Introduction
Microtubules are the principle scaffold of the mitotic spindle, serve as tracks for intracellular transport of proteins and mRNAs, and also participate in signaling functions. The repeating subunit of the microtubule is the α/β tubulin heterodimer, which polymerizes in a head-to-tail fashion to form protofilaments; typically ~13 protofilaments associate laterally to form the microtubules seen in vivo. Due to the head-to-tail assembly, the microtubule is a polar filament, with β-tubulin facing the plus end and α-tubulin at the minus end (Mitchison, 1993). In vitro experiments using purified tubulin first demonstrated that microtubules exhibit an unusual property called dynamic instability, whereby microtubules undergo prolonged periods of polymerization and depolymerization with transitions between the two states called catastrophe (from polymerization to depolymerization) and rescue (from depolymerization to polymerization)(Desai and Mitchison, 1997). In vitro, plus and minus ends both undergo dynamic instability over the same range of tubulin concentrations but display small quantitative differences.
As a result of interactions with specific binding proteins, the dynamic behavior of microtubules in vivo can differ dramatically from that described in vitro. A long list of proteins has been identified that bind at microtubule plus ends and regulate their dynamics. For example, MAP215 accelerates tubulin subunit addition at the plus end, EB1 promotes plus end growth and dynamicity, and Clip170 increases rescue frequency (Akhmanova and Steinmetz, 2008). Opposing these growth-promoting proteins are the depolymerizing Kinesin-13 motors, which use ATP hydrolysis to induce a conformational change at plus ends to promote catastrophe (Moores and Milligan, 2006). The antagonistic actions of different +TIP proteins account for the more pronounced dynamic instability of microtubules in vivo compared to microtubules composed of pure tubulin in vitro (Kinoshita et al., 2001).
In contrast to the wealth of information on the microtubule plus end, the regulation of the microtubule minus end in vivo is poorly understood. In many cell types, the minus ends are clustered and anchored at a central microtubule-organizing center (MTOC). This organization has hindered visualization of their dynamics, in contrast with plus ends which are more easily viewed at the cell periphery by microscopy. Even in organisms and cell types that lack a central MTOC (e.g. S. pombe, D. melanogaster, A. thailinia, neurons, epithelial cells, and myotubes), the microtubule minus ends appear to be embedded in poorly characterized anchoring sites around the cell (Bartolini and Gundersen, 2006;Rusan and Rogers, 2009).
Occasionally, in animal cells, microtubules are released from an MTOC or break due to actomyosin forces, thereby allowing minus ends to be observed free from any nucleating material (Rodionov and Borisy, 1997;Vorobjev et al., 1999;Yvon and Wadsworth, 1997;Waterman-Storer and Salmon, 1997;Keating et al., 1997). The conclusion from these studies is that the vast majority (80–90%) of free microtubule minus ends are stable, neither visibly growing nor shrinking. A similar stability of minus ends has been observed in cytoplasmic extracts (Rodionov et al., 1999;Vorobjev et al., 1997). Some minus ends, however, transition to rapid depolymerization resulting in the disappearance of the microtubule, and a very small percentage of microtubules treadmill through the cytoplasm (caused by simultaneous minus end shrinkage and plus end growth)(Rodionov and Borisy, 1997). Microtubule elongation from minus ends has not been reported in vivo. Thus, in contrast to the pronounced dynamic instability of plus ends, minus ends are mostly static and are indeed less dynamic than minus ends composed of pure tubulin in vitro. These results suggest that microtubule minus ends might be capped by some unknown protein(s) that suppresses subunit dynamics.
In a whole-genome RNAi screen for spindle morphology defects in Drosophila S2 cells, we identified a previously uncharacterized protein (short spindle phenotype 4 (ssp4)), whose depletion caused short spindles in mitosis and microtubule fragments in interphase (Goshima et al., 2007). Three homologues exist in humans (Baines et al., 2009), one of which localizes at microtubule minus ends located close to adherens junctions in epithelial cells (Meng et al., 2008). In this study, we show that Drosophila ssp4, which we have renamed Patronin for the Latin patronus (protector), protects microtubule minus ends in vivo against depolymerization by Kinesin-13. In the absence of Patronin, microtubules release from their nucleating sites and treadmill through the cytoplasm, a result of unhindered minus end depolymerization. Purified Patronin selectively binds to and protects minus ends against Kinesin-13-induced depolymerization in vitro, demonstrating that Patronin alone is sufficient to confer minus end stability. Thus, Patronin is the first protein demonstrated to cap and stabilize the microtubule minus end in vivo. We also show that microtubule minus end dynamics are regulated by competing actions of destabilizing and stabilizing proteins, as has been shown previously for the plus end.
Results
Depletion of Patronin results in free microtubules that move through the cytoplasm
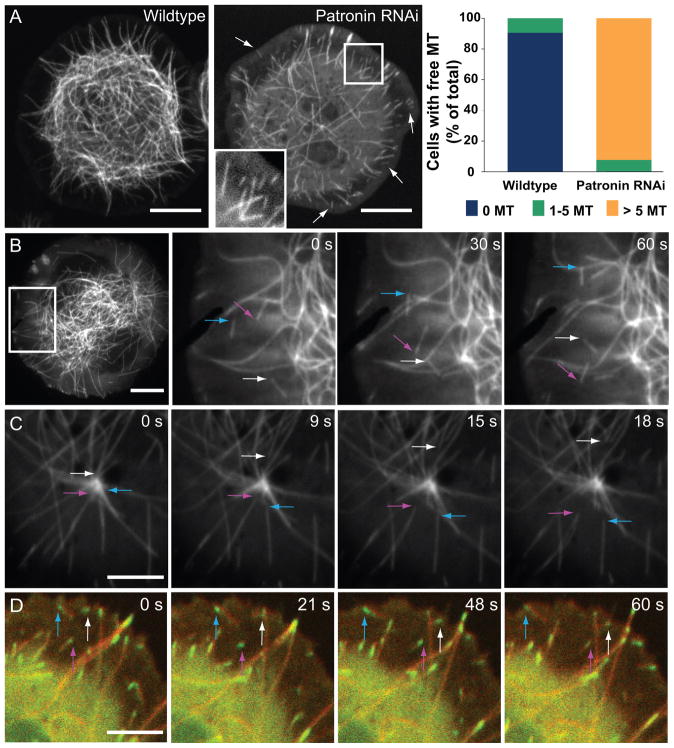
Drosophila S2 cells do not have a central MTOC in interphase, but rather generate microtubules from multiple small nucleating sites, with microtubule plus ends generally visible at the cell periphery, while minus ends lies more centrally (Rogers et al., 2008;Rusan and Rogers, 2009). In wildtype cells, free microtubules (where both the plus and minus end of the same microtubule are both clearly observed) are rarely found in the periphery (Figure 1A). In striking contrast, when Patronin was depleted by RNAi (Figure S1A), the interphase microtubule cytoskeleton became less dense (Figure 1A)(45% polymer decrease; Figure S1B) and the majority of cells had >5 free microtubules visible at the cell periphery (Figure 1A, Movie S1). Previously, we speculated that free microtubules might arise from increased severing after RNAi of Patronin (Goshima et al., 2007). However, we did not observe microtubule severing events in Patronin RNAi cells, and RNAi knockdown of microtubule severing proteins did not suppress the number of free microtubules seen after Patronin RNAi (Figure S1F).
Figure 1. Depletion of Patronin results in free microtubules that move through the cytoplasm.
Time-lapse microscopy of wildtype and Patronin-depleted GFP-tubulin Drosophila S2 cells. (A) Patronin-depleted cells have numerous free microtubules (both the plus and minus end of the same microtubule are clearly visible, arrows) which are rarely seen in wildtype cells and also have a sparser microtubule network (insert shows a region with several free microtubules). The right side shows the quantitation of free microtubules per cell from two independent experiments; colored bars indicate the percentage of cells with the number of indicated free microtubules observed (n = 200 cells per experiment; SEM <6%). Scale bars, 10 μm. See Movie S1. (B) Time-lapse TIRF microscopy of Patronin-depleted GFP-tubulin cells demonstrates that free microtubules move throughout the cytoplasm (colored arrows follow the motion of the leading end of three microtubules). Scale bar, 10 μm. See Movie S2. (C) In Patronin-depleted cells, microtubules (arrows) release and move away from the centrosome (prophase cell). Scale bar, 5 μm. See Movie S2. (D) In cells co-expressing EB1-GFP (green) and mCherry-tubulin (red), EB1 localizes to the leading end of moving microtubules (arrows), indicating that this is the microtubule plus end. See Movie S3. Brightness was adjusted in each color channel separately. Scale bar, 5 μm. See also Figures S1 and S2.
Time-lapse observation of GFP-tubulin cells provided new insight into how Patronin affects microtubules. Free microtubules appeared to move in a linear manner within the cytoplasm (Figure 1B, Movie S2). In many cases, we observed microtubules releasing from sites of nucleation and moving towards the cell periphery, which might explain the appearance of free microtubules near the cell boundary (Figure 1A, 1C, S1C, Movie S2). Since microtubules are nucleated at their minus ends, these observations indicated the free microtubules were moving with their plus ends leading and their minus ends trailing. This conclusion is further supported by observations of EB1-GFP, which always localized to the leading end of the translocating microtubule in Patronin RNAi cells (Figure 1D, Movie S3).
Free microtubules move by treadmilling in Patronin-depleted cells
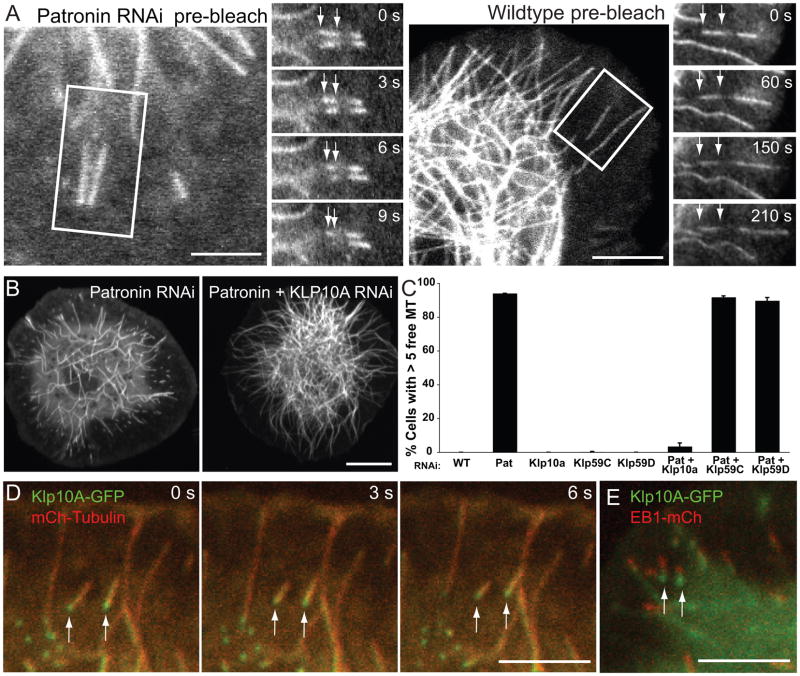
The movement of microtubules in the cytoplasm in Patronin depleted cells could result from either: 1) transport by an anchored minus end-directed motor protein (e.g. cytoplasmic dynein), or 2) microtubule treadmilling caused by tubulin addition at the plus end at a similar rate as tubulin loss at the minus end. To distinguish between these two mechanisms, we photobleached a section of a free GFP-labeled microtubule and observed how the bleach mark moved relative to the two microtubule ends. If the free microtubule is actively transported, the bleach mark should remain stationary relative to the plus and minus ends of the moving microtubule. Conversely, if the microtubule is treadmilling, the bleach mark should appear to move away from the plus end and get closer to the minus end. In Patronin-depleted cells, we observed the latter result; all plus ends moved away from the bleach mark (3.3 ± 0.3 μm/min; n = 20)(mean ± S.D.) while the minus ends moved closer (3.2 ± 0.3 μm/min; n = 20) and eventually passed through the bleached area (Figure 2A). These results indicate that microtubules move through the cytoplasm by treadmilling.
Figure 2. Free microtubules move by Klp10A-mediated treadmilling in Patronin-depleted cells.
(A) Photobleaching a mark in the middle of moving microtubules in Patronin RNAi cells reveal that the bleach mark is stationary and the trailing minus end moves towards the bleach mark (see arrows)(n=20). This indicates that the apparent motion of microtubules occurs through simultaneous tubulin polymerization at the plus end and depolymerization at the minus end. In wildtype cells, the bleach mark in a rare free microtubule remains stationary relative to the minus end, indicating that it is neither polymerizing nor depolymerizing (n=10). Scale bars, 10 μm. (B) Comparison of GFP-tubulin cells depleted of Patronin alone or both Patronin and Klp10A. Cells co-depleted of Patronin and Klp10A have a wildtype-like microtubule network and rarely have free microtubules. Scale bar, 10 μm. (C) Quantitation of the percentage of cells with >5 free microtubules shows that co-depletion of Patronin and Klp10A, but not Klp59C or Klp59D, rescues the Patronin RNAi phenotype. The mean and SEM is shown from two independent experiments (n=200 cells per experiment). (D) In Patronin-depleted cells co-expressing Klp10A-GFP (green) and mCherry-tubulin (red), Klp10A localizes to and tracks along the depolymerizing minus end of treadmilling microtubules (arrows). Scale bar, 5 μm. See Movie S3. (E) In Patronin-depleted cells co-expressing Klp10A-GFP (green) and EB1-mCherry (red), Klp10A localizes to the trailing end, while EB1 localizes to the leading end of treadmilling free microtubules (frame from a time-lapse sequence). Scale bar, 5 μm. Brightness was adjusted in each color channel separately. See also Figures S1 and S2.
We next wanted to determine whether microtubule treadmilling occurs for any free microtubule or if this phenomenon requires the depletion of Patronin. In wildtype cells, it was possible to find an occasional free microtubule, but these did not translocate in the cytoplasm. When we photobleached a free microtubule from a wildtype cell, the bleach mark remained at a constant distance from the minus end (0.01 ± 0.07 μm/min; n = 10), while the plus end continued to polymerize (3.25 ± 0.24 μm/min; n = 10)(Figure 2A). This finding suggests that free microtubule minus ends are stable in wildtype cells, as has been observed in other cell types (Dammermann et al., 2003) and that the minus end depolymerization that gives rise to microtubule treadmilling requires the depletion of Patronin. We also examined whether minus end depolymerization occurred after RNAi depletion of γ-tubulin, γ–TuRC and γ-TuSC components, since the γ-TuRC complex has been shown to bind to microtubule minus ends in vitro (Moritz et al., 1995;Zheng et al., 1995;Wiese and Zheng, 2000). However, in these RNAi cells, free microtubules were rare and did not undergo treadmilling (Figure S1D).
To learn more about microtubule behavior after Patronin depletion, we measured the plus and minus end dynamics in wildtype and Patronin depleted cells. For the microtubule plus end, the rates of growth and shrinkage and the frequencies of catastrophe and rescue were similar under Patronin-depletion and wildtype conditions (Table 1). Thus, Patronin appears to have negligible effects on plus end dynamics. In contrast, minus ends displayed very different dynamics after Patronin depletion. In Patronin RNAi cells, minus ends of treadmilling microtubules often depolymerized at a rate of 3.9 ± 0.9 μm/min (mean ± S.D.), which is similar to the plus end polymerization rate of 4.2 ± 1.3 μm/min (Table 1). The similarity in the rates of tubulin addition at the plus end and dissociation from the minus end explains why the lengths of treadmilling microtubules often remain relatively constant, with occasional shortening or lengthening when either the plus end or minus end pauses (Movie S1, S2). We also observed a more rapid minus end depolymerization rate of 10.2 ± 2.2 μm/min, and occasionally individual microtubules would transition between the slow and fast depolymerization rates (Figure S1E). Interestingly, minus end depolymerization often halted when it reached the EB1-enriched microtubule plus end tip (Figure S2A, 20 of 30 depolymerizing microtubules paused for an average of 35.8 ± 13.1 sec), indicating that +TIP proteins might help the microtubule resist continued minus end depolymerization. After such a pause, the microtubule would either continue to depolymerize and disappear (11 of 20 microtubules) or resume plus end growth and increase in length (Figure S2A; 9 of 20 microtubules). In summary, microtubule minus ends can depolymerize at two rates in vivo: one similar to plus end growth (resulting in treadmilling) and a second more rapid rate that can lead to complete microtubule disappearance and may account for the sparser microtubule network after Patronin RNAi.
Table 1. Quantitation of dynamic instability parameters in wildtype and Patronin depleted GFP-tubulin cells.
Polymerization and depolymerization rates were measured for 25 individual microtubules (per type of measurement) from 8–16 cells over 3 different experiments. The number reported is the mean and S.D. from the 25 measurements. Polymerization and depolymerization rates were measured by kymograph analysis using ImageJ. For Patronin RNAi cells, “free” microtubules were measured (both ends clearly visualized, see Movie S2). The exception (noted by an *) is the minus end dynamics in wildtype cells. Because of the high degree of stability and possible movement of the microtubule in the wildtype cytoplasm over long measurement times, we measured the microtubule minus end relative to a photobleach mark as in Fig. 2 (n = 10); the value shown is within the error of our measurement and indicates that the minus end is very stable. “N.D.”, indicates that a second rate was not detected. A comparable measurement of a minus end relative to a bleach mark in Patronin RNAi cells yielded two shrinkage rates (3.21 ± 0.31 and 10.81 ± 0.94; n = 20 for each rate), similar to that observed for tracking the minus end in microtubules without photobleach marks (shown in the Table). The microtubules scored for this table exhibited a single, constant minus end shrinkage rate. However, these two different rates of minus end shrinkage occasionally were observed for individual microtubules (Figure S1E). Catastrophe and rescue frequencies were calculated for 10 cells per condition. In each cell, 10 microtubules were observed and the frequency of catastrophe and rescue calculated over the course of 3 min. The number reported is the mean and S.D. of the frequencies calculated for each cell.
| Wildtype | Patronin RNAi | |
|---|---|---|
| Microtubule Plus End | ||
| Growth (μm/min) | 3.58 ± 1.10 | 4.22 ± 1.31 |
| Shrinkage (μm/min) | 10.21 ± 2.12 | 10.93 ± 1.56 |
| Catastrophe (min−1) | 0.12 ± 0.06 | 0.11 ± 0.05 |
| Rescue (min−1) | 0.16 ± 0.08 | 0.21 ± 0.08 |
| Microtubule Minus End | ||
| Shrinkage I (μm/min) | 0.01 ± 0.07* | 3.93 ± 0.87 |
| Shrinkage II (μm/min) | N.D. | 10.20 ± 2.21 |
Depletion of the Kinesin-13 microtubule depolymerase, Klp10A, suppresses the Patronin phenotype in interphase and mitosis
The above results reveal that Patronin protects the microtubule minus end against depolymerization in vivo. We next wanted to determine if the depolymerization was an intrinsic property of the minus end or whether another protein was actively involved. Kinesin-13s are microtubule depolymerizers that localize to both plus and minus ends in vitro, and in vivo bind to microtubule plus ends during interphase and promote their depolymerization (Desai et al., 1999; Hunter et al., 2003; Mennella et al., 2005). Kinesin-13s also promote the poleward flux of tubulin subunits towards the spindle pole during mitosis, a process that involves minus end tubulin turnover (Kwok and Kapoor, 2007; Rogers et al., 2004). To determine whether a Drosophila Kinesin-13 family member is involved in depolymerizing the microtubule minus ends after Patronin depletion, we performed double RNAi of Patronin with the three Drosophila Kinesin-13 genes (Klp10A, Klp59C, Klp59D) and examined the effect on interphase microtubule dynamics. Strikingly, co-depletion of Klp10A rescued the Patronin RNAi phenotype; the microtubule array was denser and free microtubules were no longer observed in the majority of the cells (Figure 2B, 2C). In contrast, double RNAi of either Klp59C or Klp59D with Patronin did not affect the number of free, treadmilling microtubules (Figure 2C). When a rare, free microtubule was found in a Patronin and Klp10A co-depleted cell, the minus end either remained stationary or appeared to grow, resulting in an increase in microtubule length. Interestingly, EB1-GFP localized to both ends of these growing microtubules, although it appeared more abundant at the presumed plus end at the cell periphery (Figure S2B). We also occasionally observed a transient localization of EB1-GFP at free microtubule minus ends in cells depleted of Patronin alone, which was accompanied by a pause in minus end depolymerizing and increase in microtubule length (Figure S2B). This, to our knowledge, is the first observation of in vivo minus end polymerization.
We also examined the interphase localization of Klp10A-GFP in Patronin-depleted cells. Previous studies showed that Klp10A-GFP localizes to microtubule plus ends prior to their catastrophe/depolymerization; the loading of Klp10A to growing plus ends is mediated by an interaction with EB1 (Mennella et al., 2005). In Patronin-depleted cells, we observed a prominent puncta of Klp10A-GFP tracking along the depolymerizing minus ends of treadmilling microtubules (Figure 2D, Movie S3). In cells co-expressing Klp10A-GFP and EB1-mCherry, we confirmed that more Klp10A is found at the depolymerizing end while EB1 is at the growing plus end (Figure 2E). This localization data supports the conclusion of the Klp10A rescue experiments indicating that Klp10A is actively depolymerizing minus ends in the absence of Patronin, and suggests this minus end localization is not dependent on EB1.
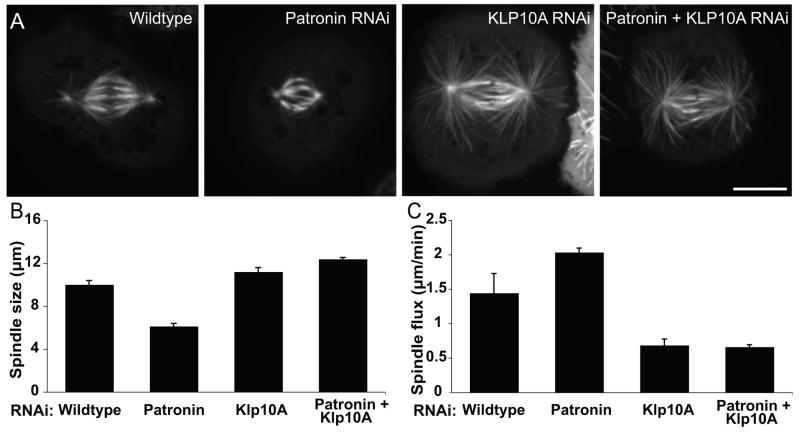
We next examined whether Klp10A is involved in producing the short spindle phenotype observed after Patronin depletion (Goshima et al., 2007). Wildtype spindles have a pole-to-pole length of 10.1 ± 1.7 μm (mean ± SD), which was reduced to 6.1 ± 1.3 μm after Patronin depletion (Figure 3A, 3B). A similar reduction was observed in acentrosomal mitotic spindle produced by centrosomin (Cnn) RNAi (Li and Kaufman, 1996)(9.6 ± 1.9 μm in Cnn RNAi cells and 6.7 ± 1.3 μm after Cnn/Patronin double RNAi (n = 35)), suggesting Patronin s function is not limited to the centrosome. Interestingly, we observed two distinct classes of short, bipolar spindles after Patronin RNAi: one in which the spindle had normal morphology with a clearly aligned metaphase plate, and another where the spindle appeared “collapsed” and the bipolar array penetrated across the metaphase plate (Figure S3B). Co-depletion of Klp10A and Patronin restored normal morphology (Figure 3A, B) and produced longer spindles (12.4 ± 2.6 μm) than wildtype cells, a length comparable to Klp10A depletion alone (11.2 ± 2.2 μm). Conversely, co-depletion of Klp59C or Klp59D and Patronin, produced shorter spindles than wildtype cells (Figure S3A). These results suggest that Patronin protects microtubule minus ends against Klp10A-induced depolymerization during mitosis and that the balance of counteracting stabilizing and destabilizing forces at the minus ends governs spindle length (see Discussion).
Figure 3. Depletion of Klp10A suppresses the Patronin phenotype in mitosis.
(A) Co-depletion of Patronin and Klp10A rescues the short spindle phenotype observed in Patronin-depleted cells and results in elongated spindles similar to those seen in Klp10A-depleted cells. Scale bar, 10 μm. (B) The mean pole-to-pole metaphase spindle length under each condition was quantified for two independent experiments (n>60 spindles per condition; error bar, SEM; p<0.001 for each reported condition). (C) The flux of tubulin towards the spindle poles was measured by photobleaching a ~1 μm stripe in the GFP-tubulin spindle and tracking its movement. The mean flux rates were quantified under each condition from two independent experiments (n=20 spindles per condition; error bar, SEM; p< 0.001 for each reported condition except the pair of Klp10A RNAi and Klp10A/Patronin RNAi flux (p<0.9)). Thus poleward flux is increased after Patronin depletion and decreased below wildtype levels when Patronin and Klp10A are co-depleted. See also Figure S3.
Poleward flux of tubulin subunits during metaphase has been associated with minus end depolymerization by Klp10A and linked to the regulation of spindle length; less poleward flux results in longer spindle and vice versa (Rath et al., 2009). Depletion of Patronin resulted in an increased flux (2.03 ± 0.06 μm/min) over wildtype (1.44 ± 0.28 μm/min), thus explaining the shorter spindle. As previously reported, Klp10A RNAi caused a dramatic reduction in flux (0.68 ± 0.09 μm/min)(Laycock et al., 2006; Rath et al., 2009). Co-depletion of Patronin and Klp10A produced a flux (0.66 ± 0.03 μm/min) similar to Klp10A alone (Figure 3C), thus explaining the long spindle phenotype.
Taken together, our results suggest that Klp10A is actively depolymerizing free microtubule minus ends in interphase and mitosis and that the presence of Patronin is able to suppress this depolymerization activity.
GFP-Patronin localizes to microtubule nucleation centers
To learn more about Patronin s functions, we determined its intracellular localization. A polyclonal antibody made against the C-terminal region of Patronin, while having considerable background staining, showed that endogenous protein localizes to centrosomes in prophase, the midbody during cytokinesis, throughout the metaphase spindle, and to punctae in interphase that often overlap with microtubules (Figure S4C).
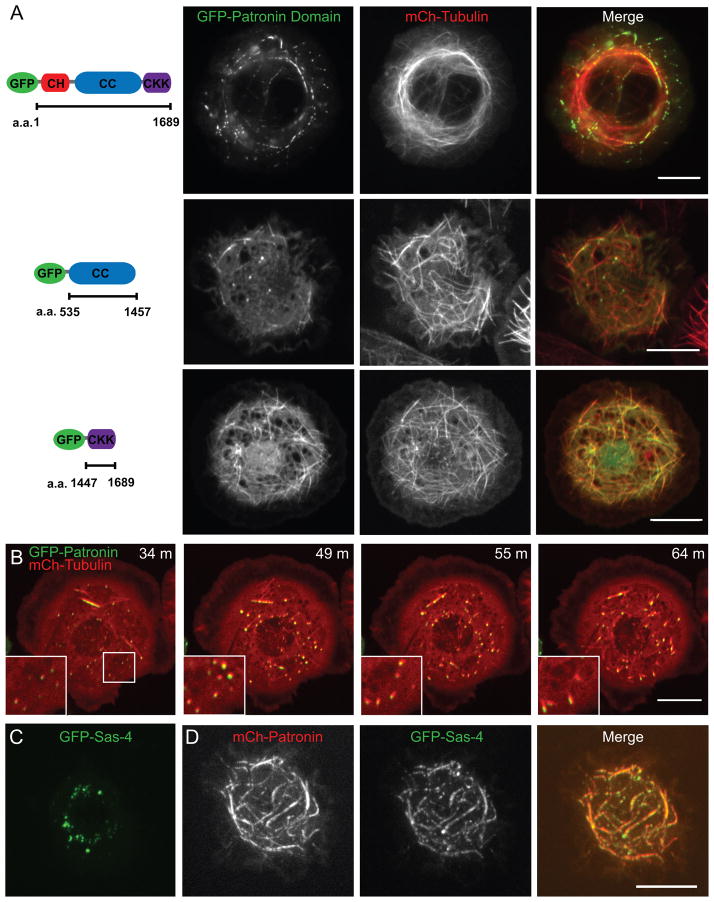
A GFP-Patronin fusion protein, which rescued the Patronin phenotype and thus is functional (Figure S4A), localized in punctae along microtubules in interphase, bundling them at moderate to high expression levels, and localized throughout the mitotic spindle (Figure 4A, Figure S4B). We also examined the localization of Patronin s three major domains: an N-terminal calponin homology domain (CH), a middle domain containing 3 predicted coiled-coils (CC), and a C-terminal microtubule binding domain (CKK domain (Baines et al., 2009)). The CH domain appeared diffuse throughout the cytoplasm (Figure S5A), while the CKK domain localized along all microtubules as previously reported (Baines et al., 2009)(Figure 4A). Interestingly, the central CC domain localized to small microtubule nucleating foci (Figure 4A, S5F) and occasionally along short stretches of microtubules.
Figure 4. GFP-Patronin localization and domain analysis.
(A) Co-expression GFP-fusions of full-length Patronin (TIRF microscopy) or Patronin domains with mCherry-tubulin (merge: GFP-Patronin in green and mCherry-tubulin in red). Localization patterns are discussed in the text. Scale bars, 10 μm. (B) Time-lapse microscopy of GFP-Patronin (green) and mCherry-tubulin (red) expressing cells re-growing their microtubule network after washout of the microtubule depolymerizing drug colcemid (time after washout is indicated). The inserts correspond to the box at 34 min. Patronin and tubulin localize to small foci, which serve as points of microtubule nucleation during the reformation of the cytoskeleton. (C) Cells expressing GFP-Sas-4 alone form cytoplasmic foci, but when GFP-Sas-4 is co-expressed with mCherry-Patronin (D), Sas-4 is recruited to sites of mCherry-Patronin along microtubules. Brightness was adjusted in each color channel separately in the merged images. See also Figures S4 and S5.
We also visualized GFP-Patronin during the depolymerization and reformation of the microtubule cytoskeleton using the microtubule depolymerizing drug colcemid. After complete microtubule depolymerization, small foci containing both GFP-Patronin and mCherry-tubulin were observed throughout the cytoplasm (data not shown). Sas-4 and γ-tubulin, established markers of microtubule nucleating centers, localized to similar foci in GFP-tubulin cells under the same conditions (data not shown). In the initial phase of microtubule regrowth, microtubules elongated out from these foci, eventually reforming the interphase microtubule array (Figure 4B). Therefore Patronin localizes to sites of new microtubule formation. A connection between Patronin and microtubule nucleating centers was also suggested by co-expression studies of mCherry-Patronin with GFP-Sas-4 (Figure 4C, 4D) or GFP-SAK (Figure S5B, S5C). GFP-Sas-4 and SAK normally are distributed as discrete cytoplasmic punctae (Figure 4C, S5B). However, when full-length Patronin (Figure 4D) or its CC domain (Figure S5F) were overexpressed along with Sas-4 and SAK, these proteins co-localized with Patronin. However, Sas-6, a centriolar protein did not co-localize with Patronin (Figure S5D, S5E). Thus, Patronin may directly or indirectly interact with a subset of proteins associated with microtubule nucleating centers.
Purified Patronin specifically binds to and protects microtubule minus ends against depolymerization in vitro
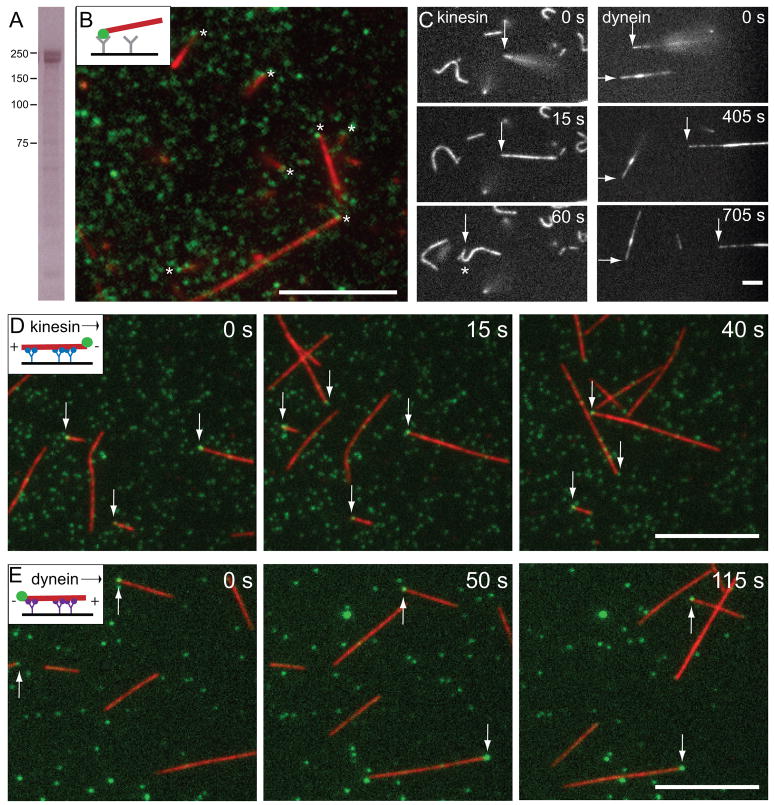
Our in vivo studies revealed that Patronin stabilizes microtubule minus ends and protects them against Kinesin-13 depolymerization. To determine if Patronin alone is sufficient for such protection, we expressed and purified full-length GFP-Patronin-6xHis (224 kDa) from baculovirus infected Sf9 cells (Figure 5A) to test its activity in vitro.
Figure 5. Purified Patronin selectively binds to microtubule minus ends in vitro.
(A) Purified GFP-Patronin-6xHis analyzed by SDS polyacrylamide gel electrophoresis and stained with Coomassie blue. Immunoblot analysis reveals that lower band of the doublet is Patronin lacking the GFP (not shown). (B) When GFP-Patronin is attached to a coverslip with anti-GFP antibody, it binds GMP-CPP stabilized rhodamine-labeled microtubules by one end (see Movie S4). Asterisks indicate the site of microtubule anchoring, which often overlaps with a GFP-Patronin spot. Scale bar, 10 μm. (C) To reveal which microtubule end was anchored to GFP-Patronin, kinesin or dynein was added after microtubule anchoring. Arrows follow a microtubule that was initially anchored by one end and then bound along its length to the motor-covered surface. With kinesin, the formerly anchored end is leading moving end (until the leading end reattaches and the microtubule buckles (asterisk, 60 s); with dynein, the formerly anchored end is trailing. See Movie S5. These assays reveal that microtubules are anchored to surface-bound Patronin selectively at their minus end (see statistics from three independent experiments in the text). Scale bar, 5 μm. Conventional kinesin (D) or dynein (E) microtubule gliding assays in the presence of GFP-Patronin (6 nM; green) demonstrate that GFP-Patronin binds selectively to the minus end. In the kinesin assay, GFP-Patronin (green) is most frequently observed at the leading end of gliding microtubules, while in the dynein assay, it resides at the trailing end. The results from three independent experiments indicate that GFP-Patronin binds selectively to the minus end. See Movie S6. Scale bars, 10 μm. Brightness was adjusted in each color channel separately.
We first wanted to establish how Patronin interacted with microtubules made from purified tubulin. We attached GFP-Patronin to a coverslip using a surface adsorbed anti-GFP antibody and then added GMP-CPP stabilized, rhodamine-labeled microtubules. Strikingly, the microtubules attached to the coverslip by only one end, resulting in filaments that swiveled in space while anchored at a single point (Figure 5B, Movie S4). In most cases, a clear spot of GFP-Patronin co-localized with the anchored end of the microtubule (asterisks, Figure 5B). Microtubules did not bind to the coverslip surface in the absence of Patronin and attached along their length when bound by anti-tubulin antibody or kinesin (data not shown). To determine if Patronin preferentially bound to the microtubule plus or minus end, microtubule gliding was induced by introducing kinesin or dynein to the assay. With kinesin, the Patronin-bound end became the leading end as kinesin moved the microtubule across the glass (128 out of 130 pre-anchored microtubules exhibited this polarity)(Figure 5C). The leading end of gliding microtubules also frequently stopped, presumably due to rebinding to Patronin, causing the microtubule to buckle due to the pushing force of kinesin (asterisk in Figure 5C, Movie S5). Conversely, when dynein was added, the Patronin-bound end now became the trailing end of the gliding microtubule (138 out of 139 microtubules)(Figure 5C, Movie S5). These results show that Patronin binds highly selectively to the microtubule minus end in vitro.
To further confirm these conclusions, we sought to visualize GFP-Patronin bound to the microtubule. In this assay, we first attached kinesin or dynein to the coverslip and then added GMP-CPP rhodamine-labeled microtubules along with purified GFP-Patronin. By TIRF microscopy, GFP-Patronin most often bound at only one end of the microtubule. With kinesin pushing the microtubule, GFP-Patronin was on the leading microtubule end (of 84 microtubules with bound GFP-Patronin, 80 had a GFP-Patronin spot at the minus end, 1 was at the plus end, and 3 appeared internal)(Figure 5D, Movie S6). With dynein transporting the microtubule, GFP-Patronin was at the trailing end (of 101 microtubules with bound GFP-Patronin, 91 were at the minus end, 4 were at the plus end, and 6 appeared internal)(Figure 5E, Movie S6). Thus, by direct observation, GFP-Patronin binds selectively to the minus end.
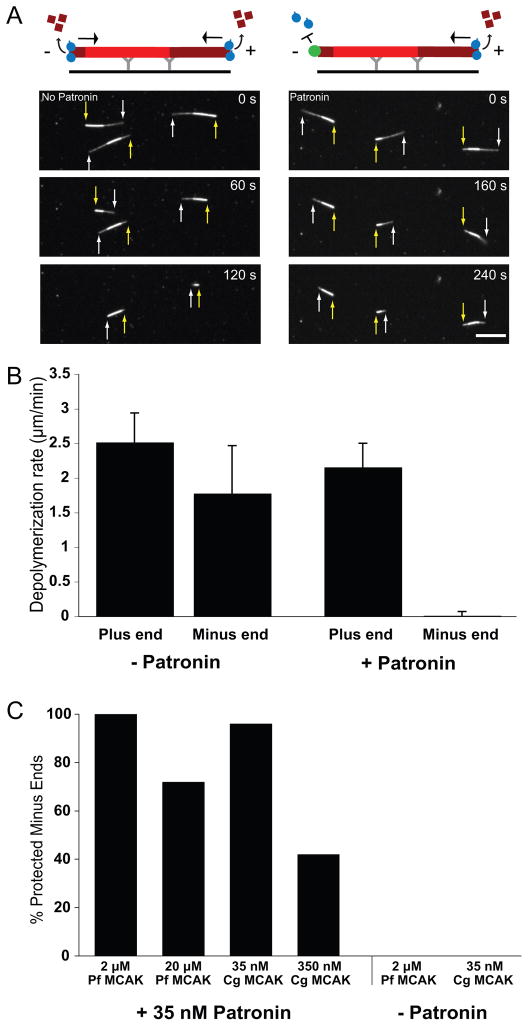
We next tested whether purified Patronin is sufficient to protect minus ends against Kinesin-13 induced depolymerization using a reconstituted assay with purified MCAK motor domain from P. falciparum (P.f. MCAK) (homologue of Klp10A, Moores et al., 2002)(Figure 6A, 6B). GMP-CPP polarity marked microtubules were adhered to the coverslip via anti-rhodamine antibody, and P.f. MCAK was added in the presence or absence of Patronin. Without Patronin, both ends of the microtubule depolymerized (plus end: 2.5 ± 0.4 μm/min, minus end: 1.8 ± 0.7 μm/min; comparable to rates reported previously in vitro (Hunter et al., 2003;Desai et al., 1999; Cooper et al., 2009). In the presence of purified Patronin, however, depolymerization from the plus end still occurred (2.2 ± 0.3 μm/min) while depolymerization from the minus end was negligible (0.01 ± 0.06 μm/min)(Figure 6A, 6B, Movie S7). We also observed selective minus end stabilization with Patronin and full-length hamster MCAK (C.g. MCAK)(Figure 6C). Higher concentrations of P.f. or C.g. MCAK, lead to the depolymerization of a portion of minus ends, suggesting that there is a competition between Patronin and MCAK for minus end binding (Figure 6C). The full-length MCAK competed more effectively than the motor domain, likely because of its higher association rate (Cooper et al., 2009). We also performed an alternative assay in which the microtubule minus end was anchored to surface adhered Patronin and a solution of P.f. MCAK was added. Once again, MCAK depolymerized the plus end rapidly, whereas the Patronin-anchored minus end did not shorten at our level of detection (Figure S6A, S6B, S6C). In summary, our in vitro studies reveal that purified Patronin binds selectively to the microtubule minus end and this binding confers protection against Kinesin-13-induced microtubule depolymerization.
Figure 6. GFP-Patronin protects microtubule minus ends from Kinesin-13-induced depolymerization in vitro.
(A) Polarity marked GMP-CPP-stabilized rhodamine-labeled microtubules were attached to the coverslip by an anti-rhodamine antibody. The minus end is closest to the region of higher fluorescence intensity in the microtubule. In the absence of Patronin, purified Kinesin-13 motor domain from P. falciparum (3 μM) depolymerizes both ends of the microtubule. In contrast, in the presence of GFP-Patronin (30 nM), Kinesin-13 only depolymerizes the dimmer, plus end (white arrows) while the minus end (yellow arrows) is stable. See Movie S7. (Note: the higher concentration of Patronin precludes imaging of individual Patronins at microtubule ends as in Fig. 5). Scale bar, 10 μm. (B) Quantitation of Kinesin-13-induced depolymerization rates at the plus and minus ends (n = 30 microtubules for each condition; mean and SD). Data is representative of three independent experiments with different microtubule preparations. (C) Patronin was mixed with the indicated concentration of either full-length Kinesin-13 from hamster (C.g.) or the motor domain from P. falciparum (P.f.) and added to polarity marked microtubules. Minus ends that showed no detectable depolymerization by the time the plus end depolymerized by >50% of the microtubule length was scored as protected. Higher concentrations of the motors are able to compete with Patronin to depolymerize a subset of minus ends. Percentages are representative of two independent experiments. See also Figure S6.
Discussion
Microtubule minus end dynamics has remained one of the least well understood properties of the microtubule cytoskeleton. Here, through in vivo and in vitro approaches, we have demonstrated that Patronin binds with high selectivity to microtubule minus ends and acts as a “cap”, stabilizing these ends and protecting them against the actions of microtubule depolymerases. The consequence of losing Patronin-mediated capping in S2 cells is dramatic. During interphase, the microtubule density decreases and microtubules released from nucleating sites treadmill through the cytoplasm. During mitosis, the spindle becomes significantly shorter and in some cases collapses to a shape that more resembles a monopolar spindle. In addition to clarifying the role of Patronin, our studies also provide new insight into the regulation of microtubule minus end dynamics. We demonstrate that minus ends are substrates for capping (Patronin), destabilizing (Kinesin-13), and possibly growth promoting or stabilizing (EB1) activities, as has been demonstrated for the microtubule plus end. The behavior of minus ends reflects a net balance of these actions, which plays an important role in the overall organization of the microtubule cytoskeleton.
Patronin mechanism
Patronin binds with high selectivity to the minus end of microtubules (>92% from our in vitro experiments), suggesting that it recognizes some unique, exposed feature at this end. In the polar microtubule, α-tubulin faces the minus end while β-tubulin faces the plus end (Mitchison, 1993). Thus we speculate that Patronin recognizes features of α-tubulin that are normally buried at the α/β interface but are exposed at the end of the microtubule. Consistent with this possibility, an anti-α-tubulin antibody has been produced that binds selectively to the microtubule minus end (Fan et al., 1996). Interestingly, selective minus end binding appears to require the cooperation of multiple regions of the Patronin protein, since the C-terminal CKK domain alone binds uniformly along the microtubule surface (Figure 4A; Baines et al., 2009).
An important functional consequence of Patronin binding to minus ends is protection against Kinesin-13 depolymerization. Kinesin-13 destabilizes microtubule ends by bending microtubule protofilaments, causing them to lose lateral interactions (Moores and Milligan, 2006). Patronin might sterically block Kinesin-13 binding and/or strengthen the lateral interactions of protofilaments, rendering minus ends resistant to depolymerization. A better understanding of how Patronin caps and protects minus ends will require higher resolution structural information of the Patronin-microtubule minus end complex.
In addition to its capping and protecting activity, Patronin might act as a scaffolding protein at microtubule nucleation centers in S2 cells. Full-length as well as Patronin s central coiled-coil region localizes to foci that nucleate microtubules, even when microtubule growth is inhibited by colcemid, and overexpression of these constructs results in the recruitment of Sas-4 and SAK, two proteins that are associated with centrioles/centrosomes (Bornens, 2002). These results raise questions of whether a scaffolding activity of Patronin might be involved in minus end capping/protection and possibly microtubule nucleation in vivo. Our in vitro data showing that purified Patronin can protect the minus end reveals that Patronin alone is sufficient for this activity, although minus end capping might be more complex and augmented by additional proteins within the cell. We thus far have not found that purified Patronin stimulates microtubule assembly from purified tubulin in vitro and the initial regrowth of microtubules after colcemid washout was similar in wildtype and Patronin RNAi cells (data not shown). However, the current experiments cannot exclude some role in nucleation. Thus, possible roles of Patronin as a scaffolding factor involved in the assembly of other proteins at microtubule minus ends awaits further investigation.
Regulation of microtubule minus end dynamics in vivo
A large and still growing list of proteins have been discovered that associate with microtubule plus ends and many exhibit opposing effects on microtubule dynamics (Akhmanova and Steinmetz, 2008;Howard and Hyman, 2007), which gives rise to the high dynamicity of plus ends in vivo and enables cells to rapidly restructure their microtubule cytoskeleton. The dynamics of microtubule minus ends in vivo has not been as well studied as plus ends, particularly at the level of single microtubules. In the few studies where minus ends have been observed in animal cells, they have been reported to be mostly stable (neither growing or shrinking; Rodionov and Borisy, 1997;Yvon and Wadsworth, 1997;Waterman-Storer and Salmon, 1997b). Minus end shrinkage and microtubule treadmilling, however, is common in Arabidopsis (Shaw et al., 2003;Ehrhardt, 2008). In contrast, microtubule minus ends composed of pure tubulin grow, shorten, and exhibit dynamic instability (Desai and Mitchison, 1997). The discrepancy between such in vitro dynamicity and in vivo stability suggests the presence of a minus end capping factor. γ-TuRC interacts directly with the microtubule minus end (Moritz et al., 1995;Zheng et al., 1995;Wiese and Zheng, 2000). However, while γ-TuRC has a clear role in microtubule nucleation in vivo, it is uncertain whether it remains bound to and stabilizes minus ends after the microtubule is nucleated. Indeed, we feel that this may not be the case, at least in Drosophila, since γ-TuRC RNAi knockdown does not greatly alter the appearance of the interphase array (Bouissou et al., 2009), produces elongated rather than short mitotic spindles (Vérollet et al., 2006), and does not generate free, treadmilling microtubules (Figure S1D, S3A). Another protein, ninein, plays a role in anchoring microtubules to MTOCs and other sites within cells (Delgehyr et al., 2005); however this interaction appears to be facilitated by γ-TuRC and ninein has not been shown yet to interact directly with minus ends. RNAi of other genes that produced a short spindle phenotype (Goshima et al., 2007) or centrosomal proteins (Sas-4, SAK, Asp, and Cnn, data not shown) did not give rise to a microtubule treadmilling phenotype indicative of minus end instability. Thus, Patronin is the only protein for which minus end capping activity as been demonstrated in vivo.
Our experiments also demonstrate that in the absence of Patronin-mediated capping, microtubule minus ends in vivo exhibit the range of behaviors seen in vitro (polymerization, depolymerization, catastrophe, and rescue) and also are acted upon by previously identified plus end binding proteins. EB1 has been used as canonical marker of microtubule plus ends in vivo. Here, we show that EB1-GFP can interact with microtubule minus ends during episodes of subunit addition (Figure S2B). Kinesin-13, which binds to plus ends and induces their catastrophe, has been suggested to depolymerize microtubule minus ends during mitosis based upon its role in spindle flux (Rogers et al., 2004), but has not been directly visualized at microtubule minus ends. Here, we show that in the absence of Patronin, the Kinesin-13 Klp10A-GFP binds to and tracks along depolymerizing minus ends and is also required for this depolymerization (Figure 2). In Patronin-depleted cells, the actions of Klp10A appear to dominate over any minus end growth promoting factors, as most microtubule minus ends undergo depolymerization and only rarely display brief periods of growth. In summary, microtubule minus ends can grow, depolymerize or be capped in vivo and the balance of proteins that promote these activities govern the behavior of microtubule minus ends in cells.
The importance of balancing stabilizing and destabilizing activities on microtubule ends is illustrated in the mitotic spindle. Net polymerization occurs at microtubule plus ends near the kinetochore and net depolymerization occurs at minus ends at the poles, which results in a poleward flux of tubulin subunits within the microtubule lattice (Kwok and Kapoor, 2007;Rogers et al., 2004). The overall balance of polymerizing and depolymerizing activities of microtubule-associated proteins governs the size and shape of the spindle (Goshima et al., 2005;Dumont and Mitchison, 2009). Studies in several organisms have implicated Kinesin-13s as major regulators of mitotic microtubule length, spindle size, and poleward flux (Mitchison et al., 2005;Rath et al., 2009;Kwok and Kapoor, 2007). Our results suggest that Patronin provides a “brake” rather than a full block on the minus end depolymerizing actions of Kinesin-13. In the absence of Patronin, Kinesin-13 is unchecked, resulting in a higher flux rate and shorter, sometimes collapsed spindles. With the depletion of both Patronin and Kinesin-13, flux is low and spindle length is longer than normal. These results imply that microtubule minus ends are not completely protected by Patronin but are subject to competing activities of Patronin and Kinesin-13, as we also demonstrate in vitro (Figure 6C). Thus, a balance of Patronin and Kinesin-13 actions on microtubules minus ends governs the length of the mitotic spindle.
The Patronin family and minus end capping in acentrosomal microtubule arrays
A single Patronin gene is found in invertebrate genomes and clear homologues do not exist or are difficult to identify in non-metazoan organisms. After Patronin (then named ssp4) was first described in Drosophila (Goshima et al., 2007), three vertebrate homologues with the same domain organization and sequence identity were reported and have been called the CAMSAP/ssp4 family of proteins (the three vertebrate branches are referred to as CAMSAP1, CAMSAP2, and CAMSAP3; (Baines et al., 2009)). All Patronin-related genes have a characteristic domain organization of an N-terminal calponin homology (CH) domain, a long central domain with interspersed predicted coiled-coil regions, and a C-terminal microtubule-binding domain (termed the CKK domain), which is the most highly conserved region of the polypeptide (Baines et al., 2009). While this work was in progress, vertebrate CAMSAP1 and a CAMSAP3 member, Nezha, were reported to interact with microtubules (Baines et al., 2009;Meng et al., 2008). Meng et. al. (2008) found that Nezha localizes specifically at microtubule minus ends located close to adherens junctions in epithelial cells and bound preferentially to the minus end in vitro (67% of microtubule-associated Nezha). However, their study did not explore whether Nezha affected the dynamics of microtubules or influenced the organization of the microtubule cytoskeleton. If the vertebrate homologues also are found to protect microtubule minus ends as shown here for Drosophila Patronin, we suggest that the currently named CAMSAP/ssp4 family be renamed as the Patronin family, retaining the phylogenetic classification of the three vertebrate branches (Patronin 1, 2, and 3)(Baines et al., 2009).
Minus end capping has been proposed to be particularly important for the formation and organization of non-radial, acentrosomal interphase microtubule arrays (Dammermann et al., 2003; Bartolini and Gundersen, 2006). The roles of the three Patronin family members in vertebrates are not yet defined, but they may have evolved to interact with distinct partners for localizing microtubule minus end capping/anchoring activities to distinct subcellular regions in epithelial cells (Meng et al., 2008), neuronal cells (Berglund et al., 2008), and other cell types with acentrosomal arrays. Thus, the three Patronin family members might provide new molecular tools for probing the organization and function of microtubules in different vertebrate cells types.
Experimental Procedures
Cell Culture and RNAi
Drosophila S2 cells (UCSF) were cultured and incubated with dsRNA as previously described (Goshima and Vale, 2003). Unless noted, cells were treated with dsRNA for 4 days, and when indicated were treated additional dsRNA at day 4 and analyzed at day 8. Plasmids and cell lines are described in the Supplemental Methods.
Live cell imaging
Cells were plated on Con A (Sigma) coated MatTek dishes for 1 hr unless noted. Live cell imaging was performed by spinning disk confocal microscopy or occasionally by TIRF microscopy (noted in the legends). Microscope equipment is described in the Suppl. Methods. For the photobleaching experiments, GFP-tubulin cells were imaged on an LSM 510 or 710 (Carl Zeiss, Inc.)(63x 1.4 NA objective). Two or three imaging scans were performed with a 488 nm laser at 1.1% power before a selected area was bleached. On the LSM 510, bleaching was achieved with a 488 nm Argon laser at 100% laser power for 4 iterations, while on the LSM710 a 405 nm laser at 45% power was used for 2 iterations. After the photobleach, scans were taken at 488 nm (1.1% power) every 3 sec. The position of a bleach mark relative to microtubule ends or within a spindle (flux measurements) was measured over time using ImageJ.
In vitro assays
GFP-Patronin with a C-terminus 6xHIS tag was expressed using the BaculoDirect system (Invitrogen). Sf9 cells were infected with P3 virus for 3 days and harvested. GFP-Patronin-6xHis was purified on a NiNTA column (Qiagen); the eluted protein was dialyzed overnight into 50 mM Tris-HCl pH 8, 150 mM KAcetate, 1 mM DTT and 10% glycerol and stored in LN2.
Flow cells were used for all in vitro assays. For the anchoring assay, anti-GFP antibody was adhered to the coverslip and 150 nM GFP-Patronin was added for 5 min. Coverslips were blocked with 1 mg/ml casein solution, after which a solution of GMP-CPP stabilized rhodamine-microtubules (See Supp. Methods), an oxygen scavenging mixture (catalase, glucose oxidase, and glucose), and 1 mg/ml casein in BRB80 was added (referred to as the microtubule solution). To determine the polarity of the anchored microtubule, the experiment was repeated with the following changes: a mixture of anti-GFP and anti-GST antibody was adhered to the coverslip, and after microtubules were anchored by Patronin, K560 kinesin (Woehlke et al., 1997) or GST-D4.4 dynein (Reck-Peterson et al., 2006), an oxygen scavenger mix and 5 mM ATP was added.
For the motility assays, a coverslip with immobilized K560 kinesin or GST-D4.4 dynein (via anti-GST) was blocked with 1 mg/ml casein and the microtubule mixture plus 6 nM GFP-Patronin and 5 mM ATP was added.
For the Kinesin-13 depolymerization assay, polarity marked GMP-CPP rhodamine-microtubules (See Supp. Methods) were anchored to the coverslip with an anti-rhodamine antibody. The indicated concentration of Kinesin-13 (either the MCAK motor domain from P. falciparum (purified as described in (Moores et al., 2002)) or full length hamster MCAK obtained from Linda Wordeman (Cooper et al., 2009)) was added with 5 mM ATP in BRB80 with an oxygen scavenger mix. Images were taken at 20 s intervals on the TE2000U Nikon microscope using a 40x 1.3 NA objective and Nikon intensilight. Microtubule lengths were measured using ImageJ software.
Supplementary Material
Acknowledgments
We thank E. Griffis, N. Stuurman and G. Goshima for guidance, discussions, and advice. G. Goshima, M. Sirajuddin, A. Carter, A. Yildiz, and A. Karunakaran contributed reagents. L. Wordeman and M. Wagenbach (U. of Washington) generously provided full-length MCAK, and J. Raff (U. of Cambridge) kindly provided a Sas-4 antibody. We thank the Physiology Course and the Cell Division Group at the MBL, Woods Hole for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhmanova A, Steinmetz M. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–22. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Baines AJ, Bignone PA, King MD, Maggs AM, Bennett PM, Pinder JC, Phillips GW. The CKK domain (DUF1781) domain binds microtubules and defines the CAMSAP/ssp4 family of animal proteins. Mol Biol Evol. 2009;26:2005–14. doi: 10.1093/molbev/msp115. [DOI] [PubMed] [Google Scholar]
- Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119:4155–63. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- Berglund L, Björling E, Oksvold P, Fagerberg L, Asplund A, Al-Khalili Szigyarto C, Persson A, Ottosson J, Wernérus H, Nilsson P, et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics. 2008;7:2019–27. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Bouissou A, Vérollet C, Sousa A, Sampaio P, Wright M, Sunkel CE, Merdes A, Raynaud-Messina B. {gamma}-Tubulin ring complexes regulate microtubule plus end dynamics. J Cell Biol. 2009;187:327–34. doi: 10.1083/jcb.200905060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JR, Wagenbach M, Asbury CL, Wordeman L. Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nature Structural & Molecular Biology. 2009;17:77–82. doi: 10.1038/nsmb.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Desai A, Oegema K. The minus end in sight. Curr Biol. 2003;13:R614–24. doi: 10.1016/s0960-9822(03)00530-x. [DOI] [PubMed] [Google Scholar]
- Delgehyr N, Sillibourne J, Bornens M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci. 2005;118:1565–75. doi: 10.1242/jcs.02302. [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Dumont S, Mitchison TJ. Force and length in the mitotic spindle. Curr Biol. 2009;19:R749–61. doi: 10.1016/j.cub.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt DW. Straighten up and fly right: microtubule dynamics and organization of non-centrosomal arrays in higher plants. Curr Opin Cell Biol. 2008;20:107–16. doi: 10.1016/j.ceb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Fan J, Griffiths AD, Lockhart A, Cross RA, Amos LA. Microtubule minus ends can be labelled with a phage display antibody specific to alpha-tubulin. J Mol Biol. 1996;259:325–30. doi: 10.1006/jmbi.1996.0322. [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–16. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–21. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Length control of the metaphase spindle. Curr Biol. 2005;15:1979–88. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19:31–5. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell. 2003;11:445–57. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating TJ, Peloquin JG, Rodionov VI, Momcilovic D, Borisy GG. Microtubule release from the centrosome. Proc Natl Acad Sci USA. 1997;94:5078–83. doi: 10.1073/pnas.94.10.5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Arnal I, Desai A, Drechsel DN, Hyman AA. Reconstitution of physiological microtubule dynamics using purified components. Science. 2001;294:1340–3. doi: 10.1126/science.1064629. [DOI] [PubMed] [Google Scholar]
- Kwok BH, Kapoor TM. Microtubule flux: drivers wanted. Curr Opin Cell Biol. 2007;19:36–42. doi: 10.1016/j.ceb.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Laycock JE, Savoian MS, Glover DM. Antagonistic activities of Klp10A and Orbit regulate spindle length, bipolarity and function in vivo. J Cell Sci. 2006;119:2354–61. doi: 10.1242/jcs.02957. [DOI] [PubMed] [Google Scholar]
- Li K, Kaufman T. The homeotic target gene centrosomin encodes an essential centrosomal component. Cell. 1996;85:585–96. doi: 10.1016/s0092-8674(00)81258-1. [DOI] [PubMed] [Google Scholar]
- Meng W, Mushika Y, Ichii T, Takeichi M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 2008;135:948–59. doi: 10.1016/j.cell.2008.09.040. [DOI] [PubMed] [Google Scholar]
- Mennella V, Rogers GC, Rogers SL, Buster DW, Vale RD, Sharp DJ. Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat Cell Biol. 2005;7:235–45. doi: 10.1038/ncb1222. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ. Localization of an exchangeable GTP binding site at the plus end of microtubules. Science. 1993;261:1044–7. doi: 10.1126/science.8102497. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Maddox P, Gaetz J, Groen A, Shirasu M, Desai A, Salmon ED, Kapoor TM. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol Biol Cell. 2005;16:3064–76. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores CA, Milligan RA. Lucky 13-microtubule depolymerisation by kinesin-13 motors. J Cell Sci. 2006;119:3905–13. doi: 10.1242/jcs.03224. [DOI] [PubMed] [Google Scholar]
- Moores CA, Yu M, Guo J, Beraud C, Sakowicz R, Milligan RA. A mechanism for microtubule depolymerization by KinI kinesins. Mol Cell. 2002;9:903–9. doi: 10.1016/s1097-2765(02)00503-8. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld M, Sedat J, Alberts B, Agard D. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 1995;378:638–40. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Rath U, Rogers GC, Tan D, Gomez-Ferreria MA, Buster DW, Sosa HJ, Sharp DJ. The Drosophila Kinesin-13, KLP59D, Impacts Pacman and Flux-based Chromosome Movement. Mol Biol Cell. 2009;20:4696–705. doi: 10.1091/mbc.E09-07-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–48. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov V, Nadezhdina E, Borisy G. Centrosomal control of microtubule dynamics. Proc Natl Acad Sci USA. 1999;96:115–20. doi: 10.1073/pnas.96.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov VI, Borisy GG. Microtubule treadmilling in vivo. Science. 1997;275:215–8. doi: 10.1126/science.275.5297.215. [DOI] [PubMed] [Google Scholar]
- Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, Scholey JM, Sharp DJ. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–70. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Peifer M, Rogers SL. A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol Biol Cell. 2008;19:3163–78. doi: 10.1091/mbc.E07-10-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan NM, Rogers GC. Centrosome Function: Sometimes Less Is More. Traffic. 2009;10:472–81. doi: 10.1111/j.1600-0854.2009.00880.x. [DOI] [PubMed] [Google Scholar]
- Shaw SL, Kamyar R, Ehrhardt DW. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 2003;300:1715–8. doi: 10.1126/science.1083529. [DOI] [PubMed] [Google Scholar]
- Vérollet C, Colombié N, Daubon T, Bourbon HM, Wright M, Raynaud-Messina B. Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation. J Cell Biol. 2006;172:517–28. doi: 10.1083/jcb.200511071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev IA, Rodionov VI, Maly IV, Borisy GG. Contribution of plus and minus end pathways to microtubule turnover. J Cell Sci. 1999;112(Pt 14):2277–89. doi: 10.1242/jcs.112.14.2277. [DOI] [PubMed] [Google Scholar]
- Vorobjev I, Svitkina T, Borisy G. Cytoplasmic assembly of microtubules in cultured cells. J Cell Sci. 1997;110(Pt 21):2635–45. doi: 10.1242/jcs.110.21.2635. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C, Salmon E. Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J Cell Biol. 1997;139:417–34. doi: 10.1083/jcb.139.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2:358–64. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD. Microtubule interaction site of the kinesin motor. Cell. 1997;90:207–16. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- Yvon AM, Wadsworth P. Non-centrosomal microtubule formation and measurement of minus end microtubule dynamics in A498 cells. J Cell Sci. 1997;110(Pt 19):2391–401. doi: 10.1242/jcs.110.19.2391. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wong M, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–83. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.