Abstract
It is expected that the attendant structural heterogeneity of human high density lipoprotein (HDL) complexes is a determinant of its varied metabolic functions. To determine structural heterogeneity of HDL, major apolipoprotein stoichiometry profiles in human HDL were determined. First, HDL was separated into two main populations, with and without apolipoprotein (apo) A-II, LpA-I and LpA-I/A-II respectively. Each main population was further separated into six individual subfractions using size exclusion chromatography (SEC). Protein proximity profiles (PPP) of major apolipoproteins in each individual subfraction was determined by optimally cross-linking apolipoproteins within individual particles with bis(sulfosuccinimidyl)suberate (BS3), a bifunctional cross linker, followed by molecular weight determination by MALDI-MS. The PPPs of LpA-I subfractions indicated that the number of apoA-I molecules increased from two to three to four upon increase in the LpA-I particle size. On the other hand, the entire population of LpA-I/A-II demonstrated the presence of only two proximal apoA-I molecules per particle, while the number of apoA-II molecules varied from one dimeric apoA-II to two and then to three. For most of the above PPP profiles, an additional population that contained a single molecule of apoC-III in addition to apoA-I and/or apoA-II was detected. Upon composition analyses of individual subpopulations, LpA-I/A-II displayed comparable proportions for total protein (~58%), phospholipids (~21%) total cholesterol (~16%), triglycerides (~5%) and free cholesterol (~4%) across subfractions. LpA-I components, on the other hand, showed significant variability. This novel information on HDL subfractions will form a basis for better understanding particle specific functions of HDL.
The anti-atherogenic properties of high density lipoproteins (HDL) have made them an attractive target for new therapies against cardiovascular disease (CVD) (1;2). HDL has antioxidative (3) as well as anti-inflammatory properties (2) and is a vehicle for reverse cholesterol transport (RCT)–cholesterol transport from peripheral tissue to the liver for disposal (1;4). RCT begins with cellular cholesterol efflux to lipid-free apolipoprotein (apo ) A-I via ATP-binding cassette transporter A1 (ABCA1) thereby generating nascent HDL, or to more mature forms of HDL via ABCG1 (5–8). The compositions and particle sizes of early forms of HDL are modified by plasma lipases, lipid transfer proteins and lecithin:cholesterol acyltransferase (LCAT), which converts nascent discoidal HDL to spherical HDL (6;9–12). Neutral lipid content in HDL is modified by cholesteryl ester transfer protein (CETP) mediated exchange of their cholesteryl esters (CE) for triglycerides (TG) in the very low density lipoprotein (VLDL) (10;13). This remodeling generates a heterogeneous mixture of HDL particles with different sizes, shapes and apolipoprotein/lipid compositions.
Although recent studies have identified more than 100 proteins associated with human HDL (14;15), and these HDL-associated proteins have been shown to be subfraction-specific (16), the majority of HDL-proteins comprise of apoA-I (~70%) and apoA-II (~20%) (17). The remainder of the proteins are mainly composed of apoC’s (mainly apoC-III, ~2–4%), apoD and apoE (<2%) (17;18). From these compositional data, one can infer that many HDL-associated proteins are minor components, perhaps only transiently present during functional interactions and remodeling processes. Thus, the major HDL apolipoproteins are most likely the most important, but not the sole determinants of HDL compositions, particle sizes and functionality. Moreover, the distinct apolipoprotein compositions of HDL subfractions are expected to control their metabolic fates.
HDL occurs as LpA-I, which contains apoA-I but not apoA-II, and LpA-I/A-II, that contains both apoA-I and apoA-II (19;20). The interactions of LpA-I/A-II and LpA-I with plasma factors are different indicating that apoA-II modulates the effects of apoA-I (21–23). Although high plasma levels of apoA-II are associated with reduction in the risk of CVD events (24;25), there is little understanding on beneficial functions of apoA-II. Furthermore, structurally, even the molecular stoichiometry of apoA-I and apoA-II in individual HDL particles or segregation of other apolipoproteins into specific native HDL particles is not known. Such information would provide a basis for further insights into the functional consequences of these individual particles. It is clear that the LpA-I and LpA-I/A-II main populations undergo different metabolic processes. These include their sites of generation (26;27), different interaction capabilities with various plasma factors (21–23) and catabolism (28). In the context of their heterogeneity, several methods have been used to determine HDL-apolipoprotein stoichiometry including immuno-nephelometry (29;30) and cross-linking followed by gel based interpretations (31). The former method measures scattered light intensity as a function of particle size and the stoichiometry estimations are then carried out comparative to standards of known composition (29;30). The latter method determines the migration distance of cross-linked HDL particles by SDS-PAGE, and the molecular weight (Mr) determinations of these complexes are carried out with respect to a set of Mr standards. This method has been widely used to determine the number of apolipoprotein molecules in HDL (26;32–34). Streaking of the gel bands, due to variability in cross-linker addition, and unpredictable gel band migration due to location of the cross-linker formation in the protein sequence, render this method less than optimal for determining apolipoprotein stoichiometry (33). We developed a method combining cross-linking chemistry with MALDI-MS that, unambiguously, determines the number of apolipoprotein molecules in reconstituted (r) HDL particles (35). The stoichiometry of proximal apolipoproteins can be determined using this method based on the absolute masses of the cross-linked apolipoproteins. The method uniquely qualifies for this purpose, has moderate sensitivity and hence capable of capturing only the most abundant apolipoproteins present in HDL. To date, methods used to detect HDL apolipoproteins, including liquid chromatography-mass spectrometry (LC-MS), have not demonstrated the capability to determine molecular stoichiometry of apolipoproteins in HDL.
Here, we report, for the first time, apolipoprotein Protein Proximity Profiling (PPP), average particle diameters, apoprotein/lipid compositions in native LpA-I and LpA-I/A-II isolated from human plasma. We have used a unique set of chromatographic techniques to first separate HDL into LpA-I and LpA-I/A-II followed by the separation of each of these main pools into six individual subfractions (Figure 1). Cross linking, followed by the application of MALDI-MS technique optimized for native HDL, demonstrated clear differences in size, composition and the number of major apolipoproteins in LpA-I vs LpA-I/A-II. Our studies have also highlighted a unique distribution of apoC-III among different HDL subfractions.
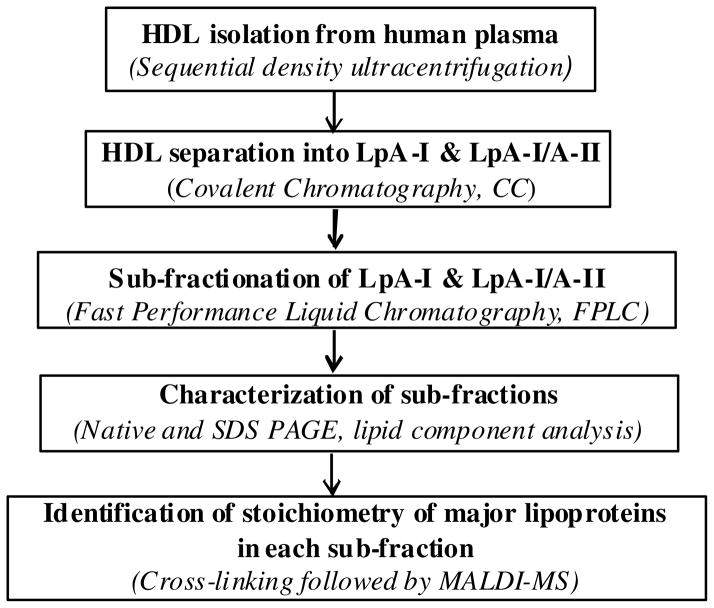
Figure 1.
Main experimental steps and techniques used in the study.
Experimental Methods
Isolation of HDL from human plasma
HDL was isolated from pooled-fresh human plasma upon sequential flotation at d = 1.063 g/ml and 1.21 g/ml as described previously (36;37). Plasma was obtained within two days of blood draw, from the Hoxworth Blood Bank, Cincinnati, OH.
HDL separation into LpA-I and LpA-I/A-II
Total HDL was separated into LpA-I and LpA-I/A-II subpopulations with the use of covalent chromatography (CC) incorporating thiopropyl sepharose beads (TPS, GE Healthcare, Piscataway, NJ) (38). Please see Supporting Information (SI) for method details.
LpA-I and LpA-I/A-II subfractionation
Individual LpA-I and LpA-I/A-II pools were further separated based on the average particle size by Size Exclusion Chromatography (SEC) using two Superose HR 6 columns (GE Healthcare, Piscataway, NJ) connected in tandem. Particle separation was carried out by introducing 10 mg of total protein in 250 μl into the column set up, operated at 0.4 ml/min flow rate. Each elution profile was divided into six sub-fractions, based on the elution volume, with the fraction number 1 (fr. 1) having the shortest elution time (hence the largest average particle size) and the fr. 6 having the largest elution time (hence the smallest average particle size). LpA-I/A-II and LpA-I subfractions with a common fraction number correspond to the same elution volume in the chromatographic profiles. In a specific case (see Discussion), 0.5 mg of total HDL protein was separated on four sizing columns connected in tandem (3 Superdex 200 followed by a single Superose 6 column).
Cross-linking of HDL subfractions and processing for MALDI-MS
All subfractions were cross-linked with 1:100 total protein to cross-linker bis(sulfosuccinimidyl) suberate (BS3) molar ratio at a total protein concentration of 1 mg/ml. Please see SI for method details including additional cross-linker use, delipidation and sample processing for MALDI.
MALDI-MS measurements and spectral processing
The measurements were carried out as before with few modifications (35). Please see SI for details. Spectral comparisons were carried out using MoverZ program (Genomic Solutions, Ann Arbor, MI). Spectra were subjected to 50% Fourier Smoothing and normalized to the same height using the most intense cross-linked mass peak in the spectrum within 50–130 kDa range, using Grams 32 software (Galactic Industries, Salem, NH).
Lipid component analysis
Total cholesterol (TC), free cholesterol (FC), total protein (TP), triglycerides (TG) and phospholipids (PL) in each subfraction were determined enzymatically (Wako Diagnostics, Richmond, VA) according to the manufacturer’s instructions on a Bio-Tech synergy HT micro-plate reader.
Gel electrophoresis
8–25% Phast Gradient gels were purchased from GE Healthcare (Piscataway, NJ ) along with the high and low molecular weight standards (Cat. Numbers 17-0445-01 and 17-0446-01); 4–30% gradient gels were cast in our laboratory. The diameter or the Mr of the cross linked complexes was calculated by comparing their migration distances with those of the standards.
Electron Microscopy Measurements
Please see SI for methods used in EM measurements.
Results
Native HDL subfractionation and characterization
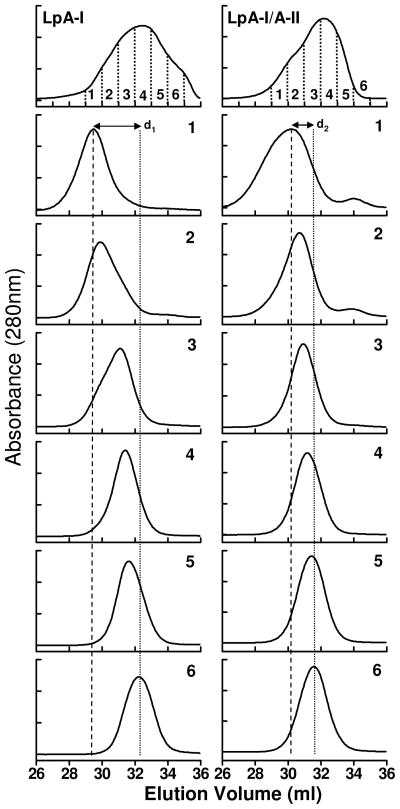
The SEC profiles of LpA-I, LpA-I/A-II and of the six individual subfractions generated there of are shown in Figure 2. LpA-I and LpA-I/A-II fractions, with same numbers, have the same elution volumes. As expected, these subfractions clearly indicated a decrease in the average particle diameters from fr. 1 to fr. 6. The shift in the average particle diameter for LpA-I (indicated by d1) is larger than for LpA-I/A-II (d2). This observation indicates that variation in the particle size was larger in LpA-I compared to that of LpA-I/A-II. This observation was confirmed with other independent HDL pools (data not shown).
Figure 2. Size exclusion chromatography of LpA-I and LpA-I/A-II.
Chromatographic traces of LpA-I and LpA-I/A-II upon separation on two Superose 6 columns in tandem (top panel). Each main pool was divided into six individual subfractions based on the elution volume and was numbered 1–6. The individual chromatography profile of each subfraction is shown below the corresponding main pool. The distances between the largest average particle size and the smallest average particle size are shown by d1 for LpA-I and d2 for LpA-I/A-II respectively. The average size of the largest and the smallest particles are shown by vertical short dash and long dash lines respectively.
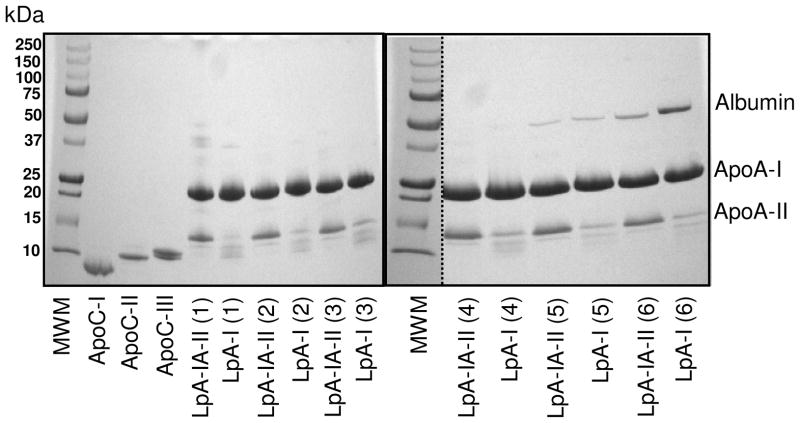
Based on SDS-PAGE (4–30%) performed under non reducing conditions, all LpA-I/A-II subfractions contained predominantly high amounts of apoA-II (greater than 90%) compared to LpA-I, thereby confirming effective separation of LpA-I from LpA-I/A-II (Figure 3). In addition to apoA-I and apoA-II, additional bands corresponding to proteins with lower Mrs were observed and were prominent in LpA-I (Figure 3). Comparison with apoC standards (apoC-I, apoC-II and apoC-III) indicated that these proteins were, likely, apoC-III1 and apoC-III2, the two major isomeric forms of apoC-III (39). Others have reported the third most abundant HDL apolipoprotein to be apoC-III, making up about 2–4 % of total HDL proteins (17;39). Moreover, serum albumin was specifically present in the smallest subfractions of LpA-I and LpA-I/A-II (fr. 6). The identities of apoA-I, apoA-II, albumin and apoC-III were confirmed by MALDI-Tof/Tof analysis, upon gel band extraction followed by tryptic digestion (data not shown). Moreover, based on Mr and MALDI/Tof/Tof, we were able to confirm the presence of apoD (29 kDa-major, 38 kDa-minor) and apoE (35 kDa) in LpA-I/A-II fr. 1 [Figure 3, Lane 1 (LpA-I/A-II)] with the identification of protein specific 2 and 7 peptides respectively from extracted gel bands. However, sequence evidence for the minor Mr band ~37 kDa on Lane 1 of LpA-I was not identified.
Figure 3. Major apolipoproteins in LpA-I and LpA-I/A-II subfractions as visualized by SDS-PAGE.
4–30% SDS-PAGE of isolated LpA-I, LpA-I/A-II subfractions (separated by FPLC, Figure 2; adjusted to 1 mg/ml total protein). ApoA-I, apoA-II band locations as well as albumin contamination (prevalent in LpA-I fractions 6) are indicated. ApoC-I, apoC-II and apoC-III standards were included. Lanes with molecular weight standards are denoted as MWM. Gels were stained with Coomassie Blue.
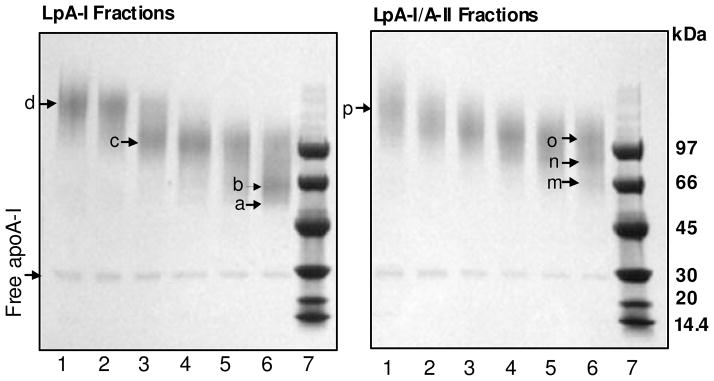
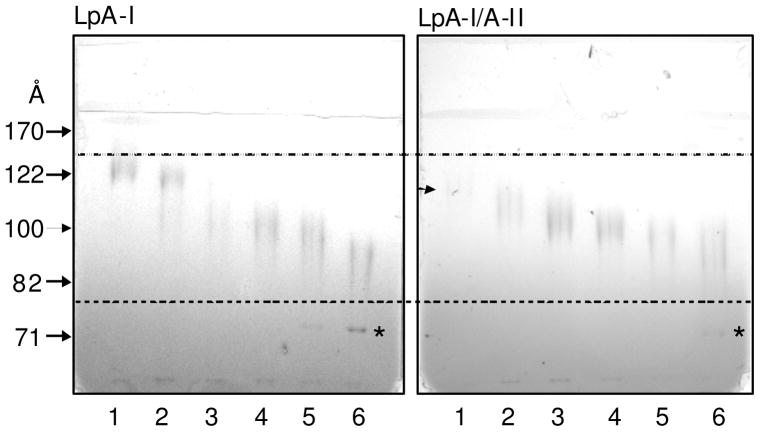
SDS-PAGE (4–15%) of cross-linked LpA-I and LpA-I/A-II subfractions revealed products with different migration patterns (Figure 4). The protein: cross-linker ratio of 1:100 was previously shown to be sufficient to almost fully cross-link proximal proteins within the cross-linker spacer arm length, 11.4 Ǻ, in discoidal rHDL particles (35). However, in native HDL a small fraction of apoA-I did not form intermolecular cross-links, implying that a fraction of apoA-I might be in an orientation not cross-linkable. Even though cross-linked subfractions appeared as smeared bands on the SDS-PAGE, a few prominent condensed bands could be pin-pointed for LpA-I and LpA-I/A-II (indicated by arrows, Figure 4). For LpA-I these bands are located ~ 52 kDa (a), 69 kDa (b), 97 kDa (c) and 152 kDa (d) while, for LpA-I/A-II they are located near 70 kDa (m), 88 kDa (n), 97 kDa (o) and 145 (p). The 69–70 kDa (b and m) bands correspond to internally cross-linked albumin and is outside the range of cross-linked complexes of LpA-I/A-II, but within the cross-linked masses for LpA-I. The apparent range of cross-linked masses across all six subfractions is larger for LpA-I compared to LpA-I/A-II. Unfortunately, exact mass ranges cannot be evaluated due to the broad streaking nature of the gel bands. Albumin migrated as a free protein, as seen by 8–25% Native PAGE (Figure 5, Lane 6 of LpA-I and LpA-I/A-II). This confirmed that albumin was present as a free contaminant protein and was not associated with HDL, contrary to what was predicted before (40). Based on the migration distances of cross-linked subfractions on Native PAGE, the average particle diameters for LpA-I and LpA-I/A-II ranged from 78–126 Å and 85–110 Å respectively (Figure 5). These dimensions are likely smaller than actual hydrodynamic diameters because cross-linking is known to reduce the apparent particle diameters. Native PAGE of non cross-linked subfractions at 1 mg/ml total protein concentration did not provide useful information due to complexes falling apart during electrophoresis (data not shown). Even though this could have been avoided by running samples at high total protein concentrations at or above 4 mg/ml) (16), we decided to maintain 1 mg/ml (standard physiological concentration for apoA-I) throughout all experiments.
Figure 4. SDS-PAGE of cross-linked LpA-I and LpA-I/A-II sub-fractions.
Individual subfractions of LpA-I and LpA-I/A-II were cross-linked at 1:100 total protein to BS3 cross-linker ratio at 1 mg/ml protein concentration and were subjected to 4–15% SDS-PAGE. Band “b” on Lane 6 (Panel LpA-I) and band “m” on Lane 6 (Panel LpA-I/A-II) correspond to albumin contamination (~66kDa). The small amount of apoA-I that did not involve in intermolecular cross-linking is shown as “free apoA-I”. Labeling of fractions are as in Figures 2 and 3. Low Molecular Weight (LMW) markers are on Lane 7. Gels were stained with Coomassie Blue.
Figure 5. Native PAGE characterization of cross-linked LpA-I and LpA-I/A-II.
Individual cross-linked LpA-I and LpA-I/A-II subfractions (at 1 mg/ml total protein concentration), were used in Figure 4 were subjected to 8–25% Native PAGE analysis. The locations of the hydro-dynamic diameters of the High Molecular Weight markers are indicated by arrows. Numbering of the sub-fractions is as in Figures 2, 3 and 4. Location of the LpA-I/A-II fraction 1, which is not clear on the gel, is marked with an arrow. Location of the internally cross-linked albumin in fraction 6 of LpA-I and LpA-I/A-II pools are marked by asterisks. Gels were stained with Coomassie Blue.
PPP of LpA-I and LpA-I/A-II subfractions
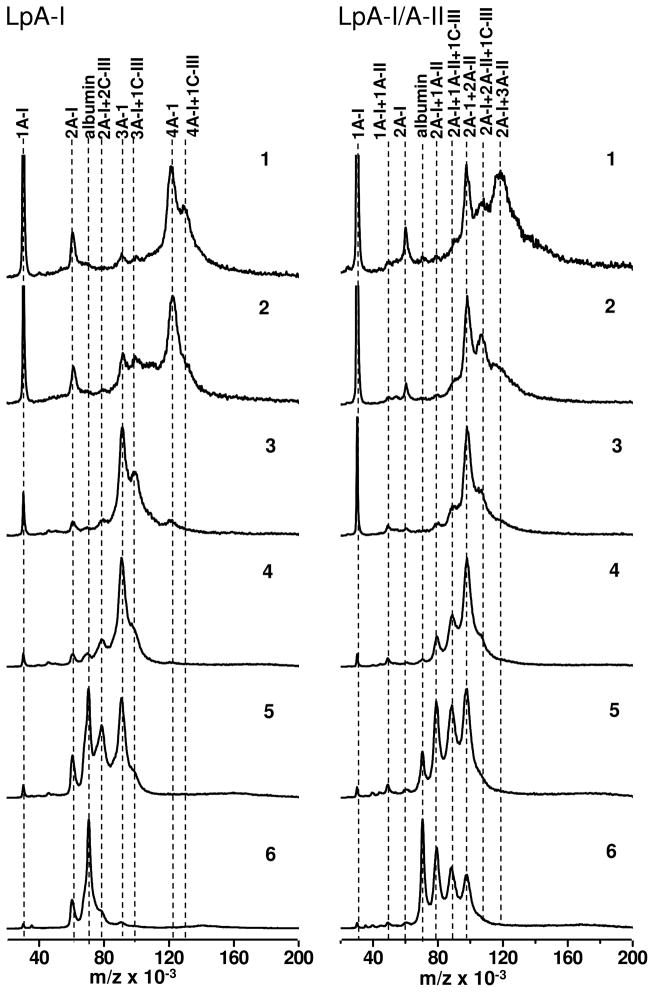
MALDI-MS of cross-linked LpA-I and LpA-I/A-II subfractions are shown in Figure 6. This MS technique has the ability to ionize and generate singly charged ions which are assignable directly to the Mr of the cross-linked complexes. All spectra were normalized to the peak height of one, using the most intense cross-linked peak in the spectrum. One of the major observations seen in MALDI spectra is that subfractions from both main pools show an average Mr shift towards higher masses when going from the smallest (fr. 6) to the largest (fr. 1) subfraction, implying that the larger particles have more apolipoprotein molecules per particle as seen by PAGE (Figure 4). However, unlike SDS-PAGE, each MALDI spectrum consisted of a set of resolved multiple peaks for which Mr can be determined within ~50 Da. Mass peaks in these spectra have originated from optimally cross-linked subfractions and hence consisted of intermolecularly cross-linked apolipoproteins within each HDL particle in a subfraction. Multiple peaks in each spectrum indicated the presence of multiple particle populations within a subfraction.
Figure 6. MALDI mass spectra of cross-linked LpA-I and LpA-I/A-II subfractions.
Individual cross-linked subfractions (shown in Figs. 4 and 5) were subjected to MALDI-MS measurements in 10–200 kDa range. Possible combinations of apolipoproteins attributable to major peaks in the MALDI spectrum is indicated above the vertical dotted lines that connects similar Mr in each spectrum. The peak location corresponds to the albumin contamination is indicated as “albumin”.
Under our cross-linking condition—1:100 total protein: BS3 at 1 mg/ml protein concentration—small LpA-I and LpA-I/A-II subfractions (frs. 6, 5 and 4) were fully cross-linked as seen by the diminution or the absence of a peak corresponding to free apoA-I that escaped intermolecular cross-linking (1A-I). However, when going from smaller to larger subfractions, the peak corresponding to monomeric apoA-I (1A-I) and cross-linked dimeric apoA-I (2A-I) were more prominent in the spectrum compared to the cross-linked complex (frs. 2 and 1). Since the cross-linking conditions were optimized for rHDL particles (35), we took steps to confirm that partial cross-linking was not due to insufficient protein: cross-linker ratio used in native HDL. To test this, we cross-linked total HDL as a function of increasing protein: BS3 ratio from 1:100 to 1:400 and recorded the spectra (data not shown). Three fold increment in the amount of BS3 did not change the peak height of the monomeric apoA-I that escaped intermolecular cross-linking, meaning, the peak corresponding to apoA-I was not due to insufficient cross linking (data not shown). Moreover, increasing the spacer arm length from 7.7 Ǻ (DSG) to 16.1 Ǻ (Sulfo-EGS) did not affect the MS peak height for monomeric apoA-I.
The increase in the amounts of monomeric apoA-I in larger subfractions seen by MALDI was not apparent on PAGE (Fig. 4). To test whether monomeric apoA-I ionization would be suppressed when dimeric apoA-I was present (a situation representative of small particles), we cross-linked rHDL that contained exactly two molecules of apoA-I at different ratios of protein to cross-linker (1:10 to 1:100). The presence of apoA-I dimer did not suppress monomeric apoA-I ionization (data not shown). Based on this observation, it can be suggested that the amount of lipid free apoA-I was very low in smaller subfractions despite less variability seen on the gel.
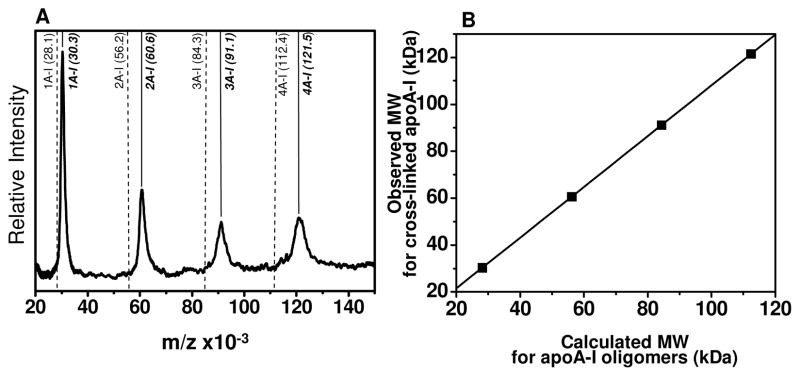
We assigned the major MALDI-MS peaks to a single HDL apolipoprotein or combinations of the most prevalent HDL apolipoproteins, whose presence and abundance were confirmed by SDS-PAGE of non-cross linked subfractions (Figure 3). The observed MALDI-MS peaks for cross-linked subfractions shifted from the calculated masses generated by the addition of molecular weights of apolipoproteins, due to the presence of cross-links in the complex (Table I). For example, every addition of BS3 cross-link resulted in 138.1 Da increment in the mass of the complex. The mass shift for different apolipoprotein combinations were predicted based on the Mr shifts observed for the cross-linked lipid-free apoA-I peaks in MALDI-MS (Figure 7). Lipid free apoA-I was processed in a way that was identical to that of native HDL subfractions by cross-linking in PBS at 1 mg/ml concentration with 1:100 molar ratio of protein: BS3 (Figure 7A). The linear regression between theoretical Mr for apoA-I oligomers and experimentally observed apoA-I MALDI-MS peaks correlated with a regression coefficient of 0.99. The equation representing the linear fit (Figure 7B) was used to predict the MALDI-MS Mr for major apolipoprotein combinations (Table 1, Predicted MALDI-MS). Theoretical addition of Mr for individual apolipoproteins (Calculated Mr), their predicted MALDI-MS, and the observed MALDI-MS peaks have been reported in Table 1. Except for two peaks, which could not be assigned to a single apolipoprotein combination due to the presence of two possibilities, clear assignments could be given to all the other peaks.
Table 1.
MALDI-MS peak assignments to most abundant HDL apolipoproteins and their combinations
| Apolipoprotein Combinations | Calculated Mr (kDa)a | Predicted MALDI-MS (kDa)b | Presence in MALDI-MSc | Clarity by MALDI-MS | Clarity by MALDI- MS and PAGE |
|---|---|---|---|---|---|
| 1A-I | 28.1 | 30.3d | YES (30.3) | YES | YES |
| 1A-I+1C-III | 36.9 | 39.8 | NO | N/A | N/A |
| 1A-I+1A-II | 45.5 | 49.1 | YES (49.1) | YES | YES |
| 2A-I | 56.2 | 60.6 | YES (60.6) | YES | YES |
| 1A-I+2A-II | 62.9 | 67.9 | NO | N/A | N/A |
| 2A-I+1C-III | 65.0 | 70.2 | YES (70.4) | NO | PARTIAL |
| Albumin | 66.4 | 71.7 | YES (70.4) | NO | PARTIAL |
| 2A-I+1A-II | 73.6 | 79.5 | YES (78.9) | NO | PARTIAL |
| 2A-I+2C-III | 73.6 | 79.5 | YES (78.3) | NO | PARTIAL |
| 2A-I+1A-II+1C-III | 82.4 | 88.9 | YES (88.7) | YES | YES |
| 3A-I | 84.3 | 91.1 | YES (91.1) | YES | YES |
| 2A-I+2A-II | 91.0 | 98.3 | YES (97.8) | YES | YES |
| 3A-I+1C-III | 93.0 | 100.5 | YES (98.9) | YES | YES |
| 2A-I+2A-II+1C-III | 99.8 | 107.8 | YES (105.6) | YES | YES |
| 3A-I+1A-II | 101.7 | 109.9 | NO | N/A | N/A |
| 3A-I+2C-III | 101.7 | 109.9 | NO | N/A | N/A |
| 2A-I+3A-II | 108.4 | 117.4 | YES (117.6) | YES | YES |
| 4A-I | 112.4 | 121.5 | YES (121.1) | YES | YES |
| 3A-I+2A-II | 119.1 | 128.7 | NO | N/A | N/A |
| 4A-I+1C-III | 121.15 | 130.9 | YES (129.5) | YES | YES |
| 5A-I | 140.5 | 151.9 | NO | N/A | N/A |
| 4A-I+1A-II | 147.2 | 159.1 | NO | N/A | N/A |
Masses of apolipoprotein combinations were calculated by addition of theoretical Mr of corresponding apolipoproteins.
Predicted MALDI-MS peak locations for cross-linked apolipoproteins in LpA-I and LpA-I/A-II subfractions were estimated based on the linear regression obtained for lipid free apoA-I measurements (Figure 7). A linear regression was generated between theoretical masses calculated for apoA-I monomer, dimer, trimer, tetramer and observed MALDI-MS masses for optimally cross-linked apoA-I oligomers, which resulted in a 0.99 correlation coefficient.
Presence of the predicted masses in MALDI-MS spectra are listed as “YES” or “NO” along with the values observed for masses in parenthesis.
Predicted masses for the rows highlighted in grey were directly taken from the cross-linked apoA-I oligomer measurements in Figure 7.
Figure 7. Linear regression between theoretical masses for oligomeric apoA-I vs observed masses for cross-linked apoA-I.
A. MALDI-MS of self associated lipid free apoA-I upon cross-linking with BS3 at 1:100 protein to BS3 molar ratio with the peak locations marked by solid lines and masses given in parentheses in Italics. Predicted Mr for non cross-linked oligomers are shown by dashed lines along with corresponding Mr in parentheses. B. Linear regression of predicted vs observed Mrs that are on panel A.
The PPPs of small subfractions of LpA-I indicated two apoA-Is (2A-Is, frs. 6 & 5) and three apoA-Is (3A-Is, frs. 5 & 4) per particle. Since there is no peak corresponding to 1A-I in these spectra, we suggest that the peak for 2A-I is not a result of partial cross-linking. Moreover, LpA-I/A-II, lacking a peak corresponding to 2A-I, further supported this interpretation and confirmed high quality of LpA-I/A-II separation from LpA-I. With increasing subfraction size, (frs. 3, 2, 1) free apoA-I (1A-I), due to incomplete cross-linking was seen. These PPPs indicated that LpA-I had mainly 3A-Is and 4A-Is per particle (frs. 2 and 1). While LpA-I had a quantized increase in the number of apoA-I molecules with increasing particle size, namely 2A-I (fr. 6, 5), 3A-I (fr. 4, 3) and 4A-I (fr. 2, 1), the combination (2A-I+2A-II) dominated all six LpA-I/A-II subfractions (Figure 6). In addition to this predominant particle, the smallest LpA-I/A-II subfraction had an additional peak, representing (2A-I+1A-II) (frs 6, 5) while the largest subfractions had (2A-I+3A-II) (fr. 1). In large LpA-I/A-II subfractions, the spectra indicated incomplete cross-linking, as seen for LpA-I large fractions (frs. 3-1).
There were cross-linked species containing one apoC-III molecule in each population including (3A-I+1C-III) and (4A-I+1C-III). However, the presence of (2A-I+1C-III) was precluded by overlap with an albumin contamination (Table I and Figure 6). Nevertheless, the Native PAGE of this subfraction indicated presence of a HDL complex, not just free albumin, hinting at the possible presence of (2A-I+1C-III). As observed for LpA-I, PPPs of each of the LpA-I/A-II subfractions were indicative of having particles with (2A-I+1A-II+1C-III), (2A-I+2A-II+1C-III). Due to the trailing of the spectral peaks for LpA-I/A-II (fr. 1) above 120 kDa, the presence of a particle population with (2A-I+3A-II+1C-III) was not clearly detectable (Figure 6). The presence of apoD and apoE preferentially in this LpA-I/A-II subfraction added to the complications of our interpretations [Figure 3, Lane 1 (LpA-I/A-II)].
Lipid Composition
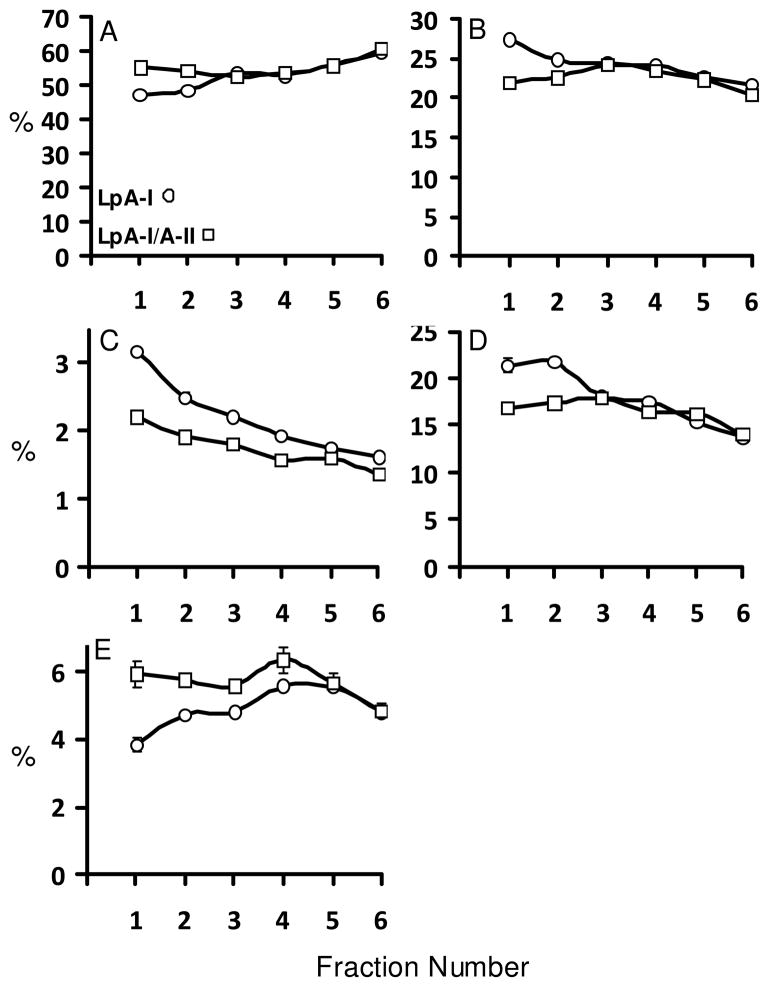
The lipid compositions, [ie. phospholipids (PL), total cholesterol (TC), free cholesterol (FC) and triglycerides (TG)], and the percentage of total protein of LpA-I and LpA-I/A-II were comparable to those reported for total HDL (41). The smaller LpA-I and LpA-I/A-II subfractions had similar lipid compositions (Figure 8). However, in larger subfractions, there were statistically significant differences between LpA-I and LpA-I/A-II for all lipid components and total protein. Consistently, the percentages of PL, FC and TC were higher in larger LpA-I fractions compared to LpA-I/A-II. On the other hand, the percentages of total protein and TG contents were significantly lower in the larger LpA-I subfraction compared to LpA-I/A-II. Another remarkable difference between LpA-I vs Lp-I/A-II subfractions was the compositional uniformity of LpA-I/A-II compared to LpA-I, across all six subfractions. This agreed with the more uniform particle sizes for the former (Figure 5) and less variability in composition in LpA-I/A-II.
Figure 8. Main lipid and total protein compositional analysis of individual LpA-I and LpA-I/A-II subfractions.
Total protein and lipid component analyses were carried out using WAKO chemical kits. The components are presented as mass percentages for individual subfractions. Subfraction labeling is as in Figures 2–6. Panels A, total protein; B, phospholipids; C, free cholesterol; D, total cholesterol; E, triglycerides.
Discussion
Major outcomes
Our measurements on LpA-I and LpA-I/A-II separated by size, unambiguously supported the presence of LpA-I with two, three and four molecules of apoA-I per particle. In contrast, all LpA-I/A-II subfractions contained only two molecules of apoA-I per particle supporting the concept that apoA-II had the ability to displace apoA-I from HDL (42;43). The number of dimeric apoA-II molecules in LpA-I/A-II subfractions varied from one to two to three. Prior reports indicated that up to two thirds of total HDL was LpA-I/A-II (20;38;44). Our data, collected on independent plasma batches, not only confirmed that approximately two thirds of HDL was LpA-I/A-II, but also revealed that the predominant LpA-I/A-II particle was (2A-I+2A-II). All LpA-I/A-II subfractions, having nearly uniform particle sizes, protein contents and major lipid components compared to quantized LpA-I particles, indicated extensive size rearrangements of large and small LpA-I upon apoA-II incorporation or direct incorporation of apoA-II onto mid sized LpA-I particles. Moreover, our data indicated a uniform distribution of apoC-III across all subfractions irrespective of particle sizes, opposing prior interpretations (Figure 6) (45). On the other hand, our analyses indicated predominant presence of apoD and apoE on a specific subfraction of LpA-I/A-II (Figure 3). Subfractions containing larger LpA-I particle populations consisted of higher TC and FC as well as estimated amounts of CE than LpA-I/A-II (Figure 8) indicating that these particles could be better mediators of the RCT pathway.
Method Optimization
The major obstacle, in compositional analysis of native HDL, is the presence of multiple particle populations with different combinations of apolipoproteins (19). Application of MALDI-MS, on cross-linked total HDL generated broad overlapping peaks with the most intense peak consistently centered ~98 kDa, originating mainly from (2A-I+2A-II) (data not shown). Our separation techniques reduced the complexity of total HDL and generated subfractions with manageable apolipoprotein heterogeneity for downstream analyses. Another challenge was the delipidation of native cross-linked subfractions, for which stringent delipidation conditions were required compared to rHDL, for obtaining clean MS peaks (see Experimental Procedures). Furthermore, we took precautions to prevent falsely detecting non covalent mass assemblies and aggregates of apolipoprotein molecules, which could be misinterpreted as a single cross-linked oligomeric entity by MALDI-MS. This was carried out by optimizing the laser power during spectral acquisitions on bovine serum albumin (BSA, Mr= 66.0 kDa), unmodified apoA-I (Mr=28.1 kDa) and cross-linked apoA-I until there were no observable oligomeric aggregates (i.e., two, three and four non-covalently bound molecules carrying a single charge, 2MH+, 3MH+ and 4MH+).
Molecular Stoichiometry
The presence of HDL particles containing 2–3 molecules of apoA-I in the highest density subfraction (HDL3c) and 4–5 molecules of apoA-I in the lowest density subfraction (HDL2b) has been reported (41). In addition, prior studies on HDL subfractions isolated by density followed by cross-linking and gel based methods, have estimated the presence of LpA-I particles with at least two, three and four molecules of apoA-I per particle (40;46;47). Supporting these findings, in vitro spherical rHDL generated by incubation of discoidal rHDL with LCAT alone and LCAT followed by CETP, were reported to contain three and two apoA-I molecules per particle respectively (32;48). Former studies on LpA-I/A-II, isolated by density subfractionation/immuno-affinity chromatography have reported an average molar ratio that ranges from 1:1 to 2:1 of apoA-I:apoA-II in LpA-I/A-II (20;47;49). For LpA-I/A-II we detected three different particle types (2A-I+1A-II), (2A-I+2A-II) and (2A-I+3A-II) with (2A-I+2A-II) being the most prominent.
Apo C-III also has been shown to possess the capability to displace apoA-I from HDL but not apoA-II implying perhaps a greater abundance of apoC-III in LpA-I. However, a prior study reported prominent presence of apoC-III in LpA-I/A-II (45). Our data showed that apoC-III was present in both LpA-I and LpA-I/A-II subfractions and in all the particle sizes. Moreover, our PPP indicated that most particle populations, that carried apoC-III contained a single inter molecularly cross-linkable apoC-III molecule, implying, most probably, these particle contained only one apoC-III. It was reported that both apoE (20;44) and apoD (45) were more abundant in LpA-I/A-II than in LpA-I. PAGE analysis [Figure 3, Lane 1 (LpA-I/A-II)], followed by MALDI-Tof/Tof carried out in our experiments indicated that apoE and apoD may be predominantly present in the largest subfraction of LpA-I/A-II.
Particle Size
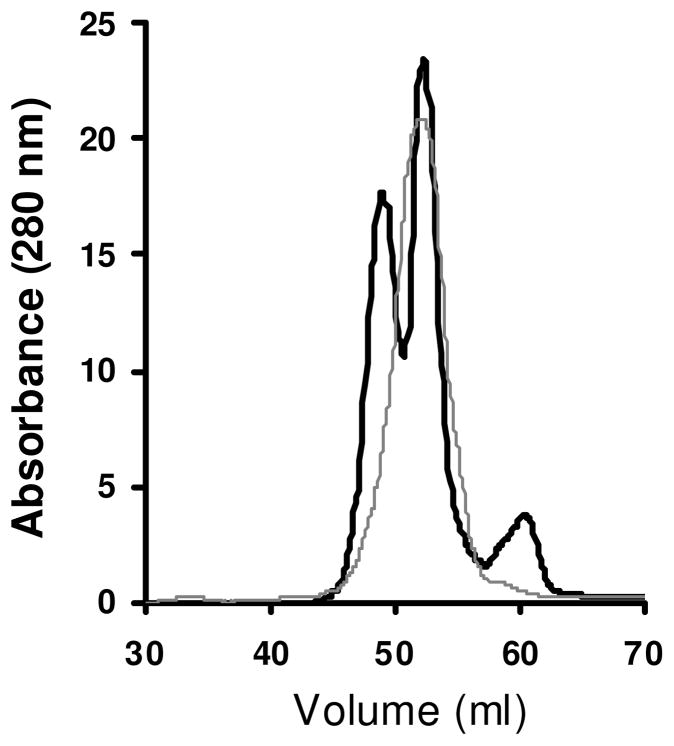
LpA-I and LpA-I/A-II separation on a two Superose 6 column set up resulted in very similar elution profiles for both pools (Fig. 1). However, PAGE analysis of different subfractions within FPLC profiles indicated different quantized particle sizes for LpA-I, while for LpA-I/A-II the variation in the particle diameters was smaller (Fig. 2 and 5). Hence we increased the resolution of chromatographic set up to broaden the elution profile for LpA-I/A-II and attempted to capture different particle sizes. LpA-I and LpA-I/A-II, separated on a set of four sizing columns in tandem (0.5 mg, 100 μl), further confirmed that LpA-I/A-II comprised of particle sizes that had less variability, while LpA-I was clearly composed of at least three distinctively different particle populations (Figure 9). While LpA-I/A-II eluted as a single peak, it overlapped with the mid peak of LpA-I (Figure 9). It was reported that LpA-I and LpA-I/A-II mainly associated with HDL2 and HDL3 respectively (44;47;49). Our FPLC data, upon comparison for the particle diameters reported for different density subfractions (47), implied that LpA-I/A-II would be associated with the higher density subfraction HDL3, while LpA-I covers both the HDL2 and HDL3 density range. The average particle diameters of cross-linked subfractions at that correspond to three FPLC peaks for LpA-I as seen by native PAGE were 71Ǻ, 82Ǻ and 105Ǻ respectively. These diameters agreed closely with the particle diameters 75–76Ǻ, 85Ǻ and 108Ǻ reported for LpA-I characterized upon immunoaffinity chromatographic separation of HDL (20;40). The reported diameter range for the entire population of LpA-I is 72–129Ǻ (47). Upon separation of LpA-I into six subfractions and subjecting to PAGE analysis with 1 mg/ml protein concentration, we were able to visualize minor population with ~125 Ǻ particle diameters (Figure 5, LpA-I, Lanes 1&2). However, we were not able to detect distinct particle sizes for LpA-I/A-II even under high resolution conditions. The PAGE analysis of FPLC peak in Figure 9 indicated that the average LpA-I/A-II particle size was ~85 Ǻ and size range of these particles was 85–108 Ǻ (Figure 5). This is in general agreement with a previous report which captured three particle populations within 78–97 Ǻ for LpA-I/A-II (47).
Figure 9. Size exclusion chromatography profiles of LpA-I and LpA-I/A-II.
LpA-I and LpA-I/A-II were separated on four sizing columns connected in tandem (three Superdex 200 columns followed by a Superose 6 column). LpA-I: black; LpA-I/A-II: grey.
Electron microscopic (EM) measurements performed on LpA-I and LpA-I/A-II main populations, averaging 500 random particles, indicated slightly different average particle diameters for LpA-I (80.9 ± 0.7 Ǻ) compared to LpA-I/A-II (88.2 ± 0.8 Ǻ). A prior study indicated that LpA-I/A-II particles were smaller (76.5 Ǻ) than LpA-I particles (99.8 Ǻ) (44). Notably, the average diameter could be dependent on the extent of abundance of the individual LpA-I particle populations of different diameters. In a situation in which the largest LpA-I particles were the most abundant, as was seen for HDL isolated from women, the average particle size could be shifted to a larger average particle diameter (46). It is also worth noting that EM produced less variability in the particle sizes compared to FPLC and Native PAGE techniques. This could be due to EM capturing only the most abundant particles in each population with near similar diameters. The width to length ratio (W/L) ratio of LpA-I (0.90 ± 0.05) and LpA-I/A-II (0.90 ± 0.05) indicated that both types of particles were spherical, a unique piece of evidence that was obtained by EM.
Limitations
The validity of our compositional analyses of HDL subfractions relies on ultracentrifugally isolated total HDL. While some reports have indicated density ultracentrifugation stripping off loosely bound proteins (45;50), studies have confirmed minimal perturbation to major apolipoproteins in HDL (20;44) including recovery of more than 90% of apoA-I and 100% apoA-II, under the salt and spin times used in our study (51). Data on apoC-III is expected to be similar to that of apoA-II because both apoA-II and apoC-III have the capability to displace apoA-I from HDL due to their higher lipophilic nature (43;52). Since apoE seemed to dissociate from HDL up to 50%, at least from rodent HDL, during ultracentrifugation (53), we cautiously report that there could be other subfractions that would contain apoE other than the largest subfraction of LpA-I/A-II. The success of the detection method we used in the study, MALDI-MS, solely relies on the optimal intermolecular cross-linking of all major apolipoproteins with in an HDL particle. Due to the presence of apoA-I in larger fractions, that escaped intermolecular cross-linking we could not rule out the presence of LpA-I particles containing more than four apoA-I molecules per particle. Experiments performed on independent batches of HDL confirmed the presence of a fraction of apoA-I in large particles not in a proper orientation for cross-linking with the rest of the proteins. This observation may also indicate protein molecules having moved apart in large particles due to a larger ncore. A recent study on HDL utilizing surface plasmon resonance spectroscopy revealed a relatively labile pool of apoA-I in HDL particles (54). Two different types of apoA-I pools in HDL was also reported based on ultracentrifugation (51). Despite optimal cross-linking of small subfractions, our data did not capture the presence of very small HDL particles with perhaps one apoA-I per particle (55).
While MALDI-MS qualifies as a unique technique for determining major apolipoprotein molecular stoichiometry, to determine stoichiometry of relatively minor apolipoproteins as apoE and apoD, further sub-fractionations followed by specific immuno-affinity isolation of these particles would be necessary. However, limitations due to stripping off apoE during ultra-centrifugation, will remain a problem. Use of FPLC in the place of ultracentrifugation could not be an alternative due to the minute amounts of sample obtainable overlap of HDL elution profile with other abundant proteins as albumin and immunoglobulins (15). Use of lipid capture reagents to bind HDL proteins and washing off abundant proteins would not be a solution for our experiments due to irreversible binding of HDL and questionable elution of native HDL complexes from lipid binding reagents (15).
Future Studies
The most important implication of PPP of different HDL subfractions would be getting insights into their functional properties. Functional assays could be carried out to determine selective cholesteryl ester uptake (via SR-B1), phospholipid hydrolysis (by Endothelial Lipase), triglyceride hydrolysis (by Hepatic Lipase) and to determine the ability to sequester cholesterol (via ABCG1). Such experiments could yield critical information as to which subfractions of HDL are more active in certain functions and would generate information to correlate functional properties with major apolipoprotein PPP. Subfraction specific major enzyme residence and their activity would be another important area of investigation. However, further improvements of gentler techniques in isolating HDL other than ultracentrifugation would be necessary prior to such analyses.
Supplementary Material
Acknowledgments
We thank Drs. Kerry Anne-Rye and Nataraja Sarma Vaitinadin for their insightful comments and Ms. Cali Smith for her excellent administrative assistance.
Abbreviations
- apo
apolipoprotein
- ABCA1
ATP-binding cassette transporter A1
- βME
β mercaptoethanol
- BS3
bis[sulfosuccinimidyl] suberate
- CC
covalent chromatography
- CE
cholesteryl ester
- CETP
cholesteryl ester transfer protein
- DTT
dithiothreitol
- FC
free cholesterol
- HDL
high density lipoproteins
- fr
fraction number
- LCAT
lecithin:cholesterol acyltransferase
- LpA-I
HDL with apoA-I but without apoA-II
- LpA-I/A-II
HDL with apoA-I and apoA-II
- Mr
Molecular weight
- PBS
Phosphate Buffered Saline
- PC
phosphatidylcholine
- PL
phospholipids
- PPP
protein proximity profile
- RCT
reverse cholesterol transport
- rHDL
reconstituted HDL
- SEC
size exclusion chromatography
- STB
Standard Tris Buffer
- TBS
Tris-Buffered Saline
- TFA
trifluoro acetic acid
- TC
total cholesterol
- TG
triglycerides
- Tof
Time of Flight
- TP
total protein
- TPS
thiopropyl sepharose
- VLDL
very low density lipoprotein
Footnotes
The work was supported by the grants from the National Institutes of Health to RAGDS (NHLBI: HL087561) and to HJP (HL030914 and HL056865).
Details of the Experimental Methods are available free of charge via the internet at http://pubs.acs.org
Reference List
- 1.Barter P, Kastelein J, Nunn A, Hobbs R. High density lipoproteins (HDLs) and atherosclerosis; the unanswered questions. Atherosclerosis. 2003;168:195–211. doi: 10.1016/s0021-9150(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 2.Ansell BJ, Watson KE, Fogelman AM, Navab M, Fonarow GC. High-density lipoprotein function recent advances. J Am Coll Cardiol. 2005;46:1792–1798. doi: 10.1016/j.jacc.2005.06.080. [DOI] [PubMed] [Google Scholar]
- 3.Navab M, Ananthramaiah GM, Reddy ST, Van Lenten BJ, Ansell BJ, Fonarow GC, Vahabzadeh K, Hama S, Hough G, Kamranpour N, Berliner JA, Lusis AJ, Fogelman AM. The oxidation hypothesis of atherogenesis: the role of oxidized phospholipids and HDL. J Lipid Res. 2004;45:993–1007. doi: 10.1194/jlr.R400001-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Curtiss LK, Valenta DT, Hime NJ, Rye KA. What is so special about apolipoprotein AI in reverse cholesterol transport? Arterioscler Thromb Vasc Biol. 2006;26:12–19. doi: 10.1161/01.ATV.0000194291.94269.5a. [DOI] [PubMed] [Google Scholar]
- 5.Arakawa R, Tsujita M, Iwamoto N, Ito-Ohsumi C, Lu R, Wu CA, Shimizu K, Aotsuka T, Kanazawa H, Abe-Dohmae S, Yokoyama S. Pharmacological inhibition of ABCA1 degradation increases HDL biogenesis and exhibits antiatherogenesis. J Lipid Res. 2009;50:2299–2305. doi: 10.1194/jlr.M900122-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zannis VI, Chroni A, Krieger M. Role of apoA-I, ABCA1, LCAT, and SR-BI in the biogenesis of HDL. Journal of Molecular Medicine. 2006;84:276–294. doi: 10.1007/s00109-005-0030-4. [DOI] [PubMed] [Google Scholar]
- 7.Yvan-Charvet L, Wang N, Tall AR. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler Thromb Vasc Biol. 2010;30:139–143. doi: 10.1161/ATVBAHA.108.179283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith JD. Insight into ABCG1-mediated cholesterol efflux. Arterioscler Thromb Vasc Biol. 2006;26:1198–1200. doi: 10.1161/01.ATV.0000221217.86465.66. [DOI] [PubMed] [Google Scholar]
- 9.Asztalos BF, Schaefer EJ, Horvath KV, Yamashita S, Miller M, Franceschini G, Calabresi L. Role of LCAT in HDL remodeling: investigation of LCAT deficiency states. J Lipid Res. 2007;48:592–599. doi: 10.1194/jlr.M600403-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Klerkx AH, El Harchaoui K, van der Steeg WA, Boekholdt SM, Stroes ES, Kastelein JJ, Kuivenhoven JA. Cholesteryl ester transfer protein (CETP) inhibition beyond raising high-density lipoprotein cholesterol levels: pathways by which modulation of CETP activity may alter atherogenesis. Arterioscler Thromb Vasc Biol. 2006;26:706–715. doi: 10.1161/01.ATV.0000205595.19612.c9. [DOI] [PubMed] [Google Scholar]
- 11.Huuskonen J, Wohlfahrt G, Jauhiainen M, Ehnholm C, Teleman O, Olkkonen VM. Structure and phospholipid transfer activity of human PLTP: analysis by molecular modeling and site-directed mutagenesis. J Lipid Res. 1999;40:1123–1130. [PubMed] [Google Scholar]
- 12.Clay MA, Newnham HH, Forte TM, Barter PI. Cholesteryl ester transfer protein and hepatic lipase activity promote shedding of apo A-I from HDL and subsequent formation of discoidal HDL. Biochim Biophys Acta. 1992;1124:52–58. doi: 10.1016/0005-2760(92)90125-f. [DOI] [PubMed] [Google Scholar]
- 13.Lagrost L. Regulation of cholesteryl ester transfer protein (CETP) activity: review of in vitro and in vivo studies. [Review] Biochim Biophys Acta. 1994;1215:209–236. doi: 10.1016/0005-2760(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 14.Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, Byun J, Vuletic S, Kassim S, Singh P, Chea H, Knopp RH, Brunzell J, Geary R, Chait A, Zhao XQ, Elkon K, Marcovina S, Ridker P, Oram JF, Heinecke JW. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon SM, Deng J, Lu LJ, Davidson WS. Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography. J Proteome Res. 2010;9:5239–5249. doi: 10.1021/pr100520x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson WS, Silva RA, Chantepie S, Lagor WR, Chapman MJ, Kontush A. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function. Arterioscler Thromb Vasc Biol. 2009;29:870–876. doi: 10.1161/ATVBAHA.109.186031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Havel PJ, Goldstein JL, Brown MS. Lipoproteins and Lipid Transport. In: Bondy PK, Rosenburg LE, editors. The Metabolic Control of Disease. Philadelphia, PA: 1980. pp. 393–494. [Google Scholar]
- 18.Bergeron N, Kotite L, Verges M, Blanche P, Hamilton RL, Krauss RM, Bensadoun A, Havel RJ. Lamellar lipoproteins uniquely contribute to hyperlipidemia in mice doubly deficient in apolipoprotein E and hepatic lipase. Proc Natl Acad Sci U S A. 1998;95:15647–15652. doi: 10.1073/pnas.95.26.15647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonas A. Lipoprotein Structure. In: Vance DE, Vance JE, editors. Biochemistry of Lipids, Lipoproteins and Membranes. Elsevier Science; 2002. pp. 483–504. [Google Scholar]
- 20.Cheung MC, Albers JJ. Characterization of lipoprotein particles isolated by immunoaffinity chromatography. Particles containing A-I and A-II and particles containing A-I but no A-II. J Biol Chem. 1984;259:12201–12209. [PubMed] [Google Scholar]
- 21.de Beer MC, Durbin DM, Cai L, Mirocha N, Jonas A, Webb NR, de Beer FC, van der Westhuyzen DR. Apolipoprotein A-II modulates the binding and selective lipid uptake of reconstituted high density lipoprotein by scavenger receptor BI. J Biol Chem. 2001;276:15832–15839. doi: 10.1074/jbc.M100228200. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, von Eckardstein A, Wu S, Assmann G. Cholesterol efflux, cholesterol esterification, and cholesteryl ester transfer by LpA-I and LpA-I/A-II in native plasma. Arteriosclerosis Thrombosis & Vascular Biology. 1995;15:1412–1418. doi: 10.1161/01.atv.15.9.1412. [DOI] [PubMed] [Google Scholar]
- 23.Hime NJ, Barter PJ, Rye KA. The influence of apolipoproteins on the hepatic lipase-mediated hydrolysis of high density lipoprotein phospholipid and triacylglycerol. J Biol Chem. 1998;273:27191–27198. doi: 10.1074/jbc.273.42.27191. [DOI] [PubMed] [Google Scholar]
- 24.Birjmohun RS, Dallinga-Thie GM, Kuivenhoven JA, Stroes ES, Otvos JD, Wareham NJ, Luben R, Kastelein JJ, Khaw KT, Boekholdt SM. Apolipoprotein A-II is inversely associated with risk of future coronary artery disease. Circulation. 2007;116:2029–2035. doi: 10.1161/CIRCULATIONAHA.107.704031. [DOI] [PubMed] [Google Scholar]
- 25.Roselli della RG, Lapolla A, Sartore G, Rossetti C, Zambon S, Minicuci N, Crepaldi G, Fedele D, Manzato E. Plasma lipoproteins, apoproteins and cardiovascular disease in type 2 diabetic patients. A nine-year follow-up study. Nutr Metab Cardiovasc Dis. 2003;13:46–51. doi: 10.1016/s0939-4753(03)80167-9. [DOI] [PubMed] [Google Scholar]
- 26.Clay MA, Pyle DH, Rye KA, Barter PJ. Formation of spherical, reconstituted high density lipoproteins containing both apolipoproteins A-I and A-II is mediated by lecithin:cholesterol acyltransferase. J Biol Chem. 2000;275:9019–9025. doi: 10.1074/jbc.275.12.9019. [DOI] [PubMed] [Google Scholar]
- 27.Gillard BK, Lin HY, Massey JB, Pownall HJ. Apolipoproteins A-I, A-II and E are independently distributed among intracellular and newly secreted HDL of human hepatoma cells. Biochim Biophys Acta. 2009;1791:1125–1132. doi: 10.1016/j.bbalip.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Beer MC, van der Westhuyzen DR, Whitaker NL, Webb NR, de Beer FC. SR-BI-mediated selective lipid uptake segregates apoA-I and apoA-II catabolism. J Lipid Res. 2005;46:2143–2150. doi: 10.1194/jlr.M500068-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Heuck CC, Erbe I, Frech K, Lohse P, Munscher G. Immunonephelometry of apolipoprotein A-II in hyperlipoproteinemic serum. Clin Chem. 1983;29:1385–1388. [PubMed] [Google Scholar]
- 30.Heuck CC, Erbe I, Flint-Hansen P. Immunonephelometric determination of apolipoprotein A-1 in hyperlipoproteinemic serum. Clin Chem. 1983;29:120–125. [PubMed] [Google Scholar]
- 31.Swaney JB, O’Brien K. Cross-linking studies of the self-association properties of apo- A-I and apo-A-II from human high density lipoprotein. J Biol Chem. 1978;253:7069–7077. [PubMed] [Google Scholar]
- 32.Silva RA, Huang R, Morris J, Fang J, Gracheva EO, Ren G, Kontush A, Jerome WG, Rye KA, Davidson WS. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc Natl Acad Sci U S A. 2008;105:12176–12181. doi: 10.1073/pnas.0803626105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhat S, Sorci-Thomas MG, Alexander ET, Samuel MP, Thomas MJ. Intermolecular contact between globular N-terminal fold and C-terminal domain of ApoA-I stabilizes its lipid-bound conformation: studies employing chemical cross-linking and mass spectrometry. J Biol Chem. 2005;280:33015–33025. doi: 10.1074/jbc.M505081200. [DOI] [PubMed] [Google Scholar]
- 34.Rye KA, Garrety KH, Barter PJ. Preparation and characterization of spheroidal, reconstituted high-density lipoproteins with apolipoprotein A-I only or with apolipoprotein A-I and A-II. Biochim Biophys Acta. 1993;1167:316–325. doi: 10.1016/0005-2760(93)90235-2. [DOI] [PubMed] [Google Scholar]
- 35.Massey JB, Pownall HJ, Macha S, Morris J, Tubb MR, Silva RA. Mass spectrometric determination of apolipoprotein molecular stoichiometry in reconstituted high density lipoprotein particles. J Lipid Res. 2009;50:1229–1236. doi: 10.1194/jlr.D800044-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lund-Katz S, Phillips MC. Packing of cholesterol molecules in human low-density lipoprotein. Biochemistry. 1986;25:1562–1568. doi: 10.1021/bi00355a016. [DOI] [PubMed] [Google Scholar]
- 37.Durbin DM, Jonas A. The effect of apolipoprotein A-II on the structure and function of apolipoprotein A-I in a homogeneous reconstituted high density lipoprotein particle. Journal of Biological Chemistry. 1997;272:31333–31339. doi: 10.1074/jbc.272.50.31333. [DOI] [PubMed] [Google Scholar]
- 38.Rosales C, Gillard BK, Courtney HS, Blanco-Vaca F, Pownall HJ. Apolipoprotein modulation of streptococcal serum opacity factor activity against human plasma high-density lipoproteins. Biochemistry. 2009;48:8070–8076. doi: 10.1021/bi901087z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito Y, Breslow JL, Chait BT. Apolipoprotein C-III0 lacks carbohydrate residues: use of mass spectrometry to study apolipoprotein structure. J Lipid Res. 1989;30:1781–1787. [PubMed] [Google Scholar]
- 40.Duverger N, Rader D, Duchateau P, Fruchart JC, Castro G, Brewer HB., Jr Biochemical characterization of the three major subclasses of lipoprotein A-I preparatively isolated from human plasma. Biochemistry. 1993;32:12372–12379. doi: 10.1021/bi00097a014. [DOI] [PubMed] [Google Scholar]
- 41.Kontush A, Chapman MJ. Functionally defective high-density lipoprotein: a new therapeutic target at the crossroads of dyslipidemia, inflammation, and atherosclerosis. Pharmacol Rev. 2006;58:342–374. doi: 10.1124/pr.58.3.1. [DOI] [PubMed] [Google Scholar]
- 42.Lagocki PA, Scanu AM. In vitro modulation of the apolipoprotein composition of high density lipoprotein. Displacement of apolipoprotein A-I from high density lipoprotein by apolipoprotein A-II. J Biol Chem. 1980;255:3701–3706. [PubMed] [Google Scholar]
- 43.Edelstein C, Halari M, Scanu AM. On the mechanism of the displacement of apolipoprotein A-I by apolipoprotein A-II from the high density lipoprotein surface. Effect of concentration and molecular forms of apolipoprotein A-II. J Biol Chem. 1982;257:7189–7195. [PubMed] [Google Scholar]
- 44.James RW, Hochstrasser D, Tissot JD, Funk M, Appel R, Barja F, Pellegrini C, Muller AF, Pometta D. Protein heterogeneity of lipoprotein particles containing apolipoprotein A-I without apolipoprotein A-II and apolipoprotein A-I with apolipoprotein A-II isolated from human plasma. J Lipid Res. 1988;29:1557–1571. [PubMed] [Google Scholar]
- 45.Nestruck AC, Niedmann PD, Wieland H, Seidel D. Chromatofocusing of human high density lipoproteins and isolation of lipoproteins A and A-I. Biochim Biophys Acta. 1983;753:65–73. doi: 10.1016/0005-2760(83)90099-1. [DOI] [PubMed] [Google Scholar]
- 46.Duverger N, Rader D, Brewer HB., Jr Distribution of subclasses of HDL containing apoA-I without apoA- II (LpA-I) in normolipidemic men and women. Arteriosclerosis & Thrombosis. 1994;14:1594–1599. doi: 10.1161/01.atv.14.10.1594. [DOI] [PubMed] [Google Scholar]
- 47.Cheung MC, Nichols AV, Blanche PJ, Gong EL, Franceschini G, Sirtori CR. Characterization of A-I-containing lipoproteins in subjects with A-I Milano variant. Biochim Biophys Acta. 1988;960:73–82. doi: 10.1016/0005-2760(88)90011-2. [DOI] [PubMed] [Google Scholar]
- 48.Jonas A, Wald JH, Toohill KL, Krul ES, Kezdy KE. Apolipoprotein A-I structure and lipid properties in homogeneous, reconstituted spherical and discoidal high density lipoproteins. J Biol Chem. 1990;265:22123–22129. [PubMed] [Google Scholar]
- 49.Atmeh RF, Shepherd J, Packard CJ. Subpopulations of apolipoprotein A-I in human high-density lipoproteins. Their metabolic properties and response to drug therapy. Biochim Biophys Acta. 1983;751:175–188. doi: 10.1016/0005-2760(83)90172-8. [DOI] [PubMed] [Google Scholar]
- 50.McVicar JP, Kunitake ST, Hamilton RL, Kane JP. Characteristics of human lipoproteins isolated by selected-affinity immunosorption of apolipoprotein A-I. Proc Natl Acad Sci U S A. 1984;81:1356–1360. doi: 10.1073/pnas.81.5.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kunitake ST, Kane JP. Factors affecting the integrity of high density lipoproteins in the ultracentrifuge. J Lipid Res. 1982;23:936–940. [PubMed] [Google Scholar]
- 52.Pownall HJ, Pao Q, Rohde M, Gotto AM. Lipoprotein-apoprotein exchange in aqueous systems: relevance to the occurrence of apoA-I and apoC proteins in a common particle. Biochem Biophys Res Commun. 1978;85:408–414. doi: 10.1016/s0006-291x(78)80057-6. [DOI] [PubMed] [Google Scholar]
- 53.van’t HF, Havel RJ. Metabolism of apolipoprotein E in plasma high density lipoproteins from normal and cholesterol-fed rats. J Biol Chem. 1982;257:10996–11001. [PubMed] [Google Scholar]
- 54.Lund-Katz S, Nguyen D, Dhanasekaran P, Kono M, Nickel M, Saito H, Phillips MC. Surface plasmon resonance analysis of the mechanism of binding of apoA-I to high density lipoprotein particles. J Lipid Res. 2010;51:606–617. doi: 10.1194/jlr.M002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Atmeh RF, bd Elrazeq IO. Small high density lipoprotein subclasses: some of their physico-chemical properties and stability in solution. Acta Biochim Pol. 2005;52:515–525. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.