Abstract
The rapid formation of numerous tissues during development is highly dependent on the swift activation of key developmental regulators. Recent studies indicate that many key regulatory genes are repressed in embryonic stem cells (ESCs), yet poised for rapid activation due to the presence of both activating (H3K4 trimethylation) and repressive (H3K27 trimethylation) histone modifications (bivalent genes). However, little is known about bivalent gene regulation. In this study, we investigated the regulation of the bivalent gene Sox21, which is activated rapidly when ESCs differentiate in response to increases in Sox2. Chromatin immunoprecipitation demonstrated that prior to differentiation, the Sox21 gene is bound by a complex array of repressive and activating transcriptional machinery. Upon activation, all identified repressive machinery and histone modifications associated with the gene are lost, but the activating modifications and transcriptional machinery are retained. Notably, these changes do not occur when ESCs differentiate in response to retinoic acid. Moreover, ESCs lacking a functional PRC2 complex fail to activate this gene, apparently due to its association with other repressive complexes. Together, these findings suggest that bivalent genes, such as Sox21, are silenced by a complex set of redundant repressive machinery, which exit rapidly in response to appropriate differentiation signals.—Chakravarthy, H., Ormsbee, B. D., Mallanna, S. K., Rizzino, A. Rapid activation of the bivalent gene Sox21 requires displacement of multiple layers of gene-silencing machinery.
Keywords: H3K4me3, H3K27me3, PRC, HDAC, Sox2
Mammalian development is a highly complex set of processes involving the formation of >200 cell types with unique gene expression profiles. Key events in these processes are the large-scale and rapid cell fate decisions that occur via expeditious activation or repression of crucial developmental regulators. Given the rapidity with which cell lineages are formed, understanding how developmental regulatory genes are activated rapidly is fundamental to our understanding of development. However, the key to proper embryogenesis lies not only in the rapid activation of lineage-specific regulators during differentiation, but also in their precise temporal and spatial regulation in response to appropriate differentiation signals. Therefore, a formidable challenge for stem cells is to ensure that key developmental regulators remain silent until specific differentiation signals appear. This delicately nuanced interplay between gene activation and repression is controlled by complex and interrelated regulatory mechanisms that are poorly understood.
Embryonic stem cells (ESCs) provide a powerful model system for dissecting the molecular mechanisms that control mammalian embryogenesis. Recent studies have shown that ESCs achieve the necessary balance between self-renewal and differentiation via a plethora of regulatory mechanisms, including the modulation of chromatin structure. Changes in chromatin structure during the differentiation of ESCs are achieved, in part, by post-translational modifications of histones, including acetylation, methylation, and ubiquitination, which alter chromatin to render it more, or less, accessible to transcriptional machinery. Several studies have examined global changes in histone modifications during ESC differentiation (1, 2). The gain or loss of specific histone modifications associated with a given gene can trigger its activation or repression depending on the location and context of histone modifications. For example, the presence of histone 3 lysine 27 trimethylation (H3K27me3) is generally associated with gene silencing, whereas the presence of histone 3 lysine 4 trimethylation (H3K4me3) is associated with gene activation (3). H3K4me3 is believed to promote gene expression by recruiting postinitiation factors and members of the splicing machinery via its interaction with the chromatin remodeler Chd1 (4, 5). Interestingly, H3K4me3 has also been shown to help recruit Jmjd2a, a histone demethylase that functions to demethylate the repressive histone modification H3K9me3 (6). H3K27me3, on the other hand, helps recruit specific repressive complexes, which cause compaction of chromatin and silence gene expression (7, 8). In this context, recent studies have suggested that >2000 genes are associated with chromatin domains enriched in H3K27me3 (negative) as well as H3K4me3 (positive) modifications in ESCs. These regions have been termed “bivalent,” and it has been suggested that genes residing within bivalent domains exist in a state “poised” for rapid transcriptional activation when specific differentiation signals appear (1,9). The mechanistic details of how these genes are regulated remains to be determined.
Several lines of evidence suggest that the polycomb repressive complex 2 (PRC2), which is responsible for catalyzing the repressive H3K27me3 modification, collaborates with ESC-specific transcription factors to globally regulate bivalent genes in ESCs (1, 2). In particular, the master regulators Sox2 and Oct4 (also known as Oct3, Oct-3/4, and Pou5f1), which control a large network of Sox2:Oct4 target genes (10, 11), have been shown to cooccupy several hundred PRC2 target genes. Notably, many Sox2:Oct4 target genes, which possess adjacent binding sites for Sox2 and Oct4, referred to as an HMG/POU cassette (11, 12), are essential for normal embryogenesis (13–18).
Previous studies have shown that small increases in the levels of Sox2 cause down-regulation of Sox2:Oct4 target genes in ESCs (19). Moreover, a 2-fold increase in the expression of Sox2 causes ESCs to differentiate into cells that exhibit markers expressed by ectoderm, mesoderm, and trophectoderm (20). RNA analysis revealed that lineage-specific genes are turned on in response to elevated levels of Sox2. One of these genes, Sox21, which is not expressed in ESCs, is strongly up-regulated within 3 h after Sox2 levels begin to rise in ESCs. Sox21 was previously reported to be a transcriptional repressor in neural stem cells, and it belongs to the same family as Sox1, 2, and 3 (21, 22). Interestingly, previous genome-wide chromatin immunoprecipitation-promoter microarray (ChIP-chip) analyses have revealed that the Sox21 gene is marked by chromatin modifications that are indicative of bivalency in ESCs (1, 2). Together, these findings led us to hypothesize that Sox21 is a poised, bivalent developmental regulator in ESCs, which is rapidly activated in response to the differentiation signal induced by elevated levels of Sox2. To test this hypothesis, we utilized an ESC model system engineered for inducible expression of Sox2 (20) that is well suited to investigating the mechanisms by which the bivalent Sox21 gene is rapidly activated.
In this study, we investigated two pertinent questions: how bivalent genes are activated during ESC differentiation, and how bivalent genes are restrained from being inappropriately activated in the absence of the appropriate differentiation signal. Our results demonstrate that crucial developmental regulators, such as Sox21, are tightly regulated by multiple, redundantly functioning regulatory mechanisms. We suggest that the complexity of Sox21 gene regulation provides new insights into the mechanisms that are likely to control other key bivalent genes during development.
MATERIALS AND METHODS
Cell culture
Sox2-inducible ESCs have been described previously (20). In this study, they are referred to as i-Sox2-ESCs. The i-Sox2-ESCs and Eed-null ESCs were maintained in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad CA, USA; http://www.invitrogen.com) containing 15% fetal bovine serum (Hyclone, Logan, UT, USA; http://www.hyclone.com) supplemented with 100 μM β-mercaptoethanol and 10 ng/ml leukemia inhibitory factor (LIF; Chemicon, Temecula, CA, USA; http://www.chemicon.com) on gelatin-coated tissue culture plastic. Stock cultures and all experimental cultures were maintained at 37°C in a moist atmosphere of 95% air and 5% CO2. Eed-null ESCs were obtained from Dr. Terry Magnuson (University of North Carolina, Chapel Hill, NC, USA; ref. 23).
RNA isolation and cDNA synthesis
i-Sox2-ESCs or Eed-null ESCs were grown for 9, 16, or 24 h in the presence or absence of 4 μg/ml Dox or 10 μM retinoic acid. RNA was isolated by Tri-Reagent (Molecular Research Center, Inc., Cincinnati, USA; http://www.mrcgene.com) and 1-bromo-3-chloropropane phase separation according to the manufacturer's protocol, with the exception of increasing the precipitation step to overnight at −20°C and performing a second precipitation with ethanol-sodium acetate before washing with ethanol and drying the RNA pellet. RNA pellets were resuspended in 200 μl of HPLC H2O. The concentration of RNA was determined by UV spectrophotometry. RNA (0.5 μg) was treated with amplification-grade DNase I (Invitrogen). cDNA was synthesized using the SuperScript III First-Strand Synthesis SuperMix (Invitrogen).
Quantitative polymerase chain reaction (qPCR)
cDNA generated from i-Sox2-ESCs or Eed-null ESCs was subjected to SYBR Green qPCR on the Cepheid SmartCycler using software version 2.0c (Cepheid, Sunnyvale, CA, USA; http://www.cepheid.com). Gene expression was assayed using RT2 Real-Time SYBR Green PCR master mix (SuperArray Bioscience Corp., Frederick, MD, USA; http://www.superarray.com) according to the manufacturer's protocol, using previously described gene-specific primers for Sox21 (20). Relative gene expression in the untreated and treated i-Sox2-ESCs and Eed-null ESCs was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Gene expression for treated cells and Eed-null ESCs is reported as the average difference in Ct value (cycle number at which significant increase in the fluorescence is detected above the background value) relative to expression of the gene in the untreated (control) i-Sox2-ESCs. Primer curves were generated for the Sox21 primers and the GAPDH primers, and the results are displayed as fold enrichment.
ChIP
ChIP was performed as described previously (24), utilizing i-Sox2-ESCs and Eed-null ESCs. In brief, i-Sox2-ESCs were plated at 1.5 × 106 cells/100 mm plate. After allowing the cells to grow for 24 h, they were refed with either 4 μg/ml Dox, or 10 μM retinoic acid. Eed-null cells were plated at 1.5 × 106 cells/100 mm plate. After 24 h, the Eed-null cells were refed with ESC medium as described above. Nine or 24 h later, the proteins present in the induced or uninduced i-Sox2-ESCs and the Eed-null ESCs were cross-linked to chromatin using 1% formaldehyde in 1× PBS for 10 min at room temperature. Glycine was used at a final concentration of 1× to quench the unreacted formaldehyde. Cells were washed twice with 10 ml of ice-cold 1× PBS and harvested by scraping in 1 ml of 1× PBS (with 0.5 mM PMSF), followed by washing once with 500 μl of 1× PBS (with 0.5 mM PMSF). Nuclei were isolated by resuspending the cells in cell lysis buffer and incubating on ice for 15 min. Lysis of nuclei was brought about by addition of 800 μl of SDS lysis buffer and incubating on ice for 10 min. The chromatin was sheared to a length of ∼500 bp by sonication. Sonicated DNA from ∼3 × 106 cells was diluted using ChIP dilution buffer and precleared by incubating with 60 μl of protein G agarose/salmon sperm DNA bead slurry (Upstate, Lake Placid, NY, USA) for 1 h at 4°C. One percent of the sheared chromatin was removed to be processed as input DNA prior to immunoprecipitation. Overnight immunoprecipitation was carried out on the remainder by incubating the sheared, precleared chromatin with 10 μg of the specific antibody, or 3 μg of normal mouse IgG, respectively. Immune complexes were collected by incubating with 60 μl of protein G agarose/salmon sperm DNA slurry for 1 h at 4°C. The remainder of the protocol was followed as described previously, except for the exclusion of the LiCl wash step, and extension of the wash time for each wash step to 10 min instead of 5 min (25). The following antibodies were utilized for ChIP experiments: RNA polymerase (05-623; Millipore, Billerica, MA, USA), H3Acetyl (06-599; Millipore), H3K4me3 (ab1012 and ab8580; Abcam, Cambridge, MA, USA), H3K27me3 (07-449; Millipore,), H2Aub (05-678; Millipore), Suz12 (ab12073; Abcam), HDAC1 (ab7028; Abcam), HDAC2 (ab7029; Abcam), Sox2 Y17 (sc-17320; Santa Cruz Biotechnology, Santa Cruz, CA, USA), Oct4 N19 (sc-8628; Santa Cruz Biotechnology), and normal mouse IgG (sc-2025; Santa Cruz Biotechnology).
qPCR for ChIP DNA
The enrichment of the specific Sox21 promoter regions over a control region located (∼3 kb) upstream of the Oct4 gene in the ChIP study was monitored using qPCR via the SYBR Green method. Relative enrichment of a protein at a specific region over the control region was calculated by comparing the normalized Ct values (normalized to input) for the respective regions. Enrichment of IgG at specific regions was determined and calculated in a similar manner. (A representative data set for enrichment at the Sox21 gene using a control IgG antibody is displayed in Fig. 3B.) Primer curves were generated for all sets of primers, and results are displayed as fold enrichment values. PCR reactions were carried out in a reaction volume of 25 μl with SYBR Green master mix (SuperArray Bioscience), and the reactions were performed with a Cepheid Smart Cycler (software version 2.0c) detection system. Statistical analysis (a single-tailed, paired Student's t test) was conducted for all the ChIP analyses that showed a difference in enrichment between the uninduced and induced states. Values of P < 0.05 were considered statistically significant. Primers utilized in the ChIP experiments are listed in Table 1.
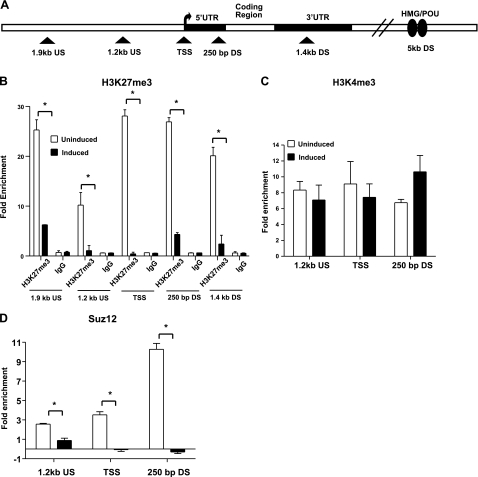
Figure 3.
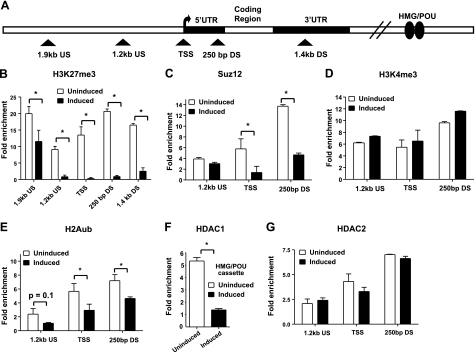
Sox21 loses its inhibitory bivalent domain after i-Sox2-ESCs are induced to differentiate by elevating Sox2 levels. A) Schematic diagram of the Sox21 gene indicating approximate location of primers (triangle) used in ChIP analysis. B–D) ChIP analysis to determine enrichment of H3K27me3 and IgG (B), H3K4me3 (C), and the PRC2 component Suz12 (D) on the Sox21 promoter region in i-Sox2-ESCs before and 24 h after induction of differentiation via treatment with 4 μg/ml Dox. Primers flanking the regions located ∼1.2 kb upstream and ∼250 bp downstream relative to the Sox21 TSS, as well as the Sox21 TSS, were utilized to determine enrichment of H3K4me3 and Suz12. Additional primers amplifying regions ∼1.9 kb upstream and ∼1.46 kb downstream of the Sox21 TSS were utilized to measure enrichment of H3K27me3 on the Sox21 gene. Representative dataset for enrichment at the Sox21 gene using a control IgG antibody is displayed in panel B. All ChIP analyses were repeated, and similar results were obtained. *P < 0.05.
Table 1.
Primers utilized for ChIP qPCR
| Primer | Sequence |
|---|---|
| 1.9 kb US of TSS | |
| Upper | TCAAAAATCAAATAGCACCC |
| Lower | CCAATTCATGTTAATGCCC |
| 1.2 kb US of Sox21 TSS | |
| Upper | CTTTGAGTGCAGGTTTAGGC |
| Lower | TGTACCGCTTTCATCCTCC |
| Sox21 TSS | |
| Upper | AAAATCCTCTCCGGGACCCT |
| Lower | CTCCGCCGCTCAACTTTCG |
| 250 bp DS of Sox21 TSS | |
| Upper | CGGTACTTGTAGTCGGGATG |
| Lower | GAAGATGGCCCAGGAGAACCC |
| 1.4 kb DS of Sox21 TSS | |
| Upper | TCATCTCTCATATACAGGCCG |
| Lower | TTGCTTTTTGCGTCTCATCC |
| Sox21 HMG/POU cassette primers | |
| Upper | CTTCATCCTTACCTTTCCCAT |
| Lower | AAACTGAATGACAAAGCGG |
| Oct4 control region primers (∼3 kb US of Oct4 TSS) | |
| Upper | ATAGGTGTGGCATTCCGCAT |
| Lower | ACCACCTGTATTTTAGAACCA |
RESULTS
Sox21 gene is bound by Sox2 and Oct4
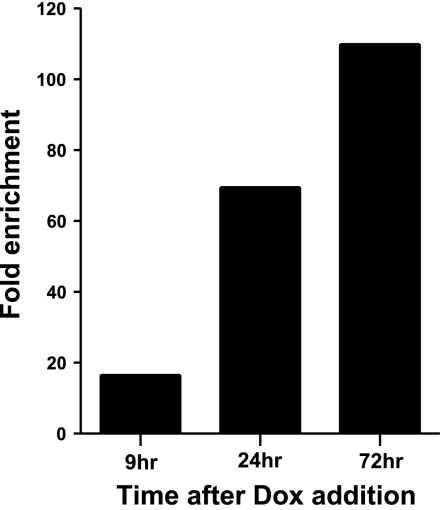
Recently, we demonstrated that elevating the levels of Sox2 ∼2-fold in ESCs triggers their differentiation into multiple cell lineages and alters the expression of >100 genes, including the Sox21 gene (20). These studies employed i-Sox2-ESCs, engineered for inducible expression of Sox2 from a doxycycline (Dox)-inducible promoter (tet-on). Treatment of the cells with Dox (induced cells) elevates Sox2 levels from a transgene. In this system, the Sox21 gene is activated ∼15-fold 9 h after Dox addition (just 3 h after Sox2 levels begin to rise), and its expression is even more strongly elevated 24 and 72 h after the initial exposure to Dox (Fig. 1). In this context, the Sox21 gene has recently been shown to possess the hallmarks of silent, bivalent developmental regulators in ESCs. Given that many bivalent genes in ESCs are associated with Sox2 and Oct4 (10), we hypothesized that the Sox21 gene may be a direct target of these transcription factors. To test this hypothesis, we initially performed in silico analysis and determined that the Sox21 gene possesses a putative HMG/POU cassette (CATTCTA/ATGCTAAT), which has been shown to bind Sox2 and Oct4 in other genes. Like the HMG/POU cassette of the Sox2 gene (26), the putative Sox21 HMG/POU cassette is located within a highly conserved region that is ∼5 kb downstream of the putative transcription start site (TSS) of the Sox21 gene (Fig. 2A). Closer examination of the putative Sox21 HMG/POU cassette revealed that it strongly resembles the consensus sequence for an HMG/POU cassette (11).
Figure 1.
Expression of Sox21 in mouse ESCs undergoing differentiation in response to elevated levels of Sox2. qPCR was performed on cDNA prepared from RNA isolated from untreated i-Sox2-ESCs or i-Sox2-ESCs treated with 4 μg/ml Dox for 9, 24, or 72 h, as described in Materials and Methods. Results are presented as fold-enrichment relative to GAPDH in Dox-induced cells vs. uninduced cells.
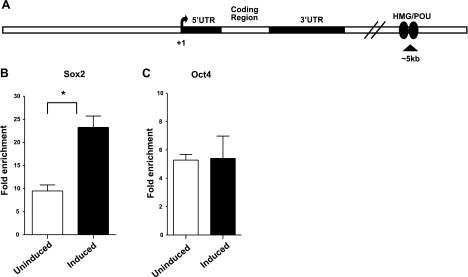
Figure 2.
Sox2 and Oct4 are associated with a region of the Sox21 gene that contains a putative HMG/POU cassette of the Sox21 gene in i-Sox2-ESCs. A) Schematic diagram of the Sox21 gene, including a putative enhancer containing an HMG/POU cassette that is located ∼5 kb downstream of the Sox21 promoter. B, C) ChIP analysis of Sox2 association (B) or Oct4 association (C) with a region within the Sox21 gene containing the putative HMG/POU cassette. ChIP was carried out before and 24 h after induction of differentiation in i-Sox2-ESCs via treatment with 4 μg/ml Dox. ChIP analyses were repeated and similar results were obtained. *P < 0.05.
To determine whether Sox2 and Oct4 are associated with the region containing the putative Sox21 HMG/POU cassette in i-Sox2-ESCs before and after differentiation, we performed ChIP analyses. We determined that both Sox2 and Oct4 are associated with the HMG/POU cassette region of the Sox21 gene in the untreated i-Sox2-ESCs (Fig. 2B, C). Moreover, we determined that the association of Sox2 with the region containing the putative Sox21 HMG/POU cassette is increased 24 h after inducible expression of Sox2 (Fig. 2B), whereas Oct4 association with the gene remains unaltered (Fig. 2C). As noted earlier, the Sox21 gene is strongly activated at this time point. These results suggest that Sox21 is a poised, bivalent, Sox2:Oct4 target gene in ESCs. Furthermore, as discussed below, Sox2 association with the Sox21 gene is elevated even at an earlier time point.
Sox21 is a bivalent gene: bivalent features disappear upon Sox21 expression
Next, we examined whether activation of the Sox21 gene, which is induced by elevating the levels of Sox2, is accompanied by a change in bivalent histone modifications. For these studies, we initially utilized ChIP analysis to examine the presence of the positive histone modification, H3K4me3, on the Sox21 gene promoter. We examined regions within 1 kb of the TSS of the gene for the presence of H3K4me3 (Fig. 3A), which is generally observed at and around the TSS of most genes. We determined that the Sox21 gene is indeed associated with H3K4me3 in uninduced i-Sox2-ESCs. Notably, this association is largely unchanged 24 h after the induction of differentiation, and it is consistent with the robust turn-on of the gene observed at this time point (Fig. 3C). Similarly, we determined that RNA polymerase II, which is often present at the promoters of poised genes in ESCs (2, 27), is also associated with the Sox21 gene before and after the induction of differentiation in the i-Sox2-ESCs (data not shown).
To extend these studies, we examined a larger region around the Sox21 TSS for the association of the negative histone modification H3K27me3 (Fig. 3A), which is reported to be more widespread in its occupancy of gene promoters. We determined that the Sox21 promoter region is extensively occupied by H3K27me3 (Fig. 3B) in the uninduced i-Sox2-ESCs. Moreover, Suz12, a core component of PRC2, which is responsible for catalyzing the repressive H3K27me3 modification, is also associated with the Sox21 promoter (Fig. 3D) in the uninduced i-Sox2-ESCs. Notably, 24 h after the induction of differentiation, the association of both H3K27me3 and Suz12 with the promoter region of the Sox21 gene is greatly diminished (Fig. 3C, D). Together, these results indicate that the rapid activation of the Sox21 gene during ESC differentiation coincides with the loss of the repressive bivalent histone modification, H3K27me3, whereas the positive bivalent feature, H3K4me3, is retained. In fact, as discussed later, these changes are initiated at a much earlier time point.
Multiple repressive complexes occupy the Sox21 gene in ESCs
Several reports suggest that bivalent, poised genes in ESCs are cooccupied by both PRC1 and PRC2 (2, 28). PRC1 catalyzes the ubiquitination of histone 2A (H2Aub), a repressive modification that is thought to result in chromatin compaction, which is not conducive to gene transcription (29,30). This led us to examine the association of H2Aub with the Sox21 gene promoter (Fig. 4A). We determined that H2Aub is associated with the promoter region of the Sox21 gene in the uninduced i-Sox2-ESCs. Upon differentiation of i-Sox2-ESCs, there was a small decrease in its association with the Sox21 gene (Fig. 4B). Although the P value was >0.05, this small decrease was reproducibly observed. Similar reductions were observed at the 9 h time point (see below).
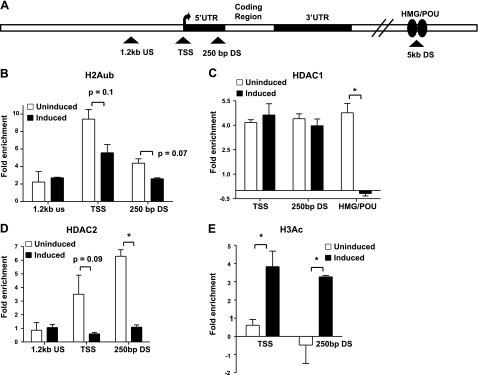
Figure 4.
Multiple repressive complexes are associated with the Sox21 gene in mouse ESCs. A) Schematic diagram of the Sox21 gene indicating approximate location of primers (triangle) used in ChIP analysis. B–E) ChIP analysis of H2Aub (B), HDAC1 (C), HDAC2 (D), and H3Ac (E) on the Sox21 gene in i-Sox2-ESCs before and 24 h after i-Sox2-ESCs were treated with 4 μg/ml Dox. Sox21 TSS, and regions ∼1.2 kb upstream and ∼250 bp downstream relative to the Sox21 TSS, were monitored for occupancy of H2Aub, HDAC2, and H3Ac. HDAC1 association was monitored at the Sox21 TSS, the region ∼250 bp downstream of the Sox21 TSS, and the region containing the putative Sox21 HMG/POU cassette. ChIP analyses were repeated with similar results. Values of P > 0.05 are indicated next to specific bar graphs in panel B. *P < 0.05.
Recent findings have shown that Sox2 is associated with histone deacetylases, HDAC1 and HDAC2 (31, 32). Given our finding that Sox2 is associated with the region of the Sox21 gene that contains a putative HMG/POU cassette, we examined whether HDACs associate with the Sox21 gene in i-Sox2-ESCs. Initially, we examined whether HDAC1 is associated with the region of the Sox21 gene that is bound by Sox2 in i-Sox2-ESCs (Fig. 4A). We determined that HDAC1 is strongly associated with this region, and that this association is greatly diminished 24 h after the gene is activated (Fig. 4C). This dramatic change in the association of HDAC1 with the Sox21 gene coincides with high expression of the gene seen at the 24 h time point. In addition, we determined that HDAC1 is also associated with the promoter region of the Sox21 gene, but that this association does not change upon the activation of the Sox21 gene.
The finding that HDAC1 is associated with Sox21 led us to examine whether another Sox2-associated protein, HDAC2, is also associated with this gene. ChIP analysis determined that HDAC2 is associated with the promoter region of the Sox21 gene in i-Sox2-ESCs (Fig. 4D), but not the region of the Sox21 gene that contains the putative HMG/POU cassette (data not shown). As in the case of HDAC1, the association of HDAC2 is diminished 24 h after the gene is activated (Fig. 4D). We extended these studies by examining whether acetylation of histone 3 (H3Ac), a chromatin modification that is associated with transcriptional activation, changes upon activation of the Sox21 gene. Indeed, H3Ac is substantially increased at the promoter region when the Sox21 gene is activated (Fig. 4E). Together, these results indicate that Sox21 is associated with multiple repressive complexes in mouse ESCs, and these complexes exit from the gene when it is activated in response to elevated levels of Sox2.
Repressive bivalent features associated with the Sox21 gene disappear early during its activation
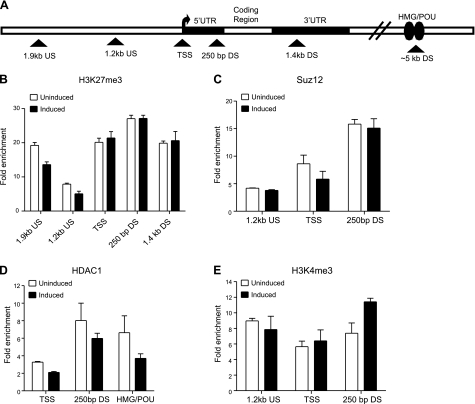
As noted above, Sox21 expression is activated within 3 h after Sox2 levels are elevated in i-Sox2-ESCs (20), which occurs 9 h after Dox addition (Fig. 1). This prompted us to examine the changes in bivalent features associated with the Sox21 gene at this early time point. More specifically, we examined whether the changes that we observed 24 h after Dox treatment were initiated by the 9 h time point. We performed ChIP analysis to determine the association of various bivalent features with the Sox21 gene at this time point. We observed the loss of most of the repressive machinery from the Sox21 gene within 9 h of the addition of Dox, coinciding with the rapid turn on of the gene seen at this time point. More specifically, we observed decreased association of the repressive features H3K27me3, Suz12, HDAC1, and to a small extent, H2Aub, with the Sox21 gene, while HDAC2 remains associated with the gene (Fig. 5A–C, E–G). Notably, the positive regulatory feature H3K4me3 remains associated with the Sox21 gene before and 9 h after the induction of differentiation by elevating Sox2 (Fig. 5D). Further, we determined that the association of Sox2 with the Sox21 gene is beginning to increase at this time point; whereas, Oct4 association with the Sox21 gene is unchanged (data not shown). These findings indicate that the loss of the repressive transcriptional machinery is tightly associated with the rapid activation of Sox21, and is likely to be a key part of the process by which this gene is rapidly activated.
Figure 5.
Sox21 loses its bivalent domains 9 h after i-Sox2-ESCs are induced to differentiate by elevating Sox2. A) Schematic diagram of the Sox21 gene indicating approximate location of primers (triangle) used in ChIP analysis. B–G). ChIP analysis of H3K27me3 (B), Suz12 (C), H3K4me3 (D), H2Aub (E), HDAC1 (F), and HDAC2 (G) on the Sox21 gene in i-Sox2-ESCs before and 9 h after i-Sox2-ESCs were treated with 4 μg/ml Dox by ChIP analyses. H3K4me3, Suz12, H2Aub, and HDAC2 association was monitored at the Sox21 TSS and at regions 1.2 kb upstream and ∼250 bp downstream relative to the Sox21 TSS; H3K27me3 association was determined at 2 additional regions, ∼1.9 kb upstream and ∼1.4 kb downstream relative to the Sox21 TSS. HDAC1 association was determined utilizing primers flanking the region containing the putative Sox21 HMG/POU cassette. ChIP analyses were repeated with similar results. Values of P > 0.05 are indicated next to specific bar graphs in panel E. *P < 0.05.
Bivalent features remain associated with the Sox21 gene when ESCs are induced to differentiate along a pathway that does not activate Sox21 expression
We also examined whether the loss of the repressive machinery associated with the Sox21 gene occurs when i-Sox2-ESCs are induced to differentiate by a completely different mechanism. To address this question, we induced the differentiation of i-Sox2-ESCs with retinoic acid (RA). Under these conditions, the Sox21 gene does not turn on (data not shown). Notably, ChIP analysis revealed that the Sox21 gene remains associated with both PRC2 components Suz12 and H3K27me3 before and after treatment of i-Sox2-ESCs with RA (Fig. 6A–C). In addition, we determined that the association of the histone deacetylase HDAC1 with the Sox21 gene is only slightly decreased under these conditions (P>0.05; Fig. 6D). As expected, the positive transcriptional features associated with the Sox21 gene, including H3K4me3 and RNA polymerase II, remain largely unchanged upon treatment with RA (Fig. 6E and data not shown). Although there was a small increase in H3K4me3 observed 250 bp downstream of the TSS after treatment with RA, this was not statistically significant (P>0.05). Together, these findings argue that loss of the repressive machinery from the Sox21 gene is tightly associated with its activation and not merely an indirect result of differentiation.
Figure 6.
Sox21 gene remains bivalent when i-Sox2-ESCs are induced to differentiate using retinoic acid. A) Schematic diagram of the Sox21 gene indicating approximate location of primers (triangle) used in the ChIP analysis. B–E) ChIP analyses of H3K27me3 (B), PRC2 component Suz12 (C), HDAC1 (D), and H3K4me3 (E) were performed in i-Sox2-ESCs before and 24 h after treatment with 10 μM RA. Primers amplifying the Sox21 TSS and the regions ∼1.2 kb upstream and ∼250 bp downstream relative to the Sox21 TSS were utilized to determine the association of Suz12 and H3K4me3 with the Sox21 promoter region. Additional primers amplifying regions ∼1.9 kb upstream and 1.4 kb downstream relative to the Sox21 TSS were utilized to monitor enrichment of H3K27me3. HDAC1 enrichment was monitored at the putative Sox21 HMG/POU cassette region, the Sox21 TSS, and 250 bp downstream of the Sox21 TSS. ChIP analyses were repeated with similar results.
Sox21 is regulated by multiple repressor complexes that function redundantly in ESCs to prevent its inappropriate expression
Thus far, our data suggest a role for several repressive complexes in the regulation of the Sox21 gene. To extend these studies, we examined the functional role of these complexes in controlling Sox21 expression. For this purpose, we used Eed-null ESCs. These cells lack one of the core components of the repressive PRC2 complex, Eed, which is essential for the functional integrity as well as the stability of the PRC2 complex (33, 34). Eed-null ESCs do not contain a functional PRC2 complex, and are therefore unable to catalyze the formation of H3K27me3 (23). Remarkably, these cells continue to express ESC-specific genes, such as Sox2 and Oct4, while also expressing a small number of lineage-specific developmental genes. They maintain ESC-like morphology, as evidenced by compact colonies, but are reported to display a tendency to differentiate (23).
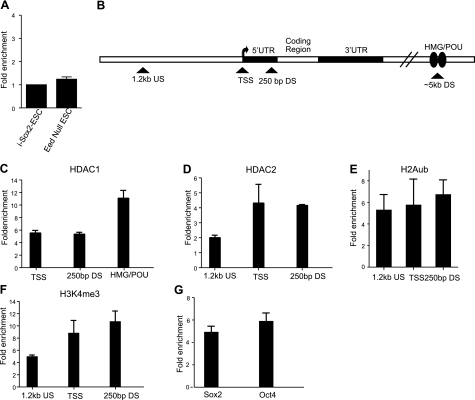
We used Eed-null ESCs to examine whether the loss of PRC2 is sufficient to activate the Sox21 gene. As reported by others (23), we confirmed that Sox21 is not activated in these cells (Fig. 7A). More specifically, the Sox21 gene does not appear to be expressed in Eed-null ESCs, and its level of expression at the RNA level is similar to that of uninduced i-Sox2-ESCs. Moreover, RNA levels of Sox21 in Eed-null ESCs are far lower than those observed in Dox treated i-Sox2-ESCs (Fig. 1). This argues that the Sox21 gene is not repressed solely by its association with PRC2. Therefore, we examined the association of HDAC1 and HDAC2 with the Sox21 gene in Eed-null ESCs. We determined that both HDAC1 and HDAC2 are associated with the inactive Sox21 gene at levels similar to those in our uninduced i-Sox2-ESCs (Fig. 7B–D; also see Fig. 4B, C). We also examined the association of PRC1 with the Sox21 gene in Eed-null ESCs. In this regard, earlier reports suggested that PRC1 is recruited to its target genes via PRC2, implying that PRC1 would not be recruited to its target genes in the absence of a functional PRC2 (35, 36). However, others have demonstrated that PRC1 can be recruited to its target genes independent of PRC2 and H3K27me3 (37). Indeed, we determined that H2Aub, a histone modification catalyzed by PRC1, is associated with the Sox21 gene promoter region in Eed-null ESCs (Fig. 7E). Equally important, we determined that the Sox21 gene is also associated with the positive histone modification H3K4me3 and with RNA polymerase II in these cells (Fig. 7F and data not shown). Finally, we determined that Sox2 and Oct4 are associated with the region of the Sox21 gene that contains the putative Sox21 HMG/POU cassette (Fig. 7G). These data indicate that the Sox21 gene remains in the bivalent state in Eed-null ESCs. Therefore, removal of a portion of the repressive machinery from the Sox21 gene is not sufficient to activate this gene.
Figure 7.
Sox21 gene remains silent in Eed-null ESCs due to the presence of redundantly functioning repressive machinery. A) qPCR was performed on cDNA prepared from RNA isolated from Eed-null ESCs or untreated i-Sox2-ESCs to measure Sox21 expression. Results are presented as fold-enrichment relative to GAPDH in Eed-null ESCs and compared to i-Sox2-ESCs, which was arbitrarily set to 1. B) Schematic diagram of the Sox21 gene indicating approximate location of primers (triangle) used in ChIP analysis. C–G) ChIP analysis of HDAC1 (C), HDAC2 (D), H2Aub (E), H3K4me3 (F), and Sox2 and Oct4 (G) was performed in Eed-null ESCs. Primers amplifying the region containing the putative HMG/POU cassette of Sox21 were utilized to determine the association of HDAC1, Sox2, and Oct4 with the Sox21 gene. Enrichment of HDAC2, H2Aub, and H3K4me3 at the Sox21 promoter was monitored at the Sox21 TSS and the regions ∼1.2 kb upstream and ∼250 bp downstream relative to the Sox21 TSS.
DISCUSSION
The work in this study investigated the poorly understood mechanisms by which bivalent, poised developmental regulators are activated rapidly during the early stages of differentiation. Our findings demonstrate, for the first time, that the inactive lineage-specific bivalent gene Sox21 is associated with a complex array of repressive machinery in ESCs. Notably, we determined that the Sox21 gene is occupied by Suz12, H3K27me3, H2Aub, HDAC1, and HDAC2 in i-Sox2-ESCs. However, nearly all the repressive machinery disappears from the Sox21 gene in response to elevated levels of Sox2. Moreover, 3 findings argue strongly that the activation of the Sox21 gene is tightly linked to the removal of this set of repressive transcriptional machinery. First, the removal of the repressive transcriptional machinery and the activation of the gene both occur just 3 h after Sox2 levels begin to rise. Second, we determined that the Sox21 gene is not activated, and does not shed its repressive machinery in response to differentiation induced by RA. Third, the lack of a single repressive complex (PRC2) is insufficient to relieve the repression of the Sox21 gene. Together, these data argue that bivalent gene regulation occurs at multiple levels; that is, genes are silenced in ESCs by multiple redundant layers of repression. This complexity of gene regulation is likely to play a key role in suppressing spurious gene activation. However, when differentiation is induced along a specific pathway, genes such as Sox21 undergo rapid activation due to the expeditious loss of repressive bivalent histone modifications and other repressive machinery.
Previous genome-wide studies determined that the transcription factors Sox2 and Oct4 cooccupy hundreds of genes associated with PRC1 and PRC2 (2, 28). Given that Sox21 was identified as a gene occupied by PRC1 and PRC2 (2, 28), we hypothesized that it may be coregulated by Sox2 and Oct4. Notably, we identified a putative Sox2:Oct4 binding site (HMG/POU cassette) located ∼5 kb downstream from the TSS of the Sox21 gene, and we determined that both Sox2 and Oct4 are associated with this region of the gene in our i-Sox2-ESC model system, both before and after the Sox21 gene is activated. These data argue that the Sox21 gene is a bivalent, Sox2:Oct4 target gene in mouse ESCs. The precise function of Sox2 and Oct4 in the modulation of Sox21 gene expression is unclear. They may perform the traditional role of a transcription factor and activate the Sox21 gene when differentiation is triggered. Alternatively, they may serve as recruitment factors for specific repressor complexes, such as HDACs, and function to silence the Sox21 gene in ESCs. Moreover, one cannot exclude the attractive model in which Sox2 and Oct4 exhibit a context-dependent dual role encompassing a silencing function in ESCs, and a gene-activating role during the onset of differentiation. Indeed, Sox2 is believed to activate specific Sox2:Oct4 target genes in ESCs, but repress Sox2:Oct4 target genes when overexpressed in ESCs (19, 20).
We determined that the activation of the Sox21 gene occurs in conjunction with the loss of its repressive, bivalent features. In particular, we demonstrated that the negative histone modification, H3K27me3, disappears from the Sox21 gene during its activation, while the positive bivalent features (H3K4me3 and RNA polymerase II) are retained. Upon the removal of the repressive machinery, rapid activation of the gene is permitted due to the continued presence of the positive transcriptional machinery, which is likely to include Sox2 and Oct4. The rapidity by which this occurs is underlined by our observation that Sox21 is turned on within 3 h of Sox2 overexpression. Thus, our results argue that bivalent gene activation is tightly coupled with loss of bivalency, which suggests that loss of the repressive, bivalent signature is responsible, at least in part, for the transcriptional activation of the gene. Moreover, the loss of repressive, bivalent features at a very early time point after the induction of differentiation reveals a probable mechanism by which rapid cell-fate decisions occur in response to specific differentiation signals throughout development.
The association of the Sox21 gene with several repressive complexes leads to speculation that there is a convergence of multiple regulatory pathways during development, which function either cooperatively or redundantly to control poised gene expression. In this regard, the H3K27me3 modification catalyzed by PRC2 is thought to act as a recognition mark to recruit PRC1, which then ubiquitinates H2Aub, causing chromatin compaction (29, 35). Notably, we observed a modest decrease in the association of H2Aub with the Sox21 gene in response to elevation of Sox2. While this decrease was not as large as that seen with other repressive features, such as the loss of H3K27me3, it was nonetheless reproducible, and it was observed both 9 and 24 h after the induction of differentiation by elevating Sox2 levels. PRC complexes have also been reported to cooperate with HDACs to control gene expression (38, 39). Moreover, Sox2 and Oct4 associate with HDACs in ESCs and during ESC differentiation (40, 41), giving rise to speculation that these repressors may be recruited to specific target genes via their association with TFs, such as Sox2 and Oct4. Indeed, we observe that HDAC1 and HDAC2 are associated with the Sox21 gene in i-Sox2-ESCs. Notably, this association is diminished upon the induction of differentiation when Sox2 is elevated. PRC2 also collaborates with H3K4 demethylases—enzymes that catalyze the removal of the activating H3K4me3 histone modification. Interestingly, this collaboration is thought to fine-tune the expression of lineage-specific developmental regulators in ESCs (42, 43). Thus, ESCs appear to utilize crosstalk between various regulatory complexes to modulate gene expression.
Notably, our study indicates that loss of the repressive machinery associated with the Sox21 gene is tightly linked to its activation. In this regard, we observed that the Sox21 gene is not activated when i-Sox2-ESCs are induced to differentiate with RA. Under these conditions, all of the repressive machinery, as well as the positive transcription machinery, remains associated with the gene. Thus, the nature of the differentiation signal appears to be critical for determining whether specific poised genes undergo activation. For example, while LIF withdrawal causes differentiation of mouse ESCs, it is not sufficient to activate certain bivalent genes (44). Hence, differentiation during embryogenesis must involve activation of highly specific transcriptional programs that up-regulate the relevant lineage-specific genes, whereas other genes remain unresponsive to these signals.
Our finding that Sox21 is occupied by multiple repressive complexes in ESCs led us to hypothesize that these complexes function in a redundant fashion. In this regard, we observed that removal of PRC2 from mouse ESCs does not appear to be sufficient to trigger the activation of Sox21. More specifically, Eed-null ESCs lack the repressive H3K27me3 modification (23), but display the continued presence of other activating features, such as Sox2, Oct4, H3K4me3, and RNA polymerase II at the Sox21 gene. This argues strongly that the gene remains repressed in these cells by additional repressive machinery (PRC1, HDAC1, and HDAC2). However, it is conceivable that other compensatory mechanisms activated in Eed-null ESCs, in order to limit their spontaneous differentiation, would also prevent the activation of the Sox21 gene if the levels of Sox2 were increased in these cells. Addressing this specific point would require engineering Eed-null ESCs for inducible Sox2 expression, since elevating Sox2 in ESCs triggers their differentiation (20). Interestingly, other studies argue that there is a strong need for repression of the Sox21 gene in ESCs. The importance of preventing inappropriate expression of developmental regulators, such as Sox21, is highlighted by a related study in our laboratory, which has determined that ectopic expression of Sox21 in mouse ESCs causes these cells to undergo differentiation (41). Thus, it appears that ESCs go to great lengths to prevent the spurious expression of key lineage markers and utilize several redundant mechanisms to ensure that key lineage-specific genes are kept silenced in ESCs, thereby protecting ESC self-renewal and pluripotency (Fig. 8).
Figure 8.
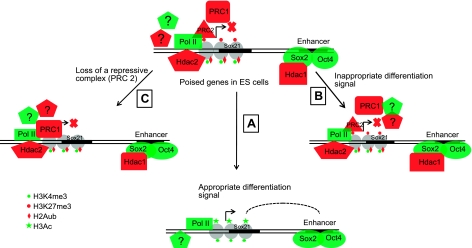
Model for bivalent gene regulation in ESCs: Bivalent, lineage-specific developmental regulatory genes remain silent in ESCs due to the presence of multiple, redundantly functioning repressive complexes, which likely collaborate to maintain a repressive environment to prevent gene activation. However, the coexistence of positive regulatory features at these genes allows for rapid activation during differentiation when appropriate signals for specific bivalent genes appear. This is facilitated by rapid exit of the repressive complexes from bivalent genes undergoing activation (A). The bivalent features remain associated with the gene when ESCs are induced to differentiate along a different pathway, and the gene remains inactive under these circumstances, which argues that the spurious activation of poised genes is prevented due to the presence of bivalent features which continue to allow the gene to remain in the poised state while preventing its inappropriate activation (B). Finally, crucial lineage-specific developmental regulators are kept under tight control via the existence of multiple layers of redundantly functioning mechanisms, and, in this way, loss of a single repressive complex is insufficient to allow gene activation due to the presence of other repressive mechanisms (C).
Currently, the molecular mechanisms responsible for the displacement of the repressive machinery during the activation of the Sox21 gene are unknown. Recent work by Duncan et al. (45) demonstrated that proteolytic cleavage of histone H3 by cathepsin L during ESC differentiation decreases the binding of PRC1 component Cbx3 to H3K27me3. Thus, it is tempting to speculate that H3 cleavage during differentiation facilitates the release of PRC components from H3K27me3 associated with the Sox21 gene. Another possible mechanism by which the PRC2 complex is displaced from the Sox21 gene is provided by Cha et al. (46), who determined that a target of PI3 kinase, Akt, phosphorylates the PRC2 component Ezh2. Intriguingly, this phosphorylation decreases the affinity of Ezh2 for H3, and inhibition of Ezh2 phosphorylation results in the repression of the PRC2 target gene, HoxA9. Another attractive possibility that could explain the disappearance of PRC from bivalent genes during differentiation is the “binary switch” model. In this scenario, phosphorylation of H3 at Ser-28 is expected to cause dissociation of the PRC2 complex occupying the adjacent H3K27 residue, and to destabilize the nucleosome, allowing access of the transcriptional machinery (47). Finally, recent work with noncoding RNAs suggests that they can collaborate with PRC proteins to control silencing of genes on the HoxD locus (48). Hence, it is conceivable that noncoding RNAs are involved in both the recruitment and removal of the PRC proteins from poised developmental regulators, such as the Sox21 gene.
In summary, our findings argue that the Sox21 gene and its chromatin structure are tightly regulated in ESCs by the presence of both activating machinery and redundant repressive machinery. This complex bivalent signature serves as a powerful mechanism to temper the rapid activation of lineage-specific genes with tight regulation. We speculate that similar complex control mechanisms operate throughout development and are indispensable for embryogenesis to take place rapidly and with great specificity.
Acknowledgments
The authors thank Dr. Terry Magnuson (University of North Carolina, Chapel Hill, NC, USA) for the gift of Eed-null ESCs, and Heather Rizzino for editorial assistance.
This work was supported by a grant from the National Institute of General Medicinal Sciences (GM 080751). B.D.O. was supported in part by a Cancer Biology Training Grant from the National Cancer Institute (CA009476). Core facilities of the University of Nebraska Medical Center Eppley Cancer Center were supported in part by a Cancer Center Support Grant from the National Cancer Institute (CA36727).
REFERENCES
- 1. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 2. Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., Jaenisch R. (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 [DOI] [PubMed] [Google Scholar]
- 3. Zhang Y., Reinberg D. (2001) Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes Dev. 15, 2343–2360 [DOI] [PubMed] [Google Scholar]
- 4. Sims R. J., 3rd, Millhouse S., Chen C. F., Lewis B. A., Erdjument-Bromage H., Tempst P., Manley J. L., Reinberg D. (2007) Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 28, 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703–717 [DOI] [PubMed] [Google Scholar]
- 6. Huang Y., Fang J., Bedford M. T., Zhang Y., Xu R. M. (2006) Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312, 748–751 [DOI] [PubMed] [Google Scholar]
- 7. Francis N. J., Kingston R. E., Woodcock C. L. (2004) Chromatin compaction by a polycomb group protein complex. Science 306, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 8. Ringrose L., Ehret H., Paro R. (2004) Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol. Cell 16, 641–653 [DOI] [PubMed] [Google Scholar]
- 9. Pasini D., Bracken A. P., Hansen J. B., Capillo M., Helin K. (2007) The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 27, 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chakravarthy H., Boer B., Desler M., Mallanna S. K., McKeithan T. W., Rizzino A. (2008) Identification of DPPA4 and other genes as putative Sox2:Oct-3/4 target genes using a combination of in silico analysis and transcription-based assays. J. Cell. Physiol. 216, 651–662 [DOI] [PubMed] [Google Scholar]
- 12. Boer B., Bernadt C. T., Desler M., Wilder P. J., Kopp J. L., Rizzino A. (2006) Differential activity of the FGF-4 enhancer in F9 and P19 embryonal carcinoma cells. J. Cell. Physiol. 208, 97–108 [DOI] [PubMed] [Google Scholar]
- 13. Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N., Lovell-Badge R. (2003) Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Scholer H., Smith A. (1998) Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 [DOI] [PubMed] [Google Scholar]
- 15. Feldman B., Poueymirou W., Papaioannou V. E., DeChiara T. M., Goldfarb M. (1995) Requirement of FGF-4 for postimplantation mouse development. Science 267, 246–249 [DOI] [PubMed] [Google Scholar]
- 16. Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. (2003) Functional expression cloning of nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655 [DOI] [PubMed] [Google Scholar]
- 17. Nakatake Y., Fukui N., Iwamatsu Y., Masui S., Takahashi K., Yagi R., Yagi K., Miyazaki J., Matoba R., Ko M. S., Niwa H. (2006) Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol. Cell. Biol. 26, 7772–7782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van den Boom V., Kooistra S. M., Boesjes M., Geverts B., Houtsmuller A. B., Monzen K., Komuro I., Essers J., Drenth-Diephuis L. J., Eggen B. J. L. (2007) UTF1 is a chromatin-associated protein involved in ES cell differentiation. J. Cell. Biol. 178, 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boer B., Kopp J., Mallanna S., Desler M., Chakravarthy H., Wilder P. J., Bernadt C., Rizzino A. (2007) Elevating the levels of Sox2 in embryonal carcinoma cells and embryonic stem cells inhibits the expression of Sox2 : Oct-3/4 target genes. Nucleic Acids Res. 35, 1773–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kopp J., Ormsbee B., Desler M., Rizzino A. (2008) Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells 26, 903–911 [DOI] [PubMed] [Google Scholar]
- 21. Sandberg M., Kallstrom M., Muhr J. (2005) Sox21 promotes the progression of vertebrate neurogenesis. Nat. Neurosci. 8, 995–1001 [DOI] [PubMed] [Google Scholar]
- 22. Chakravarthy H., Rizzino A. (2010) Sox2 (SRY-box containing gene 2). TFe Retrieved April 21, 2010 from http://www.cisreg.ca/cgi-bin/tfe/articles.pl?tfid=531
- 23. Chamberlain S. J., Yee D., Magnuson T. (2008) Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26, 1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mallanna S. K., Boer B., Desler M., Rizzino A. (2008) Differential regulation of the oct-3/4 gene in cell culture model systems that parallel different stages of mammalian development. Mol. Reprod. Dev. 75, 1247–1257 [DOI] [PubMed] [Google Scholar]
- 25. Bernadt C. T., Nowling T., Wiebe M. S., Rizzino A. (2005) NF-Y behaves as a bifunctional transcription factor that can stimulate or repress the FGF-4 promoter in an enhancer-dependent manner. Gene Expression 12, 193–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tomioka M., Nishimoto M., Miyagi S., Katayanagi T., Fukui N., Niwa H., Muramatsu M., Okuda A. (2002) Identification of sox-2 regulatory region which is under the control of oct-3/4-sox-2 complex. Nucleic Acids Res. 30, 3202–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stock J. K., Giadrossi S., Casanova M., Brookes E., Vidal M., Koseki H., Brockdorff N., Fisher A. G., Pombo A. (2007) Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell. Biol. 9, 1428–1435 [DOI] [PubMed] [Google Scholar]
- 28. Lee T. I., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S., Kumar R. M., Chevalier B., Johnstone S. E., Cole M. F., Isono K., Koseki H., Fuchikami T., Abe K., Murray H. L., Zucker J. P., Yuan B., Bell G. W., Herbolsheimer E., Hannett N. M., Sun K., Odom D. T., Otte A. P., Volkert T. L., Bartel D. P., Melton D. A., Gifford D. K., Jaenisch R., Young R. A. (2006) Control of developmental regulators by polycomb in human embryonic stem cells. Cell 125, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., Zhang Y. (2004) Role of histone H2A ubiquitination in polycomb silencing. Nature 431, 873–878 [DOI] [PubMed] [Google Scholar]
- 30. De Napoles M., Mermoud J. E., Wakao R., Tang Y. A., Endoh M., Appanah R., Nesterova T. B., Silva J., Otte A. P., Vidal M., Koseki H., Brockdorff N. (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell. 7, 663–676 [DOI] [PubMed] [Google Scholar]
- 31. Gontan C., Guttler T., Engelen E., Demmers J., Fornerod M., Grosveld F. G., Tibboel D., Gorlich D., Poot R. A., Rottier R. J. (2009) Exportin 4 mediates a novel nuclear import pathway for sox family transcription factors. J. Cell. Biol. 185, 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baltus G. A., Kowalski M. P., Tutter A. V., Kadam S. (2009) A positive regulatory role for the mSin3A-HDAC complex in pluripotency through nanog and Sox2. J. Biol. Chem. 284, 6998–7006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Montgomery N. D., Yee D., Chen A., Kalantry S., Chamberlain S. J., Otte A. P., Magnuson T. (2005) The murine polycomb group protein eed is required for global histone H3 lysine-27 methylation. Curr. Biol. 15, 942–947 [DOI] [PubMed] [Google Scholar]
- 34. Montgomery N. D., Yee D., Montgomery S. A., Magnuson T. (2007) Molecular and functional mapping of EED motifs required for PRC2-dependent histone methylation. J. Mol. Biol. 374, 1145–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Min J., Zhang Y., Xu R. M. (2003) Structural basis for specific binding of polycomb chromodomain to histone H3 methylated at lys 27. Genes Dev. 17, 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang L., Brown J. L., Cao R., Zhang Y., Kassis J. A., Jones R. S. (2004) Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell. 14, 637–646 [DOI] [PubMed] [Google Scholar]
- 37. Schoeftner S., Sengupta A. K., Kubicek S., Mechtler K., Spahn L., Koseki H., Jenuwein T., Wutz A. (2006) Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 25, 3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Van der Vlag J., Otte A. P. (1999) Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genetics 23, 474–478 [DOI] [PubMed] [Google Scholar]
- 39. Fujii S., Ito K., Ito Y., Ochiai A. (2008) Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J. Biol. Chem. 283, 17324–17332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S. Y., Qin J., Wong J., Cooney A. J., Liu D., Songyang Z. (2008) Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell. Biol. 10, 731–739 [DOI] [PubMed] [Google Scholar]
- 41. Mallanna S. K., Ormsbee B. D., Iacovino M., Gilmore J. M., Cox J. L., Kyba M., Washburn M. P., Rizzino A. (2010) Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. [E-pub ahead of print] Stem Cells doi: 10.1002/stem.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pasini D., Hansen K. H., Christensen J., Agger K., Cloos P. A., Helin K. (2008) Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and polycomb-repressive complex 2. Genes Dev. 22, 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pasini D., Cloos P. A., Walfridsson J., Olsson L., Bukowski J. P., Johansen J. V., Bak M., Tommerup N., Rappsilber J., Helin K. (2010) JARID2 regulates binding of the polycomb repressive complex 2 to target genes in ES cells. Nature 464, 306–310 [DOI] [PubMed] [Google Scholar]
- 44. Lee E. R., Murdoch F. E., Fritsch M. K. (2007) High histone acetylation and decreased polycomb repressive complex 2 member levels regulate gene specific transcriptional changes during early embryonic stem cell differentiation induced by retinoic acid. Stem Cells 25, 2191–2199 [DOI] [PubMed] [Google Scholar]
- 45. Duncan E. M., Muratore-Schroeder T. L., Cook R. G., Garcia B. A., Shabanowitz J., Hunt D. F., Allis C. D. (2008) Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell 135, 284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cha T. L., Zhou B. P., Xia W., Wu Y., Yang C. C., Chen C. T., Ping B., Otte A. P., Hung M. C. (2005) Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310, 306–310 [DOI] [PubMed] [Google Scholar]
- 47. Fischle W., Wang Y., Allis C. D. (2003) Binary switches and modification cassettes in histone biology and beyond. Nature 425, 475–479 [DOI] [PubMed] [Google Scholar]
- 48. Rinn J. L., Kertesz M., Wang J. K., Squazzo S. L., Xu X., Brugmann S. A., Goodnough L. H., Helms J. A., Farnham P. J., Segal E., Chang H. Y. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]