Abstract
The HFR1, a basic helix-loop-helix protein, is required for a subset of phytochrome A-mediated photoresponses in Arabidopsis. Here, we show that overexpression of the HFR1-ΔN105 mutant, which lacks the N-terminal 105 amino acids, confers exaggerated photoresponses even in darkness. Physiological analysis implied that overexpression of HFR1-ΔN105 activated constitutively a branch pathway of light signaling that mediates a subset of photomorphogenic responses, including germination, de-etiolation, gravitropic hypocotyl growth, blocking of greening, and expression of some light-regulated genes such as CAB, DRT112, PSAE, PSBL, PORA, and XTR7, without affecting the light-responsiveness of anthocyanin accumulation and expression of other light-regulated genes such as CHS and PSBS. Although the end-of-day far-red light response and petiole elongation were suppressed in the HFR1-ΔN105-overexpressing plants, flowering time was not affected by HFR1-ΔN105. In addition, the HFR1-ΔN105-overexpressing plants showed hypersensitive photoresponses in the inhibition of hypocotyl elongation, dependently on phytochrome A, FHY1, and FHY3 under FR light or phyB under R light, respectively. Moreover, our double mutant analysis suggested that the hypersensitive photoresponse is due to functional cooperation between HFR1-ΔN105 and other light-signaling components including HY5, a basic leucine zipper protein. Taken together, our results of gain-of-function approach with HFR1-ΔN105 suggest the existence of a complex and important basic helix-loop-helix protein-mediated transcriptional network controlling a branch pathway of light signaling and provide a useful framework for further genetic dissection of light-signaling network in Arabidopsis.
Plants sense light not only as an energy source but also to collect information about their surrounding environments, such as seasonal changes and proximity of neighboring plants (Kendrick and Kronenberg, 1994). Being sessile and photoautotrophs, plants have evolved sophisticated photosensory and regulatory systems to optimize their fitness in response to changing light conditions (Smith, 2000). Light affects various aspects of growth and development in higher plants throughout life cycles, from germination to flowering (Fankhauser and Chory, 1997). The early stage of seedling development clearly illustrates such light-dependent development. Seedlings grown in the dark undergo skotomorphogenesis, characterized by elongated hypocotyls and yellow, closed cotyledons. In response to light, seedlings follow the photomorphogenic developmental program; hypocotyls cease elongating, cotyledons become green and unfolded, and the seedlings become photosynthesis competent. Dramatic changes in gene expression underlie these marked developmental changes. Multiple genes encoding transcription factors are up- or down-regulated by light within 1 h after irradiation (Tepperman et al., 2001). Such changes in transcription factors are presumed to regulate the orchestrated expression of various downstream light-regulated genes, including CAB, CHS, XTR7, and DRT112 (Kuno et al., 2000; Ma et al., 2001).
A number of photoreceptors controlling light-dependent development, including red (R) and far-red (FR) light-absorbing phytochromes, blue (B) light receptors, cryptochromes, and phototropins have been characterized (Furuya, 1993; Lin, 2000). Among these, the phytochromes are the best characterized. Phytochromes exist as two photo-interconvertible forms, Pr and Pfr, depending on light conditions (Butler et al., 1959). In higher plants, phytochrome apoproteins are encoded by a small gene family, such as PHYA-E in Arabidopsis (Sharrock and Quail, 1989). Mutational and transgenic approaches have revealed that individual phytochromes have overlapping but distinct functions (Reed et al., 1994; Quail et al., 1995; Furuya and Schäfer, 1996; Whitelam and Devlin, 1997).
The downstream components of phytochrome signaling have been extensively characterized. Light-dependent posttranslational modifications and nuclear translocation of phytochromes have been implicated to play a role in phytochrome down-stream signaling (Lapko et al., 1997; Yeh and Lagarias, 1998; Kircher et al., 1999; Yamaguchi et al., 1999; Kim et al., 2002a). Several phytochrome-interacting molecules have been identified, implying that phytochrome may utilize multiple interacting partners to induce various photoresponses (Quail, 2002). Mutant screening using light-dependent seedling development has been fruitful to reveal a number of phytochrome-signaling components, including photoreceptors (Neff et al., 2000). One class of mutants includes the ones that exhibit altered photoresponses under different light conditions, unveiling light-dependent positive and negative regulators. Several of these are transcription factors, which regulate not only distinct but also overlapping subsets of photoresponses, including two basic helix-loop-helix (bHLH) proteins, HFR1 and PIF4 (Fairchild et al., 2000; Fankhauser and Chory, 2000; Soh et al., 2000; Huq and Quail, 2002); a basic leucine zipper (bZIP) protein, HY5 (Oyama et al., 1997); and an MYB protein, LAF1 (Ballesteros et al., 2001). EID1 and SPA1, phytochrome A (phyA)-dependent negative regulators, have been implicated to control protein stability in the nucleus (Dieterle et al., 2001; Hoecker et al. 1999; Hoecker and Quail, 2001). The other class of mutants revealed a group of repressors of photomorphogenesis, COP/DET/FUS. The cop/det/fus mutations confer photomorphogenic development even in the absence of light, including shortened hypocotyls, expanded cotyledons, and increased expression of light-inducible genes (Chory et al., 1989; Wei and Deng, 1996). Recent studies proposed that DET1, a nuclear protein, regulates gene expression via chromatin remodeling, which could control the accessibility of a promoter to specific transcription factors, for example (Benvenuto et al., 2002; Schroeder et al., 2002). COP1 encodes a RING finger protein with WD 40 repeats whose nuclear localization is negatively regulated by light (Deng et al., 1992; von Arnim and Deng, 1994). In darkness, COP1 interacts with and down-regulates several transcription factors that act as positive components in light signaling (Ang et al., 1998; Hardtke et al., 2000; Osterlund et al., 2000; Yamamoto et al., 1998, 2001; Seo et al., 2003). Other cop/det/fus loci encode an ubiquitin-conjugating enzyme or components of the COP9 signalosome complex, which was proposed to function in the proteasome-mediated protein degradation (Suzuki et al., 2002; Serino et al., 2003). Together, these findings led to the hypothesis that the primary mode of phytochrome signaling for seedling development involves posttranslational regulation on the nuclear transcription (Nagy and Schäfer, 2002). Despite an extensive list of phytochrome-signaling components, the molecular mechanisms by which these components mediate phytochrome downstream signaling mechanism are still poorly understood (Nagy and Schäfer, 2002). In particular, it is notable that no molecular components have been identified to mediate phytochrome-dependent germination as yet.
Previously, HFR1, a bHLH protein, was shown to be required for a subset of phyA-dependent responses (Fairchild et al., 2000; Fankhauser and Chory, 2000; Soh et al., 2000). To further explore the possible function of HFR1, we took gain-of-function approaches by constructing transgenic plants that overexpress full-length HFR1, C-terminal-lacking HFR1, or N-terminal-lacking HFR1 mutant. Although the transgenic plants overexpressing full-length HFR1 or C-terminal-lacking HFR1 were more or less similar to wild-type, transgenic overexpresser of mutant HFR1 lacking N-terminal 105 amino acids (HFR1-ΔN105) exhibited somewhat constitutive light responses in a subset of photomorphogenic responses in the dark. In addition to the constitutive photoresponse, the overexpression of HFR-ΔN105 also caused hypersensitive photoresponse in the inhibition of hypocotyl elongation. The results of double mutant analysis suggested that the hypersensitive photoresponse is due to the functional cooperation between HFR1-ΔN105 and other light-signaling positive elements such as HY5. Taken together, our results suggested the existence of complex and important bHLH protein-mediated transcriptional network, regulating a specific branch of light signaling in Arabidopsis.
RESULTS
Generation of Transgenic Lines That Overexpress HFR1 or Mutant HFR1 (HFR1-ΔN105)
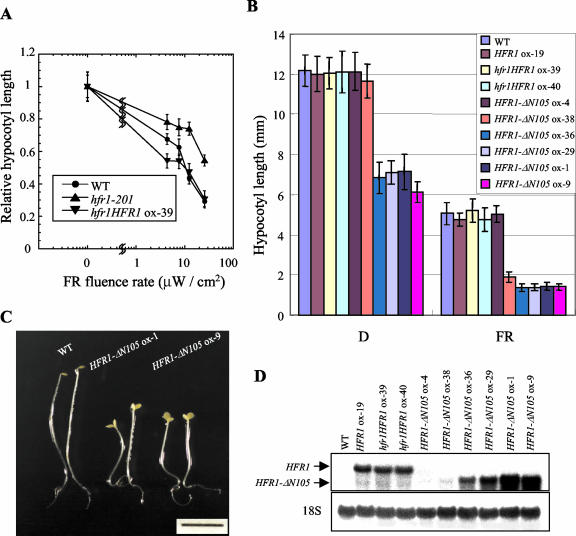
HFR1, a bHLH protein, is critical for proper photomorphogenic development in response to FR and B light (Fairchild et al., 2000; Fankhauser et al., 2000; Soh et al., 2000; Duek and Fankhauser, 2003). To further investigate the function of HFR1 in light signaling, we generated transgenic plants overexpressing full-length HFR1. When we grew the resulting independent transgenic plants overexpressing HFR1 in the hfr1-201 mutant or wild-type background, the hypocotyl phenotypes of transgenic lines were more or less similar to those of wild type (Fig. 1,A and B). Noticeably, only under low fluence rate of FR light did the transgenic plants overexpressing HFR1 show slight hypersensitivity. This suggests that the HFR1 is necessary but may not be sufficient for light signaling.
Figure 1.
Constitutive photomorphogenic phenotypes of HFR1-ΔN105-overexpressing transgenic Arabidopsis. A, Fluence rate responses of inhibition of hypocotyl elongation under FR light. The data are expressed as average relative hypocotyl length from at least 20 seedlings, normalized to their respective hypocotyl length in darkness ± sd. The average length of dark-grown seedlings was 9.8, 8.9, and 10.2 mm for wild-type, hfr1HFR1 ox-39, and hfr1-201 seedlings, respectively. Error bars = sds. WT, Wild type. B, Hypocotyl elongation phenotypes in wild-type and transgenic plants. The seedlings were grown for 4 d under FR light (21 μW cm-2) or in darkness. Each measurement was performed on at least 20 seedlings. The data are expressed as average hypocotyl length ± sd. C, Morphology of representative seedlings. The seedlings were grown for 5 d in darkness. Scale bar = 5 mm. D, RNA gel-blot analysis of wild-type and transgenic plants overexpressing full-length HFR1 or HFR1-ΔN105. Total RNA (10 μg) was loaded and subject to RNA gel-blot analysis. The blot was hybridized with a 32P-labeled HFR1-ΔN105 probe. The 18S rRNA was used as a loading control. The signals were visualized with a phosphor imager (FLA2000, Fuji, Tokyo).
Considering that HFR1 would function as a bHLH homo- or heterodimer-mediated transcriptional complex (Fairchild et al., 2000), overexpression of a subunit, full-length HFR1 may not affect significantly function of the complex. Moreover, in the case of the transcriptional regulator, it is often under various controls at posttranscriptional level (Riechmann and Ratcliffe, 2000). Deletion of a specific domain of transcriptional regulators has been effective in producing dominant negative or dominant positive phenotypes, revealing its genetic functions (Ang et al., 1998; Wang and Deng, 2002). To explore the possibilities, we conducted transgenic analysis with deletion constructs of HFR1. For the purpose, two HFR1 mutants were generated and stably introduced unto wild type: HFR1-ΔN105 lacking the N-terminal region 105 amino acids and HFR1-ΔC45 without the C-terminal region 45 amino acids, both of which have the bHLH domain (141–191 amino acids). The results showed that transgenic lines overexpressing HFR-ΔN105 showed shortened hypocotyls under both light and darkness (Fig. 1B), whereas transgenic plants overexpressing HFR1-ΔC45 did not exhibit any differences in the photoresponses, as compared with wild type (data not shown). In addition to the shortened hypocotyls, the HFR1-ΔN105 transgenic seedlings exhibited cotyledon opening/expansion and apical hook opening in darkness (Fig. 1C), implying that photomorphogenic responses were triggered in the transgenic plants even in the absence of light. The phenotypic severity of the transgenic lines appeared to correlate with expression level of HFR-ΔN105 transgene (Fig. 1, B and D). To test whether the exaggerated photoresponses observed in the HFR1-ΔN105-overexpressing lines are due to the higher levels of expression of the transgene, we compared expression levels of the full-length HFR1 and HFR1-ΔN105 transgene by RNA-blot analysis. The HFR1-ΔN105 ox-36 line exhibited shortened hypocotyls in darkness, although the level of transgene expression was lower than that of the lines expressing full-length HFR1, indicating that the exaggerated photoresponse of the HFR1-ΔN105-overexpressing lines might not be simply due to the higher level of expression of transgene. These results suggest that HFR1-ΔN105 is a gain-of-function hyperactive bHLH protein, triggering photomorphogenic responses even in darkness.
Effects of HFR1-ΔN105 Overexpression on Various Photoresponses
Phytochromes induce various photoresponses throughout development. Recent studies implicated that distinct combinations of different signaling intermediates control diverse aspects of light-dependent development (Nagy and Schäfer, 2002). For example, our recent results showed that HFR1 and HY5, a bZIP protein, might act additively to control not only overlapping but distinct subsets of the photoresponses (Kim et al., 2002b). To further understand the function of HFR1-ΔN105 in light signaling, we tested various photoresponses in the HFR1-ΔN105-overexpressing plants compared with those in the wild type.
Seed Germination
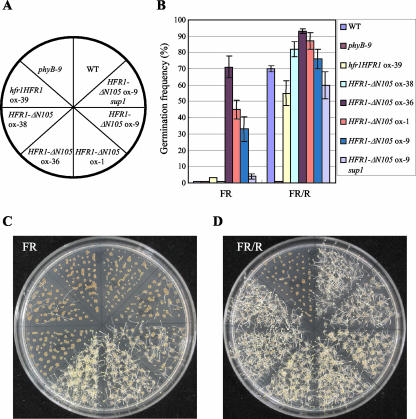
Phytochromes mediate the induction of germination by light. Treatment with a pulse of FR light just after imbibition inhibits germination, whereas subsequent irradiation with R light results in phyB-mediated seed germination (Shinomura et al., 1996). Thus, the phyB mutant seeds do not germinate even after treatment with R light. So far, other than mutations in the photoreceptor itself, no mutations that mediate phytochrome-dependent seed germination have been identified. To our surprise, the HFR1-ΔN105-overexpressing seeds germinated even after FR light treatment, suggesting that they were able to undergo light-independent germination (Fig. 2). It is notable that the transgenic lines with phytochrome-independent germination exhibited shortened hypocotyl phenotype in the dark (Fig. 1B), indicative of functional correlation between HFR1-ΔN105 and phenotypes of both germination and de-etiolation. The intragenic suppressor of HFR1-ΔN105 overexpresser (sup1) could restore the altered germination phenotype of HFR1-ΔN105 ox-9, further supporting that the constitutive germination was due to transgenic overexpression of HFR1-ΔN105. The intragenic suppressor mutation changed the Leu-173 that is highly conserved among bHLH proteins in the second helix (Heim et al., 2003) into Phe, implicating functional importance of the helix-loop-helix domain for HFR1-ΔN105. The results imply that HFR1-ΔN105 affect phytochrome signaling that leads to germination.
Figure 2.
Phytochrome-dependent germination response. A, Summary of seeds used in the germination experiment. WT, Wild type. B, Germination frequencies of wild-type or various transgenic plant lines overexpressing HFR1 or HFR1-ΔN105 seeds were measured. The seeds were treated with FR light (21 μW cm-2) for 15 min just after imbibition and then transferred to darkness without or with exposure to R light (33 μW cm-2) for 10 min. Seeds were then incubated in darkness for an additional 5 d. Each experiment was performed with at least 150 seeds. Similar results were obtained from three independent experiments. C, Representative plates from the germination experiments described in B. The plates were given a pulse of FR light and kept in darkness. D, Representative plates from the germination experiments described in B. The plates were given a pulse of R light after a pulse of FR light and then kept in further darkness.
Gravitropic Hypocotyl Growth
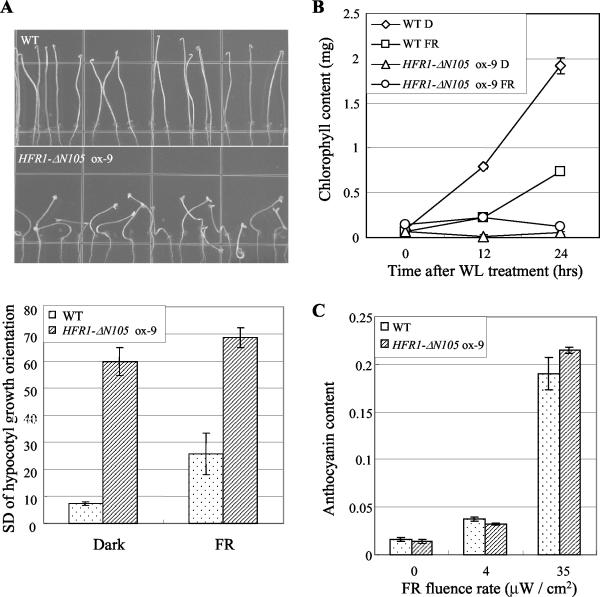
Phytochromes reduce negative gravitropism of the hypocotyls (Hangarter, 1997). The hypocotyls of wild-type seedlings grow in a randomized orientation under FR light. In contrast, hypocotyls of phyA mutants exhibit upwardly oriented growth, in a negative direction against gravity, under FR light, respectively. On the contrary, cop1 mutant seedlings show randomized hypocotyl growth even in darkness (Cao et al., 2000). The HFR1-ΔN105-overexpressing seedlings exhibited partially randomized hypocotyl growth in the dark (Fig. 3A). Thus, these results indicate that HFR1-ΔN105 may function in gravitropic hypocotyl growth.
Figure 3.
Gravitropic response of hypocotyl growth, greening response, and anthocyanin accumulation of transgenic plants overexpressing HFR1-ΔN105. A, Gravitropic response of hypocotyl growth. The seedlings were vertically grown in darkness or FR light (21 μW cm-2) for 4 d. Upper, Representative seedlings. Lower, Bars = sds of the hypocotyl growth orientations from at least 70 seedlings. The higher scores indicate more reduced gravitropism of hypocotyl growth. Error bars = ses from three independent experiments. WT, Wild type. B, FR-preconditioned blocking of greening. The seedlings were grown on Murashige and Skoog medium for 5 d in FR light (21 μW cm-2) and then irradiated with W light for indicated times. Data are expressed as average chlorophyll content (in milligrams) from 50 seedlings ± sd and were derived from three independent measurements. C, Accumulation of anthocyanin under different fluence rate of FR light. Anthocyanin measurement was performed on seedlings grown for 3 d under FR light at the indicated fluence rates. Bars = sds from three independent measurements.
Greening
Wild-type seedlings grown under FR light show reduced greening of cotyledons when exposed to W light, whereas dark-grown wild type seedlings exhibit fast and substantial accumulation of chlorophyll (Barnes et al., 1996). In contrast, cop/det/fus mutants grown in darkness show defective greening under W light (Cao et al., 2000). The HFR1-ΔN105-overexpressing plants showed reduced greening of cotyledons, even in seedlings grown in darkness (Fig. 3B), indicating that HFR1-ΔN105 affects the blocking of greening.
Anthocyanin Accumulation
Anthocyanin, a photoprotective pigment, accumulates in response to light exposure. PhyA, in particular, is a specific photoreceptor that mediates anthocyanin accumulation in response to FR light (Kunkel et al., 1996). When we measured the anthocyanin contents from seedlings grown either in darkness or under FR light, we found no obvious defects in light-dependent anthocyanin accumulation in the HFR1-ΔN105 ox-9 line, as compared with wild type (Fig. 3C). These findings suggest that HFR1-ΔN105 has no effect on the light-signaling pathways that mediate anthocyanin accumulation.
Expression of Light-Regulated Genes
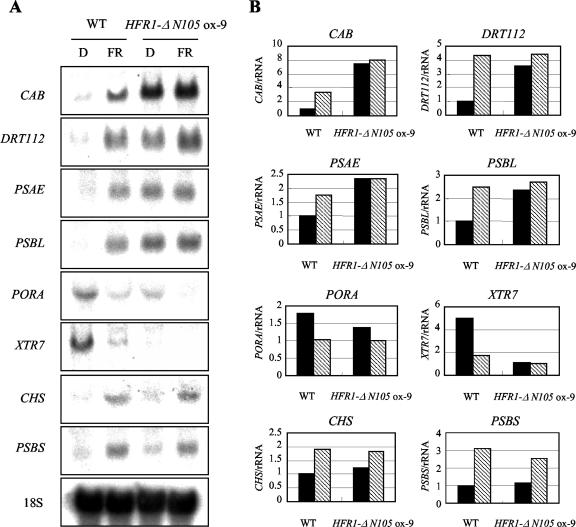
A number of genes are regulated by light, representing the molecular basis for light-dependent developmental changes (Kuno et al., 2000; Ma et al., 2001; Tepperman et al., 2001). In particular, phyA-dependent changes in gene expression in response to FR light have been useful for delineating the networks of downstream signaling components. To examine the effects of HFR1-ΔN105 on light-dependent gene expression, we used RNA gel-blot analysis to compare the effects of light on a number of light-regulated genes in transgenic and wild-type plants (Fig. 4). Compared with wild type, HFR1-ΔN105-overexpressing plants exhibited constitutive expression of CAB, PSAE, PSBL, DRT112, PORA, and XTR7. In contrast, the light-dependent expression of CHS and PSBS of HFR1-ΔN105 transgenic plants was similar to that of wild-type plants. Thus, overexpression of HFR1-ΔN105 constitutively activated a subset of genes involved in photomorphogenic responses, not affecting the expression of other subsets of light-regulated genes. These findings suggest that HFR1-ΔN105 affects a branch pathway for light-regulated gene expression.
Figure 4.
phyA-dependent gene expression of light-regulated genes in the transgenic plants overexpressing HFR1-ΔN105. A, RNA gel-blot analysis. Seedlings were grown on Murashige and Skoog-Suc medium (2% w/v) for 4 d in darkness and then transferred to FR light (14 μW cm-2) or kept in darkness (D) for an additional 12 h before extraction of total RNA. Total RNA (10 μg) was loaded and subject to RNA gel-blot analysis. The 18S rRNA was used as a loading control. Similar results were obtained in two independent experiments. WT, Wild type. B, Quantitative measurement of individual transcripts shown in A from seedlings kept in darkness (black bars) or given by 12 h of FR light (hatched bars). The values denotes relative expressions and were calculated by first normalizing each signal against 18S rRNA and then against the lowest amount of expression for each particular gene. The signals were visualized and quantified with a phosphor imager (FLA2000, Fuji). A similar trend was repeated in another independent experiment.
Photoresponses in W Light-Grown Plants
In the case of plants grown under W light, stable phytochromes, primarily phyB, mediate various photoresponses, such as hypocotyl/petiole elongation (Whitelam and Devlin, 1997). To test whether HFR1-ΔN105 affects the low fluence response, which is primarily regulated by phyB, we examined end-of-day (EOD)-FR light response. Although wild-type seedlings exhibited longer hypocotyls under EOD-FR light condition, as compared with control short-day (SD) conditions, phyB mutant plants exhibit a constitutive EOD-FR light response. The overexpression of HFR1-ΔN105 significantly suppressed hypocotyl elongation in response to EOD-FR light treatment (Fig. 5A). The results suggest that HFR1-ΔN105 ox-9 is hypersensitive to residual active phytochromes after irradiation of FR light at the EOD.
Figure 5.
Photoresponses of transgenic plants overexpressing HFR1-ΔN105 under W. A, EOD FR responses of wild-type and transgenic plants. The average hypocotyl lengths ± sds are shown from at least 20 seedlings in each group. White bars, No EOD-FR treatments; black bars, EOD-FR treatments. WT, Wild type. B, Morphology in adult wild type and transgenic plants overexpressing HFR1-ΔN105. The plants were grown for 20 d under long-day (LD; 16 h of light/8 h of darkness) or for 24 d under SD (8 h of light/16 h of darkness) conditions. Scale bar = 5 mm. C, Flowering time responses of wild-type and transgenic plants overexpressing HFR1-ΔN105. The flowering time was defined as the number of days from seed sowing until opening of the first flower. LD consists of 16 h of fluorescent lighting and 8 h of darkness. SD consists of 8 h of fluorescent lighting and 16 h of darkness.
In adult plants, petiole elongation was inhibited in HFR1-ΔN105-overexpressing plants under both LD and SD conditions (Fig. 5B). However, the flowering time of the HFR1-ΔN105-overexpressing plants was not different from that of wild type (Fig. 5C), implying that HFR1-ΔN105 may not function in floral induction. Taken together, these results indicate that HFR1-ΔN105 enhanced a subset of phytochrome-signaling pathways in response to W light that regulate hypocotyl/petiole elongation.
Hypersensitive Photoresponse of the HFR1-ΔN105 Overexpresser
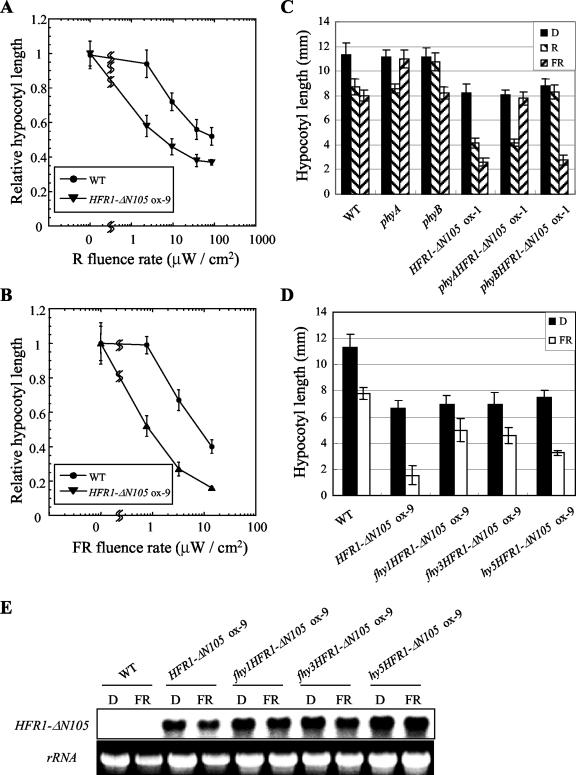
Physiological analysis of HFR1-ΔN105 transgenic plants revealed that HFR1-ΔN105 constitutively activated a branch pathway of light signaling. Interestingly, the HFR1-ΔN105 overexpresser also displayed exaggerated photoresponse in the inhibition of hypocotyl elongation (Fig. 1B). To examine the photoresponse of HFR1-ΔN105-overexpressing plants in detail, we grew seedlings under various fluence rates of R or FR light. Compared with wild type, HFR1-ΔN105 transgenic plants were hypersensitive to both R and FR light (Fig. 6, A and B). To determine which photoreceptor mediates the hypersensitive photoresponse of HFR1-ΔN105 transgenic plants, we constructed phyAHFR1-ΔN105 ox and phyBHFR1-ΔN105 ox double mutants. As shown in Figure 6C, the enhanced photoresponse of HFR1-ΔN105 ox plants under FR or R light were absent in plants in the phyA or phyB mutant backgrounds, respectively. Thus, the enhanced photoresponse of HFR1-ΔN105 transgenic plants under FR or R light required functional phyA or phyB, respectively. To test whether the hypersensitivity of HFR1-ΔN105-overexpressing plants is due to elevated expression of photoreceptor, we performed immunoblot analysis with seedlings grown in darkness or under FR light using antibody against phyA. The result showed that the phyA protein level of HFR1-ΔN105-overexpressing plants was comparable with that of wild type, irrespective of light conditions (data not shown). Thus, the hypersensitivity was not due to elevated level of photoreceptor, implying that downstream signaling after photoreceptor is responsible for the hypersensitivity of HFR1-ΔN105 overexpresser. To examine the dependence of HFR1-ΔN105 on the downstream signaling components of phyA for its enhanced photoresponse, we constructed double mutants between the HFR1-ΔN105 overexpresser and two mutants, fhy1 and fhy3. FHY1 and FHY3 are upstream components in phyA signaling and define distinct signaling branches (Desnos et al., 2001; Okamoto et al., 2001; Wang and Deng, 2002). The results showed that both FHY1 and FHY3 are necessary for the shortened hypocotyls of HFR1-ΔN105-overexpressing plants under FR light (Fig. 6D).
Figure 6.
Hypocotyl elongation responses of transgenic plants overexpressing HFR1-ΔN105 and double mutant analysis. A, Fluence rate responses of inhibition of hypocotyl elongation under R light. The data are expressed as average relative hypocotyl length from at least 20 seedlings, normalized to their respective hypocotyl length in darkness ± sd. The average length of dark-grown seedlings was 10.6 and 7.1 mm for the wild-type and HFR1-ΔN105 ox-9 seedlings, respectively. Error bars = sds. WT, Wild type. B, Fluence rate responses of inhibition of hypocotyl elongation under FR light. The data are expressed as average relative hypocotyl length from at least 20 seedlings, normalized to their respective hypocotyl length
(Legend continues on facing page.) (Legend continued from facing page) in darkness ± sd. The average length of dark-grown seedlings was 9.9 and 7.1 mm for the wild-type and HFR1-ΔN105 ox-9 seedlings, respectively. C, Hypocotyl elongation phenotypes in wild-type and transgenic plants. The seedlings were grown for 4 d under R light (33 μW cm-2), FR light (14 μW cm-2), or in darkness (D). Each measurement was performed on at least 20 seedlings. The data are expressed as average hypocotyl length ± sd. D, Hypocotyl elongation phenotypes in wild-type and transgenic plants. The seedlings were grown for 4 d under FR light (14 μW cm-2) or in darkness (D). Each measurement was performed on at least 20 seedlings. The data are expressed as average hypocotyl length ± sd. E, RNA gel-blot analysis. Seedlings were grown on Murashige and Skoog-Suc medium (2% w/v) for 4 d in darkness (D) or under FR light (14 μWcm-2) before extraction of total RNA. Total RNA (10 μg) was loaded and subject to RNA gel-blot analysis using HFR1-ΔN105 probe.
Because HFR1 and HY5 were shown to regulate not only distinct photoresponses but also overlapping response such as inhibition of hypocotyl elongation (Kim et al., 2002b), we examined the relationship between HFR1-ΔN105 and HY5. The results showed that the enhanced light responses of HFR1-ΔN105-overexpressing plants were decreased, if not completely suppressed, by the absence of HY5, implicating the functional cooperation between HY5 and HFR1-ΔN105 (Fig. 6D). The fhy1, fhy3, and hy5 mutations did neither affect the shortened hypocotyl phenotype of HFR1-ΔN105-overexpressing plants in darkness nor the expression of the transgene HFR1-ΔN105 under FR light (Fig. 6E). These results imply that HFR1-ΔN105 act synergistically with other light-signaling components such as HY5 and possibly other factors to mediate inhibition of hypocotyl elongation in response to light.
DISCUSSION
Previously, a bHLH protein, HFR1 was shown to be critical for certain aspects of photomorphogenic responses under FR and B light (Fairchild et al., 2000; Fankhauser et al., 2000; Soh et al., 2000; Duek and Fankhauser, 2003). To gain further insight into the function of HFR1 in light signaling, we exploited gain-of-function approaches by generating full-length HFR1 or mutant HFR1 lacking N- or C-terminal regions. Gain-of-function approaches using transgenic plants or dominant mutant lines have been useful to reveal genetic functions, especially in the case of transcription factors of which function have been partially understood (Sakai et al., 2001; Zhang et al., 2003). Here, we showed that overexpression of HFR1-ΔN105, but not full-length HFR1, conferred partial photomorphogenic responses in darkness and exaggerated photoresponses under light, implying that HFR1-ΔN105 may be a gain-of-function hyperactive mutant. Physiological analyses implied that a subset of photoresponses including inhibition of hypocotyl elongation, gravitropic hypocotyl growth, greening defect, and seed germination were constitutively activated, to some extent, in darkness by HFR1-ΔN105. The transgenic plants of HFR1-ΔN105 also exhibited altered light-dependent development under W light conditions. The overexpression of HFR1-ΔN105 significantly suppressed hypocotyl elongation in response to EOD-FR light treatment and petiole elongation under SD conditions. In contrast, unlike in cop/det/fus mutants, other light-dependent responses such as anthocyanin accumulation and flowering were not affected by overexpression of HFR1-ΔN105. This is consistent with our previous results that HFR1 act downstream of COP1, mediating a subset of cop1-triggered photoresponses (Kim et al., 2002b). Moreover, it should be noted that HFR1-ΔN105 is the first genetic element that controls phytochrome-dependent seed germination, other than the photoreceptor itself. Together, these results implicated that HFR1-ΔN105 activated a branch pathway of light signaling.
The activation of a specific light-signaling pathway by overexpression of HFR1-ΔN105 also could be exemplified at the level of light-dependent changes in gene expression. In HFR1-ΔN105-overexpressing seedlings grown in darkness, some light-inducible genes, such as CAB, DRT112, PSAE, and PSBL, were constitutively expressed at high levels, and some light-repressible genes including PORA and XTR7 were expressed at low levels. However, other light-regulated genes, including CHS and PSBS, exhibited normal light-responsiveness in HFR1-ΔN105 transgenic plants. The defect in PORA expression and normal induction of CHS in the HFR1-ΔN105-overexpressing line are consistent with the defective greening response and normal anthocyanin accumulation, respectively, observed in the HFR1-ΔN105 transgenic plants. It is interesting to note that the CHS and PSBS genes encode proteins implicated in the photoprotective functions upon exposure to light, whereas other genes affected by HFR1-ΔN105 are involved in the photosynthesis, greening, or cell elongation. Previous reports proposed that light signaling pathway for induction of photoprotectants might involve different intermediates from the ones for building up photosynthetic apparatus (Bowler et al., 1994; Cho et al., 2003). Our results appear to support the hypothesis and imply that the bHLH protein-mediated transcriptional network involving HFR1-ΔN105 may define distinct signaling branch, not affecting signaling pathway for photoprotective functions.
In addition to the constitutive photoresponses in the inhibition of hypocotyl elongation, overexpression of HFR1-ΔN105 enhanced the sensitivity to R and FR light. Using photoreceptor mutants, we showed that the hypersensitivity of HFR1-ΔN105-overexpressing plants was dependent on phyA, FHY1, and FHY3 under FR light and dependent on phyB under R light. Our results of western-blot analysis and the normal photosensitivity of HFR1-ΔN105-overexpressing plants in the anthocyanin accumulation suggested that the hypersensitivity is not due to overexpression of photoreceptor itself (Fig. 3D; data not shown). Double mutant analysis indicates that HY5 is, if partially, necessary for the hypersensitivity of the HFR1-ΔN105 overexpresser but not for the photomorphogenic development that occurs in darkness (Fig. 6D). The result is consistent with our previous reports that HFR1 acts additively with the bZIP transcription factor HY5 to inhibit hypocotyl elongation under FR light (Kim et al., 2002b). Together, these results suggest HY5 may work cooperatively with HFR1-ΔN105 to inhibit hypocotyl elongation in response to light, thereby accounting for the hypersensitive responses of HFR1-ΔN105 transgenic plants.
It is noteworthy that the phenotype affected by HFR1-ΔN105 overexpression is more pleiotropic than expected from the phenotype of the hfr1 mutant. The hfr1 mutation abrogated photoresponses under FR light, including inhibition of hypocotyl elongation, gravitropic hypocotyl growth, and induction of CAB gene expression (Fairchild et al., 2000; Fankhauser et al., 2000; Soh et al., 2000). In contrast, HFR1-ΔN105 overexpression induced somewhat constitutive photoresponses in regard to seed germination and greening defects, in addition to those responses affected by the hfr1 mutation. Anthocyanin accumulation and flowering were not altered in both hfr1 mutant and HFR1-ΔN105-overexpressing plants. The apparent discrepancy may reflect the existence of functional redundancy between HFR1 and unidentified bHLH proteins. In this scenario, HFR1 may function in seed germination and in greening defects, but other functionally redundant bHLH proteins can mediate germination or greening response, complementing the hfr1 mutation. Alternatively, the phenotypes of HFR1-ΔN105-overexpressing plants might represent the functions of other bHLH proteins that could dimerize with HFR1-ΔN105 and HFR1 itself. In fact, HFR1 could dimerize with other bHLH proteins (Fairchild et al., 2000; Supplemental Fig. 1). The ectopic overexpression of HFR1-ΔN105 might enhance the activity of unidentified bHLH proteins that control seed germination or greening defects. Thus, the phenotype of the HFR1-ΔN105-overexpressing plants may reflect the cumulative functions of multiple bHLH transcription factors, including HFR1, rather than solely the in vivo function of HFR1 itself. Also, it cannot be excluded that HFR1-ΔN105 may interact promiscuously with unrelated bHLH proteins to mediate various photoresponses. However, it seems to be less likely, based on the yeast (Saccharomyces cerevisiae) two-hybrid experiments, that HFR1-ΔN105 could interact with HFR1-interacting bHLH proteins and could not interact with HFR1-non interacting bHLH proteins (Supplemental Fig. 1). Moreover, HFR1-ΔN105 transgenic plants did not show any noticeable alterations in epidermal development, floral development, or fruit dehiscence that are regulated by other members of bHLH proteins (Heim et al., 2003). Nevertheless, our results suggest that transcriptional network involving bHLH proteins mediates a branch pathway of light signaling, resulting in germination, de-etiolation, gravitropic response of hypocotyl, greening defect, and expression of several light-regulated genes.
It would be an intriguing question why the full-length HFR1-overexpressing plants did not show significant alterations in photoresponses, compared with the HFR1-ΔN105-overexpressing lines. Our results of RNA gel-blot analysis showed that the N-terminal region did not affect the transcript level of transgene, indicative of posttranscriptional regulation for the function of HFR1. There are several possibilities that are not mutually exclusive. First, the N-terminal region of HFR1 may restrict the amount of HFR1 by controlling translational efficiency or protein stability. Unfortunately, the expression level of HFR1 or HFR1-ΔN105 proteins in transgenic plants was below the detection level in our experimental conditions (data not shown). Second, the N-terminal region of HFR1 may function as an inhibitory domain affecting functional activity of HFR1. In this model, N-terminal region of HFR1 may have inhibitory function for DNA-binding or productive protein complex formation, maintaining HFR1 at an inactive state in the absence of light, and light-dependent posttranslational modification on the N-terminal region might activate HFR1 to trigger a subset of photoresponses. Thus, the HFR1-ΔN105 may function constitutively even in the dark. Third, the N-terminal region of HFR1 may mediate nonproductive protein-protein interaction between HFR1 and other proteins. Thus, the mutant HFR1-ΔN105 may not form nonproductive protein complex with inhibitory proteins, becoming constitutively hyperactive. Further studies delineating the regulatory mechanisms in which the N-terminal region of HFR1 is involved will provide valuable information for the molecular processes underlying regulation of HFR1 function by light.
In summary, our studies not only unveiled functional importance of bHLH-mediated transcriptional network in a light-signaling pathway for a subset of photoresponses but also provided a useful framework for further genetic dissection of light signaling network. Further studies on the functional characterization of HFR1-interacting proteins and identification of extragenic enhancer/suppressor of HFR1-ΔN105 transgenic line would shed light on the transcriptional network that mediates various photoresponses and how light controls a complex array of photoresponses in Arabidopsis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The seeds of wild-type (Columbia [Col]), phyB-9, and phyA-211 mutants lines were obtained from the Arabidopsis Biological Resources Center (Columbus, OH). The fhy3-1 seed was kindly provided by Dr. Garry Whitelam (Leicester University, UK), and the hy5-221 seed was obtained from Dr. Xing-Wang Deng (Yale University, New Haven, CT). The fhy1-311 mutant was derived from our mutant screening with ethyl methanesulfonate-mutagenized seeds and was shown to be a null mutation (M.-S. Soh, unpublished data). The intragenic suppressor of HFR1-ΔN105 ox-9 (sup1) was isolated by suppressor screening using ethyl methanesulfonate-mutagenized M2 seeds of HFR1-ΔN105 ox-9. The mutant showed long hypocotyl and closed cotyledon in darkness, resembling wild-type seedlings. Genetic analysis showed that the mutation occurred in the transgene HFR1-ΔN105 itself. Sequencing analysis revealed that Leu-173, which is highly conserved among bHLH proteins in the second helix (Heim et al., 2003), was changed into Phe. All mutants used are from the Col background. Light conditions used were same as previously described (Soh et al., 2000). For measurement of hypocotyl lengths, seeds were surface sterilized for 5 min in commercial bleach and rinsed with sterile distilled water at least five times. Seeds were then sown onto Murashige and Skoog medium containing 0.8% (w/v) agar. After incubation at 4°C for 3 d, the plates were placed in W light for 12 h at 23°C to improve germination and then transferred to the appropriate light conditions. Data were collected from 40% of the longest seedlings to minimize variation in hypocotyl lengths among the seedlings as described previously (Soh et al., 1998). For the determination of gravitropic response, the seeds were sown in a row onto 0.8% (w/v) agar media containing 1.2% (w/v) agar. The seedlings were grown vertically and photographed to measure the angles of hypocotyl growth as described by Soh et al. (2000). For measurement of anthocyanin content, seedlings were grown on Murashige and Skoog media containing 2% (w/v) Suc in either darkness or in FR light for 3 d after W light irradiation for 12 h to induce germination. Samples of 50 seedlings were harvested. Extraction and quantitation of anthocyanin was performed as previously described (Mancinelli, 1990). For FR-preconditioned blocking of greening experiment, seedlings were grown on Murashige and Skoog medium for 5 d in either darkness or in FR light (21 μW cm-2), and then transferred to W light. Samples of 50 seedlings were harvested and homogenized. Chlorophyll was extracted in 95% (v/v) ethanol at 4°C. Chlorophyll content was estimated by spectrophotometry (Kim et al., 1996). Germination tests were performed as described by Shinomura et al. (1996). Seeds were surface-sterilized and sown on aqueous medium containing 0.7% (w/v) agar. Seeds were irradiated with FR light (21 μW cm-2) for 15 min and then kept in darkness with or without a single pulse of R light (33 μW cm-2) for 10 min. After 5 d, germination frequency was determined.
RNA Gel-Blot Analysis
Seedlings were grown on Murashige and Skoog-Suc medium (2% w/v) for 4.5 d in darkness and then transferred to FR light for the indicated times before harvesting under dim-green light. Total cellular RNA was extracted from whole seedlings using the RNeasy Miniprep kit (Qiagen, Valencia, CA). RNA gel-blot analysis was performed as described (Soh et al., 1998). The CAB2 gene probe was obtained from Dr. Joanne Chory (The Salk Institute, La Jolla, CA), and the CHS gene probe from the Arabidopsis Biological Resources Center. The Brassica napus 18S rRNA probe has been described previously (Soh et al., 1998). The PORA gene probe was generated by PCR using primers CGCGACTTCAACTCCATCAG and GGATCCAACAATGATG as described (Wang and Deng, 2002). The DRT112, XTR7, PSAE, PSBL, and PSBS gene probes were genomic fragments generated by PCR as described previously (Kuno et al., 2000).
Analysis of Arabidopsis Transgenic Lines
Full-length HFR1 was amplified by PCR with HFR1 cDNA (Soh et al., 2000) using the following primers: HFR1F4-2, 5′-CGAGAATTCATGTCGAATAATCAAGCTTTC-3′; and HFR1R8, 5′-CCTAATTTGGAATTCTTTTCTCTC-3′. The mutant HFR1 (HFR1-ΔN105) lacking the N-terminal 105 amino acids was generated by PCR using the following primers: HFR1F5, 5′-CGAGAATTCATGAGAAACAAACATGAG-3′; and HFR1R8. The EcoRI sites introduced are underlined, and the ATG start codon introduced for HFR1-ΔN105 is shown in italics. The PCR products were digested with EcoRI and then cloned into binary vector pNB96, obtained from Dr. Hong-Gil Nam (POSTECH, Pohang, Republic of Korea), in which the transgene is driven by the 35S dual promoter. The constructs were sequence verified. The resulting binary vector was introduced into Agrobacterium tumefaciens GV3101 and used to transform wild-type Arabidopsis, Col, or hfr1-201 mutant (Bechtold et al., 1993). More than 40 independent T1 transgenic plants were selected. Phenotypic analysis was performed with single T-DNA insertion lines of at least 20 independent lines.
Double Mutant Construction
To construct double mutants, we crossed HFR1-ΔN105 transgenic plants with light-signaling mutants and allowed the F1 progeny to self-pollinate to produce the F2 seeds. Basta-resistant plants were identified among the F2 seedlings and then grown for setting F3 seeds. The resulting F3 lines were tested for heterozygous basta resistance and homozygous long-hypocotyl phenotypes under appropriate light conditions. From these, basta-resistant seedlings were selected and further grown to the F4 generation; then, plants were screened for homozygous basta resistance. The resulting homozygous basta-resistant lines were designated as double mutants and used for phenotypic analysis.
Yeast (Saccharomyces cerevisiae) Two-Hybrid Experiments
The HFR1 gene and HFR1-ΔN105 were fused to the LexA DNA-binding domain of the pGilda vector (CLONTECH Laboratories, Palo Alto, CA). The PIF3 (At1g09530), AtbHLH15 (At2g20180), AtbHLH121 (At3g19860), and AtbHLH47 (At3g47640) genes were fused to the B42 activation domain of the pYESTrp2 vector (Invitrogen, Carlsbad, CA). The genes were cloned by PCR using cDNA library DNA. The yeast strain EGY48[p8op-lacZ] was used for transformation. Both bait and prey constructs were transformed into a yeast strain EGY48 [ura3, his3, trp1, and LexAop(x6)-LEU2], following the manufacturer's instructions. The transformants were selected with Glu/complete supplement mixture-His-Trp medium. Protein-protein interaction was assayed by comparing growth on Gal/Raf/complete supplement mixture-His-Trp-Leu dropout plates.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to requisite permission from any third party owners of all or parts of material. Obtaining any permission will be the responsibility of the requester.
Acknowledgments
We thank Drs. Dana Beitner-Johnson and Sumin Lee for critically proofreading this manuscript.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.029751.
This work was supported in part by Kumho Petrochemical Co., Ltd. and by the BioGreen21 program (grant to M.-S.S.).
References
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213-222 [DOI] [PubMed] [Google Scholar]
- Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada JP, Grossniklaus U, Chua NH (2001) LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev 15: 2613-2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes SA, Nishizawa NK, Quaggio RB, Whitelam GC, Chua NH (1996) Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell 8: 601-615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci 316: 1194-1196 [Google Scholar]
- Benvenuto G, Formiggini F, Laflamme P, Malakhov M, Bowler C (2002) The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr Biol 12: 1529-1534 [DOI] [PubMed] [Google Scholar]
- Bowler C, Yamagata H, Neuhaus G, Chua NH (1994) Phytochrome signal transduction pathways are regulated by reciprocal control mechanisms. Genes Dev 8: 2188-2202 [DOI] [PubMed] [Google Scholar]
- Butler WL, Norris KH, Siegelman HW, Hendricks SB (1959) Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Proc Natl Acad Sci USA 45: 1703-1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Lin Y, Cheng C-L (2000) Genetic interactions between the chlorate-resistant mutant cr88 and the photomorphogenic mutants cop1 and hy5. Plant Cell 12: 199-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DS, Hong SH, Nam HG, Soh MS (2003) FIN5 positively regulates far-red light responses in Arabidopsis thaliana. Plant Cell Physiol 44: 565-572 [DOI] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F (1989) Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58: 991-999 [DOI] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH (1992) COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβ homologous domain. Cell 71: 791-801 [DOI] [PubMed] [Google Scholar]
- Desnos T, Puente P, Whitelam GC, Harberd NP (2001) FHY1: a phytochrome A-specific signal transducer. Genes Dev 15: 2980-2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Zhou YC, Schäfer E, Funk M, Kretsch T (2001) EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev 15: 939-944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C (2003) HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J 34: 827-836 [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14: 2377-2391 [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J (1997) Light control of development. Annu Rev Cell Dev Biol 13: 203-229 [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Chory J (2000) RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiol 124: 39-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya M (1993) Phytochromes-Their molecular species, gene families, and functions. Annu Rev Plant Physiol Plant Mol Biol 44: 617-645 [Google Scholar]
- Furuya M, Schäfer E (1996) Photoperception and signaling of induction reactions by different phytochromes. Trends Plant Sci 1: 301-307 [Google Scholar]
- Hangarter RP (1997) Gravity, light and plant form. Plant Cell Environ 20: 796-800 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng X-W (2000) HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 19: 4997-5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basis helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735-747 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Quail PH (2001) The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem 276: 38173-38178 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH (1999) SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284: 496-499 [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441-2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM (1994) Photomorphogenesis in Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Kim BC, Soh MS, Kang BJ, Furuya M, Nam HG (1996) Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2. Plant J: 9: 441-456 [DOI] [PubMed] [Google Scholar]
- Kim DH, Kang JG, Yang SS, Chung KS, Song PS, Park CM (2002a) A phytochrome-associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell 14: 3043-3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Woo JC, Song PS, Soh MS (2002b) HFR1, a phytochrome A-signaling component, acts in a separate pathway from HY5, downstream of COP1 in Arabidopsis thaliana. Plant J 30: 711-719 [DOI] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bogner L, Kim L, Adam E, Harter K, Schäfer E, Nagy F (1999) Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445-1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T, Neuhaus G, Batschauer A, Chua NH, Schäfer E (1996) Functional analysis of yeast-derived phytochrome A and B phycocyanobilin adducts. Plant J 10: 625-636 [DOI] [PubMed] [Google Scholar]
- Kuno N, Muramatsu T, Hamazato F, Furuya M (2000) Identification by large-scale screening of phytochrome-regulated genes in etiolated seedlings of Arabidopsis using a fluorescent differential display technique. Plant Physiol 122: 15-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapko VN, Jiang XY, Smith DL, Song PS (1997) Posttranslational modification of oat phytochrome A: phosphorylation of a specific serine in a multiple serine cluster. Biochemistry 36: 10595-10599 [DOI] [PubMed] [Google Scholar]
- Lin C (2000) Plant blue-light receptors. Trends Plant Sci 5: 337-342 [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng X-W (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589-2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancinelli AL (1990) Interaction between light quality and light quantity in the photoregulation of anthocyanin production. Plant Physiol 92: 1191-1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Schäfer E (2002) Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu Rev Plant Biol 53: 329-355 [DOI] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J (2000) Light: an indicator of time and place. Genes Dev 14: 257-271 [PubMed] [Google Scholar]
- Okamoto H, Matsui M, Deng X-W (2001) Overexpression of the heterotrimeric G protein a-subunit enhances phytochrome-mediated inhibition of hypocotyl elongation in Arabidopsis. Plant Cell 13: 1639-1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng X-W (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462-466 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983-2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH (2002) Phytochrome photosensory signaling networks. Nat Rev Mol Cell Biol 3: 85-93 [DOI] [PubMed] [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D (1995) Phytochromes: photosensory perception and signal transduction. Science 268: 675-680 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J (1994) Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol 104: 1139-1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Ratcliffe OJ (2000) A genomic perspective on plant transcription factors. Curr Opin Plant Biol 3: 423-434 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519-1521 [DOI] [PubMed] [Google Scholar]
- Schroeder DF, Gahrtz M, Maxwell BB, Cook RK, Kan JM, Alonso JM, Ecker JR, Chory J (2002) De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr Biol 12: 1462-1472 [DOI] [PubMed] [Google Scholar]
- Seo HS, Yang J-Y, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995-999 [DOI] [PubMed] [Google Scholar]
- Serino G, Su H, Peng Z, Tsuge T, Wei N, Gu H, Deng XW (2003) Characterization of the last subunit of the Arabidopsis COP9 signalosome: implications for the overall structure and origin of the complex. Plant Cell 15: 719-731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev 3: 1745-1757 [DOI] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M (1996) Action spectra for phytochrome A- and B-specific photoinhibition of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 8129-8133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H (2000) Phytochromes and light signal perception by plants-an emerging synthesis. Nature 407: 585-591 [DOI] [PubMed] [Google Scholar]
- Soh MS, Hong SH, Hanzawa H, Furuya M, Nam HG (1998) Genetic identification of FIN2, a far red light-specific signaling component of Arabidopsis thaliana. Plant J 16: 411-419 [DOI] [PubMed] [Google Scholar]
- Soh MS, Kim YM, Han SJ, Song PS (2000) REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell 12: 2061-2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G, Yanagawa Y, Kwok SF, Matsui M, Deng XW (2002) Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev 16: 554-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98: 9437-9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035-1045 [DOI] [PubMed] [Google Scholar]
- Wang H, Deng XW (2002) Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J 21: 1339-1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW (1996) The role of the COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol 112: 871-878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Devlin PF (1997) Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ 20: 752-758 [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A (1999) Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol 3: 437-445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Deng X-W, Matsui M (2001) CIP4, a new COP1 target, is a nucleus-localized positive regulator of Arabidopsis photomorphogenesis. Plant Cell 13: 399-411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Matsui M, Ang A-H, Deng X-W (1998) Role of a COP1 interactive protein in mediating light-regulated gene expression in Arabidopsis. Plant Cell 10: 1083-1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KC, Lagarias JC (1998) Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA 95: 13976-13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859-4869 [DOI] [PubMed] [Google Scholar]