Abstract
The capacity of human poly(ADP-ribose) polymerase-1 (PARP-1) to interact with intact apurinic/apyrimidinic (AP) sites in DNA has been demonstrated. In cell extracts, sodium borohydride reduction of the PARP-1/AP site DNA complex resulted in covalent cross-linking of PARP-1 to DNA; the identity of cross-linked PARP-1 was confirmed by mass spectrometry. Using purified human PARP-1, the specificity of PARP-1 binding to AP site-containing DNA was confirmed in competition binding experiments. PARP-1 was only weakly activated to conduct poly(ADP-ribose) synthesis upon binding to AP site-containing DNA, but was strongly activated for poly(ADP-ribose) synthesis upon strand incision by AP endonuclease 1 (APE1). By virtue of its binding to AP sites, PARP-1 could be poised for its role in base excision repair, pending DNA strand incision by APE1 or the 5′-dRP/AP lyase activity in PARP-1.
Keywords: Schiff base, apurinic/apyrimidinic site-binding protein
The apurinic/apyrimidinic (AP) site is considered to be a common lesion in genomic DNA, arising at a frequency of 10,000 to 50,000 lesions per mammalian cell per day (1). If unrepaired, AP sites present mutagenic and cytotoxic consequences to the cell (2). The loss of DNA bases and attendant formation of AP sites in DNA occurs spontaneously as a result of hydrolytic cleavage of N-glycosylic bonds. AP sites are also generated through glycosylase-catalyzed removal of damaged bases during the early stage of base excision repair (BER) (3). The number of AP sites can increase dramatically under stressful conditions such as X-ray or UV light irradiation and alkylating agent exposure (4). AP sites in isolated DNA are stable enough for their use as substrates in enzymatic assays, but AP sites can be converted to single-strand breaks by alkali treatment, heating, or nucleophilic attack at the aldehydic C1′ group (5). Intact AP sites in vivo can be stable enough to be mutagenic (6) during replication. Measurements of steady-state levels of AP sites in mammalian cells have varied somewhat, but are in the range of approximately 1 site per 106 nucleotides (4, 7). Nevertheless, repair of AP sites is thought to be extremely rapid, thus minimizing the steady-state AP site level (8). In mammalian cells, the repair of AP sites is generally initiated through strand incision by AP endonuclease 1 (APE1), the second enzyme of the canonical BER pathway (9, 10). AP sites also can be incised through β-elimination via the activity of DNA glycosylases and other enzymes with associated AP lyase activity (3, 11, 12). In this case, strand incision forms a single-nucleotide gap with the AP site sugar phosphate at the 3′ margin and phosphate at the 5′ margin, and this intermediate is further processed by polynucleotide kinase, APE1, or other BER enzymes (12).
During AP site cleavage by the AP lyase activity of DNA glycosylases, an aldimine intermediate between the C1′ atom of deoxyribose and a primary amine group in the protein is formed (3, 13, 14). This intermediate, termed the Schiff base, can be reduced in vitro by sodium borohydride (NaBH4) treatment, forming an irreversible covalent complex between the enzyme and DNA (3, 13, 14). Several of the mammalian enzymes known to form the Schiff base intermediate with the AP site appear to belong to the BER system. On the other hand, an interaction of AP site DNA with proteins not considered to be involved in BER has been well documented. For example, human ribosomal protein S3 and nucleoside diphosphate kinase (NM23-H2/NDP) have been shown to form Schiff base intermediates with AP sites (15, 16). In addition, the NaBH4 trapping technique in combination with mass spectrometry has been used in a proteomic approach to identify a host of Escherichia coli and baker’s yeast proteins reactive at AP sites (17); multiple proteins belonging to a range of metabolic pathways were identified. Because the AP site in DNA appears to be promiscuous in its binding to many cellular factors, it may be important for the BER system to sequester the AP site immediately upon formation. With this idea in mind, we screened cell extracts for proteins capable of binding AP site-containing DNA and then participating in the subsequent steps in BER.
Using a 32-bp DNA duplex with an AP site in the middle of the DNA sequence, we recently identified the p80 subunit of Ku antigen (Ku) as the protein forming the predominant product of NaBH4 cross-linking in HeLa cell extract (18). Ku recently was shown to be a 5′-dRP/AP lyase involved in double-strand break repair (19). In further screening for proteins that are reactive at AP sites, we used circular AP site-containing DNA to exclude interference by Ku80. Circular double-stranded DNA was synthesized, using single-stranded M13 DNA as a template, in the presence of dUTP; then, AP sites were generated by uracil-DNA glycosylase (UDG) treatment. Unlike short duplex DNA with an AP site that predominantly cross-linked Ku80 in HeLa cell extract (18), the use of circular AP site-containing DNA allowed us to detect a unique cross-linked protein with molecular mass of ∼120 kDa. This cross-linked protein–DNA complex was isolated and examined by mass spectrometry. The analysis identified the protein as poly(ADP-ribose) polymerase-1 (PARP-1). Further characterization of the PARP-1 interaction with AP site-containing DNA and its implications are discussed.
Results and Discussion
Cross-Linking of Cell Extract Proteins to AP Site-Containing DNA and Poly(ADP-Ribose) Modification.
To study the interaction of cultured cell and tissue extract proteins with AP site-containing DNA, two types of DNA probe-bearing natural AP sites were used. One probe was a linear DNA duplex with an AP site in the middle of the 32P-5′ end-labeled strand. The second probe was a circular duplex DNA containing randomly distributed AP sites in one of the strands; double-stranded circular 32P-labeled DNA with randomly incorporated dUMP in one strand was synthesized using M13 single-stranded DNA as a template, and AP sites were created by UDG treatment immediately before use. In the case of circular AP site-containing DNA, prior to SDS-PAGE, the products of protein–DNA cross-linking were treated with nuclease to remove DNA outside of the protein–DNA complex.
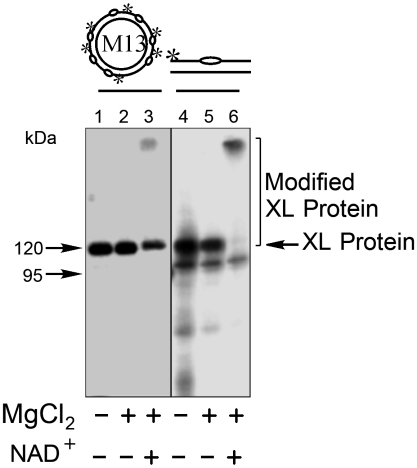
AP sites are recognized by cell extract proteins and, in particular, proteins that bind to the C1′ of deoxyribose and form the Schiff base. These proteins are studied after irreversible cross-linking by NaBH4 reduction (13, 14). Schiff base formation does not require a divalent metal, and in the initial experiments, incubations with both DNA probes contained EDTA to prevent AP site cleavage by endonucleases in the extracts. The results were compared with those from incubations with magnesium ions in excess over EDTA. Protein cross-linking in the bovine testis nuclear extract (BTNE) is shown in Fig. 1. Similar results were obtained with both incubation conditions (Fig. 1, lanes 1, 2 and 4, 5) and there was remarkable specificity for cross-linking of one extract protein. A prominent cross-linked (XL) labeled protein–DNA complex was observed at ∼120 kDa. A similar pattern of protein-circular AP site DNA cross-linking was obtained in HeLa cell extract (Fig. S1). Based on its size and the known PARP-1 recognition of BER intermediates (20–23), we suspected this XL protein might be PARP-1. To identify the protein in the ∼120-kDa complex, we first tested for poly(ADP-ribosyl)ation of the complex. Reaction mixtures with magnesium and the AP site-containing DNA probe were incubated for 15 min at 37 °C, as usual. The reaction mixture was then supplemented with NAD+, the substrate for synthesis of poly(ADP-ribose). After further incubation to allow poly(ADP-ribose) synthesis, the reaction mixture was treated with NaBH4. In the presence of NAD+, the amount of the ∼120-kDa protein–DNA complex was reduced and slower migrating material was now observed (Fig. 1, lanes 3 and 6). These results were consistent with poly(ADP-ribose) modification of the protein interacting with the DNA probe, suggesting that this protein was either automodified or modified by PARP in the extract.
Fig. 1.
Cross-linking of BTNE proteins to AP site-containing DNA probe and effect of poly(ADP-ribose) modification. The reaction conditions and product analysis were as described under Materials and Methods. 32P-labeled AP site-containing circular DNA probe (20 nM) or linear DNA probe (100 nM) was incubated with BTNE (1.25 mg/mL (lanes 2 and 5) for 15 min at 37 °C. In lanes 3 and 6, the reaction mixtures were supplemented with NAD+ and then further incubated for 5 min at 37 °C prior to NaBH4 treatment. NaBH4 was then added, and the mixtures were further incubated at 0–1 °C for 30 min. Then in lane 1, the reaction mixture was supplemented with 10 mM MgCl2. The reaction products in lanes 1–3 (proteins cross-linked with circular AP DNA) were treated with benzonase (12 units) for 15 min at 37 °C prior to SDS-PAGE. Covalently cross-linked DNA-protein complexes (XL Protein) were separated by SDS-PAGE. A representative phosphorimage illustrating cross-linking of BTNE proteins is shown. The migration positions of protein markers (M), designated in kDa, XL Protein, and poly(ADP-ribose) Modified XL Protein are indicated.
Identification of the ∼120-kDa XL Protein by Mass Spectrometry.
Next, to prove the identity of the XL protein, we performed large-scale cross-linking with the BTNE and a biotin-containing DNA probe. The cross-linked protein was prepared, purified using streptavidin-coupled paramagnetic beads, and resolved by SDS-PAGE. A well-defined stained protein band precisely corresponding to the labeled ∼120-kDa band (Fig. S2) was excised from the gel, and the protein was subjected to in-gel trypsin digestion and mass spectrometry (MS). Results from the MS analyses were searched against a database, and PARP-1 was identified. Data for the PARP-1 peptides that were identified are summarized in Table S1.
Comparison of NaBH4 Cross-Linking of Purified PARP-1 with Intact and Incised AP Site-Containing DNA.
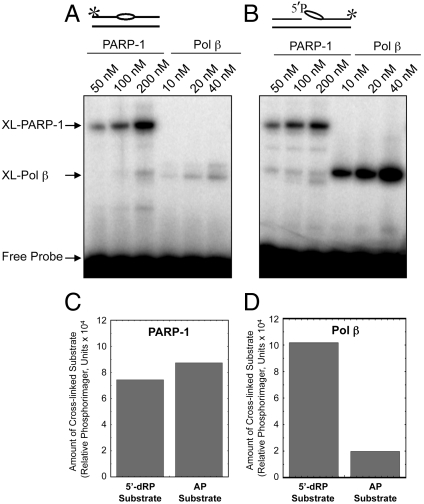
In light of the results described above, we tested for and found AP site cross-linking by purified PARP-1. We next examined purified PARP-1 cross-linking with a linear DNA containing either an intact AP site or preincised AP site; in addition to cross-linking probes, these DNAs are substrates for 5′-dRP and AP lyase enzymatic activities (to be described below). Cross-linking of PARP-1 was compared with that of DNA polymerase β (Pol β). PARP-1 and Pol β cross-linked to both of these DNA substrates in a concentration-dependent manner (Fig. 2). Pol β had a preference for the preincised AP site-containing DNA, as compared to the intact AP site (Fig. 2A, compare lanes 46 and 1012). Conversely, PARP-1 failed to show a similar preference, yielding similar cross-linking with both probes.
Fig. 2.
Comparison of cross-linking of purified PARP-1 and Pol β with 5′-dRP lyase substrate DNA and AP site-containing DNA. Cross-linking of purified PARP-1 to AP site-containing linear DNA was performed as described under Materials and Methods. Schematic representations of DNA probes are shown at Top. The * symbol denotes the position of the 32P label in the DNA. The bubble-like symbol denotes the presence of the AP site in the DNA. Representative results of PARP-1 and Pol β cross-linking to intact AP site-containing DNA (A) or preincised AP site-containing DNA (B) as a function of protein concentration. Quantification showing the yield of XL-PARP-1 (C) and XL-Pol β (D). Relative phosphorimager units of cross-linked products formed in the presence of 200 nM PARP-1 (C) and 40 nM Pol β (D) are shown.
Purified PARP-1 Cross-Linking to AP Site-Containing DNA.
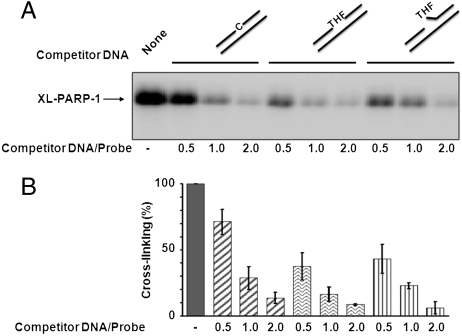
The cross-linking of PARP-1 with the AP site-containing probe raised the question of preferential PARP-1 binding to the AP site. We examined the specificity of PARP-1 interaction with AP site-containing DNA by competition experiments using three types of competitor DNA. A labeled DNA duplex with a “natural” AP site was used for PARP-1 cross-linking, and the cross-linking was competed either with a control DNA duplex (without an AP site), a synthetic AP site-containing DNA, i.e., with THF mimicking the AP site, or incised THF AP site-containing DNA. The results shown in Fig. 3 revealed that cross-linking of PARP-1 was competed with control DNA and both AP site-containing DNAs. However, the reduction was stronger in the cases of the AP site-containing DNAs and incised AP site-containing DNA than with control DNA. These results suggested that PARP-1 had greater affinity for the AP site-containing DNA than for the control DNA.
Fig. 3.
Specificity of the PARP-1 cross-linking with AP site-containing DNA, as revealed by competition with control and synthetic (THF) AP site-containing DNAs. Cross-linking of purified PARP-1 to linear natural AP site-containing DNA and the competition experiments were performed as described under Materials and Methods. (A) A representative phosphorimage showing the results of PARP-1 cross-linked to 32P-labeled AP site-containing DNA in the absence (−) or presence of competitor DNA, as indicated. The competitor-to-probe ratio is indicated. Competitor DNA contained either cytosine (C) or synthetic AP site (THF) at the same position as the natural AP site in the labeled DNA probe. A schematic representation of competitor DNAs is at Top. (B) Quantification summarizing the yield of XL-PARP-1 in three experiments similar to the experiment shown in A. In each independent experiment, the cross-linking in the absence of competitor DNA was taken as 100%. The data are the mean ± SD, n = 3.
Enzymatic Activities and PARP-1 Interaction with AP Site-Containing DNA.
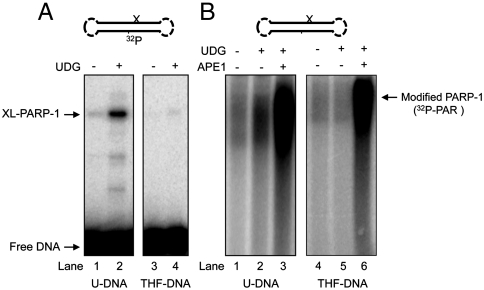
The next question regarding PARP-1’s interaction with AP sites was whether the enzyme is activated for poly(ADP-ribose) synthesis upon binding to the intact AP site. PARP-1 is well known to become activated by binding to DNA strand breaks (24, 25), and to avoid the presence of confounding DNA ends, we prepared a double-hairpin DNA for use as probe (Figs. S3 and S4). First, using this hairpin DNA with an internal 32P label, we confirmed the ability of purified PARP-1 to cross-link to the natural AP site. The results showed that double-hairpin DNA bearing the natural AP site (Fig. 4A, lane 2) was able to cross-link upon NaBH4 reduction, whereas DNA without the AP site (uracil-DNA) failed to yield any cross-linked product (Fig. 4A, lane 1). As expected, the synthetic AP site (THF)-containing DNA (without the C1′ aldehydic group) failed to cross-link (Fig. 4A, lanes 4 and 5). Next, using similar but unlabeled double-hairpin DNA and 32P-labeled NAD+ as substrate for poly(ADP-ribose) synthesis, we examined the poly(ADP-ribose) synthesis activity of PARP-1. The natural AP site was generated by UDG treatment, as in Fig. 4A. Strong PARP-1 automodification was observed only in reaction mixtures containing APE1 (Fig. 4B). PARP-1 automodification in reaction mixtures with the natural AP site, but without APE1, was only weak; this level, however, was more than the background level (Fig. 4B, compare lanes 1 and 2). Under similar conditions, the THF AP site-containing DNA failed to support poly(ADP-ribose) synthesis (Fig. 4B, lanes 4 and 5), but strong synthesis was observed when APE1 was added. These results were consistent with PARP-1 interaction with the intact AP site resulting in weak activation for autopoly(ADP-ribosyl)ation. This activation involved much less poly(ADP-ribose) synthesis than that observed with APE1-induced strand incision. Among other points, these results confirmed PARP-1’s ability to interact with the intact AP site. The stability of AP sites under these experimental conditions is demonstrated in Fig. S5.
Fig. 4.
Cross-linking and poly(ADP-ribosyl)ation activities of purified PARP-1. Experiments in A and B were performed with double-hairpin DNA substrates containing a uracil base or THF synthetic AP site, as specified under the gel images. A schematic representation of the hairpin DNA is at Top. The symbol “X” denotes the position of uracil (i.e., natural AP site) or the synthetic AP site in the DNA. (A) Results of purified PARP-1 cross-linking, as a function of UDG treatment, to internal 32P-labeled double-hairpin DNA with a uracil residue or a THF synthetic AP site. Cross-linking of PARP-1 was performed either without (lanes 1 and 3) or with (lanes 2 and 4) UDG treatment, as illustrated and described under Materials and Methods. The positions of XL-PARP-1 and free DNA are indicated. (B) Results of a poly(ADP-ribosyl)ation assay with intact (lanes 1, 2, 4, and 5) and APE1 incised (lanes 3 and 6) AP site DNAs. The presence (+) and absence (−) of UDG is indicated. Poly(ADP-ribosyl)ation was measured after incubations in the presence of 32P-labed NAD. Reaction conditions are described under Materials and Methods.
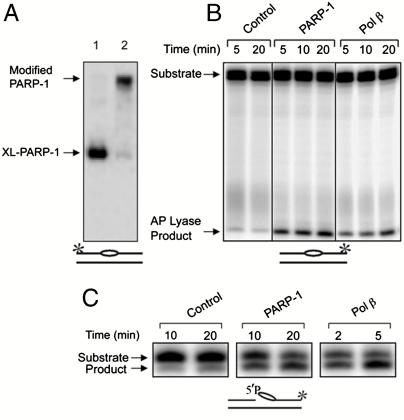
Next, to further examine PARP-1 automodification, we conducted experiments with purified PARP-1 (Fig. 5 A and B). PARP-1 was first preincubated with labeled intact linear AP site-containing DNA. The reaction mixture was then supplemented with NAD+ to allow poly(ADP-ribose) synthesis. Then, the reaction mixture was treated with NaHB4 and analyzed. The results indicated that the poly(ADP-ribose) modified enzyme was cross-linked (Fig. 5A, lane 2). In light of this result, we were curious to test PARP-1’s capacity to conduct strand incision at the AP site via AP lyase activity. As shown in Fig. 5B, PARP-1 was capable of incising AP site-containing DNA, and the activity was similar to that of Pol β. In light of PARP-1’s AP lyase activity, we also tested for 5′-dRP lyase activity. PARP-1 conducted 5′-dRP lyase activity against a preincised AP site (Fig. 5C), yet the activity appeared to be much lower than that of Pol β. These results suggested that the endogenous AP lyase activity in PARP-1 was sufficient to provide activation of poly(ADP-ribose) synthesis at the natural AP site.
Fig. 5.
Cross-linking, poly(ADP-ribosyl)ation and 5′-dRP/AP lyase activities of purified PARP-1. (A) Cross-linking (lane 1) and poly(ADP-ribosyl)ation (lane 2) were performed as in Fig. 1 using purified PARP-1 and 32P-labeled linear natural AP site-containing DNA. The positions of XL-PARP-1 and poly(ADP-ribose) modified XL-PARP-1 are indicated, and the DNA is illustrated at the bottom. (B) AP lyase activity of purified PARP-1. Reaction mixtures containing 32P-labeled intact AP site-containing DNA were assembled on ice with PARP-1 (200 nM), Pol β (200 nM), or buffer (control). Aliquots were withdrawn at the indicated intervals and analyzed for the strand incision AP lyase activity product as described under Materials and Methods. Lanes 1 and 2 represent AP site stability in the absence of PARP-1. (C) 5′-dRP lyase activity of purified PARP-1. Reaction mixtures with PARP-1 (100 nM) and Pol β (5 nM) were incubated as described. The positions of the substrates and products are indicated, and the DNA is illustrated at the bottom. The * symbol illustrates the position of the 32P label in the DNA, as in Fig. 2. In B, all eight lanes shown were from the same gel; some lanes from this gel are not shown because they are not relevant to the interpretation of this experiment, and the order of the original lanes was adjusted for use in B.
Stability of the AP Site/PARP-1 Complex.
Schiff base formation is considered a reversible process, but a PARP-1 molecule bound at the AP site may represent a rather long-lived complex. This idea raised the question of the stability of the PARP-1/AP site-containing DNA complex. To test this idea, we incubated PARP-1 in the presence of AP site-containing DNA to allow binding and formation of the Schiff base. Then, the mixture was divided into portions: One was processed immediately; the other was supplemented with an excess of THF-containing DNA to trap PARP-1 liberated from the initial complex. This mixture was then further incubated. Aliquots were withdrawn at intervals, treated with NaBH4, and analyzed by SDS-PAGE. In the presence of excess DNA trap, the amount of cross-linked material declined only ∼20% after 1 h, indicating significant stability of the PARP-1 and AP site-containing DNA complex (Fig. S6).
Interaction of APE1 and PARP-1 at the AP Site.
AP sites are considered to arise abundantly in mammalian cell and tissue genomic DNA. Therefore, understanding coordination for the various enzymes and accessory factors recognizing the AP site is an important challenge. From the results described above, PARP-1 is among the proteins interacting with the intact AP site, and we were interested in evaluating the possibility of competition between purified APE1 and PARP-1 at an intact AP site.
First, we tested for APE1 inhibition of the PARP-1 cross-linking to AP site-containing DNA in the absence of magnesium ions to prevent AP site cleavage by APE1. In these experiments, PARP-1 (200 nM) cross-linking with AP site-containing linear DNA (100 nM) was studied in the absence or presence of APE1 (20 nM). APE1 decreased the PARP-1 cross-linking (Fig. S7A, compare lanes 1 and 3), demonstrating competition of these proteins for AP DNA binding. However, when PARP-1 was first allowed to bind the AP site DNA and form the Schiff base intermediate (preincubation for 15 min at 37 °C), followed by addition of APE1 and further incubation (for 15 min), the level of PARP-1 cross-linking was unchanged by the presence of APE1 (Fig. S7A, compare lanes 1 and 2). This illustrated the inability of APE1 to displace PARP-1 from the preformed Schiff base intermediate.
A competition experiment in the presence of 10 mM Mg2+ showed that the level of PARP-1 cross-linking was decreased by factors of 3.45- and 4.0-fold by 20 nM and 100 nM APE1, respectively (Fig. S7B, compare lanes 1 and 6, 7). Analysis of AP site-containing DNA cleavage by APE1 also was conducted (Fig. S7C). PARP-1 was preincubated with the AP site-containing DNA substrate and then APE1 (20 nM or 100 nM) was added to the mixture. The incubation was continued for 2 or 15 min. At 2-min incubation with 20 nM APE1, PARP-1 inhibited the incision activity (Fig. S7C, lanes 1 and 3). However, with the longer incubation or the higher level of APE1, no significant inhibition was observed. Taken together, the data support the notion that PARP-1 and APE1 compete for AP site DNA binding.
Conclusions
PARP-1 is a regulatory enzyme involved in many different processes of DNA and RNA metabolism, including DNA base excision repair. The role of PARP-1 in BER is still under investigation, although numerous studies show an important contribution of PARP-1 in modulation of the ability of the BER enzymes to process DNA strand breaks that arise following DNA damage (25–29). Using various BER assays, evidence for important PARP-1 regulatory functions has been obtained: PARP-1 was among the core BER enzymes cross-linked to a photoreactive BER intermediate in crude extracts (20) along with APE1, Pol β, and flap endonuclease 1; studies with purified enzymes showed that PARP-1 was involved in BER along with APE1 (21, 22), Pol β, and FEN1 (21–23, 30); and PARP inhibition was found to block BER in a plasmid-based assay in vivo (31). Here, by virtue of PARP-1’s ability to interact with the intact AP site via Schiff base formation, we demonstrated a previously undescribed role for PARP-1 in regulation of the BER process. PARP-1’s interaction at the AP site could recruit this key enzyme and protect the site until APE1 becomes available to initiate strand incision and BER. Alternatively, PARP-1’s 5′-dRP/AP lyase activity could perform strand incision and trigger poly(ADP-ribosyl)ation leading to recruitment of other BER factors.
Materials and Methods
Chemicals.
Full antiprotease cocktail was from Roche Applied Science. Radioactive [γ-32P]ATP, [α-32P]dATP, and [32P]NAD+ were obtained from the Biotechnological Laboratory (Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences) and PerkinElmer Life Sciences, respectively.
Oligonucleotides.
Synthetic oligonucleotides were obtained from the Laboratory of Medicinal Chemistry (Institute of Chemical Biology and Fundamental Medicine, SB RAS) and Oligos Etc., Inc. Sequences of oligonucleotides are detailed in SI Text.
Proteins and Cell Extracts.
T4 polynucleotide kinase, E. coli UDG, Stoffel fragment of Taq DNA polymerase, DNA ligase from Methanobacterium thermoautotrophicum, and dUTP were from Biosan. Nuclease (Benzonase) was from Novagen. Human Pol β was overexpressed and purified as described (30). M. Satoh (Laval University, Quebec City, QC, Canada) kindly provided a human PARP-1 expression vector; the His-tagged full-length protein was overexpressed in E. coli and purified as described previously (32). Whole cell extract from HeLa cells was prepared as described (33). Bovine testis nuclear extract (BTNE) was prepared as described (34). All cell extracts were stored in aliquots at -70 °C.
Preparation of Linear Duplex DNA.
Linear DNA duplexes were prepared as described (18).
Preparation of M13 Circular DNA Substrate.
Uniformly 32P-labeled circular double-stranded DNA was synthesized using single-stranded M13 mp19 DNA as a template in the presence of dUTP, essentially as described previously (35). Briefly, the reaction mixture (100 μL) contained 0.27 μM single-strand M13 mp19 DNA with three annealed primers, 200 units Stoffel fragment of Taq DNA polymerase, 0.3 μCi [α-32P]dATP, 200 μM each dATP, dCTP, and dGTP, 10 μM dUTP and 190 μM dTTP, 50 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 50 mM NaCl, 1 mM ATP, and 1 pmol of thermophilic DNA ligase Methanobacterium thermoautotrophicum. DNA synthesis proceeded for 60 min at 55 °C. The reaction was stopped by addition of EDTA to a final concentration of 40 mM. Then, potassium acetate, pH 5.5, was added to a final concentration of 0.3 M. This was followed by DNA precipitation with ethanol. Precipitated DNA was dissolved in TE buffer and purified using a spin column (Sephadex G-25).
Preparation of Double-Hairpin DNA Substrates.
32P-labeled or unlabeled oligonucleotides that contained a double-hairpin sequence were annealed, ligated, and purified as detailed in SI Text.
Cross-Linking of Protein-DNA Complexes with Sodium Borohydride.
The standard reaction mixture (10 μL) contained 50 mM Hepes-KOH, pH 7.8, 5 mM EDTA, 20 mM KCl, 2 mM DTT, 32P-labeled M13 AP site-containing DNA (20 nM), linear natural AP site-containing DNA (100 nM) and hairpin DNA (100 nM) and protein extracts (1.25 mg/mL) or human recombinant PARP-1 (100/200 nM). The AP sites were generated by treating DNA with UDG immediately before the experiments. The reaction mixtures were assembled on ice and incubated for 15 min at 37 °C. After incubation, the reaction mixtures were supplemented with NaBH4 to a final concentration of 20 mM and further incubated for 30 min at 0–1 °C. In some cases, the reaction mixtures, before NaBH4 treatment, were supplemented with 10 mM MgCl2 or 400 μM NAD+ and 10 mM MgCl2 and incubated for 5 min at 37 °C. The products of covalent protein cross-linking with circular AP DNA were treated with benzonase (12 units) for 15 min at 37 °C prior to SDS-PAGE. In the competition experiments (Fig. 3), the reaction mixtures additionally contained control or THF-containing linear DNAs (intact or incised) at concentrations of 50, 100, or 200 nM, as indicated. After incubation, the reaction mixtures were supplemented with NaBH4 to a final concentration of 20 mM and further incubated for 30 min at 0–1 °C.
The reaction mixtures were quenched with Laemmli sample buffer and heated for 5 min at 95 °C. Proteins were separated by denaturing electrophoresis in a 10% polyacrylamide gel (36). The gels were dried and subjected to phosphorimaging for quantification using Molecular Imager and Quantity One software (Bio-Rad).
In experiments with double-hairpin DNA (Fig. 4A), either a uracil base or THF synthetic AP site was identically positioned, as described in SI Text. UDG, PARP-1, and DNA were incubated in the standard reaction mixture and processed as described above.
PARP-1 Autopoly(ADP-Ribosyl)ation.
The reaction mixture (10 μL) for autpoly(ADP-ribosyl)ation of PARP-1 contained 50 mM Hepes-KOH, pH 7.8, 0.5 mM EDTA, 20 mM KCl, 2 mM DTT, 5 mM MgCl2, 100 nM double-hairpin DNA 100 μM [32P]NAD+, 200 nM PARP-1, 20 nM UDG, and/or 20 nM APE1, as indicated in the figure legends. The reaction mixtures, after incubation for 20 min at 30 °C, were terminated by the addition of 10 μL SDS-PAGE buffer and heating for 5 min at 95 °C. The reaction mixtures were analyzed by 4–12% SDS-PAGE with subsequent phosphorimaging.
AP and 5′-dRP Lyase Assays.
AP and 5′-dRP and AP lyase activities of PARP-1 were determined essentially as described previously (37). Briefly, the reaction mixture (30 μL) contained 50 mM Hepes, pH 7.5, 20 mM KCl, 2 mM dithiothreitol, 1 mM EDTA, and 50 nM UDG pretreated 3′ end-labeled AP site-containing DNA or preincised AP site-containing DNA. The reaction was initiated by adding purified PARP-1 or Pol β, as indicated, followed by incubation at 37 °C. Aliquots (9 μL each) were transferred at the indicated intervals into tubes that contained 1 μL freshly prepared 200 nM NaBH4. Reaction mixtures were shifted to 0–1 °C (on ice), and incubation was continued for 30 min. After the incubation, reactions were stopped by the addition of 10 μL formamide gel-loading dye and heating to 75 °C for 2 min. Reaction products were separated by electrophoresis in a 15% polyacrylamide gel containing 8 M urea in 89 mM Tris-HCl, 89 mM boric acid, and 2 mM EDTA, pH 8.8 (38). Imaging and data analysis were performed with a Typhoon PhosphorImager and the ImageQuant software (GE HealthCare).
MALDI-TOF-MS Analysis.
Preparative cross-linking was conducted essentially as described above. The volume of the reaction mixture was increased to 1.3 mL. Samples for MALDI-TOF-MS and data acquisition were as described (18). Peptide analysis was performed using the Mascot software and National Center for Biotechnology Information protein database at http://www.expasy.org.
Assessment of the Stability of the PARP-1–AP DNA Complex.
Reaction mixtures (50 μL) were prepared as described above. PARP-1 (200 nM) was preincubated with 32P-labeled AP site-containing DNA (100 nM) for 15 min at 37 °C. Then, a portion of the reaction mixture was supplemented with unlabeled THF-containing DNA at final concentration of 1 μM to trap and prevent the reassociation of PARP-1, liberated from the complex, with the labeled DNA probe. Aliquots were taken at different times, followed by sodium borohydride treatment and analysis as above.
Supplementary Material
Acknowledgments.
We thank Dr. Yu. Gerasimova (Institute of Chemical Biology and Fundamental Medicine, SB RAS) for recording the MALDI-TOF-MS spectra. The authors also thank Bonnie Mesmer for assistance in manuscript preparation. This work was supported in part by the Russian Foundation for Basic Research [Projects 10-04-01083, 09-04-93106, and 09-04-91320 (HRJRG-102)], the Program of RAS “Molecular and Cellular Biology,” and State Contract 02.740.11.0079. This work also was supported in part by Research Projects Z01-ES050158 and Z01-ES050159 in the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009182107/-/DCSupplemental.
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Wilson DM, 3rd, Thompson LH. Life without DNA repair. Proc Natl Acad Sci USA. 1997;94:12754–12757. doi: 10.1073/pnas.94.24.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCullough AK, Dodson ML, Lloyd RS. Initiation of base excision repair: Glycosylase mechanisms and structures. Annu Rev Biochem. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 4.Atamna H, Cheung I, Ames BN. A method for detecting abasic sites in living cells: Age-dependent changes in base excision repair. Proc Natl Acad Sci USA. 2000;97:686–691. doi: 10.1073/pnas.97.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burrows CJ, Muller JG. Oxidative nucleobase modifications leading to strand scission. Chem Rev. 1998;98:1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 6.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 7.Mohsin Ali M, et al. Detection of endonuclease III- and 8-oxoguanine glycosylase-sensitive base modifications in gamma-irradiated DNA and cells by the aldehyde reactive probe (ARP) assay. J Radiat Res. 2004;45:229–237. doi: 10.1269/jrr.45.229. [DOI] [PubMed] [Google Scholar]
- 8.Sokhansanj BA, Wilson DM., 3rd Oxidative DNA damage background estimated by a system model of base excision repair. Free Radical Bio Med. 2004;37:422–427. doi: 10.1016/j.freeradbiomed.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: Definition of a family of DNA repair enzymes. Proc Natl Acad Sci USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robson CN, Hickson ID. Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E. coli xth (exonuclease III) mutants. Nucleic Acids Res. 1991;19:5519–5523. doi: 10.1093/nar/19.20.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Summer H, et al. HMGA2 exhibits dRP/AP site cleavage activity and protects cancer cells from DNA-damage-induced cytotoxicity during chemotherapy. Nucleic Acids Res. 2009;37:4371–4384. doi: 10.1093/nar/gkp375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476:73–77. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- 13.David SS, Williams SD. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem Rev. 1998;98:1221–1262. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 14.Piersen CE, McCullough AK, Lloyd RS. AP lyases and dRPases: Commonality of mechanism. Mutat Res. 2000;459:43–53. doi: 10.1016/s0921-8777(99)00054-3. [DOI] [PubMed] [Google Scholar]
- 15.Hegde V, Wang M, Deutsch WA. Human ribosomal protein S3 interacts with DNA base excision repair proteins hAPE/Ref-1 and hOGG1. Biochemistry. 2004;43:14211–14217. doi: 10.1021/bi049234b. [DOI] [PubMed] [Google Scholar]
- 16.Postel EH, Abramczyk BM, Levit MN, Kyin S. Catalysis of DNA cleavage and nucleoside triphosphate synthesis by NM23-H2/NDP kinase share an active site that implies a DNA repair function. Proc Natl Acad Sci USA. 2000;97:14194–14199. doi: 10.1073/pnas.97.26.14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rieger RA, et al. Proteomic approach to identification of proteins reactive for abasic sites in DNA. Mol Cell Proteomics. 2006;5:858–867. doi: 10.1074/mcp.M500224-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Ilina ES, Lavrik OI, Khodyreva SN. Ku antigen interacts with abasic sites. Biochim Biophys Acta. 2008;1784:1777–1785. doi: 10.1016/j.bbapap.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Roberts SA, et al. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavrik OI, et al. Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate. Evidence for the role of poly(ADP-ribose) polymerase-1 in DNA repair. J Biol Chem. 2001;276:25541–25548. doi: 10.1074/jbc.M102125200. [DOI] [PubMed] [Google Scholar]
- 21.Sukhanova MV, et al. Human base excision repair enzymes apurinic/apyrimidinic endonuclease1 (APE1), DNA polymerase beta and poly(ADP-ribose) polymerase 1: Interplay between strand-displacement DNA synthesis and proofreading exonuclease activity. Nucleic Acids Res. 2005;33:1222–1229. doi: 10.1093/nar/gki266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cistulli C, Lavrik OI, Prasad R, Hou E, Wilson SH. AP endonuclease and poly(ADP-ribose) polymerase-1 interact with the same base excision repair intermediate. DNA Repair. 2004;3:581–591. doi: 10.1016/j.dnarep.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Sukhanova M, Khodyreva S, Lavrik O. Poly(ADP-ribose) polymerase 1 regulates activity of DNA polymerase beta in long patch base excision repair. Mutat Res. 2010;685:80–89. doi: 10.1016/j.mrfmmm.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Dantzer F, et al. Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006;409:493–510. doi: 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- 25.Dantzer F, et al. Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1. Biochemistry. 2000;39:7559–7569. doi: 10.1021/bi0003442. [DOI] [PubMed] [Google Scholar]
- 26.Woodhouse BC, Dianova II, Parsons JL, Dianov GL. Poly(ADP-ribose) polymerase-1 modulates DNA repair capacity and prevents formation of DNA double strand breaks. DNA Repair. 2008;7:932–940. doi: 10.1016/j.dnarep.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Woodhouse BC, Dianov GL. Poly ADP-ribose polymerase-1: An international molecule of mystery. DNA Repair. 2008;7:1077–1086. doi: 10.1016/j.dnarep.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Jelezcova E, et al. (Parp1 activation in mouse embryonic fibroblasts promotes Pol beta-dependent cellular hypersensitivity to alkylation damage. Mutat Res. 686:57–67. doi: 10.1016/j.mrfmmm.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pachkowski BF, et al. Cells deficient in PARP-1 show an accelerated accumulation of DNA single strand breaks, but not AP sites, over the PARP-1-proficient cells exposed to MMS. Mutat Res. 2009;671:93–99. doi: 10.1016/j.mrfmmm.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sukhanova MV, Khodyreva SN, Lavrik OI. Poly(ADP-ribose) polymerase-1 inhibits strand-displacement synthesis of DNA catalyzed by DNA polymerase beta. Biochemistry-Moscow. 2004;69:558–568. doi: 10.1023/b:biry.0000029855.68502.fa. [DOI] [PubMed] [Google Scholar]
- 31.Masaoka A, Horton JK, Beard WA, Wilson SH. DNA polymerase beta and PARP activities in base excision repair in living cells. DNA Repair. 2009;8:1290–1299. doi: 10.1016/j.dnarep.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drachkova IA, et al. [Reagents for modification of protein-nucleic acids complexes. II. Site-specific photomodification of DNA-polymerase beta complexes with primers elongated by the dCTP exo-N-substituted arylazido derivatives] Bioorg Khim. 2001;27:197–204. doi: 10.1023/a:1011329220632. [DOI] [PubMed] [Google Scholar]
- 33.Biade S, Sobol RW, Wilson SH, Matsumoto Y. Impairment of proliferating cell nuclear antigen-dependent apurinic/apyrimidinic site repair on linear DNA. J Biol Chem. 1998;273:898–902. doi: 10.1074/jbc.273.2.898. [DOI] [PubMed] [Google Scholar]
- 34.Prasad R, et al. Specific interaction of DNA polymerase beta and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J Biol Chem. 1996;271:16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- 35.Zhang K, Taylor JS. Phototriggered formation and repair of DNA containing a site-specific single strand break of the type produced by ionizing radiation or AP lyase activity. Biochemistry. 2001;40:153–159. doi: 10.1021/bi001781j. [DOI] [PubMed] [Google Scholar]
- 36.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Prasad R, Beard WA, Strauss PR, Wilson SH. Human DNA polymerase beta deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J Biol Chem. 1998;273:15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.