Abstract
Bartonellae were detected in a total of 152 (23.7%) of 642 tissues from 108 (48.4%) of 223 small mammals trapped in several urban areas of Nepal. Based on rpoB and gltA sequence analyses, genotypes belonging to seven known Bartonella species and five genotypes not belonging to previously known species were identified in these animals.
Bartonella species are important zoonotic and vector-borne bacteria associated with an increasing array of emerging infections in humans and animals (6, 8, 21). Identification of new bartonellae, especially in animals that have possible contact with humans, can help to identify new human pathogens (12). Moreover, the close association between rodents and humans throughout the world makes the study of rodent-borne Bartonella essential to determine the extent to which rodents may serve as a source of human infections (7). We conducted a study on the molecular detection of Bartonella in different tissues of Nepalese small mammals.
In 1996, a total of 223 small mammals (38 Bandicota bengalensis, 3 Mus musculus castaneus, 90 Rattus rattus brunneusculus, and 92 Suncus murinus animals) were trapped in densely human populated urban areas in Nepal. Details on the trapped animals are given in Table 1. Lung, kidney, and liver tissue samples were collected and shipped to the Centers for Disease Control and Prevention (CDC), Fort Collins, CO. A total of 642 tissue samples were assessed for the presence of Bartonella DNA by rpoB and gltA sequencing as described previously (5, 23).
TABLE 1.
Prevalence of Bartonella in small mammals trapped in different districts of Nepal
| Districta | Trap areab | No. of Bartonella-infected animals/no. of animals examined (%) |
|||||
|---|---|---|---|---|---|---|---|
| Bandicota bengalensis | Mus musculus castaneus | Rattus rattus brunneusculus | Suncus murinus | Subtotal | Total | ||
| Bhaktapur | Bhelukhel | 3/5 (60) | 3/5 (60) | ||||
| Bhelukhel | 1/1 (100) | 1/1 (100) | |||||
| Pode Tole | 0/2 (0) | 4/8 (50) | 0/1 (0) | 4/11 (36.4) | |||
| NK | 4/12 (33.3) | 4/9 (44.4) | 8/21 (38.1) | ||||
| Subtotal | 0/2 (0) | 0/0 (0) | 12/26 (46.2) | 4/10 (40) | 16/38 (42.1) | ||
| Kathmandu | Hyumata | 2/2 (100) | 2/3 (66.7) | 4/5 (80) | |||
| Inakha Tole | 3/3 (100) | 3/3 (100) | |||||
| Kalimati | 2/6 (33.3) | 0/1 (0) | 1/5 (20) | 7/12 (58.3) | 10/24 (41.7) | ||
| Lagan Tole | 0/4 (0) | 2/3 (66.7) | 3/5 (60) | 5/12 (41.7) | |||
| Sabalbahal | 7/21 (33.3) | 0/1 (0) | 2/6 (33.3) | 7/14 (50) | 16/42 (38.1) | ||
| Teku | 1/4 (25) | 0/1 (0) | 4/13 (30.8) | 1/1 (100) | 6/19 (31.6) | ||
| Tokha (suburb) | 1/1 (100) | 1/1 (100) | |||||
| NK | 1/2 (50) | 15/25 (60) | 16/27 (59.3) | ||||
| Subtotal | 10/35 (28.6) | 0/3 (0) | 13/32 (40.6) | 38/63 (60.3) | 61/133 (45.9) | ||
| Lalitpur | Bakhar | 4/4 (100) | 4/4 (100) | ||||
| Bakhar Thati | 0/3 (0) | 0/3 (0) | |||||
| Lagankhel | 1/1 (100) | 1/1 (100) | |||||
| Patan | 2/2 (100) | 2/2 (100) | |||||
| Sabalbahal | 0/1 (0) | 0/1 (0) | |||||
| NK | 9/24 (37.5) | 15/17 (88.2) | 24/41 (58.5) | ||||
| Subtotal | 0/1 (0) | 0/0 (0) | 14/32 (43.8) | 17/19 (89.5) | 31/52 (59.6) | ||
| Total | 10/38 (26.3) | 0/3 (0) | 39/90 (43.3) | 59/92 (64.1) | 108/223 (48.4) | ||
The following Bartonella species were detected: in Bhaktapur, B. coopersplainsensis, B. elizabethae, B. queenslandensis, and a novel Bartonella species; in Kathmandu, B. coopersplainsensis, B. elizabethae, B. queenslandensis, B. rochalimae, B. tribocorum, and a novel Bartonella species; and in Lalitpur, B. coopersplainsensis, B. elizabethae, B. phoceensis, B. queenslandensis, B. rattimassiliensis, B. tribocorum, and a novel Bartonella species.
NK, not known.
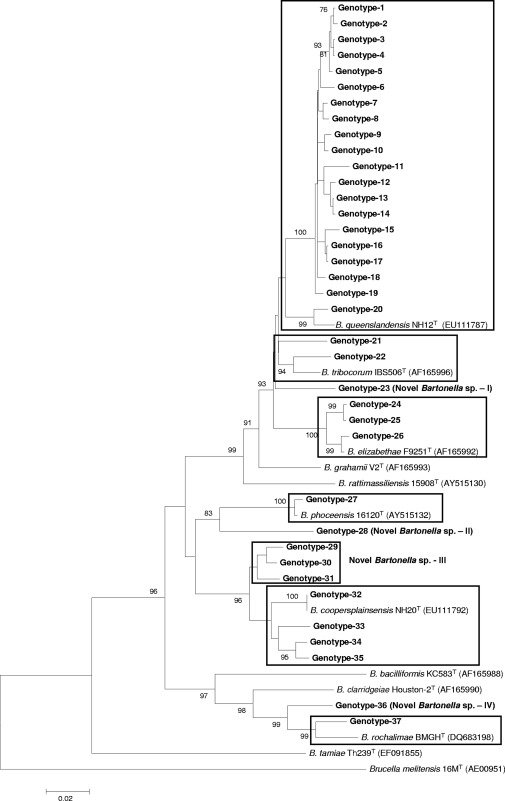
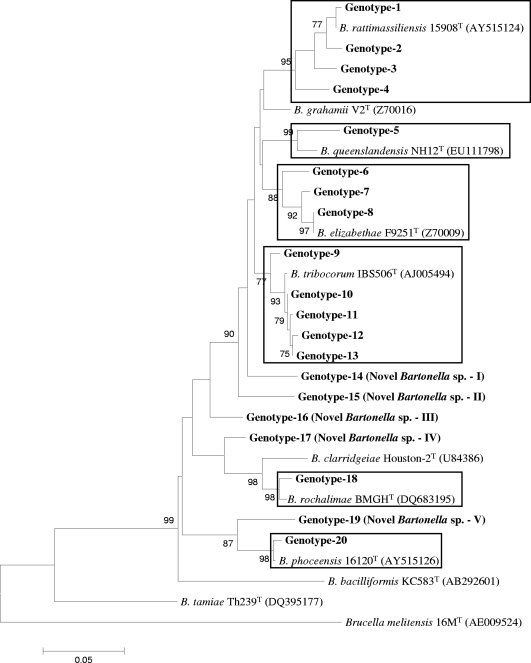
Bartonella was detected in 108 (48.4%) of 223 animals in Nepal: 10 (26.3%) of 38 B. bengalensis rats, 39 (43.3%) of 90 R. rattus brunneusculus rats, and 59 (64.1%) of 92 S. murinus shrews were infected with Bartonella (Table 1). The infection rate varied from 42 to 60% by province. The highest numbers of infected animals were in the Kathmandu and Lalitpur districts. This was because S. murinus was the most commonly infected species and the highest numbers of animals of this species were trapped in these areas. Bandicoots were the most commonly infected subjects in Kathmandu, but fewer numbers of these animals (n = 2) were trapped in Bhaktapur and Lalitpur. Only three M. musculus mice were collected (only in Kathmandu) in this study, and none of these animals were positive for Bartonella. A total of 152 (23.7%) of 642 tissue samples were positive for Bartonella. Of the three tissue types, kidney samples (29.6%) and liver samples (25.6%) were more frequently infected with Bartonella than the lung samples (17.3%; P = 0.03 and 0.04, respectively) (Table 2). DNA sequencing revealed 37 and 20 rpoB and gltA genotypes, respectively, of which 31 and 15 belonged to B. rattimassiliensis, B. queenslandensis, B. elizabethae, B. tribocorum, “B. rochalimae,” or “B. phoceensis” and the other 6 and 5 genotypes, respectively, were not genetically related to known bartonellae (Fig. 1 and 2). These novel genetic groups were found in S. murinus and R. rattus brunneusculus animals. The human pathogen B. elizabethae was detected in R. rattus brunneusculus rats (n = 5) and an S. murinus shrew (n = 1), and the human pathogen B. rochalimae was found in a B. bengalensis bandicoot (n = 1) and an S. murinus shrew (n = 1).
TABLE 2.
Detection of Bartonella DNA in different tissues of small mammals in Nepal
| Species | No. of animals infected/no. of animals tested (%) | No. of Bartonella-infected tissue samples/no. of tissue samples examined (%) |
|||
|---|---|---|---|---|---|
| Kidney | Liver | Lung | Total | ||
| Bandicota bengalensis | 10/38 (26.3) | 5/38 (13.2) | 2/38 (5.3) | 5/18 (27.8) | 12/94 (12.8) |
| Mus musculus castaneus | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/9 (0) |
| Rattus rattus brunneusculus | 39/90 (43.3) | 12/90 (13.3) | 32/90 (35.6) | 15/89 (16.9) | 59/269 (21.9) |
| Suncus murinus | 59/92 (64.1) | 49/92 (53.3) | 18/92 (19.6) | 14/86 (16.3) | 81/270 (30) |
| Total | 108/223 (48.4) | 66/223 (29.6) | 52/203 (25.6) | 34/196 (17.3) | 152/642 (23.7) |
FIG. 1.
Phylogenetic classification of Bartonella genotypes detected in small mammals of Nepal based on rpoB gene sequences. The phylogram was constructed by the neighbor-joining method with the Kimura 2-parameter model. Only the bootstrap values above 70% obtained from 1,000 replicates are given. The 76 different sequences of rpoB from small mammals of Nepal were classified into seven clusters. The GenBank accession numbers for reference sequences are given in parentheses. Only known reference Bartonella sequences without gaps and missing data are included in the phylogram. The rpoB gene sequence of Brucella melitensis 16MT was included as an out-group.
FIG. 2.
Phylogenetic classification of Bartonella genotypes found in small mammals of Nepal based on gltA gene sequences. The phylogram was constructed by the neighbor-joining method with the Kimura 2-parameter model. Only the bootstrap values above 70% obtained from 1,000 replicates are given. The 37 different sequences of gltA from small mammals of Nepal were classified into seven clusters. The GenBank accession numbers for reference sequences are given in parentheses. Only known reference Bartonella sequences without gaps and missing data are included in the phylogram. The gltA gene sequence of Brucella melitensis 16MT was included as an out-group.
Our study reports the first molecular detection and findings on the prevalence of Bartonella in small mammals of Nepal and is also the first study to compare different organ tissues from the same animal for Bartonella detection. Previously, several reports were made on the occurrence of bartonellae in rodents and other small mammals in Asian countries, including Bangladesh (3), China (28), Indonesia (27), Japan (16), Laos (1), Taiwan (19), and Thailand (2, 7, 24). Recently, Bai et al. (3) reported that 63.2% of B. bengalensis, 32.3% of R. rattus, and 42.9% S. murinus animals collected in Dhaka, Bangladesh, were infected with Bartonella, and the isolates from these animals were genetically related to Bartonella isolates found in America and Europe. Our study produced similar results in terms of the infectivity of the animals and the specificity of bartonellae. It is evident from all these studies that B. elizabethae, B. phoceensis, B. rattimassiliensis, B. rochalimae, and B. tribocorum are being circulated among small animals in Asia. Furthermore, B. coopersplainsensis and B. queenslandensis, found initially in Australia, were also detected in Nepal (this study) and Bangladesh (3), suggesting that the origin of these species might be Australasia.
The prevalence of Bartonella in Nepal was comparable with prevalence rates on other continents: in Africa, 44% in South Africa (22) and 24% in the Democratic Republic of Congo and Tanzania (V. A. K. B. Gundi et al., unpublished data); 42% in North America (18); and in Europe, 17% in France (12), 17% in central Sweden (14), 28% in southwestern Spain (20), 28% in Denmark (11), 31% in Greece (25), 31% in northeast Poland (26), 40% in Slovenia (17), and 64% in the United Kingdom (4). In a recent study, 29% prevalence was recorded in Australia (13). Such high rates of Bartonella prevalence are significant in the context of human health, because the synanthropic mammals harboring pathogenic microorganisms are often found in biotopes where they can come into close contact with humans, who might therefore be at some risk of exposure.
In this study, specificity of some bartonellae for host animals was observed (Table 3). The R. rattus brunneusculus rats carried almost all the rat-origin Bartonella species detected in this study, except B. rochalimae, which was isolated recently from R. norvegicus in Taiwan (19). In our study, S. murinus shrews harbored mostly B. queenslandensis. Studies by Bai et al. conducted in Dhaka, Bangladesh (3), also yielded evidence of the host specificity of B. queenslandensis for S. murinus. In addition, S. murinus carried some novel bartonellae in our study. On the other hand, S. murinus did not harbor other bartonellae, such as B. coopersplainsensis, B. phoceensis, and B. rattimassiliensis. These findings appear to suggest specificity of the bacteria for the host.
TABLE 3.
Bartonella coinfections in Nepalese small mammals
| Host species | No. of animals | District | Areaa | Cosurviving Bartonella combination | Genotype(s) for: |
|
|---|---|---|---|---|---|---|
| rpoB | gltA | |||||
| Bandicota bengalensis | 1 | Kathmandu | Kalimati | B. coopersplainsensis and B. queenslandensis | 35, 11 | |
| Rattus rattus brunneusculus | 1 | Lalitpur | NK | B. coopersplainsensis and B. phoceensis | 33, 27 | 20 |
| 1 | Lalitpur | NK | Novel Bartonella sp. and B. rattimassiliensis | 29, 30, 31 | 1, 2 | |
| 1 | Lalitpur | NK | B. coopersplainsensis and B. rattimassiliensis | 34 | 3 | |
| 1 | Lalitpur | NK | B. elizabethae and B. tribocorum | 24 | 9 | |
| Suncus murinus | 10 | Kathmandu | Hyumata | B. queenslandensis and novel Bartonella sp. | 13 | 14 |
| Lalitpur | Patan | 1 | 15, 17 | |||
| Kathmandu | Sabalbahal | 10 | 17 | |||
| Kathmandu | Sabalbahal | 13 | 14 | |||
| Kathmandu | NK | 15, 36 | 17 | |||
| Kathmandu | NK | 1 | 16 | |||
| Kathmandu | NK | 2 | 15 | |||
| Kathmandu | NK | 2 | 15 | |||
| Kathmandu | NK | 13 | 14 | |||
| Lalitpur | NK | 16 | 17 | |||
| 1 | Kathmandu | Sabalbahal | B. queenslandensis and B. rochalimae | 13 | 18 | |
| 1 | Lalitpur | NK | B. elizabethae and novel Bartonella sp. | 23, 36 | ||
| Total | 17 | |||||
NK, not known.
The prevalence and diversity of Bartonella species in animals or other hosts are controlled by geographical location, environmental conditions, the presence of vectors, and host species and habitats. In our study, 17 animals (15.7%) were infected with multiple Bartonella species and most of these animals had dual infections with different combinations of species (Table 4). This may be due to vector transmission between different animal hosts. According to Ellis and others, Bartonella species associated with hosts native to the Old World are phylogenetically distinct from those associated with hosts native to the New World (10). In this study, however, genotypes belonging to B. elizabethae, B. phoceensis, B. rattimassiliensis, and B. tribocorum were closely related to the isolates found in America and Europe (Tables 5 and 6), suggesting that carriers of these species might have migrated from Asia to other continents. Moreover, some novel genomic groups detected in this study were 98.3 to 100% identical to Bartonella strains found in Africa (Gundi et al., unpublished data) (Table 5). Furthermore, some genotypes, including novel genomic groups, from Nepalese rodents and shrews clustered well with other well-known rodent-associated species and human pathogens, such as B. coopersplainsensis, B. elizabethae, B. phoceensis, B. queenslandensis, B rattimassiliensis, B. rochalimae, and B. tribocorum (Fig. 1 and 2). These findings suggest that small mammals in Nepal, like those in other Asian countries, might be a potential source of Bartonella infections. Furthermore, these results suggest the need to conduct further studies to verify whether these agents might be responsible for human cases of febrile illness of unknown etiology and to determine the evolutionary, genetic, and pathogenic relationships among Nepal isolates and other isolates in Asia and on other continents.
TABLE 4.
Association between Bartonella genotypes and mammalian hosts
| Bartonella species | No. of animals associated with specific Bartonella organism(s) |
||||
|---|---|---|---|---|---|
| Bandicota bengalensis | Rattus rattus brunneusculus | Suncus murinus | Mus musculus castaneus | Total | |
| B. coopersplainsensis | 1 | 3 | 4 | ||
| B. elizabethae | 5 | 1 | 6 | ||
| B. phoceensis | 1 | 1 | |||
| B. rattimassiliensis | 3 | 3 | |||
| B. rochalimae | 1 | 1 | 2 | ||
| B. tribocorum | 1 | 4 | 2 | 7 | |
| B. queenslandensis | 1 | 2 | 47 | 50 | |
| Novel Bartonella sp. | 3 | 12 | 15 | ||
| Combination of species | 1 | 4 | 10 | 15 | |
| Total no. of associated animals/total no. of animals | 5/38 | 25/90 | 73/92 | 0/3 | 103/223 |
TABLE 5.
Genetic relationships between Bartonella species detected in Nepal and those from other geographical regions based on rpoB sequence analysis
| Bartonella sp. identified | Genotype(s) determined in this study | Host animal(s) in this studya | Accession no. of related GenBank sequence | % Sequence similarity | Host(s) of related sequence | Country of host(s) | Referenceb |
|---|---|---|---|---|---|---|---|
| B. coopersplainsensis | 32 | RRB | EU714973 | 100 | Rattus rattus | Laos | 1 |
| EU111792 | 100 | Rattus leucopus | Australia | 13 | |||
| B. phoceensis | 27 | RRB | AY515132 | 99.5 | Rattus norvegicus | France | 12 |
| AB290278 | 99.5 | Rattus rattus | Japan | 16 | |||
| Novel Bartonella sp. | 28 | RRB | FJ667575 | 98.8 | Unknown | Taiwan | D. D. Ji et al., unpublished data |
| B. queenslandensis | 1-19 | SM | EF204538 | 98.1-99.3 | Suncus murinus | Bangladesh | 3 |
| EF204539 | 97.2-100 | 3 | |||||
| AB290268 | 97.2-100 | Unknown | Japan | 16 | |||
| AB290269 | 97.1-99.9 | Unknown | |||||
| AB290270 | 97.6-98.8 | Rattus rattus | |||||
| 20 | RRB | AB290266 | 100 | Rattus rattus | Japan | 16 | |
| EU111787 | 98.4 | Melomys sp. | Australia | 13 | |||
| EU111788 | 99.2 | Rattus tunneyi | |||||
| EU111789 | 98.8 | Rattus fuscipes | |||||
| EU111790 | 99.5 | Rattus conatus | |||||
| EU111791 | 99.3 | Rattus leucopus | |||||
| Novel Bartonella sp. | 23 | SM | FJ851123 | 98.3 | Lophuromys rita | Democratic Republic of Congo | Gundi et al., unpublished |
| FJ851124 | 100 | Mus minutoides | |||||
| FJ851125 | 100 | Mus minutoides | |||||
| FJ851126 | 100 | Mus minutoides | |||||
| FJ851129 | 100 | Mus minutoides | |||||
| FJ851144 | 98.3 | Lophuromys sp. (Lophuromys dudui related) | Tanzania | Gundi et al., unpublished | |||
| FJ851146 | 98.2 | Lophuromys sp. (Lophuromys dudui related) | |||||
| B. tribocorum | 22 | BB, RRB | EU714974 | 99.9 | Rattus rattus, Mus cervicolor, Cannomys badius, Rattus exulans | Laos | 1 |
| EU714975 | 99 | Unknown | |||||
| B. elizabethae | 24 | RRB | AF165992 | 98.3 | Unknown | France | 23 |
| FJ851128 | 98.1 | Arvicanthis neumanni | Democratic Republic of Congo | Gundi et al., unpublished | |||
| B. rochalimae | 37 | BB | EU551156 | 99.6 | Rattus norvegicus | Taiwan | 19 |
BB, Bandicota bengalensis; RRB, Rattus rattus brunneusculus; SM, Suncus murinus.
The unpublished data cited are found in the GenBank database.
TABLE 6.
Genetic relationships between Bartonella species detected in Nepal and those from other geographical regions based on gltA sequence analysis
| Bartonella sp. identified | Genotype determined in this study | Host animal(s) in this studya | Accession no. of related GenBank sequence | % Sequence similarity | Host of related sequence | Country of host | Referenceb |
|---|---|---|---|---|---|---|---|
| B. rattimassiliensis | 1 | RRB | FJ179375 | 100 | Rattus norvegicus | Taiwan | 19 |
| AY515125 | 100 | Rattus norvegicus | France | 12 | |||
| 3 | RRB | AB290283 | 100 | Rattus rattus | Japan | 16 | |
| AF342933 | 100 | Rattus fuscipes | China | 28 | |||
| FJ464242 | 100 | Rattus sp. | China | D. Li et al., | |||
| FJ464244 | 100 | Rattus sp. | unpublished data | ||||
| FJ492787 | 100 | Rattus tanezumi | |||||
| FJ492792 | 100 | Rattus tanezumi | |||||
| FJ589047 | 100 | Rattus tanezumi | |||||
| B. elizabethae | 7 | RRB | AY589561 | 100 | Bandicota bengalensis | Bangladesh | 3 |
| 8 | RRB | AB445000 | 100 | Acomys cahirinus | Japan | 15 | |
| DQ884386 | 100 | Rattus norvegicus | China | Li et al., unpublished | |||
| DQ884390 | 100 | Rattus norvegicus | China | Li et al., unpublished | |||
| FJ655404 | 100 | Rattus exulans | Thailand | 2 | |||
| FJ946849 | 100 | Dog | Thailand | Y. Bai et al., unpublished data | |||
| U28072 | 100 | Unknown | Unknown | D. C. Jones et al., unpublished data | |||
| Z70009 | 100 | Human | United States | 9 | |||
| Z70020 | 100 | Rattus sp. | Peru | 5 | |||
| 6 | RRB | AF329679 | 100 | Rattus fuscipes | China | 28 | |
| FJ179377 | 100 | Taiwan | J. W. Hsieh et al., unpublished data | ||||
| FJ179391 | 100 | Taiwan | Hsieh et al., unpublished | ||||
| FJ492786 | 100 | Rattus tanezumi | China | Li and Liu, unpublished | |||
| FJ492791 | 100 | Rattus tanezumi | |||||
| FJ492793 | 100 | Rattus tanezumi | |||||
| FJ492797 | 100 | Rattus tanezumi | |||||
| FJ589045 | 100 | Rattus tanezumi | China | Li et al., unpublished | |||
| FJ589046 | 100 | Rattus tanezumi | |||||
| FJ589050 | 100 | Rattus tanezumi | |||||
| FJ589052 | 100 | Rattus tanezumi | |||||
| FJ589053 | 100 | Rattus tanezumi | |||||
| FJ589058 | 100 | Rattus tanezumi | |||||
| FJ589061 | 100 | Rattus tanezumi | |||||
| FJ589062 | 100 | Rattus tanezumi | |||||
| B. queenslandensis | 5 | RRB | AB290280 | 100 | Rattus rattus | Japan | 16 |
| AY589566 | 100 | Rattus rattus | Bangladesh | 3 | |||
| FJ179376 | 100 | Taiwan | 19 | ||||
| FJ179384 | 100 | ||||||
| FJ946846 | 100 | Dog | Thailand | Bai et al., unpublished | |||
| B. tribocorum | 13 | RRB, BB | AF086636 | 100 | Rattus rattus | Portugal | 10 |
| 10 | RRB, BB | AF075164 | 100 | Rattus norvegicus | United States | 10 | |
| AJ583111 | 100 | Unknown | South Africa | 22 | |||
| AY902183 | 100 | Rattus tanezumi | Indonesia | 27 | |||
| AY902188 | 100 | ||||||
| AY902189 | 100 | ||||||
| AY902191 | 100 | ||||||
| DQ884383 | 100 | Rattus norvegicus | China | Li et al., unpublished | |||
| DQ884387 | 100 | ||||||
| DQ884388 | 100 | ||||||
| DQ884389 | 100 | ||||||
| DQ884391 | 100 | ||||||
| DQ884392 | 100 | ||||||
| EF051466 | 100 | Rattus norvegicus | China | Li et al., unpublished | |||
| FJ464203 | 100 | Rattus norvegicus | China | Li et al., unpublished | |||
| FJ464205 | 100 | ||||||
| FJ464206 | 100 | ||||||
| FJ492782 | 100 | Rattus tanezumi | China | Li and Liu, unpublished | |||
| FJ492783 | 100 | Rattus tanezumi | |||||
| FJ492785 | 100 | Rattus tanezumi | |||||
| FJ492789 | 100 | Rattus tanezumi | |||||
| FJ492794 | 100 | Rattus tanezumi | |||||
| FJ492795 | 100 | Rattus tanezumi | |||||
| FJ492798 | 100 | Rattus tanezumi | |||||
| FJ492800 | 100 | Rattus tanezumi | |||||
| FJ589051 | 100 | Rattus tanezumi | China | Li et al., unpublished | |||
| FJ589057 | 100 | Rattus tanezumi | |||||
| B. rochalimae | 18 | SM | EU551154 | 100 | Rattus norvegicus | Taiwan | 19 |
BB, Bandicota bengalensis; RRB, Rattus rattus brunneusculus; SM, Suncus murinus.
The unpublished data cited are found in the GenBank database.
Nucleotide sequence accession numbers.
The DNA sequences obtained in this study were deposited in GenBank under accession numbers GU143433 to GU143508 (for rpoB) and GU143509 to GU143549 (for gltA) (see Tables S1 and S2 in the supplemental material).
Supplementary Material
Acknowledgments
This research was supported in part by the appointment of Vijay A. K. B. Gundi to the Emerging Infectious Diseases (EID) Fellowship Program, administered by the Association of Public Health Laboratories (APHL) and funded by the Centers for Disease and Control and Prevention (CDC).
We thank Ying Bai and Hidenori Kabeya for their help.
The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting true views of the Department of the Army or the Department of Defense.
Footnotes
Published ahead of print on 29 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Angelakis, E., K. Khamphoukeo, D. Grice, P. N. Newton, V. Roux, K. Aplin, D. Raoult, and J. M. Rolain. 2009. Molecular detection of Bartonella species in rodents from the Lao PDR. Clin. Microbiol. Infect. 15(Suppl. 2):95-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai, Y., M. Y. Kosoy, K. Lerdthusnee, L. F. Peruski, and J. H. Richardson. 2009. Prevalence and genetic heterogeneity of Bartonella strains cultured from rodents from 17 provinces in Thailand. Am. J. Trop. Med. Hyg. 81:811-816. [DOI] [PubMed] [Google Scholar]
- 3.Bai, Y., S. P. Montgomery, K. W. Sheff, M. A. Chowdhury, R. F. Breiman, H. Kabeya, and M. Y. Kosoy. 2007. Bartonella strains in small mammals from Dhaka, Bangladesh, related to Bartonella in America and Europe. Am. J. Trop. Med. Hyg. 77:567-570. [PubMed] [Google Scholar]
- 4.Birtles, R. J., S. M. Hazel, M. Bennett, K. Bown, D. Raoult, and M. Begon. 2001. Longitudinal monitoring of the dynamics of infections due to Bartonella species in UK woodland rodents. Epidemiol. Infect. 126:323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birtles, R. J., and D. Raoult. 1996. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int. J. Syst. Bacteriol. 46:891-897. [DOI] [PubMed] [Google Scholar]
- 6.Breitschwerdt, E. B., and D. L. Kordick. 2000. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin. Microbiol. Rev. 13:428-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castle, K. T., M. Kosoy, K. Lerdthusnee, L. Phelan, Y. Bai, K. L. Gage, W. Leepitakrat, T. Monkanna, N. Khlaimanee, K. Chandranoi, J. W. Jones, and R. E. Coleman. 2004. Prevalence and diversity of Bartonella in rodents of northern Thailand: a comparison with Bartonella in rodents from southern China. Am. J. Trop. Med. Hyg. 70:429-433. [PubMed] [Google Scholar]
- 8.Chomel, B. B., H. J. Boulouis, S. Maruyama, and E. B. Breitschwerdt. 2006. Bartonella spp. in pets and effect on human health. Emerg. Infect. Dis. 12:389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daly, J. S., M. G. Worthington, D. J. Brenner, C. W. Moss, D. G. Hollis, R. S. Weyant, A. G. Steigerwalt, R. E. Weaver, M. I. Daneshvar, and S. P. O'Connor. 1993. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J. Clin. Microbiol. 31:872-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis, B. A., R. L. Regnery, L. Beati, F. Bacellar, M. Rood, G. G. Glass, E. Marston, T. G. Ksiazek, D. Jones, and J. E. Childs. 1999. Rats of the genus Rattus are reservoir hosts for pathogenic Bartonella species: an Old World origin for a New World disease? J. Infect. Dis. 180:220-224. [DOI] [PubMed] [Google Scholar]
- 11.Engbaek, K., and P. A. Lawson. 2004. Identification of Bartonella species in rodents, shrews and cats in Denmark: detection of two B. henselae variants, one in cats and the other in the long-tailed field mouse. APMIS 112:336-341. [DOI] [PubMed] [Google Scholar]
- 12.Gundi, V. A., B. Davoust, A. Khamis, M. Boni, D. Raoult, and B. La Scola. 2004. Isolation of Bartonella rattimassiliensis sp. nov. and Bartonella phoceensis sp. nov. from European Rattus norvegicus. J. Clin. Microbiol. 42:3816-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gundi, V. A., C. Taylor, D. Raoult, and B. La Scola. 2009. Bartonella rattaustraliani sp. nov., Bartonella queenslandensis sp. nov. and Bartonella coopersplainsensis sp. nov., identified in Australian rats. Int. J. Syst. Evol. Microbiol. 59:2956-2961. [DOI] [PubMed] [Google Scholar]
- 14.Holmberg, M., J. N. Mills, S. McGill, G. Benjamin, and B. A. Ellis. 2003. Bartonella infection in sylvatic small mammals of central Sweden. Epidemiol. Infect. 130:149-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue, K., S. Maruyama, H. Kabeya, K. Hagiya, Y. Izumi, Y. Une, and Y. Yoshikawa. 2009. Exotic small mammals as potential reservoirs of zoonotic Bartonella spp. Emerg. Infect. Dis. 15:526-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue, K., S. Maruyama, H. Kabeya, N. Yamada, N. Ohashi, Y. Sato, M. Yukawa, T. Masuzawa, F. Kawamori, T. Kadosaka, N. Takada, H. Fujita, and H. Kawabata. 2008. Prevalence and genetic diversity of Bartonella species isolated from wild rodents in Japan. Appl. Environ. Microbiol. 74:5086-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knap, N., D. Duh, R. Birtles, T. Trilar, M. Petrovec, and T. Avsic-Zupanc. 2007. Molecular detection of Bartonella species infecting rodents in Slovenia. FEMS Immunol. Med. Microbiol. 50:45-50. [DOI] [PubMed] [Google Scholar]
- 18.Kosoy, M. Y., R. L. Regnery, T. Tzianabos, E. L. Marston, D. C. Jones, D. Green, G. O. Maupin, J. G. Olson, and J. E. Childs. 1997. Distribution, diversity, and host specificity of Bartonella in rodents from the southeastern United States. Am. J. Trop. Med. Hyg. 57:578-588. [DOI] [PubMed] [Google Scholar]
- 19.Lin, J. W., C. Y. Chen, W. C. Chen, B. B. Chomel, and C. C. Chang. 2008. Isolation of Bartonella species from rodents in Taiwan including a strain closely related to ‘Bartonella rochalimae’ from Rattus norvegicus. J. Med. Microbiol. 57:1496-1501. [DOI] [PubMed] [Google Scholar]
- 20.Marquez, F. J., J. J. Rodriguez-Liebana, M. E. Pachon-Ibanez, F. Docobo-Perez, A. Hidalgo-Fontiveros, M. Bernabeu-Wittel, M. A. Muniain, and J. Pachon. 2008. Molecular screening of Bartonella species in rodents from south western Spain. Vector Borne Zoonotic Dis. 8:695-700. [DOI] [PubMed] [Google Scholar]
- 21.Mogollon-Pasapera, E., L. Otvos, Jr., A. Giordano, and M. Cassone. 2009. Bartonella: emerging pathogen or emerging awareness? Int. J. Infect. Dis. 13:3-8. [DOI] [PubMed] [Google Scholar]
- 22.Pretorius, A. M., L. Beati, and R. J. Birtles. 2004. Diversity of bartonellae associated with small mammals inhabiting Free State province, South Africa. Int. J. Syst. Evol. Microbiol. 54:1959-1967. [DOI] [PubMed] [Google Scholar]
- 23.Renesto, P., J. Gouvernet, M. Drancourt, V. Roux, and D. Raoult. 2001. Use of rpoB gene analysis for detection and identification of Bartonella species. J. Clin. Microbiol. 39:430-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saisongkorh, W., W. Wootta, P. Sawanpanyalert, D. Raoult, and J. M. Rolain. 2009. “Candidatus Bartonella thailandensis”: a new genotype of Bartonella identified from rodents. Vet. Microbiol. 139:197-201. [DOI] [PubMed] [Google Scholar]
- 25.Tea, A., S. Alexiou-Daniel, A. Papoutsi, A. Papa, and A. Antoniadis. 2004. Bartonella species isolated from rodents, Greece. Emerg. Infect. Dis. 10:963-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welc-Faleciak, R., A. Paziewska, A. Bajer, J. M. Behnke, and E. Sinski. 2008. Bartonella spp. infection in rodents from different habitats in the Mazury Lake District, Northeast Poland. Vector Borne Zoonotic Dis. 8:467-474. [DOI] [PubMed] [Google Scholar]
- 27.Winoto, I. L., H. Goethert, I. N. Ibrahim, I. Yuniherlina, C. Stoops, I. Susanti, W. Kania, J. D. Maguire, M. J. Bangs, S. R. Telford III, and C. Wongsrichanalai. 2005. Bartonella species in rodents and shrews in the greater Jakarta area. Southeast Asian J. Trop. Med. Public Health 36:1523-1529. [PubMed] [Google Scholar]
- 28.Ying, B., M. Y. Kosoy, G. O. Maupin, K. R. Tsuchiya, and K. L. Gage. 2002. Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am. J. Trop. Med. Hyg. 66:622-627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.