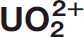Abstract
The uranyl cation ( ) can be suspected to interfere with the binding of essential metal cations to proteins, underlying some mechanisms of toxicity. A dedicated computational screen was used to identify
) can be suspected to interfere with the binding of essential metal cations to proteins, underlying some mechanisms of toxicity. A dedicated computational screen was used to identify  binding sites within a set of nonredundant protein structures. The list of potential targets was compared to data from a small molecules interaction database to pinpoint specific examples where
binding sites within a set of nonredundant protein structures. The list of potential targets was compared to data from a small molecules interaction database to pinpoint specific examples where  should be able to bind in the vicinity of an essential cation, and would be likely to affect the function of the corresponding protein. The C-reactive protein appeared as an interesting hit since its structure involves critical calcium ions in the binding of phosphorylcholine. Biochemical experiments confirmed the predicted binding site for
should be able to bind in the vicinity of an essential cation, and would be likely to affect the function of the corresponding protein. The C-reactive protein appeared as an interesting hit since its structure involves critical calcium ions in the binding of phosphorylcholine. Biochemical experiments confirmed the predicted binding site for  and it was demonstrated by surface plasmon resonance assays that
and it was demonstrated by surface plasmon resonance assays that  binding to CRP prevents the calcium-mediated binding of phosphorylcholine. Strikingly, the apparent affinity of
binding to CRP prevents the calcium-mediated binding of phosphorylcholine. Strikingly, the apparent affinity of  for native CRP was almost 100-fold higher than that of Ca2+. This result exemplifies in the case of CRP the capability of our computational tool to predict effective binding sites for
for native CRP was almost 100-fold higher than that of Ca2+. This result exemplifies in the case of CRP the capability of our computational tool to predict effective binding sites for  in proteins and is a first evidence of calcium substitution by the uranyl cation in a native protein.
in proteins and is a first evidence of calcium substitution by the uranyl cation in a native protein.
Keywords: uranium, uranyl ion, computational screening, molecular docking, modeling, C-reactive protein, surface plasmon resonance
Introduction
Metal cations are involved in many biochemical processes, and are common when focusing on proteins. Divalent cations in particular are found in many mechanisms, playing either structural or catalytic roles, or acting as mediators in protein-ligand complexes.1–4
However, some naturally abundant cations such as the uranyl ion ( ) have never been reported as integral parts in any biochemical system. Considering the large industrial use of uranium,
) have never been reported as integral parts in any biochemical system. Considering the large industrial use of uranium,  has been the subject of many toxicological studies that revealed its ability to interact with a large variety of biochemical targets. Recent investigations included analysis of specific metal carriers,5–8 of human serum proteins,9,10 and both proteomic and transcriptomic studies to identify proteins related to cellular impacts.11,12
has been the subject of many toxicological studies that revealed its ability to interact with a large variety of biochemical targets. Recent investigations included analysis of specific metal carriers,5–8 of human serum proteins,9,10 and both proteomic and transcriptomic studies to identify proteins related to cellular impacts.11,12
 is reported to deposit into bone with a metabolism similar to that of calcium,13 making ionic substitution a plausible hypothesis for some biochemical features of uranium. Le Clainche and Vita14 used the calcium binding Site I of the protein calmodulin as a template to design a peptide binding the uranyl cation. However, the direct competition of the uranyl cation at a calcium binding site of a native protein has to our knowledge not yet been reported. To probe this hypothesis, a computational tool was mandatory. Methods have been developed to detect calcium or zinc binding sites in proteins.15–18 For the uranyl ion, we used in this study the only
is reported to deposit into bone with a metabolism similar to that of calcium,13 making ionic substitution a plausible hypothesis for some biochemical features of uranium. Le Clainche and Vita14 used the calcium binding Site I of the protein calmodulin as a template to design a peptide binding the uranyl cation. However, the direct competition of the uranyl cation at a calcium binding site of a native protein has to our knowledge not yet been reported. To probe this hypothesis, a computational tool was mandatory. Methods have been developed to detect calcium or zinc binding sites in proteins.15–18 For the uranyl ion, we used in this study the only  -docking tool available, which we described previously.19
-docking tool available, which we described previously.19
Our computational method relies on rigid mainchain and full sidechain flexibility to predict  interaction sites in protein of known 3D-structures. While a few metalloproteins bind metal ions with large conformational changes, most metalloproteins impose constrained metal site geometries, with marginal fluctuation of their overall structure between metal-free and metal-bound states.20,21 In such cases, protein ligands are often observed to interact in a metal-dependent mode, that is, with direct coordination with the metal ions. Metal substitution is then expected to change protein-ligand binding properties.
interaction sites in protein of known 3D-structures. While a few metalloproteins bind metal ions with large conformational changes, most metalloproteins impose constrained metal site geometries, with marginal fluctuation of their overall structure between metal-free and metal-bound states.20,21 In such cases, protein ligands are often observed to interact in a metal-dependent mode, that is, with direct coordination with the metal ions. Metal substitution is then expected to change protein-ligand binding properties.
Our screening methodology was applied to a subset of the Protein Data Bank (PDB).22 Results were then compared to data from the Small Molecule Interaction Database (SMID),23 to pinpoint specific calcium dependant interactions between proteins and small molecules likely to be inhibited by  substitution. The C-reactive protein (CRP), a predicted template, was experimentally characterized to demonstrate this novel mode of action.
substitution. The C-reactive protein (CRP), a predicted template, was experimentally characterized to demonstrate this novel mode of action.
Results
Screening of PDB for  sites near protein ligands and metal cations
sites near protein ligands and metal cations
Sequences from protein sequence databases, such as UniProtKB/Swiss-Prot,24 and from the PDB usually differ.25,26 Missing signal peptides, missing domains or unresolved exposed loops, and redundancies due to site mutations are common discrepancies.
A simple selection scheme was used to select structures representative of native proteins and to reduce the computer screening time as detailed in Figure 1. This led to the selection of 12837 PDB unique IDs as the database subset. PDB entries corresponding to the same protein were kept in the subset despite nearly identical chain sequences and length, as long as existing or missing ligands caused slight structural variations. In such cases, the Cα root mean square deviations (RMSD) threshold used was 0.5814, corresponding to 80% similarity according to Lesk and Chothia.27
Figure 1.
Selection of PDB subset, clustering, and docking. Selection of the least mutated PDB protein chains matching each portion of wild type protein sequences, after discarding chains of near identical structure. 12837 PDB unique IDs were selected as the database subset. A two-stage screening method identified and scored  chelation sites in these structures.
chelation sites in these structures.
Our search algorithm19 was then applied to each of those structures, resulting in a set of possible chelation sites with corresponding scores based on energy minimization using the Amber force-field. The score distribution of the raw screening results is displayed in Figure 2. The calculated score [Eq. (1)] took into account both the deformation energy of the protein (Edef, difference of internal energy between the complexed and uncomplexed forms), the protein- interaction energy (EP-IUM, where IUM is the PDB identifier for
interaction energy (EP-IUM, where IUM is the PDB identifier for  ) plus nonbonded terms related to water-protein and water-uranium interactions (EPUW).
) plus nonbonded terms related to water-protein and water-uranium interactions (EPUW).
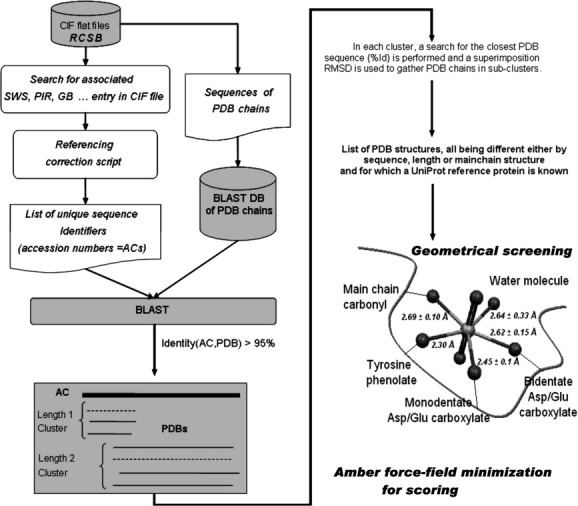
Figure 2.
Distribution of screening raw scores, fit with the sum of three Gaussian functions (thin black line). Black arrows indicate the mean of each fit and one standard deviation around the mean. The first fit from the left (bold black line, round black mark) is attributed to a population with high affinity for  (mean = −647, SD = 137); the center fit (dashed grey line) is attributed to a low affinity population (mean = −291, SD = 160); and the last fit on the right (dashed grey line, round grey mark) is attributed to a population with unfavorable deformation energy for binding such a ligand (mean = 160, SD = 179).
(mean = −647, SD = 137); the center fit (dashed grey line) is attributed to a low affinity population (mean = −291, SD = 160); and the last fit on the right (dashed grey line, round grey mark) is attributed to a population with unfavorable deformation energy for binding such a ligand (mean = 160, SD = 179).
| (1) |
This score is the sum of enthalpic energy terms, expressed in kcal/mol. Being similar to an energy function, the lower the value, the higher the propensity for interaction between the protein and the uranyl ion at each particular site. The best fit for the score distribution was obtained with a mixture of 3 Gaussian curves (Fig. 2).
Selection of an experimental target
12% of the proteins sites detected in our scheme were already populated with divalent cations: Ca2+ (8.1%), Mg2+ (1.6%), Mn2+ (1.0%), Zn2+ (0.4%), Co2+ (0.2%), and so forth. Proteins interacting with small molecules in a metal-dependent mode were also numerous. Table I reports such target proteins with high potential for  binding, that is, with a SMID ligand detected at less than 3 Å from the position of the uranyl cation.
binding, that is, with a SMID ligand detected at less than 3 Å from the position of the uranyl cation.
Table I.
Subset List of Target Proteins
| Swiss-Prot accession number | Protein name | Sequence length | Pfam families | Total #PDBs | Post clustering #PDBs | PDB files with at least one accepted scorea | SMID molecules close to uranyl hit locations | Mean of gaussian fit of scores | SD of gaussian fit of scores | Percentage of hits with score higher than mean 1SD |
|---|---|---|---|---|---|---|---|---|---|---|
| P09803 | Epithelial cadherin | 884 | Cadherin_pro; Cadherin; Cadherin_C | 7 | 6 | 1Q1P; 1FF5; 1EDH | Ca | −841 | 41 | 8% |
| P0A6A8 | Acyl carrier protein | 78 | PP-binding | 7 | 3 | 1L0H; 1L0I | Zn | −828 | 63 | 21% |
| P42566 | Epidermal growth factor receptor substrate 15 | 896 | efhand | 4 | 4 | 1F8H;1EH2;1FF1; 1C07 | Ca | −821 | 56 | 30% |
| P0A5N0 | Aminoglycoside 2′-N-acetyltransferase | 181 | Acetyltransf_1 | 4 | 2 | 1M4I; 1M44 | Kanamycin | −780 | 60 | 8% |
| P02619 | Parvalbumin beta | 107 | efhand | 6 | 5 | 3PAL; 1PVB; 4PAL; 1PAL; 2PVB | Ca | −761 | 88 | 8% |
| P02741 | C-reactive protein | 224 | Pentraxin | 3 | 3 | 1GNH; 1LJ7; 1B09 | Ca; PC | −755 | 60 | 20% |
| P37819 | Proclavaminate amidinohydrolase | 313 | Arginase | 2 | 2 | 1GQ6; 1GQ7 | Mn | −749 | 89 | 29% |
| P09860 | Troponin C, slow skeletal and cardiac muscles | 161 | efhand | 8 | 6 | 2CTN; 1DTL; 1FI5; 1AJ4; 3CTN; 1LA0 | Ca | −728 | 98 | 9% |
| P00582 | DNA polymerase I | 928 | 5_3_exonuc_N; 3_5_exonuc; DNA_pol_A | 14 | 4 | 1KFD; 2KFZ | Zn | −700 | 49 | 17% |
| Q9HYN5 | Fucose-binding lectin PA-IIL | 115 | PA-IIL (lectinII) | 12 | 2 | 1OUS; 1OXC | Ca; FUC | −673 | 59 | 10% |
| P20942 | Phosducin | 246 | Phosducin | 3 | 3 | 2TRC; 1B9X | Gd | −669 | 37 | 0% |
| Q60364 | 3,4-dihydroxy-2-butanone 4-phosphate synthase | 227 | DHBP_synthase | 3 | 2 | 1PVW; 1PVY | Zn; Ca; Pi; R5P | −666 | 38 | 0% |
| Q64537 | Calpain small subunit 1 | 270 | efhand | 5 | 4 | 1AJ5; 1DVI; 1NP8 | Ca | −659 | 51 | 23% |
| P08661 | Mannose-binding protein C | 244 | Collagen; Lectin_C | 13 | 4 | 1KZA; 1RDO; 1RDL; 1BV4 | Ca; man | −651 | 41 | 26% |
PDB entries in bold were found with hetero atoms (excluding water) less than 3 Å away from uranyl hit locations.
Four proteins attracted our attention since they all display a putative  binding site in the binding pocket for both an organic and a metallic ligand: CRP, fucose binding lectin II, DHBP synthase, and MBP-C. They represented testable models for our initial hypothesis that the uranyl ion might be a potent interferent of the formation of physiological complex.
binding site in the binding pocket for both an organic and a metallic ligand: CRP, fucose binding lectin II, DHBP synthase, and MBP-C. They represented testable models for our initial hypothesis that the uranyl ion might be a potent interferent of the formation of physiological complex.
Based on its score and availability, this study will focus on CRP. It forms a ring-shaped structure homopentamer (pCRP, 115kDa), which can interact with various types of molecules. One side of the ring can interact with polycationic compounds, carbohydrates (D-galactose related structures), or with phosphorylcholine (PC) in a calcium dependent manner. Attachment to the surface of pathogens, or binding to lyso-phosphatidylcholine of apoptotic/necrotic cells results in the activation of the classical complement cascade, through C1q binding on the other side of the molecule.28–30
The analysis of several structures revealed that the binding of Ca2+ and PC to pCRP involves only minor and local conformational rearrangements. The Cα RMSD between calcium-free (PDB 1LJ731) versus calcium-bound (1GNH32) structures, and of calcium-free versus Ca2+/PC-bound (1B0933) structures were measured at 0.33 and 0.44 Å, respectively, using 199 amino acid residues from monomer A. This was also true at the level of the quaternary structures, with Cα RSMD of 0.87 and 0.81 Å, respectively, over the 998 amino acid residues of the five monomers.
On the other hand, monomerization of pCRP to mCRP is associated with significant conformational changes.34 mCRP does not spontaneously bind PC,34–36 and was thus used as a control for nonspecific binding experiments in some of our following experiments.
Reported inhibitors of CRP–PC interaction are phosphorylcholine derivatives, including phosphate containing molecules and detergents37,38. Mg2+ was also documented as a marginal CRP ligand.35,39,40
Our results suggest that  is a potent ligand. Its mean score of −783 for all the selected sites corresponds to 85% probability of belonging to the “High affinity” population according to Bayes' inversion rule (see Fig. 2, distribution of scores). All predicted sites were located in the calcium-binding area of one of the chains, in a geometry which prevents phosphorylcholine from sterically coexisting with the uranyl ion.
is a potent ligand. Its mean score of −783 for all the selected sites corresponds to 85% probability of belonging to the “High affinity” population according to Bayes' inversion rule (see Fig. 2, distribution of scores). All predicted sites were located in the calcium-binding area of one of the chains, in a geometry which prevents phosphorylcholine from sterically coexisting with the uranyl ion.
The binding of  to CRP was demonstrated by dialysis at equilibrium in the absence of Ca2+. A
to CRP was demonstrated by dialysis at equilibrium in the absence of Ca2+. A  -loaded protein solution was extensively dialyzed for 5 h. The
-loaded protein solution was extensively dialyzed for 5 h. The  content was then quantified by inductively coupled plasma mass spectrometry (ICP-MS) in two experiments. A ratio of 0.9 ± 0.07 (at 3 SD) equivalents of
content was then quantified by inductively coupled plasma mass spectrometry (ICP-MS) in two experiments. A ratio of 0.9 ± 0.07 (at 3 SD) equivalents of  per protein subunit was measured (see Supporting Information Table SI).
per protein subunit was measured (see Supporting Information Table SI).
Supporting information Table SII lists all binding sites detected on the three PDB entries associated with CRP (Swiss-Prot AC P02741). CRP calcium binding sites display incomplete coordination spheres, as visible in PDB entry 1GNH where only calcium atoms and no PC are present in the crystal. Half of the first coordination shells of Ca2+ are populated with protein atoms, with labile water or solvent molecules not clearly visible at 3 Å resolution. These solvent-exposed half spheres of coordination are important for PC binding.
Pronase assay:  protects CRP better than Ca2+
protects CRP better than Ca2+
Sidechains involved in calcium binding are predicted to be also involved in  binding. Kinoshita et al. reported how Ca2+ binding protects the loop involving residues 141 to 148 against proteolytic cleavage by pronase (cleavage at Glu147).41
binding. Kinoshita et al. reported how Ca2+ binding protects the loop involving residues 141 to 148 against proteolytic cleavage by pronase (cleavage at Glu147).41  capability of preventing digestion was thus tested.
capability of preventing digestion was thus tested.
The protease concentration was adjusted by monitoring CRP digestion at various pronase/CRP ratios (1, 5 and 10% w/w), in the presence of 1 mM Ca2+ [Fig. 3(A)]. A 5% ratio was selected for subsequent experiments, as a trade-off between intact protein at 23 kDa, and the 16 kDa and 6.5 kDa fragments. In accordance with Kinoshita et al.,41 there was a clear decrease in proteolysis susceptibility when calcium concentrations increased [Fig. 3(B)].
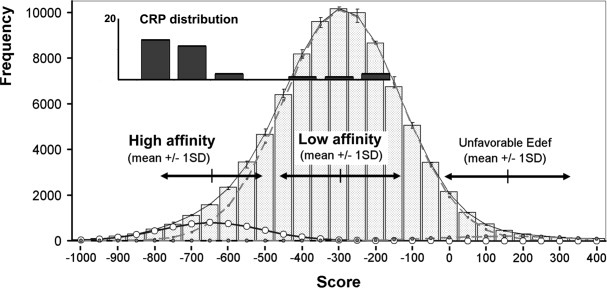
Figure 3.
Conformational sensitivity of CRP to digestion with pronase. A: Top: without bound calcium atoms, the CRP138–148 loop is largely exposed to solvent (IGNH chain D). Bottom: electrophoresis gel of CRP (200 μg/mL) digestion at a fixed CaCl2 concentration (1 mM) and an increasing concentration of pronase, which promotes CRP (23 kDa) digestion. B: Top: in the presence of two bound calcium atoms, the CRP138–148 loop is less exposed to the solvent (1GNH chain C). Bottom: electrophoresis gel of CRP (200 μg/mL) digestion at a fixed concentration of pronase (5%W/W) and an increasing concentration of CaCl2. Calcium precludes CRP digestion by pronase41. C: Top: predicted binding site geometry of chelated  geometry on CRP chain C (IGNH). The side chains responsible for calcium chelation in CRP are also involved in
geometry on CRP chain C (IGNH). The side chains responsible for calcium chelation in CRP are also involved in  binding. Bottom: protection against pronase digestion of CRP (200 μg/mL) with increasing concentrations of the uranyl ion. Electrophoresis results represent at least two independent silver staining experiments. D: MALDI-TOF mass spectra of CRP. CRP was incubated without metal (black), with 1 mM CaCl2 (dark grey) and 100 μM
binding. Bottom: protection against pronase digestion of CRP (200 μg/mL) with increasing concentrations of the uranyl ion. Electrophoresis results represent at least two independent silver staining experiments. D: MALDI-TOF mass spectra of CRP. CRP was incubated without metal (black), with 1 mM CaCl2 (dark grey) and 100 μM  (light grey) prior to pronase digestion. Relative intensity was set at 100% for the [M+H]+ peak. [M+H]+, [M+2H]2+, and [M+3H]3+ are assigned to mono, doubly, and triply charged intact CRP peaks, fragments A and B are pronase digestion products. Calcium and
(light grey) prior to pronase digestion. Relative intensity was set at 100% for the [M+H]+ peak. [M+H]+, [M+2H]2+, and [M+3H]3+ are assigned to mono, doubly, and triply charged intact CRP peaks, fragments A and B are pronase digestion products. Calcium and  spectra show very similar peak distribution, with near disappearance of digestion products in agreement with electrophoresis results, implying that the conformation of the metal-bound CRP loop is similar for both metals.
spectra show very similar peak distribution, with near disappearance of digestion products in agreement with electrophoresis results, implying that the conformation of the metal-bound CRP loop is similar for both metals.
Pronase was neither inactivated by calcium nor by  , as verified using human albumin as a control under various concentrations of CaCl2 (1 to 5 mM) and
, as verified using human albumin as a control under various concentrations of CaCl2 (1 to 5 mM) and  (0.1 to 1 mM). Nonspecific electrostatic interactions in electrophoresis experiments were also tested using Mg2+. CRP was not protected from proteolysis at Mg2+ concentration up to 10 mM (data not shown).
(0.1 to 1 mM). Nonspecific electrostatic interactions in electrophoresis experiments were also tested using Mg2+. CRP was not protected from proteolysis at Mg2+ concentration up to 10 mM (data not shown).
Figure 3(C) showed that  protects CRP against pronase more efficiently than Ca2+, since digestion profiles appeared similar at 1 mM Ca2+ and 0.1 mM
protects CRP against pronase more efficiently than Ca2+, since digestion profiles appeared similar at 1 mM Ca2+ and 0.1 mM  . The masses of fragments obtained in MALDI-TOF-MS experiments confirmed the expected cleavage site in the calcium binding loop.41 Spectra obtained were similar whenever Ca2+ or
. The masses of fragments obtained in MALDI-TOF-MS experiments confirmed the expected cleavage site in the calcium binding loop.41 Spectra obtained were similar whenever Ca2+ or  was used [Fig. 3(D)], indirectly confirming identical binding sites for both cations.
was used [Fig. 3(D)], indirectly confirming identical binding sites for both cations.
Interaction of Ca and PC with CRP
Surface plasmon resonance (SPR) was used to study the binding of PC to CRP. The setup involved immobilized PC, under the form of a PC-BSA conjugate covalently linked to the sensor chip, and circulating pCRP, and the reciprocal setting. Under both conditions the absence of Ca2+ gave no measurable signal, while increasing its concentration proportionally enhanced the signal to reach a plateau at 2 mM CaCl2, as previously reported.39 Observed signals were consistent with a maximal binding of one PC-BSA molecule per CRP pentamer. The absence of calcium in the dissociation buffer caused the immediate release of bound molecules.
Binding levels were on average and consistently seven times lower at 50 μM CaCl2 compared to those measured in the presence of 2 mM CaCl2 (data not shown). Because both calcium cations are needed for PC binding, a rough estimate of KD (Ca-CRP) can be drawn from this ratio, supposing equal affinity41 and independency of calcium binding to both sites. A 1/7 ratio for PC binding implies that individual calcium site occupancy is (1/7)0.5, relating to the Y/Rmax value in the first term of Eq. (2) depicting the law of mass action. At X = 50 μM CaCl2, we deduced a KD value of 82 μM. This agrees with reported KD, in the range of 50–78.1 μM.39,41,42
We also estimated the binding affinity of PC to CRP. The published KD values are from 5 to 18 μM for human CRP.39,40,43,44 We consistently obtained KD values between 0.4 and 1 μM for CRP interaction with PC-BSA in the presence of 50 μM Ca2+, whatever the settings and the sensor chips. Since the functionalization ratio is 10 PC molecules per BSA protein, avidity for the five binding sites of pCRP could account for the 10-fold difference with the reported affinity for free PC/CRP. Binding saturation was reached when CRP concentration was raised to 10 μM (Supporting Information Fig. S1).
Inhibition experiments:  as a potent inhibitor
as a potent inhibitor
As expected, EDTA displayed a marked inhibition of PC binding. Free PC was also found to inhibit CRP binding to immobilized PC-BSA, with an IC50 below 0.1 μM at 50 μM Ca2+ (data not shown). MgCl2 was also assayed at concentrations up to 1 mM (as a reminder, body concentration is ∼ 0.5 mM45). Neither inhibition nor enhancement of CRP-PC interaction was observed despite a molar excess of 20 over Ca2+. This result suggested that Mg2+ cannot replace Ca2+ cations in the CRP calcium binding sites. The previously reported magnesium binding to CRP35,40 is thus assumed to occur either in the calcium site with a KD value in the millimolar range at best, or in different binding sites.
 was tested in SPR experiments as an inhibitor of circulating PC-BSA binding on a CRP surface [Fig. 4(A)], along with the reversed experiment, in the presence of 50 μM CaCl2 in the buffer. This calcium concentration was considered as a good trade-off between sufficient binding signal and
was tested in SPR experiments as an inhibitor of circulating PC-BSA binding on a CRP surface [Fig. 4(A)], along with the reversed experiment, in the presence of 50 μM CaCl2 in the buffer. This calcium concentration was considered as a good trade-off between sufficient binding signal and  concentrations preserving solubility. In both cases, a strong inhibition was observed, with a half-inhibitory concentration (IC50) of 0.16 μM
concentrations preserving solubility. In both cases, a strong inhibition was observed, with a half-inhibitory concentration (IC50) of 0.16 μM  in the first setup, and 10 μM in the later setup.
in the first setup, and 10 μM in the later setup.
Figure 4.
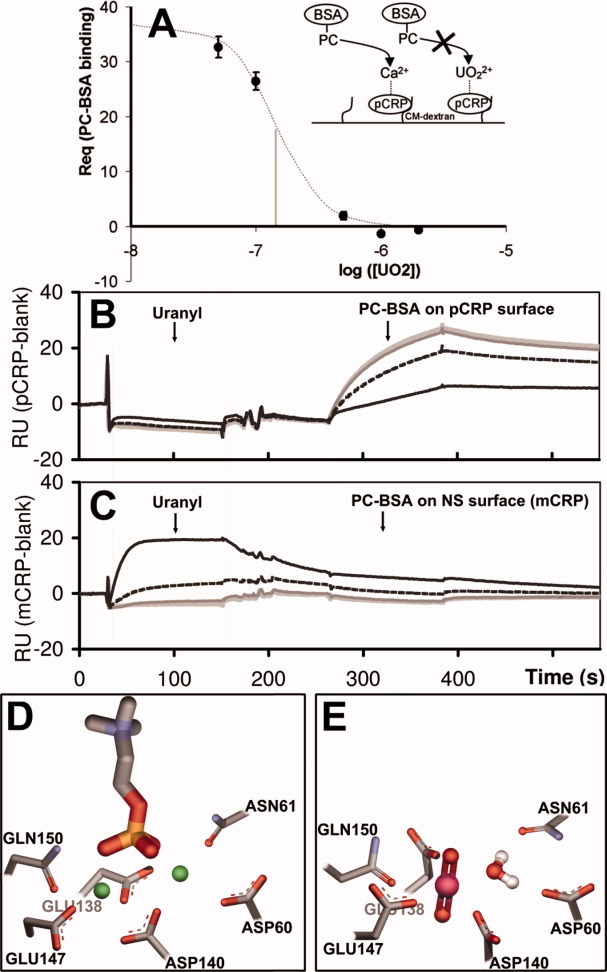
SPR binding inhibition assays. (A) Schematic and SPR data plot of  inhibition (IC50 = 0.16 μM) of PC-BSA ([PC-BSA] =100nM) binding to CRP (838 immobilized RU). B,C: SPR sensorgrams showing the effect of a
inhibition (IC50 = 0.16 μM) of PC-BSA ([PC-BSA] =100nM) binding to CRP (838 immobilized RU). B,C: SPR sensorgrams showing the effect of a  preinjection on the capacity to bind PC-BSA for pCRP (B) and mCRP (C). An increasing concentration of captured
preinjection on the capacity to bind PC-BSA for pCRP (B) and mCRP (C). An increasing concentration of captured  (light grey: 0 μM; dark grey: 0.1 μM; dashed black: 0.5 μM; black: 2 μM) only added at a capture stage results in the clear inhibition of PC-BSA (100 nM) binding on native CRP (838RU of immobilized pCRP). mCRP used for nonspecific monitoring shows no
(light grey: 0 μM; dark grey: 0.1 μM; dashed black: 0.5 μM; black: 2 μM) only added at a capture stage results in the clear inhibition of PC-BSA (100 nM) binding on native CRP (838RU of immobilized pCRP). mCRP used for nonspecific monitoring shows no  dependant signal modulation. In supporting information Figure S2,
dependant signal modulation. In supporting information Figure S2,  is also added with PC-BSA, resulting in complete inhibition of PC-BSA binding. (D) CRP calcium binding site from PDB structure 1B09 chain C (sticks) showing calcium atoms (green spheres) and the phosphorylcholine molecule (sticks). E: Predicted site geometry of the U-CRP complex upon uranyl cation and water molecule binding. Once bound, the uranyl ion prevents the phosphorylcholine molecule from docking with CRP. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
is also added with PC-BSA, resulting in complete inhibition of PC-BSA binding. (D) CRP calcium binding site from PDB structure 1B09 chain C (sticks) showing calcium atoms (green spheres) and the phosphorylcholine molecule (sticks). E: Predicted site geometry of the U-CRP complex upon uranyl cation and water molecule binding. Once bound, the uranyl ion prevents the phosphorylcholine molecule from docking with CRP. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The estimation of the refractive index increment (RII) for  diacetate diluted in water using SPR was 0.12 cm3/g instead of 0.18 cm3/g for proteins, accounting for a possible one third error in stoichiometry estimations (data not shown). The corrected formula used for the prediction of Rmax for
diacetate diluted in water using SPR was 0.12 cm3/g instead of 0.18 cm3/g for proteins, accounting for a possible one third error in stoichiometry estimations (data not shown). The corrected formula used for the prediction of Rmax for  was: (RUpred)max = RUM × (MWL/MWM) × (RIIL/RIIM).46
was: (RUpred)max = RUM × (MWL/MWM) × (RIIL/RIIM).46
Two Ca2+ atoms are needed for the binding of PC to a CRP protomer. Figure 4(D,E) show side-by-side the crystal structure of protomer C containing two calcium atoms and a molecule of PC, and the predicted location of the uranyl cation, presumably replacing the calcium atoms. One of the apical oxygen atoms is pointing in the direction normally occupied by the phosphorus atom of PC.
Specificity of the inhibition
Because of the above mentioned differences in IC50, the possibility that binding inhibition could result from either the association of  with the phosphate group of PC, or the monomerization of pCRP, was a concern. To rule them out, we used a setup where immobilized pCRP was subjected to various concentrations of
with the phosphate group of PC, or the monomerization of pCRP, was a concern. To rule them out, we used a setup where immobilized pCRP was subjected to various concentrations of  (capture step), followed by a dissociation step with running buffer. PC-BSA was then injected, either at the same uranyl ion concentration (Supporting Information Fig. S2) or with no
(capture step), followed by a dissociation step with running buffer. PC-BSA was then injected, either at the same uranyl ion concentration (Supporting Information Fig. S2) or with no  [Fig. 4(B,C)]. Regular association–dissociation stages were recorded. The corresponding binding capacity (RUpred)max was 6.5 RU of
[Fig. 4(B,C)]. Regular association–dissociation stages were recorded. The corresponding binding capacity (RUpred)max was 6.5 RU of  for a 1:1 CRP subunit/
for a 1:1 CRP subunit/ stoichiometry after RII correction. The observed binding level for
stoichiometry after RII correction. The observed binding level for  on pCRP was in the range 2.5–5 RU, that is, 40 to 80% of available calcium sites. PC-BSA maximal binding level was 65 RU in the association stage, corresponding to 13% of CRP pentamer.
on pCRP was in the range 2.5–5 RU, that is, 40 to 80% of available calcium sites. PC-BSA maximal binding level was 65 RU in the association stage, corresponding to 13% of CRP pentamer.
At a stoichiometry of 2 to 4 uranyl ions per pCRP an inhibition of PC-BSA binding between 60% and 80% at 2 μM  was observed [Fig. 4(B)]. The PC-BSA binding level was in any case fully restored in the cycle without
was observed [Fig. 4(B)]. The PC-BSA binding level was in any case fully restored in the cycle without  immediately following the 2 μM
immediately following the 2 μM  cycle.
cycle.
This showed that immobilized pCRP did not monomerize into mCRP upon  binding, since this conformational change is irreversible and mCRP is known not to bind PC. Additionally, absolute variation of baseline on the pCRP surface was within 3 RU indicating stability of the immobilized material. Since PC-BSA was never in direct contact with
binding, since this conformational change is irreversible and mCRP is known not to bind PC. Additionally, absolute variation of baseline on the pCRP surface was within 3 RU indicating stability of the immobilized material. Since PC-BSA was never in direct contact with  in Figure 4(B) experiment, it can be concluded that this inhibition was due solely to the uranyl ion interfering at the CRP calcium binding site rather than any interaction with the phosphate group of PC.
in Figure 4(B) experiment, it can be concluded that this inhibition was due solely to the uranyl ion interfering at the CRP calcium binding site rather than any interaction with the phosphate group of PC.
Direct  binding to CRP
binding to CRP
The availability of high sensitivity SPR instrument, such as Biacore T100 has opened the way to the direct measurement of small ligand interaction. The direct binding of  to CRP could be directly assessed on immobilized CRP.
to CRP could be directly assessed on immobilized CRP.
A strong nonspecific interaction for CM-dextran was observed on all sensorchips. As shown in Figure 5, the signal for a pCRP-grafted surface (3190RU of immobilized pCRP) was comparable to that of a nonfunctionalized CM-dextran surface. Nonspecific signal thus originated largely from the interaction of the uranyl ion with negatively charged side-chains or nonfunctionalized dextran carboxylate groups. Nonspecific binding was best fit using a four-parameter logistic curve, with an EC50 of 20 μM and a HillSlope of 1.9, indicating positive binding cooperativity.
Figure 5.
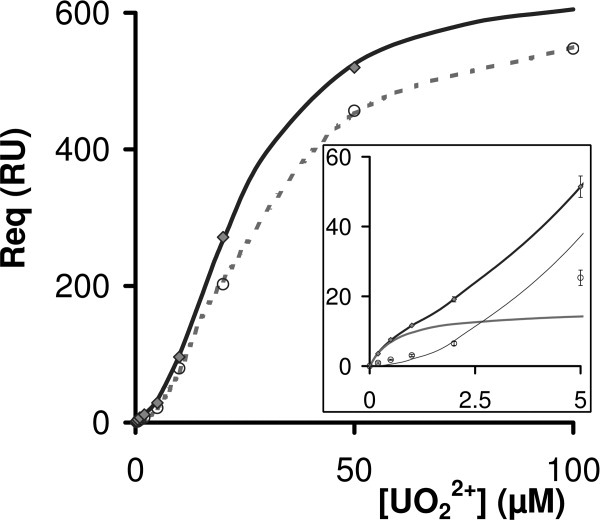
 binding SPR signal. SPR signal for
binding SPR signal. SPR signal for  binding on a pCRP surface (grey diamonds) compared to the nonspecific signal (NS) measured on a non-functionalized CM-dextran surface (open circles). Both signals are blank subtracted. The best fit of NS was obtained with a four-parameter logistic function (dashed grey line). An additional one-site binding function was necessary [Eq. (2)] for nonlinear fitting of the
binding on a pCRP surface (grey diamonds) compared to the nonspecific signal (NS) measured on a non-functionalized CM-dextran surface (open circles). Both signals are blank subtracted. The best fit of NS was obtained with a four-parameter logistic function (dashed grey line). An additional one-site binding function was necessary [Eq. (2)] for nonlinear fitting of the  binding signal on pCRP (black line). Inset: Focus on the fitted
binding signal on pCRP (black line). Inset: Focus on the fitted  binding signal on pCRP. The nonspecific contribution modeled with a four-parameter logistic curve (thin black line), the specific binding signal (dark grey line) and the fitted sum (black line) are shown.
binding signal on pCRP. The nonspecific contribution modeled with a four-parameter logistic curve (thin black line), the specific binding signal (dark grey line) and the fitted sum (black line) are shown.
The overall signal was modeled as the sum of both a standard one-site binding curve and a nonspecific logistic curve as in Eq. (2).
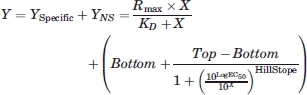 |
(2) |
By subtracting the nonspecific component from the overall signal, we could access to the specific feature of  -pCRP binding. The resulting fit is shown in Figure 5 inset, with a very good absolute sum of squares at 0.08. The corresponding KD value for the one site binding model was (6.8 ± 0.9) × 10−7 M. The fitted Rmax value at 16.2 RU was in agreement with the expected Rmax [(RUpred)max], given the quantity of CRP immobilized on the sensor chip, at 25 RU (CRP subunits, RII corrected), indicating that
-pCRP binding. The resulting fit is shown in Figure 5 inset, with a very good absolute sum of squares at 0.08. The corresponding KD value for the one site binding model was (6.8 ± 0.9) × 10−7 M. The fitted Rmax value at 16.2 RU was in agreement with the expected Rmax [(RUpred)max], given the quantity of CRP immobilized on the sensor chip, at 25 RU (CRP subunits, RII corrected), indicating that  binds about 65% of CRP calcium sites with high affinity in buffer containing 50 μM CaCl2.
binds about 65% of CRP calcium sites with high affinity in buffer containing 50 μM CaCl2.
Discussion
Experimental validation of computational prediction is unfortunately seldom achieved for a number of reasons including the difficulty in accessing biological materials. We had this requirement in mind in the design of this study. Our docking tool for  proposed several targets, and multiple binding sites in proteins. The performance of the method had to be confirmed and we selected a demonstrative example. The fact that
proposed several targets, and multiple binding sites in proteins. The performance of the method had to be confirmed and we selected a demonstrative example. The fact that  could interfere with the Ca2+ binding pocket of CRP, and consequently inhibit PC binding to CRP is original and pertinent from a toxicological point of view. Experimental results supported the predicted binding of
could interfere with the Ca2+ binding pocket of CRP, and consequently inhibit PC binding to CRP is original and pertinent from a toxicological point of view. Experimental results supported the predicted binding of  at the specific binding site for calcium, with an unexpectedly high affinity, about 100-fold higher than that of calcium.
at the specific binding site for calcium, with an unexpectedly high affinity, about 100-fold higher than that of calcium.
The colocalization of calcium and uranyl cations was confirmed by examining the selective inhibition of the pronase-sensitive calcium binding loop of CRP. Electrophoresis experiments showed that  was at least 10-fold more efficient than calcium in protecting CRP from digestion. Maldi-TOF results were also in agreement with this finding. Magnesium did not inhibit proteolysis, which rules out a nonspecific electrostatic mechanism. As seen in Figure 3(C), CRP showed slightly delayed band migration, but only at higher uranyl ion concentrations. This was considered an artifact due to silver staining since it was not observed on coomassie blue staining gels performed in parallel (Invitrogen NuPAGE Novex 10% Bis-Tris Mini gels, Supporting Information, Fig. S3). The stoichiometry of 0.9
was at least 10-fold more efficient than calcium in protecting CRP from digestion. Maldi-TOF results were also in agreement with this finding. Magnesium did not inhibit proteolysis, which rules out a nonspecific electrostatic mechanism. As seen in Figure 3(C), CRP showed slightly delayed band migration, but only at higher uranyl ion concentrations. This was considered an artifact due to silver staining since it was not observed on coomassie blue staining gels performed in parallel (Invitrogen NuPAGE Novex 10% Bis-Tris Mini gels, Supporting Information, Fig. S3). The stoichiometry of 0.9  per CRP subunit observed in ICP-MS experiments further indicated that the uranyl ion binds in the calcium binding site, one uranyl cation replacing both calcium cations.
per CRP subunit observed in ICP-MS experiments further indicated that the uranyl ion binds in the calcium binding site, one uranyl cation replacing both calcium cations.
We measured a dissociation parameter (KD) of 0.68 μM for the interaction between CRP and  in the presence of 50 μM Ca2+. Thus,
in the presence of 50 μM Ca2+. Thus,  binds to native pCRP with an affinity one-hundred fold higher than calcium, since the KD for calcium was reported to be in the range of 50–78.1 μM. Ionized calcium concentration in interstitial fluid or plasma is about 1.2 mM,45 so a competition with the uranyl ion could be expected as soon as its local concentration exceeds 10 μM. Moderate
binds to native pCRP with an affinity one-hundred fold higher than calcium, since the KD for calcium was reported to be in the range of 50–78.1 μM. Ionized calcium concentration in interstitial fluid or plasma is about 1.2 mM,45 so a competition with the uranyl ion could be expected as soon as its local concentration exceeds 10 μM. Moderate  intoxication might be sufficient to hinder CRP adhesion to apoptotic or pathogen cells. Internal contamination with uranium is often associated with inflammation47,48 and CRP levels might thus increase up to 1000 fold. CRP could then constitute a biochemical cargo for uranium in the serum. However,
intoxication might be sufficient to hinder CRP adhesion to apoptotic or pathogen cells. Internal contamination with uranium is often associated with inflammation47,48 and CRP levels might thus increase up to 1000 fold. CRP could then constitute a biochemical cargo for uranium in the serum. However,  concentrations in interstitial fluids are probably as transient as blood concentrations, so biological relevance of these dynamic processes is difficult to estimate without in vivo experiments.
concentrations in interstitial fluids are probably as transient as blood concentrations, so biological relevance of these dynamic processes is difficult to estimate without in vivo experiments.
In addition to these biochemical investigations, we examined atoms appearing in the first coordination shell of uranium. Existing data are extracted from resolved protein structures where the uranyl cation was used for phase resolution in multiple isomorphous replacement,19,49 and have also been documented for transferrin.6 We propose here the direct binding of a uranyl cation with protein side-chain atoms only as first shell ligands, apart from one water molecule, as observed in almost all  sites found in the Protein DataBank. Interestingly, Gln150 is reported here among coordinating groups in
sites found in the Protein DataBank. Interestingly, Gln150 is reported here among coordinating groups in  first-sphere. The glutamine amide oxygen atom had not been reported before as a direct uranyl ion ligand, although amide oxygen atoms are likely
first-sphere. The glutamine amide oxygen atom had not been reported before as a direct uranyl ion ligand, although amide oxygen atoms are likely  chelators since they are reported as hard Lewis bases according to the hard/soft acid/base theory (HSAB).
chelators since they are reported as hard Lewis bases according to the hard/soft acid/base theory (HSAB).
Our detection methodology was used with a particular focus on molecular interactions. Strikingly, the first selected and tested target has a calcium site prone to  competition. Such calcium binding sites favoring pentagonal or hexagonal planar chelation geometry, as needed for the uranyl ion complexation, may account for the chemical toxicity of
competition. Such calcium binding sites favoring pentagonal or hexagonal planar chelation geometry, as needed for the uranyl ion complexation, may account for the chemical toxicity of  and its distribution in tissues.
and its distribution in tissues.
Material and Methods
The screening methodology was detailed previously.19 Briefly, a first shell environment of the uranyl cation is drawn from the statistical analysis of structures of complexes from the Cambridge Structural Databank (CSD) and the PDB.  is placed at each node of a grid laid over the PDB structure in a first geometrical screening stage, and all possible binding oxygen atoms from the protein are tentatively moved towards the expected
is placed at each node of a grid laid over the PDB structure in a first geometrical screening stage, and all possible binding oxygen atoms from the protein are tentatively moved towards the expected  first shell positions, using side-chain rotamers for maximum flexibility. When this stage is favorable, a second stage involving energy minimization in the Amber force-field is applied, allowing the calculation of a score estimating the total energy of the protein-metal complex [Fig. 1 Top right].
first shell positions, using side-chain rotamers for maximum flexibility. When this stage is favorable, a second stage involving energy minimization in the Amber force-field is applied, allowing the calculation of a score estimating the total energy of the protein-metal complex [Fig. 1 Top right].
This method was applied to a nonredundant subset of the PDB, prepared to focus on the most biologically pertinent protein structures and reduce computer screening time. PDB sequences aligning with the same areas of native Swiss-Prot sequences were gathered by performing a hierarchical clustering of PDB chains based on a UniProtKB-Swiss-Prot versus PDB blast. A C-alpha RMSD was then calculated in each cluster, relative to the least mutated PDB chain compared with the accession number (AC), to retain as many alternative conformation structures as needed. The least mutated PDB entry, compared with the native sequence, was then kept in each subcluster. 12837 PDB unique IDs were selected as the database subset [Fig. 1 Top].
An automated version of our INTERALIGN program50 was used for sequence manipulations and C-alpha RMSD calculations for sequence alignments originating in the Basic Local Alignment Search Tool (BLAST). The computing time to process the PDB subset files was equivalent to 625 days on a single 3.2 GHz Pentium IV with 1024M RAM running with Linux. Database collection and processing was performed early in this work, using December 2005 version for the PDB database and March 2005 version for the SMID database (Biomolecular Object Network Databank,51).
Choice of C-reactive protein
The selection of proteins of interest [Table I] was based solely on scores and redundancy. Proteins with at least one predicted  site with a score below −500 were selected. This threshold corresponds to the mean +1SD value of the specific population distribution (84% of specific scores). Score spreading of sites per PDB entry was followed to control the number of nonspecific hits, since detected chelation areas involving common residues predominantly yield similar score levels. Proteins selected in Table I counted at least two associated structures, showed a mean value of scores below −600, a standard deviation (SD) below 100, less than 31.6% of scores higher than mean + 1SD (the expected percentage for a normal score distribution is 15.8%, 2 times as a safety coefficient), and at least one SMID ligand less than 3 Å form the uranyl ion site. We then focused on proteins interacting with molecules in a metal-dependent fashion, with an apo-form of the protein structure available to check on conformational stability compared to the metal loaded structure. This led to the selection of the calcium mediated CRP/phosphorylcholine interaction.
site with a score below −500 were selected. This threshold corresponds to the mean +1SD value of the specific population distribution (84% of specific scores). Score spreading of sites per PDB entry was followed to control the number of nonspecific hits, since detected chelation areas involving common residues predominantly yield similar score levels. Proteins selected in Table I counted at least two associated structures, showed a mean value of scores below −600, a standard deviation (SD) below 100, less than 31.6% of scores higher than mean + 1SD (the expected percentage for a normal score distribution is 15.8%, 2 times as a safety coefficient), and at least one SMID ligand less than 3 Å form the uranyl ion site. We then focused on proteins interacting with molecules in a metal-dependent fashion, with an apo-form of the protein structure available to check on conformational stability compared to the metal loaded structure. This led to the selection of the calcium mediated CRP/phosphorylcholine interaction.
Reagents
 speciation and solubility were carefully monitored for the experimental setup since neutral pH tends to form precipitates of
speciation and solubility were carefully monitored for the experimental setup since neutral pH tends to form precipitates of  -hydroxyl. In accordance with Jang et al. who studied schoepite [UO2(OH)2.H2O(c)] formation at pH 7,52 we kept
-hydroxyl. In accordance with Jang et al. who studied schoepite [UO2(OH)2.H2O(c)] formation at pH 7,52 we kept  concentration below 1 and 0.1 mM for electrophoresis and SPR experiments, respectively. All the experiments were performed within 1 day, from extemporaneous dilutions of the stock solution. The
concentration below 1 and 0.1 mM for electrophoresis and SPR experiments, respectively. All the experiments were performed within 1 day, from extemporaneous dilutions of the stock solution. The  stock solution was 100 mM
stock solution was 100 mM  diacetate, pH = 4.
diacetate, pH = 4.
Pronase from Streptomyces griseus (Calbiochem, La Jolla #537088) was freeze-dried, and CRP (Biospacific, Emeryville, #J81600135) was solubilized in 20 mM Tris-HCl (pH = 7.5), 2 mM CaCl2, 140 mM NaCl, and 0.05% NaN3. Monomeric CRP (mCRP) was prepared according to the urea method from Potempa et al.34
Unless otherwise indicated all chemical products were from Sigma-Aldrich (St-Quentin Fallavier, France) and used without further treatment. Pure water was provided by a DirectQ purification system (Millipore, Billerica).
Pronase assay
pronase/CRP ratios: 24 μg of a CRP solution at 1 mg/mL were dialyzed against 10 mM Tris-HCl and 150 mM NaCl, pH 7, using a Micro DispoDialyzer (Harvard Apparatus, Holliston). Four 30 μL CRP samples (200 μg/mL, 10 mM Tris-HCl, 150 mM NaCl, pH 7) were incubated with 1mM CaCl2 (1 h at RT), then digested (2 h at 37°C) with pronase [1 mg/mL in Tris-Buffered Saline (TBS)] at various protease/substrate ratios (1% to 10%, w/w).
Calcium protection: 1 mg/mL CRP solution was dialyzed in 25 mM Hepes, 75 mM NaCl, pH 7, then adjusted to 200 μg/mL. The protein solution was distributed (30 μL) and incubated (1 h at RT) with CaCl2 concentrations ranging from 1 to 10 mM. The samples were digested by pronase (5% w/w, 2 h, 37°C).
 protection: the preparation was identical to the previous calcium experiments, with
protection: the preparation was identical to the previous calcium experiments, with  concentrations ranging from 0.1 to 1 mM to prevent
concentrations ranging from 0.1 to 1 mM to prevent  hydrolysis and precipitation in pH 7 buffer.
hydrolysis and precipitation in pH 7 buffer.
Pronase reactions were stopped by cooling in ice and the digests were immediately analysed by SDS-PAGE, using a PhastSystem (GE Healthcare, Buc, France) with precast gradient gels PhastGel™ 8-25 and SDS buffer strips, according to the manufacturer's instructions. SeeBlue® Plus2 prestained standards were from Invitrogen (Cergy Pontoise, France).
SDS-PAGE analyses were carried out on duplicates from the different digests, and the gels were silver stained. SDS was added to each sample before heating (90°C, 10 min), but no DTT since it induces a smear on gels in the presence of  .
.
MALDI-TOF-MS assays
MALDI-TOF-MS spectra were recorded on a MALDI-TOF Biflex IV mass spectrometer (Bruker Daltonics) in positive ionization mode. Sinapinic acid prepared as saturated solution in 30% acetonitrile, 70% milli-Q water, and 0.1% trifluoroacetic acid was used as matrix for desalted unproteolyzed proteins. A CRP solution at 1 mg/mL was extensively dialyzed against 10 mM Tris-HCl and 150 mM NaCl, pH 7.4, using a Micro DispoDialyzer. Protein samples (500 μg/mL) were distributed in three glass vials (27 μL), with an added volume of 3 μL of water, CaCl2 (1 mM final concentration) or  (100 μM final concentration) before incubation (1 h, RT). Digestion (2 h at 37°C) with pronase (1 mg/mL solution in TBS) was at a protease/substrate ratio of 5% w/w. Finally, 30 μL of ethanol were added in each sample followed by sonication. Samples of 0.5 μL were spotted onto a Scout MALDI 384 target (Bruker Daltonics) using dried droplet method, with the addition of 0.5 μL of sinapinic acid. Spectra were acquired in linear mode (150–250 laser shots) in the m/z range from 5000 to 30,000. Higher masses were also scrutinized to check for the absence of multimeric CRP. Calibration was achieved using standard I from Bruker (insulin, ubiquitin, cytochrom C, myoglobin peaks), and the expected average mass of CRP (23047.32 Da).
(100 μM final concentration) before incubation (1 h, RT). Digestion (2 h at 37°C) with pronase (1 mg/mL solution in TBS) was at a protease/substrate ratio of 5% w/w. Finally, 30 μL of ethanol were added in each sample followed by sonication. Samples of 0.5 μL were spotted onto a Scout MALDI 384 target (Bruker Daltonics) using dried droplet method, with the addition of 0.5 μL of sinapinic acid. Spectra were acquired in linear mode (150–250 laser shots) in the m/z range from 5000 to 30,000. Higher masses were also scrutinized to check for the absence of multimeric CRP. Calibration was achieved using standard I from Bruker (insulin, ubiquitin, cytochrom C, myoglobin peaks), and the expected average mass of CRP (23047.32 Da).
ICP-MS assay
To load CRP with  , a 1 mg/mL CRP sample was first dialyzed against 200 mL 150 mM NaCl, 10 mM acetate buffer using a Micro DispoDialyzer (Harvard Apparatus, Holliston). Then the sample was diluted in the same buffer with added
, a 1 mg/mL CRP sample was first dialyzed against 200 mL 150 mM NaCl, 10 mM acetate buffer using a Micro DispoDialyzer (Harvard Apparatus, Holliston). Then the sample was diluted in the same buffer with added  diacetate to reach a final concentration before dialysis (10000 daltons cutoff) of 10 μM protein and 100 μM
diacetate to reach a final concentration before dialysis (10000 daltons cutoff) of 10 μM protein and 100 μM  . A sample with 100 μM
. A sample with 100 μM  without protein was also dialyzed in the same buffer as a reference. The buffer pH was adjusted to five to insure
without protein was also dialyzed in the same buffer as a reference. The buffer pH was adjusted to five to insure  solubility at a concentration up to 100 μM for an overnight run (see Supporting Information Figure S4 and Table SIII). Two 100 μL samples were then dialyzed (10000 daltons cutoff) overnight with 200 mL buffer without
solubility at a concentration up to 100 μM for an overnight run (see Supporting Information Figure S4 and Table SIII). Two 100 μL samples were then dialyzed (10000 daltons cutoff) overnight with 200 mL buffer without  . The CRP concentration was readjusted using a CRP extinction coefficient (mg/mL) of 1.818 using a Carry 300 spectrophotometer (Varian) for 280 nm absorbance measurements. Two hundred microliters of
. The CRP concentration was readjusted using a CRP extinction coefficient (mg/mL) of 1.818 using a Carry 300 spectrophotometer (Varian) for 280 nm absorbance measurements. Two hundred microliters of  loaded samples were diluted in 5% HNO3 before ICP-MS analysis (PQ Excell Option S, Thermoptek)
loaded samples were diluted in 5% HNO3 before ICP-MS analysis (PQ Excell Option S, Thermoptek)
SPR assays
Experiments were performed on a Biacore T100 instrument (GE Healthcare).  mass is above the reported sensitivity of this instrument, making
mass is above the reported sensitivity of this instrument, making  binding measurements possible. However, nonspecific binding and refractive index increment need to be taken into account for accurate measurements as detailed in this work.
binding measurements possible. However, nonspecific binding and refractive index increment need to be taken into account for accurate measurements as detailed in this work.
Free phosphorylcholine was from Sigma-Aldrich and phosphorylcholine conjugated to BSA (10 PC per BSA, functionalized on Tyr residues) was from Biosearch Technologies (Novato). PC-BSA, pCRP, and mCRP were immobilized on CM5 sensor chips using standard amine-coupling EDC/NHS chemistry followed by ethanolamine deactivation of activated CM-dextran groups. A blank flow cell was used as a reference on all chips.
The running buffer was a 10 mM Tris, 150 mM NaCl, and 50 μM CaCl2 solution without EDTA. No surfactant, such as Tween 20, was used in the running buffer since binding levels of CRP to PC-BSA in the presence of calcium were 10 times higher in the absence of detergent, with no measurable adverse effect on nonspecific binding. Calcium concentration was adjusted to 50 μM to maintain a CRP/calcium ratio slightly higher than that of the CRP long storage manufacturer solution (1 mg/mL CRP with 2 mM calcium) at CRP concentrations up to 1000 nM.
The regeneration buffer was 10 mM NaOH, 0.05% SDS, 0.5% Tween20 and 2mM EDTA, ensuring stable baseline, reproducible binding levels and minimal nonspecific binding, under all conditions.
The full SPR cycle of the uranyl ion capture experiments is given in Figure 4(B,C) and Supporting Information Figure S2. The stages were:  diacetate capture (120 sec, 10 μL/min.), dissociation/wash in running buffer (115 sec, 30 μL/min), then injection of 100 nM PC-BSA diluted in the running buffer with or without
diacetate capture (120 sec, 10 μL/min.), dissociation/wash in running buffer (115 sec, 30 μL/min), then injection of 100 nM PC-BSA diluted in the running buffer with or without  , dissociation and regeneration.
, dissociation and regeneration.
All SPR data were analyzed using Biacore T100 evaluation software version 1.1 (GE Healthcare). Fits were performed using an MW of 115 kDa for pCRP, 70 kDa for PC-BSA, and 23 kDa for mCRP.
The predicted maximum instrument responses for binding of a single ligand (RUpred)max were estimated using the standard formula (RUpred)max = RUM× (MWL/MWM) where RUM is the signal corresponding to the immobilized molecule, MWL is the molecular weight of the ligand or analyte (i.e., the circulating molecule) and MWM is the molecular weight of the immobilized molecule. Davis and Wilson46 report that correction of this formula to account for RII differences between immobilized molecule and ligand might be needed for avoid errors reaching a factor of 2 on stoichiometry predictions. We used a bare gold surface to estimate  RII value using SPR.53
RII value using SPR.53
Equation 2 was entered in Graphpad Prism version 4.00 (GraphPad Software, San Diego California, http://www.graphpad.com), which was used for nonlinear fitting of data.
Structures were drawn with DS Visualizer 2.0 (Accelrys Software) and rendered with POV-Ray 3.6 under Linux (Persistence of Vision Pty. 2004).
Acknowledgments
This work was partly supported by the programme "Toxicologie Nucléaire Environnementale" of CEA.
Glossary
Abbreviations:
- BSA-PC
phosphorylcholine hapten conjugated to bovin serum albumin
- CRP
C-reactive protein
- pCRP
pentameric CRP
- mCRP
monomeric CRP
- IUM
uranyl ion PDB identifier
- PC
phosphorylcholine (or phosphocholine)
- PDB
Protein Data Bank
- SMID
Small Molecule Interaction Database
- SPR
Surface Plasmon Resonance
References
- 1.Yamashita MM, Wesson L, Eisenman G, Eisenberg D. Where metal ions bind in proteins. Proc Natl Acad Sci USA. 1990;87:5648–5652. doi: 10.1073/pnas.87.15.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nayal M, Di Cera E. Predicting Ca(2+)-binding sites in proteins. Proc Natl Acad Sci USA. 1994;91:817–821. doi: 10.1073/pnas.91.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dudev T, Lim C. Principles governing Mg, Ca, and Zn binding and selectivity in proteins. Chem Rev. 2003;103:773–787. doi: 10.1021/cr020467n. [DOI] [PubMed] [Google Scholar]
- 4.Harding MM. The architecture of metal coordination groups in proteins. Acta Crystallogr D. 2004;60:849–859. doi: 10.1107/S0907444904004081. [DOI] [PubMed] [Google Scholar]
- 5.Hartsock WJ, Cohen JD, Segal DJ. Uranyl acetate as a direct inhibitor of DNA-binding proteins. Chem Res Toxicol. 2007;20:784–789. doi: 10.1021/tx600347k. [DOI] [PubMed] [Google Scholar]
-
6.Vidaud C, Gourion-Arsiquaud S, Rollin-Genetet F, Torne-Celer C, Plantevin S, Pible O, Berthomieu C, Quemeneur E. Structural consequences of binding of
 to apotransferrcan this protein account for entry of uranium into human cells? Biochemistry. 2007;46:2215–2226. doi: 10.1021/bi061945h. [DOI] [PubMed] [Google Scholar]
to apotransferrcan this protein account for entry of uranium into human cells? Biochemistry. 2007;46:2215–2226. doi: 10.1021/bi061945h. [DOI] [PubMed] [Google Scholar] - 7.Teulon JM, Parot P, Odorico M, Pellequer JL. Deciphering the energy landscape of the interaction uranyl-DCP with antibodies using dynamic force spectroscopy. Biophys J. 2008;95:L63–L65. doi: 10.1529/biophysj.108.141937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wegner SV, Boyaci H, Chen H, Jensen MP, He C. Engineering a uranyl-specific binding protein from NikR. Angew Chem Int Ed. 2009;48:2339–2341. doi: 10.1002/anie.200805262. [DOI] [PubMed] [Google Scholar]
- 9.Vidaud C, Dedieu A, Basset C, Plantevin S, Dany I, Pible O, Quemeneur E. Screening of human serum proteins for uranium binding. Chem Res Toxicol. 2005;18:946–953. doi: 10.1021/tx050038v. [DOI] [PubMed] [Google Scholar]
- 10.Basset C, Dedieu A, Guerin P, Quemeneur E, Meyer D, Vidaud C. Specific capture of uranyl protein targets by metal affinity chromatography. J Chromatogr A. 2008;1185:233–240. doi: 10.1016/j.chroma.2008.01.081. [DOI] [PubMed] [Google Scholar]
- 11.Dedieu A, Berenguer F, Basset C, Prat O, Quemeneur E, Pible O, Vidaud C. Identification of uranyl binding proteins from human kidney-2 cell extracts by immobilized uranyl affinity chromatography and mass spectrometry. J Chromatogr A. 2009;1216:5365–5376. doi: 10.1016/j.chroma.2009.05.023. [DOI] [PubMed] [Google Scholar]
- 12.Prat O, Berenguer F, Steinmetz G, Ruat S, Sage N, Quemeneur E. Alterations in gene expression in cultured human cells after acute exposure to uranium salt: involvement of a mineralization regulator. Toxicol in Vitro. 2010;24:160–168. doi: 10.1016/j.tiv.2009.07.035. [DOI] [PubMed] [Google Scholar]
- 13.Leggett RW. Basis for the ICRP's age-specific biokinetic model for uranium. Health Phys. 1994;67:589–610. doi: 10.1097/00004032-199412000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Le Clainche L, Vita C. Selective binding of uranyl cation by a novel calmodulin peptide. Environ Chem Lett. 2006;4:45–49. [Google Scholar]
- 15.Schymkowitz JW, Rousseau F, Martins IC, Ferkinghoff-Borg J, Stricher F, Serrano L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc Natl Acad Sci USA. 2005;102:10147–10152. doi: 10.1073/pnas.0501980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng H, Chen GT, Yang W, Yang JJ. Predicting calcium-binding sites in proteins—A graph theory and geometry approach. Proteins-Struct Funct Bioinformatics. 2006;64:34–42. doi: 10.1002/prot.20973. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Kirberger M, Qiu FS, Chen GT, Yang JJ. Towards predicting Ca2+-binding sites with different coordination numbers in proteins with atomic resolution. Proteins-Struct Funct Bioinformatics. 2009;75:787–798. doi: 10.1002/prot.22285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Vernon R, Lange O, Tyka M, Baker D. Prediction of structures of zinc-binding proteins through explicit modeling of metal coordination geometry. Protein Sci. 2010;19:494–506. doi: 10.1002/pro.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pible O, Guilbaud P, Pellequer JL, Vidaud C, Quemeneur E. Structural insights into protein-uranyl interaction: towards an in silico detection method. Biochimie. 2006;88:1631–1638. doi: 10.1016/j.biochi.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Barondeau DP, Getzoff ED. Structural insights into protein-metal ion partnerships. Curr Opin Struc Biol. 2004;14:765–774. doi: 10.1016/j.sbi.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Babor M, Greenblatt HM, Edelman M, Sobolev V. Flexibility of metal binding sites in proteins on a database scale. Proteins. 2005;59:221–230. doi: 10.1002/prot.20431. [DOI] [PubMed] [Google Scholar]
- 22.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snyder KA, Feldman HJ, Dumontier M, Salama JJ, Hogue CW. Domain-based small molecule binding site annotation. BMC Bioinformatics. 2006;7:152. doi: 10.1186/1471-2105-7-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bairoch A, Bougueleret L, Altairac S, Amendolia V, Auchincloss A, Puy GA, Axelsen K, Baratin D, Blatter MC, Boeckmann B, Bollondi L, Boutet E, Quintaje SB, Breuza L, Bridge A, Saux VBL, deCastro E, Ciampina L, Coral D, Coudert E, Cusin I, David F, Delbard G, Dornevil D, Duek-Roggli P, Duvaud S, Estreicher A, Famiglietti L, Farriol-Mathis N, Ferro S, Feuermann M, Gasteiger E, Gateau A, Gehant S, Gerritsen V, Gos A, Gruaz-Gumowski N, Hinz U, Hulo C, Hulo N, Innocenti A, James J, Jain E, Jimenez S, Jungo F, Junker V, Keller G, Lachaize C, Lane-Guermonprez L, Langendijk-Genevaux P, Lara V, Le Mercier P, Lieberherr D, Lima TD, Mangold V, Martin X, Michoud K, Moinat M, Morgat A, Nicolas M, Paesano S, Pedruzzi I, Perret D, Phan I, Pilbout S, Pillet V, Poux S, Pozzato M, Redaschi N, Reynaud S, Rivoire C, Roechert B, Sapsezian C, Schneider M, Sigrist C, Sonesson K, Staehli S, Stutz A, Sundaram S, Tognolli M, Verbregue L, Veuthey AL, Vitorello C, Yip L, Zuletta LF, Apweiler R, Alam-Faruque Y, Barrell D, Bower L, Browne P, Chan WM, Daugherty L, Donate ES, Eberhardt R, Fedotov A, Foulger R, Frigerio G, Garavelli J, Golin R, Horne A, Jacobsen J, Kleen M, Kersey P, Laiho K, Legge D, Magrane M, Martin MJ, Monteiro P, O'Donovan C, Orchard S, O'Rourke J, Patient S, Pruess M, Sitnov A, Whitefield E, Wieser D, Lin Q, Rynbeek M, di Martino G, Donnelly M, van Rensburg P, Wu C, Arighi C, Arminski L, Barker W, Chen YX, Crooks D, Hu ZZ, Hua HK, Huang HZ, Kahsay R, Mazumder R, McGarvey P, Natale D, Nikolskaya AN, Petrova N, Suzek B, Vasudevan S, Vinayaka CR, Yeh LS, Zhang J. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin ACR. Mapping PDB chains to UniProtKB entries. Bioinformatics. 2005;21:4297–4301. doi: 10.1093/bioinformatics/bti694. [DOI] [PubMed] [Google Scholar]
- 26.Domingues FS, Rahnenfuhrer J, Lengauer T. Conformational analysis of alternative protein structures. Bioinformatics. 2007;23:3131–3138. doi: 10.1093/bioinformatics/btm499. [DOI] [PubMed] [Google Scholar]
- 27.Lesk AM, Chothia C. How different amino-acid-sequences determine similar protein structures—Structure and evolutionary dynamics of the globins. J Mol Biol. 1980;136:225–270. doi: 10.1016/0022-2836(80)90373-3. [DOI] [PubMed] [Google Scholar]
- 28.Gewurz H, Zhang XH, Lint TF. Structure and function of the pentraxins. Curr Opin Immunol. 1995;7:54–64. doi: 10.1016/0952-7915(95)80029-8. [DOI] [PubMed] [Google Scholar]
- 29.Volanakis JE. Human C-reactive proteexpression, structure, and function. Mol Immunol. 2001;38:189–197. doi: 10.1016/s0161-5890(01)00042-6. [DOI] [PubMed] [Google Scholar]
- 30.Black S, Kushner I, Samols D. C-reactive protein. J Biol Chem. 2004;279:48487–48490. doi: 10.1074/jbc.R400025200. [DOI] [PubMed] [Google Scholar]
- 31.Ramadan MA, Shrive AK, Holden D, Myles DA, Volanakis JE, DeLucas LJ, Greenhough TJ. The three-dimensional structure of calcium-depleted human C-reactive protein from perfectly twinned crystals. Acta Crystallogr D Biol Crystallogr. 2002;58:992–1001. doi: 10.1107/s0907444902005693. [DOI] [PubMed] [Google Scholar]
- 32.Shrive AK, Cheetham GM, Holden D, Myles DA, Turnell WG, Volanakis JE, Pepys MB, Bloomer AC, Greenhough TJ. Three dimensional structure of human C-reactive protein. Nat Struct Biol. 1996;3:346–354. doi: 10.1038/nsb0496-346. [DOI] [PubMed] [Google Scholar]
- 33.Thompson D, Pepys MB, Wood SP. The physiological structure of human C-reactive protein and its complex with phosphocholine. Structure. 1999;7:169–177. doi: 10.1016/S0969-2126(99)80023-9. [DOI] [PubMed] [Google Scholar]
- 34.Potempa LA, Siegel JN, Fiedel BA, Potempa RT, Gewurz H. Expression, detection and assay of a neoantigen (Neo-Crp) associated with a free, human C-Reactive protein subunit. Mol Immunol. 1987;24:531–541. doi: 10.1016/0161-5890(87)90028-9. [DOI] [PubMed] [Google Scholar]
- 35.Dong AC, Caughey WS, Duclos TW. Effects of calcium, magnesium, and phosphorylcholine on secondary structures of human C-reactive protein and serum amyloid-P component observed by infrared-spectroscopy. J Biol Chem. 1994;269:6424–6430. [PubMed] [Google Scholar]
- 36.Wu Y, Ji SR, Wang HW, Sui SF. Study of the spontaneous dissociation of rabbit C-reactive protein. Biochemistry-Moscow. 2002;67:1377–1382. doi: 10.1023/a:1021862027061. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka T, Robey FA. A new sensitive assay for the calcium-dependent binding of C-Reactive protein to phosphorylcholine. J Immunol Methods. 1983;65:333–341. doi: 10.1016/0022-1759(83)90128-x. [DOI] [PubMed] [Google Scholar]
- 38.Lee RT, Takagahara I, Lee YC. Mapping the binding areas of human C-reactive protein for phosphorylcholine and polycationic compounds—relationship between the two types of binding sites. J Biol Chem. 2002;277:225–232. doi: 10.1074/jbc.M106039200. [DOI] [PubMed] [Google Scholar]
- 39.Heegaard NHH, Robey FA. A capillary electrophoresis-based assay for the binding of Ca2+ and phosphorylcholine to human C-Reactive protein. J Immunol Methods. 1993;166:103–110. doi: 10.1016/0022-1759(93)90333-3. [DOI] [PubMed] [Google Scholar]
- 40.Christopeit T, Gossas T, Danielson UH. Characterization of Ca2+ and phosphocholine interactions with C-reactive protein using a surface plasmon resonance biosensor. Anal Biochem. 2009;391:39–44. doi: 10.1016/j.ab.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 41.Kinoshita CM, Ying SC, Hugli TE, Siegel JN, Potempa LA, Jiang H, Houghten RA, Gewurz H. Elucidation of a protease-sensitive site involved in the binding of calcium to C-Reactive protein. Biochemistry. 1989;28:9840–9848. doi: 10.1021/bi00451a044. [DOI] [PubMed] [Google Scholar]
- 42.Gotschli Ec, Edelman GM. Binding properties and specificity of C-reactive protein. Proc Natl Acad Sci USA. 1967;57:706–712. doi: 10.1073/pnas.57.3.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson JK, Stroud RM, Volanakis JE. Studies on binding specificity of human C-reactive protein for phosphorylcholine. Fed Proc. 1978;37:1495. [Google Scholar]
- 44.Agrawal A, Xu YY, Ansardi D, Macon KJ, Volanakis JE. Probing the phosphocholine-binding site of human C-Reactive protein by site-directed mutagenesis. J Biol Chem. 1992;267:25352–25358. [PMC free article] [PubMed] [Google Scholar]
- 45.Foghandersen N, Altura BM, Altura BT, Siggaardandersen O. Composition of Interstitial Fluid. Clin Chem. 1995;41:1522–1525. [PubMed] [Google Scholar]
- 46.Davis TM, Wilson WD. Determination of the refractive index increments of small molecules for correction of surface plasmon resonance data. Anal Biochem. 2000;284:348–353. doi: 10.1006/abio.2000.4726. [DOI] [PubMed] [Google Scholar]
- 47.Gazin V, Kerdine S, Grillon G, Pallardy M, Raoul H. Uranium induces TNF alpha secretion and MAPK activation in a rat alveolar macrophage cell line. Toxicol Appl Pharm. 2004;194:49–59. doi: 10.1016/j.taap.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 48.Monleau M, De Meo M, Paquet F, Chazel V, Dumenil G, Donnadieu-Claraz M. Genotoxic and inflammatory effects of depleted uranium particles inhaled by rats. Toxicol Sci. 2006;89:287–295. doi: 10.1093/toxsci/kfj010. [DOI] [PubMed] [Google Scholar]
- 49.Van Horn JD, Huang H. Uranium(VI) bio-coordination chemistry from biochemical, solution and protein structural data. Coord Chem Rev. 2006;250:765–775. [Google Scholar]
- 50.Pible O, Imbert G, Pellequer JL. INTERALIGN: interactive alignment editor for distantly related protein sequences. Bioinformatics. 2005;21:3166–3167. doi: 10.1093/bioinformatics/bti474. [DOI] [PubMed] [Google Scholar]
- 51.Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, Bantoft K, Betel D, Bobechko B, Boutilier K, Burgess E, Buzadzija K, Cavero R, D'Abreo C, Donaldson I, Dorairajoo D, Dumontier MJ, Dumontier MR, Earles V, Farrall R, Feldman H, Garderman E, Gong Y, Gonzaga R, Grytsan V, Gryz E, Gu V, Haldorsen E, Halupa A, Haw R, Hrvojic A, Hurrell L, Isserlin R, Jack F, Juma F, Khan A, Kon T, Konopinsky S, Le V, Lee E, Ling S, Magidin M, Moniakis J, Montojo J, Moore S, Muskat B, Ng I, Paraiso JP, Parker B, Pintilie G, Pirone R, Salama JJ, Sgro S, Shan T, Shu Y, Siew J, Skinner D, Snyder K, Stasiuk R, Strumpf D, Tuekam B, Tao S, Wang Z, White M, Willis R, Wolting C, Wong S, Wrong A, Xin C, Yao R, Yates B, Zhang S, Zheng K, Pawson T, Ouellette BFF, Hogue CWV. The biomolecular interaction network database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jang JH, Dempsey BA, Burgos WD. Solubility of schoepite: comparison and selection of complexation constants for U(VI) Water Res. 2006;40:2738–2746. doi: 10.1016/j.watres.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 53.Tumolo T, Angnes L, Baptista MS. Determination of the refractive index increment (dn/dc) of molecule and macromolecule solutions by surface plasmon resonance. Anal Biochem. 2004;333:273–279. doi: 10.1016/j.ab.2004.06.010. [DOI] [PubMed] [Google Scholar]