Abstract
More than a hundred biochemical species, activated by neurotransmitters binding to transmembrane receptors, are important in long-term potentiation (LTP) and long-term depression (LTD). To investigate which species and interactions are critical for synaptic plasticity, many computational postsynaptic signal transduction models have been developed. The models range from simple models with a single reversible reaction to detailed models with several hundred kinetic reactions. In this study, more than a hundred models are reviewed, and their features are compared and contrasted so that similarities and differences are more readily apparent. The models are classified according to the type of synaptic plasticity that is modeled (LTP or LTD) and whether they include diffusion or electrophysiological phenomena. Other characteristics that discriminate the models include the phase of synaptic plasticity modeled (induction, expression, or maintenance) and the simulation method used (deterministic or stochastic). We find that models are becoming increasingly sophisticated, by including stochastic properties, integrating with electrophysiological properties of entire neurons, or incorporating diffusion of signaling molecules. Simpler models continue to be developed because they are computationally efficient and allow theoretical analysis. The more complex models permit investigation of mechanisms underlying specific properties and experimental verification of model predictions. Nonetheless, it is difficult to fully comprehend the evolution of these models because (1) several models are not described in detail in the publications, (2) only a few models are provided in existing model databases, and (3) comparison to previous models is lacking. We conclude that the value of these models for understanding molecular mechanisms of synaptic plasticity is increasing and will be enhanced further with more complete descriptions and sharing of the published models.
Keywords: computational model, kinetic model, long-term depression, long-term potentiation, plasticity, postsynaptic signal transduction model
1. Introduction
Synaptic plasticity is an activity-dependent change in the strength or efficacy of the synaptic connection between a pre- and postsynaptic neuron. It is induced with brief periods of synaptic activity, for example, using tetanic, high-frequency neuronal activity. Changes in synapses, in general, can last from milliseconds into years. These long-lasting changes, which require protein synthesis and gene transcription, are suggested to lead to learning and formation of memories.
The long-term activity-dependent strengthening and weakening of synapses are known as long-term potentiation (LTP; Bliss and Gardner-Medwin, 1973; Bliss and Lømo, 1973) and long-term depression (LTD; Ito et al., 1982; Ito, 1989; Dudek and Bear, 1992), respectively. Frequency-dependent LTP and LTD in the cornu ammonis 1 (CA1) region of the hippocampus, triggered by activation of N-methyl-d-aspartate (NMDA) receptors (NMDARs), are the most studied forms of long-term plasticity (see, e.g., Malenka and Bear, 2004; Citri and Malenka, 2008). In addition to hippocampal NMDAR-dependent LTP and LTD, diverse forms of LTP and LTD have been discovered in different brain regions. One example of non-NMDAR-dependent plasticity is cerebellar LTD. Some forms of LTP require neither the NMDA nor the non-NMDA ionotropic glutamate receptors (non-NMDARs include kainate receptors and α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptors, AMPARs), but do require activation of metabotropic glutamate receptors (mGluRs). This form is found, for example, in the CA1 region of the hippocampus (Lanté et al., 2006). Despite the variation in NMDAR dependence, all forms of synaptic plasticity are calcium ion (Ca2+)-dependent; only the mechanisms for Ca2+ elevation vary.
Two broad types of computational models, phenomenological and biophysical models, have been developed to understand the pre- and postsynaptic events in LTP and LTD. Phenomenological models use abstract equations to describe a relationship between neuronal activity and synaptic plasticity. Biophysical models include electrophysiological models, biochemical models, and models that include both electrophysiological properties and biochemical reactions (signaling pathways) underlying the relationship between neuronal activity and synaptic plasticity, though even these include simplifications because all the mechanisms cannot be modeled in detail. The focus of the present study is on biophysical models which concentrate on postsynaptic biochemical reactions.
This review presents an overview of 117 postsynaptic signal transduction models, categorizes them so that similarities and differences are more readily apparent, and explains how these models can be used to identify key molecules and address questions related to mechanisms underlying LTP and LTD. Section 2 presents the biological background of synaptic plasticity, Section 3 classifies the computational postsynaptic signal transduction models, and Section 4 summarizes the directions and trends of this field.
2. Synaptic Plasticity
Many different classification schemes for synaptic plasticity exist. Synaptic potentiation can be classified into three main types: short-term potentiation (STP), which lasts as long as 30–45 min; early phase LTP (E-LTP), which lasts for 1–2 h; and late phase LTP (L-LTP), which persists for considerably more than 2 h (Sweatt, 1999; Soderling and Derkach, 2000; Citri and Malenka, 2008). Synaptic depression, on the other hand, is typically classified into two types: short-term depression (STD) and LTD (Ito, 2001); though there appears to be an early and late phase LTD (E-LTD, L-LTD) also (Kauderer and Kandel, 2000). In addition, all types of plasticity involve three processes: induction, in which the mechanisms leading to plasticity are engaged; expression, which involves mechanisms allowing the plasticity to be exhibited and measured; and maintenance, which involves processes occurring after the induction phase is complete and allowing the plasticity to persist for long periods of time (Sweatt, 1999).
2.1. Mechanisms to trigger synaptic plasticity
Many different plasticity induction protocols have been developed. In general, potentiation is induced by a high-frequency stimulation and depression by a low-frequency stimulation of a chemical synapse, but there are variations in the experimental procedures depending on the cell type. Short-term plasticity is triggered typically by short trains of stimulation (Citri and Malenka, 2008). LTP is typically triggered with longer 1 s trains of high-frequency (100 Hz) stimulation (Citri and Malenka, 2008). One train triggers only E-LTP, whereas repetitive trains trigger L-LTP (Citri and Malenka, 2008). L-LTD is typically triggered with prolonged repetitive low-frequency (1 Hz) stimulation (Citri and Malenka, 2008). Theta stimulation consists of short bursts of trains repeated with 200 ms intervals and produces L-LTP, even though the number of pulses is more similar to that producing E-LTP. Spike-timing-dependent plasticity (STDP) is another protocol to trigger LTP as well as LTD. In STDP, pre- and postsynaptic neurons are stimulated independently and the timing between pre- and postsynaptic spikes determines whether potentiation or depression occurs (Markram et al., 1997; Bi and Poo, 1998; Bi and Rubin, 2005; Dan and Poo, 2006).
2.2. Molecular mechanisms of synaptic plasticity
There are various mechanisms, both pre- and postsynaptic, that lead to changes in synaptic strength, for example changes in neurotransmitter release, conductance of receptors, numbers of receptors, numbers of active synapses, and structure of synapses (Hayer and Bhalla, 2005). Several reviews about the molecular mechanisms underlying synaptic plasticity have been published (see, e.g., Bliss and Collingridge, 1993; Malenka and Nicoll, 1999; Sweatt, 1999; Soderling and Derkach, 2000; Ito, 2002; Lisman et al., 2002; Malenka and Bear, 2004; Blitzer et al., 2005; Cooke and Bliss, 2006; Wang et al., 2006; Bruel-Jungerman et al., 2007; Citri and Malenka, 2008; Santos et al., 2009). Cytosolic Ca2+ is inarguably the most critical factor: chemical buffering of Ca2+ or pharmacological blocking of Ca2+ influx prevents both potentiation and depression. There are several sources of Ca2+, depending on the brain region and the cell type. Influx through NMDARs is the most common source for LTP; influx through Ca2+-permeable AMPARs, voltage-gated Ca2+ channels, or release from intracellular stores (triggered by mGluRs which are G protein-coupled receptors) are important in many cell types. Ca2+ can activate, both directly and indirectly, protein kinases and phosphatases leading to phosphorylation–dephosphorylation cycles and, ultimately, to LTP and LTD. The next paragraphs focus on the molecular mechanisms behind NMDAR-dependent LTP and LTD, as well as cerebellar LTD, because these forms of plasticity have been studied the most both experimentally and computationally.
NMDAR-dependent potentiation is triggered by release of the neurotransmitter glutamate from the presynaptic neuron and subsequent binding to NMDARs on the postsynaptic neuron (Bliss and Collingridge, 1993; Malenka and Nicoll, 1999; Sweatt, 1999; Malenka and Bear, 2004; Citri and Malenka, 2008). After NMDARs are activated, Ca2+ can flow into the cell if the postsynaptic membrane is sufficiently depolarized to relieve the magnesium ion block from NMDAR. NMDAR-dependent LTP requires a large increase in postsynaptic Ca2+ concentration which triggers several events inside the cell. One of the most important events is Ca2+ binding to calmodulin, which then activates Ca2+/calmodulin-dependent protein kinase II (CaMKII), leading to phosphorylation of AMPARs, increase in single-channel conductance of AMPARs, and incorporation of additional AMPARs into the postsynaptic density (Citri and Malenka, 2008). Ca2+ also binds to protein kinase C (PKC) which is involved in E-LTP in some cell types (Malinow et al., 1989; Klann et al., 1993). In the hippocampus, the calmodulin-4Ca2+ complex (CaMCa4) further activates adenylyl cyclase, leading to activation of cyclic adenosine monophosphate (cAMP)-dependent protein kinase (PKA) which is required for some forms of L-LTP (Woo et al., 2003).
Transcription and also somatic and dendritic protein synthesis are required for induction of L-LTP (Bradshaw et al., 2003b), but it is unclear whether protein synthesis is required for induction of E-LTP. These nuclear and somatic events involve Ca2+/calmodulin-dependent protein kinase IV (CaMKIV), mitogen-activated protein kinase (MAPK, ERK), and PKA. For maintenance of L-LTP, the atypical PKC isozyme (PKMζ), which is an autonomously active form of PKC, is required in addition to local dendritic protein synthesis (Serrano et al., 2005).
NMDAR-dependent LTD needs only a modest increase in Ca2+ concentration (instead of the large Ca2+ increase for LTP). This modest increase in Ca2+ concentration leads to preferential activation of protein phosphatase 2B also known as calcineurin, because it has a much higher affinity for CaMCa4 than CaMKII has. Activation of protein phosphatases leads to dephosphorylation and endocytosis of AMPARs located on the plasma membrane (Citri and Malenka, 2008), and thereby the expression of LTD. Protein translation may be needed for expression and maintenance of L-LTD (Citri and Malenka, 2008), but otherwise mechanisms behind maintenance of NMDAR-dependent LTD have not been studied extensively. Some forms of LTD also require Ca2+-dependent production of endocannabinoids which travel retrogradely to produce changes in presynaptic release of neurotransmitters (Gerdeman and Lovinger, 2003).
Cerebellar LTD, the best studied form of non-NMDAR-dependent LTD, is observed at the parallel fiber to Purkinje cell synapse. Purkinje cells form synapses with several thousand parallel fibers and also receive many synaptic contacts from a single climbing fiber (Ito, 2002; Citri and Malenka, 2008). Cerebellar LTD is induced when parallel fibers and a climbing fiber are activated simultaneously. Glutamate released by parallel fibers activates mGluRs which in turn activate phospholipase C (Ito, 2002). Phospholipase C catalyzes the reaction producing diacylglycerol and inositol trisphosphate (IP3). Diacylglycerol activates PKC, and IP3 causes the release of Ca2+ from endoplasmic reticulum through IP3 receptors (IP3Rs). Phospholipase A2, which is activated by an elevation in Ca2+ concentration, produces arachidonic acid which more persistently activates PKC that is transiently activated by diacylglycerol. PKC phosphorylates AMPARs and this leads to endocytosis of AMPARs from the plasma membrane. As in hippocampal LTP, protein synthesis is needed for L-LTD (Ito, 2001).
Given that Ca2+ activates multiple processes and enzymes, such as endocannabinoid production, calcineurin, and CaMKII, it is still not clear why some stimulation protocols produce depression and some produce potentiation. Non-linear interactions between multiple pathways make a quantitative understanding difficult solely from experiments. Computer modeling synthesizes information from myriad studies ranging from plasma membrane level phenomena to intracellular phenomena. Simulations therefore provide deeper insight into mechanisms underlying plasticity and this is why modeling studies have become more and more popular during the last 10 years.
3. Computational Models
Many computational models have been developed to understand pre- and postsynaptic events in LTP and LTD. Several focused reviews that include models of a specific neural system or type of plasticity have appeared during the last 20 years (Brown et al., 1990; Neher, 1998; Hudmon and Schulman, 2002a,b; Bi and Rubin, 2005; Holmes, 2005; Wörgötter and Porr, 2005; Ajay and Bhalla, 2006; Klipp and Liebermeister, 2006; Zou and Destexhe, 2007; Morrison et al., 2008; Ogasawara et al., 2008; Bhalla, 2009; Ogasawara and Kawato, 2009; Tanaka and Augustine, 2009; Urakubo et al., 2009; Castellani and Zironi, 2010; Gerkin et al., 2010; Graupner and Brunel, 2010; Hellgren Kotaleski and Blackwell, 2010; Shouval et al., 2010); however, a comprehensive review on postsynaptic signal transduction models for LTP and LTD is lacking.
In this study, an analysis of altogether 117 postsynaptic signal transduction models published through the year 2009 is presented (see Table 1). We limit the present analysis to models of postsynaptic signal transduction pathways that are defined using several characteristics. First, the output of the model needs to be a postsynaptic aspect of the neuron. Second, some part of intracellular signaling is explicitly modeled. Thus, models in this review are required to include at least mechanisms for postsynaptic Ca2+ dynamics, Ca2+ buffers, phosphorylation–dephosphorylation cycles, LTP and LTD related enzymes, retrograde signals, or synaptic strength that depends on Ca2+ concentration. Alternatively, models that explicitly include the kinases and phosphatases underlying changes in AMPAR phosphorylation or synthesis of plasticity-related proteins are included. Models which have intracellular signaling pathways in neurons but do not address plasticity are excluded. Models of AMPAR and NMDAR activation alone, or models including only anchoring and scaffolding proteins as intracellular molecules are excluded. Lastly, purely phenomenological models of plasticity are excluded. These strict criteria are needed because of the large number of models. In addition, a few models published during 2010 are excluded (see, e.g., Clopath et al., 2010; Kim et al., 2010; Kubota and Kitajima, 2010; Nakano et al., 2010; Pepke et al., 2010; Qi et al., 2010; Rackham et al., 2010; Santamaria et al., 2010; Tolle and Le Novère, 2010a).
Table 1.
List of postsynaptic signal transduction models published each year.
| Year | Models | No. |
|---|---|---|
| 1985 | Lisman (1985) | 1 |
| 1987 | Gamble and Koch (1987) | 1 |
| 1988 | Lisman and Goldring (1988a,b) | 2 |
| 1989 | Lisman (1989) | 1 |
| 1990 | Holmes (1990), Holmes and Levy (1990), Kitajima and Hara (1990), Zador et al. (1990) | 4 |
| 1993 | De Schutter and Bower (1993), Migliore and Ayala (1993) | 2 |
| 1994 | Gold and Bear (1994), Kötter (1994), Michelson and Schulman (1994) | 3 |
| 1995 | Matsushita et al. (1995), Migliore et al. (1995), Schiegg et al. (1995) | 3 |
| 1996 | Dosemeci and Albers (1996), Fiala et al. (1996) | 2 |
| 1997 | Coomber (1997), Holmes and Levy (1997), Kitajima and Hara (1997), Migliore et al. (1997) | 4 |
| 1998 | Coomber (1998a,b), Markram et al. (1998), Murzina and Silkis (1998) | 4 |
| 1999 | Bhalla and Iyengar (1999), Kötter and Schirok (1999), Kubota and Bower (1999), Migliore and Lansky (1999a,b), Volfovsky et al. (1999) | 6 |
| 2000 | Holmes (2000), Kitajima and Hara (2000), Li and Holmes (2000), Okamoto and Ichikawa (2000a,b), Zhabotinsky (2000) | 6 |
| 2001 | Castellani et al. (2001), Franks et al. (2001), Kubota and Bower (2001), Kuroda et al. (2001), Yang et al. (2001) | 5 |
| 2002 | Abarbanel et al. (2002), Bhalla (2002a,b), Hellgren Kotaleski and Blackwell (2002), Hellgren Kotaleski et al. (2002), Holthoff et al. (2002), Karmarkar and Buonomano (2002), Karmarkar et al. (2002), Saftenku (2002), Shouval et al. (2002a,b) | 11 |
| 2003 | Abarbanel et al. (2003), Bradshaw et al. (2003a), d'Alcantara et al. (2003), Dupont et al. (2003), Kikuchi et al. (2003) | 5 |
| 2004 | Ajay and Bhalla (2004), Holcman et al. (2004), Ichikawa (2004), Murzina (2004), Steuber and Willshaw (2004), Yeung et al. (2004) | 6 |
| 2005 | Abarbanel et al. (2005), Castellani et al. (2005), Doi et al. (2005), Hayer and Bhalla (2005), Hernjak et al. (2005), Miller et al. (2005), Naoki et al. (2005), Rubin et al. (2005), Saudargiene et al. (2005), Shouval and Kalantzis (2005) | 10 |
| 2006 | Badoual et al. (2006), Lindskog et al. (2006), Miller and Wang (2006), Shah et al. (2006), Smolen et al. (2006), Zhabotinsky et al. (2006) | 6 |
| 2007 | Ajay and Bhalla (2007), Cai et al. (2007), Cornelisse et al. (2007), Delord et al. (2007), Gerkin et al. (2007), Graupner and Brunel (2007), Ichikawa et al. (2007), Kubota et al. (2007), Ogasawara et al. (2007), Schmidt et al. (2007), Smolen (2007), Tanaka et al. (2007) | 12 |
| 2008 | Achard and De Schutter (2008), Brown et al. (2008), Canepari and Vogt (2008), Clopath et al. (2008), Helias et al. (2008), Keller et al. (2008), Kubota and Kitajima (2008), Kubota et al. (2008), Pi and Lisman (2008), Santucci and Raghavachari (2008), Smolen et al. (2008), Stefan et al. (2008), Urakubo et al. (2008), Yu et al. (2008) | 14 |
| 2009 | Aslam et al. (2009), Byrne et al. (2009), Castellani et al. (2009), Jain and Bhalla (2009), Kalantzis and Shouval (2009), Kitagawa et al. (2009), Ogasawara and Kawato (2009), Schmidt and Eilers (2009), Smolen et al. (2009) | 9 |
| All | 117 |
Altogether 117 models have been published between the years 1985 and 2009. For chosen criteria, see the beginning of Section 3.
3.1. Main characteristics of models
The lists of LTP models (Table 2), LTD models (Table 3), and dual LTP and LTD models (Table 4) order the models alphabetically by the first author and by the publication month and year. Dual LTP and LTD models are able to simulate both forms of plasticity. Characteristics listed under the methods include the computational techniques: either deterministic ordinary and partial differential equations (Det.) or stochastic techniques (Stoch.) which include, for example, reaction algorithms such as the Gillespie stochastic simulation algorithm (Gillespie, 1976, 1977) and diffusion algorithms such as Brownian dynamics. A few studies also use so-called hybrid methods where different techniques are combined. The models are further classified according to the biochemical phenomena that are modeled: some models only describe reactions between chemical species (Reac.) and some also take into account the diffusion of at least some chemical species (Diff.). In addition to biochemical models, there are models which not only describe intracellular events associated with synaptic plasticity, but also take into account the associated plasma membrane and ion channel level phenomena by modeling the membrane voltage; these models are referred to as electrophysiological (Elect.). Tables 2–4 indicate the simulation tool or programing language used when known, but this piece of information is not always given in the publications. Other characteristics included in Tables 2–4 are the cell type of the model, which process of synaptic plasticity is modeled [induction (Ind.), expression (Expr.), or maintenance (Maint.)] according to the publications, time required for the dynamics of the model to reach a steady state, the model outputs used to demonstrate the change in synaptic strength, and the size of the model [less than 20 different chemical species or other model variables is defined as small (S), between 20 and 50 is medium (M), and more than 50 is large (L)]. If several different types of models are used in one publication, the size of the largest model is given. The time required for the dynamics of the model to reach a steady state is suggestive and it is not possible to compare all the models according to the time because different models use, for example, different inputs.
Table 2.
List of LTP models.
| Model | Methods | Cell type | Phases | Time | Outputs | Size |
|---|---|---|---|---|---|---|
| Ajay and Bhalla (2004) | Det. Reac./GENESIS/Kinetikita | Hippocampal CA1 N | Ind./Maint. LTP | 60–80 min | ERKII | L |
| Ajay and Bhalla (2007) | Det. Reac. Diff. Elect./GENESIS/Kinetikita | Hippocampal CA1 PN | Ind./Maint. LTP | 1–4 h | ERKII | L |
| Aslam et al. (2009) | Det. Reac./MATLAB® | Generic | Ind./Maint. L-LTP | 100 min to 40 d | CaMKII | S |
| Bhalla and Iyengar (1999) | Det. Reac. Elect./GENESIS/Kinetikita | Hippocampal CA1 N | Ind. E-LTP | 30 min | CaMKII | L |
| Bhalla (2002a) | Det. Reac. Diff. Elect./GENESIS/Kinetikita | Hippocampal CA1 N | Ind. E-LTP | 50 min | CaMKII | L |
| Bhalla (2002b) | Det. Reac./GENESIS/Kinetikita | Hippocampal CA1 N | Ind. E-LTP | 15–60 min | CaMKII | L |
| Bradshaw et al. (2003a) | Det. Reac. | Hippocampal CA1 N | Ind. LTP | CaMKII | M | |
| Canepari and Vogt (2008) | Det. Reac. | Cerebellar PC | Ind. LTP | 0.01–0.25 s | Ca2+ | S |
| Cornelisse et al. (2007) | Det. Reac. Diff./CalCb | Visual cortical layer V PN | Ind. LTP | 0.06–0.1 s | CaMCa1 | S |
| De Schutter and Bower (1993) | Det. Reac. Diff. Elect./GENESISc | Hippocampal N | Ind. LTP | 0.2 s | Ca2+ | L |
| Dupont et al. (2003) | Det. Reac. | Generic | LTP | 10–100 s | CaMKII | S |
| Franks et al. (2001) | Det. Stoch. Reac. Diff. Elect./MCelld, NEURONe | Neocortical PN | Ind. LTP | 0.2–2 s | CaMCa4 | L |
| Gamble and Koch (1987) | Det. Reac. Diff. Elect. | Hippocampal PN | Ind. LTP | 0.3 s | CaMCa4 | M |
| Gold and Bear (1994) | Det. Reac. Diff. Elect. | Hippocampal N | Ind. LTP | 0.2–0.3 s | Ca2+ | M |
| Holmes and Levy (1990) | Det. Reac. Diff. Elect. | Hippocampal DGC | Ind. LTP | 0.05–0.3 s | Ca2+ | L |
| Holmes (1990) | Det. Reac. Diff. Elect. | Hippocampal DGC | Ind. LTP | 2 s | Ca2+ | L |
| Holmes and Levy (1997) | Det. Reac. Diff. Elect. | Hippocampal DGC | Ind. LTP | 0.2 s | Ca2+, CaMCa4 | L |
| Holmes (2000) | Det. Stoch. Reac. Diff. Elect./MCelld | Hippocampal DGC | Ind. LTP | 2 s to 2 h | CaMKII | L |
| Kikuchi et al. (2003) | Det. Reac./E-Cellf | Hippocampal N | Ind. E-LTP | 10–100 min | AMPAR | L |
| Kitagawa et al. (2009) | Det. Reac./GENESIS/Kinetikita | Cerebellar PC | Ind./Expr./Maint. LTP | 2–60 min | CaMKII | L |
| Kitajima and Hara (1990) | Det. Stoch. Reac. Elect. | Hippocampal PN | Ind./Maint. LTP | 0.3 s | Ca2+ | S |
| Kubota and Bower (1999) | Stoch. Reac. | Generic | Ind. LTP | 0.02 s | CaMKII | M |
| Kubota and Bower (2001) | Det. Reac./XPPAUTg, MATLAB® | Generic | Ind. LTP | CaMKII | L | |
| Kötter (1994) | Det. Reac. | Striatal MSN | LTP | DARPP, MAP2 | S | |
| Kötter and Schirok (1999) | Det. Reac./XPPg | Striatal MSN | LTP | 1–2 s | cAMP | S |
| Li and Holmes (2000) | Det. Stoch. Reac. Diff. Elect./MCelld | Hippocampal DGC | Ind. LTP | 1–35 s | CaMKII | L |
| Lindskog et al. (2006) | Det. Reac./XPPAUTg | Striatal MSN | Ind. E-LTP | 3–30 min | DARPP32, PKA | L |
| Lisman (1985) | Det. Reac. | Generic | LTP | Kinase | S | |
| Lisman and Goldring (1988b) | Det. Stoch. Reac. | Generic | LTP | CaMKII | M | |
| Lisman and Goldring (1988a) | Det. Stoch. Reac. | Generic | LTP | CaMKII | M | |
| Lisman (1989) | Det. Reac. | Hippocampal N | LTP | CaMKII | S | |
| Markram et al. (1998) | Det. Reac. Diff. | Neocortical layer V PN | STP/LTP | 0.002–2 s | Buffered Ca2+ | L |
| Matsushita et al. (1995) | Det. Reac. | Generic | LTP | 20 s to 60 min | CaMKII | M |
| Michelson and Schulman (1994) | Stoch. Reac. | Generic | LTP | 10 s to 3 min | CaMK | L |
| Migliore and Ayala (1993) | Det. Reac. | Generic | Ind./Expr./Maint. STP/LTP | Postsyn. signal | S | |
| Miller et al. (2005) | Det. Stoch. Reac. | Generic | Ind./Maint. LTP | 2 s to 100 y | CaMKII | L |
| Miller and Wang (2006) | Stoch. Reac. | Generic | Ind./Maint. LTP | 1–50 y | CaMKII | L |
| Okamoto and Ichikawa (2000b) | Det. Reac. | Generic | Ind. LTP | CaMKII | M | |
| Okamoto and Ichikawa (2000a) | Det. Reac. Diff. | Hippocampal CA1 N | Ind. LTP | 1–10 s | CaMKII | L |
| Santucci and Raghavachari (2008) | Det. Stoch. Reac. Diff. Elect. | Hippocampal CA1 PN | Ind. LTP | 0.5–1 s | CaMKII | L |
| Schiegg et al. (1995) | Det. Reac. Diff. Elect. | Hippocampal CA1 PN | Ind. LTP | 0.1–1.5 s | Ca2+ | L |
| Smolen et al. (2006) | Det. Reac./Java | Hippocampal CA1 N | Ind./Expr. L-LTP | 2–4 h | Synaptic strength | M |
| Smolen (2007) | Det. Reac. | Hippocampal CA1 N | Maint. L-LTP | 10 h to 3 mo | Synaptic strength | M |
| Smolen et al. (2008) | Det. Stoch. Reac./Java | Hippocampal CA1 or neocortical PN | Ind./Maint. L-LTP | 2 h to 8 d | MAPK | M |
| Smolen et al. (2009) | Det. Stoch. Reac./Java | Generic | Ind./Maint. LTP | 1–6 h | CaMKII or MAPK | S |
| Volfovsky et al. (1999) | Det. Reac. Diff. Elect./FIDAPh | Hippocampal N | LTP | 0.1–1.2 s | Ca2+ | L |
| Zador et al. (1990) | Det. Reac. Diff. Elect. | Hippocampal CA1 N | Ind. LTP | 0.2–0.3 s | CaMCa4 | L |
| Zhabotinsky (2000) | Det. Reac. | Hippocampal N | Ind./Maint. LTP | 2 s to 2 y | CaMKII | S |
Models are in alphabetical order by the first author and according to the publication month and year. Tabulated characteristics are the method and model types (Det., Stoch., Reac., Diff., Elect., and simulation environment), cell type, phases of LTP, time required for the dynamics of the model to reach a steady state, model outputs, and size of the model based on the number of different chemical species or other model variables [less than 20 different chemical species or other model variables is defined as small (S), between 20 and 50 is medium (M), and more than 50 is large (L)]. All abbreviations are given in the list of abbreviations.
aGENESIS/Kinetikit (http://www.genesis-sim.org/GENESIS/; http://www.ncbs.res.in/index.php?option=com_content&task=view&id=307; Bower and Beeman, 1998; Bhalla, 2002c).
bCalC (http://web.njit.edu/~matveev/calc.html; Matveev et al., 2002).
cGENESIS (http://www.genesis-sim.org/GENESIS/; Bower and Beeman, 1998).
dMCell (http://www.mcell.cnl.salk.edu/; Stiles and Bartol, 2001).
eNEURON (http://www.neuron.yale.edu/neuron/; Carnevale and Hines, 2006).
fE-Cell (http://www.e-cell.org; Tomita et al., 1999).
gXPP, XPPAUT (http://www.math.pitt.edu/~bard/xpp/xpp.html; Ermentrout, 2002).
Table 3.
List of LTD models.
| Model | Methods | Cell type | Phases | Time | Outputs | Size |
|---|---|---|---|---|---|---|
| Achard and De Schutter (2008) | Det. Reac. Elect./GENESIS/Kinetikita | Cerebellar PC | Ind. LTD | 1 s | Ca2+ | L |
| Brown et al. (2008) | Det. Reac. Diff./Virtual Cellb | Cerebellar PC | LTD | 0.4–2 s | IP3 | M |
| Doi et al. (2005) | Det. Reac./GENESIS/Kinetikita | Cerebellar PC | Ind. LTD | 0.2–1 s | Ca2+ | L |
| Fiala et al. (1996) | Det. Reac. Elect. | Cerebellar PC | Ind. LTD | M | ||
| Hellgren Kotaleski and Blackwell (2002) | Det. Reac. Diff./XPPc | Cerebellar PC | LTD | 1–5 s | Ca2+ | S |
| Hellgren Kotaleski et al. (2002) | Det. Reac. Diff./XPPc | Cerebellar PC | Ind. LTD | 5–30 s | PKC | M |
| Hernjak et al. (2005) | Det. Reac. Diff./Virtual Cellb | Cerebellar PC | Ind. LTD | 0.1–4 s | Ca2+ | M |
| Holthoff et al. (2002) | Det. Reac. Diff. Elect./MATLAB® | Neocortical layer V PN | Ind. LTD | 0.5 s | Ca2+ | S |
| Kuroda et al. (2001) | Det. Reac./GENESIS/Kinetikita | Cerebellar PC | Ind. STD/E-,L-LTD | 15–100 min | AMPAR | L |
| Murzina (2004) | Det. Reac. Diff. Elect. | Cerebellar PC | Ind. LTD | Kinase, receptor | M | |
| Ogasawara et al. (2007) | Det. Reac. Diff. Elect. | Cerebellar PC | Ind./Expr./Maint. LTD | 20–60 min | AMPAR | L |
| Ogasawara and Kawato (2009) | Det. Stoch. Reac. | Cerebellar PC | Ind./Maint. LTD | 10 s to 70 min | Kinase | S |
| Schmidt et al. (2007) | Det. Reac. Diff./Mathematica, FEMLAB | Cerebellar PC | Ind. LTD | 0.2–4 s | Ca2+, CaM | L |
| Schmidt and Eilers (2009) | Det. Reac. Diff./Mathematica | Cerebellar PC | Ind. LTD | 0.04–3 s | Ca2+, CaM | S |
| Steuber and Willshaw (2004) | Det. Reac. Elect. | Cerebellar PC | Ind. LTD | S | ||
| Tanaka et al. (2007) | Det. Reac. | Cerebellar PC | Ind. LTD | AMPAR | M | |
| Yang et al. (2001) | Det. Reac. Elect./GENESIS/Chemesisd | Cerebellar PC | Ind. LTD | 10–100 s | PKC | L |
Models are in alphabetical order by the first author and according to the publication month and year. Tabulated characteristics are the method and model types (Det., Stoch., Reac., Diff., Elect., and simulation environment), cell type, phases of LTD, time required for the dynamics of the model to reach a steady state, model outputs, and size of the model based on the number of different chemical species or other model variables (S, M, L). All abbreviations are given in the list of abbreviations.
aGENESIS/Kinetikit (http://www.genesis-sim.org/GENESIS/; http://www.ncbs.res.in/index.php?option=com_content&task=view&id=307; Bower and Beeman, 1998; Bhalla, 2002c).
bVirtual Cell (http://vcell.org; Schaff et al., 1997; Slepchenko et al., 2003).
cXPP (http://www.math.pitt.edu/~bard/xpp/xpp.html; Ermentrout, 2002).
dGENESIS/Chemesis (http://www.genesis-sim.org/GENESIS/; http://krasnow.gmu.edu/CENlab/software.html; Bower and Beeman, 1998; Blackwell and Hellgren Kotaleski, 2002).
Table 4.
List of dual LTP and LTD models.
| Model | Methods | Cell type | Phases | Time | Outputs | Size |
|---|---|---|---|---|---|---|
| Abarbanel et al. (2002) | Det. Reac. Elect. | Hippocampal GluN | Ind. LTP/LTD | Synaptic strength | S | |
| Abarbanel et al. (2003) | Det. Reac. Elect. | Hippocampal CA1 PN | Ind. LTP/LTD | Synaptic strength | S | |
| Abarbanel et al. (2005) | Det. Reac. Elect. | Hippocampal CA1 PN | Ind. LTP/LTD | Synaptic strength | M | |
| Badoual et al. (2006) | Det. Reac. Diff. Elect./NEURONa | Cortical PN | Ind. LTP/LTD | 0.05–0.25 s | Enzyme | S |
| Byrne et al. (2009) | Stoch. Reac. Diff./Java | Hippocampal CA1 PN | Ind. LTP/LTD | 1–5 s | Ca2+, CaM | L |
| Cai et al. (2007) | Det. Stoch. Reac. Elect./Java | Hippocampal or visual cortical N | Ind. LTP/LTD | 100 s | Synaptic strength | S |
| Castellani et al. (2001) | Det. Reac. Elect. | Generic | Ind. LTP/LTD | AMPAR | S | |
| Castellani et al. (2005) | Det. Reac. | Cortical N | Ind. LTP/LTD | AMPAR | M | |
| Castellani et al. (2009) | Det. Stoch. Reac. | Generic | Ind./Maint. LTP/LTD | AMPAR | S | |
| Clopath et al. (2008) | Det. Stoch. Reac. Elect./Python | Hippocampal CA1 PN | Ind./Maint. E-, L-LTP/LTD | 3–5 h | Synaptic strength | L |
| Coomber (1997) | Det. Reac. Diff. Elect./GENESISb | Neocortical PN | Ind./Maint. LTP/LTD | 1 s | gAMPAR | L |
| Coomber (1998a) | Det. Reac./C | Generic | Ind. LTP/LTD | 5 s to 15 min | CaMKII | L |
| Coomber (1998b) | Det. Reac. | Generic | Ind. LTP/LTD | 2–60 min | CaMKII | L |
| d'Alcantara et al. (2003) | Det. Reac./MATLAB® | Cerebral cortical or hippocampal CA1 N | Ind. LTP/LTD | 20 s to 10 min | AMPAR | S |
| Delord et al. (2007) | Det. Stoch. Reac. | Generic | Ind./Maint. LTP/LTD | 4 s to 4 mo | Substrate | S |
| Dosemeci and Albers (1996) | Stoch. Reac./FutureBASIC | Generic | Ind. LTP/LTD | 20 s to 6 min | CaMKII | L |
| Gerkin et al. (2007) | Det. Reac. | Hippocampal N | Ind. LTP/LTD | 5 s | Synaptic strength | S |
| Graupner and Brunel (2007) | Det. Reac. Elect./C++, XPPAUTc | Hippocampal N | Ind./Maint. LTP/LTD | 1–3.5 min | CaMKII | M |
| Hayer and Bhalla (2005) | Det. Stoch. Reac. Diff./GENESIS/Kinetikitd, GENESIS 3/MOOSEe | Generic | LTP/LTD | 200 s to 30 h | AMPAR, CaMKII | L |
| Helias et al. (2008) | Det. Stoch. Reac. Elect./NESTf | Cortical N | Ind. LTP/LTD | CaMKII | L | |
| Holcman et al. (2004) | Stoch. Reac. Diff. | Generic | Ind. LTP/LTD | 0.4–0.6 s | Ca2+ | L |
| Ichikawa (2004) | Det. Reac. Diff./A-Cellg | Generic | Ind. LTP/LTD | CaMKII | L | |
| Ichikawa et al. (2007) | Det. Reac. Diff. Elect./A-Cellg | Hippocampal CA1 PN | Ind./Expr. LTP/LTD | CaMKII, CaN | M | |
| Jain and Bhalla (2009) | Det. Reac./GENESIS/Kinetikitd, GENESIS 3/MOOSEe | Hippocampal N | Ind. LTP/LTD | 3 h | Protein | L |
| Kalantzis and Shouval (2009) | Det. Stoch. Reac. Diff. Elect. | Hippocampal CA1 PN | Ind. LTP/LTD | 0.15 s | Synaptic strength | L |
| Karmarkar and Buonomano (2002) | Det. Reac. Elect./NEURONa | Hippocampal N | Ind. LTP/LTD | Synaptic strength | S | |
| Karmarkar et al. (2002) | Det. Reac. Elect./NEURONa | Auditory cortical layer II/III PN | Ind. LTP/LTD | Synaptic strength | S | |
| Keller et al. (2008) | Det. Stoch. Reac. Diff. Elect./MCellh, NEURONa | Hippocampal CA1 PN | Ind. LTP/LTD | 0.01–0.2 s | CaM | L |
| Kitajima and Hara (1997) | Det. Reac. Elect. | Generic | Ind./Expr. LTP/LTD | 0.04–0.05 s | Vm | M |
| Kitajima and Hara (2000) | Det. Reac. Elect. | Generic | Ind. LTP/LTD | gAMPAR | M | |
| Kubota and Kitajima (2008) | Det. Stoch. Reac. Elect./C | Cortical PN | Ind. LTP/LTD | 100 s to 80 min | Synaptic strength | L |
| Kubota et al. (2007) | Det. Stoch. Reac. Diff. | Hippocampal CA1 PN | Ind. LTP/LTD | 0.05 s | CaM | L |
| Kubota et al. (2008) | Det. Reac. Elect. | Hippocampal CA1 PN | Ind. LTP/LTD | 0.05–1 s | Synaptic strength | M |
| Migliore et al. (1995) | Det. Reac. | Hippocampal N | Ind./Expr./Maint. LTP/LTD | Postsyn. signal | S | |
| Migliore et al. (1997) | Det. Reac. | Hippocampal N | Ind./Maint. LTP/LTD | Postsyn. signal | S | |
| Migliore and Lansky (1999b) | Det. Reac. Elect./FORTRAN | Neocortical PN | Ind./Maint. LTP/LTD | 20 s | Postsyn. signal | S |
| Migliore and Lansky (1999a) | Det. Reac./FORTRAN | Hippocampal N | Ind./Maint. LTP/LTD | Postsyn. signal | S | |
| Murzina and Silkis (1998) | Det. Reac. Elect. | Hippocampal CA3 PN | Ind. LTP/LTD | 0.1 s | Vm | M |
| Naoki et al. (2005) | Det. Reac. Diff./MATLAB® | Generic | Ind./Expr. LTP/LTD | 0.5–10 s | CaMCa4 | L |
| Pi and Lisman (2008) | Det. Reac./MATLAB® | Generic | Ind./Maint. LTP/LTD, depotentiation, dedepression | 3–8 s | AMPAR | S |
| Rubin et al. (2005) | Det. Reac. Diff. Elect./XPPAUTc | Hippocampal CA1 PN | Ind. LTP/LTD | 10 s | Synaptic strength | M |
| Saftenku (2002) | Det. Reac. Elect./NEURONa | Cerebellar GrC | Ind. LTP/LTD | 100 s | Postsyn. signal | L |
| Saudargiene et al. (2005) | Det. Reac. Elect. | Generic | Ind. LTP/LTD | 0.06–0.1 s | Synaptic strength | S |
| Shah et al. (2006) | Det. Reac. Elect./Java, MATLAB® | Generic | Ind. LTP/STD/LTD | Synaptic strength | S | |
| Shouval et al. (2002a) | Det. Reac. Elect. | Generic | Ind. LTP/LTD | Synaptic strength | S | |
| Shouval et al. (2002b) | Det. Reac. Elect. | Generic | Ind. LTP/LTD | AMPAR | S | |
| Shouval and Kalantzis (2005) | Det. Stoch. Reac. Elect. | Generic | Ind. LTP/LTD | Synaptic strength | S | |
| Stefan et al. (2008) | Det. Reac./COPASIi | Generic | LTP/LTD | CaMKII, CaN | L | |
| Urakubo et al. (2008) | Det. Reac. Diff. Elect./GENESIS/Kinetikitd | Visual cortical layer II/III PN | Ind. LTP/LTD | 20 min | gsyn | L |
| Yeung et al. (2004) | Det. Reac. Elect. | Generic | Ind. LTP/LTD | 2 h | Synaptic strength | L |
| Yu et al. (2008) | Det. Stoch. Reac. Elect. | Hippocampal place N | Ind. LTP/LTD | Synaptic strength | L | |
| Zhabotinsky et al. (2006) | Det. Reac. Diff./XPPAUTc | Hippocampal CA1 N | Ind./Maint. E-, L-LTP/LTD | 10 s to 60 min | AMPAR | L |
Models are in alphabetical order by the first author and according to the publication month and year. Tabulated characteristics are the method and model types (Det., Stoch., Reac., Diff., Elect., and simulation environment), cell type, phases of LTP/LTD, time required for the dynamics of the model to reach a steady state, model outputs, and size of the model based on the number of different chemical species or other model variables (S, M, L). All abbreviations are given in the list of abbreviations.
aNEURON (http://www.neuron.yale.edu/neuron/; Carnevale and Hines, 2006).
bGENESIS (http://www.genesis-sim.org/GENESIS/; Bower and Beeman, 1998).
cXPP, XPPAUT (http://www.math.pitt.edu/~bard/xpp/xpp.html; Ermentrout, 2002).
dGENESIS/Kinetikit (http://www.genesis-sim.org/GENESIS/; http://www.ncbs.res.in/index.php?option=com_content&task=view&id=307; Bower and Beeman, 1998; Bhalla, 2002c).
eGENESIS 3/MOOSE (http://www.genesis-sim.org/GENESIS/; http://moose.sourceforge.net/).
fNEST (http://www.nest-initiative.org/; Gewaltig and Diesmann, 2007).
gA-Cell (http://www.fujixerox.co.jp/crc/cng/A-Cell/; Ichikawa, 2001).
hMCell (http://www.mcell.cnl.salk.edu/; Stiles and Bartol, 2001; Kerr et al., 2008).
iCOPASI (http://www.copasi.org/; Hoops et al., 2006).
3.2. Categorization of models
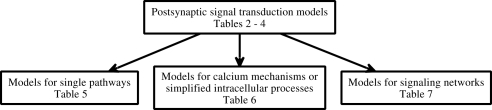
In this study, models are further categorized (Figure 1) into models for single pathways (Table 5), models for calcium mechanisms or simplified intracellular processes (Table 6), and models for signaling networks (Table 7). Models for single pathways involve at most one kinase as a model variable and do not include any receptors, ion channels, or pumps on the plasma membrane. Typically single pathways contain a pathway involving calmodulin and CaMKII and sometimes also phosphatases. Models for calcium mechanisms or simplified intracellular processes include postsynaptic Ca2+ buffers together with ion channels, receptors, or pumps, or simplified intracellular processes. The last group of models, consisting of signaling networks, takes into account interactions between at least two pathways and thus often have several protein kinases and phosphatases. These models can also include ion channels, receptors, and pumps. Several characteristics, such as model inputs, number and types of morphological compartments, molecules, ion channels, and receptors, are described for the models in the following sections. In some cases it is difficult to determine the model inputs based on the information given in the publications. For detailed biophysical models, the input is typically coupled with the plasma membrane level phenomena, such as membrane voltage. In these cases, we have indicated the change in membrane current (ΔIm) or membrane voltage (ΔVm) as the input. For more simplified models, a variety of mathematical equations are used to describe the model and the input. In these cases, we have indicated which physical property the input equation represents, such as synaptic stimulus (causing elevation in Ca2+ concentration). See also Section 4 for further comments on the presentation of input for models.
Figure 1.
Categorization of postsynaptic signal transduction models models.
Table 5.
Characteristics of models for single pathways.
| Type | Model | Inputs | Subunits/States/Residues | Ions and molecules |
|---|---|---|---|---|
| LTP | Bradshaw et al. (2003a) | Ca2+ | 6/3a/Thr-286 | Ca2+, CaM, CaMKII, PP1 |
| LTP | Dupont et al. (2003) | Ca2+, CaM, CaMCa4 | b/5c/Thr-286 | Ca2+, CaM, CaMKII |
| LTP | Kubota and Bower (2001) | Ca2+ | 2–4/5d/Thr-286, Thr-305/306 | Ca2+, CaM, CaMKII, PP1 |
| LTP | Kötter and Schirok (1999) | Ca2+ | No | AC, ATP, Ca2+, CaM, cAMP, PDE |
| LTP | Lisman (1985) | Kinase | 1/2e | 2 kinases, phosphatasef |
| LTP | Lisman and Goldring (1988b) | Ca2+ | b/3g | Ca2+, CaMKII, phosphate ion |
| LTP | Lisman and Goldring (1988a) | Ca2+ | b/3g | Ca2+, CaMKII, phosphate ion |
| LTP | Matsushita et al. (1995) | CaMCa4 | 10/5d/Thr-286, Thr-305, Ser-314 | ATP, Ca2+, CaM, CaMKII, phosphatase, phosphate ion |
| LTP | Michelson and Schulman (1994) | Ca2+ | 10/5d/Thr-286, Thr-305/306 | Ca2+, CaM, CaMK |
| LTP | Miller et al. (2005) | Ca2+ | 12/2e/Thr-286/287 | Ca2+, CaM, CaMKII, CaN, I1, PKA, PP1 |
| LTP | Miller and Wang (2006) | Ca2+ | 12/2e/Thr-286/287 | Ca2+, CaM, CaMKII, PP1 |
| LTP | Okamoto and Ichikawa (2000b) | Ca2+ | b/4h/Thr-286/287 | Ca2+, CaM, CaMKII |
| LTP | Okamoto and Ichikawa (2000a) | Ca2+ | 10/4h/Thr-286/287 | Ca2+, CaMi, CaMCa4-binding protein, CaMKII |
| LTP | Smolen et al. (2009) | Ca2+ | 1/2e | Ca2+, CaMKII or MAPK |
| LTP | Zhabotinsky (2000) | Ca2+ | 10/3j/Thr-286 | Ca2+, CaM, CaMKII, CaN, I1, PKA, PP1 |
| Dual | Byrne et al. (2009) | Ca2+ | 12/6k | Ca2+, CaM, CaMKIIl |
| Dual | Coomber (1998a) | Ca2+ | 5/7m/Thr-286 | ATP, Ca2+, CaM, CaMKII, phosphatase (CaN) |
| Dual | Coomber (1998b) | Ca2+ | 4/12/Thr-286, Thr-305/306 | ATP, Ca2+, CaM, CaMKII, phosphatase (PP1) |
| Dual | Delord et al. (2007) | Ca2+ | 1/2e | Ca2+, kinase, phosphatase, substrate |
| Dual | Dosemeci and Albers (1996) | Ca2+ | 10/4n/Thr-286, Thr-305/306 | Ca2+, CaM, CaMKII, phosphatase |
| Dual | Kubota et al. (2007) | Ca2+ | No | Ca2+, CaMo, Ng |
| Dual | Stefan et al. (2008) | Ca2+ | 1/5p | Ca2+, CaM, CaMKII, CaN |
Models are in alphabetical order by the first author and according to the publication month and year. First all LTP models are listed and then all dual LTP and LTD models. Tabulated characteristics are the model inputs, number of CaMKII or kinase subunits, number of states for each subunit, specified threonine (Thr) and serine (Ser) residues of CaMKII that are phosphorylated, as well as ions and molecules whose interactions are modeled. Note that it is not always clear if all the subunits and number of states mentioned in the publications are actually modeled and simulated. Molecules that are modeled as constants are also listed. All abbreviations are given in the list of abbreviations.
aFirst three states of those mentioned under d below are modeled.
bIt is not clearly stated in the publication how many CaMKII subunits are modeled.
cInactive, bound with CaMCa4, bound with CaMCa4 and autophosphorylated, Ca2+ dissociated from CaM bound to the phosphorylated form (trapped), and CaM dissociated from the trapped form but remains phosphorylated (autonomous).
dInactive, bound with CaMCa4, bound with CaMCa4 and autophosphorylated (trapped), CaMCa4 dissociated from the trapped form but remains phosphorylated (autonomous), and autonomous state secondary autophosphorylated (capped).
eInactive and phosphorylated.
fCa2+ is not included in the model.
gInactive, bound with Ca2+ and autophosphorylated, and Ca2+ dissociated but remains phosphorylated.
hFirst four states of those mentioned under d above are modeled.
i1-D CaM diffusion is modeled to five spines connected by a dendrite.
jInactive, bound with CaMCa4, and bound with CaMCa4 and phosphorylated or autophosphorylated.
kInactive and bound with CaM, CaMCa1, CaMCa2, CaMCa3, or CaMCa4.
l3-D CaM and CaMKII diffusion are modeled in a spine.
mInactive, bound with CaMCa4, bound with CaMCa4 and autophosphorylated, and autophosphorylated on any 1–4 sites.
nInactive, bound with CaMCa4 and autophosphorylated, autophosphorylated, and secondary phosphorylated.
o3-D CaM diffusion is modeled in a spine.
pInactive and bound with CaMCa1, CaMCa2, CaMCa3, or CaMCa4.
Table 6.
Characteristics of models for calcium mechanisms or simplified intracellular processes.
| Type | Model | Inputs | Compartments | VGICs | LGICs | Molecules and mechanisms |
|---|---|---|---|---|---|---|
| LTP | Canepari and Vogt (2008) | ICa | 1 dendritic | No | No | CD28k, FF, and PV buffers, PMCA pump |
| LTP | Cornelisse et al. (2007) | JVGCC | Several dendritic and spine compartments | No | No | CaM, CD28k, OGB-1, and PV buffers, 1-D diffusion of Ca2+ and some of the buffers, PMCA pump |
| LTP, Elect. | De Schutter and Bower (1993) | ΔIm or ΔVm | Neuron with 1192 compartments | No | NMDAR, non-NMDAR | Buffer, 1-D Ca2+ diffusion, PMCA pump |
| LTP, Elect. | Franks et al. (2001) | ΔIm or ΔVm | 1 spine | CaL, CaT | NMDAR | CaM and other buffers, 3-D Ca2+ diffusion, PMCA pump |
| LTP, Elect. | Gamble and Koch (1987) | Isyn | 1 dendritic, 2 spine-head, 2 spine-neck | Ca2+, KM | No | CaM buffer, CaN, 1-D Ca2+ diffusion, PMCA pump |
| LTP, Elect. | Gold and Bear (1994) | ΔIm or ΔVm | 1 dendritic, 4 spine-head, 3 spine-neck | No | NMDAR | Buffer, 1-D Ca2+ diffusion, PMCA pump |
| LTP, Elect. | Holmes and Levy (1990) | ΔIm or ΔVm | Neuron with several 4-compartment dendrites, 4304 spines with 4 spine-head and 3 spine-neck, 1–115 synapses | No | NMDAR, non-NMDAR | Buffer, 1-D Ca2+ diffusion, PMCA pump |
| LTP, Elect. | Holmes (1990) | ΔIm or ΔVm | Neuron with several 4-compartment dendrites, 3 spines with 5 spine-head and 3 spine-neck, 96 synapses | No | NMDAR, non-NMDAR | Buffer, 1-D Ca2+ diffusion, PMCA pump |
| LTP, Elect. | Holmes and Levy (1997) | ΔIm or ΔVm | Neuron with several 12-compartment dendrites, several spines with 4 spine-head and 4 spine-neck, several synapses, 1 axonal, 1 somatic | Ca2+, KA, KCa, Nafast | GABAAR, NMDAR, non-NMDAR | CaM and other buffers, 1-D Ca2+ diffusion, PMCA pump |
| LTP, Elect. | Holmes (2000) | ΔIm or ΔVm | Neuron with several 12-compartment dendrites, several spines with 4 spine-head and 4 spine-neck, several synapses, 1 axonal, 1 somatic | Ca2+, KA, KCa, Nafast | NMDAR, non-NMDAR | CaM buffer, CaMKIIa, CaN, 1-D Ca2+ diffusion, PMCA pump |
| LTP, Elect. | Kitajima and Hara (1990) | ΔIm or ΔVm | 1 somatic, 1 spine-head, 1 spine-neck | No | NMDAR, non-NMDAR | CaM buffer, CaMKIIb |
| LTP, Elect. | Li and Holmes (2000) | ΔIm or ΔVm | Neuron with several 12-compartment dendrites, several spines with 4 spine-head and 4 spine-neck, several synapses, 1 axonal, 1 somatic | Ca2+, KA, KCa, Nafast | NMDAR, non-NMDAR | CaM buffer, CaMKIIa, CaN, 1-D–3-D Ca2+ and Glu diffusion, PMCA pump |
| LTP | Markram et al. (1998) | ICa | 1 or 25 dendritic | No | No | Buffer, 1-D Ca2+ diffusion, PMCA pump |
| LTP | Migliore and Ayala (1993) | Presyn. stimulus | 1 pre-, 1 postsynaptic | No | No | Simplified intracellular processesc |
| LTP, Elect. | Santucci and Raghavachari (2008) | ΔIm or ΔVm | 1 pre-, 1 postsynaptic | No | AMPAR, NMDAR | CaM buffer, CaMKIId, CaN, 3-D Glu diffusion, I1, PKA, PP1, 2 vesicles |
| LTP, Elect. | Schiegg et al. (1995) | ΔIm or ΔVm | Neuron with 8 dendritic, 1 somatic, 3 spine-head, 3 spine-neck | No | AMPAR, NMDAR | CaM buffer, CaN, CICR, 1-D Ca2+ diffusion, Na+/Ca2+ exchanger, PMCA pump, Ca2+ store |
| LTP, Elect. | Volfovsky et al. (1999) | JCa, ΔIm or ΔVm | Several multi-compartment spines and dendrites | Ca2+ | No | CaM and CG-1 buffers, CaN, CICR, 3-D Ca2+ and CG-1 diffusion, PMCA and SERCA pumps, Ca2+ store |
| LTP, Elect. | Zador et al. (1990) | ΔIm or ΔVm | Neuron with 28 compartments | No | NMDAR, non-NMDAR | CaM buffer, 1-D Ca2+ diffusion, 2 PMCA pumps |
| LTD | Hellgren Kotaleski and Blackwell (2002) | Ca2+ | 1 spine | No | IP3R | Buffer, 1-D Ca2+ diffusion, IP3, PMCA pump |
| LTD | Hernjak et al. (2005) | JCa | 1–32 1-compartment spines, 2 dendritic | No | IP3R | CD28k, CG-1, and PV buffers, 1-D and 2-D diffusion of all molecules, IP3, PMCA and SERCA pumps, Ca2+ store |
| LTD, Elect. | Holthoff et al. (2002) | ΔIm or ΔVm | 1 dendritic, 1 spine-head, 1 spine-neck | CaL | No | CG-1 and other buffers, 1-D Ca2+ diffusion, PMCA and SERCA pumps |
| LTD | Schmidt et al. (2007) | ICa | 1 or 7 1-compartment spines, 1 or 7 dendritic | No | No | CaM, CD28k, OGB-1, and PV buffers, 1-D–3-D diffusion of all molecules, PMCA pump |
| LTD | Schmidt and Eilers (2009) | ICa | 1 spine, 1 dendritic | No | No | CaM, CD28k, OGB-1, and PV buffers, 1-D diffusion of all molecules, PMCA pump |
| Dual, Elect., STDP | Abarbanel et al. (2002) | Synaptic stimulus | 1 pre-, 1 postsynaptic | Noe | Simplified processes | Simplified intracellular processesc |
| Dual, Elect., STDP | Abarbanel et al. (2003) | ΔIm or ΔVm | Neuron with 1 compartment | CaT, K+, Na+ | AMPAR, NMDAR | Phosphorylation, dephosphorylation |
| Dual, Elect., STDP | Abarbanel et al. (2005) | ΔIm or ΔVm | 2 neurons with 1 presynaptic and 1 2-compartment postsynaptic | Ca2+, K+, KA, KM, Na+ | AMPAR, NMDAR | Phosphorylation, dephosphorylation |
| Dual, Elect., STDP | Badoual et al. (2006) | ΔIm or ΔVm | Neuron with 1 spine, 1 axonal, 1 dendritic, 1 somatic | CaL, KCa, KDR, KM, Na+ | AMPAR, NMDAR | 1-D Ca2+ diffusion, PMCA pump, 3 enzymes |
| Dual, Elect., STDP | Cai et al. (2007) | Synaptic stimulus | 1 pre-, 1 postsynaptic | No | NMDAR | Simplified intracellular processes, vesicle |
| Dual, Elect. | Castellani et al. (2001) | ΔIm or ΔVm | 1 spine | No | AMPAR, NMDAR | 2 kinases, 2 phosphatases |
| Dual | Castellani et al. (2009) | CaMKII | 1 postsynaptic | No | AMPAR | CaMKII, PKA, PP1c |
| Dual, Elect. | Clopath et al. (2008) | ΔIm | Neuron with 1 compartment, 100 synapses | Nof | Simplified processes | Protein synthesisc |
| Dual, Elect. | Coomber (1997) | ΔIm or ΔVm | Neuron with 149 compartments | CaL, KA, KAHP, KCa, KDR, KM, Na+ | AMPAR, NMDAR | Buffer, 1-D Ca2+ diffusion, PMCA pump |
| Dual, STDP | Gerkin et al. (2007) | Synaptic stimulus | 1 pre-, 1 postsynaptic | No | No | Simplified intracellular processesc |
| Dual, Elect., STDP | Helias et al. (2008) | Synaptic stimulus | Neuron with 1 compartment, max 10000 synapses | Nog | NMDAR | CaMKII |
| Dual | Holcman et al. (2004) | JNMDAR | 4-compartment spine | No | No | CaM buffer, CaN, 2-D Ca2+ diffusion, PMCA pump |
| Dual | Ichikawa (2004) | JNMDAR | 3112-compartment spine | No | No | CaM buffer, CaMKII, CaN, 3-D diffusion of all molecules |
| Dual, Elect. | Ichikawa et al. (2007) | ΔIm or ΔVm | 1 spine, 1 dendritic | No | AMPAR, NMDAR | CaM and other buffers, CaMKII, CaN, 1-D Ca2+ diffusion, PMCA pump |
| Dual, Elect., STDP | Kalantzis and Shouval (2009) | ΔVm | 6 spine-head, 10 spine-neck | No | NMDAR | Buffer, 1-D Ca2+ diffusion, PMCA pump |
| Dual, Elect., STDP | Karmarkar and Buonomano (2002) | Synaptic stimulus | 2 1-compartment neurons | Ca2++h | AMPAR, NMDAR | Simplified intracellular processes |
| Dual, Elect., STDP | Karmarkar et al. (2002) | Synaptic stimulus | 2 1-compartment neurons | Noh | AMPAR, NMDAR | Simplified intracellular processes |
| Dual, Elect. | Keller et al. (2008) | ΔIm or ΔVm | 1 dendritic, 1 extracellular, 1 presynaptic, 1 spine-head | Ca2+ | AMPAR, NMDAR | CaM, CD28k, OGB-1, and other buffers, 3-D diffusion of all molecules, Na+/Ca2+ exchanger, PMCA pump |
| Dual, Elect. | Kitajima and Hara (1997) | Presyn. stimulus | Several spines with 1 spine-head and 1 spine-neck, 3 dendritic, 1 presynaptic | Ca2+ | AMPAR, GABAR, NMDAR | Kinase, phosphatase, PMCA pump, vesicle |
| Dual, Elect. | Kitajima and Hara (2000) | ΔIm or ΔVm | Neuron with 2 1-8-compartment dendrites, 1 spine, 1 axonal, 1 somatic | CaL, CaN, CaT, KA, KDR, Na+ | AMPAR, NMDAR | Phosphorylation, dephosphorylation |
| Dual, Elect., STDP | Kubota and Kitajima (2008) | ΔIm or ΔVm | Neuron with 2 4-7-compartment dendrites, 1 spine, 4800 synapses, 1 somatic | KA, KAHP, Nafast' | AMPAR, GABAR, NMDAR | Simplified intracellular processes |
| Dual, Elect. | Kubota et al. (2008) | ΔIm or ΔVm | 1 spine | No | NMDAR | CaM buffer, Ng |
| Dual | Migliore et al. (1995) | Presyn. stimulus | 1 pre-, 1 postsynaptic | No | No | Simplified intracellular processesc |
| Dual | Migliore et al. (1997) | Presyn. stimulus | Several synapses with 1 pre- and 1 postsynaptic | No | No | Simplified intracellular processesc |
| Dual, Elect. | Migliore and Lansky (1999b) | Presyn. stimulus | 1 pre-, 1 postsynaptic | Noj | No | Simplified intracellular processesc |
| Dual | Migliore and Lansky (1999a) | Presyn. stimulus | 1 pre-, 1 postsynaptic | No | No | Simplified intracellular processesc |
| Dual | Naoki et al. (2005) | INMDAR | 15-compartment spine | No | No | CaM and other buffers, 1-D diffusion of all molecules, Na+/Ca2+ exchanger, PMCA and SERCA pumps |
| Dual | Pi and Lisman (2008) | JNMDAR | 1 spine | No | AMPAR | Buffer, CaMKII, PP2A, AMPAR trafficking |
| Dual, Elect., STDP | Rubin et al. (2005) | ΔIm or ΔVm | Neuron with 1 spine (dendritic), 1 somatic | CaL, KA, KAHP, KDR, Na+ | AMPAR, NMDAR | Buffer, Ca2+ detectors, 1-D Ca2+ diffusion |
| Dual, Elect. | Saftenku (2002) | ΔIm or ΔVm | Neuron with several compartments | BKCa, CaN, KA, KDR, KIR, Kslow, Nafast, Nar, Naslow | AMPAR, NMDAR | Simplified intracellular processes |
| Dual, Elect., STDP | Saudargiene et al. (2005) | ΔIm or ΔVm | 1 dendritic | No | AMPAR, NMDAR | Simplified intracellular processes |
| Dual, Elect., STDP | Shah et al. (2006) | Synaptic stimulus | 1 pre-, 1 postsynaptic | No | NMDAR | Simplified intracellular processes |
| Dual, Elect., STDP | Shouval et al. (2002a) | Synaptic stimulus | 1 synaptic | No | NMDAR | Simplified intracellular processes |
| Dual, Elect., STDP | Shouval et al. (2002b) | Synaptic stimulus | 1 pre-, 1 postsynaptic | No | AMPAR, NMDAR | 2 kinases, 2 phosphatases |
| Dual, Elect., STDP | Shouval and Kalantzis (2005) | Synaptic stimulus | 1 synaptic | No | NMDAR | Simplified intracellular processes |
| Dual, Elect., STDP | Yeung et al. (2004) | Synaptic stimulus | Neuron with 1 compartment, 120 synapses | Nog | NMDAR | Simplified intracellular processes |
| Dual, Elect., STDP | Yu et al. (2008) | Synaptic stimulus | Neuron with 1 compartment, 1000 synapses | Noi | NMDAR | Simplified intracellular processes |
Models are in alphabetical order by the first author and according to the publication month and year. First all LTP models are listed, then all LTD models, and finally all dual LTP and LTD models. Furthermore, electrophysiological (Elect.) models taking into account membrane voltage and spike-timing-dependent plasticity (STDP) models are indicated in the first column. Tabulated characteristics are the model inputs, compartments, voltage-gated ion channels (VGICs), ligand-gated ion channels (LGICs), as well as molecules and Ca2+ mechanisms modeled. ICa denotes in this study the Ca2+ current but dependency in membrane voltage is not modeled. INMDAR denotes in this study the Ca2+ current via NMDARs but dependency in membrane voltage and NMDAR kinetics are not modeled. Isyn denotes the synaptic current. JCa denotes the Ca2+ influx, JVGCC denotes the Ca2+ influx via VGCC, and JNMDAR denotes the Ca2+ influx via NMDARs. For complex CaMKII models, number of CaMKII subunits, number of states for each subunit, and specified threonine (Thr) residues of CaMKII that are phosphorylated are given. Molecules that are modeled as constants are also listed. All abbreviations are given in the list of abbreviations.
aTen CaMKII subunits/Thr-286, Thr-305/306 with five states: inactive, bound with CaMCa4, bound with CaMCa4 and autophosphorylated (trapped), CaMCa4 dissociated from the trapped form but remains phosphorylated (autonomous), and autonomous state secondary phosphorylated (capped).
bIt is not clearly stated in the publication how many CaMKII subunits are modeled but they have two states: inactive and phosphorylated.
cCa2+ is not included in the model.
dModel is by Miller et al. (2005), 12 CaMKII subunits/Thr-286/287 with two states: inactive and phosphorylated.
ePre- and postsynaptic membrane voltage are modeled.
fPostsynaptic neuron is described using adaptive exponential IF neuron model.
gPostsynaptic neuron is described using IF neuron model.
hPre- and postsynaptic neurons are described using IF neuron model.
iPostsynaptic neuron is described using LIF neuron model.
jPostsynaptic membrane voltage is modeled.
Table 7.
Characteristics of models for signaling networks.
| Type | Model | Inputs | Compartments | VGICs | LGICs | Other | Mechanisms | Pathways |
|---|---|---|---|---|---|---|---|---|
| LTP | Ajay and Bhalla (2004) | Glu, JNMDAR | 1 postsynaptic | No | No | EGFR, mGluR | CaM and other buffers | AC, CaM, CaMKIIa, CaN, Gq, MAPK, MKP, PKA, PKC, PKMζ, PLA2, PLC, PP1, Ras, SoS |
| LTP, Elect. | Ajay and Bhalla (2007) | Ca2+, ΔIm or ΔVm, JCa | Neuron with 1–324 compartments | Ca2+, KA, KAHP, KCa, KDR, Na+ | AMPAR, NMDAR | No | CaM buffer, 1-D diffusion of all molecules, PMCA pump, transport of all molecules | CaM, MAPK, PKC, PKM, PLA2, Ras |
| LTP | Aslam et al. (2009) | CaMCa4 | 1 postsynaptic | No | No | No | CaM buffer | CaMKII, CPEB1 |
| LTP, Elect. | Bhalla and Iyengar (1999) | ΔIm or ΔVm, EGF, Glu | Neuron with several compartments | Ca2+, KA, KAHP, KCa, KDR, Na2+ | AMPAR, IP3R, NMDAR | EGFR, mGluR | CaM buffer, PMCA pump, Ca2+ store | AC, CaM, CaMKIIa, CaN, Gq, MAPK, PKA, PKC, PLA2, PLC, PP1, Ras, SoS |
| LTP, Elect. | Bhalla (2002a) | ΔIm or ΔVm, EGF, Glu, hormone | Neuron with 24 dendritic, 1 somatic, 4 spine-head, 3 spine-neck | Ca2+, KA, KAHP, KCa, KDR, Na+ | AMPAR, IP3R, NMDAR | EGFR, mGluR | CaM and other buffers, 1-D Ca2+ diffusion, PMCA and SERCA pumps, Ca2+ store | AC, CaM, CaMKIIa, CaN, Gq, Gs, MAPK, PKA, PKC, PLA2, PLC, PP1, Ras, SoS |
| LTP | Bhalla (2002b) | EGF, Glu, hormone, JCa | 1 extracellular, 1 intracellular, 1 store | No | IP3R | EGFR, mGluR | CaM buffer, PMCA and SERCA pumps, Ca2+ store | AC, CaM, CaMKIIa, CaN, Gq, Gs, MAPK, PKA, PKC, PLA2, PLC, PP1, Ras, SoS |
| LTP | Kikuchi et al. (2003) | Glu, JNMDAR | 1 postsynaptic | No | AMPAR, IP3R | mGluR | CaM buffer, Ca2+ store | AC, CaM, CaMKII, CaN, Gq, I1, MAPK, MEK, MKP, PKA, PKC, PLA2, PLC, PP1, PP2A, Raf, Ras |
| LTP | Kitagawa et al. (2009) | Ca2+, GABABR | 1 postsynaptic | No | GABAAR | GABABR | CaM buffer | AC, CaM, CaMKIIb, cAMP, CaN, DARPP32, PDE1, PDE4, PKA, PP1 |
| LTP | Kubota and Bower (1999) | Ca2+ | 1 spine-head | No | AMPAR | No | CaM buffer, Ca2+ transport | AC, CaM, CaMKIIc, cAMP, CaN, I1, MAPK, PDE, PKA, PP1, Ras |
| LTP | Kötter (1994) | Ca2+, DA | 1 postsynaptic | No | No | No | Buffer | AC, CaMKII, cAMP, CaN, DARPP, MAP2, PDE, PKA, PP1 |
| LTP | Lindskog et al. (2006) | Ca2+, DA | 1 spine | No | No | D1R | CaM buffer | AC, CaM, CaMKII, CaN, DARPP32, PDE1, PDE4, PKA, PP1, PP2A |
| LTP | Lisman (1989) | Ca2+ | 1 postsynaptic | No | No | No | CaM buffer | AC, CaM, CaMKII, cAMP, CaN, I1, PDE, PKA, PP1 |
| LTP | Smolen et al. (2006) | Ca2+, cAMP, kf, Raf | 1 nucleus, 1 somatic, 1 synaptic | No | No | No | Buffer | CaMKII, CaMKIV, MAPK, PKA, gene expression |
| LTP | Smolen (2007) | Ca2+ | 1–5 synapses | No | No | No | Buffer | CaMKII, CaMKIV, MAPK, PKA, gene expression |
| LTP | Smolen et al. (2008) | Raf | 1 spine | No | No | No | No | ERK, MEK, MKKP, MKP, Rafd |
| LTD, Elect. | Achard and De Schutter (2008) | ΔIm or ΔVm | Neuron with 1600 compartments, 1 cytosolic, 1 ER, 1 PSD | BKCa, CaP, CaT, K2Ca, KA, KDR, KIR, KM, Nafast, Naslow | AMPAR, IP3R | mGluR | CD28k, MgGreen, PV, and other buffers, Na+/Ca2+ exchanger, PMCA and SERCA pumps, Ca2+ store | Gq, IP3 3-kinase, IP3 5-phosphatase, PLC |
| LTD | Brown et al. (2008) | PIP2, PLC | 1 or several 1-compartment spines, 1 dendritic | No | No | No | Buffers, 1-D and 3-D diffusion of all molecules | PIP2, PLC |
| LTD | Doi et al. (2005) | Glu, JCa | 1 cytosolic, 1 ER, 1 PSD | No | IP3R | mGluR | CD28k, MgGreen, PV, and other buffers, Na+/Ca2+ exchanger, PMCA and SERCA pumps, Ca2+ store | Gq, IP3 3-kinase, IP3 5-phosphatase, PLC |
| LTD, Elect. | Fiala et al. (1996) | cGMP, Glu | 1 cytosolic, 1 ER, 1 extracellular | KCa | IP3R | mGluR | Na+/Ca2+ exchanger, SERCA pump, Ca2+ store | CaN, G, PKC, PLC |
| LTD | Hellgren Kotaleski et al. (2002) | Ca2+, Glu | 1 spine-head, 2 spine-neck | No | IP3R | mGluR | 2 buffers, 1-D Ca2+ diffusion, Ca2+ store | G, PKC, PLA2, PLC |
| LTD | Kuroda et al. (2001) | Ca2+, Glu, NO | 1 postsynaptic | No | AMPAR | CRHR, mGluR | No | cGMP, Gq, Lyn, MAPK, MEK, PKC, PLA2, PLC, Raf |
| LTD, Elect. | Murzina (2004) | ΔVm, Glu | Neuron with 2 1-compartment spines, 5 dendritic, 1 somatic | Ca2+, K+, KCa, K Na+ | AMPAR, GABAAR | GABABR, mGluR | CaM buffer, 1-D diffusion of NO | CaM, CaMKII, CaN, cGMP, G, GC, PKC, PKG, PP1 |
| LTD, Elect. | Ogasawara et al. (2007) | ΔIm or ΔVm, Glu, NO | 1350 1-compartment spines, 30 dendritic | BKCa, CaP | AMPAR, IP3R | mGluR | CD28k, MgGreen, PV, and other buffers, 3-D diffusion of NO, PMCA and SERCA pumps, Ca2+ store | cGMP, Gq, MAPK, MEK, PKC, PLA2, PLC, Raf |
| LTD | Ogasawara and Kawato (2009) | Generic | 1 postsynaptic | No | No | No | No | 4 kinasesd |
| LTD, Elect. | Steuber and Willshaw (2004) | cGMP, Glu | 0 or 10 dendritic, 1 somatic | KCa | IP3R | mGluR | Buffer, Na+/Ca2+ exchanger, SERCA pump, Ca2+ store | CaN, G, PKC, PLC |
| LTD | Tanaka et al. (2007) | Ca2+ | 1 postsynaptic | No | AMPAR | No | No | MAPK, MEK, PKC, PLA2, Raf |
| LTD, Elect. | Yang et al. (2001) | Ca2+ | Neuron with 1600 compartments | BKCa, CaP, CaT, K2Ca, KA, KDR, KIR, KM, Nafast, Naslow | AMPAR, IP3R | mGluR | Ca2+ store | Gq, PKC, PLA2, PLC |
| Dual | Castellani et al. (2005) | Ca2+ | 1 postsynaptic | No | AMPAR | No | CaM buffer | CaM, CaMKII, cAMP, CaN, I1, PKA, PP1 |
| Dual | d'Alcantara et al. (2003) | Ca2+ | 1 postsynaptic | No | AMPAR | No | CaM buffer | CaM, CaMKII, CaN, I1, PP1 |
| Dual, Elect., STDP | Graupner and Brunel (2007) | ΔIm | 1 spine | CaL, KDR, Na+ | AMPAR, NMDAR | No | Simplified, CaM and other buffers | CaM, CaMKIIe, I1, PP1 |
| Dual | Hayer and Bhalla (2005) | Ca2+, cAMP, JNMDAR | 1 dendritic, 1 PSD, 1 spine-head | No | AMPAR | No | CaM buffer, 1-D diffusion of some of the molecules | AC, CaM, CaMKIIa, CaN, PKA, PP1 |
| Dual | Jain and Bhalla (2009) | BDNF, JNMDAR, MAPK | 1 postsynaptic | No | No | TrkB | CaM buffer | 40S, 4E-BP, AKT, CaM, CaMKIII, MAPK, mTOR, PKC, Ras, S6K, SoS |
| Dual, Elect. | Murzina and Silkis (1998) | ΔIm or ΔVm | Neuron with several compartments | Ca2+, K+, , Na+ | AMPAR, GABAAR, NMDAR | GABABR, mGluR | Buffer, Ca2+ store | AC, CaMKII, cAMP, PKA, PKC |
| Dual, Elect., STDP | Urakubo et al. (2008) | ΔIm or ΔVm | Neuron with 2-compartment spine, 20 dendritic, 1 somatic | CaL, KA, KDR, Na+, Naslow | AMPAR, NMDAR | No | CaM buffer, 1-D diffusion of most of the molecules, PMCA pump, AMPAR trafficking | CaM, CaMKIIf, CaN, cAMP, I1, PKA, PP1, PP2A |
| Dual | Zhabotinsky et al. (2006) | JNMDAR | 1 spine, 1 dendritic, 1 cell body | No | AMPAR | No | CaM buffer, 1-D diffusion of some of the molecules, AMPAR trafficking | CaM, CaMKIIg, CaN, I1, Ng, PKA, PP1, PP2A |
Models are in alphabetical order by the first author and according to the publication month and year. First all LTP models are listed, then all LTD models, and finally all dual LTP and LTD models. Furthermore, electrophysiological (Elect.) models taking into account membrane voltage and spike-timing-dependent plasticity (STDP) models are indicated in the first column. Tabulated characteristics are the model inputs, compartments, voltage-gated ion channels (VGICs), ligand-gated ion channels (LGICs), other receptors, Ca2+ mechanisms, and signaling pathways modeled. JCa denotes the Ca2+ influx and JNMDAR denotes the Ca2+ influx via NMDARs. For complex CaMKII models, number of CaMKII subunits, number of states for each subunit, and specified threonine (Thr) residues of CaMKII that are phosphorylated are given. All abbreviations are given in the list of abbreviations.
aOne CaMKII subunit/Thr-286, Thr-306 with six states: inactive, bound with CaMCa4, bound with CaMCa4 and autophosphorylated (trapped), CaMCa4 dissociated from the trapped form but remains phosphorylated (autonomous), autonomous state secondary phosphorylated (capped), and capped state dephosphorylated.
bIt is not clearly stated in the publication how many CaMKII subunits are modeled. CaMKII subunits/Thr-286/287, Thr-305/306 with six states: inactive, bound with CaMCa3, bound with CaMCa3 and autophosphorylated (trapped), CaMCa3 dissociated from the trapped form but remains phosphorylated (autonomous), autonomous state secondary phosphorylated (capped), and capped state dephosphorylated.
cIt is not clearly stated in the publication how many CaMKII subunits are modeled. CaMKII subunits/Thr-286, Thr-305/306 with five states: inactive, bound with CaMCa4, bound with CaMCa4 and autophosphorylated (trapped), CaMCa4 dissociated from the trapped form but remains phosphorylated (autonomous), and autonomous state secondary phosphorylated (capped).
dCa2+ is not included in the model.
eTwo to eight CaMKII subunits/Thr-286 with four states: inactive, bound with CaMCa4, bound with CaMCa4 and autophosphorylated, and autophosphorylated only.
fOne CaMKII subunit/Thr-286 with several states: inactive, bound with CaM, CaMCa1, CaMCa2, CaMCa3, or CaMCa4, bound and phosphorylated, and dissociated but remains phosphorylated.
gTwo models. Model 1 is one CaMKII subunit/Thr-286 with seven states: inactive, bound with CaMCa4, bound with two CaMCa4, bound with two CaMCa4 and auto-phosphorylated, CaMCa4-dissociated but remains phosphorylated, two CaMCa4 dissociated but remains phosphorylated, and autophosphorylated. Model 2 is by Miller et al. (2005), 12 CaMKII subunits/Thr-286/287 with two states: inactive and phosphorylated.
3.2.1. Models for single pathways
The models for single pathways typically focus on CaMKII (e.g., Dosemeci and Albers, 1996; Okamoto and Ichikawa, 2000a; Smolen et al., 2009), though one model for cAMP production (Kötter and Schirok, 1999) exists and several models are focused on calmodulin activation (e.g., Kubota et al., 2007; Stefan et al., 2008). Most of these models use Ca2+ concentration as the input and include reaction kinetics of CaMCa4 binding and unbinding to CaMKII subunits. Many of the models do not take into account the dodecameric structure of the CaMKII holoenzyme nor the spatial aspect of CaMCa4-dependent autophosphorylation of CaMKII between adjacent subunits. Because of the importance of CaMKII in LTP, most of these single pathway models address the same issues of amplitude and frequency dependence of Ca2+-bound calmodulin or CaMKII activation; subsequent models usually build on previous models and then advance the simulation technique (e.g., stochastic instead of deterministic simulations), or incorporate new experimental details on the CaMKII molecule.
Lisman (1985) presents one of the first models for LTP, which shows that a simple switch model has two stable states, one in which the kinase is dephosphorylated and the other in which it is almost completely phosphorylated. Switch-like behavior, important for memory formation, can be created even when reactions occur stochastically (Smolen et al., 2009), using fast and slow feedback loops. Another stochastic model (Miller et al., 2005) shows that the highly phosphorylated state of CaMKII can remain stable for years, another property which could be important for memory storage.
Okamoto and Ichikawa (2000a) demonstrate the crucial role of competition for calmodulin between spines by modeling several morphological compartments. They model CaMKII in a set of five spines connected to a dendrite and show that after autophosphorylation of CaMKII in a spine, calmodulin in the dendrite can diffuse into that spine for CaMCa4 trapping, which leads to competition since there is a limited concentration of calmodulin. Most of calmodulin is taken by those spines that experience relatively large increases in Ca2+ concentration.
A few of the models contribute to understanding of CaMKII activation though they do not explicitly model CaMKII. Delord et al. (2007) use simple models for Ca2+-controlled phosphorylation–dephosphorylation cycles with non-specific phosphoprotein substrates. Despite the simplicity of these models, the fraction of phosphorylated protein remains elevated for prolonged time periods after Ca2+ concentration returns to its basal level, representing a form of memory storage. Furthermore, the substrate phosphorylation persists in the presence of substrate turnover. Kubota et al. (2007) demonstrate that neurogranin regulates the spatiotemporal pattern of Ca2+-bound calmodulin, which has important implications for CaMKII activation and spatial specificity, by modeling diffusion of single molecules in a spine using 3-D Brownian dynamics.
Several studies show the importance of phosphatases for persistence of synaptic plasticity. Kubota and Bower (2001) show that asymptotic Ca2+ frequency sensitivity of CaMKII depends on both CaMKII and protein phosphatase 1 (PP1). Matsushita et al. (1995) show that phosphatase concentration not only controls whether CaMKII remains phosphorylated, but also controls the intensity of the input required to switch on the persistently phosphorylated state. Lisman and Zhabotinsky (2001) revisit this issue, and show that the CaMKII and PP1 bistable switch activated during the induction of LTP remains active despite the protein turnover. The bistable switch allows CaMKII autophosphorylation to be maintained at low Ca2+ concentrations, even after considering the effect of phosphatases and protein turnover. On the other hand, Bradshaw et al. (2003a) show that the presence of PP1 transforms the CaMKII bistable switch into a reversible (ultrasensitive) switch because PP1 dephosphorylates CaMKII when Ca2+ concentration is lowered to a basal level. Coomber (1998a) studies autophosphorylation and dephosphorylation of CaMKII and includes autophosphorylation of an inhibitory site caused by low-frequency stimulation. In this manner, either LTP or LTD can occur. Though using different mechanisms, both Dosemeci and Albers (1996) and Coomber (1998a,b) show that the phosphorylation of CaMKII can be sensitive to the temporal pattern of Ca2+ pulses, and this may allow CaMKII in the postsynaptic density to act as synaptic frequency detectors. The large allosteric model for calmodulin activation in the postsynaptic density by Stefan et al. (2008) explains how different Ca2+ concentrations can trigger the activation of either CaMKII or calcineurin.
3.2.2. Models for calcium mechanisms or simplified intracellular processes
Models for calcium mechanisms or simplified intracellular processes are a diverse group of models which typically address the role of Ca2+ in producing changes in synaptic strength. Most of these models focus on mechanisms controlling Ca2+ dynamics, such as Ca2+ buffers, pumps, glutamate receptors, or Ca2+-permeable ion channels. Another set of these models use more abstract equations representing intracellular processes and include an equation describing the Ca2+-dependent change in synaptic strength, in order to evaluate whether LTP or LTD occurs with repeated patterns of stimulation.
One of the most compelling questions in the field of LTP is whether high-frequency stimulation increases the spine Ca2+ concentration more than low-frequency stimulation. This has been addressed using models of Ca2+ dynamics in spines alone (see, e.g., Gamble and Koch, 1987; Kitajima and Hara, 1990; Gold and Bear, 1994; Volfovsky et al., 1999; Franks et al., 2001) or spines that include NMDAR activation by electrical activity in models of an entire neuron (see, e.g., Holmes and Levy, 1990; Zador et al., 1990; Koch and Zador, 1993). Zador et al. (1990) further demonstrate that spines compartmentalize Ca2+ (i.e., the Ca2+ signal is limited to those spines that are stimulated), thus providing a mechanism for spatial specificity. Holmes and Levy (1990) show that the frequency sensitivity of LTP requires Ca2+ buffers in addition to NMDAR properties.
A variation of this question is the effect of spine geometry on Ca2+ concentration and synaptic plasticity. Both Volfovsky et al. (1999) and Schmidt and Eilers (2009) test different spine-neck lengths and show that a long neck isolates Ca2+ signaling and calmodulin activation to the spine while stubby spines have a strong coupling between spines and the dendrite. Cornelisse et al. (2007) investigate the role of spine geometry compared to the dendrite. In particular, they demonstrate that the surface area to volume does not completely explain the difference in Ca2+ decay between a spine and dendrite. Instead, a lower buffer capacity of the spine is required to explain the experimental data.
Another important question is the role of various Ca2+ buffers in controlling Ca2+ dynamics. Many models of Ca2+ dynamics have only one or two Ca2+-binding proteins, instead of the many types found in real neurons. Markram et al. (1998) show that competition among Ca2+-binding proteins of various speeds and affinities influences the differential activation of intracellular targets. Models of Ca2+ dynamics permit tight coupling between experiments and models, but require the use of both intrinsic buffers, such as calbindin and parvalbumin, as well as Ca2+ indicators, such as Fura-FF, which themselves are fast, highly diffusible buffers. Other models have shown that buffer saturation is a crucial factor producing supralinear increases in Ca2+ concentration (Hellgren Kotaleski and Blackwell, 2002; Hernjak et al., 2005; Canepari and Vogt, 2008).
Improvements in Ca2+ imaging techniques have been accompanied by the development of sophisticated models that investigate mechanisms underlying Ca2+ microdomains. Naoki et al. (2005) take into account buffering by Ca2+-binding proteins and show that the diffusion coefficient of calmodulin has a strong effect on calmodulin activation in the microdomain near NMDARs. Kubota et al. (2008) investigate the Ca2+-binding protein neurogranin which increases Ca2+ dissociation from calmodulin. Their results show that with no Ca2+ extrusion mechanism, neurogranin increases the steady state concentration of Ca2+; however, in the presence of Ca2+ extrusion mechanisms, neurogranin instead enhances the decay rate of Ca2+. Keller et al. (2008) use MCell (Stiles and Bartol, 2001; Kerr et al., 2008) to develop one of the most advanced models of Ca2+ dynamics in a spine, including Ca2+ pumps, and both voltage-gated Ca2+ channels and NMDA-type of glutamate receptors. The voltage-dependent activation of the channels is coupled to a NEURON (Carnevale and Hines, 2006) simulation of membrane voltage. Keller et al. (2008) show that the Ca2+ gradient and calmodulin activation in the postsynaptic density depend on the order of glutamate release and action potential, and thus may explain the results of STDP experiments.
Just as recent models of Ca2+ dynamics include additional biophysical details, other models explore how biophysical processes related to, for example, glutamate receptors modulate LTP induction. Santucci and Raghavachari (2008) study the role of different types of NMDAR NR2 subunits on subsequent CaMKII activation. They show that though NR2B subunits have a more prolonged time course, the higher open probability of NR2A subunits leads to greater Ca2+ influx and CaMKII activation. The model of Li and Holmes (2000) shows that the variability in NMDAR opening, the spine-head Ca2+ concentration, and levels of CaMKII activation can play an important role in LTP induction. The spine model by Schiegg et al. (1995) includes calcineurin and Ca2+ release from stores, for example through IP3Rs, in the spine head. This study shows that the inclusion of calcineurin alone, which is a Ca2+ sensitive protein phosphatase important for synaptic depression, eliminates LTP; further inclusion of Ca2+ release from stores is required to restore LTP induction. Pi and Lisman (2008) study the role of AMPAR trafficking, modeled by inserting and removing AMPARs in the postsynaptic membrane with a rate that depends on phosphorylated CaMKII and dephosphorylated protein phosphatase 2A (PP2A). Pi and Lisman (2008) show that CaMKII activity is high during LTP, PP2A activity remains high during LTD, and neither activity is high during a basal state; thus, LTD is not a reversal of previous LTP, rather a distinct phenomenon. Clopath et al. (2008) focus on synaptic tagging, an experimental concept important for synaptic specificity of protein synthesis-dependent LTP. The model includes production of plasticity-related proteins which can be captured by tagged synapses. Non-tagged synapses can be tagged stochastically in either a high or low state. They show that synapses share protein synthesis processes which have an effect on the stabilization of potentiated synapses during the transition from E-LTP to L-LTP.
As with all computational models, verification by direct comparison with experimental data strengthens the ability to make experimental predictions and resolve conflicting experimental evidence. The study by Santucci and Raghavachari (2008) is an excellent example on developing a computationally realistic model from good quality data, using the model to resolve conflicting experimental evidence, and then making further experimental predictions. Other examples of direct comparison with experiments include studies by Markram et al. (1998), Volfovsky et al. (1999), Cornelisse et al. (2007), and Schmidt and Eilers (2009). In addition, the prediction that PP2A is critical for LTD induction has been confirmed experimentally (Nicholls et al., 2008). Cai et al. (2007) demonstrate that including the stochastic properties of synaptic transmission significantly affects the form of STDP curves, and indeed is required to explain the experimental data.
3.2.3. Models for signaling networks
Many LTP models for signaling networks are extensions of the single pathway CaMKII models. The model by Lisman (1989) is a landmark because it is one of the first to show that synaptic strength stored by CaMKII could be bidirectionally modified by physiological activity according to the postsynaptic Ca2+ concentration. Kubota and Bower (1999) predict that the CaMKII activity can be sensitive to small changes in the timing of presynaptic signal to the spine head and that CaMKII can exhibit temporal sensitivity even in the presence of PP1. Kitagawa et al. (2009) evaluate the effect of inhibitory G protein-coupled gamma-aminobutyric acid (GABA) B receptor (GABABR) activation on LTP. They show that a transient increase in Ca2+ concentration induces long-term activation of CaMKII, which is attenuated by GABABR activation due to inhibition of PKA. They further show a role for a novel positive feedback loop – one involving CaMKII-mediated downregulation of phosphodiesterase type 1.
Bhalla and Iyengar (1999), Bhalla (2002a,b), Ajay and Bhalla (2004, 2007), and Hayer and Bhalla (2005) have modeled pathways for several protein kinases and phosphatases to investigate information processing. The first study (Bhalla and Iyengar, 1999) uses synaptic stimulation of a compartmental neuron model (Holmes and Levy, 1990; Traub et al., 1991; De Schutter and Bower, 1993) to determine the Ca2+ concentration that is the input to signaling network models. Simulations show that several properties not present in individual pathways, such as feedback loops, thresholds, and sensitivity to signal strength and duration, can emerge from the interaction of pathways. Feedback loops and thresholds can give rise to bistability, offering the possibility that information can be stored within biochemical reactions in the signaling network. The role of temporal sensitivity is further explored (Bhalla, 2002a). This study shows that different input patterns are processed differently by the signaling network, thus giving rise to different outputs (input pattern discrimination). The role of the feedback loop involving MAPK and PKC is further explored in additional studies that integrate experiments and modeling (Bhalla, 2002b). The signaling network models are further refined to include PKMζ (Ajay and Bhalla, 2004, 2007), diffusional processes (Ajay and Bhalla, 2007), and electrical activity (Ajay and Bhalla, 2007) to explore mechanisms underlying MAPK activation in LTP. Ajay and Bhalla (2007) show that extracellular signal-regulated kinase (ERK, MAPK) type II (ERKII) activation after an LTP-inducing stimuli is not explained with reaction–diffusion alone but requires a distributed synaptic input and activation of voltage-gated Ca2+ channels. The model by Hayer and Bhalla (2005) shows that CaMKII and AMPAR phosphorylation form distinct bistable switches, allowing for multiple stable states of the system.
The models of striatal medium spiny neurons (Kötter, 1994; Lindskog et al., 2006) focus on integration of dopamine and glutamate signals, and explore mechanisms which are important for striatal learning. The model by Kötter (1994) is the first to investigate signaling pathways underlying plasticity in the striatum, and shows that, with Ca2+-activated adenylyl cyclase, dopamine and Ca2+ synergistically activate PKA. The model by Lindskog et al. (2006) includes the striatal adenylyl cyclase type 5, which is inhibited by Ca2+, and shows that separate transient dopamine or Ca2+ elevations each may increase the phosphorylation of cAMP-regulated phosphoprotein (DARPP32), due to Ca2+ activation of PP2A. Through this mechanism, paired stimuli yield increased PKA activation and DARPP32 phosphorylation compared to dopamine alone, in contrast to the effect of prolonged stimuli in which Ca2+ decreases DARPP32 phosphorylation. Fernandez et al. (2006) study the functions of DARPP32 with a detailed signaling network model but they do not address plasticity, thus this study is not included in Table 7. However, their study may be used as a valuable model to build on for future modeling efforts studying plasticity.
More recently models have been constructed to investigate mechanisms underlying L-LTP, by incorporating molecules such as CaMKIV, transcription factors, or the translation factor cytoplasmic polyadenylation element binding protein (CPEB1). Smolen (2007) shows that long periods of decreased activity reset synaptic strength to a low value, whereas episodic activity with short inactive periods maintains strong synapses. Smolen et al. (2008) implement a stochastic model to show that the feedback loop from MAPK to MAPK kinase kinase (Raf) increases the robustness of both stable states of MAPK activity to stochastic fluctuations. Aslam et al. (2009) show that the positive feedback loop between CaMKII and CPEB1 forms a bistable switch accounting for the protein synthesis dependence of L-LTP. In addition, Jain and Bhalla (2009) are interested in protein synthesis dependence of L-LTP, and thus investigate how the synaptic input pattern affects dendritic protein synthesis. These types of models are likely to increase because behavioral memories require protein synthesis.
Long-term depression is predominant for synapses in the cerebellum; thus, most models of LTD describe signaling networks in cerebellar Purkinje cells. Kuroda et al. (2001) investigate the mechanism producing persistent phosphorylation of AMPARs, required for LTD. Simulations show that the initial phase of phosphorylation of AMPARs depends on the activation of PKC by arachidonic acid, Ca2+, and diacylglycerol, whereas a later phase depends on the activation of a positive feedback loop and especially phospholipase A2 and arachidonic acid. Tanaka et al. (2007) further demonstrate that disrupting the positive feedback loop between several protein kinases can affect Ca2+ triggering of LTD. Brown et al. (2008) present an elaborate three-dimensional model of a Purkinje cell dendrite with spines to investigate the issue of whether sufficient phosphatidylinositol biphosphate (PIP2) is available in a single spine to achieve the experimentally estimated concentrations of IP3 required for Ca2+ release and subsequent LTD. They elegantly show that a relatively novel mechanism, namely stimulated synthesis of PIP2, is required to account for experimental results. Three of the LTD models (Yang et al., 2001; Ogasawara et al., 2007; Achard and De Schutter, 2008) use the multi-compartment, multi-channel Purkinje cell model by De Schutter and Bower (1994a,b) to simulate electrical activity leading to Ca2+ influx through synaptic and voltage-gated ion channels. Ogasawara et al. (2007) show that the nitric oxide concentration is critical for induction of LTD and for its input specificity. Achard and De Schutter (2008) re-evaluate the importance of conjunctive parallel fiber and climbing fiber inputs. They show that both inputs are required to produce a sufficient Ca2+ elevation to trigger LTD.
Because of the role of the cerebellum in eyeblink classical conditioning, several signaling network models investigate whether temporal characteristics of classical conditioning can be explained by temporal characteristics of LTD in single Purkinje cells. Fiala et al. (1996) have developed the first model to explain adaptive timing of the eyeblink response in classical conditioning. They use a biochemical variant of spectral timing for their parallel fiber inputs, and also include the effect of Ca2+-gated potassium channel activation on membrane voltage. They show that the phosphorylation state of target proteins responsible for LTD depends on the timing between climbing fiber and parallel fiber stimulation. Hellgren Kotaleski et al. (2002) include production of PKC activators by parallel fiber and climbing fiber stimulation in order to evaluate the relationship between LTD and behavior. Both Hellgren Kotaleski et al. (2002) and Doi et al. (2005) show that IP3-dependent Ca2+ dynamics are sensitive to temporal interval between parallel fiber and climbing fiber stimulation. Hellgren Kotaleski et al. (2002) further demonstrate that PKC activation is sensitive to temporal interval between parallel fiber and climbing fiber inputs (which is analogous to classical conditioning being sensitive to temporal interval). The importance of conjunctive parallel fiber and climbing fiber inputs for Ca2+ elevation is confirmed using a multi-compartment, multi-channel Purkinje cell model by Ogasawara et al. (2007) which more accurately simulates Ca2+ influx through synaptic and voltage-gated ion channels. Steuber and Willshaw (2004) show that replacing the spectral timing mechanism with Ca2+-dependent phosphorylation of mGluRs allows a single Purkinje cell to learn the adaptive timing of the eyeblink response.
More recent dual LTP and LTD models evaluate signaling network activation using spike-timing-dependent protocols (Graupner and Brunel, 2007; Urakubo et al., 2008). Urakubo et al. (2008) show that Ca2+ influx through NMDARs does not vary with spike timing (contrary to expectations) without suppression of NMDARs by Ca2+-bound calmodulin. Graupner and Brunel (2007) have constructed models for Ca2+/CaM-dependent autophosphorylation of CaMKII and PP1-dependent dephosphorylation of CaMKII. Graupner and Brunel (2007) show that CaMKII plays a central role in LTD because it is dephosphorylated during induction of LTD. More importantly, their bistable model can reproduce plasticity in response to STDP and high-frequency stimulation, without requiring abnormally low Ca2+ concentrations for dephosphorylation.
4. Analysis and Discussion
This study provides an extensive overview of 117 computational models for postsynaptic signal transduction pathways in synaptic plasticity developed over the past 25 years through 2009. Our purpose is to categorize the models so that similarities and differences are more readily apparent. Due to the large number of models, many models, though valuable, are excluded since they do not reach our criteria given in the beginning of Section 3. Some of the models included in this study are very simplified biochemical models meaning that a specific phenomenon is expressed using only a couple of reactions (see, e.g., Delord et al., 2007; Pi and Lisman, 2008). In the other extreme are the complex biophysical models that include detailed reaction–diffusion systems coupled to neuronal electrical activity (see, e.g., Bhalla, 2002a; Urakubo et al., 2008). Though model complexity has been increasing (Figures 2 and 3), the simpler biochemical models remain a valuable approach. They are relatively easy to construct, and the number of parameters to be fine-tuned is small. Not only are they computationally efficient, but they allow theoretical analysis and identification of which pathway, or combination of pathways, produces which property. On the other hand, models with detailed mechanisms are ideal for investigating which of several candidate molecules and mechanisms control or modulate a particular response. Furthermore, the direct correspondence between a detailed model and real neuron allows specific model predictions to be tested experimentally.
Figure 2.
Evolution of postsynaptic signal transduction models from 1985 to 2009. The starting point of an arrow represents the model which is used by the latter model indicated as the arrowhead. A dotted line in the arrow means that the two studies use exactly the same model (the latter study is not presented in Tables 1–9).
Figure 3.
Numbers of published postsynaptic signal transduction models per year from 1985 to 2009. (A) Numbers of LTP, LTD, and dual LTP and LTD models. (B) Numbers of reaction, reaction and diffusion, reaction and electrophysiological, as well as reaction, diffusion, and electrophysiological models. (C) Numbers of different size (S, M, and L) models. (D) Numbers of deterministic, stochastic, and deterministic and stochastic models.
In our study, the emphasis is more on evaluating the model components and on the significance of the models rather than on comparison of the actual model responses. The comparison of model responses is not trivial because all models would need to be implemented and simulated before a comparative analysis could be performed (see also Pettinen et al., 2005). Indeed, this is not only time consuming, but impossible since many of the models are neither described in sufficient detail nor provided in model databases or by other open-access means (see Table 8). Even qualitative comparison is difficult since only a few publications provide a graphical illustration of the model components and in many cases it is difficult to interpret the model input or stimulus. These observations serve also as guidelines for reviewers evaluating future publications and models: (1) all models should be described in sufficient detail including equations, inputs, outputs, compartments, variables, constants, parameters, and initial conditions; (2) graphical illustration of the model should include only those model components that actually participate in simulations; (3) the simulation tool or programing language should be specified; and (4) the model should be provided in a model database. Nordlie et al. (2009) propose a good model description practice for neuronal network models. A similar description practice is needed for signal transduction models and our study is one step toward this, as is the BioModels Database project (Le Novère et al., 2006).
Table 8.
Models provided in databases or by other open-access means.
| Model | Simulation environment | Databases |
|---|---|---|
| Ajay and Bhalla (2004) | GENESIS/Kinetikita, MATLAB®, SBMLb | DOQCSc |
| SBMLb | BioModels Databased | |
| Ajay and Bhalla (2007) | GENESIS/Kinetikita, MATLAB®, SBMLb | DOQCSc |
| SBMLb | BioModels Databased | |
| Aslam et al. (2009) | MATLAB® | Supplementary material by Aslam et al. (2009) |
| Badoual et al. (2006) | NEURONe | ModelDBf |
| Bhalla and Iyengar (1999) | GENESIS/Kinetikita, MATLAB®, SBMLb | DOQCSc |
| SBMLb | BioModels Databased | |
| SBMLb | CellMLg | |
| Bhalla (2002b) | GENESIS/Kinetikita, MATLAB®, SBMLb | DOQCSc |
| Brown et al. (2008) | Virtual Cellh | Virtual Cellh |
| Clopath et al. (2008) | Python | ModelDBf |
| Cornelisse et al. (2007) | CalCi | ModelDBf |
| d'Alcantara et al. (2003) | SBMLb | BioModels Databased |
| Doi et al. (2005) | GENESIS/Kinetikita | ModelDBf |
| Gerkin et al. (2007) | IGOR Proj | ModelDBf |
| Graupner and Brunel (2007) | XPPAUTk | ModelDBf |
| Hayer and Bhalla (2005) | GENESIS/Kinetikita, GENESIS 3/MOOSEl, MATLAB®, SBMLb | DOQCSc |
| Hernjak et al. (2005) | MathSBMLm | Virtual Cellh |
| MathSBMLm | BioModels Databased | |
| Ichikawa (2004) | A-Celln | http://www.his.kanazawa-it.ac.jp/∼ichikawa/ |
| EnglishTop.html | ||
| Ichikawa et al. (2007) | A-Celln | http://www.his.kanazawa-it.ac.jp/∼ichikawa/ |
| EnglishTop.html | ||
| Jain and Bhalla (2009) | GENESIS/Kinetikita, GENESIS 3/MOOSEl | DOQCSc |
| XML | Supplementary material by Jain and Bhalla (2009) | |
| Kitagawa et al. (2009) | SBMLb | Supplementary material by Kitagawa et al. (2009) |
| Kuroda et al. (2001) | GENESIS/Kinetikita, MATLAB®, SBMLb | DOQCSc |
| GENESIS/Kinetikita | http://www.cns.atr.jp/neuroinfo/kuroda/ | |
| SBMLb | BioModels Databased | |
| Lindskog et al. (2006) | XPPAUTk | ModelDBf |
| Migliore and Lansky (1999b) | QuickBASIC | ModelDBf |
| Saftenku (2002) | NEURONe | ModelDBf |
| Schmidt and Eilers (2009) | Mathematica | Supplementary material by Schmidt and Eilers (2009) |
| Stefan et al. (2008) | BioPAXo, CellMLg, SBMLb, Scilabp, Virtual Cellh, XPPk | BioModels Databased |
| Urakubo et al. (2008) | GENESIS/Kinetikita | ModelDBf |
| GENESIS/Kinetikita | http://www.bi.s.u-tokyo.ac.jp/kuroda-lab/info/ | |
| STDP/index.html |
aGENESIS/Kinetikit (http://www.genesis-sim.org/GENESIS/; http://www.ncbs.res.in/index.php?option=com_content&task=view&id=307; Bower and Beeman, 1998; Bhalla, 2002c).
bSBML (http://sbml.org/).
cDOQCS (http://doqcs.ncbs.res.in/; Sivakumaran et al., 2003).
dBioModels Database (http://www.biomodels.net/; Le Novère et al., 2006).
eNEURON (http://www.neuron.yale.edu/neuron/; Carnevale and Hines, 2006).
fModelDB (http://senselab.med.yale.edu/modeldb/; Migliore et al., 2003; Hines et al., 2004).
gCellML (http://www.cellml.org; Lloyd et al., 2008).
hVirtual Cell (http://vcell.org; Schaff et al., 1997; Slepchenko et al., 2003).
iCalC (http://web.njit.edu/~matveev/calc.html; Matveev et al., 2002).
jIGOR Pro (http://www.wavemetrics.com/).
kXPP, XPPAUT (http://www.math.pitt.edu/~bard/xpp/xpp.html; Ermentrout, 2002).
lGENESIS 3/MOOSE (http://www.genesis-sim.org/GENESIS/; http://moose.sourceforge.net/).
mMathSBML (http://sbml.org/Software/MathSBML).
nA-Cell (http://www.fujixerox.co.jp/crc/cng/A-Cell/; Ichikawa, 2001, 2005).
oBioPAX (http://www.biopax.org/; Luciano and Stevens, 2007).
pScilab (http://www.scilab.org/; Gomez, 1999).
Every computational model needs to be stimulated to study evoked activity even though this aspect is not always clearly indicated in the publications. In other words, an input similar to the one given in experimental wet-lab studies or as in the physiological in vivo state is required. In many cases, however, it is a challenge to mimic the input used in experiments. The construction of input stimulus is quite straightforward in cases where biophysically detailed models and a high-frequency stimulation protocol are used. In the other extreme are the models which use some function mimicking synaptic stimulus. This input type is not adequately described in many of the publications analyzed in the present study. This makes the reproduction of simulation results and the comparison of the models impossible. Therefore, the description of input stimuli should be taken into account when developing specific description language solutions for computational neuroscience and neuroinformatics.
Testing sensitivity to changes in parameter values is very important because many of the model parameters are not sufficiently constrained by experimental data. Table 9 highlights the models that evaluate whether the simulation results are sensitive to changes in parameter values. In this study, small-scale testing means that values for 10 parameters or less (for example rate constants) are varied, and large-scale testing means that values for greater than 10 parameters are varied. Table 9 shows that only a few models employ the large-scale testing of sensitivity to changes in parameter values. Publications that only test sensitivity to changes in input parameter values or do parameter estimation to fit experimental data, without analyzing the different model responses, are not included in Table 9.
Table 9.
Models testing sensitivity to changes in parameter values.
| Testing | Models |
|---|---|
| Small-scale | Holmes (1990, 2000), Holmes and Levy (1990), Gold and Bear (1994), Matsushita et al. (1995), Migliore et al. (1995), Schiegg et al. (1995), Dosemeci and Albers (1996), Fiala et al. (1996), Coomber (1998a,b), Volfovsky et al. (1999), Okamoto and Ichikawa (2000b), Zhabotinsky (2000), Kuroda et al. (2001), Hellgren Kotaleski et al. (2002), Karmarkar and Buonomano (2002), Shouval et al. (2002a,b), Abarbanel et al. (2003, 2005), d'Alcantara et al. (2003), Kikuchi et al. (2003), Hayer and Bhalla (2005), Hernjak et al. (2005), Miller et al. (2005), Naoki et al. (2005), Rubin et al. (2005), Lindskog et al. (2006), Smolen et al. (2006, 2008), Zhabotinsky et al. (2006), Cai et al. (2007), Cornelisse et al. (2007), Delord et al. (2007), Graupner and Brunel (2007), Ogasawara et al. (2007), Smolen (2007), Brown et al. (2008), Kubota and Kitajima (2008), Urakubo et al. (2008), Yu et al. (2008), Aslam et al. (2009), Castellani et al. (2009), Jain and Bhalla (2009), Kalantzis and Shouval (2009) |
| Large-scale | Bhalla and Iyengar (1999), Doi et al. (2005), Achard and De Schutter (2008), Kitagawa et al. (2009) |
Small-scale testing means that values for 10 parameters or less (for example rate constants) are varied, and large-scale testing means that values for greater than 10 parameters are varied.
In order to predict the future direction of the field, trends regarding the development of models of postsynaptic signal transduction pathways underlying LTP and LTD are illustrated (Figures 2 and 3). Figure 2 shows how different models reviewed in this study have evolved from each other. Two models are connected in Figure 2 if the publication either states directly that other models are used or the publication uses a subset of the exact same equations appearing in the older publications by the same authors. Models are excluded from Figure 2 if there is no clear evidence that they have used some other model as the basis, or if they are only based on models not reviewed in this study. Figure 2 shows that the models by Holmes and Levy (1990), Bhalla and Iyengar (1999), and Shouval et al. (2002a) are most often used as a starting point when developing new models. Zhabotinsky et al. (2006) and Graupner and Brunel (2007) cite the largest number of models when developing their models, but, on the other hand, they do not clearly state which parts of their model are taken from which other models.
Though LTP models appeared first, most of the new models are dual LTP and LTD models (Figure 3A), suggesting that these are being developed to investigate which characteristics of synaptic input patterns lead to LTP versus LTD. Despite limiting the review to models of signaling pathways, the models are extremely diverse in scope, with less than half including only reactions. Other models combine reactions and diffusion, or reactions and electrophysiological phenomena; about one-fifth have all three (Figure 3B). About one-third of the models are size small, meaning that there are less than 20 different chemical species or other model variables, and about half of the models are size large meaning that there are more than 50 different chemical species or other model variables (Figure 3C). The trend is toward increasing numbers of large models, reflecting both the increase in computational power and increasing knowledge of the biochemical pathways. Nonetheless, the continued development of small models reflects their utility in theoretical analysis. Most of the models are still deterministic even though stochastic methods have been developed more and more recently (Figure 3D). The scarcity of stochastic models compared to large models may reflect the availability of software modeling tools and analytic tools. However, several stochastic reaction–diffusion simulation tools have appeared recently (see, e.g., Kerr et al., 2008; Wils and De Schutter, 2009; Andrews et al., 2010; Byrne et al., 2010; Oliveira et al., 2010; Tolle and Le Novère, 2010b). Stochastic methods are important because very small numbers of molecules can have a dramatic effect on either strengthening or weakening the synapses and these effects should be taken into account. Another possibility is to develop and use so-called hybrid simulation methods where specific events are modeled as stochastic and others as deterministic. Though not illustrated graphically, only about one-fourth of the reviewed publications specify the simulation tool or programing language used. Most often the simulation tool used is GENESIS/Kinetikit (Bower and Beeman, 1998; Bhalla, 2002c), XPPAUT (Ermentrout, 2002), and NEURON (Carnevale and Hines, 2006). Programing languages most often used are Java and MATLAB®.
The trends in Figure 3 lead to several predictions about the future of signaling pathway modeling. The first prediction is that both the number of large models and the size of the largest model will continue to increase. Thus, existing models will be expanded to include additional signaling pathways, in parallel with the increase in experimental data of additional molecular mechanisms. Second, the trend in Figure 3D suggests that increasing number of models will be implemented stochastically or using hybrid deterministic–stochastic methods. The stochastic part of the models in particular may focus on events in the postsynaptic density and other multi-protein complexes. The third prediction is that the scope of the models will expand, with more models of dual LTP and LTD phenomena, in part because both phenomena have been measured in most cell types, and in part because the increase in size of the models is expanding to include signaling pathways for both phenomena. Related to the increase in scope of the models, more will blend reactions with diffusion or electrophysiological phenomena in order to study spatial aspects of signaling and also to better relate to experiments. In particular, modeling reactions alone is not sufficient for understanding synaptic plasticity but also electrophysiological phenomena needs to be taken into account by modeling neuronal networks (Hellgren Kotaleski and Blackwell, 2010). Further development of simulation tools (Pettinen et al., 2005; Alves et al., 2006) together with improvements in parallel computing should help in this endeavor.
Though the trend is toward larger and more complex models, this does not imply that all larger models are better than simpler models. As explained above, the quality of a model depends on many factors. Probably the most important criteria is whether the model can address a question of general scientific interest. For this reason, we have tried to organize our description of the models in order to highlight the questions addressed. Another related criteria is whether a model can make verifiable, i.e. falsifiable, predictions. Using these two criteria, models incorporating more biochemical details often appear superior, but only if the parameters can be adequately constrained. However, models which simplify the equations describing intracellular signaling pathways are more easily integrated with whole neuron electrophysiological models or able to simulate longer time frames. From this perspective they may excel for investigating whether different stimulation patterns change synaptic strength differently. It is important to note that earlier models may have been groundbreaking at the time of publication, yet their perceived quality decreases as more is learned about the interactions of intracellular molecules. Only a couple of studies reduce complex models to simpler ones and show comparative simulation results between the models (see, e.g., Hayer and Bhalla, 2005; Smolen, 2007). The reduction of model complexity will be an important research area in the future because simplified models that can capture relevant aspects of dynamics could be embedded, for example, into biologically-inspired neuronal network models when the activity of individual neurons is modeled in more detail.
To fully understand synaptic plasticity, many different characteristics of signaling pathways need to be considered. Temporal and spatial aspects of signaling are crucially important because they relate the cellular phenomenon of plasticity to the behavioral phenomenon of learning. Not only do theoreticians and modelers need to incorporate experimental findings, but also experimental progress can be enhanced by using model simulations to select the most promising experiments. Careful attention to these issues should improve the utility of modeling approaches for investigating molecular mechanisms of synaptic plasticity. The ultimate future goal of LTP and LTD modeling is to find such models for different brain regions and cells that can explain all the phases of synaptic plasticity, and then use these models to explain the differences in plasticity between brain regions or cell types. Many of the modeling studies have so far concentrated on only one type of synaptic plasticity. We believe that an analysis like the one provided by us will help in this endeavor to make more predictive models for synaptic plasticity in the future.
Acknowledgments
This work was partly supported by research project grants from Academy of Finland [106030 and 124615 (Marja-Leena Linne), 126556 (Tiina Manninen), and 129657 (Finnish Programme for Centres of Excellence in Research 2006–2011)], Swedish Research Council (Jeanette Hellgren Kotaleski), the Parkinson's Foundation (Jeanette Hellgren Kotaleski), HFSP programme (Kim T. Blackwell), and the joint NSF-NIH CRCNS programme through NIH grant R01 AA16022 and R01 AA18060 (Kim T. Blackwell). Additional support was obtained from Finnish Foundation for Economic and Technology Sciences – KAUTE (Tiina Manninen), Otto A. Malm Foundation (Tiina Manninen and Katri Hituri), Emil Aaltonen Foundation (Katri Hituri), Finnish Foundation for Technology Promotion (Katri Hituri), and two graduate schools (Tampere University of Technology Graduate School and Tampere Doctoral Programme in Information Science and Engineering) (Katri Hituri).
Abbreviations
4E-BP, 4E-binding protein; AC, adenylyl cyclase; AKT, serine/threonine kinase; AMPAR, α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor; ATP, adenosine triphosphate; BDNF, brain-derived neurotrophic factor; BKCa, high-threshold Ca2+- and voltage-gated K+ channel; CA1, cornu ammonis 1; Ca2+, calcium ion; CA3, cornu ammonis 3; CaL, high-threshold L-type Ca2+ channel; CaM, calmodulin; CaMCa1, CaM–1Ca2+ complex; CaMCa2, CaM–2Ca2+ complex; CaMCa3, CaM–3Ca2+ complex; CaMCa4, CaM–4Ca2+ complex; CaMK, Ca2+/CaM-dependent protein kinase; CaMKII, CaMK type II; CaMKIII, CaMK type III; CaMKIV, CaMK type IV; cAMP, cyclic adenosine monophosphate; CaN, high-threshold N-type Ca2+ channel; CaN, calcineurin; CaP, high-threshold P-type Ca2+ channel; CaT, low-threshold T-type Ca2+ channel; CD28k, calbindin; CG-1, Calcium-Green 1; cGMP, cyclic guanosine monophosphate; CICR, Ca2+-induced Ca2+ release; CPEB1, cytoplasmic polyadenylation element binding protein; CRHR, corticotropin-releasing hormone receptor; ΔIm, change in membrane current; ΔVm, change in Vm; D, dimensional; D1R, dopamine receptor; DA, dopamine; DARPP, cAMP-regulated phosphoprotein; DARPP32, DARPP of 32 kDa; DGC, dentate granule cell; DOQCS, Database of Quantitative Cellular Signaling; EGF, epidermal growth factor; EGFR, EGF receptor; E-LTD, early phase LTD; E-LTP, early phase LTP; ER, endoplasmic reticulum; ERK, extracellular signal-regulated kinase; ERKII, ERK type II; FF, Fura-FF; G, G protein; GABA, gamma-aminobutyric acid; GABAAR, GABA receptor A; GABABR, GABA receptor B; GABAR, GABA receptor; gAMPAR, AMPAR conductance; GC, guanylate cyclase; KCa channel conductance; Glu, glutamate; GluN, glutamatergic neuron; Gq, G protein type q; GrC, granule cell; Gs, G protein type s; gsyn, synaptic conductance; I1, inhibitor 1; ICa, Ca2+ current; IF, integrate-and-fire; INMDAR, Ca2+ current via NMDAR; IP3, inositol trisphosphate; IP3R, IP3 receptor; Isyn, synaptic current; JCa, Ca2+ influx; JNMDAR, Ca2+ influx via NMDAR; JVGCC, Ca2+ influx via VGCC; K+, potassium ion; K2Ca, low-threshold K2-type Ca2+-gated K+ channel; KA, transient A-type K+ channel; KAHP, after-hyperpolarization K+ channel; KCa, Ca2+- and voltage-gated K+ channel; KDR, delayed-rectifier K+ channel; kf, Raf, activation rate for Raf; GABAAR-activated K+ channel; GABABR-activated K+ channel; KIR, inward-rectifier K+ channel; KM, muscarine-sensitive K+ channel; Kslow, slow Ca2+-independent tetraethylammonium-insensitive K+ channel; L, large; LGIC, ligand-gated ion channel; LIF, leaky IF; L-LTD, late phase LTD; L-LTP, late phase LTP; LTD, long-term depression; LTP, long-term potentiation; Lyn, Lyn tyrosine kinase; M, medium; MAP2, microtubule-associated protein 2; MAPK, mitogen-activated protein kinase; MEK, MAPK kinase; MgGreen, Magnesium Green 1; mGluR, metabotropic glutamate receptor; MKKP, MEK phosphatase; MKP, MAPK phosphatase; MSN, medium spiny neuron; mTOR, mammalian target of rapamycin; N, neuron; Na+, sodium ion; Nafast, fast Na+ channel; Nar, recurrent Na+ channel; Naslow, non- or slowly inactivating Na+ channel; Ng, neurogranin; NMDA, N-methyl-d-aspartate; NMDAR, NMDA receptor; NO, nitric oxide; OGB-1, Oregon Green BAPTA-1; PC, Purkinje cell; PDE, phosphodiesterase; PDE1, PDE type 1; PDE4, PDE type 4; PIP2, phosphatidylinositol biphosphate; PKA, cAMP-dependent protein kinase; PKC, protein kinase C; PKG, protein kinase G; PKM, atypical PKC isozyme; PKMζ, atypical PKC isozyme; PLA2, phospholipase A2; PLC, phospholipase C; PMCA, plasma membrane Ca2+-ATPase; PN, pyramidal neuron; PP1, protein phosphatase 1; PP2A, protein phosphatase 2A; PSD, postsynaptic density; PV, parvalbumin; Raf, MEK kinase; S, small; S6K, 40S ribosomal protein S6 kinase; SBML, Systems Biology Markup Language; Ser, serine; SERCA, sarco/ER Ca2+-ATPase; SoS, son of sevenless; STD, short-term depression; STDP, spike-timing-dependent plasticity; STP, short-term potentiation; Thr, threonine; TrkB, tropomyosin-receptor kinase B; VGCC, voltage-gated Ca2+ channel; VGIC, voltage-gated ion channel; Vm, membrane voltage.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Abarbanel H. D. I., Gibb L., Huerta R., Rabinovich M. I. (2003). Biophysical model of synaptic plasticity dynamics. Biol. Cybern. 89, 214–226 10.1007/s00422-003-0422-x [DOI] [PubMed] [Google Scholar]
- Abarbanel H. D. I., Huerta R., Rabinovich M. I. (2002). Dynamical model of long-term synaptic plasticity. Proc. Natl. Acad. Sci. U.S.A. 99, 10132–10137 10.1073/pnas.132651299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abarbanel H. D. I., Talathi S. S., Gibb L., Rabinovich M. I. (2005). Synaptic plasticity with discrete state synapses. Phys. Rev. E 72, 031914. 10.1103/PhysRevE.72.031914 [DOI] [PubMed] [Google Scholar]
- Achard P., De Schutter E. (2008). Calcium, synaptic plasticity and intrinsic homeostasis in Purkinje neuron models. Front. Comput. Neurosci. 2: 8. 10.3389/neuro.10.008.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajay S. M., Bhalla U. S. (2004). A role for ERKII in synaptic pattern selectivity on the time-scale of minutes. Eur. J. Neurosci. 20, 2671–2680 10.1111/j.1460-9568.2004.03725.x [DOI] [PubMed] [Google Scholar]
- Ajay S. M., Bhalla U. S. (2006). Synaptic plasticity in vitro and in silico: insights into an intracellular signaling maze. Physiology 21, 289–296 10.1152/physiol.00009.2006 [DOI] [PubMed] [Google Scholar]
- Ajay S. M., Bhalla U. S. (2007). A propagating ERKII switch forms zones of elevated dendritic activation correlated with plasticity. HFSP J. 1, 49–66 10.2976/1.2721383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves R., Antunes F., Salvador A. (2006). Tools for kinetic modeling of biochemical networks. Nat. Biotechnol. 24, 667–672 10.1038/nbt0606-667 [DOI] [PubMed] [Google Scholar]
- Andrews S. S., Addy N. J., Brent R., Arkin A. P. (2010). Detailed simulations of cell biology with Smoldyn 2.1. PLoS Comput. Biol. 6, e1000705. 10.1371/journal.pcbi.1000705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam N., Kubota Y., Wells D., Shouval H. Z. (2009). Translational switch for long-term maintenance of synaptic plasticity. Mol. Syst. Biol. 5, 284. 10.1038/msb.2009.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badoual M., Zou Q., Davison A. P., Rudolph M., Bal T., Fregnac Y., Destexhe A. (2006). Biophysical and phenomenological models of multiple spike interactions in spike-timing dependent plasticity. Int. J. Neural Syst. 16, 79–97 10.1142/S0129065706000524 [DOI] [PubMed] [Google Scholar]
- Bhalla U. S. (2002a). Biochemical signaling networks decode temporal patterns of synaptic input. J. Comput. Neurosci. 13, 49–62 10.1023/A:1019644427655 [DOI] [PubMed] [Google Scholar]
- Bhalla U. S. (2002b). Mechanisms for temporal tuning and filtering by postsynaptic signaling pathways. Biophys. J. 83, 740–752 10.1016/S0006-3495(02)75205-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla U. S. (2002c). “Use of Kinetikit and GENESIS for modeling signaling pathways,” in Methods in Enzymology, Vol. 345, eds Hildebrandt J. D., Iyengar R. (San Diego: Academic Press; ), 3–23 [DOI] [PubMed] [Google Scholar]
- Bhalla U. S. (2009). “Molecules, networks, and memory,” in Systems Biology: The Challenge of Complexity, 1st Edn, eds Nakanishi S., Kageyama R., Watanabe D. (Tokyo: Springer; ), 151–158 [Google Scholar]
- Bhalla U. S., Iyengar R. (1999). Emergent properties of networks of biological signaling pathways. Science 283, 381–387 10.1126/science.283.5400.381 [DOI] [PubMed] [Google Scholar]
- Bi G.-Q., Poo M.-M. (1998). Synaptic modifications in cultured hippocampal neurons: dependence on spike timing, synaptic strength, and postsynaptic cell type. J. Neurosci. 18, 10464–10472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi G.-Q., Rubin J. (2005). Timing in synaptic plasticity: from detection to integration. Trends Neurosci. 28, 222–228 10.1016/j.tins.2005.02.002 [DOI] [PubMed] [Google Scholar]
- Blackwell K. T., Hellgren Kotaleski J. (2002). “Modeling the dynamics of second messenger pathways,” in Neuroscience Databases: A Practical Guide, ed. Kötter R. (Norwell, MA: Kluwer Academic Publishers; ), 63–80 [Google Scholar]
- Bliss T. V. P., Collingridge G. L. (1993). A synaptic model of memory: long-term potentiation in the hippocampus. Nature 361, 31–39 10.1038/361031a0 [DOI] [PubMed] [Google Scholar]
- Bliss T. V. P., Gardner-Medwin A. R. (1973). Long-lasting potentiation of synaptic transmission in the dentate area of the unanaesthetized rabbit following stimulation of the perforant path. J. Physiol. 232, 357–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss T. V. P., Lømo T. (1973). Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J. Physiol. 232, 331–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitzer R. D., Iyengar R., Landau E. M. (2005). Postsynaptic signaling networks: cellular cogwheels underlying long-term plasticity. Biol. Psychiatry 57, 113–119 10.1016/j.biopsych.2004.02.031 [DOI] [PubMed] [Google Scholar]
- Bower J. M., Beeman D. (1998). The Book of GENESIS: Exploring Realistic Neural Models with the GEneral NEural SImulation System, 2nd Edn New York: Telos/Springer-Verlag [Google Scholar]
- Bradshaw J. M., Kubota Y., Meyer T., Schulman H. (2003a). An ultrasensitive Ca2+/calmodulin-dependent protein kinase II-protein phosphatase 1 switch facilitates specificity in postsynaptic calcium signaling. Proc. Natl. Acad. Sci. U.S.A. 100, 10512–10517 10.1073/pnas.1932759100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw K. D., Emptage N. J., Bliss T. V. P. (2003b). A role for dendritic protein synthesis in hippocampal late LTP. Eur. J. Neurosci. 18, 3150–3152 10.1111/j.1460-9568.2003.03054.x [DOI] [PubMed] [Google Scholar]
- Brown S.-A., Morgan F., Watras J., Loew L. M. (2008). Analysis of phosphatidylinositol-4,5-bisphosphate signaling in cerebellar Purkinje spines. Biophys. J. 95, 1795–1812 10.1529/biophysj.108.130195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T. H., Kairiss E. W., Keenan C. L. (1990). Hebbian synapses: biophysical mechanisms and algorithms. Annu. Rev. Neurosci. 13, 475–511 10.1146/annurev.ne.13.030190.002355 [DOI] [PubMed] [Google Scholar]
- Bruel-Jungerman E., Davis S., Laroche S. (2007). Brain plasticity mechanisms and memory: a party of four. Neuroscientist 13, 492–505 10.1177/1073858407302725 [DOI] [PubMed] [Google Scholar]
- Byrne M. J., Putkey J. A., Waxham M. N., Kubota Y. (2009). Dissecting cooperative calmodulin binding to CaM kinase II: a detailed stochastic model. J. Comput. Neurosci. 27, 621–638 10.1007/s10827-009-0173-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M. J., Waxham M. N., Kubota Y. (2010). Cellular dynamic simulator: an event driven molecular simulation environment for cellular physiology. Neuroinformatics 8, 63–82 10.1007/s12021-010-9066-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Gavornik J. P., Cooper L. N., Yeung L. C., Shouval H. Z. (2007). Effect of stochastic synaptic and dendritic dynamics on synaptic plasticity in visual cortex and hippocampus. J. Neurophysiol. 97, 375–386 10.1152/jn.00895.2006 [DOI] [PubMed] [Google Scholar]
- Canepari M., Vogt K. E. (2008). Dendritic spike saturation of endogenous calcium buffer and induction of postsynaptic cerebellar LTP. PLoS ONE 3, e4011. 10.1371/journal.pone.0004011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevale T., Hines M. (2006). The NEURON Book, 1st Edn Cambridge, UK: Cambridge University Press; 10.1017/CBO9780511541612 [DOI] [Google Scholar]
- Castellani G. C., Bazzani A., Cooper L. N. (2009). Toward a microscopic model of bidirectional synaptic plasticity. Proc. Natl. Acad. Sci. U.S.A. 106, 14091–14095 10.1073/pnas.0905988106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani G. C., Quinlan E. M., Bersani F., Cooper L. N., Shouval H. Z. (2005). A model of bidirectional synaptic plasticity: from signaling network to channel conductance. Learn. Mem. 12, 423–432 10.1101/lm.80705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani G. C., Quinlan E. M., Cooper L. N., Shouval H. Z. (2001). A biophysical model of bidirectional synaptic plasticity: dependence on AMPA and NMDA receptors. Proc. Natl. Acad. Sci. U.S.A. 98, 12772–12777 10.1073/pnas.201404598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani G. C., Zironi I. (2010). “Biophysics-based models of LTP/LTD,” in Hippocampal Microcircuits: A Computational Modeler's Resource Book, eds Cutsuridis V., Graham B., Cobb S., Vida I. (New York: Springer; ), 555–570 [Google Scholar]
- Citri A., Malenka R. C. (2008). Synaptic plasticity: multiple forms, functions, and mechanisms. Neuropsychopharmacology 33, 18–41 10.1038/sj.npp.1301559 [DOI] [PubMed] [Google Scholar]
- Clopath C., Büsing L., Vasilaki E., Gerstner W. (2010). Connectivity reflects coding: a model of voltage-based STDP with homeostasis. Nat. Neurosci. 13, 344–352 10.1038/nn.2479 [DOI] [PubMed] [Google Scholar]
- Clopath C., Ziegler L., Vasilaki E., Büsing L., Gerstner W. (2008). Tag-trigger-consolidation: a model of early and late long-term-potentiation and depression. PLoS Comput. Biol. 4, e1000248. 10.1371/journal.pcbi.1000248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke S. F., Bliss T. V. P. (2006). Plasticity in the human central nervous system. Brain 129, 1659–1673 10.1093/brain/awl082 [DOI] [PubMed] [Google Scholar]
- Coomber C. (1997). A model of associative long-term potentiation and long-term depression in a compartmental reconstruction of a neuron. Neurocomputing 16, 189–205 10.1016/S0925-2312(97)00027-1 [DOI] [Google Scholar]
- Coomber C. (1998a). Current theories of neuronal information processing performed by Ca2+/calmodulin-dependent protein kinase II with support and insights from computer modelling and simulation. Comput. Chem. 22, 251–263 10.1016/S0097-8485(97)00002-8 [DOI] [PubMed] [Google Scholar]
- Coomber C. J. (1998b). Site-selective autophosphorylation of Ca2+/calmodulin-dependent protein kinase II as a synaptic encoding mechanism. Neural Comput. 10, 1653–1678 10.1162/089976698300017070 [DOI] [PubMed] [Google Scholar]
- Cornelisse L. N., van Elburg R. A. J., Meredith R. M., Yuste R., Mansvelder H. D. (2007). High speed two-photon imaging of calcium dynamics in dendritic spines: consequences for spine calcium kinetics and buffer capacity. PLoS ONE 2, e1073. 10.1371/journal.pone.0001073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Alcantara P., Schiffmann S. N., Swillens S. (2003). Bidirectional synaptic plasticity as a consequence of interdependent Ca2+-controlled phosphorylation and dephosphorylation pathways. Eur. J. Neurosci. 17, 2521–2528 10.1046/j.1460-9568.2003.02693.x [DOI] [PubMed] [Google Scholar]
- Dan Y., Poo M.-M. (2006). Spike timing-dependent plasticity: from synapse to perception. Physiol. Rev. 86, 1033–1048 10.1152/physrev.00030.2005 [DOI] [PubMed] [Google Scholar]
- Delord B., Berry H., Guigon E., Genet S. (2007). A new principle for information storage in an enzymatic pathway model. PLoS Comput. Biol. 3, e124. 10.1371/journal.pcbi.0030124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter E., Bower J. M. (1993). Sensitivity of synaptic plasticity to the Ca2+ permeability of NMDA channels: a model of long-term potentiation in hippocampal neurons. Neural Comput. 5, 681–694 10.1162/neco.1993.5.5.681 [DOI] [Google Scholar]
- De Schutter E., Bower J. M. (1994a). An active membrane model of the cerebellar Purkinje cell. I. Simulation of current clamps in slice. J. Neurophysiol. 71, 375–400 [DOI] [PubMed] [Google Scholar]
- De Schutter E., Bower J. M. (1994b). An active membrane model of the cerebellar Purkinje Cell. II. Simulation of synaptic responses. J. Neurophysiol. 71, 401–419 [DOI] [PubMed] [Google Scholar]
- Doi T., Kuroda S., Michikawa T., Kawato M. (2005). Inositol 1,4,5-trisphosphate-dependent Ca2+ threshold dynamics detect spike timing in cerebellar Purkinje cells. J. Neurosci. 25, 950–961 10.1523/JNEUROSCI.2727-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosemeci A., Albers R. W. (1996). A mechanism for synaptic frequency detection through autophosphorylation of CaM kinase II. Biophys. J. 70, 2493–2501 10.1016/S0006-3495(96)79821-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek S. M., Bear M. F. (1992). Homosynaptic long-term depression in area CA1 of hippocampus and effects of N-methyl-d-aspartate receptor blockade. Proc. Natl. Acad. Sci. U.S.A. 89, 4363–4367 10.1073/pnas.89.10.4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont G., Houart G., De Koninck P. (2003). Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations: a simple model. Cell Calcium 34, 485–497 10.1016/S0143-4160(03)00152-0 [DOI] [PubMed] [Google Scholar]
- Engelman M. S. (1982). FIDAP (A Fluid Dynamics Analysis Program). Adv. Eng. Softw. (1978) 4, 163–166 [Google Scholar]
- Engelman M. S. (1996). FIDAP Theoretical Manual, Version 7.5. Evanston, IL: Fluid Dynamics Inc. [Google Scholar]
- Ermentrout B. (2002). Simulating, Analyzing, and Animating Dynamical Systems: A Guide to XPPAUT for Researchers and Students, 1st Edn Philadelphia: Society for Industrial and Applied Mathematics (SIAM) [Google Scholar]
- Fernandez É., Schiappa R., Girault J.-A., Le Novère N. (2006). DARPP-32 is a robust integrator of dopamine and glutamate signals. PLoS Comput. Biol. 2, e176. 10.1371/journal.pcbi.0020176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala J. C., Grossberg S., Bullock D. (1996). Metabotropic glutamate receptor activation in cerebellar Purkinje cells as substrate for adaptive timing of the classically conditioned eye-blink response. J. Neurosci. 16, 3760–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks K. M., Bartol T. M., Sejnowski T. J. (2001). An MCell model of calcium dynamics and frequency-dependence of calmodulin activation in dendritic spines. Neurocomputing 38–40, 9–16 10.1016/S0925-2312(01)00415-5 [DOI] [Google Scholar]
- Gamble E., Koch C. (1987). The dynamics of free calcium in dendritic spines in response to repetitive synaptic input. Science 236, 1311–1315 10.1126/science.3495885 [DOI] [PubMed] [Google Scholar]
- Gerdeman G. L., Lovinger D. M. (2003). Emerging roles for endocannabinoids in long-term synaptic plasticity. Br. J. Pharmacol. 140, 781–789 10.1038/sj.bjp.0705466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerkin R. C., Bi G.-Q., Rubin J. E. (2010). “A phenomenological calcium-based model of STDP,” in Hippocampal Microcircuits: A Computational Modeler's Resource Book, eds Cutsuridis V., Graham B., Cobb S., Vida I. (New York: Springer; ), 571–591 [Google Scholar]
- Gerkin R. C., Lau P.-M., Nauen D. W., Wang Y. T., Bi G.-Q. (2007). Modular competition driven by NMDA receptor subtypes in spike-timing-dependent plasticity. J. Neurophysiol. 97, 2851–2862 10.1152/jn.00860.2006 [DOI] [PubMed] [Google Scholar]
- Gewaltig M. O., Diesmann M. (2007). NEST (neural simulation tool). Scholarpedia 2, 1430. 10.4249/scholarpedia.1430 [DOI] [Google Scholar]
- Gillespie D. T. (1976). A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 22, 403–434 10.1016/0021-9991(76)90041-3 [DOI] [Google Scholar]
- Gillespie D. T. (1977). Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361 10.1021/j100540a008 [DOI] [Google Scholar]
- Gold J. I., Bear M. F. (1994). A model of dendritic spine Ca2+ concentration exploring possible bases for a sliding synaptic modification threshold. Proc. Natl. Acad. Sci. U.S.A. 91, 3941–3945 10.1073/pnas.91.9.3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez C. (Ed.). (1999). Engineering, and Scientific Computing with Scilab. Boston: Birkhäuser [Google Scholar]
- Graupner M., Brunel N. (2007). STDP in a bistable synapse model based on CaMKII and associated signaling pathways. PLoS Comput. Biol. 3, e221. 10.1371/journal.pcbi.0030221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graupner M., Brunel N. (2010). Mechanisms of induction and maintenance of spike-timing dependent plasticity in biophysical synapse models. Front. Comput. Neurosci. 4: 136. 10.3389/fncom.2010.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer A., Bhalla U. S. (2005). Molecular switches at the synapse emerge from receptor and kinase traffic. PLoS Comput. Biol. 1, e20. 10.1371/journal.pcbi.0010020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helias M., Rotter S., Gewaltig M.-O., Diesmann M. (2008). Structural plasticity controlled by calcium based correlation detection. Front. Comput. Neurosci. 2: 7. 10.3389/neuro.10.007.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellgren Kotaleski J., Blackwell K. T. (2002). Sensitivity to interstimulus interval due to calcium interactions in the Purkinje cell spines. Neurocomputing 44–46, 13–18 10.1016/S0925-2312(02)00361-2 [DOI] [Google Scholar]
- Hellgren Kotaleski J., Blackwell K. T. (2010). Modelling the molecular mechanisms of synaptic plasticity using systems biology approaches. Nat. Rev. Neurosci. 11, 239–251 10.1038/nrn2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellgren Kotaleski J., Lester D., Blackwell K. T. (2002). Subcellular interactions between parallel fibre and climbing fibre signals in Purkinje cells predict sensitivity of classical conditioning to interstimulus interval. Integr. Physiol. Behav. Sci. 37, 265–292 10.1007/BF02734249 [DOI] [PubMed] [Google Scholar]
- Hernjak N., Slepchenko B. M., Fernald K., Fink C. C., Fortin D., Moraru I. I., Watras J., Loew L. M. (2005). Modeling and analysis of calcium signaling events leading to long-term depression in cerebellar Purkinje cells. Biophys. J. 89, 3790–3806 10.1529/biophysj.105.065771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines M. L., Morse T., Migliore M., Carnevale N. T., Shepherd G. M. (2004). ModelDB: a database to support computational neuroscience. J. Comput. Neurosci. 17, 7–11 10.1023/B:JCNS.0000023869.22017.2e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcman D., Schuss Z., Korkotian E. (2004). Calcium dynamics in dendritic spines and spine motility. Biophys. J. 87, 81–91 10.1529/biophysj.103.035972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes W. R. (1990). Is the function of dendritic spines to concentrate calcium? Brain Res. 519, 338–342 10.1016/0006-8993(90)90098-V [DOI] [PubMed] [Google Scholar]
- Holmes W. R. (2000). Models of calmodulin trapping and CaM kinase II activation in a dendritic spine. J. Comput. Neurosci. 8, 65–86 10.1023/A:1008969032563 [DOI] [PubMed] [Google Scholar]
- Holmes W. R. (2005). “Calcium signaling in dendritic spines,” in Modeling in the Neurosciences: From Biological Systems to Neuromimetic Robotics, 2nd Edn., eds Reeke G. N., Poznanski R. R., Lindsay K. A., Rosenberg J. R., Sporns O. (Boca Raton: CRC Press; ), 25–60 [Google Scholar]
- Holmes W. R., Levy W. B. (1990). Insights into associative long-term potentiation from computational models of NMDA receptor-mediated calcium influx and intracellular calcium concentration changes. J. Neurophysiol. 63, 1148–1168 [DOI] [PubMed] [Google Scholar]
- Holmes W. R., Levy W. B. (1997). Quantifying the role of inhibition in associative long-term potentiation in dentate granule cells with computational models. J. Neurophysiol. 78, 103–116 [DOI] [PubMed] [Google Scholar]
- Holthoff K., Tsay D., Yuste R. (2002). Calcium dynamics of spines depend on their dendritic location. Neuron 33, 425–437 10.1016/S0896-6273(02)00576-7 [DOI] [PubMed] [Google Scholar]
- Hoops S., Sahle S., Gauges R., Lee C., Pahle J., Simus N., Singhai M., Xu L., Mendes P., Kummer U. (2006). COPASI – a complex pathway simulator. Bioinformatics 22, 3067–3074 10.1093/bioinformatics/btl485 [DOI] [PubMed] [Google Scholar]
- Hudmon A., Schulman H. (2002a). Neuronal Ca2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annu. Rev. Biochem. 71, 473–510 10.1146/annurev.biochem.71.110601.135410 [DOI] [PubMed] [Google Scholar]
- Hudmon A., Schulman H. (2002b). Structure-function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochem. J. 364 (Pt 3), 593–611 10.1042/BJ20020228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa K. (2001). A-Cell: graphical user interface for the construction of biochemical reaction models. Bioinformatics 17, 483–484 10.1093/bioinformatics/17.5.483 [DOI] [PubMed] [Google Scholar]
- Ichikawa K. (2004). Localization of activated Ca2+/calmodulin-dependent protein kinase II within a spine: modeling and computer simulation. Neurocomputing 58–60, 443–448 10.1016/j.neucom.2004.01.079 [DOI] [Google Scholar]
- Ichikawa K. (2005). A modeling environment with three-dimensional morphology, A-Cell-3D, and Ca2+ dynamics in a spine. Neuroinformatics 3, 49–63 10.1385/NI:3:1:049 [DOI] [PubMed] [Google Scholar]
- Ichikawa K., Hoshino A., Kato K. (2007). Induction of synaptic depression by high-frequency stimulation in area CA1 of the rat hippocampus: modeling and experimental studies. Neurocomputing 70, 2055–2059 10.1016/j.neucom.2006.10.131 [DOI] [Google Scholar]
- Ito M. (1989). Long-term depression. Annu. Rev. Neurosci. 12, 85–102 10.1146/annurev.ne.12.030189.000505 [DOI] [PubMed] [Google Scholar]
- Ito M. (2001). Cerebellar long-term depression: characterization, signal transduction, and functional roles. Physiol. Rev. 81, 1143–1195 [DOI] [PubMed] [Google Scholar]
- Ito M. (2002). The molecular organization of cerebellar long-term depression. Nat. Rev. Neurosci. 3, 896–902 10.1038/nrn962 [DOI] [PubMed] [Google Scholar]
- Ito M., Sakurai M., Tongroach P. (1982). Climbing fiber induced depression of both mossy fiber responsiveness and glutamate sensitivity of cerebellar Purkinje cells. J. Physiol. 324, 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain P., Bhalla U. S. (2009). Signaling logic of activity-triggered dendritic protein synthesis: an mTOR gate but not a feedback switch. PLoS Comput. Biol. 5, e1000287. 10.1371/journal.pcbi.1000287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantzis G., Shouval H. Z. (2009). Structural plasticity can produce metaplasticity. PLoS ONE 4, e8062. 10.1371/journal.pone.0008062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmarkar U. R., Buonomano D. V. (2002). A model of spike-timing dependent plasticity: one or two coincidence detectors? J. Neurophysiol. 88, 507–513 [DOI] [PubMed] [Google Scholar]
- Karmarkar U. R., Najarian M. T., Buonomano D. V. (2002). Mechanisms and significance of spike-timing dependent plasticity. Biol. Cybern. 87, 373–382 10.1007/s00422-002-0351-0 [DOI] [PubMed] [Google Scholar]
- Kauderer B. S., Kandel E. R. (2000). Capture of a protein synthesis-dependent component of long-term depression. Proc. Natl. Acad. Sci. U.S.A. 97, 13342–13347 10.1073/pnas.97.24.13342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller D. X., Franks K. M., Bartol T. M., Jr., Sejnowski T. J. (2008). Calmodulin activation by calcium transients in the postsynaptic density of dendritic spines. PLoS ONE 3, e2045. 10.1371/journal.pone.0002045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr R. A., Bartol T. M., Kaminsky B., Dittrich M., Chang J.-C. J., Baden S. B., Sejnowski T. J., Stiles J. R. (2008). Fast Monte Carlo simulation methods for biological reaction–diffusion systems in solution and on surfaces. SIAM J. Sci. Comput. 30, 3126–3149 10.1137/070692017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S., Fujimoto K., Kitagawa N., Fuchikawa T., Abe M., Oka K., Takei K., Tomita M. (2003). Kinetic simulation of signal transduction system in hippocampal long-term potentiation with dynamic modeling of protein phosphatase 2A. Neural Netw. 16, 1389–1398 10.1016/j.neunet.2003.09.002 [DOI] [PubMed] [Google Scholar]
- Kim M. S., Huang T., Abel T., Blackwell K. T. (2010). Temporal sensitivity of protein kinase A activation in late-phase long term potentiation. PLoS Comput. Biol. 6, e1000691. 10.1371/journal.pcbi.1000691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa Y., Hirano T., Kawaguchi S.-Y. (2009). Prediction and validation of a mechanism to control the threshold for inhibitory synaptic plasticity. Mol. Syst. Biol. 5, 280. 10.1038/msb.2009.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima T., Hara K. (1990). A model of the mechanisms of long-term potentiation in the hippocampus. Biol. Cybern. 64, 33–39 10.1007/BF00203628 [DOI] [PubMed] [Google Scholar]
- Kitajima T., Hara K. (2000). A generalized Hebbian rule for activity-dependent synaptic modifications. Neural Netw. 13, 445–454 10.1016/S0893-6080(00)00028-9 [DOI] [PubMed] [Google Scholar]
- Kitajima T., Hara K.-I. (1997). An integrated model for activity-dependent synaptic modifications. Neural Netw. 10, 413–421 10.1016/S0893-6080(96)00095-0 [DOI] [PubMed] [Google Scholar]
- Klann E., Chen S. J., Sweatt J. D. (1993). Mechanism of protein kinase C activation during the induction and maintenance of long-term potentiation probed using a selective peptide substrate. Proc. Natl. Acad. Sci. U.S.A. 90, 8337–8341 10.1073/pnas.90.18.8337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipp E., Liebermeister W. (2006). Mathematical modeling of intracellular signaling pathways. BMC Neurosci. 7 (Suppl. 1), S10. 10.1186/1471-2202-7-S1-S10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C., Zador A. (1993). The function of dendritic spines: devices subserving biochemical rather than electrical compartmentalization. J. Neurosci. 13, 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötter R. (1994). Postsynaptic integration of glutamatergic and dopaminergic signals in the striatum. Prog. Neurobiol. 44, 163–196 10.1016/0301-0082(94)90037-X [DOI] [PubMed] [Google Scholar]
- Kötter R., Schirok D. (1999). Towards an integration of biochemical and biophysical models of neuronal information processing: a case study in the nigro-striatal system. Rev. Neurosci. 10, 247–266 [DOI] [PubMed] [Google Scholar]
- Kubota S., Kitajima T. (2008). A model for synaptic development regulated by NMDA receptor subunit expression. J. Comput. Neurosci. 24, 1–20 10.1007/s10827-007-0036-8 [DOI] [PubMed] [Google Scholar]
- Kubota S., Kitajima T. (2010). Possible role of cooperative action of NMDA receptor and GABA function in developmental plasticity. J. Comput. Neurosci. 28, 347–359 10.1007/s10827-010-0212-0 [DOI] [PubMed] [Google Scholar]
- Kubota Y., Bower J. M. (1999). Decoding time-varying calcium signals by the postsynaptic biochemical network: computer simulations of molecular kinetics. Neurocomputing 26–27, 29–38 10.1016/S0925-2312(99)00085-5 [DOI] [Google Scholar]
- Kubota Y., Bower J. M. (2001). Transient versus asymptotic dynamics of CaM kinase II: possible roles of phosphatase. J. Comput. Neurosci. 11, 263–279 10.1023/A:1013727331979 [DOI] [PubMed] [Google Scholar]
- Kubota Y., Putkey J. A., Shouval H. Z., Waxham M. N. (2008). IQ-motif proteins influence intracellular free Ca2+ in hippocampal neurons through their interactions with calmodulin. J. Neurophysiol. 99, 264–276 10.1152/jn.00876.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y., Putkey J. A., Waxham M. N. (2007). Neurogranin controls the spatiotemporal pattern of postsynaptic Ca2+/CaM signaling. Biophys. J. 93, 3848–3859 10.1529/biophysj.107.106849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda S., Schweighofer N., Kawato M. (2001). Exploration of signal transduction pathways in cerebellar long-term depression by kinetic simulation. J. Neurosci. 21, 5693–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanté F., de Jésus Ferreira M.-C., Guiramand J., Récasens M., Vignes M. (2006). Low-frequency stimulation induces a new form of LTP, metabotropic glutamate (mGlu5) receptor- and PKA-dependent, in the CA1 area of the rat hippocampus. Hippocampus 16, 345–360 10.1002/hipo.20146 [DOI] [PubMed] [Google Scholar]
- Le Novère N., Bornstein B., Broicher A., Courtot M., Donizelli M., Dharuri H., Li L., Sauro H., Schilstra M., Shapiro B., Snoep J. L., Hucka M. (2006). BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res. 34, D689–D691 10.1093/nar/gkj092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Holmes W. R. (2000). Comparison of CaMKinase II activation in a dendritic spine computed with deterministic and stochastic models of the NMDA synaptic conductance. Neurocomputing 32–33, 1–7 10.1016/S0925-2312(00)00137-5 [DOI] [Google Scholar]
- Lindskog M., Kim M., Wikström M. A., Blackwell K. T., Hellgren Kotaleski J. (2006). Transient calcium and dopamine increase PKA activity and DARPP-32 phosphorylation. PLoS Comput. Biol. 2, e119. 10.1371/journal.pcbi.0020119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J. (1989). A mechanism for the Hebb and the anti-Hebb processes underlying learning and memory. Proc. Natl. Acad. Sci. U.S.A. 86, 9574–9578 10.1073/pnas.86.23.9574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J., Goldring M. A. (1988a). Evaluation of a model of long-term memory based on the properties of the Ca2+/calmodulin-dependent protein kinase. J. Physiol. (Paris) 83, 187–197 [PubMed] [Google Scholar]
- Lisman J. E., Goldring M. A. (1988b). Feasibility of long-term storage of graded information by the Ca2+/calmodulin-dependent protein kinase molecules of the postsynaptic density. Proc. Natl. Acad. Sci. U.S.A. 85, 5320–5324 10.1073/pnas.85.14.5320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J., Schulman H., Cline H. (2002). The molecular basis of CaMKII function in synaptic and behavioural memory. Nat. Rev. Neurosci. 3, 175–190 10.1038/nrn753 [DOI] [PubMed] [Google Scholar]
- Lisman J. E. (1985). A mechanism for memory storage insensitive to molecular turnover: a bistable autophosphorylating kinase. Proc. Natl. Acad. Sci. U.S.A. 82, 3055–3057 10.1073/pnas.82.9.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J. E., Zhabotinsky A. M. (2001). A model of synaptic memory: a CaMKII/PP1 switch that potentiates transmission by organizing an AMPA receptor anchoring assembly. Neuron 31, 191–201 10.1016/S0896-6273(01)00364-6 [DOI] [PubMed] [Google Scholar]
- Lloyd C. M., Lawson J. R., Hunter P. J., Nielsen P. F. (2008). The CellML model repository. Bioinformatics 24, 2122–2123 10.1093/bioinformatics/btn390 [DOI] [PubMed] [Google Scholar]
- Luciano J. S., Stevens R. D. (2007). e-Science and biological pathway semantics. BMC Bioinformatics 8 (Suppl. 3), S3. 10.1186/1471-2105-8-S3-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka R. C., Bear M. F. (2004). LTP and LTD: an embarrassment of riches. Neuron 44, 5–21 10.1016/j.neuron.2004.09.012 [DOI] [PubMed] [Google Scholar]
- Malenka R. C., Nicoll R. A. (1999). Long-term potentiation – a decade of progress? Science 285, 1870–1874 10.1126/science.285.5435.1870 [DOI] [PubMed] [Google Scholar]
- Malinow R., Schulman H., Tsien R. W. (1989). Inhibition of postsynaptic PKC or CaMKII blocks induction but not expression of LTP. Science 245, 862–866 10.1126/science.2549638 [DOI] [PubMed] [Google Scholar]
- Markram H., Lübke J., Frotscher M., Sakmann B. (1997). Regulation of synaptic efficacy by coincidence of postsynaptic APs and EPSPs. Science 275, 213–215 10.1126/science.275.5297.213 [DOI] [PubMed] [Google Scholar]
- Markram H., Roth A., Helmchen F. (1998). Competitive calcium binding: implications for dendritic calcium signaling. J. Comput. Neurosci. 5, 331–348 10.1023/A:1008891229546 [DOI] [PubMed] [Google Scholar]
- Matsushita T., Moriyama S., Fukai T. (1995). Switching dynamics and the transient memory storage in a model enzyme network involving Ca2+/calmodulin-dependent protein kinase II in synapses. Biol. Cybern. 72, 497–509 10.1007/BF00199892 [DOI] [PubMed] [Google Scholar]
- Matveev V., Sherman A., Zucker R. S. (2002). New and corrected simulations of synaptic facilitation. Biophys. J. 83, 1368–1373 10.1016/S0006-3495(02)73907-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson S., Schulman H. (1994). CaM kinase: a model for its activation and dynamics. J. Theor. Biol. 171, 281–290 10.1006/jtbi.1994.1231 [DOI] [Google Scholar]
- Migliore M., Alicata F., Ayala G. F. (1995). A model for long-term potentiation and depression. J. Comput. Neurosci. 2, 335–343 10.1007/BF00961444 [DOI] [PubMed] [Google Scholar]
- Migliore M., Alicata F., Ayala G. F. (1997). Possible roles of retrograde messengers on LTP, LTD, and associative memory. Biosystems 40, 127–132 10.1016/0303-2647(96)01638-3 [DOI] [PubMed] [Google Scholar]
- Migliore M., Ayala G. F. (1993). A kinetic model of short- and long-term potentiation. Neural Comput. 5, 636–647 10.1162/neco.1993.5.4.636 [DOI] [Google Scholar]
- Migliore M., Lansky P. (1999a). Computational model of the effects of stochastic conditioning on the induction of long-term potentiation and depression. Biol. Cybern. 81, 291–298 10.1007/s004220050563 [DOI] [PubMed] [Google Scholar]
- Migliore M., Lansky P. (1999b). Long-term potentiation and depression induced by a stochastic conditioning of a model synapse. Biophys. J. 77, 1234–1243 10.1016/S0006-3495(99)76975-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliore M., Morse T. M., Davison A. P., Marenco L., Shepherd G. M., Hines M. L. (2003). ModelDB: making models publicly accessible to support computational neuroscience. Neuroinformatics 1, 135–139 10.1385/NI:1:1:135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P., Wang X.-J. (2006). Stability of discrete memory states to stochastic fluctuations in neuronal systems. Chaos 16, 026109. 10.1063/1.2208923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P., Zhabotinsky A. M., Lisman J. E., Wang X.-J. (2005). The stability of a stochastic CaMKII switch: dependence on the number of enzyme molecules and protein turnover. PLoS Biol. 3, e107. 10.1371/journal.pbio.0030107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Diesmann M., Gerstner W. (2008). Phenomenological models of synaptic plasticity based on spike timing. Biol. Cybern. 98, 459–478 10.1007/s00422-008-0233-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina G. B. (2004). Mathematical simulation of the induction of long-term depression in cerebellar Purkinje cells. Neurosci. Behav. Physiol. 34, 115–121 10.1023/B:NEAB.0000009205.08547.10 [DOI] [PubMed] [Google Scholar]
- Murzina G. B., Silkis I. G. (1998). Studies of long-term potentiation and depression of inhibitory transmission by mathematical modeling of post-synaptic processes. Neurosci. Behav. Physiol. 28, 121–129 10.1007/BF02461957 [DOI] [PubMed] [Google Scholar]
- Nakano T., Doi T., Yoshimoto J., Doya K. (2010). A kinetic model of dopamine- and calcium-dependent striatal synaptic plasticity. PLoS Comput. Biol. 6, e1000670. 10.1371/journal.pcbi.1000670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoki H., Sakumura Y., Ishii S. (2005). Local signaling with molecular diffusion as a decoder of Ca2+ signals in synaptic plasticity. Mol. Syst. Biol. 1, 2005.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher E. (1998). Usefulness and limitations of linear approximations to the understanding of Ca2+ signals. Cell Calcium 24, 345–357 10.1016/S0143-4160(98)90058-6 [DOI] [PubMed] [Google Scholar]
- Nicholls R. E., Alarcon J. M., Malleret G., Carroll R. C., Grody M., Vronskaya S., Kandel E. R. (2008). Transgenic mice lacking NMDAR-dependent LTD exhibit deficits in behavioral flexibility. Neuron 58, 104–117 10.1016/j.neuron.2008.01.039 [DOI] [PubMed] [Google Scholar]
- Nordlie E., Gewaltig M.-O., Plesser H. E. (2009). Towards reproducible descriptions of neuronal network models. PLoS Comput. Biol. 5, e1000456. 10.1371/journal.pcbi.1000456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara H., Doi T., Doya K., Kawato M. (2007). Nitric oxide regulates input specificity of long-term depression and context dependence of cerebellar learning. PLoS Comput. Biol. 3, e179. 10.1371/journal.pcbi.0020179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara H., Doi T., Kawato M. (2008). Systems biology perspectives on cerebellar long-term depression. Neurosignals 16, 300–317 10.1159/000123040 [DOI] [PubMed] [Google Scholar]
- Ogasawara H., Kawato M. (2009). “Computational models of cerebellar long-term memory,” in Systems Biology: The Challenge of Complexity, 1st Edn., eds Nakanishi S., Kageyama R., Watanabe D. (Tokyo: Springer; ), 169–182 [Google Scholar]
- Okamoto H., Ichikawa K. (2000a). A model for molecular mechanisms of synaptic competition for a finite resource. Biosystems 55, 65–71 10.1016/S0303-2647(99)00084-2 [DOI] [PubMed] [Google Scholar]
- Okamoto H., Ichikawa K. (2000b). Switching characteristics of a model for biochemical-reaction networks describing autophosphorylation versus dephosphorylation of Ca2+/calmodulin-dependent protein kinase II. Biol. Cybern. 82, 35–47 10.1007/PL00007960 [DOI] [PubMed] [Google Scholar]
- Oliveira R. F., Terrin A., Di Benedetto G., Cannon R. C., Koh W., Kim M., Zaccolo M., Blackwell K. T. (2010). The role of type 4 phosphodiesterases in generating microdomains of cAMP: large scale stochastic simulations. PLoS ONE 5, e11725. 10.1371/journal.pone.0011725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepke S., Kinzer-Ursem T., Mihalas S., Kennedy M. B. (2010). A dynamic model of interactions of Ca2+, calmodulin, and catalytic subunits of Ca2+/calmodulin-dependent protein kinase II. PLoS Comput. Biol. 6, e1000675. 10.1371/journal.pcbi.1000675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettinen A., Aho T., Smolander O.-P., Manninen T., Saarinen A., Taattola K.-L., Yli-Harja O., Linne M.-L. (2005). Simulation tools for biochemical networks: evaluation of performance and usability. Bioinformatics 21, 357–363 10.1093/bioinformatics/bti018 [DOI] [PubMed] [Google Scholar]
- Pi H. J., Lisman J. E. (2008). Coupled phosphatase and kinase switches produce the tristability required for long-term potentiation and long-term depression. J. Neurosci. 28, 13132–13138 10.1523/JNEUROSCI.2348-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Miller G. W., Voit E. O. (2010). The internal state of medium spiny neurons varies in response to different input signals. BMC Syst. Biol. 4, 26. 10.1186/1752-0509-4-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham O. J. L., Tsaneva-Atanasova K., Ganesh A., Mellor J. R. (2010). A Ca2+-based computational model for NMDA receptor-dependent synaptic plasticity at individual post-synaptic spines in the hippocampus. Front. Syn. Neurosci. 2: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin J. E., Gerkin R. C., Bi G.-Q., Chow C. C. (2005). Calcium time course as a signal for spike-timing-dependent plasticity. J. Neurophysiol. 93, 2600–2613 10.1152/jn.00803.2004 [DOI] [PubMed] [Google Scholar]
- Saftenku E. E. (2002). A simplified model of long-term plasticity in cerebellar mossy fiber-granule cell synapses. Neurophysiology 34, 216–218 10.1023/A:1020723921430 [DOI] [Google Scholar]
- Santamaria F., Gonzalez J., Augustine G. J., Raghavachari S. (2010). Quantifying the effects of elastic collisions and non-covalent binding on glutamate receptor trafficking in the post-synaptic density. PLoS Comput. Biol. 6, e1000780. 10.1371/journal.pcbi.1000780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos S. D., Carvalho A. L., Caldeira M. V., Duarte C. B. (2009). Regulation of AMPA receptors and synaptic plasticity. Neuroscience 158, 105–125 10.1016/j.neuroscience.2008.02.037 [DOI] [PubMed] [Google Scholar]
- Santucci D. M., Raghavachari S. (2008). The effects of NR2 subunit-dependent NMDA receptor kinetics on synaptic transmission and CaMKII activation. PLoS Comput. Biol. 4, e1000208. 10.1371/journal.pcbi.1000208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudargiene A., Porr B., Wörgötter F. (2005). Synaptic modifications depend on synapse location and activity: a biophysical model of STDP. Biosystems 79, 3–10 10.1016/j.biosystems.2004.09.010 [DOI] [PubMed] [Google Scholar]
- Schaff J., Fink C. C., Slepchenko B., Carson J. H., Loew L. M. (1997). A general computational framework for modeling cellular structure and function. Biophys. J. 73, 1135–1146 10.1016/S0006-3495(97)78146-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiegg A., Gerstner W., Ritz R., van Hemmen J. L. (1995). Intracellular Ca2+ stores can account for the time course of LTP induction: a model of Ca2+ dynamics in dendritic spines. J. Neurophysiol. 74, 1046–1055 [DOI] [PubMed] [Google Scholar]
- Schmidt H., Eilers J. (2009). Spine neck geometry determines spino-dendritic cross-talk in the presence of mobile endogenous calcium binding proteins. J. Comput. Neurosci. 27, 229–243 10.1007/s10827-009-0139-5 [DOI] [PubMed] [Google Scholar]
- Schmidt H., Kunerth S., Wilms C., Strotmann R., Eilers J. (2007). Spino-dendritic cross-talk in rodent Purkinje neurons mediated by endogenous Ca2+-binding proteins. J. Physiol. 581, 619–629 10.1113/jphysiol.2007.127860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P., Yao Y., Sacktor T. C. (2005). Persistent phosphorylation by protein kinase Mζ maintains late-phase long-term potentiation. J. Neurosci. 25, 1979–1984 10.1523/JNEUROSCI.5132-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N. T., Yeung L. C., Cooper L. N., Cai Y., Shouval H. Z. (2006). A biophysical basis for the inter-spike interaction of spike-timing-dependent plasticity. Biol. Cybern. 95, 113–121 10.1007/s00422-006-0071-y [DOI] [PubMed] [Google Scholar]
- Shouval H. Z., Bear M. F., Cooper L. N. (2002a). A unified model of NMDA receptor-dependent bidirectional synaptic plasticity. Proc. Natl. Acad. Sci. U.S.A. 99, 10831–10836 10.1073/pnas.152343099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shouval H. Z., Castellani G. C., Blais B. S., Yeung L. C., Cooper L. N. (2002b). Converging evidence for a simplified biophysical model of synaptic plasticity. Biol. Cybern. 87, 383–391 10.1007/s00422-002-0362-x [DOI] [PubMed] [Google Scholar]
- Shouval H. Z., Kalantzis G. (2005). Stochastic properties of synaptic transmission affect the shape of spike time-dependent plasticity curves. J. Neurophysiol. 93, 1069–1073 10.1152/jn.00504.2004 [DOI] [PubMed] [Google Scholar]
- Shouval H. Z., Wang S. S. H., Wittenberg G. M. (2010). Spike timing dependent plasticity: a consequence of more fundamental learning rules. Front. Comput. Neurosci. 4: 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran S., Hariharaputran S., Mishra J., Bhalla U. S. (2003). The Database of Quantitative Cellular Signaling: management and analysis of chemical kinetic models of signaling networks. Bioinformatics 19, 408–415 10.1093/bioinformatics/btf860 [DOI] [PubMed] [Google Scholar]
- Slepchenko B. M., Schaff J. C., Macara I., Loew L. M. (2003). Quantitative cell biology with the Virtual Cell. Trends Cell Biol. 13, 570–576 10.1016/j.tcb.2003.09.002 [DOI] [PubMed] [Google Scholar]
- Smolen P. (2007). A model of late long-term potentiation simulates aspects of memory maintenance. PLoS ONE 2, e445. 10.1371/journal.pone.0000445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen P., Baxter D. A., Byrne J. H. (2006). A model of the roles of essential kinases in the induction and expression of late long-term potentiation. Biophys. J. 90, 2760–2775 10.1529/biophysj.105.072470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen P., Baxter D. A., Byrne J. H. (2008). Bistable MAP kinase activity: a plausible mechanism contributing to maintenance of late long-term potentiation. Am. J. Physiol. Cell Physiol. 294, C503–C515 10.1152/ajpcell.00447.2007 [DOI] [PubMed] [Google Scholar]
- Smolen P., Baxter D. A., Byrne J. H. (2009). Interlinked dual-time feedback loops can enhance robustness to stochasticity and persistence of memory. Phys. Rev. E 79, 031902. 10.1103/PhysRevE.79.031902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling T. R., Derkach V. A. (2000). Postsynaptic protein phosphorylation and LTP. Trends Neurosci. 23, 75–80 10.1016/S0166-2236(99)01490-3 [DOI] [PubMed] [Google Scholar]
- Stefan M. I., Edelstein S. J., Le Novère N. (2008). An allosteric model of calmodulin explains differential activation of PP2B and CaMKII. Proc. Natl. Acad. Sci. U.S.A. 105, 10768–10773 10.1073/pnas.0804672105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuber V., Willshaw D. J. (2004). A biophysical model of synaptic delay learning and temporal pattern recognition in a cerebellar Purkinje cell. J. Comput. Neurosci. 17, 149–164 10.1023/B:JCNS.0000037678.26155.b5 [DOI] [PubMed] [Google Scholar]
- Stiles J. R., Bartol T. M. (2001). “Monte Carlo methods for simulating realistic synaptic microphysiology using MCell,” in Computational Neuroscience: Realistic Modeling for Experimentalists, ed. De Schutter E. (Boca Raton: CRC Press; ), 87–127 [Google Scholar]
- Sweatt J. D. (1999). Toward a molecular explanation for long-term potentiation. Learn. Mem. 6, 399–416 10.1101/lm.6.5.399 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Augustine G. J. (2009). “Systems biology meets single-cell physiology: role of a positive-feedback signal transduction network in cerebellar long-term synaptic depression,” in Systems Biology: The Challenge of Complexity, 1st Edn, eds Nakanishi S., Kageyama R., Watanabe D. (Tokyo: Springer; ), 159–168 [Google Scholar]
- Tanaka K., Khiroug L., Santamaria F., Doi T., Ogasawara H., Ellis-Davies G. C. R., Kawato M., Augustine G. J. (2007). Ca2+ requirements for cerebellar long-term synaptic depression: role for a postsynaptic leaky integrator. Neuron 54, 787–800 10.1016/j.neuron.2007.05.014 [DOI] [PubMed] [Google Scholar]
- Tolle D. P., Le Novère N. (2010a). Brownian diffusion of AMPA receptors is sufficient to explain fast onset of LTP. BMC Syst. Biol. 4, 25. 10.1186/1752-0509-4-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolle D. P., Le Novère N. (2010b). Meredys, a multi-compartment reaction–diffusion simulator using multistate realistic molecular complexes. BMC Syst. Biol. 4, 24. 10.1186/1752-0509-4-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita M., Hashimoto K., Takahashi K., Shimizu T. S., Matsuzaki Y., Miyoshi F., Saito K., Tanida S., Yugi K., Venter J. C., Hutchison C. A., III (1999). E-CELL: software environment for whole-cell simulation. Bioinformatics 15, 72–84 10.1093/bioinformatics/15.1.72 [DOI] [PubMed] [Google Scholar]
- Traub R. D., Wong R. K., Miles R., Michelson H. (1991). A model of a CA3 hippocampal pyramidal neuron incorporating voltage-clamp data on intrinsic conductances. J. Neurophysiol. 66, 635–650 [DOI] [PubMed] [Google Scholar]
- Urakubo H., Honda M., Froemke R. C., Kuroda S. (2008). Requirement of an allosteric kinetics of NMDA receptors for spike timing-dependent plasticity. J. Neurosci. 28, 3310–3323 10.1523/JNEUROSCI.0303-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakubo H., Honda M., Tanaka K., Kuroda S. (2009). Experimental and computational aspects of signaling mechanisms of spike-timing-dependent plasticity. HFSP J. 3, 240–254 10.2976/1.3137602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volfovsky N., Parnas H., Segal M., Korkotian E. (1999). Geometry of dendritic spines affects calcium dynamics in hippocampal neurons: theory and experiments. J. Neurophysiol. 82, 450–462 [DOI] [PubMed] [Google Scholar]
- Wang H., Hu Y., Tsien J. Z. (2006). Molecular and systems mechanisms of memory consolidation and storage. Prog. Neurobiol. 79, 123–135 10.1016/j.pneurobio.2006.06.004 [DOI] [PubMed] [Google Scholar]
- Wils S., De Schutter E. (2009). STEPS: modeling and simulating complex reaction–diffusion systems with Python. Front. Neuroinform. 3: 15. 10.3389/neuro.11.015.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo N. H., Duffy S. N., Abel T., Nguyen P. V. (2003). Temporal spacing of synaptic stimulation critically modulates the dependence of LTP on cyclic AMP-dependent protein kinase. Hippocampus 13, 293–300 10.1002/hipo.10086 [DOI] [PubMed] [Google Scholar]
- Wörgötter F., Porr B. (2005). Temporal sequence learning, prediction, and control: a review of different models and their relation to biological mechanisms. Neural Comput. 17, 245–319 10.1162/0899766053011555 [DOI] [PubMed] [Google Scholar]
- Yang K.-H., Hellgren Kotaleski J., Blackwell K. T. (2001). The role of protein kinase C in the biochemical pathways of classical conditioning. Neurocomputing 38–40, 79–85. 10.1016/S0925-2312(01)00488-X [DOI] [Google Scholar]
- Yeung L. C., Shouval H. Z., Blais B. S., Cooper L. N. (2004). Synaptic homeostasis and input selectivity follow from a calcium-dependent plasticity model. Proc. Natl. Acad. Sci. U.S.A. 101, 14943–14948 10.1073/pnas.0405555101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Shouval H. Z., Knierim J. J. (2008). A biophysical model of synaptic plasticity and metaplasticity can account for the dynamics of the backward shift of hippocampal place fields. J. Neurophysiol. 100, 983–992 10.1152/jn.01256.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zador A., Koch C., Brown T. H. (1990). Biophysical model of a Hebbian synapse. Proc. Natl. Acad. Sci. U.S.A. 87, 6718–6722 10.1073/pnas.87.17.6718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhabotinsky A. M. (2000). Bistability in the Ca2+/calmodulin-dependent protein kinase-phosphatase system. Biophys. J. 79, 2211–2221 10.1016/S0006-3495(00)76469-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhabotinsky A. M., Camp R. N., Epstein I. R., Lisman J. E. (2006). Role of the neurogranin concentrated in spines in the induction of long-term potentiation. J. Neurosci. 26, 7337–7347 10.1523/JNEUROSCI.0729-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q., Destexhe A. (2007). Kinetic models of spike-timing dependent plasticity and their functional consequences in detecting correlations. Biol. Cybern. 97, 81–97 10.1007/s00422-007-0155-3 [DOI] [PubMed] [Google Scholar]