Abstract
The DosR regulon in Mycobacterium tuberculosis is involved in respiration-limiting conditions, its induction is controlled by two histidine kinases, DosS and DosT, and recent experimental evidence indicates DosS senses either molecular oxygen or a redox change. Under aerobic conditions, induction of the DosR regulon by DosS, but not DosT, was observed after the addition of ascorbate, a powerful cytochrome c reductant, demonstrating that DosS responds to a redox signal even in the presence of high oxygen tension. During hypoxic conditions, regulon induction was attenuated by treatment with compounds that occluded electron flow into the menaquinone pool or decreased the size of the menaquinone pool itself. Increased regulon expression during hypoxia was observed when exogenous menaquinone was added, demonstrating that the menaquinone pool is a limiting factor in regulon induction. Taken together, these data demonstrate that a reduced menaquinone pool directly or indirectly triggers induction of the DosR regulon via DosS. Biochemical analysis of menaquinones upon entry into hypoxic/anaerobic conditions demonstrated the disappearance of the unsaturated species and low-level maintenance of the mono-saturated menaquinone. Relative to the unsaturated form, an analog of the saturated form is better able to induce signaling via DosS and rescue inhibition of menaquinone synthesis and is less toxic. The menaquinone pool is central to the electron transport system (ETS) and therefore provides a mechanistic link between the respiratory state of the bacilli and DosS signaling. Although this report demonstrates that DosS responds to a reduced ETS, it does not rule out a role for oxygen in silencing signaling.
Mycobacteria are strict aerobes, but Mycobacterium tuberculosis encounters microaerobic to anaerobic environments during the course of infection. Oxygen-limited microenvironments occur in mature granulomas, which are known to be avascular, inflammatory, and necrotic (1, 5, 20, 40, 56). Recent reports have detected mycobacterial DNA in visibly normal lung tissue (17), as well as adipose tissue (32). Interestingly, adipose tissue has been associated with hypoxia as well as the presence of nitric oxide (NO), both of which inhibit respiration (38, 51, 57, 60). Further, M. tuberculosis is able to survive for long periods of time in a nonreplicating and nonrespiring state (15, 55).
The DosR regulon is expressed in response to hypoxia, NO, and carbon monoxide (CO) and is thought to be important for early adaptation to these stimuli as well as long-term survival in the host (2, 25, 26, 46, 47, 53, 58). Despite induction by other gases, the presence of oxygen itself inhibits induction of the regulon (26, 41, 43, 48). The DosR regulon is regulated by the response regulator DosR (DevR; Rv3133c) and was recently shown to be positively regulated by PhoP (Rv0757) to a basal level during aerobic growth (16). DosR is activated by two sensor histidine kinases, DosT (Rv2027c) and DosS (DevS; Rv3132c). The activation of DosR occurs through autophosphorylation of either sensor followed by transfer of the phosphate to DosR (39).
DosS and DosT are modular proteins that contain C-terminal histidine kinase domains, an ATP binding domain, and two tandem GAF (cGMP, adenylyl cyclase, and FhlA) domains (GAF A and GAF B). GAF domains are small molecule-binding regulatory domains found throughout prokaryotes and eukaryotes. Within these domains, both DosT and DosS bind heme, and in DosS the location of heme binding within the GAF A domain is known. The heme moiety itself binds the divalent gases (NO, CO, and O2) responsible for modulating activity of the sensor (6, 22, 26, 41, 43, 48), and specifics of the hydrogen bond network required for ligand discrimination have been described recently (59).
The current biochemical literature on DosS is in disagreement regarding its signal for activation. One body of work indicates that DosS directly binds oxygen, suppressing the sensor's activity. In this way DosS functions as an oxygen sensor: when oxygen becomes limited and is no longer bound, the sensor autophosphorylates. Several hydrogen bond specifics of the oxygen interaction have been described, and the different hydrogen bond networks involved in discrimination between NO/CO and O2 have been delineated, as have aspects of the kinetics of oxygen binding as well as bond affinity (21, 22, 43, 48, 59). Two recent reports proposed a second model for DosT/S signaling, wherein DosS is a redox sensor (6, 26). This conclusion was based in part on in vitro oxidation rates and the stability of the oxy complex reported by these groups. Further evidence was derived from the interaction of DosS with cyanide as well as anaerobic autokinase activity assays (26). However, similar experiments resulting in contrary data have been published in two other reports by proponents of oxygen as the direct signal (21, 48). Support for the redox sensor hypothesis comes from data that demonstrate that under aerobic conditions DosS exists in an inactive Fe3+ form as a result of its rapid oxidation (but not direct binding) by O2 (6, 26); however, when DosS is reduced to Fe2+, its autokinase activity is greatly increased; thus, it uses its oxidation state as a signal for activation (6, 26). Lastly, it was demonstrated that flavin nucleotides are capable of directly reducing the heme iron of DosS (6).
Despite these various biochemical reports, little work has been conducted in vivo. In this report we investigate the stimulus involved in DosS signaling within bacterial cells.
MATERIALS AND METHODS
Growth conditions and strains.
All cultures were grown in Dubos Tween albumin medium (Becton Dickinson). For anaerobic container experiments, BD BBL GasPak Plus envelopes were used to achieve anaerobic conditions, and anaerobic indicators were used to ensure anaerobiosis. Vented culture flasks were placed in sealed GasPak containers (BD, Franklin Lakes, NJ) and stirred. All experiments had a starting culture optical density at 600 nm (OD600) of 0.2. The anaerobic dormancy model was performed as previously described (18). CFU were calculated as previously described (19). Additives were given at day 12 of the anaerobic dormancy model, 6 days after anaerobiosis (29). Tubes were opened for the addition of treatments in an anaerobic chamber to maintain the anaerobic status of the media and then further incubated for 8 days, after which they were plated. Additive concentrations were the same as those used for real-time analysis. Statistical significance was determined by a paired two-tailed Student t test. For direct oxygen concentration measurements, a 2-mm O2 sensor and TBR4100 free radical analyzer (World Precision Instruments) were used. Culture conditions were identical to those used for the experimental data, with the exception of the strain, which was an unmarked derivative of 6030 (42). Data represent the percent oxygen of untreated media and are the averages of four experiments. Oxyrase was used according to the manufacturer's guidelines.
Culture chemical treatments.
For aerobic experiments, treatments were added for 1 h. For hypoxic conditions, cultures were placed in anaerobic GasPak containers for 4 h. Treatments were then added to the media anaerobically (in an anaerobic chamber) and incubated for two additional hours for all treatments, with the exception of chlorpromazine and CSU-20. For these experiments, cultures were aerobically pretreated for 4 h and then placed in GasPak containers for four additional hours. Four hours in an anaerobic container was previously demonstrated to create a hypoxic medium environment (18). Treatment concentrations were as follows, with their respective vehicle in parentheses: 0.1 mM NO consisting of NO donor diethylenetriamine/NO adduct (water), 20 mM ascorbate (no vehicle; powder alone added), 400 μM vitamin K1 (ethanol), 400 μM vitamin K2 (DMSO), 50 μg/ml chlorpromazine (ethanol), 40 μM CSU-20 (ethanol), 1 mM 2,4-dinitrophenol (DMSO), and 10 mM nitrate (water). Vehicles for each treatment were added separately as controls. All chemicals were from Sigma, with the exception of CSU-20, which was synthesized as previously described (27).
Real-time PCR.
RNA was extracted, and cDNA prepared and labeled as previously described (53). To help stabilize RNA and avoid harvesting-induced artifacts, a solution of 5 M guanidine thiocyanate, 0.5% sodium N-lauryl sarcosine, 50 mM tri-sodium citrate, and 0.1 M 2-mercaptoethanol was used to harvest cultures at a 1:1 ratio with bacterial culture (44). Reverse transcription and quantitative real-time PCR were performed with gene-specific primers. Fluorescent probes were designed with the program FastPCR (23) and synthesized by Integrated DNA technologies. Primer sequences were as follows: Rv0239 forward (TGATTCGAACGCAAGTCCAGCTC), reverse (AGATCCGCACCATGTGCTCCA), and probe (TCGCGCACGAGCACGAAATG); Rv2626c forward (CCGCGACATTGTGATCAAAG), reverse (GCTCTGAGATGACCGGAACAC), and probe (CGAACGCAAGCATCCAGGAGATGC); and Rv1738 forward (CACTGGACCGTCGACATATCG), reverse (CGGTCGGCCGGATTG), and probe (CCAACGCAGCCGTGCCTTCG). Real-time PCR was performed on the Roche LightCycler 480. A reverse transcriptase negative reaction was used to account for residual DNA, and copy numbers were normalized to the level of Rv0239 transcript (18). These copy numbers were then used to determine the copy number per bacterium.
Microarrays.
Oligonucleotide-based microarrays were printed, microarrays were processed, RNA was extracted, and cDNA was prepared and labeled as previously reported (18, 53). Microarrays were scanned with a GenePix 4000b scanner (Molecular Devices), and spot intensities were obtained by using GenePix Pro 6.0 (Molecular Devices.) Aerobic cultures for microarray analysis were treated for 1 h with 20 mM ascorbate; the reference was untreated aerobic culture. Experiments were repeated in triplicate for the ΔdosT mutant and in duplicate for the ΔdosS mutant. Data analysis was performed as previously described (53). Significance Analysis of Microarrys (SAM) (http://www-stat.stanford.edu/∼tibs/SAM/) was used to determine statistically significant regulated genes in the ΔdosT mutant experiments (52). Genes in the ΔdosT strain included in Table 1 conformed to highly stringent selection criteria, exhibiting at least a 4-fold induction ratio and a corresponding SAM false discovery q value of zero. Microarray data are available at the NCBI Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/).
TABLE 1.
Induction ratios for ΔdosT and ΔdosS
| Gene no.a | Gene | Induction ratio |
Gene product | |
|---|---|---|---|---|
| ΔdosT strain | ΔdosS strain | |||
| DosR | ||||
| Rv0079 | 27.6 | 0.4 | Hypothetical protein | |
| Rv0080 | 12.5 | 1.2 | Conserved hypothetical protein | |
| Rv0081 | 3.1 | 1.0 | Probable transcriptional regulatory protein | |
| Rv0082 | 7.8 | 1.4 | Probable oxidoreductase | |
| Rv0569 | 14.9 | 0.8 | Conserved hypothetical protein | |
| Rv0570 | nrdZ | 5.9 | 0.5 | Probable ribonucleoside-diphosphate reductase |
| Rv0572c | 6.7 | 0.7 | Hypothetical protein | |
| Rv1733c | 5.8 | 0.7 | Probable conserved transmembrane protein | |
| Rv1736c | narX | 11.8 | 0.9 | Probable nitrate reductase |
| Rv1737c | narK2 | 18.6 | 0.5 | Possible nitrate/nitrite transporter |
| Rv1738 | 32.4 | 0.7 | Conserved hypothetical protein | |
| Rv1813c | 7.7 | 4.3 | Conserved hypothetical protein | |
| Rv1996 | 6.4 | 0.7 | Conserved hypothetical protein | |
| Rv2003c | 6.6 | 0.7 | Conserved hypothetical protein | |
| Rv2004c | 7.1 | 0.9 | Conserved hypothetical protein | |
| Rv2005c | 12.1 | 0.5 | Conserved hypothetical protein | |
| Rv2007c | fdxA | 82.0 | 0.9 | Probable ferredoxin |
| Rv2028c | 13.2 | 0.7 | Conserved hypothetical protein | |
| Rv2029c | pfkB | 20.8 | 0.9 | Probable phosphofructokinase |
| Rv2030c | 67.1 | 0.8 | Conserved hypothetical protein | |
| Rv2031c | hspX | 76.2 | 0.5 | 14-kD antigen, heat shock protein Hsp20 family |
| Rv2032 | acg | 30.8 | 0.5 | Conserved hypothetical protein |
| Rv2623 | 1.7 | 0.7 | Conserved hypothetical protein | |
| Rv2624c | 6.2 | 1.2 | Conserved hypothetical protein | |
| Rv2626c | 36.5 | 0.8 | Conserved hypothetical protein | |
| Rv2627c | 16.3 | 0.3 | Conserved hypothetical protein | |
| Rv2628 | 8.9 | 0.8 | Hypothetical protein | |
| Rv2629 | 8.1 | 0.9 | Conserved hypothetical protein | |
| Rv2630 | 4.2 | 1.6 | Hypothetical protein | |
| Rv3127 | 44.3 | 1.2 | Conserved hypothetical protein | |
| Rv3130c | tgs1 | 45.7 | 1.7 | Triacylglycerol synthase |
| Rv3131 | 41.0 | 0.9 | Conserved hypothetical protein | |
| Rv3132c | dosS | 16.7 | 0.4 | Sensor histidine kinase |
| Rv3133c | dosR | 11.1 | 0.6 | Two-component response regulator |
| Rv3134c | 26.0 | 0.7 | Conserved hypothetical protein | |
| Non-DosR | ||||
| Rv0467 | icl | 7.6 | 4.5 | Isocitrate lyase |
| Rv0791c | 5.0 | 3.5 | Conserved hypothetical protein | |
| Rv0792c | 4.4 | 3.2 | Probable GNTR family regulatory protein | |
| Rv0793 | 4.9 | 2.6 | Conserved hypothetical protein | |
| Rv2025c | 7.4 | 2.7 | Possible conserved membrane protein | |
| Rv2108 | PPE36 | 4.1 | 2.4 | PPE family protein |
| Rv2172c | 4.4 | 2.2 | Conserved hypothetical protein | |
| Rv3083 | 27.7 | 20.4 | Probable monooxygenase (hydroxylase) | |
| Rv3084 | lipR | 10.3 | 9.9 | Probable acetyl-hydrolase/esterase |
| Rv3086 | adhD | 7.3 | 8.8 | Probable zinc-type alcohol dehydrogenase |
| Rv3087 | 6.7 | 4.0 | Possible triacylglycerol synthase | |
| Rv3088 | 6.9 | 13.4 | Possible triacylglycerol synthase | |
| Rv3089 | fadD13 | 7.1 | 4.3 | Probable fatty-acid coenzyme A (CoA) ligase |
| Rv3841 | bfrB | 17.7 | 15.3 | Possible bacterioferritin |
Genes in the ΔdosT strain that were included in the table had an induction ratio of at least 4-fold and a corresponding SAM q value of 0.
Menaquinone analysis.
Bacterial cultures were concentrated by centrifugation, and 6 ml chloroform-methanol (2:1) containing 5 μg of vitamin K1 (Sigma) were added; the sample was stored in the dark at −20°C until processed. Samples were then placed on a rotating mixer (Glas-Col) for 3 h, and 1 ml of water was added to generate a phase separation. The lower, organic phase was washed once with water-methanol-chloroform (48:47:3 [vol/vol/vol]) and transferred to new tubes. The solvent was evaporated under a stream of N2, and the sample was dissolved in chloroform, loaded on a silicic acid column, and eluted with chloroform to remove the polar lipids. The elution solvent was removed from the resulting material under a N2 stream, and the sample was dissolved in ethanol in a volume normalized to the original culture OD600 and subjected to high-pressure liquid chromatography (HPLC) mass spectrometry (MS) on an HP1100 series HPLC connected to a 2000 Finnigan LCQ-DUO ion trap mass spectrometer with an APPI interface. HPLC separation was achieved using an XBridge column (Waters) and a gradient running from 100% methanol, for 2 min, to methanol-isopropanol (50:50 [vol/vol]) over 50 min at 0.4 ml min−1 with a column temperature of 40°C. Eluted molecules were subjected to positive ion MS using APPI as the ionization interface. Capillary temperature was 150°C, and APPI vaporizer temperature was 350°C.
RESULTS
DosR regulon genes respond to a reduced electron transport system.
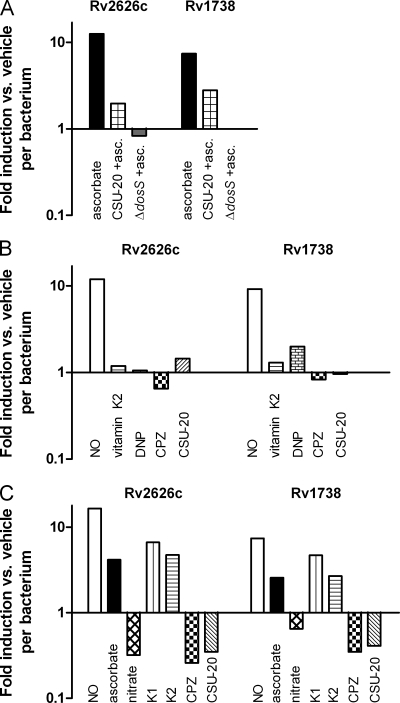
To examine DosR regulon induction, two representative regulon genes, Rv2626c and Rv1738, were selected for real-time PCR analysis. A dosT mutant was used for all experiments, except when noted, to ensure all regulation observed was solely from DosS. Ascorbate, a reductant of cytochrome c, was added to elucidate the role in DosR signaling via a reduced electron transport system (ETS). Robust induction of the two DosR regulon genes was observed when ascorbate was added to the ΔdosT strain maintained under aerobic conditions; however, no induction of the genes was observed with a ΔdosS strain (Fig. 1A). This indicated that a reductant of the ETS is capable of inducing the DosR regulon even in the presence of oxygen via DosS, but not DosT. When ascorbate was added to hypoxic cultures, increased induction of the regulon genes was also observed (Fig. 1C). However, the induction ratio was attenuated compared to that observed during aerobic conditions due to normal hypoxic induction of the regulon in the absence of ascorbate. The ETS is relatively more reduced during hypoxic conditions than during aerobic conditions due to the lack of oxygen and resulting diminution of aerobic respiration. To ensure observed signaling was not the result of hypoxia caused by the addition of ascorbate, oxygen levels were directly measured with an oxygen probe; ascorbate did not alter medium oxygen levels (Fig. 2). Oxyrase was used as a control for decreased oxygen levels, and a reduced amount of oxygen was observed. Furthermore, the lack of induction via DosT, a known oxygen sensor, indicates that oxygen levels were not responsible for induction via DosS by ascorbate.
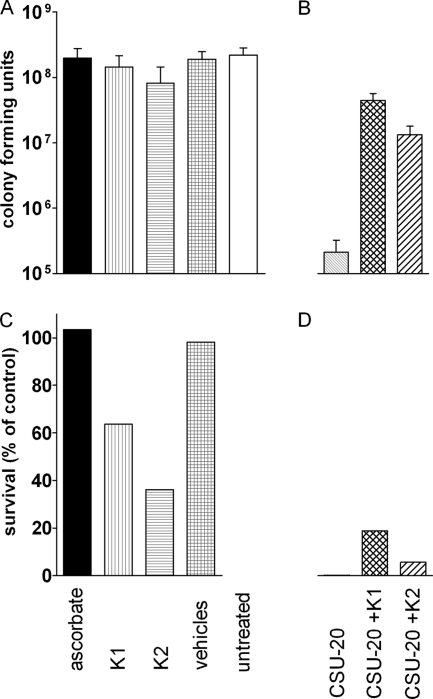
FIG. 1.
Induction of DosR regulon genes during aerobic cultures (A and B) and hypoxic conditions (C). The data shown represent fold induction of untreated or vehicle-treated cultures of the transcript copy numbers of two DosR regulon genes, Rv2626c and Rv1738, normalized to levels of Rv0239. All experiments were performed with a ΔdosT strain except when noted.
FIG. 2.
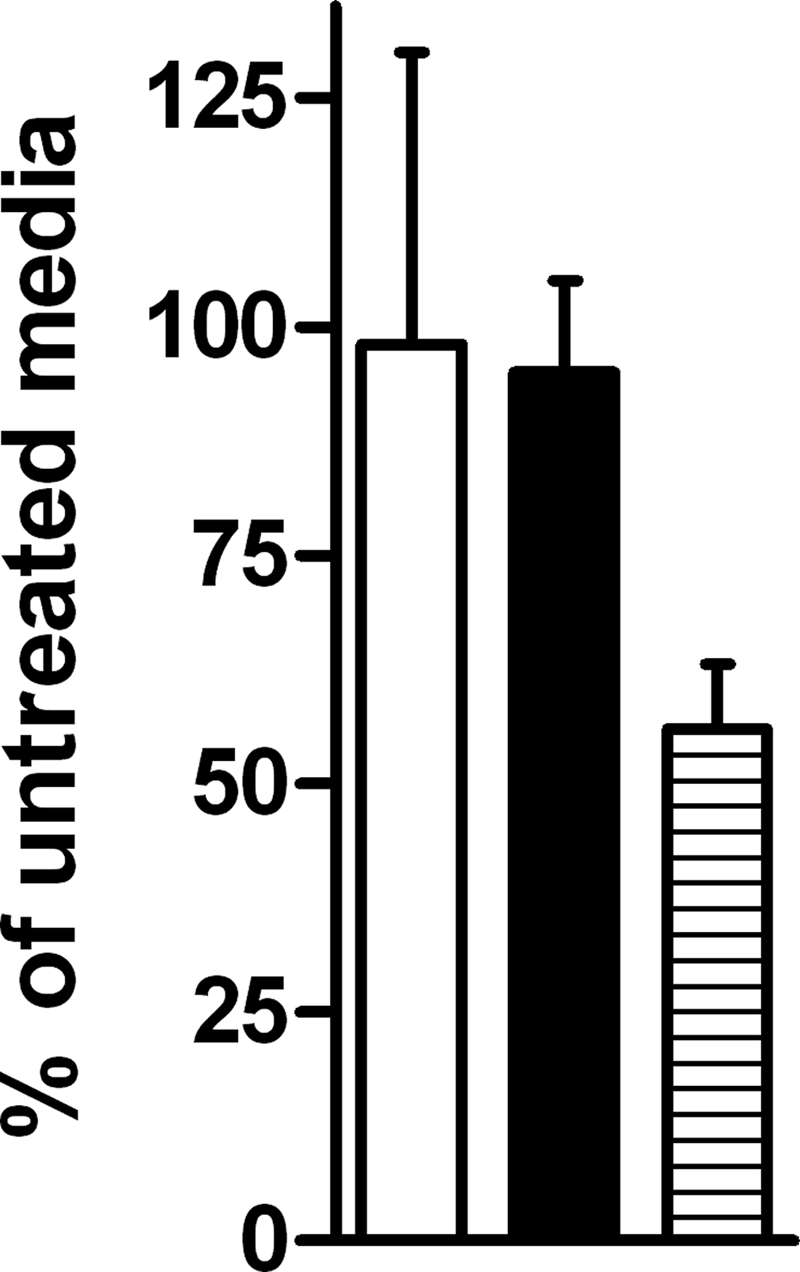
Oxygen levels of ascorbate-treated media. To ensure that observed transcriptional findings were not the result of diminished oxygen levels from the addition of ascorbate, an oxygen sensor was used for direct measurement of cultures. Results represent percent oxygen detected of untreated media. Bacterial culture is white, bacterial culture plus ascorbate is black, and media treated with Oxyrase is striped.
Additional experiments were conducted to further delineate the evident role of the ETS in DosS signaling. Chlorpromazine (CPZ), a type II oxidoreductase inhibitor, and CSU-20, a menaquinone synthesis inhibitor (27), were added to hypoxic cultures (Fig. 1C). Inhibition of oxidoreductases attenuates electron flow through the ETS, as does decreasing the size of the menaquinone pool by inhibiting menaquinone synthesis. The addition of both CPZ and CSU-20 to hypoxic cultures resulted in a marked reduction in signaling, and no effect on aerobic cultures was observed (Fig. 1B and C). When nitrate was added to hypoxic cultures, a corresponding decrease in signaling occurred (Fig. 1C). This was expected, as nitrate addition should result in oxidation of the menaquinone pool because of the ability of nitrate to function as an alternative electron acceptor. These findings corroborate the ascorbate data by correlating a decrease in electron flow into and through the menaquinone pool of the ETS (CPZ and CSU-20) or a decrease in reduction of the menaquinone pool (nitrate) with a decline in signaling. They also imply that either the menaquinone pool itself or a reduced component of the ETS is the signal monitored by DosS.
To test this further, vitamin K2, a functional unsaturated menaquinone analog, was added to cultures. Aerobically no effect was observed, but hypoxically an increased induction of the DosR regulon was observed, indicating that menaquinone levels were limiting for regulon induction during hypoxic conditions (Fig. 1B and C). When CSU-20 was added simultaneously with ascorbate to again correlate menaquinone levels and signaling, a decrease in signaling was observed compared to ascorbate-treated cultures, as expected (Fig. 1A).
Hypoxia could diminish overall proton motive force (PMF). Decreased PMF may also occur as a result of excess electrons in the ETS from the addition of ascorbate. To ensure that a decreased PMF is not the signal for induction of the DosR regulon, a protonophore, 2,4-dinitrophenol (DNP), was added to aerobic cultures. Addition of DNP resulted in no induction of the DosR regulon genes (shown in Fig. 1B and described previously in reference 3). Finally, nitric oxide was added to both aerobic and hypoxic conditions as a positive control for DosR regulon induction (53), and a 10-fold induction was observed in both conditions.
Microarray analysis of ascorbate-treated aerobic cultures.
To confirm the real-time PCR data as well as determine the overall effect of ascorbate on aerobic cultures, microarray analysis was performed on aerobic ΔdosT and ΔdosS mutant cultures treated with ascorbate. In the ΔdosT strain, 49 genes were significantly induced and had induction ratios greater than 4-fold (Table 1). Of these 49 genes, 35 were part of the DosR regulon and had an average induction ratio of 21-fold. Contrarily, the dosS strain had an average induction ratio of the DosR genes of 0.9-fold. Fourteen additional genes were induced in both mutants.
Change in menaquinone pool during entry into anaerobiosis.
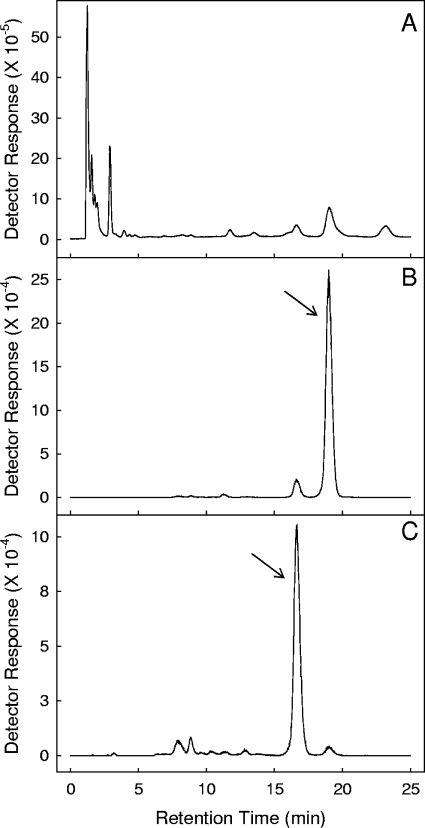
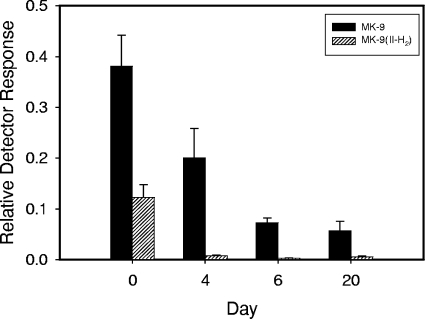
To elucidate the kinetics of the various species of menaquinone during this model as well as to show a correlation between menaquinone levels and signaling, the status of the menaquinone pool at various points during adaptation to anaerobic dormancy was next examined. The presence of oxygen in this system was as follows: time zero represents aerobic growth, day 4 represents hypoxic growth, and by day 6 the cultures were anaerobic (29). A rapid decrease in both species, MK-9(II-H2) as well as MK-9 (see Fig. 5 for structure references), was observed by the time the cultures became hypoxic (Fig. 3 and 4). However, at that point, MK-9(II-H2) ceased to decrease as rapidly and reached a fairly stable concentration, while MK-9 declined to below the limit of detection.
FIG. 3.
Representative mass spectral analysis of neutral lipids extracted from M. tuberculosis. (A) Total ion chromatogram. (B) Extracted ion chromatogram for 787.6315 Da, which corresponds to the molecular ion of MK-9(II-H2). (C) Extracted ion chromatogram for 785.6158 Da, which corresponds to the molecular ion of MK-9. Arrows indicate the peaks assigned to MK-9 (B) and MK-9(II-H2) (C).
FIG. 4.
Menaquinone content of M. tuberculosis H37Rv cells in the RAD dormancy model. Cultures were terminated at the indicated time point (days). Day 0 corresponds to logarithmic aerobic growth. Harvested cells were extracted with chloroform-methanol (2:1), and the nonpolar lipids were isolated using silicic acid column chromatography eluted with chloroform. Samples were normalized to the culture OD, subjected to LC-MS, and quantitated using vitamin K1 as an internal recovery standard to derive the relative detector response. MK-9(II-H2) is in black, and MK-9 is diagonally striped. By day 4, the levels of MK-9 had fallen to near the limit of detection.
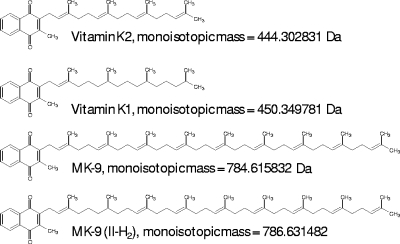
To examine the possible differential effect of menaquinone saturation status on DosS signaling, vitamin K1 and vitamin K2 were added to cultures (Fig. 1B). These compounds differ in the saturation status from the second isoprenoid group (from the head group) to the end (Fig. 5); vitamin K1 is saturated, and vitamin K2 is unsaturated. Therefore, they were added as surrogates for MK-9 and MK-9(II-H2), respectively, because the saturation status at the second isoprenoid group (from the head group) is equivalent (Fig. 5). Increased transcript levels for Rv2626 and Rv1738 were detected after the addition of vitamin K1 and were 2-fold higher relative to the addition of vitamin K2 (induction levels of the two genes versus those of the uninduced were 6.7/4.7 for K1 and 4.7/2.7 for K2) (Fig. 1B).
FIG. 5.
Structures and calculated monoisotopic masses of menaquinone analogs.
Differential survival effects of supplemental menaquinone analogs.
Under aerobic conditions it was previously demonstrated that vitamin K2 is better able than K1 (Fig. 5, structural information) to prevent killing caused by the menaquinone synthesis inhibitor CSU-20 (8). Changes in the saturation state of the menaquinone pools during anaerobic dormancy led us to determine the effects of these two menaquinones in rescuing killing by CSU-20 during anaerobic conditions. All treatments were added at day 12 of the anaerobic dormancy model, after the cultures were anaerobic for approximately 1 week, and were plated 8 days later. When treated with CSU-20, a decrease in survival of three orders of magnitude was observed (Fig. 6). Vitamin K1 had a statistically significant increased ability to rescue killing by CSU-20 compared to vitamin K2 (P < 0.0001), converse to what was reported under aerobic conditions (8), further indicating a preferred role for MK-9(II-H2) during dormancy conditions. In addition, supplementation with either menaquinone alone resulted in decreased survival of the anaerobic cultures. Vitamin K2 addition resulted in a statistically significant increase in killing compared to vitamin K1 (P < 0.05). As shown in Fig. 1, the addition of either vitamin K1 or K2 also resulted in increased DosR regulon expression. Therefore, ascorbate was tested as a control to determine if overexpression of the DosR regulon resulted in killing, which it did not (Fig. 6).
FIG. 6.
Vitamin K rescue of CSU-20-treated cultures. Cultures were treated on day 12 of the anaerobic dormancy model and incubated for 8 days. Values shown are CFU (A and B) and percent survival (C and D) derived from CFU of treated cultures divided by untreated or vehicle-treated cultures. (A and C) Ascorbate is black, vitamin K1 is vertically striped, vitamin K2 is horizontally striped, vehicles are gridded, and untreated is white. (B and D) CSU-20 is thinly diagonally striped, CSU-20 plus vitamin K1 is double diagonally striped, and CSU-20 plus vitamin K2 is thickly diagonally striped.
DISCUSSION
The findings in this report demonstrate a role for the ETS in DosS control of the DosR regulon. Data herein indicate that DosS, but not DosT, initiates signaling when treated with ascorbate, a cytochrome c reductant, even in the presence of oxygen. Interestingly, the effect of ascorbate is quite specific to the DosR regulon, although a small group of other genes is moderately affected by ascorbate in a DosS-independent manner (Table 1). This provides a link between electron flow and DosS signaling, a link further supported by the observation that inhibition of electron flow decreased signaling, as did the inhibition of menaquinone synthesis.
Regardless of the numerous routes of electron flow through the ETS, the menaquinone pool is always utilized, and thus, monitoring its redox status provides a mechanism for the bacillus to accurately sense the state of respiration. The concentration of menaquinones decreased during hypoxic entry into anaerobiosis. The unsaturated form disappeared, and the concentration of the saturated form initially declined but was maintained at a lower level. Experiments with saturated and unsaturated analogs of these species demonstrated differences in their role in signaling and survival and indicate that specific concentrations are maintained for optimal performance.
This work demonstrates that DosS responds to the redox status of the ETS; however, the direct interaction between the sensor and the ETS is as yet unknown. Recent biochemical work provides some intriguing possibilities. In vitro a ferredoxin/ferredoxin reductase pair was demonstrated to activate DosS (21). Ferredoxins are often reduced by electrons from the quinone pool; thus, there is a possible link to the ETS. The genome of M. tuberculosis contains several annotated ferredoxins, including fdxA, which is part of the DosR regulon and is thus upregulated during conditions in which DosS activates the DosR regulon (7, 53). A second report demonstrated that flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), cofactors used by many respiratory and ETS enzymes, were able to reduce DosS in vitro (6). The in vivo relevance of these interactions is as yet unknown, but their involvement with the ETS makes them interesting candidates for interaction with DosS (6). Interestingly, this same report did not observe a direct interaction between reduced menaquinone and DosS, implying that an intermediate is required. It is also worth mentioning that it was previously demonstrated with Mycobacterium smegmatis that DevS, which is a homolog to DosT (not to DosS), was insensitive to changes in redox and the menaquinone pool (28).
Several bacterial systems are known to sense components of the ETS in order to respond to a changing respiratory environment. In Escherichia coli, Aer responds to redox changes in the ETS to facilitate aerotaxis, although the mechanism of interaction is still unknown (11, 50). Also in E. coli, ArcB senses the redox state of the quinone pool (14, 30). In Helicobacter pylori, TlpD senses a decrease in electron flow and responds by altering its taxis away from electron-flow limiting conditions (45). Rhodobacter sphaeroides uses PrrBA to sense and respond to electron flow through the cbb3 oxidase (33, 34). PhoR in Bacillus subtilis responds to the reduced quinones, and several other bacterial sensors containing redox-active cysteine residues have been reported (12). Thus, it is apparent that monitoring the state of the ETS is an important function in many bacteria.
In M. tuberculosis, the DosR regulon is important for conditions that inhibit respiration, and several of these conditions result in its induction. Aside from reduction of the ETS, the slowing of aerobic respiration affects other systems which could function as signals for DosR regulon induction. Proton motive force may decrease under conditions that inhibit aerobic respiration; however, the addition of a protonophore does not induce the DosR regulon (shown in Fig. 1B and described previously in reference 3). A decreased level of ATP is likely not the signal, because while hypoxia does lower the overall amount of ATP, other additives that decrease ATP do not induce the regulon (3). Changes in respiration may have an effect on the NADH/NAD ratio; however, no correlation was shown between this ratio and DosR regulon induction via the addition of a wide variety of different additives (3). Cyanide blocks regulon induction by hypoxia and NO (53), likely by directly binding the heme moieties in DosT and DosS, an interaction shown for DosS (22). However, cyanide, a cytochrome c oxidase inhibitor, does not induce the regulon itself (3, 53). Azide, another cytochrome oxidase inhibitor, also does not affect regulon induction (22); these results imply that reduced cytochrome oxidase is not directly sensed by DosS.
Interestingly, W/Beijing strains of M. tuberculosis constitutively express the DosR regulon, and some lineages have numerous duplications and deletions, including a duplication of dosR as well as a portion of the DosR regulon (9, 13, 37). These strains also have an inactivating frameshift mutation in DosT (13, 37). It was observed in these reports that neither the DosT mutation nor the duplication of the regulon was responsible for the constitutive expression levels. In light of the findings in the present work, it seems feasible that these strains may have a metabolic abnormality which results in a constitutively reduced ETS, which could result in overexpression of the regulon via DosS. The plausibility of this is corroborated by the finding of additional gene duplications in these strains, specifically of the nuo gene cluster, responsible for encoding the type I oxidoreductase, as well as duplication of the succinate dehydrogenase gene (9). Both of the enzyme clusters encoded by these genes are involved in the ETS. We hypothesize that the mutation in DosT may be an evolutionary attempt to attenuate DosR regulon expression in certain conditions in order to counteract its overexpression.
Oxygen-limited, nonreplicating bacilli require electron flow via menaquinones, an intact proton motive force, and ATP production for survival (8, 24, 36). The major menaquinone species in mycobacteria is MK-9(II-H2), a 1,4-naphtoquinone derivative which has nine isoprene units, the second of which is saturated (Fig. 5) (31). Mono-saturated menaquinones of this type have been described for various organisms (35). Several other species of menaquinone exist in mycobacteria, although they are less prevalent, including a fully unsaturated form of the major species, MK-9 (8, 10). Functional differences between these two types of quinone species are presently unclear in the literature. Treatment with CSU-20 was shown to decrease the amount of both species of menaquinone [MK-9(II-H2) and MK-9] via menA inhibition; it was also shown to inhibit oxygen consumption (8). In this report we observed that the level of menaquinone correlated with the amount of DosR regulon expression. When oxygen became limited, the levels of menaquinone decreased, while restoring menaquinone to the system resulted in increased signaling. The decrease in menaquinone levels during anaerobic dormancy appears to result in the gradual decrease in DosR regulon expression observed in long-term models (54).
Aerobically, it was previously reported that the menaquinone analogs vitamin K1 and vitamin K2 [analogous to MK-9(II-H2) and MK-9, respectively, because of the saturation status of the second isoprenoid group; Fig. 5] were both able to rescue killing and decreased oxygen consumption caused by CSU-20; further, K2 rescued more effectively than did K1 (8). Interestingly, results reported herein demonstrate that the opposite rescuing effect occurs during anaerobic conditions: K1 rescued more effectively than K2 (Fig. 6). Differential effects of vitamin K1 and K2 as well as other related naphthoquinones have previously been reported for bacterial extracts from Mycobacterium phlei, further corroborating biological significance of the saturation status (4).
Overall, the survival data indicate that the bacilli maintain an optimal level of menaquinones during anaerobiosis, given that inhibition of synthesis results in death, but supplementation does as well. Also, it appears that not only is the amount important, but the species is as well, since addition of vitamin K2 resulted in more death than vitamin K1. The role of MK-9(II-H2) appears to be specifically important during anaerobiosis. This is based on the observed decrease of MK-9 to the level of detection and the maintenance of MK-9(II-H2) during the hypoxic adaptation period (Fig. 4). The increased anaerobic rescuing effect by K1 could be partially due to the fact that the DosR regulon is more induced by K1 than K2 (Fig. 1), and previously we reported a correlation between regulon induction and anaerobic survival (18). Thus, to summarize the menaquinone results, MK-9(II-H2) is maintained at a low level. Its analog, vitamin K1, induces the DosR regulon more effectively, rescues killing by CSU-20 more effectively, and is less toxic relative to vitamin K2, the analog of MK-9.
A recent report examined the effects of ascorbate-induced hypoxia on the DosR regulon (49). As previously reported (18), differential effects and response to oxygen levels were observed for single-sensor mutants of DosS and DosT. However, the results in our current work clearly separate the effects of oxygen and redox sensing by the two sensors. Ascorbate experiments in this work were performed at a time point when oxygen was not sufficiently limited to induce either sensor (Fig. 1 and 2 and Table 1), unlike the previously mentioned report. This enabled the differences between DosS and DosT sensing to be observed, differences which were confirmed by a variety of other additives which modulate the ETS.
This report demonstrates that induction of the DosR regulon via DosS is in response to a reduced ETS. Despite this role of DosS in redox monitoring, under aerobic conditions oxygen maintains DosS in an inactive, oxidized form. Thus, it appears that both bodies of research on DosS signaling are correct: DosS functions as both an oxygen sensor (off switch) and redox sensor (on switch). The menaquinone pool is explicitly implicated in this sensing. Menaquinone levels in general decrease, and the saturated form is preferentially maintained at low but functional levels under anaerobic conditions. Relative to the unsaturated form, this saturated form is less toxic, better able to induce signaling via DosS during hypoxia, and better able to rescue a drug-mediated decrease in the menaquinone pool. These data indicate that the saturated form of menaquinone is best utilized during oxygen-limited conditions. The menaquinone pool is dynamic, its composition changes with the respiratory environment, and its centrality in the ETS makes it a viable target for monitoring the redox status of the ETS.
Acknowledgments
This research project was funded by NIH grants RO1 AI061505, entitled the Mycobacterium tuberculosis Dormancy Program and awarded to M.I.V., AI049151 awarded to D.C.C., and T32 AI052066-06 awarded to R.W.H.
Footnotes
Published ahead of print on 15 October 2010.
REFERENCES
- 1.Anonymous. 1953. Report of panel discussion on survival and revival of tubercle bacilli in healed tuberculous lesions. Am. Rev. Tuberc. 68:477-495. [DOI] [PubMed] [Google Scholar]
- 2.Balázsi, G., A. P. Heath, L. Shi, and M. L. Gennaro. 2008. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 4:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boshoff, H. I., T. G. Myers, B. R. Copp, M. R. McNeil, M. A. Wilson, and C. E. Barry III. 2004. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J. Biol. Chem. 279:40174-40184. [DOI] [PubMed] [Google Scholar]
- 4.Brodie, A. F., and J. Ballantine. 1960. Oxidative phosphorylation in fractionated bacterial systems. III. Specificity of vitamin K reactivation. J. Biol. Chem. 235:232-237. [PubMed] [Google Scholar]
- 5.Canetti, G. 1955. The tubercle bacillus in the pulmonary lesion of man. Springer, New York, NY.
- 6.Cho, H. Y., H. J. Cho, Y. M. Kim, J. I. Oh, and B. S. Kang. 2009. Structural insight into the heme-based redox sensing by DosS from Mycobacterium tuberculosis. J. Biol. Chem. 284:13057-13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Tuberculist. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Dhiman, R. K., S. Mahapatra, R. A. Slayden, M. E. Boyne, A. Lenaerts, J. C. Hinshaw, S. K. Angala, D. Chatterjee, K. Biswas, P. Narayanasamy, M. Kurosu, and D. C. Crick. 2009. Menaquinone synthesis is critical for maintaining mycobacterial viability during exponential growth and recovery from non-replicating persistence. Mol. Microbiol. 72:85-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domenech, P., G. S. Kolly, L. Leon-Solis, A. Fallow, and M. B. Reed. 2010. Massive gene duplication event among clinical isolates of the Mycobacterium tuberculosis W/Beijing family. J. Bacteriol. 192:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunphy, P. J., P. G. Phillips, and A. F. Brodie. 1971. Separation and identification of menaquinones from microorganisms. J. Lipid Res. 12:442-449. [PubMed] [Google Scholar]
- 11.Edwards, J. C., M. S. Johnson, and B. L. Taylor. 2006. Differentiation between electron transport sensing and proton motive force sensing by the Aer and Tsr receptors for aerotaxis. Mol. Microbiol. 62:823-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eldakak, A., and F. M. Hulett. 2007. Cys303 in the histidine kinase PhoR is crucial for the phosphotransfer reaction in the PhoPR two-component system in Bacillus subtilis. J. Bacteriol. 189:410-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fallow, A., P. Domenech, and M. B. Reed. 2010. Strains of the East Asian (W/Beijing) lineage of Mycobacterium tuberculosis are DosS/DosT-DosR two-component regulatory system natural mutants. J. Bacteriol. 192:2228-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292:2314-2316. [DOI] [PubMed] [Google Scholar]
- 15.Gomez, J. E., and J. D. McKinney. 2004. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb.) 84:29-44. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalo-Asensio, J., S. Mostowy, J. Harders-Westerveen, K. Huygen, R. Hernandez-Pando, J. Thole, M. Behr, B. Gicquel, and C. Martin. 2008. PhoP: a missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS One 3:e3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernández-Pando, R., M. Jeyanathan, G. Mengistu, D. Aguilar, H. Orozco, M. Harboe, G. A. Rook, and G. Bjune. 2000. Persistence of DNA from Mycobacterium tuberculosis in superficially normal lung tissue during latent infection. Lancet 356:2133-2138. [DOI] [PubMed] [Google Scholar]
- 18.Honaker, R. W., R. L. Leistikow, I. L. Bartek, and M. I. Voskuil. 2009. Unique roles of DosT and DosS in DosR regulon induction and Mycobacterium tuberculosis dormancy. Infect. Immun. 77:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honaker, R. W., A. Stewart, S. Schittone, A. Izzo, M. R. Klein, and M. I. Voskuil. 2008. Mycobacterium bovis BCG vaccine strains lack narK2 and narX induction and exhibit altered phenotypes during dormancy. Infect. Immun. 76:2587-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imboden, P., and G. K. Schoolnik. 1998. Construction and characterization of a partial Mycobacterium tuberculosis cDNA library of genes expressed at reduced oxygen tension. Gene 213:107-117. [DOI] [PubMed] [Google Scholar]
- 21.Ioanoviciu, A., Y. T. Meharenna, T. L. Poulos, and P. R. Ortiz de Montellano. 2009. DevS oxy complex stability identifies this heme protein as a gas sensor in Mycobacterium tuberculosis dormancy. Biochemistry 48:5839-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ioanoviciu, A., E. T. Yukl, P. Moenne-Loccoz, and P. R. de Montellano. 2007. DevS, a heme-containing two-component oxygen sensor of Mycobacterium tuberculosis. Biochemistry 46:4250-4260. [DOI] [PubMed] [Google Scholar]
- 23.Kalendar, R. 2006. FastPCR, PCR primer design, DNA and protein tools, repeats and own database searches program. Helsinki, Finland. http://www.biocenter.helsinki.fi/bi/programs/fastpcr.htm.
- 24.Koul, A., L. Vranckx, N. Dendouga, W. Balemans, I. V. Den Wyngaert, K. Vergauwen, H. W. Goehlmann, R. Willebrords, A. Poncelet, J. Guillemont, D. Bald, and K. Andries. 2008. Diarylquinolines are bactericidal for dormant mycobacteria as a result of disturbed ATP homeostasis. J. Biol. Chem. 283:25273-25280. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, A., J. S. Deshane, D. K. Crossman, S. Bolisetty, B. S. Yan, I. Kramnik, A. Agarwal, and A. J. Steyn. 2008. Heme oxygenase-1 derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J. Biol. Chem. 283:18032-18039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, A., J. C. Toledo, R. P. Patel, J. R. Lancaster, Jr., and A. J. Steyn. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. U. S. A. 104:11568-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurosu, M., P. Narayanasamy, K. Biswas, R. Dhiman, and D. C. Crick. 2007. Discovery of 1,4-dihydroxy-2-naphthoate [corrected] prenyltransferase inhibitors: new drug leads for multidrug-resistant gram-positive pathogens. J. Med. Chem. 50:3973-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, J. M., H. Y. Cho, H. J. Cho, I. J. Ko, S. W. Park, H. S. Baik, J. H. Oh, C. Y. Eom, Y. M. Kim, B. S. Kang, and J. I. Oh. 2008. O2 and NO sensing mechanism through the DevSR two-component system in Mycobacterium smegmatis. J. Bacteriol. 190:6795-6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leistikow, R. L., R. A. Morton, I. L. Bartek, I. Frimpong, K. Wagner, and M. I. Voskuil. 2010. The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from non-respiring dormancy. J. Bacteriol. 192:1662-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malpica, R., B. Franco, C. Rodriguez, O. Kwon, and D. Georgellis. 2004. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. U. S. A. 101:13318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minnikin, D. E., S. M. Minnikin, M. Goodfellow, and J. L. Stanford. 1982. The mycolic acids of Mycobacterium chelonei. J. Gen. Microbiol. 128:817-822. [DOI] [PubMed] [Google Scholar]
- 32.Neyrolles, O., R. Hernandez-Pando, F. Pietri-Rouxel, P. Fornes, L. Tailleux, J. A. Payan, E. Pivert, Y. Bordat, D. Aguilar, M. C. Prevost, C. Petit, and B. Gicquel. 2006. Is adipose tissue a place for Mycobacterium tuberculosis persistence? PLoS One 1:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh, J. I., and S. Kaplan. 2000. Redox signaling: globalization of gene expression. EMBO J. 19:4237-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh, J. I., I. J. Ko, and S. Kaplan. 2004. Reconstitution of the Rhodobacter sphaeroides cbb3-PrrBA signal transduction pathway in vitro. Biochemistry 43:7915-7923. [DOI] [PubMed] [Google Scholar]
- 35.Phillips, P. G., P. J. Dunphy, K. L. Servis, and A. F. Brodie. 1969. A new menaquinone series differing in the degree of unsaturation of the side chain. Biochemistry 8:2856-2861. [DOI] [PubMed] [Google Scholar]
- 36.Rao, S. P., S. Alonso, L. Rand, T. Dick, and K. Pethe. 2008. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 105:11945-11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed, M. B., S. Gagneux, K. Deriemer, P. M. Small, and C. E. Barry III. 2007. The W-Beijing lineage of Mycobacterium tuberculosis overproduces triglycerides and has the DosR dormancy regulon constitutively upregulated. J. Bacteriol. 189:2583-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ribiere, C., A. M. Jaubert, N. Gaudiot, D. Sabourault, M. L. Marcus, J. L. Boucher, D. Denis-Henriot, and Y. Giudicelli. 1996. White adipose tissue nitric oxide synthase: a potential source for NO production. Biochem. Biophys. Res. Commun. 222:706-712. [DOI] [PubMed] [Google Scholar]
- 39.Roberts, D. M., R. P. Liao, G. Wisedchaisri, W. G. Hol, and D. R. Sherman. 2004. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 279:23082-23087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russel, W. F., S. H. Dressler, G. Middlebrook, and J. Denst. 1955. Implications of the phenomenon of open cavity healing for the chemotherapy of pulmonary tuberculosis. Am. Rev. Tuberc. 71:441-446. [PubMed] [Google Scholar]
- 41.Saini, D. K., V. Malhotra, and J. S. Tyagi. 2004. Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett. 565:75-80. [DOI] [PubMed] [Google Scholar]
- 42.Sambandamurthy, V. K., S. C. Derrick, T. Hsu, B. Chen, M. H. Larsen, K. V. Jalapathy, M. Chen, J. Kim, S. A. Porcelli, J. Chan, S. L. Morris, and W. R. Jacobs, Jr. 2006. Mycobacterium tuberculosis DeltaRD1 DeltapanCD: a safe and limited replicating mutant strain that protects immunocompetent and immunocompromised mice against experimental tuberculosis. Vaccine 24:6309-6320. [DOI] [PubMed] [Google Scholar]
- 43.Sardiwal, S., S. L. Kendall, F. Movahedzadeh, S. C. Rison, N. G. Stoker, and S. Djordjevic. 2005. A GAF domain in the hypoxia/NO-inducible Mycobacterium tuberculosis DosS protein binds haem. J. Mol. Biol. 353:929-936. [DOI] [PubMed] [Google Scholar]
- 44.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweinitzer, T., T. Mizote, N. Ishikawa, A. Dudnik, S. Inatsu, S. Schreiber, S. Suerbaum, S. Aizawa, and C. Josenhans. 2008. Functional characterization and mutagenesis of the proposed behavioral sensor TlpD of Helicobacter pylori. J. Bacteriol. 190:3244-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc. Natl. Acad. Sci. U. S. A. 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiloh, M. U., P. Manzanillo, and J. S. Cox. 2008. Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe 3:323-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sousa, E. H., J. R. Tuckerman, G. Gonzalez, and M. A. Gilles-Gonzalez. 2007. DosT and DevS are oxygen-switched kinases in Mycobacterium tuberculosis. Protein Sci. 16:1708-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taneja, N. K., S. Dhingra, A. Mittal, M. Naresh, and J. S. Tyagi. 2010. Mycobacterium tuberculosis transcriptional adaptation, growth arrest and dormancy phenotype development is triggered by vitamin C. PLoS One 5:e10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor, B. L. 2007. Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Mol. Microbiol. 65:1415-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trayhurn, P., and I. S. Wood. 2004. Adipokines: inflammation and the pleiotropic role of white adipose tissue. Br. J. Nutr. 92:347-355. [DOI] [PubMed] [Google Scholar]
- 52.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voskuil, M. I., K. C. Visconti, and G. K. Schoolnik. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb.) 84:218-227. [DOI] [PubMed] [Google Scholar]
- 55.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
- 56.Wolstenholme, G. E. W., M. P. Cameron, and C. M. O'Connor (ed.). 1955. Ciba Foundation symposium on experimental tuberculosis, bacillus and host. Little, Brown, and Company, Boston, MA.
- 57.Yan, H., E. Aziz, G. Shillabeer, A. Wong, D. Shanghavi, A. Kermouni, M. Abdel-Hafez, and D. C. Lau. 2002. Nitric oxide promotes differentiation of rat white preadipocytes in culture. J. Lipid Res. 43:2123-2129. [DOI] [PubMed] [Google Scholar]
- 58.Yuan, Y., D. D. Crane, R. M. Simpson, Y. Q. Zhu, M. J. Hickey, D. R. Sherman, and C. E. Barry III. 1998. The 16-kDa alpha-crystallin (Acr) protein of Mycobacterium tuberculosis is required for growth in macrophages. Proc. Natl. Acad. Sci. U. S. A. 95:9578-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yukl, E. T., A. Ioanoviciu, M. M. Nakano, P. R. Ortiz de Montellano, and P. Moenne-Loccoz. 2008. A distal tyrosine residue is required for ligand discrimination in DevS from Mycobacterium tuberculosis. Biochemistry 47:12532-12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yun, Z., H. L. Maecker, R. S. Johnson, and A. J. Giaccia. 2002. Inhibition of PPAR gamma 2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev. Cell 2:331-341. [DOI] [PubMed] [Google Scholar]