Abstract
Plants respond to the proximity of neighboring vegetation by elongating to prevent shading. Red-depleted light reflected from neighboring vegetation triggers a shade avoidance response leading to a dramatic change in plant architecture. These changes in light quality are detected by the phytochrome family of photoreceptors. We analyzed global changes in gene expression over time in wild-type, phyB mutant, and phyA phyB double mutant seedlings of Arabidopsis in response to simulated shade. Using pattern fitting software, we identified 301 genes as shade responsive with patterns of expression corresponding to one of various physiological response modes. A requirement for a consistent pattern of expression across 12 chips in this way allowed more subtle changes in gene expression to be considered meaningful. A number of previously characterized genes involved in light and hormone signaling were identified as shade responsive, as well as several putative, novel shade-specific signal transduction factors. In addition, changes in expression of genes in a range of pathways associated with elongation growth and stress responses were observed. The majority of shade-responsive genes demonstrated antagonistic regulation by phyA and phyB in response to shade following the pattern of many physiological responses. An analysis of promoter elements of genes regulated in this way identified conserved promoter motifs potentially important in shade regulation.
Competition for sunlight is one of the most important aspects regulating plant development. Plants can detect the presence of neighboring vegetation by sensing changes in the quality of the light reflected from these neighbors. In shade-avoiding species, a pronounced elongation response is triggered, altering the whole architecture of the plant, and in response to prolonged vegetative shading, flowering is dramatically accelerated (Smith and Whitelam, 1997). In seedlings established in an open canopy, the shade avoidance response is vital to prevent overtopping by competitors growing alongside. However, this response can be detrimental in agriculture because it limits planting density. If a shade avoidance response is induced, this reallocation of resources into elongation growth often results in a decrease in crop yield from seeds or storage organs (Ballaré et al., 1997).
Light reflected from neighboring vegetation is depleted in red (R) wavelengths because of absorption of these wavelengths by chlorophyll but remains rich in far-red (FR) light. This change in light quality is detected by the phytochrome photoreceptors: R/FR reversible photochromic pigments (Quail, 1998). Phytochrome exists in two photo-interconvertible forms, an inactive, R-absorbing Pr form and an active, FR-absorbing Pfr form. A dynamic equilibrium is established between the two forms corresponding to the R to FR ratio (R:FR) of light incident upon the plant (Smith and Holmes, 1977). A high R:FR is characteristic of open canopies and causes the equilibrium to favor the Pfr form of phytochrome. This active Pfr form acts to suppress elongation growth and flowering, resulting in a normal robust growth habit (Whitelam and Devlin, 1997). Conversely, low R:FR light reflected from neighboring vegetation pushes the phytochrome photo-equilibrium toward Pr, removing the Pfr-mediated inhibition of elongation growth and triggering the classical shade avoidance response. Small changes in elongation growth in response to shade have been observed in Sinapis alba within a few minutes using a transducer (Smith, 1994), demonstrating that this change in phytochrome photo-equilibrium is very rapidly translated into a change in the growth of the plant.
The phytochrome photoreceptors mediating the shade avoidance response have been well characterized (Quail, 1998). Five phytochromes exist in Arabidopsis, phyA to phyE, the products of a divergent gene family. PhyA is light labile, the Pfr form being rapidly degraded. PhyB to phyE are light-stable phytochromes. PhyA is the predominant phytochrome in etiolated seedlings, where it accumulates to high levels. It acts as an antenna, being very sensitive to low levels of light, and can initiate de-etiolation in response to the briefest flash of light (Johnson et al., 1994). However, the labile nature of phyA Pfr means that phyA is rapidly degraded in white light to much lower levels. Arabidopsis phyA mutants, therefore, appear wild type in bright-white light (Whitelam et al., 1993).
PhyB predominates in light-grown seedlings. phyB mutant seedlings of Arabidopsis seedlings fail to properly de-etiolate in white light and display a constitutive shade-avoiding phenotype characterized by an elongated hypocotyl, elongated petioles, and accelerated flowering (Reed et al., 1993). Furthermore, the phyB mutant shows a greatly reduced response to a reduction in R:FR. Thus, phyB is implicated as the major photoreceptor involved in the control of elongation growth in light-grown plants and the major phytochrome acting in the shade avoidance response. (The absence of any phyB Pfr in the phyB mutant has an equivalent effect to the removal of Pfr by low R:FR light.)
Therefore, in seedling establishment, phyA and phyB have contrasting effects in response to FR-rich light. PhyA interprets FR as a de-etiolation signal, whereas phyB interprets FR as a signal of shade. This antagonism between phyA and phyB can be seen clearly in etiolated seedlings that have been transferred to low R:FR light, where white light has been enriched with supplementary FR. Although phyA mutant seedlings of Arabidopsis show a clear promotion of hypocotyl elongation, in response to 24-h low R:FR treatment, wild-type seedlings show no response in this time period. The high levels of functional phyA in etiolated wild-type seedlings acts to completely counteract the shade avoidance response under these conditions (Johnson et al., 1994). A shade avoidance response can be observed in these wild-type seedlings after 5 d of low R:FR treatment. Nonetheless, phyA mutant seedlings still show an enhanced response in comparison, indicating that the role of phyA in moderating the shade avoidance response persists in de-etiolated seedlings (Johnson et al., 1994).
PhyD and phyE have also been shown to act in shade avoidance. Double and triple mutant studies have revealed roles for both phyD and phyE acting redundantly with phyB (Devlin et al., 1998,). Although phyD or phyE mutants alone show little phenotype, the phyB phyD or phyB phyE double mutants constitutively show a more extreme elongated phenotype than the phyB monogenic and show an even more attenuated response to a reduction in R:FR. Thus, phyB is the major player in shade avoidance, with minor roles for phyA, phyD, and phyE.
Although the photoreceptors involved in shade avoidance have been well characterized, little is known of the signaling events downstream or of the extent of changes occurring at the molecular level as a result of shade avoidance. A number of phytochrome-interacting factors, including one transcription factor, have been isolated recently, but no clear role for these factors has been demonstrated in shade avoidance (Ni et al., 1998; Choi et al., 1999; Fankhauser et al., 1999). Considering the dramatic changes in development resulting in shade avoidance, a large number of steps in signal transduction are predicted beyond the genes that form the direct targets of phytochrome signaling. Thus far in Arabidopsis, the only gene that has been demonstrated to show a change in transcript abundance in response to a change R:FR is HAT4 (ATHB2), a homeodomain-Leu zipper gene that shows a very rapid increase in transcript abundance in response to a reduction in R:FR. HAT4 has been demonstrated to be induced by just 1 h of simulated shade (Carabelli et al., 1993). Overexpression of HAT4 has been demonstrated to result in a constitutively shade-avoiding phenotype in Arabidopsis, demonstrating that this transcription factor plays an important role in the shade avoidance response (Schena et al., 1993).
Here, we have used DNA microarrays to analyze global changes in gene expression in response to simulated vegetative shade in wild-type and phytochrome mutant seedlings. One-week-old light-grown seedlings of Arabidopsis show a pronounced hypocotyl elongation response after just 24 h of simulated shade. These conditions formed the basis for our analysis. We demonstrate that under these conditions, phyB plays a major role as the photoreceptor mediating this promotion of hypocotyl elongation in response to shade, whereas phyA acts to moderate this response. We analyzed changes in gene expression in wild-type, phyB mutant, and phyA phyB double mutant seedlings under these conditions. We collected tissue at both 1- and 24-h time points to examine both early and late changes in gene expression in response to simulated shade.
RESULTS
Elongation Growth Response of 7-d-Old Seedlings of Arabidopsis to 24 h of Simulated Shade
Wild-type, phyB mutant, and phyA phyB double mutant seedlings of Arabidopsis were grown for 7 d in constant white light (at which point they were producing their first true leaves) before being transferred to experimental treatments. Seedlings were then either maintained in white light (high R:FR) or transferred to white light supplemented with FR (low R:FR). Hypocotyl length was measured after 24 h in these treatments.
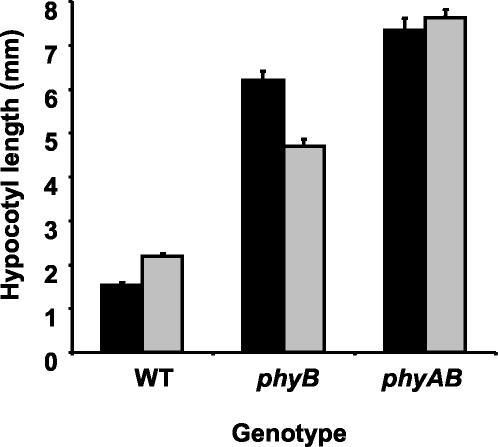
Wild-type seedlings of Arabidopsis transferred to 24-h low R:FR conditions displayed elongated hypocotyls relative to those maintained in high R:FR, evidence of a strong shade avoidance response (Fig. 1). Under normal, high R:FR conditions, phyB mutant seedlings show an elongated hypocotyl phenotype relative to wild-type seedlings. Transfer of phyB mutant seedlings to low R:FR conditions resulted in a clear inhibition of hypocotyl elongation, the reverse of the normal shade avoidance response (Fig. 1). Although phyA phyB double mutant seedlings maintained in high R:FR also displayed an elongated hypocotyl phenotype relative to wild type in seedlings under the same high R:FR conditions, the phyA phyB double mutant seedlings demonstrated no response to low R:FR (Fig. 1).
Figure 1.
Effect of simulated shade on hypocotyl length. Changes in hypocotyl length in response to simulated vegetative shade were analyzed in wild-type (WT), phytochrome B mutant (phyB), and phytochrome A phytochrome B double mutant (phyAB) seedlings. Seven-day-old white light-grown seedlings were either maintained in white light (solid bars) or transferred to white + FR light (gray bars) for 24 h, after which mean (±se) hypocotyl lengths were measured.
These data are consistent with previous finding of phyB being the major photoreceptor for the shade avoidance response, with phyA acting to moderate the degree of response.
Analysis of Gene Expression Patterns in Response to Shade
To analyze changes in gene expression in response to vegetative shade in the seedlings used above, the global gene expression profile of each of these genotypes grown in high R:FR was compared with that of seedlings grown in low R:FR. Seven-day-old wild-type, phyB mutant and phyA phyB double mutant seedlings, grown in constant light of high R:FR, were maintained in high R:FR or transferred to low R:FR conditions for 24 h. RNA was harvested after 1 and 24 h in each treatment and used to challenge Affymetrix 8K gene chips to monitor early and delayed responses to shade.
Classification of genes as shade responsive was based upon two criteria. Genes showing at least 1.5-fold change in expression level in wild-type seedlings in response to shade and showing a consistent fit across the 12 chips to a predicted pattern based upon physiological observations in the wild-type and mutant seedlings used in the experiment were categorized as shade responsive. Ten distinct patterns of expression were searched for among the genes represented on the chip.
The Pearson correlation was used to assign a P value for closeness of fit for each gene to each predicted pattern (Pavlidis and Noble, 2001). Genes that fit to a pattern with a P value < 0.01 were selected. In the rare event of a gene closely fitting to more than one pattern, the fit with the lowest P value was selected.
Genes Rapidly Differentially Regulated by Shade Solely under phyB Control
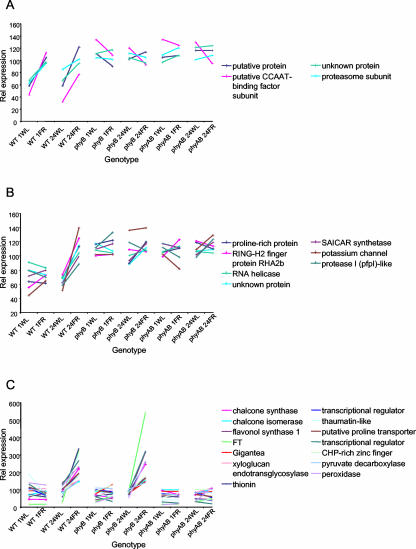
Genes rapidly up-regulated by shade under phyB control would be expected to show an increase in expression in response to both 1- and 24-h low R:FR treatments in wild-type seedlings. They would be constitutively up-regulated relative to wild type grown in white light in the phyB and phyA phyB double mutants. Conversely, genes rapidly down-regulated by shade under phyB control would show the inverse of this pattern. Genes for which the transcript levels fit to one or other of these two patterns of expression were identified by closeness of fit, across the 12 chips used in the experiment, to patterns predicted for a hypothetical gene showing such regulation (for details, see “Materials and Methods”). Genes fitting with a P value < 0.01 and showing a minimum 1.5-fold change in expression level in response to shade in wild-type seedlings were selected. Seventeen genes were classified as rapidly up-regulated by shade under phyB control using this method. Figure 2A shows those genes for which the transcript levels fit to this pattern with a P value < 0.001. Among them are the genes encoding a putative CCAAT-binding transcription factor subunit (At1g08970), and the 20S proteasome beta subunit C, PBC1 (At1g21720). Full details of all 16 genes in this list can be seen in Supplemental Table I. Nine genes were classified as rapidly down-regulated by shade under phyB control. Full details of genes in this list can also be seen in Supplemental Table II.
Figure 2.
Changes in gene expression in response to shade. Global changes in gene expression in response to simulated vegetative shade were analyzed in wild-type (WT), phytochrome B mutant (phyB), and phytochrome A phytochrome B double mutant (phyAB) seedlings. Seven-day-old white light-grown seedlings were transferred to either 1 h of white light (1WL), 1 h of white + FR light (1FR), 24 h of white light (24WL), or 24 h of white + FR light (24FR). Graphs A to C show expression patterns of transcripts matching predicted patterns of expression based upon known physiological responses with P < 0.001. Pattern fitting is described in “Materials and Methods.” Globally scaled expression values have been normalized to a mean expression of 100. A, Genes showing rapid up-regulation by shade under the control of phyB. B, Genes showing delayed up-regulation by shade under the control of phyB. C. Genes showing delayed up-regulation by shade under the control of phyA.
Genes Showing Delayed Differential Regulation by Shade Solely under phyB Control
Genes showing delayed up-regulation by shade under phyB control would be expected to show no increase in expression in response to 1-h low R:FR treatment in wild-type seedlings but to display an increase in expression in response to 24-h low R:FR treatment. They would be constitutively up-regulated relative to wild type grown in white light in the phyB and phyA phyB double mutants. Figure 2B shows those genes for which the transcript levels fit to this pattern with a P value < 0.001. Among them are the genes encoding a putative Pro-rich protein (At2g21140), the RING-H2 finger protein, RHA2b (At2g01150), an RNA helicase (At3g26560), a SAICAR synthetase (At3g21110), the potassium channel protein, AKT3 (At4g22200), the protease I (pfpI)-like protein (At2g38860). Full details of all 30 genes in this list can be seen in Supplemental Table III. Thirty-four genes were classified as showing delayed down regulation by shade under phyB control. Full details of all genes in this list can also be seen in Supplemental Table IV.
Genes Showing Differential Regulation by Shade Solely under phyA Control
Responses mediated by phyA in FR-rich light are most pronounced after prolonged irradiation (Johnson et al., 1994). Little phyA-mediated effect would be expected after only 1 h of low R:FR, but a pronounced phyA-mediated effect would be expected at the 24-h time point. Genes showing up-regulation by shade under phyA control, therefore, would be expected to show effectively zero increase in expression in response to 1 h of low R:FR treatment in wild-type seedlings but to display an increase in expression in response to 24 h low R:FR treatment. They would be expected to show a similar response in the phyB mutant. However, this response would be expected to be absent in the phyA phyB double mutant. Figure 2C shows those genes for which the transcript levels fit to this pattern with a P value < 0.001. Among them are the genes encoding thionin (At5g36910), chalcone synthase (At5g13930), FT (At1g65480), chalcone isomerase (At3g55120), flavonol synthase 1 (At5g08640), a putative Pro transporter (At2g36590), a putative transcriptional regulator (At4g04840), a putative transcriptional regulator (At4g00050), a thaumatin-like protein (At1g75040), a CHP-rich zinc finger protein (At2g19650), gigantea (At1g22770), a pyruvate decarboxylase (At5g54960), a xyloglucan endotransglycosylase (At4g30270) and peroxidase, ATP24a (At5g39580). Full details of all 36 genes in this list can be seen in Supplemental Table V. Nineteen genes were classified as showing down-regulation by shade under phyA control. Full details of all genes in this list can also be seen in Supplemental Table VI.
Genes Rapidly Differentially Regulated by Shade under phyC, D, or E Control
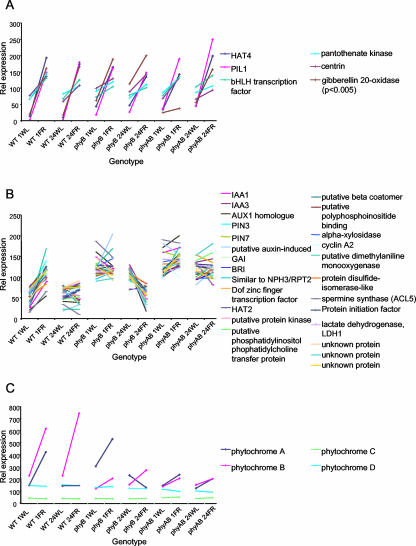
Genes rapidly up-regulated by shade under phyC, D, or E control would be expected to show an increase in expression in response to both 1 and 24 h of low R:FR treatments in wild-type, phyB mutant, and phyA phyB mutant seedlings. Eleven genes were classified as rapidly up-regulated by shade under phyC, D, or E control using this method. Figure 3A shows those genes for which the transcript levels fit to this pattern with a P value < 0.001. Among them are the genes encoding homeobox-Leu zipper, HAT4 (At4g16780), PIF3-like protein, PIL1 (At2g46970), pantothenate kinase (At1g60440), a putative bHLH transcription factor (At3g57795), and a putative centrin (At4g27280). Notably, the gene encoding GA 20-oxidase 3 (At5g07200) was found to fit to this pattern with P < 0.005. Full details of all 11 genes in this list can be seen in Supplemental Table VII. Seven genes were classified as rapidly down-regulated by shade under phyC, D, or E control. Full details of all genes in this list also can be seen in Supplemental Table VIII.
Figure 3.
Changes in gene expression in response to shade. Global changes in gene expression in response to simulated vegetative shade were analyzed in wild-type (WT), phytochrome B mutant (phyB), and phytochrome A phytochrome B double mutant (phyAB) seedlings. Seven-day-old white light-grown seedlings were transferred to either 1 h of white light (1WL), 1 h of white + FR light (1FR), 24 h of white light (24WL), or 24 h of white + FR light (24FR). Graphs A and B show expression patterns of transcripts matching predicted patterns of expression based upon known physiological responses with P < 0.001. Pattern fitting is described in “Materials and Methods.” Globally scaled expression values have been normalized to a mean expression of 100. A, Genes showing rapid up-regulation by shade under the control of phyC, D, or E. B, Genes showing rapid up-regulation by shade under the control of phyB, where phyA moderates this response in prolonged shade. C, Expression patterns for transcripts of the phytochrome genes included in the analysis.
Genes Showing Antagonistic Regulation by phyA and phyB in Response to Shade
Many physiological responses to shade are regulated primarily by phyB but are moderated by the action of phyA under prolonged low R:FR irradiation. Genes showing such regulation by shade would demonstrate a differential regulation by shade under phyB control after 1 h of low R:FR treatment, but under prolonged irradiation, an antagonistic action of phyA would be revealed, reducing this differential regulation. Genes up-regulated by shade in this way would demonstrate an up-regulation after 1 h of low R:FR treatment in wild-type seedlings but would display a reduced change in expression in response to 24 h of low R:FR treatment. In phyB mutants grown in white light, these genes would be constitutively up-regulated relative to wild type grown in white light. In the absence of phyB, these genes would show no response to 1 h of low R:FR treatment, but the moderating effect of phyA would be made apparent in response to 24 h of low R:FR treatment in that these genes would display a down-regulation after 24 h of low R:FR treatment. In phyA phyB double mutants grown in white light, these genes would be, again, constitutively up-regulated relative to wild type grown in white light. However, in the absence of both phyA and phyB, these genes would show no response to either 1 or 24 h of low R:FR treatment. Conversely, genes down-regulated by shade in this way would demonstrate the inverse of this pattern. Genes for which the transcript levels fit to one or the other of these two patterns of expression were selected as previously described. These groupings formed by far the largest groupings of shade-responsive genes. Ninety-nine genes were classified as showing up-regulation by shade under the control phyB, moderated by phyA using this method. Figure 3B shows those genes whose transcript levels fit to this pattern with a P value < 0.001. Among them are the genes encoding HAT2 (At5g47370), a putative dimethylaniline monooxygenase (At4g28720), the auxin transport protein, PIN3 (At1g70940), IAA3 (At1g04240), a putative polyphosphoinositide-binding protein (At3g51670), a putative beta coatomer (At4g31480), BRI1 (At4g39400), cyclin A2 (At1g15570), GAI (At1g14920), a putative phosphatidylinositol phophatidylcholine transfer protein (At2g21540), a putative auxin-induced protein (At4g36110), a putative protein kinase (At2g47060), IAA1 (At4g14560), a protein similar to NPH3/RPT2 (At4g37590), alpha-xylosidase (At1g68560), PIN7 (At1g23080), a dof zinc finger transcription factor (At3g61850), a protein disulfide-isomerase-like protein (At3g54960), lactate dehydrogenase, LDH1 (At4g17260), spermine synthase, ACL5 (At5g19530), an AUX1 homolog (At5g49630), and a protein initiation factor (At4g18300). Notably, the gene encoding phototropin 1, PHOT1 (At3g45780) was found to fit to this pattern with P < 0.006. Full details of all 99 genes in this list can be seen in Supplemental Table IX.
Forty-one genes were classified as showing down-regulation by shade under the control phyB, moderated by phyA. Full details of all genes in this list can also be seen in Supplemental Table X. Notably, this list includes the gene encoding transcription factor HY5 (At5g11260), previously demonstrated to be involved in light signaling.
Phytochrome A and B Are Strongly Up-Regulated by Shade
The transcript encoding phytochrome A showed a rapid up-regulation in response to shade after 1 h of low R:FR treatment in both wild-type and phyB mutant seedlings (Fig. 3C). The expression of PHYA, however, showed no increase relative to white light levels after 24 h of low R:FR treatment. The absence of any PHYA transcript in the phyA phyB double mutant meant that no PHYA expression was observed in this line. The retention of a response to shade in the phyB mutant suggests the involvement of phytochromes C, D, or E. The response to 1 but not 24 h of low R:FR treatment, however, suggests that phyA acts antagonistically to phyC, D, or E in prolonged supplementary FR.
The transcript encoding phytochrome B showed a rapid up-regulation in response to both 1 and 24 h shade in wild-type seedlings (Fig. 3C). The absence of any PHYB transcript in the phyB and phyA phyB double mutants meant that no PHYB expression was observed in this line. The two other phytochrome genes represented on the chip, PHYC and PHYD, showed no response to shade (Fig. 3C).
Confirmation of Microarray Data
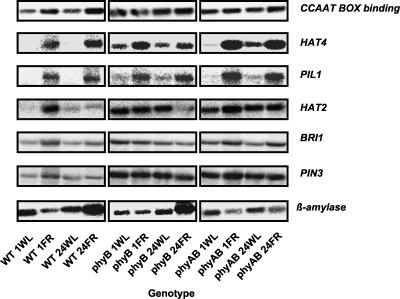
Northern blotting was used to confirm the expression profiles of a selection of genes showing a fit to one of several of the predicted patterns above. Transcript levels of the following genes were measured across the same 12 genotypes and treatments that were used in generating the chip data (Fig. 4). The transcripts encoding the putative CCAAT transcription factor (At1g08970), the homeodomain-Leu zipper transcription factor, HAT4 (At4g16780), the PIF3-like basic helix loop helix transcription factor, PIL1 (At2g46970), the homeodomain-Leu zipper transcription factor, HAT2 (At5g47370), and the Leu-rich repeat receptor kinase brassinosteroid insensitive 1, BRI1 (At4g39400), the auxin transport protein, PIN3 (At1g70940), and the transcript-encoding β-amylase (At4g17090) were tested. In each case, the transcript levels showed a close qualitative correlation to the to the microarray data (Fig. 4).
Figure 4.
Confirmation of patterns of gene expression by northern blotting. Changes in gene expression in response to simulated vegetative shade were analyzed in wild-type (WT), phytochrome B mutant (phyB), and phytochrome A phytochrome B double mutant (phyAB) seedlings. Seven-day-old white light-grown seedlings were transferred to either 1 h of white light (1WL), 1 h of white + FR light (1FR), 24 h of white light (24WL), or 24 h of white + FR light (24FR). Northern-blot analysis confirmed both the patterns of expression observed in the microarray data for the genes indicated.
Functional Analysis of Genes Differentially Regulated by Shade
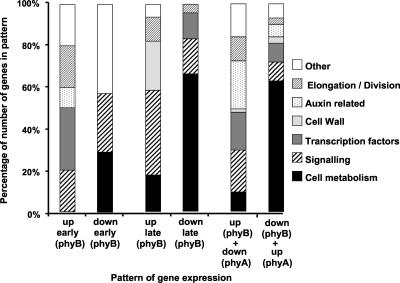
A functional analysis of genes in each of the patterns of gene expression revealed a number of noteworthy trends. Seven functional categories were used to catalogue gene function: “cell metabolism related,” “signaling molecules,” “transcription factors,” “cell wall related,” “auxin related,” “cell elongation/division related,” and “others.” Function was assigned based on the Affymetrix annotation provided by the Salk Institute Genomic Analysis Library. Those genes categorized as, simply, “expressed protein,” “hypothetical protein,” or “putative protein” were omitted from this functional analysis. For each of the patterns of gene expression, the percentage of the total number of differentially regulated genes that were allotted to each of seven functional categories was calculated. Results for patterns showing differential regulation by shade either under the control of phyB alone or where the action of phyB is moderated by phyA are shown in Figure 5.
Figure 5.
Functional analysis of genes differentially regulated by shade. Transcripts fitting to the following patterns of expression in response to shade: rapid up-regulation by shade under the control of phyB (up early [phyB]); rapid down-regulation by shade under the control of phyB (down early [phyB]); delayed up-regulation by shade under the control of phyB (up late [phyB]); delayed down-regulation by shade under the control of phyB (down late [phyB]);rapid up-regulation by shade under the control of phyB, where phyA moderates this response in prolonged shade (up [phyB] down [phyA]); and rapid down-regulation by shade under the control of phyB, where phyA moderates this response in prolonged shade (down [phyB] up [phyA]) were manually assigned to one of the indicated functional categories. Genes annotated as “unknown protein,” “hypothetical protein,” “putative protein,” or “expressed protein” were omitted from the functional classification. The graphs shows the percentage of the total number of genes fitting each pattern that were assigned to each functional category.
A significantly greater percentage of the total number of genes down-regulated by shade, as opposed to up-regulated, were associated with cell metabolism. This was true for both early and late shade-regulated patterns and for those patterns representing regulation by phyB alone or by both phyB and phyA. In particular genes associated with photosynthesis and fatty acid metabolism and redox metabolism were highly represented among those down-regulated.
A significant percentage of the total number of genes displaying rapid up-regulation in response to shade were transcription factors. Again, this was true both for patterns representing regulation by phyB alone or by both phyB and phyA.
A number of cell wall-associated genes displayed late up-regulation in response to shade under phyB control. Of these, the majority were pectinesterases and pectate lyases, associated with cell wall loosening.
It is also noteworthy that a substantial number of genes associated with cell elongation or division were up-regulated by shade. Again, this was true both for patterns representing regulation by phyB alone or by where phyA moderates the action of phyB.
A large number or auxin-related genes were found to fit to the pattern corresponding to genes up-regulated by shade under the control of phyB but where the action of phyA moderates the response under prolonged low R:FR irradiation. In particular, a number of the indole-3-acetic acid (IAA) family of auxin-regulated transcription factors were regulated in this manner. This forms the largest functional grouping of genes fitting to this pattern. Coupled with the fact that this pattern represents the majority of shade-regulated genes, this represents a significant finding.
Analysis of Promoter Elements Overrepresented among Shade-Regulated Genes
The first 500 bp of the promoters of all of the genes fitting with a P value < 0.001 to the largest shade-regulated grouping (those up-regulated by shade under the control of phyB but where the action of phyA moderates the response under prolonged low R:FR irradiation) was examined for overrepresented hexamer sequences. A number of hexamers corresponding to recognized eukaryotic transcription factor-binding motifs were identified as overrepresented, that is present at a significantly higher frequency among these sequences than among the promoter sequences of the Arabidopsis genome as a whole (for full list of overrepresented hexamers, see Supplemental Table XI). A P value was assigned to represent the significance of this overrepresentation (For details see “Materials and Methods”).
The palindromic sequence ACTAGT, shown to be involved in the control of stromelysin gene expression in mammals, was present with a frequency of 40% among this group as opposed to a frequency of 11% throughout the whole genome (P = 0.00015). The sequence TTATTA, a classical T-box, involved in regulation of genes involved in development in a range of species, though not in plants, was present with a frequency of 88% among this group as opposed to a frequency of 60% throughout the whole genome (P = 0.00206). The sequence TAATTA representing the recognized core-binding motif for homeodomain proteins was present with a frequency of 68% among this group as opposed to a frequency of 40% throughout the whole genome (P = 0.00306). Notably, this frequency rises to 94% among this group if 1,000 bp of promoter sequence is considered. The sequence TGTGGT, the mammalian AML/CB-Falpha transcription factor family recognition site, was present with a frequency of 44% among this group as opposed to a frequency of 21% throughout the whole genome (P = 0.00542). Finally, the canonical E-box cis-acting sequence CATGTG was present with a frequency of 40% among this group as opposed to a frequency of 19% throughout the whole genome (P = 0.00792). This frequency rises to 64% among this group if 1,000 bp of promoter sequence is considered.
DISCUSSION
The shade avoidance response is a major determinant in planting density in many economically important crops. Crop yield severely reduces as planting density increases with increasingly more resources being directed toward elongation growth (Ballaré et al., 1997). Likewise, in many crop species such as cabbage (Brassica oleraceo var. capitata), where a tight rosette habit is desirable, or root crops, where allocation of reserves to storage is required, acceleration of flowering due to shade avoidance is detrimental.
We analyzed changes in gene expression in response to shade in young seedlings previously established for 7 d in constant white light. Seedlings were exposed to either 1 or 24 h of constant low R:FR light before being harvested. Changes in gene expression in response to shade under these conditions were examined in seedlings of the phyB mutant and the phyA phyB double mutant and wild-type seedlings. PhyA and phyB represent the most abundant phytochromes in established seedlings, and both have been demonstrated to be involved to some degree in the shade avoidance response. A clear elongation response was seen in 7-d-old wild-type seedlings transferred to low R:FR for 24 h. Analysis of 7-d-old phyB mutant and phyA phyB double mutant seedlings confirmed that phyB acts as the major photoreceptor for this shade avoidance response, whereas phyA acts as a moderator in this response. One- and 24-h time points were chosen in an attempt to identify both early and late shade-regulated genes in these seedlings.
Seven-day-old seedlings were chosen for this assay because seedlings of this age are not yet competent to flower. In more mature seedlings grown under such conditions, prolonged low R:FR light would cause an acceleration of flowering, meaning that changes in gene expression involved in the flowering process would be detected in addition to those directly involved in the shade avoidance response. Furthermore, this made cultivation on petri dishes possible, generating very uniform growth conditions and allowing harvest of a large number of whole seedlings from which RNA was pooled.
We used pattern fitting software to search for genes responding to shade in a manner consistent with the physiological changes observed in response to shade. It was predicted that some genes would show a response mediated by phyB alone, some by phyA alone, and others by the antagonistic actions of both. One further category was searched for: those genes still showing a clear response to shade in the absence of both phyA and phyB. Genes showing at least 1.5-fold change in expression level in wild-type seedlings in response to shade and showing a consistent fit across the 12 chips to a predicted pattern based upon physiological observations (P < 0.01); were categorized as shade responsive.
Shade-responsive genes (301) were identified using these criteria fitting to one of 10 distinct, predicted patterns. This method for analysis of chip data allows more subtle changes in gene expression to be considered meaningful. Only 92 genes among the 301 fitting to these patterns of expression showed a greater than 2-fold change in expression level. The majority showed only modest changes in expression level but showed such tight fits to physiological patterns that we are confident that they are genuinely shade regulated. This is consistent with the difficulty, up until now, of identifying shade-responsive genes despite numerous efforts. The only previously identified shade-responsive gene is that encoding the HAT4 homeodomain transcription factor (Carabelli et al., 1993), a gene also identified as shade responsive using our method.
Of particular interest were genes with a putative role in signaling or transcriptional regulation that could form part of a shade-specific signal transduction pathway as opposed to those genes that are end point targets of shade avoidance in affecting plant architecture. Manipulation of such end point genes is likely to have a constitutive effect on plant architecture that could be detrimental in terms of agricultural yield. Conversely, a shade-specific signal transduction pathway component could be manipulated as a method of engineering out the shade avoidance response without affecting plant growth under non-shade conditions. Two possible approaches could be used. The most obvious approach would involve manipulation of genes that show a change in expression in response to shade, specifically under the control of phyB, C, D, or E. Reducing the response of key genes regulated in this way would be predicted to reduce shade avoidance. Alternatively, genes showing change in expression in shade specifically under the control of phyA could be manipulated to enhance the phyA-mediated moderation of shade avoidance (Robson et al., 1996).
A number of interesting genes encoding potential shade signal transduction components were identified, including a number of transcription factors, some of which already have been implicated as having a role in light signaling. The gene encoding homeobox-Leu zipper, HAT4 (Schena et al., 1993; Steindler et al., 1999) fit closely to the predicted pattern for genes showing a rapid up-regulation of expression in shade specifically under the control of phyC, D, or E. This agrees with previous observations (Franklin et al., 2003) that phyB, D, and E act redundantly to regulate HAT4 transcript levels and provides a good positive control for the current experimental results. HAT4 encodes a developmental regulator (Schena et al., 1993), and its involvement in shade avoidance has been demonstrated clearly. Plants overexpressing HAT4 show a constitutive shade-avoiding phenotype, thus demonstrating that manipulation of shade avoidance responsive genes does have the potential to control plant architecture (Steindler et al., 1999). Notably, the homeobox-Leu zipper HAT2, similar in sequence to HAT4, although not previously identified as being involved in shade signaling, also showed strong shade responsiveness, fitting closely to the predicted pattern for genes showing antagonistic regulation by phyA and phyB in response to shade.
The transcript encoding PIF3-like protein, PIL1, also fit closely to the predicted pattern for genes showing a rapid up-regulation of expression in shade specifically under the control of phyC, D, or E. PIL1 has been demonstrated to interact with TOC1, a protein playing a role both in the circadian clock and in the regulation of elongation growth by light (Makino et al., 2002; Mas et al., 2003), whereas PIF3 has been shown to interact directly with phyA and phyB (Ni et al., 1998), making PIL1 a very interesting candidate potential shade-specific signal transduction component.
The transcript encoding the DNA-binding protein, LHY (Schaffer et al., 1998), fits closely to the predicted pattern for genes showing down-regulation of expression in shade specifically under the control of phyA. LHY plays a key role in the circadian oscillator in Arabidopsis (Schaffer et al., 1998). It has been demonstrated that loss of LHY leads to early flowering, although this phenotype is specific to short days and, therefore, is not consistent with an involvement of LHY in shade avoidance (Mizoguchi et al., 2002).
Finally, the transcript encoding the bZIP protein HY5 (Oyama et al., 1997) shows a strong down regulation by shade regulated primarily under the control of phyB, but this is moderated by the action of phyA under prolonged low R:FR irradiation. The HY5 transcription factor has been demonstrated to be involved in the up-regulation of light-responsive genes in seedling establishment, and this is consistent with its down-regulation in response to shade (Chattopadhyay et al., 1998).
A number of recognized components involved in protein degradation also form potential shade signal transduction components. The genes encoding 20S proteasome beta subunit C (PBC1) and the RING-H2 finger proteins (RHA2b and RMA1, both E3 ubiquitin-protein ligases) all show strong shade responsiveness. This suggests that protein degradation may play a role in the shade avoidance response. Protein degradation has been demonstrated to play a key role in regulation of light responses during seedling establishment. In particular, a key step in maintaining skotomorphogenesis in darkness involves the degradation of HY5 via the COP9 signalosome. The COP9 signalosome has similarity to the lid of the 19S regulatory particle of the 26S proteasome (which also contains a 20S proteolytic core) and has been shown to interact with E3 ubiquitin ligases (Schwechheimer and Deng, 2001).
Two other intracellular signaling components, showing a response to shade, have been implicated previously in light signaling. The transcript encoding gigantea, a nucleoplasmically localized protein with a proposed role in phyB signaling (Huq et al., 2000), fits closely to the predicted pattern for genes showing an up-regulation of expression in shade, specifically under the control of phyA. The transcript encoding a PHOT1 photoreceptor-interacting protein essential for phototropism (Motchoulski and Liscum, 1999) fit closely to the predicted pattern for genes showing antagonistic regulation by phyA and phyB in response to shade.
One further group of interesting genes encoding potential shade-specific signal transduction components include a number of hormone-related factors. In particular, a number of other auxin-related factors also fit to the predicted pattern for genes showing antagonistic regulation by phyA and phyB in response to shade. These include the transcripts encoding two of the PIN family of auxin transport proteins (Friml et al., 2002), PIN3 and PIN7; a number of early auxin-induced transcription factors (Liscum and Reed, 2002), IAA1, IAA3, IAA5, IAA11, and IAA19; a GH3-like protein; an AUX1 homolog; and a number of other auxin-regulated proteins.
Links between light and auxin signaling have been proposed on several occasions. The shy2/iaa3 mutant was isolated a suppressor of the hy2 phytochrome chromophore-deficient mutant (Kim et al., 1996). The shy2/iaa3 monogenic mutant displays a constitutively photomorphogenic phenotype (Tian and Reed, 1999). More significantly, it was demonstrated subsequently that phytochrome is capable of phosphorylating SHY2/IAA3 (Colon-Carmona et al., 2000).
The genes encoding brassinosteoid insensitive 1 (BRI1; Nam and Li, 2002), GA insensitive (GAI; Peng et al., 1997), and GA 20-oxidase 3 (Coles et al., 1999) also show a strong shade responsiveness.
It is most likely, however, that moderation of hormone signaling by light is an downstream event in light signaling, rather than an early step in shade-specific signal transduction.
It is significant that the pattern representing the largest grouping of shade-responsive genes was indicative of genes showing antagonistic regulation by phyA and phyB in response to shade. Thus, the majority of shade-responsive genes respond in the same manner as that seen for the whole-plant gross physiological response to shade. It remains to be seen whether any of the genes in this grouping are primary transcription-regulated targets of shade signaling, in which case this antagonistic action of phyA and phyB must concur at the promoters of these genes. It would seem quite likely that phyA and phyB action can converge on the same promoter. Evidence for the convergence of phyA and phyB on a single promoter element is provided by the fact that both phyA and phyB can bind the bHLH transcription factor, PIF3, known to bind to and activate transcription via G-box elements in the promoters light-regulated genes (Martinez-Garcia et al., 2000). The antagonistic action of phyA and phyB in shade avoidance is, most likely, purely a property of the manner of light activation of the two photoreceptors, phyA being active in FR light and phyB being activated by R light and inactivated by FR light. It does not, therefore, require the proposition of different positive and negative sites of action at the promoters of genes showing antagonistic regulation. Alternatively, the possibility of separate primary transcription-regulated targets for phyA and phyB in the promoters of some genes cannot be ruled out given that some genes were shown to be solely regulated by one or other of these phytochromes.
The overrepresentation of certain recognized transcription factor-binding sites in the promoters of genes rapidly up-regulated by shade under the control of phyB, where phyA moderates this response in prolonged shade, could provide clues to key transcription factors acting specifically in early shade signaling. Two potential cis-acting sequences are of particular interest. The prevalence of the sequence, TAATTA, representing the recognized core-binding motif for homeodomain proteins (Ades and Sauer, 1994) is consistent with the documented involvement of homeodomain-containing transcription factors in the shade avoidance. The well-characterized rapidly shade up-regulated HAT4 transcription factor has been demonstrated to bind to the sequence TAATMATTAA (where M = A or C). However, HAT4 has, thus far, only been shown to act as a suppressor of gene expression. This is, therefore, not consistent with HAT4 being responsible for the shade up-regulation of genes in this category. It is, however, possible that HAT4 could also act as a positive regulator of gene expression. Alternatively, the identification of another shade-up-regulated homeodomain transcription factor, HAT2, in this study, raises the possibility that this could be responsible for the up-regulation of genes containing this TAATTA promoter sequence.
The canonical E-box CIS-acting sequence CATGTG, which represents a binding site for a number of bHLH transcription factors in mammals (Berberich et al., 1992; Yasumoto et al., 1994; Liu et al., 1997), was also overrepresented among this group of genes. The bHLH transcription factor PIF3 and the bZip transcription factor HY5 involved in light regulation have been demonstrated to be capable of activating transcription from closely related E-box elements (G-boxes, CACGTG; Chattopadhyay et al., 1998; Martinez-Garcia et al., 2000). As yet, no plant transcription factors have been demonstrated to bind to the sequence CATGTG, although it is tempting to speculate that, if this were a genuine CIS-acting element in plants, it also may be bound be a bHLH transcription factor. This is particularly exciting given the identification of a number of shade-regulated bHLH transcription factors, including the PIF3-like protein PIL1.
Another interesting finding was the rapid up-regulation of both PHYA and PHYB transcripts in response to shade. It is potentially very significant that the transcripts encoding the major photoreceptors involved in the shade avoidance response are, themselves, shade up-regulated. In particular, the shade up-regulation of PHYB transcript is intriguing because it is suggestive of a negative feedback mechanism. Such changes in PHYB transcript levels in response to shade have been observed elsewhere. Hall et al. (2002) demonstrated that expression of a firefly luciferase reporter driven by the PHYB promoter (PHYB::LUC) is dramatically increased by end-of-day FR light treatment, a treatment that also simulates shade. Furthermore, they showed that this response is under phyB control. Similarly, Somers and Quail (1995) demonstrated that phyB acts to negatively regulate its own expression in etiolated seedlings of Arabidopsis.
It must be made clear, however, that changes in transcript level are not necessarily indicative of changes in protein level. Circadian regulation of PHYB::LUC expression does not appear to be backed up by changes in the level of total phyB protein. Bognar et al. (1999) observed little variation in total phyB protein level over the circadian day. However, when the same authors analyzed expression of a translational PHYB::PHYB::LUC fusion, they demonstrated that synthesis of new phyB protein does follow the circadian pattern displayed by the PHYB::LUC fusion (Bognar et al., 1999). The authors propose that posttranslational mechanisms affecting the phyB protein mask the effects of the circadian rhythm of phyB synthesis.
If the changes in phyB transcript level observed in response to shade are translated into equally significant changes at the protein level, this would lead a suppression of the shade avoidance response. Although increased levels of phyB protein would not change the ratio of Pr to Pfr established under shade, it would lead to an increase in the absolute amount of both forms. The increase in amount of the active PfrB form would lead to an increased inhibition of elongation growth relative to that seen initially in response to shade.
In conclusion, we have identified a number of potential shade-specific signaling components. A key step, now, will be to test the effect of mutation in the genes encoding these factors. It would be expected that mutants deficient in key up-regulated signaling components would fail to show aspects of the shade avoidance response. As such, this work offers a number of potential targets that could be used to beneficially manipulate both crop plants and ornamentals to allow an increase in planting density without affecting plant architecture and flowering time.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of wild type, the phyB-1 mutants, and the phyA-201 phyB-1 double mutant Arabidopsis of the Landsberg erecta ecotype were sown on Murashige and Skoog agar plates covered with discs of Whatman No. 1 filter paper (Whatman, Clifton, NJ). Seeds were stratified in darkness at 4°C for 3 d and then placed in continuous white light (photosynthetically active radiation, 60 μmol m-2 s-1; R:FR, 8.99) at 22°C for exactly 7 d. Seedlings were then either maintained in these conditions or transferred to white light supplemented with FR light (photosynthetically active radiation, 60 μmol m-2 s-1; R:FR, 0.05). Approximately 200 seedlings of each genotype were harvested after 1 and 24 h from each treatment by scraping whole seedlings into aluminum foil packets and flash freezing in LN2.
Light Sources
White light was provided by Philips (New York) cool-white 20-W F20T12/CW tubes. Supplementary FR light was provided by two QB1310CS-670-735, LED hybrid lamps (Quantum Devices Inc., Barneveld, WI). All light measurements were made using an LI-1800 portable spectroradiometer (LI-COR, Lincoln, NE).
Measurement of Hypocotyl Length
Hypocotyl lengths were measured using Scion Image software (Scion, Frederick, MD) to analyze digital images of seedlings laid out flat on agar plates. Data represent the mean (±se) of at least 29 seedlings for each treatment.
Hybridization of Affymetrix Genome Arrays
Total RNA was prepared from the staged tissue samples using the Qiagen RNeasy Plant Mini Kit (Qiagen, Valencia, CA). Double-stranded cDNA was synthesized from 5 μg of total RNA using Superscript II reverse transcriptase (Life Technologies/Gibco-BRL, Cleveland). This cDNA was used as a template to synthesize biotin-labeled cRNA by in vitro transcription using an ENZO BioArray High Yield RNA transcript labeling kit (ENZO, Farmingdale, NY). Amplified cRNA was fragmented and hybridized to GeneChip microarrays (Affymetrix, Santa Clara, CA) overnight at 45°C. The hybridized arrays were washed and stained with biotinylated anti-streptavidin antibody and a phycoerythrin streptavidin conjugate and then scanned using a Hewlett-Packard GeneArray Scanner (Hewlett-Packard, Palo Alto, CA). Affymetrix GeneChip software was used to determine the average difference values between perfectly matched oligonucleotide probes and 1-bp mismatches for each probe set. Data were then scaled globally such that the average intensity of each microarray equaled a target intensity of 200 (Wodicka et al., 1997). The resulting hybridization intensity values reflect the abundance of a given mRNA relative to the total mRNA population and were used in all subsequent analyses.
Statistical Analysis
Pattern fitting was performed by a feature selection method known as template matching (Pavlidis and Noble, 2001). In brief, the expression pattern of each gene across all experimental conditions was compared with a template representing the pattern being sought. Each template was a binary vector of 12 values (one for each experimental condition), with a value of 0 corresponding to a low expression level and a value of 1 corresponding to a contrasting high expression level. The 12 values within each template correspond to the following experimental conditions: wild type, 1 h of white light; wild type, 1 h of white light + FR; wild type, 24 h of white light; wild type, 24 h of white light + FR; phyB, 1 h of white light; phyB, 1 h of white light + FR; phyB, 24 h of white light; phyB, 24 h of white light + FR; phyA phyB, 1 h of white light; phyA phyB, 1 h of white light + FR; phyA phyB, 24 h of white light; and phyA phyB, 24 h of white light + FR, respectively. The templates used were as follows: 010111111111 (rapid up-regulation by shade under the control of phyB), 101000000000 (rapid down-regulation by shade under the control of phyB), 000111111111 (delayed up-regulation by shade under the control of phyB), 111000000000 (delayed down-regulation by shade under the control of phyB), 000100010000 (delayed up-regulation by shade under the control of phyA), 111011101111 (delayed down-regulation by shade under the control of phyA), 010101010101 (rapid up-regulation by shade under the control of phyC, D, or E), 101010101010 (rapid down-regulation by shade under the control of phyC, D, or E), 010011101111 (rapid up-regulation by shade under the control of phyB, where phyA moderates this response in prolonged shade), and 101100010000 (rapid down-regulation by shade under the control of phyB, where phyA moderates this response in prolonged shade). The Pearson correlation coefficient of the expression profile of each gene with the template was calculated and used as a measure of the agreement of the profile with the predicted pattern. A P value was calculated for each correlation coefficient using a Student's t test, and only those genes with P values less than 0.01, mean expression value of 10 or more across the 12 chips, and a minimum of 1.5-fold change in expression level in response to shade were considered for further analysis. It is important to note that the correlation coefficient only depends on the shape of the profile, so all profiles having a pattern similar to that of the template will be given small P values regardless of fold change and expression levels.
Northern Blotting
Total RNA was extracted using RNeasy (Qiagen) according to the manufacturer's instructions, and 10 μg was analyzed by northern blot as previously described (Alabadí et al., 2001). Equal loading was confirmed by ethidium bromide staining (data not shown). The transcripts were detected by hybridizing the blots with radiolabeled cDNAs, which were obtained by reverse transcriptase-PCR using the following primers: PIL1for gagtcagtctaagccacaac, PIL1rev gcaacatcgtaggtggtctt; HAT4for tcatccgaggaatcgacgtg, HAT4rev gccgtgagatatcctcgtcg; HAT2for aacgtcgaggaagaagctca, HAT2rev acctgaccagcacaagcaac; CCAATfor tcaagcattttgggagaacc, CCAATrev ttttcctggtcaggttggtc; BRI1for gaatctatccgcttcgttgc, BRI1rev gaatgttcccggagaatgaa; PIN3for aatgaatctccggttcatcg, PIN3rev tctcagctccggtgagattt; and B-amylfor gggaccttgtggagaattga, B-amylrev tccatgcaggtgaagttgag.
Analysis of Promoter Elements Overrepresented among Shade-Regulated Genes
Stretches of sequence 500 bp upstream of the start codon were obtained for each gene encoding the group of 25 transcripts fitting with a P value < 0.001 to the pattern: rapid up-regulation by shade under the control of phyB, where phyA moderates this response in prolonged shade. Analysis for overrepresented hexamer “words” was performed using the statistical motif analysis tool available at The Arabidopsis Information Resource Web site (http://www.arabidopsis.org). The motif analysis program (originally written by Dr. Robert Ewing, Arabidopsis Functional Genomics Consortium, Carnegie Institute, Stanford, CA) compares the frequencies of 6-mer words in groups of sequences (on both strands) with the frequencies of these words in the current genomic sequence set of 27,117 sequences, also consisting of 500 bp upstream of the start codon of each gene (The Institute for Genomic Research, July 2002 release).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Supplementary Material
Acknowledgments
The authors are grateful to Dr. Marty Straume (University of Virginia Center for Biomathematical Technology, Charlottesville) for advice on pattern fitting analysis and to Dr. John Hogenesch (Genomics Institute of the Novartis Research Foundation, San Diego, CA) for help with regulatory element analysis.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.034397.
This work was supported by the National Institutes of Health (grant no. GM56006), by the Torrey Mesa Research Institute (funding to S.A.K.), by EMBO (long-term fellowship to P.F.D.), and by Pew Charitable Trusts (fellowship to M.J.Y.). This is manuscript no. 15909–CB of The Scripps Research Institute.
The online version of this article contains Web-only data.
References
- Ades SE, Sauer RT (1994) Differential DNA-binding specificity of the engrailed homeodomain: the role of residue 50. Biochemistry 33: 9187-9194 [DOI] [PubMed] [Google Scholar]
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880-883 [DOI] [PubMed] [Google Scholar]
- Ballaré CL, Scopel AL, Sánchez RA (1997) Foraging for light: photosensory ecology and agricultural implications. Plant Cell Environ 20: 820-825 [Google Scholar]
- Berberich S, Hyde-DeRuyscher N, Espenshade P, Cole M (1992) max encodes a sequence-specific DNA-binding protein and is not regulated by serum growth factors. Oncogene 7: 775-779 [PubMed] [Google Scholar]
- Bognar LK, Hall A, Adam E, Thain SC, Nagy F, Millar AJ (1999) The circadian clock controls the expression pattern of the circadian input photoreceptor, phytochrome B. Proc Natl Acad Sci USA 96: 14652-14657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M, Sessa G, Baima S, Morelli G, Ruberti I (1993) The Arabidopsis Athb-2 and -4 genes are strongly induced by far-red-rich light. Plant J 4: 469-479 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673-683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G, Yi H, Lee J, Kwon YK, Soh MS, Shin B, Luka Z, Hahn TR, Song PS (1999) Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 401: 610-613 [DOI] [PubMed] [Google Scholar]
- Coles JP, Phillips AL, Croker SJ, Garcia-Lepe R, Lewis MJ, Hedden P (1999) Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J 17: 547-556 [DOI] [PubMed] [Google Scholar]
- Colon-Carmona A, Chen DL, Yeh KC, Abel S (2000) Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol 124: 1728-1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC (1998) Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10: 1479-1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Robson PR, Patel SR, Goosey L, Sharrock RA, Whitelam GC (1999) Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol 119: 909-915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Yeh KC, Lagarias JC, Zhang H, Elich TD, Chory J (1999) PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284: 1539-1541 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Praekelt U, Stoddart WM, Billingham OE, Halliday KJ, Whitelam GC (2003) Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol 131: 1340-1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Wisniewska J, Benkova E, Mendgen K, Palme K (2002) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806-809 [DOI] [PubMed] [Google Scholar]
- Hall A, Kozma-Bognar L, Bastow RM, Nagy F, Millar AJ (2002) Distinct regulation of CAB and PHYB gene expression by similar circadian clocks. Plant J 32: 529-537 [DOI] [PubMed] [Google Scholar]
- Huq E, Tepperman JM, Quail PH (2000) GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci USA 97: 9789-9794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E, Bradley M, Harberd NP, Whitelam GC (1994) Photoresponses of light-grown phyA mutants of Arabidopsis: phytochrome A is required for the perception of daylength extensions. Plant Physiol 105: 141-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Soh MS, Kang BJ, Furuya M, Nam HG (1996) Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2. Plant J 9: 441-456 [DOI] [PubMed] [Google Scholar]
- Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387-400 [PubMed] [Google Scholar]
- Liu Y, Nifuji A, Tamura M, Wozney JM, Olson EN, Noda M (1997) Scleraxis messenger ribonucleic acid is expressed in C2C12 myoblasts and its level is down-regulated by bone morphogenetic protein-2 (BMP2). J Cell Biochem 67: 66-74 [DOI] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T (2002) The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol 43: 58-69 [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859-863 [DOI] [PubMed] [Google Scholar]
- Mas P, Alabadi D, Yanovsky MJ, Oyama T, Kay SA (2003) Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carre IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629-641 [DOI] [PubMed] [Google Scholar]
- Motchoulski A, Liscum E (1999) Arabidopsis NPH3: a NPH1 photoreceptor-interacting protein essential for phototropism. Science 286: 961-964 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203-212 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657-667 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983-2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Noble WS (2001) Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol 2: Research 0042.1–0042.15 [DOI] [PMC free article] [PubMed]
- Peng JR, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194-3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH (1998) The phytochrome family: dissection of functional roles and signalling pathways among family members. Philos Trans R Soc Lond Biol 353: 1399-1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson PRH, McCormac AC, Irvine AS, Smith H (1996) Genetic engineering of harvest index in tobacco through overexpression of a phytochrome gene. Nat Biotechnol 14: 995-998 [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219-1229 [DOI] [PubMed] [Google Scholar]
- Schena M, Lloyd AM, Davis RW (1993) The HAT4 gene of Arabidopsis encodes a developmental regulator. Genes Dev 7: 367-379 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Deng XW (2001) COP9 signalosome revisited: a novel mediator of protein degradation. Trends Cell Biol 11: 420-426 [DOI] [PubMed] [Google Scholar]
- Smith H (1994) Sensing the light environment: the functions of the phytochrome family. In RE Kendrick, GHM Kronenberg, eds, Photomorphogenesis in Plants, Ed 2, Vol 2. Kluwer, Dordrecht, The Netherlands, pp 377-416 [Google Scholar]
- Smith H, Holmes MG (1977) The function of phytochrome in the natural environment: III. Measurement and calculation of phytochrome photo-equilibrium. Photochem Photobiol 25: 547-550 [Google Scholar]
- Smith H, Whitelam GC (1997) The shade avoidance syndrome: multiple responses mediated by multiple phytochromes. Plant Cell Environ 20: 840-844 [Google Scholar]
- Somers DE, Quail PH (1995) Phytochrome-mediated light regulation of PHYA- and PHYB-GUS transgenes in Arabidopsis thaliana seedlings. Plant Physiol 107: 523-534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I (1999) Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126: 4235-4245 [DOI] [PubMed] [Google Scholar]
- Tian Q, Reed JW (1999) Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126: 711-721 [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Devlin PF (1997) Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ 20: 752-758 [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757-768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodicka L, Dong H, Mittmann M, Ho MH, Lockhart DJ (1997) Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol 15: 1359-1367 [DOI] [PubMed] [Google Scholar]
- Yasumoto K, Yokoyama K, Shibata K, Tomita Y, Shibahara S (1994) Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol Cell Biol 14: 8058-8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.