Abstract
Voltage-clamp techniques are typically used to study the plasma membrane proteins, such as ion channels and transporters that control bioelectrical signals. Many of these proteins have been cloned and can now be studied as potential targets for drug development. The two approaches most commonly used for heterologous expression of cloned ion channels and transporters involve either transfection of the genes into small cells grown in tissue culture or the injection of the genetic material into larger cells. The standard large cells used for the expression of cloned cDNA or synthetic RNA are the egg progenitor cells (oocytes) of the African frog, Xenopus laevis.
Until recently, cellular electrophysiology was performed manually, one cell at a time by a single operator. However, methods of high-throughput electrophysiology have been developed which are automated and permit data acquisition and analysis from multiple cells in parallel. These methods are breaking a bottleneck in drug discovery, useful in some cases for primary screening as well as for thorough characterization of new drugs. Increasing throughput of high-quality functional data greatly augments the efficiency of academic research and pharmaceutical drug development. Some examples of studies that benefit most from high-throughput electrophysiology include pharmaceutical screening of targeted compound libraries, secondary screening of identified compounds for subtype selectivity, screening mutants of ligand-gated channels for changes in receptor function, scanning mutagenesis of protein segments, and mutant-cycle analysis. We describe here the main features and potential applications of OpusXpress, an efficient commercially available system for automated recording from Xenopus oocytes. We show some types of data that have been gathered by this system and review realized and potential applications.
Keywords: channelopathies, voltage clamp, mutant-cycle analysis, Alzheimer’s disease, unnatural amino acids
Introduction
Electrophysiology has long been considered one of the more esoteric aspects of neuroscience; invisible ion channels are probed with miraculously selective drugs to determine their effects on the ephemeral electrical signals of brain cells. We know that this bioelectricity is the very essence of the nervous system but, studied with only the most sophisticated equipment capable of measuring small numbers of charged particles as they traverse cell membranes at time scales faster than a blink of an eye, we see only the ghosts of its true dynamic vitality, frozen in small snapshots on oscilloscopes or computer screens. However, that perspective is changing as our accumulated understanding of cellular function and disease is allowing us to identify specific ion channels and transporters as therapeutic targets. Advances in molecular biology have permitted us to draw these target molecules out of the brain and other tissues and study them in new ways. Ion channel and transporter genes that have been cloned can be expressed heterologously in other cells and then studied in detail, combining electrophysiological and pharmacological approaches. This has been a tremendous boon to both basic research and drug development.
Not so long ago, fully automated electrophysiology experiments were thought to be impossible because of the interactive and highly technical nature of the experiments. The goal of developing instruments that could record the ionic currents of channels or electrogenic transporters, deliver solutions, and analyze data for multiple experiments without operator intervention, was first conceived for the pharmaceutical industry as a way to break a logjam in ion channel drug discovery. While other high-throughput screening methods for ion channel targets have been developed, these measurement methods suffered from high rates of false positive and negative findings. Traditionally, verification of positive hits required highly trained scientists doing experiments that were very labor-intensive and time-consuming. This, one cell and one channel type at a time, approach created a logjam in the discovery pipeline and was a disincentive to use ion channels as targets for drug discovery on a larger scale.
We know from the human genome project that there are around 300-400 ion channel genes [1]. Yet, at present, only about 30 ion channels are screened as drug targets by the pharmaceutical industry, leaving a huge untapped source of possible ion channel drug targets. While ion channels were first discovered and studied in neuronal and muscle cells, they are found in all cells of the body and have been implicated in a wide range of neurological and muscular disorders, including migraine, epilepsy, myotonia, and cardiac rhythm problems. Ion channels have also been implicated in disorders of other tissues such as cystic fibrosis. Also, it is worth noting that many drugs that target gene products other than ion channels also interact with ion channels, which means that pharmaceutical companies need to test all compounds for cross-reactivity, especially in the area of targets involved in the cardiac action potential and contractility.
The cloning of ion channels and transporters has made it possible to study the functional properties of identified genes in a small number of suitable expression systems. This approach utilizes cell types that lack endogenous proteins which would confound results and are amenable to genetic transfection or are large enough to be directly injected with genetic material.
The Xenopus oocyte was one of the first heterologous expression systems proven to be useful [2] and is still widely used today, due to its large size, its faithful expression of channel proteins in the cell membrane, and the relative absence of endogenous channels which might complicate analysis of electrophysiology measurements. Ricardo Miledi and coworkers first used Xenopus oocytes to study nicotinic acetylcholine receptors in 1982 by injecting mRNA isolated from cat muscle [2]. Within five years Miledi’s group alone had used Xenopus oocytes to study more than a dozen types of ion channels and receptors, by injecting RNA prepared from various tissues. They also made the important discovery that for many neurotransmitter receptor subtypes that were not themselves ion channels (e.g. G-protein coupled receptors), the Xenopus oocyte carried the necessary transduction mechanisms to convert receptor activation to an easily-measured current mediated by calcium-dependent chloride channels [3, 4]. As cloned receptors and channels became available in the 1980s, the Xenopus oocyte system became even more valuable, since the cloning revealed the existence of multiple receptor subtypes, each of which could be individually characterized by injecting single clones into the oocytes. This made it possible to identify drugs with selectivity for specific receptor subtypes [5].
The large uniform size of Xenopus oocytes (approximately 1 mm in diameter) made it seem feasible to create an automated electrophysiology recording system for Xenopus oocytes. Realization of this dream has required the development of computer-driven systems that fully integrate software and hardware. In this article we outline the salient features of the oocyte expression system for the study of cloned ion channels, and we describe a recently developed automated multichannel high throughput electrophysiology system for oocyte recording.
Voltage Clamp Measurements from Xenopus oocytes
The African clawed frog, Xenopus laevis, has a long history as an experimental animal, going back to endocrinology and embryology studies in the early 1950s. Xenopus oocytes were first studied in the late sixties and early seventies, but have been most widely used as an expression system since the seminal work by the Miledi group in the early 1980s. Why Xenopus oocytes and why voltage-clamp recordings?
The most direct and quantitative measures of ion channel and transporter function are made with electrical recording of the actual ion flow across the cell membrane due to activation of the channels or transporter proteins. This is best achieved using one of several voltage-clamp techniques, where the voltage across the membrane is controlled and the currents measured provide information about gating function and pore properties on a millisecond or faster time-scale. The basic method of voltage-clamp involves the use of microelectrodes and an amplifier to measure a cell’s membrane potential and inject current, so that the cells membrane potential stays fixed (i.e. is clamped) at a potential set by the experimentalist. In this way, voltage-clamping controls the membrane voltage reliably and records the currents due to channel activation evoked by an experimental stimulus. This has the function of isolating channel-mediated currents from capacitive currents or other ion channel currents that might be stimulated if the cell membrane potential is allowed to change. Additionally, voltage clamp is the most valuable method for the study of the voltage-dependence of gating in voltage-gated ion channels, since it allows the experimentalist to separate out the effects of voltage and time as channels open and close in response to voltage jumps. Voltage-clamp also permits the study the voltage-dependence of compounds that bind to ion channels and alter their function.
The true beauty of voltage clamping for pharmacological studies comes from the fact that for a given cell there is a direct linear relationship between the response measured, as percent of the cell’s maximum possible response, and the fraction of the total population of ion channels that opened to create the current measured. This linear relationship is derived from Ohm’s law, I=(Em−Erev)G, which states that current will be the product of the electrical driving force (the difference between the membrane potential (Em)and the zero current potential, or “reversal potential”, (Erev) for the channels of interest) and the conductance (i.e. of the ion channels opened by the stimulus). The experimentalist controls the driving force by choosing solutions to establish the desired reversal potential and by selecting the voltages for the voltage-clamp to maintain. The conductance in response to a given stimulus is equal to the total number of channels (N), the fraction that respond to the stimulus (Popen) and the conductance of a single open channel (γ). Thus Ohm’s law for voltage-clamp recordings can be written I = (Em − Erev)NPopenγ. For a given cell (Em − Erev), N, and γ are constants so that I/Imax is directly proportional to Popen. This linear relationship makes voltage clamp an ideal method for quantitative concentration-response studies.
Voltage-clamp can be conducted either with two electrodes, one used to measure membrane voltage and the other to inject current to keep the voltage at the command voltage, or alternatively, with one electrode that carries out both functions. Single electrode voltage clamp is most often done with patch-clamp electrodes [6], which provide a tight seal to the cell’s membrane and a relatively low resistance connection to the cell interior. Such “whole-cell patch” voltage clamp recording is best conducted on relatively small cells, since with larger cells, or elaborately branched neurons, it becomes impossible to control voltage over the entire surface of the cell with a single electrode. Two electrode voltage clamp (TEVC) is the most common approach for voltage-clamping a large cell and the method ideally suited for cells the size of an oocyte. Xenopus oocytes are extremely robust cells and will tolerate the use of relatively blunt, low-resistance electrodes for passing current. Additionally, when amplifiers are used that can create a virtual ground reference in the bath, current necessary to control the voltage across the cell membrane can also be supplied through the very low resistance bath electrode. This use of a virtual ground (the Axon CNS guide [7]) allows currents as large as several tens of microamps to be recorded without losing control of the cell’s membrane potential.
Molecular targets
All kinds of ion channels can be studied with electrophysiology. One classification of ion channels separates them by how they open and close (i.e. gate). Based on this classification, ion channels can be ligand-gated, voltage-gated, or spontaneously-gated. The ligand-gated channels include those gated by extracellular ligands such as neurotransmitters (e.g. ACh, 5HT, GABA and glutamate) and by intracellular ligands such as cAMP and Ca2+ ions. Among the voltage-gated channels, channels are often identified by their selectivity for specific ions (K+, Na+, Ca2+, Cl−); although channels that are relatively non-selective have also been described.
Many kinds of ion transporters can also be studied with these methods, as long as there is a net movement of charge from one side of the membrane to the other. These so-called electrogenic transporters can be cotransporters or antiporters, depending on whether the transported ions and other chemicals move across the membrane in the same or opposite direction (for review see [8]). For example, many transporters couple movement of chemicals into or out of the cell to the passive movement of sodium and/or potassium ions down their electrochemical gradient into or out of the cell. Examples of this type of transporter include the sodium-coupled glucose transporter and sodium-coupled amino acid transporters, for example those that transport glutamate, aspartate, or other amino acids into cells along with Na ions. Examples of antiporters include the Na/Ca or Na/H exchangers, which transport Ca or H ions in the opposite direction that Na ions move. Other electrogenic transporters that can be studied using electrophysiology techniques include those driven by ATP hydrolysis such as the ubiquitous Na/K ATPase, a transporter that moves 3 Na ions out of the cell and 2 K ions into the cell, setting up the ionic gradient that drives many of the other transport processes. Because cloned ion channels and transporters can be expressed in the oocyte plasma membrane, it becomes possible to record ionic currents through them with voltage-clamp techniques and to study their function on a millisecond, and even fraction of a millisecond, time scale.
Generally speaking, the oocyte expression system is particularly useful for studying these ion channels and transporters due to both its ease-of-use and its reliability. Having relatively few endogenous ion channels (that are expressed at relatively low levels), Xenopus oocytes have been used to express a wide variety of ion channels and transporters in plasma membranes by injecting RNA into the cytoplasm [9] or DNA into the nucleus [10]. RNA can be in the form of mRNA isolated from tissues of interest [2] or cRNA transcribed from cloned or mutated channel DNA [9]. DNA injected into the nucleus can also be from cloned wild-type or mutant channels [11]. Xenopus laevis oocytes can be readily harvested in plentiful numbers . The cells are hardy, require rather simple culture conditions, can survive in vitro for up to several weeks, and are easy to inject. Expression levels can be easily regulated by the amount of RNA or DNA injected. Exogenously expressed channels tend to be so well expressed they easily swamp out most underlying endogenous conductances that are present.
While we normally focus on the heterologously-expressed ion channels in the Xenopus oocytes, there is one type of endogenous ion channel which cannot be ignored, calcium-dependent chloride channels. Fortuitously, these channels not only serve to amplify small signals through calcium-permeable channels such as the nicotinic α7 receptor [12], their presence also allows the oocytes to serve as a useful expression system for many G-protein receptors. The oocytes have all the necessary cellular machinery to connect the activation of receptors that link to either GQ or G11 to generate large, easily recorded, electrophysiological responses. Based on this approach, Xenopus oocytes have been used to study such diverse G-protein coupled receptors as odorant detectors [13], parathyroid hormone [14] and CRF receptors [15], as well as multiple subtypes of metabotropic serotonin, melatonin, dopamine, adrenergic, glutamate, ACh, and GABA receptors [16-26].
Caveats for oocyte recording
It is important to keep in mind that, as with any biological preparation, low levels of a variety of ion channels have been reported to exist in oocytes, and experimentalists should bear this in mind and conduct appropriate controls. Moreover, although the calcium-dependent chloride channels can often be seen as an advantage, their contribution to the current response can sometimes be problematical for some sorts of studies where receptor-mediated currents need to be studied in strict isolation, for example to determine current-voltage relationships or ionic selectivity. While there may be technical limitations to electrophysiological recordings from oocytes, especially with large currents recorded following voltage steps [27], for most experiments, these limitations are not a problem. Additionally, for receptors composed of multiple subunits, oocytes may support subunit combinations not commonly found in nature, or may take natural subunit combinations and configure them differently than they might typically be configured in mammalian cells (e. g different ratios of α and β subunits, [28]). However, such anomalies are relatively rare and, in fact, oocytes have been useful to show how novel subunits that do not function in isolation can coassemble with other functional subunit combinations and so modify the properties of the receptor complexes found in vivo [29].
There are a few differences in the environment for a channel in Xenopus oocytes compared to mammalian cells. Post-translational processing of proteins in oocytes can be a little different from that in mammalian cell lines and the composition of the cell membrane may also be somewhat different. This can be an issue for those most interested in studying channels or transporters derived from human tissue and who want to understand the functional mechanisms and drug interactions strictly in the context of native human tissue. Also, it has been noted that there is a shift in the dose-response curve of many compounds when assayed with TEVC recordings, leading to an increase in the IC50. The mechanism of this decrease in potency has not been clearly identified but has been proposed to be due to non-specific binding to the yolk and vitelline membrane of the oocyte [30]. While it is true that some channels express much better in oocytes than in mammalian cells, it is also true that there are channels that express better in mammalian cells [31].
While TEVC is almost universally used to record current responses in oocytes, additional methods can be used to supplement that approach and address specific questions. One such alternative oocyte approach is the “cut-open oocyte” recording method. With this technically challenging approach, the cell is internally perfused so that drugs and different ionic solutions can be applied to the inside of the oocyte [32]. More frequently, patch-clamp methods are used to supplement TEVC recording from oocytes. Macropatches with macroscopic currents can be recorded from a section of the oocyte membrane, providing better recording bandwidth. Single-channel records from smaller membrane patches can also be used, to tease out the molecular events that underlie the easily recorded macroscopic currents from TEVC. Both macropatch and single channel patches can be made from any of the standard patch clamp orientations, including cell-attached, inside-out, or outside-out patches [33].
Relative advantages of Xenopus oocytes compared to other expression systems
The most common alternative expression system, and the only other one easily amenable to electrophysiological studies, relies on either stably or transiently expressed cloned genes in tissue culture cell lines such as human embryonic kidney (HEK) or Chinese hamster ovary (CHO) cells. Typically, non-stable transfection leads to expression in only a small percentage of the cell population, as low as 20%, and for only a short time. Several strategies have been used to increase the probability that the cells chosen for recording have been transfected. One strategy is ‘panning’ for transfected cells by using antibodies or other means to enrich the percentage of mammalian cells that have the channel of interest. Another is co-transfection of the channel with a marker of transfection like green fluorescent protein (GFP), to make it easy to identify which cells are most likely to have been transfected with the exogenous channels. These and other methods are used to increase the chances of finding transfected mammalian cells in the limited window of time of, typically a few days, over which the channels are expressed. In order to create stably transfected cell lines, cells are grown for several passages in selective medium and then have to be sub-cloned, and then each clone has to be screened for expression levels. While creating mammalian cell lines that stably express the channel of interest often works well, for some channels this method can be problematic and take much longer than even the standard month or two. Once a stably transfected cell line has been established, standard tissue culture techniques are required to maintain the cell line. With mammalian cell lines, the voltage-clamp measurement of choice is patch clamping. While manual patch-clamping of mammalian cells is relatively laborious and technically difficult compared to oocyte recording, automated patch clamp systems have increased the practicality of this approach [34-36].
Chief among the advantages of expressing channels and transporters in oocytes is that it takes a very short turn-around time to go from isolating the DNA from a new channel clone, or from creating mutations for structure-function studies in established clones, to being able to study channel function. To obtain a quick answer about channel function, cDNA or in vitro-transcribed cRNA is injected into the oocyte. Many channels express within 1 to 2 days in oocytes. Even though for other channels, expression can take a few days longer, this rapid transition from gene product to data is an attractive aspect of oocytes as an alternative to expression in mammalian cells. Another major advantage of using oocytes is that virtually every oocyte injected will express the channel of interest, unlike mammalian cells.
As noted above, expression of ion channels in oocytes is particularly useful for studying newly cloned or mutated channels due to the rapid turnaround time between cloning or mutating a channel and making the functional measurement. Oocytes also provide an excellent alternative expression system when channels don’t express in mammalian expression systems. Additionally, oocytes are particularly useful when you need to co-express several channel subunits at once, something that is more difficult to do in mammalian cells. Oocytes are also easily amenable to simple perfusion of drugs acting on the extracellular surface of cells. It is far more challenging to make efficient solution delivery to small tissue-cultured cells.
Oocyte recording of native mammalian receptors, channels, and transporters
As an alternative to studying cloned receptors, Miledi and coworkers have shown that it is possible to reconstitute native mammalian receptors directly in oocyte membranes [37, 38]. This is accomplished by injecting vesicles into oocytes that have been prepared from tissues with channels of interest. The vesicles have fully formed channel proteins that can incorporate into the cell membrane. This technique, which has so far only been shown possible in the oocyte expression system, is particularly useful for studying differences in channel function, for example, between diseased and normal tissues [39, 40]. This latter capability could be of significant use in helping to determine the roles for ion channels in various disease states.
High throughput approaches for the study of ion channels and transporters
Electrophysiological study of cloned ion channels, in particular using voltage-clamp, is clearly a method of great strength and utility for both basic science and the pharmaceutical industry. However, while an academic scientist may base a successful career on publishing a few papers a year, drug discovery and pharmaceutics are driven by the need to conduct hundreds of studies and evaluate thousands of drug leads every year. This defines the challenge of high throughput approaches to the study of ion channels and transporters. Traditionally, drug companies have resorted to non-electrophysiological approaches in order to achieve high throughput screening of drug candidates.
Until recently, high throughput screening relied either on binding studies or flux measurements with radioisotopes. Now methods using the FLIPR and VIPR [34] systems, for example, have been developed that can give fluorescent readouts from multiwell plates based on either calcium or voltage-sensitive indicators. These approaches can all be used to rapidly screen large numbers of compounds, but each has limitations compared to voltage-clamp recording. For example, while binding studies can identify compounds with affinity for a molecular target, they cannot discriminate agonists from antagonists nor indeed if the compounds have any functional effect. Likewise, since for both flux studies and the fluorescent readout systems, it may be possible to obtain a maximal response with submaximal receptor occupancy, these approaches may not discriminate partial agonists from full agonists. This can be related to the presence of “spare receptors” or to nonlinear aspects of the relationship between receptor occupancy and response. For example, when a membrane potential indicator dye is used to study cells that are not voltage-clamped, small amounts of receptor activation can produce large changes in membrane voltage due to the large initial driving force. However, once the cell has become depolarized, large amounts of additional receptor activation produce negligible increases in the voltage signal, and the cell will never become more depolarized than the reversal potential of the channel being studied. This is in marked contrast to the linear relationship between the measured response (i.e. current) and receptor activation, and points to one of the great strengths of voltage-clamp studies. Measurements based on calcium indicators can also show non-linear properties since, frequently, voltage-dependent calcium channels can convert a small depolarization into a large calcium response.
High throughput electrophysiology (HTEP) recording
With all of the advantages provided by voltage-clamp recording, clearly an ideal for drug discovery would be high throughput electrophysiology, such as might be achieved by developing integrated and automated systems for either patch-clamp or TEVC recording. Ideally, direct electrical recordings for high throughput electrophysiology should provide efficient and sensitive screening with low numbers of false positives or negatives. Systems like PatchXpress [35, 36] have taken on that challenge for recording from small cells, and likewise multichannel automated systems such as OpusXpress have made high throughput electrophysiology an obtainable goal for oocyte recording.
Traditionally, oocyte recording was a relatively low throughput process, performed one oocyte at a time by a trained technician requiring an hour or more of set-up time each day and similar clean up time. Often, traditional perfusion systems used large volumes of experimental drugs with plumbing cobbled together from various syringes, valves and tubing, often controlled manually and therefore not synchronized to the data acquisition systems, except by manual intervention. Each laboratory, and indeed each investigator within a laboratory, might have their own system for organizing (or disorganizing as the case might be) data and taking notes. Even so, the contributions to science from such low throughput systems continue to be enormous, almost making up for the trouble that a given investigator might have trying to find a specific single raw data file, recorded by a long-since departed part-time student technician on an obsolete computer five years ago. Clearly though, the traditional approach is one with large room for improvement.
Goals for high throughput electrophysiology systems
In trying to define how an ideal HTEP system might be configured, we can begin with a discussion of how such a system would be used, and what sorts of experimental goals it would be directed toward. One of the most common applications for HTEP would be to define the concentration-response relationships of drugs with known activity for specific channels or transporters. For example, by applying varying concentrations of a putative agonist for a ligand-gated ion channel to oocytes expressing the channel of interest, the concentration-dependence of the response can be used to define an EC50 value. In order to determine if the experimental drug is a full agonist or partial agonist, all that is required is to make comparisons in single oocytes between the responses to the experimental drug and a reference agonist with known efficacy and potency. Likewise putative antagonists could be characterized across concentrations for their ability to block the response of ligand-gated channels to agonists in co-application experiments, or to block the activity of voltage-dependent channels when the compounds are present during the voltage steps used to activate the channels. For both of these sorts of experiments, responses obtained in the absence of the antagonist would serve as the internal control so that IC50 values can be easily obtained. Various ancillary questions would also need to be addressable with an HTEP system, such as determinations of recovery rates, the voltage-dependence and state-dependence of drug interaction with the channel, and whether the effect occurred through competitive or noncompetitive mechanisms. For these kinds of studies, the great advantage of an HTEP system, aside from whatever intrinsic increase in efficiency the system might bring, would be the ability to record from multiple cells in parallel. In this way, a single run might generate all the necessary replicates to complete a publishable data set.
The other most important potential application for HTEP is drug screening. Oocytes expressing a target gene could be exposed to a large number of different compounds and monitored for their responses, again as compared to responses to reference compounds or control conditions. Looking at multiple oocytes in parallel, several sets of oocytes could be tested in duplicate with many different compounds so that dozens of different drugs could be screened in a single one hour experiment. Important for this approach would be the ability to monitor the health and responsiveness of individual cells throughout the course of each experiment. This is important because if cells fail, due to unexpected effects of the drugs or other reasons, they should be taken off-line so that the drug solutions could be saved for testing on new cells.
The utility of HTEP is obvious, particularly for projects that require screening a large number of compounds or require a large number of cloned or mutated channels to be tested. Examples of the latter include alanine scanning mutagenesis [41], cysteine accessibility scanning [42], and mutant-cycle analysis [43]. HTEP would also expedite projects that require a large number of solution changes to characterize a single response, as with ligand-gated channels. An HTEP system that is optimized for oocytes would come with all of the advantages of traditional oocyte recording discussed above and, additionally, would be cost effective since it is likely that technology for automated handling of large cells such as oocytes would be less expensive than automated patch-clamp systems. Also, a single parallel recording system is likely to be far less expensive than multiple manual systems and would also make effective use of technical staff.
With these sorts of scenarios in mind we identify key features needed for an HTEP oocyte system to be useful (see Table I).
Table 1.
Key Features for HTEP
| Generate large amounts of high quality data |
| Automation of multiple experiments |
| Accurate and well-timed drug delivery coordinated with data acquisition |
| Research quality voltage control to evaluate voltage-dependent gating, permeation, compound binding |
| Easy to set up and run |
| Cost effective |
| Flexible fluidics and voltage protocols for studying a variety of channels and transporters |
| Multiple additions of different fluids to each cell |
| Automated real-time monitoring |
| For experiment quality and termination |
| For analysis of experiment progress |
| Integrated and intuitive methods for storing, organizing, analyzing, archiving, retrieving data |
OpusXpress: an example of High-Throughput Electrophysiology optimized for oocytes
OpusXpress is a high-throughput electrophysiology system in which voltage control, data acquisition, fluid delivery, and real-time analysis are all automated and are coordinated based on user specifications. With the OpusXpress system these functions are also performed in parallel, enabling a single operator to run an experiment on up to 8 oocytes at a time while simultaneously preparing for the next set of experiments, greatly increasing the rate of data acquisition by a single operator and enhancing operational efficiency.
For automated electrophysiology to realize its full potential, it must not compromise on data quality. The OpusXpress system has many features, both in hardware and software, that ensure optimal data quality. OpusXpress recordings have the low noise, high bandwidth, and excellent voltage control of conventional manual TEVC oocyte recordings, allowing recording on a microsecond time scale. This is due to the optimal approach angle of the electrodes that minimizes capacitive coupling yet readily impales the oocytes, placement of the bath electrode near the oocyte but downstream of fluid inflow, high output compliance research quality amplifiers for rapid charging of the cell membrane, ultra-high DC gain to maintain voltage clamp, and virtual ground control of the bath electrode to prevent electrode drift. When HTEP is chosen for screening instead of much higher throughput plate-based assays, it is either because HTEP is the only way to screen a particular target or because HTEP gives a more accurate report of the true effect of a compound on function, and tends not to give the false results of the higher throughput methods. OpusXpress provides the data quality necessary to permit comparison of data with conventional electrophysiology recordings. Compromising on quality with HTEP would result in the worst of both worlds, throughput lower than multiwell plate-based systems and less-reliable data.
The flexibility built into OpusXpress enhances its capability by allowing easy transition between parallel and independent operation and by providing tools to create a wide variety of experimental protocols. While the OpusXpress system operates mostly in parallel, for most of its functions users can isolate a single element to work independently, if desired. For example, if 8 oocytes are impaled and in voltage-clamp mode before starting an experiment, and one or more oocytes become leaky, all good oocytes can be left in voltage-clamp mode while the bad ones are replaced. In another example, if one or more cells fail in the middle of an experiment and will no longer provide good data, OpusXpress can automatically terminate the experiment on only the bad cells while continuing experiments on the remaining good cells.
OpusXpress makes it easy for users to set up and run customized experiments for testing a wide range of ion channels and electrogenic ion transporters. The system can be used not only for simple screening procedures, but also for mechanistic studies of channel function and compound interaction. The system can run procedures with complex user-defined voltage protocols that are coordinated with fluid delivery from two different and complementary fluidics systems. One fluidics system allows continuous flow of buffer from one of two buffer reservoirs, driven by peristaltic pumps, while the other allows robotic delivery of fluids to each oocyte from up to 24 different wells of a multiwell plate, all with user-defined flow rates.
For automated real-time monitoring, OpusXpress software allows users to distinguish responses to reference compounds from responses to experimental compounds and generate statistics reports in real time, so that parameters such as response to reference compounds can be used to monitor and control experiments in a dynamic manner. For example, if individual cells do not show responses to reference compounds above some threshold value, such cells can be taken off-line and not used for drug testing. Note that these designations are useful for off-line analysis as well.
The pharmaceutical industry especially benefits from HTEP, as it advances the drug discovery process, screening targeted chemical libraries for hits and misses, and facilitating rapid determinations of potency and efficacy series. Although OpusXpress was designed with the needs of the pharmaceutical industry in mind, it has also proven to be an invaluable asset to numerous academic labs and has helped generate many peer-reviewed publications [12, 44-83].
Alternative HTEP systems
Alternative HTEP systems for oocytes have been developed, though only one (Roboocyte by MultiChannel Systems, Reutlingen, Germany) is commercially available at the time of this writing. Others have been created as in-house projects by technology development groups within large pharmaceutical companies, but their use has typically been restricted to those within the company and to partners. These alternative systems have made significantly different choices in system design that affect system capability and throughput.
Examples of experiments Performed with OpusXpress
Rapidly desensitizing α7-type nAChR
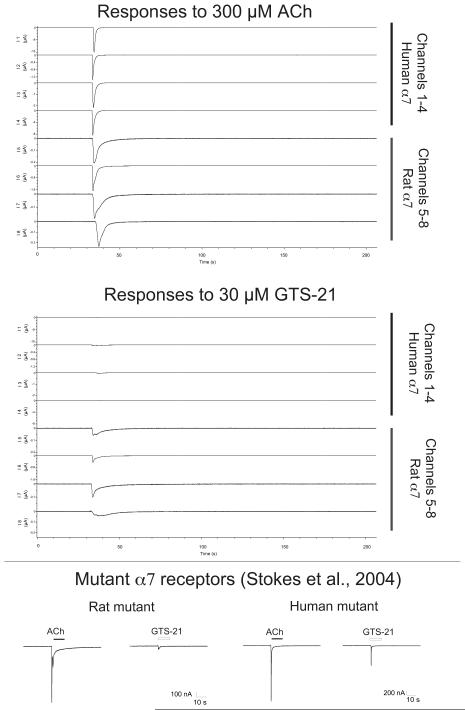
OpusXpress has been used to study many different types of ion channels in the Papke laboratory, including nicotinic acetylcholine receptors (nAChR), and especially the α7 subtype nAChR. The α7 nAChR shows a unique form of rapid desensitization that is dependent on agonist concentration [84]. This phenomenon has been studied extensively with α7 expressed in oocytes and also with the native α7-type nAChR of acutely-dissociated hypothalamic neurons, with essentially the same results and conclusions in both systems. For example, the calculated EC50s estimated for α7 receptor activation by ACh, choline and the experimental agonist 4OH-GTS-21 are essentially the same for both the oocyte experiments and the acutely dissociated neurons [12, 85]. Sample oocyte records of α7 nAChR are shown in Figure 1. These recordings came from a large study which used OpusXpress to determine the structural differences between rat and human forms of α7 that accounted for why the potential therapeutic agent GTS-21 preferentially activates rat α7 compared to human α7 receptors [75]. As shown in the figure, a combination of 3 single point mutations were necessary and sufficient to reverse this difference between rat and human α7 receptors. The data shown represents a final conclusion of what was, in fact, a very large study made possible by OpusXpress. In the paper, Stokes et al. report findings based on concentration response studies of over 15 different agonists on both wild-type human and rat receptors, as well as studies of ACh and GTS-21 on 30 mutants or chimeric receptors. In each case, the concentration-dependence of both activation and desensitization were examined, so in total the entire study encompassed over 150 concentration-response analyses conducted with OpusXpress. A large fraction of this body of work was conducted side by side with other projects in the laboratory, over the course of less than a month.
Figure 1.
Relative responses of human and rat α7 receptors to ACh and GTS-21 are regulated by 3 amino acids. The upper panels compare the responses of wild-type rat and human α7 receptors, expressed in oocytes and currents recorded with OpusXpress, to control applications of 300 μM ACh and applications of 100 μM of the experimental partial agonist GTS-21. The control response amplitudes are scaled to the same size, and the experimental responses are normalized to their respective controls. Note the relatively low efficacy of GTS-21 for human wild-type receptors compared to rat. As part of an extensive study conducted with OpusXpress (see text), it was ultimately shown that this difference in GTS-21 efficacy was due to 3 specific differences in the sequence of rat and human receptors in the agonist binding site [75]. As shown in the lower panels, GTS-21 has reduced efficacy for the rat α7 mutant N184S, K186R, S167G and increased efficacy for the reciprocal human mutant α7 S184N, R186K, G167S.
The nicotinic α7 receptor has been widely acknowledged as a potentially important therapeutic target for the treatment of Alzheimer’s disease [86-96]. Therefore, the Papke laboratory [5, 49, 51-53, 59-62, 64, 75, 97-102], along with others, including numerous drug company teams [103-113] have been working toward developing new α7-selective agonists and positive allosteric modulators of α7 [114, 115]. The assessment of functional properties is a crucial step in the screening of potential new drug candidates, and the development of automated electrophysiological recording systems such as OpusXpress has facilitated the process of testing new drugs to a large degree. However, while the simple screening of multiple drugs at a single concentration identifies “hits” and “misses”, the generation of full concentration-response studies is still a bottleneck in drug development. This impasse too can be lessened by making the most out of the large amounts of data which are generated by systems such as OpusXpress.
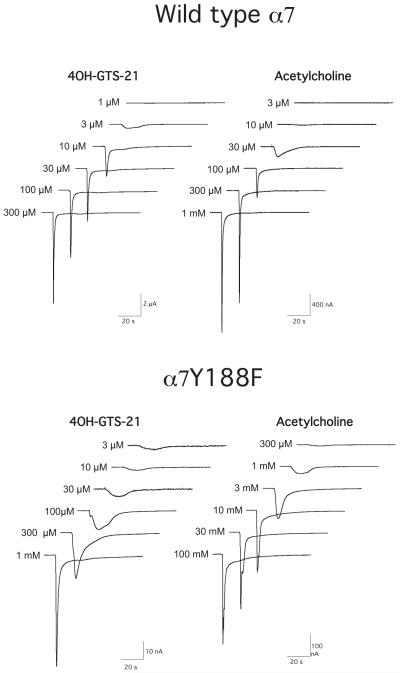
For example, we have noted that the α7 nicotinic acetylcholine receptor displays a unique concentration-dependence of response kinetics [12, 116]. This feature is surprisingly consistent regardless of whether the drugs tested are high or low potency, full agonists or partial agonist, whether the α7 clone is from human or other vertebrate species and is usually even a feature of α7 mutants, unless the mutations are within the transmembrane domain [11, 68, 69]. This character of α7 nAChR responses is illustrated in Figure 2, which shows the characteristic concentration-dependent changes in α7 receptor responses in wild-type and mutant (Y188F) α7 receptors to the partial agonist 4OH-GTS-21 and to ACh. The Y188F mutation has a large effect on the potency of ACh [49], with little effect on the potency of 4OH-GTS-21. Hence the concentration-dependent effects on response waveforms in the mutant are shifted for ACh, but not for 4OH-GTS-21.
Figure 2.
Families of agonist-evoked responses recorded with OpusXpress from oocytes expressing wild-type α7 nAChR or an α7 mutant (Y188F), that shows a selective reduction in ACh potency [49]. The reduction in ACh potency is reflected in the shift in ACh concentration required to evoke transient synchronous activation of receptors rather than more sustained low amplitude responses.
Based on the systematic analysis of many different concentration-response studies utilizing either human or rat α7 nAChR we developed a method that permits estimates of EC50 and Imax values for experimental drugs to be generated from single concentration responses [66]. This method involves the normalization of each experimental response to an ACh control response from the same cell, and then analyzing that normalized response with transformation function based on the relationship between the net charge and peak current to their respective EC50 values, derived from our large α7 agonist data base. This transform then defines the “functional concentration” (the test concentration relative to the estimated EC50) at which each experimental agonist was tested. At low functional concentrations net charge is large relative to peak current amplitude and at higher functional concentrations this relationship reverses, so that for any single concentration response the ratio of net charge to peak current can be used to estimate functional concentration. Efficacy can then be estimated by comparing the observed (net charge) response to the expected value for a full agonist at the estimated functional concentration. This extended analysis, combined with automated recording methods greatly increases the efficiency with which promising new drug candidates can be characterized for α7 receptor activity.
Automated recording systems like OpusXpress, especially when applied with methods such as our waveform analysis described above, can be a major boon for drug screening. However, such systems are arguably an even greater asset for hypothesis-driven research requiring the generation and testing of multiple mutant receptor subtypes, since the oocyte methodology allows new mutants to be tested almost immediately. Moreover, the retesting of previously generated mutants can be done on the spur of the moment, since it only requires finding the RNA in the freezer and injecting it into a fresh lot of cells. Any mutant can be worked up in just a matter of a day or two. In contrast, if transfected cell lines, of the sorts used for automated patch clamping, have been placed in storage, it often takes weeks to grow the cells up in sufficient number for experiments. Even so, cells brought up from frozen stocks sometimes have lost the phenotypic expression of interest. However, not only is RNA able to be kept in storage for up to several years at a time, but the cDNAs themselves last indefinitely and can be used to make fresh RNA in a day. Such flexibility allows an oocyte lab the ability to study literally hundreds of different receptor subtypes in an ordinary year, including various mutants, chimeras, and receptors with differing subunit composition and species of origin.
Neurotransmitter receptor expression in oocytes following injection of hippocampal membranes
Along with drug development and hypothesis testing, the oocyte expression system, powered by an acquisition engine like OpusXpress, can also be a tool for discovery and insight into the underpinnings of ion channel-related disease. This is because diseased tissue can be harvested and membranes from that tissue prepared and injected into oocytes. The oocytes incorporating those membranes then reconstitute the ion channel profile of the tissues from which the membranes were harvested, making it easier to study differences due to the disease state.
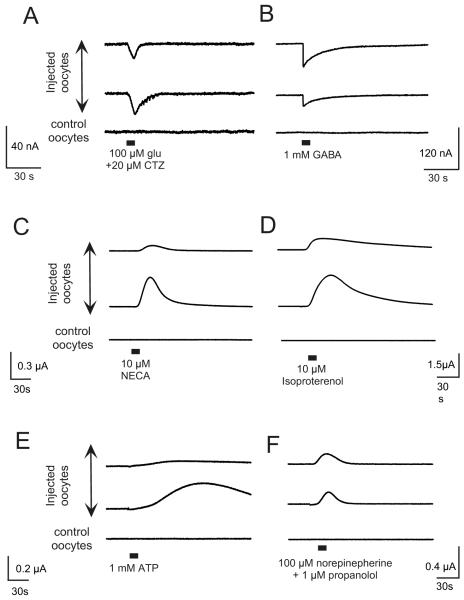
Figure 3 shows data obtained with OpusXpress utilizing this method of injecting brain membranes into Xenopus oocytes [38]. In such experiments, with membranes from rat hippocampi, we have been able to detect responses to GABA, glutamate, isoproterenol (i. e beta adrenergic agonist), norepinepherine + propranolol (i. e. alpha adrenergic receptor activation), NECA (adenosine receptor agonist), and dopamine. The responses varied in magnitude as well as direction of current (inward vs. outward). In panel A, typical responses to 100 μM glutamate are compared between 2 oocytes injected with hippocampal membranes and a control sham-injected oocyte. In order to improve the detection of ionotropic glutamate receptor responses, the data shown were obtained in the presence of the AMPA-receptor potentiating agent, cyclothiazide (CTZ). In panel B, responses to 1 mM of the nonselective agonist GABA are shown. However, through the use of selective agonists such as muscimol (not shown), we have been able determine that both GABAa and GABAb receptors contribute to these responses. Additionally, important pharmacological properties are preserved in these preparations. For example, responses to the GABAa agonist muscimol were enhanced 49 ±16 % when muscimol was co-applied with the benzodiazepines, flunitrazepam (data not shown).
Figure 3.
Functional receptors expressed in Xenopus oocytes from the injection of hippocampal membranes. Responses of oocytes injected with hippocampal membranes (upper 2 traces) to the applications of A) 100 μM glutamate plus 20 μM cyclothiazide (CTZ), B) 1 mM GABA, C) 10 μM NECA, D) 10 μM isoproterenol, E) 1mM ATP, and F) 100 μM nor-epinephrine plus 1 μM propranolol In each case the responses of oocytes injected with hippocampal membranes are compared to sham-injected ‘control oocytes’ recorded at the same time.
Analysis of ion channel function with unnatural amino acids
Another area in which a remarkable potential for scientific discovery has been realized with the oocyte expression system, in combination OpusXpress, has been to investigate novel theories about ion channel function through the incorporation of unnatural amino acids [46]. Mutations can be introduced into genes of interest that create codons for which no naturally occurring transfer RNA exists. Artificial tRNAs, linked to custom-designed unnatural amino acids, can then be injected into the oocytes along with the mutant cRNA to create proteins that can address the very most detailed probative questions about ion channels and other pharmacological targets. This approach has been used to demonstrate the importance of a Cis-trans isomerization at a proline for opening the pore of a 5HT3a receptor [54], to probe the Mg2+ blockade site of an N-methyl-D-aspartate (NMDA) receptor [55], and to investigate the agonist activation of GABA(A) receptors [57].
Conclusions
The advent of systems such as OpusXpress for HTEP study of ion channels and transporters in Xenopus oocytes marks a real breakthrough for both research and therapeutics. It represents another critical technology taken through the rite of passage from where it was available only to the pioneers with primitive plumbing and homemade recording chambers, to a truly modern piece of laboratory equipment. It is not an exaggeration to say that with a machine like OpusXpress available in their lab, an investigator can formulate a new hypothesis over coffee in the morning and be sending a publication-quality test of that hypothesis to his or her collaborators by e-mail at lunch time. It allows investigators to embrace larger challenges. It will no longer be appropriate to dismiss projects involving large amounts of mutant testing or mutant cycle analysis as “overly ambitious”. The automation of the process will translate into rapid acquisition of large amounts of uniformly high quality data and the efficient use of time spent in preparation and data analysis. This new level of efficiency allows users to focus on the science rather than on the technical details.
It seems likely that for scientists in both academia and industry, in the very near future tools such as OpusXpress will be seen as ways not only to do old things better, but also to do entirely new things. For example, by being able to directly assay ion channel function in healthy and diseased tissue by injecting membranes into oocytes and screening those cells rapidly with multiple ion channel probes using OpusXpress, we may gain new insights into diseases as diverse as epilepsy and Alzheimer’s disease. As these and other diseases give up their secrets, new high throughput drug screening will also help tell us how to target underlying ion channel and/or neurotransmitter anomalies.
Acknowledgements
We thank Dr. Henry Lester (Cal Tech) and David Yamane (MDS Analytical Technologies) for helpful comments. For technical assistance with the oocyte experiments in the Papke lab since the beginning of the OpusXpress era, Dr. Papke thanks Julia K. Porter Papke, Brian Jack, Lisa Jacobs, Jenny Hughes, Chad Brodbeck, Dolan AbuAouf, Sara Braley, Adrienne Argenio, Irena Garic, Bernadette Schoneburg, Amanda Waber, Theresa Kappel, Hillary Schiff, Josh Buhr, Andon Placzek, Chris Coverdill, Jeff Garris, and Clare Stokes. Research in the Papke lab is supported by NIH grants, GM57481, DA 017548, AG10485, and T32 AG00196, the McKnight foundation, and collaborators in the pharmaceutical industry. An OpusXpress 6000 was provided to the Papke lab by Axon Instruments, now part of MDS Analytical Technologies.
Footnotes
Although the authors were on the original development team for OpusXpress neither is now affiliated with MDS Analytical Technologies or has any direct financial interest in OpusXpress.
References
- 1.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, et al. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 2.Miledi R, Parker I, Sumikawa K. Embo J. 1982;1:1307–12. doi: 10.1002/j.1460-2075.1982.tb01315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gundersen CB, Miledi R, Parker I. Proc R Soc Lond B Biol Sci. 1983;219:103–9. doi: 10.1098/rspb.1983.0062. [DOI] [PubMed] [Google Scholar]
- 4.Miledi R, Parker I. J. Physiol. 1984;357:173–183. doi: 10.1113/jphysiol.1984.sp015495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Fiebre CM, Meyer EM, Zoltewicz J, Henry JC, Muraskin S, Kem WR, Papke RL. Mol Pharm. 1995;47:164–171. [PubMed] [Google Scholar]
- 6.Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ. Pflüegers Arch. Eur. J. Physiol. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 7.MolecularDevices . The Axon CNS Guide. Molecular Devices; Foster City, CA: 2006. pp. 38–42. [Google Scholar]
- 8.Dubyak GR. Adv Physiol Educ. 2004;28:143–54. doi: 10.1152/advan.00046.2004. [DOI] [PubMed] [Google Scholar]
- 9.Connolly J, Boulter J, Evans K, Mason P, Gardner PD, Patrick J, Heinemann S. Society for Neuroscience. 1986 [Google Scholar]
- 10.Ballivet M, Nef P, Couturier S, Rungger D, Bader CR, Bertrand D, Cooper E. Neuron. 1988;1:847–852. doi: 10.1016/0896-6273(88)90132-8. [DOI] [PubMed] [Google Scholar]
- 11.Bertrand D, Devillers-Thiéry A, Revah F, Galzi J-L, Hussy N, Mulle C, Bertrand S, Ballivet M, Changeux J-P. Proc. Natl. Acad. Sci. USA. 1992;89:1261–1265. doi: 10.1073/pnas.89.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papke RL, Papke JKP. Br J of Pharm. 2002;137:49–61. doi: 10.1038/sj.bjp.0704833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahmen N, Wang HL, Margolis FL. J Neurochem. 1992;58:1176–9. doi: 10.1111/j.1471-4159.1992.tb09379.x. [DOI] [PubMed] [Google Scholar]
- 14.Racke FK, Hammerland LG, Dubyak GR, Nemeth EF. FEBS Lett. 1993;333:132–6. doi: 10.1016/0014-5793(93)80390-g. [DOI] [PubMed] [Google Scholar]
- 15.Moriarty TM, Gillo B, Sealfon S, Landau EM. Brain Res. 1988;464:201–5. doi: 10.1016/0169-328x(88)90026-5. [DOI] [PubMed] [Google Scholar]
- 16.Singer D, Boton R, Moran O, Dascal N. Pflugers Arch. 1990;416:7–16. doi: 10.1007/BF00370215. [DOI] [PubMed] [Google Scholar]
- 17.Fraser SP, Barrett P, Djamgoz MB, Morgan PJ. Neurosci Lett. 1991;124:242–5. doi: 10.1016/0304-3940(91)90104-2. [DOI] [PubMed] [Google Scholar]
- 18.Sumikawa K, Parker I, Miledi R. Proc R Soc Lond B Biol Sci. 1984;223:255–60. doi: 10.1098/rspb.1984.0093. [DOI] [PubMed] [Google Scholar]
- 19.Miledi R, Parker I, Sumikawa K. J Physiol. 1987;383:213–29. doi: 10.1113/jphysiol.1987.sp016405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bahouth SW, Hadcock JR, Malbon CC. J Biol Chem. 1988;263:8822–6. [PubMed] [Google Scholar]
- 21.Blitzer RD, Omri G, De Vivo M, Carty DJ, Premont RT, Codina J, Birnbaumer L, Cotecchia S, Caron MG, Lefkowitz RJ, et al. J Biol Chem. 1993;268:7532–7. [PubMed] [Google Scholar]
- 22.Nathanson MH, Burgstahler AD, Orloff JJ, Mani A, Moyer MS. Am J Physiol. 1994;267:C94–103. doi: 10.1152/ajpcell.1994.267.1.C94. [DOI] [PubMed] [Google Scholar]
- 23.Birnbaum AK, Wotta DR, Law PY, Wilcox GL. Brain Res Mol Brain Res. 1995;28:72–80. doi: 10.1016/0169-328x(94)00185-h. [DOI] [PubMed] [Google Scholar]
- 24.Taniyama K, Takeda K, Ando H, Kuno T, Tanaka C. FEBS Lett. 1991;278:222–4. doi: 10.1016/0014-5793(91)80121-i. [DOI] [PubMed] [Google Scholar]
- 25.Wolff MA, Wingate VP. Invert Neurosci. 1998;3:305–15. doi: 10.1007/BF02577690. [DOI] [PubMed] [Google Scholar]
- 26.Schwarz DA, Barry G, Eliasof SD, Petroski RE, Conlon PJ, Maki RA. J Biol Chem. 2000;275:32174–81. doi: 10.1074/jbc.M005333200. [DOI] [PubMed] [Google Scholar]
- 27.Sigworth FJ, Zhou J. Methods Enzymol. 1992;207:746–62. doi: 10.1016/0076-6879(92)07054-r. [DOI] [PubMed] [Google Scholar]
- 28.Nelson ME, Kuryatov A, Choi CH, Zhou Y, Lindstrom J. Mol Pharmacol. 2003;63:332–41. doi: 10.1124/mol.63.2.332. [DOI] [PubMed] [Google Scholar]
- 29.Gerzanich V, Kuryatov A, Anand R, Lindstrom J. Mol. Pharmacol. 1996;51:320–7. [PubMed] [Google Scholar]
- 30.Kiehn J, Lacerda AE, Wible B, Brown AM. Circulation. 1996;94:2572–9. doi: 10.1161/01.cir.94.10.2572. [DOI] [PubMed] [Google Scholar]
- 31.Grinevich VP, Letchworth SR, Lindenberger KA, Menager J, Mary V, Sadieva KA, Buhlman LM, Bohme GA, Pradier L, Benavides J, Lukas RJ, Bencherif M. J Pharmacol Exp Ther. 2005;312:619–26. doi: 10.1124/jpet.104.075069. [DOI] [PubMed] [Google Scholar]
- 32.Costa AC, Patrick JW, Dani JA. Biophys. J. 1994;67:395–401. doi: 10.1016/S0006-3495(94)80494-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Methfessel C, Witzemann V, Takahashi T, Mishina M, Numa S, Sakmann B. Pflüegers Arch. Eur. J. Physiol. 1986;407:577–588. doi: 10.1007/BF00582635. [DOI] [PubMed] [Google Scholar]
- 34.Dunlop J, Roncarati R, Jow B, Bothmann H, Lock T, Kowal D, Bowlby M, Terstappen GC. Biochem Pharmacol. 2007 doi: 10.1016/j.bcp.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 35.Ly JQ, Shyy G, Misner DL. Clin Lab Med. 2007;27:201–8. doi: 10.1016/j.cll.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 36.Dubin AE, Nasser N, Rohrbacher J, Hermans AN, Marrannes R, Grantham C, Van Rossem K, Cik M, Chaplan SR, Gallacher D, Xu J, Guia A, Byrne NG, Mathes C. J Biomol Screen. 2005;10:168–81. doi: 10.1177/1087057104272295. [DOI] [PubMed] [Google Scholar]
- 37.Palma E, Trettel F, Fucile S, Renzi M, Miledi R, Eusebi F. Proc Natl Acad Sci U S A. 2003;100:2896–900. doi: 10.1073/pnas.0438006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miledi R, Palma E, Eusebi F. Methods Mol Biol. 2006;322:347–55. doi: 10.1007/978-1-59745-000-3_24. [DOI] [PubMed] [Google Scholar]
- 39.Palma E, Amici M, Sobrero F, Spinelli G, Di Angelantonio S, Ragozzino D, Mascia A, Scoppetta C, Esposito V, Miledi R, Eusebi F. Proc Natl Acad Sci U S A. 2006;103:8465–8. doi: 10.1073/pnas.0602979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ragozzino D, Palma E, Di Angelantonio S, Amici M, Mascia A, Arcella A, Giangaspero F, Cantore G, Di Gennaro G, Manfredi M, Esposito V, Quarato PP, Miledi R, Eusebi F. Proc Natl Acad Sci U S A. 2005;102:15219–23. doi: 10.1073/pnas.0507339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panchenko VA, Glasser CR, Mayer ML. J Gen Physiol. 2001;117:345–60. doi: 10.1085/jgp.117.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karlin A, Akabas MH. Methods Enzymol. 1998;293:123–145. doi: 10.1016/s0076-6879(98)93011-7. [DOI] [PubMed] [Google Scholar]
- 43.Penzotti JL, Lipkind G, Fozzard HA, Dudley SC., Jr. Biophys J. 2001;80:698–706. doi: 10.1016/S0006-3495(01)76049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abaffy T, Matsunami H, Luetje CW. J Neurochem. 2006;97:1506–18. doi: 10.1111/j.1471-4159.2006.03859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abaffy T, Malhotra A, Luetje CW. J Biol Chem. 2007;282:1216–24. doi: 10.1074/jbc.M609355200. [DOI] [PubMed] [Google Scholar]
- 46.Beene DL, Dougherty DA, Lester HA. Curr Opin Neurobiol. 2003;13:264–70. doi: 10.1016/s0959-4388(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 47.Beene DL, Price KL, Lester HA, Dougherty DA, Lummis SC. J Neurosci. 2004;24:9097–104. doi: 10.1523/JNEUROSCI.2429-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dwoskin LP, Joyce BM, Zheng G, Neugebauer NM, Manda VK, Lockman P, Papke RL, Bardo MT, Crooks PA. Biochem Pharmacol. 2007;74:1271–82. doi: 10.1016/j.bcp.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horenstein NA, McCormack TJ, Stokes C, Ren K, Papke RL. J Biol Chem. 2006;282:5899–909. doi: 10.1074/jbc.M609202200. [DOI] [PubMed] [Google Scholar]
- 50.Hsiao B, Mihalak KB, Repicky SE, Everhart D, Mederos AH, Malhotra A, Luetje CW. Mol Pharmacol. 2006;69:27–36. doi: 10.1124/mol.105.015164. [DOI] [PubMed] [Google Scholar]
- 51.Kem WR, Mahnir VM, Prokai L, Papke RL, Cao X, Le Francois S, Wildeboer K, Prokai-Tatrai K, Papke JKP, Soti F. Mol Pharm. 2004;65:56–67. doi: 10.1124/mol.65.1.56. [DOI] [PubMed] [Google Scholar]
- 52.Leonik FM, Papke RL, Horenstein NA. Bioorg Med Chem Lett. 2007 doi: 10.1016/j.bmcl.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 53.Lopez-Hernandez G, Placzek AN, Thinschmidt JS, Lestage P, Trocme-Thibierge C, Morain P, Papke RL. Neuropharmacology. 2007;53:134–44. doi: 10.1016/j.neuropharm.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 54.Lummis SC, Beene DL, Lee LW, Lester HA, Broadhurst RW, Dougherty DA. Nature. 2005;438:248–52. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 55.McMenimen KA, Dougherty DA, Lester HA, Petersson EJ. ACS Chem Biol. 2006;1:227–34. doi: 10.1021/cb6000944. [DOI] [PubMed] [Google Scholar]
- 56.Mihalak KB, Carroll FI, Luetje CW. Mol Pharmacol. 2006;70:801–5. doi: 10.1124/mol.106.025130. [DOI] [PubMed] [Google Scholar]
- 57.Padgett CL, Hanek AP, Lester HA, Dougherty DA, Lummis SC. J Neurosci. 2007;27:886–92. doi: 10.1523/JNEUROSCI.4791-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papke RL. J Pharmacol Exp Ther. 2002;301:765–73. doi: 10.1124/jpet.301.2.765. [DOI] [PubMed] [Google Scholar]
- 59.Papke RL, Meyer EM, Lavieri S, Bollampally S, Papke T, Horenstein B, Papke JKP. J Neuropharm. 2004;46:1023–1038. doi: 10.1016/j.neuropharm.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 60.Papke RL, Papke JKP, Rose GM. BioOrg Med Chem Lett. 2004;14:1849–53. doi: 10.1016/j.bmcl.2003.09.104. [DOI] [PubMed] [Google Scholar]
- 61.Papke RL, Schiff HC, Jack BA, Horenstein NA. Neurosci. let. 2005;378:140–144. doi: 10.1016/j.neulet.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 62.Papke RL, McCormack TJ, Jack BA, Wang D, Bugaj-Gaweda B, Schiff HC, Buhr JD, Waber AJ, Stokes C. Eur. J. of Pharm. 2005;524:11–18. doi: 10.1016/j.ejphar.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 63.Papke RL. Life Sci. 2005 [Google Scholar]
- 64.Papke RL, Zheng G, Horenstein NA, Dwoskin LP, Crooks PA. Bioorg Med Chem Lett. 2005;15:3874–80. doi: 10.1016/j.bmcl.2005.05.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Papke RL, Buhr JD, Francis MM, Choi KI, Thinschmidt JS, Horenstein NA. Mol Pharmacol. 2005;67:1977–90. doi: 10.1124/mol.105.011676. [DOI] [PubMed] [Google Scholar]
- 66.Papke RL. Life Sci. 2006;78:2812–9. doi: 10.1016/j.lfs.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 67.Papke RL, Dwoskin LP, Crooks PA. J Neurochem. 2007;101:160–7. doi: 10.1111/j.1471-4159.2006.04355.x. [DOI] [PubMed] [Google Scholar]
- 68.Placzek AN, Grassi F, Papke T, Meyer EM, Papke RL. Mol Pharmacol. 2004;66:169–177. doi: 10.1124/mol.66.1.169. [DOI] [PubMed] [Google Scholar]
- 69.Placzek AN, Grassi F, Meyer EM, Papke RL. Mol Pharmacol. 2005;68:1863–76. doi: 10.1124/mol.105.016402. [DOI] [PubMed] [Google Scholar]
- 70.Rodrigues-Pinguet N, Jia L, Li M, Figl A, Klaassen A, Truong A, Lester HA, Cohen BN. J Physiol. 2003;550:11–26. doi: 10.1113/jphysiol.2003.036681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodriguez EA, Lester HA, Dougherty DA. Proc Natl Acad Sci U S A. 2006;103:8650–5. doi: 10.1073/pnas.0510817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodriguez EA, Lester HA, Dougherty DA. Rna. 2007;13:1703–14. doi: 10.1261/rna.666807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodriguez EA, Lester HA, Dougherty DA. Rna. 2007;13:1715–22. doi: 10.1261/rna.667607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shiembob DL, Roberts RL, Luetje CW, McIntosh JM. Biochemistry. 2006;45:11200–7. doi: 10.1021/bi0611715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stokes C, Papke JKP, McCormack T, Kem WR, Horenstein NA, Papke RL. Mol. Pharm. 2004;66:14–24. doi: 10.1124/mol.66.1.14. [DOI] [PubMed] [Google Scholar]
- 76.Wanner KW, Nichols AS, Walden KK, Brockmann A, Luetje CW, Robertson HM. Proc Natl Acad Sci U S A. 2007;104:14383–8. doi: 10.1073/pnas.0705459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiu X, Hanek AP, Wang J, Lester HA, Dougherty DA. J Biol Chem. 2005;280:41655–66. doi: 10.1074/jbc.M508635200. [DOI] [PubMed] [Google Scholar]
- 78.Clark RJ, Fischer H, Dempster L, Daly NL, Rosengren KJ, Nevin ST, Meunier FA, Adams DJ, Craik DJ. Proc Natl Acad Sci U S A. 2005;102:13767–72. doi: 10.1073/pnas.0504613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clark RJ, Fischer H, Nevin ST, Adams DJ, Craik DJ. J Biol Chem. 2006;281:23254–63. doi: 10.1074/jbc.M604550200. [DOI] [PubMed] [Google Scholar]
- 80.Armishaw CJ, Daly NL, Nevin ST, Adams DJ, Craik DJ, Alewood PF. J Biol Chem. 2006;281:14136–43. doi: 10.1074/jbc.M512419200. [DOI] [PubMed] [Google Scholar]
- 81.Loughnan M, Nicke A, Jones A, Schroeder CI, Nevin ST, Adams DJ, Alewood PF, Lewis RJ. J Biol Chem. 2006;281:24745–55. doi: 10.1074/jbc.M603703200. [DOI] [PubMed] [Google Scholar]
- 82.Jin AH, Brandstaetter H, Nevin ST, Tan CC, Clark RJ, Adams DJ, Alewood PF, Craik DJ, Daly NL. BMC Struct Biol. 2007;7:28. doi: 10.1186/1472-6807-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nevin ST, Clark RJ, Klimis H, Christie MJ, Craik DJ, Adams DJ. Mol Pharmacol. 2007 doi: 10.1124/mol.107.040568. [DOI] [PubMed] [Google Scholar]
- 84.Papke RL, Meyer E, Nutter T, Uteshev VV. Eur J Pharmacol. 2000;393:179–95. doi: 10.1016/s0014-2999(00)00009-1. [DOI] [PubMed] [Google Scholar]
- 85.Uteshev VV, Meyer EM, Prokai L, Papke RL. 31st Annual Meeting of the Society for Neuroscience; 2001.p. 144.2. [Google Scholar]
- 86.Levin ED, Rezvani AH. Eur J Pharmacol. 2000;393:141–6. doi: 10.1016/s0014-2999(99)00885-7. [DOI] [PubMed] [Google Scholar]
- 87.Court J, Martin-Ruiz C, Piggott M, Spurden D, Griffiths M, Perry E. Biol Psychiatry. 2001;49:175–84. doi: 10.1016/s0006-3223(00)01116-1. [DOI] [PubMed] [Google Scholar]
- 88.Nordberg A. Biol Psychiatry. 2001;49:200–10. doi: 10.1016/s0006-3223(00)01125-2. [DOI] [PubMed] [Google Scholar]
- 89.Perry EK, Martin-Ruiz CM, Court JA. Alcohol. 2001;24:63–8. doi: 10.1016/s0741-8329(01)00130-6. [DOI] [PubMed] [Google Scholar]
- 90.Yu WF, Guan ZZ, Bogdanovic N, Nordberg A. Exp Neurol. 2005;192:215–25. doi: 10.1016/j.expneurol.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 91.D’Andrea MR, Nagele RG. Curr Pharm Des. 2006;12:677–84. doi: 10.2174/138161206775474224. [DOI] [PubMed] [Google Scholar]
- 92.Sapronov NS, Fedotova YO, Kuznetsova NN. Bull Exp Biol Med. 2006;142:700–2. doi: 10.1007/s10517-006-0455-y. [DOI] [PubMed] [Google Scholar]
- 93.Liu Q, Zhang J, Zhu H, Qin C, Chen Q, Zhao B. Faseb J. 2007;21:61–73. doi: 10.1096/fj.06-5841com. [DOI] [PubMed] [Google Scholar]
- 94.Wang HY, Lee DH, D’Andrea MR, Peterson PA, Shank RP, Reitz AB. J. Biol. Chem. 2000;275:5626–32. doi: 10.1074/jbc.275.8.5626. [DOI] [PubMed] [Google Scholar]
- 95.Wevers A, Witter B, Moser N, Burghaus L, Banerjee C, Steinlein OK, Schutz U, de Vos RA, Steur EN, Lindstrom J, Schroder H. Acta Neurol. Scand. Suppl. 2000;176:42–8. doi: 10.1034/j.1600-0404.2000.00306.x. [DOI] [PubMed] [Google Scholar]
- 96.Hurst RS, Hajos M, Raggenbass M, Wall TM, Higdon NR, Lawson JA, Rutherford-Root KL, Berkenpas MB, Hoffmann WE, Piotrowski DW, Groppi VE, Allaman G, Ogier R, Bertrand S, Bertrand D, Arneric SP. J Neurosci. 2005;25:4396–405. doi: 10.1523/JNEUROSCI.5269-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hunter BH, Papke RL, de Fiebre CM, Meyer EM. Neurosci. Lett. 1994;168:130–134. doi: 10.1016/0304-3940(94)90433-2. [DOI] [PubMed] [Google Scholar]
- 98.Papke RL, Bencherif M, Lippiello P. Neurosci. Lett. 1996;213:201–204. doi: 10.1016/0304-3940(96)12889-5. [DOI] [PubMed] [Google Scholar]
- 99.Papke RL, Meyer EM, de Fiebre CM. 26th Annual Meeting of the Society for Neuroscience; 1996.p. 602.4. [Google Scholar]
- 100.Meyer EM, Tay ET, Papke RL, Meyers C, Huang G, de Fiebre CM. Brain Res. 1997;768:49–56. doi: 10.1016/s0006-8993(97)00536-2. [DOI] [PubMed] [Google Scholar]
- 101.Meyer EM, Tay ET, Zoltewicz JA, Papke RL, Meyers C, King M, Fiebre C. M. d. J Pharmacol Exp Ther. 1998;284:1026–1032. [PubMed] [Google Scholar]
- 102.Marrero MB, Papke RL, Bhatti BS, Shaw S, Bencherif M. J. Pharm. Exp. Ther. 2004;309:16–27. doi: 10.1124/jpet.103.061655. [DOI] [PubMed] [Google Scholar]
- 103.Sullivan JP, Donnelly-Roberts D, Briggs CA, Anderson DJ, Gopalakrishnan M, Xue IC, Piattoni-Kaplan M, Molinari E, Campbell JE, McKenna DG, Gunn DE, Lin NH, Ryther KB, He Y, Holladay MW, Wonnacott S, Williams M, Arneric SP. J Pharmacol Exp Ther. 1997;283:235–46. [PubMed] [Google Scholar]
- 104.Macor JE, Gurley D, Lanthorn T, Loch J, Mack RA, Mullen G, Tran O, Wright N, Gordon JC. Bioorg Med Chem Lett. 2001;11:319–21. doi: 10.1016/s0960-894x(00)00670-3. [DOI] [PubMed] [Google Scholar]
- 105.Macor JE, Mullen G, Verhoest P, Sampognaro A, Shepardson B, Mack RA. J Org Chem. 2004;69:6493–5. doi: 10.1021/jo049404q. [DOI] [PubMed] [Google Scholar]
- 106.Marrero MB, Papke RL, Bhatti BS, Shaw S, Bencherif M. J Pharmacol Exp Ther. 2004;309:16–27. doi: 10.1124/jpet.103.061655. [DOI] [PubMed] [Google Scholar]
- 107.Rueter LE, Anderson DJ, Briggs CA, Donnelly-Roberts DL, Gintant GA, Gopalakrishnan M, Lin NH, Osinski MA, Reinhart GA, Buckley MJ, Martin RL, McDermott JS, Preusser LC, Seifert TR, Su Z, Cox BF, Decker MW, Sullivan JP. CNS Drug Rev. 2004;10:167–82. doi: 10.1111/j.1527-3458.2004.tb00011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Walker DP, Wishka DG, Piotrowski DW, Jia S, Reitz SC, Yates KM, Myers JK, Vetman TN, Margolis BJ, Jacobsen EJ, Acker BA, Groppi VE, Wolfe ML, Thornburgh BA, Tinholt PM, Cortes-Burgos LA, Walters RR, Hester MR, Seest EP, Dolak LA, Han F, Olson BA, Fitzgerald L, Staton BA, Raub TJ, Hajos M, Hoffmann WE, Li KS, Higdon NR, Wall TM, Hurst RS, Wong EH, Rogers BN. Bioorg Med Chem. 2006;14:8219–48. doi: 10.1016/j.bmc.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 109.Biton B, Bergis OE, Galli F, Nedelec A, Lochead AW, Jegham S, Godet D, Lanneau C, Santamaria R, Chesney F, Leonardon J, Granger P, Debono MW, Bohme GA, Sgard F, Besnard F, Graham D, Coste A, Oblin A, Curet O, Vige X, Voltz C, Rouquier L, Souilhac J, Santucci V, Gueudet C, Francon D, Steinberg R, Griebel G, Oury-Donat F, George P, Avenet P, Scatton B. Neuropsychopharmacology. 2007;32:1–16. doi: 10.1038/sj.npp.1301189. [DOI] [PubMed] [Google Scholar]
- 110.Boess FG, De Vry J, Erb C, Flessner T, Hendrix M, Luithle J, Methfessel C, Riedl B, Schnizler K, van der Staay FJ, van Kampen M, Wiese WB, Koenig G. J Pharmacol Exp Ther. 2007;321:716–25. doi: 10.1124/jpet.106.118976. [DOI] [PubMed] [Google Scholar]
- 111.Hashimoto K, Ishima T, Fujita Y, Matsuo M, Kobashi T, Takahagi M, Tsukada H, Iyo M. Biol Psychiatry. 2007 doi: 10.1016/j.biopsych.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 112.Kristensen SE, Thomsen MS, Hansen HH, Timmermann DB, Hay-Schmidt A, Mikkelsen JD. Neurosci Lett. 2007;418:154–8. doi: 10.1016/j.neulet.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 113.Wishka DG, Walker DP, Yates KM, Reitz SC, Jia S, Myers JK, Olson KL, Jacobsen EJ, Wolfe ML, Groppi VE, Hanchar AJ, Thornburgh BA, Cortes-Burgos LA, Wong EH, Staton BA, Raub TJ, Higdon NR, Wall TM, Hurst RS, Walters RR, Hoffmann WE, Hajos M, Franklin S, Carey G, Gold LH, Cook KK, Sands SB, Zhao SX, Soglia JR, Kalgutkar AS, Arneric SP, Rogers BN. J Med Chem. 2006;49:4425–36. doi: 10.1021/jm0602413. [DOI] [PubMed] [Google Scholar]
- 114.Timmermann DB, Gronlien JH, Kohlhaas KL, Nielsen EO, Dam E, Dyhring T, Ahring PK, Peters D, Holst D, Christensen JK, Malysz J, Briggs CA, Gopalakrishnan M, Olsen GM. J Pharmacol Exp Ther. 2007 doi: 10.1124/jpet.107.120436. [DOI] [PubMed] [Google Scholar]
- 115.Gronlien JH, Haakerud M, Ween H, Thorin-Hagene K, Briggs CA, Gopalakrishnan M, Malysz J. Mol Pharmacol. 2007 doi: 10.1124/mol.107.035410. [DOI] [PubMed] [Google Scholar]
- 116.Papke RL, Thinschmidt JS. Neurosci. Let. 1998;256:163–166. doi: 10.1016/s0304-3940(98)00786-1. [DOI] [PubMed] [Google Scholar]