Abstract
Root-produced organic compounds in xylem sap, such as hormones and amino acids, are known to be important in plant development. Recently, biochemical approaches have revealed the identities of several xylem sap proteins, but the biological functions and the regulation of the production of these proteins are not fully understood. XYLEM SAP PROTEIN 30 kD (XSP30), which is specifically expressed in the roots of cucumber (Cucumis sativus), encodes a lectin and is hypothesized as affecting the development of above-ground organs. In this report, we demonstrate that XSP30 gene expression and the level of XSP30 protein fluctuate in a diurnal rhythm in cucumber roots. The rhythmic gene expression continues for at least two or three cycles, even under continuous light or dark conditions, demonstrating that the expression of this gene is controlled by a circadian clock. Removal of mature leaves or treatment of shoots with uniconazole-P, an inhibitor of gibberellic acid (GA) biosynthesis, dampens the amplitude of the rhythmic expression; the application of GA negates these effects. These results suggest that light signals perceived by above-ground organs, as well as GA that is produced, possibly, in mature leaves, are important for the rhythmic expression of XSP30 in roots. This is the first demonstration of the regulation of the expression of a clock-controlled gene by GA.
The higher plant body consists of functionally specialized organs such as the leaf, stem, flower, and root. Because plants grow in changing environments, it is essential for different organs to interact to ensure that the plant body develops and functions properly. Information transfer between organs is essential for synchronized plant development (Bernier, 1988; Kolek and Kozinka, 1991). A major route for the transfer of materials between organs consists of the vascular bundles, which are composed mainly of the xylem and phloem. Numerous materials are translocated over long distances through these bundles. The xylem is composed mainly of xylem vessels, which form a type of apoplastic space. Organic nutrients, such as amino acids, sugars, and organic acids, as well as water and inorganic nutrients, are transported through the xylem to the aboveground organs (Schurr and Schulze, 1995; Zornoza et al., 1996; Satoh et al., 1998). Interestingly, the roots control aspects of the development of aerial organs, possibly acting via growth-related compounds in the xylem sap (Kinet et al., 1993; Satoh, 1996). For example, cytokinin, abscisic acid, and other growth-related compounds that are synthesized in root tissues are involved in stomatal responses (Else et al., 1995; Liang et al., 1997), leaf senescence (Nooden et al., 1990; Soejima et al., 1992), lateral bud development (Bangerth, 1994; Beveridge et al., 1997), flower bud formation (Kinet et al., 1993), leaf greening (Kato et al., 2002), and adventitious root formation (Kuroha et al., 2002).
Recently, macromolecules have been found in xylem sap, including oligo- and polysaccharides (Satoh et al., 1992; Campbell et al., 1995) and proteins such as peroxidase (Biles and Abeles, 1991), chitinase (Masuda et al., 2001), a cucumber (Cucumis sativus) root-specific Gly-rich protein (CRGRP; Sakuta et al., 1998; Sakuta and Satoh, 2000), pathogenesis-related (PR) proteins (Rep et al., 2002), a Cys-rich protein (Rep et al., 2003), and a novel 30-kD protein (XSP30; Masuda et al., 1999). However, the regulation of the production of these xylem sap proteins and their physiological functions are not well understood.
A major environmental factor for plants, the diurnal light cycle, is perceived mainly in leaves, but signal transduction resulting from this cue is controlled in many organs (McClung, 2001). In plants, photoreceptors that perceive light signals, such as phytochromes and cryptochromes, influence the expression of biological clock component genes, such as TIMING OF CAB EXPRESSION 1 (Strayer et al., 2000), LATE ELONGATED HYPOCOTYL (Schaffer et al., 1998), and CIRCADIAN CLOCK ASSOCIATED 1 (Wang and Tobin, 1998) via factors that interact with photoreceptors and adjust the biological clock in 24-h periods (Makino et al., 2002; Mizoguchi et al., 2002; Mouradov et al., 2002). The biological clock signals that are distributed to each organ regulate many aspects of development, such as flowering, leaf movement, and hypocotyl elongation (McClung, 2001). The expression of numerous genes is controlled by circadian rhythms (Somers, 1999; Harmer et al., 2000). In addition, the diurnal light cycle affects the levels of plant hormones produced in the shoot. The expression of the gene that encodes gibberellic acid (GA) 20-oxidase, a key regulatory enzyme in the gibberellin biosynthetic pathway in leaves, is controlled by the diurnal light cycle and is correlated to tuber formation in potato (Solanum tuberosum; Carrera et al., 1999). Endogenous levels of indole-3-acetic acid (IAA) fluctuate in a circadian rhythm and are involved in the growth of the internode (Jouve et al., 1999). However, the effects of circadian rhythms on the root have not been fully studied; these effects are less intuitive because roots are spatially separated from leaves, where the circadian rhythms are adjusted. Information on clock-controlled gene expression and protein accumulation in roots is quite limited.
In our previous study of cucumber xylem sap, a novel 30-kD protein (XSP30) was identified (Masuda et al., 1999). The sequence of the protein encoded by the XSP30 cDNA has significant homology to the B chain of ricin, a toxic protein with Gal-specific lectin activity from the seeds of castor bean (Ricinus communis; Lord et al., 1994). Ricin superfamily lectins are characterized by a ribosome-inactivating domain (A chain) and a Gal-binding lectin domain (B chain; Lord et al., 1994). Although XSP30 lacks a ribosome-inactivating domain (A chain), it binds to the free (N-acetylglucosamine)2 in N-linked glycans (Oda et al., 2003). XSP30 is expressed only in roots, but the glycoprotein-conjugated N-linked glycans recognized by XSP30 are most abundant in leaves (Masuda et al., 1999; Oda et al., 2003), leading to speculation that XSP30 is a signaling molecule that is transported from the root to the shoot.
In this report, we show that XSP30 expression in cucumber roots follows a diurnal pattern, and that the oscillation in the expression of its gene is controlled by a circadian clock, which is, in turn, possibly affected by leaf gibberellins. The specific expression of XSP30 in root vascular tissues is also shown. This is the first report of a circadian control of a root-specific gene that is affected by GA.
RESULTS
Root-Specific Expression of the XSP30 Gene
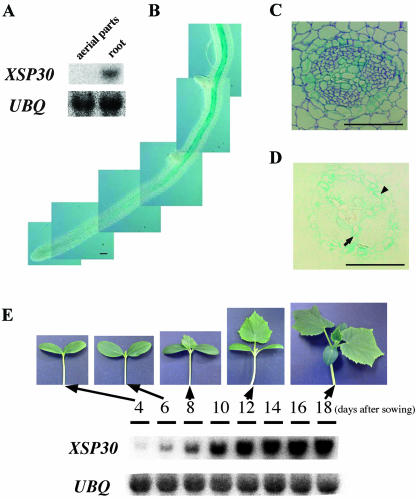
We have found that XSP30 is expressed only in root tissue (Masuda et al., 1999), as no transcripts can be detected in the aerial organs of cucumber (Fig. 1A). To further analyze this expression pattern, we constructed a chimeric gene (PXSP30::GUS), consisting of the XSP30 promoter fused to a GUS reporter gene, and detected the distribution of the GUS activity driven by the XSP30 promoter in transgenic cucumber plants (Fig. 1B). GUS activity was analyzed in the hairy roots that emerged from the cut surface of cotyledons transformed with PXSP30::GUS. Strong GUS staining was observed in the central cylinder of mature roots (Fig. 1B), and the staining intensity gradually decreased in younger portions of the roots. No GUS staining was detected at the root tip or in emerging lateral roots. Cross sections revealed that GUS expression was restricted to the xylem parenchyma and pericycle cells in the central cylinder, and no GUS staining was observed in the phloem (Fig. 1, C and D).
Figure 1.
Root-vascular tissue-specific XSP30 expression. A, Root-specific expression of XSP30. Total RNA (10 μg lane-1) extracted from aboveground organs or roots of 30-d-old cucumber plants was subjected to RNA gel-blot analysis. The transcripts were probed using a full-length XSP30 cDNA and ubiquitin fragments as probes. The ubiquitin cDNA was isolated from cucumber roots, where it is expressed constitutively. B through D, β-Glucuronidase (GUS) activity in PXSP30::GUS-transgenic hairy roots. GUS activity was observed in the central cylinder of mature roots, but not in the root tip shown in B. GUS-stained transgenic roots were embedded in Technovit 7100 (Kulzer and Co., Werheim, Germany), and thin serial sections were stained with toluidine blue (C) or left un-stained (D). The central cylinder is shown. Scale bars correspond to 100 μm. The arrow and arrowhead indicate GUS staining in the xylem parenchyma and pericycle cells, respectively. E, Seedling development-dependent expression of XSP30 in roots. Total RNA (10 μg lane-1) extracted at dusk from roots of seedlings at 4, 6, 8, 10, 12, 14, 16, and 18 d after sowing were subjected to RNA gel-blot analysis. The transcripts were probed using a full-length XSP30 cDNA and a ubiquitin fragment as probes.
Gel-blot analysis of total RNA prepared from the roots of cucumber seedlings of different ages showed that XSP30 expression was barely detectable in the roots of 4-d-old seedlings, but increased gradually with maturity, concurrent with leaf development, after 6 d (Fig. 1E).
Diurnal Oscillation of XSP30 Gene Expression and Protein Accumulation
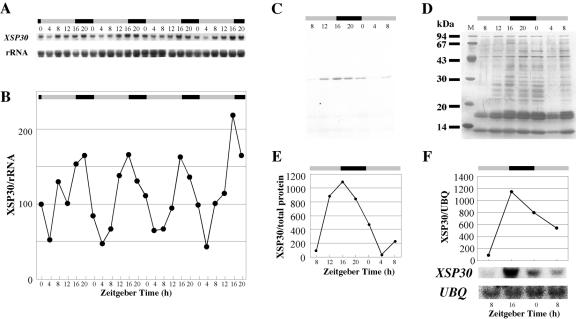
To test whether environmental signals affect XSP30 gene expression in cucumber roots, we examined XSP30 mRNA levels in plants under various conditions. In the course of this study, we found that XSP30 gene expression showed a diurnal pattern under a photoperiod of 16 h of light/8 h of dark (Fig. 2, A and B). XSP30 gene expression peaked at time 16 and decreased to trough level at around time 4. Peaks in expression levels occurred at time 16, except for during the first cycle.
Figure 2.
Diurnal regulation of XSP30 gene expression and XSP30 content of xylem sap. A and B, Time course of XSP30 expression. Cucumber plants were grown under a 16-h light/8-h dark photoperiod. Total RNA (10 μg lane-1) was extracted from the roots of seedlings every 4 h, beginning on the 11th d after sowing. RNA samples were subjected to RNA gel-blot analysis with XSP30 cDNA and rRNA probes. Shown are the original autoradiograph in A and the ratio of the intensity of hybridization of XSP30 and rRNA in B. C through E, Detection of XSP30 protein in xylem sap. Xylem sap was collected at 4-h intervals from stems of cucumber plants 30 d after sowing. The proteins in equal volumes of sap (5 μL lane-1) were separated by SDS-PAGE and detected with anti-XSP30 serum on nylon membranes (C) or silver stained (D). The ratio of the intensity of the XSP30 signal and staining of the total proteins is shown in E. The XSP30 gene expression in cucumber roots, 30 d after sowing, is shown in F. Total RNA (10 μg lane-1) was extracted from the roots of seedlings every 8 h, beginning on d 30 after sowing. RNA samples were subjected to RNA gel-blot analysis with an XSP30 cDNA and a ubiquitin fragment as probes. The original autoradiograph and the ratio of the intensity of hybridization to the XSP30 and ubiquitin transcripts are shown. Dawn was defined as zeitgeber time 0. The periods of light and dark are indicated as shaded and black bars, respectively.
The amount of XSP30 protein in root xylem sap was evaluated by immunoblotting with anti-XSP30 serum (Masuda et al., 1999). A diurnal rhythm in XSP30 protein abundance occurred with a peak at dusk (time 16 in this experiment; Fig. 2, C-E). This diurnal pattern in protein abundance was quite similar to the fluctuation of the abundance of the XSP30 mRNA (Fig. 2, A, B, and F).
Circadian Clock-Controlled Expression of the XSP30 Gene
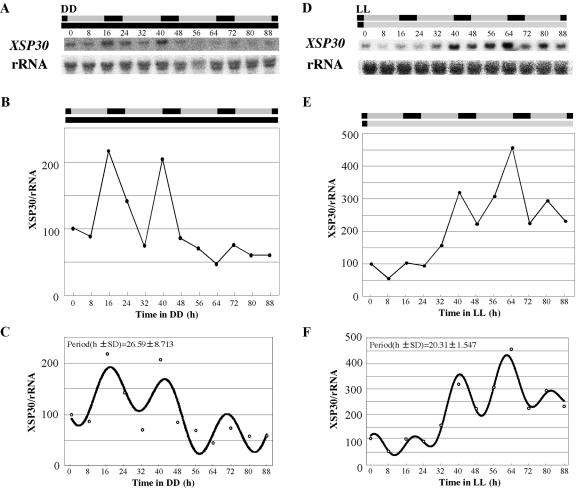
The XSP30 mRNA level began to increase before dusk and to decrease before dawn (Fig. 2, A and B). This anticipation of the diurnal rhythm is a common feature in circadian rhythms. When seedlings grown under a 16-h light/8-h dark photoperiod for 13 days were transferred to continuous dark (DD) or continuous light (LL), the XSP30 gene showed a rhythmic pattern of expression for at least two cycles under DD (Fig. 3, A and B) and three cycles under LL (Fig. 3, D and E). The data were Fourier transformed and the periods were estimated as 26.59 (±8.713) hours in DD and 20.31 (±1.547) hours in LL (Fig. 3, C and F). To confirm the strength of rhythms, the relative amplitude errors (RAE, see “Material and Methods”) were calculated as 0.308 (0.6 > RAE) in DD and 0.731 (1 > RAE > 0.6) in LL. A perfect sine wave gives an RAE of 0 and RAE = 1 defines the limit of statistical significance of rhythm, suggesting substantial and weak rhythms of XSP30 expression in DD and LL, respectively. The experiments of Figure 3 have been performed three times. Statistically significant rhythms were detected twice, and in the third replicate (data not shown), the time courses were suggestive, although not statistically significant. Therefore, we conclude that a circadian clock contributes to the regulation of expression of XSP30.
Figure 3.
Expression of XSP30 under DD or LL. Thirteen-day-old seedlings grown under a 16-h light/8-h dark photoperiod were transferred to DD (A-C) or LL (D-F). Total RNA was extracted every 8 h from roots of treated seedlings. RNA samples were subjected to RNA gel-blot analysis with an XSP30 cDNA and rRNA as probes. The original autoradiograph is shown (A and D), as well as the ratio of the intensity of hybridization to XSP30 and rRNA (B and E). The data were Fourier transformed and the period were estimated using Fast Fourier Transform-Non-Linear Least Squares program as described (Plautz et al., 1997; C and F). The shaded and black bars indicate the original periods of light and dark, respectively.
Decapitation Eliminates Rhythmic Expression of XSP30 in Roots
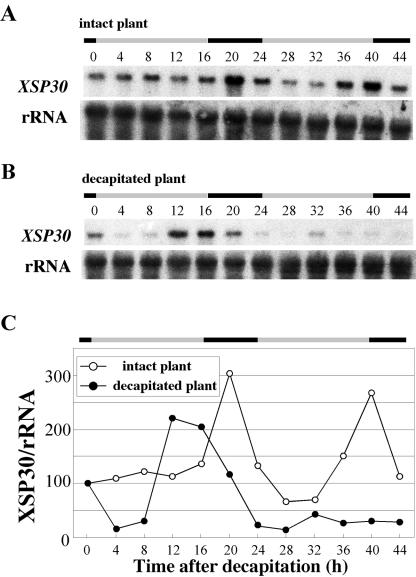
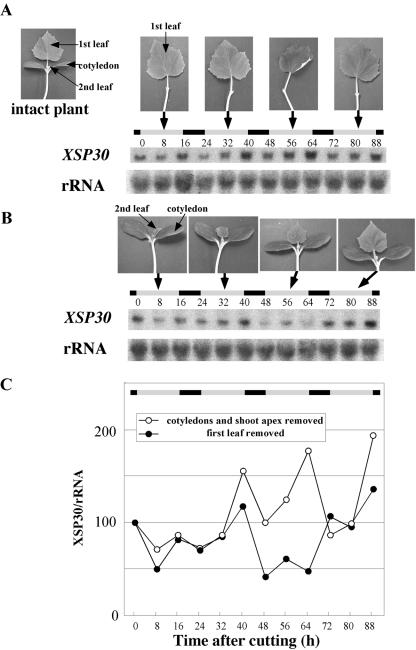
Light signals are important for the entrainment of a circadian clock. Aboveground organs are thought to receive the light signals that control a variety of biological processes in plants. We have recently demonstrated that cotyledons are required for cell division during tissue reunion in the cortex of cut cucumber (Asahina et al., 2002). This led us to test whether the diurnal expression of XSP30 in roots requires the aboveground organs. When the first leaf, the cotyledons, and the shoot apex were removed from cucumber seedlings at the middle of the hypocotyl, the XSP30 mRNA abundance peaked at around dusk (time 12-16) after decapitation. However, this increase in XSP30 expression at dusk was not observed on the 2nd d after organ removal (Fig. 4, B and C). In contrast, intact control plants showed a normal diurnal rhythm, and XSP30 expression peaked at times 20 and 40 (Fig. 4, A and C). The results suggest that these aboveground organs are involved in the control of the rhythmic expression of XSP30 in cucumber roots.
Figure 4.
Effect of decapitation of aboveground organs on XSP30 expression in roots. Total RNA (10 μg lane-1) was extracted every 4 h from the roots of 15-d-old cucumber plants grown under a 16-h light/8-h dark photoperiod from intact (A) or decapitated plants (B). Root RNA samples were subjected to RNA gel-blot analysis with an XSP30 cDNA and rRNA as probes. The original autoradiograph is shown (A and B), as well as the ratio of the intensity of hybridization to XSP30 and rRNA (C). The periods of light and dark are indicated as shaded and black bars, respectively.
Involvement of the First Leaf in Diurnal Gene Expression
Gene expression in roots was examined after removal of the first leaf or the cotyledons and shoot apex from 13-d-old seedlings. Rhythmic expression of XSP30 was observed during at least the 2nd, 3rd, and 4th d after removal of the cotyledon and shoot apex (Fig. 5, A and C). XSP30 expression peaked at times 40, 64, and 88 on the 2nd, 3rd, and 4th d after removal, respectively. In contrast, the XSP30 mRNA level was significantly lower in plants from which only the first leaf was removed (Fig. 5, B and C). These results suggest that mature leaves are required for the high amplitude of the diurnal expression of XSP30 in cucumber roots, and that translocatable signals, possibly including plant hormones such as GAs, auxins, and brassinosteroids may be involved in this process.
Figure 5.
Effect of leaf removal on XSP30 expression in roots. The cotyledons plus the shoot apex (A), or the mature first leaf (B), were removed from 13-d-old seedlings, and the roots were collected every 8 h for RNA extraction. Root RNA samples were subjected to RNA gel-blot analysis with an XSP30 cDNA and rRNA as probes. The original autoradiograph is shown (A and B), as well as the ratio of the intensity of hybridization to XSP30 and rRNA (C). The periods of light and dark are indicated as shaded and black bars, respectively.
Oscillating XSP30 Expression Is Possibly Related to Leaf Gibberellins
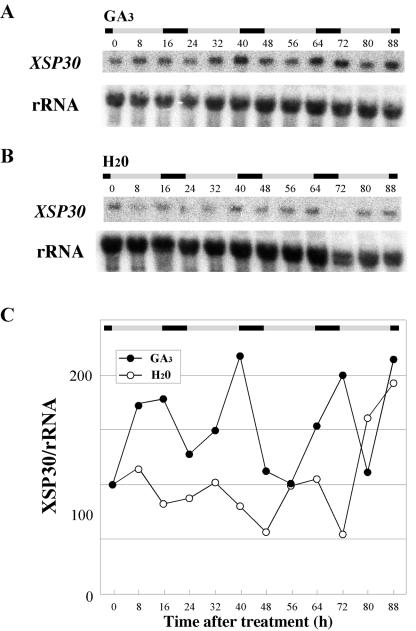
A gibberellin biosynthesis inhibitor (uniconazole-P), an auxin polar transport inhibitor (2,3,5-triiodobenzoic acid), or a brassinosteroid biosynthesis inhibitor (brassinazole; Asami et al., 2000) were applied to leaves, and their effects on XSP30 expression in roots were analyzed. Only uniconazole-P significantly affected the expression of XSP30 in roots (data not shown). Therefore, we examined the relationship between gibberellin and XSP30 gene expression in roots. GA was sprayed on the cotyledons and shoot apices of cucumber seedlings after removal of the first leaf. Treatment with GA fully restored the high-amplitude rhythmic expression of XSP30 in the roots of these seedlings (Fig. 6, A and C). In contrast, in control seedlings treated with water, XSP30 expression failed to return to this expression pattern (Figs. 6, B and C, and 5, B and C). The data in Figure 6 were Fourier transformed and the periods and the RAE were estimated as 26.74 (±0.962) hours and 0.311 in plants treated with water and 27.32 (±0.564) hours and 0.256 in plants treated with GA.
Figure 6.
Effects of gibberellin on XSP30 expression in roots. Mature first leaves were removed from 13-d-old seedlings, and 2 × 10-4 m GA3 (A) or water (B) was applied every 2 d to the cotyledons and shoot apex. Roots were collected every 8 h for RNA extraction. Root RNA samples were subjected to RNA gel-blot analysis with an XSP30 cDNA and rRNA as probes. The original autoradiograph is shown (A and B), as well as the ratio of the intensity of hybridization to XSP30 and rRNA (C). The periods of light and dark are indicated as shaded and black bars, respectively.
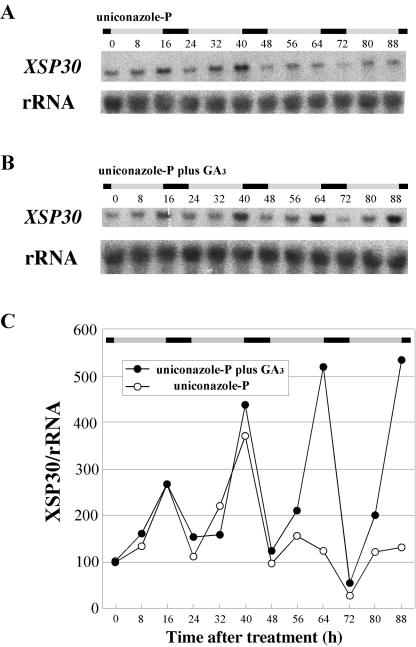
To examine the involvement of endogenous gibberellins in activating XSP30 expression, uniconazole-P was applied to the shoots of intact plants. This resulted in a dramatic decrease in XSP30 expression within the 48 h after the uniconazole-P treatment (Fig. 7, A and C), but rhythmic expression with a lower amplitude was observed during the 3rd and 4th d. Simultaneous application of GA3 with uniconazole-P resulted in no change in the oscillation pattern, with maximum expression at time 64 and 88 on the 3rd and 4th d, respectively (Fig. 7, B and C). The data in Figure 7 were Fourier transformed and the periods and the RAE were estimated as 22.82 (±0.909) hours and 0.366 in plants treated without GA and 23.79 (±1.224) hours and 0.341 in plants treated with GA. To elucidate whether gibberellin transported from the shoot affects the expression of XSP30 in roots, 10-4 m GA3 was supplied to the roots of uniconazole-P-treated plants. The application of GA3 to roots did not reverse the effects of uniconazole-P (data not shown). These results suggest that gibberellins produced in leaves are involved in the control of the diurnally oscillating pattern of XSP30 gene expression in cucumber roots.
Figure 7.
Effect of uniconazole-P and gibberellin on XSP30 expression in roots. Uniconazole-P (10-4 m; A) or 10-4 m uniconazole-P plus 2 × 10-4 m GA3 (B) was applied to the shoot, including the cotyledons, first leaf, and shoot apex, of 13-d-old seedlings. Roots were collected every 8 h for RNA extraction. Root RNA samples were subjected to RNA gel-blot analysis with an XSP30 cDNA and rRNA as probes. The original autoradiograph is shown (A and B), as well as the ratio of the intensity of hybridization to XSP30 and rRNA (C). The periods of light and dark are indicated as shaded and black bars, respectively.
DISCUSSION
Diurnal Control of XSP30 Gene Expression in Roots
In plants, leaf movements, hypocotyl elongation, stomatal opening, and floral induction are controlled by a circadian clock (McClung, 2001). These effects are likely the result of the circadian rhythm-dependent expression of numerous plant genes (Somers, 1999). Among 8,200 genes examined using microarrays of Arabidopsis genes, approximately 6% exhibited circadian expression patterns in steady-state mRNA levels (Harmer et al., 2000). Clock-controlled expression of these genes was tested using young seedlings 7 to 10 d after germination. Because roots are spatially separated from leaves, where the circadian rhythms are adjusted, the effects of circadian rhythms on roots have not been fully studied. However, a few examples of diurnally oscillating gene expression in roots have been reported. The genes that encode certain enzymes involved in nitrogen fixation are expressed in a diurnal pattern (Lejay et al., 1999; Abd-el Baki et al., 2000), as are the histone H1 and chalcone synthase transcripts (Corlett et al., 1998; Thain et al., 2002). Apart from the report that the diurnal expression pattern of the nitrate transporter gene Nrt in roots is caused by photoassimilates (Lejay et al., 1999), there has been no report on diurnal control mechanisms in roots. In this report, we have demonstrated diurnal oscillations in the expression of XSP30 in cucumber roots (Fig. 2). XSP30 protein levels also oscillated after the expression pattern of the gene (Fig. 2). The oscillation in gene expression was sustained for two or three cycles under continuous DD and LL conditions (Fig. 3). This result suggests that the expression of XSP30 in roots is controlled by a circadian clock.
Possible Fine Tuning of XSP30 Expression by GA
The regulation of the period, which is the time after which a definite phase of the oscillation reoccurs, and the amplitude, which is the difference between maximum value and mean value in a sinusoidal oscillation, is quite important for the proper functioning of clock-controlled genes in various biological processes. For example, the expression of the floral activator gene CONSTANS (CO) is controlled by a circadian clock. The diurnal pattern of CO mRNA accumulation is different in inductive (long days) versus noninductive (short days) photoperiods (Suarez-Lopez et al., 2001). Several circadian rhythm mutants affect CO expression in ways that correlate with their early- or late-flowering phenotypes. For example, in short day-grown elf3 and elf4 mutants, the level of CO mRNA is increased at all time points as compared with wild-type plants (Suarez-Lopez et al., 2001; Doyle et al., 2002). Increased CO expression is consistent with the early flowering phenotypes of elf3 and elf4.
We report evidence that gibberellin is involved in the expression of XSP30 in roots (Figs. 6 and 7). One of the major sites of production of gibberellin, which controls various aspects of plant development, is the leaf (Yamaguchi and Kamiya, 2000). Gibberellin production in pea (Pisum sativum) and pumpkin (Cucurbita pepo) is thought to be greatest in young expanded leaves (Smith et al., 1992, 1998). It is possible that this may also be true in cucumber seedlings, and that the decrease in XSP30 expression after removal of the first leaf is caused by the decrease in the endogenous levels of gibberellin (Figs. 4 and 5).
The effect of gibberellin on the regulation of XSP30 gene expression appears to be independent of the possible circadian regulation. In other words, the diurnal oscillation pattern of XSP30 expression does not appear to be controlled by the oscillating level of endogenous gibberellin, because application of high concentrations of GA did not disrupt the diurnal expression pattern of XSP30 (Figs. 6 and 7). GA is likely involved in the regulation of the amplitude or maximum expression level of XSP30, but not the periodicity, which might be controlled by a circadian clock (Figs. 2 and 3).
It is possible to interpret our results based on this hypothesis. The XSP30 expression cycles because the clock, if it is involved, may be gating the responsiveness of XSP30 expression to GA, because GA (or some signal downstream of GA) transport to the roots may oscillate over the diurnal cycle, or both. Diurnal GA synthesis in sorghum (Sorghum bicolor) partly supports this idea (Foster and Morgan, 1995). The overall expression pattern of XSP30 seems to be fine-tuned by GA and also, possibly, by a circadian rhythm.
We tested whether inhibitors of gibberellin, auxin, and brassinosteroid biosynthesis or transport affected the diurnal oscillation pattern of XSP30 gene expression, and we found that only the GA biosynthesis inhibitor, uniconazole-P, had a significant effect in our experimental conditions (Fig. 7 and A. Odah and S. Satoh, unpublished data). However, there is a possibility that signals other than gibberellin also participate in the diurnal expression pattern of XSP30 in roots. One signaling molecule candidate is IAA, which controls the rate of internodal elongation, and the endogenous levels of which fluctuate in a circadian rhythm in Arabidopsis (Jouve et al., 1999). Although no circadian rhythm in IAA levels has been demonstrated in cucumber, we postulate that such a rhythm might occur. In pea, removal of the apical bud inhibits certain GA-controlled processes in the remaining stem tissue, such as elongation, and the level of GA1 is also dramatically reduced. The effect of the apical bud can be replaced by applying auxin to the cut stump of the decapitated stem; this procedure restores the endogenous GA1 level and the elongation (Ross et al., 2000). It is possible that auxin may act similarly in cucumber seedlings, with the effects of GA replaced by applying auxin. IAA, or other signals produced in the shoot in a diurnal pattern, may be translocated to roots to establish a diurnal rhythm in underground tissues. Further analysis using inhibitors of IAA and other plant hormones at different concentrations will give us more information on this process.
The overall expression pattern of XSP30 may be fine-tuned by GA and by the circadian clock. This is quite similar to the growth pattern of the Arabidopsis inflorescence stem in which the circadian clock modulates auxin transport, auxin sensitivity, or both, to yield a rhythm in the rate of elongation (Jouve et al., 1998, 1999). The Lhc (also called CAB) mRNA levels exhibit a robust circadian pattern under free-running conditions in LL, despite the continuous presence of phytochrome in the active form (Millar and Kay, 1996). This observation led to the conclusion that the circadian clock must negatively regulate (or “gate”) the light-induced transcription of the Lhc and other clock-regulated plant genes, and this is called the gating hypothesis (Millar and Kay, 1996). Rhythmic alteration in the sensitivity of stomata to extracellular signals are also reported (Webb, 1998). The response of the stomata to these signals (the acute response) depends on the phase of the circadian cycle at which the signal is applied. Thus, the circadian clock can modulate (or gate) the acute responses of stomata (Webb, 1998).
Possible Physiological Function(s) of XSP30
Several examples of the effects of gibberellins applied directly to roots have been reported (Tanimoto, 1994; Yaxley et al., 2001). In the present study, gibberellins applied to the shoot affected XSP30 gene expression in roots (Figs. 6 and 7), suggesting that gibberellins are translocated to the roots or that gibberellins induce the production of a mediator in the shoot that causes a response in the roots. Supplying 10-4 m GA directly to the roots of uniconazole-P-treated plants did not affect XSP30 expression (data not shown), suggesting that mediators are involved in the transmission of gibberellin signals to roots. An alternative possibility is that GA applied directly to roots does not produce a physiological reaction detectable under our experimental conditions. However, in vegetative pea plants, grafting studies have indicated that endogenous GA1 is essentially immobile. In contrast, applied GA1 is very mobile, moving readily throughout the plant. In this sense, GA1 applied to pea plants behaves quite differently from endogenous GA1 (Proebsting et al., 1992). A detailed analysis to assess whether the effect(s) of the endogenous and applied GAs are similar will be performed using genetic and transgenic approaches.
The XSP30 promoter directed GUS expression specifically in the xylem parenchyma and pericycle cells in the central cylinder of mature transgenic hairy roots (Fig. 1). Thus, XSP30 is likely to be produced in the vascular tissues of mature roots. CRGRP-1 and CRGRP-2, genes that encode other xylem sap proteins, are also expressed in xylem parenchyma cells in the central cylinder of cucumber roots, and these proteins are transported to aboveground organs via xylem vessels (Sakuta et al., 2000). Production of proteins in the central cylinder of the root is probably necessary for the loading of macromolecules into xylem vessels. For example, the gene encoding the putative phosphate channel PHO1, which is hypothesized to load phosphate into the xylem, is expressed preferentially in parenchyma cells in the central cylinder of Arabidopsis roots (Hamburger et al., 2002). Recently, the gene for a transporter involved in the xylem loading of boron was reported to be expressed in the pericycle cells of Arabidopsis roots (Takano et al., 2002).
In summary, our results suggest that GA originating in the leaf, or some signal downstream of GA, provides important inducer(s) to generate high amplitude diurnal oscillations of the XSP30 gene expression. A circadian clock might be involved in the rhythmic expression of the gene, and both of these factors may optimize and fine-tune XSP30 gene expression in xylem parenchyma and pericycle cells of mature cucumber roots (Fig. 2). Any reciprocal regulation between circadian clocks and GA functions is still unclear. Possible clues include the expression of the GA20 oxidase gene being controlled by daily light/dark cycles, and the involvement of PHOR1, a GA-signaling mediator, in the photoperiodic control of tuber formation in potatoes (Carrera et al., 1999). Red light controls GA turnover by up-regulating the gene expression of GA2ox2, which converts GA1 to GA8 in pea (Reid et al., 2002). Therefore, there is an alternative possibility that the clock might affect GA turnover as well as synthesis. Analysis of endogenous GA level and XSP30 expression under the light cycle condition will elucidate the possible regulation between circadian clocks and GA functions. Recently, we have found that XSP30 has lectin activity and recognizes the N-linked glycans of glycoproteins in cucumber leaves (Oda et al., 2003). One attractive possibility is that the XSP30 protein is produced in underground roots and is delivered to leaves, where it could regulate biological processes through direct binding to the N-acetylglucosamine sugar chains of glycoproteins. In this sense, XSP30 may play a role in the communication between roots and shoots.
MATERIALS AND METHODS
Plant Materials
Seeds of cucumber (Cucumis sativus cv Shimoshirazu-jibai) were obtained from the Sakata Seed Co. (Kanagawa, Japan). Cucumber plants were grown in artificial soil (Kurehakagaku, Tokyo) under white fluorescent light (40 μmol m-2 s-1) with a 16-h light/8-h dark photoperiod at 28°C.
RNA Gel-Blot Analysis
Full-length cDNA probes for XSP30 (DDBJ; AB025717), ubiquitin, and rDNA were prepared using the PCR. Total RNA (10 μg lane-1) was isolated as described previously (Sakuta et al., 1998) from the combined roots of 10 treated plants. RNA gel-blot analysis was performed using a 32P-labeled probes and a BioImaging Analyzer (BAS 5000; Fuji Photo Film, Tokyo). Signal intensity was quantified using Science Lab 98 Image Gauge software (version 3.1; Fuji Photo Film). Values were represented relative to the values of samples at time 0 after normalization to the rRNA controls. Values at time 0 were shown as 100. All RNA gel blots were performed at least twice and usually with independent samples (see Supplemental Figs. 2 and 3). Fourier transforms, period, and RAE estimates were obtained using the Fast Fourier Transform-Non-Linear Least Squares program as described (Plautz et al., 1997). The RAE is the value of the amplitude error estimate divided by the value of the most probable amplitude estimate. RAE can range from a value of zero for an infinitely well-determined rhythmic component (zero error) to a value of one, theoretically, for a minimally determined rhythmic component (error in the amplitude equals the amplitude value itself).
Cloning of the XSP30 Promoter
To analyze the XSP30 promoter, XSP30 genomic DNA was cloned by thermal asymmetric interlaced PCR (Liu et al., 1995). The primers used, from the 3′ to the 5′ end, were: 5′-CTTTCCGTGACTTCCCAACTTTGTG-3′, 5′-TCTTCTGGGTCCATTTCTTGTTAGG-3′ and, 5′-CGTCACATGGTGATAATCGAGTTGG-3′. Short arbitrary degenerate primers were designed to have melting points (Tm) of 63°C to 68°C and 44°C to 46°C (Liu et al., 1995).
Plant Transformation and Induction of Hairy Roots
An 820-bp DNA fragment of the XSP30 genomic sequence upstream of the putative initiation codon (PXSP30) was amplified by PCR. The product was cloned upstream of, and in frame with, the GUS coding sequence in a modified pBI121 vector, which contains the cauliflower mosaic virus 35S promoter fused upstream of the hygromycin phosphotransferase gene (Iwai et al., 2001). This plasmid was introduced into Agrobacterium tumefaciens R1000 (Kamada et al., 1995) by the freeze-thaw method (Zahm et al., 1984). Transgenic hairy roots were obtained by inoculating the excised surface of 7-d-old cucumber plant cotyledons with A. tumefaciens harboring the above plasmid. The cotyledons were cultured on Murashige and Skoog agar-solidified medium under continuous light at 28°C.
Histochemical Analysis of GUS Activity
For GUS staining, hairy roots that emerged from the cotyledons were immersed in 1 mg mL-1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide in 50 mm sodium phosphate, pH 7.0. Samples were then subjected to a vacuum for 5 min and incubated at 37°C for 8 h (Jefferson et al., 1987). Stained samples were fixed in 2% (w/v) paraformaldehyde and 0.5% (w/v) glutaraldehyde in 50 mm sodium phosphate buffer (pH 7.0) at room temperature for 4 h and were then dehydrated by passage through an ethanol series. Dehydrated samples were embedded in Technovit 7100. Thin serial sections were cut with a tungsten knife using a microtome (RM-2415; Leica, Wetzlar, Germany). The sections were expanded in drops of water on glass slides (APS-coated microglass slides; Matsunami Glass Industries, Kishiwada, Japan) and were dried at 50°C.
SDS-PAGE of Xylem Sap Proteins
Xylem sap was collected, as described previously (Sakuta et al., 1998), at 4-h intervals from the cut stems of 1-month-old cucumber plants, at 4 h before dusk, at dusk, at midnight, at dawn, at 4 h after dawn, and at midday. The samples were immediately frozen in liquid nitrogen and stored at -20°C. Xylem sap samples (5 μL lane-1) were mixed with an equal volume of buffer (120 mm Tris-HCl, pH 6.8, 20% [v/v] glycerol, 4% [w/v] SDS, 0.01% [w/v] bromphenol blue, and 10% [v/v] β-mercaptoethanol), boiled for 10 min in a water bath, and then centrifuged at 15,000g for 5 min. The resulting supernatants were subjected to SDS-PAGE, as described by Laemmli (1970). After electrophoresis, the gels were stained with silver (Sil-Best stain; Nacalai Tesque, Kyoto) or subjected to immunological detection (Sakuta et al., 1998). To determine the amount of total protein, the entire intensity of each lane staining was quantified using Science Lab 98 Image Gauge software (version 3.1; Fuji Photo Film).
Immunological Detection of XSP30
Proteins were transferred from SDS-PAGE gels to nitrocellulose filters (ADVANTEC, Tokyo) in 25 mm Tris, 192 mm Gly, and 20% (v/v) methanol at 40 V for 2 h (Gershoni and Palade, 1982). The filters were incubated in phosphate-buffered saline (PBS) with 2% (w/v) bovine serum albumin overnight at 4°C, and then for 1 h in a 0.1% (v/v) solution of the XSP30-specific antiserum prepared from a rat immunized with an XSP30 fusion protein (Masuda et al., 1999). The filter was washed with 0.1% (v/v) Tween 20 in PBS and was agitated in a solution of 1,000-fold diluted goat anti-rat immunoglobulin G horseradish peroxidase-conjugated antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) in PBS for 1 h. Proteins were visualized by incubating the filter in PBS containing 0.03% (w/v) 3,3-diaminobenzidine and 0.003% (v/v) H2O2. The intensity of the signals was quantified using Science Lab 98 Image Gauge software (version 3.1; Fuji Photo Film).
Decapitation and Elimination of Aboveground Organs
The shoots of seedlings were removed at dawn with a razor blade at the middle of the hypocotyl, 15 d after sowing. Roots were collected from these seedlings every 4 h for RNA preparation. The first leaf or the cotyledons plus the shoot apex of the cucumber plants was cut off, 13 d after sowing, at dawn with a razor blade, and the roots were collected every 8 h for RNA preparation.
Gibberellin and Uniconazole-P Treatments
GA3 and uniconazole-P were dissolved in ethanol and the concentration of the solution was diluted 1,000-fold in 0.1% (v/v) Tween 20. The leaves and the shoot apex of 13-d-old plants were sprayed with 2 × 10-4 m GA3, 10-4 m uniconazole-P, or, as a control, 0.1% (v/v) ethanol, each with 0.1% (v/v) Tween 20. Roots were collected every 8 h for RNA preparation.
Acknowledgments
We thank Dr. Seiyei Yamakawa, Taichi Oguchi, Masashi Asahina, and Madoka Shimizu of the University of Tsukuba (Tsukuba, Japan) for their valuable suggestions.
This work was supported in part by the “Research for the Future” Program of the Japan Society for the Promotion of Science (grant no. JSPS-RFTF97L00601).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.030742.
References
- Abd-el Baki GK, Siefritz F, Man HM, Weiner H, Kaldenhoff R, Kaiser WM (2000) Nitrate reductase in Zea mays L. under salinity. Plant Cell Environ 23: 515-521 [Google Scholar]
- Asahina M, Iwai H, Kikuchi A, Yamaguchi S, Kamiya Y, Kamada H, Satoh S (2002) Gibberellin produced in the cotyledon is required for cell division during tissue-reunion in the cortex of cut cucumber and tomato hypocotyls. Plant Physiol 129: 201-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123: 93-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangerth F (1994) Response of cytokinin concentration in the xylem exudate of bean (Phaseolus vulgaris L.) plants to decapitation and auxin treatment, and relationship to apical dominance. Planta 194: 439-442 [Google Scholar]
- Bernier G (1988) The control of floral evocation and morphogenesis. Annu Rev Plant Physiol Plant Mol Biol 39: 175-219 [Google Scholar]
- Beveridge CA, Murfet IC, Kerhoas L, Sotta B, Miginiac E, Rameau C (1997) The shoot controls zeatin riboside export from pea roots: evidence from the branching mutant rms4. Plant J 11: 339-345 [Google Scholar]
- Biles CL, Abeles FB (1991) Xylem sap proteins. Plant Physiol 96: 597-601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Jackson SD, Prat S (1999) Feedback control and diurnal regulation of gibberellin 20-oxidase transcript level in potato. Plant Physiol 119: 765-773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JA, Loveys BR, Lee VWK, Strother S (1995) Growth-inhibiting properties of xylem exudate from Vitis vinifera. Aust J Plant Physiol 22: 7-13 [Google Scholar]
- Corlett JE, Wilkinson S, Thompson AJ (1998) Diurnal control of the drought-inducible putative histone H1 gene in tomato. J Exp Bot 49: 945-952 [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74-77 [DOI] [PubMed] [Google Scholar]
- Else MA, Hall KC, Arnold GM, Davies WJ, Jackson MB (1995) Export of abscisic acid, 1-aminocyclopropane-1-carboxylic acid phosphate, and nitrate from roots to shoots of flooded tomato plants. Plant Physiol 107: 377-384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KR, Morgan PW (1995) Genetic regulation of development in Sorghum bicolor. Plant Physiol 108: 337-343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershoni JM, Palade GE (1982) Electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to a positively charged membrane filter. Anal Biochem 124: 396-405 [DOI] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, Peteot JM, Somerville C, Poirier Y (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889-902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathway in Arabidopsis by the circadian clock. Science 290: 2110-2113 [DOI] [PubMed] [Google Scholar]
- Iwai H, Ishii T, Satoh S (2001) Absence of arabinan in the side chains of the pectic polysaccharides strongly associated with cell walls of Nicotiana plumbaginifolia non-organogenic callus with loosely attached constituent cells. Planta 213: 907-915 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusion: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901-3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouve L, Gaspar T, Kevers C, Greppin H, Agosti RD (1999) Involvement of indole-3-acetic acid in the circadian growth of the first internode of Arabidopsis. Planta 209: 136-142 [DOI] [PubMed] [Google Scholar]
- Jouve L, Greppin H, Agosti RD (1998) Arabidopsis thaliana floral stem elongation: evidence for an endogenous circadian rhythm. Plant Physiol Biochem 36: 469-472 [Google Scholar]
- Kamada H, Tachikawa Y, Saitou T, Harada H (1995) Effects of light and growth-regulators on adventitious bud formation in horseradish (Armoracia-rusticana). Plant Cell Rep 14: 611-615 [DOI] [PubMed] [Google Scholar]
- Kato C, Kato H, Asami T, Yoshida S, Noda H, Kamada H, Satoh S (2002) Involvement of xylem sap zeatin-O-glucoside in cucumber shoot greening. Plant Physiol Biochem 40: 949-954 [Google Scholar]
- Kinet JM, Lejeune P, Bernier G (1993) Shoot-root interactions during floral transition: a possible role for cytokinins. Environ Exp Bot 33: 459-469 [Google Scholar]
- Kolek J, Kozinka V (1991) Physiology of the plant root system. In, eds, Development in Plant and Soil Sciences. Kluwer Academic Press, London, pp 130-202
- Kuroha T, Kato H, Asami T, Yoshida S, Kamada H, Satoh S (2002) A trans-zeatin riboside in root xylem sap negatively regulates adventitious root formation on cucumber hypocotyls. J Exp Bot 53: 2193-2200 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685 [DOI] [PubMed] [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive FD, Filleur S, Daniel-Vedele F, Gojon A (1999) Molecular and functional regulation of two NO3- uptake systems by N- and C-status of Arabidopsis plants. Plant J 18: 509-519 [DOI] [PubMed] [Google Scholar]
- Liang J, Zhang J, Wong MH (1997) How do roots control xylem sap ABA concentration in response to soil drying? Plant Cell Physiol 38: 10-16 [Google Scholar]
- Liu Y, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457-463 [DOI] [PubMed] [Google Scholar]
- Lord JM, Roberts LM, Robertus JD (1994) Ricin: structures, mode of action, and some current applications. FASEB J 8: 201-208 [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T (2002) The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: characterization with APRR1-overexpressing plants. Plant Cell Physiol 43: 58-69 [DOI] [PubMed] [Google Scholar]
- Masuda S, Kamada H, Satoh S (2001) Chitinase in cucumber xylem sap. Biosci Biotechnol Biochem 65: 1183-1885 [DOI] [PubMed] [Google Scholar]
- Masuda S, Sakuta C, Satoh S (1999) cDNA cloning of a novel lectin-like xylem sap protein and its root-specific expression in cucumber. Plant Cell Physiol 40: 1177-1181 [DOI] [PubMed] [Google Scholar]
- McClung CR (2001) Circadian clock in plants. Annu Rev Plant Physiol Plant Mol Biol 52: 139-162 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Kay SA (1996) Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci USA 93: 15491-15496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carré IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629-641 [DOI] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell Suppl 2002: 111-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooden LD, Singh S, Letham DS (1990) Correlation of xylem sap cytokinin levels with monocarpic senescence in soybean. Plant Physiol 93: 33-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda A, Sakuta C, Kamada H, Satoh S (2003) Xylem sap lectin, XSP30, recognizes GlcNAc sugar chains of glycoproteins in cucumber leaf. Plant Biotechnol 20: 67-74 [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12: 204-217 [DOI] [PubMed] [Google Scholar]
- Proebsting WM, Hedden P, Lewis MJ, Crocker SJ, Proebsting LN (1992) Gibberellin concentration and transport in genetic lines of pea. Plant Physiol 100: 1354-1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB, Botwright NB, Smith JJ, O'Neill DP, Kerckhoffs LHJ (2002) Control of gibberellin levels and gene expression during de-etiolation in pea. Plant Physiol 128: 734-741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Dekker HL, Vossen JH, de Boer AD, Houterman PM, de Koster CG, Cornelissen BJC (2003) A tomato xylem sap protein represents a new family of small cysteine-rich proteins with structural similarity to lipid transfer proteins. FEBS Lett 543: 82-86 [DOI] [PubMed] [Google Scholar]
- Rep M, Dekker HL, Vossen JH, de Boer AD, Houterman PM, Speijer D, Back JW, de Koster CG, Cornelissen BJC (2002) Mass spectrometric identification of PR protein in xylem sap of fungus-infected tomato. Plant Physiol 130: 904-917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JJ, O'Neill DP, Smith JJ, Kerckhoffs LHJ, Elliott RC (2000) Evidence that auxin promotes gibberellin A(1) biosynthesis in pea. Plant J 21: 547-552 [DOI] [PubMed] [Google Scholar]
- Sakuta C, Oda A, Yamakawa S, Satoh S (1998) Root-specific expression of genes for glycine-rich proteins cloned by use of an antiserum against xylem sap proteins of cucumber. Plant Cell Physiol 39: 1330-1336 [DOI] [PubMed] [Google Scholar]
- Sakuta C, Satoh S (2000) Root-specific gene expression of novel glycine-rich proteins and their systemic delivery to the wall of metaxylem vessels via the xylem sap in cucumber. Plant Cell Physiol 41: 627-638 [DOI] [PubMed] [Google Scholar]
- Satoh S (1996) Inhibition of flowering of cucumber grafted on rooted squash stock. Physiol Plant 97: 440-444 [Google Scholar]
- Satoh S, Iizuka C, Kikuchi A, Nakamura N, Fujii T (1992) Protein and carbohydrates in xylem sap from squash root. Plant Cell Physiol 33: 841-847 [Google Scholar]
- Satoh S, Kuroha T, Wakahoi T, Inouye Y (1998) Inhibition of the formation of adventitious roots on cucumber hypocotyls by the fractions and methoxybenzylglutamine from xylem sap of squash root. J Plant Res 111: 541-546 [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219-1229 [DOI] [PubMed] [Google Scholar]
- Schurr U, Schulze ED (1995) The concentration of xylem sap constituents in root exudate, and in sap from intact, transpiring castor bean plants (Ricinus communis L.). Plant Cell Environ 18: 409-420 [Google Scholar]
- Smith MW, Yamaguchi S, Tahar AA, Kamiya Y (1998) The first step of gibberellin biosynthesis in pumpkin is catalyzed by at least two copalyldiphosphate synthases encoded by differentially regulated genes. Plant Physiol 118: 1411-1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith VA, Knatt CJ, Gaskin P, Reid JB (1992) The distribution of gibberellins in vegetative tissues of Pisum sativum L. Plant Physiol 99: 368-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soejima H, Sugiyama T, Ishihara K (1992) Changes in cytokinin activities and mass spectrometric analysis of cytokinins in root exudates of rice plant (Oryza sativa L.). Plant Physiol 100: 1724-1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE (1999) The physiology and molecular basis of the plant circadian clock. Plant Physiol 121: 9-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock cone TOC1, an autoregulatory response regulator homolog. Science 289: 768-771 [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116-1120 [DOI] [PubMed] [Google Scholar]
- Takano J, Noguchi K, Yasumori M, Kobayashi M, Gajdos Z, Miwa K, Hayashi H, Yoneyama T, Fujiwara T (2002) Arabidopsis boron transporter for xylem loading. Nature 420: 337-340 [DOI] [PubMed] [Google Scholar]
- Tanimoto E (1994) Interaction of gibberellin A3 and ancymidol in the growth and cell wall extensibility of dwarf pea roots. Plant Cell Physiol 35: 1019-1028 [Google Scholar]
- Thain SC, Murtas G, Lynn JR, McGrath RB, Millar AJ (2002) The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol 130: 102-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207-1217 [DOI] [PubMed] [Google Scholar]
- Webb AAR (1998) Stomatal rhythms. In PJ Lumsden, AJ Millar, eds, Biological Rhythms and Photoperiodism in Plants. BIOS Scientific Publishers, Oxford, pp 69-80
- Yamaguchi S, Kamiya Y (2000) Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol 41: 251-257 [DOI] [PubMed] [Google Scholar]
- Yaxley JR, Ross JJ, Sherriff LJ, Reid J (2001) Gibberellin biosynthesis, mutation and root development in pea. Plant Physiol 125: 627-633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahm P, Hohmeyer C, Geider K (1984) Site specific mutagenesis of the Ti plasmid by transformation of Agrobacterium tumefaciens with mutagenized T-DNA fragments cloned in E. coli plasmids. Mol Gen Genet 194: 188-194 [Google Scholar]
- Zornoza P, Gonzalez M, Serrano S, Carpena O (1996) Intervarietal differences in xylem exudate composition and growth under contrasting forms of N supply in cucumber. Plant Soil 178: 311-317 [Google Scholar]