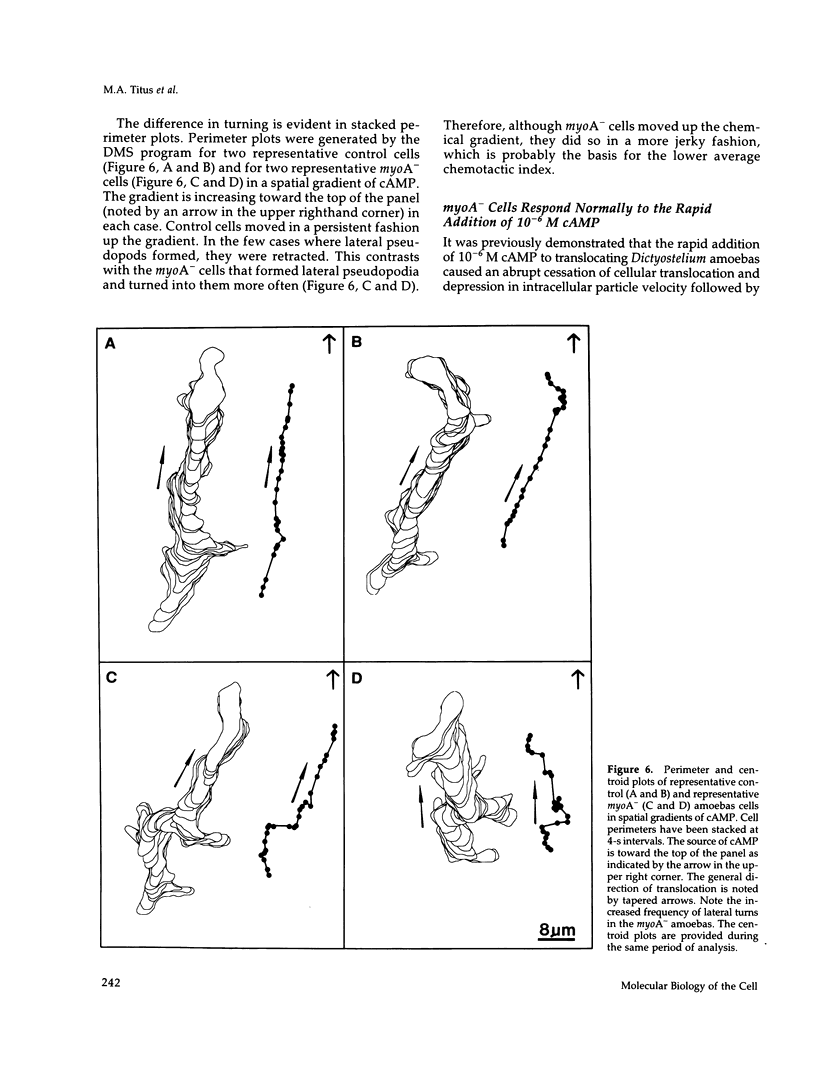
Abstract
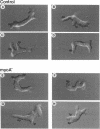
The myoA gene of Dictyostelium is a member of a gene family of unconventional myosins. The myosin Is share homologous head and basic domains, but the myoA gene product lacks the glycine-, proline-, alanine-rich and src homology 3 domains typical of several of the other myosin Is. A mutant strain of Dictyostelium lacking a functional myoA gene was produced by gene targeting, and the motility of this strain in buffer and a spatial gradient of the chemoattractant cyclic AMP was analyzed by computer-assisted methods. The myoA- cells have a normal elongate morphology in buffer but exhibit a decrease in the instantaneous velocity of cellular translocation, an increase in the frequency of lateral pseudopod formation, and an increase in turning. In a spatial gradient, in which the frequency of pseudopod formation is depressed, myoA- cells exhibit positive chemotaxis but still turn several times more frequently than control cells. These results demonstrate that the other members of the unconventional myosin family do not fully compensate for the loss of functional myoA gene product. Surprisingly, the phenotype of the myoA- strain closely resembles that of the myoB- strain, suggesting that both play a role in the frequency of pseudopod formation and turning during cellular translocation.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. J., Pollard T. D. Binding of myosin I to membrane lipids. Nature. 1989 Aug 17;340(6234):565–568. doi: 10.1038/340565a0. [DOI] [PubMed] [Google Scholar]
- Albanesi J. P., Fujisaki H., Hammer J. A., 3rd, Korn E. D., Jones R., Sheetz M. P. Monomeric Acanthamoeba myosins I support movement in vitro. J Biol Chem. 1985 Jul 25;260(15):8649–8652. [PubMed] [Google Scholar]
- Barylko B., Wagner M. C., Reizes O., Albanesi J. P. Purification and characterization of a mammalian myosin I. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):490–494. doi: 10.1073/pnas.89.2.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carboni J. M., Conzelman K. A., Adams R. A., Kaiser D. A., Pollard T. D., Mooseker M. S. Structural and immunological characterization of the myosin-like 110-kD subunit of the intestinal microvillar 110K-calmodulin complex: evidence for discrete myosin head and calmodulin-binding domains. J Cell Biol. 1988 Nov;107(5):1749–1757. doi: 10.1083/jcb.107.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. H., Borysenko C. W. The 110,000-dalton actin- and calmodulin-binding protein from intestinal brush border is a myosin-like ATPase. J Biol Chem. 1984 Nov 25;259(22):14128–14135. [PubMed] [Google Scholar]
- Coluccio L. M., Bretscher A. Mapping of the microvillar 110K-calmodulin complex: calmodulin-associated or -free fragments of the 110-kD polypeptide bind F-actin and retain ATPase activity. J Cell Biol. 1988 Feb;106(2):367–373. doi: 10.1083/jcb.106.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelman K. A., Mooseker M. S. The 110-kD protein-calmodulin complex of the intestinal microvillus is an actin-activated MgATPase. J Cell Biol. 1987 Jul;105(1):313–324. doi: 10.1083/jcb.105.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté G. P., Albanesi J. P., Ueno T., Hammer J. A., 3rd, Korn E. D. Purification from Dictyostelium discoideum of a low-molecular-weight myosin that resembles myosin I from Acanthamoeba castellanii. J Biol Chem. 1985 Apr 25;260(8):4543–4546. [PubMed] [Google Scholar]
- De Lozanne A., Spudich J. A. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987 May 29;236(4805):1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- DeLozanne A., Lewis M., Spudich J. A., Leinwand L. A. Cloning and characterization of a nonmuscle myosin heavy chain cDNA. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6807–6810. doi: 10.1073/pnas.82.20.6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerec M., Adelberg E. A., Clark A. J., Hartman P. E. A proposal for a uniform nomenclature in bacterial genetics. Genetics. 1966 Jul;54(1):61–76. doi: 10.1093/genetics/54.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doberstein S. K., Pollard T. D. Localization and specificity of the phospholipid and actin binding sites on the tail of Acanthamoeba myosin IC. J Cell Biol. 1992 Jun;117(6):1241–1249. doi: 10.1083/jcb.117.6.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhoff T. T., Titus M. A., Manstein D. J., Ruppel K. M., Spudich J. A. Molecular genetic tools for study of the cytoskeleton in Dictyostelium. Methods Enzymol. 1991;196:319–334. doi: 10.1016/0076-6879(91)96029-q. [DOI] [PubMed] [Google Scholar]
- Fukui Y., De Lozanne A., Spudich J. A. Structure and function of the cytoskeleton of a Dictyostelium myosin-defective mutant. J Cell Biol. 1990 Feb;110(2):367–378. doi: 10.1083/jcb.110.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y., Lynch T. J., Brzeska H., Korn E. D. Myosin I is located at the leading edges of locomoting Dictyostelium amoebae. Nature. 1989 Sep 28;341(6240):328–331. doi: 10.1038/341328a0. [DOI] [PubMed] [Google Scholar]
- Hammer J. A. Novel myosins. Trends Cell Biol. 1991 Aug;1(2-3):50–56. doi: 10.1016/0962-8924(91)90089-r. [DOI] [PubMed] [Google Scholar]
- Hayden S. M., Wolenski J. S., Mooseker M. S. Binding of brush border myosin I to phospholipid vesicles. J Cell Biol. 1990 Aug;111(2):443–451. doi: 10.1083/jcb.111.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howley P. M., Israel M. A., Law M. F., Martin M. A. A rapid method for detecting and mapping homology between heterologous DNAs. Evaluation of polyomavirus genomes. J Biol Chem. 1979 Jun 10;254(11):4876–4883. [PubMed] [Google Scholar]
- Jung G., Hammer J. A., 3rd Generation and characterization of Dictyostelium cells deficient in a myosin I heavy chain isoform. J Cell Biol. 1990 Jun;110(6):1955–1964. doi: 10.1083/jcb.110.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G., Saxe C. L., 3rd, Kimmel A. R., Hammer J. A., 3rd Dictyostelium discoideum contains a gene encoding a myosin I heavy chain. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6186–6190. doi: 10.1073/pnas.86.16.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht D. A., Cohen S. M., Loomis W. F., Lodish H. F. Developmental regulation of Dictyostelium discoideum actin gene fusions carried on low-copy and high-copy transformation vectors. Mol Cell Biol. 1986 Nov;6(11):3973–3983. doi: 10.1128/mcb.6.11.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht D. A., Loomis W. F. Antisense RNA inactivation of myosin heavy chain gene expression in Dictyostelium discoideum. Science. 1987 May 29;236(4805):1081–1086. doi: 10.1126/science.3576221. [DOI] [PubMed] [Google Scholar]
- Korn E. D., Hammer J. A., 3rd Myosins of nonmuscle cells. Annu Rev Biophys Biophys Chem. 1988;17:23–45. doi: 10.1146/annurev.bb.17.060188.000323. [DOI] [PubMed] [Google Scholar]
- Kuspa A., Maghakian D., Bergesch P., Loomis W. F. Physical mapping of genes to specific chromosomes in Dictyostelium discoideum. Genomics. 1992 May;13(1):49–61. doi: 10.1016/0888-7543(92)90201-3. [DOI] [PubMed] [Google Scholar]
- Lynch T. J., Albanesi J. P., Korn E. D., Robinson E. A., Bowers B., Fujisaki H. ATPase activities and actin-binding properties of subfragments of Acanthamoeba myosin IA. J Biol Chem. 1986 Dec 25;261(36):17156–17162. [PubMed] [Google Scholar]
- Lynch T. J., Brzeska H., Miyata H., Korn E. D. Purification and characterization of a third isoform of myosin I from Acanthamoeba castellanii. J Biol Chem. 1989 Nov 15;264(32):19333–19339. [PubMed] [Google Scholar]
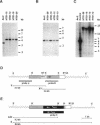
- Manstein D. J., Titus M. A., De Lozanne A., Spudich J. A. Gene replacement in Dictyostelium: generation of myosin null mutants. EMBO J. 1989 Mar;8(3):923–932. doi: 10.1002/j.1460-2075.1989.tb03453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta H., Gadasi H., Collins J. H., Korn E. D. Multiple forms of Acanthamoeba myosin I. J Biol Chem. 1979 May 10;254(9):3624–3630. [PubMed] [Google Scholar]
- Mayer B. J., Hamaguchi M., Hanafusa H. A novel viral oncogene with structural similarity to phospholipase C. Nature. 1988 Mar 17;332(6161):272–275. doi: 10.1038/332272a0. [DOI] [PubMed] [Google Scholar]
- Miyata H., Bowers B., Korn E. D. Plasma membrane association of Acanthamoeba myosin I. J Cell Biol. 1989 Oct;109(4 Pt 1):1519–1528. doi: 10.1083/jcb.109.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., Rubin G. M. The Drosophila ninaC locus encodes two photoreceptor cell specific proteins with domains homologous to protein kinases and the myosin heavy chain head. Cell. 1988 Mar 11;52(5):757–772. doi: 10.1016/0092-8674(88)90413-8. [DOI] [PubMed] [Google Scholar]
- Mooseker M. S., Coleman T. R. The 110-kD protein-calmodulin complex of the intestinal microvillus (brush border myosin I) is a mechanoenzyme. J Cell Biol. 1989 Jun;108(6):2395–2400. doi: 10.1083/jcb.108.6.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellen W., Datta S., Reymond C., Sivertsen A., Mann S., Crowley T., Firtel R. A. Molecular biology in Dictyostelium: tools and applications. Methods Cell Biol. 1987;28:67–100. doi: 10.1016/s0091-679x(08)61637-4. [DOI] [PubMed] [Google Scholar]
- Pasternak C., Spudich J. A., Elson E. L. Capping of surface receptors and concomitant cortical tension are generated by conventional myosin. Nature. 1989 Oct 12;341(6242):549–551. doi: 10.1038/341549a0. [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Doberstein S. K., Zot H. G. Myosin-I. Annu Rev Physiol. 1991;53:653–681. doi: 10.1146/annurev.ph.53.030191.003253. [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Korn E. D. Acanthamoeba myosin. I. Isolation from Acanthamoeba castellanii of an enzyme similar to muscle myosin. J Biol Chem. 1973 Jul 10;248(13):4682–4690. [PubMed] [Google Scholar]
- Porter J. A., Hicks J. L., Williams D. S., Montell C. Differential localizations of and requirements for the two Drosophila ninaC kinase/myosins in photoreceptor cells. J Cell Biol. 1992 Feb;116(3):683–693. doi: 10.1083/jcb.116.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D. R. "DMS," a computer-assisted system for quantitating motility, the dynamics of cytoplasmic flow, and pseudopod formation: its application to Dictyostelium chemotaxis. Cell Motil Cytoskeleton. 1988;10(1-2):91–106. doi: 10.1002/cm.970100114. [DOI] [PubMed] [Google Scholar]
- Soll D. R. Methods for manipulating and investigating developmental timing in Dictyostelium discoideum. Methods Cell Biol. 1987;28:413–431. doi: 10.1016/s0091-679x(08)61660-x. [DOI] [PubMed] [Google Scholar]
- Soll D. R., Voss E., Varnum-Finney B., Wessels D. "Dynamic Morphology System": a method for quantitating changes in shape, pseudopod formation, and motion in normal and mutant amoebae of Dictyostelium discoideum. J Cell Biochem. 1988 Jun;37(2):177–192. doi: 10.1002/jcb.240370205. [DOI] [PubMed] [Google Scholar]
- Spudich J. A. Dictyostelium discoideum: methods and perspectives for study of cell motility. Methods Cell Biol. 1982;25(Pt B):359–364. doi: 10.1016/s0091-679x(08)61433-8. [DOI] [PubMed] [Google Scholar]
- Spudich J. A. In pursuit of myosin function. Cell Regul. 1989 Nov;1(1):1–11. doi: 10.1091/mbc.1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M. L., Ferenz C. R., Kelleher K. L., Kriz R. W., Knopf J. L. Sequence similarity of phospholipase C with the non-catalytic region of src. Nature. 1988 Mar 17;332(6161):269–272. doi: 10.1038/332269a0. [DOI] [PubMed] [Google Scholar]
- Titus M. A., Warrick H. M., Spudich J. A. Multiple actin-based motor genes in Dictyostelium. Cell Regul. 1989 Nov;1(1):55–63. doi: 10.1091/mbc.1.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnum-Finney B. J., Voss E., Soll D. R. Frequency and orientation of pseudopod formation of Dictyostelium discoideum amebae chemotaxing in a spatial gradient: further evidence for a temporal mechanism. Cell Motil Cytoskeleton. 1987;8(1):18–26. doi: 10.1002/cm.970080104. [DOI] [PubMed] [Google Scholar]
- Varnum B., Edwards K. B., Soll D. R. Dictyostelium amebae alter motility differently in response to increasing versus decreasing temporal gradients of cAMP. J Cell Biol. 1985 Jul;101(1):1–5. doi: 10.1083/jcb.101.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnum B., Soll D. R. Effects of cAMP on single cell motility in Dictyostelium. J Cell Biol. 1984 Sep;99(3):1151–1155. doi: 10.1083/jcb.99.3.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M. C., Barylko B., Albanesi J. P. Tissue distribution and subcellular localization of mammalian myosin I. J Cell Biol. 1992 Oct;119(1):163–170. doi: 10.1083/jcb.119.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick H. M., Spudich J. A. Myosin structure and function in cell motility. Annu Rev Cell Biol. 1987;3:379–421. doi: 10.1146/annurev.cb.03.110187.002115. [DOI] [PubMed] [Google Scholar]
- Wessels D., Murray J., Jung G., Hammer J. A., 3rd, Soll D. R. Myosin IB null mutants of Dictyostelium exhibit abnormalities in motility. Cell Motil Cytoskeleton. 1991;20(4):301–315. doi: 10.1002/cm.970200406. [DOI] [PubMed] [Google Scholar]
- Wessels D., Schroeder N. A., Voss E., Hall A. L., Condeelis J., Soll D. R. cAMP-mediated inhibition of intracellular particle movement and actin reorganization in Dictyostelium. J Cell Biol. 1989 Dec;109(6 Pt 1):2841–2851. doi: 10.1083/jcb.109.6.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels D., Soll D. R., Knecht D., Loomis W. F., De Lozanne A., Spudich J. Cell motility and chemotaxis in Dictyostelium amebae lacking myosin heavy chain. Dev Biol. 1988 Jul;128(1):164–177. doi: 10.1016/0012-1606(88)90279-5. [DOI] [PubMed] [Google Scholar]
- Wessels D., Soll D. R. Myosin II heavy chain null mutant of Dictyostelium exhibits defective intracellular particle movement. J Cell Biol. 1990 Sep;111(3):1137–1148. doi: 10.1083/jcb.111.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond S. H. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. J Cell Biol. 1977 Nov;75(2 Pt 1):606–616. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zot H. G., Doberstein S. K., Pollard T. D. Myosin-I moves actin filaments on a phospholipid substrate: implications for membrane targeting. J Cell Biol. 1992 Jan;116(2):367–376. doi: 10.1083/jcb.116.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]