Abstract
Transferrin receptor 1 (TfR) plays a critical role in cellular iron import for most higher organisms. Cell surface TfR binds to circulating iron-loaded transferrin (Fe-Tf) and transports it to acidic endosomes, where low pH promotes iron to dissociate from transferrin (Tf) in a TfR-assisted process. The iron-free form of Tf (apo-Tf) remains bound to TfR and is recycled to the cell surface, where the complex dissociates upon exposure to the slightly basic pH of the blood. Fe-Tf competes for binding to TfR with HFE, the protein mutated in the iron-overload disease hereditary hemochromatosis. We used a quantitative surface plasmon resonance assay to determine the binding affinities of an extensive set of site-directed TfR mutants to HFE and Fe-Tf at pH 7.4 and to apo-Tf at pH 6.3. These results confirm the previous finding that Fe-Tf and HFE compete for the receptor by binding to an overlapping site on the TfR helical domain. Spatially distant mutations in the TfR protease-like domain affect binding of Fe-Tf, but not iron-loaded Tf C-lobe, apo-Tf, or HFE, and mutations at the edge of the TfR helical domain affect binding of apo-Tf, but not Fe-Tf or HFE. The binding data presented here reveal the binding footprints on TfR for Fe-Tf and apo-Tf. These data support a model in which the Tf C-lobe contacts the TfR helical domain and the Tf N-lobe contacts the base of the TfR protease-like domain. The differential effects of some TfR mutations on binding to Fe-Tf and apo-Tf suggest differences in the contact points between TfR and the two forms of Tf that could be caused by pH-dependent conformational changes in Tf, TfR, or both. From these data, we propose a structure-based model for the mechanism of TfR-assisted iron release from Fe-Tf.
Differences in the contact points between the transferrin receptor and the two forms of transferrin (with or without iron) are consistent with pH-dependent conformational changes in transferrin, its receptor, or both
Introduction
Transferrin receptor 1 (TfR) is a homodimeric type II membrane protein that plays a critical role in the primary iron acquisition mechanism for all iron-requiring cell types in vertebrates (Enns 2002). TfR binds the serum iron-carrier protein transferrin (Fe-Tf) and imports it to acidic endosomes, where iron is released and transported to the cytosol. The complex between TfR and iron-free transferrin (apo-Tf) is then recycled to the cell surface where apo-Tf dissociates and returns to circulation (reviewed in Enns et al. 1996). TfR also binds the hereditary hemochromatosis protein HFE (Parkkila et al. 1997; Feder et al. 1998). HFE is a class I major histocompatibility complex (MHC)-related protein that is mutated in patients with hereditary hemochromatosis (Feder et al. 1996), an iron-storage disease characterized by excessive iron absorption leading to an accumulation of iron principally in the liver, heart, pancreas, parathyroid, and pituitary gland, leading to tissue damage (Cullen et al. 1999).
The X-ray crystal structures of the human TfR ectodomain, both alone (Lawrence et al. 1999) and in complex with HFE (Bennett et al. 2000), have been reported. The homodimeric TfR ectodomain contains three domains on each polypeptide chain: a protease-like domain resembling amino- and carboxypeptidases (residues 121–188 and 384–606), an apical domain (residues 189–383), and a helical domain involved in TfR homodimerization (residues 607–760). Intact TfR also includes a glycosylated stalk region (residues 90–120), a transmembrane domain (residues 62–89), and an N-terminal cytoplasmic domain (residues 1–61) that includes a tyrosine-based endosomal sorting sequence (YTRF) (Enns 2002). The structure of a 2:1 HFE/TfR complex (two HFEs bound to a homodimeric TfR) shows that each HFE interacts with helices 1 and 3 of the TfR helical domain (Bennett et al. 2000) (Figure 1A and 1B). The central portion of the interface includes a hydrophobic core consisting of TfR residues Leu619, Val622, and Tyr643 packed against hydrophobic residues from the α1 domain helix of HFE.
Figure 1. TfR Structure.
(A) Ribbon diagram of TfR homodimer derived from the 3.2 Å structure of TfR (Lawrence et al. 1999). The HFE-binding site (deduced from an analysis using the HFE/TfR co-crystal structure [Bennett et al. 2000]) on the TfR helical domain closest to the viewer is highlighted in cyan.
(B) Space-filling representation of one chain from the TfR homodimer, with the HFE structural epitope residues highlighted as in (A). The location of the interdomain cleft is indicated by an orange asterisk.
(C–E) Summary of effects of TfR substitutions for binding HFE (C), Fe-Tf (D), and apo-Tf (E). Color-coding of the TfR sidechains designates the effects of the substitutions on binding affinities as indicated.
Figures were made with Molscript (Kraulis 1991) or GRASP (Nicholls et al. 1993) and rendered with Raster3D (Merritt and Bacon 1997).
The structures of various transferrins (Tfs) and related proteins such as lactoferrin have been studied extensively by X-ray crystallography (Bailey et al. 1988; Anderson et al. 1989; Gerstein et al. 1993; Zuccola 1993; Kurokawa et al. 1995, 1999; Baker et al. 1998; Karthikeyan et al. 1999). Tf and its relatives are single-chain molecules consisting of two similarly folded lobes (the N- and C-lobes), each of which contains two domains (NI and NII in the N-lobe; CI and CII in the C-lobe). Diferric Tf (Fe-Tf) contains two iron atoms, each held in a cleft between the domains of each lobe. Transition between the ferric and iron-free states of Tf involves significant conformational changes (Grossmann et al. 1992, 1993). Specifically, loss of iron results in a 54°–63° rotation between the two domains that comprise each lobe (Gerstein et al. 1993). Additionally, the interface between the lobes repacks, exposing previously buried residues and burying previously exposed residues (Kurokawa et al. 1999). In vivo, these conformational changes presumably take place while Tf is bound to TfR, as the two proteins remain complexed throughout endocytosis and recycling (Dautry-Varsat et al. 1983).
Free Fe-Tf releases iron at acidic pH, but binding to TfR affects the iron release at both basic and acidic pH (Bali and Aisen 1991, 1992; Bali et al. 1991). At pH 7.4, iron release from Fe-Tf bound to TfR is slower than from free Fe-Tf. At low pH, the opposite effect is observed, such that binding to TfR significantly increases the iron-release rate (Bali and Aisen 1991, 1992; Bali et al. 1991; Sipe and Murphy 1991). Attempts to determine the mechanism by which TfR mediates these effects on the iron release rate have been hampered by a lack of detailed knowledge of the binding footprints of Fe-Tf and apo-Tf on TfR and by the unavailability of crystal structures of Fe-Tf or apo-Tf bound to TfR.
Although the structural details of the interaction between Tf and TfR remain unknown, early studies established that two Tf molecules bind to each TfR homodimer (Enns and Sussman 1981) by primarily interacting with what is now structurally defined as the TfR helical domain (Buchegger et al. 1996). A subsequent mutagenesis study further localized the binding site to include a conserved RGD sequence (residues 646–648) within the TfR helical domain (Dubljevic et al. 1999). The HFE/TfR co-crystal structure revealed that HFE directly contacts TfR residues 646 and 648 (Bennett et al. 2000), which is consistent with biochemical inhibition studies that suggested that HFE and Tf bind to the same or an overlapping site on TfR (Lebrón et al. 1999). As Fe-Tf is a large protein (approximately 90Å × 50Å × 40 Å, measured using the structure of iron-bound ovo-Tf [Kurokawa et al. 1995]), the remainder of the Tf contact site on TfR could include other TfR domains, the TfR interdomain cleft, or both (Figure 1B), as previously suggested (Lawrence et al. 1999). A subsequent mutagenesis study sought to identify other Tf-contacting residues on TfR (West et al. 2001). In that study, residues identified from the HFE/TfR co-crystal structure as involved in contacting HFE were mutated, and their effects on binding to HFE and Fe-Tf were quantitatively evaluated. These experiments identified several residues within the TfR helical domain that are involved in binding to each protein (defined as a substitution producing a greater than or equal to 5-fold reduction in binding affinity) and confirmed that the Fe-Tf- and HFE-binding sites on TfR overlap. However, the larger size of Tf relative to the HFE ectodomain (679 amino acids in Tf compared with 374 for the HFE/β2-microglobulin ectodomain) suggested that Fe-Tf could contact residues outside of the TfR helical domain. Also, the effects of the TfR substitutions on binding to apo-Tf were not evaluated; thus, the question of whether Fe-Tf and apo-Tf bind differently to TfR was not addressed.
We therefore sought to expand the library of TfR mutants to more extensively map the Fe-Tf interface and to compare the effects of TfR mutants for binding to Fe-Tf versus apo-Tf. Here we report the affinities of 30 mutants of human TfR for binding to HFE and Fe-Tf at pH 7.5 and to apo-Tf at pH 6.3. As expected, the most important residues for Tf binding are located in the center of the TfR helical domain in the vicinity of critical residues for HFE binding. However, we also identified residues within the TfR protease-like domain that make significant contributions to binding of Fe-Tf, but not apo-Tf, to TfR. Conversely, substitution of residues at the edge of the TfR helical domain affects binding of apo-Tf, but not Fe-Tf. This information, together with the identification of common Fe-Tf- and apo-Tf-contacting residues within the helical domain, constrains the possible positions of Fe-Tf and apo-Tf on TfR, allowing for construction of structural models for the placement of the two forms of Tf on TfR. Our data also suggest a structural mechanism to explain TfR's role in the pH-dependent modulation of iron release rates from Fe-Tf.
Results
Design of TfR Mutants
Our choice of TfR residues to substitute was guided by the crystal structures of TfR alone and bound to HFE (Lawrence et al. 1999; Bennett et al. 2000) and by a previous binding study involving ten TfR point mutants (West et al. 2001) (§ symbol in Table 1). Four substitutions at the HFE-binding site on the TfR helical domain (L619A, R629A, Y643A, and F650A) were found to significantly reduce (greater than or equal to 5-fold) the binding affinity for both HFE and Fe-Tf at pH 7.5, giving a first-order map of the Fe-Tf-binding site on TfR (West et al. 2001). In order to identify additional TfR residues critical for Fe-Tf binding and to evaluate their effects on apo-Tf binding, we extended our TfR mutant library to include an additional 20 mutants. The new set of mutations were chosen using three different strategies: (1) an alanine scan involving solvent-exposed residues on helix 3 of the helical domain (R651A, S654A, T658A, N662A, E664A) (classified as H3 in Table 1); (2) substitution of residues in the TfR interdomain cleft (see Figure 1B), suggested to be part of the Fe-Tf-binding site (Lawrence et al. 1999), for the residues of chicken TfR, which does not bind human Tf (Buchegger et al. 1996) (P710R, K717Q) (classified as IDC in Table 1); (3) mutation of large solvent-exposed hydrophobic residues, which often provide much of the free energy of binding in protein–protein interactions (Jones and Thornton 1996; Tsai et al. 1997; Lo Conte et al. 1999), throughout the remaining TfR surface area (Y123S, F187A, F396A, F521A, Y523S, W528A, W702A, F760A) (classified as Hφ in Table 1). A second generation of mutants was subsequently made to further define newly identified binding sites (W124A, D125K, E606K, D610A) and to test the effect of combining substitutions (Y123S/G647A). Mutants involving TfR residues known from the HFE/TfR crystal structure (Bennett et al. 2000) to contact HFE are denoted as part of the HFE structural epitope in Table 1.
Table 1. Comparison of the Binding Affinities of TfR Mutants for HFE and Fe-Tf at pH 7.5 and Apo-Tf at pH 6.3.
For each substituted position, residues at that position in human and six other species of TfR—feline, canine, hamster, mouse, rat, and chicken—and two species of TfR2 (human and mouse) are listed in the first column
The second column (HFE Structural Epitope) identifies TfR residues that are part of the crystallographically identified HFE structural epitope. Yes indicates that the residue contains an atom within 4 Å of HFE and No that all atoms of the residue are >4 Å from HFE. The asterisk indicates a glycine residue that is covered by HFE, but which lacks a sidechain and therefore does not contain atoms within 4 Å of HFE (Bennett et al. 2000)
The third column classifies substitutions as to their location or chemical character: HΦ, solvent-exposed hydrophobic residue; H1, H2, or H3, part of the HFE structural epitope on helix 1, 2, or 3 of the TfR helical domain; IDC, part of the TfR interdomain cleft
The fourth column lists the name of the mutant. The § symbol denotes mutants that were also evaluated for HFE and Fe-Tf binding in previous experiments (West et al. 2001). Current results for these mutants agree closely with those reported previously (West et al. 2001). The KDs for wild-type TfR are averages derived from 22 independent measurements, and the numbers after the plus/minus sign represent standard deviations. Mutants with significant relative affinity reductions are highlighted (green, between 5-fold and 30-fold; red, greater than 30-fold). Mutants that showed no binding are labeled N.B. ΔΔG values were calculated from the mutant to wild-type affinity ratios and plotted for each mutant in the form of a histogram, shown in Figure S1
TfR mutants were expressed as N-terminally 6x-His-tagged soluble ectodomains in baculovirus-infected cells, as previously described (Lebrón et al. 1998; West et al. 2001). In a previous TfR mutagenesis study, it was shown that mutants that had a strong effect on binding were properly folded as determined by comparison of their far-UV circular dichroism spectra and gel filtration profiles to that of wild-type TfR (West et al. 2001). In this study, we note that all of the newly made mutants retain wild-type or near wild-type binding affinities for at least one of the three TfR ligands tested (HFE, Fe-Tf, or apo-Tf) (Table 1), confirming their structural integrity.
Affinity Measurements and Analyses
Each of the TfR mutants designed in the current screen, plus the mutants from the previous study (West et al. 2001), were tested in a surface plasmon resonance-based assay for binding to either a soluble form of HFE at pH 7.5, Fe-Tf at pH 7.5, or apo-Tf at pH 6.3 (Table 1; Figure 2). For these experiments, filtered insect cell supernatants containing secreted recombinant TfR mutants were injected over a biosensor chip to which an anti-pentaHis antibody had been immobilized. The antibody captures TfR by binding to its two 6x-His tags, thereby allowing oriented coupling of the receptors to the biosensor chip. HFE, Fe-Tf, or apo-Tf was then injected over the antibody/TfR-coupled sensor chip, and binding data were fit to a bivalent ligand model in which equilibrium dissociation constants (KD1 and KD2) were derived for binding to the first and the second binding sites on homodimeric TfR (see Table 1) (West et al. 2001).
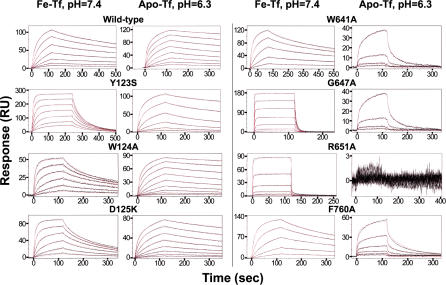
Figure 2. Biosensor Analyses of Tf Binding to Immobilized Wild-Type and Selected Mutant TfR Molecules.
Sensorgrams (black lines) of injected Fe-Tf or apo-Tf binding to wild-type TfR (top left) or the indicated TfR mutants are shown with best-fit binding curves (red lines) derived from a bivalent ligand model (see Materials and Methods) superimposed. The sensorgrams demonstrate that the binding responses are concentration dependent, and the superimposed binding curves demonstrate the close fit of the binding model to the experimental data. Concentrations of injected proteins for each sensorgram are given below as two numbers: the first is the highest injected concentration (nM), and the second is the dilution factor, either 2-fold (2×) or 3-fold (3×), that relates successive injections. For each TfR sample, there are two sets of numbers, the first being for Fe-Tf and the second for apo-Tf. Wild-type (31, 2×; 200, 2×), Y123S (250, 2×; 330, 3×), W124A (2,000, 3×; 2,000, 2×), D125K (2,000, 3×; 1,000, 2×), W641A (110, 3×; 1,000, 3×), G647A (6,000, 3×; 780, 3×), R651A (5,000, 3×; 1,000, 3×), F760A (110, 3×; 270, 3×).
Our previous mutagenesis study established that both the HFE- and Fe-Tf-binding sites on TfR include residues within helices 1 and 3 of the TfR helical domain (West et al. 2001) (see Figure 1A and 1B). In the present study, we tested the previously prepared TfR mutants for binding apo-Tf at pH 6.3 and re-evaluated their binding to HFE and Fe-Tf at pH 7.5. In agreement with the previous results, we found that two mutants, L619A and Y643A, showed no detectable HFE binding and a significant (greater than or equal to 5-fold) decrease in Fe-Tf binding. These substitutions also significantly reduced apo-Tf binding at acidic pH. Two other mutants, R629A and Q640A, were again found to significantly reduce HFE binding and to have a relatively minor effect on Fe-Tf binding (R629A) or no significant effect (Q640A). The effects of these substitutions on apo-Tf binding correlated with their effects on Fe-Tf binding. Likewise, the F650A mutant, which shows a moderate reduction in binding affinity for both HFE and Fe-Tf, also shows a reduced affinity for binding apo-Tf. Only one of the previously analyzed mutants, G647A, exhibited a major (greater than 100-fold) reduction in Fe-Tf binding affinity, and the present analysis reveals that it has a similar effect on apo-Tf binding. Interestingly, one of the previously analyzed mutants, W641A, which does not significantly affect HFE or Fe-Tf binding at pH 7.5, exerted a significant reduction in the binding affinity for apo-Tf at pH 6.3 (see Table 1; Figure 1E; Figure 2), suggesting that it might be possible to find additional substitutions with differential effects on binding of the two forms of Tf.
Our first strategy for finding additional residues critical for Tf binding involved substitution of solvent-exposed residues C-terminal to the Tf-binding epitope residues Gly647 and Phe650 on helix 3 of the TfR helical domain. Of the five new TfR mutants constructed (R651A, S654A, T658A, N662A, E664A), only one (R651A) affected Tf binding, resulting in a greater than 2,800-fold reduction in binding of Fe-Tf and apo-Tf. Having identified a “hot spot” for Tf binding involving TfR helical domain residues Gly647 and Arg651, we then searched for residues affecting Tf binding that were distant from this site, which would allow approximate positioning of the bi-lobed Tf structure on TfR. Two residues within the cleft formed by portions of the three TfR domains were changed to their chicken TfR counterparts to test the prediction that Tf binds to the TfR interdomain cleft (Lawrence et al. 1999). There were no significant differences in Tf binding affinity for either the P710R or the K717Q mutants, suggesting that at least this region of the interdomain cleft is not critical for binding to either form of Tf. Consistent with this interpretation, we found a second binding site at the base of the TfR protease-like domain that is distant from the interdomain cleft (approximately 46 Å). The Y123S mutant, which was constructed as part of a screen to test the effects of changing large solvent-exposed hydrophobic residues, shows a significantly reduced affinity for Fe-Tf, but not to apo-Tf or HFE. To confirm that Tyr123 forms part of the Fe-Tf-binding site, three additional mutants were constructed: the double mutant Y123S/G647A and the two single mutants W124A and D125K. The double mutant showed an increased effect on Fe-Tf binding compared to the G647A alone, consistent with the involvement of Tyr123 in Fe-Tf binding. In addition, the W124A and D125K single mutants, which change residues adjacent to Tyr123, also reduced TfR's affinity for Fe-Tf, but not apo-Tf. Thus, the base of the protease-like domain in the vicinity of Tyr123 is involved in differential binding to the iron-loaded form of Tf, but not apo-Tf. None of the other substitutions constructed in the screen of solvent-exposed hydrophobic residues significantly affected binding to either form of Tf or to HFE.
As Tf is a bi-lobed structure, it should be possible to evaluate the binding of isolated lobes to wild-type and mutant TfRs to gain information regarding the positions of the two Tf lobes on TfR. Isolated iron-loaded Tf C-lobe (Fe-C-lobe) binds to TfR with an affinity of approximately 650 nM (Zak et al. 1994; Zak and Aisen 2002). Assuming independent binding of the two Tf lobes without effects of cooperativity, the affinity increase to a KD of approximately 1 nM for intact Fe-Tf binding to TfR suggests the KD for binding isolated N-lobe would be approximately 1.5 mM. This affinity is too weak to be detected by most binding assays. Consistent with this assumption, isolated Tf N-lobe neither binds detectably to TfR nor donates iron to TfR-expressing cells (Zak et al. 1994; Mason et al. 1997). We therefore tested purified Fe-C-lobe (Zak and Aisen 2002) for binding to wild-type TfR and selected TfR mutants.
Fe-C-lobe was injected over wild-type TfR and TfR mutants (Y123S, D125K, R651A, F760A) in a biosensor binding assay as described for Fe-Tf above. Binding data were analyzed using an equilibrium-based approach because the rapid kinetics of the Fe-C-lobe interaction with wild-type TfR do not allow accurate derivation of kinetic rate constants (Figure 3). No significant changes in affinity were observed for Fe-C-lobe binding to two mutants in the TfR protease-like domain (Y123S, D125K), but a mutation in the central portion of the Fe-Tf functional epitope on TfR, R651A, eliminated detectable binding (Figure 3). In addition, binding of Fe-C-lobe was not significantly affected by the F760A mutation in the TfR helical domain, which reduces the affinity of apo-Tf, but not Fe-Tf (see Figure 2; Table 1). These results suggest that Tf C-lobe contacts the TfR helical domain, but not the protease-like domain.
Figure 3. Biosensor Analyses of Fe-C-Lobe Binding to Immobilized Wild-Type and Selected Mutant TfR Molecules.
Plots of the equilibrium binding response, normalized to the Rmax value (the ligand immobilization value) derived from fitting, versus concentration of injected Fe-C-lobe, are shown for the indicated TfR mutants along with the wild-type TfR control that was present in an adjacent flow cell on the same biosensor chip. Best-fit binding curves derived from a bivalent ligand model are shown as solid lines connecting the datapoints (squares for wild-type TfR and triangles for TfR mutants). The R651A mutant exhibited no binding and was not fit. A summary of derived binding constants is shown in the lower right panel. The KDs for wild-type TfR are averages derived from three independent measurements, and the number after the plus/minus sign represents the standard deviation.
Discussion
Despite many years of investigation of the Tf/TfR pathway for iron uptake, molecular details about the interaction between TfR and Tf have been limited largely due to a lack of structural information for a Tf/TfR complex. In the absence of a three-dimensional structure, site-directed mutagenesis can be used to map out a protein–protein interaction. To narrow down a subset of residues for mutageneis from the 639 residues in a soluble TfR monomer, we used the crystal structures of TfR alone (Lawrence et al. 1999) and TfR bound to HFE (Bennett et al. 2000) to locate solvent-exposed residues in the vicinity of the HFE-binding site, which was suggested from competition studies to overlap with the Tf-binding site on TfR (Lebrón et al. 1999). We identified residues within the TfR helical domain whose substitution affected binding of both HFE and Fe-Tf at pH 7.5 in a previous mutagenesis study involving ten human TfR mutants (West et al. 2001). These results established that HFE and Fe-Tf bind to the same or an overlapping site on TfR. In the present study, we have expanded the library of TfR mutants to more precisely map the Tf-binding site on TfR and compared binding of Fe-Tf and apo-Tf to TfR. From a survey of 29 point mutants of human TfR, we identified 11 residues, which, when substituted, reduce the affinity of TfR for either human Fe-Tf, apo-Tf, or both (see Table 1). Six of the 11 residues are completely conserved in different species of TfR and in a more recently identified Tf-binding receptor, TfR2, which shares 45% sequence identity with TfR (Kawabata et al. 1999). Most notably, four of the residues exerting the largest effects on Fe-Tf binding, apo-Tf binding, or both (Leu619, Trp641, Gly647, and Arg651) are completely conserved across all currently known TfR and TfR2 sequences (see Table 1). Others, such as the tyrosines at positions 123 and 643, are either conserved or conservatively substituted for phenylalanine in some TfR species. By contrast, of the 18 positions at which substitutions did not significantly affect Tf binding, 16 are not conserved, and two (Phe187 and Glu664) are conservatively substituted (see Table 1). These results suggest that our conclusions about the mode of binding between human Tf and human TfR can be generalized to include Tf/TfR complexes from other species and the interaction between TfR2 and Tf.
From a quantitative analysis of the affinities of the different TfR mutants for Fe-Tf and apo-Tf, we can classify the residues we mutated using the criteria of Wells and colleagues (Cunningham and Wells 1993), which categorize the structural and functional epitope residues in a protein–protein interaction. The functional epitope is defined as residues exerting a major effect on the binding affinity (a ΔΔG value, ≥2 kcal/mol after substitution of a single residue, corresponding to an affinity reduction of at least 30-fold at room temperature). The structural epitope on a protein is all residues at the contact interface with the binding partner, which can be deduced from a co-crystal structure (Cunningham and Wells 1993). Substitution of some, but not all, of the residues at the structural epitope of a protein–protein interface will result in affinity changes (Cunningham and Wells 1993). This is illustrated in our study by comparing the crystallographically-defined structural epitope on TfR for binding HFE (Bennett et al. 2000) (see Figure 1A and 1B; Table 1; Figure S1) with the results of mutagenic mapping of residues affecting HFE binding (see Figure 1C). In the absence of a Tf/TfR co-crystal structure, we can use our mutagenesis results to predict the functional and structural epitope residues (affinity reductions of greater than or equal to 30-fold or between 5- and 30-fold, respectively) on TfR for binding to Fe-Tf and apo-Tf. From the comparison of wild-type and mutant TfR binding affinities, Arg651 was identified as a functional epitope residue for binding both Fe-Tf at pH 7.5 and binding apo-Tf at pH 6.3, as substitution of this single residue to alanine greatly reduces binding to either form of Tf (see Table 1). In combination with the previously studied G647A mutant (Dubljevic et al. 1999; West et al. 2001), which reduces affinity for both Fe-Tf and apo-Tf by over 100-fold, these residues define a functional epitope for Fe-Tf and apo-Tf binding located in the bottom central portion of the TfR helical domain (see Figure 1D and 1E). Two other nearby residues, Leu619 and Trp641, can be considered part of the functional epitope for binding apo-Tf. The HFE/TfR crystal structure shows that these residues are at the contact interface with HFE (see Figure 1A and 1B) (Bennett et al. 2000), but with the exception of Leu619, their substitutions do not significantly affect HFE binding (see Table 1). Instead, the functional epitope for HFE binding is shifted slightly upwards on the TfR helical domain from the Fe-Tf functional epitope to include residues Leu619 and Tyr643 (see Figure 1C). Thus, although most of the functional epitope residues for binding of HFE and Tf are physically separated, they are close enough that binding of either HFE or Fe-Tf to TfR would sterically preclude binding of the other species (see Figure 1C and 1D). In addition, some substitutions in TfR significantly lower the affinity for both HFE and Tf (L619A, R629A, Y643A, and F650A) (see Table 1).
Since Tf is a larger molecule than HFE, we reasoned that Tf could also interact with residues not contained in the HFE binding footprint on TfR. We therefore tested substitutions of residues outside of the TfR helical domain for their effects on binding to Tf. To narrow down the search, we chose to substitute solvent-exposed hydrophobic residues, which are often found in protein–protein interfaces (Jones and Thornton 1996; Lo Conte et al. 1999). We also restricted the search to residues within approximately 90 Å (the longest dimension of Fe-Tf) of the Fe-Tf functional-binding epitope for substitution. Using this strategy, we identified a region at the base of the protease-like domain involving residues Tyr123, Trp124, and Asp125, where substitutions showed significant effects on binding to Fe-Tf at pH 7.5, but not to HFE at pH 7.5 or to apo-Tf at pH 6.3 (see Figure 1D and 1E; Table 1). Having defined two predicted Fe-Tf contact areas on TfR that are separated by approximately 33 Å (measured between TfR residues Arg651 and Tyr123) constrains the ways in which Tf can interact with TfR. In particular, computer modeling suggests that a single Tf lobe cannot make productive contacts with both regions of TfR (A. M. Giannetti, unpublished data); thus both lobes of Fe-Tf are likely to be involved in the interface with TfR. Previous studies of the binding of isolated Fe-N- and Fe-C-lobes of Tf suggested that the majority of the binding energy in the Tf/TfR interaction comes from the C-lobe (Zak et al. 1994; Zak and Aisen 2002). It has also been observed that mixing purified N- and C-lobes results in a significant enhancement of TfR binding over that of C-lobe alone (Mason et al. 1997; Zak and Aisen 2002). These observations are consistent with a Tf orientation on TfR in which the C-lobe contacts the Tf functional epitope on the TfR helical domain and the N-lobe contacts the Tyr123 area at the base of the TfR protease-like domain (Figure 4). In this model, allosteric effects need not be invoked to explain the increased affinity of the N-lobe/C-lobe mixture over C-lobe alone (Zak and Aisen 2002). Instead, the observed increase in affinity is predicted to arise from direct contacts between the N-lobe and TfR. To test the predicted orientation of Tf on TfR (Figure 4), we compared the affinities of isolated Fe-C-lobe (Zak and Aisen 2002) to wild-type TfR and to TfR mutants with substitutions in the helical domain (R651A, F760A) and the protease-like domain (Y123S, D125K) (see Figure 3). As predicted, substitutions in the protease-like domain do not affect binding of Fe-C-lobe, whereas a functional epitope substitution (R651A) in the TfR helical domain eliminates detectable binding of Fe-C-lobe to TfR.
Figure 4. Model for the Binding of Fe-Tf and Apo-Tf to TfR.
The figures representing each molecule are drawn to scale as an outline around the known structures of TfR (Lawrence et al. 1999), Fe-ovo-Tf (Kurokawa et al. 1995), and apo-ovo-Tf (Kurokawa et al. 1999). Membrane-bound TfR includes a stalk region that places the TfR ectodomain about 30 Å above the cell surface (Fuchs et al. 1998), which would allow the Tf molecule to extend below the plane of the TfR ectodomain. At basic pH, Fe-Tf (orange, with the iron atom positions shown as black dots) and TfR (blue) associate to make a complex containing one TfR homodimer and two Fe-Tf molecules, one bound to each polypeptide chain of the TfR homodimer. Fe-Tf makes energetically favorable contacts at basic pH to residues identified by mutagenesis in the TfR helical domain (red) and the protease-like domain (green). Acidification results in iron release and large conformational changes in the Tf structure as it becomes apo-Tf (gray). Apo-Tf does not make energetically favorable contacts with the protease-like domain, but retains binding to the helical domain-binding site (red) and makes new contacts to the helical domain (yellow), thereby stabilizing the complex. Upon return to basic pH, the apo-Tf molecules dissociate from TfR. This is also illustrated in Video S1.
Our binding data also allow us to assess potential differences in the binding of Fe-Tf versus apo-Tf to TfR. Two prior observations are consistent with differences in the binding footprints of Fe-Tf and apo-Tf on TfR. First, Fe-Tf undergoes a large conformational change upon acidification and release of iron, as deduced by comparison of crystal structures of ferric and iron-free forms of Tf and Tf-related molecules such as the lactoferrins (Bailey et al. 1988; Anderson et al. 1989; Gerstein et al. 1993; Zuccola 1993; Kurokawa et al. 1995, 1999; Baker et al. 1998; Karthikeyan et al. 1999) (see Figure 4). Second, TfR has been suggested to undergo a pH-dependent conformational change resulting in aggregation at pH <6 in the absence of Tf (Turkewitz et al. 1988). Our finding of differential effects of TfR substitutions for binding Fe-Tf at pH 7.5 versus apo-Tf at pH 6.3 is consistent with conformational changes in Tf,TfR, or both at acidic pH. We find one TfR region that affects binding of Fe-Tf, but not apo-Tf (the region near Tyr123 involving TfR residues 123–125 at the base of the protease-like domain), and another region that affects binding of apo-Tf, but not Fe-Tf or Fe-C-lobe (the region defined by Trp641 and Phe760, two spatially proximal residues [10.2 Å apart] at the edge of the TfR helical domain) (see Figure 1E and 1F). The apo-Tf-specific binding site may be important for TfR's ability to significantly accelerate iron release from receptor-bound Fe-Tf (Bali and Aisen 1991). Taking all of our data into account, we propose the following structure-based mechanism to explain TfR-assisted iron release from Fe-Tf (see Figure 4; Video S1).
First, Fe-Tf binds to TfR at pH 7.5, with the C-lobe making critical contacts to the TfR region defined by Arg651 in the helical domain and the N-lobe making additional favorable contacts with the second Tf-binding site defined by Tyr123, Trp124, and Asp125. The interaction of the complementary surfaces on each protein presumably limits Fe-Tf's freedom to sample more open states, thereby favoring the closed iron-bound state and lowering the iron release rate from both Tf lobes. This is consistent with the experimental evidence (Navati et al. 2003) suggesting that iron release is reduced by an order of magnitude under conditions favoring a closed Tf conformation relative to conditions favoring an open conformation.
Second, as the pH is lowered, protonation of key residues in Fe-Tf allows it to sample open conformations that facilitate iron release (Navati et al. 2003). A conformational change in Tf, TfR, or both allows additional Tf interactions between Tf and the hydrophobic binding surface defined by TfR residues Trp641 and Phe760. These new interactions stabilize Tf in an open conformation exposing the iron-binding site and thereby enhancing the rate of iron release.
The extensive mutagenic mapping of ligand binding to TfR reported here has revealed residues responsible for functional binding to HFE, Fe-Tf, and apo-Tf. These data confirm that HFE and Tf bind to a physically and functionally overlapping site on the TfR helical domain, although the most important receptor residues for binding are different for the two proteins. Thus, HFE and Fe-Tf must compete with each other for binding to cell surface TfR, which may have functional significance in HFE's role in maintaining iron homeostasis (Townsend and Drakesmith 2002). Additionally, we have found that Fe-Tf makes specific contacts not only to the TfR helical domain through its C-lobe, but also to the TfR protease-like domain, which implies that there are specific N-lobe/TfR contacts contributing to Tf binding. Finally, our demonstration that Fe-Tf and apo-Tf have different binding footprints on the surface of TfR provides insight into the mechanism by which TfR binding differentially affects iron release rates from Fe-Tf at acidic and basic pH.
Materials and Methods
Preparation of TfR ligands
A soluble form of human HFE (residues 1–275 of the mature protein noncovalently associated with the light chain β2-microglobulin) was expressed and purified as previously described (Lebrón et al. 1998). Human Fe-Tf was prepared from apo-Tf (Sigma, St. Louis, Missouri, United States) by incubation with bicarbonate and excess ferric ammonium sulfate. Free iron was removed by dialysis, and the protein was further purified by gel-filtration chromatography. Iron saturation was 100% as determined spectrophotometrically (A465/A280, ∼0.05) (He and Mason 2002). Purified recombinant Fe-C-lobe was cleaved from a full-length Fe-Tf in which the loop that connects the N- and C-lobes was replaced with a Factor Xa site (Zak and Aisen 2002). Concanavalin A chromatography was used to separate the glycosylated C-lobe from unglycosylated N-lobe (Zak and Aisen 2002). Protein concentrations were determined from the A280- value using extinction coefficients of 52,200 M−1 cm−1 (Fe-C-lobe) (O. Zak, personal communication), 83,360 M−1 cm−1 (Tf), and 96,570 M−1 cm−1 (HFE/β2-microglobulin) (Lebrón et al. 1998).
Production of wild-type TfR and TfR mutants
Soluble human TfR and TfR mutants were expressed in a lytic baculovirus/insect cell expression system as previously described (Lebrón et al. 1998). Mutations were introduced through PCR mutagenesis (Quickchange, Strategene, La Jolla, California, United States) into a baculovirus expression vector (pACGP67A; Pharmingen, San Diego, California, United States) containing a hydrophobic leader sequence, 6x-His tag, Factor Xa site, and residues 121–760 of human TfR. All mutations were confirmed by DNA sequencing of the protein-coding region of the vector. The Y123S mutation was further confirmed by N-terminal sequencing of the purified mutant protein, yielding the sequence ADPHHHHHHSSGIEGRGEFRLSWDD (the serine substitution for tyrosine is underlined), corresponding to residual leader sequence residues (A), vector-encoded sequence (DP), the 6x-His tag, spacer residues (SSG), a Factor Xa site (IEGR), a spacer segment (GEF), and residues 121–126 of the mutant TfR (RLSWDD). The double mutant Y123S/G647A was constructed by introducing the Y123S substitution into the G647A–TfR expression construct, after which the protein-coding region of the expression plasmid was again sequenced. Recombinant viruses were generated by cotransfection of a transfer vector with linearized viral DNA (Baculogold, Pharmingen). Supernatants of baculovirus-infected High 5 cells were used as the source of wild-type TfR and TfR mutants for surface plasmon resonance-based affinity measurements.
Affinity measurements
We used a BIACORE 2000 biosensor system (Pharmacia, LKB Biotechnology, Uppsalla, Sweden) to assay the interaction between TfR and HFE, Fe-Tf, and apo-Tf as described (West et al. 2001). Binding of injected proteins (the analytes were HFE, Fe-Tf, or apo-Tf) to a protein immobilized on the sensor chip (the ligand was TfR) results in changes in surface plasmon resonance that are read out in real time as resonance units (RUs) (Fägerstam et al. 1992; Malmqvist 1993).
For each experiment, the four flow cells of a CM5 biosensor chip (Pharmacia) were prepared by covalently attaching an anti-His-tag antibody (anti-PentaHis; Qiagen, Valencia, California, United States) to a coupling density of 2,000–4,000 RUs through standard amine coupling chemistry (BIACORE manual). Insect cell supernatants (50–300 μl) containing secreted 6x-His-tagged wild-type or mutant TfR were passed through a 0.2 μm filter and injected over one of the four flow cells of a biosensor chip at a flow rate of 30 μl/min, resulting in stable binding of TfR to density of 200–400 RUs. In a typical experiment, a small amount of TfR immediately dissociates from the anti-His antibody, but most TfR protein (>85%) remains bound during the course of the injection of the TfR ligands, resulting in a negligible baseline drift. On each biosensor chip, one flow cell containing only the immobilized antibody was used as the reference cell, one cell containing wild-type TfR served as an internal control for binding of the three TfR binding partners, and TfR mutants were coupled to the other two flow cells. HFE or Fe-Tf was injected over the flow cells at 50 μl/min or 70 μl /min, respectively, at 25° C in 50 mM PIPES (pH 7.5), 150 mM NaCl, and 0.005% surfactant P20 (v/v). All analyte injections were made as serial 2- or 3-fold dilutions. The HFE concentration series ranged from 30 nM to 10 μM, and the Fe-Tf and apo-Tf injections typically spanned from 1 nM to 200 nM, except for experiments involving low-affinity mutants requiring higher concentrations to properly derive affinities (see legend to Figure 2). In test experiments, the sensorgrams from duplicate injections could be overlaid to within the experimental noise; thus, single injections were done for each concentration of injected protein in a binding experiment. Between successive injections of analytes, the chips were regenerated to preinjection response levels by either flowing with running buffer until baseline was achieved (in the case of HFE) or by a 12-second injection of the injection buffer containing 0.5 M MgCl2 (in the case of Fe-Tf). This treatment did not cause dissociation of TfR from the anti-His-tag antibody. Apo-Tf was injected in 50 mM PIPES (pH 6.3), 150 mM NaCl, 0.005% surfactant P20 (v/v), including 50 μM desferrioxamine as an iron chelator, and chip regeneration was achieved with an injection of the same buffer at pH 7.5.
Raw sensorgram data were preprocessed using the Scrubber software package (BioLogic Software, Campbell, Australia; http://www.biologic.com.au). The response from the reference flow cell was subtracted from the experimental flow cells to eliminate bulk refractive index changes. The response from the average of at least three buffer-only injections was then subtracted to correct for potential systematic instrument artifacts. Kinetic constants were obtained by simultaneous fitting of the association and dissociation phases of all curves in the working set using the program Clamp99 (Morton and Myszka 1998). The data were fit to a bivalent ligand model, which describes the two sequential binding events for either Tf or HFE binding to homodimeric TfR. A simple 1:1 binding model did not account for the observed data as judged from large residuals in the fits (data not shown). Equilibrium dissociation constants (KDs) were calculated from the ratio of the dissociation and association rate constants, koff (s−1) and kon (M−1·s−1), respectively, yielding KDs for the first and second binding events (KD1 and KD2) in the following reaction mechanism:
 |
where A is either HFE, Fe-Tf, or apo-Tf. For independent binding sites, the apparent stepwise equilibrium dissociation constants (KD1 and KD2) are related to the intrinsic binding constants for the first and second binding events to TfR (KD,intrinsic and KD,intrinsic), as follows:
Hence, if the binding of a TfR ligand is independent of whether a ligand is bound on the other face of the TfR homodimer, KD2 = 4KD1.
For each mutant, the relative effect on HFE, Fe-Tf, or apo-Tf binding was calculated as a ratio between the mutant KD and the average of 22 independent determinations of the wild-type KD (see Table 1) and as a ratio between the mutant KD and the wild-type KD derived from wild-type protein coupled to a flow cell on the same sensor chip as the mutant (data not shown). No significant differences were found for the two methods of calculating the ratios. All mutants were evaluated for HFE and Fe-Tf binding in at least two independent experiments. For apo-Tf binding, those mutants that showed a significant difference in binding compared to wild-type TfR were reevaluated in a separate, independent experiment. No significant differences in KDs were observed in independent determinations of mutant affinities. When accurate affinities could not be derived in a duplicate experiment due to problems with baseline drift, visual inspection of the sensorgrams demonstrated that each mutant exerted the same relative effects compared with wild-type TfR in independent binding experiments. Table 1 presents affinities derived from one binding experiment per mutant/ligand pair. The reproducibility of the binding experiments can be assessed by the standard deviation of the wild-type TfR affinity for each of the ligands (derived from 22 independent binding experiments) and from the fact that the affinities of many of the mutants are not significantly changed compared to wild-type TfR.
For binding interactions involving the Fe-C-lobe, which reach equilibrium quickly, we derived KDs using an equilibrium-based approach. In these experiments, KDs were derived by non-linear regression analysis of plots of Req (the equilibrium binding response) versus the log of the analyte concentration. The data were fit to a binding model assuming a bivalent ligand in BIAevaluation 3.0 (BIACORE). We were unable to detect significant amounts of binding between apo-C-lobe and wild-type TfR at pH 6.3, presumably due to an intrinsically weak binding affinity.
Supporting Information
Histogram of ΔΔG values for the change relative to wild-type TfR in TfR mutant affinities for HFE (blue), Fe-Tf (pink), and apo-Tf (gray). ΔΔG values (the difference in binding energy for a mutant TfR compared to wild-type TfR) were calculated using the KD1 values from Table 1 as ΔΔG = −RTln(KD1,mut/KD1,wild-type), where R is the gas constant (1.99 × 10−3 kcal mol−1 K−1), and T is the temperature in degrees Kelvin (298 K). The dashed green line represents the cutoff for TfR mutants with a greater than or equal to 5-fold affinity reduction in ligand binding, and the dashed red line indicates a greater than or equal to 30-fold affinity reduction. An orange star indicates non-binding mutants and mutants with a greater than 160-fold affinity reduction whose ΔΔG values exceed the y-axis limit of the histogram (L619A and Y643A, ≥4 kcal/mol; G647A = 3.2 kcal/mol; and R651A, ≥4.6 kcal/mol).
(1.86 MB TIFF).
(12 MB MOV).
Accession Numbers
The SwissProt accessions numbers for the proteins discussed in this paper are β2-microglobulin (P01884), Fe-Tf (P02787), HFE (Q30201), TfR canine (Q9GLD3), TfR chicken (Q90997), TfR feline (Q9MYZ3), TfR hamster (Q07891), TfR human (P02786), TfR mouse (Q62351), TfR rat (Q99376), TfR2 human (Q9UP52), and TfR2 mouse (Q62351).
Acknowledgments
This work was supported by grants from the National Institutes of Health (1-R01-DK60770 to PJB and DK-15056 to Dr. Philip Aisen) and an National Research Service Award predoctoral training grant (5T32-GM-7616 to AMG). We are grateful to Inderjit Nangiana and Cynthia Jones (Caltech Protein Expression Facility) for assistance in expressing TfR proteins and to the Caltech Protein/Peptide Micro-Analytical Laboratory for protein sequencing. We thank Drs. Anthony West, Andy Herr, Caroline Enns, and Anne B. Mason for helpful discussions and members of the Björkman lab for critical reading of the manuscript. We give a special thanks to D. G. Myszka for beta versions of Clamp and Scrubber and discussions of BIACORE experimental details.
Abbreviations
- apo-Tf
iron-free transferrin
- Fe-C-lobe
iron-loaded transferrin C-lobe
- Fe-Tf
diferric transferrin
- HFE
hereditary hemochromatosis protein
- KD
equilibrium dissociation constant
- MHC
major histocompatibility complex
- PIPES
piperazine-1,4-bis(2-ethanesulphonic) acid
- RU
resonance unit
- Tf
transferrin
- TfR
transferrin receptor
Conflicts of interest. The authors have declared that no conflicts of interest exist.
Author contributions. AMG and PJB conceived and designed the experiments. AMG performed the experiments. AMG and PJB analyzed the data. AMG, PMS, and OZ contributed reagents/materials/analysis tools. AMG and PJB wrote the paper. AMG developed and produced the animated figure.
Academic Editor: Janet Thornton, European Bioinformatics Institute
References
- Anderson BF, Baker HM, Norries GE, Rice DW, Baker EN. Structure of human lactoferrin: Crystallographic structure analysis and refinement at 2.8 Å resolution. J Mol Biol. 1989;209:711–734. doi: 10.1016/0022-2836(89)90602-5. [DOI] [PubMed] [Google Scholar]
- Bailey S, Evans RW, Garratt RC, Gorinsky B, Hasnain S, et al. Molecular structure of serum transferrin at 3.3 Å resolution. Biochemistry. 1988;27:5804–5812. doi: 10.1021/bi00415a061. [DOI] [PubMed] [Google Scholar]
- Baker EN, Anderson BF, Baker HM, MacGillivray RTA, Moore SA, et al. Three-dimensional structure of lactoferrin: Implications for function, including comparisons with transferrin. Adv Exp Med Biol. 1998;443:1–14. [PubMed] [Google Scholar]
- Bali PK, Aisen P. Receptor-modulated iron release from transferrin: Differential effects on N- and C-terminal sites. Biochemistry. 1991;30:9947–9952. doi: 10.1021/bi00105a019. [DOI] [PubMed] [Google Scholar]
- Bali PK, Aisen P. Receptor-induced switch in site-site cooperativity during iron release by transferrin. Biochemistry. 1992;31:3963–3967. doi: 10.1021/bi00131a011. [DOI] [PubMed] [Google Scholar]
- Bali PK, Zak O, Aisen P. A new role for the transferrin receptor in the release of iron from transferrin. Biochemistry. 1991;30:324–328. doi: 10.1021/bi00216a003. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Lebrón JA, Bjorkman PJ. Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature. 2000;403:46–53. doi: 10.1038/47417. [DOI] [PubMed] [Google Scholar]
- Buchegger F, Trowbridge IS, Liu L-FS, White S, Collawn JF. Functional analysis of human/chicken transferrin receptor chimeras indicates that the carboxy-terminal region is important for ligand binding. Eur J Biochem. 1996;235:9–17. doi: 10.1111/j.1432-1033.1996.0009u.x. [DOI] [PubMed] [Google Scholar]
- Cullen LM, Anderson GJ, Ramm GA, Jazwinska EC, Powell LW. Genetics of hemochromatosis. Annu Rev Med. 1999;50:87–98. doi: 10.1146/annurev.med.50.1.87. [DOI] [PubMed] [Google Scholar]
- Cunningham BC, Wells JA. Comparison of a structural and a functional epitope. J Mol Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]
- Dautry-Varsat A, Ciechanover A, Lodish HF. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc Natl Acad Sci U S A. 1983;80:2258–2262. doi: 10.1073/pnas.80.8.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubljevic V, Sali A, Goding A. A conserved RGD (Arg–Gly–Asp) motif in the transferrin receptor is required for binding to transferrin. Biochem J. 1999;341:11–14. [PMC free article] [PubMed] [Google Scholar]
- Enns CA. The transferrin receptor. In: Templeton DM, editor. Molecular and cellular iron transport. New York: Marcel Dekker; 2002. pp. 71–94. [Google Scholar]
- Enns CA, Sussman HH. Physical characterization of the transferrin receptor in human placentae. J Biol Chem. 1981;256:9820–9823. [PubMed] [Google Scholar]
- Enns CA, Rutledge EA, Williams AM. Biomembranes. New York: JAI Press; 1996. The transferrin receptor; pp. 255–287. [Google Scholar]
- Fägerstam LG, Frostell-Karlsson A, Karlsson R, Persson B, Rönnber I. Biospecific interaction analysis using surface plasmon resonance detection applied to kinetic, binding site and concentration analysis. J Chromatogr. 1992;597:397–410. doi: 10.1016/0021-9673(92)80137-j. [DOI] [PubMed] [Google Scholar]
- Feder JN, Gnirke A, Thomas W, Zsuchihashi Z, Ruddy DA, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- Feder JN, Penny DM, Irrinki A, Lee VK, Lebrón JA, et al. The hemochromatosis gene product complexes with the transferrin receptor, and lowers its affinity for ligand binding. Proc Natl Acad Sci U S A. 1998;95:1472–1477. doi: 10.1073/pnas.95.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs H, Lüchken U, Tauber R, Engel A, Gessner R. Structural model of phospholipid-reconstituted human transferrin receptor derived by electron microscopy. Structure. 1998;6:1235–1243. doi: 10.1016/s0969-2126(98)00124-5. [DOI] [PubMed] [Google Scholar]
- Gerstein M, Anderson BF, Norris GE, Baker EN, Lesk AM, et al. Domain closure in lactoferrin: Two hinges produce a see-saw motion between alternative close-packed interfaces. J Mol Biol. 1993;234:357–372. doi: 10.1006/jmbi.1993.1592. [DOI] [PubMed] [Google Scholar]
- Grossmann JG, Neu M, Pantos E, Schwab FJ, Evans RW, et al. X-ray solution scattering reveals conformational changes upon iron uptake in lactoferrin, serum and ovo-transferrins. J Mol Biol. 1992;225:811–819. doi: 10.1016/0022-2836(92)90402-6. [DOI] [PubMed] [Google Scholar]
- Grossmann JG, Neu M, Evans RW, Lindley PF, Appel H, et al. Metal-induced conformational changes in transferrins. J Mol Biol. 1993;229:585–590. doi: 10.1006/jmbi.1993.1063. [DOI] [PubMed] [Google Scholar]
- He Q-Y, Mason AB. Molecular aspects of release of iron from transferrin. In: Templeton DM, editor. Molecular and cellular iron transport. New York: Marcel Dekker; 2002. pp. 95–124. [Google Scholar]
- Jones S, Thornton JM. Principles of protein–protein interaction. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan S, Paramasivam M, Yadav S, Srinivasan A, Singh TP. Structure of buffalo lactoferrin at 2.5 Å resolution using crystals grown at 303 K shows different orientations of the N and C lobes. Acta Crystallogr D Biol Crystallogr. 1999;55:1805–1813. doi: 10.1107/s0907444999010951. [DOI] [PubMed] [Google Scholar]
- Kawabata H, Yang R, Hirama T, Vuong PT, Kawno S, et al. Molecular cloning of transferrin receptor 2. J Biol Chem. 1999;274:20826–20832. doi: 10.1074/jbc.274.30.20826. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. MolScript: A program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Kurokawa H, Mikami B, Hirose M. Crystal structure of diferric hen ovotransferrin at 2.4 Å resolution. J Mol Biol. 1995;254:196–207. doi: 10.1006/jmbi.1995.0611. [DOI] [PubMed] [Google Scholar]
- Kurokawa H, Dewan JC, Mikami B, Sacchettini JC, Hirose M. Crystal structure of hen apo-ovotransferrin: Both lobes adopt an open conformation upon loss of iron. J Biol Chem. 1999;274:28445–28452. doi: 10.1074/jbc.274.40.28445. [DOI] [PubMed] [Google Scholar]
- Lawrence CM, Ray S, Babyonyshev M, Galluser R, Borhani DW, et al. Structure of the ectodomain of human transferrin receptor. Science. 1999;286:779–782. doi: 10.1126/science.286.5440.779. [DOI] [PubMed] [Google Scholar]
- Lebrón JA, Bennett MJ, Vaughn DE, Chirino AJ, Snow PM, et al. Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell. 1998;93:111–123. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- Lebrón JA, West AP, Bjorkman PJ. The hemochromatosis protein HFE competes with transferrin for binding to the transferrin receptor. J Mol Biol. 1999;294:239–245. doi: 10.1006/jmbi.1999.3252. [DOI] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, Janin J. The atomic structure of protein–protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- Malmqvist M. Biospecific interaction analysis using biosensor technology. Nature. 1993;361:186–187. doi: 10.1038/361186a0. [DOI] [PubMed] [Google Scholar]
- Mason AB, Tam BM, Woodworth RC, Oliver RW, Green BN, et al. Receptor recognition sites reside in both lobes of human serum transferrin. Biochem J. 1997;326:77–85. doi: 10.1042/bj3260077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Morton TA, Myszka DG. Kinetic analysis of macromolecular interactions using surface plasmon resonance. Methods Enzymol. 1998;295:268–294. doi: 10.1016/s0076-6879(98)95044-3. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Bharadwaj R, Honig B. GRASP: Graphical representation and analysis of surface properties. Biophys J. 1993;64:A166–A166. [Google Scholar]
- Parkkila S, Waheed A, Britton RS, Bacon BR, Zhou XY, et al. Association of the transferrin receptor in human placenta with HFE, the protein defective in hereditary hemochromatosis. Proc Natl Acad Sci U S A. 1997;94:13198–13202. doi: 10.1073/pnas.94.24.13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipe DM, Murphy RF. Binding to cellular receptor results in increased iron release from transferrin at mildly acidic pH. J Biol Chem. 1991;266:8002–8007. [PubMed] [Google Scholar]
- Townsend A, Drakesmith H. Role of HFE in iron metabolism, hereditary haemochromatosis, anaemia of chronic disease, and secondary iron overload. Lancet. 2002;359:786–790. doi: 10.1016/S0140-6736(02)07885-6. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Lin SL, Wolfson HJ, Nussinov R. Studies of protein–protein interfaces: A statistical analysis of the hydrophobic effect. Protein Sci. 1997;6:53–64. doi: 10.1002/pro.5560060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkewitz AP, Schwartz AL, Harrison SC. A pH-dependent reversible conformational transition of the human transferrin receptor leads to self-association. J Biol Chem. 1988;263:16309–16315. [PubMed] [Google Scholar]
- West AP, Giannetti AM, Herr AB, Bennett MJ, Nangiana JS, et al. Mutational analysis of the transferrin receptor reveals overlapping HFE and transferrin binding sites. J Mol Biol. 2001;313:385–397. doi: 10.1006/jmbi.2001.5048. [DOI] [PubMed] [Google Scholar]
- Zak O, Aisen P. A new method for obtaining human transferrin C-lobe in the native conformation: Preparation and properties. Biochemistry. 2002;41:1647–1653. doi: 10.1021/bi015868w. [DOI] [PubMed] [Google Scholar]
- Zak O, Trinder D, Aisen P. Primary receptor-recognition site of human transferrin is in the C-terminal lobe. J Biol Chem. 1994;269:7110–7114. [PubMed] [Google Scholar]
- Zuccola HJ. Atlanta, Georgia: Georgia Institute of Technology; 1993. The crystal structure of monoferric human serum transferrin. [dissertation] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Histogram of ΔΔG values for the change relative to wild-type TfR in TfR mutant affinities for HFE (blue), Fe-Tf (pink), and apo-Tf (gray). ΔΔG values (the difference in binding energy for a mutant TfR compared to wild-type TfR) were calculated using the KD1 values from Table 1 as ΔΔG = −RTln(KD1,mut/KD1,wild-type), where R is the gas constant (1.99 × 10−3 kcal mol−1 K−1), and T is the temperature in degrees Kelvin (298 K). The dashed green line represents the cutoff for TfR mutants with a greater than or equal to 5-fold affinity reduction in ligand binding, and the dashed red line indicates a greater than or equal to 30-fold affinity reduction. An orange star indicates non-binding mutants and mutants with a greater than 160-fold affinity reduction whose ΔΔG values exceed the y-axis limit of the histogram (L619A and Y643A, ≥4 kcal/mol; G647A = 3.2 kcal/mol; and R651A, ≥4.6 kcal/mol).
(1.86 MB TIFF).
(12 MB MOV).