Coulombe R, Macheroux P, Chisla S, Vrielink A. The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site. EMBO J. 2000;19:4204–4215. doi: 10.1093/emboj/19.16.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Diggins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Chen W, Shi W, Qi F, Dong X. SA-LAAO, a novel L-amino acid oxidase that enables Streptococcus oligofermentans to outcompete Streptococcus mutans by generating H2O2 from peptone. J Bacteriol. 2008;190:4716–4721. doi: 10.1128/JB.00363-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Johnson PM, Ko K-C, Kamio M, Germann MW, Derby CD, Tai PC. Cloning, characterization and expression of escaping, a broadly antimicrobial FAD-containing L-amino acid oxidase from ink of the sea hare Aplysia californica. J Exp Biol. 2005;208:3609–3622. doi: 10.1242/jeb.01795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.