Abstract
Gene duplication is supposed to be the major source for genetic innovations. However, how a new duplicate gene acquires functions by integrating into a pathway and results in adaptively important phenotypes has remained largely unknown. Here, we investigated the biological roles and the underlying molecular mechanism of the young kep1 gene family in the Drosophila melanogaster species subgroup to understand the origin and evolution of new genes with new functions. Sequence and expression analysis demonstrates that one of the new duplicates, nsr (novel spermatogenesis regulator), exhibits positive selection signals and novel subcellular localization pattern. Targeted mutagenesis and whole-transcriptome sequencing analysis provide evidence that nsr is required for male reproduction associated with sperm individualization, coiling, and structural integrity of the sperm axoneme via regulation of several Y chromosome fertility genes post-transcriptionally. The absence of nsr-like expression pattern and the presence of the corresponding cis-regulatory elements of the parental gene kep1 in the pre-duplication species Drosophila yakuba indicate that kep1 might not be ancestrally required for male functions and that nsr possibly has experienced the neofunctionalization process, facilitated by changes of trans-regulatory repertories. These findings not only present a comprehensive picture about the evolution of a new duplicate gene but also show that recently originated duplicate genes can acquire multiple biological roles and establish novel functional pathways by regulating essential genes.
Author Summary
Gene duplication has long been appreciated as a major source for new genes and new functions. Nevertheless, it is still a fascinating mystery how new duplicate genes are functionally integrated into the existing gene network and how they contribute to the novel functions of organisms at the pathway level. By studying the recently originated kep1 gene family in Drosophila melanogaster, we show that one of the young duplicate genes, nsr, has evolved important biological functions associated with male reproduction by regulating several essential fertility genes in the short evolutionary period after its birth. The evolutionary dynamics, biological roles, and the underlying molecular mechanism of nsr revealed in this study present a vivid and comprehensive example of how new genes acquire important biological functions and demonstrate that recently originated new genes can regulate pre-existing essential genes and create novel architectures of genetic pathways.
Introduction
Gene duplication is a fundamental evolutionary process and provides a major source for genetic novelties [1]–[3]. The usual fate of a gene duplicate is pseudogenization, but some duplicates can fortuitously survive through neofunctionalization, in which one copy retains its ancestral function while the other copy acquires a novel function, or subfunctionalization, in which the duplicate and the ancestral copies subdivide the ancestral functions [4], [5]. The two processes, especially neofunctionalization, should have contributed greatly to the biological diversity by providing genetic innovations.
However, how a new duplicate gene acquires functions by integrating into a pathway and results in adaptively important phenotypes has remained largely unknown. Studying the recently originated young genes could be a very informative way to illustrate these processes, as genes at the early stage of evolution should have retained their original features well, which could have changed with time [3]. Currently, a number of young duplicate genes with potential biological functions have been reported [6]–[13]. Among them, three young Drosophila duplicate genes, arisen by retroposition, were reported to have male-related functions: K81 was proposed to be a testes-expressed paternal effect gene [6], mojoless is required for male germline survival [7], and sphinx is an RNA-coding gene responsible for male courtship behavior [8], [14]. Nevertheless, little is known about how these young duplicate genes have been integrated into the molecular pathways and thereby have realized their functions in the host species.
In this study, we systematically characterized a young Drosophila gene of the kep1 gene family, which originated recently in the Drosophila melanogaster (D. melanogaster) species complex (including D. melanogaster, D. simulans, D. mauritiana, and D. sechellia) about 5.4–12.8 million years ago through the duplications of the kep1 gene locus, mediated by the transposon DNAREP1_DM [15]. We performed a comprehensive investigation of its functions within an evolutionary context and successfully revealed its biological roles as well as the underlying molecular mechanism. The results shed novel light on the functional origin of new genes at the pathway level.
Results
Evolutionary Analysis of the kep1 Gene Family
There are 7 members in the kep1 gene family, and their phylogenetic distributions are illustrated in Figure 1A. The parental gene kep1 is present in all Drosophila species. Through the duplications of the kep1 gene locus, the new genes nsr (novel spermatogenesis regulator, CG3875), CG3927, CR9337, and CG4021 originated in the common ancestor of the D. melanogaster species complex, and CR9337-r and CR33318 occurred after the sibling species in the complex diverged [15]. In this study, we focused on the intact new duplicates nsr, CG3927, and CG4021 in D. melanogaster, in which the genetic manipulations are feasible.
Figure 1. Evolutionary analysis of kep1 family.
(A) Origination events of the newly originated kep1 gene family. The phylogeny of the Drosophila species and the divergence time are indicated [18]. On the phylogenetic tree, the blue box represents the parental gene kep1, and green and grey boxes represent the intact new genes and pseudogenes of the kep1 family, respectively. The approximate starting point of the origination of the kep1 family is depicted as a red circle. (B) Multiple alignments for protein sequences of kep1 family genes in D. melanogaster. The asterisks denote the positions of identical amino acids. The blue line under the alignment shows the KH RNA-binding domain. (C) Distribution of dN/dS throughout the kep1-nsr pair. With 120-bp windows and 6-bp slides, dN/dS ratios were estimated using the maximum likelihood method [53] and plotted. Blue and red spots represent dN/dS ratios that are statistically significantly lower and higher than the neutral expectation (p-value <0.05, two-tailed Fisher's exact test), respectively. Regions embedded in the KH domain, as depicted by the grey block, are enriched with signals of purifying selection. (D) Likelihood values of nucleotide substitutions for nsr in Drosophila lineages. Numbers of nonsynonymous and synonymous substitutions for the entire coding sequences are labeled above the lineages, and ω values (dN/dS) are labeled beneath the lineages. A ω value representing the lineage that shows significant evidence of positive selection is highlighted in red. Abbreviations: D. melanogaster (mel); D. simulans (sim); D. sechellia (sec); D. yakuba (yak); D. erecta (ere), and D. ananassae (ana).
The kep1 family copies are located dispersedly on the second chromosome. D. melanogaster kep1 is a pre-mRNA splicing factor, influencing female fertility, eye development, and immune responses to bacterial infection [16]. Consistent with that, the coding sequences of kep1 are conserved throughout the Drosophila phylogeny (Table S1). Multiple alignments of the protein sequences of kep1 family members show that the three intact new genes have a well-retained KH RNA-binding domain but possess highly diverged C-termini (Figure 1B). By sliding window analysis, the ratio of nonsynonymous changes (dN) over synonymous changes (dS) for each kep1-new gene pair was estimated and tested for selection. For all gene pairs, significant purifying selection signals are enriched in the KH domain region (Figure 1C and Figure S1A), revealing functional constraint on the new genes. Most interestingly, the C-termini between the kep1-nsr pair shows significant positive selection signal (dN/dS = 6.11, p-value <0.05) (Figure 1C), which probably arose from accelerated evolution in the nsr as a result of adaptive evolution.
We analyzed the evolutionary patterns along the phylogenetic branches for nsr (Figure 1D), CG3927, and CG4021 (Figure S1B), based on the maximum likelihood estimates of ω values (dN/dS) [17]. If we assume that the duplication events happened when D. melanogaster and D. yakuba diverged 7.4 million years ago [18], even using the most conservative estimate of the synonymous substitution rate for Drosophila [19]–[21], 24.3, 17.9, and 22.6 synonymous substitutions are expected to occur in the ancestral lineage of the D. melanogaster species complex for nsr, CG3927, and CG4021, respectively. These numbers are far beyond our observations, which are 2.6 for nsr, 0 for CG3927, and 9.3 for CG4021 (Figure 1D and Figure S1B). Therefore, the three new duplicate copies must have originated very late in the ancestral lineage, probably close to the split point of the sibling species in the D. melanogaster species complex. In the ancestral lineage, there are many nonsynonymous substitutions in the new genes, and the estimated ω values are 3.192 for nsr (Figure 1D), infinite for CG3927 (there are no synonymous mutations), and 1.149 for CG4021 (Figure S1B), in which the ones for nsr and CG3927 are significantly larger than the neutral expectation (Table S2), indicating that positive selection should have shaped the two new genes, especially nsr. On the branches leading to individual species, the ω values decline, possibly because the new genes might have evolved functions that are under selective constraint.
Tissue-Specific Expression and Subcellular Localization of New Duplicate Genes in the kep1 Family
In D. melanogaster, the kep1 copy is ubiquitously expressed [22], but the new duplicate copies display a male-specific expression pattern, according to our RT-PCR results (Figure S2A). To provide clues for the biological functions of new kep1 family genes, GFP was fused to the coding sequences of each gene to designate their detailed expression patterns in D. melanogaster (Figure S2B). Since the uniform male-specific expression pattern for all of the new duplicate genes is more likely a consequence of a shared regulatory region rather than independently evolved genetic mutations, we used the homologous upstream regulatory sequences of all kep1 family genes as the driving promoter (Figure S2D). As expected, the shared regulatory region is sufficient to drive similar male-specific expression for each of the GFP-tagged kep1 family proteins, which are unexceptionally enriched in the primary spermatocytes of testes (Figure 2A–2D). Previous large-scale profiling of gene expression patterns in D. melanogaster testes demonstrated that all kep1 family genes showed a high level of mitosis and meiosis expression, followed by much-reduced post-meiosis expression [23]. This result is consistent with our observation and also suggests that the kep1 family genes may be expressed in the spermatogonial stage as well.
Figure 2. Expression analysis of kep1 family proteins by GFP transgene in D. melanogaster.
(A–D) Low-magnification fluorescent images of transgenic GFP-fused Kep1 (A), Nsr (B), CG3927 (C), and CG4021 (D) proteins (green) in testes of D. melanogaster strain w1118. All of the kep1 family genes are enriched in the primary spermatocytes. (E–H) High-magnification fluorescent images of transgenic GFP-fused Kep1 (E), Nsr (F), CG3927 (G), and CG4021 (H) proteins in the primary spermatocytes. Proteins of kep1 family genes are located in the nuclear regions of the primary spermatocytes, which are distinguishable from the three diffusely staining nuclear regions (three white arrows), corresponding to the three main chromosome bivalents. Note that all kep1 family proteins are distributed in speckles (yellow arrow). (I and J) Comparisons of subcellular localization between Kep1 and new gene proteins. Kep1 is tagged with GFP (green); while Nsr and CG4021 are tagged with TAP (red), respectively. Nsr protein shows a wider expression region than Kep1 protein (arrow) (I), and CG4021 protein is completely co-localized with Kep1 protein (J). Scale bars: 200 µm for A–D; 10 µm for E–H; 50 µm for I and J.
In the primary spermatocytes, kep1 family proteins are localized in a specked nuclear pattern (Figure 2E–2H), a highly diagnostic feature for spliceosomal components [24], [25]. Considering that D. melanogaster kep1 is a splicing factor responsible for the alternative splicing of the Drosophila caspase molecule dredd [16], the observation above led us to speculate that new kep1 family genes might regulate the pre-mRNA processing of genes required for spermatogenesis and sperm function.
Evolution of novel subcellular localization after duplication is thought to be an important evolutionary mechanism for the origins of genes with novel functions [26]. Though both are distributed in punctuate nuclear structures of primary spermatocytes, the localization of Nsr protein is much broader than the Kep1 protein (Figure 2I). RNase A treatment of testes could lead to the ectopic accumulation and dispersal of GFP-tagged Nsr protein (Figure S2E, S2F, S2G, 2H), indicating that the Nsr protein is localized in an RNA-dependent manner, and its expanded nuclear localization might imply a novel RNA-binding property. CG4021 protein is localized, completely overlapping with the Kep1 protein, in primary spermatocyte nuclei (Figure 2J), and CG3927 protein was found to have a lack of a significant fluorescent signal for the comparison.
Loss-of-Function Analysis for the kep1 Family Genes
To comprehensively understand the biological functions of the kep1 family genes, we have generated null mutants for all four intact gene copies in D. melanogaster by either gene targeting knockout [27] or imprecise P-element excision [28] (Figure 3A and 3B). The wild-type (WT) control flies of the mutants are WT recombinants created by targeted mutagenesis or precisely excised strains of P-element excision, for the sake of an identical genetic background between the mutant and the WT flies. The null males of nsr display significantly reduced fecundity when compared to the WT males (p-value <0.001, Mann-Whitney U test) (Figure 3C). This phenotype can be fully restored by introducing the genomic sequences of nsr back into the genome (Figure 3C). Heterozygous flies of nsr mutants are equally fertile as the WT flies (Figure 3C). We found that the sperm storage tissue (seminal vesicle) of nsr male mutants was empty or contained little sperm, if any (Figure 4A and 4B). During D. melanogaster spermatogenesis, germ cells from gonial precursors differentiate into cysts of 64 syncytial spermatids, which will undergo an actin-based individualization process, in which a bulk of unneeded cytoplasm is eliminated from the spermatids through remodeling of the cyst membrane. Extrusion of the cytoplasm along sperm bundles can form visible cystic bulges, which will migrate to the distal ends and are detached as waste bags. An actin structure, termed the “investment cone (IC),” is formed at the site where each spermatid develops its own membrane [29], [30]. We labeled the sperm bundles together with the cystic bulges and waste bags with GFP under control of the don juan (dj) gene promoter [31], and the ICs are visualized by FITC-conjugated phalloidin. The testis of nsr mutant male contains comparable amounts of spermatids as their WT controls; however, the structures of cystic bulges and waste bags are largely absent (Figure 4C and 4D). In WT flies, ICs in the same cyst move coordinately in clusters (Figure 4E), while they are scattered along the sperm bundles in the nsr mutants (Figure 4F). The phenotypes above are typical features of an impaired individualization process [30]. Electron microscopy examination further confirmed that the spermatids of nsr mutants are unindividualized, with substantial amounts of residual cytoplasm (Figure 4G and 4H). As the final step of spermatogenesis, the spermatids are assembled by coiling at the base of the testis to facilitate their transport into seminal vesicles [29]. Under a phase-contrast microscope, the sperm bundles of nsr mutants are twisted at the distal ends of testis, instead of regular coiling (Figure 4I and 4J). Therefore, nsr is functionally involved in both sperm individualization and coiling.
Figure 3. Generation of null mutants for each kep1 family gene and the male fertility test.
(A) Schematic maps of mutant alleles of kep1 family genes generated by P-element excision or targeted mutagenesis. The exons (black block), start codon (ATG), stop codon (TAA/TGA/TAG), and deleted genomic regions are indicated. For kep1 and CG4021, the P-element insertion for further excision is shown by a triangle, with a Bloomington stock number given underneath. (B) RT-PCR examination of null mutants (Null) for each kep1 family gene relative to WT flies (WT). Negative control (NC) is the reaction without reverse transcriptase, and the expression of rp49 is used as internal control. (C) Fertility test for nsr WT and mutant males. nsr WT: WT controls with identical genetic background with nsr mutants; nsr -/-: homozygous nsr mutants; nsr -/CyO: heterozygous nsr mutants; nsr Rescue: flies with a copy of WT nsr transgene in the nsr mutant background. Error bars indicate standard deviation. (D) Fertility test for kep1, CG3927, and CG4021 WT and mutant (-/-) males. Error bars indicate standard deviation.
Figure 4. Morphological characterization of nsr mutants.
(A and B) Seminal vesicles (sv) from nsr WT (A) and mutant males (B) stained by Hoechst 33342. A shriveled seminal vesicle was observed in nsr mutants. (C and D) dj-GFP labeled elongated spermatids from WT and nsr mutant testes. The elongated spermatids from WT testis (C) are tightly organized in bundles with visible cystic bulges (cb) (arrowhead) and waste bags (wb) (arrow). In contrast, the elongated spermatids from nsr mutants (D) are much looser; the structures of cb and wb can hardly be observed. (E and F) Phalloidin-stained ICs in testes of WT and nsr mutants (inset shows a region at higher magnification). In WT testis, the ICs progress syncytially (E), while the syncytial movement of ICs is abnormal in nsr mutant testis (F). (G and H) Electron microscopic images of cyst from WT and nsr mutant testes at the late stage of individualization (inset shows a region at higher magnification). Individualized spermatids in WT cyst contain highly ordered organelles, including the major mitochondrial derivative (mj), the minor mitochondrial derivative (mi), and the axoneme (ax) (G), while the spermatid individualization is abolished in the nsr mutant cyst, as revealed by excess cytoplasmic remnants (arrow) and poorly assembled organelles (H). (I and J) Testis bases of WT and nsr mutant flies under a phase contrast microscope. Unlike regular coiling in the WT flies (I), the spermatids are tangled at the testis base of nsr mutant (J). Scale bars: 200 µm for A–D; 100 µm for E and F; 1 µm for G and H.
In contrast, though kep1 is required for female fertility in D. melanogaster [16], no significant difference in male fertility was detected between kep1 mutant males and their WT controls (Figure 3D). Also, we did not observe reduced fertility (Figure 3D) or other obvious defects for the CG3927 and CG4021 mutants. Considering that only nsr exhibits a robust signature of positive selection, this result may not be surprising. Either CG3927 and CG4021 have not acquired new functions or their phenotypic effects are not strong enough to be detected in our phenotyping assay.
Requirement of nsr for the Integrity of Sperm Axoneme Structure by Regulating Several Y-Linked Male Fertility Genes
Microarray comparison of the transcription profiles between nsr WT and mutant testes only identified 14 genes that exhibited at least a 2-fold difference at the expression level, but none of them seemed to be male fertility-related (Table S3). Considering that the background hybridization noise and lack of probes for some genes might limit the power of microarray, we further implemented whole transcriptome shortgun sequencing (RNA-Seq), which is regarded as a more precise way for measurements of transcript levels [32]. Using the Illumina paired-end sequencing platform, we generated 16.3 million reads (75-bp) for WT testes and 9.6 million for nsr mutant testes. Based on these transcriptome data, we identified 10 genes that were significantly differentially expressed (>5-fold) between WT and mutants. Among them, kl-2, kl-3, and kl-5 are known male fertility genes, and the others are either not correlated with male fertility or functionally unknown (Table S4).
The kl-2, kl-3, and kl-5 genes are 12.4-fold, 10.0-fold, and 6.84-fold down-regulated in the mutants, respectively (Table S4), and their sharp reductions in expression were validated by real-time PCR (Figure 5A). Interestingly, the three genes were located adjacently on the Y chromosome, and all encode dynein heavy chain polypeptides of the sperm axoneme [33]–[35]. The phenotypic defect associated with the sterility of kl-2 mutants is not very clear [36], [37], while kl-3 or kl-5 mutations by P-element insertions result in loss of the outer dynein arm of the sperm axoneme and irregular coiling of spermatid tails, and complete deletion of either locus causes defects in sperm individualization [37]–[39]. Electron microscope examination of the spermatid flagellum showed that the outer dynein arms of sperm axonemes were also missing in the nsr mutants (Figure 5B–5F). The deficiencies of nsr mutants, including sperm individualization, coiling, and axonemal structures, fit well with the phenotypes of the kl-3 and kl-5 mutants. This substantial agreement of the loss-of-function phenotypes between the Y-linked genes kl-3, kl-5, and nsr indicates that nsr is involved in male functions by regulating kl-3, kl-5, and, possibly, kl-2 as well. Moreover, it is very likely that nsr regulates the kl-2, kl-3, and kl-5 genes post-transcriptionally, because their primary transcript levels are largely unaltered between the mutants and WT flies, as shown by real-time PCR results (Figure 5A). This is also in accordance with the conserved RNA-binding domain (Figure 1B and 1C) and the splicing factor-like distribution pattern (Figure 2F) of the Nsr protein. More importantly, our co-immunoprecipitation experiment demonstrated that the pre-mRNA cleavage stimulatory factor CstF-64 [40] can be specifically immunoprecipitated by TAP-tagged Nsr protein from testis extracts (Figure S3A and S3B). This result fortifies the idea that nsr might function as an RNA processing factor, although future studies are needed to explore how nsr and CstF-64 collaboratively process the primary transcripts of these male genes.
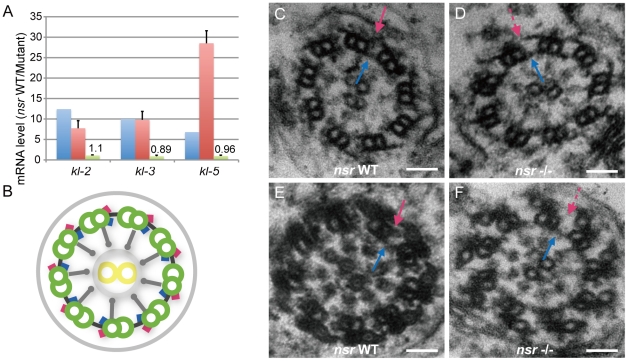
Figure 5. Identification of the Y chromosome genes kl-2, kl-3, and kl-5 as downstream targets of nsr.
(A) Histogram of kl-2, kl-3, and kl-5 RNA level changes in testes of nsr mutants for mature transcripts, estimated by RNA-Seq (blue) and quantitative real-time PCR (red), and for primary transcripts estimated by quantitative real-time PCR (green). Error bars indicate standard deviation. Both RNA-Seq and real-time PCR results show that the mature transcripts are down-regulated in the mutant testes. The difference between the estimations of kl-5 by RNA-Seq and real-time PCR is possibly due to the larger variations of genes with lower abundance in RNA-Seq [62]. The real-time PCR result shows that the levels of primary transcripts are largely unaltered between the WT and mutant testes. (B) Graphic illustration of axoneme structure [63], [64]. The axoneme is composed of a central pair of singlet microtubules (yellow circle) surrounded by nine doublet microtubules (green circle), anchored by outer (red) and inner (blue) dynein arms that can mediate axoneme motility. Radical spokes (grey spoke) pass from each doublet fiber toward the central singlets. (C–F) Electron microscopic images of early- (C and D) and late-stage (E and F) spermatid axonemes for WT and nsr mutants. The late-stage axoneme is distinct from early-stage axoneme by extensive accessory structures. For both stages, the inner dynein arms of the axoneme are normal in nsr mutants (blue arrow), but the outer dynein arms (red arrow) are constantly missing (dashed red arrow). Scale bars: 50 nm.
Functional Status of Ancestral kep1 in the Pre-Duplication Species D. yakuba
We traced the functional status of kep1 in the pre-duplication species D. yakuba by detecting its expression pattern using Kep1 antibody (Figure S3C). Surprisingly, immunocytochemistry with Kep1 antibody showed only background staining of D. yakuba testis (Figure 6D), whereas it is capable of yielding a robust staining pattern in the primary spermatocytes of D. melanogaster (Figure 6B and 6C), exactly as revealed by transgenic GFP localization (Figure 2A). The antibody worked well in detecting Kep1 proteins in ovary extracts from both D. yakuba and D. melanogaster by Western blot (Figure S3D), ruling out the possibility that the antibody sensitivity is not equally sufficient for detecting Kep1 protein of D. yakuba.
Figure 6. Evolutionary history of the expression patterns of kep1 family proteins.
(A) Phylogenetic tree of Drosophila species. The pre- and post-duplication lineages are denoted by blue lines and red lines, respectively. (B–F) Immunostaining of testes with different genotypes using Kep1 antibody shows fluorescent signals in primary spermatocytes for the D. melanogaster species complex (B, E and F) but only background staining for the pre-duplication species D. yakuba (D). This suggests that the primary spermatocyte-biased expression patterns of kep1 family genes should have been established in the common ancestor of the D. melanogaster species complex after the split of D. yakuba. (G) Transgenic GFP, regulated by cis-elements (including promoter, 5′ UTR and coding sequences) of D. yakuba kep1 in D. melanogaster, is also enriched in primary spermatocytes, indicating that the cis-elements of kep1 have not changed between D. melanogaster and D. yakuba, and thus, trans-regulatory changes should have contributed to the observed testicular expression patterns of kep1 family genes in D. melanogaster. The abbreviations of Drosophila species are the same as in Figure 1. Scale bars: 200 µm.
Absence of Kep1 protein in D. yakuba testis suggests that the kep1 gene should not be ancestrally required for male fertility, and it also raises the questions of when and how the novel testicular expression patterns of the kep1 family in D. melanogaster has been evolved. The immunofluorescent signals of Kep1 proteins in the sibling species of D. melanogaster, D. simulans (Figure 6E) and D. sechellia (Figure 6F), suggest that this novel pattern has been established in the common ancestor of the D. melanogaster species complex. This interspecies difference of expression pattern between D. yakuba and D. melanogaster may arise from either cis-acting or trans-acting regulatory changes. The two genetic factors can be distinguished by testing the transcriptional activity of D. yakuba's cis-elements of kep1 in D. melanogaster. Controlled by D. yakuba's cis-elements of kep1, GFP was also found to accumulate in the primary spermatocytes in D. melanogaster (Figure 6G) with the same subcellular localization as with the control of the cis-elements of D. melanogaster kep1 (Figure S3E). This means that the activity of the cis-elements has not been differentiated between D. yakuba and D. melanogaster, and it is the changes in trans-regulatory repertoires that most likely have enabled all kep1 family genes to obtain novel testicular expression patterns.
Discussion
There are two possible scenarios to explain the current functional roles of nsr in D. melanogaster: neofunctionalization and subfunctionalization [4], [5]. Our results tend to support the neofunctionalization scenario, although we cannot completely exclude the possibility of subfunctionalization.
Several pieces of evidence support the neofunctionalization scenario. Firstly, the parental gene kep1 is under strict purifying selection across the Drosophila phylogeny (Table S1). The significant conservation of kep1 and its inessentiality for male fertility in the pre-duplication species D. yakuba is consistent with the reported functions of kep1 in female fertility, eye development, and immune response [16] but not male fertility (Figure 3D) in D. melanogaster. These results suggest that kep1 possibly has retained its ancestral functions without evolving novel male functions after the duplication events, and nsr is free to evolve new functions. Secondly, nsr shows a robust signal of positive selection (Figure 1C and 1D), especially in the C-termini (Figure 1C). As we know, RNA recognition is a complex biological process that may need the collaboration of multiple factors; the RNA-binding domain alone possibly does not contain sufficient information for specific targeting [41], [42]. Thus, the rapidly evolving C-termini of nsr could have contributed to novel RNA-binding ability by mediating co-option with different cofactors, and this idea is further strengthened by the specific immunoprecipitation of the pre-mRNA cleavage stimulatory factor CstF-64 by the Nsr protein (Figure S3B). The subcellular localization pattern of the Nsr protein is also different from the Kep1 protein by displaying a larger localization range in the nuclei of primary spermatocytes (Figure 2I), and cell type-specific expression or subcellular localization is regarded as one of the strategies for RNA-binding proteins to regulate specific splicing events [42]. Although it is still not clear what is the concrete molecular process that the novel distribution pattern of nsr has contributed to its roles in spermatogenesis, it is possible that this novel distribution might allow the spatial-specific assembling between nsr and its cofactors, and the subsequent specific regulation of mRNA substrates. Thirdly, our antibody did not detect obvious expression of Kep1 protein in D. yakuba testis, and thus, the parental gene kep1 should not be ancestrally required for male fertility. After the split of D. yakuba, trans-regulatory changes possibly occurred prior to or accompanied by the duplications of kep1, which enabled the kep1 family genes to obtain novel testicular expression patterns and thereby lend them an opportunity to evolve novel male functions, as nsr has done.
Nevertheless, the alternative subfunctionalization scenario cannot be completely excluded if a recent “gain and loss” turnover of male functions for kep1 did happen or if kep1 has lost its male functions in the D. yakuba lineage for some reason. In the recent “gain and loss” turnover, the parental gene kep1 could have acquired an essential role in spermatogenesis after the split of D. melanogaster and D. yakuba but prior to the duplication events, whereas the new copy nsr has taken over the spermatogenesis role from kep1 after its origination.
The new duplicate gene nsr displays tremendous divergence from kep1 at the levels of biological function and molecular pathway. The kep1 gene participates in female fertility by regulating the apoptosis molecule dredd [16], whereas the new gene nsr is integrated into the spermatogenesis pathway by regulating Y-linked male fertility genes; thus, our findings also provide an unusual case, showing a functional transition in a new gene from a female role to male role. It is interesting that the newly originated genes are often expressed primarily in male reproductive tissues in diverse organisms [43]–[47], and most of the new Drosophila genes with known functions [6]–[8], together with nsr, are associated with male reproduction. This phenomenon pronounces that new genes may tend to be functionally male-biased and suggests a significant role of natural selection and sexual selection in the fixation of beneficial mutations for male reproductive success.
Our study reveals that nsr has been integrated into fundamental developmental processes by regulating pre-existing essential genes. Interestingly, the sperm maturation aspects that nsr participates in are conserved during evolution [48]. For example, the failure of eliminating sperm cytoplasm and loss of the outer axonemal dynein arm can also cause many types of human infertility [49]–[51]. The functional mechanism of nsr indicates that new genes could contribute to the evolutionary turnover of molecular pathways governing essential and conserved developmental processes, which partially explains the phenomenon that the same developmental processes in different organisms are sometimes achieved by a different set of genes. The positive selection signal and biological functions of nsr together strongly suggest that nsr might have contributed to the adaptive evolution of male reproductive pathways in the D. melanogaster species complex.
Materials and Methods
Evolutionary Analysis
Protein sequences of nsr, CG3927, CG4021, and kep1 in D. melanogaster are downloaded from FlyBase (http://flybase.org) and aligned by ClustalW (http://www.ebi.ac.uk/Tools/clustalw). Orthologous coding sequences of kep1 family genes in other Drosophila species (http://flybase.org) were predicted using a combination of BLAT (http://genome.ucsc.edu) and GeneWise (http://www.ebi.ac.uk/Tools/Wise2) and manually checked. Alignments of coding sequences mentioned below are performed by MEGA 3.2 [52], considering the coding structures. To estimate the selective constraint on kep1 through the Drosophila phylogeny, alignments of kep1 coding sequences from different Drosophila species were tested for purifying selection by MEGA 3.2 pairwisely. To detect the selective pressure on the new genes of the kep1 family, alignments of the coding sequences between kep1 and each new gene were performed and calculated for the dN/dS ratio with 120-bp windows and 6-bp slides. For each window, the maximum likelihood method [53] was used to test if the dN/dS ratio was significantly different from one (two-tailed Fisher's exact test).
The ω (dN/dS) values in the phylogeny of new kep1 family genes were estimated using the maximum likelihood approach, implemented by the codeml free-ratio model in the PAML4.2 package (http://abacus.gene.ucl.ac.uk/software/paml.html) [17]. To test if the ω ratio in the ancestral lineage of the D. melanogaster species complex was significantly different from one, the likelihood of the two-ratio model with an estimated ω was compared to an alternative two-ratio model, with ω constrained to be one for this lineage.
Fly Strains
All Drosophila strains were maintained at 25°C using standard cornmeal medium. The transgenic strains were produced by microinjection of w1118 embryos following standard P-element-mediated germline transformation [54]. P-element insertion stocks DG20303 and KG07486 were ordered from Bloomington Stock Center. Strains for P-element excision (Sp/CyO; Δ2-3, Sb/TM6B and Sp/CyO; MKRS/TM6B) are kindly provided by Dr. Yongqing Zhang. Strains for targeted mutagenesis (70FLP70I-SceI, 70FLP and 70I-CreI) were previously described by Xie and Golic (2004).
Transgenic Constructs of kep1 Family Genes
For GFP-tagged vectors, the pH-Stinger plasmid [55] was modified by excision with SpeI/NheI and re-ligation to remove its Hsp70 promoter and nuclear GFP. Gene promoter sequences (plus 5′ UTR) and GFP coding sequences were then cloned into XbaI/EcoRI and EcoRI/KpnI sites of the modified plasmid. Coding sequences of each gene were added into EcoRI sites and selected for correct insertion orientation (Figure S2B). TAP-tagged transgenic vectors were constructed similarly but had GFP replaced with a TAP tag, which consists of two IgG-binding domains of protein A (ProtA) and a calmodulin-binding peptide (CBP) separated by a TEV protease cleavage site [56] (Figure S2C). For all the vectors above, a homologous upstream region of kep1 family genes (including D. yakuba kep1) was adopted as the promoter sequence (Figure S2D). A rescue construct of nsr was prepared by inserting a 2.8-kb DNA fragment, ranging from the end of the upstream gene to the start of the downstream gene, into the NotI site of the pW8 transformation vector (FlyBase). The primer information is available in Table S5.
Generation of Null Flies of Each kep1 Family Gene and Male Fertility Test
P-element excision: The fly strains DG20303 (with a P-element at the 5′ UTR of kep1) and KG07486 (with a P-element annotated to locate at the nsr locus but found to be inserted at the 5′ UTR of CG3927 after PCR validation) were mobilized with Δ2-3 transposase by standard P-element excision, respectively [28]. Excision lines were screened by PCR, and the endpoints were determined by sequencing.
Gene knockout by ends-in targeting: The targeting vectors were designed to create a deletion, spanning from 42-bp downstream of the transcriptional start site to a site within the 3′ UTR of nsr, and a deletion spanning from the start codon to a site within the 3′ UTR of CG4021, respectively. Targeted mutagenesis was performed as previously described [27]. Donor flies bearing the targeting vector were generated and crossed with flies carrying heat shock-activated FLP recombinase and I-SceI endonuclease (70FLP70I-SceI). The 0-3 day old progeny were heat-shocked at 38.5°C for 1 hour, and the enclosed white-eye virgins were crossed with males constitutively expressing FLP recombinase (70FLP). In total, at least 1000 vials were screened for nonmosaic red-eye individuals with successful insertions of the targeted allele at the site of the endogenous allele. Stocks of the recombinant flies were established and crossed with flies carrying heat shock-activated I-CreI endonuclease (70I-CreI). We heat-shocked 0-3 day old progeny at 38.5°C for 1 hour and screened for white-eye adults with recombinant reduction events at the targeted site. The reduction events will lead to either removal of the allele or maintenance of the WT allele. Strains of both genotypes were established to serve as knockout and WT lines, respectively.
For the male fertility test, an individual male of each genotype (<1 day) was placed with three w1118 virgin females, which were collected within 5 hours of enclosure and aged for 2 days. The progeny were counted on the 18th day after the mating and compared between the mutant and WT lines using Mann-Whitney U test.
Antibody Preparation and Immunofluorescence Assay
A polyclonal antibody was raised against the glutathione-S-transferase-Kep1 (amino acids 233–313) recombinant protein in guinea pigs. Testis squashes and immunostaining were performed as previously described [57]. The primary antibodies used are guinea pig anti-Kep1 serum (1∶200 dilution) for Kep1 protein and rabbit peroxidase-antiperoxidase complex (PAP) (1∶1000 dilution, Sigma) for ProtA. The secondary antibodies are Alexa 555-conjugated anti-guinea pig and Alexa 594-conjugated anti-rabbit (Molecular Probes). Testes were co-stained with Hoechst 33342 (1 µg/ml, Molecular Probes) to visualize nuclear DNA if needed. FITC-conjugated phalloidin (1∶100 dilution) was used for IC staining. RNase A treatment was performed as previously described [58] by a 10-min incubation of TBS with 50 µg/ml RNase A (Fermentas), and the controls were incubated in the same buffer, but free of RNase A.
Western Analysis
For sample preparation, adult testes or ovaries from 0–5 day old flies were dissected in PBS, transferred to RIPA buffer, ground, and boiled at 95°C for 10 min for lysis. The primary antibodies used were PAP (1∶2000 dilution, Sigma), mouse anti-β-actin (1∶3000 dilution, Abcam), and guinea pig anti-Kep1 (1∶500 dilution). Peroxidase-conjugated secondary antibodies were used for signal detection (1∶10000 dilution, Santa Cruz).
Immunoprecipitation Assay
Six hundred testes of 0–3 day old flies carrying TAP-tagged Kep1 protein, TAP-tagged Nsr protein, or TAP-tagged CG4021 protein were used for affinity purification, respectively. Testes were ground in 100 µl RIPA buffer plus protease inhibitor cocktail (Roche) with the Sample Grinding Kit (GE Healthcare). The cell suspension was centrifuged at 4°C for 5 min, the supernatant was pre-cleared by 5 µl protein G plus-agarose beads (Santa Cruz), and incubated with 2 µl PAP at 4°C overnight. Then, 10 µl protein G plus-agarose beads were added to the mixture and incubated at 4°C for 1 hour. Complexes of TAP-tagged proteins were liberated from the beads by cleavage of TEV protease as previously described [56], subjected to SDS-PAGE, and visualized by Coomassie blue staining. The protein band of interest was cut out and identified by MALDI-ToF mass spectrometry.
Electronic Microscopy
The dissected testes from WT controls and nsr mutants were fixed in 2.5% glutaraldehyde, washed twice with PBS, post-fixed with OsO4, and dehydrated in an ascending series of ethanol. The resultant specimens were embedded in Araldite, sliced into ultrathin sections (50–100 nm), stained with 1% uranyl acetate, and examined with a JEOL electron microscope.
RT-PCR and Real-Time RT-PCR
Total RNA was isolated from adult testes with Trizol reagent (Invitrogen) and treated with DNase I (Fermentas). Reverse-transcription was performed using the RevertAid First Strand cDNA Synthesis kit (Fermentas) with a no-reverse-transcriptase reaction as the negative control. Real-time PCR was performed in triplicate with SYBR Green PCR Mix (Bio-Rad) and subjected to the ABI 7000 Sequence Detection System. Oligo-dT primer was used to synthesize the cDNA templates for detecting mature transcripts and random hexamer primer for primary transcripts. Information on the PCR primers is available in Table S5. The relative concentration of genes was calculated by analyzing their dissociation curves using the constitutively expressed gene rp49 as the internal control.
Microarray Analysis
With Trizol reagent (Invirtrogen), total RNA was extracted from testes of 0–2 day old nsr mutant and WT flies, respectively. After amplification, mRNA was fluorescently labeled by GeneChip One-Cycle Target Labeling (Affymetrix) and subjected to GeneChip Drosophila Genome 2.0 Arrays (Affymetrix) in duplicate. Image collection was performed in accordance with standard Affymetrix protocols. The significance of gene expression change was estimated using the Significance Analysis of Microarrays (SAM) algorithm, which assigns a score to each gene on the basis of change in gene expression relative to the standard deviation of repeated measurements [59]. The microarray data have been deposited in Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE22289.
Paired-End cDNA Library Construction for Illumina Genome Analyzer 2 (GA2) Sequencing
With Trizol reagent (Invirtrogen), 5 µg total RNA was extracted from testes of 0-1 day old nsr mutant and WT flies, respectively. The first-strand cDNA was synthesized with oligo-dT primer by Superscripts II reverse transcriptase (Invitrogen), and second strand cDNA synthesis was followed according to the standard protocol. Then, the double-stranded cDNA was purified with the Qiaquick PCR purification kit (Qiagen) and fragmented with a nebulizer (Invirtrogen), resulting in an average size of 150–250-bp. Overhangs of resultant fragmented cDNAs were blunted with T4 DNA polymerase (NEB) and Klenow polymerase (NEB) and treated with 3′-5′ exonuclease-deficient Klenow polymerase (NEB) to generate 3′ overhangs. After that, cDNA was ligated to an Illumina PE adapter oligo mix by the Quick ligation kit (Qiagen). The adapter-modified cDNA within 200-bp was isolated by agarose gel, extracted with the QIAquick Gel Extraction Kit (NEB), and amplified by PCR reaction. Finally, the library products were sequenced using the Illumina GA2 sequencing machine. Sequence data from this study have been submitted to the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession number SRA020074.
Measurement of Gene Expression Using Data of Illumina GA2 Sequencing
The generated 75-bp raw reads were mapped to the genomic sequences of D. melanogaster (Ensembl release 55: ftp://ftp.ensembl.org/pub/release-55/fasta/drosophila_melanogaster) using SOAP2 software (http://www.soapmaker.ca) [60]. The count of covering reads for each annotated transcript (Ensembl release 55: ftp://ftp.ensembl.org/pub/release-55/gtf/drosophila_melanogaster) was calculated as the index of their expression level. The alteration of transcript level between nsr mutants and WT flies was estimated and normalized for the variation of the total data size of transcript reads. The significance of expression difference (p-value) for each gene (the longest transcript) was further computed according to the R package “DEGseq” using the MA-plot-based method with a random sampling model and followed by an adjustment with q-values for multiple testing corrections [61].
Supporting Information
Evolutionary analysis of CG3927 and CG4021. (A) Distributions of dN/dS throughout kep1-CG3927 and kep1-CG4021 pairs. With 120-bp windows and 6-bp slides, dN/dS ratios were estimated using the maximum likelihood method and plotted. Blue spots represent dN/dS ratios that are statistically significantly lower than the neutral expectation (p-value <0.05, two-tailed Fisher's exact test). Regions embedded in the KH domain, as depicted by the grey block, are enriched with signals of purifying selection. The shorter alignment of the kep1-CG3927 pair than the kep1-nsr and kep1-CG4021 pairs is mostly due to less well-aligned sequences caused by indels. (B) Likelihood values of nucleotide substitutions for CG3927 and CG4021 in Drosophila lineages. Numbers of nonsynonymous and synonymous substitutions for the entire coding sequences are labeled above the lineages, and ω values (dN/dS) are labeled beneath the lineages. Abbreviations: D. melanogaster (mel); D. simulans (sim); D. sechellia (sec); D. yakuba (yak), and D. erecta (ere).
(0.48 MB TIF)
Expression analysis of kep1 family proteins. (A) Male-specific expression pattern of new kep1 family genes shown by RT-PCR. Total RNA was extracted from the whole body of 0–5 day old adults. F: female; M: male. Negative control (NC) was reaction without reverse transcriptase. The expression of rp49 was used as the internal control. (B and C) Schematic representations of GFP (B) and TAP (C) transgene constructs. CDS: coding sequences. (D) Alignments of the homologous promoter sequences of D. melanogaster kep1 family genes and D. yakuba kep1. The asterisks denote the position of identical nucleotides. (E–H) The localization sensitivity of GFP-tagged Nsr protein to RNase A treatment. The localization of GFP-tagged Nsr protein exhibits ectopic diffusion and accumulation after RNase A treatment (E, G) compared with the mock-treated control (F, H). Scale bars: 200 µm for E and F; 20 µm for G and H.
(1.42 MB TIF)
Immunoprecipitation results of TAP-tagged kep1 family proteins and pre-duplication ancestral subcellular localization of Kep1 protein. (A) Western blot of testis extracts from 0–5 day old flies, with the indicated genotypes. The blot was probed with PAP (to recognize the ProtA of TAP) and β-Actin antibody (loading control). The absence of ProtA in w1118 lane indicates the specificity of PAP antibody. (B) Immunoprecipitation of TAP-tagged kep1 family proteins with testis extracts from 0–3 day flies. A 50-72-kDa band corresponding to CstF-64 (identified by mass spectrometry) was immunoprecipitated by TAP-tagged Nsr protein (lane 2) but not in TAP-tagged Kep1 protein (lane 1) or TAP-tagged CG4021 protein (lane 3). (C) Western blot of testis extracts from 0–5 day old WT flies showing that Kep1 antibody recognizes a band of about 43-kDa (arrow), which is consistent with the reported size of Kep1 protein, and that this band is absent in kep1 mutants. (D) Western blot of testis (lane 1 and 2) and ovary (lane 3 and 4) extracts of 0–5 day old D. melanogaster and D. yakuba probed with Kep1 antibody. Kep1 protein is detectable in ovary but not testis of D. yakuba (arrow). (E) Overlapping subcellular localization between the D. melanogaster TAP-tagged Kep1 (mel-Kep1-TAP) and the transgenic GFP-tagged Kep1 protein regulated by the cis-elements (including promoter, 5′ UTR and coding sequences) of D. yakuba kep1 (yak-Kep1-GFP) in D. melanogaster. Scale bars: 20 µm.
(1.23 MB TIF)
dN/dS ratios of kep1 among Drosophila species (Kumar method). All the pairwise species comparisons show that the parental gene kep1 is subjected to significant purifying selection (p-value <0.001, MEGA Z-test, kumar method). Abbreviations: D. melanogaster (mel); D. simulans (sim); D. sechellia (sec); D. yakuba (yak); D. erecta (ere) and D. ananassae (ana); D. pseudobscura (pse).
(0.03 MB DOC)
Maximum likelihood tests of positive selection for new kep1 family genes. The dN/dS ratios were set to be ω0 for the background branches and ω1 for the foreground branch, which is the phylogenetic lineage from D. yakuba to the common ancestor of the D. melanogaster species complex.
(0.03 MB DOC)
List of genes identified by microarray to be >2-fold differentially expressed between testes of nsr WT and mutant flies. Significance Analysis of Microarrays (SAM) identified 14 genes that changed at least 2-fold with a q-value cutoff of 0.01 for significance.
(0.02 MB XLS)
List of genes identified by RNA-seq to be >3-fold differentially expressed between testes of nsr WT and mutant flies. The significance of expression difference (p-value) for each gene (longest transcript) was computed using MA-plot-based method with a random sampling model and followed by an adjustment with q-values for multiple testing corrections. With a p-value cutoff of 0.001, 10 genes (red) and 43 genes are identified to be significantly changed at least 5-fold and 3-fold, respectively.
(0.04 MB XLS)
Primer Information. F: forward primer; R: reverse primer.
(0.03 MB XLS)
Acknowledgments
We thank the three anonymous reviewers' suggestions for the manuscript. We are grateful to Zhaohui Wang and her lab members for providing fly strains and immunizing guinea pigs, Yongqing Zhang for sharing protocols and fly strains for P-element excision, and Chi-Fa Huang for technical support in establishing the Drosophila transgenic platform. We also thank Renjie Jiao, Hongzheng Dai, and Sha Sun for advice on targeted mutagenesis.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by a CAS key grant (KSCX2-YW-N-060), a NSFC key grant (No. 30930056), the 100 Talents Program of Chinese Academy of Sciences, and a CAS-Max Planck Society Fellowship to WW and by a NSFC grant (No. 30871342) and a grant (GREKF08-02) from the State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, to HY. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ohno S. New York: Springer-Verlag; 1970. Evolution by gene duplication. [Google Scholar]
- 2.Zhou Q, Wang W. On the origin and evolution of new genes–a genomic and experimental perspective. J Genet Genomics. 2008;35:639–648. doi: 10.1016/S1673-8527(08)60085-5. [DOI] [PubMed] [Google Scholar]
- 3.Long M, Betran E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 4.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 6.Loppin B, Lepetit D, Dorus S, Couble P, Karr TL. Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr Biol. 2005;15:87–93. doi: 10.1016/j.cub.2004.12.071. [DOI] [PubMed] [Google Scholar]
- 7.Kalamegham R, Sturgill D, Siegfried E, Oliver B. Drosophila mojoless, a retroposed GSK-3, has functionally diverged to acquire an essential role in male fertility. Mol Biol Evol. 2007;24:732–742. doi: 10.1093/molbev/msl201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai H, Chen Y, Chen S, Mao Q, Kennedy D, et al. The evolution of courtship behaviors through the origination of a new gene in Drosophila. Proc Natl Acad Sci U S A. 2008;105:7478–7483. doi: 10.1073/pnas.0800693105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li D, Dong Y, Jiang Y, Jiang H, Cai J, et al. A de novo originated gene depresses budding yeast mating pathway and is repressed by the protein encoded by its antisense strand. Cell Res. 2010;20:408–420. doi: 10.1038/cr.2010.31. [DOI] [PubMed] [Google Scholar]
- 10.Rogers RL, Bedford T, Lyons AM, Hartl DL. Adaptive impact of the chimeric gene Quetzalcoatl in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2010;107:10943–10948. doi: 10.1073/pnas.1006503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Dean AM, Brunet F, Long M. Evolving protein functional diversity in new genes of Drosophila. Proc Natl Acad Sci U S A. 2004;101:16246–16250. doi: 10.1073/pnas.0407066101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nurminsky DI, Nurminskaya MV, De Aguiar D, Hartl DL. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature. 1998;396:572–575. doi: 10.1038/25126. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Zhang YP, Rosenberg HF. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat Genet. 2002;30:411–415. doi: 10.1038/ng852. [DOI] [PubMed] [Google Scholar]
- 14.Wang W, Brunet FG, Nevo E, Long M. Origin of sphinx, a young chimeric RNA gene in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2002;99:4448–4453. doi: 10.1073/pnas.072066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S, Arguello JR, Li X, Ding Y, Zhou Q, et al. Repetitive element-mediated recombination as a mechanism for new gene origination in Drosophila. PLoS Genet. 2008;4:e3. doi: 10.1371/journal.pgen.0040003. doi: 10.1371/journal.pgen.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Fruscio M, Styhler S, Wikholm E, Boulanger MC, Lasko P, et al. Kep1 interacts genetically with dredd/caspase-8, and kep1 mutants alter the balance of dredd isoforms. Proc Natl Acad Sci U S A. 2003;100:1814–1819. doi: 10.1073/pnas.0236048100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 18.Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- 19.Li W. Sunderland: Sinauer Associates; 1997. Molecular Evoltuion. [Google Scholar]
- 20.Pritchard JK, Schaeffer SW. Polymorphism and divergence at a Drosophila pseudogene locus. Genetics. 1997;147:199–208. doi: 10.1093/genetics/147.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heger A, Ponting CP. Evolutionary rate analyses of orthologs and paralogs from 12 Drosophila genomes. Genome Res. 2007;17:1837–1849. doi: 10.1101/gr.6249707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Fruscio M, Chen T, Bonyadi S, Lasko P, Richard S. The identification of two Drosophila K homology domain proteins. Kep1 and SAM are members of the Sam68 family of GSG domain proteins. J Biol Chem. 1998;273:30122–30130. doi: 10.1074/jbc.273.46.30122. [DOI] [PubMed] [Google Scholar]
- 23.Vibranovski MD, Lopes HF, Karr TL, Long M. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 2009;5:e1000731. doi: 10.1371/journal.pgen.1000731. doi: 10.1371/journal.pgen.1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 25.Spector DL. Nuclear organization of pre-mRNA processing. Curr Opin Cell Biol. 1993;5:442–447. doi: 10.1016/0955-0674(93)90009-f. [DOI] [PubMed] [Google Scholar]
- 26.Byun-McKay SA, Geeta R. Protein subcellular relocalization: a new perspective on the origin of novel genes. Trends Ecol Evol. 2007;22:338–344. doi: 10.1016/j.tree.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Xie HB, Golic KG. Gene deletions by ends-in targeting in Drosophila melanogaster. Genetics. 2004;168:1477–1489. doi: 10.1534/genetics.104.030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, et al. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindsley DL, Tokuyasu KT. Spermatogensis. In: Ashburner M, Wright TR, editors. Genetics and Biology of Drosophila. New York: Academic Press; 1980. pp. 225–294. [Google Scholar]
- 30.Fabrizio JJ, Hime G, Lemmon SK, Bazinet C. Genetic dissection of sperm individualization in Drosophila melanogaster. Development. 1998;125:1833–1843. doi: 10.1242/dev.125.10.1833. [DOI] [PubMed] [Google Scholar]
- 31.Santel A, Winhauer T, Blumer N, Renkawitz-Pohl R. The Drosophila don juan (dj) gene encodes a novel sperm specific protein component characterized by an unusual domain of a repetitive amino acid motif. Mech Dev. 1997;64:19–30. doi: 10.1016/s0925-4773(97)00031-2. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldstein LS, Hardy RW, Lindsley DL. Structural genes on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1982;79:7405–7409. doi: 10.1073/pnas.79.23.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gepner J, Hays TS. A fertility region on the Y chromosome of Drosophila melanogaster encodes a dynein microtubule motor. Proc Natl Acad Sci U S A. 1993;90:11132–11136. doi: 10.1073/pnas.90.23.11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carvalho AB, Lazzaro BP, Clark AG. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc Natl Acad Sci U S A. 2000;97:13239–13244. doi: 10.1073/pnas.230438397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardy RW, Lindsley DL, Livak KJ, Lewis B, Siversten AL, et al. Cytogenetic analysis of a segment of the Y chromosome of Drosophila melanogaster. Genetics. 1984;107:591–610. doi: 10.1093/genetics/107.4.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardy RW, Tokuyasu KT, Lindsley DL. Analysis of spermatogenesis in Drosophila melanogaster bearing deletions for Y-chromosome fertility genes. Chromosoma. 1981;83:593–617. doi: 10.1007/BF00328522. [DOI] [PubMed] [Google Scholar]
- 38.Zhang P, Stankiewicz RL. Y-Linked male sterile mutations induced by P element in Drosophila melanogaster. Genetics. 1998;150:735–744. doi: 10.1093/genetics/150.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Timakov B, Zhang P. Genetic analysis of a Y-chromosome region that induces triplosterile phenotypes and is essential for spermatid individualization in Drosophila melanogaster. Genetics. 2000;155:179–189. doi: 10.1093/genetics/155.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kenan DJ, Query CC, Keene JD. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 42.Singh R, Valcarcel J. Building specificity with nonspecific RNA-binding proteins. Nat Struct Mol Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- 43.Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci U S A. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature. 2007;450:233–237. doi: 10.1038/nature06323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vibranovski MD, Zhang Y, Long M. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 2009;19:897–903. doi: 10.1101/gr.088609.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Krester DM, Kerr JB. The Physiology of Reproduction. New York: Raven Press Ltd; 1994. The cytology of testis. pp. 1177–1290. [Google Scholar]
- 49.Jouannet P, Escaller D, Serres C, David G. Motility of human sperm without outer dynein arms. J Submicrosc Cytol. 1983;15:67–71. [PubMed] [Google Scholar]
- 50.Wolf JP, Feneux D, Escalier D, Rodrigues D, Frydman R, et al. Pregnancy after subzonal insemination with spermatozoa lacking outer dynein arms. J Reprod Fertil. 1993;97:487–492. doi: 10.1530/jrf.0.0970487. [DOI] [PubMed] [Google Scholar]
- 51.Keating J, Grundy CE, Fivey PS, Elliott M, Robinson J. Investigation of the association between the presence of cytoplasmic residues on the human sperm midpiece and defective sperm function. J Reprod Fertil. 1997;110:71–77. doi: 10.1530/jrf.0.1100071. [DOI] [PubMed] [Google Scholar]
- 52.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 53.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 54.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 55.Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726, 728, 730, 732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- 56.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 57.Wang Z, Mann RS. Requirement for two nearly identical TGIF-related homeobox genes in Drosophila spermatogenesis. Development. 2003;130:2853–2865. doi: 10.1242/dev.00510. [DOI] [PubMed] [Google Scholar]
- 58.Heatwole VM, Haynes SR. Association of RB97D, an RRM protein required for male fertility, with a Y chromosome lampbrush loop in Drosophila spermatocytes. Chromosoma. 1996;105:285–292. doi: 10.1007/BF02524646. [DOI] [PubMed] [Google Scholar]
- 59.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li R, Yu C, Li Y, Lam TW, Yiu SM, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 61.Wang L, Feng Z, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 62.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 63.Kiefer BI. Development, organization, and degeneration of the Drosophila sperm flagellum. J Cell Sci. 1970;6:177–194. doi: 10.1242/jcs.6.1.177. [DOI] [PubMed] [Google Scholar]
- 64.Mitchison TJ, Mitchison HM. How cilia beat. Nature. 2010;463:308–309. doi: 10.1038/463308a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Evolutionary analysis of CG3927 and CG4021. (A) Distributions of dN/dS throughout kep1-CG3927 and kep1-CG4021 pairs. With 120-bp windows and 6-bp slides, dN/dS ratios were estimated using the maximum likelihood method and plotted. Blue spots represent dN/dS ratios that are statistically significantly lower than the neutral expectation (p-value <0.05, two-tailed Fisher's exact test). Regions embedded in the KH domain, as depicted by the grey block, are enriched with signals of purifying selection. The shorter alignment of the kep1-CG3927 pair than the kep1-nsr and kep1-CG4021 pairs is mostly due to less well-aligned sequences caused by indels. (B) Likelihood values of nucleotide substitutions for CG3927 and CG4021 in Drosophila lineages. Numbers of nonsynonymous and synonymous substitutions for the entire coding sequences are labeled above the lineages, and ω values (dN/dS) are labeled beneath the lineages. Abbreviations: D. melanogaster (mel); D. simulans (sim); D. sechellia (sec); D. yakuba (yak), and D. erecta (ere).
(0.48 MB TIF)
Expression analysis of kep1 family proteins. (A) Male-specific expression pattern of new kep1 family genes shown by RT-PCR. Total RNA was extracted from the whole body of 0–5 day old adults. F: female; M: male. Negative control (NC) was reaction without reverse transcriptase. The expression of rp49 was used as the internal control. (B and C) Schematic representations of GFP (B) and TAP (C) transgene constructs. CDS: coding sequences. (D) Alignments of the homologous promoter sequences of D. melanogaster kep1 family genes and D. yakuba kep1. The asterisks denote the position of identical nucleotides. (E–H) The localization sensitivity of GFP-tagged Nsr protein to RNase A treatment. The localization of GFP-tagged Nsr protein exhibits ectopic diffusion and accumulation after RNase A treatment (E, G) compared with the mock-treated control (F, H). Scale bars: 200 µm for E and F; 20 µm for G and H.
(1.42 MB TIF)
Immunoprecipitation results of TAP-tagged kep1 family proteins and pre-duplication ancestral subcellular localization of Kep1 protein. (A) Western blot of testis extracts from 0–5 day old flies, with the indicated genotypes. The blot was probed with PAP (to recognize the ProtA of TAP) and β-Actin antibody (loading control). The absence of ProtA in w1118 lane indicates the specificity of PAP antibody. (B) Immunoprecipitation of TAP-tagged kep1 family proteins with testis extracts from 0–3 day flies. A 50-72-kDa band corresponding to CstF-64 (identified by mass spectrometry) was immunoprecipitated by TAP-tagged Nsr protein (lane 2) but not in TAP-tagged Kep1 protein (lane 1) or TAP-tagged CG4021 protein (lane 3). (C) Western blot of testis extracts from 0–5 day old WT flies showing that Kep1 antibody recognizes a band of about 43-kDa (arrow), which is consistent with the reported size of Kep1 protein, and that this band is absent in kep1 mutants. (D) Western blot of testis (lane 1 and 2) and ovary (lane 3 and 4) extracts of 0–5 day old D. melanogaster and D. yakuba probed with Kep1 antibody. Kep1 protein is detectable in ovary but not testis of D. yakuba (arrow). (E) Overlapping subcellular localization between the D. melanogaster TAP-tagged Kep1 (mel-Kep1-TAP) and the transgenic GFP-tagged Kep1 protein regulated by the cis-elements (including promoter, 5′ UTR and coding sequences) of D. yakuba kep1 (yak-Kep1-GFP) in D. melanogaster. Scale bars: 20 µm.
(1.23 MB TIF)
dN/dS ratios of kep1 among Drosophila species (Kumar method). All the pairwise species comparisons show that the parental gene kep1 is subjected to significant purifying selection (p-value <0.001, MEGA Z-test, kumar method). Abbreviations: D. melanogaster (mel); D. simulans (sim); D. sechellia (sec); D. yakuba (yak); D. erecta (ere) and D. ananassae (ana); D. pseudobscura (pse).
(0.03 MB DOC)
Maximum likelihood tests of positive selection for new kep1 family genes. The dN/dS ratios were set to be ω0 for the background branches and ω1 for the foreground branch, which is the phylogenetic lineage from D. yakuba to the common ancestor of the D. melanogaster species complex.
(0.03 MB DOC)
List of genes identified by microarray to be >2-fold differentially expressed between testes of nsr WT and mutant flies. Significance Analysis of Microarrays (SAM) identified 14 genes that changed at least 2-fold with a q-value cutoff of 0.01 for significance.
(0.02 MB XLS)
List of genes identified by RNA-seq to be >3-fold differentially expressed between testes of nsr WT and mutant flies. The significance of expression difference (p-value) for each gene (longest transcript) was computed using MA-plot-based method with a random sampling model and followed by an adjustment with q-values for multiple testing corrections. With a p-value cutoff of 0.001, 10 genes (red) and 43 genes are identified to be significantly changed at least 5-fold and 3-fold, respectively.
(0.04 MB XLS)
Primer Information. F: forward primer; R: reverse primer.
(0.03 MB XLS)