Abstract
Several hundred Arabidopsis genes, transcriptionally regulated by phytochrome A (phyA), were previously identified using an oligonucleotide microarray. We have now identified, in silico, conserved sequence motifs in the promoters of these genes by comparing the promoter sequences to those of all the genes present on the microarray from which they were sampled. This was done using a Perl script (called Sift) that identifies over-represented motifs using an enumerative approach. The utility of Sift was verified by analysis of circadian-regulated promoters known to contain a biologically significant motif. Several elements were then identified in phyA-responsive promoters by their over-representation. Five previously undescribed motifs were detected in the promoters of phyA-induced genes. Four novel motifs were found in phyA-repressed promoters, plus a motif that strongly resembles the DE1 element. The G-box, CACGTG, was a prominent hit in both induced and repressed phyA-responsive promoters. Intriguingly, two distinct flanking consensus sequences were observed adjacent to the G-box core sequence: one predominating in phyA-induced promoters, the other in phyA-repressed promoters. Such different conserved flanking nucleotides around the core motif in these two sets of promoters may indicate that different members of the same family of DNA-binding proteins mediate phyA induction and repression. An increased abundance of G-box sequences was observed in the most rapidly phyA-responsive genes and in the promoters of phyA-regulated transcription factors, indicating that G-box-binding transcription factors are upstream components in a transcriptional cascade that mediates phyA-regulated development.
Transcriptional control is of critical importance in mediating the responses of eukaryotic cells to external stimuli. The promoter of a gene (the regulatory DNA sequence upstream of the transcribed region) is centrally important in determining if and when transcription will be initiated. The nucleotide sequence of the promoter specifies the recruitment of DNA-binding proteins, including the transcription factors that regulate gene expression. The short DNA sequence motifs that specify protein binding are therefore the essential functional components of the promoter. The level of interest in the mechanisms of transcriptional regulation has led to a number of advances in the computational analysis of regulatory DNA sequence. Databases of known transcription factor binding sites can detect the presence of protein-recognition elements in a given promoter, but only when the binding site of the relevant DNA-binding protein and its tolerance to mismatches in vivo is already known. Because this knowledge is currently limited to a small subset of transcription factors, much effort has been devoted to discovery of regulatory motifs by comparative analysis of the DNA sequences of promoters. By finding conserved regions between multiple promoters, motifs may be identified with no prior knowledge of transcription factor-binding sites. The promoters of coregulated genes are likely to be responsive to the same pathway and therefore to share common regulatory motifs, providing a potential way to discover new mechanisms of transcriptional regulation. The currently used computational approaches can be grouped into those using sequence alignment (alignment methods) and those where statistical analysis of the abundance of short sequences is used to detect over-represented motifs (enumerative methods). The previous implementations of these algorithms are reviewed by Ohler and Niemann (2001).
Alignment methods (e.g. Roth et al., 1998) have the disadvantage that they make no distinction between core promoter elements (e.g. TATA box) and regulatory sequences specific to a coregulated gene set. The researcher is faced with a list of many potential sequence motifs, the most high-scoring of which are usually core promoter motifs. In addition, the models used to predict likelihood of elements occurring at random are usually optimized for yeast or mammalian genomes, making these programs of little use for plant promoter analysis. These issues are now being addressed (Thijs et al., 2002). Alignment type approaches typically do not provide any information to the user that allows the degree of sequence similarity between related elements to be interpreted in a biological context. Similar elements, bound by related proteins but with divergent functions, are thus difficult to distinguish from each other using this method. In addition, the computational (both memory and CPU) requirements for alignment scale exponentially with the number of promoter sequences analyzed. Thus, using this method, analysis of more than 20 to 30 intergenic regions from Arabidopsis is intractable using the computer facilities currently accessible to most biologists.
The “enumerative” method has been previously used by van Helden et al. (1998), Jensen and Knudsen (2000), and Vanet et al. (2000). This method consists of counting small sequence motifs, and looking for sequences enriched to statistically significant levels. Such an approach has recently been used for investigation of 5′-untranslated region sequences in plants (Hulzink et al., 2003) but has not to our knowledge been used before for promoter sequence analysis in plants.
Complex regulatory networks are revealed by microarray experiments, in which large numbers of transcripts are assayed simultaneously (Futcher, 2002). Most microarray experiments reveal several hundred coregulated genes that may share common promoter motifs. Harmer et al. (2000) used an Align-Ace (Roth et al., 1998) analysis of a subset of the promoters of their circadian-regulated gene list, together with biological analysis of putative elements, to discover the single element described in their paper. Detailed re-analysis of this data by Michael and McLung (2003) has revealed a number of potentially important motifs in the promoters of these genes, using existing alignment programs. However, alignment techniques are not well suited to the comparative analysis of hundreds of promoters, because of their CPU-intensive nature. In addition, prior knowledge of the biological system is needed for the interpretation of numerous potential motifs identified, most of which are related to the core functions of the promoter.
In this paper, we investigate the control of transcription in Arabidopsis in response to light stimuli via the phytochrome photoreceptor. Phytochromes are central to the responses of higher plants to light (Smith, 2000). Until recently, however, the number of known light-regulated genes has been limited to a few dozen, most of which respond to signals from the phytochromes. The response of transcription of this limited set of genes to light has been extensively studied (Terzaghi and Cashmore, 1995). Several small DNA motifs in the promoter region of the gene, for example the G-box and I-box (Giuliano et al., 1988), have been shown to be both necessary and sufficient for the induction of transcription of certain genes in response to light. Analysis of promoters of orthologous, light-induced genes from a range of species can locate motifs conserved across large evolutionary distances (Arguello-Astorga and Herrera-Estrella, 1996).
Many genetic mutants in Arabidopsis that show aberrant responses to stimuli have been shown to inactivate genes encoding proteins that bind DNA or are associated with the regulation of transcription (e.g. Putterill et al., 1995; Oyama et al., 1997; Stockinger et al., 1997; Sakai et al., 2001). In some cases, the binding specificities of these proteins have been elucidated by DNA footprinting studies and/or random binding site selection experiments (Chattopadhyay et al., 1998). Furthermore, such binding sites have been identified as regions critical to the function of promoters showing strong transcriptional regulation by external stimuli (Menkens et al., 1995). However, the number of known transcription factor-binding sites in plants remains small, and it is likely that many critical regulatory promoter elements are not amenable to discovery by these methods.
Computational analysis of promoter sequences provides an alternative means of investigation, now that the sequences of large groups of coregulated genes are available. Several hundred far-red-light-responsive genes have been identified by Tepperman et al. (2001), which are responsive to signals transmitted by the phytochrome A (phyA) photoreceptor pathway. These genes were grouped into functional classes and categorized into “early”- and “late”-responsive transcripts, according to whether a 2-fold response was observed within 1 h of the commencement of the far-red-light stimulus or within the subsequent 24 h of illumination. The analysis of the promoters of these genes was not attempted by Tepperman et al., in common with most investigations of transcriptional regulation using microarray data. The reason for this may be that analysis of promoters for large gene-lists, such as that described by Tepperman et al., is intractable using existing, publicly available programs. In the work described here, a computational method was developed to discover potential new DNA regulatory motifs by their over-representation in the promoters of this set of coregulated genes. Because several light- or phytochrome-responsive elements have already been characterized (Terzaghi and Cashmore, 1995), this data set provides an opportunity to test a new approach and to investigate the potential existence of uncharacterized regulatory pathways alongside the established framework.
RESULTS
Computational Approach
The approach we describe here is a modification of the standard enumerative method, which examines a promoter set for sequences over-represented with respect to the remainder of the genome (van Helden et al., 1998). Here, we compare the abundance of a motif in a set of coregulated promoters with the abundance of the same sequence in the promoters of all the genes on the microarray with which the coregulated gene set was identified. By this means, we compare the sequences identified as coregulated directly with the distribution from which they were sampled. In most previous implementations of the enumerative approach, promoter sequences are compared with the genome as a whole. By comparing one set of promoters with another, we internally eliminate sequences that are over-represented in all promoters. Such sequences can be over-represented because they are involved in functions common to all promoters or because of the different nucleotide composition of promoters from the rest of the genome because of the absence of coding sequence.
By comparing coregulated promoters to the rest of the microarray, rather than all of the promoters in the genome, we sample from the distribution from which the coregulated gene set was originally determined. When the genes represented on the microarray are a subset of the genome, as is the case with the Arabidopsis array used in the studies described here, this subset is rarely randomly sampled from the genome. Often the genes present are the most highly expressed, which can cause false results in a promoter comparison analysis with the promoters of the total genome.
Another general problem with the enumerative method is that it is vulnerable to false positives, caused by the presence of multimeric repetitive sequences in one or more of the promoters. If motifs are simply counted, and the overall abundance is compared, repeats lead to the components of the repeat being the most statistically significant hits. We overcame this problem by also requiring that promoters containing one or more motifs are significantly more common in the coregulated gene set than they are in the set of promoters of genes represented on the microarray. Only those motifs statistically over-represented by both raw counting and on a per-promoter basis are reported. We call the program Sift, and we have made the data and source code we used available on-line at http://www.pgec.usda.gov/Quail/Hudson-promoter/.
Analysis of Circadian Gene Promoters
Motifs such as the evening element (Harmer et al., 2000) are totally conserved in many well-characterized light- and circadian-regulated promoter sequences in plants. The evening element has already been shown to be biologically significant in the circadian regulation of Arabidopsis genes. The promoters of the circadian-regulated genes identified by Harmer et al. therefore make a useful “training data set” to verify whether a method is capable of detecting regulatory promoter elements from a set of upstream genomic sequences.
Using the method described here to find conserved sequences on the sense strand of the promoters of this subset, our best hit by a considerable margin was the “evening element” described and verified by in vivo analysis by Harmer et al. (2000), followed by the G-box, found to be over-represented in circadian-regulated promoters by Michael and McClung (2003). We then investigated which other promoter motifs were significantly over-represented in the circadian-regulated gene set. Figure 1 shows the results of the complete analysis of this set of promoters. Note that after the different versions of the evening element, the next best hit, at P = 2.9 × 10-8, was the G-box, CACGTG. A G-box-related sequence, CCACGT, was found to be an even more significant hit when both the sense and reverse complement of the promoters were searched. The evening element, on the other hand, was less significantly enriched when both strands were considered. The “core” evening element AAATATCT was found on the sense strand of 103 of 419 circadian-regulated promoters and on the antisense strand of 83 of 419. This may indicate that the evening element is preferentially present on the sense strand.
Figure 1.
Elements over-represented in circadian-regulated promoters. These are elements found in the promoters of circadian-regulated genes identified by Harmer et al. (2000). Each sequence shown was individually identified as a statistically significant over-represented sequence in circadian gene promoters. The sequences in each aligned group are those we determined to represent the same motif. The P value was determined using the binomial distribution to find the likelihood of the observed number of elements occurring in a randomly chosen set of promoters. The letters bs or ss designate whether the element was detected as over-represented on both strands (bs) or just one strand (ss).
Only elements where enrichment was such that the binomial distribution gave P < 10-5 are shown. Several other elements met this criterion. Most are sub-types or variants of the evening element or the G-box (Fig. 1). The evening element is a type of GATA or I-box element (Giuliano et al., 1988), because it contains the antisense GATA sequence TATC. Other GATA-type elements not corresponding to perfectly conserved evening elements were found to be over-represented in these promoters, particularly the sequence GGATAAA. However, the elements CTAAAATA, AAAGTCCTAA, CACTAACCAC, and ATCACATAT do not fit into any previously described categories of light- or circadian-regulatory promoter elements in plants, to our current knowledge.
Analysis of phyA-Regulated Promoters
Perhaps the best characterized of the environmental transcriptional responses in plants is the induction of transcription of nuclear genes in response to light signals, particularly those from phytochrome. The well-known elements defined by previous analyses include GT-1 sites, GATA or I-box elements, G-boxes, and some basal promoter elements such as the CCAAT box (Terzaghi and Cashmore, 1995). We set out to use the phyA-regulated gene sets described by Tepperman et al. (2001) to investigate which of these elements were over-represented in the set of genes regulated by phyA in response to continuous far-red light. This not only gave us the opportunity to define those promoter elements with a role in mediating the responses of a large number of phyA-regulated genes, but also the potential to discover new elements, over-represented in these promoters, that may be involved in the regulation of a significant subset of the phyA-responsive transcripts.
phyA-Induced Genes
We first addressed the upstream sequences of all of the genes known to be transcriptionally induced by the phyA pathway, which were defined previously (Tepperman et al., 2001). The genes in question were divided by these authors into two groups, early and late, according to whether or not a 2-fold increase in transcript abundance was seen within 1 h of the beginning of far-red-light stimulus. For the present analysis, we considered the promoters of the early and late genes together as a single group. This larger sample size increased the signal-to-noise ratio for the detection of elements. We were able to unambiguously identify the upstream 2-kb “promoter” sequences for 514 of these genes.
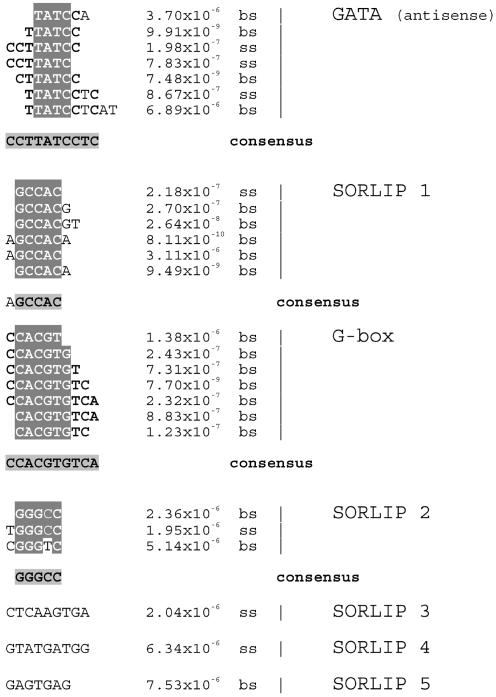
Figure 2 shows the over-represented motifs on either the sense strand or both strands of the set of phyA-induced genes. These motifs include the well-characterized element GATA box or I-box, in this case as TATC on the sense strand (and therefore on the antisense strand as GATA, as described by Giuliano et al. [1988]). TATC, the antisense GATA element (also known as the LAMP element) has previously been found to be strongly conserved between phytochrome-regulated promoters (Grob and Stuber, 1987). Another clearly recognizable, previously characterized element, known to be involved in light regulation of gene expression, is the G-box (Chattopadhyay et al., 1998; Giuliano et al., 1988; Menkens et al., 1995; Martinez-Garcia et al., 2000). Note that certain flanking bases are over-represented next to these core elements, notably TTATCC/GGATAA for the GATA element, compatible with the Box II sequences from the rbcS gene of Arabidopsis, pea (Pisum sativum), and others (Arguello-Astorga and Herrera-Estrella, 1998). These results may provide insight into how an element such as G-box, which is over-represented in rapidly phyA-induced genes, phyA-repressed genes, and circadian-regulated genes, may be specifically targeted by multiple trans-acting factors by means of different conserved flanking sequences.
Figure 2.
Elements over-represented in phyA-induced promoters. These are elements found in the promoters of phytochrome-A-induced genes identified by Tepperman et al. (2001). Each sequence shown was individually identified as a statistically significant over-represented sequence in phyA-induced gene promoters. The sequences in each aligned group are those we determined to represent the same motif. The P value was determined using the binomial distribution to find the likelihood of the observed number of elements occurring in a randomly chosen set of promoters. The letters bs or ss designate whether the element was detected as over-represented on both strands (bs) or just one strand (ss).
A number of sequences were identified that are over-represented in the phyA-induced promoters but have not been previously characterized. We refer to these sequences as sequences over-represented in light-induced promoters (SORLIPs). SORLIP 1 is the most over-represented of these and the most statistically significant hit; the core sequence is GCCAC with an A at the 5′ end and A or G at the 3′ end as conserved flanking bases. It appears to be strand-independent, because it is the strongest hit when both strands are considered (Fig. 2). The other significantly over-represented sequences we found, SOR-LIPs 2 though 5, are detailed in Figure 2.
phyA-Repressed Genes
We then applied the same approach to the study of the promoters of 259 genes identified by Tepperman et al. (2001) as transcriptionally repressed, again in response to continuous far-red light acting through the phyA photoreceptor. The repression of transcription in response to light has been less well studied than induction of transcription. Certain elements have been identified as important for repression of specific genes, such as DE1 for PRA2 (Inaba et al., 1999; Inaba et al., 2000) and RE1 for PHYA itself (Bruce et al., 1991; Dehesh et al., 1994).
We found a number of strongly conserved sequences over-represented in the promoters of the phyA-repressed genes. The most significant of these sequences was, again, the G-box. The G-box was more enriched in the promoters of phyA-repressed genes than phyA-induced genes or circadian-regulated genes (see “Distribution of Motifs in the Genome”). However, when the multiple, significantly enriched, totally conserved G-box motifs in this set of promoters are aligned (Fig. 3), it is clear that although the core CACGTG sequence is the same as that which is abundant in phyA-induced and circadian-regulated promoters, the flanking nucleotides of the most over-represented elements are distinctly different. The consensus sequence of the phyA-induced promoter G-box (CCACGTGTCA) is strongly supported, with all of the over-represented fragments showing 100% identity to one another where they overlap (Fig. 2). The G-box motifs over-represented in phyA-repressed genes have the consensus CTTCACGTGG, or because the over-representation of most of these sequences is strand independent, CCACGTGAAG. The three flanking nucleotides at one end of the palindrome therefore form a distinct signature.
Figure 3.
Elements over-represented in phyA-repressed promoters. These are elements found in the promoters of phytochrome-A-repressed genes identified by Tepperman et al. (2001). Each sequence shown was individually identified as a statistically significant over-represented sequence in light-repressed gene promoters. The sequences in each aligned group are those we determined to represent the same motif. The P value was determined using the binomial distribution to find the likelihood of the observed number of elements occurring in a randomly chosen set of promoters. The letters bs or ss designate whether the element was detected as over-represented on both strands (bs) or just one strand (ss).
We also found other motifs that were very strongly over-represented in the phyA-repressed set of promoters, which we term sequences over-represented in light-repressed promoters (SORLREPs; Fig. 3). The consensus sequence of the most common motif of this group, TTTTACTAGT, is very close to the DE1 sequence, GGATTTTACAGT (Inaba et al., 1999). DE1 has been shown by Inaba et al. (2000) to be sufficient to confer light repression on a minimal 35S promoter. The GC-rich RE1 element (Bruce et al., 1991), although present in the Arabidopsis PHYA promoter (Dehesh et al., 1994), does not appear to be strongly enriched in the large set of repressed gene promoters investigated here, although it does resemble the SORLIP 2 sequence found in the phyA-induced promoter set. In addition, we detected four more elements that are over-represented in phyA-repressed promoters SORLREP 2 through 5 (Fig. 3).
Distribution of Motifs in the Genome
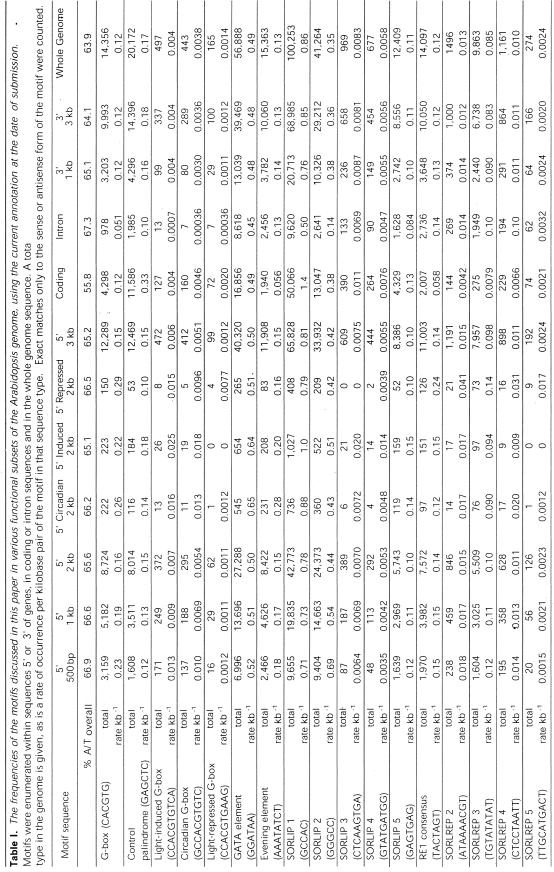
To investigate the information obtainable from the detection of an element within a promoter sequence, we enumerated exact matches to the elements described here in the sequence of the whole of the Arabidopsis genome, and subsets of that sequence. Note that palindromic sequences, such as G-box, were treated differently, in that only one strand of the genome was searched (to prevent each motif being counted twice). In the case of non-palindromic sequences, both strands were searched. The subsets of the genome analyzed were introns, intergenic regions, regions 5′ and 3′ of coding regions, coding regions themselves, and the subsets of 2-kb 5′ promoter sequences used in this paper, regulated by circadian rhythms or phytochrome signaling. The results of this analysis are given in Table I.
Table 1.The frequencies of the motifs discussed in this paper in various functional subsets of the Arabidopsis genome, using the current annotation at the date of submission.
It can be seen from Table I that the frequency of finding a “core” G-box sequence, CACGTG, in the 500 bp upstream of any gene is 0.23 per kilobase—i.e. roughly one-tenth of all genes have a G-box in the immediate 5′ 500 bp. G-boxes are therefore almost twice as common in this region of promoter sequence as in coding regions or the genome as a whole, despite the increased A/T content of promoter sequences. This distribution was not shared by a control palindromic sequence with the same A/T content (GAGCTC), which, as expected from a GC rich region without termination codons, was much more abundant in coding sequence than in the 5′ upstream promoter regions.
In the 2 kb upstream from the ATG start codon, the abundance of the G-box drops to 0.16 per kilobase pair—i.e. the G-box is more common closer to the translation start point. In the whole “induced” gene set, this rate is increased to 0.22 per kilobase pair; the G-box is more common, significantly (as shown earlier in this section), but far from ubiquitous in the promoters of phyA-induced genes. The G-box is clearly over-represented in the promoters of circadian-regulated and phyA-repressed genes. However, of the 14,356 times that CACGTG occurs in the current version of the Arabidopsis genome sequence, only 595 occur in the promoters of a gene regulated 2-fold by phyA signaling or by circadian rhythms. It is likely that many other genes are influenced by these environmental stimuli, but to an extent below the resolution of the microarray experiments. However, 4,298 G-boxes occur in regions that are annotated as coding for proteins. Therefore, G-boxes may exist in genomic sequence but may not be signals for light- or circadian-regulated transcription. The positional context of the G-box must therefore be necessary for its function as a designator of phyA-regulated transcription.
When the most common flanking bases from the G-boxes of promoters in the induced, repressed, and circadian gene lists are considered, the sequences become more specific both to promoters and to the conditions described. Only four of the phyA-repressed promoter G-box flanking sequences contain all of the most favored bases for this group. However, none of the phyA-induced promoters contained that sequence. The phyA-induced consensus G-box was present in the phyA-repressed promoters, but was 40% less frequent. The flanking bases of the G-box sequence may therefore confer specificity to certain functions and help to provide a positional context for the core sequence.
The GATA element GGATAA was, as expected, more frequent in the phyA-induced and circadian-regulated promoters. The evening element (also a GATA element) was also over-represented in promoters of both the phyA-induced and circadian-regulated genes. Of the new sequences we identified, the SORLIPs all were more common in the phyA-induced promoters than elsewhere in the genome. The SORLREP elements were also all most common in phyA-repressed genes, although interestingly SORLREP4 was also common in circadian-regulated promoters.
Different Distributions of Elements Are Present in Early- and Late-Responsive Promoters
Tepperman et al. (2001) placed the transcripts responding to phyA signaling into early and late categories; early indicates a 2-fold change in level within 1 h, late a 2-fold change within 24 h. We investigated the distribution of the elements described in Table I within these categories of the phyA-regulated genes. The results are shown in Figure 4.
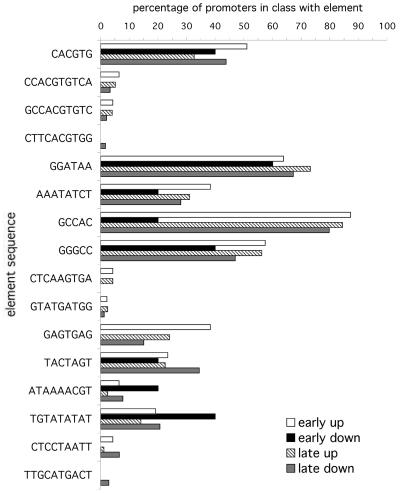
Figure 4.
Distribution of motifs by promoter response. The graph shows the percentage of promoters containing putative and known light-regulatory sequence motifs. The promoters are grouped by their response to far-red light (early, transcripts that respond 2-fold within 1 h; late, transcripts that respond 2-fold within 24 h; up or down indicate the direction of the response to the stimulus). The bars show the percentage of promoters in each list containing an exact sequence match to the sequence given by the vertical axis.
The majority of the elements investigated here are of roughly equal abundance between the early and late categories and are therefore likely to generally specify the response of promoters to the phyA pathway. Some elements (e.g. GGATAA) seem to be more abundant in the late category, which indicates that the signal transduction pathway to these elements may contain more steps. The G-box, CACGTG, which is over-represented in both phyA-induced and -repressed promoters (Figs. 2 and 3), is more common in the early-induced than the late-induced promoters; it shares this distribution with the evening element, AAATATCT. The same is true of the “induced” (CCACGTGTCA) and “circadian” (GCCACGTGTC) consensus G-boxes with flanking sequences and the SORLIP5 element, GAGTGAG. These elements, which predominate in the early-responsive promoters, are more likely to have the fewest steps in the signal transduction cascade to gene expression. Note that there are few elements that predominate in the early-repressed promoters; however there are very few genes that show 2-fold repression within 1 h (Tepperman et al., 2001) because a very short transcript half-life is required for this to be observed. It is likely that many of the genes in the “late-repressed” class are very rapidly responsive in terms of the reduction of transcription from the promoter, but that this change takes more than 1 h to manifest itself in the mRNA concentration. Certain elements are rare, but unique to a particular class. For example, SORLIP3, which contains the elements of the G-box separated by two adenines (CACAAGTGA) is found in just over 4% of early- or late-induced promoters (21 elements in all [Table I]) but is not found in any of the phyA-repressed promoters.
Elements Are Differentially Distributed According to the Functional Class of the Gene Product
The data of Tepperman et al. (2001) indicate that genes encoding transcription factors predominate among the transcripts that respond the most rapidly to light signals. The distribution of elements among the promoters of the functional classes of genes described by Tepperman et al. was investigated, and the results are given in Figure 5. These functional categories were defined by Tepperman et al. to differentiate genes likely to be involved in different aspects of the changes associated with photomorphogenic development. The individual elements and where they are contained in each promoter are listed in the supplemental data, available in the online version of this article at http://www.plantphysiol.org.
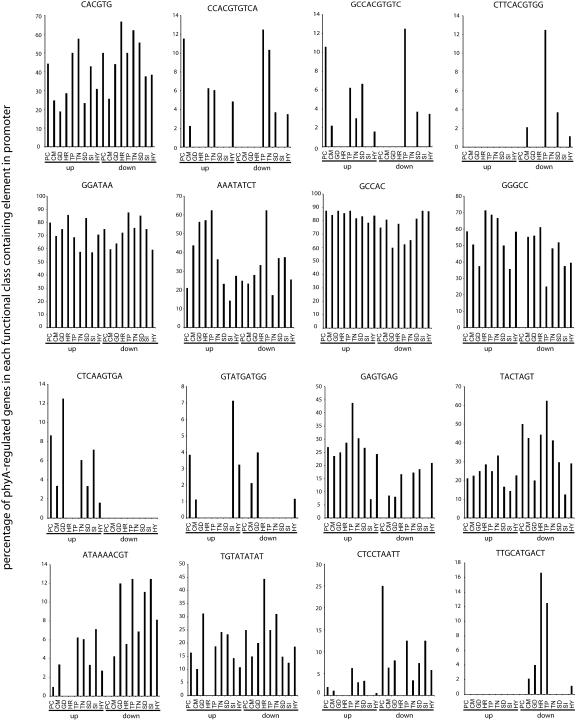
Figure 5.
Distribution of motifs by functional class of the downstream gene. The graphs each show the percentage of phyA-regulated genes in a given functional category containing an exact sequence motif in the promoter. The genes are grouped by the far-red-light response (induced [up] or repressed [down]) and by the functional category of the gene product. Functional categories are: PC, photosynthesis and chloroplast; CM, cellular metabolism related; GD, growth and development; HR, hormone related; TP, transmembrane transporters; TN, transcription; SD, stress and defense; SI, signaling; HY, hypothetical or unknown function (categories as defined by Tepperman et al. [2001]).
For some elements, this approach does not yield easily interpretable results, because the elements are rare and so the graph is noisy when the elements are spread into so many categories (an extreme example is the “repressed” G-box consensus, CCACGTGAAG, which is present in only four promoters, exactly one of each of the functional categories in which it is found in Fig. 5). In other cases, such as the GGATAA element, the abundance of the element (it occurs over 27,000 times in promoter regions of the genome; Table I) means that it is present in a large percentage of promoters of every functional category, although it is more abundant in induced than repressed promoters.
For some elements, however, the functional category distribution may give information about their in vivo function. The evening element AAATATCT has an intriguing distribution, being extremely common in the promoters of genes in the cellular metabolism, growth and development, hormone-related, and transporter categories of phyA-induced genes and less common in the promoters of photosynthesis-related genes and genes encoding transcription factors. This relationship is close to the inverse of that observed for the G-box, CACGTG, which is more common in the promoters of phyA-induced transcription factor genes than in any other functional category and is also relatively common in photosynthesis-related, phyA-induced genes. This pattern does not hold for phyA-repressed genes, where the G-box is common in most categories. However, the G-box is highly abundant in phyA-repressed transcription factor genes, whereas the evening element is unusually rare in promoters of this functional class. The G-boxes with perfectly conserved flanking regions may be too rare for this distribution to be meaningful.
DISCUSSION
We have identified new DNA sequence elements that are conserved between promoters regulated by the phyA pathway. The tool we used was designed specifically to allow us to analyze large numbers of coregulated promoters, identified using the large, oligonucleotide microarrays currently available. It may be of value to other researchers using microarrays to identify cis-regulatory elements, and for that reason, we provide the Perl script and the data files at http://www.pgec.usda.gov/Quail/Hudson-promoter/. Using appropriate data files, this approach could be applied to any organism where microarray data and upstream sequence information are available.
Using this tool, we have identified a number of totally conserved elements that are enriched in circadian-regulated promoters to a statistically significant level. These elements are disproportionately common (i.e. present in a larger number of promoter regions) with respect to the promoters of expressed, non-circadian-regulated genes. These motifs therefore do not represent constitutive, ubiquitous promoter elements, because they are not present in all promoters and are significantly more common in those promoters regulated by circadian rhythms. The fact that the most significant hits are sequences previously known to be important for circadian transcriptional regulation demonstrates that our approach can provide biologically relevant motif data from coregulated sets of promoters.
We have identified a number of conserved elements upstream of the phyA-induced and -repressed genes, and we believe these elements are likely to be involved in the function of phyA-regulated promoters, because they are enriched with respect to the background set of expressed gene promoters. Many of these elements have been described previously. Not only are GATA/I-box and G-box well established (Terzaghi and Cashmore, 1995) and DE1 also previously described (Inaba et al., 2000), but the element we refer to as SORLIP 1 has been previously interpreted as an inverted Box II-like motif in the Arabidopsis GAPB promoter (Arguello-Astorga and Herrera-Estrella, 1996). We believe, however, that it is not part of the same transcriptional system as the G-box, because to our knowledge, few basic-helix-loop-helix (bHLH) transcription factors would bind an element divergent from the E-box consensus CANNTG (Toledo-Oritz et al., 2003). Nor does it contain the b-zip core motif ACGT, required for binding HY5, for example (Chattopadhyay et al., 1998). Its strong conservation and abundance in the phyA-regulated promoters described here lead us to believe that it is not an irregular Box II element but rather a separate entity. Of the remaining elements described, none directly resemble known light-inducible elements, but SORLIP 2 resembles the RE1 element (Bruce et al., 1991), and SORLIP 5 strongly resembles the maize (Zea mays) endosperm box (Lohmer et al., 1991). Although the endosperm box and RE1 elements have not been previously shown to have a role in light-induced transcription, they may be involved in mediating responses downstream of phyA, perhaps in combination with other elements.
Because the elements we describe here are not in any case unique to the promoters in which they are over-represented, none of them can be considered to be diagnostic indicators of the ability of a promoter sequence to respond transcriptionally to a particular environmental stimulus. However, we do believe that we have shown that over-representation of sequence motifs is a useful tool for the discovery of new regulatory pathways, given large sets of coregulated genes. Our results suggest that the presence of a motif in the 5′-untranscribed region of a gene is not in itself sufficient to confer strong transcriptional responses on all of the genes sharing such a motif. The distinctive characteristics of strongly responding promoters are probably both the presence of conserved flanking regions to cis-regulatory elements and conserved combinations of different promoter elements.
The evening element is known to be involved in circadian regulation of transcription (Harmer et al., 2000), and the G-box known to be important for phytochrome-regulated transcriptional induction (Menkens et al., 1995) as well as stress and defense responses (e.g. Arias et al., 1993). The work of Michael and McClung (2003) has linked the G-box with circadian-regulated genes in plants, and the related E-box core element (CANNTG), frequently in full G-box form, is known to be important in mammalian circadian promoters (Darlington et al., 1998; Gekakis et al., 1998; Hogenesch et al., 1998). The distribution of the G-box and the evening element between the promoters of different functional classes of genes is very intriguing. This may be an indication that these two elements control distinct subsets of genes by function, possibly due to differing requirements at different times in the day/night cycle.
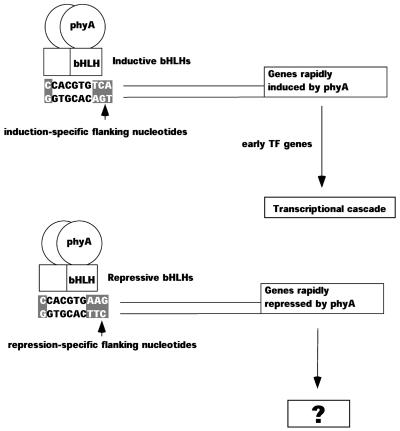
To our knowledge, over-representation of G-box sequences in light-repressed genes has not been reported previously. It has been previously established that alteration of flanking sequences around the CACGTG core of the G-box can markedly affect the transcriptional behavior of the downstream gene (Salinas et al., 1992). It is shown in Figure 3 and Table I that there is a highly significant enrichment of G-boxes in phyA-repressed promoter sequences. The G-boxes of phyA-repressed promoters show different conserved flanking nucleotides than the G-boxes of phyA-induced promoters. The different flanking sequence between the conserved G-box motifs of light-induced and light-repressed genes may specify which of these two opposing transcriptional responses to light occurs in a given promoter (Fig. 6).
Figure 6.
Distinct conserved flanking nucleotides of the G-boxes in phyA-induced and -repressed promoters may indicate a branch point in a transcriptional cascade. G-boxes are common in the promoters of the “first wave” of phyA-responsive genes. The active form of phyA is known to bind to bHLH proteins, which then bind G-box sequences. Genes encoding transcriptional regulators are prominent among these early-responding genes. The G-box/bHLH system is therefore correlated with both rapid induction and rapid repression of transcriptional regulators, making it the first step in a transcriptional cascade. Induction or repression is specified according to the flanking nucleotides around the CACGTG core. The consensus sequence of the G-boxes in phyA-induced promoters is CCACGTGTCA. The repressed promoters may be bound by yet to be discovered bHLH repressor proteins, specific to the conserved nucleotides CCACGTGAAG. The downstream events from the rapidly phyA-repressed transcriptional regulators remain to be characterized.
Importantly, the G-box is very abundant in the most rapidly responding, phyA-induced genes. It is less abundant in the promoters of genes that respond more slowly (Fig. 4). The G-box core sequence is also abundant in phyA-repressed promoters, which may be rapidly responsive, although the time resolution of our measurement of this response is limited by the half-life of the relevant transcripts. The G-box is disproportionately common in the promoters of genes that encode transcription factors, in both phyA-induced and phyA-repressed promoters (Fig. 5). The combination of these two observations is strong evidence that the G-box mediated responses are upstream in a transcriptional cascade leading to transcriptional regulation of genes with other elements. In other words, G-box-regulated genes are responding to far-red light after fewer intervening steps. This fits the observation that the transcription factor PIF3 can bind directly to phytochrome in a photoreversible fashion while bound to a DNA duplex with a G-box sequence (Martinez-Garcia et al., 2000).
Changes in expression of rapidly responding transcripts, particularly those encoding transcription factors, may be a prerequisite for the more global changes in gene expression that happen downstream. The predominance of G-boxes in the promoters of these genes indicates that the G-box, and proteins that bind it, may be involved in these critical early steps. This is illustrated in Figure 6. Rapidly responding genes, encoding transcriptional regulators, may be induced or repressed by phytochrome according to the specificity of different bHLH DNA-binding proteins. More than one bHLH has been shown to be involved in phytochrome signaling (e.g. PIF3 [Ni et al., 1999], HFR1 [Fairchild et al., 2000] and PIF4 [Huq and Quail, 2002]). Yet more members of the large family of bHLHs in Arabidopsis may be involved combinatorially in phytochrome-regulated gene expression (Toledo-Oritz et al., 2003). Sub-families of bHLH proteins within this large gene family are likely to have different specificities for the flanking nucleotides around the G-box core sequence. The products of this rapidly responding “first wave” of transcription factor genes may then go on to produce the later, genomic-scale changes in transcription observed by Tepperman et al. (2001). These downstream transcription factors are likely to bind the GATA element, SORLIPs and SORLREPs, and other cis-regulatory motifs in the downstream promoters. The amplification of the cellular signal by this transcriptional cascade could be sufficient to explain the global developmental changes that result from a conformational change in a photoreceptor such as phyA.
MATERIALS AND METHODS
Data Sources
The gene lists used for the analysis of phyA-regulated promoters, derived from the data of Tepperman et al. (2001) are available at http://www.pgec.usda.gov/Quail/phyB_tables/downloads/Table.1.xls.hqx. The circadian-regulated Arabidopsis gene lists of Harmer et al. (2000) are available at http://www.sciencemag.org/feature/data/1055592files/1055592.shl. Both of these data sets were obtained using the original Affymetrix Arabidopsis genome chip, an oligonucleotide array containing sequences representing approximately 8,200 genes, described by Zhu and Wang (2000). In the case of Tepperman et al. (2001), the coregulated gene list comprises genes with transcripts that respond 2-fold or more to far-red light, in wild type but not in the phyA mutant. This was determined at one or more time points across a 24-h time course of RNA samples from dark-grown Arabidopsis seedlings exposed to far-red light from time zero. In the case of Harmer et al. (2000), the coregulated genes have transcripts with expression patterns that show a 0.95 or higher Pearson correlation coefficient to one of several thousand computer-generated cosine curves. This was determined across a detrended 44-h time course of RNA samples from light-grown plants entrained under circadian conditions.
The analysis shown, including all of that in Table I, was performed using the total genomic, upstream, downstream, intergenic, intron, and coding sequences available by ftp download, accessible from The Arabidopsis Information Resource (http://www.arabidopsis.org).
Sequence Analysis
The coregulated gene clusters used to recover sets of coregulated promoters were identified by Tepperman et al. (2001) or by Harmer et al. (2000). Transcriptional start sites in Arabidopsis are currently not well annotated, so annotated translation start sites were used to define the start of “promoter” regions of 2 kb of 5′ sequence. The approach uses an exact pattern-match to enumerate each one-through 10-mer in the coregulated set of promoters. For each motif in the coregulated promoter sequences, the number of occurrences of that motif was compared with an expected value derived from the frequency of that element in the sequence of the promoters for the whole microarray. A version of the one-degree-of-freedom chisquared test was used to produce a value that was then compared with a table of critical values.
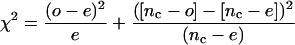 |
1 |
where o is the total number of elements in the promoters of the gene cluster, ob is the total number of elements in the baseline data (all of the promoters for the microarray), nc is the number of 2-kb promoters of the gene cluster, and nb is number of 2-kb promoters in the baseline data.
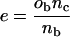 |
2 |
The preliminary chi-squared step provides a few-hundred candidate sequences for the next step. This step is to compare the number of promoters with each motif, in the coregulated promoter subset of interest, to the number with the same motif in the set of all of the promoters for genes on the microarray. The list of over-represented DNA oligomers with P < 0.001 (after a Bonferroni correction for multiple testing) from the chi-squared test is fed into the secondary analysis. The promoters in the coregulated subset and for all of the genes on the microarray are each evaluated for the presence or absence of the search motif, again by an exact match. The probability of the element being present in the number of promoters in the query set by random sampling of the promoters on the microarray is estimated using the binomial distribution.
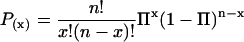 |
3 |
where x is the number of 2-kb promoters in the gene cluster positive for the element, n is the number of promoters in the gene cluster and the probability Π of each promoter containing one or more elements is approximated by dividing the number of promoters positive for the element on the entire microarray xb by the number of genes on the microarray nb.
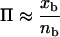 |
4 |
Elements meeting the critical P value required were aligned using the Multalign algorithm (Corpet, 1988) with manual curation, and the assignment of sections of the alignment to individual motifs was performed manually. Correlation with previously described cis-regulatory elements was performed using plant CARE (https://oberon.rug.ac.be:8443/care-bin/qfm_querycare.htpl), PLACE (http://www.dna.affrc.go.jp/htdocs/PLACE/), and literature searches. Source code and other files required for this analysis are available at http://www.pgec.usda.gov/Quail/Hudson-promoter/.
Acknowledgments
We thank Jim Tepperman for valuable discussions of gene lists affected by far-red light and assistance with the submission process, Stacey Harmer for further information on circadian-regulated genes, and Nick Kaplinsky for helpful discussions.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.030437.
This work was supported by the National Institutes of Health (grant no. GM47475), by the U.S. Department of Agriculture-Agricultural Research Service Current Research Information Service (grant no. 5335–2100–017–00D), and by Torrey Mesa Research Institute (San Diego).
The online version of this article contains Web-only data.
References
- Arguello-Astorga GR, Herrera-Estrella LR (1996) Ancestral multipartite units in light-responsive plant promoters have structural features correlating with specific phototransduction pathways. Plant Physiol 112: 1151-1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello-Astorga GR, Herrera-Estrella LR (1998) Evolution of light-regulated plant promoters. Annu Rev Plant Physiol Plant Mol Biol 49: 525-555 [DOI] [PubMed] [Google Scholar]
- Arias JA, Dixon RA, Lamb CJ (1993) Dissection of the functional architecture of a plant defense gene promoter using a homologous in vitro transcription initiation system. Plant Cell 5: 485-496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce WB, Deng XW, Quail PH (1991) A negatively acting DNA sequence element mediates phytochrome-directed repression of phyA gene transcription. EMBO J 10: 3015-3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang L-H, Puente P, Deng X-W, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673-683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881-10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA (1998) Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280: 1599-1603 [DOI] [PubMed] [Google Scholar]
- Dehesh K, Franci C, Sharrock RA, Somers DE, Welsch JA, Quail PH (1994) The Arabidopsis phytochrome A gene has multiple transcription start sites and a promoter sequence motif homologous to the repressor element of monocot phytochrome A genes. Photochem Photobiol 59: 379-384 [DOI] [PubMed] [Google Scholar]
- Futcher B (2002) Transcriptional regulatory networks and the yeast cell cycle. Curr Opin Cell Biol 14: 676-683 [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280: 1564-1569 [DOI] [PubMed] [Google Scholar]
- Giuliano G, Pechersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR (1988) An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA 85: 7089-7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob U, Stuber K (1987) Discrimination of phytochrome dependent light inducible from non-light inducible plant genes: prediction of a common light-responsive element (LRE) in phytochrome dependent light inducible plant genes. Nucleic Acids Res 15: 9957-9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110-2113 [DOI] [PubMed] [Google Scholar]
- Hogenesch JB, Gu YZ, Jain S, Bradfield CA (1998) The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci USA 95: 5474-5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulzink RJM, Weerdesteyn H, Croes AF, Gerats T, Antonius van Herpen MM, van Helden J (2003) In silico identification of putative regulatory sequence elements in the 5′-untranslated region of genes that are expressed during male gametogenesis. Plant Physiol 132: 75-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441-2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14: 2377-2391 [PMC free article] [PubMed] [Google Scholar]
- Inaba T, Nagano Y, Reid JB, Sasaki Y (2000) DE1: a 12 bp cis-regulatory element sufficient to confer dark-inducible and light down-regulated expression to a minimal promoter in pea. J Biol Chem 275: 19723-19727 [DOI] [PubMed] [Google Scholar]
- Inaba T, Nagano Y, Sakakibara T, Sasaki Y (1999) Identification of a cis-regulatory element involved in phytochrome down-regulated expression of the pea small GTPase gene pra2. Plant Physiol 120: 491-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Knudsen S (2000) Automatic discovery of regulatory patterns in promoter regions based on whole cell expression data and functional annotation. Bioinformatics 16: 326-333 [DOI] [PubMed] [Google Scholar]
- Lohmer S, Maddaloni M, Motto M, Di Fonzo N, Hartings H, Salamini F, Thompson RD (1991) The maize regulatory locus Opaque-2 encodes a DNA-binding protein which activates the transcription of the b-32 gene. EMBO J 10: 617-624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859-863 [DOI] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Cashmore AR (1995) The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci 20: 506-510 [DOI] [PubMed] [Google Scholar]
- Michael TP, McClung CR (2003) Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol 132: 629-639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1999) Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400: 781-784 [DOI] [PubMed] [Google Scholar]
- Ohler U, Niemann H (2001) Identification and analysis of eukaryotic promoters: recent computational approaches. Trends Genet 17: 56-60 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983-2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847-857 [DOI] [PubMed] [Google Scholar]
- Roth FP, Hughes JD, Estep PW, Church GM (1998) Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol 16: 939-945 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519-1521 [DOI] [PubMed] [Google Scholar]
- Salinas J, Oeda K, Chua NH (1992) Two G-box-related sequences confer different expression patterns in transgenic tobacco. Plant Cell 4: 1485-1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H (2000) Phytochromes and light signal perception by plants: an emerging synthesis. Nature 407: 585-591 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035-1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98: 9437-9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR (1995) Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol 46: 445-474 [Google Scholar]
- Thijs G, Marchal K, Lescot M, Rombauts S, De Moor B, Rouze P, Moreau Y (2002) A gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J Comput Biol 9: 447-464 [DOI] [PubMed] [Google Scholar]
- Toledo-Oritz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749-1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J, Andre B, Collado-Vides J (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J Mol Biol 281: 827-842 [DOI] [PubMed] [Google Scholar]
- Vanet A, Marsan L, Labigne A, Sagot MF (2000) Inferring regulatory elements from a whole genome. An analysis of Helicobacter pylori sigma(80) family of promoter signals. J Mol Biol 297: 335-353 [DOI] [PubMed] [Google Scholar]
- Zhu T, Wang X (2000) Large-scale profiling of the Arabidopsis transcriptome. Plant Physiol 124: 1472-1476 [DOI] [PMC free article] [PubMed] [Google Scholar]