Abstract
To define the functions of genes previously identified by expression profiling as being rapidly light induced under phytochrome (phy) control, we are investigating the seedling de-etiolation phenotypes of mutants carrying T-DNA insertional disruptions at these loci. Mutants at one such locus displayed reduced responsiveness to continuous red, but not continuous far-red light, suggesting a role in phyB signaling but not phyA signaling. Consistent with such a role, expression of this gene is induced by continuous red light in wild-type seedlings, but the level of induction is strongly reduced in phyB-null mutants. The locus encodes a novel protein that we show localizes to the nucleus, thus suggesting a function in light-regulated gene expression. Recently, this locus was identified as EARLY FLOWERING 4, a gene implicated in floral induction and regulating the expression of the gene CIRCADIAN CLOCK-ASSOCIATED 1. Together with these previous data, our findings suggest that EARLY FLOWERING 4 functions as a signaling intermediate in phy-regulated gene expression involved in promotion of seedling de-etiolation, circadian clock function, and photoperiod perception.
Arabidopsis has developed elaborate photosensory systems to optimize its growth and development to daily and seasonal changes in the light environment. The five phytochromes (phyA–E), the two cryptochromes (cry1 and cry2), and the two phototropins (phot1 and phot2) confer sensitivity to light quality, quantity, duration, and direction (for review, see Quail, 2002). Seedlings deficient in phyA-mediated-signaling pathways are tall in continuous far-red light (FRc), and ones deficient in phyB pathways are tall in continuous red light (Rc). In addition to these influences on cell elongation and shoot morphogenesis, light also controls the transition to flowering. phy and cry have been implicated in the entrainment of the Arabidopsis circadian clock (Somers et al., 1998; also see Fankhauser and Staiger, 2002), triggering flowering under the appropriate photoperiods. For example, Arabidopsis flowers earlier under long-day than short-day conditions. This relationship between light input and clock output is important for synchronization to the environmental changes.
Microarray-based analysis has revealed that phyA regulates the expression of a master set of transcription factor genes early on during the de-etiolation process (Tepperman et al., 2001). Several of these early-response transcription regulators have G-box (CACGTG) sequences in their promoters (Hudson and Quail, 2003). The phy-interacting factor 3 (PIF3), a basic helix-loop-helix type transcription factor is capable of binding the G-box sequences in these promoters. Furthermore, light induces nuclear translocation of the biologically active (Pfr) form of phyB, which can bind to the DNA-bound PIF3 (Ni et al., 1998, 1999; Martinéz-García et al., 2000), suggesting regulation of the expression of a set of specific target genes (see Quail, 2000). Among others, the group of early light-induced genes, with G-box sequences in their promoters, includes CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), CONSTANS (CO), and TIMING OF CHLOROPHYLL A/B BINDING PROTEIN 1 (TOC1)-LIKE genes (Tepperman et al., 2001). CCA1 and LHY are two closely related MYB-like transcription factors that when overexpressed, cause photoperiod-insensitive early flowering and circadian arhythmicity (Schaffer et al., 1998; Wang and Tobin, 1998; Green and Tobin, 1999). CCA1, LHY, and TOC1, which is a pseudoresponse regulator, function in an autoregulatory feedback loop and are integral components of the central oscillator in Arabidopsis (Harmer et al., 2001; Devlin, 2002). The evening element (EE; AAATATCT) is a known binding site for the CCA1 and LHY proteins in the TOC1 promoter, repressing its expression (Harmer et al., 2000; Mizoguchi et al., 2002). The transcriptional feedback loop is completed by TOC1 activating the expression of CCA1 and LHY late in the day (Alabadi et al., 2001). Mutations in the TOC1 gene result in long hypocotyl phenotypes in Rc and FRc (Mas et al., 2003), indicating reduced sensitivity to Rc and FRc.
Several other mutations that affect light-dependent circadian processes in Arabidopsis also exhibit altered light sensitivities in various developmental processes. EARLY FLOWERING 3 (ELF3) function is required for the light sensitivity of the central oscillator in the circadian cycle (McWatters et al., 2000). This gene encodes a nuclear-localized putative transcriptional regulator that interacts with phyB (Hicks et al., 2001; Liu et al., 2001), and loss-of-function elf3 mutant plants have a long hypocotyl phenotype (Zagotta et al., 1996). PSEUDO RESPONSE REGULATOR 7, related to TOC1, functions as a signaling intermediate in phy-regulated phasing of the circadian clock, as well as in seedling de-etiolation (Kaczorowski and Quail, 2003). ZEITLUPE encodes a clock-associated F-box containing protein, and the zeitlupe seedlings are hypersensitive to Rc (Somers et al., 2000). GIGANTEA (GI) and CO promote flowering in long-day conditions without any effect in short-day conditions (see Simpson et al., 1999). GI is a novel protein (Fowler et al., 1999) that is localized to the nucleus and is involved in seedling Rc sensitivity (Huq et al., 2000). CO encodes a putative zinc finger transcription factor (Putterill et al., 1995) that is induced early in a phyA-dependent manner (Tepperman et al., 2001). It promotes flowering by activating the expression of FLOWERING LOCUS T and SUPPRESSOR OF OVER EXPRESSION OF CO1 (Kabayashi et al., 1999; Kardailsky et al., 1999; Samach et al., 2000). Hence, CO expression is light sensitive, and it underlies the photoperiodic control of flowering (Suarez-Lopez et al., 2001).
To examine the global patterns of gene expression early on during the de-etiolation process and to identify new genes involved in the early phy-signaling pathways, we performed oligonucleotide microarray analysis of the wild-type and phyA-null mutant Arabidopsis seedlings grown in FRc (Tepperman et al., 2001). To determine the functional relevance of these rapidly regulated genes to phy signaling, we have initiated a systematic analysis using reverse-genetic approaches to disrupt their activities. One of the genes identified as 5293 (protein accession number AAB95293) belonging to the hypothetical and unknown group (see Tepperman et al., 2001) was selected for further analysis of its potential role in early phyA-regulated transcriptional events. We identified insertional mutants at this locus and evaluated their photomorphogenic phenotypes. Toward the completion of this study, a report appeared identifying this locus as EARLY FLOWERING 4 (ELF4), involved in photoperiod perception and circadian regulation (Doyle et al., 2002). Mutations in elf4 were shown to affect the expression patterns of CCA1, COLD-CIRCADIAN RHYTHM-RNA BINDING 2, and CHLOROPHYLL A/B BINDING PROTEIN and to cause early flowering under short-day conditions, which was attributed to the observed elevated expression of CO (Doyle et al., 2002). Here, we report that ELF4 is involved in controlling phyB-mediated seedling deetiolation and is localized to the nucleus, suggesting an intermediary signaling function between the photoreceptor and downstream light-dependent pathways, including the circadian clock.
RESULTS
ELF4 Is a phyA-Regulated Early-Induced Gene Belonging to a Small Arabidopsis Gene Family
A group of phyA-regulated early response genes of yet unknown function were identified by comparative oligonucleotide microarray analysis of expression in wild-type and phyA-null Arabidopsis 4-d-old seedlings grown in FRc for up to 24 h (Tepperman et al., 2001). One of the genes in this group, designated as 5293 by the last four digits of its protein accession number (AAB95293), was induced rapidly in FRc, exhibiting robust responsiveness within 1 h, only in the wild-type seedlings, and relatively insignificant expression levels in the phyA seedlings. This gene 5293 was recently identified in a screen for early flowering mutations by Doyle et al. (2002) and is therefore designated hereafter by its given name, ELF4.
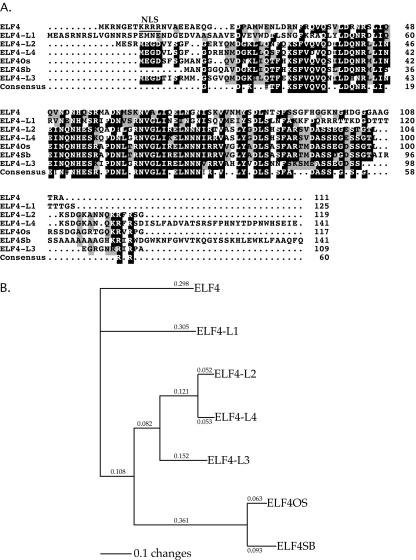
The ELF4 gene is predicted to encode a novel 111-amino acid protein with no significant homology to proteins of known function. ELF4 belongs to a small but highly conserved Arabidopsis gene family (Fig. 1A; also see Doyle et al., 2002). Comparison of the predicted amino acid sequences of the family members showed that they share a high percentage of identity with ELF4 and are designated here as ELF4-Like (ELF4-L). Our database searches here have identified a new member of this family, ELF4-L3 (At2g06255) not reported previously (see Fig. 1, A and B). The ELF4 family now consists of seven members, including one from rice (ELF4Os), one from sorghum (ELF4Sb), and five from Arabidopsis (percent identities to ELF4: ELF4-L1, 53%; ELF4-L2, 47%; and ELF4-L3 and ELF4-L4, 44%). Furthermore, 26 of the 111 amino acid residues of ELF4 (about 23%) are invariant (D22, F33, Q37, L40, D41, N43, R44, L46, I47, N51, N53, H54, S56, D60, N65, V66, L68, I69, E71, N73, N75, V79, Y83, D85, L86, and F90) within the entire ELF4 protein family (Fig. 1A). Phylogenetic tree analysis of the family members (Fig. 1B) shows that ELF4 and ELF4-L1 form a separate phylogenetic subgroup. ELF4-L2 and ELF4-L4 are very closely related to each other, and they are more closely related to the other family members than to the ELF4 subgroup (Fig. 1, A and B). There are an additional 27 amino acid residues that are invariant among ELF4-L2, ELF4-L3, ELF4-L4, ELF4OS, and ELF4SB (Fig. 1A). Hence, within these other family members, a total of 53 amino acid residues are invariant. Our analysis of the ELF4 gene structure revealed three EEs in its promoter within 600 nucleotides upstream of the ATG start site (Fig. 2A), consistent with circadian regulation of its expression (Doyle et al., 2002).
Figure 1.
Comparison of predicted amino acid sequences of the ELF4 family. A, Amino acid sequence alignments of ELF4 protein family members, including ELF4 (At2g40080), ELF4-L1 (At2g29950), ELF4-L2 (At1g72630), ELF4-L3 (At2g06255), ELF4-L4 (At1g17455) from Arabidopsis, ELF4Os (AAD27669) from rice (Oryza sativa) and ELF4Sb (AAD27564) from sorghum (Sorghum bicolor). Reverse font, Identical residues; gray boxes, similar residues. Numbers at the right indicate amino acid residues. All sequences shown are full length, except for ELF4Sb (1–141 of the 438 amino acids shown). The alignments were performed using MultiAlign (Corpet, 1988). The putative nuclear localization signal (NLS) in ELF4 is underlined. B, Phylogenetic neighbor-joining tree of the aligned sequences. The unrooted tree was constructed using PAUP 4.0 software, showing the putative evolutionary relationships of the ELF4 family members. The branch lengths are proportional to the indicated distance values (changes) between sequences.
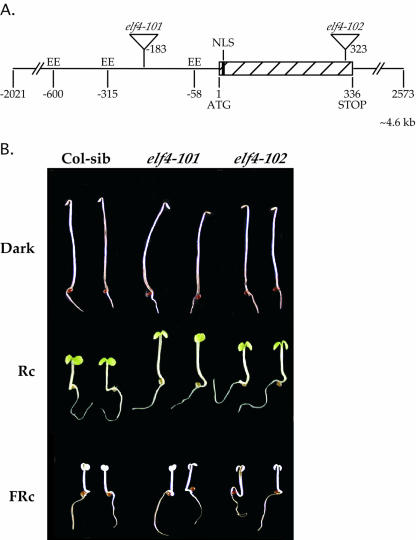
Figure 2.
elf4 mutant seedlings have reduced responsiveness to Rc. A, Structure of the ELF4 gene, showing the positions of the T-DNA insertion sites in the elf4-101 and the elf4-102 mutants, three EEs in the promoter, and the putative NLS (black rectangle). The numbers indicate nucleotide positions relative to the first nucleotide of the ATG start codon. B, Four-day-old wild-type sibling (Col-sib), elf4-101, and elf4-102 mutant seedlings grown in the dark, Rc (9.5 μmol m-2 s-1), or FRc (2.7 μmol m-2 s-1).
elf4 Mutant Seedlings Have Reduced Sensitivity to Rc during De-Etiolation
We obtained two independent T-DNA insertion lines from Syngenta Biotechnology, Inc. (Research Triangle Park, NC). elf4-101 is predicted to have a T-DNA insertion 183 nucleotides upstream of the ATG start codon, whereas elf4-102 is predicted to have a T-DNA insertion at 323 bases downstream of the start codon, near the C terminus (Fig. 2A). Homozygous T-DNA lines of elf4-101 and elf4-102 were selected and back-crossed (Columbia [Col] ecotype). A wild-type sibling (lacking the elf4-102 insertion, designated as Col-sib) was selected as a control wild-type line for phenotypic comparisons. As shown in Figure 2B, the elf4-101 and elf4-102 seedlings have longer hypocotyls than the Col-sib seedlings when grown in Rc for 4 d. There was no difference in the hypocotyl lengths between the mutant and wild-type seedlings when grown in darkness or FRc for 4 d (Fig. 2B).
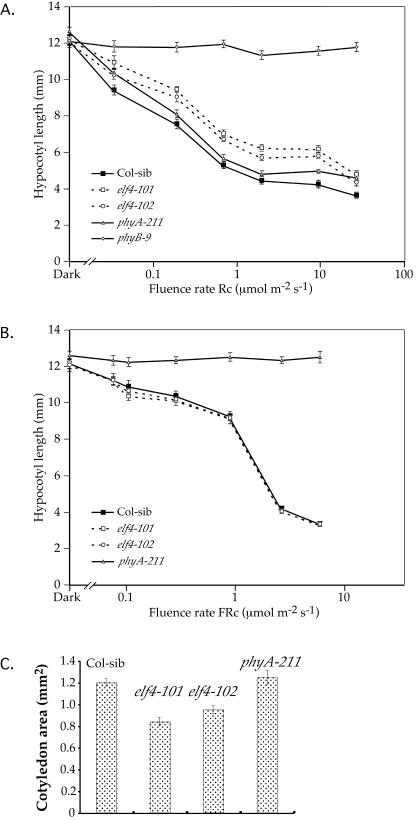
The elf4 mutant seedlings were subjected to Rc and FRc fluence rate response analysis to quantitatively characterize their sensitivities to the two wavelengths. elf4-101 and elf4-102 seedlings are taller than the wild-type (Col-sib) seedlings at all fluence rates tested in Rc (Fig. 3A), indicating reduced sensitivity to Rc. In contrast, the elf4 mutant seedlings are indistinguishable from the Col-sib seedlings in the FRc fluence rates tested (Fig. 3B). A second index of reduced Rc sensitivity in the de-etiolation process is cotyledon expansion. The elf4-101 and elf4-102 mutant seedlings have reduced expansion of cotyledon area, in comparison with the Col-sib and the phyA-211 (Fig. 3C). In addition, the adult elf4 mutant plants have slightly elongated petioles (data not shown). Overall, these results indicate that elf4 mutant seedlings are hyposensitive to Rc and are impaired in phyB-mediated seedling de-etiolation.
Figure 3.
elf4 mutants have reduced sensitivity to Rc but not to FRc. Fluence rate response curves for Col-sib, elf4-101, and elf4-102 mutants under Rc (A) and FRc (B). The phyB-9 mutant in Rc and phyA-211 mutant in Rc and FRc, are included for hypocotyl length comparisons. C, elf4 mutant seedlings show reduced cotyledon expansion. Four-day-old seedlings grown in Rc (9.5 μmol m-2 s-1).
PhyB Is Required to Induce Normal Levels of ELF4 Gene Expression in Rc
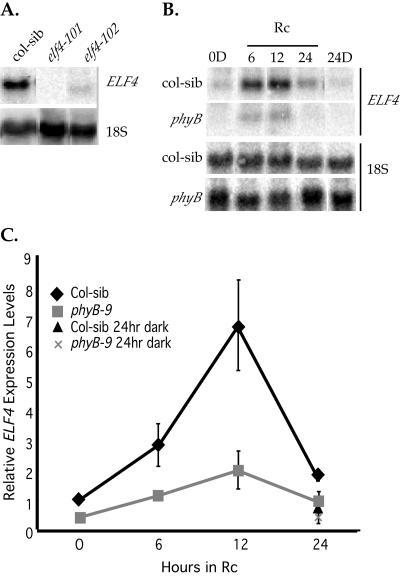
The ELF4 transcript levels peak at 12 h in FRc (Tepperman et al., 2001), so we used this time point to assay the ELF4 transcript levels in the Col-sib and the elf4 mutant seedlings. As seen in Figure 4A, the ELF4 transcript is present in Col-sib seedlings grown in FRc for 12 h, but the transcript was undetectable in the elf4-101 mutant (T-DNA insertion in the promoter) seedlings (Fig. 4A). The ELF4 transcript was also undetectable in all of the elf4-101 mutant seedlings exposed to any of the Rc or other FRc treatments (data not shown). Hence, elf4-101 appears to be a null allele. The elf4-102 mutant (T-DNA insertion near the C terminus) seedlings express a smaller sized transcript at very low levels (Fig. 4A).
Figure 4.
Rc-induced expression of the ELF4 gene is phyB dependent. Representative northern blots probed with ELF4 riboprobes. A, ELF4 transcript levels in Col-sib, elf4-101, and elf4-102 mutant seedlings grown in FRc for 12 h. B, ELF4 expression in wild-type, Col-sib, and phyB-9 mutant seedlings grown in Rc (7 μmol m-2 s-1) for 6, 12, or 24 h. RNA from dark controls (0D and 24D) are also included. C, Quantitation of the ELF4 transcript levels from four independent replicates. The values were normalized to the 18S rRNA signal, and the mean values for each time point were plotted with ses.
ELF4 gene expression is regulated by phyA (Tepperman et al., 2001), but the elf4 mutant seedlings show reduced Rc sensitivity (Figs. 2 and 3). To test the involvement of phyB in ELF4 gene expression, we examined the ELF4 transcript levels in 4-d-old Colsib and phyB-9 mutant seedlings grown in darkness or exposed to 6, 12, or 24 h of Rc and compared them with those observed in FRc. The pattern of expression in FRc was similar to that previously reported (Tepperman et al., 2001), with expression peaking at 12 h of FRc exposure (data not shown). Rc also induced expression in wild-type, Col-sib seedlings with a temporal pattern very similar to FRc (Fig. 4, B and C). By contrast, the phyB-9 mutation has a considerable affect on ELF4 expression levels in Rc (Fig. 4, B and C). Although the temporal pattern of ELF4 expression remains similar in Col-sib and phyB-9 mutant seedlings, ELF4 expression is reduced in the phyB-9 null seedlings by 3- to 4-fold (Fig. 4C). This indicates that phyB is necessary for normal induction of ELF4 expression in response to the Rc signal. This observation is consistent with the ELF4 expression profile observed in microarray-based analysis of wild-type and phyB mutant seedlings grown in Rc (J.M. Tepperman and P.H. Quail, unpublished data).
ELF4 Is Localized to the Nucleus
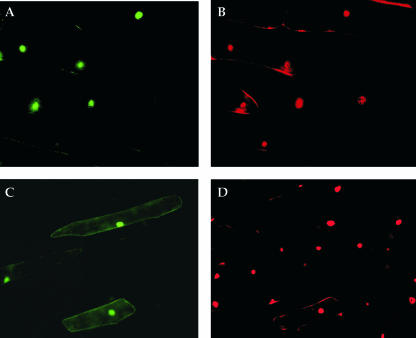
The predicted amino acid sequence of ELF4 contains a cluster of basic residues (amino acids 8–11, KRRR) that can function as a potential NLS (marked in Figs. 1A and 2A). To investigate the subcellular localization of ELF4, an enhanced green fluorescent protein (EGFP)-ELF4 fusion driven by a 35S promoter was transiently expressed in onion epidermal cells. The green fluorescent protein (GFP) fluorescence in the transformed onion epidermal cells expressing this fusion protein is observed exclusively in the nucleus (Fig. 5, A and B). In contrast, a parallel assay with EGFP alone showed distribution throughout the cell in addition to the nucleus (Fig. 5, C and D). The passive distribution of the EGFP-alone control into the nucleus in addition to the cytoplasm is expected, due to its small size. The EGFP-ELF4 fusion, however, is present only in the nucleus and is not detectable in the cytoplasm. These results provide evidence that the ELF4 protein is likely to be targeted to the nucleus, consistent with a potential function in gene regulation.
Figure 5.
ELF4 localizes to the nucleus. Transient transfection assays in onion epidermal cells using EGFP-ELF4 constructs. A and B, Cells expressing the EGFP-ELF4 fusion protein; C and D, cells expressing the EGFP-protein control. GFP florescence (A) and propidium iodide (PI)-stained (B) nuclei show that all of the detectable EGFP-ELF4 fusion protein is nuclear localized. GFP fluorescence (C) and PI-stained (D) nuclei show that the EGFP protein control is distributed throughout the cell, including the cytoplasm and nucleus.
DISCUSSION
The expression of several genes of as yet unknown function is rapidly regulated by phyA in response to FRc light signals, as reported in our previous microarray-based analysis of wild-type and phyA mutant seedlings (Tepperman et al., 2001). We speculated that these genes might function in phyregulated networks. The results presented here indicate that one of the phyA-regulated early-induced “unknown” genes, recently identified as ELF4 (Doyle et al., 2002), is involved in the phyB-mediated Rc-induced seedling de-etiolation process (Fig. 6). The reason for the absence of a phenotype in elf4 mutant seedlings under FRc is unknown, but the data suggest a bifurcation between phyA- and phyB-signaling pathways.
Figure 6.
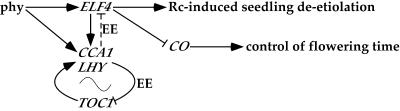
Simplified model of ELF4 function in phy-regulated seedling de-etiolation, circadian rhythms, and flowering time. ELF4 expression is regulated by phy in response to light signals, as is the expression of CCA1 and LHY. ELF4 functions as a positive regulator of phyB-mediated seedling de-etiolation. ELF4 function is closely linked to the central oscillator, thereby functioning in clock maintenance and regulating circadian rhythmicity. Like TOC1, ELF4 has EE in its promoter, and it induces the expression of CCA1. We propose that the ELF4 expression is regulated in a negative manner, possibly by CCA1/LHY binding to the EE in its promoter, similar to the regulation of TOC1. ELF4 represses CO expression exerting control on flowering time.
The elf4 mutant phenotype observed in Rc appears to be attributable to the lack of normal phyB-induced expression of ELF4 (Fig. 4). phyB-null mutant seedlings have a 3- to 4-fold reduction in ELF4 expression levels in Rc compared with wild type (Fig. 4E). These data indicate that phyB is required for the normal high levels of expression in Rc. Evidently, this phyB-induced expression in Rc is important for ELF4 function, because elf4 mutant seedlings lacking expression exhibit a Rc-specific phenotype. The Rc specificity of the phenotype suggests that ELF4 functions downstream in a phyB-regulated pathway.
There is compelling evidence that upon photocon-version to the biologically active form, the phy molecule is translocated into the nucleus (Nagy and Schäfer, 2000a, 2000b). PIF3, a basic helix-loop-helix-class transcriptional regulator, interacts specifically with the Pfr form of phy while bound to its DNA target site and appears to be involved in Rc-regulated gene expression (for review, see Quail, 2002). There is a growing list of other proteins that are also nuclear localized and implicated in phy-mediated signaling, including some that are involved in the circadian clock and photoperiod sensitivity, like ELF3, GI, TOC1, and PSEUDO RESPONSE REGULATOR 7 (Huq et al., 2000; Strayer et al., 2000; Liu et al., 2001; Kaczorowski and Quail, 2003). As we show here, ELF4 is also localized to the nucleus (Fig. 5) and may function to coregulate the expression of a subset of phyB-regulated genes. Our preliminary attempts using in vitro immunoprecipitation assays do not show any evidence for direct interactions between ELF4 and the phys (data not shown).
We isolated two T-DNA insertion lines, elf4-101 and elf4-102. The insertion in elf4-101 is in the promoter region, and the ELF4 mRNA was not detected in the mutant seedlings, indicating that this is likely to be an elf4-null allele. The elf4-102 insertion is near the C terminus, and a smaller sized transcript was observed in the elf4-102 seedlings. elf4-101 and elf4-102 are defective in phyB-mediated Rc responses in the hypocotyl and cotyledons. The inhibition of these Rc responses is more subtle in elf4-102 compared with the inhibition in elf4-101 seedlings. This difference may be attributed to reduced ELF4 functionality due to the short, lower abundance transcript and potentially truncated protein present in the elf4-102 mutant seedlings. On the basis of these findings, it appears that ELF4 functions in a positive manner downstream in the phyB-signaling pathway, regulating Rc-induced seedling photomorphogenesis (Fig. 6). elf4 mutants flower earlier than the wild type under short-day, noninductive conditions (Doyle et al., 2002). The elf4-101 and elf4-102 mutants also flower early (data not shown) and are evidently photoperiod insensitive. It has been proposed that the early-flowering phenotype is a result of the elevated levels of CO in the elf4 mutants (Doyle et al., 2002). As depicted in our proposed simplified model shown in Figure 6, ELF4 might function to repress the expression of CO. ELF4 plays a crucial role in photoperiod perception and the control of flowering time by regulating the expression of this key gene in the flowering pathway (Doyle et al., 2002).
Arabidopsis plants lacking ELF4 function have lower levels of CCA1 transcripts (Doyle et al., 2002), indicating that ELF4 controls CCA1 expression (Fig. 6). CCA1 and LHY are also proposed direct targets of the phy-dependent light-induction pathway (Fig. 6), possibly via PIF3 bound to their G-box promoter elements (Martinéz-García et al., 2000; Tepperman et al., 2001; Quail, 2002). CCA1 and LHY bind to an EE in the TOC1 promoter, repressing its expression as part of the mechanism by which the circadian oscillator functions (Fig. 6; Harmer et al., 2000; Mizoguchi et al., 2002). Our searches for promoter sequences revealed three EE in the ELF4 promoter (Fig. 2A), suggesting that it may also be a target for repression by CCA1 and LHY (Fig. 6). Lower levels of CCA1 and LHY in elf4 mutants suggests that like TOC1, ELF4 may function to activate the transcription of these genes, thereby perhaps being closely linked to the feedback loop, which is essential for the free-running clock and consistent with the view that ELF4 function is closely associated with the central oscillator (Eriksson and Miller, 2003). Together, these and previous data suggest that ELF4 functions in a phyB-mediated signaling pathway, possibly by regulating gene expression, and that ELF4 is closely associated with the central oscillator and is essential in clock maintenance and photoperiod perception.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Measurements
The Arabidopsis T-DNA insertion lines in Col ecotype background were identified by searching the database of the Syngenta Arabidopsis Insertion Library Project (formerly GARLIC), through an Academic collaboration with Syngenta Biotechnology. Homozygous T-DNA insertion lines were isolated using PCR with T-DNA- and gene-specific primers flanking the T-DNA insertion sites. The homozygous lines were crossed back once to Col and then isolated as elf4-101 and elf4-102. One of the wild-type siblings (Col-sib), lacking the elf4-102 T-DNA insertion was used thereon as a control.
Sterilized seeds were plated on growth medium plates without Suc (Hoecker et al., 1999), stratified for 3 d at 4°C, synchronized by a 3-h white-light (WL) treatment followed by a 21-h dark treatment at 21°C and then transferred to various light conditions as specified. The light sources used were as described (Wagner et al., 1991) and the fluence rates were monitored by using a spectroradiometer (model L1–1800, LI-COR, Lincoln, NE). Hypocotyl and cotyledon measurements were performed 96 h after the WL treatment using a digital camera (Coolpix 990, Nikon, Tokyo) and NIH Image software (National Institutes of Health, Bethesda, MD).
RNA Isolation and Hybridization
Seeds were plated as described above. After the synchronization with WL, seeds were transferred back to the dark at 21°C for 4 d. The 4-d-old seedlings were irradiated with Rc (7 μmol m-2 s-1) or FRc (2 μmol m-2 s-1) for 6, 12, or 24 h. Dark control seedlings were harvested before (0 D) or after (24 D) the light treatments. Tissue was harvested in the dark and frozen immediately in liquid nitrogen. RNA was isolated using the Plant RNeasy kit according to the manufacturer's instructions (Qiagen,Valencia, CA). Ten micrograms of total RNA was separated on 1.2% (w/v) agarose gels containing 0.67% (w/v) formaldehyde and blotted on to Magna nylon transfer membranes (Osmonics, Westborough, MA) by capillary action in 20× SSC. Membranes were cross-linked using a UV-Crosslinker 1800 (Stratagene, La Jolla, CA). A full-length cDNA clone (expressed sequence tag, 118M24) was obtained (Arabidopsis Biological Resource Center, Columbus, OH), was sequenced to confirm the ELF4 sequence, and was linearized at the 5′ end before being used as a template for riboprobes. The ELF4 antisense-RNA probe was synthesized in vitro in the presence of [α-32P]UTP (3,000 Ci mmol-1; Amersham Biosciences, Piscataway, NJ) using the Riboprobe Transcription System (Promega, Madison, WI). Blots were prehybridized for 2 h at 60°C, hybridized overnight, and then washed, as described (Khanna et al., 1999). Blots were then exposed overnight and visualized using a PhosphorImager (Storm 860, Molecular Dynamics, Sunnyvale, CA). The 18S probe was labeled using the Redi-Prime II kit (Amersham Biosciences) and hybridized according to Church and Gilbert (1984). The ELF4 expression levels were quantified using ImageQuant for Mac v.1.2 (Molecular Dynamics) and were normalized to the 18S rRNA signal. Four independent replicates were performed, and the mean values for each time point were plotted with ses.
Subcellular Localization
The expressed sequence tag clone (118M24) was used as a template for PCR, using PFU Turbo polymerase (Stratagene) and primers containing internal restriction sites (EcoRI and XbaI) to amplify the full-length open reading frame of ELF4, which was inserted into the pEZS-CL vector (S. Cutler and D. Ehrhardt, Carnegie Institute of Washington, Stanford, CA) to create a 35S:EGFP-ELF4 construct. The construct was sequenced and used for bombarding onion epidermal peels for transient transfection assays as described (Huq et al., 2000). The nuclei were stained with 0.1 μg mL-1 PI. A 35S:EGFP construct was used as control.
Acknowledgments
We thank C.L. Lanzatella for Arabidopsis seed plating and for taking excellent care of plants in the greenhouse, J.M. Tepperman for providing critical data, A. Smith for solution preparation, and members of our laboratory for helpful discussions and comments.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.030007.
This work was supported by the National Institutes of Health (grant no. GM47475), by the Department of Energy (grant no. DE–FGO3–87ER13742), by the U.S. Department of Agriculture/Agricultural Research Service Current Research Information System (CRIS; grant no. 5335–2100–017–00D), and by the Torrey Mesa Research Institute (San Diego).
References
- Alabadi D, Oyama T, Yanovski MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1. Science 293: 880-883 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991-1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881-10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF (2002) Signs of the time: environmental input to the circadian clock. J Exp Biol 53: 1535-1550 [DOI] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognár L, Nagy F, Miller AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74-77 [DOI] [PubMed] [Google Scholar]
- Eriksson ME, Miller A (2003) The circadian clock: a plant's best friend in a spinning world. Plant Physiol 132: 732-738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Staiger D (2002) Photoreceptors in Arabidopsis thaliana in light perception, signal transduction and entrainment of the endogenous clock. Planta 216: 1-16 [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Somach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679-4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tobin EM (1999) Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA 96: 4176-4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110-2113 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Panda S, Kay SA (2001) Molecular bases of circadian rhythms. Annu Rev Cell Dev Biol 17: 215-253 [DOI] [PubMed] [Google Scholar]
- Hicks KA, Albertson TM, Wagner DR (2001) EARLY FLOWERING 3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH (1999) SPA1: a WD-repeat protein specific to phytochrome A signal transduction. Science 284: 496-499 [DOI] [PubMed] [Google Scholar]
- Hudson ME, Quail PH (2003) Identification of key promoter motifs involved in the network of phyA-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol 133: 1605-1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Tepperman JM, Quail PH (2000) GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci USA 97: 9789-9794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabayashi Y, Kaya H, Goto K, Iwabuchi M (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960-1962 [DOI] [PubMed] [Google Scholar]
- Kaczorowski KA, Quail PH (2003) Arabidopsis PSEUDO RESPONSE REG ULATOR 7 (PRR7) is a signaling intermediate in phytochrome regulated de-etiolation and phasing of the circadian clock. Plant Cell (in press) [DOI] [PMC free article] [PubMed]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer. FT Sci 286: 1962-1965 [DOI] [PubMed] [Google Scholar]
- Khanna R, Lin X, Watson JC (1999) Photoregulated expression of the PsPK3 and PsPK5 genes in pea seedlings. Plant Mol Biol 39: 231-242 [DOI] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR (2001) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293-1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinéz-García JF, Huq E, Quail PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859-863 [DOI] [PubMed] [Google Scholar]
- Mas P, Alabadi D, Yanovsky MJ, Oyama T, Kay SA (2003) Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWatters HG, Bastow RM, Hall A, Miller AJ (2000) The ELF3 zeitnehmer regulates light signaling to the circadian clock. Nature 408: 716-720 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatly K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carré IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629-641 [DOI] [PubMed] [Google Scholar]
- Nagy F, Schäfer E (2000a) Nuclear and cytosolic events of light-induced, phytochrome-regulated signaling in higher plants. EMBO J 19: 157-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Schäfer E (2000b) Control of nuclear import and phytochromes. Curr Opin Plant Biol 3: 450-454 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657-667 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1999) Binding of phytochrome B to its nuclear signaling partner PIF3 is reversibly induced by light. Nature 400: 781-784 [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847-857 [DOI] [PubMed] [Google Scholar]
- Quail PH (2000) Phytochrome interacting factors. Semin Cell Dev Biol 11: 457-466 [DOI] [PubMed] [Google Scholar]
- Quail PH (2002) Photosensory perception and signaling in plant cells: new paradigms? Curr Opin Cell Biol 14: 180-188 [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613-1616 [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219-1229 [DOI] [PubMed] [Google Scholar]
- Simpson GG, Gendall AR, Dean C (1999) When to switch to flowering. Annu Rev Cell Dev Biol 15: 519-550 [DOI] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488-1490 [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319-329 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768-771 [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatly K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116-1120 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang H-S, Wand X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98: 9437-9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DR, Tepperman JM, Quail PH (1991) Overexpression of phytochrome B induces a short hypocotyl phenotype in transgenic Arabidopsis. Plant Cell 3: 1275-1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207-1217 [DOI] [PubMed] [Google Scholar]
- Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR (1996) The Arabidopsis ELF3 gene regulates negative photomorphogenesis and the photoperiodic induction of flowering. Plant J 10: 691-702 [DOI] [PubMed] [Google Scholar]