Abstract
Genomic imprinting results in preferential expression of the paternal, or maternal allele of certain genes. We have performed a genome-wide characterization of imprinting in the mouse embryonic and adult brain. This approach uncovered parent-of-origin allelic effects in over 1300 loci. We identified parental bias in the expression of individual genes and of specific transcript isoforms, with differences between brain regions. Many imprinted genes are expressed in neural systems associated with feeding and motivated behaviors, and parental biases preferentially target genetic pathways governing metabolism and cell adhesion. We observed a preferential maternal contribution to gene expression in the developing brain and a major paternal contribution in the adult brain. Thus, parental expression bias emerges as a major mode of epigenetic regulation in the brain.
Parent-of-origin effects influence gene expression and trait inheritance in offspring. Genomic imprinting is a form of epigenetic regulation that results in the preferential expression of the paternally or maternally inherited allele of certain genes (1). Currently, fewer than 100 imprinted genes have been identified, and the evolutionary pressures that underlie imprinting are debated (2, 3). Clinical and experimental data suggest roles for imprinting in regulating brain development and function (4). In humans, Prader-Willi Syndrome (PWS) and Angelman Syndrome (AS) result from a deletion of the paternal or maternal copy of 15q11-13, respectively. PWS is associated with hyperphagia, stubbornness and compulsive traits (5), whereas AS is associated with absent speech, happy affect and inappropriate laughter (6). Further, studies of parthenogenetic (PG)- and androgenetic (AG)- chimeras in the mouse have suggested preferential maternal contribution to the development of the cortex, but preferential paternal contribution to the hypothalamus (7, 8). Such biased roles have yet to be clearly demonstrated. Moreover, despite tantalizing reports, our understanding of the neural systems governed by imprinted genes, and of the scope and features of imprinted loci expressed in the brain is very limited.
Imprinting refers to functional differences between the maternal and paternal chromosomes or alleles (9), and is also used more strictly to define complete allele-specific silencing (10). Known imprinted genes have been shown to display all-or-none and biased allelic expression according to the gene and tissue considered (11, 12). We report here a genome-wide analysis of parental allelic effects involving complete silencing or parental biases in gene expression in the murine embryonic day 15 (E15) brain, and in the adult male and female cortex (medial prefrontal cortex (mPFC)) and hypothalamus (preoptic area (POA)). Together with a companion study (13), our data suggests that substantial maternal and paternal biases in gene expression originate from the X chromosomes and autosomes, respectively. These results may shed light on gene regulatory processes underlying brain function, evolution and disease.
Imprinted Gene Expression in the Adult CNS
To gain insight into neural systems affected by imprinting, we performed an in silico study of the expression pattern of known imprinted genes in the adult brain. The expression pattern of 45 known imprinted genes was investigated across 118 distinct adult brain regions in the Allen Brain Atlas (Fig. 1 and fig. S1). A heatmap based on the relative number of known imprinted genes expressed in a given brain region identified 26 out of 118 brain regions as hotspots for the expression of imprinted genes, whereas the expression hotspots of 20 randomly selected control genes with known biallelic expression were located mainly in cortical and olfactory regions and appeared entirely distinct from that of imprinted genes (Fig. 1 and fig. S1). Brain regions predicted from earlier studies to be enriched for imprinted gene expression indeed emerged as hotspots, such as the medial preoptic area (MPOA), which regulates mating, maternal behavior and thermoregulation (14). From our data, aminergic systems and neural systems associated with feeding and motivated behaviors constituted the largest source of imprinting hotspots. These included: the arcuate nucleus (ARC), dorsal raphe (DR), substantia nigra pars compacta (SNc), ventral tegmental area (VTA), dorsal hypothalamic area, locus ceruleus (LC) and nucleus accumbens (NA) (15, 16). These findings enticed us to perform a more detailed and large-scale analysis to characterize and compare parent-of-origin effects governing gene expression in distinct brain regions.
Fig. 1.
A map of imprinted gene expression in the adult CNS identifies distinctive hot- and cold-spots. Presence (colored squares) versus absence (dark grey squares) of imprinted gene expression was mapped in a representative subset of brain regions (full map in Fig. S1). Randomly selected biallelic control genes are indicated by green squares. The heatmap was assigned for each brain region according to the number of standard deviations from the mean for the number of imprinted genes expressed (Cooler to Warmer (standard deviations): <−2, <−1.5, <−1, >+1, >+1.5, >+2).
A High Resolution Approach to Analyze Imprinting
We used Illumina RNA-Seq technology to characterize the transcriptome of brain tissues from F1 hybrids resulting from reciprocal crosses of CAST/EiJ (CAST) and C57BL/6J (C57) mice (F1 initial cross (F1i): CAST mother X C57 father; F1 reciprocal cross (F1r): C57 mother X CAST father). Single nucleotide polymorphisms (SNPs) were identified by separately sequencing the CAST and C57 transcriptomes of the original parents (except for E15 brains), and the subsequent base calls were used to distinguish transcription from maternal and paternal alleles in F1i and F1r (Table S1, fig. S2, S3 and supplementary online material (SOM)). We characterized parent-of-origin effects governing gene expression in the E15 brain, as well as the adult male and female mPFC and POA. For the current study, male and female samples were treated as biological replicates. This approach is appropriate for the detection of parental effects that are independent of the sex of the offspring.
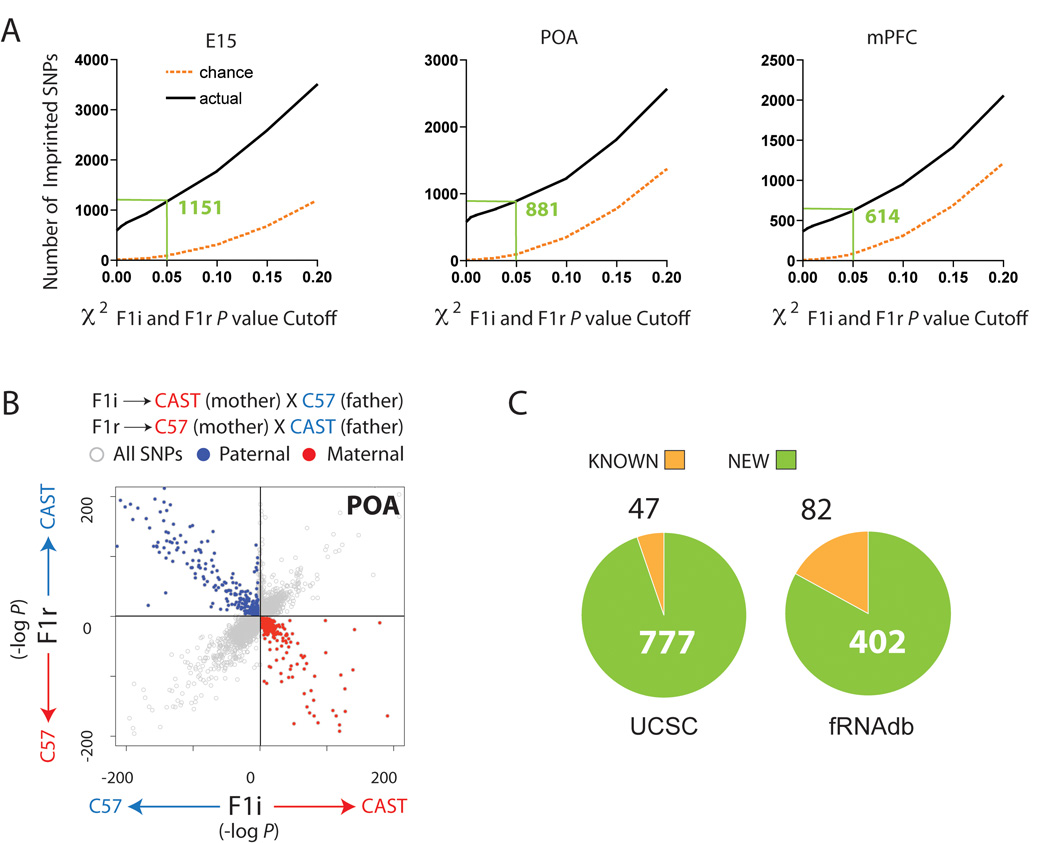
Imprinting was assessed by χ2 tests in both initial and reciprocal crosses as described in the SOM. The total number of SNP sites exhibiting a significant parent-of-origin effect was determined for a range of χ2 P value cutoffs (0.001 to 0.2) and compared to the number expected by chance (Fig. 2A). We selected a cutoff of P<0.05 for each cross (E15 False Discovery Rate (FDR) = 0.06, POA FDR = 0.1, mPFC FDR = 0.1). Our approach yields highly accurate and reproducible results, as demonstrated by multiple controls detailed in the SOM. Scatterplots of the −log (P) for the F1i and F1r data for each SNP site clearly indicated exclusive selection of paternally and maternally expressed loci relative to the total dataset (Fig. 2B and fig. S4). Overall, SNPs identified by our approach (excluding mitochondrial and X chromosome SNP sites) exhibited a robust parental expression bias with a mean of 87±15% (mean±SD). Parent-specific biases emerged as a continuum from the dataset, suggesting that imprinting may manifest as relative allele-specific expression bias rather than strict monoallelic transcription, or that allelic bias is cell-type specific and is partially masked by cellular heterogeneity in brain samples (Fig. 2B and fig. S4). Our approach that includes sequencing of transcriptomes from parents and hybrid offspring, as well as increased sequence depth, likely contribute to differences in results between our and previous studies (17, 18).
Fig. 2.
Identification of loci exhibiting parent-of-origin allelic effects in the embryonic and adult CNS using Illumina sequencing. (A) Plots of the number of SNP sites exhibiting parental expression bias identified by sequencing (black) compared to chance expectations (orange) at various χ2 P value cutoffs. Green values indicate number of imprinted SNPs detected at P<0.05. (B) Scatterplot of the -log (P) of the two-tailed χ2 probability (P) for individual SNP sites for the F1i versus the F1r cross (POA shown). SNP sites identified by P<0.05 cutoff in each cross are indicated by red and blue dots. (C) Numbers of known and uncovered genes associated with parental allelic effects.
Genome-wide Analysis of Imprinting
Imprinted genes and genes with imprinted features were identified by the presence of one or more SNP sites exhibiting a significant paternal or maternal expression bias, as described above. This approach enabled us to identify 1308 candidate imprinted loci, among which 824 genes annotated in the University of California Santa Cruz genome database (UCSC) (5.7% of the ~14520 genes assessed) (Fig. 2C, Table S2) and 484 putative ncRNAs annotated in the functional RNA database (fRNAdb) (4.1% of the 11545 ncRNAs assessed) (Fig. 2C, Table S3). Of these, 604 have known human orthologs. Of the 86 previously known imprinted genes, 72 were expressed in one or more brain regions and contained SNPs above the 10 read minimum cutoff. Among those, 47 were called imprinted, whereas the remaining 25 exhibited biallelic expression in all brain regions tested. Of the 484 ncRNAs associated with parental allelic effects based on alignments to the fRNAdb, we classified 82 as “known” based on genomic positions directly or closely associated with previously known imprinted ncRNAs, including Apeg3, Copg2as, Air, Nespas, H19, Peg12, Snurf/Snrpn/Ube3aas, Gtl2, and Rian (19).
A gene ontology analysis revealed that biological processes associated with parental allelic effects are mostly related to metabolic processes in the developing brain (eg. primary metabolic process, FDR = 4.11E-14), and to cell-adhesion in the adult brain (eg. cell adhesion, FDR=1.45E-8) (Table S4). These findings are striking in light of previous work that identified roles for imprinted genes in growth, feeding, metabolism and thermoregulation (2). We report here and in our companion study (13), parental allelic effects at key conserved regulators of metabolism such as Interleukin-18 (Il18) (13) and the mitochondrial ribosomal protein Mrpl48 (20), as well as cell-adhesion, such as cadherin 15 (cdh15).
Characterization of Gene Clusters with Parent-of-Origin Allelic Effects
Analysis of the genomic distribution of all loci identified in our study shows a scattered distribution across all chromosomes (fig. S5). An algorithm was applied that searched for >2 imprinted genes and/or ncRNAs residing within a 1Mb window. This window size correctly identified previously characterized imprinted gene clusters (eg: H19-IGF2, Mest-Copg2, Dlk1-Mest), with the exception of the 4MB long PWS-AS cluster that splits into two clusters (Table S5). This analysis identified 204 putative imprinted gene clusters, which encompass 65% of the genes and ncRNAs identified in our study. The presence of imprinted ncRNAs has been demonstrated to play a critical role in the regulation of imprinting for many known imprinted gene clusters (1) and 106 (52%) of these candidate clusters contained both coding and putative noncoding loci (Table S5). For a summary of data for known imprinted gene clusters, see fig. S6.
Our approach identified features in imprinted gene clusters known to be associated with brain functions and disorders. For example, Peg13 and Kcnk9 (linked to Birk-Barel mental retardation (21)) were found to be part of a larger cluster that includes 1810044A24Rik (also called Trappc9 and linked to mental retardation (22)), several maternally expressed ncRNAs, and a MEG, Eif2c2 (argonaute2) (Fig. 3A and B). From our data, it appears that 1810044A24Rik undergoes isoform-specific imprinting, which is revealed by SNPs within the unique exon and 3’UTR of the uc007wbn.1 isoform that are all paternally expressed. SNPs located in the exons shared by all other isoforms (uc007wbp.1, uc007wbm.1 and uc007wbl.1) are maternally expressed.
Fig. 3.
Features of imprinted gene clusters associated with neurological disorders and diseases. (A) UCSC browser tracks indicate all reads aligning uniquely to the genome (Expression, black) and the percentage of transcriptome aligned reads at SNP sites that were assigned to the paternal (blue, positive) versus maternal allele (red, negative). Analysis of 1810044A24Rik isoforms (1: uc007wbn.1, 2: uc007wbl.1 (uc007wbm.1 not shown), 3: uc007wbp.1) revealed mixed isoform specific imprinting at this locus. A paternally expressed isoform (uc007wbn.1) was identified by SNPs located in the unique 3’ exon and UTR. (B) Bar graphs indicate parental bias for SNP sites located in a 1810044A24Rik shared exon (SNP_ID: uc007wbl.1_1339), the 3’UTR of the uc007wbn.1 isoform (SNP_ID: uc007wbn.1_4207), and Eif2c2 (SNP_ID: uc007wbu.1_1944). Sequenom analysis validated maternal expression of Eif2c2 (E15 brain). Highlighted regions in browser tracks (A; red: Eif2c2; blue: 1810044A24Rik) indicate regions detailed in bar graphs. χ2 analysis (***P<0.001; **P< 0.01). (C) A large region of paternally-biased transcription was uncovered in the PWS-AS gene cluster between the SNRPN/SNURF locus and Ndn. (D) Sequenom validation of paternally-biased expression by DOKist4 in the PWS-AS cluster (SNP_ID: uc009hex.1_3057).
In the PWS-AS cluster we uncovered a large region between Snrpn and Ndn that hosts numerous paternally expressed imprinted ncRNAs, including two predicted microRNAs (mir-344 and mir-344-2) (Fig. 3C). Sequenom analysis of allele-specific expression with an independent cohort of animals replicated the Illumina results and clearly revealed strict paternal expression of the DOKist4 gene within this region (Fig. 3D).
Brain Region- and Developmental Stage- Specific Parent-of-origin Allelic Effects
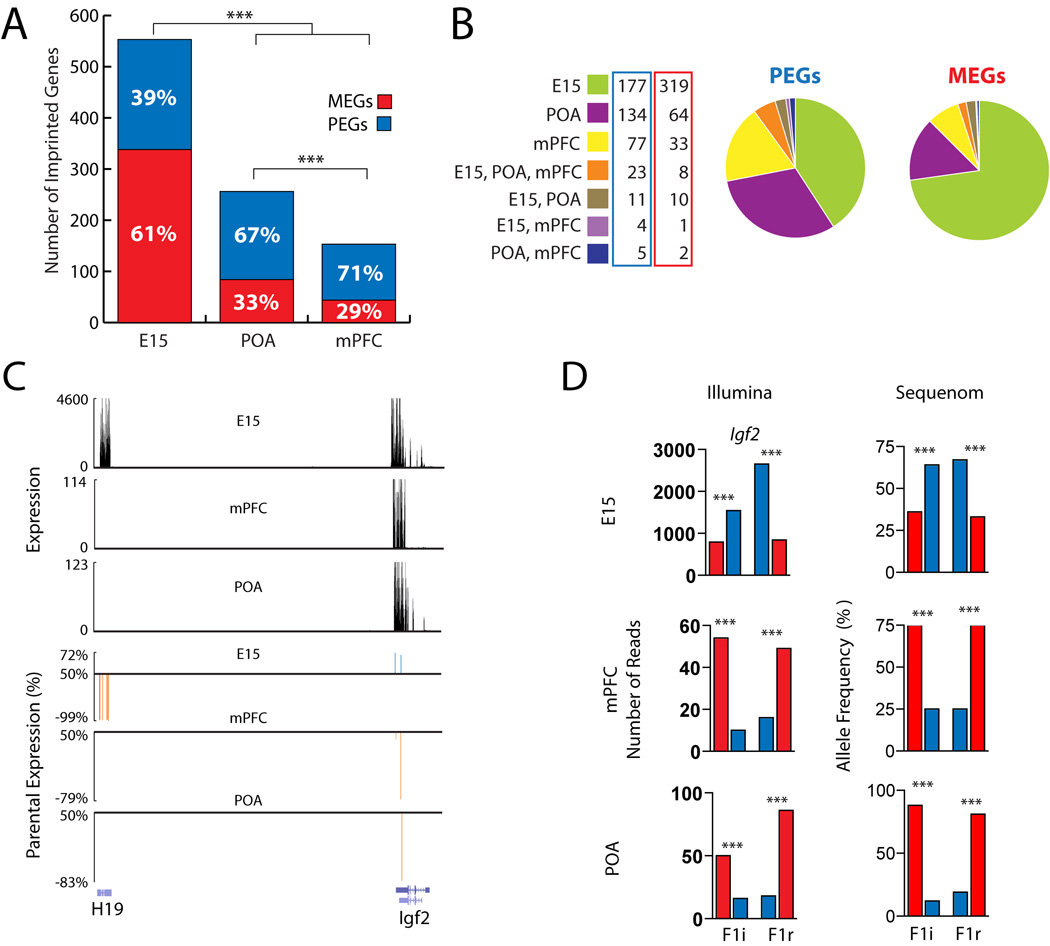
A total of 553 UCSC genes associated with parental allelic effects were uncovered in the E15 brain, compared to 256 in the adult POA (P<0.001; χ2 Analysis) and 153 in the adult mPFC (P<0.0001; χ2 Analysis) (Fig. 4A and B).
Fig. 4.
Parent-of-origin allelic effects influence gene expression in a developmental and region specific manner in the CNS. (A) Comparison of the total numbers of UCSC annotated genes with parent-of-origin allelic effects in the E15 brain, adult POA and mPFC. Red and blue bars indicate MEGs and PEGs identified in each sample, respectively. (B) Pie charts comparing the relative numbers of PEGs and MEGs identified in the E15 brain, mPFC, and POA. (C) Spatiotemporal regulation of imprinting at the H19-Igf2 locus revealed by UCSC Browser tracks of raw expression data (black) and parental expression bias (blue: paternal, red: maternal) at identified SNP sites in H19 and Igf2. (D) Igf2 allele-specific expression inversion confirmed by Illumina and Sequenom (SNP_ID: uc009kod.1_2313). Raw expression tracks of reads uniquely aligning to the genome are shown below in black. χ2 analysis (***P<0.001; **P< 0.01; *P< 0.05).
61% of genes identified in the E15 brain were MEGs, revealing a significant maternal bias in the developing brain (PEGs: 215, MEGs: 338, P<0.0001; χ2 Analysis) (Fig 4A). In contrast, a paternal bias was observed in both the adult POA (PEGs: 172, MEGs: 84, P<0.0001; χ2 Analysis) and the adult mPFC (PEGs: 109, MEGs: 44; P<0.0001; χ2 Analysis), such that ~70% of genes identified in the adult brain were PEGs. The observed parental allelic biases were statistically significant through a range of different P value cutoffs (P<0.03, P<0.05 and P<0.1) that increased the total number of genes by over three-fold, thus indicating a robust signal-to-noise ratio in the data. The biases were not present at higher P value cutoffs (P<0.9).
Of the 824 UCSC annotated genes associated with parental allelic effects in the E15 brain, POA or mPFC, 769 (93%) were expressed and had SNP site read depths above the cutoff of 10 in all of the three target brain tissues. However, most demonstrated a significant parental expression bias in only one of the target tissues (Fig. 4B). A majority was found exclusively in the E15 brain, including 73% of all MEGs. Further, only 5 PEGs were shared between the adult POA and mPFC. 74% of the genes imprinted in all three samples were PEGs. These results suggest that parental influence over gene expression is highly spatially and temporally regulated in the brain.
Two examples of this phenomenon are detailed here and in the SOM (fig. S7 and S8). The Igf2-H19 locus has been linked to colorectal and other forms of cancers (23), Beckwith-Wiedemann syndrome (BWS) (23) and Silver-Russell Syndrome (24). H19 is a maternally expressed ncRNA (25) and Igf2 is a canonical PEG that promotes placental and embryonic growth (2). In endodermal and mesodermal cell lineages, the reciprocal parental expression of the two genes is due to a competition for promoter access to a shared set of enhancers located downstream of H19 (26, 27). Maternal H19 expression is directly involved in regulating the paternal expression of Igf2 (28). Previous studies have suggested that imprinting at this locus is more complex in the brain (28–30).
Our data document maternal expression of H19 and paternal bias of Igf2 in the E15 brain (Fig. 4C and D). H19 is not expressed in the adult mPFC or POA, and 80% of Igf2 transcription in the adult male and female POA and mPFC originates from the maternal allele (Fig. 4C and D). These data were confirmed by Sequenom on a distinct cohort of animals (Fig. 4D). Similarly, a gene cluster encompassing Grb10 and Dopa Decarboxylase (Ddc) displays spatiotemporally regulated parental allelic effects (fig. S7 and S8).
These examples, and the reproducibility of the parental allelic biases in independent male, female samples, and by Sequenom, highlight the extraordinary complexity of parental influence over transcription in the CNS.
Complex Parent-of-origin Allelic Effects in the Brain
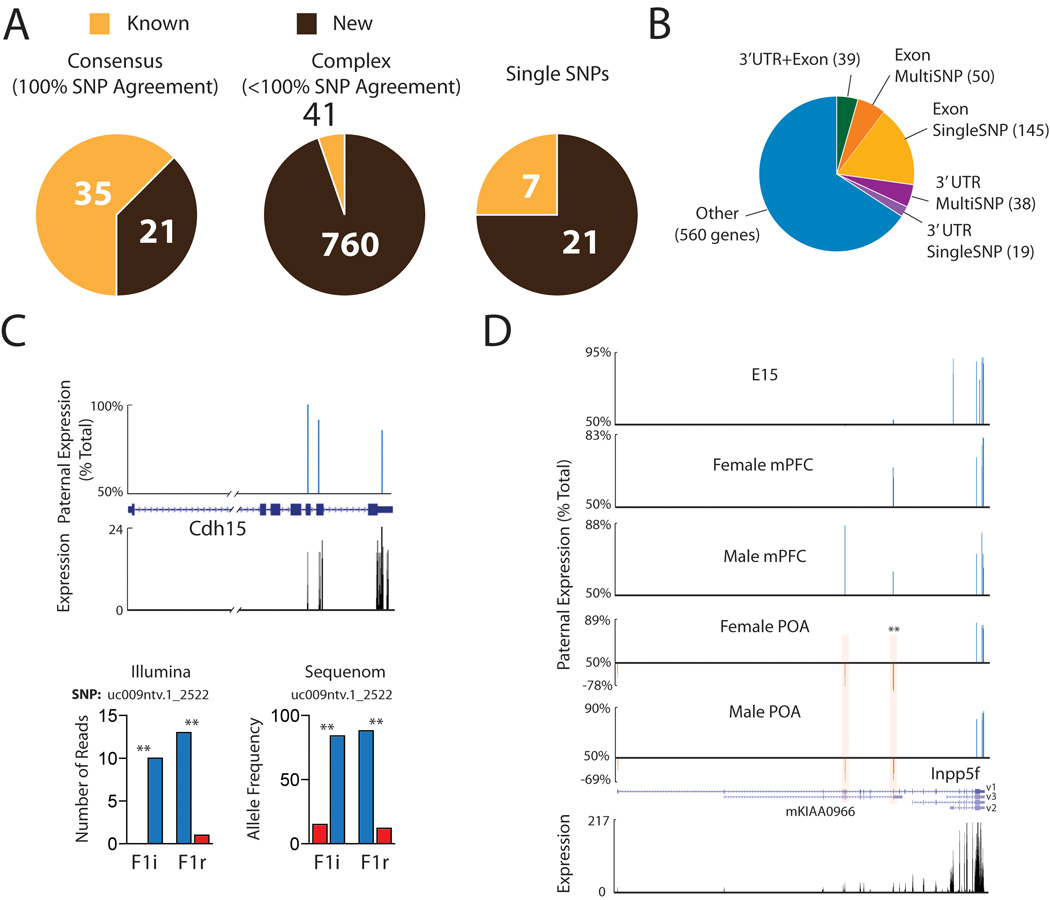
Three general categories of genes with parent-of-origin expression bias emerged from our analysis in known, as well as newly identified loci, which we term consensus, complex, and single SNP loci (Fig. 5A, Table S6–S12). Consensus loci have multiple SNPs, at least one SNP site above the P value cutoff of 0.05 in each cross and 100% of SNPs agree with the direction of the parental expression bias in both the F1i and F1r cross. Complex loci have multiple SNPs, with one or more SNPs above the P value cutoff and one or more SNP sites that differ (ie. biallelic expression, strain or opposite parental bias). Finally, a subset of genes had a single SNP site or multiple SNPs within 32 bp of each other (the size of a single read). Thirty-five of the 47 previously known imprinted genes displayed consensus imprinting in at least one brain region (Table S6). However, several of these same genes also exhibited complex imprinting in other samples, such that 41 known imprinted genes were identified as complex in one or more brain regions. Seven known imprinted genes were identified based on a single SNP site (Fig. 5A). Detailed analysis of the positions of SNPs with parental allelic bias within complex loci revealed genes in which monoallelic SNPs are confined to a specific exon (195 genes), to the 3’UTR (57 genes), or to the final exon and 3’UTR region (39 genes) (Fig. 5B), suggesting that, in these genes, the parental allelic effect is restricted to only one or a few transcript isoforms. In a subset of these cases, the same parental bias is confirmed by multiple SNPs in the exon or 3’UTR (Fig. 5B). A large proportion of the genes exhibited parental effects in the last exon (including 3’UTR region), but involved disagreements between SNP sites in the same region of the gene (classified as “other” (560 genes)). In some cases, as detailed below, these disagreements appear to be related to the fact that only a subset of the SNPs for a given complex gene are able to distinguish a specific imprinted isoform from other overlapping transcripts arising from the same locus.
Fig. 5.
Characterization of complex genes associated with parental allelic effects in the CNS transcriptome. (A) Numbers of previously known and newly uncovered consensus, complex, or single SNP imprinted genes. (B) Proportions of complex genes with parent-of-origin allelic effects localized to one or more exons (Exon), the 3’UTR (last exon), or 3’UTR+Exons, or other outcomes (ie. disagreements between SNPs in the same exon or 3’UTR). Exons or 3’ UTRs with more than one SNP for evidence are indicated separately (MultiSNP). (C) UCSC browser tracks at the cdh15 locus indicate preferential expression of the paternal allele (paternal allele expression bias: blue, POA data shown). Illumina and Sequenom analysis confirmed preferential expression of the paternal allele for cdh15 (SNP_ID: uc009ntv.1_2522). (D) Complex spatiotemporal and isoform-specific imprinting at the Inpp5f locus. A significant maternal bias was observed specifically in the region of Inpp5f_v1 that overlaps with mKIAA0966 in adult POA. Highlighted SNP sites of particular interest are statistically significant in both crosses by χ2 analysis (***P<0.001; **P< 0.01; *P< 0.05).
Cadherin 15 (cdh15), a gene prospectively linked to intellectual disability in humans (31), emerges as a consensus imprinted locus, in which all three SNPs display preferential expression of the paternal allele in independent male and female samples (Fig. 5C) and by Sequenom on an independent cohort of animals (Fig. 5C). Other notable consensus imprinted genes include Bcl2l1, a major regulator of apoptosis linked to cancer (32), and Eif2c2 (also called argonaute2), involved in microRNA and short-interfering RNA (siRNA)-mediated gene silencing (33) (Table S6).
Detailed analysis of complex loci revealed remarkable and so far unsuspected features of parent-of-origin transcription bias (Table S7–11). In the Inpp5f locus, three isoforms have previously been described (Fig. 5D) with preferential paternal expression of Inpp5f_v2 and Inpp5f_v3, while Inpp5f_v1 was reported biallelic (34). In our analysis, SNP sites aligning to the Inpp5f_v2 and Inpp5f_v3 isoforms confirmed strict paternal expression. Four SNP sites located in exons shared by the Inpp5f_v1 isoform and an overlapping UCSC annotated transcript (mKIAA0966) indicated a modest and nonsignificant paternal bias in expression in the adult mPFC and E15 brain. However, in the adult male and female POA 73% of transcription at these sites (P<0.01, in F1i and F1r cross) originated from the maternal allele. A single SNP found in the first exon of Inpp5f_v1, indicated a modest, nonsignificant maternal expression bias in POA. Thus, our approach resolved complex regional-, developmental stage- and isoform- specific parental bias in the transcriptome.
Recently, a highly complex form of imprinting has been described for the gene H13, such that some H13 isoforms are maternally expressed, whereas others are paternally expressed (35). Our analysis confirmed these results (fig. S9). Here we find that Herc3, a host gene for the known PEG, Nap1l5 showed features indicative of isoform specific imprinting in a manner very similar to H13 (fig. S10). Additional examples of complex parent-of-origin effects in the CNS transcriptome are presented in the SOM for Lsm14a, Pafah1b3, and Ndel1 (fig. S11). Other notable complex loci include cdh2 (neuronal-cadherin), which plays a central role in brain morphogenesis (36), as well as arnt2 (aryl hydrocarbon receptor nuclear transclocator 2), a gene with multiple isoforms that regulates hypothalamic development in concert with other imprinted genes, such as Ndn (37). Many genes identified in our analysis exhibited complex patterns of parental allelic effects for which the underlying mechanism and functional significance are not yet clear.
Finally, several loci in our dataset did not display the classical pattern associated with parent-of-origin expression biases, but instead displayed significant differences in the relative expression of the maternal and paternal alleles in F1i versus F1r offspring, which we refer to as cross-effects. These effects were analyzed separately and the findings are detailed in the supplemental data (fig. S12).
Discussion
Our study documents over ~1300 protein coding genes and putative ncRNAs associated with parental allelic effects in expression in the brain. The resolution and reproducibility of our approach is highlighted by the correct detection of maternally-inherited mtDNA and male X-linked loci, highly correlated parental bias among male and female samples from the same adult brain regions, and finally, by independent confirmation using sequenom for select examples. From our study, parent-of-origin effects in the brain emerges as a very complex and widespread form of epigenetic regulation characterized by brain region-, developmental stage-, and isoform-specific parental allelic effects. These findings build substantially upon earlier studies that identified imprinted genes in which monoallelic expression is restricted to a developmental stage (31), tissue (39, 40), or cell type (41). Such complex regulation is likely to involve the combined effects of specific parent-or origin allelic DNA methylation patterns and histone-modifications, as well as tissue- and cell-type specific promoters and enhancers (40, 42). Recent work suggests that alternative polyadenylation sites may also contribute to the generation of distinct maternal and paternal isoforms (35). It will be of interest to determine whether other emerging epigenetic mechanisms that appear to influence the expression of alternative exons and 3’UTRs in the transcriptome, such as nucleosome positioning and histone modifications (43, 44), might be relevant to the complex parent-of-origin effects uncovered in our data.
Early studies of imprinting gave rise to the concept of a maternal influence centered in the cortex and a paternal influence centered in the hypothalamus (7, 8). A slightly different picture emerged from our study, such that significant maternal influence was uncovered in the embryonic brain, whereas a robust paternal bias was observed in both adult cortex (mPFC) and hypothalamus (POA). Our companion study suggests maternal control over adult brain gene expression residing on the X chromosome (13). Our findings may provide insights into brain evolution, function and neurological disease, due to the prominent involvement of X-linked genes in neurological function (45) and the unique susceptibility of imprinted loci to mutation and dysregulation (46).
Supplementary Material
Acknowledgments
We thank L. Luo, T. Maniatis, R. Losick, S. Hippenmeyer, and members of the Maniatis and Dulac labs for critical comments on the manuscript. We thank R. Hellmiss for help with figures and R. Jaenisch for Xegfp mice. This work was supported by the Klarman Foundation for Eating Disorders and the Howard Hughes Medical Institute (HHMI). CG is supported by a Human Frontiers long-term fellowship and Alberta Heritage Foundation for Medical Research Incentive Award. CD is an HHMI investigator. All data can be found in GEO: GSE22131
References and Notes
- 1.Reik W, Walter J. Nat Rev Genet. 2001;2:21. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 2.Haig D. Annu Rev Genet. 2004;38:553. doi: 10.1146/annurev.genet.37.110801.142741. [DOI] [PubMed] [Google Scholar]
- 3.Keverne EB. Hormones and behavior. 2001;40:146. doi: 10.1006/hbeh.2001.1685. [DOI] [PubMed] [Google Scholar]
- 4.Wilkinson LS, Davies W, Isles AR. Nat Rev Neurosci. 2007;8:832. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- 5.Cassidy SB, Driscoll DJ. Eur J Hum Genet. 2009;17:3. doi: 10.1038/ejhg.2008.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Buggenhout G, Fryns JP. Eur J Hum Genet. 2009;17:1367. doi: 10.1038/ejhg.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen ND, et al. Proc Natl Acad Sci USA. 1995;92:10782. doi: 10.1073/pnas.92.23.10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keverne EB, Fundele R, Narasimha M, Barton SC, Surani MA. Brain Res Dev Brain Res. 1996;92:91. doi: 10.1016/0165-3806(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 9.Hall JG. Am J Hum Genet. 1990;46:857. [PMC free article] [PubMed] [Google Scholar]
- 10.Bartolomei MS, Tilghman SM. Annu Rev Genet. 1997;31:493. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez A, Fiering S, Martinez E, Galton VA, St Germain D. Endocrinology. 2002;143:4483. doi: 10.1210/en.2002-220800. [DOI] [PubMed] [Google Scholar]
- 12.Ono R, et al. Genome Res. 2003;13:1696. doi: 10.1101/gr.906803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregg C, Jiangwen Z, Butler JE, Haig D, Dulac C. companion study. 2010 [Google Scholar]
- 14.Keverne EB. Adv Genet. 2007;59:217. doi: 10.1016/S0065-2660(07)59008-5. [DOI] [PubMed] [Google Scholar]
- 15.Gao Q, Horvath TL. Annu Rev Neurosci. 2007;30:367. doi: 10.1146/annurev.neuro.30.051606.094324. [DOI] [PubMed] [Google Scholar]
- 16.Hyman SE, Malenka RC, Nestler EJ. Annu Rev Neurosci. 2006 doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- 17.Babak T, et al. Curr Biol. 2008;18:1735. doi: 10.1016/j.cub.2008.09.044. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, et al. PLoS ONE. 2008;3:e3839. doi: 10.1371/journal.pone.0003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulz R, et al. Epigenetics. 2008;3:89. doi: 10.4161/epi.3.2.5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koc EC, et al. J Biol Chem. 2001;276:43958. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 21.Barel O, et al. Am J Hum Genet. 2008;83:193. doi: 10.1016/j.ajhg.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mochida GH, et al. Am J Hum Genet. 2009;85:897. doi: 10.1016/j.ajhg.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chao W, D'Amore PA. Cytokine Growth Factor Rev. 2008;19:111. doi: 10.1016/j.cytogfr.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delaval K, Wagschal A, Feil R. Bioessays. 2006;28:453. doi: 10.1002/bies.20407. [DOI] [PubMed] [Google Scholar]
- 25.Bartolomei MS, Zemel S, Tilghman SM. Nature. 1991;351:153. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 26.Bartolomei MS, Webber AL, Brunkow ME, Tilghman SM. Genes Dev. 1993;7:1663. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- 27.Webber AL, Ingram RS, Levorse JM, Tilghman SM. Nature. 1998;391:711. doi: 10.1038/35655. [DOI] [PubMed] [Google Scholar]
- 28.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Nature. 1995;375:34. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 29.Hu JF, Vu TH, Hoffman AR. Mol Endocrinol. 1995;9:628. doi: 10.1210/mend.9.5.7565809. [DOI] [PubMed] [Google Scholar]
- 30.Hemberger M, et al. Dev Genes Evol. 1998;208:393. doi: 10.1007/s004270050195. [DOI] [PubMed] [Google Scholar]
- 31.Bhalla K, et al. Am J Hum Genet. 2008;83:703. doi: 10.1016/j.ajhg.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beroukhim R, et al. Nature. 2010;463:899. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, et al. Science. 2004;305:1437. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 34.Wood AJ, Bourc'his D, Bestor TH, Oakey RJ. Nucleic Acids Res. 2007;35:7031. doi: 10.1093/nar/gkm742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood AJ, et al. Genes Dev. 2008;22:1141. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadowaki M, et al. Dev Biol. 2007;304:22. doi: 10.1016/j.ydbio.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 37.Caqueret A, Yang C, Duplan S, Boucher F, Michaud JL. Horm Res. 2005;64:222. doi: 10.1159/000088977. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, et al. Mol Cell Biol. 2004;24:270. doi: 10.1128/MCB.24.1.270-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smit MA, et al. Mamm Genome. 2005;16:801. doi: 10.1007/s00335-004-2415-z. [DOI] [PubMed] [Google Scholar]
- 40.Arnaud P, et al. Hum Mol Genet. 2003;12:1005. doi: 10.1093/hmg/ddg110. [DOI] [PubMed] [Google Scholar]
- 41.Yamasaki Y, et al. Hum Mol Genet. 2005;14:2511. doi: 10.1093/hmg/ddi255. [DOI] [PubMed] [Google Scholar]
- 42.McEwen KR, Ferguson-Smith AC. In: Genomic Imprinting - A Model for Roles of Histone Modifications in Epigenetic Control. Ferguson-Smith AC, Greally JM, Martienssen RA, editors. Springer; 2009. pp. 235–258. [Google Scholar]
- 43.Tilgner H, et al. Nat Struct Mol Biol. 2009;16:996. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 44.Spies N, Nielsen CB, Padgett RA, Burge CB. Mol Cell. 2009;36:245. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Disteche Nguyen. Brain Res. 2006;1126:46. doi: 10.1016/j.brainres.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 46.Skinner Jirtle. Nat Rev Genet. 2007;8:253. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.