Abstract
Increasing evidence suggests that parentally supplied RNA plays crucial roles during eukaryotic development. This epigenetic contribution may regulate gene expression from the earliest stages. Although present in a variety of eukaryotes, maternally inherited characters are especially prominent in ciliated protozoa, in which parental noncoding RNA molecules instruct whole-genome reorganization. This includes removal of nearly all noncoding DNA and sorting the remaining fragments, producing extremely gene-rich somatic genomes. Chromosome fragmentation and extensive replication produce variable DNA copy numbers in the somatic genome. Understanding the forces that drive and regulate copy number change is fundamental. We show that RNA molecules present in parental cells during sexual reproduction can regulate chromosome copy number in the developing nucleus of the ciliate Oxytricha. Experimentally induced changes in RNA abundance can both increase and decrease the levels of corresponding DNA molecules in progeny, demonstrating epigenetic inheritance of chromosome copy number. These results suggest that maternal RNA, in addition to controlling gene expression or DNA processing, can also program DNA amplification levels.
Keywords: copy number variation, genome rearrangement
DNA copy number profoundly influences evolution (1–4), permitting changes in gene and protein expression (5–7) that may be acted upon by positive selection (7), permit adaptation (8–11), or have links to human genetic disease (12) and disease susceptibility (13–15), including autism-spectrum disorders (16) and several cancers (5–6, 17–22).
Because most of Oxytricha’s more than 20,000 unique “nanochromosomes” in its somatic nucleus bear only a single gene (23, 24) and copy number can vary independently (25), Oxytricha provides a unique opportunity to study copy number variation. The absence of physical linkage potentially releases most constraints on gene copy number, encouraging gene regulation at the level of DNA copy number, in addition to transcription. Although longer, germline chromosomes participate in conventional mitosis during vegetative growth, the tiny somatic macronuclear chromosomes that differentiate from the germline chromosomes undergo amitosis, permitting both drift and regulation of copy number (26, 27). Observed levels vary over four orders of magnitude (23, 25, 27–31). With such high copy numbers, a typical macronucleus may contain tens of millions of nanochromosomes, averaging 2.2 kb bound by short telomeres (23). Such high copy numbers may permit high rates of transcription and translation to sustain Oxytricha’s large (∼100 μm) cells (23).
Our previous work revealed that noncoding template RNAs that originate from the parental macronucleus guide a highly elaborate process of DNA deletion, unscrambling, and assembly to form the macronuclear genome (32). Disruption of specific RNA templates disables rearrangement of the corresponding gene, and injection of artificial templates reprograms the DNA rearrangement pathway. Here we investigate whether the same maternal RNA templates that guide genome rearrangement in Oxytricha can also control the level of chromosome amplification in the developing nucleus.
Results
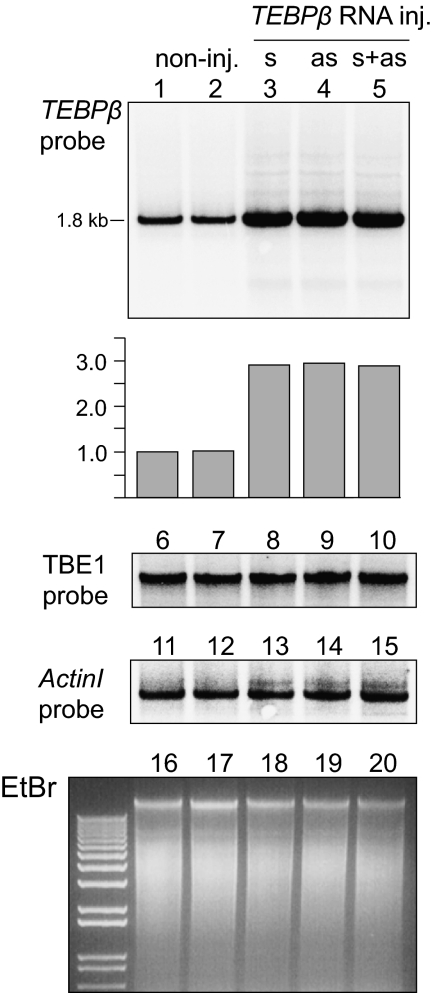
To test whether the level of maternal transcripts available during sexual reproduction has an influence on DNA copy number in the sexual progeny, we independently microinjected full-length RNA copies of two different gene-sized chromosomes, carrying Telomere-End-Binding Protein-β (TEBPβ) or ActinI genes, into the cytoplasm of mating cells. TEBPβ represents an example of a conventional gene, requiring only DNA deletion but no reordering during nuclear development, whereas ActinI represents a scrambled gene that requires both DNA inversions and permutations to produce a functional copy (23). Injections occurred between 10 to 15 h after mixing of complementary mating types, before pairs separate, around the time of fertilization. The progeny from mating pairs injected with TEBPβ RNA demonstrated normal growth rate but 2.9-fold greater levels (i.e., 290% increase) of TEBPβ nanochromosomes, regardless of whether they were injected with sense, antisense, or both RNA strands (Fig. 1). Therefore, subsequent experiments used only the sense strand. The uniform ActinI hybridization (Fig. 1, lanes 11–15) suggests that the effects of TEBPβ RNA microinjection were sequence-specific. In addition, there was no effect of RNA microinjection on the overall amount of macronuclear DNA, based on ethidium bromide staining (Fig. 1, lanes 16–20), supporting the sequence specificity of the effect. PCR and DNA sequencing confirmed that the DNA copy number increase is not a result of any contaminating DNA that may have been coinjected with RNA, because the progeny DNA contains multiple alleles, whereas the synthetic templates derive from a single PCR clone that also contains additional point substitutions (SI Text). The presence of multiple TEBPβ alleles also supports their zygotic origin.
Fig. 1.
Microinjection of RNA increases chromosome copy number in sexual progeny. Southern analysis of whole-cell DNA isolated from pooled progeny of 40 cells (20 conjugating pairs) in each experiment injected (inj.) with TEBPβ sense (s) or antisense (as) RNA or both strands. Hybridization to a TEBPβ probe (lanes 1–5) shows increased levels of the WT TEBPβ chromosome in progeny of injected cells (lanes 3–5). Gray bars indicate relative amount of TEBPβ probe signal. Lanes 6 to 10 and 11 to 15 are hybridization to a TBE1 transposase and ActinI probe, respectively, as DNA loading controls, in addition to ethidium bromide staining (lanes 16–20) on a 0.9% agarose gel. ActinI hybridization and ethidium bromide staining indicate that the effect of TEBPβ RNA microinjection is sequence-specific, as it does not increase the levels of ActinI chromosomes or of macronuclear DNA in general.
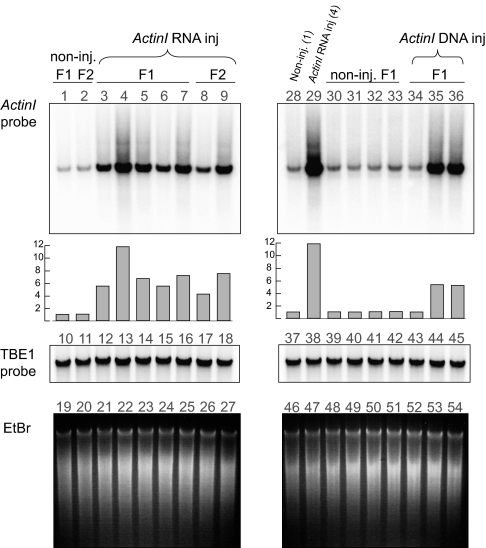
To confirm the effect and examine its epigenetic inheritance, we injected synthetic sense RNA for the complete ActinI chromosome into the cytoplasm of five mating pairs. These individual pairs were isolated after injection and their progeny cells were cultured separately. Southern analysis revealed 5.5- to 11.8-fold higher ActinI chromosome copy levels in the five independent experiments. To test whether the RNA-induced DNA copy number increase transferred epigenetically to the next generation, we induced sexual conjugation by starving the progeny of ActinI-injected cells after 2 wk of vegetative growth. Conjugating pairs that were separated and grown vegetatively for another 2 wk also demonstrated increased ActinI nanochromosome copy number in F2 cells (4.3–7.5-fold; Fig. 2), supporting epigenetic inheritance of this acquired trait.
Fig. 2.
Microinjection of RNA or DNA increases chromosome copy number in sexual progeny. Southern analysis of total DNA from progeny of cells injected with sense RNA copies of the complete ActinI nanochromosome. Lanes 1 to 9 and 28 to 36 contain DNA from the progeny of individually injected mating pairs hybridized to an ActinI probe. F1 cells (lanes 3–7) show increased levels of the ActinI nanochromosome. F2 cells (lanes 8 and 9) that were derived from the progeny of individual mating pairs isolated from F1 cells also show higher amounts of the ActinI chromosome, although slightly less than their parents (lanes 3 and 4). Lanes 30 to 33 contain DNA from four different clonal cultures derived from noninjected parents. They do not exhibit differences in ActinI chromosome copy number. Lanes 34 to 36 contain DNA from the progenies of cells injected with the full-length synthetic DNA chromosome. Two show increased levels of ActinI chromosomes. To compare relative signal intensities on both blots, samples 1 and 4 (Left) were duplicated in lanes 28 and 29. Gray bars indicate relative levels of ActinI probe signal. TBE1 transposase probe provided a loading control (lanes 10–18 and 37–45) in addition to ethidium bromide staining (lanes 19–27 and 46–54). The ethidium staining also confirms that ActinI injection did not influence the overall levels of macronuclear DNA, helping to exclude non-sequence-specific effects.
We also tested whether increased DNA copy number in the parental nucleus can induce increased amplification of the corresponding chromosome in sexual progeny. We injected full-length DNA copies of a cloned ActinI nanochromosome containing additional point substitutions into the somatic nuclei of mating pairs 6 to 8 h after mixing, approximately 1 to 3 h after pair formation. Of the three pairs that survived nuclear injection to allow examination of progeny after 12 d of growth, two showed an approximate 5.4-fold increase levels of ActinI nanochromosome copy number (Fig. 2). Cloning and sequencing revealed the absence of the artificial substitutions in the injected DNA (sites marked with an asterisk in SI Text), as well as the presence of multiple alleles, confirming the amplification of endogenous ActinI chromosomes and not simply the added presence of injected DNA in the progeny.
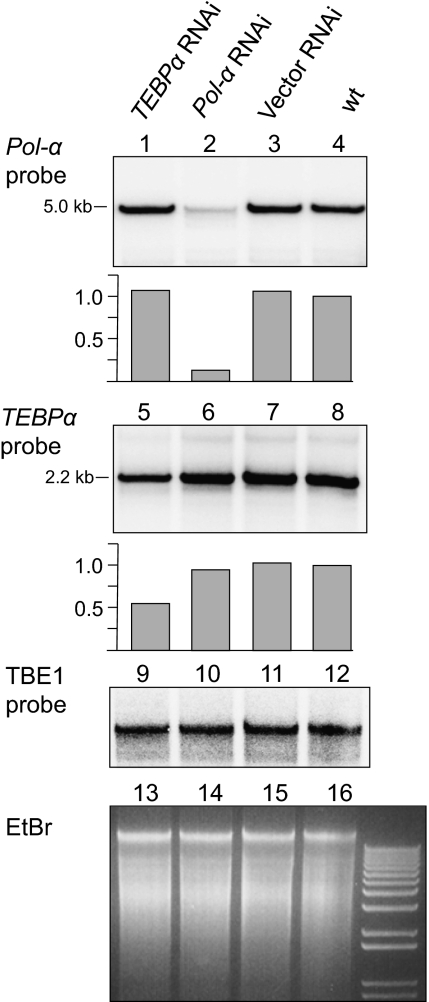
Reciprocally, we used RNAi to test whether reduced availability of RNA during conjugation has a negative effect on chromosome amplification. Pooled progenies of conjugating cells that were treated with RNAi against TEBPα and DNA Polymerase-α (pol-α), both scrambled genes, previously displayed down-regulation of corresponding mRNA levels during conjugation and aberrant rearrangement of the corresponding DNA loci (32). Here we report that these cells also display 1.8-fold (i.e., 180%) and 7.7-fold reduced levels, respectively, of the mature DNA nanochromosomes (Fig. 3). The stronger effect for pol-α is consistent with its more severe effect on DNA rearrangements (32). The two independent RNAi experiments have a sequence-specific effect on the respective gene copy numbers, with no effect on the nonhomologous gene (Fig. 3, lanes 1 and 6), reinforcing the conclusion that RNA level during conjugation influences DNA copy number in the progeny in a sequence-specific manner.
Fig. 3.
RNAi during conjugation reduces chromosome copy number in progeny. Southern analysis of whole-cell DNA from progeny of cells treated with RNAi against pol-α (lanes 1–4) and TEBPα (lanes 5–8). RNAi during conjugation reduces the levels of corresponding chromosomes in sexual progeny (lanes 2 and 5). Gray bars indicate relative levels of pol-α and TEBPα probe signals. TBE1 transposase probe signal (lanes 9–12), as well as ethidium bromide staining of whole-cell DNA (lanes 13–16), provided a loading control.
Discussion
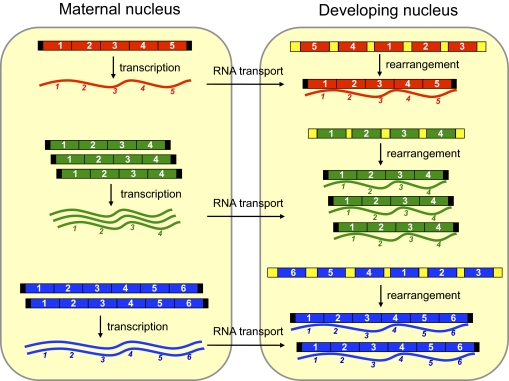
Our data support a model of template RNA-mediated epigenetic inheritance of DNA copy number in the Oxytricha macronucleus (Fig. 4) involving chromosome replication during development. As chromosome copy number varies in the somatic nucleus, cells should produce RNA templates during conjugation in approximately the same proportion as their DNA concentration in a mature parent nucleus. After template RNAs guide DNA rearrangements, the level of correctly processed chromosomes may also correlate with available template RNA concentration, suggesting a quantitative relationship from maternal DNA to maternal RNA to progeny DNA. This permits establishment of chromosome copy number very early during development (23). Furthermore, the effect of RNA injection on chromosome copy number is not dependent on the type of DNA rearrangement during chromosomal maturation, influencing both scrambled and nonscrambled genes.
Fig. 4.
Model for RNA-guided epigenetic inheritance of chromosome copy number in Oxytricha. During sexual conjugation, all DNA in the maternal somatic nucleus produces transcripts in approximately similar abundance to the chromosome copy number. After transport to the developing nucleus, the maternal RNAs guide DNA rearrangements, yielding a quantity of mature chromosomes proportional to the level of available templates, or alternatively, the template RNAs may protect correctly processed DNA molecules from degradation. Numbered DNA and RNA segments represent regions that are retained during genome rearrangement, whereas yellow boxes are deleted. Black boxes represent telomeres.
A previous observation in Paramecium noted that the presence of siRNA during conjugation leads to local DNA deletions within the targeted region (33). This is consistent with a “scan-RNA” model proposed for Paramecium and Tetrahymena (34, 35) wherein injected siRNA cause degradation of homologous maternal transcripts, which subsequently are unable to sequester the germline scan RNAs, resulting in deletion of corresponding DNA in the developing nucleus. It is unknown, however, whether maternal RNA in these and other organisms can influence DNA copy number, as we show for Oxytricha.
The combination of imprecise nuclear division during vegetative growth and high ploidy allows individual chromosome copy number to vary widely (26, 27). In addition, most reports of DNA copy number in ciliates reflect population averages that mask individual variation, if copy number is not measured in individual cells. Although somatic variation can be acted upon by selection during asexual growth, after approximately 80 divisions, Oxytricha trifallax cells cannot produce viable progeny (36), and extreme departures in chromosome copy number from initial, postconjufation levels may contribute to senescence (30). The possibility that chromosome copy number is not regulated during vegetative growth underscores the need to regulate it during conjugation. RNA-mediated epigenetic control of DNA copy number offers both robustness and plasticity to genomes, because they may transmit somatically acquired changes in copy number across generations if the new copy number confers a selective advantage, yet still retain the ability to reset DNA copy number to previous levels upon mating with a “wild-type” cell or by further somatic change in the opposite direction. It remains to be determined whether individual exconjugant cells in a mating pair adopt the chromosome copy numbers of one parent or acquire the average of both parents’ RNA template levels. The potential reversibility of RNA-mediated epigenetic phenomena facilitates adaptation in a changing environment. The expanding roles of RNA-mediated inheritance underscore the inadequacy of the germline genome and the need for the parental cell to equip its daughters with an RNA cache bearing all of the additional information necessary for proper development.
Methods
Microinjection of DNA and RNA.
Synthetic RNA and DNA were generated by PCR, followed by restriction digestion, ligating segments, and cloning into pCR2.1-TOPO vector (Invitrogen). DNA versions were prepared by telomere-to-telomere PCR from the cloned DNA, and RNA versions of each strand were prepared by in vitro transcription of PCR products with T7 promoter appended to the forward or the reverse primer. RNA samples were DNase-treated after in vitro transcription and before injection. Approximately 5 pL DNA or 10 pL RNA (5 μg/μL) was injected (IM 300; Narishige) into the macronucleus or cytoplasm, respectively, of each cell in a mating pair visualized by phase-contrast inverted microscopy (Axiovert 200; Zeiss) as described previously (32). After injection, the pairs were pooled together or cultured separately. The same number of noninjected pairs were always treated identically as a control. Whole-cell DNA was isolated 12 to 14 d later for analysis. Individual exconjugants were observed under the microscope and each cell displayed morphological features characteristic of sexual reproduction in Oxytricha (also called Sterkiella), such as rounded cell shape or a large macronuclear anlagen. Thus, as described earlier (32), on the basis of cell morphology, the analyzed F2 cells were the progeny of F1.
RNAi.
Double-stranded RNA feeding was performed by using a modified Paramecium protocol (37) that we described previously (32), in which ciliates were fed live bacteria, supplemented with algae. Silencing plasmids contained 1.5 kb of TEBPα or 1.8 kb of the pol-α macronuclear sequence (details and RNAi-feeding sequences provided in ref. 32). Pooled progeny cells were analyzed.
Cells.
O. trifallax (Sterkiella histriomuscorum) strains JRB310 and JRB510 (38) were cultured in inorganic salts medium (39) with the algae Chlorogonium elongatum as a food source and Klebsiella oxytoca as a food supplement. To induce mating, JRB310 and JRB510 cells were mixed in Pringsheim salts (40) at 2,000 cells/mL, with only Klebsiella as food source. Early pairs appear 4 to 6 h after mixing of mating types, and maximum pairing (>60%) occurs within 24 h after mixing.
DNA and RNA isolation.
Cells were harvested after 12 to 14 d of recovery in vegetative culture by addition of 50 mM EDTA and centrifugation at 2,500 × g for 10 min. DNA was extracted with the NucleoSpin Tissue kit (Macherey-Nagel). RNA was extracted with NucleoSpin RNA II (Macherey-Nagel).
PCR After RNAi.
All PCR products were amplified with FastStart High-Fidelity PCR systems (Roche Applied Science).
Southern Hybridization.
DNA (approximately 1 μg) was electrophoresed through an ethidium bromide-stained 0.9% agarose gel, depurinated in gel (0.25% HCl 15 min; washed in 0.4 M NaOH for 15 min), and transferred to Hybond XL membrane in 0.4 M NaOH with a Nytran TurboBlotter (GE Healthcare). Probes were generated by random priming [Prime-It (Stratagene) with (α-32P)-dATP 3,000 Ci/mmol (Perkin-Elmer)] of WT Oxytricha strain JRB310 TEBPα, TEBPβ, Pol-α, ActinI, and TBE1 transposase PCR products and purified (NucTrap; Stratagene). After overnight hybridization at 60 °C (0.5 M NaPO4, pH 7.2, 1% BSA, 1 mM EDTA, 7% SDS) the membrane was washed in 0.2× SSC with 0.1% SDS (30 min, 60 °C), and visualized on a Storm 860 PhosphorImager.
Supplementary Material
Acknowledgments
We thank E. Swart, J. Wang, T. Doak, and Y. Zhou for valuable assistance and discussion. This study was supported by National Science Foundation Grant 0923810 and National Institutes of Health Grant GM59708.
Footnotes
See Commentary on page 21951.
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012236107/-/DCSupplemental.
References
- 1.Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 2008;320:1629–1631. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- 2.Cooper GM, Nickerson DA, Eichler EE. Mutational and selective effects on copy-number variants in the human genome. Nat Genet. 2007;39(7 suppl):S22–S29. doi: 10.1038/ng2054. [DOI] [PubMed] [Google Scholar]
- 3.Korbel JO, et al. The current excitement about copy-number variation: How it relates to gene duplications and protein families. Curr Opin Struct Biol. 2008;18:366–374. doi: 10.1016/j.sbi.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dopman EB, Hartl DL. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:19920–19925. doi: 10.1073/pnas.0709888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyman E, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–6245. [PubMed] [Google Scholar]
- 6.Pollack JR, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perry GH, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1188–1190. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullis CA. Molecular aspects of the environmental induction of heritable changes in flax. Heredity. 1977;38:129–154. [Google Scholar]
- 9.Riehle MM, Bennett AF, Long AD. Genetic architecture of thermal adaptation in Escherichia coli. Proc Natl Acad Sci USA. 2001;98:525–530. doi: 10.1073/pnas.021448998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunham MJ, et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Infante JJ, Dombek KM, Rebordinos L, Cantoral JM, Young ET. Genome-wide amplifications caused by chromosomal rearrangements play a major role in the adaptive evolution of natural yeast. Genetics. 2003;165:1745–1759. doi: 10.1093/genetics/165.4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estivill X, Armengol L. Copy number variants and common disorders: Filling the gaps and exploring complexity in genome-wide association studies. PLoS Genet. 2007;3:1787–1799. doi: 10.1371/journal.pgen.0030190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 14.Aldred PMR, Hollox EJ, Armour JA. Copy number polymorphism and expression level variation of the human alpha-defensin genes DEFA1 and DEFA3. Hum Mol Genet. 2005;14:2045–2052. doi: 10.1093/hmg/ddi209. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 16.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein CA, et al. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc Natl Acad Sci USA. 1999;96:4494–4499. doi: 10.1073/pnas.96.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucito R, et al. Detecting gene copy number fluctuations in tumor cells by microarray analysis of genomic representations. Genome Res. 2000;10:1726–1736. doi: 10.1101/gr.138300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain AN, et al. Quantitative analysis of chromosomal CGH in human breast tumors associates copy number abnormalities with p53 status and patient survival. Proc Natl Acad Sci USA. 2001;98:7952–7957. doi: 10.1073/pnas.151241198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snijders AM, et al. Genome-wide-array-based comparative genomic hybridization reveals genetic homogeneity and frequent copy number increases encompassing CCNE1 in fallopian tube carcinoma. Oncogene. 2003;22:4281–4286. doi: 10.1038/sj.onc.1206621. [DOI] [PubMed] [Google Scholar]
- 21.van de Wiel MA, et al. Expression microarray analysis and oligo array comparative genomic hybridization of acquired gemcitabine resistance in mouse colon reveals selection for chromosomal aberrations. Cancer Res. 2005;65:10208–10213. doi: 10.1158/0008-5472.CAN-05-0760. [DOI] [PubMed] [Google Scholar]
- 22.Langdon JA, et al. Combined genome-wide allelotyping and copy number analysis identify frequent genetic losses without copy number reduction in medulloblastoma. Genes Chromosomes Cancer. 2006;45:47–60. doi: 10.1002/gcc.20262. [DOI] [PubMed] [Google Scholar]
- 23.Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman DC, Anderson RC, DuBois ML, Prescott DM. Macronuclear gene-sized molecules of hypotrichs. Nucleic Acids Res. 1995;23:1279–1283. doi: 10.1093/nar/23.8.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harper DS, Song K, Jahn CL. Overamplification of macronuclear linear DNA molecules during prolonged vegetative growth of Oxytricha nova. Gene. 1991;99:55–61. doi: 10.1016/0378-1119(91)90033-8. [DOI] [PubMed] [Google Scholar]
- 26.Baird SE, Klobutcher LA. Differential DNA amplification and copy number control in the hypotrichous ciliate Euplotes crassus. J Protozool. 1991;38:136–140. doi: 10.1111/j.1550-7408.1991.tb06033.x. [DOI] [PubMed] [Google Scholar]
- 27.Baird SE, Klobutcher LA. Genetic characterization and use of a restriction fragment length variant in the hypotrichous ciliate Euplotes crassus. J Protozool. 1988;35:459–465. doi: 10.1111/j.1550-7408.1988.tb04130.x. [DOI] [PubMed] [Google Scholar]
- 28.Gaunitz C, Witte H, Gaunitz F. Primary structure of a gene-sized DNA encoding calmodulin from the hypotrichous ciliate Stylonychia lemnae. Gene. 1992;119:191–198. doi: 10.1016/0378-1119(92)90271-p. [DOI] [PubMed] [Google Scholar]
- 29.Steinbruck G. Overamplification of genes in macronuclei of hypotrichous ciliates. Chromosoma. 1983;88:156–163. [Google Scholar]
- 30.Duerr HP, Eichner M, Ammermann D. Modeling senescence in hypotrichous ciliates. Protist. 2004;155:45–52. doi: 10.1078/1434461000163. [DOI] [PubMed] [Google Scholar]
- 31.Herrick GS, Cartinhour SW, Williams KR, Kotter KP. Multiple sequence versions of the Oxytricha fallax 81-MAC alternate processing family. J Protozool. 1987;34:429–434. doi: 10.1111/j.1550-7408.1987.tb03207.x. [DOI] [PubMed] [Google Scholar]
- 32.Nowacki M, et al. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garnier O, Serrano V, Duharcourt S, Meyer E. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol Cell Biol. 2004;24:7370–7379. doi: 10.1128/MCB.24.17.7370-7379.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mochizuki K, Gorovsky MA. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev. 2004;18:2068–2073. doi: 10.1101/gad.1219904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lepère G, Bétermier M, Meyer E, Duharcourt S. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 2008;22:1501–1512. doi: 10.1101/gad.473008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adl SM, Berger JD. Timing of life cycle morphogenesis in synchronous samples of Sterkiella histriomuscorum. II. The sexual pathway. J Eukaryot Microbiol. 2000;47:443–449. doi: 10.1111/j.1550-7408.2000.tb00073.x. [DOI] [PubMed] [Google Scholar]
- 37.Galvani A, Sperling L. RNA interference by feeding in Paramecium. Trends Genet. 2002;18:11–12. doi: 10.1016/s0168-9525(01)02548-3. [DOI] [PubMed] [Google Scholar]
- 38.Williams K, Doak TG, Herrick G. Developmental precise excision of Oxytricha trifallax telomere-bearing elements and formation of circles closed by a copy of the flanking target duplication. EMBO J. 1993;12:4593–4601. doi: 10.1002/j.1460-2075.1993.tb06148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang W-J, Stover NA, Addis VM, Landweber LF. A micronuclear locus containing three protein-coding genes remains linked during macronuclear development in the spirotrichous ciliate Holosticha. Protist. 2004;155:245–255. doi: 10.1078/143446104774199628. [DOI] [PubMed] [Google Scholar]
- 40.Laughlin TJ, Henry JM, Phares EF, Long MV, Olins DE. Methods for the large-scale cultivation of an Oxytricha (Ciliophora: Hypotricha) J Eukaryot Microbiol. 1983;30:63–64. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.