Abstract
Peroxisomes participate in many important functions in plants, including seed reserve mobilization, photorespiration, defense against oxidative stress, and auxin and jasmonate signaling. In mammals, defects in peroxisome biogenesis result in multiple system abnormalities, severe developmental delay, and death, whereas in unicellular yeasts, peroxisomes are dispensable unless required for growth of specific substrates. PEX10 encodes an integral membrane protein required for peroxisome biogenesis in mammals and yeast. To investigate the importance of PEX10 in plants, we characterized a Ds insertion mutant in the PEX10 gene of Arabidopsis (AtPEX10). Heterozygous AtPEX10::dissociation element mutants show normal vegetative phenotypes under optimal growth conditions, but produce about 20% abnormal seeds. The embryos in the abnormal seeds are predominantly homozygous for the disruption allele. They show retarded development and some morphological abnormalities. No viable homozygous mutant plants were obtained. AtPEX10 fused to yellow fluorescent protein colocalized with green fluorescent protein-serine-lysine-leucine, a well-documented peroxisomal marker, suggesting that AtPEX10 encodes a peroxisomal protein that is essential for normal embryo development and viability.
Peroxisomes are involved in a diverse repertoire of functions in plant cells. In addition to the well-established metabolic roles in mobilization of seed lipid via the peroxisomal pathways of β-oxidation and the glyoxylate cycle, and the salvage of carbon via the photorespiratory cycle, new functions are still being discovered. Much of this information has come from the study of mutants. In recent years it has become apparent that peroxisomes play important roles in reactive oxygen metabolism, including the formation and turnover of the signaling molecules nitric oxide and H2O2 (Corpas et al., 2001), as well as in the biosynthesis of the major auxin indole acetic acid (Zolman et al., 2000) and the oxylipin jasmonic acid (Stintzi and Browse, 2000). Both of the latter are formed by peroxisomal β-oxidation from the precursor molecules indole butyric acid (IBA) and 3-oxo-2(2′[Z]-pentenyl)-cyclopentane-1-octanoic acid), respectively. Some mutants in peroxisome biogenesis or function have a profound effect on plant development. The comatose mutant (cts-1,2), which is defective in transport of fatty acids or acyl coenzyme A (CoA) into peroxisomes, fails to activate the developmental switch from dormancy to germination (Footitt et al., 2002). The ted3 mutant, which is a gain-of-function mutation in the peroxisomal protein PEX2, was isolated as a suppressor of the de-etiolated 1 (det1-1) mutant (Hu et al., 2002).
Peroxisomes have no DNA, therefore all peroxisomal proteins are encoded by nuclear genes and the products imported posttranslationally. Twenty-five genes required for this process have been isolated from various yeast species. Many of them have homologs in mammals, plants, and invertebrates, although the functions that peroxisomes perform differs in these diverse organisms. These genes required for peroxisome biogenesis have been given the acronym “PEX” and the corresponding encoded protein “peroxin” (Distel et al., 1996).
Pex10p is an integral peroxisome membrane protein that has been characterized in mammals and various yeast species. Mutants in Pex10p have a defect in transporting matrix proteins containing peroxisomal-targeting signals one (PTS1) and two (PTS2) into peroxisomes. Import receptors for PTS1 or PTS2 proteins are encoded by PEX5 and PEX7, respectively. Pex5p and Pex7p are cycling receptors that bind their cargo in the cytosol, and subsequently dock at the peroxisome membrane at a complex that includes Pex13p and Pex14p. (See Sparkes and Baker, 2002 for a recent review of peroxisome protein import.) In a human fibroblast line deficient in PEX10 (PBD100), the PTS1 receptor Pex5p accumulates on the surface of the peroxisome (Chang et al., 1999) and, consistent with this, an epistatic analysis of PEX mutants in the yeast Pichia pastoris placed the function of Pex10p downstream of receptor docking (Collins et al., 2000). Pex10p is believed to span the membrane twice with N- and C-termini exposed to the cytosol (Okumoto et al., 1998; Faber et al., 2001). The C terminus has a C3HC4 RING finger motif that has been shown to be essential for function (Kalish et al., 1995). Two other peroxins, Pex2p and Pex12p, have RING finger motifs and these three RING finger peroxins interact with one another (Okumoto et al., 2000). These data have led to the suggestion (although direct proof is still lacking) that the RING peroxins may form (part of) a translocation channel for import of matrix proteins. All Pex10 proteins have a conserved motif “TLGEEY” of unknown function in the amino-terminal part of the protein.
In humans, mutations that affect peroxisome biogenesis lead to disease. Mutations that disturb import of PTS1- and PTS2-targeted proteins result in the Zellweger spectrum of peroxisome biogenesis disorders. The most severe clinical symptoms are Zellweger syndrome itself, which results in neurological, hepatic, and renal dysfunction, as well as facial abnormalities and muscle weakness. Patients with this syndrome suffer multiple biochemical abnormalities arising from the loss of peroxisomal functions, and rarely survive their first year (Wanders, 1999). Mutations in several different genes, including PEX10, can give rise to Zellweger syndrome (Okumoto et al., 1998; Warren et al., 1998).
With the completion of the Arabidopsis genome sequence, 15 possible homologs of mammalian and fungal PEX genes have been identified (Mullen et al., 2001; Charlton and Lopez-Huertas, 2002). Genetic screens have identified mutants in four of them, AtPEX2 (TED3; Hu et al., 2002), AtPEX5 (Zolman et al., 2000), AtPEX14 (Hayashi et al., 2000), and AtPEX16 (SSE1; Lin et al., 1999). We show that null Arabidopsis pex10 (Atpex10) mutants display delayed embryo development, which accounts for the high frequency of seed abortion in the progeny of heterozygous individuals. Possible reasons for this phenotype in the context of the known functions of plant peroxisomes and alternative explanations are discussed.
RESULTS
Subcellular Localization of AtPEX10-YFP and Expression of AtPEX10 in Arabidopsis Plants
Previously, we have isolated a cDNA encoding a protein with 47% to 56% sequence similarity to the product of the PEX10 gene from mammals and fungi, and we have shown that it is encoded by a single genomic locus, At2g26350 (Baker et al., 2000; Sparkes and Baker, 2002). To investigate whether AtPEX10 is a true ortholog of the PEX10 gene of mammals and fungi, we examined its subcellular localization by fusing it to the yellow fluorescent protein (YFP; Fig. 1).
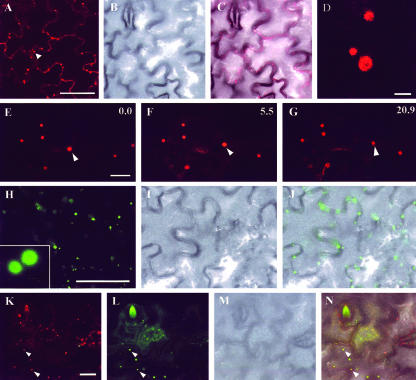
Figure 1.
AtPEX10-YFP fusion localizes to peroxisomes in tobacco (Nicotiana tabacum) leaf epidermal cells. A, Tobacco leaf epidermal cells transiently expressing a AtPEX10-YFP fusion. Scale bar = 50 μm. B, Same cells as in A visualized with transmitted light to show cell boundaries. C, Merged image of A and B. D, Higher magnification of structures highlighted with AtPEX10-YFP. Note the absence of fluorescence in the center of these structures (scale bar = 2 μm). E through G, Time-lapse sequence of epidermal cells expressing an AtPEX10-YFP fusion. An arrowhead indicates one of these structures covering a greater distance in comparison with other AtPEX10-YFP bodies present in the same field of view. Time is expressed in seconds at the top right of each frame. Scale bar = 10 μm. H through J, Leaf epidermal cells transiently expressing a GFP-SKL fusion imaged with GFP settings of the confocal microscope (H; scale bar = 50 μm). Inset in H represents a higher magnification comparable with that in D. I, Same cells as in H imaged with transmitted light. The two images are merged in J. K through N, Colocalization of GFP and YFP fusions in epidermal cells of a leaf in which a mixed population of Agrobacterium tumefaciens bearing AtPEX10-YFP and GFP-SKL constructs was inoculated. K, AtPEX10-YFP expression. Scale bar = 20 μm. L, Same cells as in K showing GFP-SKL expression. M, Transmitted light image of cells pictured in K and L. N, In a merged image of K through M, it is possible to identify cells that are coexpressing AtPEX10-YFP and GFP-SKL. In these cells, AtPEX10-YFP colocalizes with peroxisomes highlighted with GFP-SKL (arrowheads).
AtPEX10-YFP was expressed transiently in tobacco leaf epidermal cells and was visualized using confocal microscopy. AtPEX-YFP located to punctate spherical structures similar in size to plant peroxisomes (0.5-1.5 μm; Fig. 1, A and D). Low laser settings of the microscope allowed us to identify the distribution of the YFP fluorescence toward the rim of some of these spherical structures (Fig. 1D, compare with inset in H from GFP-SKL expression). Tobacco leaf peroxisomes generally contain catalase crystals, which may explain the pattern of fluorescence. These structures showed distinct behaviors being stationary or moving over a few microns or more (Fig. 1, E-G), all of which are characteristic of peroxisome movement in Arabidopsis and onion (Allium cepa; Mano et al., 2002; Mathur et al., 2002; Muench and Mullen, 2003).
To investigate the nature of the structures highlighted by AtPEX10-YFP, we transiently expressed in tobacco leaf epidermal cells a green fluorescent protein (GFP) fused to the C-terminal tripeptide Ser-Lys-Leu (SKL), a common signal sequence for peroxisomal matrix proteins. A YFP-SKL fusion has already been used in Arabidopsis and onion as an in vivo marker for peroxisomes (Mathur et al., 2002). In leaf epidermal cells, GFP-SKL fluorescence highlighted spherical structures similar to those visualized with a AtPEX10-YFP fusion (Fig. 1, H-J). To confirm the identity of the AtPEX10-YFP structures with peroxisomes, we cotransformed tobacco leaf epidermal cells with cultures of A. tumefaciens containing binary vectors AtPEX10-YFP and GFP-SKL. Confocal observation of cells coexpressing both constructs revealed colocalization of fluorescence due to expression of the AtPEX10-YFP with GFP-SKL (Fig. 1, K-N). Similar results were also obtained when the AtPEX10-YFP construct was transiently expressed in Arabidopsis plants that had been stably transformed with a peroxisomal targeted GFP (the A5 line described by Cutler et al., 2000; data not shown). These results allowed us to conclude that the AtPEX10-YFP fusion localizes to peroxisomes.
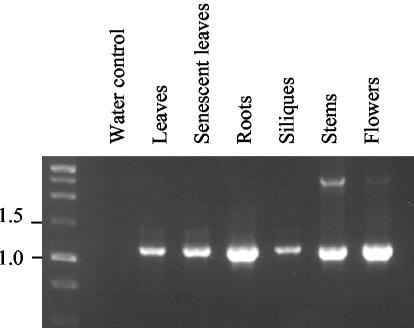
The tissue expression profile of AtPEX10 was determined by reverse transcriptase (RT)-PCR (Fig. 2). Total RNA was extracted from green leaves, senescent leaves, roots, siliques, stems, and flowers and was reverse transcribed and amplified with PEX10 gene-specific primers P4 and P11 (see Fig. 3A). AtPEX10 transcript was detected in all tissues tested. AtPEX10 expression was also detected in seedlings by northern blotting, with maximal expression occurring at d 1 and 2 after imbibition (data not shown). This is in good agreement with the expression profile of other PEX genes from Arabidopsis (Lopez-Huertas et al., 2000).
Figure 2.
Arabidopsis PEX10 is expressed in a range of tissues. RT-PCR of AtPEX10 from Arabidopsis tissues (indicated above lanes) shows that AtPEX10 is widely expressed. The primers used were P4 and P11, and the expected size of AtPEX10 amplified product is 1.2 kbp.
Figure 3.
Location and segregation of the dissociation element (Ds) in the Atpex10 line. A, Schematic of genomic sequence of AtPEX10, boxes refer to exons. The Ds element is located in exon 4, and primers used to determine location and orientation of the Ds element in AtPEX10 are displayed. B, Arabidopsis PEX10 (AJ276134) is predicted to have two transmembrane domains (underlined) and a conserved zinc RING finger at the carboxy terminus (boxed residues). The section in bold italics represents the part of the protein that could theoretically be translated from the mutant allele. Five PCR products derived from independent PCR reactions were sequenced. All five contained the substitution M184K relative to the previously published sequences AC002505 and AF119572. In addition, the substitution P199L was found in four out of the five independent PCR products, but the E357K mutation was found in only one PCR product. Therefore, it seems likely that M184K and P199L are genuine polymorphisms, but E357K is a PCR-induced mutation. These three substitutions are shown in bold type. C, Genomic extracts from seven progeny from selfed Atpex10 heterozygotes were genotyped. Two PCR reactions per plant were performed with primer combinations P11 and P18 (black arrowhead) and P11 and DS3 (white arrowhead). P11 and P18 amplifies a 910-bp product from the wild-type allele, and P18 and DS3 amplifies a product of 453 bp from the disrupted allele. Two segregating wild-type (WT) and five heterozygotes (Hh) are shown. Water controls and control PCR reactions from genomic DNA from wild-type plants are also shown.
Characterization of an Arabidopsis pex10 Disruption Mutant
A potential Ds insertion mutant in At2g26350 (AtPEX10) was reported among a population of sequenced insertion sites (Parinov et al., 1999). Seed from this line (SGT3777; NASC stock no. N100156 we refer to here as the T0 generation) were ordered and selected on kanamycin. The site of insertion was confirmed by sequencing PCR products amplified from genomic DNA from T0 plants, using AtPEX10-and Ds-specific primers (Fig. 3A). The Ds insert in AtPEX10 is located in exon four, 599 bp downstream of the ATG (Fig. 3A), and is in the opposite orientation to the AtPEX10 gene. This orientation should result in an AtPEX10-β-glucuronidase (GUS) transcriptional fusion, as the Ds element carries a GUS gene trap with splice acceptor and donor sites in all three reading frames (Parinov et al., 1999). Unfortunately, no GUS activity could be detected in any tissues tested (data not shown). The Ds insertion is predicted to result in a severely truncated protein that lacks the RING domain and both predicted transmembrane domains (Fig. 3B). Therefore, it is unlikely that the truncated protein will be correctly inserted into the peroxisome membrane.
Southern-blot analysis of the Atpex10 line showed a single Ds insertion within the genome in AtPEX10 (data not shown). Three hundred and fifty-five seeds from a selfed heterozygous Atpex10 plant selected on kanamycin resulted in 240 kanamycin-resistant and 105 kanamycin-sensitive seedlings. Therefore, the ratio of kanamycin-resistant to -sensitive seed approximates 2:1 instead of the 3:1 ratio expected for the segregation of a single dominant marker. Forty T1 progeny from two selfed heterozygous T0 plants were genotyped by PCR. Examples of seven individuals are shown along with a wild-type control (Fig. 3C). In total, 14 wild-type and 26 heterozygous plants were obtained (1:2 ratio). No Atpex10 plants homozygous for the Atpex10 allele containing the Ds insert were present in the 40 plants analyzed.
Peroxisomal β-oxidation is an essential metabolic pathway. It produces the energy and carbon skeletons required for seedling establishment by mobilizing stored lipids (triacylglycerol) in the seed. A number of β-oxidation mutants require Suc for seedling establishment (Hayashi et al., 1998; Zolman et al., 2000, 2001; Footitt et al., 2002). To determine whether homozygous mutant plants simply fail to germinate, seed from a selfed heterozygous Atpex10 plant were compared with wild type for differences in germination levels (as defined by radicle emergence) in the presence and absence of 1% (w/v) Suc. No significant differences (at the 50% level of significance) were found between wild type and seed from the heterozygous Atpex10 mutant in the presence or absence of Suc (data not shown). Establishment, defined as the ability to produce true foliage leaves within 18 d of plating, was also tested. The percentage of seedlings establishing without Suc from a selfed heterozygote was low, but not significantly different (10% level of significance) compared with wild type (34.6% established for WT without Suc compared with 48.8% [χ2 = 2.19; P > 0.1] and 35.2% [χ2 = 0.95; P > 0.1] for seedlings from two selfed heterozygous plants). Therefore, the lack of homozygous mutant plants cannot be explained by failure to germinate or establish in the absence of Suc.
Heterozygous plants grew normally and did not differ in appearance from wild-type plants grown along side them. Similarly, transmission electron micrographs of transverse sections through mesophyll cells treated with a peroxisomal cytochemical stain diaminobenzidine revealed no obvious difference in the ultrastructure of cotyledon cells of heterozygous Atpex10 and wild-type seedlings (data not shown). Additionally, the morphology of the peroxisomes, mitochondria, and chloroplasts appear unaltered from wild type, and are all closely associated. Heterozygote and wild-type seedlings showed similar sensitivity to 2,4-dichlorophenoxybutyric acid (2,4-DB; data not shown), a herbicide that is bioactivated by peroxisomal β-oxidation and inhibits root growth (Hayashi et al., 1998), suggesting that the heterozygous mutants have no detectable defect in β-oxidation.
Selfed Heterozygous pex10 Plants Produce a High Frequency of Abnormal Seed
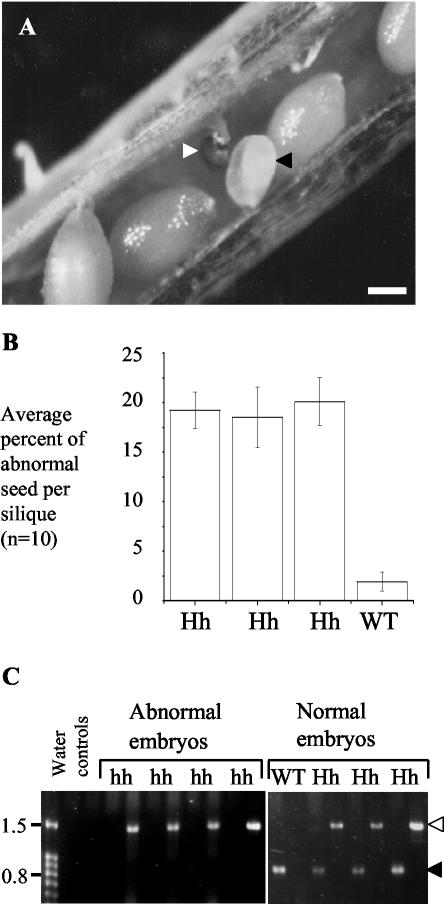
Developing seed from three heterozygous and one wild-type selfed plant were analyzed. During Arabidopsis seed development, the external appearance of the immature seed changes from white to green, and then browns before seed release. Immature seed within a single silique develop at approximately the same rate, therefore, abnormal seed appear white, or brown and shriveled in comparison with normal green developing seed (Fig. 4A; Meinke, 1994). Ten siliques were removed from each plant, and the developing seeds were removed and scored for abnormalities in external morphology. Approximately 300 seeds per plant were counted. The average frequency of abnormal seeds per silique for a heterozygous plant was 19.4%, whereas only 1.9% of seed from siliques from wild-type plants were abnormal (Fig. 4B). The levels of abnormal seed seen in the three heterozygous Atpex10 mutant plants were found to be significantly different from wild-type levels at the 0.1% level of significance (χ2 = 386.3, 253.5, and 357.1; P > 0.001).
Figure 4.
Homozygous Atpex10 mutants are embryo lethal. A and B, Developing seed from siliques from selfed heterozygotes show a higher level of abnormal seeds per silique than seen in wild type. The abnormal seed are white (black arrowhead) or brown and shriveled (white arrowhead). Scale bar = 0.2 mm. C, Embryos were dissected from the developing seed and were genotyped by PCR. Two PCR reactions per embryo were performed using primers GUS1 and GUS2 (white arrowhead) or EM1 and EM2 (black arrowhead). GUS1 and GUS2 amplifies a 1.5-kbp product from the disrupted Atpex10 allele; EM1 and EM2 amplifies a 755-bp product from the wild-type allele.
Homozygous Atpex10 Is Embryo Lethal
As no homozygous Atpex10 plants were identified in PCR genotype screens, and the segregation of wild-type to heterozygous plants was 1:2, it seemed likely that the missing homozygous mutants might be found among the embryos in the abnormal seed. Seeds were classified as normal or abnormal on the basis of morphological appearance, and the embryos cleanly dissected away from the surrounding endosperm tissue. Single-embryo PCR was carried out with two primer pairs, EM1 and EM2, and GUS1 and GUS2 (Figs. 3A and 4C). Results from four morphologically abnormal embryos and four normal embryos are shown in Figure 4C. All of the abnormal embryos contain only the disruption allele, demonstrating that they are homozygous for the Atpex10 gene. One of the normal embryos contains only the wild-type allele-specific band of 755 bp, whereas the remaining three contain 755 bp and 1.4 kbp bands and are therefore heterozygous. Forty embryos were genotyped in total; 31 classified as morphologically abnormal and nine as normal. Twenty-nine of the 31 abnormal embryos were homozygous mutants, the remaining two were heterozygotes. Of the nine embryos that appeared normal, three were wild type, four were heterozygotes, and two were homozygous mutants. These results indicated that a large proportion of the homozygous mutant population are morphologically abnormal, although the presence of two homozygous mutants among the morphologically normal embryos suggest that some may develop further than others. However, the results from germination tests and genotyping adult plants indicate that these embryos do not give rise to viable seed.
To investigate the developmental abnormalities associated with the homozygous Atpex10 mutation, developing seeds were examined by light microscopy using Normarski optics. Developing seeds from siliques from selfed heterozygous Atpex10 plants were removed. Morphologically abnormal seeds were compared with seeds of normal appearance from the same silique. In all cases, abnormal embryos were at an earlier developmental stage than the normal embryos from the same silique. Figure 5B shows an embryo from a normal seed at the torpedo stage of development (about 4.5 d after flowering under our growth conditions), whereas Figure 5C shows an abnormal embryo from the same silique at the earlier globular stage normally seen around 2.5 d after flowering. Figure 5D shows a normal embryo from a different silique at the curled cotyledon stage (more than 5 d after flowering), whereas abnormal embryos from this silique (Fig. 5E) and staged siliques from the same time point (Fig. 5, F-H) ranged from globular to heart stages. These and further observations from staged siliques demonstrate that the abnormal embryos do not arrest at a single developmental time point, but appear to develop more slowly and eventually abort. Genotyping of embryos by PCR indicated that some homozygous mutant embryos could develop at least as far as the torpedo stage. Seed-containing embryos at the torpedo stage of development begin to green. Embryos from green seed were removed and genotyped, but their developmental stage was not recorded, and could therefore have been more advanced than the torpedo stage. This result suggests that the phenotype of Atpex10 homozygous mutants should be classified as variable using the classification of McElver et al. (2001). Some evidence of developmental abnormalities was seen in some embryos. Comparison between Figure 5, E and G, indicates Figure 5G is an enlarged globular stage embryo. Similar comparison between Figure 5, F and H, indicates the embryo in Figure 5H is an enlarged heart stage embryo. Additional observations made during dissection of embryos from their seed coat for PCR analysis indicated some also displayed unevenly sized cotyledons (data not shown). Enlarged or abnormal embryos are a fairly common occurrence in embryo-lethal mutants, for example, acc1-1, defective in a cytosolic acetyl CoA carboxylase (Baud et al., 2003). Likewise, a homozygous insertion in AtCSLA7, a cellulose synthase-like putative glycosyl transferase, was embryo lethal and resulted in slower development compared with wild type (Goubet et al., 2003).
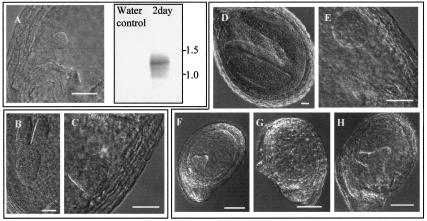
Figure 5.
Mutants show developmental delay. A, RT-PCR with AtPEX10-specific primers was performed on whole seed from wild-type plants 2 d postflowering. Normarski image of developing embryo shows that at 2 d postflowering, embryos are at early globular stage of development. The amplified products were blotted and probed with an AtPEX10-specific probe. B through H, Developing seed from selfed heterozygous Atpex10 plants were removed from the silique and viewed using Normarski optics. Seeds were separated based on normal (B and D) or abnormal (C, and E through H) external appearance, where B and C are from the same silique and D and E are from the same silique. F through H are from siliques containing morphologically normal embryos at the same developmental stage as D. Embryos in normal seeds are at a later developmental stage to embryos in abnormal seeds from the same silique, and some of the abnormal embryos appear to be larger (compare E with G, and F with H). Scale bar: A through E = 10 μm; F through H = 50 μm.
Wild-type developing seeds were studied to determine whether AtPEX10 expression early in embryogenesis occurred. Total RNA was extracted from whole wild-type seed-containing embryos at the early globular stage of development (Fig. 5A). RTPCR with AtPEX10-specific primers followed by Southern blotting with an AtPEX10-specific probe showed that AtPEX10 is expressed at this developmental stage (Fig. 5A). This is consistent with a lack of AtPEX10 leading to defects in development. To determine whether abnormal embryos still had peroxisomes, cleanly dissected embryos at the heart-torpedo stage were fixed and embedded for electron microscopy. No peroxisomes could be identified even after cytochemical staining with diaminobenzidine. However, the cellular ultrastructure was highly abnormal. In some cases, nuclei were visible with prominent nucleoli. The nuclei were clearly surrounded by a double membrane but this was highly distended with gaps of up to 0.27 μm between the two membranes. The cytoplasm was packed with heterogeneous vesicles, and no other organelles could be identified with confidence. It seemed likely that these cells were dying and no conclusions could be drawn about the presence or absence of peroxisomes (data not shown).
The Embryo-Lethal Phenotype Can Be Complemented by a Wild-Type Copy of AtPEX10
Plants heterozygous for the Atpex10 disruption allele were transformed with a wild-type genomic copy of AtPEX10. Two plants (C1A and C4A) resistant to hygromycin, the selectable marker for the transgene, were recovered. PCR analysis indicated that they were transformed heterozygous individuals (data not shown). To determine whether the AtPEX10 transgene complemented the embryo-lethal phenotype, two criteria were tested. The first criterion was to test whether the levels of aborted seed were reduced to that expected for complemented plants, and the second criterion was whether homozygous plants could be grown from the complemented seeds. The former was tested by dissecting 10 siliques from C4A and C1A self-fertilized plants, and scoring for the number of abnormal versus normal seeds. The percentage of abnormal seeds per silique was determined and the average of 10 siliques was taken. Results indicate that the levels of abnormal seeds per silique from selfed C1A (4.47%) and C4A (1.39%) plants is similar to that expected for complemented plants carrying one (6.25%) or two (1.56%) copies of the transgene, respectively. T2 seeds harvested from self-fertilized C4A and C1A plants were selected on kanamycin, and the resistant progeny were genotyped. Genomic DNA extracted from the resistant progeny was amplified with primers specific for the transgene or for the chromosomal copy of AtPEX10 (data not shown). Out of the eight plants tested, two lacked the chromosomal copy of AtPEX10 and were kanamycin resistant. These two individuals are homozygous for the Ds insertion allele. Therefore, the embryo-lethal phenotype can be complemented by the AtPEX10 gene, giving rise to adult plants.
DISCUSSION
AtPEX10 fused at the amino terminus of YFP colocalizes with GFP-SKL when transiently expressed in tobacco (Fig. 1) or with a peroxisome-targeted GFP-MFP2 fusion protein when expressed in Arabidopsis (data not shown). The size and motility of the structures labeled by the AtPEX10-YFP fusion protein are consistent with those previously reported for plant peroxisomes (Mano et al., 2002; Mathur et al., 2002; Muench and Mullen, 2003), indicating that AtPEX10 is a peroxisomal protein. An Atpex10 Ds insertion allele (SGT3777) has a recessive embryo-lethal phenotype. Selfed heterozygote plants produced approximately 20% abnormal seeds, the majority of which were homozygous for the Atpex10 disruption allele, and no homozygous mutant adult plants were obtained. Abnormal seeds contained embryos at an earlier developmental stage than normal seeds in the same silique. Some embryos appeared abnormal or enlarged as well as developmentally delayed. Similar phenotypes have been seen in some other embryo-lethal mutants (e.g. Patton et al., 1998; Baud et al., 2003; Goubet et al., 2003). As AtPEX10 is widely expressed in plant tissues, Atpex10 is presumably an example of the class of mutants whose embryo lethality reflects an essential function that is first required during embryogenesis (Meinke, 1995; Patton et al., 1998).
Heterozygous (Atpex10/AtPEX10) plants do not display a vegetative phenotype or detectable abnormalities at the cellular level, or in β-oxidation under optimal growth conditions. Seeds from a selfed heterozygous Atpex10 plant are not resistant to 2,4-DB. This compound is bioactivated to 2,4-D (an auxin) by peroxisomal β-oxidation and has been used as a selection for mutants in β-oxidation and peroxisome biogenesis. Some resistant mutants show a long root phenotype when grown on 2,4-DB or its analog IBA even when heterozygous, presumably as they do not metabolize enough 2,4-DB to result in inhibition (Hayashi et al., 1998; Zolman et al., 2000). Therefore, these results suggest that β-oxidation is not significantly impaired in the Atpex10 heterozygote.
Seed development is a complex process and many genes when mutated can give rise to embryo lethality (Tzafrir et al., 2003; www.seedgenes.org). Recent estimates suggest that 500 to 1,000 genes may be essential for seed development in Arabidopsis (McElver et al., 2001). Known embryo-defective genes encode proteins with diverse functions, including metabolism, cell growth, transcription, translation, protein fate, protein transport and traffic, and plant defense (McElver et al., 2001). In some cases, the link between the mutation and the phenotype is obvious, but in many cases, it is unclear. Why should the Arabidopsis pex10 mutant be embryo lethal? Considering that AtPEX10 is localized to peroxisomes, and Pex10p in yeast and mammals is involved in peroxisomal matrix import, the most likely explanation is that peroxisome matrix protein import is abolished or severely impaired. Three possibilities relating to the role(s) peroxisomes may play during embryogenesis suggest themselves. First, peroxisomes may play an essential nutritional role in embryogenesis. Nutritional deficiency, for example, in biotin (Patton et al., 1998) or folate (Ishikawa et al., 2003) biosynthesis can result in an embryo-lethal phenotype. The bio2 (Patton et al., 1998) and gla1 (Ishikawa et al., 2003) mutants arrest between the globular and heart stage, when presumably the supply of nutrients from maternal and/or endosperm tissue is no longer sufficient to support embryo development. Genes encoding β-oxidation enzymes are relatively abundant in expressed sequence tag collections from developing Arabidopsis seeds (White et al., 2000), suggesting that some fatty acids synthesized in the seeds are turned over during embryogenesis, possibly providing energy for differentiation and development. Second, peroxisomes play a major role in detoxifying reactive oxygen species. Although antioxidants are present in other cellular compartments, perhaps embryos with defective peroxisomes suffer an overload of reactive oxygen species that results in cell death. Finally, peroxisomes may be a source of signal molecules that are necessary for proper development. Peroxisomes are required for the formation of jasmonate and of indole acetic acid from IBA, although precursors of these molecules (IBA and 12-oxophytodienoic acid), which are not dependent upon peroxisomes, have some biological activities (Bartel et al., 2001; Weber, 2002). The role of jasmonate or other lipid-derived signals in embryogenesis has not, to our knowledge, been addressed. Mutations in ABP1, a proposed auxin receptor, or MONOPTEROS, a transcription factor that can bind auxin-responsive elements, gives rise to defects during embryogenesis, and null abp1 mutants are embryo lethal (Hardtke and Berleth, 1998; Chen et al., 2001). Recently, double mutants in the medium-chain (acx3) and short-chain (acx4) acyl-CoA oxidase activities were reported to abort at the preheart stage, which provides direct evidence of the requirement for short chain β-oxidation activity during embryogenesis (Rylott et al., 2003). Whether this lethality is due to a disruption of lipid-based signaling, sequestration of free CoA, or accumulation of possibly toxic short-chain acyl-CoAs remains to be established.
The phenotype of the Atpex10 mutant described in this paper is very different from the phenotypes of other peroxisomal mutants such as the pex5-1 (Zolman et al., 2000) and ped2 (pex14) mutants of Arabidopsis (Hayashi et al., 2000), which are embryo lethal, although it is not known if these are null alleles. Other peroxisome mutants in Arabidopsis include sse1 (caused by a T-DNA insertion in a putative PEX16 homolog; Lin et al., 1999) and ted3 a gain-of-function allele of AtPEX2 (Hu et al., 2002). Homozygous sse1 seeds are shrunken in appearance due to an increase in starch deposition at the expense of lipids and proteins. High amounts of starch accumulate in developing seed of Arabidopsis from late-heart until curled cotyledon stage, and synthesis of triacylglycerol, the storage lipid used during germination, increases markedly at late-heart stage (Baud et al., 2002). The shrunken seeds of sse1 are not viable (Lin et al., 1999) and the embryo-defective nature of this mutant has been confirmed. A detailed analysis has shown that embryos abort at a variety of stages, although 43% attain the normal mature cotyledon shape (http://www.seedgenes.org). Although SSE1 could partially complement a Yarrowia lipolytica pex16 mutant, information on the subcellular localization of SSE1 has not been presented. Further studies are required to elucidate the link between peroxisome function, suggested by the ability of SSE1 to partially complement the Y. lipolytica pex16 mutant, and defective storage product accumulation. The Atpex10 mutant described here is clearly different from sse1, as abnormalities leading to cell death and seed abortion are seen early in embryogenesis, before the deposition of storage reserves. The expression data shown in Figure 5A and the peroxisomal location (Fig. 1) are consistent with an essential role for AtPEX10 in peroxisomes during the pattern formation phase of embryogenesis. However, our data do not exclude other roles for AtPEX10 at this or later developmental stages. The most similar mutant to Atpex10 is the ted3 (Atpex2) knockout described by Hu et al. (2002). They suggested that Atpex2 mutants are embryo lethal, based on the observation that the siliques from heterozygotes were smaller and contained fewer seeds, and, as is the case for Atpex10, no homozygous mutant adult plants could be recovered. Interestingly, TED3 (PEX2) encodes a RING finger protein that in other species has been shown to interact with Pex10p (Okumoto et al., 2000).
Other possibilities are that AtPEX10 is involved in processes not directly linked to protein import into peroxisomes. The complex phenotype of the ted3 mutant plants suggests a role for TED3 (PEX2) in peroxisome-nucleus communication, as ted3 suppressed some of the misregulated gene expression seen in the det1-1 mutant (Hu et al., 2002). While this manuscript was under review, a report appeared describing the same Arabidopsis pex10 mutant, including an ultrastructural analysis of the aborted embryos (Schumann et al., 2003). In contrast to our findings that heart stage embryos contained degenerating cells, these authors were able to observe identifiable organelles. They were unable to detect peroxisomes, but showed that there were abnormalities in endoplasmic reticulum, lipid bodies, and protein bodies. However, the interpretation of such data is problematic as these embryos eventually die, and such changes may not be a direct result of the loss of AtPEX10 function, but may be secondary effects of processes disrupted as a primary consequence of the mutation. Clearly, answering these questions concerning the primary function of AtPEX10 and the underlying biochemical basis of the embryo-lethal phenotype are of great importance, but will require the study of alleles with partial or conditional function.
MATERIALS AND METHODS
Plant Growth Conditions
Seeds were imbibed and germinated on Murashige and Skoog salts (2.36 g L-1), plant agar (0.8 g L-1; Ducheva, Haarlem, Netherlands) with kanamycin (50 μg mL-1) or hygromycin (30 μg mL-1) selection where specified, or 1% (w/v) Suc. Carbenicillin (0.5 mg mL-1) and amphotericin B (2.5 μg mL-1) were included in all seedling media except for selection with hygromycin. Seedlings were carefully removed from tissue culture, planted in compost, and kept under propagators in short-day conditions (18°C, 8-h light photoperiod). Propagators were removed after approximately 1 week and the plants were kept in short-day conditions for around 3 to 4 weeks or until flowering was required. After this time, the plants were transferred to long-day growth conditions (22°C, 16-h light photoperiod).
GFP-SKL and PEX10-YFP C-Terminal Fusion Construction
For GFP-SKL generation and subcloning into a binary vector (pVKH18En6; Batoko et al., 2000), we amplified GFP from pLL4:cGFP5 (Brandizzi et al., 2003) with ExTaq (TaKara, Kyoto) using a forward-strand primer (5′-CGATCTAGAGCAGATCGATGAGTAAAGGAGAAGAACTTTTCACTGGAG-3′) and a reverse primer containing the necessary codons encoding for Ser-Lys-Leu and a stop codon (reverse-strand 5′-GCTGAGCTCGGCTAAAGTTTTGATTTGTATAGTTCATCCATGCCATGTG-3′). The XbaI-GFP5-SKL-SacI PCR fragment was cloned into pGEM-T easy (Promega, Madison, WI) and was sequenced before being subcloned into the multiple cloning site of pVKH18En6.
The AtPEX10 CDS was amplified from AtPEX10 cDNA (accession no. AJ276134) using gateway primers (forward-strand 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGAAACGATGAGGCTTAATGGGGAT-3′, reverse-strand 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTAAAATCAGAATGATACAAACA-3′) with Pfx proof-reading DNA polymerase (Invitrogen, Paisley, UK). The PCR product was cloned into pGEM T-easy (Promega) and was sequenced before being inserted into the gateway donor vector pDONR207 (Invitrogen) via the gateway BP reaction. The fragment was then transferred into the pCAMBIA 1300-YFP (5′-35S promoter-cassette B-eYFP-nos terminator) destination vector (described below) using the gateway LR reaction.
The pCAMBIA 1300-YFP binary gateway destination vector contains the following construct: 5′-cauliflower mosaic virus (CaMV) 35S promoter-gateway cassette B-eYFP-NOS terminator. The CaMV35S promoter (800 bp) and nopaline synthase terminator (250 bp) DNA fragments in this construct originate from pBI121 (Jefferson et al., 1987). The GUS reporter gene located downstream of the 35S promoter in pBI121 was replaced by PCR-amplified enhanced YFP (eYfp, accession no. AF242870) using BamHI and SacI restriction sites. The eYFP fragment was amplified with the following oligonucleotides: forward strand, 5′-ATCGGATCCCGGGATGGTGAGCAAGGGCGAGGA, reverse strand, 5′-CTAGAGCTCACTTGTACAGCTCGTCCAT. The 5′ ends of the oligomers contain, respectively, BamHI and SacI restriction sites to enable cloning into pBI121. The Cassette B DNA fragment (Invitrogen) was then placed upstream of eYFP into a SmaI restriction site present in the forward-strand oligonucleotide and was thus introduced during the previous cloning step. The entire 5′-CaMV 35S promoter-gateway cassette B-eYFP-nopaline synthase terminator construct was then transferred into the HindIII and EcoRI sites of pCAMBIA 1300 (8,958 bp).
Transient Expression of Fluorescent Constructs and Confocal Microscopy
The DNA encoding for AtPEX10-YFP and GFP-SKL was transfected into Agrobacterium tumefaciens strain GV3101 (pMP90). Four-week-old tobacco (Nicotiana tabacum) SR1 (cv Petit Havana) greenhouse plants grown at 21°C were used for A. tumefaciens-mediated transient expression (Batoko et al., 2000). A single colony from the transformants was inoculated into 5 mL of yeast extract broth medium (per liter: 5 g of beef extract, 1 g of yeast extract, 5 g Suc, and 0.5 g of MgSO4.7H2O), supplemented with appropriate antibiotics. The bacterial culture was incubated at 30°C with agitation for 20 h. Two hundred microliters of bacterial culture was then pelleted in a 1.5-mL tube by centrifugation at 2,200g for 5 min at room temperature. The pellet was washed once with 1 mL of infiltration medium (50 mm MES, 2 mm Na3PO4.12H2O, 1 mm acetosyringone, and 5 mg mL-1 Glc) and was then resuspended in 1 mL of the same buffer. For experiments requiring coexpression of two different constructs, 100 μL of each bacterial culture was mixed before the wash in infiltration medium.
The bacterial suspension was inoculated using a 1-mL syringe without a needle by gentle pressure through the stomata on the lower epidermis surface. Transformed plants were then incubated under normal growth conditions.
Transformed leaves were analyzed 24 to 48 h after infection of lower epidermal cells. An inverted laser scanning microscope (LSM 510; Zeiss, Jena, Germany) and a 63× oil immersion objective were used for confocal imaging. For imaging expression of GFP-SKL, excitation line of an argon ion laser of 488 nm was used with a 505- to 530-nm band pass filter in the single-track facility of the microscope. For imaging, expression of AtPEX10-YFP excitation line of an argon ion laser of 514 nm was used with a 530- to 600-nm band pass filter in the single-track facility of the microscope. For imaging coexpression of YFP and GFP constructs, excitation lines of an argon ion laser of 458 nm for GFP and 514 nm for YFP were used alternately with line switching using the multitrack facility of the microscope. The fluorescence was detected using a 458/514-nm dichroic beam splitter and a 475- to 525-nm band pass filter for GFP and a 560- to 615-nm band pass filter for YFP. Time-lapse scanning was acquired with LSM 510 imaging system software (Zeiss). Postacquisition image processing was with LSM 5 Image Browser (Zeiss) and Adobe Photoshop 5.0 software (Adobe Systems, Mountain View, CA).
DNA and RNA Extraction, and PCR Analysis
Approximately 100 developing seeds from the same staged time point were dissected from siliques, placed in approximately 100 μL of diethyl pyrocarbonate (DEPC)-treated water in a 1.5-mL tube, frozen in liquid nitrogen, and stored at -80°C. RNA was extracted by grinding the frozen sample with an Eppendorf grinder after adding equal volumes of phenol: chloroform and sand. Four hundred microliters of RNA extraction buffer (25 mm Tris-HCl, pH 8, 25 mm EDTA, pH 8, 75 mm NaCl, 1% [w/v] SDS, and 7.8% [v/v] β-mercaptoethanol) followed by 400 μL of phenol:chloroform was then added and centrifuged at 20,627g at room temperature for 10 min. The aqueous layer was removed to a clean tube, an equal volume of cold isopropanol was added, and the mixture was vortexed and centrifuged at 20,627g for 10 min at room temperature. The top phase was removed to a clean tube, 10 μg of glycogen was added, and it was mixed by inversion. One-third of the volume of 8 m LiCl was added (to give 2 m final concentration) and the mixture was kept at 4°C for 16 h. The sample was centrifuged at 20,627g for 15 min, supernatant was removed, and pellet was washed with 500 μL of cold 2 m LiCl and centrifuged for 10 min at 20,627g. The supernatant was discarded and the pellet was dissolved in 500 μL of DEPC water and incubated at 70°C for 10 min. The RNA was precipitated by adding 0.1 volumes 3 m sodium acetate and 2.5 volumes of 100% (w/v) ethanol, incubating on ice for 1 h, and centrifuging at 20,627g for 30 min. The resulting pellet was washed twice with 70% (w/v) ethanol and was dried in a speed vacuum. RNA was resuspended in DEPC-treated water, and one-half of the volume was used for RT-PCR.
Genomic DNA was extracted from Arabidopsis whole plant tissue using the Nucleon phytopure plant DNA extraction kit (Amersham Life Sciences, Piscataway, NJ) and according to Edwards et al. (1991). PCR from whole plant tissue was carried out using 1× Taq polymerase buffer (Promega), 0.2 mm dNTPs, 0.2 μm primers, and 0.5 units of Taq polymerase (Promega). PCR amplification from DNA extracted from complemented plants was carried out using the ExTaq polymerase system (TaKara). Genomic DNA extraction and PCR amplification from embryos were carried out using the REDExtract-N-Amp plant PCR kit (Sigma, Poole, UK) with the following modifications. Extraction volumes were 20 μL of dilution and extraction buffers into an initial 25-μL sample volume. PCR amplification used 4 μL of embryo extract, 0.5 μm primers, and 10 μL of REDExtract-N-Amp mix in a final volume of 20 μL. Standard PCR cycling conditions were one cycle at 95°C for 1 min, five cycles at 94°C for 20 s, 63°C for 1 min, 72°C for 2 min; five cycles of 94°C for 20 s, 55°C for 1 min, 72°C for 2 min; and 30 cycles of 94°C for 20 s, 45°C for 1 min, 72°C for 2 min followed by a final extension of 72°C for 10 min. Embryo PCR and RT-PCR had an additional 10 cycles of the 45°C annealing temperature cycle. RT-PCR cycling conditions began with reverse transcription at 47°C for 40 min followed by 94°C for 2 min, which was immediately followed by the PCR cycling conditions.
RNA from mature tissues was prepared by the method of Ausubel (Ausubel et al., 1998), and 500 ng of total RNA was used for RT-PCR. RT-PCR was performed using the single-step Reverse-iT RT-PCR kit (AB-gene, Surrey, UK) as instructed by the manufacturers. Cycling conditions were one cycle for 2 min at 95°C; 30 cycles for 1 min at 94°C, for 1 min at 55°C, and for 2 min at 74°C; and one cycle for 10 min at 74°C.
The primers used in this study were: P4, 5′-CCCATTGTGCCTAAAAATCAG-3′; P11, 5′-ATGAGGCTTAATGGGGATTCG-3′; P18; 5′-CGCTGAATAGCTCGGTGCCAC-3′; EM1, 5′-CGGTACAATTTTTCCGACACC-3′; EM2, 5′-GGTATACATTACCACAGGCC-3′; DS3, 5′-ACGGTCGGGAAACTAGCTCTA-3′; GUS1, 5′-GCAAGCTTGATGGTATCGGTGTGAGCGTCGC-3′; GUS2, 5′-GCTCTAGAGTCCTGTAGAAACCCCAACCCGTG-3′; PCAMRB, 5′-CAGGTCGACTCTAGAGGATC-3′; and OP10, 5′-CGAACACCTCATATGCGTTG-3′. The position of primers is shown schematically in Figure 3A and their use is described in the relevant section of the results.
Embryo Dissection and Microscopy Techniques
Developing seeds were dissected from siliques and prepared for Normarski microscopy according to Aida (1997). After preparation, the developing seed were placed onto a glass side with concave wells from Agar Scientific (Essex, UK) with clearing solution and a glass coverslip (22 × 22 mm, 1.5 mm thick) lowered on top. Normarski images were taken with a digital camera (C4742-95; Hamamatsu, Bridgewater, NJ) attached to a microscope (Axiovert 135; Zeiss). Embryos were dissected from the developing seed according to Scott et al. (1998) except that embryos were dissected into sterile water (purified using an Elga reverse-osmosis water purifier), not 1 m KOH. Dissection and images were taken using a dissecting microscope (MZ12; Leica, Deerfield, IL) with an camera (SLR; Olympus, Melville, NY) attached. Seedlings were harvested, stained, fixed, and sectioned for electron microscopy according to Frederick and Newcomb (Frederick and Newcomb, 1969). Microscopy was performed using a electron microscope (1200ex; JOEL, Tokyo).
Complementation
Binary vector pCAMBIA 1300 (pCAMBIA, Canberra, Australia) was digested with EcoRI and was ligated with a 7-kbp genomic fragment containing AtPEX10 isolated from EcoRI digestion of BAC T9J22. A. tumefaciens strain GV3101 (mp90) was transformed by electroporation with the binary vector and was selected on kanamycin. Arabidopsis plants were transformed by the floral dip method (Clough and Bent, 1998), and transformants were selected on 30 μg mL-1 hygromycin (Sigma). Silwett l-77 was obtained from Lehle Seeds (Round Rock, TX).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank the following colleagues for their advice on various aspects of this work: Prof. Phil Gilmartin, Dr. Brendan Davies, Dr. Lesley McCartney, Dr. Mike Deeks (University of Leeds); Dr. Peter Eastmond, Prof. Ian Graham, Dr. Liz Rylott (University of York); and Dr. Greg Briarty (University of Nottingham, retired). We also thank Barbara Johnson and Fiona Moulton for their important contributions in caring for the plants and Malcolm Willis and Adrian Hick for help with the microscopy.
This work was funded by the Biotechnology and Biological Sciences Research Council (grant no. 24/P13265 to A.B. and a studentship to I.A.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.031252.
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup shaped cotyledon mutant. Plant Cell 9: 841-857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore D, Seidman JG, Smith JA, Struhl K (1998) Current Protocols in Molecular Biology. Greene Publishing/Wiley Interscience, New York
- Baker A, Charlton W, Johnson B, Lopez-Huertas E, Oh J, Sparkes I, Thomas J (2000) Biochemical and molecular approaches to understanding protein import into peroxisomes. Biochem Soc Trans 28: 499-504 [PubMed] [Google Scholar]
- Bartel B, LeClere S, Magidin M, Zolman BK (2001) Inputs to the active indole-3-acetic acid pool: de novo synthesis, conjugate hydrolysis, and indole-3-butyric acid β-oxidation. J Plant Growth Regul 20: 198-216 [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I (2000) A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus for normal Golgi movement in plants. Plant Cell 12: 2201-2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Boutin J, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151-160 [Google Scholar]
- Baud S, Guyon V, Kronenberger J, Wuilleme, S. Miquel M, Caboche M, Lepiniec L, Rochat C (2003) Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant J 33: 75-86 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Hanton S, Pinto daSilva LL, Boevink P, Evans D, Oparka K, Denecke J, Hawes C (2003) ER quality control can lead to retrograde transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant J 34: 269-281 [DOI] [PubMed] [Google Scholar]
- Chang CC, Warren DS, Sacksteder KA, Gould SJ (1999) PEX12 interacts with PEX5 and PEX10 and acts downstream of receptor docking in peroxisomal matrix protein import. J Cell Biol 147: 761-773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton W, Lopez-Huertas E (2002) PEX genes in plants and other organisms. In A Baker, IA Graham, eds, Plant Peroxisomes. Biochemistry, Cell Biology and Biotechnological Applications. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 385-426
- Chen J, Ullah H, Young JC, Sussman MR, Jones AM (2001) ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev 15: 902-911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735-743 [DOI] [PubMed] [Google Scholar]
- Collins CS, Kalish JE, Morrell JC, McCaffery JM, Gould SJ (2000) The peroxisome biogenesis factors Pex4p, Pex22p, Pex1p, and Pex6p act in the terminal steps of peroxisomal matrix protein import. Mol Cell Biol 20: 7516-7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Del Rio LA (2001) Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci 6: 145-150 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffiths JS, Somerville CR (2000) Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718-3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel B, Erdmann R et al. (1996) A unified nomenclature for peroxisome biogenesis factors. J Cell Biol 135: 1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber KN, Kram AM, Ehrmann M, Veenhuis M (2001) A novel method to determine the topology of peroxisomal membrane proteins in vivo using the tobacco etch virus protease. J Biol Chem 276: 36501-36507 [DOI] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, Larson TR, Graham IA, Baker A, Holdsworth M (2002) Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J 21: 2912-2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick SE, Newcomb EH (1969) Cytochemical localisation of catalase in leaf microbodies (peroxisomes). J Cell Biol 43: 343-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet F, Misrahi A, Park SK, Zhang Z, Twell D, Dupree P (2003) AtCSLA7, a cellulose synthase-like putative glycosyl transferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol 131: 547-557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17: 1405-1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Nito K, Toriyama-Kato K, Kondo M, Yamaya T, Nishimura M (2000) AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J 19: 5701-5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M (1998) 2, 4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10: 183-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JP, Aguirre M, Peto C, Alonso J, Ecker J, Chory J (2002) A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297: 405-409 [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Machida C, Yoshioka T, Kitano H, Machida Y (2003) The GLOBULAR ARREST 1 gene which is involved in the biosynthesis of folates is essential for embryogenesis in Arabidopsis thaliana. Plant J 33: 235-244 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) Gus fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901-3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalish JE, Theda C, Morrell J, Berg JM, Gould SJ (1995) Formation of peroxisome lumen is abolished by loss of Pichia pastoris Pas7p, a zinc-binding integral membrane protein of the peroxisome. Mol Cell Biol 15: 6406-6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Sun L, Nguyen LV, Rachubinski RA, Goodman HM (1999) The Pex16p homolog SSE1 and storage organelle formation in Arabidopsis seeds. Science 284: 328-330 [DOI] [PubMed] [Google Scholar]
- Lopez-Huertas E, Charlton WL, Johnson B, Graham IA, Baker A (2000) Stress induces peroxisome biogenesis genes. EMBO J 19: 6770-6777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S, Nakamori C, Hayashi M, Kato A, Kondo M, Nishimura M (2002) Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP:dynamic morphology and actin-dependent movement. Plant Cell Physiol 43: 331-341 [DOI] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Hulskamp M (2002) Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants. Plant Physiol 128: 1031-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushmann MA et al. (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751-1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D (1994) Seed development in Arabidopsis thaliana. In CR Somerville, EM Meyerowitz, eds, Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Meinke D (1995) Molecular genetics of plant embryogenesis. Annu Rev Plant Physiol Plant Mol Biol 46: 369-394 [Google Scholar]
- Muench DG, Mullen RT (2003) Peroxisome dynamics in plant cells: a role for the cytoskeleton. Plant Sci 164: 307-315 [Google Scholar]
- Mullen RT, Flynn CR, Trelease RN (2001) How are peroxisomes formed? The role of the endoplasmic reticulum and peroxins. Trends Plant Sci 6: 273-278 [DOI] [PubMed] [Google Scholar]
- Okumoto K, Abe I, Fujiki Y (2000) Molecular anatomy of the peroxin Pex12p-ring finger domain is essential for Pex12p function and interacts with the peroxisome-targeting signal type 1-receptor Pex5p and a ring peroxin, Pex10p. J Biol Chem 275: 25700-25710 [DOI] [PubMed] [Google Scholar]
- Okumoto K, Itoh R, Shimozawa N, Suzuki Y, Tamura S, Kondo N, Fujiki Y (1998) Mutations in PEX10 is the cause of Zellweger peroxisome deficiency syndrome of complementation group B. Hum Mol Genet 7: 1399-1405 [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang W, Kumaran M, Sundaresan V (1999) Analysis of flanking sequences from dissociation insertion lines: a database of reverse genetics in Arabidopsis. Plant Cell 11: 2263-2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton DA, Schetter AL, Franzmann LH, Nelson K, Ward ER, Meinke DW (1998) An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol 116: 935-946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylott EL, Rogers CA, Gilday AD, Edgell T, Larson TR, Graham IA (2003) Arabidopsis mutants in short- and medium-chain acyl-coA oxidase activities accumulate Acyl-CoAs and reveal that fatty acid β-oxidation is essential for embryo development. J Biol Chem 278: 21370-21377 [DOI] [PubMed] [Google Scholar]
- Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C (2003) Ath- PEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci USA 100: 9626-9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Bailey J, Dickinson HG (1998) Parent of origin effects on seed development in Arabidopsis thaliana. Development 125: 3329-3341 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Baker A (2002) Peroxisome biogenesis and protein import in plants animals and yeasts: enigma and variations? Mol Membr Biol 19: 171-185 [DOI] [PubMed] [Google Scholar]
- Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97: 10625-10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Dickrerman A, Brazhnik O, Nguyen Q, McElever J, Frye C, Patton D, Meinke D (2003) The Arabidopsis seed genes project. Nucleic Acids Res 31: 90-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders RJA (1999) Peroxisomal disorders: clinical, biochemical, and molecular aspects. Neurochem Res 24: 565-580 [DOI] [PubMed] [Google Scholar]
- Warren DS, Morrell JC, Moser HW, Valle D, Gould SJ (1998) Identification of PEX10, the gene defective in complementation group 7 of the peroxisome-biogenesis disorders. Am J Hum Genet 63: 347-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H (2002) Fatty acid-derived signals in plants. Trends Plant Sci 7: 217-224 [DOI] [PubMed] [Google Scholar]
- White JA, Todd J, Newman T, Focks N, Girke T, Martinez O, Jaworski JG, Ohlrogge JB, Benning C (2000) A new set of Arabidopsis expressed sequence tags from developing seeds: the metabolic pathway from carbohydrates to seed oil. Plant Physiol 124: 1582-1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B (2001) The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol 127: 1266-1278 [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323-1337 [DOI] [PMC free article] [PubMed] [Google Scholar]