Abstract
Plant and protozoan microtubules are selectively sensitive to dinitroanilines, which do not disrupt vertebrate or fungal microtubules. Tetrahymena thermophila is an abundant source of dinitroaniline-sensitive tubulin, and we have modified the single T. thermophila α-tubulin gene to create strains that solely express mutant α-tubulin in functional dimers. Previous research identified multiple α-tubulin mutations that confer dinitroaniline resistance in the human parasite Toxoplasma gondii, and when two of these mutations (L136F and I252L) were introduced into T. thermophila, they conferred resistance in these free-living ciliates. Purified tubulin heterodimers composed of L136F or I252L α-tubulin display decreased affinity for the dinitroaniline oryzalin relative to wild-type T. thermophila tubulin. Moreover, the L136F substitution dramatically reduces the critical concentration for microtubule assembly relative to the properties of wild-type T. thermophila tubulin. Our data provide additional support for the proposed dinitroaniline binding site on α-tubulin and validate the use of T. thermophila for expression of genetically homogeneous populations of mutant tubulins for biochemical characterization.
An extraordinary number of small molecules target the eukaryotic α-β tubulin dimer. Compounds that shift the normal equilibrium between free dimers and polymers to destabilize or stabilize microtubules are exploited for diverse applications, ranging from cancer chemotherapy to treatment of helminth infections (31, 32). Although many compounds interact with almost all tubulin isotypes, some small molecules are selectively active against phylogenetically restricted subsets of tubulins. For example, helminth and fungal tubulins are selectively sensitive to several benzimidazoles (benomyl, albendazole, and mebendazole) that require the presence of “susceptible” amino acids (E198 and F200) in β-tubulin (33, 36, 37). Dinitroanilines represent another group of selective small molecules. They are synthetic compounds that inhibit microtubules in plants and protozoa but are inactive against the microtubules of vertebrates and fungi (reviewed in references 50, 53, and 70). These molecules (e.g., oryzalin and trifluralin) have been used in commercial herbicide formulations for over 40 years (53). Dinitroaniline binding studies using plant, protozoan, and vertebrate tubulins established that only sensitive tubulins bind dinitroanilines (9, 28, 49, 72). The ability of dinitroanilines to selectively disrupt the microtubules of protozoan parasites without affecting vertebrate microtubules suggests the exciting possibility that we may be able to develop novel antiparasitic agents by understanding the mechanism of action of these compounds on sensitive tubulins.
Resistance to microtubule-disrupting or -stabilizing drugs is often associated with point mutations to α- or β-tubulin that alter polymerization or binding site properties of tubulin heterodimers. Genetic studies of a wide variety of dinitroaniline-sensitive organisms have identified mutations to α-tubulin associated with development of resistance. Studies using the unicellular green alga Chlamydomonas reinhardtii identified the Y24H mutation, and work with the higher land plants Eleusine indica (goosegrass) and Setaria viridis (green foxtail) identified the mutations T239I and M268T (goosegrass) and L136F and T239I (foxtail) (6, 11, 30, 79). Research from our group using the apicomplexan parasite Toxoplasma gondii identified 35 unique α-tubulin point mutations that confer oryzalin resistance (45, 52). The T. gondii mutations include the substitutions L136F and T239I, akin to the plant mutations. We were able to convert sensitive parasites into resistant lines by homologous integration of α-tubulin transgenes bearing individual mutations identified in our screen. Moreover, we were able to confer resistance with α-tubulin transgenes bearing the M268T or F24H (equivalent to Y24H) mutations, which we did not identify in our resistance screen but were associated with dinitroaniline resistance in other organisms (45). This suggests that resistance mechanisms are conserved in plants and protozoa and that there is most likely a common dinitroaniline binding site and mechanism of action.
Computational studies support a model in which the dinitroaniline binding site is located in the α-tubulin subunit beneath the H1-S2 loop, and compound binding disrupts protofilament contacts in the microtubule lattice (48, 52). Specifically, studies using flexible dinitroaniline docking to Toxoplasma, Leishmania, Plasmodium, and bovine tubulin structures have identified a consistent binding site on protozoan but not vertebrate α-tubulin. Molecular dynamics simulations suggest that dinitroaniline binding profoundly limits flexibility of the α-tubulin H1-S2 loop, which is drawn toward the core of the protozoan tubulin dimer (48). Based on docking studies and the location of the mutations on the α-tubulin subunit, we developed a model for how these mutations confer dinitroaniline resistance. This model predicts that (i) mutations within or near the binding site reduce tubulin dimer-dinitroaniline ligand affinity and (ii) mutations to the M or H1-S2 loops or to the GTPase-activating domain increase protofilament affinity within the microtubule lattice to compensate for the destabilizing effect of dinitroanilines (53).
In order to test our model of dinitroaniline resistance mechanisms, we required purified tubulin so that we could measure assembly properties and the dinitroaniline avidities of dimers bearing resistance mutations. Purification of biochemical quantities of tubulin from T. gondii is constrained by the fact that it is an obligate intracellular parasite with a minimal (but critically important) microtubule cytoskeleton. Traditionally, researchers have exploited Escherichia coli expression to produce proteins for biochemical and structural analysis. Tubulin folding requires the TCP-1 chaperones, which are specific to eukaryotes, and to date no one has expressed polymerization-competent α-β dimers in bacteria (67, 77). Although some researchers have used bacterially expressed tubulin monomers for drug binding studies, we strongly believe that such studies must be carried out on polymerization-competent α-β dimers. Simple eukaryotic organisms, such as Saccharomyces cerevisiae and Tetrahymena thermophila, have reduced-copy-number, genetically tractable tubulin genes and provide the opportunity to assess the effects of amino acid changes in the context of genetically homogeneous tubulin (8, 17, 23, 24, 76).
T. thermophila is a free-living ciliate that is classified as an alveolate, along with the apicomplexans and dinoflagellates (13, 38). It has a single α-tubulin gene and two identical β-tubulin genes (19, 47) that encode tubulin, which is incorporated into diverse microtubule populations. The primary source of heterogeneity in these ciliates arises from posttranslational modifications (PTMs) to α- and β-tubulins (16, 68). Since T. thermophila is amenable to bulk culture, is genetically tractable, and is related to T. gondii, we chose to exploit it to study the effects of dinitroaniline resistance mutations on the properties of protozoan tubulin.
In this study we expressed α-tubulins bearing the L136F or I252L mutation in T. thermophila as the sole source of α-tubulin. These mutations correspond to the L136F and V252L mutations in T. gondii. The L136F mutation is located in the computationally determined binding site for dinitroanilines, and we predicted that it would reduce affinity for oryzalin. The V252L mutation is located in the GTPase activating domain. It should be noted that the lethal mutation D252A in budding yeast does not correspond to this position, since S. cerevisiae has an earlier single amino acid insertion; I/V252 corresponds to yeast tubulin L253, which is located between the essential GAP residues D252 and E255 (2, 60). We hypothesized that the V252L mutation decreases activation of the β-tubulin GTPase, leading to slowed hydrolysis of GTP and consequently a larger GTP cap. However, the data presented here indicate that both the L136F and I252L point mutations alter the properties of tubulin in unanticipated ways to modify dinitroaniline affinity and its critical concentration (Cc).
MATERIALS AND METHODS
Growth curves.
T. thermophila at a starting density of 3 × 105 cells/ml was grown in 250-ml flasks with 50 ml of SPP growth medium (1% protease peptone, 0.1% yeast extract, 0.2% glucose, 0.003% EDTA-ferric sodium salt, 100 μg/ml streptomycin, 100 U/ml penicillin, 0.25 μg/ml amphotericin B [Fungizone]) and oryzalin at 0, 7.5, and 15 μM. The culture turbidity was measured in a spectrophotometer (Bio-Rad) as the optical density at 600 nm (OD600) every 4 h for 60 h.
T. thermophila tubulin mutagenesis.
To introduce mutations into Atu1p α-tubulin, we used a derivative of the pTUB100E3 plasmid (26), pTub100E3/α-His6 (J. Gaertig, unpublished data). The QuikChange site-directed mutagenesis kit (Stratagene) was used to introduce point mutations using the primers GGTCTCCAAGGTTTCTTTGTCTTCAACTCCGTCGGTGG and its reverse complement for L136F mutagenesis and GGTGCCCTTAACGTCGATCTCACTGAATTCTAAACTAACTTGG and its reverse complement for V252L mutagenesis. The constructs were verified by sequencing.
Biolistic transformation of T. thermophila.
For each sample, ∼2 μg of purified plasmid (Qiagen Maxiprep kit) was linearized by digestion with HindIII and resuspended in TE (10 mM Tris-HCl [pH 7.5], 1 mM EDTA) after phenol-chloroform precipitation. T. thermophila heterokaryon strains AAKO2.7 and AAKO5.5 were used for mating, because both have the ATU1 α-tubulin gene deleted from their germ line genomes (25, 26). To cross the strains, 100-ml cultures of each strain were starved in 10 mM Tris-HCl (pH 7.5) for 16 to 24 h at 30°C without shaking. The cells were suspended in 10 mM Tris-HCl (pH 7.5) to 1 × 107 cells/ml and were spread on moist filter paper prior to bombardment at 900 lb/in2 using the DuPont Biolistic PDS-1000/He particle delivery system (Bio-Rad). T. thermophila was bombarded with 1.0-μm gold particles coated with 2 μg of digested plasmid. Immediately following bombardment, cells were suspended in 50 ml of SPP medium, incubated at 30°C for 0.5 to 2 h, plated into 96-well plates, and selected in SPP supplemented with 80 mg/ml paromomycin sulfate (to kill cells with parental heterokaryon genomes). The surviving T. thermophila pTub100E3/α-His6 transformants were confirmed by sequencing of the amplified ATU1 gene coding region.
Immunofluorescence of T. thermophila tubulin.
T. thermophila cultures at ∼2 × 105 cells/ml were harvested by centrifugation at 225 × g for 3 min. The cells were washed in 10 ml of 10 mM Tris (pH 7.4) and spun for 3 min at 225 × g. The pellet was resuspended in ∼0.5 ml of buffer and fixed in 4.5 ml of ice-cold fixative consisting of 50% ethanol, 0.1% Triton X-100 in PHEM [10 mM EGTA, 2 mM MgCl2, 60 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)] (73). Cells were incubated at room temperature (RT) for 30 min. The cells were washed once in 3 ml of phosphate-buffered saline (PBS) for 5 min and blocked with 5 ml of blocking buffer (PBS, 0.1% Tween 20, 3% bovine serum albumin) for 30 min at RT. Cells were incubated with a 1:1,000 dilution of the primary antibody in blocking buffer (anti-α-tubulin; 1-5-2; Sigma) for 1 h at RT and then rinsed with PBS for 5 min and incubated in a 1:1,000 dilution of goat anti-mouse Alexa Fluor 488 (Invitrogen) in blocking buffer (RT for 1 h). Cells were rinsed three times with PBS for 5 min each, with the first wash containing Hoechst stain at a 1:1,000 dilution, and mounted in a polyvinyl alcohol-based medium as previously described (51).
Quantification of tubulin.
To quantify tubulin in T. thermophila, lysates were loaded onto gels at a concentration of 19,500 cells per lane along with standards consisting of known amounts of purified bovine tubulin (Cytoskeleton, Inc.). The samples were resolved by SDS-PAGE on a 12.5% gel (Bio-Rad) and blotted to nitrocellulose for immunoblot analysis. Blots were probed with a mouse anti-α-tubulin antibody (1-5-2; Sigma) and detected with an Alexa Fluor 488-conjugated goat anti-mouse secondary antibody (Invitrogen). Samples were imaged with a Typhoon Trio+ (GE Healthcare) and quantified with ImageQuant software (GE Healthcare).
Purification of T. thermophila tubulin.
The protocol for purification of T. thermophila tubulin was adapted from the existing Leishmania tarentolae tubulin isolation protocol (78). T. thermophila (∼6 × 108 cells, from 6 liters of culture) was collected by centrifugation at 3,750 × g, resuspended in 40 ml PME + P buffer (PME [0.1 M PIPES {pH 6.9}, 1 mM MgCl2, 1 mM EGTA] plus protease inhibitors [1 mM benzamidine, 0.5 mM phenylmethylsulfonyl fluoride, and 25 μg/ml leupeptin]) on ice and sonicated (Bio-Rad) with 10 30-s bursts at 25 W with a 2-min cooling interval between each burst (61). The sonicated suspensions were cooled on ice for 30 min and then centrifuged at 17,000 × g for 40 min at 4°C. The resulting supernatant was filtered through glass wool, and a peristaltic pump was used to load a 10-ml column of a previously equilibrated mixture (two volumes of PME + P buffer and DEAE-Sepharose Fast Flow matrix [GE Healthcare]) at a rate of 2.5 ml/minute. The column was washed with two column volumes of PME + P buffer followed by four column volumes of PME + P containing 0.1 M KCl and 0.25 M glutamate (pH 6.9). Tubulin was eluted with two column volumes of PME + P buffer containing 0.3 M KCl and 0.75 M glutamate (pH 6.9). Fractions (2.5 ml) were collected after the start of the 0.3 M KCl-0.75 M glutamate elution. Fractions containing tubulin were identified by SDS-PAGE and pooled, and GTP, dimethyl sulfoxide (DMSO), and additional MgCl2 were added to final concentrations of 10 mM MgCl2, 8% (vol/vol) DMSO, and 2 mM GTP. The tubulin solution was then incubated at 37°C for 30 min for assembly and then spun at 50,000 × g at 30°C for 30 min. The supernatant was removed following centrifugation, and the remaining pellet consisting of microtubules was rinsed once with warm PME (∼37°C) and resuspended in ∼1.5 ml cold PME. The pellet was further solubilized via probe sonication (30 ∼5-s bursts at 10 W) by using a Fischer Scientific sonicator. After the tubulin-rich solution was incubated on ice for 30 min, it was spun at 50,000 × g at 4°C for 30 min. The supernatant containing heterodimeric tubulin was stored at −80°C in 100-μl aliquots. We assessed the quality of tubulin from each preparation by SDS-PAGE. As assessed by Coomassie blue staining, our preparations were consistently free of detectable levels of contaminating proteins.
Tryptophan-quenching dinitroaniline binding assay and calculation of Kd values.
Quenching of intrinsic tubulin fluorescence by oryzalin was based on an established method (71, 72). Tubulin samples (0.3 or 1 μM) were mixed with various concentrations of oryzalin as indicated in the figures (0 to 80 μM). Tubulin tryptophan fluorescence was excited at 290 nm, and emission was recorded every 5 nm from 310 to 340 nm. Intensity at 325 nm was taken for calculation of quenching, following correction for a (minor) inner filter effect based on a control experiment with the same drug additions to a solution of 10 μM N-acetyl-tryptophanamide. Fractional quenching of fluorescence was fitted to a single-site binding model using the Prism software from GraphPad.
Critical concentration assays.
To determine the Cc values for the different tubulins, we carried out assembly assays for each tubulin sample. Tubulin was assembled for 30 min under the assembly conditions described above, in a 100-μl reaction volume. Assembled tubulin was pelleted for 20 min in an airfuge (Beckman) at maximal speed. A 30-μl aliquot from the middle of the tube was removed to measure the free tubulin concentration by bicinchoninic acid (BCA) assay. After the remaining supernatant was removed, the pellet was suspended in 100 μl ice-cold 1× PME and quantified by BCA assay. Polymerized tubulin in the pellet was plotted as a function of the total tubulin in the reaction mixture. Extrapolation of this plot to zero polymer gave the maximum tubulin concentration in the absence of polymer (i.e., the Cc).
Tubulin assembly assays.
Tubulin assembly assays were based on an established method (7). Reactions were carried out in 96-well half-area microplates (Costar) in a final volume of 50 μl, and reaction mixtures contained final concentrations of 7 μM tubulin (0.7 mg/ml), 0.1 mM PIPES (pH 6.9), 1 mM EGTA, 1 mM MgCl2, 10% (vol/vol) DMSO, and 1 mM GTP, with oryzalin concentrations of 0 or 5 μM. The tubulin solution containing PIPES, EGTA, MgCl2, and oryzalin was added to the microplate on ice and incubated for 10 min. Assembly was initiated by the addition of 10 μl of 5 mM GTP in 25% DMSO, with absorbance values read by using a microplate reader (SpectraMax Plus) at 351 nm at 30°C. Electron microscopy samples were adsorbed onto Formvar/carbon-coated grids and negatively stained with 1% aqueous uranyl acetate.
RESULTS
T. thermophila has abundant, dinitroaniline-sensitive tubulin.
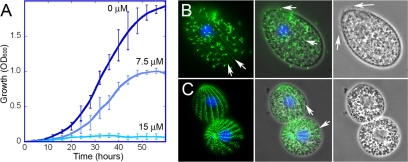
The free-living ciliate T. thermophila is an excellent source of abundant dinitroaniline-sensitive tubulin, and we investigated it as a system for expressing functional mutant tubulins for biochemical characterization. Previous work from our group had defined a large number of point mutations in α1-tubulin that confer dinitroaniline resistance in T. gondii. Since T. gondii is an obligate intracellular parasite, it is not a feasible source of material for biochemical studies. However, T. thermophila and T. gondii α-tubulins share 95% identity, which corresponds to 22 amino acid differences between T. thermophila Atu1p and T. gondii α1-tubulin, 12 of which are conservative substitutions (see Fig. S1 in the supplemental material). Wild-type T. thermophila, including the wild-type strain for these studies, is sensitive to dinitroanilines such as oryzalin (18). The wild-type (control) strain used here exclusively expresses a wild-type T. thermophila α-tubulin sequence with a carboxy-terminal His6 tag, and growth of this strain is inhibited by increasing concentrations of oryzalin (Fig. 1A). When T. thermophila from cultures grown in the presence of oryzalin were labeled with an antitubulin antibody, the abundant ciliary axonemes that typically cover the cell surface (Fig. 1B) were lost (Fig. 1C). Interestingly, some other highly stable tubulin structures, such as the cortical microtubules, were largely retained. Oryzalin-treated T. thermophila cells also appear smaller, perhaps as a function of reduced nutrient uptake due to a reduction or disruption of oral cilia.
Fig. 1.
Wild-type T. thermophila cells are dinitroaniline sensitive. (A) Growth of wild-type T. thermophila at 30°C in medium with 0, 7.5, or 15 μM oryzalin. Wild-type T. thermophila cells showed reduced growth at 7.5 μM and complete inhibition at 15 μM. (B and C) Microscopy of T. thermophila cells grown in the absence (B) or presence (C) of 7.5 μM oryzalin indicated that many microtubule structures are disrupted by dinitroaniline treatment. T. thermophila cells were labeled with antitubulin antibody (green) and 4′,6-diamidino-2-phenylindole (DAPI, which labels the nuclei blue). The left panels show merged blue and green fluorescence, the middle panels show the fluorescence image merged with a phase-contrast image, and the right panels show a phase-contrast image alone. (B, left) Normal T. thermophila cell covered with tubulin-containing cilia, but also with underlying microtubules, such as the cortical microtubules (arrows). (B, middle and right) T. thermophila cilia are visible by both tubulin immunofluorescence and phase imaging (arrows). (C, middle) Oryzalin-treated T. thermophila cells lose most or all cilia, making the underlying cortical microtubules (arrows) more visible.
Mutant T. thermophila α-tubulin genes complement the ATU1 deletion.
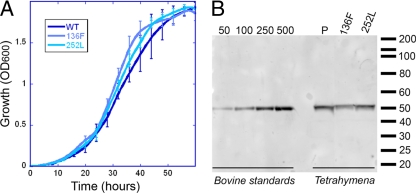
The heterokaryon strains AAKO2.7 and AAKO5.5 each contain a deletion of the single α-tubulin gene ATU1 in the “germ line” micronucleus (25, 26). Essential expression of Atu1p α-tubulin is provided by an intact copy of ATU1 in the “somatic” macronucleus. Mating the AAKO2.7 and AAKO5.5 strains triggers destruction of their macronuclei, and reconstituted macronuclei (derived from zygotic micronuclei) lack the ATU1 gene. In the absence of a complementing gene supplied in trans, loss of α-tubulin leads to cell death. After mating the ATU1 knockout heterokaryons, we attempted to rescue lethality by using T. thermophila Atu1p α-tubulin transgenes containing H28Q, L136F, R243S, T239I, or I252L mutations. These point mutations correspond to substitutions that confer dinitroaniline resistance in T. gondii. We recovered viable strains expressing the L136F or I252L mutations as the sole source of α-tubulin in T. thermophila. Curiously, these residues are located within 5 Å of each other within the tubulin dimer, although only L136 is a proposed binding site residue. The mutant strains showed comparable growth rates to the wild-type strain in the absence of oryzalin (Fig. 2A) and expressed nearly identical levels of tubulin per cell (30.2 ± 1.0 pg of protein; mean ± standard deviation [SD]) (Fig. 2B). Although we did not recover strains with the H28Q, T239I, or R243S mutations, the complementation experiments were only performed once. We do not have any evidence to suggest that tubulin bearing any of these mutations would not be functional in Tetrahymena. In fact, since the mutations were originally identified in the sole copy of an essential α-tubulin gene in Toxoplasma, they most likely represent viable substitutions in tubulin.
Fig. 2.
Complemented T. thermophila strains show normal growth and express similar levels of tubulin. (A) T. thermophila wild-type (WT) and mutant strains grow with similar kinetics at 30°C in the absence of oryzalin. (B) Immunoblot analysis of cell lysates equivalent to ∼9,750 cells per well indicated that the three T. thermophila strains (the wild-type complemented strain and strains expressing tubulin with the L136F or V252L point mutation) express essentially equivalent concentrations of tubulin per cell (30 pg). This blot was probed with the anti-α-tubulin monoclonal antibody 1-5-2.
Mutant α-tubulins alter T. thermophila growth in oryzalin.
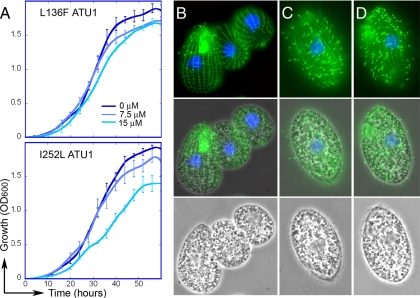
T. thermophila strains that express wild-type α-tubulin exhibited growth inhibition in 7.5 to 15 μM oryzalin (Fig. 1A). The L136F and I252L strains were less sensitive to oryzalin, as illustrated by their continued growth in increasing concentrations of this compound. The L136F strain was essentially insensitive to 15 μM oryzalin (Fig. 3A, top). Growth of the I252L strain was somewhat attenuated in 15 μM oryzalin, as the slope of log-phase growth was reduced and the cells appeared to reach stationary phase at a lower cell density (Fig. 3A, bottom). Unfortunately, due to the hydrophobicity of oryzalin and other dinitroanilines, we could not reliably assay the growth of these T. thermophila strains with higher compound concentrations, as it was apparent that the dinitroanilines precipitate out of SPP growth medium when present above ∼20 μM. When T. thermophila grown in 7.5 μM oryzalin was examined by immunofluorescence, cells expressing wild-type tubulin lost most cilia and exhibited aberrant morphology (Fig. 3B). These defects included increased numbers of cells with two micronuclei, shrunken macronuclei, a rounded cell shape, a smaller size, or loss of cilia from the somatic cortex and oral cavity. When T. thermophila cells expressing L136F and I252L mutations in α-tubulin were stained after growth in 7.5 μM oryzalin, the cell population was composed of healthy-looking, growing, and ciliated cells (Fig. 3D and E).
Fig. 3.
(A) Growth of complemented strains in 0, 7.5, and 15 μM oryzalin. (A) T. thermophila cells expressing the L136F ATU1 transgene (top panel) and T. thermophila cells expressing the I252L ATU1 transgene (bottom panel). (B to D) Immunofluorescence of T. thermophila cells cultured in oryzalin. Wild-type T. thermophila cells grown in 7.5 μM oryzalin lose most or all cilia but retain underlying cortical microtubules (B), whereas T. thermophila cells expressing the L136F (C) or I252L (D) tubulin mutations retain cilia.
Purification of wild-type and mutant tubulins.
Tetrahymena strains expressing wild-type, L136F, and I252L tubulins were grown in bulk culture (∼6 liters) for purification of tubulin. After optimizing protocols which exploit DEAE chromatography and cycles of induced polymerization and depolymerization to isolate functional tubulin dimers (61), we obtained highly purified samples that were free of any discernible high-molecular-weight microtubule-associated proteins (MAPs) or other contaminating proteins (Fig. 4A). These samples exhibited GTP-dependent assembly, which could be observed as increased light scattering (increased optical density over time at 350 nm). In order to rule out denatured protein aggregation as a source of increased light scattering, we verified that substitution of GDP for GTP prevented a change in optical density under otherwise-identical assembly conditions (Fig. 4B). Electron microscopy of negatively stained samples showed that the tubulins form microtubules and a few other polymeric forms in the presence of GTP (Fig. 4C). These samples were used to investigate differences between wild-type tubulin and L136F or I252L mutant tubulins.
Fig. 4.
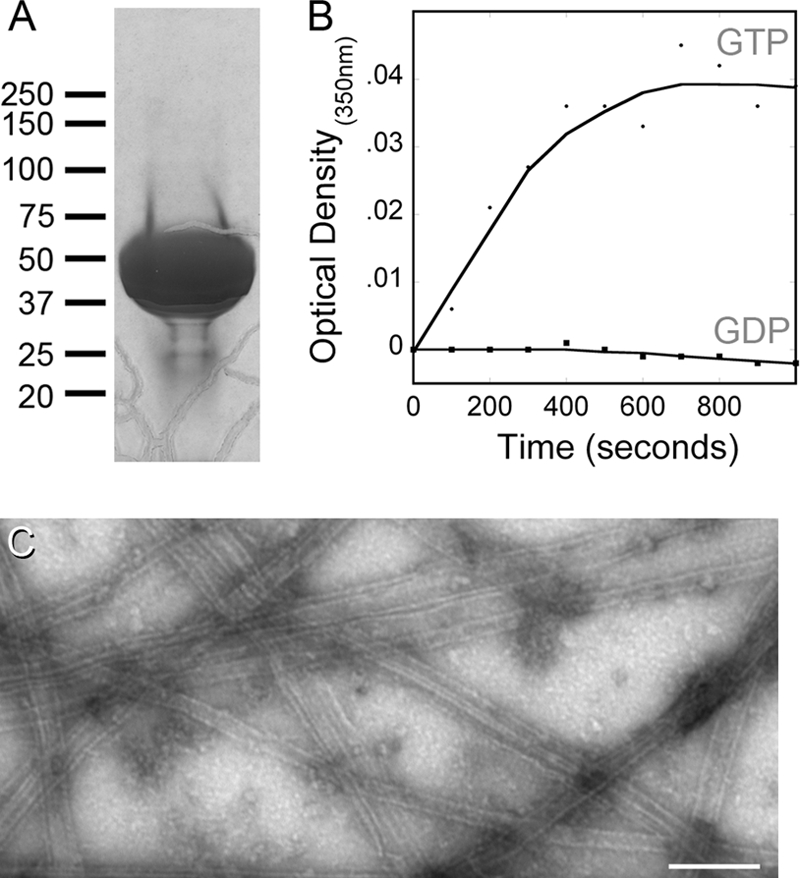
Purified tubulin is free of discernible MAPs and shows GTP-dependent assembly. (A) An overloaded Coomassie blue-stained SDS-PAGE gel illustrates the purity of DEAE-purified, cycled L136F tubulin used for biochemistry experiments; the absence of contaminating proteins (particularly any high-molecular-weight microtubule-associated proteins) is typical of all our protein purifications. (B) Assembly of I252L tubulin (as well as the wild-type and L136F samples [data not shown]) is dependent upon the presence of GTP (circles). There was no change in optical density when the assembly reaction mixture contained GDP (squares), indicating that the light scattering reflects assembly rather than aggregation of denatured protein. (C) Electron microscopy of negatively stained samples demonstrated that tubulins polymerize to microtubules and other polymeric forms in the presence of GTP. The wild-type tubulin sample is shown, but all tubulins assembled into similar polymers in this assay. Bar, 200 μm.
T. thermophila α-tubulin mutants have reduced oryzalin affinity.
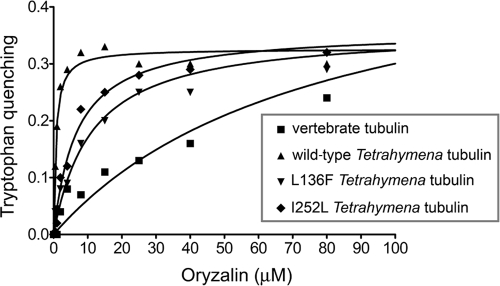
The Tetrahymena tubulin dimer has three tryptophans within the α-subunit polypeptide and four tryptophans within the β-subunit polypeptide. These confer tubulin in solution with inherent fluorescence emission at 325 nm. In cases where small-molecule ligands act as fluorescence resonance acceptors, tryptophan quenching at 325 nm is a straightforward method to measure compound binding to a protein (12, 71, 72). Oryzalin binding brings this small-molecule ligand into sufficiently close proximity to resonance quench tryptophan emission, and it therefore reduces the absolute fluorescence from the sample in proportion to oryzalin binding. While a vertebrate tubulin control exhibited minimal quenching at high concentrations of oryzalin, wild-type tubulin purified from T. thermophila showed substantial tryptophan quenching at 1 to 2 μM oryzalin, indicating avid binding. The L136F and I252L T. thermophila tubulins have dramatically reduced quenching in oryzalin. Tryptophan quenching data can be used to calculate Kd values for the interaction of tubulin with oryzalin (Fig. 5 and Table 1). It should be noted that it is possible to measure quenching at 80 μM oryzalin because its solubility is higher in the PME buffer used for this assay than in Tetrahymena SPP growth medium. The maximal quenching for all T. thermophila tubulin samples was remarkably similar, indicating that oryzalin bound in the same location to these three samples. Vertebrate tubulin has a much lower affinity for oryzalin, as reflected in a much higher Kd, and it also has a different maximal fluorescence quenching from what was observed with T. thermophila tubulin. These data indicate that wild-type T. thermophila tubulin has a Kd of 0.44 μM for oryzalin, in contrast to vertebrate tubulin, which has a Kd of 77 μM for oryzalin. Both L136F and I252L mutant tubulins have reduced oryzalin affinities: L136F tubulin has a Kd of 11 μM and I252L tubulin has a Kd of 6.7 μM.
Fig. 5.
Binding curves for association of tubulin with oryzalin. Oryzalin binding was modeled using a one-site binding, nonlinear fit. Wild-type T. thermophila tubulin, L136F T. thermophila tubulin, and I252L T. thermophila tubulin all have similar maximal quench values, indicating that oryzalin interacts in the same fashion with each of the three tubulins. The binding curve and maximal quenching are different for vertebrate tubulin. These data indicate that the Kd values for oryzalin are 77 μM for vertebrate tubulin and 0.44 μM for wild-type T. thermophila tubulin, which is reduced to 11 μM for L136F T. thermophila tubulin and 6.7 μM for I252L T. thermophila tubulin.
Table 1.
Biochemical properties of T. thermophila tubulins
| α-Tubulin type | Assembly in oryzalin | Oryzalin Kda (μM) | Critical concnb (μM) |
|---|---|---|---|
| Wild type | Sensitive | 0.44 ± 0.1 | 5.5 ± 0.9 |
| L136F | Resistant | 11.0 ± 2 | 2.5 ± 0.7 |
| I252L | Resistant | 6.7 ± 1 | 5.6 ± 0.6 |
The standard error associated with a specific Kd value was generated within the analysis program and reflects an estimate of the fit for the data set (each of which was generated from three different samples).
Variations reported for the critical concentrations reflect the standard errors of the means from three experiments.
L136F tubulin has a decreased critical concentration.
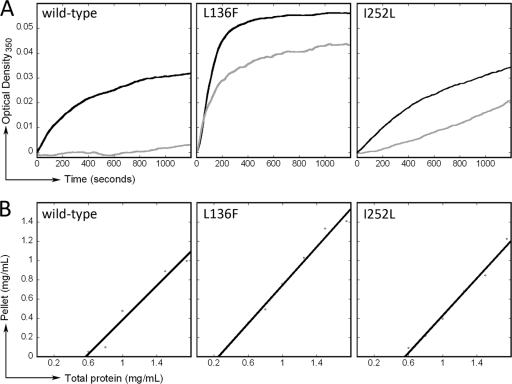
As tubulin heterodimers in solution assemble into microtubules the turbidity increases, which can be observed as a change in the optical density at 351 nm. Samples at a concentration of 7 μM tubulin were assayed for assembly in the presence or absence of 5 μM oryzalin. In this assay, microtubule assembly is defined as an increase in turbidity above 0.02 absorbance units. All three tubulin samples (wild-type, L136F, and I252L) assembled in the absence of oryzalin (Fig. 6A, black traces). However, in 5 μM oryzalin, wild-type T. thermophila tubulin did not polymerize, although both L136F and V252L mutant tubulins assembled (Fig. 6A, gray traces). Since L136F tubulin appears to avidly assemble over a range of concentrations (∼8 to 17.5 μM) (data not shown), we determined the Cc values for wild-type and mutant T. thermophila tubulins (Fig. 6B and Table 1). The tubulin Cc defines the concentration of tubulin required for equilibrium between microtubule assembly and disassembly. Above the Cc, tubulin will polymerize until the tubulin pool reaches the Cc, and below the Cc microtubules will depolymerize into free tubulin subunits. A lower Cc value indicates that subunits have greater affinity within the microtubule lattice, permitting assembly at lower tubulin concentrations. Our assays indicated that wild-type and I252L T. thermophila tubulins have essentially identical Cc values: 0.55 ± 0.09 mg/ml for wild-type tubulin and 0.56 ± 0.06 mg/ml for I252L tubulin. In contrast, L136F tubulin has a decreased Cc of 0.25 ± 0.07 mg/ml.
Fig. 6.
Mutant tubulins show altered assembly properties. (A) Polymerization of 0.7 mg/ml (7 μM) purified T. thermophila tubulin was followed at 351 nm in the presence (gray) or absence (black) of 0 or 5 μM oryzalin. Wild-type T. thermophila tubulin assembles normally in the absence of oryzalin but fails to polymerize in 5 μM oryzalin. L136F T. thermophila tubulin and I252L T. thermophila tubulin both assemble in 5 μM oryzalin. (B) Determination of Cc values for wild-type, L136F, and I252L tubulins. Wild-type T. thermophila tubulin has a Cc of 0.55 ± 0.09 mg/ml. The Cc for L136F tubulin is dramatically decreased to 0.25 ± 0.07 mg/ml, whereas I252L tubulin behaves similarly to wild-type tubulin with a Cc value of 0.56 ± 0.06 mg/ml.
DISCUSSION
Although the structure of the tubulin dimer within the microtubule lattice has been determined (20, 21, 42, 44, 54, 55, 57), precise biochemical roles for many amino acids in both α- and β-tubulins remain ill-defined. The situation is complicated by natural sequence variations: there are species-specific differences in tubulins and multiple isotypes of both α- and β-tubulins in multicellular organisms (22, 39-41, 65, 69, 75). Adding to this complexity, directed or selected mutations to α- or β-tubulin alter cellular phenotypes (cold or drug sensitivity, or more complex phenotypes such as flagellar function, leaf and petal shape, or neuronal behavior) (15, 29, 56, 59, 60, 62). Results from studies of heterogeneous populations of tubulin, such as the properties of tubulin in the presence of tubulin binding drugs, reflect the averaged properties of isotype mixtures.
Tubulin dimers can be altered by diverse PTMs, such as acetylation of K40 in α-tubulin or glycylation or glutamylation of the carboxy-terminal tails of both α- and β-tubulins (68). PTMs differentially mark distinct microtubule subpopulations within a cell, which influences their association with other proteins. For example, kinesin-1 binds preferentially to detyrosinated and acetylated microtubules (43, 58). When the enzymes responsible for acetylation (MEC-17) or glycylation (TTLL3A to -F) are deleted, Tetrahymena becomes resistant to Taxol (Paclitaxel) and sensitive to oryzalin (1, 74, 76). This suggests that these PTMs increase microtubule stability within the cell. Although purified tubulin samples unavoidably contain heterogeneous PTMs, because these modifications are likely to act by modulating association with microtubule-associated proteins rather than by directly influencing tubulin biochemistry, it is unlikely that they have an overt effect on the assembly or drug binding properties of purified tubulin.
Although it is impossible to control the tubulin PTMs, it is possible to reduce complexity in tubulin samples by exploiting genetically tractable simple organisms with single genes for α- and/or β-tubulin. These organisms provide the opportunity to prepare monospecific samples to compare the activities of distinct amino acid sequences in tubulin. For example, S. cerevisiae, which has two α-tubulin genes, TUB1 and TUB3, and a single β-tubulin gene, TUB2 (63, 64, 66), has been exploited to dissect the roles of isotype differences and binding site residues. Under wild-type transcriptional control, the TUB1 α-tubulin gene, but not the TUB3 α-tubulin gene, is sufficient for survival, although tub3 null yeast cells have increased benomyl sensitivity (63, 64). If TUB3 expression levels are increased, this α-tubulin is sufficient to support growth of a tub1 null (8). When TUB1p and TUB3p dimers are individually isolated from tub3 and tub1 null strains, it is clear that the 39-amino-acid differences make TUB3p-containing microtubules less dynamic than TUB1p-containing microtubules (8). This is consistent with the increased sensitivity of tub3 null strains to benomyl. Researchers have also introduced directed mutations into the sole β-tubulin gene (TUB2) to analyze the requirements for Taxol sensitivity. S. cerevisiae is innately insensitive to the microtubule-stabilizing drug Taxol, due both to drug efflux (plasma membrane transporters confer multidrug resistance) and to small differences in β-tubulin residues required for Taxol binding. Introduction of five substitutions to β-tubulin is sufficient to confer Taxol binding (24). When this mutant β-tubulin gene is introduced into a transporter knockout strain, Taxol exposure causes cells to develop long microtubules and arrest during cell division, similar to the effects of Taxol on vertebrate cells (14).
We are interested in understanding the basis of the selective action of dinitroanilines on plant and protozoan tubulin subsets and the influence of resistance mutations on the properties of tubulin. In both plants and protozoa, dinitroaniline resistance is associated with amino acid substitutions to the α-tubulin subunit of the tubulin dimer (3–6, 11, 30, 45, 52, 79). We identified 35 different single point mutations to the α1-tubulin gene that confer dinitroaniline resistance in T. gondii (45, 46). In some, but not all, cases the substitution makes the tubulin more like insensitive vertebrate tubulin. For example, although the V252L mutation in Toxoplasma converts valine to leucine, the residue typically seen in vertebrate tubulins, the L136F mutation converts a residue that is conserved between Toxoplasma and vertebrate tubulins into an atypical residue at this position. The resistance mutations cluster into distinct domains, which led us to propose that they act by distinct mechanisms: (i) to increase microtubule stability or (ii) to decrease tubulin affinity for dinitroanilines (53). Although in some cases we could observe overt phenotypes associated with the expression of a mutant tubulin in T. gondii, such as lengthened, hyperstabilized microtubules associated with the H28Q mutation, in most cases the only detectable phenotypes associated with mutant tubulins in parasites were dinitroaniline resistance and reduced fitness in the absence of drug selection (45, 46). In order to characterize the biochemical properties of tubulin heterodimers bearing these amino acid substitutions, we needed to exploit a genetically tractable, dinitroaniline-sensitive organism for expression of altered polymerization-competent tubulin.
We previously demonstrated that α-tubulin transgenes bearing M268T or F24H (equivalent to Y24H) mutations, which are associated with dinitroaniline resistance in other organisms, also confer resistance in T. gondii (45). This suggested that substitutions to T. thermophila α-tubulin residues would confer resistance, even in cases where the mutated residue was different between wild-type T. gondii and wild-type T. thermophila tubulin. We engineered an established T. thermophila α-tubulin gene construct to individually contain the mutations H28Q, L136F, R243S, T239I, and I252L. With the exception of residue 252, all sites of mutation occurred in amino acid residues that are conserved between T. thermophila and T. gondii (see Fig. S1 in the supplemental material). However, the residue at position 252 is a valine in T. gondii and an isoleucine in T. thermophila. Although substitution of a leucine for an isoleucine in T. thermophila is a subtler alteration than substitution of a leucine for a valine, the I252L mutation conferred dinitroaniline resistance in T. thermophila.
We determined that the Cc value for wild-type T. thermophila tubulin is 0.56 mg/ml. This is within the range of values that have been observed for tubulins isolated from other eukaryotes. Although tubulins isolated from sea urchin (Strongylocentrotus purpuratus), budding yeast, and the protozoan parasite Leishmania amazonensis have Cc values in the range of 0.1 to 0.2 mg/ml, tubulin from bovine brain has a Cc of 0.56 mg/ml and tubulin from corn (Zea mays) has a Cc of 0.83 mg/ml (8, 10, 27, 34, 35, 71). It is important to note that buffer ionic strength and additives such as Taxol or DMSO influence relative Cc values, so that a close comparison of values from different studies is not informative. However, in this research, we used identical tubulin assembly conditions to measure Cc values for T. thermophila tubulin with L136F or I252L substitutions. These results can be directly compared and indicate that I252L tubulin assembly is essentially identical to that of wild-type T. thermophila tubulin, while L136F tubulin has a dramatically reduced Cc, indicating that it assembles with greater avidity.
Previous researchers have used several different techniques to measure the affinity of plant or protozoan tubulin dimers for the dinitroaniline oryzalin. Tubulins isolated from higher land plants have Kd values of 0.095 μM (Zea mays) and 2.59 μM (suspension cultures of rose tissue) (28, 49). Tubulin isolated from L. amazonensis has a Kd of 19 μM (71). However, Leishmania and other kinetoplastid parasites are relatively insensitive to oryzalin. In comparison, they have a Kd value of 1.7 μM for the dinitroaniline analog GB-II-5, which is a much more effective inhibitor of kinetoplastid tubulin. Our measurements indicated that wild-type T. thermophila tubulin has an oryzalin Kd of 0.44 μM. Oryzalin affinity is reduced in the L136F and I252L mutant T. thermophila tubulins, which have Kd values of 11 and 6.7 μM, respectively. These data indicate that single mutations to α-tubulin can dramatically change oryzalin sensitivity, although resistant T. thermophila tubulins are still more sensitive to oryzalin than bovine brain tubulin, which has a Kd of 77 ± 25 μM for oryzalin.
The biochemical properties of wild-type tubulin and tubulins bearing the L136F and I252L substitutions indicate that we cannot precisely anticipate the mechanism of resistance in mutant tubulins by location of point mutations within the structure of the tubulin dimer. We hypothesized that I252L tubulin would produce hyperstabilized microtubules due to reduced activity of the GTPase activating domain. Our experiments indicate that it has a Cc value similar to wild-type tubulin and most likely works by allosterically altering the drug binding site conformation to reduce affinity for dinitroanilines. This mechanism is much more consistent with its ability to confer high levels of oryzalin resistance (∼40 μM) in T. gondii (45). We predicted that the L136F mutation would decrease the affinity of tubulin for dinitroanilines. Our binding data indicate that this is the case, but we also discovered an unanticipated reduction in the Cc associated with this substitution, most likely an indication that L136F tubulin also has increased subunit affinity within the microtubule lattice. Previous observations of the H28Q mutation, which is located both in the H1-S2 loop and in the proposed dinitroaniline binding site, had suggested that it is not always possible to assign a single mechanism (dinitroaniline or dimer-dimer affinity) for resistance mutations (45). The observations presented here corroborate this conclusion. In this report we demonstrate that mutations that confer dinitroaniline resistance in T. gondii (L136F and I252L) and green foxtail (L136F) confer resistance in T. thermophila. These data suggest that the drug binding site and mechanism of disruption are conserved in diverse, dinitroaniline-sensitive organisms. These studies are also proof of concept that expression of altered tubulin in T. thermophila is an excellent method for dissecting the precise role of single amino acid changes in tubulin function.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Sept (University of Michigan) and Susan Dutcher (Washington University) for commenting on the manuscript. We are grateful to Grant MacGregor for use of his airfuge and technical advice and to Tara Reed and Darany Tan for assistance with media preparation, T. thermophila culturing, and tubulin purification.
Research presented in this paper was supported by NIH grant AI067981 (N.S.M.), NSF grant MBC-0639934 (J.G.), and by funds from the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (D.L.S.).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 24 September 2010.
REFERENCES
- 1.Akella J. S., Wloga D., Kim J., Starostina N. G., Lyons-Abbott S., Morrissette N. S., Dougan S. T., Kipreos E. T., Gaertig J. 2010. MEC-17 is an alpha-tubulin acetyltransferase. Nature 467:218–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anders K. R., Botstein D. 2001. Dominant-lethal alpha-tubulin mutants defective in microtubule depolymerization in yeast. Mol. Biol. Cell 12:3973–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthony R. G., Hussey P. J. 1999. Dinitroaniline herbicide resistance and the microtubule cytoskeleton. Trends Plant Sci. 4:112–116 [DOI] [PubMed] [Google Scholar]
- 4.Anthony R. G., Hussey P. J. 1999. Double mutation in Eleusine indica alpha-tubulin increases the resistance of transgenic maize calli to dinitroaniline and phosphorothioamidate herbicides. Plant J. 18:669–674 [DOI] [PubMed] [Google Scholar]
- 5.Anthony R. G., Reichelt S., Hussey P. J. 1999. Dinitroaniline herbicide-resistant transgenic tobacco plants generated by co-overexpression of a mutant alpha-tubulin and a beta-tubulin. Nat. Biotechnol. 17:712–716 [DOI] [PubMed] [Google Scholar]
- 6.Anthony R. G., Waldin T. R., Ray J. A., Bright S. W., Hussey P. J. 1998. Herbicide resistance caused by spontaneous mutation of the cytoskeletal protein tubulin. Nature 393:260–263 [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya G., Herman J., Delfin D., Salem M. M., Barszcz T., Mollet M., Riccio G., Brun R., Werbovetz K. A. 2004. Synthesis and antitubulin activity of N1- and N4-substituted 3,5-dinitro sulfanilamides against African trypanosomes and Leishmania. J. Med. Chem. 47:1823–1832 [DOI] [PubMed] [Google Scholar]
- 8.Bode C. J., Gupta M. L., Suprenant K. A., Himes R. H. 2003. The two alpha-tubulin isotypes in budding yeast have opposing effects on microtubule dynamics in vitro. EMBO Rep. 4:94–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan M. M., Fong D. 1990. Inhibition of leishmanias but not host macrophages by the antitubulin herbicide trifluralin. Science 249:924–926 [DOI] [PubMed] [Google Scholar]
- 10.Davis A., Sage C. R., Wilson L., Farrell K. W. 1993. Purification and biochemical characterization of tubulin from the budding yeast Saccharomyces cerevisiae. Biochemistry 32:8823–8835 [DOI] [PubMed] [Google Scholar]
- 11.Delye C., Menchari Y., Michel S., Darmency H. 2004. Molecular bases for sensitivity to tubulin-binding herbicides in green foxtail. Plant Physiol. 136:3920–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dufour E., Haertle T. 1991. Binding of retinoids and beta-carotene to beta-lactoglobulin. Influence of protein modifications. Biochim. Biophys. Acta 1079:316–320 [DOI] [PubMed] [Google Scholar]
- 13.Fast N. M., Xue L., Bingham S., Keeling P. J. 2002. Re-examining alveolate evolution using multiple protein molecular phylogenies. J. Eukaryot. Microbiol. 49:30–37 [DOI] [PubMed] [Google Scholar]
- 14.Foland T. B., Dentler W. L., Suprenant K. A., Gupta M. L., Jr., Himes R. H. 2005. Paclitaxel-induced microtubule stabilization causes mitotic block and apoptotic-like cell death in a paclitaxel-sensitive strain of Saccharomyces cerevisiae. Yeast 22:971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukushige T., Siddiqui Z. K., Chou M., Culotti J. G., Gogonea C. B., Siddiqui S. S., Hamelin M. 1999. MEC-12, an alpha-tubulin required for touch sensitivity in C. elegans. J. Cell Sci. 112 (Pt 3):395–403 [DOI] [PubMed] [Google Scholar]
- 16.Gaertig J., Cruz M. A., Bowen J., Gu L., Pennock D. G., Gorovsky M. A. 1995. Acetylation of lysine 40 in alpha-tubulin is not essential in Tetrahymena thermophila. J. Cell Biol. 129:1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaertig J., Gu L., Hai B., Gorovsky M. A. 1994. High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 22:5391–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaertig J., Thatcher T. H., Gu L., Gorovsky M. A. 1994. Electroporation-mediated replacement of a positively and negatively selectable beta-tubulin gene in Tetrahymena thermophila. Proc. Natl. Acad. Sci. U. S. A. 91:4549–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaertig J., Thatcher T. H., McGrath K. E., Callahan R. C., Gorovsky M. A. 1993. Perspectives on tubulin isotype function and evolution based on the observation that Tetrahymena thermophila microtubules contain a single alpha- and beta-tubulin. Cell Motil. Cytoskel. 25:243–253 [DOI] [PubMed] [Google Scholar]
- 20.Gigant B., Curmi P. A., Martin-Barbey C., Charbaut E., Lachkar S., Lebeau L., Siavoshian S., Sobel A., Knossow M. 2000. The 4 A X-ray structure of a tubulin:stathmin-like domain complex. Cell 102:809–816 [DOI] [PubMed] [Google Scholar]
- 21.Gigant B., Wang C., Ravelli R. B., Roussi F., Steinmetz M. O., Curmi P. A., Sobel A., Knossow M. 2005. Structural basis for the regulation of tubulin by vinblastine. Nature 435:519–522 [DOI] [PubMed] [Google Scholar]
- 22.Gull K., Hussey P. J., Sasse R., Schneider A., Seebeck T., Sherwin T. 1986. Tubulin isotypes: generation of diversity in cells and microtubular organelles. J. Cell Sci. Suppl. 5:243–255 [DOI] [PubMed] [Google Scholar]
- 23.Gupta M. L., Jr., Bode C. J., Dougherty C. A., Marquez R. T., Himes R. H. 2001. Mutagenesis of beta-tubulin cysteine residues in Saccharomyces cerevisiae: mutation of cysteine 354 results in cold-stable microtubules. Cell Motil. Cytoskel. 49:67–77 [DOI] [PubMed] [Google Scholar]
- 24.Gupta M. L., Jr., Bode C. J., Georg G. I., Himes R. H. 2003. Understanding tubulin-Taxol interactions: mutations that impart Taxol binding to yeast tubulin. Proc. Natl. Acad. Sci. U. S. A. 100:6394–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hai B., Gaertig J., Gorovsky M. A. 2000. Knockout heterokaryons enable facile mutagenic analysis of essential genes in Tetrahymena. Methods Cell Biol. 62:513–531 [DOI] [PubMed] [Google Scholar]
- 26.Hai B., Gorovsky M. A. 1997. Germ-line knockout heterokaryons of an essential alpha-tubulin gene enable high-frequency gene replacement and a test of gene transfer from somatic to germ-line nuclei in Tetrahymena thermophila. Proc. Natl. Acad. Sci. U. S. A. 94:1310–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hugdahl J. D., Bokros C. L., Hanesworth V. R., Aalund G. R., Morejohn L. C. 1993. Unique functional characteristics of the polymerization and MAP binding regulatory domains of plant tubulin. Plant Cell 5:1063–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hugdahl J. D., Morejohn L. C. 1993. Rapid and reversible high-affinity binding of the dinitroaniline herbicide oryzalin to tubulin from Zea mays L. Plant Physiol. 102:725–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishida T., Kaneko Y., Iwano M., Hashimoto T. 2007. Helical microtubule arrays in a collection of twisting tubulin mutants of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 104:8544–8549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.James S. W., Silflow C. D., Stroom P., Lefebvre P. A. 1993. A mutation in the alpha 1-tubulin gene of Chlamydomonas reinhardtii confers resistance to anti-microtubule herbicides. J. Cell Sci. 106:209–218 [DOI] [PubMed] [Google Scholar]
- 31.Jordan A., Hadfield J. A., Lawrence N. J., McGown A. T. 1998. Tubulin as a target for anticancer drugs: agents which interact with the mitotic spindle. Med. Res. Rev. 18:259–296 [DOI] [PubMed] [Google Scholar]
- 32.Jordan M. A. 2002. Mechanism of action of antitumor drugs that interact with microtubules and tubulin. Curr. Med. Chem. Anticancer Agents 2:1–17 [DOI] [PubMed] [Google Scholar]
- 33.Jung M. K., Wilder I. B., Oakley B. R. 1992. Amino acid alterations in the benA (beta-tubulin) gene of Aspergillus nidulans that confer benomyl resistance. Cell Motil. Cytoskel. 22:170–174 [DOI] [PubMed] [Google Scholar]
- 34.Keller T. C., III, Jemiolo D. K., Burgess W. H., Rebhun L. I. 1982. Strongylocentrotus purpuratus spindle tubulin. II. Characteristics of its sensitivity to Ca++ and the effects of calmodulin isolated from bovine brain and S. purpuratus eggs. J. Cell Biol. 93:797–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller T. C., III, Rebhun L. I. 1982. Strongylocentrotus purpuratus spindle tubulin. I. Characteristics of its polymerization and depolymerization in vitro. J. Cell Biol. 93:788–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lacey E. 1990. Mode of action of benzimidazoles. Parasitol. Today 6:112–115 [DOI] [PubMed] [Google Scholar]
- 37.Lacey E., Prichard R. K. 1986. Interactions of benzimidazoles (BZ) with tubulin from BZ-sensitive and BZ-resistant isolates of Haemonchus contortus. Mol. Biochem. Parasitol. 19:171–181 [DOI] [PubMed] [Google Scholar]
- 38.Leander B. S., Keeling P. J. 2003. Morphostasis in alveolate evolution. Trends Ecol. Evol. 18:395–402 [Google Scholar]
- 39.Lewis S. A., Gilmartin M. E., Hall J. L., Cowan N. J. 1985. Three expressed sequences within the human beta-tubulin multigene family each define a distinct isotype. J. Mol. Biol. 182:11–20 [DOI] [PubMed] [Google Scholar]
- 40.Lewis S. A., Gu W., Cowan N. J. 1987. Free intermingling of mammalian beta-tubulin isotypes among functionally distinct microtubules. Cell 49:539–548 [DOI] [PubMed] [Google Scholar]
- 41.Lewis S. A., Lee M. G., Cowan N. J. 1985. Five mouse tubulin isotypes and their regulated expression during development. J. Cell Biol. 101:852–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H., DeRosier D. J., Nicholson W. V., Nogales E., Downing K. H. 2002. Microtubule structure at 8 Å resolution. Structure 10:1317–1328 [DOI] [PubMed] [Google Scholar]
- 43.Liao G., Gundersen G. G. 1998. Kinesin is a candidate for cross-bridging microtubules and intermediate filaments. Selective binding of kinesin to detyrosinated tubulin and vimentin. J. Biol. Chem. 273:9797–9803 [DOI] [PubMed] [Google Scholar]
- 44.Lowe J., Li H., Downing K. H., Nogales E. 2001. Refined structure of alpha beta-tubulin at 3.5 Å resolution. J. Mol. Biol. 313:1045–1057 [DOI] [PubMed] [Google Scholar]
- 45.Ma C., Li C., Ganesan L., Oak J., Tsai S., Sept D., Morrissette N. S. 2007. Mutations in alpha-tubulin confer dinitroaniline resistance at a cost to microtubule function. Mol. Biol. Cell 18:4711–4720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma C., Tran J., Li C., Ganesan L., Wood D., Morrissette N. 2008. Secondary mutations correct fitness defects in Toxoplasma gondii with dinitroaniline resistance mutations. Genetics 180:845–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGrath K. E., Yu S. M., Heruth D. P., Kelly A. A., Gorovsky M. A. 1994. Regulation and evolution of the single alpha-tubulin gene of the ciliate Tetrahymena thermophila. Cell Motil. Cytoskel. 27:272–283 [DOI] [PubMed] [Google Scholar]
- 48.Mitra A., Sept D. 2006. Binding and interaction of dinitroanilines with apicomplexan and kinetoplastid alpha-tubulin. J. Med. Chem. 49:5226–5231 [DOI] [PubMed] [Google Scholar]
- 49.Morejohn L. C., Bureau T. E., Mole-Bajer J., Bajer A. S., Fosket D. E. 1987. Oryzalin, a dinitroaniline herbicide, binds to plant tubulin and inhibits microtubule polymerization in vitro. Planta 172:252–264 [DOI] [PubMed] [Google Scholar]
- 50.Morgan R. E., Werbovetz K. A. 2008. Selective lead compounds against kinetoplastid tubulin. Adv. Exp. Med. Biol. 625:33–47 [DOI] [PubMed] [Google Scholar]
- 51.Morrissette N. S., Bedian V., Webster P., Roos D. S. 1994. Characterization of extreme apical antigens from Toxoplasma gondii. Exp. Parasitol. 79:445–459 [DOI] [PubMed] [Google Scholar]
- 52.Morrissette N. S., Mitra A., Sept D., Sibley L. D. 2004. Dinitroanilines bind alpha-tubulin to disrupt microtubules. Mol. Biol. Cell 15:1960–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morrissette N. S., Sept D. 2008. Dinitroaniline interactions with tubulin: genetic and computational approaches to define the mechanisms of action and resistance, p. 327–350InBlume Y. B., Baird W. V., Yemets A. I., Breviario D. (ed.), The plant cytoskeleton: a key tool for agro-biotechnology. Springer Dordrecht, New York, NY [Google Scholar]
- 54.Nogales E., Whittaker M., Milligan R. A., Downing K. H. 1999. High-resolution model of the microtubule. Cell 96:79–88 [DOI] [PubMed] [Google Scholar]
- 55.Nogales E., Wolf S. G., Khan I. A., Luduena R. F., Downing K. H. 1995. Structure of tubulin at 6.5 Å and location of the taxol-binding site. Nature 375:424–427 [DOI] [PubMed] [Google Scholar]
- 56.Rathinasamy K., Panda D. 2006. Suppression of microtubule dynamics by benomyl decreases tension across kinetochore pairs and induces apoptosis in cancer cells. FEBS J. 273:4114–4128 [DOI] [PubMed] [Google Scholar]
- 57.Ravelli R. B., Gigant B., Curmi P. A., Jourdain I., Lachkar S., Sobel A., Knossow M. 2004. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 428:198–202 [DOI] [PubMed] [Google Scholar]
- 58.Reed N. A., Cai D., Blasius T. L., Jih G. T., Meyhofer E., Gaertig J., Verhey K. J. 2006. Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 16:2166–2172 [DOI] [PubMed] [Google Scholar]
- 59.Reijo R. A., Cooper E. M., Beagle G. J., Huffaker T. C. 1994. Systematic mutational analysis of the yeast beta-tubulin gene. Mol. Biol. Cell 5:29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richards K. L., Anders K. R., Nogales E., Schwartz K., Downing K. H., Botstein D. 2000. Structure-function relationships in yeast tubulins. Mol. Biol. Cell 11:1887–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sackett D. L., Werbovetz K. A., Morrissette N. S. 2010. Isolating tubulin from nonneural sources. Methods Cell Biol. 95:17–32 [DOI] [PubMed] [Google Scholar]
- 62.Savage C., Hamelin M., Culotti J. G., Coulson A., Albertson D. G., Chalfie M. 1989. mec-7 is a beta-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev. 3:870–881 [DOI] [PubMed] [Google Scholar]
- 63.Schatz P. J., Pillus L., Grisafi P., Solomon F., Botstein D. 1986. Two functional alpha-tubulin genes of the yeast Saccharomyces cerevisiae encode divergent proteins. Mol. Cell. Biol. 6:3711–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schatz P. J., Solomon F., Botstein D. 1986. Genetically essential and nonessential alpha-tubulin genes specify functionally interchangeable proteins. Mol. Cell. Biol. 6:3722–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sullivan K. F., Cleveland D. W. 1986. Identification of conserved isotype-defining variable region sequences for four vertebrate beta tubulin polypeptide classes. Proc. Natl. Acad. Sci. U. S. A. 83:4327–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thomas J. H., Neff N. F., Botstein D. 1985. Isolation and characterization of mutations in the beta-tubulin gene of Saccharomyces cerevisiae. Genetics 111:715–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ursic D., Sedbrook J. C., Himmel K. L., Culbertson M. R. 1994. The essential yeast Tcp1 protein affects actin and microtubules. Mol. Biol. Cell 5:1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verhey K. J., Gaertig J. 2007. The tubulin code. Cell Cycle 6:2152–2160 [DOI] [PubMed] [Google Scholar]
- 69.Villasante A., Wang D., Dobner P., Dolph P., Lewis S. A., Cowan N. J. 1986. Six mouse alpha-tubulin mRNAs encode five distinct isotypes: testis-specific expression of two sister genes. Mol. Cell. Biol. 6:2409–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Werbovetz K. A. 2002. Tubulin as an antiprotozoal drug target. Mini Rev. Med. Chem. 2:519–529 [DOI] [PubMed] [Google Scholar]
- 71.Werbovetz K. A., Brendle J. J., Sackett D. L. 1999. Purification, characterization, and drug susceptibility of tubulin from Leishmania. Mol. Biochem. Parasitol. 98:53–65 [DOI] [PubMed] [Google Scholar]
- 72.Werbovetz K. A., Sackett D. L., Delfin D., Bhattacharya G., Salem M., Obrzut T., Rattendi D., Bacchi C. 2003. Selective antimicrotubule activity of N1-phenyl-3,5-dinitro-N4, N4-di-n-propylsulfanilamide (GB-II-5) against kinetoplastid parasites. Mol. Pharmacol. 64:1325–1333 [DOI] [PubMed] [Google Scholar]
- 73.Williams N. E., Honts J. E., Kaczanowska J. 1990. The formation of basal body domains in the membrane skeleton of Tetrahymena. Development 109:935–942 [DOI] [PubMed] [Google Scholar]
- 74.Wloga D., Webster D. M., Rogowski K., Bre M. H., Levilliers N., Jerka-Dziadosz M., Janke C., Dougan S. T., Gaertig J. 2009. TTLL3 is a tubulin glycine ligase that regulates the assembly of cilia. Dev. Cell 16:867–876 [DOI] [PubMed] [Google Scholar]
- 75.Wolf N., Regan C. L., Fuller M. T. 1988. Temporal and spatial pattern of differences in microtubule behaviour during Drosophila embryogenesis revealed by distribution of a tubulin isoform. Development 102:311–324 [DOI] [PubMed] [Google Scholar]
- 76.Xia L., Hai B., Gao Y., Burnette D., Thazhath R., Duan J., Bre M. H., Levilliers N., Gorovsky M. A., Gaertig J. 2000. Polyglycylation of tubulin is essential and affects cell motility and division in Tetrahymena thermophila. J. Cell Biol. 149:1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yaffe M. B., Farr G. W., Miklos D., Horwich A. L., Sternlicht M. L., Sternlicht H. 1992. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature 358:245–248 [DOI] [PubMed] [Google Scholar]
- 78.Yakovich A. J., Ragone F. L., Alfonzo J. D., Sackett D. L., Werbovetz K. A. 2006. Leishmania tarentolae: purification and characterization of tubulin and its suitability for antileishmanial drug screening. Exp. Parasitol. 114:289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamamoto E., Zeng L., Baird W. V. 1998. Alpha-tubulin missense mutations correlate with antimicrotubule drug resistance in Eleusine indica. Plant Cell 10:297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.