Abstract
The HIV gp41 protein catalyzes fusion between viral and host cell membranes and its apolar N-terminal region or “fusion peptide” binds to the host cell membrane and plays a key role in fusion. “HFP” is a construct containing the fusion peptide sequence, induces membrane vesicle fusion, and is an important fusion model system. Earlier solid-state NMR (SSNMR) studies showed that when HFP is associated with membranes with ~30 mol% cholesterol, the first sixteen residues have predominant β strand secondary structure and a fraction of the strands form antiparallel β sheet structure with residue 16→1/1→16 or 17→1/1→17 registries of adjacent strands. In some contrast, other SSNMR and infrared studies have been interpreted to support a large fraction of approximately in-register parallel registry of adjacent strands. However, the samples had extensive isotopic labeling and other structural models were also consistent with the data. The present SSNMR study uses sparse labeling schemes that reduce ambiguity in the determination of the fraction of HFP molecules with parallel β registry. Quantitative analysis of the data shows that the parallel fraction is at most 0.15 with a much greater fraction of antiparallel 16→1/1→16 and 17→1/1→17 registries. These data strongly support a model of HFP-induced vesicle fusion caused by antiparallel rather than parallel registries and provide insight into the arrangement of gp41 molecules during HIV/host cell fusion. This study is an example of quantitative determination of a complex structural distribution by SSNMR including experimentally-validated inclusion of natural abundance contributions to the SSNMR data.
Keywords: HIV, fusion, peptide, parallel, antiparallel, solid-state NMR
AIDS is caused by membrane-enveloped human immunodeficiency virus (HIV) which infects host cells by fusion, i.e. joining of viral and host cell membranes (1). Fusion is facilitated by gp41 which is an integral HIV membrane protein. The N-terminal ~175-residue gp41 ectodomain lies outside the virus and X-ray crystal and liquid-state nuclear magnetic resonance (LSNMR) structures of soluble regions of the ectodomain have shown molecular trimers (1–5). These structures lacked the ~25-residue N-terminal “fusion peptide” region which plays a key role in fusion and infection as evidenced by inhibition of fusion and infection when gp41 had mutations within the fusion peptide region (1, 6, 7). Peptides with the fusion peptide sequence are denoted HFPs and have been studied as model fusion systems because they induce vesicle fusion and because their mutation-fusion activity relationships are similar to those of in vivo fusion and infection (1, 8–10).
The HFP structure-function literature includes NMR data showing random coil structure for HFP in aqueous solution (11, 12). Solid-state nuclear magnetic resonance (SSNMR) has shown predominant β sheet structure for residues 1–16 of membrane-associated HFP where the membranes contained ~30 mol% cholesterol which is comparable to the mol% cholesterol of membranes of HIV and host cells of HIV (13–16). A fluorescence and infrared (IR) study reported the time-resolved courses of HFP structural changes and the intervesicle lipid mixing function following addition of a HFP solution to a membrane vesicle solution (17). The experimental rates (Rs) were ordered RHFP membrane binding > RHFP β sheet formation > Rlipid mixing and were consistent with the sequence: (1) random coil HFPs bind to a membrane vesicle; (2) HFP structure changes to oligomeric β sheet; and finally (3) vesicle fusion.
The biological relevance of HFP oligomers is further supported by the molecular trimer structure of soluble regions of the gp41 ectodomain (3–5). The region between residues T25 and G85 of each molecule was a continuous helix and the helices of the different molecules formed a parallel coiled-coil. The fusion peptide region was not included in the protein constructs for these structures but would be N-terminal of residue T25. A C-terminally cross-linked HFP trimer (HFPtr) was therefore synthesized to mimic the close proximity of the three T25 residues in the coiled-coil. Relative to HFP monomer, HFPtr induced membrane vesicle fusion with ~40-fold faster rate which supported the functional significance of the trimer (18). Although both the monomer and trimer formed β sheet oligomers in membranes with cholesterol, HFPtr is more deeply inserted which correlates with greater membrane perturbation and a reduction of the vesicle fusion activation energy (19). The in vivo importance of fusion peptide oligomers was also demonstrated by dominant inhibition of fusion and infection in viruses and cells for which a small fraction of the gp41 had the V2E point mutation in the fusion peptide region (7, 20). Analyses of these data supported the involvement of multiple gp41 trimers and fusion peptides in fusion (21). Electron micrographs of virus-cell contacts have also been interpreted to show multiple gp41 trimers at the contact site (22). Functional importance of fusion peptide trimers has also been demonstrated for fusion peptides of other viruses (23, 24).
Because of the aforementioned functional significance of HIV fusion peptide oligomers, there has been effort to elucidate the distribution of structures of membrane-associated HFP oligomers. SSNMR has played a key role in this effort in particular for samples prepared in a manner similar to that of fusion assays with addition of an aqueous fusion peptide solution to a membrane vesicle solution (14). Appendage of a C-terminal lysine tag to HFP greatly reduced HFP aggregation in aqueous solution and allowed separation of pelleted fused vesicles with bound HFP from unbound HFP in the supernatant (12, 18, 25). HFP/lipid binding was supported by SSNMR detection of a HFP A1 13CO(carbonyl)-lipid 31P distance of ~5 Å (19). For membrane-associated HFP, the 13C chemical shifts derived from an unambiguous assignment were consistent with a fully extended β strand conformation for residues between A1 and G16 (15). Detection of intermolecular 13C-13C and 13C-15N distances of ~5 Å supported β sheet oligomer/aggregate structure and the A1 13CO-lipid 31P contact and other data suggest that the number of molecules in the oligomer is small (15, 19, 26).
The present work focuses on quantitative determination of populations of specific β sheet registries. The clearest information to-date on this topic has been a SSNMR experiment on membrane-associated HFP with an A14 13CO label and a G3 15N label whose separation (rCN) was >20 Å along a single β strand (15). SSNMR can detect labeled 13CO-15N dipolar coupling (dCN) where dCN = 3109/rCN3 with d in Hz and r in Å. The minimum detectable dCN≈ 10 Hz correlates with rCN ≈ 7 Å so that detectable dCN in this sample were necessarily ascribed to interrather than intramolecular 13CO-15N proximity. SSNMR detection of d > 30 Hz strongly supported a significant fraction of molecules with intermolecular A14-G3 hydrogen bonding and labeled rCN of 4.1 and 5.5 Å, i.e. 16→1/1→16 antiparallel β sheet registry. Fig. 1c displays this registry with isotopic labeling from the present study and not the earlier study. Detection of similarly large d in an A14 13CO/I4 15N HFP sample supported a fraction of 17→1/1→17 antiparallel registry.
Figure 1.
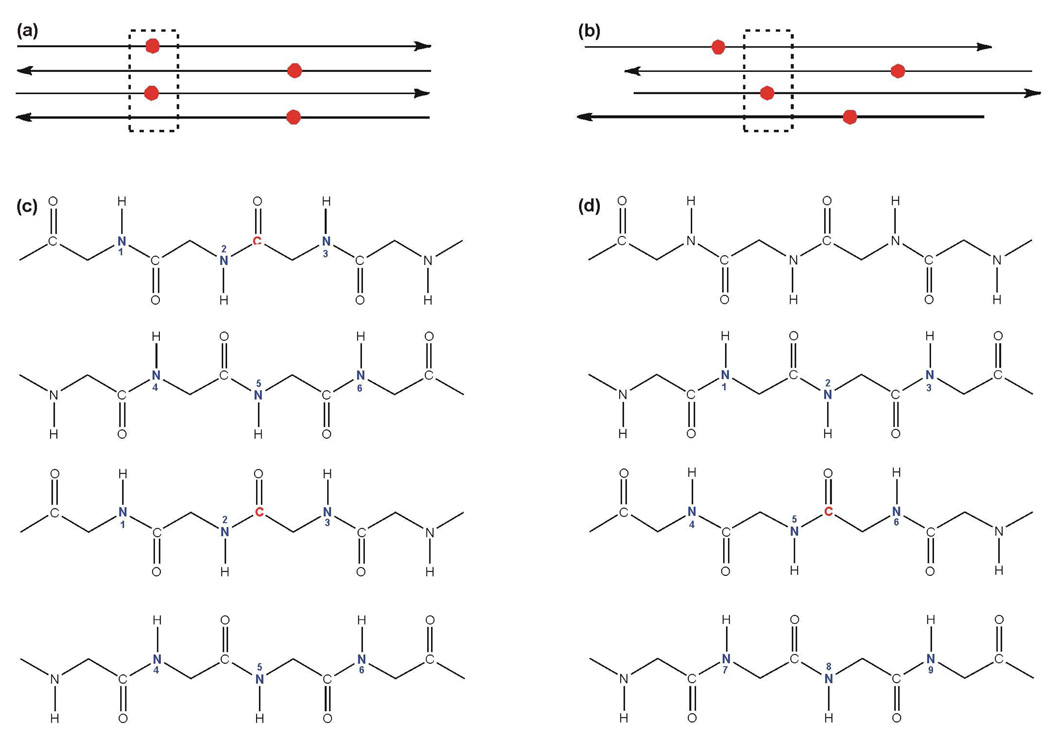
(a) HFPs where red and blue correspond to 13CO and 15N labeled residues, respectively. (b) HFP-NC, HFP-P, HFP-A, and HFP-AP were SSNMR samples which each contained a mixture of 13CO and 15N labeled peptides in a 1:2 mol ratio. The HFP-NC sample was a mixture of HFP-F8 and HFP-A6L7 that had been lyophilized separately. The other samples were membrane-associated HFPs that formed β sheet structure with a molecular mixture of 13CO and 15N labeled peptides in the sample. (c) Registries probed by the SSNMR REDOR experiments and labeled 13CO/labeled 15N proximities for the membrane-associated HFPs in these registries. Consideration of residue 1→16 or 1→17 registries is based on the fully extended conformation of this HFP region. For parallel sheets, there is CO(residue i) – HN(residue i+1) hydrogen bonding of adjacent strands.
At most half of the membrane-associated HFP molecules were in the 16→1/1→16 or 17→1/1→17 registries, i.e. a large fraction of the molecules were in registries not detected in either the A14 13CO/G3 15N or A14 13CO/I4 15N labeled samples. Because of the close proximity of the T25 residues of the three molecules of the gp41 trimer, a reasonable hypothesis for a populated HFP registry is in-register parallel β sheet, i.e. 1→17/1→17 in Fig. 1c. An earlier SSNMR study attempted to test this hypothesis using samples each containing an equimolar mixture of two labeled HFPs, one with three sequential backbone 13CO labels and the other with three sequential backbone 15N labels (27). Detection of an average dCN > 10 Hz for a G5-L7 13CO/G5-L7 15N sample and a F11-G13 13CO/F11-G13 15N sample were consistent with a fraction of in-register parallel HFP molecules. However, because the samples were extensively labeled, the data were also consistent with other parallel or antiparallel registries. In addition, the data reflected an average of many intermolecular dCNs so it was not possible to determine the fraction of molecules with a particular registry. There have also been efforts to detect in-register parallel structure using SSNMR measurement of intermolecular 13C-13C dipolar couplings (dCCs) where dCC = 7710/rCC3 with dCC in Hz and rCC in Å. For HFP with a single 13CO label and inregister parallel structure, the labeled interstrand rCC ≈ 5 Å with dCC ≈ 70 Hz (28, 29). These parameters will be independent of the residue that is 13CO labeled. For membrane-associated HFP with F8 13CO, a best-fit dCC ≈ 70 Hz was detected whereas for membrane-associated HFPtr, dCC depended on the position of the labeled 13CO residue with a range of 10–60 Hz (30, 31). This residue dependence argued against a major fraction of in-register parallel structure in HFPtr.
There was also an IR spectroscopy effort to distinguish between the 1→17/1→17 parallel and 16→1/1→16 antiparallel registries using samples that contained backbone 13CO labeling at either: (1) A1 to V3, G5 to I9; (2) F8 to G16; or (3) A1 to V3, G5 to G16 (32). The IR wavenumbers and intensities of different samples were interpreted to support a large fraction of parallel structure and little antiparallel structure. However, in our view, the extensive labeling of the IR samples precluded quantitation of specific registries and greater support for this argument is provided in the Discussion section.
The present paper reports a determination of the fraction of parallel structure in membrane-associated HFP oligomers. We were motivated to study this question because of: (1) the functional significance of HIV fusion peptide oligomers; and (2) the existing undefinitive and conflicting data and interpretations relevant to this question. As part of this effort, we developed a model to quantify effects of natural abundance 13C and 15N nuclei on SSNMR measurements of dCN and experimentally validated this model.
MATERIALS AND METHODS
Materials
9-fluorenylmethoxycarbonyl (FMOC) protected amino acids and FMOC-Ala-Wang resin were purchased from Peptides International (Louisville, KY). Isotopically labeled amino acids were purchased from Cambridge Isotopes (Andover, MA) and were FMOC-protected using literature methods (33). Fig. 1a shows the labeled HFPs. The 23 N-terminal residues (AVGIGALFLGFLGAAGSTMGARS) are a consensus sequence of the gp41 fusion peptide, non-native W24 is an A280 chromophore and the non-native lysines greatly reduced HFP aggregation in aqueous solution prior to membrane binding (12, 18). This ensured that membrane-associated β sheet oligomers/aggregates were formed after membrane binding.
HFP was manually synthesized and then cleaved from the resin for three hours in a solution of trifluoroacetic acid (TFA):water:anisole:thioanisole:ethanedithiol in a 90:5:2:2:2 volume ratio. After precipitation with cold diethyl ether, centrifugation, and dissolution of the pellet in water, crude HFP was purified by reversed-phase high-performance liquid chromatography with a semi-preparative C4 column and a water-acetonitrile gradient containing 0.1% TFA. Acetonitrile and TFA were removed with nitrogen gas and water was then removed by lyophilization. HFP purity was >95% as determined by mass spectrometry. HFP amounts were quantified using A280 of aqueous solutions of HFP with ε = 5600 M−1cm−1.
SSNMR samples
As shown in Fig. 1b, each sample contained a 13CO and a 15N labeled peptide in a 1:2 mol ratio. Three samples contained membrane-associated HFP β sheet oligomers/aggregates that were each a statistical mixture of 13CO and 15N labeled HFPs. Detection of substantial dCN by SSNMR indicated proximity of the labeled 13CO and 15N nuclei on adjacent strands and was used to estimate the fractional populations of specific registries as detailed below. As shown in Fig. 1c, the HFP-P sample was designed to detect parallel 1→17/1→17 and 2→17/1→16 registries, the HFP-A sample was designed to detect previously observed antiparallel 16→1/1→16 and 17→1/1→17 registries, and the HFP-AP sample was designed to detect both parallel 1→17/1→17 and 2→17/1→16 registries as well as antiparallel 16→1/1→16 and 17→1/1→17 registries.
In addition to the potential proximity of labeled 13CO and 15N nuclei, there will always be proximity between labeled 13CO and some natural abundance (n.a.) 15N nuclei as well as proximity between some n.a. 13CO and labeled 15N nuclei. These proximities will contribute to the dCN detected in the SSNMR experiment and should be included in the data modeling. Quantitative understanding of these proximities required a negative control (HFP-NC) sample with: (1) the same relative fractions of labeled 13CO, 15N, and n.a. sites as the HFP-P, HFP-A, and HFP-AP samples; and (2) labeled 13CO – labeled 15N rCN that are much greater than the REDOR detection limit of ~7 Å. One possibility was a sample made like HFP-P, HFP-A, and HFP-AP but with labels at sites that do not form intermolecular hydrogen bonds. This possibility was not pursued because the distribution of registries of membrane-associated HFP is not yet well-defined. Instead, the HFP-NC sample was a physical mixture of lyophilized HFP-F8 (5.0 mg) and HFP-A6L7 (10.0 mg) without any membrane. Each peptide was lyophilized separately and the two peptides were then mixed in the solid phase to form a uniform physical mixture. Water and membrane were not added to the physical mixture so that the labeled 13COs and 15Ns remained much farther apart than the 7 Å REDOR detection limit. Although there were populations of β sheet as well as α helical lyophilized peptides in the HFP-NC sample, each population yielded very similar (ΔS/S0) – see Results section for further details.
For the HFP-P, HFP-A, and HFP-AP samples, the membrane composition was 1,2-di-O-tetradecyl-sn-glycero-3-phosphocholine (DTPC) lipid, 1,2-di-O-tetradecyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (DTPG) lipid, and cholesterol in a 8:2:5 mol ratio. This composition reflected the large amount of choline lipid and fractions of negatively charged lipid and cholesterol in membranes of host cells of HIV (16). Ether- rather than more physiologically abundant ester-linked lipids were used because the latter have two COs/molecule that would contribute substantial n.a. 13CO signal. Bilayer phase is retained for ether-linked lipids with cholesterol and with HFP (34–36). In addition, membrane-associated HFP has predominant β sheet structure in either ester-linked lipid + cholesterol or ether-linked lipid + cholesterol compositions (30).
Samples were prepared by first dissolving DTPC (40 µmol), DTPG (10 µmol), and cholesterol (25 µmol) in chloroform and removing the chloroform with nitrogen gas and vacuum. The lipid film was suspended in 2 mL of 5 mM HEPES buffer at pH 7.0 with 0.01% NaN3 preservative. The suspension was homogenized with ten freeze-thaw cycles and large unilamellar vesicles were formed by extrusion through a 100 nm diameter polycarbonate filter (Avestin, Ottawa, ON). A separate solution was prepared with 13CO-labeled HFP (3.0 mg) and 15N labeled HFP (6.0 mg) in HEPES buffer (32 mL). The HFP solution was added drop-wise to the vesicle solution and the combined solution was gently stirred overnight. Ultracentrifugation at ~150000g for four hours pelleted membranes with bound HFP while unbound HFP remained in the supernatant (14). The pellet was lyophilized, transferred to the SSNMR rotor, and rehydrated with 30 µL H2O (37). The validity of the lyophilization/rehydration approach was evidenced by peak 13CO chemical shifts that were within 0.6 ppm of those of samples that were not lyophilized (15).
SSNMR experiments
Data were collected on a 9.4 T spectrometer (Varian Infinity Plus, Palo Alto, CA) using a triple resonance MAS probe equipped for 4.0 mm rotors and tuned to 13C, 1H, and 15N nuclei at respective frequencies of 100.8, 400.8, and 40.6 MHz. The 13C chemical shift was externally referenced to the methylene resonance of adamantane at 40.5 ppm and the 13C transmitter was set to 153 ppm. The 13CO-15N dipolar coupling (dCN) was probed with the rotational-echo double-resonance (REDOR) experiment with typical parameters: (1) 52 kHz 1H π/2 pulse; (2) 2.2 ms cross-polarization with 74 kHz 1H field and 83–98 kHz ramped 13C field; (3) dephasing period of duration τ for which the “S0” and “S1” acquisitions had 60 kHz 13C π pulses at the end of each rotor cycle except the last cycle, and the S1 acquisitions additionally had 59 kHz 15N π pulses in the middle of each rotor cycle; and (4) 13C detection (30, 38). The MAS frequency was 10 kHz, the recycle delay was 2 s, 88 kHz TPPM 1H decoupling was applied during the dephasing and detection periods, and XY-8 phase cycling was applied to the 13C π pulses and to the 15N π pulses (39, 40). Experiments were calibrated using a lyophilized helical peptide with sequence AEAAAKEAAAKEAAAKA, N-terminal acetylation and C-terminal amidation, and A9 13CO and A13 15N labels (30, 41). The labeled rCN ≈ 4.1 Å corresponds to dCN ≈ 45 Hz. Samples were typically cooled by nitrogen gas at −50 °C to enhance 13CO signal and reduce motional averaging of dCN (42). The typical difference between 13C shift in cooled and uncooled membrane-associated HFP samples is ≤ 0.5 ppm and indicates little variation in secondary structure with temperature (26). For each sample, data were collected for τ = 2.2, 8.2, 16.2, 24.2, 32.2, 40.2, and 48.2 ms.
The 13CO-15N dipolar coupling, dCN, is detected by reduction of 13CO signal intensity in the S1 spectrum relative to the S0 spectrum with greater reduction at increased τ. These intensities were also denoted S1 and S0 and were determined from integration over a shift range that encompassed most of the 13CO signal. A range of 8 ppm was used for HFP-NC spectra and a range of 5 ppm was used for HFP-P, HFP-A, and HFP-AP spectra. The normalized dephasing:
| (1) |
and its standard deviation:
| (2) |
where σS0 and σS1 were the experimental root-mean-squared deviations of the spectral intensities derived from 12 regions of the spectrum that did not include spectral features (43).
Experimental dephasing of a membrane-associated HFP sample was modeled as a sum of S0 and a sum of S1 signals of different spin geometries of 13CO and 15N nuclei where the geometries reflected statistical distributions of n.a. 13CO and 15N nuclei as well as geometries of (1) 1→17/1→17 and 2→17/1→16 parallel adjacent strand registries; (2) 16→1/1→16 and 17→1/1→17 antiparallel registries; and (3) other “X” registries where all labeled rCN > 7 Å and S1 = S0. The notation (ΔS/S0)sim will be generally used for simulated (ΔS/S0) and can refer to a particular spin geometry or to the population weighted sum using calculations from different spin geometries. For the former case, the (S1/S0)sim ≡ γ were calculated using the SIMPSON program with input parameters that included dCNs as well as Euler angles in a fixed crystal frame for each 13CO-15N vector and for the 13CO chemical shift anisotropy (CSA) principal axis system (44, 45). These input parameters were calculated by the SIMMOL program using 13CO and 15N coordinates from a region of a high-resolution crystal structure with the appropriate structural motif, e.g. parallel β sheet (46). Coordinates were obtained from the following Protein Data Bank (PDB) files: 1JK3, 1IGD, 1NKI, 2E4T, 1CEX, 1MNZ, and 2IWW. For each spin geometry, (S1/S0)sim was the average of ten different SIMPSON calculations and each calculation was based on input parameters from a different set of atomic coordinates. The 13CO CSA principal values of 247, 176, and 99 ppm were inputs to the SIMPSON calculations and 1Hs and relaxation were not considered.
RESULTS
Chemical shift and conformational distributions
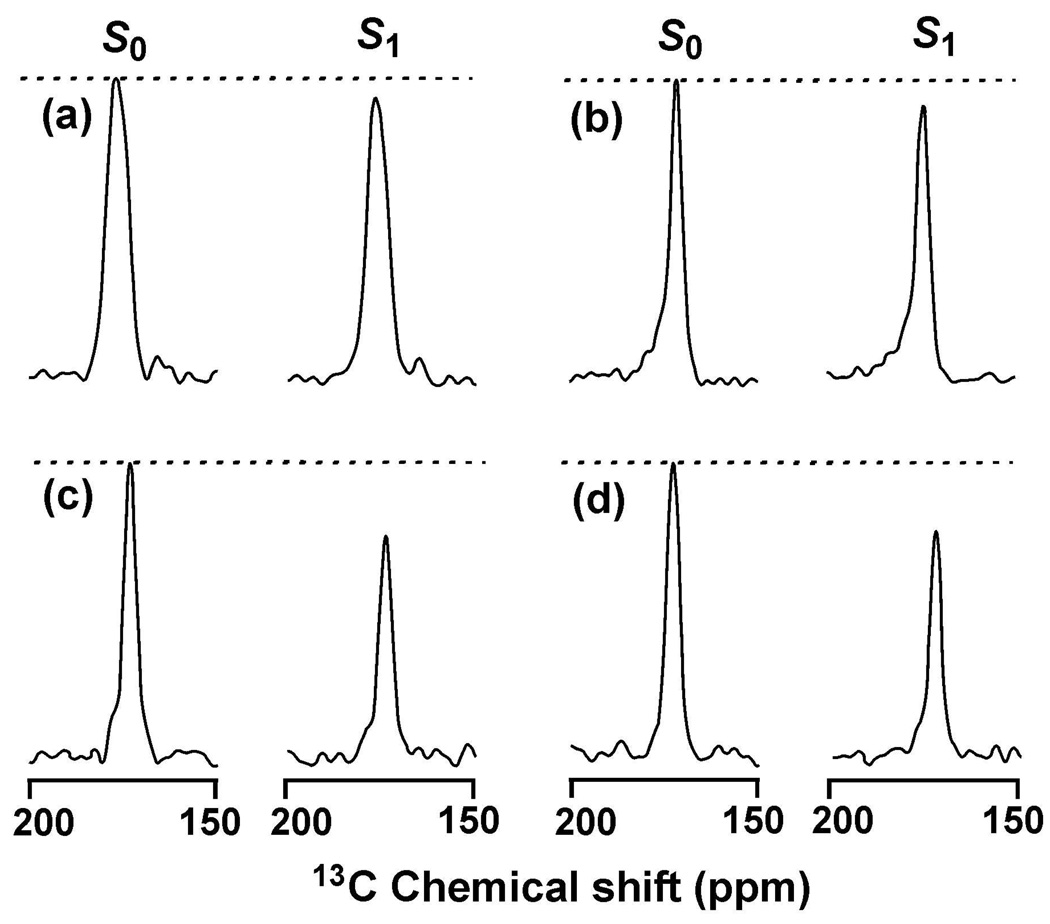
Fig. 2 displays REDOR S0 and S1 13C SSNMR spectra for τ = 32.2 ms. Each S0 spectrum has a ~50% contribution from the labeled 13CO and ~50% contribution from n.a. 13COs of the unlabeled residues. The full-width at half-maximum linewidths of the membrane-associated HFP samples in Fig. 2b–d are 3–4 ppm and indicate a distinct secondary structure. For the HFP-AP sample with F8 13CO label, the peak 13CO shift of 173 ppm is the same as was observed for F8 13CO of HFP in known β strand conformation and is very different from the 178 ppm shift observed in α helical conformation (15, 30). For the HFP-P and HFP-A samples with L12 13CO label, the 174 ppm peak shift is also the same as β strand HFP and different from the 179 ppm shift of Leu in helical HFP (30, 47). Overall, the shifts and linewidths are consistent with the fully extended conformation that has been observed for the first sixteen residues of HFP associated with membranes with biologically relevant cholesterol content (15).
Figure 2.
REDOR S0 and S1 13C SSNMR spectra at 32.2 ms dephasing time for (a) HFP-NC, (b) HFP-P, (c) HFP-A, or (d) HFP-AP. Each spectrum was processed with 200 Hz line broadening and baseline correction and was the sum of: (a) 38624; (b) 23488; (c) 24914; or (d) 14240 scans. Relatively narrow 13CO signals were observed in the HFP-P, HFP-A, and HFP-AP samples because the HFPs were membrane-associated with predominant β sheet conformation at the labeled 13CO site. A broader 13CO signal was observed in the HFP-NC sample because there was no membrane and there were populations of lyophilized HFP with either α helical or β sheet conformation at the labeled 13CO site.
The linewidth of the lyophilized HFP-NC sample with F8 13CO label is ~7 ppm and correlates with a broad distribution of secondary structures that is also evidenced by a 176 ppm peak 13CO shift that is midway between typical Phe helical and β strand shifts (48).
Qualitative analysis of REDOR data
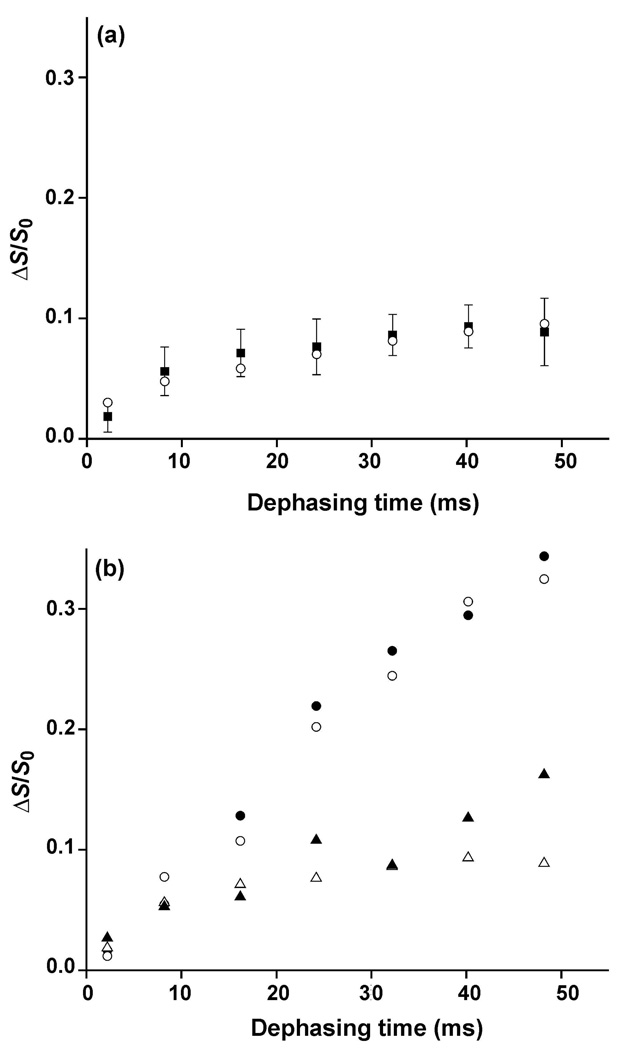
Relative to the S0 signals, there is attenuation in the S1 signals of 13COs within ~7 Å of 15Ns and the associated ΔS/S0 normalized dephasing increased with dephasing time, Fig. 2 and Fig. 3. Because of the physical separation of the 13CO and 15N labeled HFPs in HFP-NC, the S1 attenuation and (ΔS/S0)exp of this sample reflected F8 13CO-n.a. 15N and n.a. 13CO-A6,L7 15N proximities but not F8 13CO-A6,L7 15N proximity, Figs. 2a and 3a. There was similar S1 attenuation and (ΔS/S0)exp in HFP-P, Figs. 2b and 3b, which demonstrated that there was little L12 13CO-G13,A14 15N proximity in HFP-P and only a small fraction of parallel 1→17/1→17 and 2→17/1→16 registries. There was much larger S1 attenuation and (ΔS/S0)exp for the HFP-A sample, Figs. 2c and 3b, which indicated significant L12 13CO-G5,A6 15N proximity and therefore a substantial fraction of antiparallel 16→1/1→16 and 17→17/1→17 registries. Comparably large S1 attenuation and (ΔS/S0)exp were observed for the HFP-AP sample, Figs. 2d and 3b. The similarity of the HFP-NC and HFP-P data and the similarity of the HFP-A and HFP-AP data were consistent with ascribing F8 13CO-L9,G10 15N proximity in HFP-AP to antiparallel 16→1/1→16 and 17→1/1→17 registries rather than parallel 1→17/1→17 and 2→17/1→16 registries. Detection of a substantial fraction of these antiparallel registries is consistent with earlier SSNMR data for sparsely labeled HFP (15). Detection of only a small fraction of parallel registries is a new result and disagrees with previous interpretations of SSNMR and IR data for samples with extensive labeling (27, 32). These new data highlight the importance of sparse labeling to reduce interpretational ambiguity for systems with a structural distribution like membrane-associated HFP.
Figure 3.
(a) Plot of REDOR (ΔS/S0)exp (filled squares with error bars) and (ΔS/S0)sim(open circles) vs dephasing time for the lyophilized HFP-NC sample. The (ΔS/S0)sim were calculated using a mixture of n.a.d. models with fractional populations: α helical, 0.5; min β sheet, 0.25; max β sheet, 0.25. (b) Plots of (ΔS/S0)exp vs dephasing time for: HFP-NC, open triangles; HFP-P, filled triangles; HFP-A, open circles; HFP-AP, filled circles. The typical σexp is ±0.02. Variation of ±0.02 in (ΔS/S0)exp was also observed between two different preparations of the same sample type, e.g. HFP-A.
Natural abundance models
Quantitative analysis of the (ΔS/S0)exp to yield the fraction of parallel and antiparallel HFP registries requires an accurate natural abundance dephasing (n.a.d.) model, i.e. a model that accounts for effects of labeled 13CO-n.a. 15N and n.a. 13CO-labeled 15N proximities. Both types of proximities were considered but for conciseness of presentation, the discussion in this paper focuses on labeled 13CO-n.a. 15N. One measure of validity of a n.a.d. model was agreement within experimental error between (ΔS/S0)exp of HFP-NC and (ΔS/S0)sim of the model. Consideration was first given to the HFP-F8 regions of HFP-NC including the spin geometries of one or two labeled 13COs and one n.a. 15N. Geometries with two or more n.a. 15Ns were not considered because the fractional isotopic abundance of 15N is only 0.0037. For each geometry, the SIMPSON program was used to calculate (S1/S0)sim as a function of the dephasing time τ. Only geometries with rCN < 7 Å were considered because those with rCN > 7 Å do not affect (S1/S0)sim within our experimental signal-to-noise. We consider this a “long-range” n.a.d. model which is distinguished from a “short-range” model of earlier studies that only considered n.a. nuclei separated by one or two bonds from a labeled nucleus, i.e. rCN < 3 Å (15, 30). The broad spectral linewidth of HFP-NC indicated both helical and β strand conformational populations and coordinates of spin geometries for both α helical and β sheet structures were obtained from corresponding regions of high-resolution structure PDB files. For α helical structure, the rCN < 7 Å criterion resulted in geometries with a single labeled 13CO at residue i and a single n.a. 15N at a residue between i − 3 and i + 5. These nine geometries are one aspect of the α n.a.d. model.
Fig. 4 illustrates relevant labeled 13COs and n.a. 15Ns for antiparallel β sheet structure. The strands in panels a and c are “fully constrained” to a single registry with resultant six unique spin geometries. Three geometries had one labeled 13CO and one n.a. 15N within the same strand and three geometries had two labeled 13COs on vicinal strands and one n.a. 15N in the intervening strand. In panels b and d, the strands have different registries so that the labeled 13CO in the top strand was >7 Å from the nine n.a. 15N sites of the 13CO of the third strand. The structure of panels b and d has nine unique spin geometries and is denoted a maximum β sheet n.a.d. (max β n.a.d.) model while the structure of panels a and c has six geometries and is denoted a minimum β sheet n.a.d. (min β n.a.d.) model. In either structure there are nine n.a. 15N sites within 7 Å of each labeled 13CO but in the min n.a.d. model, some sites (e.g. 4–6 in Fig. 4c) are “shared” between two 13COs, i.e. within 7 Å of two 13COs. This reduces the average number of n.a. 15N sites per 13CO and the overall n.a.d.
Figure 4.
(a, b) Schematic diagrams of the HFP-F8 region of the HFP-NC sample in antiparallel β sheet structure with labeled 13COs represented as red circles. Panel a shows a model that is fully constrained to a single registry while panel b shows multiple registries. (c, d) β sheet backbone representations of the respective boxed regions of panels a and b with labeled 13COs in red and possible n.a. 15N sites in blue, i.e. sites for which a n.a. 15N is within 7 Å of a labeled 13CO. A particular spin geometry will have only one 15N. The min n.a.d. model is shown in panel c and each spin geometry will have either one labeled 13CO and one n.a. 15N (#1, 2 or 3) or two labeled 13COs and one n.a. 15N (#4, 5, or 6). The max n.a.d model is shown in panel d and each spin geometry will have one labeled 13CO and one n.a. 15N.
There are many indices and parameters in the quantitative modeling and descriptions and possible values for them are compiled in Table 1. For the α, min β, or max β n.a.d. models, the average γ = (S1/S0) for the relevant spin geometries:
| (3) |
where “ na ” refers to natural abundance, l = 0 or 1, “N ” refers to n.a. 15N, j is the index of a particular spin geometry, and J is the number of unique spin geometries of the model (J = 9 for α or max β n.a.d. and J = 6 for min β n.a.d.). The γ0Nna(τ) = 1 for all τ while γ1Nna(τ) were calculated using [S1j (τ)/S0j (τ)] sim from the SIMPSON program and generally decreased with increasing τ. After setting a total labeled 13CO population of 1.0 for HFP-F8 in HFP-NC, the relative population affected by n.a. 15N is J × 0.0037 while the remainder population, [1 − (J × 0.0037)], has S1 = S0. There is also an n.a. 13CO contribution from unlabeled residues in HFP-F8 with a relative population 30 × 0.011 and with S1 = S0. Similar analysis for 15N labeled HFP-A6L7 in HFP-NC results in:
| (4) |
where l = 0 or 1, “C” refers to n.a. 13CO, k is the index of a particular spin geometry, K is the number of unique n.a. 13CO-labeled 15N spin geometries of the model (K = 10, 8, or 12, respectively, for α, min β, or max β n.a.d. models), and γ0Cna(τ) = 1. Accounting for the 1:2 ratio of HFP-F8:HFP-A6L7, the total n.a. 13CO population of HFP-A6L7 is 2 × 31 × 0.011 with population 2K × 0.011 affected by labeled 15N and the remainder having S1 = S0. For HFP-NC in total:
| (5) |
where l = 0 or 1 and the terms in the first braces are τ- and l-dependent.
Table 1.
Indices and parameters
| Index/parameter | Description | Values |
|---|---|---|
| ft | fractional population of structure t | for membr. assoc. samples, determined by fitting |
| j, k | index for n.a. site <7 Å from a labeled site: j, n.a. 15N near labeled 13CO; k, n.a 13CO near labeled 15N |
|
| J, K | number of n.a. sites <7 Å from a labeled site: J, n.a. 15Ns near labeled 13CO; K, n.a 13COs near labeled 15N |
α helical structure, J = 9, K = 10; min β sheet structure, J = 6, K = 8; max β sheet structure, J = 9, K = 12 |
| l | REDOR data type index | 0 ≡ no dipolar evolution 1 ≡ dipolar evolution |
| m | datum index | 1, 2, 3, 4, 5, 6, or 7 |
| Sl, Slj, Slk, Slu | REDOR signal intensity | determined by experiment or calculation |
| t, t1, t2 | structural population index; for unconstrained model of membr. assoc. samples, t1 indexes the top/middle registry and t2 indexes the middle/bottom registry |
for membr. assoc. samples: 1 ≡ parallel registry 2 ≡ antiparallel registry 3 ≡ other “X” registry |
| u | membr. assoc. sample index | 1 ≡ HFP-P 2 ≡ HFP-A 3 ≡ HFP-AP |
| v | index for arrangement of three adjacent labeled HFPs in membr. assoc. samples – middle HFP has 13CO labeling and the 13CO is hydrogen bonded to HN of top HFP |
1 ≡ 13CO HFP (top), 13CO HFP (bottom) 2 ≡ 15N HFP (top), 13CO HFP (bottom) 3 ≡ 13CO HFP (top), 15N HFP (bottom) 4 ≡ 15N HFP(top),15N HFP(bottom) |
| wv | fractional population of arrangement of three adjacent labeled HFPs in membr. assoc. samples |
fully constrained model: w1 = 1/9, w2 = 2/9, w3 = 2/9, w4 = 4/9; unconstrained model: w1 = 1/81, w2 = 2/81, w3 = 2/81, w4 = 4/81 |
| γlNna(τ), γlCna(τ) |
Sl(τ)/S0: γlN(τ), labeled 13CO-n.a. 15N; γlC(τ), n.a. 13CO-labeled 15N |
γ0N(τ) = 1; γ0C(τ) = 1; γ1N(τ) and γ1C(τ) determined by calculation |
| γltuvlab(τ), γlt1t2uvlab(τ) |
Slulab(τ)/S0lab for arrangement of labeled 13CO and 15N nuclei: γ1tuvlab(τ), fully constrained model; γ1t1t2uvlab(τ), unconstrained model |
γ0tuvlab(τ) = 1; γ0t1t2uvlab(τ) = 1; γ1tuvlab(τ) and γ1t1t2uvlab(τ) determined by calculation |
| τ | REDOR dephasing time | 2.2, 8.2, 16.2, 24.2, 32.2, 40.2, or 48.2 ms |
For each n.a.d. model (α, min β, and max β), (ΔS/S0)sim for each τ was calculated with Eq. 5 and statistical comparison was then made to (ΔS/S0)exp:
| (6) |
where m is the index for an experimental datum, i.e. a particular τ. The respective χ2 for the α, min β, and max β n.a.d. models were 1.2, 3.8, and 2.0 which were all less than the number of degrees of fitting, 7, i.e. the number of data, 7, minus the number of independent fitting parameters, 0. The validity of the approach to n.a.d. calculation was supported by good fits for all models (43).
The broad 13CO linewidth of HFP-NC in Fig. 2a was consistent with two HFP populations, one with helical and one with β strand secondary structure. It was therefore reasonable to calculate (ΔS/S0)sim for “mixtures” with contributions from multiple models:
| (7) |
where l = 0 or 1, t was the index that referred to the α, min β, or max β model, ft was fractional population with Σft = 1, and each Sltsim(τ) was calculated using Eq. 5. The HFP-NC distribution of 13CO shifts indicated fα ≈ 0.5 and fminβ + fmaxβ ≈ 0.5 but did not provide information about individual fminβ or fmaxβ Fitting using fα = 0.5 and fminβ = 0.5, fmaxβ = 0.0 yielded χ2 = 1.5 while fitting using either fα = 0.5, fminβ = 0.0, fmaxβ = 0.5 or fα = 0.5, fminβ = 0.25, fmaxβ = 0.25 yielded χ2 = 1.2 and (ΔS/S0)sim in Fig. 3a were calculated with the latter distribution. The (ΔS/S0)sim from all three conformational distributions fit well to the (ΔS/S0)exp and these models are statistically similar. Together with previously described good fitting for different secondary structure models show that n.a.d. is accurately calculated with these models and only weakly dependent on secondary and tertiary structure. The key feature of all these well-fitting long-range models was consideration of the multiple n.a. sites within 7 Å of a labeled site which led to continually increasing (ΔS/S0) with τ, Fig. 3a. The (ΔS/S0)sim were also calculated using a short-range model that only considered n.a. sites separated by one or two bonds from each labeled site. The (ΔS/S0)sim were systematically less than the (ΔS/S0)exp with resultant poor fit and χ2 = 29.
Quantitative analysis of registry populations – fully constrained model
For membrane-associated HFP, there is a single distribution of registries which we model as fractions of: (1) 1→17/1→17 and 2→17/1→16 parallel registries; (2) 16→1/1→16 and 17→1/1→17 antiparallel registries; and (3) X registries not detected by any of our labeling schemes, Fig. 1c. Fraction 1 contributed to the (ΔS/S0)exp of HFP-P, fraction 2 contributed to the (ΔS/S0)exp of HFP-A, and fractions 1 and 2 contributed to (ΔS/S0)exp of HFP-AP. The overall goal was best-fit determination of these fractions based on the (ΔS/S0)exp of the three samples, Fig. 3b, and this analysis required calculation of the n.a.d. contribution to (ΔS/S0)exp. Because a 1:2 13CO-HFP:15N-HFP ratio was used for all samples, this contribution was calculated using models developed for HFP-NC and resulted in a modified Eq. 5 appropriate for HFP-P, HFP-A, and HFP-AP:
| (8) |
| (9) |
where Slna is the sum of the first two braced terms in Eq. 8 and Slulab is the third braced term. Each membrane-associated sample is labeled by the index u where u = 1, 2, or 3 respectively refers to HFP-P, HFP-A, or HFP-AP, Table 1 and Fig. 1b. The first braced term in Eq. 8 corresponds to labeled and n.a. 13CO that experience n.a.d., the second braced term corresponds to n.a. 13COs that do not experience n.a.d., and the third braced term corresponds to labeled 13COs that do not experience n.a.d. but may experience dephasing from labeled 15Ns. The secondary structure of membrane-associated HFP was predominantly β sheet, Fig. 2b–d, and the best estimates of the n.a.d. terms in the first braced term were taken to be the average of the max β and min β calculated values. In the second and third braced terms, K and J were estimated to be their respective average values of 10 and 7.5. S0ulab(τ) was calculated using γ0tuvlab(τ) = 1 while γ1tuvlab(τ) and therefore S1ulab(τ) were first calculated with a “fully constrained” model, Fig. 4a,c, in which a β sheet region contained either: (1) 1→17/1→17 or 2→17/1→16 parallel registries; or (2) 16→1/16→1 or 17→1/1→17 antiparallel registries; or (3) X registries not directly detected by any of our labeling schemes, Fig. 1c. A sample was considered to be a mixture of the three registry types each denoted by index t = 1, 2, or 3 and fractional population ft, Table 1. The S1ulab(τ) was calculated by modified Eq. 7:
| (10) |
The γ1tuvlab(τ) values depended on the labeled dCNs and therefore rCNs which in turn depended on registry type t and sample labeling u, Fig. 1c. For some combinations of t and u, all labeled rCN > 7 Å with consequent dCN ≈ 0 and γ1tuvlab(τ) = 1. Specific examples are t = 1 and u = 2, t = 2 and u = 1, and t = 3 and u = 1, 2, or 3. For other combinations of t and u, γ1tuvlab(τ) were determined from SIMPSON calculations and Fig. 5a–d displays schematic examples for t = 2, u = 2 with numerical values of γ1tuvlab(τ) and more details in the Supporting Information. Column a, b, c, or d corresponds to particular arrangements of 13CO and 15N labeled HFPs that are respectively denoted by the index v = 1, 2, 3, or 4. For each v, the typical difference between the calculated γ122vlab(τ) for the 16→1/1→16 or 17→1/1→17 registry was ≤ 0.01 and the final γ122vlab(τ) were the average for the two registries. The antiparallel γ123vlab(τ) of HFP-AP were analogously calculated and the parallel γ111vlab(τ) of HFP-P and parallel γ113vlab(τ) of HFP-AP were calculated using the 1→17/1→17 registry and had similar values to γs calculated using the 2→17/1→16 registry. Fractional weightings wv were based on the 1:2 ratio of 13CO HFP:15N HFP with w1 = 1/9, w2 = 2/9, w3 = 2/9, and w4 = 4/9. A more complete version of Eq. 10:
| (11) |
with indices and parameters summarized in Table 1.
Figure 5.
Schematics of three adjacent HFPs for HFP-A, i.e. u = 2, in (a–d) fully constrained or (e–h) unconstrained models. Red and blue correspond to 13CO and 15N labeled residues, respectively, and the labeled 13CO in the middle strand was hydrogen bonded to the HN group of the residue in the top strand. Panels a–d display antiparallel 16→1/1→16 (top) or 17→1/1→17 (bottom) registries while panels e–h display parallel 1→17/1→17, antiparallel 16→1/1→16, and X registries where X refers to a registry for which the labeled rCN > 7 Å, i.e. beyond the approximate detection limit of the SSNMR experiment, and which is not 1→17/1→17, 2→17/1→16, 16→1/1→16, or 17→1/1→17. Correspondence between columns and the index v are: a and e, v = 1; b and f, v = 2; c and g; v = 3; d and h; v = 4. Both rows of three-strand arrangements in panels a–d correspond to t = 2 and the row, t1, t2 correspondence in panels e–h is: row 1, t1 = 1, t2 = 1; row 2, t1 = 1, t2 = 2; row 3, t1 = 1, t2 = 3; row 4, t1 = 2, t2 = 1; row 5, t1 = 2, t2 = 2; row 6, t1 = 2, t2 = 3; row 7, t1 = 3, t2 = 1; row 8, t1 = 3, t2 = 2; row 9, t1 = 3, t2 = 3. For each three-strand arrangement enclosed by a box, the γ1tuvlab(τ) or γ1t1t2uvlab(τ) were calculated by SIMPSON simulation. For arrangements with t, t1, or t2= 2, fitting to experiment was based on γs that were the average of those calculated with 16→1/1→16 and 17→1/1→17 registries although the latter registry is not displayed in panels e–h. For any arrangement not enclosed by a box, γ1tuvlab(τ) = 1 or γ1t1t2uvlab(τ) = 1.
The values of f1, f2, and f3 were the same for the HFP-P, HFP-A, and HFP-AP samples with f3 = 1 – f1 – f2. Best-fit values of f1 and f2 were obtained by calculating χ2(f1, f2) using an expression analogous to Eq. 6:
| (12) |
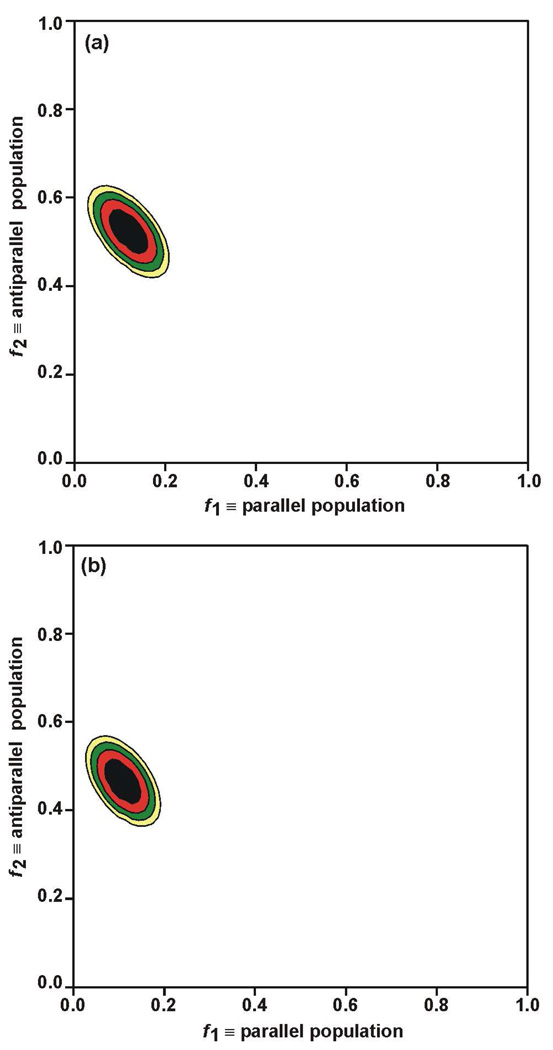
and then selecting the f1 and f2 values which corresponded to minimum χ2, i.e. χ2min. In Eq. 12, m is the index for each τ, and [ΔS(f1, f2)/S0]umsim was determined using Eqs. 8–11. For this fully constrained model, Fig. 6a displays a plot of χ2 vs f1 and f2 with best-fit f1 = 0.12 and f2 = 0.52 and χ2min = 11. The model was reasonable as evidenced by χ2min which was smaller than the number of degrees of fitting, 19, i.e. the number of data, 21, minus the number of fitting parameters, 2. The f1 fractional parallel population, Fig. 1c, was small which was consistent with qualitative analysis of the data, Fig. 3b. The f2 antiparallel population was substantially larger and also consistent with Fig. 3b. The f3 ≈ 0.35 indicated a substantial population of X registries not detected by the labeling of the three samples.
Figure 6.
Contour plots of χ2 vs f1 parallel and f2 antiparallel fractional populations for (a) fully constrained and (b) unconstrained models. In each plot, f1 is the sum of populations of 1→17/1→17 and 2→17/1→16 parallel registries and f2 is the sum of populations of 16→1/1→16 and 17→1/1→17 antiparallel registries. For plot a, the best-fit values were f1 = 0.12 ± 0.03 and f2 = 0.52 ± 0.04 with χ2min = 11, and for plot b, f1 = 0.11 ± 0.03, f2 = 0.46 ± 0.04, and χ2min = 8. Parameter uncertainties were determined by the region of χ2 within about three units of χ2min. In plot a, the black, red, green, yellow, and white regions correspond to χ2 < 14, 14 < χ2 < 17, 17 < χ2< 20, 20 < χ2< 23, and χ2 > 23 and in plot b, the regions correspond to χ2 < 11, 11 < χ2 < 14, 14 < χ2 < 17, 17 < χ2 < 20, and χ2 > 20.
The above fitting was done using a long-range n.a.d. model that considered effects of n.a. sites within 7 Å of each labeled nucleus. Fitting displayed in Fig. 6a was based on n.a.d. calculated from half min β and half max β sheet structure, Fig. 4, but the best-fit f1 and f2 and χ2min were not sensitive to the structural composition of the long-range n.a.d. model. For example, fitting done using n.a.d. for half α helical and half max β sheet structure yielded best-fit f1 and f2 and χ2min respectively within 0.01, 0.01, and 1 of the corresponding Fig. 6a values. For HFP-NC fitting, n.a.d. was underestimated by a short-range model that only considered n.a. sites separated by one or two bonds from each labeled site. This effect was also observed when fitting membrane-associated HFP data with the short-range n.a.d. model and led to best-fit f1 = 0.22 and f2 = 0.57 which were significantly higher than the Fig. 6a values. The χ2min = 20 using the short-range model was also higher than the χ2min in Fig. 6a.
Quantitative analysis of registry populations – unconstrained model
In addition to the fully constrained model for strand registries, an alternate “unconstrained” fitting model was also considered for which there was local mixing of: (1) 1→17/1→17 parallel registries; (2) 16→1/1→16 and 17→1/1→17 antiparallel registries; and (3) X registries not directly detected by any of our labeling schemes, Fig. 1c. Each pairwise registry type was labeled by t =1, 2, or 3 with fractional population ft. For this unconstrained model, Fig. 5e–h displays schematics of three-strand registries with 13CO labeled HFP in the middle strand. Each e–h row has three-strand registries that were each a combination of two registries labeled by specific t1 and t2 which denote the respective t of the top/middle and middle/bottom strands. As with the fully constrained models, the registries in each e–h column corresponded to a particular 13CO HFP/15N HFP arrangement which respective label v = 1, 2, 3, or 4. The 13CO HFP:15N HFP population ratio of 1:2 correlated with a sum weighting of 1/9 for the v = 1 registries with individual registry weighting w1 = 1/(9×9) = 1/81. The sum weightings for v = 2, 3, or 4 were respectively 2/9, 2/9, or 4/9 with respective individual weightings w2 = 2/81, w3 = 2/81, and w4 = 4/81. Eq. 11 was modified for the unconstrained model:
| (13) |
Similar to the fully constrained model, many combinations of t1, t2, u, and v have rCN > 7 Å with consequent dCN ≈ 0 and γ1t1t2uvlab(τ) = 1. In Fig. 5e–g, such registries are not enclosed by a box. Similar to results for the fully constrained model, the γ1t1t2uvlab(τ) were similar for the two antiparallel registries and an average value was used.
The values of f1, f2, and f3 in the unconstrained model were the same for the HFP-P, HFP-A, and HFP-AP samples with f3 = 1 – f1 – f2. Best-fit values of f1 and f2 were obtained with Eq. 12 and Fig. 6b displays a plot of χ2 vs f1 and f2 with best-fit f1 = 0.11 and f2 = 0.46 and corresponding χ2min = 8. The unconstrained model was reasonable as evidenced by a best-fit χ2 which was smaller than the number of degrees of fitting, 19. Similar to the results of the fully constrained model, the f1 fractional parallel population was small and the f2 antiparallel population and f3 ≈ 0.4 other population were significant.
This unconstrained model fitting was done with n.a.d. calculated with a long-range model and half min β and half max β structure. Similar to the fully constrained model, best-fit f1, f2, and χ2 for the unconstrained model were: (1) negligibly affected by the structural distribution of the long-range n.a.d. model; and (2) significantly increased by use of a short-range n.a.d. model.
DISCUSSION
This paper sets an upper limit of ~0.15 on the fraction of membrane-associated HFP in in-register parallel β sheet structure and this result is supported by both qualitative analysis of the data, Fig. 3b, as well as quantitative analyses with fully constrained and unconstrained models, Fig. 6. Both models fit the data well and yielded similar best-fit fractional population of parallel registries and similar populations of antiparallel registries. The small fractional parallel population agrees with some earlier SSNMR studies but differs from interpretations of other SSNMR and IR data which respectively reported ~0.5 and ~1.0 fractions of in-register parallel structure (27, 30–32). The present study used samples with sparse isotopic labeling while the earlier studies interpreted to support a large fraction of parallel structure used samples with extensive labeling. We think that there was ambiguity of interpretation in the studies of extensively labeled samples and that the data could also be reasonably interpreted in terms of small in-register parallel population. For example, the earlier SSNMR study also used the REDOR technique but with only a single τ (24 ms) and with samples containing equimolar amounts of HFP 13CO labeled on three sequential residues and HFP 15N labeled on three sequential residues. The typical (ΔS/S0)exp was ~0.1 and was approximately independent of the positions of the labeled residues and also had some contribution from n.a.d. It was not possible to do unambiguous quantitative analysis of registry distributions because: (1) each sample was extensively labeled so that non-zero (ΔS/S0) was expected for many different registries; (2) (ΔS/S0)exp were only measured for a single τ; and (3) a “HFP-NC”-type sample was not studied and n.a.d. was therefore not quantitatively modeled.
The samples for the IR study were also extensively labeled with backbone 13CO labeling at either: (1) A1 to V3, G5 to L9; (2) F8 to G16; or (3) A1 to V3, G5 to G16. The authors’ interpretation of their spectra to support predominant in-register parallel structure was based in part on expected effects of (13C=16O electric dipole)…(13C=16O electric dipole) coupling on 13C=O vibrational wavenumber and intensity. However, their interpretation appeared to neglect the substantial intramolecular coupling between 13C=Os on adjacent residues and we note that this coupling is independent of registry. In addition to these “undiluted” samples, three “diluted” samples were studied that had an equimolar mixture of a labeled and unlabeled peptide. The wavenumber (v) of a 13C=16O vibration is sensitive to nearby (~5 Å away) C=O vibrations and is higher with 12C=16O neighbors than with 13C=16O neighbors. If there is hydrogen bonding between 13CO labeled residues of adjacent strands in an undiluted sample, the corresponding diluted sample should have an increased fraction of 13C=O/12C=O proximities, decreased fraction of 13C=O/13C=O proximities, and Δv = vdiluted − vundiluted > 0. If there were a major fraction of parallel 1→17/1→17 structure (as claimed by the authors), dilution of (1) A1-V3, G5-L9; (2) F8-G16; or (3) A1-V3, G5-G16 labeled HFPs would have had comparable effect on proximities and resulted in similar Δv. However, the experimental ΔvA1-V3,G5-L9 ≈ ΔvF8-G16 ≈ (ΔvA1-V3,G5-G16)/2 which is inconsistent with a large fraction of in-register parallel structure. Like the earlier SSNMR study on extensively labeled samples, extensive labeling of the IR samples also meant that the IR data were consistent with many registry distributions and precluded more quantitative analysis of the distribution. Overall, the sparse labeling of the present SSNMR study allowed for much more unambiguous and quantitative determination of the populations of specific registries. This general approach can be applied in the future to determine the registry distributions of HFP constructs with very high or low fusogenicity such as HFPtr or V2E mutant, respectively.
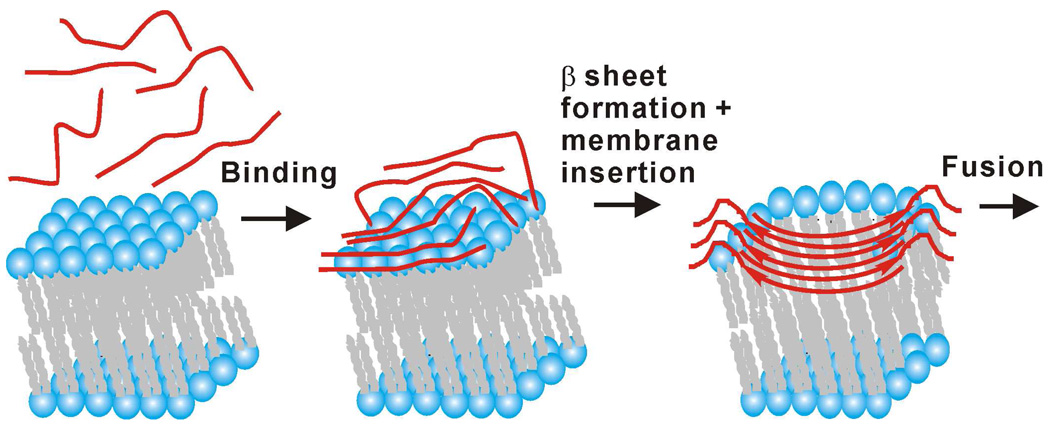
Fig. 7 displays a structure/function model for HFP based on results from this and earlier studies. Prior to membrane binding, HFP is monomeric in aqueous solution and has random coil structure (11, 12, 18). HFP sequentially: (1) binds to membranes; (2) forms β sheet oligomers with a significant fraction of 16→1/1→16 and 17→1/1→17 antiparallel registries; and (3) induces membrane fusion as monitored by intervesicle lipid mixing (12, 14, 17, 18). It is also known that the A6 and L9 residues of β sheet HFP insert shallowly into the membrane with correlation between membrane insertion depth and both membrane perturbation and fusion rate (19, 36, 49). A global structure-function model is non-transmembrane HFP insertion perturbs bilayer structure and moves the membrane on the fusion reaction coordinate towards the highly perturbed transition state with consequent reduction in fusion activation energy and increase in fusion rate.
Figure 7.
Pictorial model of HFP (red lines) binding to membranes followed by antiparallel β sheet formation and membrane insertion and then fusion. Time increases from left-to-right. For reasons of clarity, some lipids are not shown in the right-most picture. Although there are no data yet on fusion peptide structure during HIV/host cell fusion, the antiparallel β sheet structure of the right-most picture is plausible because: (1) the structure is consistent with multiple trimers at the fusion site; and (2) the structure is membrane-inserted with deeper insertion positively correlated with increased membrane perturbation and vesicle fusion rate.
There is functional and electron microscopic evidence that multiple gp41 trimers are required for fusion and Fig. 7(right) shows a β sheet HFP hexamer as would be reasonable for interleaved antiparallel fusion peptides from two gp41 trimers. While there are no data specifically supporting a HFP hexamer, HFP β sheet oligomers likely contain a small number of molecules because: (1) for ~90% of HFP molecules, there is an A1 13CO-lipid 31P distance of ~5 Å, i.e. close contact of most HFPs with the membrane; and (2) significant temperature dependence of intensities of SSNMR spectra (19, 26). Interleaved antiparallel fusion peptides from multiple gp41 trimers may also be the fusogenic structure of HIV/host cell fusion. As noted above, this structure can insert into and perturb membranes which are likely requirements for HIV/host cell fusion.
The experimentally observed membrane insertion of the central β sheet region (e.g. A6 to L12) of HFP is consistent with ΔGinsertion ≈ −6 kJ/mol for the fully constrained 17→1/1→17 registry as calculated by summing individual residue insertion energies for L12→A6/A6→L12, Fig. 1c (50). A similar calculation yielded ΔGinsertion ≈ −3 kJ/mol for L12→G5/G5→L12 of the 16→1/1→16 registry. The ΔGinsertion ≈ −6 kJ/mol for A6→L12/A6→L12 of the 1→17/1→17 parallel registry suggests that ΔGinsertion does not underlie the preference for antiparallel over parallel structure. This preference may instead be due to ΔGelectrostatic as the HFP N-terminus is located in the high water content lipid headgroup region and is therefore likely protonated. Closest intermolecular NH3+−NH3+ distance is ~5 Å for the low population 1→17/1→17 registry and ~10 Å for the significantly populated 17→1/1→17 registry. For εdielectric = 78 and hexameric HFP, ΔGelectrostatic ≈ +5.1 kJ/mol for the 1→17/1→17 registry and +1.5 kJ/mol for the 17→1/1→17 registry.
We expect that inclusion of the non-native C-terminal W(K)6A tag does not contribute to the preference for antiparallel over parallel registry because either registry would have similar minimized electrostatic repulsion energy. Such repulsion would be minimized by: (1) extensive solvation of the tag; and (2) large inter-tag distances that are possible because of random coil tag structure. Tag solvation is supported by the previously observed lack of membrane insertion of HFP beyond residue L12 and random coil tag structure is supported by broad NMR linewidths in the C-terminal region of HFP (14, 19). We also note that inclusion of the tag has minor effect on fusion activity and that similar REDOR ΔS/S0 were observed for mixtures of triply 13CO and 15N labeled HFPs with or without the tag (12, 27).
In contrast to the reasonably large distribution of membrane-associated HFP registries, i.e. significant 16→1/1→16, 17→1/1→17, and X registries, SSNMR studies of β sheet registries of protein in amyloid fibrils have typically shown a single registry which is usually in-register parallel, e.g. 1→17/1→17 (28, 29). The width of a fibril is at most a few protein molecules and the length is >200 molecules and along the intermolecular β sheet hydrogen bonding direction. The amyloid fibrils are grown in aqueous solution (without lipid) and their greater registry homogeneity may reflect ordered fibril growth from seeds (51).
One distinctive feature of the present study is the development of a quantitative n.a.d. model that was experimentally validated. Accurate fitting of the HFP-NC data and fitting of the membrane-associated HFP data relied on a long-range n.a.d. model which included effects of n.a. nuclei <7 Å from each labeled nucleus. For this model, the n.a.d. was approximately independent of secondary and tertiary structure. The n.a.d. was systematically underestimated by a short-range model which only considered n.a. nuclei separated by one or two bonds from each labeled nucleus.
Supplementary Material
ACKNOWLEDGEMENT
NMR and mass spectra were obtained at Michigan State University facilities. We acknowledge useful discussions with Dr. Wei Qiang, Dr. Kelly Sackett, and Dr. Charles Gabrys.
ABBREVIATIONS
- CO
carbonyl
- d
magnetic dipole-dipole coupling
- DTPC
1,2-di-O-tetradecyl-sn-glycero-3-phosphocholine
- DTPG
1,2-di-O-tetradecyl-sn-glycero-3-phosp-rac-(1-glycerol) sodium salt
- FMOC
9-fluorenylmethoxycarbonyl
- HEPES
N-(2-hydroxyethyl)piperazine-N'-2-ethanesulfonic acid
- HFP
HIV fusion peptide
- HFPtr
HIV fusion peptide trimer
- HIV
human immunodeficiency virus
- IR
infrared
- MAS
magic angle spinning
- max n.a.d.
maximum natural abundance dephasing
- min n.a.d.
minimum natural abundance dephasing
- n.a.
natural abundance
- n.a.d.
natural abundance dephasing
- NMR
nuclear magnetic resonance
- PDB
Protein Data Bank
- r
internuclear distance
- REDOR
rotational-echo double resonance
- SIMPSON
simulation program for solid-state NMR spectroscopy
- SSNMR
solid-state NMR
- TFA
trifluoroacetic acid
- TPPM
two-pulse phase-modulation
Footnotes
This work was supported by NIH award AI47153 to D.P.W.
SUPPORTING INFORMATION AVAILABLE
Table of (ΔS/S0)exp values and schematics of spin geometries and associated γ1tuvlab(τ) or γ1t1t2uvlab(τ) calculated by SIMPSON simulation. This information is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan K, Liu J, Wang J, Shen S, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caffrey M, Cai M, Kaufman J, Stahl SJ, Wingfield PT, Covell DG, Gronenborn AM, Clore GM. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 1998;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang ZN, Mueser TC, Kaufman J, Stahl SJ, Wingfield PT, Hyde CC. The crystal structure of the SIV gp41 ectodomain at 1.47 A resolution. J. Struct. Biol. 1999;126:131–144. doi: 10.1006/jsbi.1999.4116. [DOI] [PubMed] [Google Scholar]
- 5.Buzon V, Natrajan G, Schibli D, Campelo F, Kozlov MM, Weissenhorn W. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. Plos Pathogens. 2010;6:e1000880. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freed EO, Myers DJ, Risser R. Characterization of the fusion domain of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Proc. Natl. Acad. Sci. U.S.A. 1990;87:4650–4654. doi: 10.1073/pnas.87.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freed EO, Delwart EL, Buchschacher GL, Jr, Panganiban AT. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc. Natl. Acad. Sci. U.S.A. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durell SR, Martin I, Ruysschaert JM, Shai Y, Blumenthal R. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion. Mol. Membr. Biol. 1997;14:97–112. doi: 10.3109/09687689709048170. [DOI] [PubMed] [Google Scholar]
- 9.Nieva JL, Agirre A. Are fusion peptides a good model to study viral cell fusion? Biochim. Biophys. Acta. 2003;1614:104–115. doi: 10.1016/s0005-2736(03)00168-8. [DOI] [PubMed] [Google Scholar]
- 10.Epand RM. Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta. 2003;1614:116–121. doi: 10.1016/s0005-2736(03)00169-x. [DOI] [PubMed] [Google Scholar]
- 11.Chang DK, Chien WJ, Cheng SF. The FLG motif in the N-terminal region of glucoprotein 41 of human immunodeficiency virus type 1 adopts a type-I beta turn in aqueous solution and serves as the initiation site for helix formation. Eur. J. Biochem. 1997;247:896–905. doi: 10.1111/j.1432-1033.1997.00896.x. [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Prorok M, Castellino FJ, Weliky DP. Oligomeric β-structure of the membrane-bound HIV-1 fusion peptide formed from soluble monomers. Biophys. J. 2004;87:1951–1963. doi: 10.1529/biophysj.103.028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira FB, Goni FM, Muga A, Nieva JL. Permeabilization and fusion of uncharged lipid vesicles induced by the HIV-1 fusion peptide adopting an extended conformation: dose and sequence effects. Biophys. J. 1997;73:1977–1986. doi: 10.1016/S0006-3495(97)78228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Gabrys CM, Weliky DP. Solid-state nuclear magnetic resonance evidence for an extended beta strand conformation of the membrane-bound HIV-1 fusion peptide. Biochemistry. 2001;40:8126–8137. doi: 10.1021/bi0100283. [DOI] [PubMed] [Google Scholar]
- 15.Qiang W, Bodner ML, Weliky DP. Solid-state NMR spectroscopy of human immunodeficiency virus fusion peptides associated with host-cell-like membranes: 2D correlation spectra and distance measurements support a fully extended conformation and models for specific antiparallel strand registries. J. Am. Chem. Soc. 2008;130:5459–5471. doi: 10.1021/ja077302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krasslich HG. The HIV lipidome: A raft with an unusual composition. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buzon V, Padros E, Cladera J. Interaction of fusion peptides from HIV gp41 with membranes: A time-resolved membrane binding, lipid mixing, and structural study. Biochemistry. 2005;44:13354–13364. doi: 10.1021/bi050382r. [DOI] [PubMed] [Google Scholar]
- 18.Yang R, Prorok M, Castellino FJ, Weliky DP. A trimeric HIV-1 fusion peptide construct which does not self-associate in aqueous solution and which has 15-fold higher membrane fusion rate. J. Am. Chem. Soc. 2004;126:14722–14723. doi: 10.1021/ja045612o. [DOI] [PubMed] [Google Scholar]
- 19.Qiang W, Sun Y, Weliky DP. A strong correlation between fusogenicity and membrane insertion depth of the HIV fusion peptide. Proc. Natl. Acad. Sci. U.S.A. 2009;106:15314–15319. doi: 10.1073/pnas.0907360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang XZ, Kurteva S, Ren XP, Lee S, Sodroski J. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J. Virol. 2005;79:12132–12147. doi: 10.1128/JVI.79.19.12132-12147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnus C, Rusert P, Bonhoeffer S, Trkola A, Regoes RR. Estimating the stoichiometry of Human Immunodeficiency Virus entry. J. Virol. 2009;83:1523–1531. doi: 10.1128/JVI.01764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sougrat R, Bartesaghi A, Lifson JD, Bennett AE, Bess JW, Zabransky DJ, Subramaniam S. Electron tomography of the contact between T cells and SIV/HIV-1: Implications for viral entry. PLOS Pathogens. 2007;3:571–581. doi: 10.1371/journal.ppat.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau WL, Ege DS, Lear JD, Hammer DA, DeGrado WF. Oligomerization of fusogenic peptides promotes membrane fusion by enhancing membrane destabilization. Biophys. J. 2004;86:272–284. doi: 10.1016/S0006-3495(04)74103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan JH, Lai CB, Scott WRP, Straus SK. Synthetic fusion peptides of tick-borne Encephalitis virus as models for membrane fusion. Biochemistry. 2010;49:287–296. doi: 10.1021/bi9017895. [DOI] [PubMed] [Google Scholar]
- 25.Han X, Tamm LK. A host-guest system to study structure-function relationships of membrane fusion peptides. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13097–13102. doi: 10.1073/pnas.230212097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bodner ML, Gabrys CM, Parkanzky PD, Yang J, Duskin CA, Weliky DP. Temperature dependence and resonance assignment of 13C NMR spectra of selectively and uniformly labeled fusion peptides associated with membranes. Magn. Reson. Chem. 2004;42:187–194. doi: 10.1002/mrc.1331. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Weliky DP. Solid state nuclear magnetic resonance evidence for parallel and antiparallel strand arrangements in the membrane-associated HIV-1 fusion peptide. Biochemistry. 2003;42:11879–11890. doi: 10.1021/bi0348157. [DOI] [PubMed] [Google Scholar]
- 28.Benzinger TL, Gregory DM, Burkoth TS, Miller-Auer H, Lynn DG, Botto RE, Meredith SC. Propagating structure of Alzheimer's β-amyloid(10–35) is parallel β-sheet with residues in exact register. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Quart. Rev. Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 30.Zheng Z, Yang R, Bodner ML, Weliky DP. Conformational flexibility and strand arrangements of the membrane-associated HIV fusion peptide trimer probed by solid-state NMR spectroscopy. Biochemistry. 2006;45:12960–12975. doi: 10.1021/bi0615902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Z, Qiang W, Weliky DP. Investigation of finite-pulse radiofrequency-driven recoupling methods for measurement of intercarbonyl distances in polycrystalline and membrane-associated HIV fusion peptide samples. Magn. Reson. Chem. 2007;245:S247–S260. doi: 10.1002/mrc.2160. [DOI] [PubMed] [Google Scholar]
- 32.Sackett K, Shai Y. The HIV fusion peptide adopts intermolecular parallel β-sheet structure in membranes when stabilized by the adjacent N-terminal heptad repeat: A 13C FTIR study. J. Mol. Biol. 2005;350:790–805. doi: 10.1016/j.jmb.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 33.Lapatsanis L, Milias G, Froussios K, Kolovos M. Synthesis of N-2,2,2-(trichloroethoxycarbonyl)-L-amino acids and N-(9-fluorenylmethoxycarbonyl)-L-amino acids involving succinimidoxy anion as a leaving group in amino-acid protection. Synthesis. 1983;8:671–673. [Google Scholar]
- 34.Siminovitch DJ, Ruocco MJ, Makriyannis A, Griffin RG. The effect of cholesterol on lipid dynamics and packing in diether phosphatidylcholine bilayers. X-ray diffraction and 2H-NMR study. Biochim. Biophys. Acta. 1987;901:191–200. doi: 10.1016/0005-2736(87)90115-5. [DOI] [PubMed] [Google Scholar]
- 35.Komatsu H, Rowe ES. Effect of cholesterol on the ethanol-induced interdigitated gel phase in phosphatidylcholine: Use of fluorophore pyrene-labeled phosphatidylcholine. Biochemistry. 1991;30:2463–2470. doi: 10.1021/bi00223a024. [DOI] [PubMed] [Google Scholar]
- 36.Gabrys CM, Yang R, Wasniewski CM, Yang J, Canlas CG, Qiang W, Sun Y, Weliky DP. Nuclear magnetic resonance evidence for retention of a lamellar membrane phase with curvature in the presence of large quantities of the HIV fusion peptide. Biochim. Biophys. Acta. 2010;1798:194–201. doi: 10.1016/j.bbamem.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huster D, Xiao LS, Hong M. Solid-state NMR investigation of the dynamics of the soluble and membrane-bound colicin Ia channel-forming domain. Biochemistry. 2001;40:7662–7674. doi: 10.1021/bi0027231. [DOI] [PubMed] [Google Scholar]
- 38.Gullion T, Schaefer J. Rotational-echo double-resonance NMR. J. Magn. Reson. 1989;81:196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 39.Gullion T, Baker DB, Conradi MS. New, compensated Carr-Purcell sequences. J. Magn. Reson. 1990;89:479–484. [Google Scholar]
- 40.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J. Chem. Phys. 1995;103:6951–6958. [Google Scholar]
- 41.Long HW, Tycko R. Biopolymer conformational distributions from solid-state NMR: alpha-helix and 3(10)-helix contents of a helical peptide. J. Am. Chem. Soc. 1998;120:7039–7048. [Google Scholar]
- 42.Fowler DJ, Weis RM, Thompson LK. Kinase-active signaling complexes of bacterial chemoreceptors do not contain proposed receptor-receptor contacts observed in crystal structures. Biochemistry. 2010;49:1425–1434. doi: 10.1021/bi901565k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bevington PR, Robinson DK. Data Reduction and Error Analysis for the Physical Sciences. 2nd ed. Boston: McGraw-Hill; 1992. [Google Scholar]
- 44.Oas TG, Hartzell CJ, McMahon TJ, Drobny GP, Dahlquist FW. The carbonyl 13C chemical-shift tensors of 5 peptides determined from 15N dipole-coupled chemical shift powder patterns. J. Am. Chem. Soc. 1987;109:5956–5962. [Google Scholar]
- 45.Bak M, Rasmussen JT, Nielsen NC. SIMPSON: A general simulation program for solid-state NMR spectroscopy. J. Magn. Reson. 2000;147:296–330. doi: 10.1006/jmre.2000.2179. [DOI] [PubMed] [Google Scholar]
- 46.Bak M, Schultz R, Vosegaard T, Nielsen NC. Specification and visualization of anisotropic interaction tensors in polypeptides and numerical simulations in biological solid-state NMR. J. Magn. Reson. 2002;154:28–45. doi: 10.1006/jmre.2001.2454. [DOI] [PubMed] [Google Scholar]
- 47.Jaroniec CP, Kaufman JD, Stahl SJ, Viard M, Blumenthal R, Wingfield PT, Bax A. Structure and dynamics of micelle-associated human immunodeficiency virus gp41 fusion domain. Biochemistry. 2005;44:16167–16180. doi: 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- 48.Zhang HY, Neal S, Wishart DS. RefDB: A database of uniformly referenced protein chemical shifts. J. Biomol. NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 49.Tristram-Nagle S, Chan R, Kooijman E, Uppamoochikkal P, Qiang W, Weliky DP, Nagle JF. HIV fusion peptide penetrates, disorders, and softens T-cell membrane mimics. J. Mol. Biol. 2010;402:139–153. doi: 10.1016/j.jmb.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, Heijne G. Recognition of tranmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 51.Petkova AT, Leapman RD, Guo ZH, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.