Abstract
Summary: Pseudomonas aeruginosa strains exhibit significant variability in pathogenicity and ecological flexibility. Such interstrain differences reflect the dynamic nature of the P. aeruginosa genome, which is composed of a relatively invariable “core genome” and a highly variable “accessory genome.” Here we review the major classes of genetic elements comprising the P. aeruginosa accessory genome and highlight emerging themes in the acquisition and functional importance of these elements. Although the precise phenotypes endowed by the majority of the P. aeruginosa accessory genome have yet to be determined, rapid progress is being made, and a clearer understanding of the role of the P. aeruginosa accessory genome in ecology and infection is emerging.
INTRODUCTION
Overview of the Pseudomonas aeruginosa Genome
Pseudomonas aeruginosa is a Gram-negative bacterium notable for its ability to thrive in highly diverse ecological niches and to cause significant morbidity and mortality among compromised humans. Key to the survival of P. aeruginosa in environments ranging from soil to various living host organisms is its metabolic versatility. It can subsist on a variety of different carbon sources for energy, is able to utilize nitrogen as a terminal electron acceptor to respire under anaerobic conditions, has minimal nutrient requirements, and grows at temperatures of up to 42°C (169). Also important to the persistence of P. aeruginosa in nature is its ability to form polysaccharide-encased, surface-attached communities known as biofilms and to resist protozoan predation by directly injecting cytotoxic effector proteins into the cytosol of eukaryotic cells through a type III secretion system (2, 141, 169). The same properties that confer ecological success also allow P. aeruginosa to cause opportunistic infections in humans. Due to its metabolic diversity, P. aeruginosa can even multiply in certain disinfectants and metabolize many antibiotics (20, 50). As a result, hospital outbreaks have been traced to the use of many different contaminated solutions. Furthermore, by forming biofilms, P. aeruginosa can colonize the surfaces of medical devices such as catheters and bronchoscopes and can survive disinfection protocols. Once inside humans, P. aeruginosa injects host cells with a combination of type III secretion effector proteins, disrupting host cell signal transduction cascades and inducing host cell death (123). Other well-characterized P. aeruginosa virulence factors include secreted enzymes that degrade the host extracellular matrix (157), an exotoxin that inhibits host cell RNA translation (234), adhesins (43, 214), and a flagellum (63). Given these properties, it is not surprising that P. aeruginosa ranks among the leading causes of hospital-acquired pneumonia, urinary tract infection, bloodstream infection, and surgical site infection in intensive care units (34, 101, 160). In addition to being frequent, hospital-acquired P. aeruginosa infections are often also severe, with an attributable mortality rate of approximately 40% for mechanically ventilated patients with P. aeruginosa pneumonia (62). P. aeruginosa is also the leading cause of respiratory infections in individuals with cystic fibrosis (CF) (169), as well as a frequent cause of exacerbations in individuals with advanced chronic obstructive pulmonary disease (179). Thus, P. aeruginosa is a significant concern to medical practitioners.
Consistent with the remarkable adaptability that allows P. aeruginosa to be both a ubiquitous environmental organism and a consummate opportunistic pathogen, the P. aeruginosa genome is large and complex. The first P. aeruginosa genome to be sequenced was that of strain PAO1, a strain widely used in research studies and originally isolated from a wound (93). Sequencing studies in 2000 indicated that this strain has a genome size of 6.3 Mbp and contains 5,570 predicted open reading frames (ORFs), making it the largest bacterial genome to be sequenced fully at that time. The large size of the P. aeruginosa genome reflects the numerous and distinct gene families that it contains. This is in contrast to some other large bacterial genomes, whose size reflects gene duplication events rather than greater genetic and functional diversity. Specifically, the P. aeruginosa genome contains a disproportionately large number of genes predicted to encode outer membrane proteins involved in adhesion, motility, antibiotic efflux, virulence factor export, and environmental sensing by two-component systems. Additionally, consistent with the bacterium's metabolic versatility, the P. aeruginosa genome has a large number of genes encoding transport systems and enzymes involved in nutrient uptake and metabolism. Considering the genetic diversity of the P. aeruginosa genome, it is not surprising that it contains one of the highest percentages of predicted regulatory genes (8.4%) of all bacterial genomes (211).
Since the sequencing of PAO1, the genomes of several clinical P. aeruginosa isolates have also been sequenced, allowing comparison of the genomes of different P. aeruginosa strains (121, 140, 189, 232). Comparative genomic analysis revealed that the P. aeruginosa genome is a mosaic consisting of relatively conserved genomic sequences with interspersed accessory genetic material. Therefore, the current paradigm is that the P. aeruginosa pangenome is the sum of two components: a core genome and an accessory genome. The core genome of P. aeruginosa is defined as the genes that are present in nearly all strains of P. aeruginosa and encode a set of metabolic and pathogenic factors shared by all P. aeruginosa strains, irrespective of origin (environmental, clinical, or laboratory). The core genome constitutes approximately 90% of the total genome and is highly conserved from strain to strain. For example, a microarray-based comparison of 18 different P. aeruginosa strains found that 97% of the 267 examined PAO1 virulence-related genes were conserved across all strains (233). In contrast, the accessory genome encompasses genes that are found in some P. aeruginosa strains but not others. These segments are not scattered randomly throughout the core genome; rather, they tend to cluster in certain loci. Mathee et al. used the term “regions of genomic plasticity (RGPs)” to describe such loci (140). The genetic sequences occupying many RGPs are often referred to as genomic islands (>10 kb) or islets (<10 kb). Although the definition of a genomic island is evolving (54, 88, 104), here we use this term to refer to a horizontally acquired genetic element present in the chromosome of some strains but absent from closely related strains. Together, the genetic content of the RGPs forms a large proportion of the accessory genome. Genetic elements within the accessory genome may encode properties that contribute to the niche-based adaptation of the particular strains that harbor them. Although plasmids constitute an important part of the P. aeruginosa gene pool, we limit our discussion to genetic elements found within the P. aeruginosa chromosome.
In the absence of intergenomic comparisons, a number of features can be used to identify components of the P. aeruginosa accessory genome. Much of the accessory genome can be identified by its aberrant G+C content, codon usage, and tetranucleotide usage. Since P. aeruginosa is characterized by a high G+C content (∼66.6%), genes acquired from other species (as is the case for much of the accessory genome) generally have a lower G+C content than that of the P. aeruginosa core genome. Once integrated into the P. aeruginosa chromosome, however, foreign DNA experiences the same pressures as the rest of the P. aeruginosa genome, and thus, over time, it may lose the sequence compositional differences that once distinguished it from the P. aeruginosa core genome. Another identifying feature of the accessory gene pool is a close association with genetic elements facilitating mobility. Moreover, genomic islands are frequently mosaic, possessing features from multiple different mobile genetic elements (108). Notably, though, the mobile elements associated with genomic islands are often degenerate due to accumulated deletions and rearrangements resulting in the loss of one or more of the activities necessary for mobility. Indeed, for many accessory elements, a mechanism of transfer cannot be inferred with certainty due to insufficient similarity with known mobile elements. A final identifying feature of the accessory gene pool is a predilection for insertion at particular sites within the P. aeruginosa core genome, as described above (140). That is, there are specific loci (i.e., RGPs) within the core genome that act as hot spots for the insertion of accessory genes. In particular, core genome tRNA genes are frequently targeted for the insertion of accessory genetic elements (231).
Functional Importance of the Accessory Genome
The accessory genome is central to P. aeruginosa biology. Along with deletions, rearrangements, and mutations within the core genome, the horizontal transfer of components of the accessory genome is a primary contributor to P. aeruginosa genome evolution. Beyond shaping the content and arrangement of the P. aeruginosa genome as a whole, however, accessory genetic elements also confer specific phenotypes that are advantageous under certain selective conditions. For example, by encoding novel catabolic pathways, certain components of the P. aeruginosa accessory gene pool are thought to promote persistence in otherwise inhospitable environments containing heavy metals and toxic organic compounds (3, 33). Of particular interest in the field of bioremediation are the accessory genome elements encoding catabolic pathways capable of degrading common environmental pollutants, such as polycyclic aromatic hydrocarbons, polychlorinated biphenyls, and pesticides. Importantly, the accessory genome is also of great medical relevance. It is a source not only of genes that promote P. aeruginosa persistence within various host species by encoding virulence factors (87) but also of genes encoding resistance to multiple classes of antibiotics (144). Given the already high intrinsic resistance of P. aeruginosa conferred by multidrug efflux pumps and a β-lactamase encoded within the core genome, dissemination of antibiotic resistance by elements of the accessory genome is especially concerning. In fact, the rapid spread of multidrug-resistant strains, many of which owe their broad resistance to genes of the accessory genome, led the Infectious Diseases Society of America to declare P. aeruginosa one of the six “top-priority dangerous, drug-resistant microbes” against which novel pharmaceuticals are desperately needed (213). Knowledge of the accessory genome is therefore imperative to the application of P. aeruginosa in biotechnology and the development of effective medical interventions to treat or prevent P. aeruginosa infections in humans.
MAJOR COMPONENTS OF THE P. AERUGINOSA ACCESSORY GENOME
The majority of the P. aeruginosa accessory genome can be grouped into four broad categories: (i) integrative and conjugative elements (ICEs), (ii) replacement islands, (iii) prophages and phage-like elements, and (iv) transposons, insertion sequences (ISs), and integrons. However, it should be recognized that components of the accessory genome are often formed by a combination of different functional modules. As a result, many elements cannot be assigned unambiguously to one category or another. Moreover, some elements lack features consistent with any of these categories. Nonetheless, the use of this classification system provides a useful framework for discussing the P. aeruginosa accessory genome.
ICEs
General features.
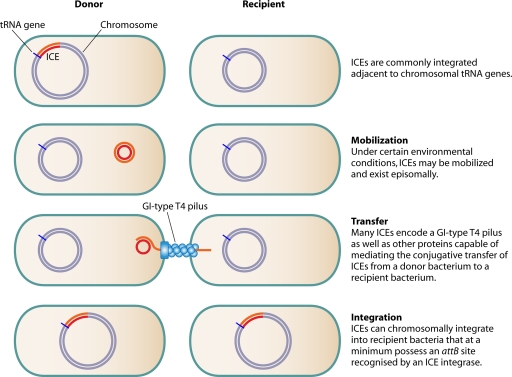
Many P. aeruginosa genomic islands are ICEs or are derived from such elements. ICE is a term coined by Burrus et al. (28, 30) to describe self-transmissible genetic elements that must integrate into an existing replicon to accomplish replication (Fig. 1). ICEs possess a combination of both plasmid and phage-associated DNA properties. Like plasmids, ICEs can exist as circular extrachromosomal elements and are transferred by self-mediated conjugation. However, in contrast to plasmids, which usually become chromosomally integrated only if they contain chromosome-like sequences that mediate RecA-dependent homologous recombination, all ICEs can undergo phage integrase-mediated chromosomal integration. Integration occurs via site-specific recombination between an ICE recombination site (attP) and a recombination site on the bacterial chromosome (attB). Within the category of ICEs, P. aeruginosa genomic islands can be subdivided further into those that have retained mobility and those that have become fixed due to degeneration of their phage or conjugative elements. Also, whereas plasmids by definition replicate autonomously, most ICEs do not replicate on their own but rather rely on duplication of the chromosome for replication. Because of their ability to move from strain to strain and to integrate into the bacterial chromosome, some ICEs were formerly referred to as conjugative transposons (28).
FIG. 1.
ICEs are self-transmissible genomic islands that can transiently exist as circular extrachromosomal elements, are transferred by self-mediated conjugation, and undergo integrase-mediated chromosomal integration.
Characterized mobilizable P. aeruginosa ICEs range in size from 81 to 108 kb and share a syntenic set of 72 ORFs with >75% sequence identity (108, 109, 235). Though the functions of many of the genes within this highly conserved backbone have yet to be demonstrated experimentally, they are predicted to mediate excision, self-transfer to a new host, and reintegration. Currently, the mechanism of excision from the P. aeruginosa chromosome is not understood, although integrases appear to be necessary for this activity (29, 124, 177, 180, 194). Once excised, ICEs circularize to reform an attP site and restore the attB site on the P. aeruginosa chromosome (30, 177, 182). Maintenance of the episomal form of the ICE is achieved through expression of an ICE-carried partitioning gene (177). Conjugative transfer is then likely mediated by a subset of the genes in the conserved backbone that encode a type IV secretion system (T4SS). Based on gene content, order, and homology, the ICE-associated T4SSs represent a novel lineage of T4SSs distinct from the classical type IV A systems exemplified by the Agrobacterium tumefaciens VirB-VirD4 DNA-protein transfer system and the type IV B systems exemplified by the Legionella pneumophila Dot/Icm protein secretion system (103). The ICE-associated T4SSs, referred to as “genomic island (GI)-type T4SSs,” are widely distributed and appear to be associated exclusively with genomic islands. Since their initial discovery on the Haemophilus influenzae antibiotic resistance island ICEHin1056, GI-type T4SSs have been identified on genomic islands in Salmonella enterica serovar Typhi, Erwinia carotovora, Methylibium petroleiphilum, Xylella fastidiosa, and Pseudomonas spp. The GI-type T4SSs identified by bioinformatics in P. aeruginosa ICEs are only putative, since in most cases they have yet to be characterized functionally. Nonetheless, based on their high degree of homology with a large subset of the 24 genes comprising the GI-type T4SS on ICEHin1056 (103), it is reasonable to assume that the corresponding P. aeruginosa genes encode factors that mediate conjugal transfer of their cognate ICEs. Following transfer, mobilizable ICEs can recognize and insert at specific chromosomal sites by utilizing an integrase that is also encoded within the conserved backbone. Specifically, these integrases catalyze site-specific recombination between the attP site of their cognate ICE and a chromosomal attB site, which is often located at the 3′ end of a tRNA gene (96, 231).
With the exception of some variability in the organization of their origins of replication, the primary difference between the various mobilizable ICEs is their “cargo” genes (235). As described above, the mobilizable P. aeruginosa ICEs share large regions with synteny and high sequence similarity that encode the functions necessary for their mobility and dissemination (149) (Fig. 2). Within certain regions of this backbone are clusters of additional ORFs not essential for mobilization or dissemination; these cargo ORFs confer diverse phenotypes on the organisms that contain the ICEs. Cargo ORFs are often grouped into modules, with little relationship between different modules. This structure suggests that ICEs evolved through multiple recombination events, with acquisition of individual modules occurring at different times and from different sources.
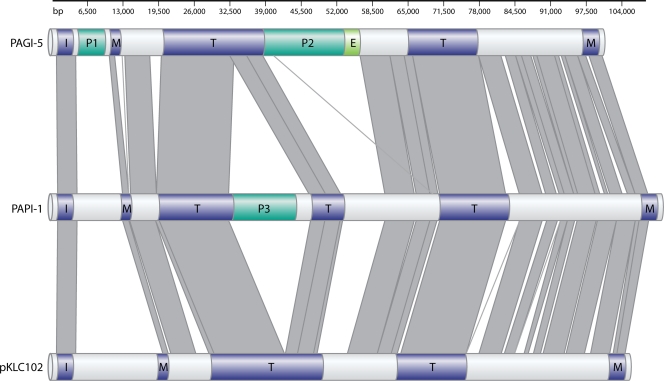
FIG. 2.
PAGI-5, PAPI-1, and pKLC102 share a similar modular structure. Gray lines indicate regions of similar sequence. Purple boxes represent putative integration (I), transfer (T), and maintenance (M) gene modules common to all three ICEs. The integration module consists of the xerC integrase gene. The maintenance modules include the genes encoding the ParE maintenance toxin and the Soj partitioning protein. The transfer modules include genes encoding a coupling protein and a conjugative relaxase as well as GI-type T4SS genes. A few select cargo regions unique to particular ICEs and shown to contribute to pathogenicity (P) are shown in dark green. A set of PAGI-5 cargo ORFs with a potential ecological (E) role in conferring mercury resistance are shown in light green. (Adapted from reference 14 with permission.)
Although mobilization and transfer of some ICEs have been demonstrated, many other genetic elements with features of ICEs appear to have lost these capacities. Deletion or disruption of genes essential for the various steps required for transfer from one bacterium to another has occurred in many of these elements. For the purposes of this review, these ICE-like elements, which retain many of their former structural features, are also referred to as ICEs.
Specific P. aeruginosa ICEs.
A number of P. aeruginosa ICEs have been identified and partially characterized (Table 1). Many of these fall into two large families: pKLC102-related ICEs and clc-like ICEs. Here we review representative members of both families and discuss their evolution and function.
TABLE 1.
Selected P. aeruginosa ICE
| ICE | Size (kb) | tRNA integration site (if applicable) | Features | NCBI GenBank accession no. | Reference(s) |
|---|---|---|---|---|---|
| PAGI-2 | 105 | tRNAGly | Cargo genes thought to function in complexing with and transport of heavy metals | AF440523 | 119 |
| PAGI-3 | 103 | tRNAGly | Cargo genes thought to confer metabolic, transport, and resistance capacities | AF440524 | 119 |
| PAGI-4 | 23 | tRNALys | Contains genes with putative metabolic functions | AY258138 | 108 |
| PAGI-5 | 99 | tRNALys | Contributes to virulence in mouse model of acute pneumonia | EF611301 | 14 |
| PAGI-8 | 18 | tRNAPhe | Predicted to encode an ATPase, a Zn-dependent transcriptional regulator, and a DotA/TraY-like protein | EF611304 | 15 |
| LESGI-1 | 46 | tRNAPro | Contains several ORFs similar to those for predicted proteins in nonpseudomonads | FM209186 (whole LESB58 sequence) | 232 |
| LESGI-3 | 111 | tRNAGly | Cargo genes thought to confer transport capacities | FM209186 (whole LESB58 sequence) | 232 |
| LESGI-5 | 29 | Contributes to virulence in a rat lung chronic infection model; high prevalence of carriage in CF patients | FM209186 (whole LESB58 sequence) | 68, 232 | |
| PAPI-1 | 108 | tRNALys | Cargo includes numerous genes shown to affect virulence in infection models; self-transmissible | AY273869 | 35, 87, 177 |
| PAPI-2 | 11 | tRNALys | Contains gene encoding the type III secreted virulence factor ExoU | AY273870 | 87, 113 |
| Dit island | 112 | tRNAGly | Putative diterpenoid metabolism island | NZ_AAKW00000000 (whole 2192 sequence) | 140 |
| pKLC102 | 104 | tRNALys | Can exist episomally; ICE progenitor | AY257538 | 108 |
| pKLK106 | 106 | tRNALys | Can exist episomally; ICE progenitor | AF285417-AF285424 | 106 |
| clc element | 105 | tRNAGly | Found in P. knackmussii strain B13; contains genes for chlorocatechol degradation; self-transmissible; ICE progenitor | AJ617740 | 181, 182 |
The pKLC102 family of ICEs includes pKLC102 itself along with Pseudomonas aeruginosa genomic island 4 (PAGI-4), PAGI-5, P. aeruginosa pathogenicity island 1 (PAPI-1), and PAPI-2. Each member of this family is thought to have evolved from pKLC102, a widely distributed 104-kb ICE first identified in P. aeruginosa clone C (108, 109). The pKLC102 XerC/XerD-like integrase recognizes a chromosomal attB site consisting of a 15- to 20-bp motif found within the 3′ ends of two distinct tRNALys genes. Thus, pKLC102 can be integrated chromosomally at either of these two loci. pKLC102 was the first P. aeruginosa genomic island shown to be mobilizable, and it is now known to have an unusually high spontaneous mobilization frequency (10%). pKLC102 also appears to be an aberration among the P. aeruginosa ICEs in that it occurs at 30 episomal copies per cell, suggesting that it is actually capable of autonomous replication and is therefore more appropriately categorized as a conjugative plasmid (109). Like plasmids, pKLC102 possesses an origin of replication (oriV). pKLC102 oriV, however, has a unique structure consisting of 16 highly conserved 57-bp direct repeats on one end and an AT-rich region preceded by four palindromes on the other end (235). An ICE that is highly similar to pKLC102 in sequence and integration site specificity and can also exist in both episomal and integrated forms is pKLK106 (106). Outside its conserved backbone, pKLC102 contains a myriad of cargo genes, including genes encoding novel fatty acid synthases, the products of a putative chemotaxis operon, a cold adaptation protein, a polyketide synthase, a phage antirepressor, four putative transcription regulators, and a synthase for a cyclic β-(1,2)-glucan (108). This last protein is a periplasmic component critical for mediating bacterium-host interactions in other prokaryotes (98). Though these putative encoded proteins suggest numerous possibilities, no specific phenotypes have yet been assigned to the pKLC102 cargo ORFs.
PAGI-4 is a pKLC102-like ICE that appears to have once possessed a full backbone of mobility genes but became immobilized through a series of deletion events. One border of PAGI-4 has nearly complete sequence identity to the integrase-encoding segment of pKLC102. Consequently, PAGI-4, like pKLC102, is found integrated at a tRNALys gene. The events that led to the immobilization of PAGI-4 have yet to be elucidated, but Klockgether et al. hypothesized that fixation in the chromosome occurred following insertion of a large composite transposon near the PAGI-4 chromosomal integration site (108). Following fixation, PAGI-4 appears to have sustained further deterioration in the form of truncation events that reduced it to its current 23-kb size.
PAPI-1 and the related element PAGI-5 are prototypical ICEs that endow enhanced pathogenic characteristics upon the strains that harbor them. PAPI-1 is a 108-kb ICE identified in the broad-host-range pathogenic strain PA14 (87). Interstrain transfer of a circular extrachromosomal form of PAPI-1 is mediated by the PAPI-1 GI-type T4SS, which utilizes a self-encoded type IV pilus for conjugation (35, 177). Interestingly, processing of the precursor of the major subunit of this pilus requires a prepilin peptidase encoded within the core genome of P. aeruginosa. The requirement for this prepilin peptidase may restrict transfer of PAPI-1 to P. aeruginosa or related bacterial species that encode a compatible prepilin peptidase (35). PAPI-1 is so named because many of its cargo ORFs appear to promote pathogenicity. Nineteen of its cargo ORFs, when mutated, resulted in diminished virulence in an Arabidopsis leaf infiltration model or a mouse thermal injury model of infection (87). Consistent with acquisition through horizontal gene transfer (HGT), many of the virulence genes identified in PAPI-1 have their closest known homologs in organisms other than P. aeruginosa. With the exception of a number of two-component regulatory genes (e.g., pvrR, which has been shown to affect biofilm formation [56]), the majority of virulence genes identified in PAPI-1 encode hypothetical proteins of unknown function. As a result, the mechanisms by which PAPI-1 cargo ORFs enhance virulence have yet to be determined. PAGI-5, identified in a highly virulent P. aeruginosa pneumonia isolate, is a 99-kb ICE that is similar to PAPI-1 (Fig. 2). Illustrating the capacity of ICEs to continually evolve through acquisition and loss of cargo ORFs, PAGI-5 carries two regions of cargo ORFs (NR-I and NR-II) that are absent in PAPI-1 (14). Among a panel of 35 P. aeruginosa isolates cultured from patients with ventilator-associated pneumonia, these cargo regions were present in only the more virulent strains (Fig. 3) (14). Additional studies confirmed that these regions of PAGI-5 contribute to pathogenicity, as deletion of either attenuated virulence in a mouse model of acute pneumonia (14). Thus, like many of the PAPI-1 cargo ORFs, the cargo ORFs of PAGI-5 are associated with pathogenic functions. However, categorizing PAGI-5 purely as a pathogenic ICE may be inappropriate, as PAGI-5 also possesses cargo ORFs with features suggesting ecological roles. Namely, PAGI-5 contains a cluster of genes with homology to a mercury ion-induced transcriptional regulator gene, a mercuric ion reductase gene, and a mercuric ion transport protein gene. Like the pKLC102 XerC/XerD-like integrase, the PAGI-5 and PAPI-1 XerC/XerD-like integrases are tyrosine recombinases. Also, the PAGI-5 and PAPI-1 integrases are specific for the same 15- to 20-bp attB motif as the pKLC102 integrase, consistent with the theory that both PAGI-5 and PAPI-1 are derived from pKLC102. As expected, all three ICEs insert into the 3′ end of tRNALys genes.
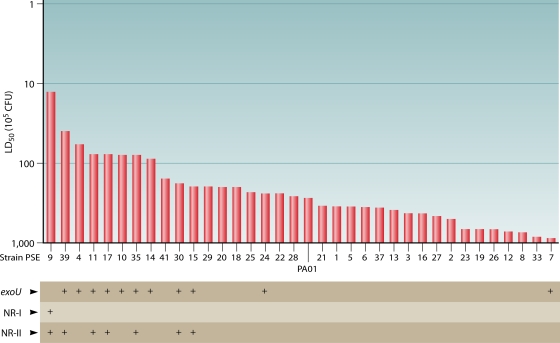
FIG. 3.
Elements of the P. aeruginosa accessory genome are associated with increased virulence in P. aeruginosa clinical isolates. The genomes of 35 P. aeruginosa isolates cultured from the airways of patients with ventilator-associated pneumonia were examined for the presence of three components of the accessory genome: exoU, NR-I, and NR-II. The laboratory strain PAO1 was included as a control. The exoU gene is part of PAPI-2, and NR-I and NR-II are cargo regions carried by PAGI-5. Virulence was quantified by measuring the LD50 in a mouse model of acute pneumonia. The y axis has been inverted such that higher bars represent increased levels of virulence. All three genetic elements are found more commonly in highly pathogenic strains. (Adapted from reference 192 with permission. Copyright 2003 Infectious Diseases Society of America.)
Similar to PAPI-1 and PAGI-5, PAPI-2 is an ICE that has demonstrated virulence properties. This island is an 11-kb element with a G+C content of 56.4%, which is substantially lower than that of the P. aeruginosa chromosome (87). PAPI-2 is most notable for belonging to a family of genomic islands that carry the genes encoding the protein ExoU and its chaperone, SpcU (113). ExoU is a phospholipase effector protein secreted by the P. aeruginosa type III secretion system that functions as a potent virulence factor in animal models and has been associated with poor clinical outcomes in human patients (66, 84, 85, 114, 190). Other PAPI-2-related islands that carry the gene encoding ExoU are ExoU islands A, B, and C. Thus, PAPI-2 and the ExoU islands form a related subset of ICEs within the large pKLC102 family (113). Acquisition of these islands appears to markedly enhance virulence, since strains that possessed them were associated with particularly severe disease in a mouse model of pneumonia (Fig. 3) (82, 192). Likewise, deletion of PAPI-2 has been associated with decreased virulence (82, 192). Interestingly, PAPI-2 acted synergistically with PAPI-1 in a pneumonia model, demonstrating that distinct accessory genomic elements can act in concert (82).
The second large family of P. aeruginosa ICEs is comprised of the clc-like elements, which include clc itself, PAGI-2, PAGI-3, and LES genomic island 3 (LESGI-3). This family of ICEs is derived from the clc element, a 105-kb genomic island first identified in Pseudomonas knackmussii strain B13 as the genetic locus responsible for the bacterium's ability to metabolize 3-chlorobenzoate (55, 181). The cargo ORFs of the clc element confer on this ICE its pronounced catabolic characteristics. In addition to the clcRABDE genes, which encode enzymes for 3- and 4-chlorocatechol degradation, the clc element cargo includes a functional operon for 2-aminophenol degradation, a putative aromatic compound transporter gene, and a gene predicted to encode an aromatic ring dioxygenase (71). In terms of mobility, the clc element appears to be transferred readily between not only gammaproteobacteria (including P. aeruginosa) but also betaproteobacteria (70, 86, 183). Its chlorocatechol degradation cargo genes are highly similar to those of Ralstonia sp. (218), and its transfer has been documented in complex microbial communities in chlorocatechol-contaminated sludge, soil, and wastewater (205). The clc element attB site used for chromosomal integration is within the 3′ end of two tandem tRNAGly genes (181, 182), and integration is mediated by a bacteriophage P4-like integrase.
Two P. aeruginosa ICEs closely related to the clc element are PAGI-2 (105 kb) and PAGI-3 (103 kb), which were first identified in strain C and strain SG17M, respectively (119). The clc element, PAGI-2, and PAGI-3 each contain bacteriophage P4-like integrase genes. Interestingly, and for unclear reasons, neither PAGI-2 nor PAGI-3 excises at a detectable rate (109). These two ICEs, along with the 111-kb ICE LESGI-3 from strain LES (232), share a similar bipartite structure: the portion of each ICE adjacent to the integration site is conserved, and the remaining ICE sequence consists of unique cargo ORFs. Many PAGI-2 cargo ORFs have close homologs in Cupriavidus (formerly Ralstonia) metallidurans. In particular, C. metallidurans CH34, a strain isolated from the sludge of a zinc decantation tank, contains an ICE with 99.9% sequence identity to PAGI-2 (109). The LESGI-3 cargo ORFs encode proteins for the complexing and transport of heavy metals. Similarly, the PAGI-3 cargo ORFs appear to encode metabolic features and transport and resistance capacities, though the extent to which they actually strengthen the metabolic versatility of P. aeruginosa has yet to be determined.
Replacement Islands
General features.
Lipopolysaccharide (LPS) O antigen, pyoverdine, pili, and flagella are critical determinants of P. aeruginosa fitness. These macromolecules mediate fundamental processes such as bacterium-bacterium interaction, iron acquisition, adhesion, and motility. Moreover, these macromolecules are surface exposed and, as such, are under selective pressure as the targets of phage predation and immune recognition (42, 72, 117, 168). The genes responsible for the synthesis and, in the case of pili and flagella, posttranslational modification of each of these macromolecules are grouped together in gene clusters known as “replacement islands” (Table 2) (201). Smith et al. (201) coined this term to describe the O antigen biosynthesis, pyoverdine, pilin, and flagellar glycosylation genetic loci because similar to genomic islands, each of these gene clusters contains horizontally acquired components and is highly divergent between different strains. Unlike other genomic islands, however, versions of these genetic elements are present and occupy the same sites in nearly all P. aeruginosa genomes. This maintenance of multiple functionally related islands in the population is thought to be a product of intense selective pressure to resist phage killing (or, in the case of the pyoverdine locus, to resist pyocin killing) and to escape detection by the host immune system.
TABLE 2.
P. aeruginosa replacement islands
| Replacement island | Size (kb) | Selected NCBI GenBank accession no. | Reference(s) |
|---|---|---|---|
| O-antigen biosynthetic locus | 14-25 | AC104719 (O1), AC104731 (O2), AC104733 (O3), AC104734 (O4), AC104735 (O5), AC104736 (O6), AC104737 (O7; derived from ATCC O7 strain), AC104738 (O8), AC104739 (O9), AC104720 (O10), AC104721 (O11), AC104722 (O12; derived from ATCC O12 strain), AC104723 (O13), AC104724 (O14), AC104725 (O15; derived from ATCC O15 strain), AC104726 (O15; Lory bank), AC104727 (O16), AC104728 (O17), AC104729 (O18), AC104730 (O19), AC104732 (O20) | 184 |
| Pyoverdine locus | 39-50 | AY765259 (pyoverdine type I; strain 10-15), AY765263 (pyoverdine type II; strain MSH), AY765261 (pyoverdine type III; strain 206-12) | 201, 204 |
| Pilin and pilin modification genes | 1-4 | AY112719 (pilAI and group 1 accessory gene tfpO; strain PA131533), AF511653 (pilAII; strain PA5235), PA14 chromosome positions 5233456 to 5233511 (www.pseudomonas.com) (pilAIIIand group III accessory gene tfpY; strain PA14), AY112720 (pilAIV and group IV accessory genes tfpW and tfpX; strain PA5196), AY112718 (pilAV and group V accessory gene tfpZ; strain PA110594) | 116 |
| Flagellin glycosylation island | 8-16 | AF332547 (long, a-type flagellin; strain PAK), AY280452 (short, a-type flagellin; strain JJ692), PAO1 chromosome positions 1175614 to 1182697 (www.pseudomonas.com) (b-type flagellin; strain PAO1) | 7, 9 |
Specific P. aeruginosa replacement islands.
The outermost part of the LPS molecule consists of repeating sugar moieties known as the O antigen/polysaccharide. Among strains that produce O antigen, significant variation exists in its structure and chain length, allowing for strain classification by serotyping. Thus far, there are 20 different P. aeruginosa O antigen serotypes recognized by the International Antigenic Typing Scheme (133). With the exception of serotype O15 and O17 strains, which use biosynthetic genes carried elsewhere in the genome, the genes encoding the major enzymes for O antigen biosynthesis are found in a single cluster that occupies a common genetic locus in all P. aeruginosa strains. These genes encode the proteins that mediate the chemical modification and sequential assembly of sugars, the translocation of the resulting polysaccharide subunits to the periplasm, and the ligation of polysaccharide subunits to form the repetitive polysaccharide chains that comprise the O antigen. Given the variability of the P. aeruginosa O antigen, it follows that the O antigen biosynthesis locus is highly divergent between different P. aeruginosa strains (184). Consistent with acquisition through HGT from an organism with a lower G+C content than that of P. aeruginosa, the G+C contents of many O antigen biosynthesis genes are significantly lower than that of the P. aeruginosa chromosome. The precise method of horizontal transfer by which these genes were acquired is unknown, but evidence suggests bacteriophage-mediated transfer from Gram-positive bacteria. For example, a number of O antigen biosynthetic genes are highly similar to Staphylococcus aureus capsule synthesis genes or contain nucleotide sequences highly similar to that of a Bacillus subtilis bacteriophage attachment site (53). The presence of a particular O antigen biosynthetic cluster could affect the fitness of a P. aeruginosa strain in several ways. Though a comparison of the virulence properties of individual O antigens has yet to be made, several studies have demonstrated that the O antigen contributes to P. aeruginosa pathogenicity (168). Strains lacking O antigen were highly attenuated in virulence in a mouse burned-skin model of P. aeruginosa infection (48) and were more sensitive to serum-mediated killing in humans (51). Given that the antipseudomonal activities of outer membrane-destabilizing cationic peptides are mediated through interaction with LPS O antigen, it is conceivable that O antigen biosynthetic clusters confer different degrees of resistance or susceptibility to these host defense molecules.
Pyoverdine is a major siderophore produced by P. aeruginosa to scavenge and transport iron(III). It has a complex structure consisting of a quinoline-derived chromophore, which gives pyoverdine its characteristic yellow-green fluorescence, and a variable peptide chain synthesized by a nonribosomal peptide synthetase (146). Any given P. aeruginosa strain may produce one of three structurally different pyoverdine types (types I to III), allowing strains to be divided into three siderovars. Each siderovar produces a specific pyoverdine and pyoverdine receptor combination, since the receptors are specific for their cognate pyoverdine type (147). In addition to intertype variation, there is significant intratype pyoverdine and pyoverdine receptor variation (201). As a result, the pyoverdine genetic locus (which contains the gene encoding the pyoverdine receptor, nonribosomal peptide synthetase genes, and a putative ABC transporter gene) is highly divergent among P. aeruginosa strains (204). While much of the intratype divergence at the pyoverdine locus can be accounted for by recombination events between different P. aeruginosa pyoverdine types, diversity in the gene encoding the pyoverdine receptor in particular is likely due to HGT (21, 171). Though the G+C content of the pyoverdine locus is not significantly different from that of the P. aeruginosa core genome, it exhibits unusual codon and tetranucleotide usage. Furthermore, the type I and type II pyoverdine receptors are similar to receptors from Azotobacter vinelandii and A. tumefaciens, respectively. Like P. aeruginosa, both of these soil bacteria have high G+C contents, perhaps explaining the similar G+C contents of the P. aeruginosa pyoverdine locus and core genome. It has been proposed that diversity in the pyoverdine receptor gene drives diversity in the nonribosomal peptide synthetase and putative ABC transporter genes, since the proteins encoded by these genes must change to functionally accommodate new pyoverdine receptors. The selective pressures driving pyoverdine receptor heterogeneity are unclear. In contrast to O antigen, pili, and flagella, the pyoverdine receptor is not a receptor for phage entry but is an entry target for pyocins (16), which are bacterially produced phage-like molecules with antibacterial properties. Alternatively, evolution of the pyoverdine receptor and the resulting generation of new pyoverdine structures may prevent “stealing” of the pyoverdine siderophore by other bacteria in the same environment (227). Thus, prevention of pyocin entry and siderophore stealing may provide powerful selective pressures that drive pyoverdine receptor diversity.
P. aeruginosa type IV pili, which are polymers of monomeric subunits of pilin, mediate adherence to host cell surfaces and are involved in biofilm formation (79, 206). Additionally, type IV pili are particularly popular receptors for P. aeruginosa phages (23, 26, 188, 224). Early studies demonstrated interstrain variation in pilin gene sequence and protein glycosylation status (38, 203). More recently, an analysis of type IV pili from hundreds of diverse P. aeruginosa isolates demonstrated that there are five distinct groups of P. aeruginosa pilin genes (groups I to V) (116). Each pilin gene group is unique in terms of the sequence and length of pilin it encodes as well as its associated accessory genes, which encode proteins mediating pilin posttranslational glycosylation (116). The presence of a tRNAThr gene adjacent to the pilin gene and the fact that some P. aeruginosa pilins are more closely related to pilins of distinct bacterial species (e.g., Dichelobacter nodosus, Eikenella corrodens, Ralstonia solanacearum, and Xanthomonas campestris) than to other P. aeruginosa pilins strongly suggest that pilin gene diversity was generated through HGT. Pilin-associated genes modify pilin in several ways. In the case of group I pili, an associated accessory gene encodes a protein that transfers an LPS O antigen unit to a specific pilin residue (116). In the case of group IV pili, an associated gene encodes an arabinosyltransferase that attaches d-arabinofuranose to several different pilin amino acid residues (115, 221). The functions of the accessory genes associated with group III and V pili are unknown, but they are thought to encode proteins that also modify pilin through posttranslational modifications such as glycosylation. Group II pili do not have associated accessory genes. Interestingly, the group I pilin gene and its corresponding glycosyltransferase gene are overrepresented in P. aeruginosa strains isolated from CF patients (116) and are thus among the few examples of horizontally acquired alleles associated with a particular type of human infection. Though pilin glycosylation in general has been shown to increase P. aeruginosa colonization and survival in the lungs of mice during acute pneumonia (200), the role of type-specific pilin modifications in P. aeruginosa virulence has yet to be determined.
P. aeruginosa produces a single polar flagellum, which mediates motility and adherence and is a potent stimulator of the innate immune response (176). The flagellar filament is a polymer of flagellin protein subunits. P. aeruginosa flagella are glycosylated, and variation of the flagellin protein and its glycosylation allow classification of P. aeruginosa strains as producing either a-type or b-type flagellin (7-9, 191). This variation results from sequence diversity in the flagellin gene itself as well as in a set of flagellin glycosylation genes known as the flagellin glycosylation island, which is inserted within the flagellar biosynthetic gene cluster. The different glycosylation islands encode proteins producing glycans of distinct sizes and targeting different flagellin amino acid residues (191, 220). The flagellin glycosylation islands are thought to have been acquired through either transformation or generalized phage transduction followed by chromosomal capture through double reciprocal recombination (178). Glycosylation does not appear to affect flagellum-mediated motility, as assessed by in vitro motility assays. However, flagellar glycosylation does play a significant role in strain virulence in vivo. Mutant P. aeruginosa strains with nonglycosylated flagellins exhibited 50% lethal dose (LD50) values that were 35- to >10,000-fold greater than those of their parental counterparts in a mouse burned-skin model of infection (8). Additionally, glycosylated flagella stimulate greater interleukin-8 (IL-8) release from epithelial cells, suggesting that glycan moieties may enhance Toll-like receptor 5 (TLR5) recognition of flagellin (219). Whether glycan modifications produced by different flagellar glycosylation islands differentially enhance virulence in specific hosts and infection types has yet to be examined.
Prophages and Phage-Like Elements
General features.
Bacteriophages are highly abundant and genetically diverse organisms composed of a protein coat surrounding a single- or double-stranded DNA or RNA genome, which at a minimum contains essential genes encoding proteins that allow the phage to parasitize the bacterial replication machinery. Bacteriophages may be either virulent or temperate. Virulent bacteriophages lyse their bacterial host cells. In contrast, temperate bacteriophages can display lysogeny, a nonlytic growth mode in which the phage genome is integrated into the bacterial host chromosome in a site-specific recombinase-dependent manner. Once integrated into a bacterial chromosome, the bacteriophage is termed a prophage and may develop mutations or undergo recombination events with other prophages in the bacterial host chromosome. As this occurs, prophages may deteriorate and become permanently fixed in the chromosome. In the case of lysogenic conversion, integrated phages confer upon their bacterial hosts novel phenotypic properties, such as toxin production. If these properties enhance fitness, lysogenized bacterial strains are at a competitive advantage compared to nonlysogenized strains. Upon exposure to DNA-damaging stimuli, prophages retaining the necessary genes may be mobilized from the bacterial chromosome. During mobilization, errors can occur in phage DNA packaging, resulting in the transfer of bacterial genomic DNA to the next bacterial host in a process known as transduction. Specifically, the transduction process can be either generalized (bacterial DNA by itself may be packaged into phage particles) or specialized (phage DNA along with adjacent bacterial DNA is packaged into phage particles). Generalized transduction in particular is a significant contributor to the horizontal transfer of virulence and antibiotic resistance genes (89).
Specific P. aeruginosa prophages and phage-like elements.
There are at least 60 different temperate P. aeruginosa phages (4), and the majority of P. aeruginosa isolates are thought to be lysogenized by at least one phage (94) (Table 3). In particular, double-stranded DNA tailed phages constitute the majority of phages infecting P. aeruginosa. These phages are divided into three families based on tail morphology: Siphoviridae, Myoviridae, and Podoviridae. Of these, the Siphoviridae family of long, noncontractile-tailed phages is the most common. P. aeruginosa phages are a reservoir for genetic diversity. A study of 18 diverse P. aeruginosa phages revealed that 82% of the predicted P. aeruginosa phage proteome is of unknown function (118). Furthermore, tailed phages are themselves genetic mosaics formed by genetic transfer from other phages and bacteria (24, 32, 224). Historically, the study of P. aeruginosa phages was limited to epidemiological typing. More recently, though, there has been renewed interest in P. aeruginosa phages as contributors to interstrain differences in virulence and as integral players in P. aeruginosa biofilm development. Illustrating the ubiquitous nature of P. aeruginosa bacteriophages, recent whole-genome sequencing of P. aeruginosa strain LES revealed six prophage clusters, each derived from one or more of the P. aeruginosa phages F10, D3112, D3, and Pf1 (232). Furthermore, mutations in genes from three of these prophage clusters significantly attenuated virulence, as determined in competition studies performed with a rat model of chronic lung infection (232). The mechanisms by which these LES prophages contribute to virulence are unclear.
TABLE 3.
Selected P. aeruginosa phages and prophage-like elements
| Element | Size (kb)a | Features | Reference(s) |
|---|---|---|---|
| LES prophage 2 | 42 | Contains regions of homology to Siphoviridae family phage F10; contributes to virulence in a rat lung chronic infection model | 118, 232 |
| LES prophage 3 | 43 | Contains regions of homology to phage F10, P. aeruginosa strain 2192, and LES prophage 5; contributes to virulence in a rat lung chronic infection model | 232 |
| LES prophage 4 | 37 | Similar in sequence to transposable phage D3122 | 224, 232 |
| LES prophage 5 | 50 | Shares regions of similarity with phage D3; contributes to virulence in a rat lung chronic infection model | 112, 232 |
| LES prophage 6 | 8 | Similar to filamentous phage Pf1 | 92, 232 |
| φCTX | 36 | Double-stranded DNA Myoviridae family phage encoding a pore-forming cytotoxin that contributes to the virulence of P. aeruginosa strains harboring it | 12, 153 |
| PAGI-6 | 44 | Exhibits a high level of sequence identity to φCTX; however, lacks genes encoding φCTX integrase and cytotoxin | 15 |
| D3 | 56 | Double-stranded DNA Siphoviridae family phage containing a “seroconverting operon” that changes lysogenized P. aeruginosa strains from serotype O5 to O16 | 112, 155 |
| Pf1 | 7 | Single-stranded DNA filamentous phage highly upregulated during P. aeruginosa biofilm development | 92, 228 |
| Pf4 | 12 | Single-stranded DNA filamentous phage affecting P. aeruginosa biofilm phenotypic variation and differentiation as well as virulence | 187, 226 |
| PT-6 | NK | Double-stranded DNA Podoviridae family bacteriophage that produces an alginase | 75 |
| R-type and F-type pyocins | 12-15 | Defective prophages related to phage P2 and phage lambda; encode phage tail particles with antibacterial activity; affect susceptibility to fluoroquinolone antibiotics | 19, 25, 154 |
| F116 | 65 | Double-stranded DNA Podoviridae family generalized transducing phage; digests alginate and encodes several putative proteins with amino acid sequence similarity to proteins from fluorescent Pseudomonas species | 32, 118 |
| D3112 | 38 | Double-stranded DNA Siphoviridae family generalized transducing phage with transposase-mediated integration; representative of one of two groups of P. aeruginosa transposable phages (D3112-like and B3-like); like other tailed P. aeruginosa phages, it exemplifies a mosaic genetic structure; predicted to encode several proteins with sequence similarity to other phage proteins as well as proteins from the bacterial plant pathogen Xanthomonas fastidiosa | 224 |
NK, not known.
Another P. aeruginosa prophage that affects virulence is φCTX (12). φCTX is a double-stranded DNA Myoviridae family phage that contains the ctx gene, which encodes a pore-forming toxin (153). This phage enhances virulence in a number of infection models (12, 161). PAGI-6, a φCTX-related genetic element that lacks ctx and an integrase gene, is an example of a prophage that has undergone multiple recombination and deletion events, resulting in permanent immobilization and altered virulence potential (15).
A fascinating example of lysogenic conversion in P. aeruginosa is LPS alteration mediated by the Siphoviridae family bacteriophage D3. Initial studies reported that serotype O5 strain PAO1 bacteria lysogenized by the LPS-specific phage D3 are resistant to subsequent infection by the same phage (112, 117). Further studies revealed that a 3.6-kb “seroconverting operon” within the D3 genome converted the PAO1 O-antigen serotype from O5 to O16 (155). Presumably, D3 evolved to carry serotype-converting genes to avoid superinfection. Notably, though, since the O antigen is a surface structure that both affects bacterial adhesion to eukaryotic cells and is an antigen recognized by host immune cells, seroconversion may potentially benefit the pathogenesis of P. aeruginosa infections by enhancing bacterial adherence or evasion of host immune responses. Indeed, PAO1 strains lysogenized with D3 display enhanced adherence to human buccal epithelial cells (217). Additionally, lysogeny with FIZ15, a phage that has similar properties to those of D3, is associated with increased resistance to phagocytosis by mouse peritoneal macrophages and increased resistance to killing by human serum (217).
In addition to modifying virulence, bacteriophages can also alter other aspects of P. aeruginosa biology. For example, the filamentous phage Pf4 mediates the appearance of small-colony variants (SCVs), a colony morphotype that is associated with poor lung function in CF patients and with enhanced antibiotic resistance (226). In P. aeruginosa biofilms, filamentous prophage genes are highly differentially upregulated and contribute to the maturation and structural integrity of these bacterial communities (187, 228). Also consistent with a function in remodeling biofilm architecture, Podoviridae family members PT-6 and F116 produce factors that digest the exopolysaccharide alginate, a primary matrix component of P. aeruginosa biofilms (75, 80).
Many P. aeruginosa strains harbor two tandem prophages, derived from phage P2 and phage lambda, that have evolutionarily specialized as narrow-spectrum antibacterial bacteriocins known as R- and F-type pyocins, respectively (154). These pyocins are defective prophages, as they contain phage tail genes but lack genes needed for phage head formation, replication, and integration. Consequently, instead of encoding full phage particles, R- and F-type pyocins encode nuclease- and protease-resistant, rod-like particles resembling phage tails (76, 212). The pyocin phage tail particles are thought to exert their bacteriocin activity by producing pores in the membranes of nonpyocinogenic strains. Spontaneous pyocin production rates are low; however, similar to the conditions stimulating prophage mobilization, pyocin activity can be induced by mutagenic stimuli such as UV irradiation (100) or treatment with the DNA cross-linking agent mitomycin C (105). Although the capacity to produce pyocins is widespread among P. aeruginosa strains (69), the physiological role of these molecules is unclear. R- and F-type pyocins may be important in interstrain competition and even, potentially, interspecies competition, since R-type pyocins are active against Neisseria gonorrhoeae (125), Neisseria meningitidis (5), Haemophilus ducreyi (65), and H. influenzae (167). Additionally, since pyocin production is increased by oxidative stress, it has been proposed that pyocin may have some activity against eukaryotic cells and that its production is a P. aeruginosa defense mechanism against oxidative attack by these cells (41).
Transposons, Insertion Sequences, and Integrons
General features.
Transposable elements are genetic entities that mediate their own translocation from one site to another, usually unrelated site on the same or a different DNA molecule (17, 18), and they are ubiquitous in P. aeruginosa and other bacteria. All functional transposable elements contain a gene or a group of genes encoding a transposase or transposase complex, which mediates transposition by binding to short (15 to 40 bp) inverted repeat (IR) sequences at the borders of the transposable element. Most P. aeruginosa transposable elements can transpose into many different sites on a DNA molecule. Some, though, display site bias for AT-rich sequences or, in the case of some transposon family members, display striking specificity for sequences within certain conserved genes (165, 166, 207). Transposable elements will generally produce a staggered cut at their insertion site; inserted sequences then become flanked by short direct repeats upon transposition. IS elements are small transposable elements that encode only the functions needed for transposition (Fig. 4 A). Larger transposable elements that generally encode functions in addition to transposition are known as transposons. Transposons are further subdivided into two classes: composite transposons and complex transposons (17). Composite transposons are composed of two IS elements with intervening stretches of DNA (Fig. 4B). The IS elements form terminal IR sequences flanking these stretches, allowing the IS-encoded transposase to mediate transposition of the DNA segments caught between them. Complex transposons are more complicated structures, delimited by short terminal IR sequences and containing a dedicated transposase gene, usually in addition to several other genes (Fig. 4C).
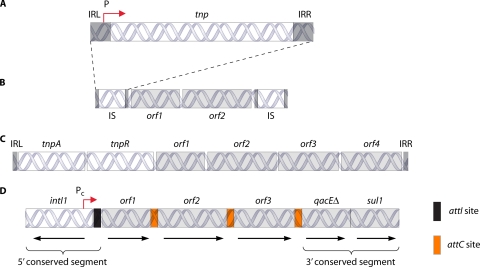
FIG. 4.
General structures of transposable elements and integrons. (A) Organization of a typical IS. Terminal inverted repeats are represented by gray bars labeled IRL (left inverted repeat) and IRR (right inverted repeat). A single open reading frame encoding the transposase tnp is between the inverted repeats. The transposase promoter (P) is partially contained within the IRL. (B) Organization of a composite transposon. One or more genes (light gray boxes labeled orf) are flanked by two usually identical insertion sequences (IS). Transposition is mediated by the IS-contained transposase recognizing the IRL of the first IS and the IRR of the second IS. (C) Organization of a Tn3-like complex transposon. Transposase and recombinase genes are represented by white boxes labeled tnpA and tnpR, respectively. Non-transposition-related coding sequences are represented by light gray boxes labeled orf. These coding sequences are frequently found in the context of integrons. (D) Organization of a typical class 1 integron. The 5′ conserved region consists of the integrase gene intI1 followed by the attI recombination site, represented by a small black box. A forward-directed promoter (Pc) within intI1 drives expression of gene cassettes. Gene cassettes (labeled orf1, orf2, and orf3) are separated by their attC sites (orange boxes). The 3′ conserved segment in most class 1 integrons consists of a truncated quaternary ammonium compound resistance gene (qacEΔ) fused with a sul1 sulfonamide resistance gene.
Integrons are genetic entities that capture exogenous gene cassettes and ensure their expression. All integrons are composed of three core components: (i) a promoter, (ii) a primary recombination site (attI) located downstream of the promoter, and (iii) a gene encoding a tyrosine recombinase family integrase (Fig. 4D). The integrase is capable of inserting gene cassettes containing a 3′ imperfect inverted repeat sequence known as an attC site or “59-base element” into the integron primary recombination site in a RecA-independent manner (142). Integrons by themselves are not mobile. Rather, they achieve mobility only when linked to an existing mobile genetic element. Common mobile genetic element carriers on which integrons can be found are conjugative plasmids and transposons. The genetic elements collected within integrons are gene cassettes, i.e., discrete units that are acquired as circularized DNA consisting of a single gene and an attC site, which is paired with attI for integrase-mediated recombination (185, 210). Following insertion of a gene cassette into an integron's attI recombination site, the attI recombination site is reformed, allowing additional cassettes to be inserted into the integron. The end-to-end gene cassettes can then be expressed as an operon from the integron promoter (17).
On the basis of integrase sequences, at least five classes of integrons have been described (142). In P. aeruginosa, the majority of integrons belong to class 1, in which antibiotic resistance gene cassettes are particularly common (142). These integrons are frequently associated with Tn402-derived transposons (142). Other classes of integrons are quite rare in P. aeruginosa. For example, to date, there has been only one report of class 2 integrons found in P. aeruginosa (236) and no published reports of class 3, 4, or 5 integrons.
Specific P. aeruginosa transposons, insertion sequences, and integrons.
A number of transposons and integrons have been identified in P. aeruginosa (Tables 4 and 5). Most of these characterized transposons and integrons carry antibiotic resistance genes. This association, however, likely represents the disproportionate amount of attention paid by researchers to the role of transposon- and integron-mediated genetic exchange in antibiotic resistance propagation rather than being an intrinsic characteristic of transposons and integrons.
TABLE 4.
Selected P. aeruginosa transposons
| Transposon | Size (kb) | Type | Features | NCBI GenBank accession no. | Reference(s) |
|---|---|---|---|---|---|
| Tn6061 | 26.6 | Complex | Tn3 family transposon; contains 10 antibiotic resistance genes against β-lactamases (blaVEB-1, blaOXA-10), aminoglycosides [ant(2′)-Ia, ant(3″)-Ia, ant(4′)-IIb], tetracycline [tet(G)], rifampin (aar-2), chloramphenicol (cmlA5, floR), and sulfonamides (sul1) | GQ388247 | 46 |
| Tn1213 | 4.1 | Composite | Flanked by ISPa12/ISPa13 insertion sequences; contains β-lactamase blaPER-1 gene expressed under the control of a promoter within insertion sequence ISPa12;associated with P. aeruginosa outbreak in Warsaw, Poland | AY779042 | 60, 174 |
| Tn4401b | 10.0 | Complex | Tn3 family transposon; contains carbapenemase blaKPC-2 gene and two insertion sequences, ISKpn7 and ISKpn6; similar sequence is found in all KPC-expressing Enterobacteriaceae | EU176013 | 150 |
| Tn6001 | 14.6 | Complex | Contains integron In450 carrying metallo-β-lactamase gene blaVIM-3, three copies of aminoglycoside acetyltransferase gene aacA4, aminoglycoside adenylyltransferase gene aadB, sulfonamide resistance gene sul1, orf2 (putative fosfomycin resistance gene), and two other ORFs of unknown function; associated with broad-spectrum antibiotic resistance among P. aeruginosa isolates in Taiwan | EF138817 | 216 |
| Tn801 | 1.5 | Complex | Tn3 family transposon; contains extended-spectrum β-lactamase gene blaTEM-21 preceded by resolvase gene interrupted by insertion sequence IS6100 containing transposase tnpA; associated with ESBL-expressing P. aeruginosa outbreak in nursing home in France | AF080442, AF466526 | 27, 57, 58 |
TABLE 5.
Selected P. aeruginosa class 1 integrons
| Integron | Size (kb) | Gene cassettes (functions) (in order of position downstream from attI site) | NCBI GenBank accession no. | Reference(s) |
|---|---|---|---|---|
| In59.2 | 3.5 | aacA29a (aminoglycoside acetyltransferase), blaVIM-2 (metallo-β-lactamase), aacA29b (aminoglycoside acetyltransferase) | EU118149 | 198 |
| In59.3 | 3.5 | aacA29a (aminoglycoside acetyltransferase), blaVIM-17 (metallo- β-lactamase), aacA29b (aminoglycoside acetyltransferase) | EU118148 | 198 |
| In70.2 | 6.6 | blaVIM-1 (metallo-β-lactamase), aacA4 (aminoglycoside acetyltransferase), aphA15 (aminoglycoside phosphotransferase), aadA1 (aminoglycoside adenylyltransferase) | AJ581664 | 186 |
| In113 | 5.3 | blaIMP-1 (metallo-β-lactamase), aac(6′)-Iae (6′-N-aminoglycoside acetyltransferase), aadA1 (aminoglycoside 3″-adenylyltransferase) | AB104852 | 193 |
| In120 | >3.0 | aac4 (aminoglycoside acetyltransferase), blaBEL-1 (extended-spectrum β-lactamase), smr2 (putative small drug resistance protein), aadA5 (aminoglycoside adenylyltransferase) | GU250439 | 173 |
| In122 | 7.2 | blaVIM-2(metallo-β-lactamase), blaOXA-2 (oxacillinase), aac(6′)-Ib (aminoglycoside-6′-N-acetyltransferase), aadB (aminoglycoside adenylyltransferase), qacG (quaternary ammonium compound resistance) | AY507153 | 44 |
| In163 | 3.9 | aacA4 (aminoglycoside acetyltransferase), blaOXA-56 (extended-spectrum β-lactamase), aadA7 (aminoglycoside adenylyltransferase), orf1 (putative transposase) | AY660529 | 36 |
| Unnamed integron | >4.9 | blaVIM-2 (metallo-β-lactamase), qacF (quaternary ammonium compound resistance), aacA4 (aminoglycoside acetyltransferase), catB3 (chloramphenicol resistance), blaOXA-30 (extended-spectrum β-lactamase), aadA1 (aminoglycoside adenylyltransferase) | DQ287356 | 102 |
| Unnamed integron | >3.2 | blaOXA-74 (extended-spectrum β-lactamase), aac(6′)-Ib-cr (fluoroquinolone-acetylating aminoglycoside acetyltransferase), cmlA7 (chloramphenicol acetyltransferase) | EU161636 | 129 |
A common family of complex transposons in P. aeruginosa consists of the Tn3-like transposons, which are characterized by similar terminal IR sequences, tnpA-encoded transposases, and frequently a second recombination enzyme gene, tnpR, encoding a site-specific recombinase (78, 197). The presence of certain members of this family can dramatically impact the ability of the host bacterial strain to cause disease in a health care setting. One example currently causing substantial concern among clinicians involves strains carrying the emerging KPC family of β-lactamases, which are now causing outbreaks worldwide (recently reviewed by Nordmann et al. [158]). Unlike most β-lactamases, KPC enzymes are capable of hydrolyzing carbapenems, one of the few remaining effective agents for the treatment of many multiresistant bacteria. Although the KPC enzymes are detected primarily among Klebsiella pneumoniae strains and other Enterobacteriaceae, they have also been observed in P. aeruginosa. The Tn3-like transposon Tn4401 was shown to carry blaKPC-2, encoding one member of the KPC family of β-lactamases, in a highly resistant P. aeruginosa isolate from Colombia (150). In fact, parts of the Tn4401 transposon have been identified with every sequence of blaKPC-like genes to date, suggesting that this transposon plays a role in the dissemination of blaKPC genes within and between species of bacteria, including P. aeruginosa (150). A number of other transposons have also been characterized and shown to contribute to antimicrobial resistance in P. aeruginosa (127, 130).
In addition to carrying gene cassettes encoding antibiotic resistance elements, transposable elements can influence gene expression in other ways. Although insertion sequences carry no genes other than those encoding the elements necessary for transposition, an insertion sequence in P. aeruginosa has been shown to modulate expression of an adjacent gene. The promoter sequence in the insertion sequence IS1999 increased expression of the extended-spectrum β-lactamase (ESBL) gene blaVEB-1 when IS1999 inserted into the integron-specific recombination site attI just upstream of this antibiotic resistance gene in P. aeruginosa (10, 152). The ubiquity of ISs, thought to represent 2 to 5% of bacterial genomic DNA (40, 138), suggests that horizontal transfer not just of genes but also of regulatory elements may play a prominent role in modulation of antibiotic resistance or other adaptive functions in P. aeruginosa (151).
As with transposons, the best-studied integrons are those that carry antibiotic resistance genes. Such integrons have the potential to dramatically enhance the fitness of the host bacterium in the health care setting (39, 135, 175). Integron-associated antibiotic resistance genes have been identified in several outbreaks of strains producing metallo-β-lactamases (MBLs) (74, 111, 163, 172, 215). Like KPC β-lactamases, MBLs hydrolyze most β-lactam antibiotics, including carbapenems (31, 134, 223). Integrons rarely carry MBL genes alone; they commonly also contain other antibiotic resistance determinants, such as genes encoding aminoglycoside acetyltransferases, phosphotransferases, and adenylyltransferases (159, 222, 223). Owing to their ability to capture and ensure the expression of multiple different resistance gene cassettes, integrons are particularly dominant contributors to the development of multidrug-resistant P. aeruginosa strains (81, 132, 144). In addition to MBLs and aminoglycoside resistance genes, other integron-associated β-lactamases have also been implicated in P. aeruginosa outbreaks in hospital settings worldwide (36, 164).
The reservoir from which gene cassettes are collected by integrons remains unclear. In several hospital outbreaks, the sources of horizontally transferred genetic material were identified as P. aeruginosa strains or other Gram-negative bacilli that are pathogenic to humans. It has been shown, however, that antibiotic resistance gene cassettes in outbreak-related strains of P. aeruginosa can, in some cases, be traced back to environmental strains of bacteria (198). Examinations of environmental sources have uncovered an immense gene cassette metagenome among bacteria and other organisms in the natural environment that can serve as a nearly inexhaustible supply of genetic diversity (95, 148, 209). Hence, the environment likely constitutes a deep reservoir from which antibiotic resistance genes can be acquired by and disseminated among P. aeruginosa strains.
Other Genetic Elements
Many of the sequences that comprise the P. aeruginosa accessory genome do not resemble ICEs, replacement islands, prophages, transposable elements, or integrons. Some of these elements have likely undergone extensive decay and rearrangements, which prevents their appropriate classification. Still others, though, likely belong to categories of genetic elements that have not yet been characterized. Here we discuss a few examples of such elements.
Rearrangement hot spot (rhs) elements were first characterized in Escherichia coli but were subsequently found in a number of Gram-negative bacteria, including P. aeruginosa and species of Salmonella, Yersinia, Vibrio, Actinobacillus, and Burkholderia (90, 99, 131). Their presence and number vary from strain to strain. For example, E. coli strain K-12 contains five rhs elements that constitute 0.8% of its entire genome, whereas other E. coli strains do not harbor any of these elements (91). rhs elements vary in structure but typically consist of at least a large ORF comprised of a conserved rhs core followed by a short highly variable core extension region (90). rhs cores encode a signature repeated peptide motif [YDxxGRL(I/T)]. Interestingly, rhs cores and core extensions often differ significantly in G+C content even though they form a single large ORF, suggesting that rhs elements are in-frame composites of two distinct elements (99). Together, these features suggest that rhs elements are acquired by HGT, but the mechanism by which this occurs is unclear. The function of rhs elements is also enigmatic, although an E. coli rhs element was implicated in transport of capsular polysaccharide (143) and a Pseudomonas savastanoi rhs element was recently shown to facilitate bacteriocin production (199). For P. aeruginosa, several rhs elements have been described. PAGI-9 and PAGI-10 are rhs-like ORFs that are 6.7 kb and 2.5 kb in size, respectively, and were identified in the hypervirulent P. aeruginosa strain PSE9 by subtractive hybridization with a less virulent strain (15). rhs elements have also been reported in P. aeruginosa isolates from patients with cystic fibrosis and in strain PAO1 (47, 86).
As mentioned previously, the exoU gene encodes an effector protein of the P. aeruginosa type III secretion system and is carried by a family of related ICEs. Another effector protein of the P. aeruginosa type III secretion system that is encoded by an accessory gene is ExoS (83). Like ExoU, it is a potent virulence determinant (196), but its origin appears to be more complex. ExoS shares 76% amino acid identity with a third P. aeruginosa type III secretion effector protein, ExoT, and the exoS and exoT genes are thought to have arisen by gene duplication (237). In contrast to the exoT gene, which is found in every P. aeruginosa strain, the exoS gene is present in only about 70% of P. aeruginosa strains (64). Thus, the exoS gene is part of the P. aeruginosa accessory genome, whereas the exoT gene is not. Interestingly, the sequences surrounding the exoS gene are conserved in nearly all P. aeruginosa strains, and the exoS gene itself is not a part of a discernible mobile genetic element. It is, however, located between 10-bp direct repeats, and sequence analysis of strains lacking exoS is consistent with a recombination event between these direct repeats facilitating the excision of exoS from the chromosome (233). It is also perplexing that nearly all strains of P. aeruginosa carry either the exoS gene or the exoU gene, but not both (64), an observation that is difficult to explain since these genes are located at different loci. Kulasekara et al. proposed a model whereby acquisition of PAPI-2 or other exoU-containing islands caused a deletion of the exoS gene from the P. aeruginosa chromosome (113). The exoU-containing islands themselves are flanked by 20-bp repeats, named TAGI repeats, that have some similarity to the 10-bp repeats flanking the exoS gene. Kulasekara and colleagues postulated that the TAGI repeats are involved in integration of the exoU islands by a site-specific recombinase and that expression of this recombinase during integration simultaneously causes excision of exoS through recognition of the related 10-bp repeats flanking exoS. This model still requires experimental verification, but if it is confirmed, it will demonstrate that interactions between distinct elements of the accessory genome are much more intricate than previously thought.
In addition to the relatively small rhs elements and the exoS gene, many larger accessory genetic elements also display no clear genetic mechanism of acquisition. PAGI-1, for example, is a 49-kb island that was the first genomic island identified in P. aeruginosa (128). Portions of this island are present in 85% of clinical isolates. Besides several proteins of unknown function, genes on PAGI-1 encode transcriptional regulators and dehydrogenases implicated in reactive oxygen species detoxification. Intriguingly, PAGI-1 lacks any sequences associated with transduction, transposition, or conjugation. More recent genomic studies have identified additional genomic islands for which no mode of transfer is apparent. For example, PAGI-7 is a 22-kb genomic island that has a G+C content of 55.8% and is found in only some P. aeruginosa strains (15). However, it is not found adjacent to tRNA genes. Rather, it is integrated within an ORF predicted to encode the ATP-dependent helicase HprB. Although the island interrupts this ORF, no portion of the ORF is deleted or repeated, and the mechanism of insertion is unknown. PAGI-7 itself contains 20 predicted ORFs, including multiple mobility-associated ORFs, predicted transcriptional regulator genes, and a ptxABCDE operon, which is predicted to encode enzymes for the oxidation of phosphite to phosphate (45, 145).
HGT AND EVOLUTION OF THE ACCESSORY GENOME
As alluded to in the preceding sections, HGT plays a major role in shaping the accessory genome of P. aeruginosa. However, it is not the only process that does so. Selective genome reduction can result in a core gene segment becoming an accessory gene segment retained only in the strains derived from progenitors that did not undergo a deletion event. This is exemplified by the observation of large chromosomal deletions resulting in pyomelanin production and loss of pyoverdine synthesis and uptake in P. aeruginosa strains isolated from the lungs of CF patients (61) and perhaps by the case of the exoS gene (113). Alternatively, strain-specific mutations, genetic rearrangements, and duplications followed by clonal expansion can transform genomic regions that were once found in all strains into unique accessory regions. For example, changes in the pyoverdine receptor drive the accumulation of strain-specific compensatory mutations in other pyoverdine locus genes (201). Together with HGT, these ongoing processes continually modify the content of the accessory genome.
By far, though, the most important process contributing to the evolution of the P. aeruginosa accessory genome is HGT. For prokaryotes, HGT can be defined as gene acquisition through one of three possible mechanisms: (i) transduction, or DNA transfer via a bacteriophage; (ii) conjugation, or DNA transfer via intimate cell-cell contact; and (iii) transformation, or transfer of naked DNA. While P. aeruginosa is known to engage in transduction and conjugation, there are no published reports of natural transformation in P. aeruginosa. Nevertheless, several lines of evidence suggest that transformation may be a means of HGT in P. aeruginosa. The type IV pilus of P. aeruginosa is functional for DNA uptake and can restore transformation in a Pseudomonas stutzeri pilA mutant (77) and an N. gonorrhoeae pilE mutant (1). Also, P. aeruginosa has homologs of many of the proteins required for DNA uptake by N. gonorrhoeae. Finally, a number of studies have demonstrated that many segments of the P. aeruginosa chromosome appear to have undergone high levels of recombination between different P. aeruginosa genotypes (107, 170, 229). Although the mechanism of this recombination is unclear, natural competence under certain environmental conditions could account for at least a portion of it. In contrast to transformation, transduction and conjugation have been well documented for P. aeruginosa (11, 97).
At present, the environmental signals that trigger the HGT process are poorly understood (6). For example, though the spontaneous excision rates for the mobilizable ICEs pKL102, clc, and PAPI-1 have been determined in vitro (110, 177, 194, 195), how these laboratory rates compare to mobilization rates in the natural environment has yet to be determined. Likewise, the in vivo growth conditions affecting ICE transfer from one bacterium to another through GI-type T4SSs are unknown, though in vitro findings suggest that ICE mobilization occurs at late growth stages and may be affected by cyclic AMP levels (35). In contrast, several well-characterized conditions, such as mitomycin C treatment, use of antibiotics, UV light, and physiological stresses (e.g., amino acid deprivation), are known to promote mobilization of prophage accessory elements. Additionally, it was recently demonstrated that phage production by P. aeruginosa in soil and groundwater can be triggered by acyl-homoserine lactone quorum-sensing signal molecules (73). Thus, phage-mediated HGT may occur in a microbial population density-dependent manner, consistent with the prevailing thought that high bacterial density niches such as biofilms are rich environments for HGT (202).
Once mobilized, accessory elements may be transferred to a new host, but the efficiency of this process depends upon a number of factors, including the degree of relatedness between the donor and recipient cells and the HGT history of the recipient cell. As expected, host background has a great impact on the transferability of mobilized accessory genetic elements. Reduced relatedness between donor and recipient cells not only is associated with metabolic and gene expression incompatibilities but also results in lower homologous recombination rates due to differences in regulatory motifs asymmetrically distributed between leading and lagging DNA strands (120). Still, the transfer process appears to be remarkably tolerant. Evidence for interspecies transfer of ICEs between P. aeruginosa and Burkholderia cepacia complex bacteria within biofilms in the CF lung (59) and between P. aeruginosa and C. metallidurans (119) in sludge waste has been found. Interestingly, HGT recipients may not be limited to other bacteria. For example, a 7-kb mobile genetic element identified in strain PAO1 encodes a phospholipase with greatest similarity to mammalian and yeast phospholipases, suggesting that transfer between P. aeruginosa and eukaryotic cells is possible (230). Metagenomic studies of complex communities such as soils, which are particularly densely populated with diverse species, will likely reveal more examples of interspecies transfer, better define the exact degree of species dissimilarity tolerated by the P. aeruginosa HGT systems, and show whether particular types of accessory genes possess qualities that make them more itinerant than others.
In addition to kinship, there is mounting evidence that HGT is affected by the particular repertoire of accessory elements already acquired by a bacterium. That is, it appears that bacteria can “remember” their HGT histories. For instance, Qiu et al. reported that the transfer efficiency of the ICE PAPI-1 is greatly reduced when recipient cells already harbor PAPI-1 (178). This observation suggests the presence of a system that prevents redundant ICE acquisition and is reminiscent of plasmid incompatibility systems, which reduce the efficiency of conjugation between bacteria carrying identical or related plasmids (137). Additionally, many bacteria possess clustered, regularly interspaced, short palindromic repeat (CRISPR) loci that inhibit the uptake of foreign DNA (139). CRISPR loci are present in 40% of eubacterial genomes, including that of P. aeruginosa (239), and encode CRISPR RNAs (crRNAs) that confer immunity against previously encountered bacteriophages and conjugative plasmids (13, 225). Specifically, CRISPR arrays are composed of 24- to 48-nucleotide DNA repeats separated by unique spacers. CRISPR-associated (CAS) genes, which encode proteins that function in CRISPR RNA processing and foreign element silencing, are located nearby. CRISPR arrays are initially transcribed as long RNA molecules that are then processed into individual repeat-spacer units. These units target foreign DNA for degradation through an unknown mechanism involving CAS proteins. New spacers corresponding to phage or plasmid sequences are integrated into the 5′ end of existing CRISPR arrays during phage infection or plasmid conjugation. These new spacers confer resistance to subsequent transduction/conjugation events with the cognate phage/plasmid or other phages/plasmids bearing the same sequence. Thus, CRISPR loci may define one of the barriers limiting DNA transfer in a particular strain of P. aeruginosa. Furthermore, the order of spacer sequences within particular CRISPR loci may provide a “log” of the temporal history of horizontal gene acquisition in a particular strain.
Just as accessory gene mobilization and transfer are regulated, so is expression of acquired accessory genes. For this reason, not all accessory genetic elements are expressed. An in vitro transcriptional profiling study of the ICE PAGI-2 found that all but 1 of the 111 PAGI-2 ORFs is transcriptionally silent (110). This was true in both rich and minimal media and during different growth phases. Given that PAGI-2 contains a putative mercury resistance operon, the authors of this study postulated that PAGI-2 transcriptional induction might be mercury exposure dependent. Nevertheless, they were unable to stimulate PAGI-2 transcription in growth medium supplemented with mercury. Thus, it is unclear whether most of the genes in this large ICE are ever expressed. In contrast, it was found that the majority of ORFs in pKLC102, including several with homology to PAGI-2 ORFs, are highly expressed in vitro. Since pKLC102 can exist episomally at multiple copy numbers, while PAGI-2 is immobile and exists as a single chromosomal copy, differences in mobility and copy number may in part account for the disparate transcript levels. Also affecting the expression of accessory genetic elements is the histone-like nucleoid structuring (H-NS) family of proteins. H-NS proteins are regulators of gene expression in numerous Gram-negative bacterial species and play a role in selectively silencing horizontally acquired genes (22, 136). Since the A+T content of horizontally acquired genes is typically higher than that of the core genome, H-NS proteins act as transcriptional silencers of foreign DNA by binding to specific sequences within AT-rich elements. Once bound to DNA, adjacent H-NS proteins oligomerize across AT-rich stretches of DNA to cause transcriptional silencing. Two H-NS family proteins that have been studied in P. aeruginosa are MvaT and MvaU, which act coordinately and preferentially bind regions of the P. aeruginosa chromosome corresponding to accessory genetic elements (37). Not surprisingly, then, double mutation of mvaT and mvaU results in activation of chromosomally integrated filamentous Pf4 prophage (126). The extent of H-NS-mediated repression of horizontally transferred genes has yet to be determined. Also, it remains unclear whether some horizontally transferred elements in P. aeruginosa encode “antisilencing” proteins that are able to overcome H-NS-mediated repression (208).
HGT may be advantageous to the recipient strain when it enhances the recipient's metabolic or pathogenic capacities; however, the existence of H-NS proteins and functionally similar factors devoted to the silencing of horizontally acquired genes suggests that foreign DNA can also have a deleterious impact on host cell fitness (122, 156). This is the case when newly acquired accessory genes result in toxicity from enhanced gene dosage or interference with global gene expression networks (49). Additionally, integration of large pieces of foreign genetic material may adversely affect genome stability. Certain bacteria are able to adapt to the acquisition of novel genetic material and negate any fitness cost. For example, in E. coli the deleterious effects of acquiring some new conjugative plasmids are ameliorated through the accumulation of mutations that repress plasmid gene expression (70). The fitness cost of HGT in P. aeruginosa has been assessed only for the specific case of the ICE clc, whose integration was found to have a minimal effect on the fitness of strain PAO1 (182). In this case, integration resulted in uncoupling of the clc integrase gene from its constitutive promoter, preventing high levels of integrase expression. Since excessive amounts of integrase are deleterious to Pseudomonas (70), Gaillard and colleagues proposed that this uncoupling may explain the observed negligible impact of clc integration (52, 67). The consequences of integration of other accessory elements will need to be examined to resolve the critical determinants affecting P. aeruginosa fitness.
FUTURE DIRECTIONS
As high-throughput sequencing technologies continue to advance and become more affordable, the number of P. aeruginosa whole-genome sequences available will dramatically increase. Correspondingly, comparisons of newly sequenced strains will expand our knowledge of the P. aeruginosa accessory genome. In particular, sequencing of large numbers of ecological isolates has the potential to reveal novel niche-specific accessory genetic elements contributing to the remarkable ability of P. aeruginosa to survive in a wide range of environmental reservoirs. Annotating and classifying novel accessory genetic elements will not be trivial, given their mosaic structure and disproportionately large percentage of ORFans (ORFs with no significant similarity to other sequences in the database) (162, 238). New bioinformatic and homology-based gene annotation programs will be invaluable in this regard. For example, many recently developed online bioinformatic tools were designed to identify and classify accessory genetic elements (14). Likewise, advances in our understanding of the mechanisms of HGT will undoubtedly define the role of many genes involved in the transfer and maintenance of elements within the accessory genome. Determining the function of proteins encoded by “cargo” genes will be more difficult. In some cases, function may be deduced by sequence comparison to the ever-increasing number of genes encoding proteins of known function. In other cases, rigorous genetic and biochemical analyses on a gene-by-gene basis will be required. Regardless of the approaches used, characterization of the accessory genome will be essential for a more complete understanding of the environmental and pathogenic potential of P. aeruginosa.
Acknowledgments
This work was supported by National Institutes of Health grants 2R01 AI053674, 5K02 AI065615, 1R01 AI075191, 1R21 AI088286 (A.R.H.), 5F30 ES016487-03 (V.L.K.), 5T32 AI007476-13, and F32 AI089068 (E.A.O.).
REFERENCES
- 1.Aas, F. E., M. Wolfgang, S. Frye, S. Dunham, C. Lovold, and M. Koomey. 2002. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46:749-760. [DOI] [PubMed] [Google Scholar]
- 2.Abd, H., B. Wretlind, A. Saeed, E. Idsund, K. Hultenby, and G. Sandstrom. 2008. Pseudomonas aeruginosa utilises its type III secretion system to kill the free-living amoeba Acanthamoeba castellanii. J. Eukaryot. Microbiol. 55:235-243. [DOI] [PubMed] [Google Scholar]
- 3.Aguilar-Barajas, E., M. I. Ramírez-Díaz, H. Riveros-Rosas, and C. Cervantes. 2010. Heavy metal resistance in pseudomonads, p. 255-282. In J. L. Ramos and A. Filloux (ed.), Pseudomonas: molecular microbiology, infection and biodiversity, vol. 6. Springer, New York, NY. [Google Scholar]
- 4.Akhverdian, V. Z., E. A. Khrenova, V. G. Bogush, T. V. Gerasimova, and N. B. Kirsanov. 1984. Wide distribution of transposable phages in natural Pseudomonas aeruginosa populations. Genetika 20:1612-1619. [PubMed] [Google Scholar]
- 5.Andersen, S. R., G. Bjune, J. Lyngby, K. Bryn, and E. Jantzen. 1995. Short-chain lipopolysaccharide mutants of serogroup B Neisseria meningitidis of potential value for production of outer membrane vesicle vaccines. Microb. Pathog. 19:159-168. [DOI] [PubMed] [Google Scholar]
- 6.Arnold, D. L., R. W. Jackson, N. R. Waterfield, and J. W. Mansfield. 2007. Evolution of microbial virulence: the benefits of stress. Trends Genet. 23:293-300. [DOI] [PubMed] [Google Scholar]
- 7.Arora, S. K., M. Bangera, S. Lory, and R. Ramphal. 2001. A genomic island in Pseudomonas aeruginosa carries the determinants of flagellin glycosylation. Proc. Natl. Acad. Sci. U. S. A. 98:9342-9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arora, S. K., A. N. Neely, B. Blair, S. Lory, and R. Ramphal. 2005. Role of motility and flagellin glycosylation in the pathogenesis of Pseudomonas aeruginosa burn wound infections. Infect. Immun. 73:4395-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arora, S. K., M. C. Wolfgang, S. Lory, and R. Ramphal. 2004. Sequence polymorphism in the glycosylation island and flagellins of Pseudomonas aeruginosa. J. Bacteriol. 186:2115-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aubert, D., T. Naas, and P. Nordmann. 2003. IS1999 increases expression of the extended-spectrum beta-lactamase VEB-1 in Pseudomonas aeruginosa. J. Bacteriol. 185:5314-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babalova, M., J. Blahova, M. Lesicka-Hupkova, K. Kralikova, V. Krcmery, and V. Schaefer. 1995. A bi-modal transfer of antibiotic resistance by conjugation and transduction from an imipenem-resistant strain of Pseudomonas aeruginosa. Zentralbl. Bakteriol. 283:61-68. [DOI] [PubMed] [Google Scholar]
- 12.Baltch, A. L., R. P. Smith, M. Franke, W. Ritz, P. Michelsen, L. Bopp, and F. Lutz. 1994. Pseudomonas aeruginosa cytotoxin as a pathogenicity factor in a systemic infection of leukopenic mice. Toxicon 32:27-34. [DOI] [PubMed] [Google Scholar]
- 13.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709-1712. [DOI] [PubMed] [Google Scholar]
- 14.Battle, S. E., F. Meyer, J. Rello, V. L. Kung, and A. R. Hauser. 2008. Hybrid pathogenicity island PAGI-5 contributes to the highly virulent phenotype of a Pseudomonas aeruginosa isolate in mammals. J. Bacteriol. 190:7130-7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battle, S. E., J. Rello, and A. R. Hauser. 2009. Genomic islands of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 290:70-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baysse, C., J. M. Meyer, P. Plesiat, V. Geoffroy, Y. Michel-Briand, and P. Cornelis. 1999. Uptake of pyocin S3 occurs through the outer membrane ferripyoverdine type II receptor of Pseudomonas aeruginosa. J. Bacteriol. 181:3849-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett, P. M. 2004. Genome plasticity: insertion sequence elements, transposons and integrons, and DNA rearrangement. Methods Mol. Biol. 266:71-113. [DOI] [PubMed] [Google Scholar]
- 18.Bennett, P. M. 1999. Integrons and gene cassettes: a genetic construction kit for bacteria. J. Antimicrob. Chemother. 43:1-4. [PubMed] [Google Scholar]
- 19.Blazquez, J., J. M. Gomez-Gomez, A. Oliver, C. Juan, V. Kapur, and S. Martin. 2006. PBP3 inhibition elicits adaptive responses in Pseudomonas aeruginosa. Mol. Microbiol. 62:84-99. [DOI] [PubMed] [Google Scholar]
- 20.Bodey, G. P., R. Bolivar, V. Fainstein, and L. Jadeja. 1983. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 5:279-313. [DOI] [PubMed] [Google Scholar]
- 21.Bodilis, J., B. Ghysels, J. Osayande, S. Matthijs, J. P. Pirnay, S. Denayer, D. De Vos, and P. Cornelis. 2009. Distribution and evolution of ferripyoverdine receptors in Pseudomonas aeruginosa. Environ. Microbiol. 11:2123-2135. [DOI] [PubMed] [Google Scholar]
- 22.Bouffartigues, E., M. Buckle, C. Badaut, A. Travers, and S. Rimsky. 2007. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 14:441-448. [DOI] [PubMed] [Google Scholar]
- 23.Bradley, D. E. 1973. A pilus-dependent Pseudomonas aeruginosa bacteriophage with a long noncontractile tail. Virology 51:489-492. [DOI] [PubMed] [Google Scholar]
- 24.Braid, M. D., J. L. Silhavy, C. L. Kitts, R. J. Cano, and M. M. Howe. 2004. Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. J. Bacteriol. 186:6560-6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brazas, M. D., and R. E. Hancock. 2005. Ciprofloxacin induction of a susceptibility determinant in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:3222-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Budzik, J. M., W. A. Rosche, A. Rietsch, and G. A. O'Toole. 2004. Isolation and characterization of a generalized transducing phage for Pseudomonas aeruginosa strains PAO1 and PA14. J. Bacteriol. 186:3270-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 29.Burrus, V., and M. K. Waldor. 2003. Control of SXT integration and excision. J. Bacteriol. 185:5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burrus, V., and M. K. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376-386. [DOI] [PubMed] [Google Scholar]
- 31.Bush, K. 2001. New beta-lactamases in gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin. Infect. Dis. 32:1085-1089. [DOI] [PubMed] [Google Scholar]
- 32.Byrne, M., and A. M. Kropinski. 2005. The genome of the Pseudomonas aeruginosa generalized transducing bacteriophage F116. Gene 346:187-194. [DOI] [PubMed] [Google Scholar]
- 33.Campos-García, J. 2010. Metabolism of acyclic terpenes by Pseudomonas, p. 235-254. In J. L. Ramos and A. Filloux (ed.), Pseudomonas: molecular microbiology, infection and biodiversity, vol. 6. Springer, New York, NY. [Google Scholar]
- 34.Carmeli, Y., N. Troillet, A. W. Karchmer, and M. H. Samore. 1999. Health and economic outcomes of antibiotic resistance in Pseudomonas aeruginosa. Arch. Intern. Med. 159:1127-1132. [DOI] [PubMed] [Google Scholar]
- 35.Carter, M. Q., J. Chen, and S. Lory. 2010. The Pseudomonas aeruginosa pathogenicity island PAPI-1 is transferred via a novel type IV pilus. J. Bacteriol. 192:3249-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carvalho, A. P., R. M. Albano, D. N. de Oliveira, D. A. Cidade, L. M. Teixeira, and A. Marques Ede. 2006. Characterization of an epidemic carbapenem-resistant Pseudomonas aeruginosa producing SPM-1 metallo-beta-lactamase in a hospital located in Rio de Janeiro, Brazil. Microb. Drug Resist. 12:103-108. [DOI] [PubMed] [Google Scholar]
- 37.Castang, S., H. R. McManus, K. H. Turner, and S. L. Dove. 2008. H-NS family members function coordinately in an opportunistic pathogen. Proc. Natl. Acad. Sci. U. S. A. 105:18947-18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castric, P. 1995. pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology 141:1247-1254. [DOI] [PubMed] [Google Scholar]
- 39.Chanawong, A., F. H. M'Zali, J. Heritage, A. Lulitanond, and P. M. Hawkey. 2001. SHV-12, SHV-5, SHV-2a and VEB-1 extended-spectrum beta-lactamases in Gram-negative bacteria isolated in a university hospital in Thailand. J. Antimicrob. Chemother. 48:839-852. [DOI] [PubMed] [Google Scholar]
- 40.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 41.Chang, W., D. A. Small, F. Toghrol, and W. E. Bentley. 2005. Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics 6:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chibeu, A., P. J. Ceyssens, K. Hertveldt, G. Volckaert, P. Cornelis, S. Matthijs, and R. Lavigne. 2009. The adsorption of Pseudomonas aeruginosa bacteriophage phiKMV is dependent on expression regulation of type IV pili genes. FEMS Microbiol. Lett. 296:210-218. [DOI] [PubMed] [Google Scholar]
- 43.Comolli, J. C., A. R. Hauser, L. Waite, C. B. Whitchurch, J. S. Mattick, and J. N. Engel. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 67:3625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corvec, S., L. Poirel, E. Espaze, C. Giraudeau, H. Drugeon, and P. Nordmann. 2008. Long-term evolution of a nosocomial outbreak of Pseudomonas aeruginosa producing VIM-2 metallo-enzyme. J. Hosp. Infect. 68:73-82. [DOI] [PubMed] [Google Scholar]
- 45.Costas, A. M., A. K. White, and W. W. Metcalf. 2001. Purification and characterization of a novel phosphorus-oxidizing enzyme from Pseudomonas stutzeri WM88. J. Biol. Chem. 276:17429-17436. [DOI] [PubMed] [Google Scholar]
- 46.Coyne, S., P. Courvalin, and M. Galimand. 2010. Acquisition of multidrug resistance transposon Tn6061 and IS6100-mediated large chromosomal inversions in Pseudomonas aeruginosa clinical isolates. Microbiology 156:1448-1458. [DOI] [PubMed] [Google Scholar]
- 47.Croft, L., S. A. Beatson, C. B. Whitchurch, B. Huang, R. L. Blakeley, and J. S. Mattick. 2000. An interactive web-based Pseudomonas aeruginosa genome database: discovery of new genes, pathways and structures. Microbiology 146:2351-2364. [DOI] [PubMed] [Google Scholar]
- 48.Cryz, S. J., Jr., T. L. Pitt, E. Furer, and R. Germanier. 1984. Role of lipopolysaccharide in virulence of Pseudomonas aeruginosa. Infect. Immun. 44:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dahlberg, C., and L. Chao. 2003. Amelioration of the cost of conjugative plasmid carriage in Escherichia coli K12. Genetics 165:1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dantas, G., M. O. Sommer, R. D. Oluwasegun, and G. M. Church. 2008. Bacteria subsisting on antibiotics. Science 320:100-103. [DOI] [PubMed] [Google Scholar]
- 51.Dasgupta, T., T. R. de Kievit, H. Masoud, E. Altman, J. C. Richards, I. Sadovskaya, D. P. Speert, and J. S. Lam. 1994. Characterization of lipopolysaccharide-deficient mutants of Pseudomonas aeruginosa derived from serotypes O3, O5, and O6. Infect. Immun. 62:809-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daubin, V., and H. Ochman. 2004. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 14:1036-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dean, C. R., C. V. Franklund, J. D. Retief, M. J. Coyne, Jr., K. Hatano, D. J. Evans, G. B. Pier, and J. B. Goldberg. 1999. Characterization of the serogroup O11 O-antigen locus of Pseudomonas aeruginosa PA103. J. Bacteriol. 181:4275-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 55.Dorn, E., M. Hellwig, W. Reineke, and H. J. Knackmuss. 1974. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol. 99:61-70. [DOI] [PubMed] [Google Scholar]
- 56.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 57.Dubois, V., C. Arpin, P. Noury, C. Andre, L. Coulange, and C. Quentin. 2005. Prolonged outbreak of infection due to TEM-21-producing strains of Pseudomonas aeruginosa and enterobacteria in a nursing home. J. Clin. Microbiol. 43:4129-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dubois, V., C. Arpin, P. Noury, and C. Quentin. 2002. Clinical strain of Pseudomonas aeruginosa carrying a bla(TEM-21) gene located on a chromosomal interrupted TnA type transposon. Antimicrob. Agents Chemother. 46:3624-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eberl, L., and B. Tummler. 2004. Pseudomonas aeruginosa and Burkholderia cepacia in cystic fibrosis: genome evolution, interactions and adaptation. Int. J. Med. Microbiol. 294:123-131. [DOI] [PubMed] [Google Scholar]
- 60.Empel, J., K. Filczak, A. Mrowka, W. Hryniewicz, D. M. Livermore, and M. Gniadkowski. 2007. Outbreak of Pseudomonas aeruginosa infections with PER-1 extended-spectrum beta-lactamase in Warsaw, Poland: further evidence for an international clonal complex. J. Clin. Microbiol. 45:2829-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ernst, R. K., D. A. D'Argenio, J. K. Ichikawa, M. G. Bangera, S. Selgrade, J. L. Burns, P. Hiatt, K. McCoy, M. Brittnacher, A. Kas, D. H. Spencer, M. V. Olson, B. W. Ramsey, S. Lory, and S. I. Miller. 2003. Genome mosaicism is conserved but not unique in Pseudomonas aeruginosa isolates from the airways of young children with cystic fibrosis. Environ. Microbiol. 5:1341-1349. [DOI] [PubMed] [Google Scholar]
- 62.Fagon, J. Y., J. Chastre, Y. Domart, J. L. Trouillet, J. Pierre, C. Carne, and C. Gibert. 1989. Nosocomial pneumonia in patients receiving continuous mechanical ventilation. Am. Rev. Respir. Dis. 139:877-884. [DOI] [PubMed] [Google Scholar]
- 63.Feldman, M., R. Bryan, S. Rajan, L. Scheffler, S. Brunnert, H. Tang, and A. Prince. 1998. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect. Immun. 66:43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feltman, H., G. Schulert, S. Khan, M. Jain, L. Peterson, and A. R. Hauser. 2001. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147:2659-2669. [DOI] [PubMed] [Google Scholar]
- 65.Filiatrault, M. J., R. S. Munson, Jr., and A. A. Campagnari. 2001. Genetic analysis of a pyocin-resistant lipooligosaccharide (LOS) mutant of Haemophilus ducreyi: restoration of full-length LOS restores pyocin sensitivity. J. Bacteriol. 183:5756-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Finck-Barbançon, V., J. Goranson, L. Zhu, T. Sawa, J. P. Wiener-Kronish, S. M. J. Fleiszig, C. Wu, L. Mende-Mueller, and D. Frank. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25:547-557. [DOI] [PubMed] [Google Scholar]
- 67.Fischer, D., and D. Eisenberg. 1999. Finding families for genomic ORFans. Bioinformatics 15:759-762. [DOI] [PubMed] [Google Scholar]
- 68.Fothergill, J. L., E. Mowat, M. J. Ledson, M. J. Walshaw, and C. Winstanley. 2010. Fluctuations in phenotypes and genotypes within populations of Pseudomonas aeruginosa in the cystic fibrosis lung during pulmonary exacerbations. J. Med. Microbiol. 59:472-481. [DOI] [PubMed] [Google Scholar]
- 69.Fyfe, J. A., G. Harris, and J. R. Govan. 1984. Revised pyocin typing method for Pseudomonas aeruginosa. J. Clin. Microbiol. 20:47-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaillard, M., N. Pernet, C. Vogne, O. Hagenbuchle, and J. R. van der Meer. 2008. Host and invader impact of transfer of the clc genomic island into Pseudomonas aeruginosa PAO1. Proc. Natl. Acad. Sci. U. S. A. 105:7058-7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gaillard, M., T. Vallaeys, F. J. Vorholter, M. Minoia, C. Werlen, V. Sentchilo, A. Puhler, and J. R. van der Meer. 2006. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 188:1999-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Geiben-Lynn, R., K. Sauber, and F. Lutz. 2001. Flagellin inhibits Myoviridae phage phiCTX infection of Pseudomonas aeruginosa strain GuA18: purification and mapping of binding site. Arch. Microbiol. 176:339-346. [DOI] [PubMed] [Google Scholar]
- 73.Ghosh, D., K. Roy, K. E. Williamson, S. Srinivasiah, K. E. Wommack, and M. Radosevich. 2009. Acyl-homoserine lactones can induce virus production in lysogenic bacteria: an alternative paradigm for prophage induction. Appl. Environ. Microbiol. 75:7142-7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gibb, A. P., C. Tribuddharat, R. A. Moore, T. J. Louie, W. Krulicki, D. M. Livermore, M. F. Palepou, and N. Woodford. 2002. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new bla(IMP) allele, bla(IMP-7). Antimicrob. Agents Chemother. 46:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Glonti, T., N. Chanishvili, and P. W. Taylor. 2010. Bacteriophage-derived enzyme that depolymerizes the alginic acid capsule associated with cystic fibrosis isolates of Pseudomonas aeruginosa. J. Appl. Microbiol. 108:695-702. [DOI] [PubMed] [Google Scholar]
- 76.Govan, J. R. 1974. Studies on the pyocins of Pseudomonas aeruginosa: morphology and mode of action of contractile pyocins. J. Gen. Microbiol. 80:1-15. [DOI] [PubMed] [Google Scholar]
- 77.Graupner, S., V. Frey, R. Hashemi, M. G. Lorenz, G. Brandes, and W. Wackernagel. 2000. Type IV pilus genes pilA and pilC of Pseudomonas stutzeri are required for natural genetic transformation, and pilA can be replaced by corresponding genes from nontransformable species. J. Bacteriol. 182:2184-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grinsted, J., F. de la Cruz, and R. Schmitt. 1990. The Tn21 subgroup of bacterial transposable elements. Plasmid 24:163-189. [DOI] [PubMed] [Google Scholar]
- 79.Hahn, H. P. 1997. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa. Gene 192:99-108. [DOI] [PubMed] [Google Scholar]
- 80.Hanlon, G. W., S. P. Denyer, C. J. Olliff, and L. J. Ibrahim. 2001. Reduction in exopolysaccharide viscosity as an aid to bacteriophage penetration through Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 67:2746-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harris, A., C. Torres-Viera, L. Venkataraman, P. DeGirolami, M. Samore, and Y. Carmeli. 1999. Epidemiology and clinical outcomes of patients with multiresistant Pseudomonas aeruginosa. Clin. Infect. Dis. 28:1128-1133. [DOI] [PubMed] [Google Scholar]
- 82.Harrison, E. M., M. E. Carter, S. Luck, H. Y. Ou, X. He, Z. Deng, C. O'Callaghan, A. Kadioglu, and K. Rajakumar. 2010. Pathogenicity islands PAPI-1 and PAPI-2 contribute individually and synergistically to the virulence of Pseudomonas aeruginosa strain PA14. Infect. Immun. 78:1437-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hauser, A. R. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7:654-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hauser, A. R., E. Cobb, M. Bodí, D. Mariscal, J. Vallés, J. N. Engel, and J. Rello. 2002. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 30:521-528. [DOI] [PubMed] [Google Scholar]
- 85.Hauser, A. R., P. J. Kang, and J. Engel. 1998. PepA, a novel secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol. Microbiol. 27:807-818. [DOI] [PubMed] [Google Scholar]
- 86.Hayden, H. S., W. Gillett, C. Saenphimmachak, R. Lim, Y. Zhou, M. A. Jacobs, J. Chang, L. Rohmer, D. A. D'Argenio, A. Palmieri, R. Levy, E. Haugen, G. K. Wong, M. J. Brittnacher, J. L. Burns, S. I. Miller, M. V. Olson, and R. Kaul. 2008. Large-insert genome analysis technology detects structural variation in Pseudomonas aeruginosa clinical strains from cystic fibrosis patients. Genomics 91:530-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.He, J., R. L. Baldini, E. Deziel, M. Saucier, Q. Zhang, N. T. Liberati, D. Lee, J. Urbach, H. M. Goodman, and L. G. Rahme. 2004. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. U. S. A. 101:2530-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hentschel, U., and J. Hacker. 2001. Pathogenicity islands: the tip of the iceberg. Microbes Infect. 3:545-548. [DOI] [PubMed] [Google Scholar]
- 89.Hertveldt, K., and R. Lavigne. 2008. Bacteriophages of Pseudomonas, p. 255-263. In B. H. A. Rehm (ed.), Pseudomonas: model organism, pathogen, cell factory. Wiley-VCH, Weinheim, Germany.
- 90.Hill, C. W. 1999. Large genomic sequence repetitions in bacteria: lessons from rRNA operons and Rhs elements. Res. Microbiol. 150:665-674. [DOI] [PubMed] [Google Scholar]
- 91.Hill, C. W., C. H. Sandt, and D. A. Vlazny. 1994. Rhs elements of Escherichia coli: a family of genetic composites each encoding a large mosaic protein. Mol. Microbiol. 12:865-871. [DOI] [PubMed] [Google Scholar]
- 92.Hill, D. F., N. J. Short, R. N. Perham, and G. B. Petersen. 1991. DNA sequence of the filamentous bacteriophage Pf1. J. Mol. Biol. 218:349-364. [DOI] [PubMed] [Google Scholar]
- 93.Holloway, B. W. 1955. Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol. 13:572-581. [DOI] [PubMed] [Google Scholar]
- 94.Holloway, B. W., J. B. Egan, and M. Monk. 1960. Lysogeny in Pseudomonas aeruginosa. Aust. J. Exp. Biol. Med. Sci. 38:321-329. [DOI] [PubMed] [Google Scholar]
- 95.Holmes, A. J., M. R. Gillings, B. S. Nield, B. C. Mabbutt, K. M. Nevalainen, and H. W. Stokes. 2003. The gene cassette metagenome is a basic resource for bacterial genome evolution. Environ. Microbiol. 5:383-394. [DOI] [PubMed] [Google Scholar]
- 96.Hou, Y. M. 1999. Transfer RNAs and pathogenicity islands. Trends Biochem. Sci. 24:295-298. [DOI] [PubMed] [Google Scholar]
- 97.Hupkova, M., J. Blahova, and V. Krcmery. 1994. Transfer, by conjugation and transduction, of resistance to ceftazidime and cefotaxime from the same clinical isolate of Pseudomonas aeruginosa. Zentralbl. Bakteriol. 281:196-200. [DOI] [PubMed] [Google Scholar]
- 98.Inon de Iannino, N., G. Briones, M. Tolmasky, and R. A. Ugalde. 1998. Molecular cloning and characterization of cgs, the Brucella abortus cyclic beta(1-2) glucan synthetase gene: genetic complementation of Rhizobium meliloti ndvB and Agrobacterium tumefaciens chvB mutants. J. Bacteriol. 180:4392-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jackson, A. P., G. H. Thomas, J. Parkhill, and N. R. Thomson. 2009. Evolutionary diversification of an ancient gene family (rhs) through C-terminal displacement. BMC Genomics 10:584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jacob, F. 1954. Induced biosynthesis and mode of action of a pyocine, antibiotic produced by Pseudomonas aeruginosa. Ann. Inst. Pasteur (Paris) 86:149-160. [PubMed] [Google Scholar]
- 101.Jarvis, W. R. 2003. Epidemiology and control of Pseudomonas aeruginosa infections in the intensive care unit, p. 153-168. In A. R. Hauser and J. Rello (ed.), Severe infections caused by Pseudomonas aeruginosa. Kluwer Academic Publishers, Boston, MA.
- 102.Jeong, J. H., K. S. Shin, J. W. Lee, E. J. Park, and S. Y. Son. 2009. Analysis of a novel class 1 integron containing metallo-beta-lactamase gene VIM-2 in Pseudomonas aeruginosa. J. Microbiol. 47:753-759. [DOI] [PubMed] [Google Scholar]
- 103.Juhas, M., D. W. Crook, I. D. Dimopoulou, G. Lunter, R. M. Harding, D. J. Ferguson, and D. W. Hood. 2007. Novel type IV secretion system involved in propagation of genomic islands. J. Bacteriol. 189:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Juhas, M., J. R. van der Meer, M. Gaillard, R. M. Harding, D. W. Hood, and D. W. Crook. 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33:376-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kageyama, M., and F. Egami. 1962. On the purification and some properties of a pyocin, a bacteriocin produced by Pseudomonas aeruginosa. Life Sci. 1:471-476. [DOI] [PubMed] [Google Scholar]
- 106.Kiewitz, C., K. Larbig, J. Klockgether, C. Weinel, and B. Tummler. 2000. Monitoring genome evolution ex vivo: reversible chromosomal integration of a 106 kb plasmid at two tRNA(Lys) gene loci in sequential Pseudomonas aeruginosa airway isolates. Microbiology 146:2365-2373. [DOI] [PubMed] [Google Scholar]
- 107.Kiewitz, C., and B. Tummler. 2000. Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J. Bacteriol. 182:3125-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Klockgether, J., O. Reva, K. Larbig, and B. Tummler. 2004. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J. Bacteriol. 186:518-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Klockgether, J., D. Wurdemann, O. Reva, L. Wiehlmann, and B. Tummler. 2007. Diversity of the abundant pKLC102/PAGI-2 family of genomic islands in Pseudomonas aeruginosa. J. Bacteriol. 189:2443-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Klockgether, J., D. Wurdemann, L. Wiehlmann, and B. Tummler. 2008. Transcript profiling of the Pseudomonas aeruginosa genomic islands PAGI-2 and pKLC102. Microbiology 154:1599-1604. [DOI] [PubMed] [Google Scholar]
- 111.Kouda, S., M. Ohara, M. Onodera, Y. Fujiue, M. Sasaki, T. Kohara, S. Kashiyama, S. Hayashida, T. Harino, T. Tsuji, H. Itaha, N. Gotoh, A. Matsubara, T. Usui, and M. Sugai. 2009. Increased prevalence and clonal dissemination of multidrug-resistant Pseudomonas aeruginosa with the blaIMP-1 gene cassette in Hiroshima. J. Antimicrob. Chemother. 64:46-51. [DOI] [PubMed] [Google Scholar]
- 112.Kropinski, A. M. 2000. Sequence of the genome of the temperate, serotype-converting, Pseudomonas aeruginosa bacteriophage D3. J. Bacteriol. 182:6066-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kulasekara, B. R., H. D. Kulasekara, M. C. Wolfgang, L. Stevens, D. W. Frank, and S. Lory. 2006. Acquisition and evolution of the exoU locus in Pseudomonas aeruginosa. J. Bacteriol. 188:4037-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kurahashi, K., O. Kajikawa, T. Sawa, M. Ohara, M. A. Gropper, D. W. Frank, T. R. Martin, and J. P. Wiener-Kronish. 1999. Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J. Clin. Invest. 104:743-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kus, J. V., J. Kelly, L. Tessier, H. Harvey, D. G. Cvitkovitch, and L. L. Burrows. 2008. Modification of Pseudomonas aeruginosa Pa5196 type IV pilins at multiple sites with D-Araf by a novel GT-C family arabinosyltransferase, TfpW. J. Bacteriol. 190:7464-7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kus, J. V., E. Tullis, D. G. Cvitkovitch, and L. L. Burrows. 2004. Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) versus non-CF patients. Microbiology 150:1315-1326. [DOI] [PubMed] [Google Scholar]
- 117.Kuzio, J., and A. M. Kropinski. 1983. O-antigen conversion in Pseudomonas aeruginosa PAO1 by bacteriophage D3. J. Bacteriol. 155:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kwan, T., J. Liu, M. Dubow, P. Gros, and J. Pelletier. 2006. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. J. Bacteriol. 188:1184-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Larbig, K. D., A. Christmann, A. Johann, J. Klockgether, T. Hartsch, R. Merkl, L. Wiehlmann, H. J. Fritz, and B. Tummler. 2002. Gene islands integrated into tRNA(Gly) genes confer genome diversity on a Pseudomonas aeruginosa clone. J. Bacteriol. 184:6665-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lawrence, J. G., and H. Hendrickson. 2003. Lateral gene transfer: when will adolescence end? Mol. Microbiol. 50:739-749. [DOI] [PubMed] [Google Scholar]
- 121.Lee, D. G., J. M. Urbach, G. Wu, N. T. Liberati, R. L. Feinbaum, S. Miyata, L. T. Diggins, J. He, M. Saucier, E. Deziel, L. Friedman, L. Li, G. Grills, K. Montgomery, R. Kucherlapati, L. G. Rahme, and F. M. Ausubel. 2006. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 7:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee, S. W., and G. Edlin. 1985. Expression of tetracycline resistance in pBR322 derivatives reduces the reproductive fitness of plasmid-containing Escherichia coli. Gene 39:173-180. [DOI] [PubMed] [Google Scholar]
- 123.Lee, V. T., R. S. Smith, B. Tummler, and S. Lory. 2005. Activities of Pseudomonas aeruginosa effectors secreted by the type III secretion system in vitro and during infection. Infect. Immun. 73:1695-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lesic, B., S. Bach, J. M. Ghigo, U. Dobrindt, J. Hacker, and E. Carniel. 2004. Excision of the high-pathogenicity island of Yersinia pseudotuberculosis requires the combined actions of its cognate integrase and Hef, a new recombination directionality factor. Mol. Microbiol. 52:1337-1348. [DOI] [PubMed] [Google Scholar]
- 125.Levin, J. C., and D. C. Stein. 1996. Cloning, complementation, and characterization of an rfaE homolog from Neisseria gonorrhoeae. J. Bacteriol. 178:4571-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li, C., H. Wally, S. J. Miller, and C. D. Lu. 2009. The multifaceted proteins MvaT and MvaU, members of the H-NS family, control arginine metabolism, pyocyanin synthesis, and prophage activation in Pseudomonas aeruginosa PAO1. J. Bacteriol. 191:6211-6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li, H., M. A. Toleman, P. M. Bennett, R. N. Jones, and T. R. Walsh. 2008. Complete sequence of p07-406, a 24,179-base-pair plasmid harboring the blaVIM-7 metallo-beta-lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrob. Agents Chemother. 52:3099-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liang, X., X. Q. Pham, M. V. Olson, and S. Lory. 2001. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J. Bacteriol. 183:843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Libisch, B., L. Poirel, Z. Lepsanovic, V. Mirovic, B. Balogh, J. Paszti, Z. Hunyadi, A. Dobak, M. Fuzi, and P. Nordmann. 2008. Identification of PER-1 extended-spectrum beta-lactamase producing Pseudomonas aeruginosa clinical isolates of the international clonal complex CC11 from Hungary and Serbia. FEMS Immunol. Med. Microbiol. 54:330-338. [DOI] [PubMed] [Google Scholar]
- 130.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lin, R. J., M. Capage, and C. W. Hill. 1984. A repetitive DNA sequence, rhs, responsible for duplications within the Escherichia coli K-12 chromosome. J. Mol. Biol. 177:1-18. [DOI] [PubMed] [Google Scholar]
- 132.Lister, P. D., D. J. Wolter, and N. D. Hanson. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22:582-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu, P. V., and S. Wang. 1990. Three new major somatic antigens of Pseudomonas aeruginosa. J. Clin. Microbiol. 28:922-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Livermore, D. M., and N. Woodford. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489-495. [DOI] [PubMed] [Google Scholar]
- 135.Lolans, K., A. M. Queenan, K. Bush, A. Sahud, and J. P. Quinn. 2005. First nosocomial outbreak of Pseudomonas aeruginosa producing an integron-borne metallo-beta-lactamase (VIM-2) in the United States. Antimicrob. Agents Chemother. 49:3538-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lucchini, S., G. Rowley, M. D. Goldberg, D. Hurd, M. Harrison, and J. C. Hinton. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lyras, D., A. W. Chan, J. McFarlane, and V. A. Stanisich. 1994. The surface exclusion system of RP1: investigation of the roles of trbJ and trbK in the surface exclusion, transfer, and slow-growth phenotypes. Plasmid 32:254-261. [DOI] [PubMed] [Google Scholar]
- 138.Mahillon, J., C. Leonard, and M. Chandler. 1999. IS elements as constituents of bacterial genomes. Res. Microbiol. 150:675-687. [DOI] [PubMed] [Google Scholar]
- 139.Marraffini, L. A., and E. J. Sontheimer. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mathee, K., G. Narasimhan, C. Valdes, X. Qiu, J. M. Matewish, M. Koehrsen, A. Rokas, C. N. Yandava, R. Engels, E. Zeng, R. Olavarietta, M. Doud, R. S. Smith, P. Montgomery, J. R. White, P. A. Godfrey, C. Kodira, B. Birren, J. E. Galagan, and S. Lory. 2008. Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. U. S. A. 105:3100-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Matz, C., A. M. Moreno, M. Alhede, M. Manefield, A. R. Hauser, M. Givskov, and S. Kjelleberg. 2008. Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. ISME J. 2:843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mazel, D. 2006. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4:608-620. [DOI] [PubMed] [Google Scholar]
- 143.McNulty, C., J. Thompson, B. Barrett, L. Lord, C. Andersen, and I. S. Roberts. 2006. The cell surface expression of group 2 capsular polysaccharides in Escherichia coli: the role of KpsD, RhsA and a multi-protein complex at the pole of the cell. Mol. Microbiol. 59:907-922. [DOI] [PubMed] [Google Scholar]
- 144.Mesaros, N., P. Nordmann, P. Plesiat, M. Roussel-Delvallez, J. Van Eldere, Y. Glupczynski, Y. Van Laethem, F. Jacobs, P. Lebecque, A. Malfroot, P. M. Tulkens, and F. Van Bambeke. 2007. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin. Microbiol. Infect. 13:560-578. [DOI] [PubMed] [Google Scholar]
- 145.Metcalf, W. W., and R. S. Wolfe. 1998. Molecular genetic analysis of phosphite and hypophosphite oxidation by Pseudomonas stutzeri WM88. J. Bacteriol. 180:5547-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Meyer, J. M. 2000. Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch. Microbiol. 174:135-142. [DOI] [PubMed] [Google Scholar]
- 147.Meyer, J. M., V. A. Geoffroy, C. Baysse, P. Cornelis, I. Barelmann, K. Taraz, and H. Budzikiewicz. 2002. Siderophore-mediated iron uptake in fluorescent Pseudomonas: characterization of the pyoverdine-receptor binding site of three cross-reacting pyoverdines. Arch. Biochem. Biophys. 397:179-183. [DOI] [PubMed] [Google Scholar]
- 148.Michael, C. A., M. R. Gillings, A. J. Holmes, L. Hughes, N. R. Andrew, M. P. Holley, and H. W. Stokes. 2004. Mobile gene cassettes: a fundamental resource for bacterial evolution. Am. Nat. 164:1-12. [DOI] [PubMed] [Google Scholar]
- 149.Mohd-Zain, Z., S. L. Turner, A. M. Cerdeno-Tarraga, A. K. Lilley, T. J. Inzana, A. J. Duncan, R. M. Harding, D. W. Hood, T. E. Peto, and D. W. Crook. 2004. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 186:8114-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Naas, T., G. Cuzon, M. V. Villegas, M. F. Lartigue, J. P. Quinn, and P. Nordmann. 2008. Genetic structures at the origin of acquisition of the beta-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Naas, T., L. Philippon, L. Poirel, E. Ronco, and P. Nordmann. 1999. An SHV-derived extended-spectrum beta-lactamase in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:1281-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Naas, T., L. Poirel, A. Karim, and P. Nordmann. 1999. Molecular characterization of In50, a class 1 integron encoding the gene for the extended-spectrum beta-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 176:411-419. [DOI] [PubMed] [Google Scholar]
- 153.Nakayama, K., S. Kanaya, M. Ohnishi, Y. Terawaki, and T. Hayashi. 1999. The complete nucleotide sequence of phi CTX, a cytotoxin-converting phage of Pseudomonas aeruginosa: implications for phage evolution and horizontal gene transfer via bacteriophages. Mol. Microbiol. 31:399-419. [DOI] [PubMed] [Google Scholar]
- 154.Nakayama, K., K. Takashima, H. Ishihara, T. Shinomiya, M. Kageyama, S. Kanaya, M. Ohnishi, T. Murata, H. Mori, and T. Hayashi. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38:213-231. [DOI] [PubMed] [Google Scholar]
- 155.Newton, G. J., C. Daniels, L. L. Burrows, A. M. Kropinski, A. J. Clarke, and J. S. Lam. 2001. Three-component-mediated serotype conversion in Pseudomonas aeruginosa by bacteriophage D3. Mol. Microbiol. 39:1237-1247. [DOI] [PubMed] [Google Scholar]
- 156.Nguyen, T. N., Q. G. Phan, L. P. Duong, K. P. Bertrand, and R. E. Lenski. 1989. Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol. Biol. Evol. 6:213-225. [DOI] [PubMed] [Google Scholar]
- 157.Nicas, T., and B. H. Iglewski. 1985. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can. J. Microbiol. 31:387-392. [DOI] [PubMed] [Google Scholar]
- 158.Nordmann, P., G. Cuzon, and T. Naas. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228-236. [DOI] [PubMed] [Google Scholar]
- 159.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in Gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 160.Obritsch, M. D., D. N. Fish, R. MacLaren, and R. Jung. 2004. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob. Agents Chemother. 48:4606-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.O'Callaghan, R. J., L. S. Engel, J. A. Hobden, M. C. Callegan, L. C. Green, and J. M. Hill. 1996. Pseudomonas keratitis. The role of an uncharacterized exoprotein, protease IV, in corneal virulence. Invest. Ophthalmol. Vis. Sci. 37:534-543. [PubMed] [Google Scholar]
- 162.Ou, H. Y., X. He, E. M. Harrison, B. R. Kulasekara, A. B. Thani, A. Kadioglu, S. Lory, J. C. Hinton, M. R. Barer, Z. Deng, and K. Rajakumar. 2007. MobilomeFINDER: web-based tools for in silico and experimental discovery of bacterial genomic islands. Nucleic Acids Res. 35:W97-W104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pagani, L., C. Colinon, R. Migliavacca, M. Labonia, J. D. Docquier, E. Nucleo, M. Spalla, M. Li Bergoli, and G. M. Rossolini. 2005. Nosocomial outbreak caused by multidrug-resistant Pseudomonas aeruginosa producing IMP-13 metallo-beta-lactamase. J. Clin. Microbiol. 43:3824-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pena, C., C. Suarez, F. Tubau, C. Juan, B. Moya, M. A. Dominguez, A. Oliver, M. Pujol, and J. Ariza. 2009. Nosocomial outbreak of a non-cefepime-susceptible ceftazidime-susceptible Pseudomonas aeruginosa strain overexpressing MexXY-OprM and producing an integron-borne PSE-1 beta-lactamase. J. Clin. Microbiol. 47:2381-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Peters, J. E., and N. L. Craig. 2001. Tn7: smarter than we thought. Nat. Rev. Mol. Cell. Biol. 2:806-814. [DOI] [PubMed] [Google Scholar]
- 166.Petrovski, S., and V. A. Stanisich. 2010. Tn502 and Tn512 are res site hunters that provide evidence of resolvase-independent transposition to random sites. J. Bacteriol. 192:1865-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Phillips, N. J., C. M. John, L. G. Reinders, B. W. Gibson, M. A. Apicella, and J. M. Griffiss. 1990. Structural models for the cell surface lipooligosaccharides of Neisseria gonorrhoeae and Haemophilus influenzae. Biomed. Environ. Mass Spectrom. 19:731-745. [DOI] [PubMed] [Google Scholar]
- 168.Pier, G. B. 2007. Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. Int. J. Med. Microbiol. 297:277-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pier, G. B., and R. Ramphal. 2005. Pseudomonas aeruginosa, p. 2587-2615. In G. Mandell, J. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases. Elsevier Churchill Livingstone, Philadelphia, PA.
- 170.Pirnay, J. P., F. Bilocq, B. Pot, P. Cornelis, M. Zizi, J. Van Eldere, P. Deschaght, M. Vaneechoutte, S. Jennes, T. Pitt, and D. De Vos. 2009. Pseudomonas aeruginosa population structure revisited. PLoS One 4:e7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pirnay, J. P., S. Matthijs, H. Colak, P. Chablain, F. Bilocq, J. Van Eldere, D. De Vos, M. Zizi, L. Triest, and P. Cornelis. 2005. Global Pseudomonas aeruginosa biodiversity as reflected in a Belgian river. Environ. Microbiol. 7:969-980. [DOI] [PubMed] [Google Scholar]
- 172.Pitout, J. D., G. Revathi, B. L. Chow, B. Kabera, S. Kariuki, P. Nordmann, and L. Poirel. 2008. Metallo-beta-lactamase-producing Pseudomonas aeruginosa isolated from a large tertiary centre in Kenya. Clin. Microbiol. Infect. 14:755-759. [DOI] [PubMed] [Google Scholar]
- 173.Poirel, L., L. Brinas, A. Verlinde, L. Ide, and P. Nordmann. 2005. BEL-1, a novel clavulanic acid-inhibited extended-spectrum beta-lactamase, and the class 1 integron In120 in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:3743-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Poirel, L., L. Cabanne, H. Vahaboglu, and P. Nordmann. 2005. Genetic environment and expression of the extended-spectrum beta-lactamase blaPER-1 gene in Gram-negative bacteria. Antimicrob. Agents Chemother. 49:1708-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Poirel, L., E. Lebessi, M. Castro, C. Fevre, M. Foustoukou, and P. Nordmann. 2004. Nosocomial outbreak of extended-spectrum beta-lactamase SHV-5-producing isolates of Pseudomonas aeruginosa in Athens, Greece. Antimicrob. Agents Chemother. 48:2277-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Prince, A. 2006. Flagellar activation of epithelial signaling. Am. J. Respir. Cell Mol. Biol. 34:548-551. [DOI] [PubMed] [Google Scholar]
- 177.Qiu, X., A. U. Gurkar, and S. Lory. 2006. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 103:19830-19835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Qiu, X., B. R. Kulasekara, and S. Lory. 2009. Role of horizontal gene transfer in the evolution of Pseudomonas aeruginosa virulence. Genome Dyn. 6:126-139. [DOI] [PubMed] [Google Scholar]
- 179.Rakhimova, E., L. Wiehlmann, A. L. Brauer, S. Sethi, T. F. Murphy, and B. Tummler. 2009. Pseudomonas aeruginosa population biology in chronic obstructive pulmonary disease. J. Infect. Dis. 200:1928-1935. [DOI] [PubMed] [Google Scholar]
- 180.Ramsay, J. P., J. T. Sullivan, G. S. Stuart, I. L. Lamont, and C. W. Ronson. 2006. Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol. Microbiol. 62:723-734. [DOI] [PubMed] [Google Scholar]
- 181.Ravatn, R., S. Studer, D. Springael, A. J. Zehnder, and J. R. van der Meer. 1998. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 180:4360-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ravatn, R., S. Studer, A. J. Zehnder, and J. R. van der Meer. 1998. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J. Bacteriol. 180:5505-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ravatn, R., A. J. Zehnder, and J. R. van der Meer. 1998. Low-frequency horizontal transfer of an element containing the chlorocatechol degradation genes from Pseudomonas sp. strain B13 to Pseudomonas putida F1 and to indigenous bacteria in laboratory-scale activated-sludge microcosms. Appl. Environ. Microbiol. 64:2126-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Raymond, C. K., E. H. Sims, A. Kas, D. H. Spencer, T. V. Kutyavin, R. G. Ivey, Y. Zhou, R. Kaul, J. B. Clendenning, and M. V. Olson. 2002. Genetic variation at the O-antigen biosynthetic locus in Pseudomonas aeruginosa. J. Bacteriol. 184:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 186.Riccio, M. L., L. Pallecchi, J. D. Docquier, S. Cresti, M. R. Catania, L. Pagani, C. Lagatolla, G. Cornaglia, R. Fontana, and G. M. Rossolini. 2005. Clonal relatedness and conserved integron structures in epidemiologically unrelated Pseudomonas aeruginosa strains producing the VIM-1 metallo-beta-lactamase from different Italian hospitals. Antimicrob. Agents Chemother. 49:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rice, S. A., C. H. Tan, P. J. Mikkelsen, V. Kung, J. Woo, M. Tay, A. Hauser, D. McDougald, J. S. Webb, and S. Kjelleberg. 2009. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 3:271-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Roncero, C., A. Darzins, and M. J. Casadaban. 1990. Pseudomonas aeruginosa transposable bacteriophages D3112 and B3 require pili and surface growth for adsorption. J. Bacteriol. 172:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Roy, P. H., S. G. Tetu, A. Larouche, L. Elbourne, S. Tremblay, Q. Ren, R. Dodson, D. Harkins, R. Shay, K. Watkins, Y. Mahamoud, and I. T. Paulsen. 2010. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS One 5:e8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Roy-Burman, A., R. H. Savel, S. Racine, B. L. Swanson, N. S. Revadigar, J. Fujimoto, T. Sawa, D. W. Frank, and J. P. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 191.Schirm, M., S. K. Arora, A. Verma, E. Vinogradov, P. Thibault, R. Ramphal, and S. M. Logan. 2004. Structural and genetic characterization of glycosylation of type a flagellin in Pseudomonas aeruginosa. J. Bacteriol. 186:2523-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Schulert, G. S., H. Feltman, S. D. P. Rabin, C. G. Martin, S. E. Battle, J. Rello, and A. R. Hauser. 2003. Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J. Infect. Dis. 188:1695-1706. [DOI] [PubMed] [Google Scholar]
- 193.Sekiguchi, J., T. Asagi, T. Miyoshi-Akiyama, T. Fujino, I. Kobayashi, K. Morita, Y. Kikuchi, T. Kuratsuji, and T. Kirikae. 2005. Multidrug-resistant Pseudomonas aeruginosa strain that caused an outbreak in a neurosurgery ward and its aac(6′)-Iae gene cassette encoding a novel aminoglycoside acetyltransferase. Antimicrob. Agents Chemother. 49:3734-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sentchilo, V., R. Ravatn, C. Werlen, A. J. Zehnder, and J. R. van der Meer. 2003. Unusual integrase gene expression on the clc genomic island in Pseudomonas sp. strain B13. J. Bacteriol. 185:4530-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sentchilo, V., A. J. Zehnder, and J. R. van der Meer. 2003. Characterization of two alternative promoters for integrase expression in the clc genomic island of Pseudomonas sp. strain B13. Mol. Microbiol. 49:93-104. [DOI] [PubMed] [Google Scholar]
- 196.Shaver, C. M., and A. R. Hauser. 2004. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect. Immun. 72:6969-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sherratt, D. 1989. Tn3 and other related transposable elements: site-specific recombination and transposition, p. 163-184. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, DC.
- 198.Siarkou, V. I., D. Vitti, E. Protonotariou, A. Ikonomidis, and D. Sofianou. 2009. Molecular epidemiology of outbreak-related Pseudomonas aeruginosa strains carrying the novel variant blaVIM-17 metallo-beta-lactamase gene. Antimicrob. Agents Chemother. 53:1325-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sisto, A., M. G. Cipriani, M. Morea, S. L. Lonigro, F. Valerio, and P. Lavermicocca. 2010. An Rhs-like genetic element is involved in bacteriocin production by Pseudomonas savastanoi pv. savastanoi. Antonie Van Leeuwenhoek 98:505-517. [DOI] [PubMed] [Google Scholar]
- 200.Smedley, J. G., III, E. Jewell, J. Roguskie, J. Horzempa, A. Syboldt, D. B. Stolz, and P. Castric. 2005. Influence of pilin glycosylation on Pseudomonas aeruginosa 1244 pilus function. Infect. Immun. 73:7922-7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Smith, E. E., E. H. Sims, D. H. Spencer, R. Kaul, and M. V. Olson. 2005. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. J. Bacteriol. 187:2138-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sorensen, S. J., M. Bailey, L. H. Hansen, N. Kroer, and S. Wuertz. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3:700-710. [DOI] [PubMed] [Google Scholar]
- 203.Spangenberg, C., R. Fislage, W. Sierralta, B. Tummler, and U. Romling. 1995. Comparison of type IV-pilin genes of Pseudomonas aeruginosa of various habitats has uncovered a novel unusual sequence. FEMS Microbiol. Lett. 127:158. [DOI] [PubMed] [Google Scholar]
- 204.Spencer, D. H., A. Kas, E. E. Smith, C. K. Raymond, E. H. Sims, M. Hastings, J. L. Burns, R. Kaul, and M. V. Olson. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:1316-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Springael, D., K. Peys, A. Ryngaert, S. Van Roy, L. Hooyberghs, R. Ravatn, M. Heyndrickx, J. R. van der Meer, C. Vandecasteele, M. Mergeay, and L. Diels. 2002. Community shifts in a seeded 3-chlorobenzoate degrading membrane biofilm reactor: indications for involvement of in situ horizontal transfer of the clc-element from inoculum to contaminant bacteria. Environ. Microbiol. 4:70-80. [DOI] [PubMed] [Google Scholar]
- 206.Sriramulu, D. D., H. Lunsdorf, J. S. Lam, and U. Romling. 2005. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54:667-676. [DOI] [PubMed] [Google Scholar]
- 207.Stanisich, V. A., R. Arwas, P. M. Bennett, and F. de la Cruz. 1989. Characterization of Pseudomonas mercury-resistance transposon Tn502, which has a preferred insertion site in RP1. J. Gen. Microbiol. 135:2909-2915. [DOI] [PubMed] [Google Scholar]
- 208.Stoebel, D. M., A. Free, and C. J. Dorman. 2008. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 154:2533-2545. [DOI] [PubMed] [Google Scholar]
- 209.Stokes, H. W., A. J. Holmes, B. S. Nield, M. P. Holley, K. M. Nevalainen, B. C. Mabbutt, and M. R. Gillings. 2001. Gene cassette PCR: sequence-independent recovery of entire genes from environmental DNA. Appl. Environ. Microbiol. 67:5240-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 211.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalk, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saler, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 212.Takeda, Y., and M. Kageyama. 1975. Subunit arrangement in the extended sheath of pyocin R. J. Biochem. 77:679-684. [DOI] [PubMed] [Google Scholar]
- 213.Talbot, G. H., J. Bradley, J. E. Edwards, Jr., D. Gilbert, M. Scheld, and J. G. Bartlett. 2006. Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Dis. 42:657-668. [DOI] [PubMed] [Google Scholar]
- 214.Tang, H., M. Kays, and A. Prince. 1995. Role of Pseudomonas aeruginosa pili in acute pulmonary infection. Infect. Immun. 63:1278-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tsakris, A., A. Poulou, I. Kristo, T. Pittaras, N. Spanakis, S. Pournaras, and F. Markou. 2009. Large dissemination of VIM-2-metallo-beta-lactamase-producing Pseudomonas aeruginosa strains causing health care-associated community-onset infections. J. Clin. Microbiol. 47:3524-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tseng, S. P., J. C. Tsai, L. J. Teng, and P. R. Hsueh. 2009. Dissemination of transposon Tn6001 in carbapenem-non-susceptible and extensively drug-resistant Pseudomonas aeruginosa in Taiwan. J. Antimicrob. Chemother. 64:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Vaca-Pacheco, S., G. L. Paniagua-Contreras, O. Garcia-Gonzalez, and M. de la Garza. 1999. The clinically isolated FIZ15 bacteriophage causes lysogenic conversion in Pseudomonas aeruginosa PAO1. Curr. Microbiol. 38:239-243. [DOI] [PubMed] [Google Scholar]
- 218.van der Meer, J. R., C. Werlen, S. F. Nishino, and J. C. Spain. 1998. Evolution of a pathway for chlorobenzene metabolism leads to natural attenuation in contaminated groundwater. Appl. Environ. Microbiol. 64:4185-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Verma, A., S. K. Arora, S. K. Kuravi, and R. Ramphal. 2005. Roles of specific amino acids in the N terminus of Pseudomonas aeruginosa flagellin and of flagellin glycosylation in the innate immune response. Infect. Immun. 73:8237-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Verma, A., M. Schirm, S. K. Arora, P. Thibault, S. M. Logan, and R. Ramphal. 2006. Glycosylation of b-type flagellin of Pseudomonas aeruginosa: structural and genetic basis. J. Bacteriol. 188:4395-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Voisin, S., J. V. Kus, S. Houliston, F. St. Michael, D. Watson, D. G. Cvitkovitch, J. Kelly, J. R. Brisson, and L. L. Burrows. 2007. Glycosylation of Pseudomonas aeruginosa strain Pa5196 type IV pilins with mycobacterium-like alpha-1,5-linked d-Araf oligosaccharides. J. Bacteriol. 189:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Walsh, T. R. 2005. The emergence and implications of metallo-beta-lactamases in Gram-negative bacteria. Clin. Microbiol. Infect. 11(Suppl. 6):2-9. [DOI] [PubMed] [Google Scholar]
- 223.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-beta-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wang, P. W., L. Chu, and D. S. Guttman. 2004. Complete sequence and evolutionary genomic analysis of the Pseudomonas aeruginosa transposable bacteriophage D3112. J. Bacteriol. 186:400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Waters, L. S., and G. Storz. 2009. Regulatory RNAs in bacteria. Cell 136:615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Webb, J. S., M. Lau, and S. Kjelleberg. 2004. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:8066-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.West, S. A., and A. Buckling. 2003. Cooperation, virulence and siderophore production in bacterial parasites. Proc. Biol. Sci. 270:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 229.Wiehlmann, L., G. Wagner, N. Cramer, B. Siebert, P. Gudowius, G. Morales, T. Kohler, C. van Delden, C. Weinel, P. Slickers, and B. Tummler. 2007. Population structure of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 104:8101-8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wilderman, P. J., A. I. Vasil, Z. Johnson, and M. L. Vasil. 2001. Genetic and biochemical analyses of a eukaryotic-like phospholipase D of Pseudomonas aeruginosa suggest horizontal acquisition and a role for persistence in a chronic pulmonary infection model. Mol. Microbiol. 39:291-303. [DOI] [PubMed] [Google Scholar]
- 231.Williams, K. P. 2002. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucleic Acids Res. 30:866-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Winstanley, C., M. G. Langille, J. L. Fothergill, I. Kukavica-Ibrulj, C. Paradis-Bleau, F. Sanschagrin, N. R. Thomson, G. L. Winsor, M. A. Quail, N. Lennard, A. Bignell, L. Clarke, K. Seeger, D. Saunders, D. Harris, J. Parkhill, R. E. Hancock, F. S. Brinkman, and R. C. Levesque. 2009. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool epidemic strain of Pseudomonas aeruginosa. Genome Res. 19:12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wolfgang, M. C., B. R. Kulasekara, X. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Woods, D. E., S. J. Cruz, and R. L. Friedman. 1982. Contribution of toxin A and elastase to virulence of Pseudomonas aeruginosa in chronic lung infections of rats. Infect. Immun. 36:1223-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Wurdemann, D., and B. Tummler. 2007. In silico comparison of pKLC102-like genomic islands of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 275:244-249. [DOI] [PubMed] [Google Scholar]
- 236.Xu, Z., L. Li, M. E. Shirtliff, M. J. Alam, S. Yamasaki, and L. Shi. 2009. Occurrence and characteristics of class 1 and 2 integrons in Pseudomonas aeruginosa isolates from patients in southern China. J. Clin. Microbiol. 47:230-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yahr, T. L., J. T. Barbieri, and D. W. Frank. 1996. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa. J. Bacteriol. 178:1412-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zaneveld, J. R., D. R. Nemergut, and R. Knight. 2008. Are all horizontal gene transfers created equal? Prospects for mechanism-based studies of HGT patterns. Microbiology 154:1-15. [DOI] [PubMed] [Google Scholar]
- 239.Zegans, M. E., J. C. Wagner, K. C. Cady, D. M. Murphy, J. H. Hammond, and G. A. O'Toole. 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J. Bacteriol. 191:210-219. [DOI] [PMC free article] [PubMed] [Google Scholar]