Abstract
Protists that live under low-oxygen conditions often lack conventional mitochondria and instead possess mitochondrion-related organelles (MROs) with distinct biochemical functions. Studies of mostly parasitic organisms have suggested that these organelles could be classified into two general types: hydrogenosomes and mitosomes. Hydrogenosomes, found in parabasalids, anaerobic chytrid fungi, and ciliates, metabolize pyruvate anaerobically to generate ATP, acetate, CO2, and hydrogen gas, employing enzymes not typically associated with mitochondria. Mitosomes that have been studied have no apparent role in energy metabolism. Recent investigations of free-living anaerobic protists have revealed a diversity of MROs with a wider array of metabolic properties that defy a simple functional classification. Here we describe an expressed sequence tag (EST) survey and ultrastructural investigation of the anaerobic heteroloboseid amoeba Sawyeria marylandensis aimed at understanding the properties of its MROs. This organism expresses typical anaerobic energy metabolic enzymes, such as pyruvate:ferredoxin oxidoreductase, [FeFe]-hydrogenase, and associated hydrogenase maturases with apparent organelle-targeting peptides, indicating that its MRO likely functions as a hydrogenosome. We also identified 38 genes encoding canonical mitochondrial proteins in S. marylandensis, many of which possess putative targeting peptides and are phylogenetically related to putative mitochondrial proteins of its heteroloboseid relative Naegleria gruberi. Several of these proteins, such as a branched-chain alpha keto acid dehydrogenase, likely function in pathways that have not been previously associated with the well-studied hydrogenosomes of parabasalids. Finally, morphological reconstructions based on transmission electron microscopy indicate that the S. marylandensis MROs form novel cup-like structures within the cells. Overall, these data suggest that Sawyeria marylandensis possesses a hydrogenosome of mitochondrial origin with a novel combination of biochemical and structural properties.
It is now widely accepted that the most recent common ancestor of extant eukaryotes possessed an endosymbiont-derived mitochondrial organelle of alphaproteobacterial ancestry (see reference 16 for a recent review). Although most well-known eukaryotes contain mitochondria that aerobically respire to produce ATP, a vast diversity of anaerobic eukaryotic lineages have been discovered that lack classical mitochondrial structures. Instead of mitochondria, biochemically diverse double-membrane-bounded organelles have been found that function under low-oxygen conditions (for recent reviews, see references 5, 6, 17, 23, 54, and 57). The discovery and recent investigations of the functions of these mitochondrion-related organelles (MROs) has greatly expanded our understanding of both the conservation and diversity of functions that these organelles can perform.
A number of protist lineages, including the parabasalids, several anaerobic ciliate groups, and anaerobic chytrid fungi, are known to possess MROs termed “hydrogenosomes,” which generate ATP and molecular hydrogen from anaerobic energy metabolism. Over the last few decades, intensive molecular and biochemical investigation of the functions of the hydrogenosomes of Trichomonas vaginalis have revealed that they possess a mitochondrion-like protein import apparatus and they carry out additional mitochondrial functions, including the synthesis of iron-sulfur (Fe-S) clusters and metabolism of some amino acids (9, 35). Other anaerobic protistan lineages contain “mitosomes,” which are typically smaller than hydrogenosomes and have no obvious energy metabolic role. Some of these mitosomes, such as those of Giardia intestinalis (54) and some Microsporidia (22), possess mitochondrion-like protein import apparatuses and seem to function in Fe-S cluster biogenesis. Other mitosomes, such as those of Entamoeba, retain mitochondrial heat shock and chaperonin proteins (2, 32) yet seem to carry out biochemical functions, such as sulfur activation (34), that are not typically associated with aerobic mitochondria.
Recent investigations of little-studied protists have further revealed a continuum of diversity of function in MROs, from organelles that are complex metabolic intermediates between mitochondria and hydrogenosomes (e.g., Nyctotherus ovalis [7] and Blastocystis sp. [51]) to apparent intermediates between hydrogenosomes and mitosomes (e.g., Mastigamoeba [21] and Trimastix [25]). The potential functional diversity of MROs could even be greater since there are a myriad of additional poorly studied anaerobic protists that, in transmission electron microscopy studies (26), have been shown to contain MRO-like structures that have yet to be intensively investigated. By studying their organelles, we can obtain important insights into their evolutionary origin, the range of their biochemical flexibility within the eukaryotic tree of life, and possibly even the origin of eukaryotes themselves (5).
Sawyeria marylandensis is an amoeba from anaerobic sediments that belongs to the class Heterolobosea (24, 39, 48). Small subunit rRNA phylogenies (SSU rRNA) show Sawyeria branching in a robust clade with the two other anaerobic heteroloboseids for which molecular data are available, namely, Psalteriomonas lanterna and Monopylocystis visvesvarai (39). Sawyeria is not a strict anaerobe but a microaerophile since it grows in cultures with trace amounts of oxygen, and to date, it has been described only as a gymnamoeba, lacking a cyst or flagellate stage (39). Sawyeria cells are uninucleate and lack classical mitochondria. However, like Psalteriomonas lanterna, transmission electron micrographs reveal that Sawyeria contains densely staining organelles that resemble mitochondria in size and shape (39). Recently an expressed sequence tag (EST) survey of Psalteriomonas lanterna revealed the existence of transcripts encoding canonical mitochondrial proteins. These findings, combined with ultrastructural arguments, were interpreted as evidence that its densely staining organelles were mitochondrion-related organelles (12). More surprisingly, the full genome sequence of the model system aerobic heteroloboseid Naegleria gruberi was reported to have genes that encode enzymes typically associated with anaerobic energy metabolism, including [FeFe]-hydrogenase and its maturase proteins (19), and the encoded proteins appeared to possess N-terminal mitochondrial targeting signals. Although experimental follow-up studies are needed to verify these biochemical predictions, it seems that the capacity for anaerobic energy metabolism may be relatively old within mitochondria of the heteroloboseid lineage.
Here we report the first detailed morphological and molecular characterization of the mitochondrion-derived organelles of Sawyeria marylandensis. By directed degenerate PCR studies and an expressed sequence tag (EST) survey of this organism, we identified several homologues of mitochondrion- and hydrogenosome-targeted proteins, including pyruvate:ferredoxin oxidoreductase (PFO), [FeFe]-hydrogenase, and the recently described hydrogenase maturase protein HydG. The proteins we have identified, as well as the presence of predicted N-terminal mitochondrion-type targeting peptides, indicate that Sawyeria marylandensis likely has MROs with hydrogenosomal functions and a unique proteome composition. Phylogenetic analyses of some of these proteins show that this organelle has evolved from an ancestral heteroloboseid mitochondrion. Our electron microscopic morphological analyses clearly demonstrate that the Sawyeria organelles have two bounding membranes and form novel cup-shaped structures. These data expand our knowledge of both the structural and functional diversity of mitochondrion-derived organelles within unicellular eukaryotes.
MATERIALS AND METHODS
Culture conditions.
Sawyeria marylandensis cells were grown at 20°C in the dark in ATCC medium 802 in 15-ml Falcon tubes previously bacterized with a mix of different anaerobic bacteria. The culture was transferred weekly. When the culture reached peak density, cells were harvested by centrifugation at 300 × g and resuspended in Tri-reagent (Molecular Research Center, Cincinnati, OH).
EST data.
Total RNA was extracted using Tri-reagent following the manufacturer's guidelines. cDNA libraries were constructed by Amplicon Express (Pullman, WA). The number of ESTs passing quality control and submitted to further analysis was 9,300. EST data were automatically clustered by tools implemented in TBestDB (http://amoebidia.bcm.umontreal.ca/pepdb/searches/login.php?bye=true) (38) and AnaBench (http://anabench.bcm.umontreal.ca/anabench/) (4). Moreover, data were also manually clustered using the Phred and Phrap software programs (18). The 9,300 sequences of Sawyeria marylandensis resulted in a total of 2,672 unique clusters.
Gene searching.
A BLASTX search of the EST clusters against mitochondrial human and yeast proteomes (3, 52) was carried out, and the best hits were then searched again by BLASTX against the GenBank nonredundant database. Sequences that consistently showed best hits to mitochondrial or hydrogenosomal homologs were further analyzed. In addition to sequence-based searches, we also conducted a text-based search using the name of mitochondrial/hydrogenosomal proteins within the automatic annotation pipeline of TBestDB (38). Trichomonas and Giardia proteomes were searched using BLASTP for homologs of the Sawyeria proteins that we had identified as putatively mitochondrial. Positive hits were then blasted back against the nr NCBI database, and in some cases, homology was further assessed by aligning the sequences into eukaryotic alignments.
RNA extraction, PCR, cloning, and sequencing.
[FeFe]-hydrogenase, PFO, Cpn60, and IscS were directly amplified by reverse transcriptase PCR (RT-PCR) using either degenerate or “exact-match” primers as described below. Total RNA and purified poly(A)+ mRNA were obtained using standard methods. First-strand cDNA was obtained via reverse transcription using an oligo(dT) primer. A portion of the gene was obtained by RT-PCR. Full cDNA and gene sequences were obtained by random amplification of cDNA ends (RACE) using the Gene Racer kit (Invitrogen Corporation, Carlsbad, CA). The different genes were amplified using different combinations of specific and degenerate primers. PCR products were purified and cloned using the Topo-TA cloning kit for sequencing (Invitrogen, Carlsbad, CA). Both coding and noncoding strands were sequenced from at least three different clones.
Phylogenetic analyses.
Alignments were constructed with the MacGDE2.3 (49) or MacClade software program. All individual gene alignments were manually revised and edited, and only those positions that were unambiguously aligned were included in the final analyses. Maximum-likelihood (ML) phylogenetic trees were constructed using the RAxML software program (50) with a WAG model of protein evolution and with a gamma distribution (WAG+Γ). Statistical support was obtained from 100 bootstrap replicates using RAxML. In the case of NuoF, an additional tree was constructed in RAxML by first recoding the amino acids into four functional categories as specified elsewhere (45) and analyzing the data with the general time reversible (GTR) model with a gamma distribution.
Prediction of putative targeting peptides.
Most proteins that localize in hydrogenosomes or mitosomes have N-terminal targeting peptides. We searched for targeting peptides in our Sawyeria proteins using two different targeting peptide prediction software tools: MITOPROT (10) and PSORT (37).
Electron microscopy.
Cells were fixed in a 0.1 M cacodylate buffer containing 2.5% gluteraldehyde at 4°C. Postfixation in 1% osmium tetroxide and 0.25% uranyl acetate was also conducted before stepwise dehydration in acetone at 4°C. Samples were embedded in Epon resin, and 60 nm-sections were made on an LKB-Huxley ultramicrotome using a Diatome diamond knife. Poststaining was conducted in an aqueous solution containing 2% uranyl acetate and lead citrate. Sections were examined on a Jeol JEM 1230 transmission electron microscope operating at 80 kV, and the images were captured using a digital camera.
3D organelle reconstruction.
A digital three-dimensional (3D) model of serial transmission electron microscope sections of Sawyeria MROs was generated using the software program TrackEM (an ImageJ plugin for three-dimensional modeling; see http://www.ini.uzh.ch/∼acardona/trakem2.html). The different sections (n = 20) were registered. Using the various modeling functions, traces of the MRO outlines were generated. The traces were rendered in the program to generate a 3D model of the Sawyeria hydrogenosome.
Nucleotide sequence accession numbers.
New sequences have been deposited in GenBank under the accession numbers listed in Table 1.
Table 1.
Mitocondrial/hydrogenosomal proteins in Sawyeria marylandensis and comparison with N. gruberi, T. vaginalis, and G. intestinalisa
| Category or S. marylandensis mitochondrial/hydrogenosomal protein | Accession no.d |
N-terminal extensione | Annotationf |
||||
|---|---|---|---|---|---|---|---|
| TBestDB | NCBI | Ng | Tv | Gi | |||
| Energy metabolism | |||||||
| [FeFe]-hydrogenaseb | EF612752 (SmHydA) | Yes | + | + | + | ||
| EF612753 (SmHydB) | |||||||
| Pyruvate:ferredoxin oxidoreductase (PFO)b | EF612751 | Yes | − | + | + | ||
| Alpha-succinyl-CoA synthetaseb | SML00000322 | EF612760 | Yes | + | + | − | |
| Beta-succinyl-CoA synthetaseb | SML00000048 | EF612761 | Yes | + | + | − | |
| 2Fe-2S Ferredoxinc | SML00003510 | EC818301 | ? | + | + | + | |
| Putative glycerol kinasec | SML00001707 | EC823814 | ? | + | − | + | |
| Fe-hydrogenase assembly | |||||||
| HydGb | SML00000047 | EF612762 | Yes | + | + | − | |
| HydFc | SML00000703 | EC826105 | ? | + | + | − | |
| Mitochondrial carrier family | |||||||
| Putative ATP/ADP Carrier (AAC)b | SML00000111 | EC824912 | No | + | + | − | |
| SML00000209 | EC825377 | ||||||
| Mitochondrial import pathway | |||||||
| Cpn60b | EU233795 | Yes | + | + | + | ||
| Hsp70b | EF612755 | Yes | + | + | + | ||
| Cpn10b | EF612758 | No | + | + | − | ||
| TOM34c | SML00000787 | EC825338 | ? | −/+ | − | − | |
| TIM 23c | SML00000812 | EC826107 | ? | + | − | − | |
| TIM 44c | SML00000628 | EC825535 | ? | + | − | − | |
| Mitochondrial respiratory complex I | |||||||
| 51 kDa (NuoF)b | EF612754 | Yes | + | + | − | ||
| 24 kDa (NuoE)c | SML00003413 | EC824286 | ? | + | + | − | |
| ISC pathway | |||||||
| IscSb | EF612756 | Yes | + | + | + | ||
| IscUb | EF612757 | ? | + | + | + | ||
| Frataxinc | SML00000479 | EC823663 | ? | + | + | + | |
| Fatty acid metabolism | |||||||
| Propionyl-CoA carboxylase alpha chainc | SML00000070 | EC826322 | ? | + | − | − | |
| Propionyl-CoA carboxylase beta chainc | SML00000244 | EC825818 | ? | + | − | − | |
| 3-Ketoacyl-CoA thiolasec | SML00000148 | EC826272 | + | − | − | ||
| Amino acid metabolism | |||||||
| Dihydrolipoamide dehydrogenase (L protein)b | SML00002324 | EF612759 | Yes | + | − | − | |
| Glycine cleavage system Hc | SML00003411 | EC819138 | ? | + | + | − | |
| Glycine cleavage system Pc | SML00003181 | EC821614 | ? | + | + | − | |
| 4-Aminobutyrate transaminasec | SML00000492 | EC825405 | ? | + | − | − | |
| Methylmalonyl-CoA mutasec | SML00000635 | EC825225 | ? | + | − | − | |
| Serine hydroxymethyltransferasec | SML00001624 | EC824484 | ? | + | + | − | |
| Branched chain ketoacid dehydrogenase E1, alpha polypeptidec | SML00000388 | EC826530 | ? | + | − | − | |
| SML00002275 | EC817751 | ||||||
| Branched-chain alpha-keto acid dehydrogenase E1-beta subunitc | SML00001068 | EC825728 | ? | + | − | − | |
| Lipoamide acyltransferase (branched-chain alpha-keto acid dehydrogenase complex) E2c | SML00000474 | EC826090 | ? | + | − | − | |
| Glutamate dehydrogenasec | SML00000006 | EC826618 | ? | + | + | + | |
| Alanine aminotransferasec | SML00000316 | EC825413 | ? | + | − | − | |
| Aspartate aminotransferasec | SML00001965 | EC817939 | ? | + | + | + | |
| Ornithine carbamoyltransferasec | SML00001073 | EC825834 | ? | −/+ | + | + | |
| Putative delta-1-pyrroline-5-carboxylate dehydrogenasec | SML00000009 | EC826523 | ? | + | − | − | |
| Putative glutaryl-CoA dehydrogenasec | SML00001691 | EC823693 | ? | + | − | − | |
| 2-Amino-3-ketobutyrate coenzyme A ligasec | SML00000023 | EC826214 | ? | + | − | − | |
| Other functions | |||||||
| Thioredoxin reductase 2c | SML00002472 | EC820868 | ? | + | + | + | |
| NADH-cytochrome b5 reductasec | SML00002452 | EC819124 | ? | + | − | − | |
| Succinate semialdehyde dehydrogenasec | SML00000871 | EC822494 | ? | −/+ | − | − | |
| Dynamin like proteinc | SML00000756 | EC822881 | ? | + | + | + | |
Hits confirmed by BLAST (E < 10−5) to NCBI and PFAM.
Phylogeny shown in Fig. 1 or the supplemental material.
Automatic annotation as obtained by the AnaBench pipeline (http://anabench.bcm.umontreal.ca/anabench/).
For those genes fully sequenced, NCBI accession numbers are provided in bold. For the rest, TBestDB accession numbers are provided for the contigs plus an additional NCBI accession number for a representative EST of the contig.
? indicates an incomplete sequence at the N terminus.
Ng, Naegleria gruberi; Tv, Trichomonas vaginalis; Gi, Giardia intestinalis. “−/+” indicates that a potential distant homolog of the protein was found but its assignment to the specific protein family named could not be confirmed by reciprocal BLAST.
RESULTS
PCR and EST survey.
From our expressed sequence tag (EST) survey of Sawyeria marylandensis, we obtained 9,300 sequences that were grouped into 2,672 unique clusters. We carried out a Basic Local Alignment Search Tool (BLAST) search of our cluster consensus sequences against human and yeast mitochondrial proteomes (3, 52), and the clusters that had strong hits in these databases were then used as queries in BLAST searches of the GenBank nonredundant (nr) database. In addition, a text-based gene/protein name search for known mitochondrion- and hydrogenosome-targeted proteins was carried out within the automatic annotation pipeline of TBestDB (38). Genes encoding other canonical mitochondrion- or hydrogenosome-targeted proteins (namely, [FeFe]-hydrogenase, PFO, chaperonin 60 [Cpn60], and cysteine desulfurase [IscS]) were directly amplified from cDNA by RT-PCR using degenerate primers. The results of our transcriptomic (EST) and degenerate PCR survey of Sawyeria marylandensis for genes whose products are typically targeted to mitochondria or hydrogenosomes are shown in Table 1. We found 43 genes that encoded proteins involved in energy metabolism, [FeFe]-hydrogenase assembly, mitochondrial import and carriers, subunits of mitochondrial respiratory complex I, components of the iron-sulfur cluster (ISC) pathway, and the metabolism of fatty acids and amino acids. Interestingly, the predicted partial proteome of the Sawyeria marylandensis mitochondrion-derived organelles based on this survey appears to contain a number of pathways/functions that are lacking in the hydrogenosomes of Trichomonas and the mitosomes of Giardia (Table 1). A search for homologs of these Sawyeria genes in the genome of the aerobic mitochondriate heteroloboseid amoeba Naegleria gruberi revealed a high degree of overlap in content (Table 1). This overlap includes a number of genes encoding enzymes involved in anaerobic “hydrogenosomal” metabolism, such as [FeFe]-hydrogenase and associated maturases, although a gene coding for the anaerobic enzyme pyruvate:ferredoxin oxidoreductase in Naegleria gruberi is apparently absent (Table 1). Other genes, such as those encoding a putative TOM34, ornithine carbamoyltransferase, and succinate semialdehyde dehydrogenase, had apparent distant homologs (possibly paralogs) in N. gruberi but were clearly more similar to other eukaryotic or bacterial homologs in the GenBank nonredundant database. Since none of these last three Sawyeria sequences had convincing organellar targeting peptides, their significance with respect to the function of its MROs remains unclear.
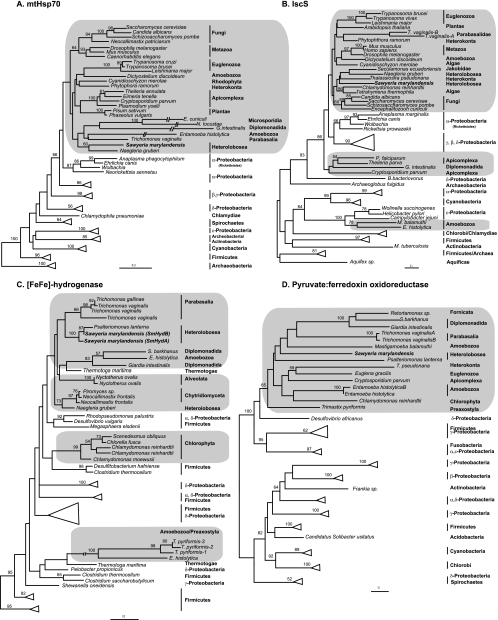
For those genes where the full coding sequence was obtained, we performed phylogenetic analyses. Figure 1 shows the estimated maximum likelihood (ML) trees of mitochondrial Hsp70, IscS, [FeFe]-hydrogenase, and PFO, while the remaining trees are presented in the supplemental material.
Fig. 1.
Maximum-likelihood (ML) phylogenetic trees of mitochondrial Hsp70 (A), cysteine desulfurase (IscS) (B), [FeFe]-hydrogenase (C), and pyruvate:ferredoxin oxidoreductase (D). Trees were constructed using RAxML with a WAG+Γ model of evolution. Bootstrap support values above 50 are shown over branches.
Like Trichomonas, Sawyeria expresses homologues of the characteristic hydrogenosomal marker proteins pyruvate:ferredoxin oxidoreductase (PFO) and [FeFe]-hydrogenase. Two genes coding for [FeFe]-hydrogenase have been identified. Both Sawyeria [FeFe]-hydrogenases are predicted to have the same protein module structure as Neocallimastix, Piromyces, and most Trichomonas copies (59); that is, they possess a [2Fe-2S] “plant ferredoxin-like” module, a His-ligated [4Fe-4S] module, 2[4Fe-4S] modules, and an H cluster. The two Sawyeria homologues (SmHydA and SmHydB) clearly group with the [FeFe]-hydrogenase of the anaerobic heteroloboseid Psalteriomonas lanterna (bootstrap value [BV] = 100% in the phylogenetic tree; Fig. 1C). SmHydB is more closely related to the heteroloboseid Psalteriomonas lanterna homologue than to the other Sawyeria copy (the SmHydA), suggesting that a gene duplication took place before Sawyeria and Psalteriomonas diverged from their last common ancestor. Further data on Psalteriomonas will be needed to confirm whether this organism encodes a second copy orthologous to SmHydA or whether it has secondarily lost it. Oddly, the Sawyeria/Psalteriomonas clade does not group specifically with the [FeFe]-hydrogenase homologue from their heteroloboseid sister Naegleria gruberi (which groups with anaerobic chytrid fungi instead), although all of the heteroloboseid homologs share the same protein module structure (not shown). Since the branches in the tree separating the two heteroloboseid groups have low bootstrap values, nothing definitive can be concluded from this branching pattern. As in previous analyses, eukaryotes as a whole do not appear as monophyletic (29). On the other hand, the PFO topology shows eukaryotes as a weakly supported (BV = 65%) monophyletic group, with Sawyeria and Psalteriomonas homologs branching off sequentially in a poorly resolved region of the tree (Fig. 1D).
At least two of the three genes encoding proteins involved in [FeFe]-hydrogenase assembly (HydG and HydF) were also detected as ESTs in Sawyeria. The HydG tree shows the five eukaryotic sequences forming a monophyletic group with moderate bootstrap support (BV = 71%) (see Fig. S4 in the supplemental material), with a Sawyeria-plus-Naegleria subgroup.
Sawyeria possesses other proteins presumably involved in energy metabolism, such as a 2Fe-2S ferredoxin, which, in trichomonad hydrogenosomes, transfers electrons from PFO to [FeFe]-hydrogenase, and the alpha and beta subunits of succinyl-coenzyme A (CoA) synthetase (SCS), the principle ATP-generating enzyme of hydrogenosomal energy metabolism (see reference 57). Both SCS alpha and beta subunit trees show all eukaryotes emerging as a group, with a monophyletic Heterolobosea (Sawyeria plus Naegleria) lineage occupying an unclear position within this group (see Fig. S1 and S2 in the supplemental material).
Also like that of Trichomonas, the Sawyeria genome encodes both the 51-kDa (NuoF) and 24-kDa (NuoE) subunits of the NADH dehydrogenase module in the mitochondrial respiratory complex I (NADH:ubiquinone oxidoreductase) (Table 1). The ML tree of 51-kDa NADH proteins (see Fig. S3 in the supplemental material) shows Sawyeria clustering strongly with a homologue from Psalteriomonas. This clade groups with Trichomonas as a sister to the main group of eukaryotes (including the Naegleria homolog) plus a subgroup of the alphaproteobacteria. To check if this is caused by phylogenetic artifacts (27), we also constructed a phylogenetic tree based on a recoding of the amino acids into functional categories (e.g., see reference 45). However, the resulting ML topology was the same as that of the nonrecoded analyses (data not shown).
Several proteins involved in protein import and maturation were detected. These include a homologue of the mitochondrial translocase of the outer membrane 34 (TOM34) and translocases of the inner membrane 23 and 44 (TIM23 and TIM44), all of which are part of the core mitochondrial protein import machinery that seems to have evolved early in eukaryotic evolution (14). Surprisingly, although we were able to identify a protein encoded by the N. gruberi genome that was distantly related to the Sawyeria TOM34 sequence, upon further database searches it became clear that this protein was more closely related to tetratricopeptide repeat (TPR)-containing proteins of other eukaryotes and was unlikely to be a member of the TOM34 family.
We also found a homolog of the mitochondrial heat shock protein 70 (mtHsp70), which is involved in protein import into the mitochondrial matrix, as well as chaperonins 60 and 10 (Cpn60 and Cpn10), which form the chaperonin complex that refolds imported mitochondrial proteins. The ML tree of mtHsp70 shows Sawyeria grouping with the other heteroloboseid sequence, that of Naegleria gruberi, with weak bootstrap support (BV = 60%) (Fig. 1A). In this tree, all eukaryotes form a weakly supported clade emerging from within the alphaproteobacteria as sisters to the Rickettsiales. Similarly, all eukaryotes form a strongly supported monophyletic group, with the alphaproteobacteria branching as their sister group in the Cpn60 tree (see Fig. S5 in the supplemental material). In this tree, Sawyeria groups with Naegleria and Psalteriomonas homologs as expected. The Cpn10 tree also shows a monophyletic group of eukaryotes in which Sawyeria and Naegleria homologs branch together (see Fig. S6).
We identified genes in the ESTs encoding proteins involved in Fe-S cluster assembly, including those encoding IscS, IscU, and frataxin. Sawyeria IscS groups with the heterolobosean Naegleria, the jakobid Seculamonas, and the dinoflagellate Thalassiosira within the eukaryotic clade, although with low statistical support (Fig. 1B). Again eukaryotes appear to emerge from within the alphaproteobacteria, weakly grouping with the Rickettsiales. The resolution in the IscU tree is poor, with low bootstrap support for many branches, and shows the eukaryotes as paraphyletic (see Fig. S7 in the supplemental material).
Interestingly, we have also found some of the same enzymes typically associated with amino acid metabolism that are apparently retained in the hydrogenosomes of Trichomonas or Neocallimastix. For example, Sawyeria appears to express a homolog of dihydrolipoamide dehydrogenase (L protein or PDH-E3). Dihydrolipoamide dehydrogenase is one of the three subunits of the pyruvate dehydrogenase complex and is also part of two multienzyme complexes of amino acid metabolism typically found in mitochondria: the glycine cleavage system complex and the branched-chain alpha-keto acid dehydrogenase. While we were unable to identify any other subunit of the PDH complex, we did detect homologs of all of the putative subunits of the glycine cleavage system complex and all of the subunits of the branched-chain alpha-keto acid dehydrogenase complex (Table 1). The phylogeny of this protein family again shows eukaryote mitochondrion-targeted sequences as monophyletic, with the Sawyeria homolog emerging within the group (see Fig. S8 in the supplemental material).
Finally, both mitochondria and hydrogenosomes export the ATP they produce to the cytosol via an ADP/ATP carrier (AAC), a unique eukaryotic protein not found in prokaryotes that is part of a larger family of mitochondrial carrier proteins. Our search for AAC identified two putative AACs in Sawyeria. The ML tree shows both copies, plus a homolog from Psalteriomonas lanterna, as the sister group to the ATP-ADP carriers (with BV equal to 89%), occupying a pivotal position between AAC and other mitochondrial carrier proteins (see Fig. S9 in the supplemental material). The Sawyeria AAC-B paralog strongly grouped with the Psalteriomonas homolog, again indicating that a possible ancestral gene duplication event has occurred within the anaerobic heteroloboseid genome. The distant relationship between these proteins and the AAC homolog from Naegleria (which emerges as part of the canonical eukaryotic mitochondrial clade) seems to indicate a distinct ancestry.
Predicted organellar targeting peptides.
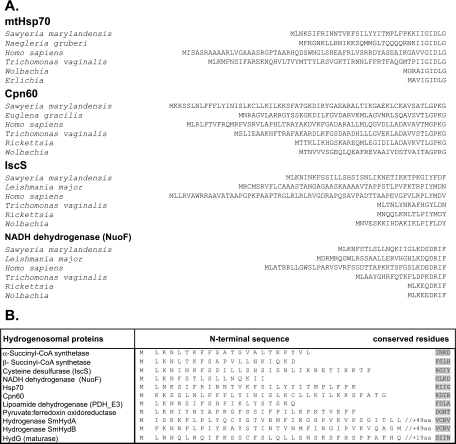
Manual alignment of the amino-terminal ends of Sawyeria proteins with those of other eukaryotes and bacterial homologues revealed the presence of extensions (of around 15 to 28 amino acids long) compared to bacterial sequences (Table 1 and Fig. 2). Of all these proteins, just two (Hsp70 and PFO) are predicted to possess an N-terminal mitochondrial targeting peptide using the targeting peptide prediction software (see Materials and Methods). In fact, arginines, which are suggested to be important for organelle import (47), are generally missing in the apparent MRO targeting peptides of Sawyeria. Given the evolutionary divergence of Sawyeria from organisms whose targeting peptides have been used to train targeting peptide prediction software tools (e.g., Saccharomyces, humans etc), it is not surprising that bona fide targeting peptides are not always recognized by these tools. Based on comparisons with other proteins, we observed that putative N-terminal targeting peptides for Sawyeria proteins most often start with a consensus motif of ML[K,N][K,N] (Fig. 2B). However, cleavable N-terminal extensions are probably not the only signals by which proteins are targeted and subsequently imported into the Sawyeria MROs, as has recently been shown for Trichomonas (33) and is well known for other mitochondria (14).
Fig. 2.
Comparison of the N termini of several different protein homologues from Sawyeria and several representative eukaryote sequences, showing the putative targeting peptides. Aligned bacterial homologues, which do not contain targeting peptides, are also shown. Most of the putative targeting peptides of Sawyeria were not predicted by automated software (see Materials and Methods and the main text).
Morphological characterization of Sawyeria MROs.
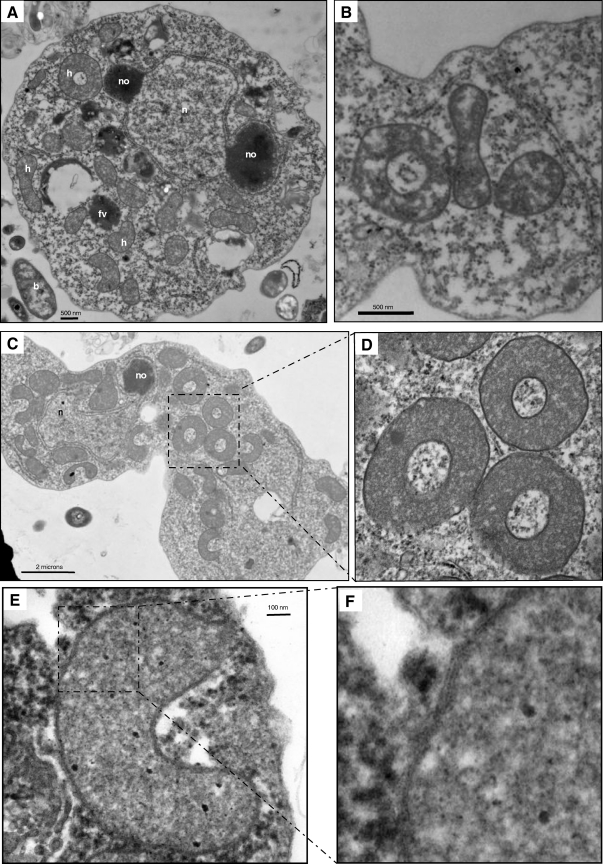
Sections from Sawyeria marylandensis cells examined in transmission electron micrographs revealed the presence of abundant densely stained organelles in the cytoplasm (Fig. 3 A to F). These organelles measure ∼1 to 1.5 μm, are clearly bounded by two membranes (Fig. 3F), and are likely to be MROs. They are not, however, surrounded by rough endoplasmic reticulum (RER), as has been described for the MROs of Psalteriomonas lanterna and the mitochondria of other members of the Heterolobosea (40). We detected two different morphologies of Sawyeria MROs. Some of the organelles appeared bean shaped, as previously described (39), and others presented as round bodies with central holes, while others were cup shaped (see Fig. 3B and D for a view of different shapes of the hydrogenosome).
Fig. 3.
Transmission electron micrographs of Sawyeria marylandensis. (A) Full view of a Sawyeria marylandensis cell. (B) Partial view of an S. marylandensis cell showing the three different shapes of the mitochondrion-related organelle. (C) S. marylandensis cell with several hydrogenosomes. (D) An inset view of picture C. (E) View of a hydrogenosome of S. marylandensis. (F) Close view showing double-membrane structure of the organelle.
The three-dimensional (3D) reconstructions of the Sawyeria MROs using serial transmission electron microscope sections confirmed the two different shapes (Fig. 4). Both types harbor an invagination, which forms a cavity. The bean-shaped MRO is about 1.8 μm tall, 1.5 μm long, and 0.840 μm wide. The cup-shaped MRO is 1.5 μm tall and 1.65 μm wide. Both MRO types display a nonhomogenous and moderately electron-dense interior. Within the nonhomogenous content, there are a few whitish compartments with diffuse borders that resemble vesicles. These vesicle-like protuberances typically contain one gray spot at the periphery or in the middle and a stack of gray filaments. The cup-shaped MROs can possess up to three vesicle-like protuberances surrounding them, while the bean-shaped organelles typically have only one of these buds, which is always located at the extreme end of the structure.
Fig. 4.
3D reconstruction of shape of Sawyeria hydrogenosomes derived from serial sectioning transmission electron microscopy. The bean-shaped MRO is about 1.8 μm tall, 1.5 μm long, and 0.840 μm wide. The cup-shaped MRO is 1.5 μm tall and 1.65 μm wide. See the main text for more details.
DISCUSSION
Our EST and PCR surveys have yielded a number of genes that encode proteins typical of mitochondria and hydrogenosomes, with putative N-terminal targeting sequences that likely direct them to the MROs of Sawyeria marylandensis. We hypothesize that these organelles are in fact hydrogenosomes with a novel proteome and biochemical functions, although biochemical studies of hydrogen production for these organelles will be needed to test this hypothesis. Phylogenetic analyses of the mitochondrial import system components (Cpn60 and mtHsp70), ISC system components, and other proteins unequivocally suggest that the Sawyeria marylandensis hydrogenosomes are related to those of Psalteriomonas lanterna and the mitochondria of Naegleria gruberi and thus must have evolved from ancestral heteroloboseid mitochondria.
Energy metabolism in Sawyeria MROs.
In most aerobic eukaryotes, conversion of pyruvate to acetyl-CoA occurs in mitochondria via the PDH enzyme complex. In anaerobic protists, this conversion is typically catalyzed by PFO and/or by a pyruvate:formate lyase (PFL). PFO transfers the electrons generated in the process to the ferredoxin protein, and a [FeFe]-hydrogenase enzyme is responsible for reoxidizing the reduced ferredoxin and coupling this with the reduction of protons to form H2 gas (28). While PFO and [FeFe]-hydrogenase activities are thought not to be associated with the mitosomes in Giardia and Entamoeba (31, 44, 56), they occur in the hydrogenosomes of Trichomonas (36). Recent studies have found homologs of these two enzymes in the anaerobes Mastigamoeba balamuthi (21), Blastocystis (51), and Trimastix pyriformis (25). A recent survey (29) suggests both of these enzymes are not only present in anaerobic taxa but are patchily distributed across the eukaryotic tree and are present in both aerobic and anaerobic lineages. Our finding of two copies of genes encoding [FeFe]-hydrogenase, genes for two of the three hydrogenase naturase proteins, and a single gene for pyruvate:ferredoxin oxidoreductase (PFO) that, where known, possess an N-terminal mitochondrial/hydrogenosomal targeting peptide (HydE is a partial sequence) in Sawyeria marylandensis suggests this organism truly possesses hydrogenosomes. The lack of an obvious phylogenetic affinity of Sawyeria/Psalteriomonas [FeFe]-hydrogenases to homologs recently identified in Naegleria (19) makes it difficult to determine how old this protein is within the heteroloboseid lineage.
Of the three described hydrogenase maturases, we have identified two (HydG and HydF) (Table 1) in Sawyeria. Since ESTs are only a partial representation of the transcriptome, it is possible that HydE is also a part of the Sawyeria proteome that has yet to be detected. HydG, HydF, and HydE have been shown to be necessary for H-cluster maturation and have been exclusively found in bacteria containing [FeFe]-hydrogenase and, recently, in Chlamydomonas reinhardtii, Trichomonas vaginalis, and Trimastix pyriformis (25, 41–43). Some of these proteins have also recently been found in aerobic protists (29), including the heterolobosean Naegleria gruberi, which has all three maturases (19), suggesting a fairly ancient origin of these proteins within the heterolobosea.
Sawyeria also expresses the two subunits (alpha and beta) of the succinyl-CoA synthetase (SCS), which typically catalyzes substrate-level phosphorylation that converts succinyl-CoA to succinate and synthesizes ATP. This enzyme is part of the tricarboxylic acid cycle in aerobic mitochondria and is the main ATP-generating enzyme in anaerobic energy metabolism of hydrogenosomes. Both SCS alpha and beta subunits have putative N-terminal extensions that again suggest that they are located in the MROs, and both are related to presumably mitochondrion-targeted homologs in Naegleria gruberi. In the absence of a canonical mitochondrial oxidative phosphorylation pathway, the SCS enzyme is likely an important source of ATP for Sawyeria. In hydrogenosomes and some anaerobic mitochondria, the acetyl-CoA generated by PFO is converted to succinyl-CoA (the substrate for the SCS enzyme) via an acetate:succinate CoA transferase (ASCT) (36, 53, 58). Although we did not detect any ASCT homologs in our survey, we suspect an enzyme with this activity exists in Sawyeria marylandensis MROs, which would complete the hydrogenosomal energy generation pathway. Finally, although we detected what may be an E3 subunit of the PDH complex, in the absence of other PDH subunits, we suspect this protein instead is part of the amino acid metabolic systems described below.
NADH dehydrogenase subunits.
In Trichomonas, both the 51-kDa (NuoF) and 24-kDa (NuoE) subunits of the NADH dehydrogenase module in mitochondrial respiratory complex I (NADH:ubiquinone oxidoreductase) have been shown to be located in hydrogenosomes, possibly functioning to oxidize the NADH produced by the hydrogenosomal malic enzyme and transfer the electrons to ferredoxin, which is in turn oxidized by [FeFe]-hydrogenase (27). Sawyeria also encodes the NuoF and NuoE proteins, which could function in exactly the same way as their homologs in Trichomonas hydrogenosomes. Our phylogenetic analyses of NuoF show that Sawyeria, Psalteriomonas, and Trichomonas sequences are basal to all other eukaryotic homologues (see Fig. S3 in the supplemental material), and in contrast to the findings of Hrdy et al. (27), a phylogenetic analysis of amino acids grouped into functional categories did not change the optimal topology inferred. However, we cannot absolutely reject the possibility that our topology is due to a phylogenetic artifact, and further investigation of this issue is beyond the scope of this article. Interestingly, Sawyeria NuoF appears to have an N-terminal extension, again consistent with a hydrogenosomal localization and function in this organism.
Iron-sulfur cluster biogenesis.
Mitochondria play a vital role in Fe-S cluster assembly in eukaryotes (30). This is one of the most conserved functions in mitochondria and mitochondrion-related organelles (55), although in Entamoeba and Mastigamoeba, ISC has apparently been replaced, by lateral gene transfer, with epsilonproteobacterium-related homologues of NifS and NifU (1, 21). Sawyeria has canonical IscS and IscU proteins with a putative N-terminal targeting peptide (Fig. 2) that group within the eukaryotes in respective phylogenetic trees (Fig. 1B; see also Fig. S7 in the supplemental material). Our survey has also detected frataxin, another key protein in iron-sulfur biogenesis that was also recently found within trichomonad hydrogenosomes (13). So far, it appears that frataxin homologs are absent from both the mitosomes of Giardia and Entamoeba (13).
Amino acid metabolism.
Recently, some evidence for amino acid metabolism in hydrogenosomes was found. In Trichomonas, the serine hydroxymethyl transferase and two proteins of the glycine decarboxylase complex, the glycine cleavage H protein and dihydrolipoamide dehydrogenase (L protein), have been demonstrated to localize in hydrogenosomes (35). Moreover, the ornithine transcarbamoylase, a mitochondrial enzyme of the arginine biosynthesis pathway, seems to localize in the hydrogenosomes of Neocallimastix frontalis (20).
The L protein, in other organisms, is a subunit of several mitochondrial 2-oxo-acid dehydrogenase complexes: PDH, the glycine cleavage system, and the branched-chain alpha-keto acid dehydrogenase. This protein carries out a flavin-mediated oxidation of the dihydrolipoyl groups of other enzymes in the complex to lipoate and ultimately transfers the electrons to NAD+ to produce NADH. Since we found all other subunits of the glycine cleavage system (GCS) and branched-chain alpha-keto acid dehydrogenase complexes but did not find any other PDH subunits (Table 1), it is likely that this protein is involved in the two former functions but not the latter.
In mitochondria, GCS functions to methylenate one carbon transferring cofactor, N5,N10-methylenetetrahydrofolate (CH2-THF), using glycine as a substrate and reducing NAD+ in the process (15). The CH2-THF generated by the mitochondrial glycine cleavage system is used in the methylenation and subsequent hydroxylation of glycine to form serine by serine hydroxymethyl transferase (SHMT) (46), a homolog of which we also found in S. marylandensis (Table 1). Note that these findings are in contrast to those for Trichomonas, where so far only two members of the glycine cleavage system (the L and H proteins) have been identified (35).
The branched-chain alpha-keto acid dehydrogenase (BCKDH) functions in the degradation of branched-chain amino acids, such as valine, isoleucine, and leucine, which ultimately will yield acyl-CoA and carbon dioxide. S. marylandensis seems to have the two subunits of the BCKDH and the dihydrolipoamide dehydrogenase, which together may function as part of the multisubunit branched-chain alpha-keto acid dehydrogenase complex. In any case, regardless of its precise function, the phylogeny shows Sawyeria dihydrolipoamide dehydrogenase grouping with the rest of eukaryotic mitochondrial homologs (see Fig. S8 in the supplemental material).
Protein import system.
Since most mitochondrial proteins are encoded in the nucleus, a protein import system that ensures their delivery into the organelle is an important feature of eukaryotes. Core components of the protein import machinery may be present in all eukaryotes, although with some differences (8, 14). Some of those components, namely, the TOM and TIM 23 complexes, do not have counterparts in bacteria and may be de novo acquisitions of the eukaryotic lineage (14) (although see reference 11 for putative homologs in proteobacteria of some components of the TIM 23 complex). Sawyeria appears to express a gene similar to that encoding the mitochondrial translocase of the outer membrane 34 (TOM34), which is a subunit of the TOM complex, as well as translocases of the inner membrane 23 and 44 (TIM23 and TIM44), which are both subunits of the TIM23 complex. Surprisingly, although we found orthologs of TIM23 and TIM44 encoded in the Naegleria gruberi genome, we were unable to confidently identify an ortholog of TOM34.
Protein import into the organelle also requires mtHsp70, and once the proteins have been imported into mitochondria, they require refolding into their native structures by the chaperonin complex formed by the Cpn60 and Cpn10 subunits. S. marylandensis expresses all three of these proteins, which are indeed quite ubiquitous among mitochondria and MROs, except in the Microsporidia (5). The phylogenetic trees (Fig. 1A; see also Fig. S5 and S6 in the supplemental material) of these proteins place Sawyeria with the rest of the eukaryotic mitochondria/hydrogenosomal homologues, although with different specific affiliations depending on the protein. In any case, a mitochondrial ancestry is clearly supported in all three cases.
Transport of other molecules.
Proteins from the mitochondrial carrier family are involved in the bidirectional transport of metabolites, nucleotides, amino acids, cofactors, carboxylic acids, and inorganic anions across the inner membrane of the organelle. The two Sawyeria carriers appear, along with a homolog in Psalteriomonas, to emerge in an intermediate position between ATP/ADP carriers (AAC) and other mitochondrial carriers (see Fig. S9 in the supplemental material), so we cannot confidently assign them to any of the members of this diverse family of proteins.
The Sawyeria marylandensis hydrogenosome has a novel shape.
Our data convincingly show for the first time that the S. marylandensis hydrogenosomes have a double membrane (Fig. 3) and often adopt a cup shape. Since we did not observe ribosomes inside of the organelle, and given its “stripped-down” hydrogenosome-like metabolic properties, it is likely that the organelle does not possess a genome. This is in agreement with the fact that we never observed an organelle signal in fluorescence microscopy using the DNA-specific dye 4′,6-diamidino-2-phenylindole (DAPI) (data not shown). The morphology of S. marylandensis hydrogenosomes is more complex than originally described (39), with two different shapes (Fig. 3 and 4). The reason for the different morphologies and the biological implications of these shapes are not yet understood. It may well be that different morphologies are due to the different metabolic conditions within different parts of these cells.
Conclusions.
We have shown that the microaerophilic amoeba Sawyeria marylandensis represents a new case of a protist that likely harbors a hydrogenosome with a unique combination of biochemical pathways, although biochemical studies would be helpful to confirm or refute the predicted metabolic capacities. The fact that this organism (as well as its sister, Psalteriomonas) seems likely to possess the canonical “hydrogenosomal”-type energy metabolism but is not closely related to other hydrogenosome-containing groups, such as parabasalids, chytrid fungi, and ciliates, indicates that this kind of metabolism is either a retained ancestral feature of some or all of these eukaryotes or represents more-recent convergent adaptations to a biochemically “optimal” state (or some combination of the two scenarios; see Hug et al. (2010) for further discussion [29]). In either case, the presence of enzymes involved in mitochondrial pathways such as branched-chain amino acid degradation that are not known to exist in other well-studied hydrogenosomes (e.g., those of Trichomonas vaginalis) is consistent with findings of recent studies (21, 25, 51) suggesting a wide spectrum of functions exist in MROs across eukaryotic diversity. The presence of some “hydrogenosomal” enzymes that are likely targeted to the mitochondria of Naegleria gruberi, an aerobic relative of Sawyeria marylandensis and Psalteriomonas lanterna, opens new possibilities for comparative studies within the Heterolobosea aimed at understanding the evolutionary transition between mitochondria and hydrogenosomes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the grant MOP-62809, awarded to A.J.R. from the Canadian Institutes of Health Research (CIHR). I.R.-T. was supported by a CIHR postdoctoral fellowship and by an ICREA contract.
We thank Jacquie de Mestral and Alastair G. Simpson for technical help. We thank Martin Kolisko, Alex Stechmann, Bill Martin, and Tudor Borza for helpful discussions. We also thank Mariona Ruiz-Barberà, Martí Ruiz-Barberà, and Olivia Ruiz-Barberà for their continuous assistance.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 29 October 2010.
REFERENCES
- 1.Ali V., Shigeta Y., Tokumoto U., Takahashi Y., Nozaki T. 2004. An intestinal parasitic protist, Entamoeba histolytica, possesses a non-redundant nitrogen fixation-like system for iron-sulfur cluster assembly under anaerobic conditions. J. Biol. Chem. 279:16863–16874 [DOI] [PubMed] [Google Scholar]
- 2.Anderson I. J., Loftus B. J. 2005. Entamoeba histolytica: observations on metabolism based on the genome sequence. Exp. Parasitol. 110:173–177 [DOI] [PubMed] [Google Scholar]
- 3.Andreoli C., Prokisch H., Hortnagel K., Mueller J. C., Munsterkotter M., Scharfe C., Meitinger T. 2004. MitoP2, an integrated database on mitochondrial proteins in yeast and man. Nucleic Acids Res. 32:D459–D462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badidi E., De Sousa C., Lang B. F., Burger G. 2003. AnaBench: a Web/CORBA-based workbench for biomolecular sequence analysis. BMC Bioinform. 4:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barberà M. J., Ruiz-Trillo I., Leigh J., Hug L. A., Roger A. J. 2007. The diversity of mitochondrion-related organelles amongst eukaryotic microbes, p. 239–275 InMartin W. F., Müller M. (ed.), Origin of mitochondria and hydrogenosomes. Springer-Verlag, Berlin, Germany [Google Scholar]
- 6.Biagini G. A., Bernard C. 2000. Primitive anaerobic protozoa: a false concept? Microbiology 146:1019–1020 [DOI] [PubMed] [Google Scholar]
- 7.Boxma B., de Graaf R. M., van der Staay G. W., van Alen T. A., Ricard G., Gabaldon T., van Hoek A. H., Moon-van der Staay S. Y., Koopman W. J., van Hellemond J. J., Tielens A. G., Friedrich T., Veenhuis M., Huynen M. A., Hackstein J. H. 2005. An anaerobic mitochondrion that produces hydrogen. Nature 434:74–79 [DOI] [PubMed] [Google Scholar]
- 8.Burri L., Williams B. A., Bursac D., Lithgow T., Keeling P. J. 2006. Microsporidian mitosomes retain elements of the general mitochondrial targeting system. Proc. Natl. Acad. Sci. U. S. A. 103:15916–15920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlton J. M., Hirt R. P., Silva J. C., Delcher A. L., Schatz M., Zhao Q., Wortman J. R., Bidwell S. L., Alsmark U. C., Besteiro S., Sicheritz-Ponten T., Noel C. J., Dacks J. B., Foster P. G., Simillion C., Van de Peer Y., Miranda-Saavedra D., Barton G. J., Westrop G. D., Muller S., Dessi D., Fiori P. L., Ren Q., Paulsen I., Zhang H., Bastida-Corcuera F. D., Simoes-Barbosa A., Brown M. T., Hayes R. D., Mukherjee M., Okumura C. Y., Schneider R., Smith A. J., Vanacova S., Villalvazo M., Haas B. J., Pertea M., Feldblyum T. V., Utterback T. R., Shu C. L., Osoegawa K., de Jong P. J., Hrdy I., Horvathova L., Zubacova Z., Dolezal P., Malik S. B., Logsdon J. M. J., Henze K., Gupta A., Wang C. C., Dunne R. L., Upcroft J. A., Upcroft P., White O., Salzberg S. L., Tang P., Chiu C. H., Lee Y. S., Embley T. M., Coombs G. H., Mottram J. C., Tachezy J., Fraser-Liggett C. M., Johnson P. J. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claros M. G. 1995. MitoProt, a Macintosh application for studying mitochondrial proteins. Comput. Appl. Biosci. 11:441–447 [DOI] [PubMed] [Google Scholar]
- 11.Clements A., Bursac D., Gatsos X., Perry A. J., Civciristov S., Celik N., Likic V. A., Poggio S., Jacobs-Wagner C., Strugnell R. A., Lithgow T. 2009. The reducible complexity of a mitochondrial molecular machine. Proc. Natl. Acad. Sci. U. S. A. 106:15791–15795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Graaf R. M., Duarte I., van Alen T. A., Kuiper J. W., Schotanus K., Rosenberg J., Huynen M. A., Hackstein J. H. 2009. The hydrogenosomes of Psalteriomonas lanterna. BMC Evol. Biol. 9:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolezal P., Dancis A., Lesuisse E., Sutak R., Hrdy I., Embley T. M., Tachezy J. 2007. Frataxin, a conserved mitochondrial protein, in the hydrogenosome of Trichomonas vaginalis. Eukaryot. Cell 6:1431–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolezal P., Likic V., Tachezy J., Lithgow T. 2006. Evolution of the molecular machines for protein import into mitochondria. Science 313:314–318 [DOI] [PubMed] [Google Scholar]
- 15.Douce R., Bourguignon J., Neuburger M., Rebeille F. 2001. The glycine decarboxylase system: a fascinating complex. Trends Plant Sci. 6:167–176 [DOI] [PubMed] [Google Scholar]
- 16.Embley T. M., Martin W. 2006. Eukaryotic evolution, changes and challenges. Nature 440:623–630 [DOI] [PubMed] [Google Scholar]
- 17.Embley T. M. 2006. Multiple secondary origins of the anaerobic lifestyle in eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ewing B., Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 19.Fritz-Laylin L. K., Prochnik S. E., Ginger M. L., Dacks J. B., Carpenter M. L., Field M. C., Kuo A., Paredez A., Chapman J., Pham J., Shu S., Neupane R., Cipriano M., Mancuso J., Tu H., Salamov A., Lindquist E., Shapiro H., Lucas S., Grigoriev I. V., Cande W. Z., Fulton C., Rokhsar D. S., Dawson S. C. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140:631–642 [DOI] [PubMed] [Google Scholar]
- 20.Gelius-Dietrich G., Ter Braak M., Henze K. 2007. Mitochondrial steps of arginine biosynthesis are conserved in the hydrogenosomes of the chytridiomycete Neocallimastix frontalis. J. Eukaryot. Microbiol. 54:42–44 [DOI] [PubMed] [Google Scholar]
- 21.Gill E. E., Diaz-Trivino S., Barbera M. J., Silberman J. D., Stechmann A., Gaston D., Tamas I., Roger A. J. 2007. Novel mitochondrion-related organelles in the anaerobic amoeba Mastigamoeba balamuthi. Mol. Microbiol. 66:1306–1320 [DOI] [PubMed] [Google Scholar]
- 22.Goldberg A. V., Molik S., Tsaousis A. D., Neumann K., Kuhnke G., Delbac F., Vivares C. P., Hirt R. P., Lill R., Embley T. M. 2008. Localization and functionality of microsporidian iron-sulphur cluster assembly proteins. Nature 452:624–628 [DOI] [PubMed] [Google Scholar]
- 23.Hackstein J. H. P., Tjaden J., Koopman W., Huynen M. 2007. Hydrogenosomes (and related organelles, either) are not the same, p. 135–159 InMartin W. F., Müller M. (ed.), Origin of mitochondria and hydrogenosomes. Springer-Verlag, Berlin, Germany [Google Scholar]
- 24.Hampl V., Hug L., Leigh J. W., Dacks J. B., Lang B. F., Simpson A. G., Roger A. J. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups.” Proc. Natl. Acad. Sci. U. S. A. 106:3859–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hampl V., Silberman J. D., Stechmann A., Diaz-Trivino S., Johnson P. J., Roger A. J. 2008. Genetic evidence for a mitochondriate ancestry in the ‘amitochondriate’ flagellate trimastix pyriformis. PLoS One 3:e1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hampl V., Simpson A. G. B. 2008. Possible mitochondria-related organelles in poorly-studied “amitochondriate” eukaryotes, p. 265–282 InTachezy J. (ed.), Hydrogenosomes and mitosomes: mitochondria of anaerobic eukaryotes. Springer Science+Business Media, Berlin, Germany [Google Scholar]
- 27.Hrdy I., Hirt R. P., Dolezal P., Bardonova L., Foster P. G., Tachezy J., Embley T. M. 2004. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature 432:618–622 [DOI] [PubMed] [Google Scholar]
- 28.Hrdy I., Muller M. 1995. Primary structure and eubacterial relationships of the pyruvate:ferredoxin oxidoreductase of the amitochondriate eukaryote Trichomonas vaginalis. J. Mol. Evol. 41:388–396 [PubMed] [Google Scholar]
- 29.Hug L. A., Stechmann A., Roger A. J. 2010. Phylogenetic distributions and histories of proteins involved in anaerobic pyruvate metabolism in eukaryotes. Mol. Biol. Evol. 27:311–324 [DOI] [PubMed] [Google Scholar]
- 30.Lill R., Muhlenhoff U. 2005. Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 30:133–141 [DOI] [PubMed] [Google Scholar]
- 31.Lloyd D., Ralphs J. R., Harris J. C. 2002. Giardia intestinalis, a eukaryote without hydrogenosomes, produces hydrogen. Microbiology (Reading, England) 148:727–733 [DOI] [PubMed] [Google Scholar]
- 32.Loftus B., Anderson I., Davies R., Alsmark U. C., Samuelson J., Amedeo P., Roncaglia P., Berriman M., Hirt R. P., Mann B. J., Nozaki T., Suh B., Pop M., Duchene M., Ackers J., Tannich E., Leippe M., Hofer M., Bruchhaus I., Willhoeft U., Bhattacharya A., Chillingworth T., Churcher C., Hance Z., Harris B., Harris D., Jagels K., Moule S., Mungall K., Ormond D., Squares R., Whitehead S., Quail M. A., Rabbinowitsch E., Norbertczak H., Price C., Wang Z., Guillen N., Gilchrist C., Stroup S. E., Bhattacharya S., Lohia A., Foster P. G., Sicheritz-Ponten T., Weber C., Singh U., Mukherjee C., El-Sayed N. M., Petri W. A., Jr., Clark C. G., Embley T. M., Barrell B., Fraser C. M., Hall N. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433:865–868 [DOI] [PubMed] [Google Scholar]
- 33.Mentel M., Zimorski V., Haferkamp P., Martin W., Henze K. 2008. Protein import into hydrogenosomes of Trichomonas vaginalis involves both N-terminal and internal targeting signals: a case study of thioredoxin reductases. Eukaryot. Cell 7:1750–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miichi F., Abu Yousuf M., Nakada-Tsukui K., Nozaki T. 2009. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc. Natl. Acad. Sci. U. S. A. 106:21731–21736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukherjee M., Brown M. T., McArthur A. G., Johnson P. J. 2006. Proteins of the glycine decarboxylase complex in the hydrogenosome of Trichomonas vaginalis. Eukaryot. Cell 5:2062–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller M. 1993. The hydrogenosome. J. Gen. Microbiol. 139:2879–2889 [DOI] [PubMed] [Google Scholar]
- 37.Nakai K., Horton P. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34–36 [DOI] [PubMed] [Google Scholar]
- 38.O'brien E. A., Koski L. B., Zhang Y., Yang L., Wang E., Gray M. W., Burger G., Lang B. F. 2007. TBestDB: a taxonomically broad database of expressed sequence tags (ESTs). Nucleic Acids Res. 35:D445–D451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Kelly C. J., Silberman J. D., Amaral Zettler L. A., Nerad T. A., Sogin M. L. 2003. Monopylocystis visvesvarai n. gen., n. sp. and Sawyeria marylandensis n. gen., n. sp.: two new amitochondrial heterolobosean amoebae from anoxic environments. Protist 154:281–290 [DOI] [PubMed] [Google Scholar]
- 40.Page F. C., Blanton R. L. 1985. The heterolobosea(Sarcodina: Rhizopoda), a new class uniting the Schizopyrenida and the Acrasidae(Acrasida). Protistologica 21:121–132 [Google Scholar]
- 41.Posewitz M. C., King P. W., Smolinski S. L., Smith R. D., Ginley A. R., Ghirardi M. L., Seibert M. 2005. Identification of genes required for hydrogenase activity in Chlamydomonas reinhardtii. Biochem. Soc Trans. 33:102–104 [DOI] [PubMed] [Google Scholar]
- 42.Posewitz M. C., King P. W., Smolinski S. L., Zhang L., Seibert M., Ghirardi M. L. 2004. Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J. Biol. Chem. 279:25711–25720 [DOI] [PubMed] [Google Scholar]
- 43.Putz S., Dolezal P., Gelius-Dietrich G., Bohacova L., Tachezy J., Henze K. 2006. Fe-hydrogenase maturases in the hydrogenosomes of Trichomonas vaginalis. Eukaryot. Cell 5:579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reeves R. E., Warren L. G., Susskind B., Lo H. S. 1977. An energy-conserving pyruvate-to-acetate pathway in Entamoeba histolytica. Pyruvate synthase and a new acetate thiokinase. J. Biol. Chem. 252:726–731 [PubMed] [Google Scholar]
- 45.Rodriguez-Ezpeleta N., Brinkmann H., Roure B., Lartillot N., Lang B. F., Philippe H. 2007. Detecting and overcoming systematic errors in genome-scale phylogenies. Syst. Biol. 56:389–399 [DOI] [PubMed] [Google Scholar]
- 46.Schirch V., Szebenyi D. M. 2005. Serine hydroxymethyltransferase revisited. Curr. Opin. Chem. Biol. 9:482–487 [DOI] [PubMed] [Google Scholar]
- 47.Schneider G., Sjoling S., Wallin E., Wrede P., Glaser E., von Heijne G. 1998. Feature-extraction from endopeptidase cleavage sites in mitochondrial targeting peptides. Proteins 30:49–60 [PubMed] [Google Scholar]
- 48.Silberman J. D., Simpson A. G., Kulda J., Cepicka I., Hampl V., Johnson P. J., Roger A. J. 2002. Retortamonad flagellates are closely related to diplomonads—implications for the history of mitochondrial function in eukaryote evolution. Mol. Biol. Evol. 19:777–786 [DOI] [PubMed] [Google Scholar]
- 49.Smith S. W., Overbeek R., Woese C. R., Gilbert W., Gillevet P. M. 1994. The genetic data environment an expandable GUI for multiple sequence analysis. Comput. Appl. Biosci. 10:671–675 [DOI] [PubMed] [Google Scholar]
- 50.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 51.Stechmann A., Hamblin K., Perez-Brocal V., Gaston D., Richmond G. S., van der Giezen M., Clark C. G., Roger A. J. 2008. Organelles in blastocystis that blur the distinction between mitochondria and hydrogenosomes. Curr. Biol. 18:580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor S. W., Fahy E., Zhang B., Glenn G. M., Warnock D. E., Wiley S., Murphy A. N., Gaucher S. P., Capaldi R. A., Gibson B. W., Ghosh S. S. 2003. Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 21:281–286 [DOI] [PubMed] [Google Scholar]
- 53.Tielens A. G., van Grinsven K. W., Henze K., van Hellemond J. J., Martin W. 2010. Acetate formation in the energy metabolism of parasitic helminths and protists. Int. J. Parasitol. 40:387–397 [DOI] [PubMed] [Google Scholar]
- 54.Tovar J. 2007. Mitosomes of parasitic protozoa: biology and evolutionary significance, p. 276–300 InMartin W. F., Müller M. (ed.), Origin of mitochondria and hydrogenosomes. Springer-Verlag, Berlin, Germany [Google Scholar]
- 55.Tovar J., Leon-Avila G., Sanchez L. B., Sutak R., Tachezy J., van der Giezen M., Hernandez M., Muller M., Lucocq J. M. 2003. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature 426:172–176 [DOI] [PubMed] [Google Scholar]
- 56.Townson S. M., Upcroft J. A., Upcroft P. 1996. Characterisation and purification of pyruvate:ferredoxin oxidoreductase from Giardia duodenalis. Mol. Biochem. Parasitol. 79:183–193 [DOI] [PubMed] [Google Scholar]
- 57.van der Giezen M., Tovar J., Clark C. G. 2005. Mitochondrion-derived organelles in protists and fungi. Int. Rev. Cytol. 244:175–225 [DOI] [PubMed] [Google Scholar]
- 58.van Grinsven K. W., Rosnowsky S., van Weelden S. W., Putz S., van der Giezen M., Martin W., van Hellemond J. J., Tielens A. G., Henze K. 2008. Acetate:succinate CoA-transferase in the hydrogenosomes of Trichomonas vaginalis: identification and characterization. J. Biol. Chem. 283:1411–1418 [DOI] [PubMed] [Google Scholar]
- 59.Voncken F. G., Boxma B., van Hoek A. H., Akhmanova A. S., Vogels G. D., Huynen M., Veenhuis M., Hackstein J. H. 2002. A hydrogenosomal [Fe]-hydrogenase from the anaerobic chytrid Neocallimastix sp. L2. Gene 284:103–112 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.